Abstract
Background
The continuous improvement and evolution of immune cell phenotyping requires periodic upgrading of laboratory methods and technology. Flow cytometry laboratories that are participating in research protocols sponsored by the NIAID are required to perform “switch” studies to validate performance before methods for T-cell subset analysis can be changed.
Methods
Switch studies were conducted among the four flow cytometry laboratories of the Multicenter AIDS Cohort Study (MACS), comparing a 2-color, lyse-wash method and a newer, 3-color, lyse no-wash method. Two of the laboratories twice failed to satisfy the criteria for acceptable differences from the previous method. Rather than repeating more switch studies, these laboratories were allowed to adopt the 3-color, lyse no-wash method. To evaluate the impact of the switch to the new method at these two sites, their results with the new method were evaluated within the context of all laboratories participating in the NIH-NIAID-Division of AIDS Immunology Quality Assurance (IQA) proficiency-testing program.
Results
Laboratory performance at these two sites substantially improved relative to the IQA standard test results. Variation across the four MACS sites and across replicate samples was also reduced.
Conclusions
Although switch studies are the conventional method for assessing comparability of laboratory methods, two alternatives to the requirement of repeating failed switch studies should be considered: (1) test the new method and assess performance on the proficiency testing reference panel, and (2) prior to adoption of the new methods, use both the old and the new method on the reference panel samples and demonstrate that performance with the new method is better according to standard statistical procedures. These alternatives may help some laboratories’ transition to a new and superior methodology more quickly than if they are required to attempt multiple, serial switch studies.
Keywords: HIV, immunophenotyping, flow cytometry, quality assessment, multicenter studies
Decline in the number of CD4+ T-cells and expansion of CD8+ T-cells are hallmarks of HIV-1 disease progression. The most widely used method for quantifying T-cells in HIV-1 infection is flow cytometry. Many technical factors affect flow cytometry-based immunophenotyping results, including flow cytometer quality control, specimen age, antibodies used, method of sample preparation, and analytical/gating procedures (1–5). Given the numerous variables, it is critical that all sites generating flow cytometry data be monitored for proficiency in terms of accuracy and precision both within and between laboratories, especially for multi-center studies. The National Institute of Allergy and Infectious Diseases, Division of AIDS (DAIDS), has developed such a program, the Immunological Quality Assessment (IQA) Program, which is sponsored by DAIDS and administered by the New Jersey School of Medicine and Dentistry (3). All laboratories generating flow cytometry data for NIH studies are required to participate in this program and to maintain satisfactory performance in it. The IQA sends matched blood samples to all participating laboratories and then analyzes and summarizes their results according to specific statistical criteria. To evaluate inter- and intralaboratory performance, the T-cell results for each sample are compared with the median T-cell value for all laboratories reporting results for that sample. Values that differ from the median by 4% or less are considered acceptable. Intralaboratory performance is tested with blinded replicates sent to each laboratory and performance is considered acceptable if the range is within 3%.
DAIDS has taken a strong, central role in directing flow cytometry laboratories to utilize improved methodologies with published guidelines for appropriate methods to perform CD4 enumeration in HIV infected individuals (2–4), and is an advocate for their adoption. In January, 2001, the DAIDS IQA Flow Cytometry Advisory Committee recommendations called for the use of CD45 gating for CD4 and CD8 immunophenotyping, based on the increased precision of lymphocyte gating strategies utilizing CD45 vs. right angle scatter rather than light scatter properties only (1,2,4). Laboratories using the CD3/CD4 and CD3/CD8 antibody combinations were required to begin using either CD3/CD4/CD45 and CD3/CD8/CD45 (3-color) or CD3/CD8/CD45/CD4 (4-color) combinations. However, laboratories could switch to the new method only after successfully completing a “switch” study involving the direct comparison of the old and new methods and obtaining approval from the IQA Advisory Committee. The switch study requires that 60 patient specimens with a CD4 percentage of ≤30% [by the 2-color method (4)] be tested by both the old and the new method. To “pass” the switch study, differences between the results must be within targeted ranges. Based on these evaluations, laboratories have been either certified to switch to the new method or, if the two methods did not agree sufficiently, required to repeat the evaluation.
