Abstract
The Rag family proteins are Ras-like small GTPases that play a critical role in amino acid-stimulated mTORC1 activation by recruiting mTORC1 to lysosome. Despite progress in the mechanistic understanding of Rag GTPases in mTORC1 activation, little is known about the physiological function of Rag GTPases in vivo. Here, we show that loss of RagA and RagB (RagA/B) in cardiomyocytes results in hypertrophic cardiomyopathy and phenocopies lysosomal storage diseases although mTORC1 activity is not substantially impaired in vivo. We demonstrate that despite upregulation of lysosomal protein expression by constitutive activation of the transcription factor EB (TFEB) in RagA/B knockout mouse embryonic fibroblasts, lysosomal acidification is compromised due to decreased v-ATPase level in the lysosome fraction. Our study uncovers RagA/B GTPases as key regulators of lysosomal function and cardiac protection.
Introduction
Rag family proteins (Rag A, B, C, D) are a unique subgroup of the Ras GTPase superfamily. Unlike other Ras family proteins, Rag GTPases are significantly larger and form heterodimers. For example, RagA or RagB forms a stable complex with RagC or RagD, with the possibility of forming four distinct complexes 1. RagA was initially identified as a functional homologue of the yeast Gtr1 that also forms a heterodimer with Gtr2 (homologous to Rag C and Rag D). Genetic studies revealed that Gtr1/Gtr2 is involved in various cellular processes such as nuclear transport, microautophagy, and intracellular trafficking 2-4.
Our group and Sabatini's lab discovered that the Rag GTPases play an essential role in amino acid-mediated mTORC1 activation 5, 6. The heterodimer formation of Rag GTPases is critical for mTORC1 activation, but RagA/B appears to be more important than RagC/D in mTORC1 activation. Thus, overexpression of the constitutively active form (GTP-bound mutant) of RagA (or B), regardless of the nucleotide status of RagC (or D), can activate mTORC1 even in the absence of amino acids 5, 6. It has been reported that amino acids can stimulate RagA/B and RagC/D to bind GTP and GDP, respectively 6, therefore leading to mTORC1 activation. However, a recent study showed that the binding of Rag GTPases to mTORC1 is regulated by amino acids without altering GTP charging of the Rag GTPase heterodimer, suggesting that additional mechanisms may be involved in amino acid-induced mTORC1 activation7.
Rag GTPases are anchored on the surface of lysosomes by a protein complex called Ragulator in close proximity to Rheb 8. The active Rag heterodimeric complex (RagA/B GTP-bound and RagC/D GDP-bound) binds directly to Raptor, a subunit of the mTORC1 complex, thereby recruiting the kinase complex to lysosome. At the lysosome, the mTORC1 is activated by Rheb, which mediates growth factor signals by acting downstream of the TSC1/TSC2 GTPase activating protein (GAP)9,10. The Ragulator also functions as a GEF (guanidine exchange factor) for RagA/B and connects the v-ATPase (vacuolar H+-adenosine triphosphatase) to the Rag GTPases for mTORC1 activation by amino acids 11, 12. Therefore, Rag GTPases mediates amino acid signals by recruiting the mTORC1 complex to lysosome.
Lysosomes are cellular organelles that hydrolyze metabolites and cellular wastes to maintain cellular homeostasis or recycling of nutrients. Lysosomal dysfunction caused by genetic mutations in lysosomal proteins causes lysosomal storage diseases (LSDs). Thus, LSDs are associated with abnormal accumulation of metabolites, and the major affected organs are the metabolic centers of the body such as liver, skeletal muscle, and heart 13, 14. Rag GTPases-mTORC1 signaling regulates lysosomal biogenesis via transcription factor EB (TFEB), which promotes lysosomal protein expression 15-17. Rag GTPases and mTORC1 regulate TFEB by direct binding and phosphorylation, respectively. Intriguingly, overexpression of TFEB can facilitate autophagy flux and ameliorate LSDs in vitro and in vivo 18-20. Therefore, it is conceivable that the downregulation of Rag GTPases-mTORC1 signaling may facilitate lysosome function due to TFEB activation.
In this study, we investigated the physiological function of Rag GTPases using conditional knockout mice. We show that loss of RagA/B in cardiomyocytes causes cardiac hypertrophy and phenocopies lysosomal storage diseases. Moreover, we demonstrate that despite constitutive activation of TFEB, the lysosomal expression of v-ATPase is decreased in Rag A/B knockout (KO) cells, resulting in compromised lysosomal acidification and dysfunction. Our study provides new insights into the critical physiological functions of RagA/B in heart and defines a novel role of Rag GTPases in lysosomal function by regulating lysosomal v-ATPase expression.
Results
Loss of RagA and RagB in heart causes cardiac hypertrophy
Formation of heterodimer is essential for Rag GTPase function and stability. Therefore, we reasoned that deletion of both RagA and RagB, which are highly homologous and functionally redundant, should be sufficient to inactivate the Rag GTPases. To investigate the physiological role of Rag GTPases in specific tissues, we generated RagA and RagB double conditional knockout mice using the Cre-loxP system (Supplementary Fig.1A).
Since Rag GTPases regulate amino acid-mediated mTORC1 activation 5, 6, and mTORC1 signaling plays a critical role in the muscle tissues 21-24, we utilized a transgenic mouse model that expresses Cre recombinases under the control of muscle creatine kinase promoter (Mck-Cre) to delete the floxed alleles in skeletal and cardiac muscle tissues 25. In the course of generating Mck-Cre/+;RagA fl/fl;RagB fl/fl (hereinafter RagA/B cKO), we first confirmed the Cre-mediated deletion of floxed alleles in RagA fl/fl or RagB fl/fl mice using genomic DNA PCR analyses (Supplementary Fig. 1B). Although the recombination of floxed alleles by Cre was detected in skeletal muscles, we were surprised that the RagA protein levels were not significantly reduced in the RagA/B cKO muscles (Supplementary Fig. 2B). Moreover, we did not find any noticeable pathological phenotypes in skeletal muscles of RagA/B cKO mice (Supplementary Fig. 2). Based on these observations, we speculate that the lack of phenotype in the skeletal muscles of RagA/B cKO mice is probably due to both the incomplete deletion (less than 100%) of the floxed alleles by the Cre recombinase and the presence of multiple nuclei (some with RagA/B deletion and some with no RagA/B deletion) in skeletal muscle fibers. Therefore muscle fibers still have some myofiber nuclei in which RagA and RagB are not deleted.
On the other hand, loss of RagA and RagB in cardiomyocytes caused severe cardiac hypertrophy (Figure 1 and 2). Unlike the skeletal muscle, the RagA protein levels were significantly reduced in the heart tissues of RagA/B cKO mice (Figure 1A). Because the anti-RagA antibody could also weakly detect RagB, we found that RagB protein was detected in the control heart tissues, but not in the RagA/B cKO hearts (arrowhead in Fig. 1A), showing that both RagA and RagB floxed alleles were efficiently deleted by Cre in the heart.
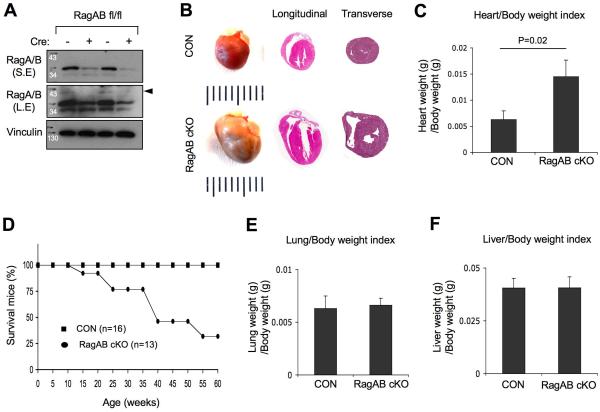
Figure 1. Loss of RagA and RagB causes cardiomegaly and premature death.
(A) RagA and RagB protein levels are decreased in RagA/B cKO hearts. Heart tissue lysates were prepared from control or RagA/B cKO mice and analyzed by immunoblotting. S.E, short exposure; L.E, long exposure. (B) Representative images of whole mount hearts (4 months old), and H&E stained longitudinal and transverse sections of the hearts from 4-month and 1-year old mice, respectively. Ruler represents 1 mm per interval. (C) Heart/Body weight index. Heart weight was divided by body weight. Values represent the mean ± SD of data (n=8 per group). P=0.02, Wilcoxon rank sum test. (D) Survival curve of control and RagA/B cKO mice. P <0.00001, Mantel-Cox (logrank) test. (E) Lung/body weight index. Lung weight was divided by body weight of the mouse. Values represent the mean ± SD of data (n=8 per group). (F) Liver/body weight index. Liver weight was divided by body weight of the mouse. Values represent the mean ± SD of data (n=8 per group).
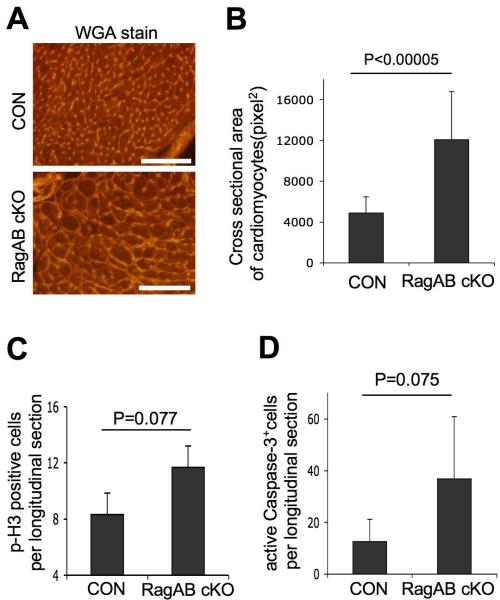
Figure 2. Cardiac hypertrophy in RagA/B cKO mice.
(A) Images of wheat germ agglutinin (WGA)-stained heart tissues. Representative images from control and RagA/B cKO tissues are shown. Scale bars, 100 μm. (B) Cross sectional area of cardiomyocytes. The area of cardiomyocytes from (A) was measured using the ImageJ software. Values represent the mean ± SD of data (n=15 per group). (C) Quantification of cell proliferation in heart tissues. Longitudinal sections of whole mount of hearts were stained with phsopho-Histone3 (p-H3) antibody to detect proliferating cardiomyocytes. The p-H3 positive cells in a whole longitudinal tissue section were counted. Values represent the mean ± SD of data (n=3 per group). (D) Quantification of apoptotic cell death in heart tissue. The serial sections of tissues from (C) were examined for apoptotic cell death by immunohistochemical staining using antibody that binds to cleaved (active) caspase-3. The active caspase-3 positive cells in a whole longitudinal tissue section were counted. Values represent the mean ± SD of data (n=5 per group). P values were determined by Wilcoxon rank sum test.
