Abstract
Response of bacterial pathogen to environmental bacteria and its host is critical for understanding of microbial adaption and pathogenesis. Here, we used RNA-Seq to comprehensively and quantitatively assess the transcriptional response of Acidovorax avenae subsp. avenae strain RS-1 cultivated in vitro, in vivo and in co-culture with rice rhizobacterium Burkholderia seminalis R456. Results revealed a slight response to other bacteria, but a strong response to host. In particular, a large number of virulence associated genes encoding Type I to VI secretion systems, 118 putative non-coding RNAs, and 7 genomic islands (GIs) were differentially expressed in vivo based on comparative genomic and transcriptomic analyses. Furthermore, the loss of virulence for knockout mutants of 11 differentially expressed T6SS genes emphasized the importance of these genes in bacterial pathogenicity. In addition, the reliability of expression data obtained by RNA-Seq was supported by quantitative real-time PCR of the 25 selected T6SS genes. Overall, this study highlighted the role of differentially expressed genes in elucidating bacterial pathogenesis based on combined analysis of RNA-Seq data and knockout of T6SS genes.
Acidovorax avenae subsp. avenae (Aaa), causes diseases in many plants with economic importance. In particular, bacterial brown stripe of rice has been reported in many countries in the world1,2,3. Although this rice disease has been proven to be economically important, very little is known about the molecular basis of pathogenesis of Aaa in rice plants. Liu et al.4 characterized pilP, which is required for twitching motility, pathogenicity, and biofilm formation of Aaa strain RS-1. However, it is crucial for elucidating bacterial pathogenesis to examine how the host and other environmental bacteria alter the global pattern of pathogen gene expression5,6. The whole genome of Aaa strain RS-1 was recently published7.
Identification of global gene expression in bacteria, and characterization of their roles in pathogen physiology, disease, and defense against the host and environmental bacteria, is an important initial step in understanding the pathogenesis. Using direct high-throughput Illumina sequencing of cDNAs, some bacterial transcriptome have been recently reported, while most of these researches focused on the transcriptome of in vitro cultivated bacteria8,9,10,11. However, it is the characterization of the bacterial transcriptome during in vivo infection of its host that eventually could provide the most significant insights into bacterial pathogenesis. Indeed, transcriptome analysis in human and animal pathogens such as Vibrio cholerae has revealed differential expression in vivo vs. in vitro conditions6. In contrast, little is known about the in vivo expression profile of plant pathogenic bacteria due to the absence of an efficient method to directly collect bacterial cells from diseased plant tissues.
Fortunately, a method has been recently developed to obtain in vivo samples by detaching the bacteria from tissues of infected leaves that were cut into pieces and put into distilled water, while in vitro samples were obtained by incubating bacteria in Luria Bertani broth. Furthermore, the differential composition of outer membrane (OM) proteome has been observed between in vivo and in vitro based on LC-MS/MS in combination with an in silico analysis of OM proteome of Aaa strain RS-11. In particular, Type VI secretion systems (T6SS) core components, such as OmpA/MotB domain containing proteins and an ATP dependent Clp protease, were identified in the OM proteome under in vivo conditions, but not under in vitro conditions1. This result highlighted that bacteria under in vivo conditions are ideal for this study of OM proteome that may be involved in the survival and pathogenicity of Aaa strain RS-1.
In addition to the pathogen-host interaction, bacterial pathogenicity is also influenced by other bacterial species in host and natural environment. This may also be true for the rice pathogen Aaa. Recent studies have revealed that bacteria alter their gene expression when confronted with another bacterial species. For example, Pseudomonas fluorescens strain Pf0-1 shows a species-specific transcriptional and metabolic response to bacterial competitors5. Therefore, it is also necessary to examine the transcriptional response of rice pathogen Aaa to other rice associated bacteria such as Burkholderia seminalis strain R456, which was isolated from rice rhizosphere and is nonpathogenic to rice. B. seminalis strain R456 protected rice seedlings from infection by Rhizoctonia solani in our previous studies1,12.