This report is based on the experience of the Multicenter AIDS Cohort Study (MACS) laboratories performing switch studies to transition from a 2-color to a 3-color T-cell immunophenotyping methodology. When two of the four MACS flow cytometry laboratories (which were certified for two-color analyses) repeatedly failed the required switch study, approval was sought and received to adopt the new (3-color, lyse no-wash) method despite the failures. This adoption resulted in more precise laboratory performance at these sites, suggesting that under certain circumstances a rigid requirement to repeat a failed switch study may be counterproductive. Based on this experience, we offer two possible alternatives to the current requirement of having to repeat a failed switch study, in an effort to increase the speed and efficiency of adopting new technologies.
MATERIALS AND METHODS
Blood Specimens
The specimens consisted of ethylenediamine tetra-acetate (EDTA) anticoagulated whole blood. Five blood samples were shipped bimonthly to participating laboratories including the MACS sites and were processed for flow cytometric analysis on the day they were received. The MACS study sites and flow cytometry laboratories are located in metropolitan Baltimore, Chicago, Los Angeles, and Pittsburgh. The samples shipped to the MACS sites during the study period were matched (i.e., aliquots of the same sample), thus permitting interlaboratory comparisons across the sites. The IQA shipments of five samples typically included two to four blinded replicate samples to assess intralaboratory reproducibility. IRB approval was in place for collection and analysis of peripheral blood specimens at the IQA and MACS laboratories, respectively, for this work.
Instrumentation
Flow cytometry was performed with a FACSCalibur® flow cytometer utilizing Cellquest® software (BDIS) at sites 1 and 2 and with an EPICS XL® flow cytometer utilizing System 2® software (Beckman Coulter, Hialeah, FL) at sites 3 and 4.
Two-Color Flow Cytometry
At sites 3 and 4 specimens were stained with Simultest® reagents (Becton Dickinson Immunocytometry Systems (BDIS), San Jose, CA), which included the following combinations of monoclonal antibodies, conjugated to fluorescein isothiocyanate (FITC) and phycoerythrin (PE): CD45/CD14 to determine the optimal lymphocyte gate, and CD3/CD4 and CD3/CD8 to define CD4+ and CD8+ T-cells, respectively. Samples were stained, lysed, and analyzed according to a whole blood staining procedure as described (6,7).
Three-Color Flow Cytometry
Specimens were stained with Tritest® reagents (BDIS), which included the following combinations of monoclonal antibodies conjugated to FITC, PE, and peridinin chlorophyll protein (PerCP): CD3/CD4/CD45 and CD3/CD8/CD45. The specimens were stained and lysed according to a lyse no-wash method, lymphocytes were gated on CD45 vs. side scatter (SSC), and T-cells (CD3+ CD4+ and CD3+CD8+) were analyzed from within this gate as described (2,4). To assess the proportion of lymphocytes that were included in the analysis (i.e., lymphocyte recovery), backgating was performed in which ungated CD3+CD4+ and CD3+CD8+ cells were displayed in an exact copy of the CD45 vs. SSC gate used to define the lymphocyte population. T-lymphocyte recovery of >95% was considered acceptable. The Tritest reagents were validated for Coulter EPICS XL instrumentation by parallel testing on the FACSCalibur and XL. The instruments performed equivalently with respect to percentage of lymphocytes, lymphocyte purity, and lymphocyte recovery (data not shown).
Statistical Analysis
Percentages of CD3+, CD4+, and CD8+ T-cells were determined by the four MACS laboratories for each sample, and differences were calculated between these and the corresponding median values obtained by all of the IQA laboratories that reported results for that sample. The average number of laboratories that reported results to the IQA for a given sample was 75, with a range of 32–83. For the present analysis, immunophenotyping data from two distinct time periods were compared. In Time Period A [November 2001 through March 2004], sites 1 and 2 of the MACS laboratories used 3-color cytometry and sites 3 and 4 used 2-color. In Time Period B [January 2005 to May 2006], all MACS sites used 3-color cytometry.