The difference of heart size between RagA/B cKO mice and control littermates was easily noticeable, and both left and right ventricles were highly dilated in RagA/B cKO hearts (Figure 1B). Consistent with the increased heart size, the heart/body weight index of RagA/B cKO mice was significantly increased compared with the control mice (Figure 1C), whereas the body weight of RagA/B cKO mice was not significantly different from the control littermates. Addtionally, we found that the RagA/B cKO mice displayed a premature sudden death phenotype (Figure 1D). 50% of RagA/B cKO mice died at 7~8 months, but did not show any noticeable signs of illness or pain like hunched position or impaired motor activity before the death. Meanwhile, the weight of lung and liver tissues was not affected in RagA/B cKO mice (Figure. 1E and F), suggesting a relatively compensated hemodynamic state with no clear signs of overt heart failure.
Because an increase of cell size and/or cell proliferation could contribute to organ size, we investigated which factors could have contributed to the heart enlargement. We compared the size of RagA/B cKO cardiomyocytes with control cardiomyocytes using heart tissue sections stained with wheat germ agglutinin (Figure 2A). The cell size measurement showed that the RagA/B cKO cardiomyocytes are almost 3 times larger when compared to control cardiomyocytes (Figure 2B). While a cell proliferation marker, phospho-Histone3 (p-H3) staining showed a slight increase (P=0.077, Wilcoxon rank sum test) of p-H3 positive cells in RagA/B cKO hearts despite few overall proliferating cells in a whole longitudinal section (Figure. 2C). Since there was a marginal increase (P=0.075) of cleaved caspase-3 positive cells in RagA/B cKO hearts (Figure. 2D) and the RagA/B cKO hearts displayed massive degeneration (see below), the increased number of proliferating cells in RagA/B cKO hearts may be due to a regeneration or repair process of the cardiac muscle 26. Therefore, we conclude that the cardiac enlargement of RagA/B cKO mice is resulted from the cellular hypertrophy of cardiomyocytes.
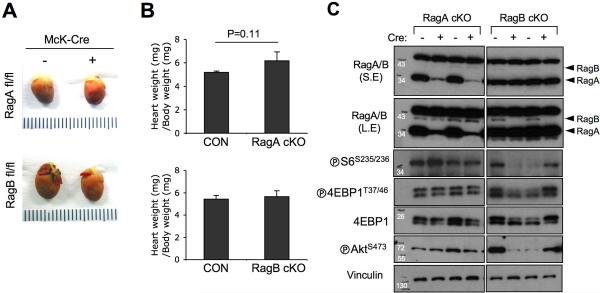
Of note, the single conditional knockout of either RagA or RagB in cardiomyocytes was not sufficient to induce cardiac enlargement (Figure. 3A), further supporting a functional redundancy of RagA and RagB in the heart. Thus, the heart/body weight index of the single conditional knockout mice was not different from the control littermates (Figure. 3B), and mTORC1 signaling was not altered in the single knockout mice (Figure. 3C). Despite the dominant RagA expression in the heart, RagB protein level was not changed upon RagA deletion (Figure. 3C).
Figure 3. RagA or RagB single conditional KO in heart does not cause cardiac hypertrophy.
(A) Representative images of whole mount hearts. Hearts were isolated from Mck-Cre/+;RagA flox/flox (2 months old) or Mck-Cre/+;RagB flox/flox (3 months old) mice, and the size of heart was compared with control littermates. (B) Heart/Body weight index. Heart weight was divided by body weight. Values represent the mean ± SD of data (n=3 per group). P values were determined by signed rank sum test. (C) Immunoblot analysis using heart tissue lysates. Tissue lysates from control, RagA, or RagB single cKO hearts were analyzed by immunoblotting using the indicated antibodies.
RagA/B cKO causes heart dysfunction and cardiomyopathy
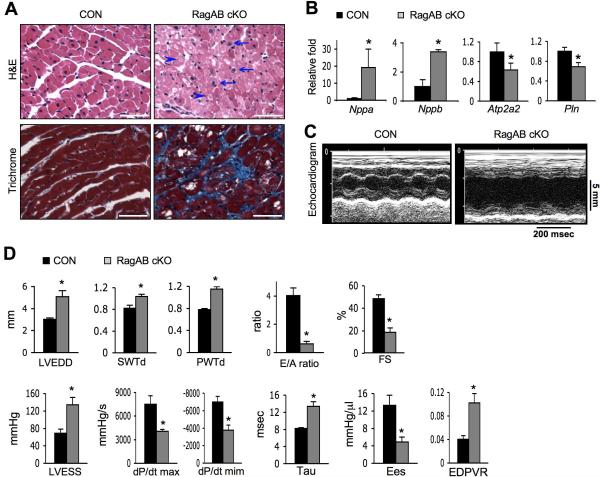
We further examined the pathologic features of the cardiac hypertrophy in RagA/B cKO mice. Histological analysis demonstrated characteristics of cardiomyopathy in RagA/B cKO hearts, including myocyte disarray and nuclear enlargement (Figure 4A, refer to arrows). Furthermore, vacuole formation was prevalent (arrow heads in Fig. 4A), and fibrosis was evident by the positive trichrome staining in 1 year old mice (Figure 4A, bottom panel).
Figure 4. Characterization of the cardiac hypertrophy in RagA/B cKO mice.
(A) Images of H&E (top, 2 month-old mice) or Masson's trichrome (bottom, 1 year-old mice) stained heart tissues. Arrows and arrowheads point enlarged nuclei and vacuole formation, respectively. Representative images are shown. Scale bars, 50 μm. (B) Molecular markers of cardiac remodeling in RagA/B cKO hearts. mRNA levels of indicated genes were determined by quantitative RT-PCR. Values represent the mean ± SD of data (n=4 per group). (C) Images of echocardiography. Representative images of tracing are shown. (D) Measurements of echocardiography and parameters of hemodynamic analyses. LVEDD, left ventricular end-diastolic diameter; SWTd, diastolic septal wall thickness; PWTd, diastolic posterior wall thickness; E/A ratio, early to late ventricular filling velocity; FS, fractional shortening; LVESS, left ventricular end-systolic stress; dP/dt max and min, the rate of left ventricle pressure rise in systole and diastole, respectively;.Ees, end-systolic elastance; EDPVR, end-diastolic pressure-volume relation slope. Values represent the mean ± S.E of data (n=4 or 5 per group). * P < 0.05, Wilcoxon rank sum test.
Functional failure of heart is associated with the up- or down-regulation of particular biomarkers. Quantitative RT-PCR analysis demonstrated a typical profile of cardiac remodeling markers in RagA/B cKO hearts (Figure 4B). Both natriuretic peptide A (Nppa) and natriuretic peptide B (Nppb) mRNA levels were upregulated, indicating increased heart wall stress. In contrast, the cardiac muscle calcium pump, sarcoplasmic reticulum calcium ATPase 2 (Atp2a2), and its regulator phospholamban (Pln), were downregulated.
Consistently, echocardiography and hemodynamic analyses showed severe left ventricular dilation and contractile dysfunction of RagA/B cKO hearts (Figure 4C and 4D). Specifically, the left ventricular end-diastolic diameter, diastolic septal/posterior wall thickness, and left ventricular-end-systolic stress were significantly increased in RagA/B cKO hearts. Meanwhile, both hemodynamic and echocardiographic parameters of systolic function were altered in RagA/B cKO mice with respect to control mice. Fractional shortening, dP/dT and the end-systolic elastance (Ees) were significantly reduced in the cKO mice. Diastolic function also appeared to be decreased in RagA/B cKO mice as compared to controls: E/A ratio and –dP/dT were significantly decreased, whereas Tau and the end-diastolic pressure-volume relation slope (EDPVR) were significantly increased. Taken together, these data show that RagA/B cKO display eccentric hypertrophy with severe reduction of cardiac function.
Defective autophagy flux in RagA/B KO cells
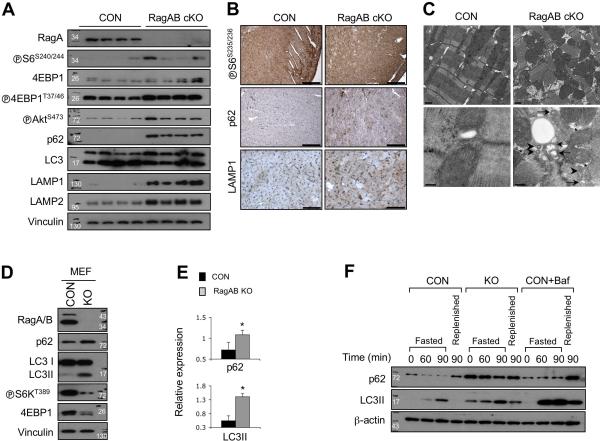
To determine the molecular mechanism of the cardiac hypertrophy, we investigated whether mTORC1 pathway was dysregulated in RagA/B cKO hearts (Figure 5A). Interestingly, the downstream targets of mTORC1, S6 and 4EBP1 phosphorylation levels were slightly increased in RagA/B cKO hearts, suggesting that mTORC1 activation was not severely impaired by loss of RagA/B in vivo. Immunohistochemical staining confirmed that the cardiomyocytes of RagA/B cKO mice maintained the level of S6 phosphorylation, suggesting that mTORC1 activity was not significantly compromised (Figure 5B). These observations also suggest an alternative mechanism in mTORC1 activation in the absence of RagA/B. Further analyses using another group of mice showed that the mTORC1 activation status in the heart varied among individual mice, but was not abnormally altered in the RagA/B cKO hearts (Supplementary Fig. 3). Interestingly the phospho-AktS473 level was increased in the RagA/B cKO hearts, suggesting that either the negative feedback loop of mTORC1 to mTORC2 was deregulated or the growth factor-Akt signaling axis was upregulated to compensate for the impaired cardiomyocytes in the RagA/B cKO hearts.
Figure 5. Defective autophagy flux in RagA/B cKO hearts and MEFs.