Here we aim to comprehensively and quantitatively investigate the transcriptional responses of Aaa strain RS-1 under in vitro culture, in vivo infection to rice plant, and under co-culture with rice rhizobacterium B. seminalis strain R456 using RNA-Seq technologies.
Results
Quality analysis of RNA-Seq data
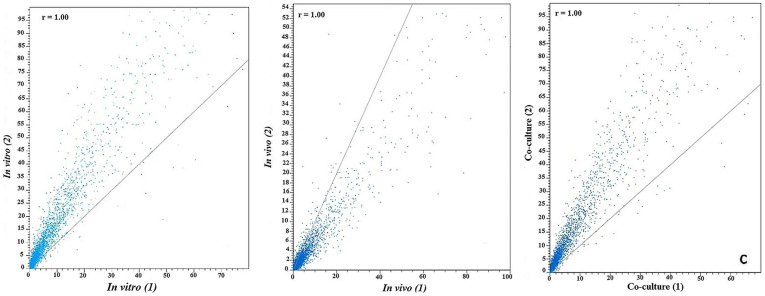
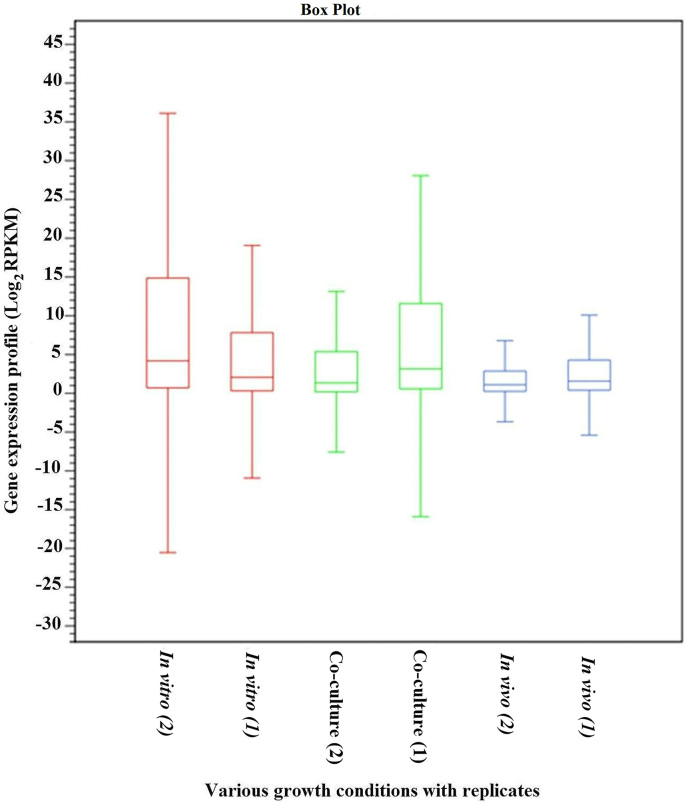
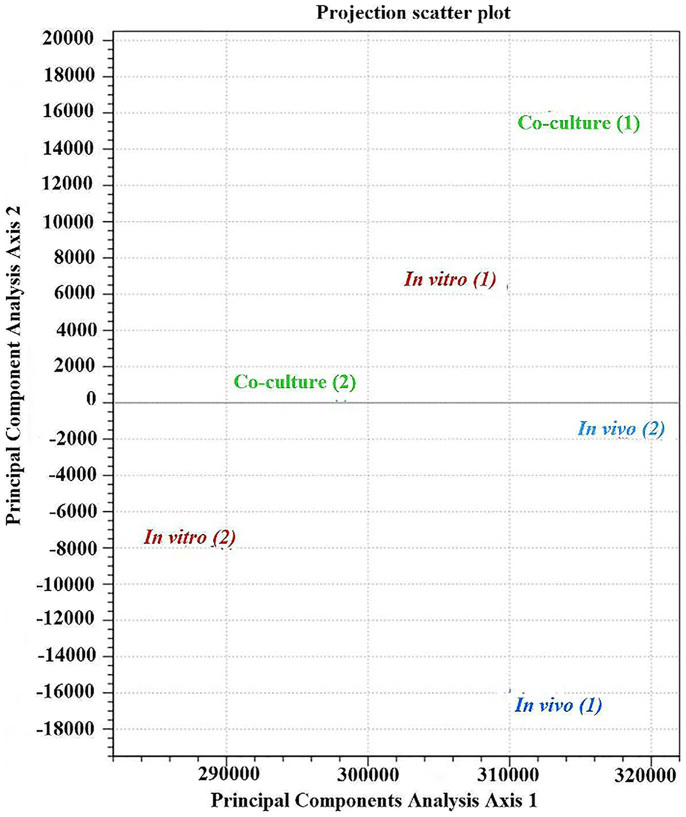
The total number of reads obtained from each sample of Aaa strain RS-1 was between 14,620,058 and 31,932,210, while the number of mapped cDNA reads varied between 13,360,933 and 26,011,042 per samples (Table 1). Furthermore, heat maps of coverage revealed numerous regions with transcript abundance that was uniformly high or uniformly low for each RNA sample (Figure 1). All data used in our analyses were highly reproducible in terms of the high correlation between two biological replicates for each condition (R = 0.95–0.98, P < 0.001; Figure 2). In addition, the box plots indicated that the locations of the distributions of the expression values in all six samples of Aaa strain RS-1 are generally similar, although there is considerable difference in the spread of the expression level (log2RPKM) of expressed annotated genes between two biological replicates of each sample (Figure 3). In agreement with hierarchical clustering, a PCA plot based on 6 digital gene expression profiles resulted in a clear separation between the growth conditions and two biological replicates (Figure 4).
Table 1. Summary of Acidovorax avenae subsp. avenae strain RS-1 cDNA samples sequenced using the Illumina genome analyze.
| Sample name | In vitro (1) | In vitro (2) | In vivo (1) | In vivo (2) | Co-culture (1) | Co-culture (2) |
|---|---|---|---|---|---|---|
| Number of reads | 21,439,332 | 14,620,058 | 28,261,598 | 31,932,210 | 27,392,918 | 28,330,180 |
| Mapped reads | 20,062,237 | 13,360,933 | 16,328,205 | 18,776,074 | 26,011,042 | 26,310,263 |
| Unique mapped | 1,552,759 | 1,983,472 | 1,267,326 | 1,246,124 | 2,080,783 | 3,546,162 |
| mRNA percent | 7.2% | 13.6% | 4.5% | 3.9% | 7.6% | 12.5% |
Figure 1. Distribution of differentially expressed genes visualized using GenomeViz software under three conditions of Acidovorax avenae subsp. avenae strain RS-1.
Outmost rings indicate all coding regions in the genomes colored according to COG designation. Red and purple rings correspond to co-culture (1) and (2); blue and light blue rings correspond to in vitro (1) and (2); orange and green rings correspond to in vivo (1) and (2). COG (color): functional designations are described below. J (gold1): translation, ribosomal structure and biogenesis; A (orange3): RNA processing and modification; K (DarkOrange1): transcription; L (DarkOrange3): DNA replication, recombination and repair; B (maroon): Chromatin structure and dynamics; D (AntiqueWhite1): Cell division and chromosome partitioning; Y (yellow): Nuclear structure; V (pink): Defense mechanisms; T (tomato1): Signal transduction mechanisms; M (PeachPuff3): Cell envelope biogenesis, outer membrane; N (MediumPurple1): Cell motility and secretion; Z (red): Cytoskeleton; W (green): Extracellular structures; U (DeepPink): Intracellular trafficking, secretion, and vesicular transport; O (PaleGreen1): Posttranslational modification, protein turnover, chaperones; C (RoyalBlue4): Energy production and conversion; G (Blue1): Carbohydrate transport and metabolism; E (DodgerBlue1): Amino acid transport and metabolism; F (SkyBlue3): Nucleotide transport and metabolism; H (LightBlue1): Coenzyme metabolism; I (Cyan3): Lipid metabolism; P (MediumPurple4): Inorganic ion transport and metabolism; Q (aquamarine4): Secondary metabolites biosynthesis, transport and catabolism; R (gray90): General function prediction only; S (gray70): Function unknown; - (gray50): Not in COGs.
Figure 2. Correlation (R = 0.95–0.98, P < 0.001) of RNA-Seq data between two biological replicates of Acidovorax avenae subsp. avenae strain RS-1 under the condition of (a) in vitro; (b) in vivo; (c) co-culture.
Figure 3. Box plot of the expression level (log2RPKM) of annotated expressed genes in all six samples of Acidovorax avenae subsp. avenae strain RS-1.
Figure 4. Principle component analysis of RPKM-based expression values of Acidovorax avenae subsp. avenae strain RS-1 transcriptome under in vivo, in vitro and co-culture conditions.
The 6 samples shown in the 2D plane spanned by their first two principal components. This type of plot is useful for visualizing the overall effect of experimental covariates and batch effects. For this data set, no batch effects besides the known effects of condition and lib Type are discernible.