Interlaboratory comparisons presented here were based on IQA samples that were analyzed by all sites. All nonreplicate samples were included in the analysis, and for replicate samples only the first of the replicates (as labeled by the IQA) was included. (This method is unbiased with respect to replicate values and in any case there was minimal variation among the replicates in all laboratories, with differences ≤2% in all but three sets of replicate analyses (out of 85 performed). The Wilcoxon Sign Rank test was used to test for differences in CD4+ and CD8+ percentages between the IQA and the four sites (SYSTAT Software Inc., Richmond, CA). Box plots were used to display the data (see legend to Fig. 1 for explanation of the box plots). For analysis of replicate samples, we used a mixed linear model (Software R, R Project for Statistical Computing, R-Project.org). The analysis fitted separate models for the two time periods. Each model consisted of a fixed effect for the sample and a random effect for the site, thus yielding estimates of the components of variation within and across sites. The residuals from these models were carefully scrutinized and no violations of the models’ assumptions were found. Analysis included all replicate sets so that exclusion of a replicate set by one site did not exclude replicate data from the other three sites (50 out of 56 replicate sets were reported for period A and 35 of 36 for period B). The models correct for differences in sample numbers across time periods and for differences in replicate numbers, which ranged from two to four replicates per IQA shipment.
Fig. 1.
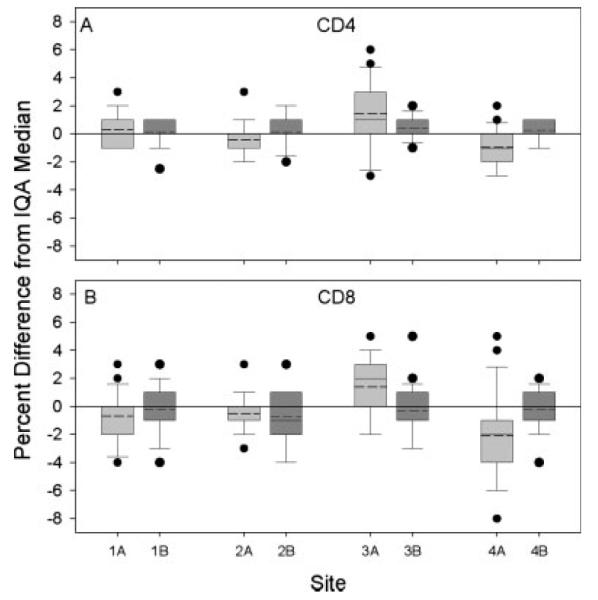
Differences between the CD4+ T-cell percentages (A) and CD8 T-cell (B) percentages obtained by the four MACS laboratories and the median IQA T-cell percentages obtained by the IQA program. Plots show results from two time periods for comparison across sites and across methods. Boxes 1A to 4A (light shading) summarize data from the time period when sites 1 and 2 used 3-color and sites 3 and 4 used 2-color (11/01–3/04; n = 23). Boxes 1B–4B (dark shading) summarize data from the time period when all sites used 3-color (1/05–5/06; n = 23). For each box, the lower and upper bounds of the box represent the 25th and 75th percentiles of the data, respectively; thus, the height of the box represents the intra-quartile range (IQR) of the data. The horizontal lines within the boxes are the mean (dashed line) and the median (solid line) of the data. The upper and lower whiskers represent the 90th and 10th percentiles, respectively. Circles represent data points that were outliers, defined as data that were >90th percentile or <10th percentile.
RESULTS
Data for this study included results for IQA shipments from November 2001 through May 2006. All MACS laboratories remained fully certified by the IQA program during the entire study period.
Tables 1 and 2 summarize the results for CD4+ and CD8+ T cells, respectively, from the IQA and from the four MACS sites for specimens received during the two distinct study periods. Tables 1 and 2 show results for Time Period A, when sites 1 and 2 used 3-color and sites 3 and 4 used 2-color, and for Time Period B, when all four sites used 3-color. The same number of samples was analyzed in each time period. Overall, the medians for all T-cell subsets for the four MACS laboratories were within 2% of the IQA medians. Using the IQA definition of an acceptable result (i.e., a value within ±4% of the median for all laboratories participating in the IQA), sites 3 and 4 had more unacceptable results (n = 9) than sites 1 and 2 (n = 0) in Time Period A, especially for CD3+CD8+ T-cells. The same was true for CD3+ T cells (data not shown), although it should be noted the IQA does not formally grade the CD3+ T-cell results. In Time Period A, the percentages of CD4+ and CD8+ T-cells reported by sites 3 and 4 were slightly but significantly different from the IQA median, but this difference was not seen during Time Period B when they used 3-color. Similarly, the number of unacceptable results at sites 3 and 4 decreased markedly in Time Period B (n = 1).