(A) Immunoblot analysis using heart tissue lysates. Tissue lysates from control or RagA/B cKO hearts were analyzed by immunoblotting using the indicated antibodies. (B) Immunohistochemical analysis of heart tissues. Heart tissue sections were analyzed by immunohistochemistry using the indicated antibodies. Representative images are shown. Scale bars, 200 μm (◅S6S235/236 and p62); 50 μm (LAMP1). (C) Transmission electron microscopy of heart tissues. The ultrastructure of control and RagA/B cKO hearts was examined by TEM. Arrows and arrowheads indicate accumulation of AVi (initial autophagic vacuoles) and AVd (late autophagic vacuoles/autolysosomes), respectively. Asterisks point the electron-lucent spaces. Scale bars, 1 μm (top panel); 200 nm (bottom panel). (D) Immunoblot analysis of control and RagA/B KO MEFs. Cells were cultured in a nutrient rich condition, and then cell lysates were analyzed by immunoblotting with the indicated antibodies. (E) Quantification of immunoblot data. Levels of p62 and LC3II were determined by densitometry. Values represent the mean ± SD of data (n=3). *P < 0.05, Wilcoxon rank sum test. (F) Immunoblot analysis of autophagy flux. Cells were cultured in a nutrient rich medium, and then incubated in HBSS to induce autophagy. For the replenished samples, cells were re-incubated for another 90 min in a nutrient rich medium after 90 min of starvation. A set of control cells was cultured in the presence of bafilomycin A1 (100 nM). Cell lysates were prepared at the indicated time points and analyzed by immunoblotting. CON, control MEFs; KO, RagA/B KO MEFs; Baf, bafilomycin A1.
Inhibition of autophagy flux is known to cause cardiac hypertrophy in mice 27, 28, and the Rag GTPases localize lysosomes that play critical roles in autophagy flux. Thus, we examined whether the autophagy flux was affected in RagA/B cKO hearts. The conversion of LC3-I to LC3-II (a lipidated form of LC3) is a commonly used as a marker for autophagy induction, whereas the degradation of p62 is used to measure protein degradation by autophagy flux. Interestingly, p62 protein was abnormally accumulated in RagA/B cKO hearts even though the level of LC3 lipidation was not significantly different from that of control hearts (Figure 5A-B). These results suggest that autophagy initiation (autophagosome formation) may not be significantly altered while autolysosomal degradation is blocked in RagA/B cKO hearts 29. In addition, the lysosomal marker proteins, lysosome-associated membrane protein-1 (LAMP1) and LAMP2, were abnormally accumulated in RagA/B cKO hearts (Figure 5A-B). We further assessed the ultrastructure of cardiomyocytes in tissue sections using transmission electron microscopy (TEM) (Figure 5C and Supplementary Fig. 4A-F). RagA/B cKO cardiomyocytes displayed degeneration and myofibril disarray. Consistent with the observed vacuoles in histological analysis (Figure 4A), there were many electron-lucent spaces in the cytoplasm of RagA/B cKO myocytes (asterisks in Figure 5C and Supplementary Fig. 4A-F). Moreover, they were associated with accumulation of initial autophagic vacuoles (AVi, arrows) and late (degradative) autophagic vacuoles/autolysosomes (AVd, arrowheads in Figure 5C and Supplementary Fig. 4A-F) 29. Accordingly, these data indicate that autophagosomes and autolysosomes are abnormally accumulated in RagA/B cKO hearts.
Because the autophagy flux is a very dynamic process and it is difficult to manipulate the process in vivo, we utilized RagA and RagB double knockout mouse embryonic fibroblast cells (RagA/B KO MEFs) for further investigation. First, we performed immunoblot analysis using cells that were cultured in nutrient rich conditions (Figure 5D). Both RagA and RagB proteins were absent in RagA/B KO MEFs, and phosphorylation of the mTORC1 substrates, S6K and 4EBP1, was decreased but not abolished. Interestingly, both p62 and LC3-II levels were increased about 2 and 3 fold, respectively, in RagA/B KO MEFs compared with control cells (Figure 5E). To compare the autophagy flux of RagA/B KO MEFs with that of control cells under different nutrient conditions, cells were fasted with the Hank's balanced salt solution (HBSS) for 60 or 90 minutes, and then cultured in a nutrient rich medium for 90 minutes (Figure 5F). Again, the basal LC3-II level was higher in RagA/B KO MEFs compared to control cells under normal conditions (time 0). LC3-II levels were further increased upon fasting in both wild type and RagA/B KO MEFs, suggesting that the LC3 lipidation and autophagy initiation were normally induced in the KO cells. Consistently, rapamycin, an mTOR inhibotor treatment further increased the LC3-II formation in the RagA/B KO MEFs under the nutrient rich condition (Supplementary Fig. 4G). These data show a tight inverse correlation between mTORC1 activity and LC3-II formation. Thus, the elevated basal LC3-II formation in the KO MEFs was probably due to, at least in part, the reduced mTORC1 activity in these cells. Meanwhile, LC3-II in RagA/B KO MEFs did not disappear after the cells were replenished with a nutrient rich medium, whereas LC3-II disappeared in control MEFs under the same treatment (Figure 5F). Taken together, these data indicate that autophagy initiation can still be stimulated by nutrient deprivation in the RagA/B KO cells. However, p62 protein level was maintained at a higher level and remained even under starvation conditions in RagA/B KO MEFs despite more LC3 lipidation, suggesting that degradation of p62 was uncoupled from LC3-II accumulation (autophagy induction) in RagA/B KO MEFs. Of note, accumulation of both p62 protein and LC3-II in RagA/B KO MEFs was similar to the control MEFs treated with bafilomycin-A1, a v-ATPase inhibitor that blocks autophagy flux by inhibiting lysosomal acidification (Figure 5F). Furthermore, we also observed that bafilomycin-A1 treatment during autophagy flux did not further increase the accumulation of p62 and LC3-II in the RagA/B KO MEFs (Supplementary Fig. 4H). Taken together, these results support a model that the late stage of autophagy flux, autolysosomal degradation is likely compromised in RagA/B KO MEFs.
Altered expression of lysosomal proteins in RagA/B KO MEFs
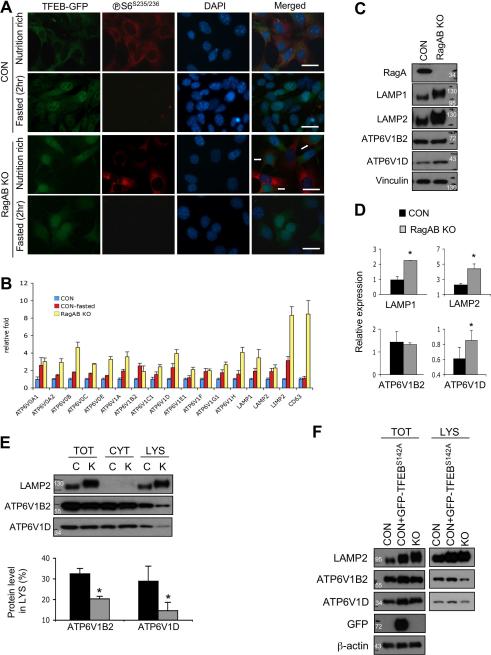
Recent studies showed that TFEB is a master regulator of lysosome biogenesis and autophagy by stimulating expression of many lysosomal and autophagy related genes15. TFEB function is regulated by nutrients and its nuclear localization is inhibited by mTORC1 and Rag GTPases 16, 17, 30. Interestingly, the levels of the lysosomal marker LAMP1 and LAMP2 were increased in RagA/B cKO hearts (Figure 5A). Accordingly, we explored the possibility that deregulation of TFEB in RagA/B KO MEFs may contribute to lysosomal dysfunction. At first, control and RagA/B KO MEFs were infected with recombinant retroviruses expressing TFEB-GFP, and then the localization of TFEB was investigated under either nutrient rich conditions or starvation conditions (Figure 6A). As expected, TFEB-GFP protein was exclusively localized in the cytoplasm under nutrient rich conditions that promoted high mTORC1 activity as indicated by the positive staining for phospho-S6S235/236. TFEB-GFP was translocated to the nucleus in control MEFs under starvation conditions (2 hours in HBB solution). In contrast, TFEB-GFP protein was localized in the nucleus in RagA/B KO MEFs regardless of nutrient conditions and even in the phospho-S6S235/236 positive cells (arrows, Figure 6A). These data show that TFEB is constitutively active in RagA/B KO cells regardless of the mTORC1 activation status.
Figure 6. Altered lysosomal protein expression in RagA/B KO MEFs.
(A) Nuclear localization of TFEB in RagA/B KO MEFs. TFEB-GFP expressing control or RagA/B KO MEFs were cultured either in a nutrient rich medium or in HBSS for 2 hours, and then cells were fixed to examine the localization of TFEB-GFP proteins and immuno-stained with phospho-S6S235/236 antibody. Representative images are shown. Scale bars, 50 μm. (B) Quantitative RT-PCR of v-ATPase subunits and lysosomal membrane proteins. Total RNAs were prepared from control, 2 hour-fasted control, or RagA/B KO MEFs, and the expression levels of indicated genes were determined. (C) Immunoblot analysis of v-ATPase subunits and lysosomal membrane proteins. The levels of lysosomal membrane proteins and v-ATPase subunits were compared by immunoblotting. (D) Quantification of immunoblot analysis by densitometry. (E) The levels of v-ATPase subunits in lysosome fraction. Cell lysates were fractionated, and the amounts of v-ATPase subunits in each fraction were compared between control and RagA/B KO MEFs. The loading amount of cell lysates was normalized with ATP6V1D, and the percentage of the v-ATPase subunit level in lysosome fraction was determined by densitometric analysis. (F) Comparison of the lysosomal v-ATPase level in TFEBS142A expressing cells. The amounts of LAMP2, ATP6V1B2, and ATP6V1D in total or lysosomal fraction of GFP- TFEBS142A expressing cells were compared with control and RagA/B KO MEFs. For the lysosomal fraction, the protein loading amounts were normalized with LAMP2 levels. C, control MEFs; K, RagA/B KO MEFs; TOT, total; CYT, cytosolic fraction; LYS, lysosome fraction. All values represent the mean ± SD of data (n=3 per group). *P < 0.05, Wilcoxon rank sum test.