Differential genes expression
According to the method of Nagalakshimi et al.13, the RNA-Seq expression values in this study were divided into four categories: (i) non transcribed (average GEI < 1), (ii) low transcript levels (average GEI ≥ 1 and <10), (iii) medium transcript levels (average GEI ≥ 10 and <25), and (iv) high transcript levels (average GEI ≥ 25). Indeed, 3137, 2972 and 3018 showed an average GEI ≥ 1.0 in vitro, in vivo and in co-culture, revealing that about 60% of the 4853 annotated genes in the genome of Aaa strain RS-1 are transcribed under the three growth conditions. Among these genes, 504, 189, and 495 had high transcript levels; 577, 312, and 534 had medium transcript levels; 2056, 2471, and 1989 had low transcript levels in vitro, in vivo and in co-culture, respectively.
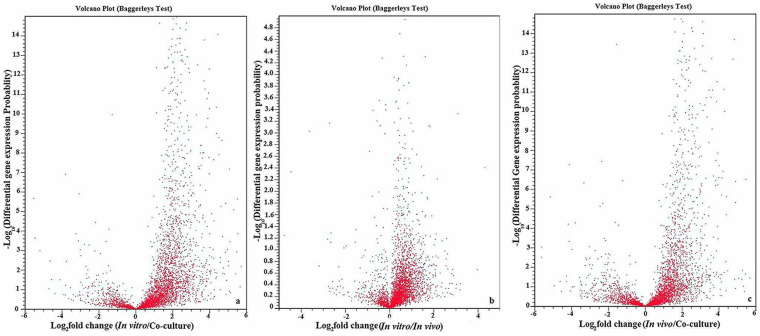
The global analysis of differentially expressed genes and their absolute and relative distributions of reads under the three conditions are shown in Figure 1. Furthermore, the transcriptional changes of two fold or higher between two conditions were illustrated in Figure 5 using the variance analysis package TopHat with statistically significant (P < 1 × 10−5) based on the normalized gene expression value. In addition, Supplementary Figure S1 revealed the Dendrograms and Heatmap of the top 50 differentially expressed genes between the three conditions of Aaa strain RS-1. Among them, the role of in vivo down-regulated clpB in pathogenicity was confirmed in this section of T6SS genes knockout.
Figure 5. Volcano plot of log2 fold-change (x-axis) versus −log10 FDR-corrected p-value (y-axis, representing the probability that the gene is differentially expressed) in RNA-Seq data of Acidovorax avenae subsp. avenae strain RS-1 under the conditions of (a) in vitro vs. co-culture; (b) in vitro vs. in vivo; (c) in vivo vs. co-culture.
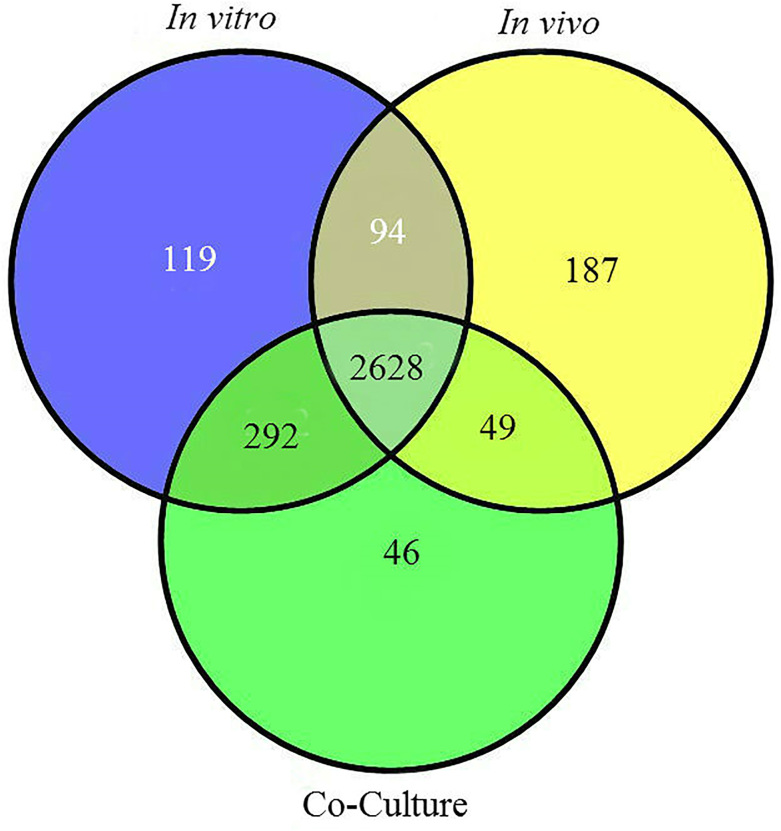
In general, 2628 genes were induced by all the three conditions, while 49 genes were specifically induced by both in vivo and co-culture (Figure 6). In addition, 292 genes expression were specifically induced by both in vitro and co-culture but not in vivo, meaning these gene were down regulated in vivo, while 94 genes were specifically induced by both in vitro and in vivo, but not co-culture, meaning these genes were down regulated in co-culture (Figure 6). The higher number in both specifically expressed genes and down regulated expressed genes revealed that in vivo resulted in a greater differential expression relative to co-culture. Notably, most T3SS and T6SS genes expression were ≥2-fold either up- or down- regulated under in vivo conditions, while only several T3SS and T6SS genes expression was differentially regulated by co-culture relative to in vitro.
Figure 6. Venn diagram representing the number of differentially expressed genes in Acidovorax avenae subsp. avenae RS-1 during in vitro (blue circle), in vivo (yellow circle) and co-culture (Green circle) conditions.
Furthermore, expression of 119, 187 and 46 genes were specifically induced by in vitro, in vivo and co-culture, respectively (Figure 6). Obviously, in vitro specifically expression means these genes were down-regulated by both in vivo and co-culture, while the number of co-culture induced genes is less than one quarter of that of in vivo induced genes relative to in vitro. Based on clusters of orthologous genes (COG) designations, in vivo specifically induced genes were classified into the 18 functional categories (Supplementary Table S1). Among these functional categories, the main focuses were 30 carbohydrate transport and metabolism, 13 intracellular trafficking, secretion, and vesicular transport, 24 signal transduction mechanisms, which have been widely reported to be involved in the virulence of bacterial pathogens. In particular, ATP-binding cassette (ABC) transporter genes that are involved in pathogenicity of a number of bacterial species14,15 account for nearly half of in vivo induced carbohydrate transport and metabolism genes. In addition, intracellular trafficking, secretion, and vesicular transport mainly consists of T3SS components, which is an essential requirement for the virulence of many bacterial pathogen of plants, animals and humans16,17.
