Abstract
Use of mRNA-based vaccines for tumour immunotherapy has gained increasing attention in recent years. A growing number of studies applying nanomedicine concepts to mRNA tumour vaccination show that the mRNA delivered in nanoparticle format can generate a more robust immune response. Advances in the past decade have deepened our understanding of gene delivery barriers, mRNA’s biological stability and immunological properties, and support the notion for engineering innovations tailored towards a more efficient mRNA nanoparticle vaccine delivery system. In this review we will first examine the suitability of mRNA for engineering manipulations, followed by discussion of a model framework that highlights the barriers to a robust anti-tumour immunity mediated by mRNA encapsulated in nanoparticles. Finally, by consolidating existing literature on mRNA nanoparticle tumour vaccination within the context of this framework, we aim to identify bottlenecks that can be addressed by future nanoengineering research.
1. Introduction
mRNA is a biomolecule built by nature and shaped through evolution as a transient messenger of genetic information. The attractiveness of mRNA delivery is founded on its potential for higher transfection efficiencies in non-dividing cells (no nuclear entry required), rapid expression, predictable kinetics as well as higher safety profile compared to plasmid DNA.1 mRNA delivery is gaining attention because of a gradual acceptance from the research community that in vitro transcribed mRNA is not as biologically labile as initially thought. Improved understanding of mRNA stability2 in the last decade has led to optimized designs of in vitro transcribed mRNA. Structural features such as 3’ globin UTR, anti-reverse cap analogue, polyA tail as well as use of modified nucleotides have all led to enhanced mRNA translation.3 Such improvements have made an impact in the clinic because dendritic cells (DCs) are transfected efficiently with in vitro transcribed mRNA and subsequently applied as a tumour vaccine. This has led to the development of mRNA-based cellular therapy approaches4, 5 as well as direct in vivo injection of mRNA in naked6, 7 and nanoparticle formats.8–13
With established clinical infrastructure for the manufacturing and quality control of GMP-grade mRNA, there is much incentive to broaden the application of mRNA through biomedical engineering approaches. The most common manipulation of mRNA is its encapsulation in nanoparticles for enhanced delivery efficiencies. While there is a handful of published work on mRNA nanoparticle-mediated tumour vaccination in preclinical studies, there is currently no mRNA nanoparticle vaccine in the clinical pipeline, thus making this a fertile direction for nanomedicine research. There is also growing interest in gene delivery researchers who are venturing into the mRNA arena, as well as mRNA vaccinologists who are searching for effective ways to deliver mRNA to antigen presenting cells in vivo. It would seem that after almost two decade of advances made in nucleic acid delivery systems of RNAi and DNA that we would have solved the problem of mRNA delivery to dendritic cells, but we have not, at least not yet. Nevertheless, a good understanding of gene delivery barriers has been achieved in the gene delivery field and many tools have also been developed to address these delivery barriers. In addition, the immunological properties of mRNA, signalling pathways through pattern-recognition receptors (e.g. TLR, RLRs) and their implications on immune responses have also been elucidated in recent years. Hence, we are at an appropriate juncture where collusion of engineering technologies and biomedical sciences can help fulfil the translational potential of mRNA.
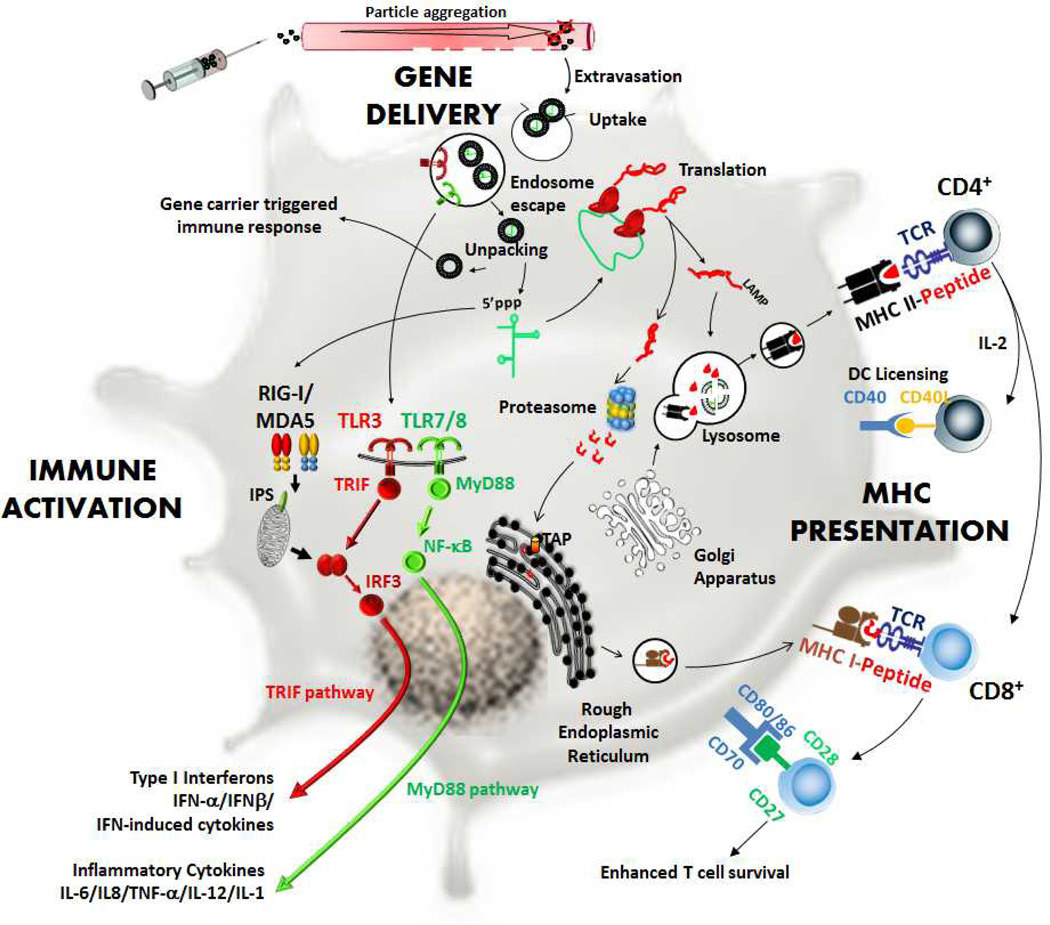
In this review we will first discuss whether mRNA is amenable for engineering manipulation. We will then draw on knowledge of gene delivery and mRNA immunotherapy to put together a frame work for mRNA nanoparticle tumour vaccination. This frame work is a set of delivery and biological barriers highlighting rate limiting steps to antigen presentation mediated by mRNA nanoparticle vaccine: gene delivery (extracellular and intracellular), immune activation, and MHC presentation (Fig. 1). We will describe and discuss these barriers within the context of genetic immunization with examples from published studies on mRNA nanoparticle tumour vaccination and DC transfection. Studies pursuing mRNA nanoparticle vaccination have often been performed in niche areas (either delivery-focused or immunology-focused). It is also the aim of this review to cast these studies together in an overall picture to help identify bottlenecks to mRNA nanoparticle-mediated immunization.
Fig. 1.
Proposed frame work for mRNA nanoparticle mediated tumour vaccination. A combination of three overlapping processes determines immune response: gene delivery, immune activation, and major histocompatibility complex (MHC) presentation. In this framework, nanoparticles administered into the body have to overcome extracellular barriers from the site of administration to antigen presenting cells. After cell uptake, nanoparticles have to escape the endosome and mRNA has to unpack from the gene carrier and enter the protein translation pathway. At the same time, through pattern recognition receptors, mRNA nanoparticle needs to mediate immune activation (via TLR3,7/RIG-I,MDA5) so that cytokines are secreted for ensuing adaptive immune responses. Protein translated from delivered mRNA molecule needs to enter both the MHC I (via proteasome) and II (via lysosome) processing pathways so that antigen peptides can be presented on both MHC I and II molecules.
2. Is mRNA stable for biomedical manipulation?
From the delivery efficiency perspective, one key advantage of using mRNA for gene therapy is that transfection can be achieved without the need for gene entry into the nucleus. Consequently transfection of non-dividing cells (e.g. DCs, T cells, neurons), which were previously considered unattractive candidates for non-viral gene therapy, are now actively being investigated1, 14, 15. This rationale and other advantages for using mRNA as an antigen encoding molecule have also been well reviewed16–20. Alongside advances in molecular biology, substantial progress has also been made to understand the regulation of mRNA biological stability,2 and improved understanding on various mechanisms of mRNA degradation has led to a focused effort to optimize the mRNA structure for enhanced intracellular stability and increased protein translation capabilities. These studies demonstrate the importance of an optimized 5’ cap analogue, a 3’UTR globin sequence and a sufficiently long poly-A tail in the structural makeup of the mRNA, leading to increased gene expression in dendritic cells.18 More recently, investigations into the TLR-mediated immunogenicity of mRNA have led to the development of modified mRNA. It was initially observed that endogenous RNA were significantly less potent in triggering TLR-mediated immune response than in vitro-transcribed RNA.21 It was then discovered that modified nucleotides (e.g. pseudouridines) contained within endogenous RNA not only suppressed immune responses, but also enhanced protein translation in mRNA transcribed with these modified nucleotides22. The mechanism of enhancement is predominantly caused by a lack of Type I interferon secretion from transfected cells and to a lesser extent by the enhanced intracellular stability of the modified mRNA22. The conclusions make sense since modified nucleotides are also building blocks of structural RNAs such as ribosomes, which are non-immunogenic and relatively resistant to degradation.
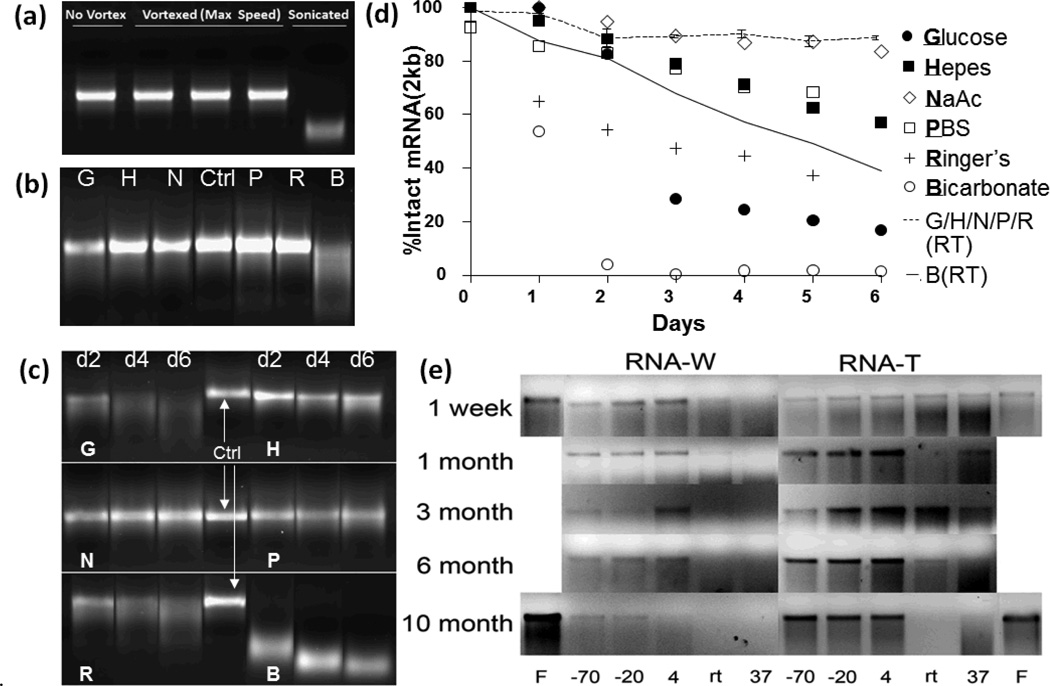
Enhanced biological stability and performance of mRNA in vivo attract the attention of biomedical engineers because of the opportunities to incorporate mRNA into biomaterials or medical devices for therapeutic applications. Many devices that have been designed for DNA delivery, such as nano-/micro- particles, transcutaneous microneedles,24, 25 hydrogels26–28 and other macroformulations,29 have not been well developed for mRNA. The question then becomes whether mRNA is physically and/or chemically stable for manipulation under fabrication conditions. As shown in Fig.2a (unpublished data), mRNA can withstand significant vortex-induced shear stress, but rapidly degrades upon sonication. Freeze-dried mRNA can remain stable for up to 10 months (Fig.2e, column “F”).23 Freeze-drying is also a feasible method to obtain highly concentrated mRNA dissolved in the desired buffer compared to column recovery (e.g. RNeasy kit). Solution stability of naked mRNA is a key piece of information currently missing in the context of the development of controlled release devices such as hydrogels. Degradation is caused by hydrolysis of the phosphodiester bond of mRNA backbone caused by nucleophilic attack from hydroxide ions or 2’-OH group present in the ribose sugar residues of mRNA itself. Notwithstanding commercially available RNA storage solutions of proprietary nature, to develop encapsulation technologies, biomedical engineers need to know the physical stability of in vitro transcribed mRNA in common defined buffers. At present, this information can only be found for distilled water and aqueous trehalose solution (Fig.2e).23 In both buffers, there is no significant compromise in structural integrity when mRNA is stored at or below 4°C. However, the effect of temperature becomes significant at or above room temperature (Fig.2e). In our hands, mRNA at 4°C is stable in all buffers including bicarbonate for up to 6 days (data not shown). This is consistent with published data shown in Fig.2e. We observe that at room temperature, mRNA stability becomes pH dependent. As shown in Fig.2b (unpublished data), mRNA diluted in sodium bicarbonate is completely degraded but remain stable in neutral and acidic buffers. At 37°C, effects of buffer become apparent (Fig.2c, unpublished data). mRNA remains relatively stable in Hepes, PBS and sodium acetate, degrades gradually over time in Ringer’s lactate and 5% glucose (unbuffered) solution, but degrades rapidly in sodium bicarbonate. Using densitometry quantification (Image J), degradation profiles in different buffers at room temperature and 37°C are quantified and plotted on Fig.2d (unpublished data).