To determine the transcriptional activity of TFEB in control and RagA/B KO MEFs, we measured the expression of TFEB target genes by quantitative RT-PCR (qPCR) (Figure 6B). We assessed a subset of known TFEB direct target genes, including some v-ATPase subunits and lysosomal membrane proteins, because they are essential for lysosomal acidification and function 31-33. As expected, most of the TFEB target genes were upregulated by starvation in control MEFs (Figure 6B). We also found that all TFEB target genes tested were significantly upregulated in RagA/B KO MEFs compared with control MEFs under nutrient rich conditions, further supporting the constitutive activation of TFEB in RagA/B KO MEFs.
We next compared the protein levels of TFEB target genes using immunoblotting analysis (Figure 6C). Consistent with the qPCR data, the protein levels of LAMP1 and LAMP2 were about 2 fold more abundant in the RagA/B KO MEFs than the control MEFs. However, the v-ATPase subunit ATP6V1B2 protein level was not changed while another v-ATPase subunit ATP6V1D protein level was marginally (about 50%) increased in RagA/B KO MEFs (Figure 6D). Thus, these data indicate that the molecular ratio between the lysosomal membrane proteins and v-ATPase subunits is altered in RagA/B KO MEFs.
v-ATPase can localize on intracellular vesicular membranes and the plasma membrane. Lysosomal localization of the v-ATPase is required to acidify lysosomes. Therefore, we investigated whether the altered expression of lysosomal proteins affects the level of v-ATPase on lysosomes in RagA/B KO MEFs. We fractionated cells into cytoplasm and lysosome fractions, and examined the v-ATPase distribution (Figure 6E, the total protein loading amounts were normalized with ATP6V1D levels). We found that the levels of both ATP6V1B2 and ATP6V1D protein in the lysosome fraction were significantly lower in RagA/B KO MEFs compared with control cells, and the relative ratio of v-ATPase subunits to the lysosomal membrane protein LAMP2 was decreased in RagA/B KO cells. Quantification analysis revealed that only about 15~20% of total ATP6V1B2 and ATP6V1D was co-isolated in the lysosome fraction of RagA/B KO MEFs, while about 30~33% of those proteins resided in the lysosome fraction of control MEFs.
These data are intriguing because it has been shown that the activation of TFEB facilitates autophagy flux and lysosomal function 18-20. Therefore, we further investigated whether constitutive activation of TFEB contributed to the decreased v-ATPase expression in the lysosome fraction. Overexpression of a constitutively active mutant TFEB (TFEBS142A) 18 in control MEFs upregulated both LAMP2 and ATP6V1D protein levels similar to those in the RagA/B KO cells, supporting the notion that RagA/B regulate lysosomal protein expression via TFEB. However, the lysosomal localization of ATP6V1B2 and ATP6V1D in the TFEBS142A expressing cells was still higher than the RagA/B KO MEFs (Figure 6F, the lysosome fractions were normalized with LAMP2 levels), indicating that RagA/B influence v-ATPase localization independent of TFEB. Taken together, our data suggest that the Rag GTPases modulate lysosomal function, at least in part by affecting lysosomal protein expression and localization.
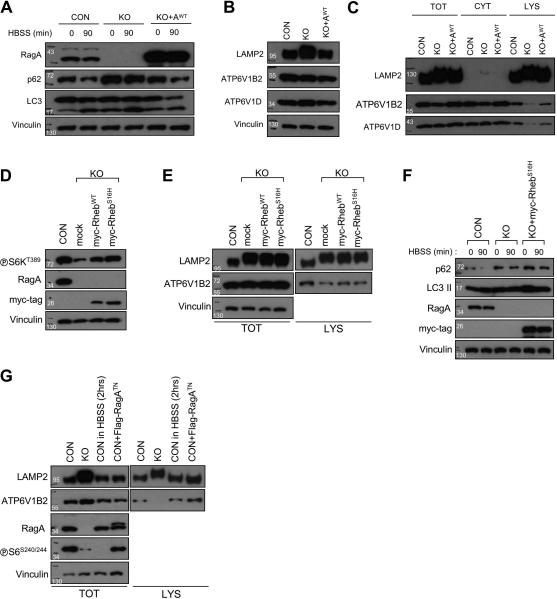
In order to determine whether the decreased lysosomal v-ATPase expression in RagA/B KO MEFs can be restored by RagA, the RagA/B KO MEFs were infected with RagA-encoding recombinant retroviruses. Re-introduction of RagA in the RagA/B KO MEFs not only restored both ATP6V1B2 and ATP6V1D levels in the lysosome fraction, but also rescued the defects of autophagy flux (Figure. 7A-C, the fractionated samples were normalized with LAMP2 level). Furthermore, to investigate whether the diminished mTORC1 activity affected the lysosomal v-ATPase localization in RagA/B KO MEFs, we also generated RagA/B KO MEF cell lines expressing either RhebWT or constitutively active Rheb (RhebS16H). Despite substantial restoration of mTORC1 activity by RhebWT or RhebS16H as indicated by increased S6K1 phosphorylation (Figure 7D), the total LAMP2 protein level and lysosomal ATP6V1B2 localization were not restored to normal (Figure 7E). Moreover, autophagy flux was not recovered in RhebS16H expressing RagA/B KO MEFs (Figure 7F). We also examined whether mTORC1 inactivation by either nutrient deprivation or dominant negative RagA (RagATN) expression in control MEFs affects lysosomal v-ATPase localization. The expression of RagATN did not completely inhibit mTORC1 probably due to insufficient expression level and had no substantial effect on the lysosomal localization of v-ATPase. Intriguingly, the nutrient deprivation also did not affect the lysosomal v-ATPase localization despite of complete mTORC1 inactivation (Figure 7G). Collectively, these data indicate that the Rag GTPases modulate the lysosomal v-ATPase localization independent of mTORC1 activity.
Figure 7. Restored autophagy flux and lysosomal v-ATPase localization in RagA/B KO MEFs by RagAWT expression, but not by RhebWT or RhebS16H expression.
(A) Restoration of autophagy flux in RagA/B KO MEFs by reintroducing RagAWT. RagA/B KO MEFs were infected with wild-type RagA (RagAWT) expressing retroviruses, and then autophagy flux was examined by immunoblotting. (B) Immunoblot analysis of TFEB target gene expression. The expression levels of LAMP2, ATP6V1B2, and ATP6V1D were restored by RagAWT expression in the RagA/B KO MEFs. (C) Immunoblot analysis of lysosomal v-ATPase subunits. Cells were fractionated, and the levels of v-ATPase subunits in total, cytoplasm, and lysosomal fraction were examined by immunoblotting. The protein loading amounts were normalized with LAMP2 protein levels. (D) RhebWT or RhebS16H expression substantially restores mTORC1 activity in RagA/B KO MEFs. Stable cell lines were generated by infecting RagA/B KO MEFs with RhebWT or RhebS16H encoding retroviruses, and then cell lysates were analyzed by immunoblotting. (E) RhebWT or RhebS16H does not restore lysosomal v-ATPase localization in RagA/B KO MEFs. Cells were fractionated, and the lysates were analyzed by immunoblotting. (F) Immunoblot analysis of autophagy flux. Cells were cultured in a nutrient rich medium followed by starvation in HBSS for 90 min, and autophagy flux markers were examined by immunoblotting. (G) Nutrient deprivation or RagATN expression in control MEFs does not alter lysosomal v-ATPase localization. Starved control MEFs (in HBSS for 2 hrs) or RagATN expressing control MEFs were fractionated and analyzed by immunoblotting. CON, control MEFs; KO, RagA/B KO MEFs; AWT, RagA wild-type; TOT, total; CYT, cytoplasm; LYS, lysosome fraction.
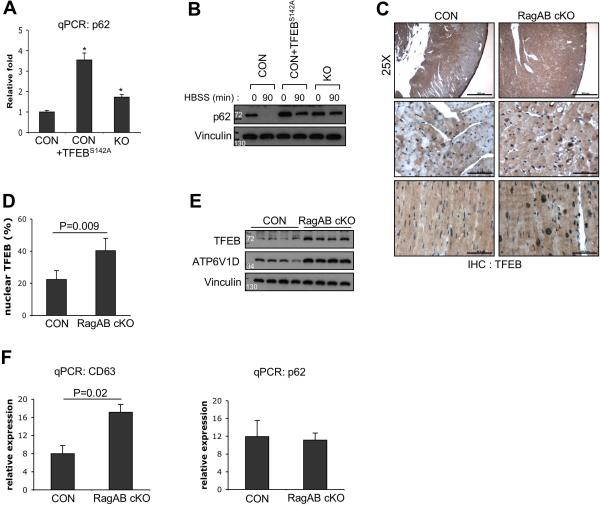
Given the fact that TFEB regulates p62 expression18 and its constitutive activation in RagA/B KO MEFs, we examined whether the amount of p62 mRNA in the KO MEFs or TFEBS142A expressing control MEFs were upregulated. qPCR analyses showed that p62 mRNA levels were about 1.7 and 3.5 fold increased in the RagAB KO MEFs and TFEBS142A expressing cells, respectively (Figure 8A). However, nutrient starvation could substantially decrease p62 protein in both the control and the TFEBS142A expressing MEFs, but not in the RagA/B KO MEFs (Figure 8B). As expected, p62 levels were higher in the TFEBS142A MEFs likely due to enhanced transcription. These data indicate that both the defective autolysosomal degradation and transcriptional upregulation contribute to the increase level of p62 in the RagA/B KO MEFs.
Figure 8. Increased TFEB activity in RagA/B KO MEFs and RagA/B cKO hearts.
(A) p62 mRNA levels were upregulated in RagA/B KO MEFs and TFEBS142A expressing control MEFs. Total RNAs were prepared from cells and p62 mRNA levels were quantified using qPCR. Values represent mean ± SD of data (n=4 per group). *P value=0.021. (B) Immunoblot analysis of p62 degradation by starvation. p62 levels were substantially decreased in control and TFEBS142A expressing control MEFs by starvation (in HBSS for 90min), but not in RagA/B KO MEFs. (C) Immunohistochemical analyses of heart tissues using TFEB antibody. Top panel, low magnification view shows uneven distribution of TFEB expression in the control hearts (scale bars=500μm). Middle panel, high magnification view of left ventricular wall areas (scale bars=50μm). Bottom panel, inter-ventricular septum areas (scale bars=50μm). (D) Quantification of TFEB positive nuclei. The number of TFEB positive nuclei were divided by total number of nuclei per microscopic field. Values represent mean ± SD of data (n=5 per group). (E) Immunobot analysis of heart tissue lysates. TFEB and its target ATP6V1D levels were upregulated in RagA/B cKO hearts compared with controls. (F) Quantitative RT-PCR analyses of TFEB target genes from heart tissues. Total RNAs were prepared from heart tissues, and mRNA levels of TFEB target genes, CD63 and p62 were quantified by qPCR. Values represent mean ± SD of data (n=4 per group). P values were determined by Wilcoxon rank sum test.