Genome-wide transcriptome analyses of secretion systems
Genome-wide comparative in silico analysis identified Type I, II, III, and type IV secretion systems in Aaa strain RS-1 (Supplementary Table S2), while type VI secretion system was identified in our previous study1 by referring the secretion systems of Aaa strain 19860 drafted by KEGG18. In addition, the analysis of secretion systems revealed two clusters for T2SS, two clusters for T3SS and one cluster for T6SS, while individual components of other identified secretion systems are distributed at various positions in Aaa strain RS-1 genome.
Among 36 T1SS genes, 4 and 5 genes were expressed specifically co-culture and in vitro, respectively, while 24 genes were unexpressed under both conditions. In contrast, 26 and 5 genes were expressed specifically in vivo and in vitro, respectively, while 4 genes were unexpressed under both conditions (Supplementary Table S2). Among 15 T2SS genes, 14 genes were common expressed while one gene was unexpressed under the three conditions. Of the common expressed genes, co-culture and in vivo caused a ≥2 fold down-regulation in expression of 1 and 6 genes, respectively (Supplementary Table S2). Among 23 T3SS genes, no gene was specifically expressed co-culture and in vitro, while 2 out of 9 common expressed genes had a ≥2-fold change of down-regulation. Furthermore, 7 and 0 gene was specifically expressed in vivo and in vitro, respectively, while 6 out of the 9 common expressed genes had a ≥2-fold change, including up-regulation of 4 genes and down-regulation of 2 genes (Supplementary Table S2). Among 10 T4SS genes, 9 genes were common expressed while one gene was unexpressed under the three conditions. Of the common expressed genes, co-culture and in vivo caused a ≥2 fold down-regulation in expression of 1 gene and 3 genes, respectively (Supplementary Table S2). Among 25 T6SS genes, 0 and 5 genes were specifically expressed co-culture and in vitro, respectively, while 3 out of the 18 common expressed genes had a ≥2-fold change of up-regulation (Supplementary Table S2). Furthermore, 0 and 5 genes were specifically expressed in vivo and in vitro, respectively, while 17 out of the 18 common expressed genes had a ≥2-fold change including up-regulation of 2 genes and down-regulation of 15 genes (Supplementary Table S2).
GIs in Aaa strain RS-1 and transcription
Comparative genomic analysis revealed 7 putative GIs that contain 89 genes in Aaa strain RS-1 genome (Supplementary Figure S2; Table S3). Furthermore, transcription profiles analysis indicated that 1 and 4 genes were expressed specifically in co-culture and in vitro, respectively, while 59 genes were unexpressed under both conditions. Of the 25 common expressed genes, there was not a ≥2-fold change in genes expression between co-culture and in vitro. Similarly, 4 and 4 genes were expressed specifically in vivo and in vitro, respectively, while 56 genes were unexpressed under both conditions. Of the 25 common expressed genes, in vivo resulted in a ≥2-fold down-regulation of 11 genes.
Genome-wide transcriptome analyses of ncRNAs
Four hundred and forty six ncRNAs were predicted in the genome of Aaa strain RS-1 using RNAspace, SIPHT, and Rfam methods, which have been applied for the discovery of ncRNAs in several plant pathogenic bacteria19,20. After removing tRNA, and rRNA as well as redundant ncRNAs, a comprehensive list of 188 ncRNAs was obtained in Aaa strain RS-1 genome while Aaa strain ATCC19860 genome was used as a reference. Furthermore, BLAST search of the putative ncRNAs resulted in several hits even though the significance of the alignment was E < 0.02, and found 55 of which coordinates with Rfam database. In addition, 118 out of 188 ncRNAs were confirmed based on transcriptomic analysis of Aaa strain RS-1 by searching the intergenic regions for differential expression between in vitro, in vivo and co-culture RNA-Seq data (Supplementary Table S4).
Among 118 Aaa strain RS-1 expressed ncRNAs, BLAST search revealed that the total 30 ncRNAs coordinates with Rfam database, while their names and functions were presented in Supplementary Table S4. In details, 7, 11, and 25 ncRNAs were expressed under in vivo, co-culture and in vitro conditions, respectively. Furthermore, the function of ncRNAs may be more associated with the specifically expressed ncRNAs in Aaa strain RS-1, which include 2 in vivo specifically expressed ncRNAs (tRNA and Glycine), 2 co-culture specifically expressed ncRNAs (6S and CRISPR-DR28), and 17 in vitro specifically expressed ncRNAs (5 CRISPR-DR28, 11 tRNAs and 1 suhB).
Validation of Illumina sequence data using qRT-PCR
Illumina sequence data were validated by comparing the gene's total transcript level estimated from the RNA-Seq data with quantitative RT-PCR results of 25 selected T6SS genes in Aaa strain RS-1 (Supplementary Table S5). Indeed, the squared correlation coefficient r2 value between the two methods was 0.78 for the in vivo vs. in vitro expression and 0.69 for co-culture vs. in vitro expression, respectively (Supplementary Figure S3). The high correlation observed in this study verified the efficiency and robustness of the RNA-Seq transcriptome of Aaa strain RS-1 cultivated under three different conditions.
Validation of in vivo differential expression using T6SS mutants
The role of in vivo differential expression from RNA-Seq data was further validated by constructing knockout mutants of the 13 selected T6SS genes and compared their rice pathogenicity with that of wild type strain RS-1. In general, the result of T6SS mutants gave a strong support for this RNA-Seq data. Indeed, 11 mutants out of 12 in vivo differentially expressed T6SS genes that include 1 up- and 11 down-regulated genes lost or reduced the pathogenicity to rice plants compared to wild type strain RS-1, while neither in vivo expression change nor virulence loss of mutant for pppA gene (Table 2). Overall, the loss of virulence for knockout mutants of T6SS genes emphasized the importance of these genes in bacterial pathogenicity.
Table 2. Validation of in vivo differential expression from transcriptional profile of Acidovorax avenae subsp. avenae strain RS-1 by the changes in pathogenicity of T6SS gene mutants constructed in this study.
| NGEVa of RNA-Seq | Validation by mutation | ||||
|---|---|---|---|---|---|
| Locus Tag | T6SS components | In vitro | In vivo | Gene | Pathogenicity |
| Acav_1267 | ClpB | 5.9 | 2 | ΔclpB | loss |
| Acav_1504 | Hcp | 123.6 | 24.85 | Δhcp | loss |
| Acav_1905 | VgrG-8 | 11.95 | 5 | ΔvgrG | loss |
| Acav_1512 | IcmF | 6.4 | 1.8 | ΔicmF | loss |
| Acav_1519 | DUF879 | 5.85 | 2.6 | ΔdotU | loss |
| Acav_1511 | DotU/OmpA/MotB | 11.05 | 4.4 | ΔompA | loss |
| Acav_2399 | VgrG-3 | 4.05 | 0 | ΔimpA | loss |
| Acav_1516 | Duf877/EvpB | 68.35 | 22.55 | ΔevpB | decrease |
| Acav_0662 | VgrG-2 | 13.7 | 0 | ΔvgrG | loss |
| Acav_0298 | VgrG-1 | 1.65 | 3.7 | ΔvgrG | loss |
| Acav_1513 | ImpM | 4.85 | 1.9 | ΔimpM | loss |
| Acav_4620 | PppA | 8.95 | 6.1 | ΔpppA | Not loss |
| Acav_1509 | Lip | 12.65 | 3.55 | Δlip | Not loss |
aNGEV: Normalized gene expression value. The detailed knockout of these T6SS genes and their pathogenic function will be published in another paper.
