Abstract
This perspective reviews the many dimensions of base excision repair from a 10,000 foot vantage point and provides one person’s view on where the field is headed. Enzyme function is considered under the lens of X-ray diffraction and single molecule studies. Base excision repair in chromatin and telomeres, regulation of expression and the role of posttranslational modifications are also discussed in the context of enzyme activities, cellular localization and interacting partners. The specialized roles that base excision repair play in transcriptional activation by active demethylation and targeted oxidation as well as how base excision repair functions in the immune processes of somatic hypermutation and class switch recombination and its possible involvement in retroviral infection are also discussed. Finally the complexities of oxidative damage and its repair and its link to neurodegenerative disorders, as well as the role of base excision repair as a tumor suppressor are examined in the context of damage, repair and aging. By outlining the many base excision repair-related mysteries that have yet to be unraveled, hopefully this perspective will stimulate further interest in the field.
Keywords: Base excision repair, DNA glycosylases, AP endonuclease, Repair enzyme structures, DNA glycosylase search, Repair in chromatin, BER subpathways, BER crosspathways, Repair in telomeres, BER and transcriptional regulation, BER in the immune system, Neurodegenerative diseases, Cancer, Aging
1. Base excision repair overview
Base excision repair (BER) is a highly conserved pathway from bacteria to humans and is responsible for repairing the vast majority of endogenous DNA damage including alkylations, oxidations, deaminations and depurinations, as well as single-strand breaks (SSBs) (for reviews see [1,2]. Thus, the primary function of BER is to remove these frequently produced lesions and maintain genomic integrity. However, as discussed below, the enzymes involved in BER have been co-opted to take part in a variety of apparently unrelated cellular functions (see Fig. 1). Although BER will be discussed in general terms, this perspective will focus on the first two steps of the process in mammals, where the enzymes involved are homologous across species. Since this article covers such a broad range of topics, recent reviews will be the primary material referenced with apologies to the investigators who originally reported the observations.
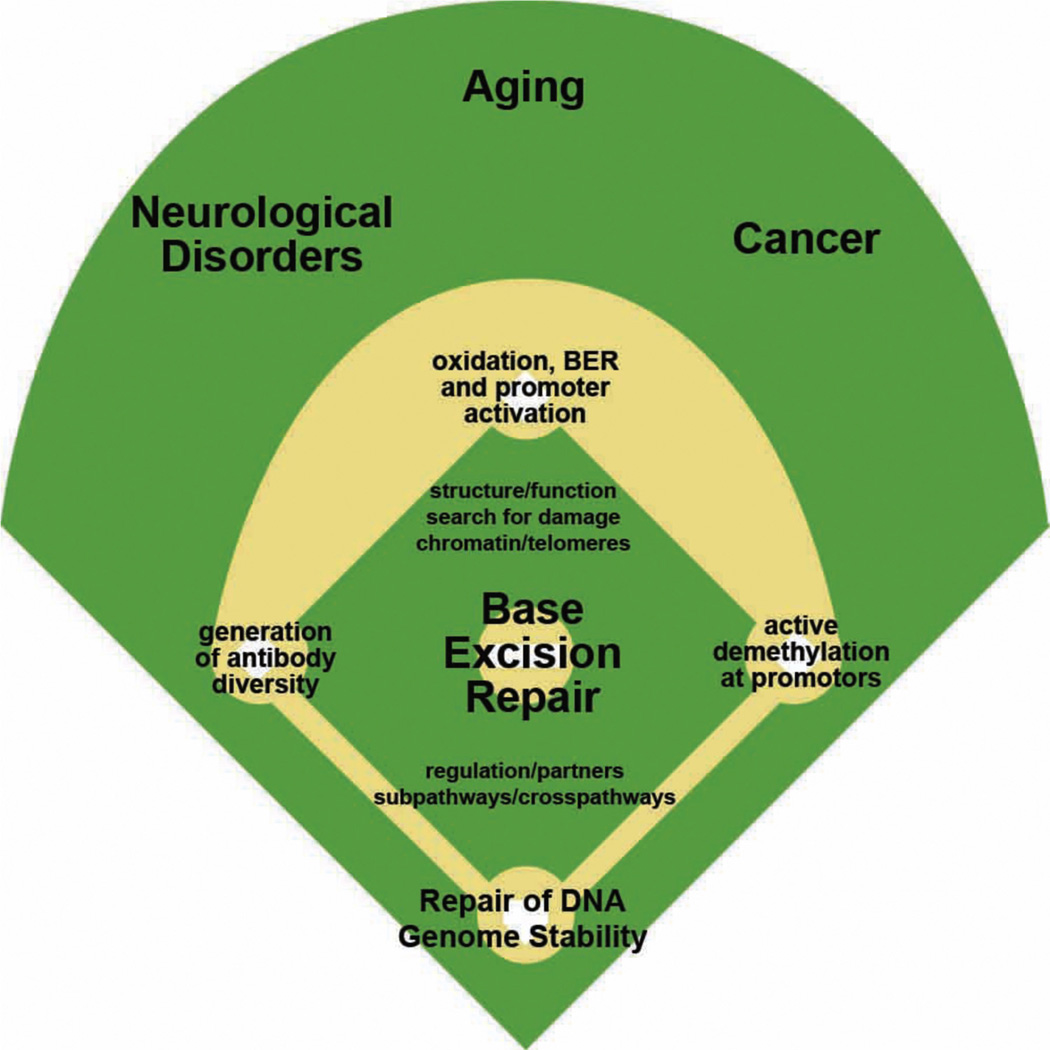
Fig. 1.