Table 1.
CD4+ T-Cell Percentages Obtained by the Four MACS Laboratories during the Study Period, Compared with the Median for Reporting Laboratories Participating in the IQA
| IQA | Site 1 | Site 2 | Site 3 | Site 4 | |
|---|---|---|---|---|---|
| Time period A | |||||
| Number of specimensa | 23 | 23 | 23 | 23 | 23 |
| Mean | 23.2 | 23.5 | 22.8 | 24.7 | 22.3 |
| Median | 23 | 24 | 22 | 23 | 22 |
| 95% CI Upper | 26.3 | 26.7 | 26.0 | 27.9 | 25.2 |
| 95% CI Lower | 20.2 | 20.3 | 19.6 | 21.5 | 19.3 |
| P valueb | 0.285 | 0.09 | 0.012+ | 0.003+ | |
| Number of unacceptable resultsc | 0 | 0 | 2 | 0 | |
| Time period B | |||||
| Number of specimens | 23 | 23 | 23 | 23 | 23 |
| Mean | 20.8 | 20.9 | 20.9 | 21.2 | 21.0 |
| Median | 20 | 20 | 22 | 21 | 20 |
| 95% CI Upper | 24.5 | 24.6 | 24.5 | 24.9 | 24.9 |
| 95% CI Lower | 17.0 | 17.2 | 17.3 | 17.5 | 17.1 |
| P valueb | 0.285 | 0.496 | 0.026 | 0.116 | |
| Number of unacceptable resultsc | 0 | 0 | 0 | 0 |
Time period A, when sites 1 and 2 used 3-color and sites 3 and 4 used 2-color, and time period B, when all sites used 3-color.
Only specimens analyzed at all four laboratories (23/66) were included in this analysis.
P-value by Wilcoxon Sign Rank test comparing each site with the IQA, significance set at P < 0.0125 after Bonferroni correction for multiple comparisons.
Values that differed from the IQA median by 5% or more.
Table 2.
CD8 Cell Percentages Obtained by the Four MACS Laboratories during the Study Period, Compared with the Median for Reporting Laboratories Participating in the IQA
| IQA | Site 1 | Site 2 | Site 3 | Site 4 | |
|---|---|---|---|---|---|
| Time period A | |||||
| Number of specimensa | 23 | 23 | 23 | 23 | 23 |
| Mean | 50.7 | 50.0 | 50.2 | 52.1 | 48.7 |
| Median | 52 | 51 | 51 | 53 | 50 |
| 95% CI Upper | 55.5 | 54.5 | 54.6 | 56.5 | 52.8 |
| 95% CI Lower | 46.3 | 45.6 | 45.8 | 47.8 | 44.5 |
| P valueb | 0.080 | 0.058 | 0.006+ | 0.005+ | |
| Number of unacceptable resultsc | 0 | 0 | 1 | 6 | |
| Time period B | |||||
| Number of specimens | 23 | 23 | 23 | 23 | 23 |
| Mean | 50.6 | 50.3 | 49.8 | 50.3 | 50.3 |
| Median | 51 | 52 | 50 | 51 | 52 |
| 95% CI Upper | 55.2 | 55.0 | 54.3 | 54.5 | 55.1 |
| 95% CI Lower | 45.9 | 45.7 | 45.3 | 46.0 | 45.5 |
| P valueb | 0.629 | 0.60 | 0.267 | 0.444 | |
| Number of unacceptable resultsc | 0 | 0 | 1 | 0 |
Time period A, when sites 1 and 2 used 3-color and sites 3 and 4 used 2-color, and time period B, when all sites used 3-color.
Only specimens analyzed at all four laboratories (23/66) were included in this analysis.
P-value by Wilcoxon Sign Rank test comparing each site with the IQA, Significance set at P < 0.0125 after Bonferroni correction for multiple comparisons.
Values that differed from the IQA median by 5% or more.
Figure 1 shows the distributions of the differences, on individual samples, between the CD4+ and CD8+ T-cell percentages obtained by the MACS laboratories, and the IQA medians for the corresponding specimens. The box plots labeled 1–4 A show data for Time Period A. It can be seen that sites 1 and 2 (which used the 3-color method during this period) were closer to the IQA medians, had a narrower inter-quartile range (IQR), and had fewer outliers than sites 3 and 4 (which used the 2-color method during this time period). This was especially evident for CD8+ T-cells. Again, similar results were observed for CD3+ T-cells (data not shown).