Discussion
This study systematically examined and compared transcriptomes of Aaa strain RS-1 cultivated under three different conditions using RNA-Seq technologies. In general, this result indicated the difference in the transcriptional response of bacterial pathogen to environmental bacteria and its host. Indeed, a slight transcriptional response was observed when co-cultured with rice rhizobacterium. However, in vivo infection caused a strong transcriptional response, while a large number of genes were differentially expressed in vivo based on comparative genomic and transcriptomic analyses. This result provided us a new clue to understand microbial adaption and pathogenesis.
These results demonstrated the reliability of using strand-specific Illumina-based RNA-Seq for the transcriptomics studies of Aaa strain RS-1 cultivated under three different conditions. Indeed, comparative analyses revealed largely consistent global profiles for each RNA sample regardless of sequencing depth, while the high percentage of reads was assignable to the genome. Furthermore, the high correlation was found between two biological replicates for each condition, which are generally considered to be required for RNA-Seq data analysis. In addition, the reliability of expression data obtained by RNA-Seq was also supported by quantitative real-time PCR of the selected T6SS genes. Taken together, these results strongly supported that our RNA-Seq data is reliable for further analysis of differential gene expressions.
Many studies have showed that bacteria adapt to the host and environmental conditions by altering their patterns of gene expression5,6,9. Although the cut-offs between low, medium, and high transcript levels were arbitrary, the in vitro gene number of each category was slightly changed by co-culture, supporting the result that the growth of Aaa strain RS-1 was unaffected by co-culture (data not shown). However, the gene number of each category in vitro was markedly changed as compared to that of in vivo, suggesting differential gene expressions between the two conditions. Therefore, it could be inferred that the RNA-Seq data in this study will provide much information for understanding of bacterial pathogenesis.
Among the in vivo differentially expressed genes, some genes such as those involved in secretion systems, GIs and ncRNAs, have been well reported for their role in pathogenicity of plants and animal as well as human bacterial pathogen20,21. Indeed, the in vivo differential expression was observed in a large number of secretion systems genes, in particular T3SS and T6SS genes. However, the expression of some T3SS genes was unaffected or down-regulated by in vivo infection in this study. This is different with previous studies, which revealed that T3SS genes are widely expressed in a variety of bacterial pathogens17,21,22. Furthermore, the down-regulation of in vivo expression for majority of T6SS genes was contrast with other studies, which revealed that T6SS genes were often in vivo induced in a variety of bacterial pathogen of animals and humans23,24,25. These results suggest the complexity of secretion systems genes expression in the adaption of bacterial pathogens to the host environment.
Of these differentially expressed GI genes, vgrG (Acav_0662), encoding a type VI secretion-associated protein, was noted. This gene was highly expressed in vitro, but was completely inhibited by both in vivo and co-culture. Furthermore, the gene mutant constructed in this study significantly reduced pathogenicity compared to the wild type strain, which was consistent with previous studies that found that GIs have been reported to be involved in bacterial pathogenicity26,27. Although it is still unclear about the potential importance of the other differentially expressed GI genes in the pathogenicity of Aaa strain RS-1 to rice, the in vivo expression change suggest that these genes in Aaa strain RS-have a role in the response to host.
Our understanding about ncRNAs has noticeably increased in the last decade although the exact function of many ncRNAs is still a mystery19,28. ncRNAs are RNAs that are transcribed, but not translated into protein. They include well-characterized transfer RNAs and ribosomal RNAs, snRNAs, snoRNAs, and miRNAs, as well as a plethora of new ncRNAs28. From our study, it could be inferred that the differentially expressed ncRNAs in Aaa strain RS-1 may have a role in the response to host and other bacterium. Furthermore, many ncRNAs were unable to be confirmed from Rfam database in this study, which may be considered as novel ncRNAs as these are undescribed by any other studies.
The role of differentially expressed T6SS genes in response to host was further justified by comparing the pathogenicity of the deletion mutants constructed in this study with the wild type strain. In general, the mutation of in vivo non-active pppA gene did not affect the pathogenicity to rice compared to the wild type strain, while the loss of pathogenicity was found for the mutants of 11 in vivo differentially expressed genes, including 10 in vitro highly expressed genes, and the vgrG-1 gene, which was up-regulated in vivo and down-regulated in vitro. However, in contrast with the above differentially expressed genes, this result also revealed that the in vivo down-regulated gene lip still display a role in pathogenicity when mutagenized.
No change in pathogenicity of the lip mutant to rice revealed the discrepancy between the in vivo expression response and the function of gene. Indeed, in vivo differential expression revealed the response of T6SS cluster genes to host, while the T6SS has been proposed to function in protein secretion, which is an essential process for a variety of biological functions. Furthermore, the loss in virulence of the mutants may be due to both a direct reduction in the secretion of effector and an indirect disruption in structure and function of the T6SS machinery, influencing the secretion of other proteins, eventually resulting in the reduction in the pathogenicity to rice.
However, it is still generally unclear about the role of each T6SS cluster gene in structure and function of the T6SS machinery, which, taken as a whole, has been proposed to be associated with the virulence of a variety of bacteria. Interestingly, this study revealed the difference in biological functions between T6SS genes. Indeed, the mutation of all differentially expressed (including up- and down-regulated) T6SS genes except lip gene caused the reduction in growth rate and biofilm formation, which may, at least partially, attribute to the reduction in pathogenicity to rice. In contrast, the mutation of differentially expressed lip gene and in vivo non-active pppA gene did not cause the reduction in growth rate and biofilm formation compared to the wild type strain (data not shown). Therefore, it could be suggested that the change in pathogenicity to rice may be more likely due to an indirect disruption in structure and function of the T6SS machinery by the mutation of differentially expressed genes, influencing the secretion of proteins involved in functions such as the growth, and biofilm formation, eventually resulting in the loss of virulence.