Fig. 2.
Solution Stability of mRNA. (a) Ctrl: control, V: mRNA vortexed at max speed for 9 minutes or sonicated (bath type) for 3 minutes and incubated at 4°C overnight (b) Agarose gel electrophoresis of mRNA (200µg/ml) in G,H,N,P,R,B buffers incubated at room temperature for 6 days. (day 6 is shown) G: 5% glucose (unbuffered solution), H: 100mM Hepes, N: 100mM sodium acetate, P: PBS, R: Ringer’s lactate, B: 0.75% sodium bicarbonate. Ctrl: control mRNA stored at −20°C. (c) Agarose gel electrophoresis of mRNA (200µg/ml) in G,H,N,P,R,B buffers incubated at 37°C over 6 days (days 2,4,6 are shown). (d) Degradation profiles of mRNA analyzed on agarose gel (quantified with densitometry analysis). mRNA (200µg/ml) is incubated at 37°C (symbols) or room temperature (lines) in G,H,N,P,R,B buffers (e) Agarose gel electrophoresis of mRNA stored in water (RNA-W) or trehalose (RNA-T, unbuffered solution) over time. “F” indicates freeze-dried RNA.23 (a) – (d) are unpublished data by the authors. (e) is reproduced with permission.23
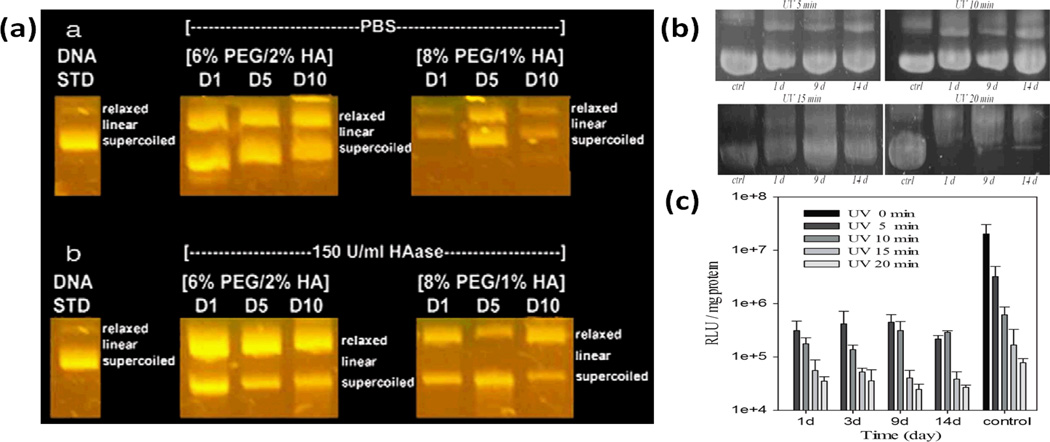
The solution stability of mRNA is indeed lower than that of plasmid DNA. Plasmid DNA’s solution stability has been demonstrated in hydrogels, which typically has over 95% water content. Agarose gel electrophoresis shows that DNA recovered from UVA cross-linked hydrogels remain intact after being left in PBS or TE buffer at 37°C for up to 10 days (Fig.3a)28 and 14 days (Fig.3b)26, respectively. The proportion of DNA in relaxed conformation increased while the supercoiled conformation decreased over time.26, 28 While UV can inactivate plasmid DNA in a dose-dependent manner, changes from coiled to relax conformation did not significantly affect the bioactivity of plasmid DNA released from hydrogels throughout the 14 day duration of the study (Fig.3c).26 Corresponding information on the UV stability of mRNA has yet to emerge.
Fig. 3.
(a) Agarose gel electrophoresis analysis of structural integrity of DNA encapsulated in UV cross-linked PEG-Hyaluronic Acid hydrogel.28 (b) Agarose gel electrophoresis analysis of structural integrity of DNA encapsulated in UV cross-linked Pluronic-Hyaluronic Acid hydrogel.28 (c) Bioactivity of DNA released from Pluronic-Hyaluronic Acid hydrogel cross-linked using different UV intensity.26 Reproduced with permission.26,28
In summary, mRNA is shear resistant and hence it can be more efficiently mixed with gene carriers during formulation to form smaller particles with better reproducibility. In addition, solution stability of mRNA is not a limiting factor in the fabrication of mRNA-based biomedical devices because it is compatible with commonly used buffers and can remain stable at room temperature during the fabrication process. However, it may be limited by hydrolysis at physiological temperature and may not be suitable for long term sustained release applications. This may limit the application of mRNA in medical devices such as micro-needle technology. Hence improving solution stability of naked mRNA at physiological temperature will be a significant advancement in this area.
3. Barriers to Non-Viral mRNA Nanoparticle Tumour Vaccination
mRNA nanoparticle tumour vaccination is a multifaceted process. The first stage implicates the gene delivery process (gene delivery, Fig.1) where nanoparticles encapsulating the mRNA need to overcome extracellular barriers (to reach antigen presenting cells), endocytic barriers (to enter the cell), followed by intracellular barriers (to release the mRNA into the cytoplasm). In the second stage, the protein translated from mRNA has to be optimally presented on major histocompatibility complexes (MHC) molecules (MHC presentation, Fig.1) to activate T cells through the recognition of MHC-peptide complexes. In the third stage, antigen presenting cells need to be immunologically primed (immune activation, Fig.1) to provide the necessary co-stimulatory signals and pro-survival cytokines to ensure the robust development and proliferation of antigen-specific T cells. These three stages constitute a network of barriers which mRNA nanoparticle vaccines have to overcome in order to achieve the desired response, and will be the subject of the following section.
3.1 Gene Delivery
3.1.1 Extracellular Gene Delivery of Nanoparticles to Target Organs
The gene delivery process is the first hurdle to mRNA nanoparticle vaccination (Fig. 1). When mRNA nanoparticles are administered, they need to reach the target organ and be efficiently taken up and expressed by antigen presenting cells. To achieve the former, nanoparticles have to overcome extracellular barriers defined as the impediments to efficient transport of nanoparticles from the point of administration to the antigen presenting cells. To achieve the latter, nanoparticles have to overcome intracellular barriers defined as the ability to escape the endosomes and efficiently unpack to release the mRNA into the cytoplasm (discussed in the next section). The route of administration undoubtedly determines the magnitude of extracellular barrier. As there are many options,30–32 we will focus our discussion only on systemic, subcutaneous and intranasal route of administration, highlighting delivery barriers and ideal nanoparticle properties.
3.1.1.1 Systemic Administration
In systemic delivery, colloidal stability of mRNA nanoparticles is critical to ensure efficient transport through the blood stream to the target organs, preferably the spleen. Colloidal instability in biological fluid is a poorly understood phenomenon. Particle aggregation can be caused by interaction between the cationic nanoparticles and the anionic components in blood. When injected into the systemic circulation, positively charged nucleic-acid nanoparticles aggregate through interaction with erythrocytes and other negatively charged serum proteins such as albumin, IgM, IgG, complement C3. Particle aggregation can also be caused by the heterogeneous zeta potential of the cationic nanoparticles. Ho et al have shown that DNA-polyplexes formulated with Turbofect© (poly(2-hydroxypropyleneimine)) in bulk mixing aggregated rapidly via the second mechanism unless its formulation is confined within picoliter volume droplets generated by a microfluidic device.33 Presumably the nanoparticles assembled in small volume are more uniform in composition and surface charge, therefore less prone to aggregation. Li et al have also shown that the particle size of non-pegylated liposome-protamine-DNA complexes (LPD) increase from 135nm to 647nm after they are mixed with serum.34 Particle aggregation creates a problem because upon reaching the target organ, they become too large to extravasate the blood capillaries through the endothelial fenestrations to get to the underlying cells (Fig. 1).