To examine the role of the RagA/B in TFEB regulation in vivo, we investigated nuclear TFEB localization in the RagA/B cKO heart tissue using immunohistochemistry (Figure 8C). TFEB expression was not distributed evenly in control hearts, specifically lower in the ventricular wall area, whereas consistent TFEB expression was observed in RagA/B cKO hearts (top panel, Figure 8C). Quantitative analysis showed that the percentage of TFEB positive nuclei per field was increased about 2 fold in the RagA/B cKO hearts compared with control hearts (Figure 8D). Consistent with this observation, immunoblotting analyses showed that TFEB protein level and its transcriptional target ATP6V1D were increased in the RagA/B cKO hearts (Figure 8E), confirming that TFEB activity was upregulated in the cKO hearts. Of note, it has been also suggested that TFEB positively regulates its own expression34. However, unlike RagA/B KO MEFs, p62 mRNA level was not significantly increased in the RagA/B cKO hearts even though another TFEB target gene, CD63 was significantly upregulated (Figure 8F). A possible explanation is that the elevated p62 protein in the RagA/B cKO hearts (Figure 5A) might indirectly suppress its own expression. Alternatively, the high basal TFEB activity in the control hearts (nuclear TFEB signals were observed in ~20% cells, Figure 8D) may mask the p62 induction by the RagA/B KO-induced further activation of TFEB. Nonetheless, our data suggest that in the RagA/B cKO hearts, impaired autolysosomal degradation contributes to p62 accumulation.
Lysosomal acidification is compromised in RagA/B KO MEFs
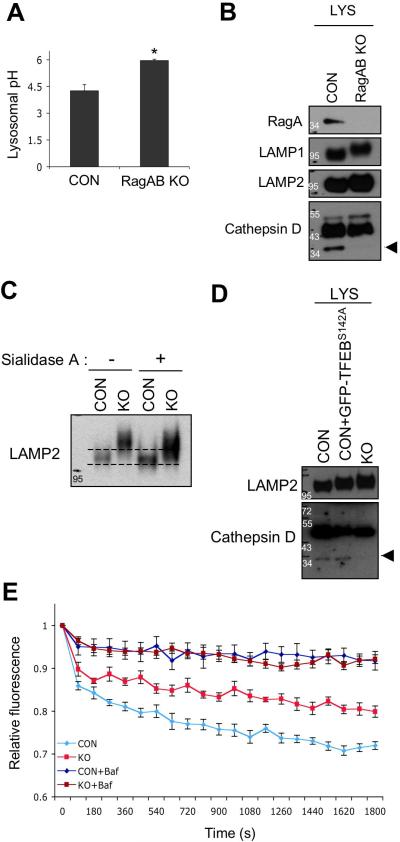
The accumulation of p62 and autophagosomes in the RagA/B KO hearts and MEFs suggests a possible defect in lysosomal degradation. Acidification is critical for lysosome function and hydrolytic degradation. Therefore, we assessed the lysosomal pH in live cells to determine whether the decreased lysosomal v-ATPase level affected the pH of lysosome. The lysosomal pH of RagA/B KO MEFs was significantly higher than that of control MEFs (Figure 9A), supporting the notion that the Rag GTPases play a role in lysosomal function. Some lysosomal hydrolytic enzymes are matured by a proteolytic process in a pH-dependent manner, like cathepsin D maturation 35. Thus, we investigated whether the maturation of cathepsin D was defective in RagA/B KO MEFs. Immunoblot analysis using the lysosome fractions (normalized with LAMP1 protein level) showed that the matured cathepsin D was significantly impaired in RagA/B KO MEFs (arrow head, Figure 9B), indicating defective acidification of lysosome and dysfunction. Moreover, we consistently observed that both LAMP1 and LAMP2 proteins present higher molecular weights in RagA/B KO MEFs compared with control MEFs. Multiple post-translational modifications occur in the lysosomes, and the sialylation of LAMP1 and 2 are modified by lysosomal N-acetyl-α-neuraminidase (sialidase) 36, 37. Thus, treatment of sialidase A increased the mobility of LAMP2 in the RagA/B KO MEFs, although not to the migration of the LAMP2 in the control MEFs (Figure 9C), indicating that LAMP2 proteins were over-sialylated in RagA/B KO MEFs. Consistent with unchanged lysosomal v-ATPase level in TFEBS142A expressing control MEFs (Figure 6F), we also observed that the maturation of cathepsin D was not impaired in those cells (Figure 9D).
Figure 9. Lysosomal acidification is compromised in RagA/B KO MEFs.
(A) Lysosomal pH measurement in live cells. Cells were fed with OG541-conjugated dextran for overnight followed by serum-starvation for 1 hour, and then trypsinized for spectrofluorimetric measurement. Values represent the mean ± SD of data (n=3 per group). *P < 0.05, Wilcoxon rank sum test. (B) Immunoblot analysis of cathepsin D maturation in lysosome fraction. Lysosome fractions from control and RagA/B KO MEFs were analyzed by immunoblotting using the indicated antibodies. The amount of protein loading was normalized with LAMP1 protein level. Arrowhead indicates the matured form of cathepsin D. (C) Sialidase A treatment increases mobility of LAMP2 on SDS-PAGE. Cell lysates were incubated with sialidase A for 3hrs followed by SDS-PAGE and immunoblotting. (D) Cathepsin D maturation in GFP-TFEBS142A expressing cells. The amounts of matured cathepsin D in lysosomal fraction of GFP-TFEBS142A expressing cells were compared with control and RagA/B KO MEFs using immunoblotting. The amount of protein loading was normalized with LAMP2 protein level. Arrowhead indicates the matured form of cathepsin D. (E) A compromised lysosomal acidification capacity in RagA/B KO MEFs. Cells were fed with a pH sensitive fluorescent dye (OG541)-conjugated dextran for overnight, and then starved in a serum-free medium for 2 hours. v-ATPase-mediated acidification of lysosome-enriched fraction was measured in the absence or in the presence of bafilomycin A1 (100nM) in vitro. The amount of lysosome fraction was normalized with LAMP2 protein level. Each point represents the mean ± S.E of three 30-second intervals from three independent measurements. CON, control MEFs; KO, RagA/B KO MEFs; Baf, bafilomycin A1.
To determine the molecular mechanism for defects in lysosomal acidification of the RagA/B KO cells, we measured v-ATPase activity in the lysosome fraction, which was loaded with pH-sensitive fluorescent dye-conjugated dextran (Figure 9E). Bafilomycin-A1, a specific v-ATPase inhibitor, was used to confirm the specificity of the v-ATPase assay. Consistent with the altered v-ATPase protein levels, the acidification rate of lysosome from RagA/B KO MEFs was significantly reduced compared with that from control MEFs (Figure 9E). These results suggest that the reduced v-ATPase activity in the lysosomes of RagA/B KO cells is responsible for the defective lysosomal acidification and dysfunction. We next accessed whether Rag GTPases might directly regulate v-ATPase activity. Addition of immuno-purified wild-type or mutant RagA proteins (and co-immunoprecipitated RagC proteins) was not able to restore the lysosomal acidification in vitro (Supplementary Fig. 5A-D). Altogether, our data indicate that lysosomal acidification is compromised in RagA/B KO MEFs due to the reduced lysosomal v-ATPase, and thus resulting in lysosomal dysfunction.
RagA/B cKO hearts resemble lysosomal storage diseases
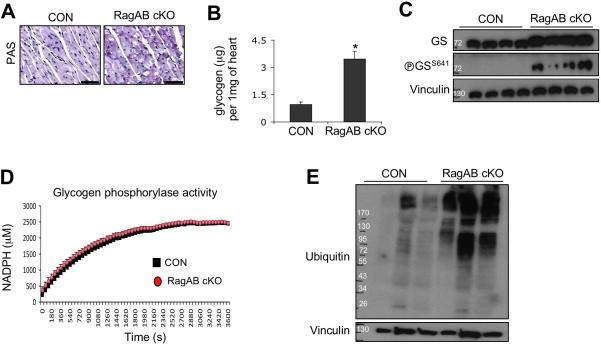
Mutations in multiple genes coding for lysosomal proteins cause lysosomal storage diseases (LSDs) in humans. Cardiac hypertrophy found in both Pompe disease (acid alpha-glucosidase defect) and Danon disease (LAMP2 defect) is associated with abnormal glycogen accumulation in cardiomyocytes due to the lysosomal dysfunction 38, 39. In fact, we noticed that glycogen was abnormally accumulated in RagA/B cKO hearts revealed by TEM (electro-dense granules (small double arrows) in Supplementary Fig. 4A-F), suggesting that the cardiac hypertrophy of RagA/B cKO hearts resembles lysosomal storage disease. We first confirmed the abnormal glycogen accumulation in RagA/B cKO heart sections by Periodic acid-Schiff staining (Figure 10A). Using a biochemical assay, we found that the amount of glycogen in the RagA/B cKO heart tissues was about 3 times more than the controls (Figure 10B).
Glycogen metabolism can be controlled by the classic glycogen synthase and phosphorylase. Immunoblot analysis using the heart tissue lysates showed that the glycogen synthase level was not different between control and RagA/B cKO hearts (Figure 10C). Glycogen synthase activity is inhibited by phosphorylation of S64140. We found that phosphorylation of S641 in glycogen synthase was higher in RagA/B cKO hearts than the control mice, indicating that the glycogen synthase activity was repressed in the cKO heart. This is probably due to a compensatory effect of cells to counter balance the elevated glycogen level in the RagA/B cKO heart. Therefore, the abnormal glycogen accumulation could not be due to hyperactivation of glycogen synthase in the RagA/B cKO hearts.
We next examined the glycogen phosphorylase, the major enzyme responsible for glycogen hydrolysis. Glycogen phosphorylase activity assays using tissue lysates demonstrated that the enzyme activity was indistinguishable between the control and the RagA/B cKO hearts (Figure 10D). Altogether, these results show that the abnormal glycogen accumulation in the RagA/B cKO heart is not due to glycogen synthase or phosphorylase, but lysosomal dysfunction may be the major contributor for the glycogen accumulation.