In summary, this study first examined the transcriptional profile of Aaa strain RS-1 cultivated under three different conditions, which provided a basis for analysis of biological function such as pathogenesis. Indeed, RNA-Seq data revealed in vivo differential expression of a large number of genes in particular many important virulence-related genes such as those involved in secretion systems, GIs and ncRNAs, while the role of differentially expressed genes was highlighted by the loss of virulence for T6SS gene mutants. Overall, this study clearly indicated that RNA-Seq-based transcriptome analysis of rice bacterial pathogen during infection and co-culture produced a robust, sensitive, and accessible data set for identification of specific cues in bacteria-host or bacteria-bacteria and novel genes for bacterial pathogenicity and commensalism.
Methods
Strains and growth conditions
Aaa strain RS-1 and B. seminalis strain R456 were isolated from diseased rice plants2,7 and rice rhizosphere12,29, respectively, and were stored in 20–30% sterile glycerol at −80°C. The samples of Aaa strain RS-1 for in vitro and in vivo analysis were prepared as described before1. The co-culture analysis was conducted according to Di Cagno et al.30 and Ruiz et al.31. Briefly, Aaa strain RS-1 and B. seminalis strain R456 were inoculated and incubated in chambers of a double culture vessel apparatus separated by a 0.22-μm membrane filter (Millipore Isopore™). In order to avoid the possible contamination during in vivo and co-culture operation, all bacterial samples were further confirmed based on the sequence analysis of 16S-rDNA2.
RNA harvesting, mRNA purification and cDNA synthesis
RNA extraction and purification were performed with RNeasy Mini Kit (Qiagen). RNA samples were treated with DNaseI and purified by phenol-chloroform-isoamyl alcohol extraction and ethanol precipitation. In order to remove rRNA and polyadenylated mRNA, ten micrograms from each total RNA sample was subsequently treated with the MICROB Express Bacterial mRNA Enrichment kit (Ambion), RiboMinus™ Transcriptome Isolation Kit (Bacteria) (Invitrogen) and the Sera-mag Magnetic Oligo(dT) Beads (Thermo) following the manufacturer's instructions. Samples were resuspended in 15 μL of RNase-free water. mRNA enriched RNAs were chemically fragmented to the size range of 150–250 bp using 1 × fragmentation solution (Ambion) for 2.5 min at 94°C. Double stranded cDNA was generated using the SuperScript Double-Stranded cDNA Synthesis Kit (Invitrogen). Briefly, each mRNA sample was mixed with 100 pmol of random hexamers (Invitrogen), incubated at 65°C for 5 min, chilled on ice, mixed with 4 μl of First-Strand Reaction Buffer (Invitrogen), 2 μl of 0.1 M DTT, 1 μl of 10 mM RNase-freed NTPmix, 1 μl of SuperScript III reverse transcriptase (Invitrogen), and incubated at 50°C for 1 h. To synthesize the second strand, the following Invitrogen reagents were added: 51.5 μl of RNase-free water, 20 μl of second-strand reaction buffer, 2.5 μl of 10 mM RNase-free dNTP mix, 50 U E. coli DNA Polymerase, 5U E. coli RNase H (Invitrogen), and incubated at 16°C for 2.5 h.
Library preparation and Illumina sequencing
Paired End Sample Prep kit (Invitrogen) was used for RNA-Seq library generation according to the manufacturer's instructions as follows: Fragmented cDNA was end-repaired, ligated to Illumina adaptors, and amplified by 18 cycles of PCR. Paired-end 100-bp reads were generated by sequencing using the Illumina Hiseq2000 Genome Analyzer instrument.
RNA-Seq data analysis
After removing the low quality reads and adaptors, RNA-Seq reads were aligned to the corresponding Aaa type strain ATCC 19860 (accession number CP002521) genome using Tophat 2.0.732,33, allowing for a maximum of two mismatch, for the genome sequence of Aaa strain RS-1 is yet not completed7. If reads mapped to more than one location, only the highest score one was kept. The reads that map to rRNA and tRNA regions were removed from further analysis. RPKM (Reads Per Kilobase per Million mapped reads) expression values were further calculated with Cufflinks 2.0.233. Principal component analysis (PCA) was done on RPKM-based expression values. In addition, according to the cut-offs of Gene Expression Index (GEI) described in Nagalakshimi et al.13, genes were classified into four categories. Cuffdiff was then used to determine the differential expression by comparing transcript abundances between pairs of duplicate experiments. Significant differential expressed genes (FDR value < 10−2 and at least two fold changes) were selected for further analysis.
Genome-wide transcriptome analysis of secretion systems
Bacterial pathogenesis relies mainly on the activity of proteins secreted by a variety of secretion systems, including the well documented type I to type V secretion systems34,35,36,37,38 and a recently discovered Type VI secretion system (T6SS)39,40. The components and locations of secretion systems homologs in Aaa strain RS-1 were determined by BLASTN, BLASTP and TBLASTX searches of strain ATCC19860's T1SS-T6SS against strain RS-1 genome. Furthermore, the role of secretion systems in host pathogenicity and bacterial commensalism were determined by comparing RNA-Seq data of Aaa strain RS-1 in vivo and co-culture conditions to that of in vitro conditions, respectively.
Genome-wide transcriptome analysis of genomic islands
Genomic islands (GIs) that related to gene horizontal transfer have been found to play an important role in pathogenicity of a variety of bacterial pathogen26,27. Gene components of GIs in Aaa strain RS-1 were identified by retrieving the pre-computed GIs of Aaa strain ATCC19860 from strain RS-1 genome. GIs of Aaa strains ATCC19860 were analyzed by using IslandViewer software web server, which utilizes two sequence composition-based approaches SIGI-HMM and IslandPath-DIMOB. Moreover, the association of GIs with host pathogenicity and bacterial commensalism was determined by comparing the expression level of each identified GI gene in Aaa strain RS-1 cultured in vivo and in co-culture conditions with that of in vitro conditions, respectively, based on the RNA-Seq data.
Genome-wide transcriptome analysis of non-coding RNAs
Noncoding (nc) RNA genes are reported to work directly as structural or regulatory RNAs instead of mRNA that encodes proteins19,28. Genes encoding these untranslated RNA molecules are present in the genomes of many Gram negative bacteria playing role in virulence, niche adaption and so on. From both genome and RNA-Seq data of Aaa strain RS-1, computational prediction analysis of ncRNAs were performed by RNAspace41, SIPHT42, and RNAz43. Final list of putative ncRNAs was made from the nucleotides with >50 bp. Comparative analysis of ncRNAs between Aaa strain ATCC19860 and Acidovorax avenae subsp. citrulli strain AAC01 was also conducted. Total no. of ncRNAs under in vivo, in vitro and co-culture was also explored as described by Yoder-Himes et al.44. Briefly, all genes with read coverage in regions having coverage higher than the least expressed 20% were further analyzed. To settle on the borders of ncRNAs within each candidate intergenic region, a sliding window of 20 bp was used to optimize subregion continuity of expression, requiring the lowest expression to be at least 30% of the highest expression in the range (or higher than the expression of the medium of the gene coverage). Candidates sized 100 bp or more were further considered. To prevent misclassification of untranslated regions (UTRs) as ncRNAs, candidates having expression similar to one of the flanking genes were discarded.