This cartoon was inspired by the 1996 Failla lecture delivered by the Author to the Radiation Research Society discussing base excision repair of radiation-induced free radical damage. The model of the baseball diamond based on Abbot and Costello’s skit “Who’s on First, What’s on Second, I Don’t Know’s on Third,” was used to describe the activities of the four sets of BER enzymes, the glycosylases (first base), AP endonucleases (second base), DNA polymerases (third base), and ligases (home plate), which was about the extent of what was known at the time. This cartoon jumps almost two decades to display in outline form our current knowledge of BER processing which has definitely come from the minor to the major leagues.
The initial step in BER is the search for the lesions in DNA by DNA glycosylases. In humans there are eleven of these enzymes, four devoted to the removal of mispaired uracil and thymine, six to the repair of oxidative damage, and one to the removal of alkylated bases. The glycosylases that recognize uracil, thymine, and alkylated bases are monofunctional, and remove the damaged base by cleaving the N-glycosyl bond between the base and the sugar. The resulting abasic site is recognized by an apurinic (AP) endonuclease (APE1), which cleaves the abasic site leaving a sugar attached to the 5′ side of the nick. The resulting 3′ hydroxyl is a substrate for the repair polymerase, DNA polymerase β (Pol β), which also has a lyase activity that removes the sugar attached to the 5′ phosphate. The gap is filled in and sealed by a DNA ligase. The glycosylases that recognize oxidative lesions are bifunctional and not only excise the damaged base but also cleave the DNA backbone, leaving either an α, β unsaturated aldehyde or a phosphate attached to the 3′ side of the nick. The sugar is removed by the phosphodiesterase activity of APE1 and the phosphate group by polynucleotide kinase (PNK).The remainder of the steps are the same as described for the pathway initiated by the monofunctional enzymes. This process is called short patch BER and in some circumstances can be redirected to a longer patch process, often because the sugar is inefficiently removed from the 5′ end of the nick. In this case a number of different polymerases can take over, including the replicative polymerases or pol λ. The damage-containing strand is displaced and removed by FEN1, and the nick is sealed by a ligase. Ligase 1 seals the nick in long patch BER and either ligase 1 or ligase IIIα/XRCC1 seals the nick in short patch BER. If the starting lesion is an abasic site, repair is initiated by APE1 but the subsequent enzymatic steps are the same. Single-strand breaks may have any one of a number of end blocks that need to be removed (see Section 6) before repair can proceed. Poly(ADP)ribose polymerase 1 (PARP1), and XRCC1, a scaffolding protein, are also important players in the BER process.