One of the hallmarks of autophagy flux defect is the accumulation of ubiquitinated proteins. Accumulation of ubiquitinated proteins is also observed in lysosomal storage diseases including a mouse model of Pompe disease 14, 41. Consistent with the defects in autophagy flux and lysosomal function in RagA/B cKO hearts, ubiquitinated proteins were massively accumulated in RagA/B cKO heart tissues compared with control hearts (Figure 10E). Collectively, these data show that the histopathological phenotype of RagA/B cKO hearts resembles the lysosomal storage diseases in humans.
Discussion
Rag GTPases are key transducers relaying amino acid signals to cellular regulation42. In response to amino acid stimulation, the Rag GTPases are activated by the Ragulator-mediated guanine nucleotide exchange and recruit mTORC1 to lysosomes via a direct binding to Raptor, a subunit of mTORC1. Rheb GTPase downstream of growth factor signaling subsequently activates the lysosomal associated mTORC1. mTORC1 has been the only well-defined physiological target of Rag GTPases in modulating cell metabolism and growth so far. In this study, we show that loss of both RagA and RagB in cardiomyocytes causes hypertrophic cardiomyopathy that resembles lysosomal storage diseases in humans. Notably, phosphorylation of mTORC1 downstream molecules was not significantly reduced in the RagA/B knockout heart, indicating alternative mechanism or compensation for mTORC1 activation in the RagA/B KO cells. Of interest, a recent study also showed that mTORC1 can be activated regardless of nutrient levels in the absence of RagA by unknown mechanism 43. Although further studies are necessary to elucidate the molecular mechanism of Rag GTPases-independent mTORC1 activation, our study suggests an mTORC1-independent function of Rag GTPases in regulating lysosome and autophagy flux. We propose that the defect in lysosome function and autophagy flux is responsible for the pathological phenotypes observed in the RagA/B cKO hearts. Constitutive activation of TFEB by the loss of Rag GTPases leads to excessive expression of lysosomal proteins with altered stoichiometry and decreased lysosomal localization. As a result, reduced lysosomal v-ATPase activity leads to the elevated lysosome pH and lysosomal dysfunction. Our study defines a new role of Rag GTPases in regulating lysosomal function in vivo.
As an integrator of nutrient levels in the cell, mTORC1 negatively regulates autophagy initiation 44. Because Rag GTPases mediate mTORC1 activation by nutrients, one may expect an increased autophagy flux by impaired mTORC1 activity due to the loss of Rag GTPases. In fact, our data show that the basal level of LC3 lipidation is increased significantly in RagA/B KO MEFs even under nutrient rich condition (Figure 5D and 5F), suggesting that autophagy initiation is increased in the RagA/B KO cells. However, p62, which is degraded in autolysosomes at the end of autophagy flux 45, 46, is accumulated in the KO cells. Therefore, the simultaneous accumulation of p62 and LC3-II demonstrates a defective autophagy flux in RagA/B KO MEFs. This is consistent with the defective lysosomal function in the KO cells. Intriguingly, it has also been shown that mTORC1 regulates the reformation of lysosomes subsequent to autolysosomal degradation 47. Currently, it is not clear whether the compromised mTORC1 activation by loss of Rag GTPases affects the reformation of lysosomes.
LSDs are associated with defective autophagy flux in most cases 48. In this study, we found that the hypertrophy of RagA/B cKO hearts displays characteristic features of LSDs: the accumulation of autophagosomes/autolysosomes, p62 protein, glycogen, and ubiquitinated proteins. Therefore, our in vitro and in vivo data strongly demonstrate that loss of Rag GTPases leads to lysosomal dysfunction. The final clearance of intracellular components by autophagy requires fusion with and degradation by lysosomes. Lysosomal dysfunction is likely the cause for the defects in autophagy flux and cardiomyopathy in the RagA/B cKO heart. Our data support a critical role of autophagy in maintaining healthy cardiac function.
Heart is one of the major organs affected in many LSDs, and infantile or childhood onset of Pompe disease is usually lethal due to heart failure 14, 39. RagA/B cKO mice also died prematurely (Figure 1D). Interestingly, the mice did not show any noticeable signs of illness before death, suggesting that death may be caused predominantly by a fatal arrhythmia, or in some cases by an acute heart failure development which immediately precedes the death. Interestingly, the lung weight was not changed in RagA/B cKO mice, suggesting that the hemodynamic state of these animals is relatively compensated despite the severe reduction of systolic function. This may be due to the activation of several known compensatory adaptations that limit the reduction of cardiac output and the increase of pulmonary vascular pressure and lung congestion in RagA/B cKO mice. RagA/B cKO mice develop extensive ventricular dilation that can partly compensate for the reduction of fractional shortening and limit the stroke volume drop. The Frank-Starling mechanism is probably still preserved in the heart of RagA/B cKO mice and it can temporarily increase the stroke volume in these animals. Chronic ventricular dilation is also known to be associated with increased ventricular compliance that would allow the ventricle of RagA/B cKO mice to have a greater end-diastolic volume without an extensive parallel increase of end-diastolic pressure. In addition, the observed increase of myocardial wall thickness and overall ventricular mass can temporarily increase ventricular contractility in RagA/B cKO mice. Finally, neurohormonal activation could also be another mechanism that would contribute to compensating the hemodynamic state of RagA/B cKO mice49.
Recent studies have shown potential roles of Rag GTPases in lysosome function. Rag GTPases interact with v-ATPases via Ragulator 8, 11. Moreover, Rag GTPases directly interact with TFEB to modulate the mTORC1-mediated TFEB cytoplasmic localization 16, 17. TFEB is constitutively activated in the RagA/B KO cells, and as a consequence, TFEB target gene expression is significantly elevated (Figure 6A-B). Despite the increased v-ATPase expression, the levels of the v-ATPase protein associated with lysosomes are actually decreased in RagA/B KO MEFs compared with control MEFs (Figure 6E). As a result, lysosomal v-ATPase activity is reduced and lysosomal pH is higher (Figure 9). Consistent with this notion, maturation of lysosomal proteins, such as cathepsin D and LAMP1 and 2, is impaired in the RagA/B KO cells. Intriguingly, overexpression of constitutively active mutant TFEB is not sufficient to recapitulate the RagA/B KO phenotypes (Figure 6F and 9D). Therefore, RagA/B modulate lysosome function in part by affecting the lysosomal localization of v-ATPase and in part by suppressing TFEB activity.
VPS15 is an essential component of the VPS34 complex that is critical for autophagy induction as well as vesicular trafficking. Of note, loss of Vps15 in skeletal muscles exhibits pathologic phenotypes similar to RagA/B cKO mice 50. The Vps15 knockout skeletal muscles develop autophagic vacuolar myopathy associated with accumulation of p62, autophagosomes, and glycogen. Intriguingly, it has been also shown that Gtr1 and Gtr2, the yeast orthologues of Rag GTPases, regulate vesicular trafficking 4. Therefore, Rag GTPases may regulate protein and vesicular trafficking, such as v-ATPase localization to lysosomes, by a mechanism independent of mTORC1. Future studies are necessary to gain the molecular insight of mTORC1 independent function of Rag GTPases.
Methods
Generation of RagA/B conditional knockout mice
RagA targeting vector was constructed by inserting PCR amplified RagA genomic DNA fragments including the promoter, exon, and 3’ UTR sequences in between two loxP sites of pflox-FRT-neo vector (Dr. Ju Chen, University of California-San Diego), and the targeting vector was injected into mouse ES cells (129 SvJ) to generate RagA conditional allele (RagAfl/+) by homologous recombination. G418-resistant ES cell clones were screened by Southern blotting and genomic DNA (gDNA) PCR. A targeted ES clone was injected into C57BL/6 blastocysts to generate chimeric mice. Germ-line transmission of RagA floxed allele of chimeric mice was confirmed by gDNA PCR genotyping, and then the chimeric mice were mated with FLPeR mice (Gt(ROSA)26Sortm1(FLP1)Dym/J; The Jackson Laboratory) to remove neomycin resistant gene from the targeted allele.
RagB conditional allele was also generated as described above, but only the exon 3 (encodes a GDP/GTP binding site) was flanked by two loxP sites. RagAfl/+;RagBfl/+ mice were generated by breeding between RagA and RagB heterozygotes, and then the double heterozygote mice were intercrossed to generate the RagAfl/fl;RagBfl/fl mice.
To delete the floxed alleles in the skeletal and cardiac muscles, the single or double conditional knockout mice were mated with transgenic mice expressing Cre recombinases under the control of muscle creatine kinase promoter (B6.FVB(129S4)-Tg(Ckmm-cre)5Khn/J; The Jackson Laboratory), and the offspring were backcrossed with the conditional knockout mice. Therefore, the genetic background of mice is 129 SvJ and C56BL/6 mixed background, and littermates from mating of Mck-Cre/+; RagAfl/fl;RagBfl/fl and RagAfl/fl;RagBfl/fl were compared in all experiments.
All animal procedures were approved by the University of California-San Diego and Rutgers New Jersey Medical School Institutional Animal Care and Use Committee and carried out in accordance with the guidelines.
Generation of RagA/B knockout MEFs and cell culture
RagAfl/fl;RagBfl/fl embryos were dissected out at E13.5 and trypsinized for 1 hr at 37°C. Primary MEFs were then immortalized with SV40 T antigen using a retroviral vector. The immortalized MEFs were infected with adenoviruses encoding GFP and Cre-GFP (1K MOI, Cell Biolabs, USA) to generate control and RagA/B KO MEFs, respectively. Cells were cultured in high glucose DMEM (Invitrogen, USA) supplemented with 10% FBS (Hyclone, USA) and antibiotics (penicillin/streptomycin).
Cell fractionation
Overnight cultured cells (1×106 cells on a 10cm dish) were trypsinized and resupended in 750μl of cold fractionation buffer (50mM KCl, 90mM Potassium gluconate, 1mM EGTA, 50mM Sucrose, 5mM glucose, protease inhibitor cocktail tablet, 20mM HEPES pH 7.4). The cells were then lysed by sonication, and nuclear fraction was removed by centrifugation at 1000g for 10min at 4°C. The supernatant was further centrifuged at 20000g for 30min at 4°C. The precipitated lysosome enriched fraction was resuspended in the fractionation buffer, and the supernatant was aliquoted as cytosolic fraction.