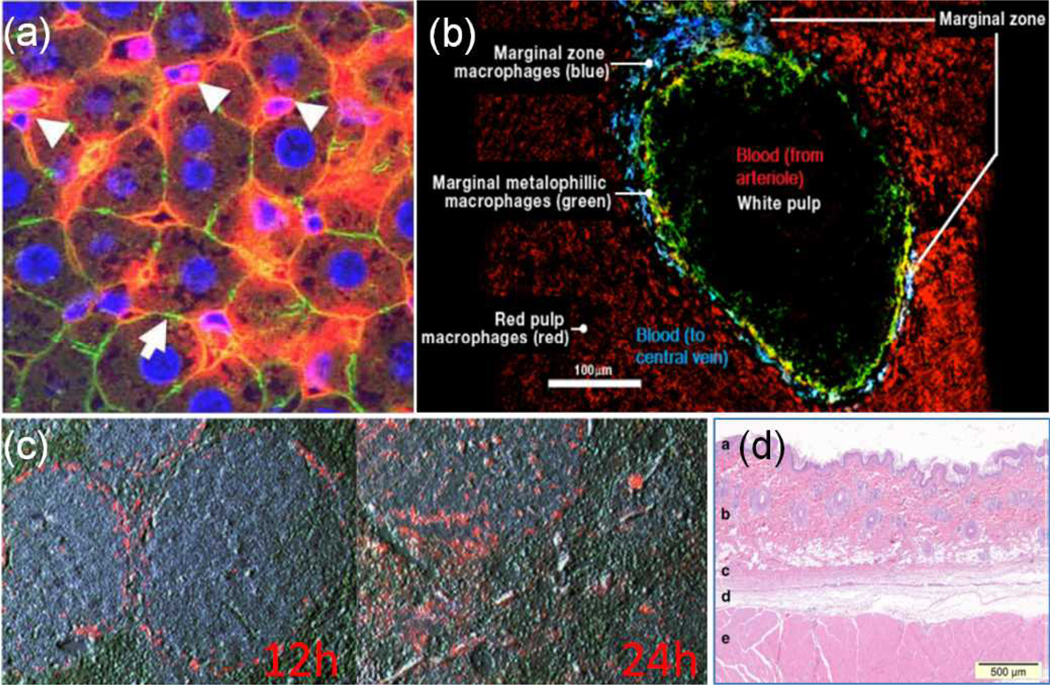
Blood vessels in the liver and spleen are organized into sinusoids, where endothelial fenestrations are wider and blood flows slower. This allows nanoparticles to extravasate into the tissues reaching the inner hepatocytes in the liver or white pulp cells in the spleen (predominantly T cells, DCs and macrophages). Aggregated particles that are too large to pass through the endothelial fenestrations, including those that have been opsonized, will be cleared from the blood stream by Kupffer cells interspersed between hepatocytes in the liver lobules (Fig. 3a) and by marginal zone macrophages (MZf) in the spleen (Fig. 3b). Gene loaded particles administered intravenously are unevenly distributed. For example, 60% and 55% of intravenously injected non-pegylated LPD lipopolyplexes30 and PEI-25K-DNA polyplexes 35 accumulate in the liver. Similarly, 80% and 10% of mRNA lipopolyplexes administered intravenously via the tail vein accumulate in the liver and spleen, respectively.11
As lipid-based nanoparticles are the most frequently evaluated formulation for mRNA immunization, we will further elaborate on its in vivo distribution. Lipid-based nanoparticles that are distributed to the spleen after intravenous administration are sequestered in the splenic marginal zone after 12 hours and gradually move out into the white pulp (T cell region) after 24 hours (Fig. 3c). At the 24 hour time point, an increased infiltration of CD11b+ and CD11c+ cells in the spleen is detected.36 At this time, about 25% of splenic CD11b+ cells and 15% of splenic CD11c+ cells have taken up lipopolyplexes (e.g. LPDs) distributed to the spleen after intravenous administration,36 but only about 4% of CD11c+ cells express the encapsulated GFP mRNA.11 The transfection efficiency of CD11c+ cells is increased (from 4% to 13% based on GFP+ cells) when mRNA lipopolyplexes are functionalized with mannose.11 As macrophages can also present antigens to T cells after being activated via a ROS-mediated pathway37 directly by themselves or indirectly through DCs, the total number of antigen presenting cells targeted by intravenous administration is actually quite reasonable. Still, this may not be an efficient strategy because a significant amount of nucleic acid is actually taken up by the liver and not the spleen. These nucleic acids that are distributed to the liver are also very poorly expressed,34, 35 presumably degraded by Kupffer cells. Similarly, splenic marginal zone macrophages and immature DCs both engender highly degradative endocytic pathways27 that break down intracellular cargoes destined for MHC class II processing through the endosome-lysosome pathways (Fig.1).38 Hence, it is important that nanoparticles can escape from the early/late endosome efficiently, otherwise a large proportion of the mRNA nanoparticles taken up by the DCs and macrophages in the spleen will be degraded. In addition, due to a lobe-sided biodistribution, large nanoparticle doses may be required to achieve adequate splenic transfection, which may lead to gene carrier-associated toxicities.
Nevertheless, many studies have reported that intravenously administered mRNA nanoparticle formulations can activate antigen specific CTLs.8, 10–12, 42, 43. These studies will be reviewed in later sections. Whether such CTL levels are robust enough to achieve therapeutic responses comparable to mRNA-DC vaccine remain to be seen. To summarize, for systemic administration of mRNA nanoparticles, it is clear that the nanoparticles can be taken up by antigen presenting cells in the liver and spleen. However, a large dose is necessary because the particles are distributed poorly to the spleen and DCs are not efficiently transfected. Hence for IV administered mRNA nanoparticle vaccine formulations, improving splenic distribution and transfection efficiency of splenic DCs are necessary measures to advance the field.
3.1.1.2 Subcutaneous Administration
In subcutaneous administration, the objective is to deliver mRNA-encapsulated nanoparticles to lymph node DCs through the lymphatic system (and transfecting them), or alternatively transfect dermal dendritic cells already present in the skin. Although intradermal injection may be more favourable due to the prevalence of dermal DCs, administering a single dose precisely within the thin layer of dermis (‘b’ in Fig. 4d) may be a challenging procedure in mice models, where most experimental vaccination studies are conducted. The subcutaneous space is a non-cellular region (‘d’ in Fig. 4d) found between the skin and skeletal muscles (‘c’ and ‘e’ in Fig. 4b) that can be easily accessed through skin folding. Consequently, extracellular barriers associated with this subcutaneous vaccination are related to poor targeting of dermal dendritic cells and trafficking efficiency of particles to the lymph nodes.
Fig. 4.
(a) Cross section of murine liver sinusoids. Systemically administered non-pegylated fluorescently labeled LPD lipopolyplexes trapped within the sinusoids of liver lobules and taken up by Kupffer cells (white arrows).39 Red: LPD, Blue: DAPI, Green: Phalloidin. (b) Immunostain of mouse spleen’s cross-section showing locations of white pulp (DCs & T cells) and marginal zone. Blood flow direction from white to red pulp. (blue: marginal zone macrophages; green: marginal metalophillic macrophages; red: red pulp macrophages). (c) Localization of non pegylated LPD in mouse spleen 12 and 24 hours after intravenous administration.40 (d) Anatomy of rat skin. (a epidermis, b dermis, c skin (panniculus) muscle, d subcutaneous connective tissue, and e skeletal muscle of trunk).41 Reproduced with permission.41,39,40
Studies conducted to investigate the determinants of lymphatic trafficking reveal that particle size, charge and colloidal stability are factors influencing the rate of transport through the subcutaneous space. Although particles in these studies were made with a wide range of materials, there is a consensus that ultra-small particles (<50nm) are the most efficiently transported44, 45, while those ranging from 100nm to 300nm can also reach the lymph nodes42, 44–50. Proteins (∼7% by mass, predominantly albumins and globulins) are also present in interstitial fluids51 but their concentrations are significantly lower compared to blood and as a result particle aggregation may not be a significant impediment in subcutaneous vaccination.
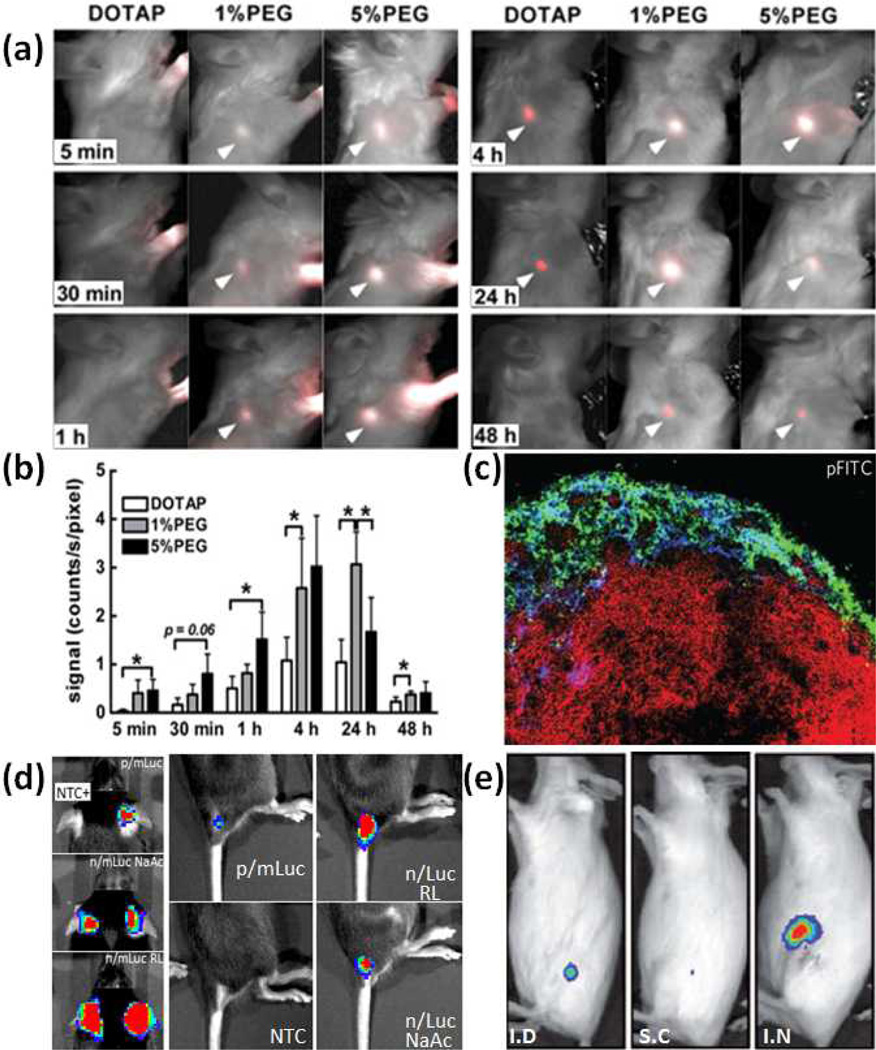
Another observation made by Moghini et al 47 using neutral DOPC liposomes is that increased particle hydrophilicity (via pegylation) enhances trafficking efficiencies through reduced non-specific interactions with interstitial proteins. In a similar study, Zhuang et al 46 show that a high cationic charge on DOTAP liposomes (ζ-potential:+43mV) significantly reduces its trafficking capacity (Fig. 5a) compared to pegylated DOTAP liposomes (ζ-potential:+15mV). Pegylated liposomes appear in the lymph node as early as 30 minutes after administration compared to 4 hours (for non pegylated liposomes), confirming a passive lymphatic transport mechanism (note: Evans Blue injected subcutaneously through the tail base labels the inguinal lymph node within 30 minutes52). Notably in both studies, increasing PEG length accelerates particle transport, but they are not well retained in the lymph node (Fig. 5b). Beside liposomes, polymeric particles formulated using chitosan and heparin (size: 200nm to 1µm, ζ-potential +25mV) can also be found in popliteal lymph nodes 45 minutes after footpad injection (Fig. 5c).48
Fig. 5.
(a) Lymph node trafficking of DOTAP liposomes (0%/1%/5% PEG) over six time periods.46 (b) Distribution of DOTAP Liposomes (0%/1%/5% PEG) in the lymph node over time measured based on radioactivity.46 (c) Popliteal lymph nodes isolated 45 minutes after injection of heparin-polylysine particles. Green: polylysine, Blue:Lyve-1 (lymphatic endothelial cells), Red: B220 (B cell marker).48 (d) C57Bl/6 mice subcutaneously administered with luciferase mRNA encapsulated in nanoparticles (p/mLuc) or in naked form (n/mLuc) diluted in sodium acetate (NaAc) or Ringer’s Lactate (RL). Bioluminescence assayed 4h post administration. NTC: Non Transfected Control.53 (e) Balb/c mice administered with naked luciferase mRNA via intradermal (ID), subcutaneous (SC) and intranodal (IN) routes. Bioluminescence from inguinal lymph node assayed 16h post administration.6 Reproduced with permission.46,6, 48, 53
Although nodal transfection mediated by subcutaneously administered mRNA nanoparticles have not been well studied, transfection efficiency and transgene expression kinetics mediated by mRNA nanoparticles at the subcutaneous site have been reported. mRNA subcutaneously administered in nanoparticle form is not only poorly expressed, but is also expressed over a shorter period of time compared to naked mRNA at the site of injection (Fig. 5d).53
The poor local transfection performance may be caused by entrapment within the extracellular matrix, while naked mRNA can diffuse through the extracellular matrix to reach muscle and dermal cells on each side.54 Although particle properties are characterized (250nm in 50% serum, ζ-potential −15mV), their transport to the lymph node has not been determined. The fact that naked mRNA can mediate high transfection efficiencies via subcutaneous administration shows that it can remain relatively stable in the subcutaneous space. However, it is not stable enough to be transported through lymphatics and expressed in the lymph node (Fig. 5e).6
Despite a lack of direct evidence of nodal DC transfection from subcutaneously administered mRNA nanoparticles, antigen specific CTL cell responses are efficiently induced8, 9, 12 and anti-tumour therapeutic responses have been reported.8 These studies indicate that dermal DCs may play a significant role in the induction of immunity for subcutaneously injected mRNA nanoparticles.