Consistency of qRT-PCR with RNA-Seq profile
RNA-Seq data was confirmed by examining the expression of 25 T6SS genes by qRT-PCR. Primers for qRT-PCR (Supplementary Table S6) of 25 T6SS genes were designed using Premier Primer 5 (Premier Biosoft Int., Palo Alto, CA, USA), while 16S-rRNA gene was used as the reference2. Total RNA of Aaa strain RS-1 under in vitro, in vivo and co-culture was extracted as described above. The cDNA was synthesized with a PrimeScript™ RT reagent Kit with gDNA Eraser (Perfect Real Time) (TaKaRa, Dalian, China) and was then used directly as the template for qRT-PCR using a SYBR® Premix Ex Taq™ (Tli RNaseHPlus) (TaKaRa, Dalian, China) following the manufacturer's instruction and qRT-PCR was performed on ABI Prism 7500 sequence detection system (Applied Biosystems, USA). Normalized expression levels of the target gene transcripts were calculated relative to the 16S-rRNA gene using the ΔΔCt method, where CT is the threshold cycle. Each result represents the average of three independent determinations. The amplification of qRT-PCR was initiated by denaturation at 95°C 30 s; followed by 40 cycles at 95°C 5 s, 60°C 34 s; with a final standard melting curve stage. After optimization, the threshold value was finally optimizing at 0.22, average relative concentrations were calculated using Microsoft Excel. In addition, the consistency of the two different techniques was determined according to the method of Lee et al.45 by calculating the Squared Pearson correlation coefficient (r2) between the RNA-Seq data and qRT-PCR results of 25 T6SS genes expression.
Virulence of in vivo differentially expressed T6SS genes
The in vivo induced differential gene expression suggests that these genes have a role in the response to its host, which could be further validated by constructing various deletion mutations to examine their role in bacterial pathogenesis. Among the differentially expressed genes, the T6SS genes were selected to be mutagenized for the newly recognized secretion system has been shown to have versatile roles in virulence, symbiosis, interbacterial interactions, and antipathogenesis, which will make it more likely to find the phenotypic change between mutants and the wild type strain. In addition, various types (up-regulation, down-regulation and no significant change) of in vivo expression responses have been observed between T6SS cluster genes, which makes it interesting to examine the function of T6SS cluster genes with different expression patterns. In brief, in-frame deletion of T6SS genes were performed by homologous recombination on the background of Aaa strain RS-1 as described by Liu et al.4. Pathogenicity of wild type and mutants to rice was evaluated by examining the emergence of rice seeds and the height of rice seedlings, which was carried out in the perlite substrate according to the method of Li et al.2. Each treatment had three replicates and each contains 10 rice seeds. The experiment was repeated twice.
Author Contributions
B.L. and M.I. contributed equally to this work. B.L. and G.C.S. supervised the work. M.I., Z.Q.C. and M.Y.G. performed the experiments. M.I., F.X. and M.K. analyzed the data. All authors contributed to the writing of manuscript.
Supplementary Material
Supplementary information
Supplementary Table S1
Supplementary Table S2
Supplementary Table S3
Supplementary Table S4
Supplementary Table S5
Supplementary Table S6
Acknowledgments
This work was supported by Zhejiang Provincial Natural Science Foundation of China (R13C140001; Y3090150), National Natural Science Foundation of China (31371904), Zhejiang Provincial Project (2014C32010), the Fundamental Research Funds for the Central Universities, the Agricultural Ministry of China (nyhyzx 201303015; 201003029; 201003066), Key Subject Construction Program of Zhejiang for Modern Agricultural Biotechnology and Crop Disease Control (2010DS700124-KF1101; - KF1203; - KF1309; - KF1410) and German Research Foundation (KU2679/2-1; BU890/21- 1).
Accession numbers: RNA-Seq raw data files are accessible through the GEO Series accession number-GPL17669: GSM1220690-In vitro Rep 1; GSM1220691-In vitro Rep 2; GSM1220692-In vivo Rep 1; GSM1220693-In vivo Rep 2; GSM1220694-Co-culture Rep 1; GSM1220695-Co-culture Rep 2.