The mammalian DNA glycosylases that recognize base damages all have structural or functional homologs in bacteria and in fact the human enzymes are able to complement repair defects in bacteria (for reviews see [3,4]). The human mono-functional glycosylases recognize a broad spectrum of lesions. Alkyl guanine glycosylase/methyl purine glycosylase (AGG/MPG) recognizes alkylated purines and ethenopurines and is primarily found in mammals. Uracil DNA glycosylase (UNG) removes uracil as well as 5-hydroxyuracil and other uracil derivatives in both single- and double-stranded DNA. Single-strand-selective monofunctional uracil glycosylase 1 (SMUG1) recognizes many of the same substrates as UNG in single- and double-stranded DNA including 5-hydroxymethyluracil (5-hmU). Thymine DNA glycosylase (TDG) removes uracil, thymine and 5-hydroxymethyluracil when paired with guanine in double-stranded DNA. Methyl CpG binding domain protein 4 (MBD4) recognizes uracil, thymine and 5-hydroxymethyluracil when paired with guanines in CpG dinucleotides as well as other substrates recognized by the uracil glycosylases. MUTYH, also a monofunctional glycosylase, removes adenine when misincorporated opposite 7,8-dihydro-8-oxoguanine (8-oxoG) or 2,6-diamino-4-hydroxy-5-formamidopyrimidine (FapyG).
Both OGG1 and NTH1 are bifunctional housekeeping glycosylases. In double- stranded DNA, the principal substrates for OGG1 are 8-oxoG and FapyG paired with cytosine while those for NTH1 are oxidized pyrimidines and formamidopyrimidines. The bifunctional Nei-like proteins NEIL1 and NEIL2 remove oxidized pyrimidines and formamidopyrimidines in both single-stranded and duplex DNA, although NEIL2 prefers single-stranded DNA. NEIL3 also recognizes a broad spectrum of oxidized pyrimidines and formamidopyrimidines but has weak glycosylase activity on duplex DNA and weak lyase activity on all substrates [5]. The best substrates for all of the NEIL proteins are the further oxidation products of 8-oxoG, spiroiminodihydantoin (Sp) and guanidinohydantoin (Gh) (for a review see [6]).
APE1, which has 3 → 5 exonuclease and 3′ phosphodiesterase activities in addition to its AP endonuclease activity, is also highly conserved from Escherichia coli to humans (for a review see [7]) The N-terminal domain of APE1 contains a redox regulatory region. A second APE family member, APE2, has only weak AP endonuclease activity but strong 3′ phosphodiesterase and 3′→5′ exonuclease activities. Different DNA polymerases and DNA ligases are used across phyla for repair synthesis and nick sealing with Pol β being the primary repair polymerase in humans.
BER reactions reconstituted in vitro strongly suggest that repair occurs by a handoff mechanism (for reviews see [8,9]). The glycosylases remain bound to their AP site product until displaced by APE1, a step which is rate-limiting for most mammalian glycosylases, and subsequent reactions are coordinated by PARP1 and/or XRCC1 with Pol β and DNA ligase completing the repair pathway. In fact, immunoprecipitates of XRCC1 are able to completely repair AP sites [10]. Other studies have shown that the glycosylases and APE1 interact with downstream enzymes and that immunoprecipitates of NEIL 1 or 2 contain PNK, Pol β, LigIIIα, XRCC1, PCNA and FEN1. Similarly UNG2 [11] has been shown to associate with APE1, Pol β, XRCC1, PNK, the replicative polymerases, ligase 1 and protein cyclin A; this complex can perform BER of uracil and AP sites. These observations have led to the hypothesis that the BER enzymes function under certain circumstances as a complex or “BERosome.” In the cell, however, many of these interactions among BER factors and enzymes are possibly transient in nature and likely influenced by post-translational modifications. Moreover, it is unlikely that the lesion search is undertaken by a BERosome. Thus, it seems more plausible that the replication or transcription associated glycosylases such as UNG2, NEIL1 and NEIL2 function in a stable BERosomal complex. But we need to know more.