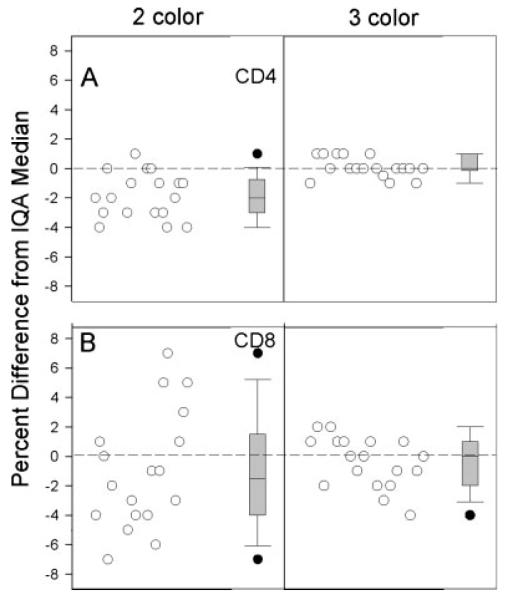
Twice during the study period, site 4 attempted to switch to the 3-color method but failed the required switch study (data not shown). As per IQA procedures, the specific statistical reasons for the failure were not shared with site 4; however, failure can occur when more than 20% of specimens differ by 4% or more across the two methods. After the second failure, in April 2004, a discussion was held with the IQA program staff as to whether this site should adopt the 3-color method. It was decided that since the 3-color method had proved superior to the 2-color method in many other laboratories, the IQA would approve this site to begin routine use of the 3-color method. Figure 2 provides a summary of the results of this change on IQA performance at site 4 for CD4+ and CD8+ T-cells. For both T-cell subsets, the percentage differences from the IQA medians were smaller, the IQRs were narrower, and the outliers were fewer. Similar results were achieved for CD3+ T-cells (data not shown).
Fig. 2.
Distributions of the differences between CD4+ (A) and CD8= (B) T-cell percentages obtained by site 4 and the IQA medians on individual specimens, using the 2-color lyse-wash method (left) and the 3-color lyse no-wash method (right). Within each plot, differences for individual samples are shown on the left, and box plots of the differences are shown on the right (box plots defined as per legend to Fig. 1). To equalize sample size across methods, only the last eighteen 2-color samples were included for comparison with the 3-color results. Plots show improved IQA performance for the 3-color lyse no-wash method.
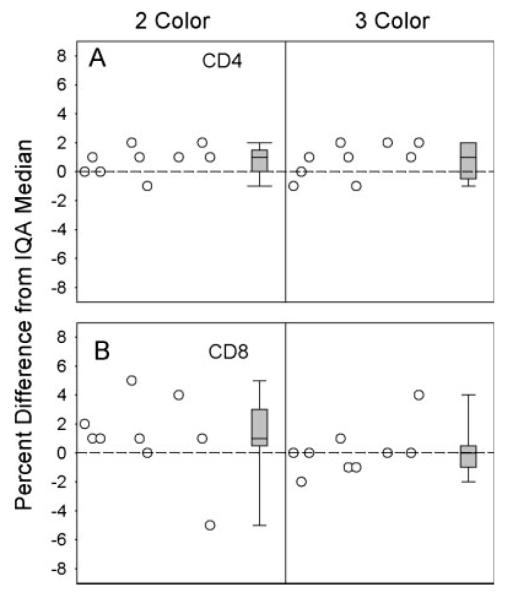
After the experience of site 4 as described above, site 3 also failed their switch study. To prepare for the possibility that it might fail a second switch study, site 3 began analyzing IQA samples using both the 2- and 3-color methods concurrently. As shown in Figure 3, the results with the 3-color method were substantially closer to the IQA median for CD8 and not much different for CD4 (although there was less room for improvement in the CD4 measurements). A second switch study was completed, and was failed. Nevertheless, the lab was allowed to adopt the 3-color method, in part because of the improved data on the IQA samples using the 3-color method, and in part because of the improvement that had been seen at site 4 after they switched methods. IQA performance of site 3 improved after the switch, just as it had in site 4. The improvement in performance of sites 3 and 4 on the IQA can also be seen in Figure 1 by comparing sites 3A versus 3B and 4A versus 4B: both the heights of the boxes and the differences from the IQA medians for these sites were reduced in Time Period B as compared to Time Period A. Figure 1 also shows that the differences from the IQA medians were more congruent across the four MACS sites in Time Period B as compared to A, i.e., after sites 3 and 4 switched to the 3-color method.