References
- Ibrahim M. et al. Differential expression of in vivo and in vitro protein profile of outer membrane of Acidovorax avenae subsp. avenae. PLoS ONE 7, e49657 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B. et al. Occurrence of bacterial brown stripe of rice in soilless culture system caused by Acidovorax oryzae in China. J. Gen. Plant Pathol. 77, 64–67 (2011). [Google Scholar]
- Wang Y. L. et al. Differentiation in MALDI-TOF MS and FTIR spectra between two closely related species Acidovorax oryzae and Acidovorax citrulli. BMC Microbiol. 12, 182 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. et al. Characterization of pilP, a gene required for twitching motility, pathogenicity, and biofilm formation of Acidovorax avenae subsp. avenae RS-1. Eur. J. Plant Pathol. 134, 551–560 (2012). [Google Scholar]
- Garbeva P., Silby M. W., Raaijmakers J. M., Levy S. B. & de Boer W. Transcriptional and antagonistic responses of Pseudomonas fluorescens Pf0-1 to phylogenetically different bacterial competitors. ISME J. 5, 973–985 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandlik A. et al. RNA-Seq-Based Monitoring of Infection-Linked Changes in Vibrio cholerae Gene Expression. Cell Host & Microbe 10, 165–174 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie G. L. et al. Genome sequence of the rice pathogenic bacterium Acidovorax avenae subsp. avenae RS-1. J. Bacteriol. 193, 5013–5014 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto A. C., Melo-Barbosa H. P., Miyoshi A., Silva A. & Azevedo V. Application of RNA-Seq to reveal the transcript profile in bacteria. Genet. Mol. Res. 10, 1707–1718 (2011). [DOI] [PubMed] [Google Scholar]
- Dotsch A. et al. The Pseudomonas aeruginosa Transcriptome in Planktonic Cultures and Static Biofilms Using RNA Sequencing. PLoS ONE 7, e31092 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas B. J., Chin M., Nusbaum C., Birren B. W. & Livny J. How deep is deep enough for RNA-Seq profiling of bacterial transcriptomes? BMC Genomics 13, 734 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R. et al. RNA-seq based transcriptional map of bovine respiratory disease pathogen “Histophilus somni 2336”. PloS ONE 7, e29435 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B. et al. Phenotypic and molecular characterization of rhizobacterium Burkholderia sp. strain R456 antagonistic to Rhizoctonia solani, sheath blight of rice. World J. Microb. Biot. 27, 2305–2313 (2011). [Google Scholar]
- Nagalakshimi U. et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320, 1344–1349 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buell C. R. et al. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. U. S. A. 100, 10181–10186 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama H., Akiba K., Hori H., Ando T. & Nakae T. Tat pathway-mediated translocation of the sec pathway substrate protein MexA, an inner membrane component of the MexAB-OprM xenobiotic extrusion pump in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 54, 1492–1497 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman D. S. et al. A functional screen for the type III (Hrp) secretome of the plant pathogen Pseudomonas syringae. Science 295, 1722–1726 (2002). [DOI] [PubMed] [Google Scholar]
- Jock S., Donat V., López M. M., Bazzi C. & Geider K. Following spread of fire blight in Western, Central and Southern Europe by molecular differentiation of Erwinia amylovora strains with PFGE analysis. Environ. Microbiol. 4, 106–114 (2002). [DOI] [PubMed] [Google Scholar]
- Kanehisa M. & Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coenye T. et al. Identification of putative noncoding RNA genes in the Burkholderia cenocepacia J2315 genome. FEMS Microbiol. Lett. 276, 83–92 (2007). [DOI] [PubMed] [Google Scholar]
- Lee K. et al. A Genome-Wide Survey of Highly Expressed Non-Coding RNAs and Biological Validation of Selected Candidates in Agrobacterium tumefaciens. PLoS ONE 8, e70720 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dow J. M. et al. Biofilm dispersal in Xanthomonas campestris is controlled by cell-cell signaling and is required for full virulence to plants. Proc. Natl. Acad. Sci. U. S. A. 100, 10995–11000 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z. M. et al. Harpin, elicitor of the hypersensitive response produced by the plant pathogen Erwinia amylovora. Science 257, 85–88 (1992). [DOI] [PubMed] [Google Scholar]
- Hood R. D. et al. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host & Microbe 7, 25–37 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz S., Hood R. D. & Mougous J. D. What is type VI secretion doing in all those bugs? Trends Microbiol. 18, 531–537 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell A. B. et al. Type VI secretion delivers bacteriolytic effectors to target cells. Nature 475, 343–349 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker J. & Kaper J. B. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54, 641–679 (2000). [DOI] [PubMed] [Google Scholar]
- Dobrindt U., Hochhut B., Hentschel U. & Hacker J. Genomic islands in pathogenic and environmental microorganisms. Nat. Rev. Microbiol. 2, 414–424 (2004). [DOI] [PubMed] [Google Scholar]
- Liu J. & Carnilli A. A broadening world of bacterial small RNAs. Curr. Opin. Microbiol. 13, 18–23 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L. X. & Xie G. L. Diversity and distribution of Burkholderia cepacia complex in the rhizosphere of rice and maize. FEMS Microbiol. Lett. 266, 231–235 (2007). [DOI] [PubMed] [Google Scholar]
- Di Cagno R., De Angelis M., Coda R., Minervini F. & Gobbetti M. Molecular adaptation of sourdough Lactobacillus plantarum DC400 under co-cultivation with other lactobacilli. Res. Microbiol. 160, 358–366 (2009). [DOI] [PubMed] [Google Scholar]
- Ruiz L., Sanchez B., de los Reyes-Gavilan C. G., Gueimonde M. & Margolles A. Coculture of Bifidobacterium longum and Bifidobacterium breve alters their protein expression profiles and enzymatic activities. Int. J. Food Microbiol. 133, 148–153 (2009). [DOI] [PubMed] [Google Scholar]
- Trapnell C., Pachter L. & Salzberg S. L. TopHat: discovering splice junctions with RNA-seq. Bioinformatics 25, 1105–1111 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L., Paulsen I. T., Tchieu J., Hueck C. J. & Saier M. H. Phylogenetic analyses of the constituents of Type III protein secretion systems. J. Mol. Microbiol. Biotechnol. 2, 125–144 (2000). [PubMed] [Google Scholar]
- Craig L., Pique M. E. & Tainer J. A. Type IV pilus structure and bacterial pathogenicity. Nat. Rev. Microbiol. 2, 363–378 (2003). [DOI] [PubMed] [Google Scholar]
- Filloux A. The underlying mechanisms of type II protein secretion. Biochim. Biophy. Acta 1694, 163–179 (2004). [DOI] [PubMed] [Google Scholar]
- Kondo M. et al. Genetic organization of the hrp gene cluster in Acidovorax avenae strain N1141 and a novel effectors protein that elicits immune responses in rice (Oryza sativa L.). Biosci. Biotechnol. Biochem. 76, 129–138 (2012). [DOI] [PubMed] [Google Scholar]
- Korotkov K. V., Sandkvist M. & Hol W. G. J. The type II secretion system: biogenesis, molecular architecture and mechanism. Nat. Rev. Microbiol. 10, 336–351 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jani A. J. & Cotter P. A. Type VI secretion. Not just for pathogenesis anymore. Cell Host & Microbe 8, 2–6 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler M. & Mekalanos J. J. Type 6 Secretion Dynamics Within and Between Bacterial Cells. Science 337, 815 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cros M. J. et al. RNAspace.org: An integrated environment for the prediction, annotation, and analysis of ncRNA. RNA 17, 1947–1956 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoo J. S., Chai S. F., Mohamed R., Nathan S. & Firdaus-Raih M. Computational discovery and RT-PCR validation of novel Burkholderia conserved and Burkholderia pseudomallei unique sRNAs. BMC Genomics 13, S13 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber A. R., Findeiss S., Washietl S., Hofacker I. L. & Stadler P. F. RNAz 2.0: improved noncoding RNA detection. Pac. Symp. Biocomput. 15, 69–79 (2010). [PubMed] [Google Scholar]
- Yoder-Himes D. R. et al. Mapping the Burkholderia cenocepacia niche response via high-throughput sequencing. Proc. Natl. Acad. Sci. U. S. A. 106, 3976 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. et al. Accurate quantification of transcriptome from RNA-Seq data by effective length normalization. Nucleic Acids Res. 39, e9 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information
Supplementary Table S1
Supplementary Table S2
Supplementary Table S3
Supplementary Table S4
Supplementary Table S5
Supplementary Table S6