3.1.1.3 Intranasal Administration
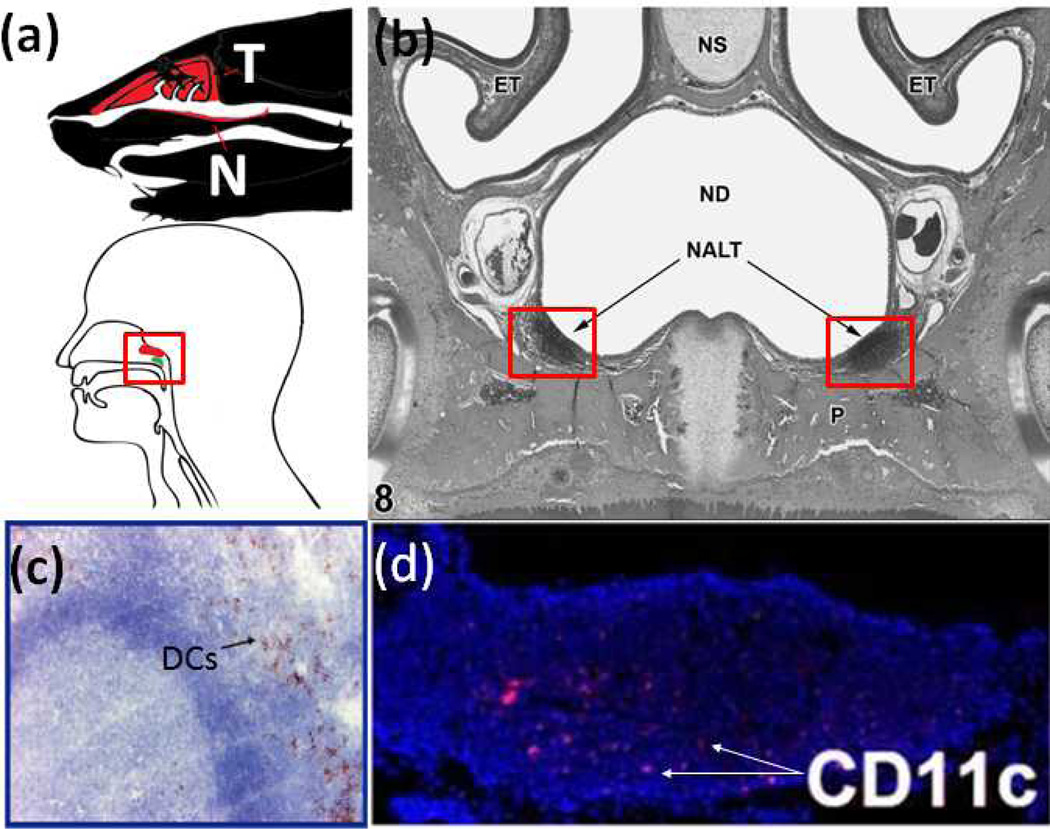
In intranasal administration, particles are directly delivered to lymphoid tissues located in the nasal cavity (Fig. 6a), avoiding barriers posed by systemic or lymphatic transport discussed earlier. Immune cells within these lymphoid tissues are arranged in an organized follicular tissue structure (nasal associated lymphoid tissue, Fig. 6b)55 found directly under the nasal epithelium. The objective is to deliver particles through the thin nasal epithelium to the immune cells composed of not only B and T cells, but also dendritic cells (Fig. 6c, 6d). The nasal epithelium above the NALT is a single layer of epithelial cells littered with microfold cells (M cells). M cells are of interest in nasal particle delivery because they translocate particles from the epithelium to the underlying NALT.56 Particles translocated by M cells may be taken up by NALT cells, drained passively to the cervical lymph nodes through lymphatic vessels or transported actively through NALT DCs. Particle size (and aggregation issues) may not be a significant barrier to transport across the nasal epithelium because M cells are known to translocate particles with sizes up to several microns.57
Fig. 6.
Nasal Associated Lymphiod Tissue (NALT). (a) Location of NALT in mouse (top; T: turbinates, N: NALT) and human (bottom; Red: adenoids; Green: tubal tonsil). (b) Cross section of nasal cavity showing location of NALT in mouse.55 (NS: nasal septum; ND: nasal cavity; ET: turbinates) (c) Cross section of human adenoid tonsil stained for plasmacytoid DCs (brown).58 (d) mouse NALT stained for CD11c+ DCs (red).59 Reproduced with permission.58,59,55
Nevertheless, many studies show that particles at or below 1 micron tend to yield better functional outcomes.57, 60, 61 As the epithelial surface is negatively charged due to the presence of the glycocalyx, particles with net positive charges (and expectedly so) mediate better immune responses. Ironically, particles with higher negative zeta potentials (at least −25mV) can also be efficiently transported by M cells.60, 61 It appears that decreased ionic concentration of the buffer used to suspend the particles can increase M cell uptake, through potentiating the particle charge density.60
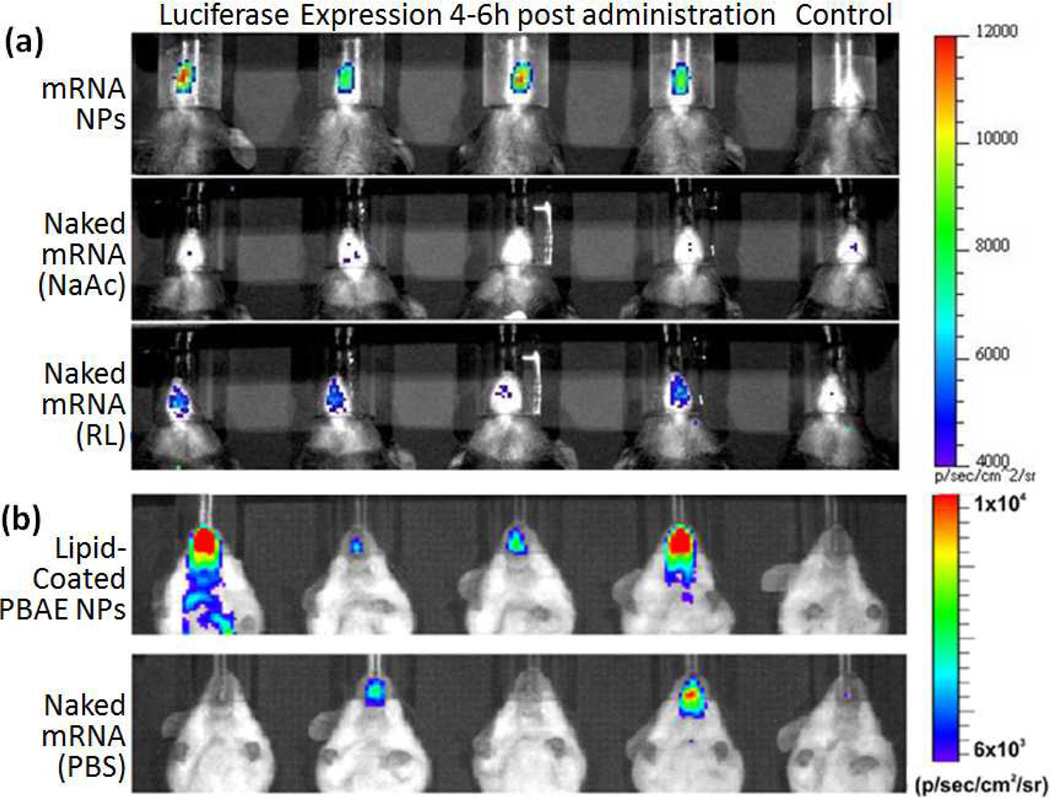
The relatively permissive transport mechanism through the nasal epithelium is undermined by the fact that fluid instilled into the nasal cavity is rapidly cleared from the nose shortly after administration (80% by mass within the first hour).62 Therefore increased particle adhesion onto the nasal walls, especially through the use of mucoadhesive materials such as chitosan can enhance immune responses albeit at a very high dose.63, 64 Although mucoadhesion increases the residence time of particles on the nasal epithelium, it can also impede their movement through the mucus, making it a property that needs to be carefully managed to achieve optimal results.65 We have reported nasal transfection with lipid-based mRNA nanoparticles (180nm, +40mV in water).53 Although these particles are cleared almost as quickly as naked mRNA, luciferase expression peaks at 4 hours and remains detectable for up to 24 hours compared to naked mRNA which is detectable only at the 4 hour time point (Fig. 7a).53 Similar expression kinetics is observed in another study where luciferase mRNA surface adsorbed on pegylated core shell nanoparticles (280nm, +40mV in water) are intranasally administered (Fig. 7b).66
Fig. 7.
Mice intranasally administered with mRNA encoding luciferase (in above mentioned formulations) followed by intraperitoneal injection of luciferin 15 minutes prior to bioluminescence imaging. (a) C57Bl/6 mice imaged 4 hours post administration.53 (b) Balb/c mice imaged 6 hours post administration.66 (PBAE: poly-beta amino ester, NaAc: sodium acetate, RL: Ringer’s Lactate). Reproduced with permission.53,66
Unlike drug-loaded particles, cellular uptake is a requirement for gene-loaded particles to exert a biological effect. As mentioned earlier, the mucus barrier is one that should not be underestimated because a mucus layer as thin as 20nm can block particle access to M cells.67 Hence particle properties that facilitate transport across the mucus will enhance nasal delivery of nanoparticle vaccine. It has been shown that pegylated (PEG length ∼5kDa) or medium sized particles (200nm and 500nm) diffuse through mucus more efficiently than non-pegylated or smaller particles (100nm).65 Since the mucus barrier of NALT in a healthy subject is relatively thin, it is uncertain if pegylation can significantly enhance delivery efficiencies.
In summary, nasal administration of mRNA nanoparticle vaccine is an attractive strategy for tumour vaccination due to its non-invasive nature. Recent studies have demonstrated that nasal transfection using mRNA encapsulated in nanoparticles is a feasible concept. Further studies are warranted to ascertain if this route of delivery can induce an anti-tumour response. M cell transport efficiencies may be enhanced by using highly negatively charged particles, increasing residence time of the particles on nasal epithelium as well as enhancing the mucus-penetrating power of the nanoparticles. The incorporation of such properties into mRNA nanoparticles will help overcome extracellular barriers associated with nasal nanoparticle vaccine delivery.
3.1.2 Cytosolic Gene Delivery of mRNA in Dendritic Cells
Upon reaching the target organ, mRNA nanoparticles need to be efficiently taken up by antigen presenting cells, escape from the endosomal compartment and unpack to release the mRNA for protein translation. Early studies showed that DCs were poorly transfected by lipoplexes.68 Based on %GFP+ cells, transfected immature and mature human monocyte derived DCs using DMRIE-C are 7.5% and 4%, respectively. In the same study, transfection mediated by electroporation was 63% and 33%, respectively. Consequently, there was little interest in chemical transfection of DCs. But significant advances in gene carrier development in the past decade have improved the prospect of DC transfection by mRNA.