Lysosomal pH measurement in live cells
Cells grown on a 6-well plate were incubated with 30μg/ml Oregon Green-514 conjugated dextran for overnight. The following day, cells were incubated in a serum-free medium for 1hr and then collected by trypsinization. Cells were resuspended in 200μl of HBSS. The fluorescence of sample was measured at 530nm by excitation at 440nm and 490nm at 30s intervals for 10min using a 96-well plate reader (Infinite M200, TECAN, USA). The average of collected values was interpolated to a pH calibration curve that was plotted by control cell aliquots in nigericin (10μg/ml) containing buffered isotonic solutions (pH 3~7).
In vitro lysosome acidification assay
This assay was performed as previously described 11 with a few modifications. Cells were grown in a culture medium supplemented with 30μg/ml Oregon Green-514 conjugated dextran (Molecular Probes, USA) for overnight, and then incubated in a serum-free medium for 2hr followed by 1μM FCCP treatment for 15min. Lysosome fraction was prepared in the presence of 1μM FCCP. The lysosome enriched fraction (LEF) was then resuspended in 180μl of fractionation buffer without FCCP and transferred to a black 96-well plate. The baseline fluorescence of sample was first measured at 530nm by excitation at 440nm and 490nm at 30s intervals for 5min using a 96-well plate reader (Infinite M200, TECAN, USA). The acidification of LEF by v-ATPase was initiated by adding 5mM ATP and 5mM MgCl2, and the fluorescence measurement was resumed for 30min. To verify the specificity of assay, 100nM bafilomycin-A1 (Cayman Chemicals, USA) was directly added to the resuspended LEF.
Generation of stable MEF cell lines
To generate the TFEB-GFP expressing cells, pEGFP-N1-TFEB plasmid (#38119, Addgene) was sub-cloned into a retroviral vector (pMxs-IRES-Puro). Recombinant retroviruses were made by transfecting the retroviral vectors into packaging cells, and then MEFs were infected with the recombinant retroviruses. Infected cells were selected using puromycin (4μg/ml).
The GFP-TFEBS142A plasmid was constructed by a quick-change mutagenesis method using the above retroviral plasmid as template.
Myc-tagged human Rheb cDNA was PCR amplified and sub-cloned into pMxs-IRES-Puro vector. The constitutively active mutant (RhebS16H) was constructed by a quick-change mutagenesis method.
Quantitative RT-PCR analysis
Total RNA was prepared from tissues or MEFs using either Trizol solution (Invitrogen, USA) or RNeasy Plus mini kit (Qiagen, Germany), and 200ng of total RNA was reverse-transcribed using iScript cDNA systhesis kit (Bio-Rad, USA). 10ng of cDNA mix was added to KAPA SYBR FAST Master Mix (KAPA Biosystems, USA) with specific primer sets (0.5μM, Supplementary Table 1) and the PCR was performed using ABI 7300. All Ct values were normalized with HPRT and the relative expression level was determined using the 2−ΔCt method (ΔCt sample = Ct sample – Ct geomean of housekeeping genes).
Immunohistochemistry and histological analysis
Surgically removed tissues were immediately fixed in 4% paraformaldehyde (Electron Microscopy Sciences, USA) + PBS solution for overnight followed by paraffin embedding. For immunohistochemistry, deparaffinized tissue sections were rehydrated and then microwaved for 5 min in 10mM Sodium Citrate buffer (pH 6.0) for the epitope retrieval. After quenching of endogenous peroxidase activity and blocking procedures, the tissue sections were incubated with primary antibodies for overnight at 4°C. For all rabbit IgG primary antibodies (listed in Supplementary Table 2), the immunostaining was performed using a Vectastain elite ABC kit and a DAB peroxidase substrate kit (Vector Laboratories, USA) according to the manufacturer's instruction. Otherwise, horseradish peroxidase-conjugated secondary antibodies were used for the staining. The antibody list can be found in the supplemental information.
Other histological stains such as H&E, Masson's trichrome, and Peridodic Schiff staining were performed by the UCSD histology core.
Immunoblot analysis
Snap-frozen tissues were homogenized in a cold tissue lysis buffer (1% SDS, 1% NP-40, 50mM NaF, 2mM EDTA, 1mM Na3VO4, protease inhibitor cocktail tablet (Roche, Germany), 100mM NaCl, 10mM Tris-HCl pH 7.5) followed by centrifugation at 13,000g for 10min at 4°C to remove undissolved materials. Protein concentration was determined using detergent compatible protein assay reagents (Bio-Rad, USA) and equal amount of total protein was used for SDS-PAGE. After the resolved proteins were transferred on PVDF membrane (Millipore, USA), the membrane was incubated with primary antibody diluted in 5% BSA + TBST buffer for overnight at 4°C. Antibody bound target proteins were detected with HRP-conjugated secondary antibodies and ECL solution.
1×105 MEFs were plated on a well of 12-well plate and cultured overnight in high-glucose DMEM medium (Invitrogen, USA) supplemented with 10% FBS (Hyclone, USA) and penicillin (100unit/ml)/streptomycin (100mg/ml, Gibco, USA). For the autophagy flux induction, cells were first replenished with a fresh medium and then incubated in Hank's balanced salt solution (HBSS, Mediatech Inc., USA) for the indicated time period in the absence or in the presence of 100~200nM bafilomycin-A1 (Cayman Chemical, USA). After a rinse with PBS, cell lysates were colleted in 1× SDS sample buffer followed by SDS-PAGE.
For the quantification of immunoblot data, the densitiometry of scanned images was measured using the Quantity One software (Bio-Rad, USA).
The un-cropped scanned images of immunoblots can be found in the Supplementary information (Supplementary Fig. 6).
Transmission electron microscopy
Mice were perfused with modified Karnovsky's fixative (2.5% glutaraldehyde and 2% paraformaldehyde in 0.15M sodium cacodylate buffer, pH 7.4), and collected tissues were cut into small pieces followed by incubation in the fixative for overnight at 4°C. Ultrathin sections were prepared and examined using a JOEL 1200EX II transmission electron microscope.
Glycogen measurement
About 10~20mg of snap-frozen tissue was homogenized in 200μl dH2O followed by boiling for 5min. The boiled homogenates were spun down at 16,000g for 5min, and 10μl of the supernatant was used for the assay. The glycogen level was determined using EnzyChrom glycogen assay kit (BioAssay System, USA) as instructed by the manufacturer.
Glycogen phosphorylase activity assay
20~80mg of snap-frozen tissues were homogenized in 200~800μl imidazole buffer (0.5mM DTT, 5mM EDTA, 20mM KF, 50mM Imidazole solution-pH 7.0). The homogenates were centrifuged at 13,000g for 10min at 4°C. 10ml of the supernatant was added to 200μl of reaction buffer (50mM imidazole solution-pH 7.0, 20mM glycogen, 20mM K2HPO4, 0.5mM MgCl2, 1mM EDTA, 0.1mM NADP+, 2μM glucose 1,6-bisphosphate, 0.5mM DTT, 0.025% BSA, 0.6U/ml P-glucomutase, 1mM 5’AMP, 1U/ml glucose 6-phosphate dehydrogenase). NADPH production by the enzymatic reaction was measured at 450nm by excitation at 350nm (1min intervals for 1hr).
Immuno-purification of RagA proteins
HEK293A cells were co-transfected with plasmids encoding Flag-RagA and RagC using PolyJet™ DNA In Vitro Tranfection reagent (SinaGen Lab.). After 48h of transfection, cells from two dishes (15cm) were collected using Mild Lysis Buffer (MLB: 10mM Tris-HCl, pH7.5, 2mM EDTA, 100mM NaCl, 1% NP-40, 50mM NaF, 1mM Na3VO4, protease inhibitor cocktail (Roche)). After centrifugation, the supernatants were transferred and then incubated with the anti-Flag resin (Sigma-Aldrich, USA). The resin was washed three times with MLB and elution was carried out with buffer containing 20mM Tris pH7.5, 5mM EDTA and 3xflag-peptide. Eluted proteins were loaded onto a column (Pierce) to remove detergent followed by dialyzation in fractionation buffer. Purified protein was visualized by Coomassie blue staining and the protein concentration was determined.
Sialidase A treatment
Cells were plated on 12-well plate and cultured for overnight. Cells were then collected in mild lysis buffer and the cell lysates were denatured using a Glyopro Enzymatic Deglycosylation Kit (Prozyme, USA). Denatured lysates were incubated with 2μl of sialidase A at 37°C for 3hrs as instructed by the manufacturer.
Echocardiography
2~6 month-old male and female mice were anesthetized by intraperitoneal injection of 2, 2, 2- tribromoethanol (Sigma-Aldrich, 300 mg/kg). After shaving the chest, the animal was placed on a warm pad, and echocardiograph examination was performed by using Acuson Sequoia C256 (Siemens Medical Solutions). A 13-MHz linear ultrasound probe was used. All the measurements of left ventricular (LV) internal diameter were taken in M-mode short-axis. LV end-diastolic diameter (LVEDD) was measured at the time of the apparent maximal LV diastolic dimension, while LV end-systolic diameter (LVESD) was measured at the time of the most anterior systolic excursion of the posterior wall. In the same positions, diastolic and systolic wall measurements were taken. LV fractional shortening (FS) expressed in percentages was calculated as follows: FS= (LVEDDLVESD)/LVEDD × 100. Mitral inflow peak E and A wave velocities were also measured.
Hemodynamic Analysis
2~6 month old male and female mice were anesthetized by intraperitoneal injection of 2, 2, 2- tribromoethanol (Sigma-Aldrich, 300 mg/kg) and the right carotid artery was cannulated with a high-fidelity microtip pressure transducer catheter (1.4 Fr, Model SPR-839; Millar Instruments, Houston, TX). The catheter was advanced into the aorta to measure aortic pressure and into the LV to measure LV function and pressures. LV pressure and +dP/dt and −dP/dt (change in pressure over time) were recorded using a chart recorder (Model MT95K2; Astro-Med, West Wanwick, RI).
Statistical analysis
P (two-sided) values were determined by the Wilcoxon rank sum or signed rank (for paired samples) test for the comparison of two experimental groups using Mstat software (Dr. Drinkwater, University of Wisconsin-Madison). For the comparison of survival curve, the Mantel-Cox (logrank) test was used.
Supplementary Material
Figure 10. Cardiac hypertrophy in RagA/B cKO mice exhibits the features of lysosomal storage diseases.