Fig. 3.
Distributions of differences between the CD4+ (A) and CD8+ (B) T-cell percentages obtained by site 3 and the IQA medians on individual specimens. Plots are from a paired analysis showing results from the 2-color method on the left and results from the 3-color method on the right side of each plot. Both individual values and box plots (defined as in the legend to Fig. 2) are shown. The nine samples include nonreplicates and only the first of the replicates as labeled by the IQA. The plots show improved IQA performance for CD8 for the 3-color method.
Table 3 shows an analysis of the variability of the replicate samples during the two time periods studied (i.e., when 2- and 3-color were used vs. when only 3-color was used). After sites 3 and 4 switched to 3-color analysis (Time Period B), the intralaboratory and interlaboratory standard deviations of replicate samples were lower than they had been in Time Period A when the four MACS sites used mixed immunophenotyping methods. The lower variation across the MACS sites helps demonstrate proficiency at using the 3-color method by sites 3 and 4 despite the failed switch studies. These analyses were made possible by the IQA program and the clear result is improved interlaboratory comparability of immunophenotyping data across the four MACS laboratories.
Table 3.
Estimated Components of Variance Expressed as Standard Deviation
| Intralaboratory variation | Interlaboratory variation | |
|---|---|---|
| CD4 time period A | 1.48 (1.31–1.68)a | 1.54 (0.68–3.51) |
| CD4 time period B | 0.85 (0.74–0.98) | 0.09 (0.00–2.67) |
| CD8 time period A | 2.20 (1.94–2.48) | 0.98 (0.43–2.24) |
| CD8 time period B | 1.52 (1.33–1.75) | 0.34 (0.08–1.45 |
Values are the estimated standard deviation and the 95% confidence interval for that estimate.
DISCUSSION
The NIAID DAIDS IQA program requires its certified flow cytometry laboratories to satisfactorily complete a switch study prior to receiving approval to adopt a newer method. Satisfactory completion of the switch study depends on the attainment of minimal statistical differences observed between the data procured by the old vs. the new method. This procedure helps maintain data integrity both within and between sites. However, when both methods do not agree sufficiently, laboratories are often required to repeat the switch study. This requires a substantial investment of time and money and may delay the adoption of the newer, improved method. If the newer method results in a substantial improvement in accuracy and precision as compared to the older method, the differences in the results may exceed the acceptability criteria and the switch study would fail. The latter would suggest that an alternative to minimum differences between the old and new methods as the criteria for switch approval should be considered.
Switch studies have been instrumental in helping DAIDS-sponsored laboratories implement new technologies. Our findings suggest, however, that two alternative methods should be considered for approval of laboratories that fail the required switch studies. The first alternative is to test the new method on proficiency samples and demonstrate that the laboratory’s performance improves relative to the IQA standard. The second alternative is to perform paired analyses of proficiency-testing specimens with both methods before adopting the new method for regular laboratory specimens, and demonstrate that the new method results in fewer failed IQA specimens and smaller differences from the IQA median than the old method. In either case prior approval from the IQA Advisory Committee would still need to be obtained before switching the laboratory samples to a new method. We highly recommend that laboratories planning to switch to a new method begin immunophenotyping the IQA samples by both the old and new method in advance of submitting switch study data. These data provide an indication of the effect of the new method on IQA testing results, and will also provide statistical evidence of potential improvement.
The failure of the switch studies in the two MACS flow laboratories was primarily due to problems with analyses of CD8+ T-cells, particularly for samples with elevated CD8+ T-cell percentages. This conclusion is supported by the fact that data generated using the 2-color method resulted in a greater number of outliers and by the direct comparison of paired analyses showing improved precision of the IQA specimens tested at site 3. In some cases, specific causes for discrepancies in the methods could be identified, such as a restricted light scatter lymphocyte gate that occasionally excluded the smaller B-cells, and loss of CD8 cells by aggregation and “escape” from the light scatter gate (data not shown).