In vitro transfection is commonly studied using DC2.469 and JAWS II (ATCC) cell lines, but also with primary murine bone marrow derived dendritic cells (BMDCs) and human monocyte derived DCs. Perche et al11 developed a targeted lipopolyplex formulation prepared by addition of mannosylated and histidylated liposomes to mRNA pre-condensed with PEG and histidine-modified polylysine.70 Lipids and polylysine, the building blocks of LPD, were custom synthesized to incorporate imidazole moieties to enhance endosome escape via the proton-sponge effect. A portion of the lipids was also conjugated with mannose to enhance targeting. Interestingly, the lipid tail was linked to its head group via a phosphoramide bond to improve biocompatibility. Transfection efficiencies based on GFP+ cells against DC2.4 in Opti-MEM using these targeted lipopolyplexes was significantly higher (60%) compared to the same lipopolyplexes without mannosylated lipids (40%).
Cheng et al.71 developed a series of “DPE” triblock co-polymers composed of a DMAEMA (dimethylaminoethyl methacrylate) segment for cationic-mediated mRNA binding, a PEGMA (polyethylene glycol methacrylate) segment to impart colloidal stability and a DEAEMA-co-BMA (copolymer of diethylaminoethyl methacrylate and butyl methacrylate) segment to achieve pH sensitivity for endosome escape. Transfection efficiencies against DC2.4 cells in serum free media reached 50% (GFP+).
Su et al.66 developed a formulation where mRNA was delivered via lipid-enveloped pH sensitive core-shell nanoparticles. Based on a similar concept reported for DNA vaccine72, mRNA was electrostatically adsorbed onto the surface of pegylated cationic nanoparticles composed of pH-sensitive core and a PEG/DOTAP/DOPC lipid shell. These particles were efficiently taken up by 80% of DC2.4 in the presence of 10% serum but only 30% were transfected. This discrepancy was attributed to degradation of mRNA, which may not be well protected via surface adsorption.
Commercially available mRNA transfection reagents are also relatively efficient in DC transfection. Mockey et al.14 used Lipofectamine to transfect JAWS II cells with luciferase mRNA to study the potentiation of mRNA translation by the length of the poly-A tail. Phua et al.53 used Stemfect mRNA transfection reagent to study the transfection efficiency and transgene expression kinetics of mRNA in naked and nanoparticle format. Based on GFP reporter gene expression, the transfection efficiencies in immature BMDC and immature human monocyte derived DCs were 63% and 52% respectively, while that in DC2.4 and JAWS II cells were 98% and 80%, respectively. Kariko et al.22 used Mirusbio Trans-IT mRNA transfection reagent to study enhancement in protein translation mediated by luciferase mRNA synthesized with various modified nucleotides. Compared to its unmodified form, luciferase expression was increased by up to four fold in immature BMDCs. Weissman et al.43 transfected human monocyte derived DCs with HIV gag encapsulated in lipofectin (DOTMA) to study the in vitro primary immune responses. DOTAP based liposomes were used for mRNA immunotherapy, although only in vivo immunotherapy data were reported.8
3.2 Major Histocompatibility Complex (MHC) Presentation
In a classical infection model, pathogens taken up by DCs via phagocytosis go through the endosome-lysosome pathway (synonymous to MHC II processing pathway in antigen presenting cells).73 DCs present antigens to CD4+ T cells through MHC class II molecules and also to the CD8+ CTLs through MHC class I molecules (via cross presentation, Fig. 8),73 activating both CD4+ T cells and CD8+ CTLs (cytotoxic T lymphocytes). To achieve a robust cellular immune response, antigens have to be presented by DCs in the context of both MHC I and II. Activated CD4+ T cells stimulate CTLs through the production of IL-2 and license DCs through CD40/CD40-ligand (L) interactions (Fig. 1). Licensed DCs up-regulate co-stimulatory molecules CD70, CD80 and CD86 needed to provide co-stimulation (i.e. second signal) to complete CTL activation (Fig.1). CTLs activated in this way are programmed for survival, while those that do not receive co-stimulation (i.e. unlicensed DCs that present antigens to CTLs via MHC I molecules but do not provide co-stimulation via CD70/80/86 are eventually deleted.74 This pathway is relevant to most synthetic peptide/protein based nanoparticle vaccine, but not for mRNA-based vaccine.
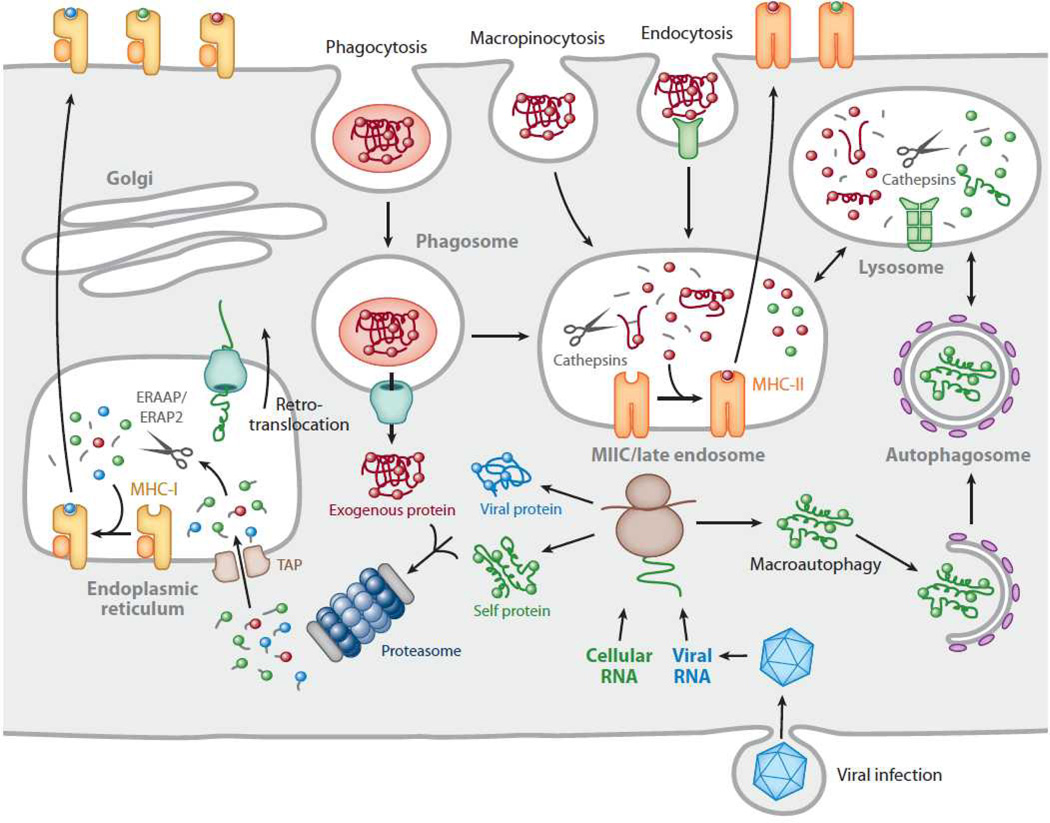
Fig. 8.
Trafficking of antigens for processing and presentation on major histocompatibility complex (MHC) molecules: Cytosolic proteins are processed primarily by the proteasome. Short peptides are then transported into the endoplasmic reticulum (ER) by the transporter associated with antigen processing (TAP) for subsequent assembly with MHC-I molecules. In certain antigen-presenting cells, particularly dendritic cells, exogenous proteins can also be fed into this pathway by retrotranslocation from phagosomes, a phenomenon known as cross-presentation. Exogenous proteins are primarily presented by MHC-II molecules. Antigens are internalized by several pathways, including phagocytosis, macropinocytosis, and endocytosis, and eventually traffic to a mature or late endosomal compartment, often called the MHC-II compartment, or MIIC, where they are processed and loaded onto MHC-II molecules. Cytoplasmic/nuclear antigens can also be trafficked into the endosomal network via autophagy for subsequent processing and presentation with MHC-II molecules. Reproduced with permission.73
mRNA vaccination is more similar to viral infection. In contrast to classical infection, antigens are instead derived within the cytoplasmic compartment (Fig. 8). Although cytoplasmic antigens are efficiently presented on MHC class I molecules, they tend to be poorly presented on MHC class II molecules, as only 10–30% of peptides bound to MHC II are derived from cytoplasmic and nuclear proteins.73 It is now known that cytoplasmic antigens are presented on MHC II via various autophagy mechanisms, in particular, the chaperone-mediated autophagy is a major pathway involved in MHC II presentation of cytosolic proteins. In this pathway, cytoplasmic chaperones Hsc70 and Hsp90, together with lysosomal transmembrane protein (LAMP) selectively shuttle epitopes into lysosomes, effectively infiltrating the MHC II pathway (Fig. 8).73 To enhance mRNA tumour vaccination, the chaperone-mediated autophagy pathway has been exploited to increase MHC II-peptide presentation. A single LAMP-1 or DC-LAMP75 sequence can be cloned into the cDNA template so that the mRNA is eventually translated as a LAMP tagged protein (Fig. 1). Co-delivering mRNA encoding both LAMP tagged and untagged protein has been shown to be effective in enhancing anti-tumour immunity in colorectal and melanoma immunotherapy models.10, 76, 77
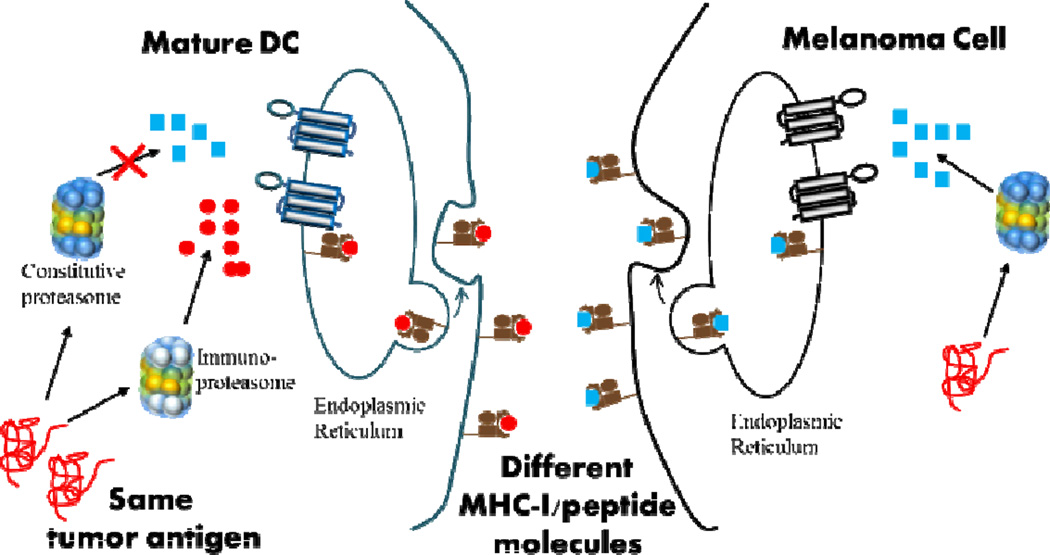
In addition to delivering antigens to the right locations on DCs’ cell surfaces, it has recently been shown that antigen peptides generated from immunologically mature DCs may not be optimal. When mRNA transfected DCs undergo maturation, constitutive subunits X(β5), Y(β1), and Z(β2) of DC’s proteasomes are replaced by the inducible subunits LMP7 (β5i), LMP2(β1i), and MECL1(β2i), leading to the formation of immunoproteasome.78 As a result, antigen peptides presented by mature DCs are exclusively generated by immunoproteasomes (iPs, triangles in Fig. 9) and CTLs predominantly recognize iP-generated peptides. In contrast, the proteasomes of tumour cells (such as melanoma) remains unmodified and MHC I-peptide complexes found on the surface of melanoma cells are exclusively generated by constitutive proteasomes (cP, squares in Fig. 9). Consequently, antigen specific CTLs recognize melanoma cells poorly even though antigen peptides are derived from the same antigen protein, resulting in a misdirected and suboptimal anti-tumour immune response. This problem has been addressed by inhibiting the conversion of cP to iP in mature DCs by knocking down the expression of inducible subunits through the use of siRNA targeting LMP7 (β5i), LMP2(β1i), and MECL1(β2i).79 This siRNA strategy has recently been proven to be effective in humans.80
Fig. 9.