2. Base excision repair enzyme structures inform function
The enzymatic mechanisms of all the BER proteins (for a review see [12]) have been significantly informed by their structures (for reviews see [13–16]). The DNA glycosylases are members of four different structural families. AAG/MPG has a unique structure while there are six subfamilies of the uracil glycosylase enzymes. Three of these subfamilies are present in eukaryotes, with UNG, SMUG1 and TDG being the mammalian representatives. NTH1, OGG1, MUTYH and the glycosylase domain of MBD4 belong to the HhH superfamily. The NEIL or Nei-like glycosylases are members of the Fpg/Nei family; Fpg (formamidopyrimidine DNA glycosylase) and Nei are their E. coli homologs. APE1 and APE2 are members of the same family and are orthologs of E. coli Xth, the major AP endonuclease present in prokaryotes.
Crystal structures have shown that all of the glycosylases that recognize damaged bases (for reviews see [13–16]) evert the damaged base out of the DNA helix into a substrate binding pocket before cleaving the N-glycosyl bond. These structures have also defined the relationships between the substrate binding pockets and the active site nucleophiles. Interestingly the same base damage can be recognized by glycosylases having different structures. Like the glycosylases, APE1 also flips the abasic site out of the DNA helix for catalysis. In all of the DNA glycosylase- and APE–DNA complexes, the DNA backbone is severely kinked; however, the overall B-form DNA structure is maintained by insertion of “void-filling” amino acid residues that take the place of the extruded base and usually interact with the orphaned base. Structural studies have also revealed the DNA footprint and enzyme/DNA interacting residues. These include a number of specific DNA binding motifs, such as the helix–hairpin–helix motif present in all members of the HhH superfamily and iron sulfur clusters present in some. DNA binding motifs in the Fpg/Nei family include the helix-two-turns-helix and zinc/zincless finger motifs. Moreover, the αF-β9/10 loop or 8-oxoG capping loop found only in Fpg proteins is involved in stabilizing 8-oxoG lesions but no other lesions in the lesion binding pocket [17]. The glycosylase structures coupled with site-directed mutagenesis have provided mechanistic insights into glycosylase function.
Structures of Pol β (for reviews see [18,19]) have shown the polymerase domain to possess the classic polymerase fingers, palm and thumb subdomains, as well as an additional N-terminal domain containing the 5′ dRP lyase activity. The crystal structures also revealed conformational changes upon Pol β binding to DNA. The crystal structures of ligase I (for reviews see [4,20,21]) bound to DNA reveal those subdomains that encircle DNA and the position of the nicked DNA ends within the ligase active site. In addition to its repair function, ligase l functions during DNA replication of Okazaki fragments. DNA ligase III (for a review see [22]) has a domain structure similar to that of Ligase I, is extended in the absence of DNA, and encircles the nicked DNA in a closed clamp structure. Ligase III partners with XRCC1 which, unlike Ligase III, has a nuclear localization signal.
Although the structures of most of the BER players have been solved in complex with DNA and in the case of glycosylases, with a lesion in the active site, there are some exceptions. As yet there are no structures of the mammalian glycosylases that recognize oxidized pyrimidines (NTH1 and the three NEIL proteins) with DNA lesions in the substrate binding pocket. However, the structures of two viral orthologs of the NEIL proteins, mimivirus Nei1 and Nei2, have been solved in complex with DNA, and in the case of mimivirus Nei1, with lesions in the active site [23,24]. The latter structures together with site-specific mutagenesis clearly show that there are no specific interactions between amino acid residues in the glycosylase and the lesion, supporting prior hypotheses that the lesion search must occur before extrusion of the DNA base. The structures of human NTH1 or even prokaryotic Nth1 with lesions in the substrate binding pocket have still eluded all attempts.