(A) Periodic acid-Schiff staining of heart tissues. Representative images are shown. Scale bars, 50 μm. (B) Biochemical measurement of glycogen level in heart tissues. Glycogen levels were determined using an enzyme-based colorimetric assay kit. Values represent the mean ± SD of data (n=4 per group). *P < 0.03, Wilcoxon rank sum test. (C) Immunoblot analysis of glycogen synthase-1 in heart. Tissue lysates were prepared from control and RagA/B cKO hearts, and levels of glycogen synthase and phosphoglycogen synthase were investigated by immunoblotting. (D) Glycogen phosphorylase activity assay. Using an enzyme-based assay, activity of glycogen phosphorylase in heart tissue was determined by measuring NADPH production in the reaction. Each point represents the mean ± SD of data (n=4 per group). (E) Accumulation of ubiquitinated proteins in RagA/B cKO hearts. Levels of ubiquitinated proteins in heart tissues were examined by immunoblotting.
Acknowledgements
We thank members of the Guan Lab for discussions and especially thank Dr. Masa Hoshijima (UCSD) for suggestions and examination of TEM data. We also thank Tracy Guo for her help and expertise on primary cardiomyocyte preparation. This work was supported by grants from the National Institutes of Health (CCT5042 and CCT2413) to K.L.G.
Footnotes
Author contributions
Y.C.K. and K.L.G. conceived the study, designed the experiments, and prepared the manuscript. Y.C.K., H.W.P., and J.S.M. performed the experiments and analyzed the data. J.L.J, R.C.R, and X.W. performed the experiments and provided critical preliminary data. S.S. and J.S. carried out the cardiac function assays, analyzed the data, and discussed the results.
Competing financial interests The authors declare no competing financial interests
References
- 1.Sekiguchi T, Hirose E, Nakashima N, Ii M, Nishimoto T. Novel G proteins, Rag C and Rag D, interact with GTP-binding proteins, Rag A and Rag B. J Biol Chem. 2001;276:7246–7257. doi: 10.1074/jbc.M004389200. [DOI] [PubMed] [Google Scholar]
- 2.Nakashima N, Noguchi E, Nishimoto T. Saccharomyces cerevisiae putative G protein, Gtr1p, which forms complexes with itself and a novel protein designated as Gtr2p, negatively regulates the Ran/Gsp1p G protein cycle through Gtr2p. Genetics. 1999;152:853–867. doi: 10.1093/genetics/152.3.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dubouloz F, Deloche O, Wanke V, Cameroni E, De Virgilio C. The TOR and EGO protein complexes orchestrate microautophagy in yeast. Mol Cell. 2005;19:15–26. doi: 10.1016/j.molcel.2005.05.020. [DOI] [PubMed] [Google Scholar]
- 4.Gao M, Kaiser CA. A conserved GTPase-containing complex is required for intracellular sorting of the general amino-acid permease in yeast. Nat Cell Biol. 2006;8:657–667. doi: 10.1038/ncb1419. [DOI] [PubMed] [Google Scholar]
- 5.Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008;10:935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sancak Y, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oshiro N, Rapley J, Avruch J. Amino acids activate mTOR complex1 without changing Rag GTPase guanyl nucleotide charging. J Biol Chem. 2013 doi: 10.1074/jbc.M113.528505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sancak Y, et al. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inoki K, Li Y, Xu T, Guan KL. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 2003;17:1829–1834. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Menon S, et al. Spatial Control of the TSC Complex Integrates Insulin and Nutrient Regulation of mTORC1 at the Lysosome. Cell. 2014;156:771–785. doi: 10.1016/j.cell.2013.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zoncu R, et al. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science. 2011;334:678–683. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bar-Peled L, Schweitzer LD, Zoncu R, Sabatini DM. Ragulator Is a GEF for the Rag GTPases that Signal Amino Acid Levels to mTORC1. Cell. 2012;150:1196–1208. doi: 10.1016/j.cell.2012.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Futerman AH, van Meer G. The cell biology of lysosomal storage disorders. Nat Rev Mol Cell Biol. 2004;5:554–565. doi: 10.1038/nrm1423. [DOI] [PubMed] [Google Scholar]
- 14.Platt FM, Boland B, van der Spoel AC. The cell biology of disease: lysosomal storage disorders: the cellular impact of lysosomal dysfunction. J Cell Biol. 2012;199:723–734. doi: 10.1083/jcb.201208152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Settembre C, Fraldi A, Medina DL, Ballabio A. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat Rev Mol Cell Biol. 2013;14:283–296. doi: 10.1038/nrm3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Settembre C, et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012;31:1095–1108. doi: 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martina JA, Puertollano R. Rag GTPases mediate amino acid-dependent recruitment of TFEB and MITF to lysosomes. J Cell Biol. 2013;200:475–491. doi: 10.1083/jcb.201209135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Settembre C, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pastore N, et al. Gene transfer of master autophagy regulator TFEB results in clearance of toxic protein and correction of hepatic disease in alpha-1-anti-trypsin deficiency. EMBO Mol Med. 2013;5:397–412. doi: 10.1002/emmm.201202046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song W, et al. TFEB regulates lysosomal proteostasis. Hum Mol Genet. 2013;22:1994–2009. doi: 10.1093/hmg/ddt052. [DOI] [PubMed] [Google Scholar]
- 21.Bentzinger CF, et al. Skeletal muscle-specific ablation of raptor, but not of rictor, causes metabolic changes and results in muscle dystrophy. Cell Metab. 2008;8:411–424. doi: 10.1016/j.cmet.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 22.Risson V, et al. Muscle inactivation of mTOR causes metabolic and dystrophin defects leading to severe myopathy. J Cell Biol. 2009;187:859–874. doi: 10.1083/jcb.200903131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang D, et al. MTORC1 regulates cardiac function and myocyte survival through 4E-BP1 inhibition in mice. J Clin Invest. 2010;120:2805–2816. doi: 10.1172/JCI43008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shende P, et al. Cardiac raptor ablation impairs adaptive hypertrophy, alters metabolic gene expression, and causes heart failure in mice. Circulation. 2011;123:1073–1082. doi: 10.1161/CIRCULATIONAHA.110.977066. [DOI] [PubMed] [Google Scholar]
- 25.Bruning JC, et al. A muscle-specific insulin receptor knockout exhibits features of the metabolic syndrome of NIDDM without altering glucose tolerance. Mol Cell. 1998;2:559–569. doi: 10.1016/s1097-2765(00)80155-0. [DOI] [PubMed] [Google Scholar]
- 26.Mercola M, Ruiz-Lozano P, Schneider MD. Cardiac muscle regeneration: lessons from development. Genes Dev. 2011;25:299–309. doi: 10.1101/gad.2018411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakai A, et al. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med. 2007;13:619–624. doi: 10.1038/nm1574. [DOI] [PubMed] [Google Scholar]
- 28.Jaber N, et al. Class III PI3K Vps34 plays an essential role in autophagy and in heart and liver function. Proc Natl Acad Sci U S A. 2012;109:2003–2008. doi: 10.1073/pnas.1112848109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sardiello M, et al. A gene network regulating lysosomal biogenesis and function. Science. 2009;325:473–477. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- 31.Forgac M. Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat Rev Mol Cell Biol. 2007;8:917–929. doi: 10.1038/nrm2272. [DOI] [PubMed] [Google Scholar]
- 32.Saftig P, Klumperman J. Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat Rev Mol Cell Biol. 2009;10:623–635. doi: 10.1038/nrm2745. [DOI] [PubMed] [Google Scholar]
- 33.Palmieri M, et al. Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum Mol Genet. 2011;20:3852–3866. doi: 10.1093/hmg/ddr306. [DOI] [PubMed] [Google Scholar]
- 34.Settembre C, et al. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat Cell Biol. 2013;15:647–658. doi: 10.1038/ncb2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zaidi N, Maurer A, Nieke S, Kalbacher H. Cathepsin D: a cellular roadmap. Biochem Biophys Res Commun. 2008;376:5–9. doi: 10.1016/j.bbrc.2008.08.099. [DOI] [PubMed] [Google Scholar]
- 36.Zanoteli E, et al. Muscle degeneration in neuraminidase 1-deficient mice results from infiltration of the muscle fibers by expanded connective tissue. Biochim Biophys Acta. 2010;1802:659–672. doi: 10.1016/j.bbadis.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yogalingam G, et al. Neuraminidase 1 is a negative regulator of lysosomal exocytosis. Dev Cell. 2008;15:74–86. doi: 10.1016/j.devcel.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van der Ploeg AT, Reuser AJ. Pompe's disease. Lancet. 2008;372:1342–1353. doi: 10.1016/S0140-6736(08)61555-X. [DOI] [PubMed] [Google Scholar]
- 39.Linhart A, Elliott PM. The heart in Anderson-Fabry disease and other lysosomal storage disorders. Heart. 2007;93:528–535. doi: 10.1136/hrt.2005.063818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Skurat AV, Roach PJ. Phosphorylation of sites 3a and 3b (Ser640 and Ser644) in the control of rabbit muscle glycogen synthase. J Biol Chem. 1995;270:12491–12497. doi: 10.1074/jbc.270.21.12491. [DOI] [PubMed] [Google Scholar]
- 41.Raben N, et al. Suppression of autophagy in skeletal muscle uncovers the accumulation of ubiquitinated proteins and their potential role in muscle damage in Pompe disease. Hum Mol Genet. 2008;17:3897–3908. doi: 10.1093/hmg/ddn292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jewell JL, Guan KL. Nutrient signaling to mTOR and cell growth. Trends Biochem Sci. 2013;38:233–242. doi: 10.1016/j.tibs.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Efeyan A, et al. RagA, but Not RagB, Is Essential for Embryonic Development and Adult Mice. Dev Cell. 2014 doi: 10.1016/j.devcel.2014.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alers S, Loffler AS, Wesselborg S, Stork B. Role of AMPK-mTOR-Ulk1/2 in the regulation of autophagy: cross talk, shortcuts, and feedbacks. Mol Cell Biol. 2012;32:2–11. doi: 10.1128/MCB.06159-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pankiv S, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 46.Komatsu M, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 47.Yu L, et al. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature. 2010;465:942–946. doi: 10.1038/nature09076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lieberman AP, et al. Autophagy in lysosomal storage disorders. Autophagy. 2012;8:719–730. doi: 10.4161/auto.19469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Klabunde RE. Cardiovascular Physiology Concepts. Edn. 2nd Lippincott Williams & Wilkins; 2011. [Google Scholar]
- 50.Nemazanyy I, et al. Defects of Vps15 in skeletal muscles lead to autophagic vacuolar myopathy and lysosomal disease. EMBO Mol Med. 2013;5:870–890. doi: 10.1002/emmm.201202057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.