Misdirected anti-tumour immune response stimulated by mature tumour associated antigen loaded DCs.
In summary, the lack of MHC II presentation in mRNA vaccination is a problem that has been overcome through co-delivery of LAMP-tagged mRNA to access the MHC II pathway. Besides LAMP-tagged mRNA, co-delivery of multiple mRNAs encoding additional adjuvants, such as the TriMix7 or GM-CSF8 has been shown to enhance anti-tumour immunity. In addition, peptides generated by immunoproteasomes of mature DCs leading to misdirected CTL specificities against tumor antigens has been overcome through co-delivery of siRNA targeting inducible subunits of immunoproteasomes. All of these suggest that the future of mRNA nanoparticle vaccine will necessarily be a co-delivery system. It is a great challenge to ensure every single nanoparticle contains every type of mRNA required by design in a particular study. Significant heterogeneity may result because bulk handling methods employed in co-encapsulation/loading of mRNA into nanoparticles may not provide a consistent formulation process. While this may not be critical in proof-of-concept studies, issues related to throughput and batch-to-batch consistency may impact the translation of nanoparticle vaccines. Hence, operator-independent nanoparticle formulation methods will be necessary to ensure controlled and consistent synthesis of nanoparticles in the future.81
3.3 Immune Activation
To achieve a robust T cell response, the mRNA nanoparticle formulation must not only ensure that dendritic cells translate and present tumour antigens encoded by the mRNA, it must also concomitantly stimulate them to maturity, which is defined as the secretion and expression of immunological factors and co-stimulatory molecules necessary for complete T cell activation. If immune activation is inadequate, transfected dendritic cells may turn tolerogenic and create immunological acceptance of the tumour instead.82
Major immune activation pathways associated with mRNA are shown in Fig. 1. mRNA itself activates toll-like receptors (TLR3, 7 and 8)21 located in the endosomes of immune cells as well as fibroblasts and epithelial cells. The signalling involved can be broadly classified into the TRIF and MyD88 pathways. The former leads to the secretion of type I interferon while the latter results in the secretion of pro-inflammatory cytokines (Interleukins and TNF family). TLR3, which senses double stranded RNA (derived from internal hairpin structures of mRNA), trigger the secretion of both type I interferon via a TRIF dependent pathway as well as pro-inflammatory cytokines via the MyD88 pathways. TLR7, which senses single stranded RNA, triggers the secretion of inflammatory cytokines via MyD88 pathway. In addition to endosome associated immune receptors, cytosolic retinoic acid inducible gene-I (RIG-I) receptor 83 can be activated by triphosphates at the 5’ end of uncapped mRNA leading to the secretion of type I interferon (Fig. 1). Although it is a common practice to cap the 5’ end of mRNA with an anti-reverse cap analogue during in vitro transcription, capping efficiency is usually about 80%, leaving exposed 5’ triphosphates from uncapped fraction available for RIG-I activation. MDA5 is another cytosolic receptor sharing the RIG-I signalling pathway that can be activated by long-double stranded RNA (such as self-replicating RNA, which has re-emerged recently in the vaccine field). 84
Pro-inflammatory cytokines induced from mRNA have been identified as TNF-α,7,85 IL-1β,6 IL-12,6,86, 87 IL-6,7,6, 85, 87, 88 IL-8.88 Similarly Type I interferons induced from mRNA are IFN-α83, 85, 86, 89 and IFN-β.83, 85 Interestingly, chemokines such as GRO (Growth-Regulated Oncogene), MCP-1 (Monocyte Chemoattractant Protein-1), RANTES (Regulated on Activation, Normal T cell Expressed and Secreted) and MDC (Macrophage-Derived Chemokine) have also been reported when peripheral blood cells are pulsed with naked mRNA.88 These secretions exert not only an immediate innate immune response, but also adaptive immune responses by facilitating the maturation of professional antigen presenting cells (dendritic cells, B cells, activated macrophages) through up-regulation of MHC II and co-stimulatory molecules (CD80, 86) as well as a change in chemokine receptors. The secretion of Type-I interferon induced by innate immune response to mRNA has recently been shown to be counterproductive in the induction of antigen specific T cells when mRNA is delivered via DOTAP/DOPE lipoplexes.12 This study confirms the idea that not all “self-adjuvant” effects derived from mRNA facilitate the development of cellular immunity.
The immunogenicity of mRNA has been highlighted in recent years due to increasing interest in their therapeutic application outside immunotherapy. A better understanding of the structural86, 91 and molecular requirements3, 22 for mRNA’s biological stability and mechanism of immune activation have led to increased translational capacity mRNA and boosted its potential for gene therapy applications. However, as there are reports describing non-modified mRNA as “non-immunogenic”76,16 the impression that mRNA is highly immunogenic has to be put into perspective. mRNA delivered by electroporation is not efficient in mediating DC maturation. DC maturation marker CD83 (<30%) and co-stimulatory molecule CD80 (<40%)68, 92 are both poorly up-regulated in human monocyte-derived DCs in vitro, even though the transfection efficiency based on GFP expression is 76%. However, the co-stimulatory molecule CD86 is relatively well up-regulated (>70%), indicating that transfected DCs can be partially matured with mRNA. CD86 up-regulation of a similar magnitude is also observed in nodal DCs, when mRNA is directly injected into inguinal lymph nodes. However this study did not report the transfection efficiency based on GFP+ cells, staining of CD83 and CD80, making it difficult to conclude the extent of DC maturation. Overall, these data suggest that partial maturation may only occur in mRNA-loaded DCs in vitro, compared to >90% maturation (based on expression of CD80, CD83, CD86) in DCs treated with cytokines.
An inherent advantage of mRNA nanoparticle vaccine is that its overall immunogenicity can be modified by the gene carrier. When delivered in nanoparticle format, CD80 and CD86 expression on human monocyte-derived DCs (pulsed with mRNA-lipofectin) were up-regulated (>90%) compared to cytokine treatment (>80%). CD83 expression, however, remained relatively low.43 Interestingly, CD80 and CD86 are both up-regulated (>50%) by lipofectin liposomes alone while CD83 expression remained low (4%). As the transfection efficiency mediated by lipofectin in human monocyte-derived DCs is unlikely to be >90%, this response is more consistent with immunostimulating effects of cationic liposomes via the ROS mechanism, a topic that has been comprehensively reviewed.50,93 In another study, Rettig et al compared the cytokine secretion profile when human peripheral blood mononuclear cells (PBMCs) are treated with β-Gal RNA either in naked form or encapsulated by protamine. β-Gal RNA nanoparticles mediated significantly higher levels of IL-6, IL-8 and MCP-1 compared to naked mRNA,88 suggesting that cytokine secretion can also be affected by the addition of protamine. Protamine-condensed mRNA also induced CD86 on DCs, but the transfection efficiency of the mRNA is abrogated. Similarly, Oliwia et al. also showed that mRNA lipoplexes induced high levels of IL-6 and IL-12 and TNF-α in murine lungs following intranasal instillation.94 In this study, cytokine secretion data from liposome alone or naked mRNA was not available. In another study, subcutaneously injected mRNA-DOTAP/DOPE liposomes but not the liposomes alone, induce high levels of IL-6, IL-1β and type I interferon in DCs.12 This lack of cytokine induction by empty liposomes is consistent with the adjuvant effects of DOTAP-based vaccines,93 and an interesting contrast between lipid and polymeric gene carriers.
In an effort to evaluate the use of modified RNA for immunotherapy, Pollard et al. investigated the effects of IFN-α on T cell response.12 Surprisingly, IFN-α knockout mice immunized subcutaneously with DOTAP/DOPE mRNA lipoplexes develop more robust antigen specific T cell responses compared to wild type mice. Although results coming from knock-out models are encouraging, a direct confirmation is needed from wild type models immunized with modified mRNA to confirm its utility in immunotherapy.
On the other hand, there is evidence that gene carriers may decrease the overall immunogenicity of the mRNA formulations. Uchida et al. showed that mRNA encapsulated in nanomicelles (PEG-polyamino acid block co-polymer) administered into the central nervous system induce lower levels of IL-6, TNF-α, IFN-α4 and IFN-β1 from neural tissues compared to naked mRNA.85 The mechanism, however, remains to be elucidated.
To summarize, immunotherapies that use mRNA encapsulated in nanoparticles may benefit from immunogenicity or lack thereof of gene carriers. Immune stimulating properties of mRNA in terms of the mechanism of activation and the extent to which it can modify DC phenotype are well characterized. But there is still a knowledge gap on how molecular structure of the gene carrier modifies the immunogenicity of mRNA. Lipid-based formulations are capable of up-regulating costimulatory molecules such as CD80/86, but are poor in inducing DC maturation and cytokine secretions necessary for a productive T cell activation. Protamine can up-regulate CD86 expression and also induce high levels of pro-inflammatory cytokines. PEG-poly amino acid micelles, on the other hand, attenuate immune responses.
4. mRNA Nanoparticle Vaccine Delivery Systems for Tumour Immunotherapy
In the previous sections, we discussed a framework for mRNA nanoparticle delivery: namely (1) gene delivery, (2) immune activation and (3) MHC processing. Despite numerous reports on mRNA nanoparticle (mRNA-NP) delivery, only a handful studied its therapeutic efficacy in terms of either immune responses (CTL assay/IFN-γ secretion) or overall survival. In this section, we review these studies (listed in Table 1) highlighting current progress in mRNA-NP mediated tumour vaccination and provide a perspective on the bottleneck of this exciting area of research.
Table 1.
mRNA Nanoparticle Tumour Vaccination Studies.
| Ref | Antigen/µg x number of doses/delivery system | Cytotoxic T Lymphocyte Response | Splenic IFN-γ |
Survival Data |
Size/ Zeta Potential |
||||
|---|---|---|---|---|---|---|---|---|---|
| IV | SC | ID | IM | IP | |||||
| Martinon et al(1993)95 | Influenza nucleoprotein/12µg x 2/ Anionic Lipoplex: DPPC/DPPS/Chol (Encapsulation efficiency 5–10%) |
+ | + | − | Not reported Extruded through 200nm membranes |
||||
| Zhou et al (1999)13 |
GP100/ 8µg x 2/ HVJ-liposomes: Egg PC/Chol/DC-Chol |
Works only if injected directly into spleen. CTL levels comparable with positive control |
|||||||
| Hoerr et al (2000)9 |
LacZ/30µg x1/Lipopolyplex: Unifectin+protamine | ++ | + | − | − | Not reported |
|||
| LacZ/30µg x1/ Lipoplex: Unifectin only | − | − | |||||||
| LacZ/30µg x1/Protamine only | − | − | + | − | |||||
| mLacZ/30µg x1/Naked format | + | ||||||||
| Hess et al (2006)8 |
OVA/5µg x 2/Lipoplex: DOTAP | + | + | ||||||
| OVA/3µg x2/Lipoplex: DOTAP/DOPE | + | − | + | ||||||
| OVA+GMCSF/3+1µg x1/Lipoplex:DOTAP-DOPE | ++ | ||||||||
| Mockey et al (2007)10 |
MART-1/25µg x2/Lipopolyplex: PEG-HpK,HDHE,Chol |
+ | + | + | 100nm/ +13mV (10mM Hepes) |
||||
| Perche et al (2011)11 |
MART-1/25µg x 2(IV)/ Targeted lipopolyplex: PEG-HpK, HDHE/HDHE-mannose,Chol) |
+ | 150nm/ +17mV (10mM Hepes) |
||||||
| Pollard et al(2013)12 | GAG/20µg x2 (IV,SC, wild type mice)/ Lipoplex: DOTAP-DOPE(IV/SC) |
+ | Not reported | ||||||
| GAG/20µg x2 (IV, IFN± −/− mice)/ Lipoplex: DOTAP/DOPE(IV) |
++ | ||||||||
IV: intravenous (tail vein); SC: subcutaneous (base of tail); ID: intradermal (ear pinnae); IM: intramuscular; IP: intraperitoneal. “+” and “−” respectively indicate statistically significant and insignificant result compared to controls. “++” indicates results are significantly higher than “+” samples reported in the same study.