All of the mammalian proteins possess disordered regions which have been shown to interact with other cooperating proteins (for reviews see [25,26]). These interactions are likely dynamic influenced by cell cycle and cellular stress conditions. The amino acids involved in these protein-protein interactions have often been elucidated by molecular techniques but the structural visualization of these interactions has been difficult. Small-angle X-ray scattering (SAXS) is currently being used to probe the nature of the repair protein/protein interacting domains (for example see [27]). Moreover, particular protein/protein interactions will most certainly be influenced by post-translational modifications. Clearly, visualization of specific protein/protein complexes is essential for understanding and clarification of the functions of any “BERosome” that may assemble in response to changes in cellular conditions.
3. The DNA glycosylase search for lesions
The primary enzymes in the BER pathway responsible for the lesion search are the DNA glycosylases, the first enzymes in the pathway (for reviews see [28–33]). As glycosylases do not use any energy while they are doing their job, they must find lesions by thermal diffusion. Although not definitive, glycosylases appear to use a combination of sliding and distributive interactions in the search for lesions. Correlated cleavage ensemble studies were able to estimate whether the DNA glycosylase dissociates from the DNA to find another DNA molecule with a lesion or continues on the same molecule to find a second lesion. At low salt concentrations bacterial Udg, MutY and Fpg glycosylases appear to slide several hundred base pairs before dissociating from the DNA [28,34,35]. Similar studies showed human AGG to be able to hop over a protein bound to the DNA to find the second lesion [36] while Udg could hop over nicks and gaps [26,27].
More recently, single molecule approaches have employed undamaged DNA molecules to visualize the glycosylase search and have provided major insights into the process [33,37–40]. The first human enzyme examined, OGG1, was shown to move along DNA in a bidirectional and random manner and no change in this behavior as a function of salt concentration was observed. Since changes in salt did not influence the diffusion constant, it was concluded that OGG1 does not hop on DNA. Furthermore, the diffusion constant was in keeping with the enzyme rotating around the DNA molecule. The three bacterial DNA glycosylases that recognize oxidative DNA damages, Fpg, Nei and Nth, were also studied using single molecule approaches. Similar to what had been observed with OGG1, the sliding behavior of these three enzymes was bidirectional, random and highly redundant. Importantly, the bacterial enzymes use a wedge residue to probe for damage since mutation of this residue to an alanine resulted in an order of magnitude increase in the rate of diffusion. The diffusive behavior of the three bacterial glycosylases was also examined on DNA containing the damages they recognize. The enzymes were observed to stop at a frequency proportional to the number of damages contained in the DNA molecule. Observations to date can be explained by a chemo-mechanical model in which DNA glycosylases rotationally scan DNA and randomly spot check for damage, slowing down substantially while doing so. Once a glycosylase finds a damage it stops to remove it.
Although a cohesive picture of the lesion search is emerging from these single molecule studies, much remains to be elucidated. The diffusive behavior of other mammalian DNA glycosylases and enzymes in the entire BER pathway need to be examined using single molecule assays. Furthermore, stringing up lambda DNA for single molecule studies is a far cry from the configuration of DNA in chromatin. It would thus be informative to examine glycosylase motion on nucleosome tightropes.
4. Base excision repair in chromatin
Several laboratories have reconstituted single nucleosomes with positioned lesions and asked how the BER enzymes recognize and remove the lesions (for a review see [41]). What is clear is that the processing of oxidatively damaged bases and abasic sites by glycosylases or AP endonucleases, respectively, depends on the helical orientation of the lesion and its position relative to the nucleosome dyad axis [42–49]. Outward facing lesions near the nucleosome edge are readily processed by glycosylases and APE1. If the lesion is facing inward toward the histone octamer, it is still processed but much less efficiently. The current hypothesis is that spontaneous reversible partial unwrapping of nucleosomal DNA allows for access by the glycosylase or AP endonuclease to the inwardly facing lesion in the nucleosome. In keeping with this idea, it was found that when the lesion was facing toward the histone octamer, removal by NTH1, NEIL1 and APE1 depended on the enzyme concentration, with effective concentrations approaching the estimated in vivo values. It was also observed that lesions closer to the dyad axis are more difficult to process than those further away, possibly due to less frequent unwrapping. Interestingly, like the glycosylases and APE1, when presented with a gap, Pol β readily fills it in when it faces away from the histone octamer but does so much less efficiently when it faces toward the octamer. In contrast, when presented with a nick, ligase IIIα/XRCC1 behaves identically regardless of whether the nick is facing outward or inward toward the histone octamer. In fact, when the entire BER system is reconstituted, complete repair is observed whether the lesion was facing outward or inward; however, nucleosome disruption appears to occur during the final ligase IIIα/XRCC1 step. It is not surprising that some disruption occurs during the ligation step given that ligase encircles the DNA molecule.