The improvement in the laboratory performance with the adoption of the 3-color, lyse no-wash method including CD45 lymphocyte gating was expected in view of previous studies demonstrating the improved precision of lyse no-wash methods incorporating the CD45 vs. SSC strategy for gating lymphocytes (1,2,4). In addition, the lyse no-wash method reduces manipulation of the cells (e.g., washing and centrifugation) and is therefore less prone to the formation of cellular aggregates and “escapees.” Given the expected improvements with the new method, the adoption of new criteria by which laboratories may complete switch studies will help laboratories adopt new methods more quickly and without the cost of unnecessarily repeating switch studies.
ACKNOWLEDGMENTS
We thank Marianne Chow, Eugene Cimino, Marisela Lua Killian, Edwin Molina, Carlos Nahas, Tricia Nilles, Nicholas J. O’Gorman and Kim Stoika for their contributions in the MACS flow cytometry laboratories; and Mr. Raul Louzao for management of the IQA project and for data review and technical assistance to MACS flow cytometry units.
Grant sponsor: National Institutes of Health; Grant number: N01–AI 95356; Grant sponsors: National Institute of Allergy and Infectious Diseases (NIAID), National Cancer Institute; Grant sponsor: National Heart, Lung and Blood Institute; Grant numbers: UO1-AI-35042, 5-MO1-RR-00722, UO1-AI-35043, UO1-AI-37984, UO1-AI-35039, UO1-AI-35040, UO1-AI-37613, UO1-AI-35041; Grant sponsors: National Institute on Drug Abuse (NIDA), National Institute of Mental Health (NIMH), Office of AIDS Research, of the NIH; Grant sponsor: DHHS; Grant number: U01-AI-068613.
LITERATURE CITED
- 1.Gelman R, Wilkening C. Analysis of quality assessment studies using CD45 for gating lymphocytes for CD3+CD4+% Cytometry. 2000;42:1–4. doi: 10.1002/(sici)1097-0320(20000215)42:1<1::aid-cyto1>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 2.Schnizlein-Bick C, Mandy F, O’Gorman M, Paxton H, Nicholson J, Hultin L, Gelman R, Wilkening C, Livnat D. Use of CD45 gating in the three and four-color flow cytometric immunophenotyping: Guideline from the National Institute of Allergy and Infectious Diseases, Division of AIDS. Cytometry. 2002;50:46–52. doi: 10.1002/cyto.10073. [DOI] [PubMed] [Google Scholar]
- 3.Cavelli T, Denny TN, Paxton H, Gelman R, Kagan J. Guidelines for flow cytometric immunophenotyping: A report from the National Institute of Allergy and Infectious Diseases, Division of AIDS. Cytometry. 1993;14:702–714. doi: 10.1002/cyto.990140703. [DOI] [PubMed] [Google Scholar]
- 4.Nicholson J, Kidd P, Mandy F, Livnat D, Kagan J. Three-color supplement to the NIAID DAIDS guideline for flow cytometric immunophenotyping. Cytometry. 1996;26:227–230. doi: 10.1002/(SICI)1097-0320(19960915)26:3<227::AID-CYTO8>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 5.Gelman R, Cheng S-C, Kidd P, Waxdal M, Kagan J. Assessment of the effects of instrumentation, monoclonal antibody, and fluoro-chrome on flow cytometric immunophenotyping: A report based on 2 years of the NIAID DAIDS flow cytometry quality assessment program. Clin Immunol Immunopathol. 1993;66:150–162. doi: 10.1006/clin.1993.1019. [DOI] [PubMed] [Google Scholar]
- 6.Schenker EL, Hultin LE, Bauer KD, Ferbas J, Margolick JB, Giorgi JV. Evaluation of a dual-color flow cytometry immunophenotyping panel in a multicenter quality assurance program. Cytometry. 1993;14:307–317. doi: 10.1002/cyto.990140311. [DOI] [PubMed] [Google Scholar]
- 7.Giorgi JV, Landay A. HIV infection: Diagnosis and disease progression evaluation. In: Darzynkiewicz Z, Robinson JP, Crissman HA, editors. Methods in Cell Biology. Academic Press; Orlando, FL: 1994. pp. 174–181. [DOI] [PubMed] [Google Scholar]