The first attempt in mRNA-NP vaccine was reported in 1993 by Martinon et al.95 using anionic liposomes composed of phosphatidylcholine(PC)/phosphatidylserine(PS)/cholesterol. mRNA encoding influenza nucleoprotein was encapsulated into liposomes by the hydration method. This method is inefficient and yields only about 10% in encapsulation efficiency.95 Even so, antigen-specific CTL response was induced if the particles were administered via IV or SC route but not IP route.95 This result is consistent with a similar study, where IP vaccination route also failed to induce CTL response.9 The authors attributed the ineffectiveness of the IP route to particle aggregation and a lack of peritoneal antigen presenting cells.95 This is an interesting study because it uses pH-insensitive anionic liposomes that not only interact poorly with negatively charged cell membranes, but they are also inefficient in endosome escape (Fig. 1). Yet they mediate antigen specific CTL responses indicating that mRNA is translated by antigen presenting cells. Nevertheless, this PS containing mRNA formulation may be phagocytosed by antigen presenting cells of the monocyte-phagocyte system via PS receptors and the anionic nature of the lipoplexes may resist opsonin-mediated aggregation in the blood upon IV administration. Also, since the liposomes were extruded with a 200nm membrane, they could be efficiently translocated to the lymph nodes upon SC administration. In summary, this formulation may be promising if the encapsulation efficiency is increased and if pH-sensitive molecules are incorporated.
Hoerr et al. demonstrated that the use of liposomes (Unifectin) protects mRNA from nuclease-mediated degradation in vivo and significantly increased CTL response via IV route, but failed to induce any CTL response via the IM and IP routes.9 In addition, they also showed that although small amounts of protamine can protect naked mRNA from nuclease degradation, mice subcutaneously immunized with mRNA formulated with protamine alone (without Unifectin) did not enhance CTL response compared to naked mRNA.
Hess et al. first reported survival data from mRNA nanoparticle tumour vaccination.8 In this detailed study, OVA mRNA encapsulated in DOTAP liposomes was intradermally administered into mice ear pinnae twice (14 days apart) in a prophylactic immunotherapy model, followed by subcutaneous tumour challenge with EG7-OVA cells. Splenic CD8+ T cells from immunized mice demonstrate lytic activities both in vitro (CTL assay) and in vivo (adoptively transfer of CFSE labeled OVA pulsed splenocytes). These mice also experienced significant delay in tumour onset and progression against EG7-OVA tumour challenge through Day 16. The formulation applied in this study was DOTAP lipoplexes prepared by mixing mRNA with DOTAP liposomes in PBS. These lipoplexes were not characterized for size and zeta potential in this study. But since DOTAP lipoplexes has been reported to aggregate when they are formulated in the presence of salt, the mRNA-DOTAP lipoplexes prepared in this study could have been aggregated.96 Particle aggregation, as mentioned earlier, reduces extracellular and intracellular gene delivery efficiencies. And this may be the reason for a higher CTL response following IV compared to ID administration and an absence thereof from SC administration (tail base). Also, aggregated particles administered IV can still be taken up by macrophages in the liver and spleen, develop into effective antigen presenting cells through ROS mediated activation (by DOTAP) and transfer the antigen to endogenous DCs or activate T cells directly. On he other hand, ID administration induced a dose dependent CTL response because a small fraction of mRNA lipoplexes remained bioactive after ID administration, which could transfect dermal DCs leading to CTL response. As this was a small fraction of the given dose, a larger dose could have led to higher dermal DC transfection and presumably higher CTL response.
In this review, we are able to correlate observed CTL responses to particle properties of lipoplexes because they have been characterized in other studies under similar conditions. As shown in Table 1, mRNA nanoparticle tumour vaccination studies are often reported without much information about particle properties. To optimize therapeutic outcome, particle characterization such as size, zeta potential measured under the applied physiological conditions should be an important aspect of future immunotherapy experiments.
In the study by Hess et al., CTL response from mRNA encapsulated in DOTAP/DOPE was also four times higher than in DOTAP liposomes. This shows that the inclusion of DOPE, a helper lipid with fusogenic property, facilitates intracellular gene transfer to antigen presenting cells.97 This indicates, at least in the DOTAP-based mRNA liposomes, that endosome escape maybe a rate-limiting step for immune modulation. In addition, CTL responses were enhanced when OVA mRNA was co-delivered with GM-CSF (to attract DCs), but not CD80 (co-stimulatory molecules) or IL-2 (T cell proliferation) mRNA. These results suggest that threshold levels of GMCSF needed to enhance immune response is much lower than CD80 and IL-2. It also re-affirms that a co-delivery system is the optimal mRNA nanoparticle tumour vaccine.
Mockey et al. also reported anti-tumour efficacy of mRNA nanoparticle vaccination using histidylated lipids optimized for mRNA delivery.10 Each phosphoramide lipid contained a single histidine head group to facilitate endosome escape via proton sponge effect. Mannosylated lipids (11% molar ratio) were later incorporated in the formulation (Man11-LPR) to enhance uptake.98 The colloidal stability of mRNA nanoparticles formulated with these lipids in physiological salt concentration and in serum is unknown although particles are about 150nm (ζ-potential +14–18mV) in 10mM Hepes. But given its almost identical in vivo biodistribution with LPD, their colloidal stability may be similar to that of unpegylated LPDs. In Mockey et al., MART-1 (Melanoma antigen recognized by T-cells 1) mRNA encapsulated in these histidylated lipopolyplexes was intravenously administered into mice via tail vein twice (7 days apart) in a prophylactic model, followed by a subcutaneous tumour challenge with B16 F10 cells. Developed tumours progressed more slowly in nanoparticle immunized mice compared to control mice vaccinated with luciferase mRNA nanoparticles. In a subsequent study by Perche et al.11 MART-1 mRNA encapsulated in Man11-LPR or non-mannosylated LPR formulations were re-evaluated in the prophylactic B16 F10 melanoma immunotherapy model. The median survival of mice immunized with MART-1 mRNA nanoparticles formulated with Man11-LPR was 5 and 10 days longer than mice immunized with non-mannosylated LPR and NaCl controls, respectively. Although the effectiveness of Man11-LPR in a therapeutic immunotherapy model remains to be determined, this study demonstrates that the presence of a targeting ligand improves overall therapeutic efficacy through enhanced uptake of nanoparticles by antigen presenting cells.
In this section, we reviewed studies that evaluate mRNA nanoparticle formulations for tumour vaccination. Although nanoparticle properties are not well characterized in many of these studies, it is clear that the use of mRNA nanoparticles consistently induce CTL responses. These studies show that nanoparticle delivery of mRNA tumour vaccination is a fertile research direction.
5. Conclusions
Advances in the past decade have deepened our understanding of mRNA’s biological stability and immunological properties and provided extensive evidence to support the need for engineering innovations to deliver mRNA for genetic vaccination. In this review we discussed the physical stability of mRNA in aqueous buffers and clarified the technical possibility of subjecting mRNA to engineering manipulations. Presently, mRNA tumour vaccines administered directly in vivo benefits from being encapsulated with gene carriers in nanoparticle format and a growing number of studies are looking at the feasibility of applying nanomedicine concepts, such as ligand decoration for APC targeting, pegylation for colloidal stability, or microfluidics synthesis for formulation, to mRNA tumour vaccination. A few deliberate attempts have been made on the rational design of mRNA gene carrier,11, 66, 71 but most have not been functionally evaluated in vivo for immune or therapeutic response.11, 71 An optimal mRNA gene carrier for tumour vaccination, therefore, has yet to emerge. Instead, off-the-shelves gene carriers are the most frequently used to encapsulate mRNA and evaluated for their ability to stimulate antigen specific T cell responses, often without adequate particle characterization. Adding to the challenge is that different routes of vaccination probably require different optimal gene carriers and formulations. Given the sustained interest in this field, the development of effective mRNA formulations will no doubt accelerate in the near future to advance the field of mRNA immunotherapy.
Acknowledgements
This research is supported by the Department of Defense (DoD) awards W81XWH-12-1-0260 (SK.N) and W81XWH-12-1-0261 (KW.L), NIH AI96305
References
- 1.Lee J, Boczkowski D, Nair S. Methods Mol. Biol. 2013;969:111–125. doi: 10.1007/978-1-62703-260-5_8. [DOI] [PubMed] [Google Scholar]
- 2.Wu XY, Brewer G. Gene. 2012;500(1):10–21. doi: 10.1016/j.gene.2012.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grudzien-Nogalska E, et al. Methods Mol. Biol. 2013;969:55–72. doi: 10.1007/978-1-62703-260-5_4. [DOI] [PubMed] [Google Scholar]
- 4.Phua KK, et al. Adv Healthc Mater. 2013 [Google Scholar]
- 5.Walch B, et al. Gene Ther. 2012;19(3):237–245. doi: 10.1038/gt.2011.121. [DOI] [PubMed] [Google Scholar]
- 6.Kreiter S, et al. Cancer Res. 2010;70(22):9031–9040. doi: 10.1158/0008-5472.CAN-10-0699. [DOI] [PubMed] [Google Scholar]
- 7.Van Lint S, et al. Cancer Res. 2012;72(7):1661–1671. doi: 10.1158/0008-5472.CAN-11-2957. [DOI] [PubMed] [Google Scholar]
- 8.Hess PR, et al. Cancer Immunol. Immunother. 2006;55(6):672–683. doi: 10.1007/s00262-005-0064-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoerr I, et al. Eur. J. Immunol. 2000;30(1):1–7. doi: 10.1002/1521-4141(200001)30:1<1::AID-IMMU1>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 10.Mockey M, et al. Cancer Gene Ther. 2007;14(9):802–814. doi: 10.1038/sj.cgt.7701072. [DOI] [PubMed] [Google Scholar]
- 11.Perche F, et al. Nanomedicine. 2011;7(4):445–453. doi: 10.1016/j.nano.2010.12.010. [DOI] [PubMed] [Google Scholar]
- 12.Pollard C, et al. Mol. Ther. 2013;21(1):251–259. doi: 10.1038/mt.2012.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou WZ, et al. Hum. Gene Ther. 1999;10(16):2719–2724. doi: 10.1089/10430349950016762. [DOI] [PubMed] [Google Scholar]
- 14.Mockey M, et al. Biochem. Biophys. Res. Commun. 2006;340(4):1062–1068. doi: 10.1016/j.bbrc.2005.12.105. [DOI] [PubMed] [Google Scholar]
- 15.Zou S, et al. Int. J. Pharm. 2010;389(1–2):232–243. doi: 10.1016/j.ijpharm.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamamoto A, et al. Eur. J. Pharm. Biopharm. 2009;71(3):484–489. doi: 10.1016/j.ejpb.2008.09.016. [DOI] [PubMed] [Google Scholar]
- 17.Tavernier G, et al. J. Controlled Release. 2011;150(3):238–247. doi: 10.1016/j.jconrel.2010.10.020. [DOI] [PubMed] [Google Scholar]
- 18.Kreiter S, et al. Curr. Opin. Immunol. 2011;23(3):399–406. doi: 10.1016/j.coi.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 19.Mitchell DA, Nair SK. J. Clin. Invest. 2000;106(9):1065–1069. doi: 10.1172/JCI11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boczkowski D, Nair S. Expert Opinion on Biological Therapy. 2010;10(4):563–574. doi: 10.1517/14712591003614749. [DOI] [PubMed] [Google Scholar]
- 21.Kariko K, et al. Immunity. 2005;23(2):165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 22.Kariko K, et al. Mol. Ther. 2008;16(11):1833–1840. doi: 10.1038/mt.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones KL, Drane D, Gowans EJ. BioTechniques. 2007;43(5):675–681. doi: 10.2144/000112593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar A, et al. J Control Release. 2012;163(2):230–239. doi: 10.1016/j.jconrel.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DeMuth PC, et al. Adv Mater. 2010;22(43):4851–4856. doi: 10.1002/adma.201001525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chun KW, et al. Biomaterials. 2005;26(16):3319–3326. doi: 10.1016/j.biomaterials.2004.07.055. [DOI] [PubMed] [Google Scholar]
- 27.Kasper FK, et al. J Control Release. 2005;104(3):521–539. doi: 10.1016/j.jconrel.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 28.Wieland JA, Houchin-Ray TL, Shea LD. J Control Release. 2007;120(3):233–241. doi: 10.1016/j.jconrel.2007.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phua K, Leong KW. Nanomedicine (Lond) 2010;5(2):161–163. doi: 10.2217/nnm.09.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tseng YC, Mozumdar S, Huang L. Adv. Drug Del. Rev. 2009;61(9):721–731. doi: 10.1016/j.addr.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Owens DE, Peppas NA. Int J Pharmaceut. 2006;307(1):93–102. doi: 10.1016/j.ijpharm.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 32.Kabanov AV. Pharmaceutical Science & Technology Today. 1999;2(9):365–372. doi: 10.1016/s1461-5347(99)00186-8. [DOI] [PubMed] [Google Scholar]
- 33.Ho YP, et al. Nano Lett. 2011;11(5):2178–2182. doi: 10.1021/nl200862n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li S, et al. Gene Ther. 1998;5(7):930–937. doi: 10.1038/sj.gt.3300683. [DOI] [PubMed] [Google Scholar]
- 35.Merdan T, et al. Bioconjug. Chem. 2005;16(4):785–792. doi: 10.1021/bc049743q. [DOI] [PubMed] [Google Scholar]
- 36.Dileo J, et al. Mol. Ther. 2003;7(5 Pt 1):640–648. doi: 10.1016/s1525-0016(03)00064-9. [DOI] [PubMed] [Google Scholar]
- 37.Maemura K, et al. Immunol. Cell Biol. 2005;83(4):336–343. doi: 10.1111/j.1440-1711.2005.01323.x. [DOI] [PubMed] [Google Scholar]
- 38.Medd PG, Chain BM. Semin. Cell Dev. Biol. 2000;11(3):203–210. doi: 10.1006/scdb.2000.0162. [DOI] [PubMed] [Google Scholar]
- 39.Li SD, Huang L. Biochim. Biophys. Acta. 2009;1788(10):2259–2266. doi: 10.1016/j.bbamem.2009.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Idoyaga J, Steinman R. Immunology Image Resource. [webpage]; Available from: http://www.cell.com/immunity/image_resource-spleen. [Google Scholar]
- 41.Richter WF, Bhansali SG, Morris ME. Aaps Journal. 2012;14(3):559–570. doi: 10.1208/s12248-012-9367-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Allen TM, Hansen CB, Guo LSS. Biochim. Biophys. Acta. 1993;1150(1):9–16. doi: 10.1016/0005-2736(93)90115-g. [DOI] [PubMed] [Google Scholar]
- 43.Weissman D, et al. J. Immunol. 2000;165(8):4710–4717. doi: 10.4049/jimmunol.165.8.4710. [DOI] [PubMed] [Google Scholar]
- 44.Reddy ST, et al. J. Controlled Release. 2006;112(1):26–34. doi: 10.1016/j.jconrel.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 45.Manolova V, et al. Eur. J. Immunol. 2008;38(5):1404–1413. doi: 10.1002/eji.200737984. [DOI] [PubMed] [Google Scholar]
- 46.Zhuang Y, et al. J. Controlled Release. 2012;159(1):135–142. doi: 10.1016/j.jconrel.2011.12.017. [DOI] [PubMed] [Google Scholar]
- 47.Moghimi SM. Biomaterials. 2006;27(1):136–144. doi: 10.1016/j.biomaterials.2005.05.082. [DOI] [PubMed] [Google Scholar]
- 48.John ALS, et al. Nat. Mater. 2012;11(3):250–257. doi: 10.1038/nmat3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Daftarian P, et al. Cancer Res. 2011;71(24):7452–7462. doi: 10.1158/0008-5472.CAN-11-1766. [DOI] [PubMed] [Google Scholar]
- 50.Chen WS, Yan WL, Huang L. Cancer Immunology Immunotherapy. 2008;57(4):517–530. doi: 10.1007/s00262-007-0390-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Witte CL, et al. Circulation. 1969;40(5):623. doi: 10.1161/01.cir.40.5.623. [DOI] [PubMed] [Google Scholar]
- 52.Harrell MI, Iritani BM, Ruddell A. J. Immunol. Methods. 2008;332(1–2):170–174. doi: 10.1016/j.jim.2007.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Phua KKL, Leong KW, Nair SK. J. Controlled Release. 2013;166(3):227–233. doi: 10.1016/j.jconrel.2012.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Probst J, et al. Gene Ther. 2007;14(15):1175–1180. doi: 10.1038/sj.gt.3302964. [DOI] [PubMed] [Google Scholar]
- 55.Cesta MF. Toxicol. Pathol. 2006;34(5):599–608. doi: 10.1080/01926230600865531. [DOI] [PubMed] [Google Scholar]
- 56.Miller H, et al. World J Gastroenterol. 2007;13(10):1477–1486. doi: 10.3748/wjg.v13.i10.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vila A, et al. Int. J. Pharm. 2005;292(1–2):43–52. doi: 10.1016/j.ijpharm.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 58.Hartmann E, et al. Clin. Vaccine Immunol. 2006;13(11):1278–1286. doi: 10.1128/CVI.00172-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nacer A, et al. Mucosal Immunol. 2013 [Google Scholar]
- 60.Rajapaksa TE, et al. J. Biol. Chem. 2010;285(31):23739–23746. doi: 10.1074/jbc.M110.126359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Matsuo K, et al. J. Controlled Release. 2011;152(2):310–316. doi: 10.1016/j.jconrel.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 62.Southam DS, et al. American Journal of Physiology-Lung Cellular and Molecular Physiology. 2002;282(4):L833–L839. doi: 10.1152/ajplung.00173.2001. [DOI] [PubMed] [Google Scholar]
- 63.Xu JH, et al. Clin. Vaccine Immunol. 2011;18(1):75–81. doi: 10.1128/CVI.00263-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Roy K, et al. Nat. Med. 1999;5(4):387–391. doi: 10.1038/7385. [DOI] [PubMed] [Google Scholar]
- 65.Lai SK, Wang YY, Hanes J. Adv. Drug Del. Rev. 2009;61(2):158–171. doi: 10.1016/j.addr.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Su X, et al. Mol. Pharm. 2011;8(3):774–787. doi: 10.1021/mp100390w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Frey A, et al. J. Exp. Med. 1996;184(3):1045–1059. doi: 10.1084/jem.184.3.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Van Tendeloo VF, et al. Blood. 2001;98(1):49–56. doi: 10.1182/blood.v98.1.49. [DOI] [PubMed] [Google Scholar]
- 69.Shen ZH, et al. J. Immunol. 1997;158(6):2723–2730. [PubMed] [Google Scholar]
- 70.Gao X, Huang L. Biochemistry (Mosc) 1996;35(3):1027–1036. doi: 10.1021/bi952436a. [DOI] [PubMed] [Google Scholar]
- 71.Cheng C, et al. Biomaterials. 2012;33(28):6868–6876. doi: 10.1016/j.biomaterials.2012.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Singh M, et al. Proc. Natl. Acad. Sci. U. S. A. 2000;97(2):811–816. doi: 10.1073/pnas.97.2.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Blum JS, Wearsch PA, Cresswell P. Annu. Rev. Immunol. 2013;31:443–473. doi: 10.1146/annurev-immunol-032712-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kurts C, Robinson BWS, Knolle PA. Nature Reviews Immunology. 2010;10(6):403–414. doi: 10.1038/nri2780. [DOI] [PubMed] [Google Scholar]
- 75.de Saint-Vis B, et al. Immunity. 1998;9(3):325–336. doi: 10.1016/s1074-7613(00)80615-9. [DOI] [PubMed] [Google Scholar]
- 76.Bonehill A, et al. J. Immunol. 2004;172(11):6649–6657. doi: 10.4049/jimmunol.172.11.6649. [DOI] [PubMed] [Google Scholar]
- 77.Nair SK, et al. Nat. Biotechnol. 1998;16(4):364–369. doi: 10.1038/nbt0498-364. [DOI] [PubMed] [Google Scholar]
- 78.Kloetzel PM. Nat Immunol. 2004;5(7):661–669. doi: 10.1038/ni1090. [DOI] [PubMed] [Google Scholar]
- 79.Dannull J, et al. Blood. 2007;110(13):4341–4350. doi: 10.1182/blood-2007-04-083188. [DOI] [PubMed] [Google Scholar]
- 80.Dannull J, et al. J. Clin. Invest. 2013;123(7):3135–3145. doi: 10.1172/JCI67544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Valencia PM, et al. Nat Nanotechnol. 2012;7(10):623–629. doi: 10.1038/nnano.2012.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bachmann MF, Kopf M. Curr. Opin. Immunol. 2002;14(4):413–419. doi: 10.1016/s0952-7915(02)00363-1. [DOI] [PubMed] [Google Scholar]
- 83.Hornung V, et al. Science. 2006;314(5801):994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 84.Geall AJ, et al. Proc. Natl. Acad. Sci. U. S. A. 2012;109(36):14604–14609. doi: 10.1073/pnas.1209367109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Uchida S, et al. Plos One. 2013;8(2) doi: 10.1371/journal.pone.0056220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lund JM, et al. Proc. Natl. Acad. Sci. U. S. A. 2004;101(15):5598–5603. doi: 10.1073/pnas.0400937101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Scheel B, et al. Eur. J. Immunol. 2004;34(2):537–547. doi: 10.1002/eji.200324198. [DOI] [PubMed] [Google Scholar]
- 88.Scheel B, et al. Eur. J. Immunol. 2005;35(5):1557–1566. doi: 10.1002/eji.200425656. [DOI] [PubMed] [Google Scholar]
- 89.Diebold SS, et al. Science. 2004;303(5663):1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- 90.Su XF, et al. Mol Pharmaceut. 2011;8(3):774–787. doi: 10.1021/mp100390w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kranz LM, D M, Holzmann M, Reuter K, Selmi A, Fritz D, Meng M. Induction of potent anti-tumoral immunity via systemic delivery of antigen-encoding RNA-lipoplexes; Germany. 11th Annual Meeting of the Association for Cancer Immunotherapy; 2013. p. 281. [Google Scholar]
- 92.Chung DJ, et al. J Transl Med. 2013;11:166. doi: 10.1186/1479-5876-11-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Watson DS, Endsley AN, Huang L. Vaccine. 2012;30(39):5799–5799. doi: 10.1016/j.vaccine.2012.01.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Andries O, et al. J Control Release. 2013;167(2):157–166. doi: 10.1016/j.jconrel.2013.01.033. [DOI] [PubMed] [Google Scholar]
- 95.Martinon F, et al. Eur. J. Immunol. 1993;23(7):1719–1722. doi: 10.1002/eji.1830230749. [DOI] [PubMed] [Google Scholar]
- 96.Yan W, Huang L. Int. J. Pharm. 2009;368(1–2):56–62. doi: 10.1016/j.ijpharm.2008.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Regelin AE, et al. Biochim. Biophys. Acta. 2000;1464(1):151–164. doi: 10.1016/s0005-2736(00)00126-7. [DOI] [PubMed] [Google Scholar]
- 98.Perche F, et al. J. Drug Target. 2011;19(5):315–325. doi: 10.3109/1061186X.2010.504262. [DOI] [PubMed] [Google Scholar]