Despite these advances, little is known about histone chaperones or nucleosome remodeling complexes that may respond to the need for BER. SW1/SNF was reported to increase the efficiency of the initial steps of BER in a model nucleosome [50] and ISW1 and ISW2 were reported to enhance the excision of uracil from nucleosomes in vitro [51]. Also, little is known about the possible impact of histone modifications on the efficiency of BER in nucleosomes, although it should be pointed out that studies with reconstituted histones from chicken erythrocytes and unmodified recombinant histones have yielded similar results [47,49]. Finally, understanding BER on single reconstituted nucleosomes in vitro, although a major advance, is still a far cry from strings of nucleosomes or condensed 30 nm chromatin fibers.
5. Base excision repair in eukaryotic cells
BER is clearly a cellular workhorse necessary not only to maintain genome stability but also for its many specialized roles. Thus, the proteins involved in these multiple different processes must be regulated at the transcriptional, posttranscriptional, post-translational and targeted degradation levels, and likely by small non-coding RNA as well. Regulation includes a number of signal transduction pathways and in addition, regulation of BER is complicated by the fact that BER has been co-opted for other duties (see Fig. 1). Moreover, there are external stimuli to contend with as well as variations in internal stimuli moderated by the fact that ROS, whose resulting DNA lesions are repaired by BER, are also essential for a variety of signaling pathways (for reviews see [52,53]). For example, Boldogh’s group [54–56] has shown that OGG bound to its product, 8-oxoG, binds to Rac1-GDP, which results in GTP exchange and an increase in ROS levels involved in redox signaling. Furthermore, upon removal of 8-oxoG by OGG, the released 8-oxoG base activates Ras GPTase, resulting in phosphorylation of its downstream targets. More recently [57] APE2 was shown to be required for ATR-Chk1 checkpoint activation in response to hydrogen peroxide treatment of Xenopus egg extracts. It is clear that our understanding of the involvement of BER in signal transduction pathways is in its infancy and much needs to be learned. Many of the cellular issues surrounding BER that are discussed in this section should be able to be addressed in the near future using high resolution microscopy such as STORM as well as live cell imaging.
A consistent picture of the transcriptional regulation of the DNA repair enzymes has yet to be determined (for a review see [58]). The promoters of the housekeeping glycosylases, OGG1 and NTH1 have been cloned and analyzed and BRCA1 has been shown to stimulate the expression of both enzymes [59] (and for a review see [60]). There are a number of conflicting studies on the upregulation of the DNA glycosylases and other BER enzymes in response to stress. For example, OGG1 expression has been reported to increase following exposure to a variety of insults but other studies show no such changes. Oxidative stress upregulates expression of MUTYH and NEIL1 although no increase in transcription of any of the bifunctional glycosylases was observed following low doses of ionizing radiation [61]. Expression of BER enzymes is clearly an area that needs further work possibly by using microarray expression analysis in a coordinated fashion on several model systems.
BER glycosylases also show tissue-specific expression as determined primarily by Northern blot analyses, with much of the data coming from mouse, rat and adult human tissues. There is considerable variation in the expression of all the glycosylases among different tissues with NEIL3 being unique insofar as it is only expressed in stem cells, blood forming organs and during embryonic development (for a review see [62]).
The BER glycosylases are also differentially regulated during the cell cycle [58]. In the case of the monofunctional enzymes, UNG2 is upregulated in S phase, repairs postreplicative U opposite A, and interacts with PCNA (which likely is required given its postreplication duty). UNG1, a splice variant synthesized from the same gene, is localized to the mitochondria. SMUG1 is not cell cycle regulated while TDG expression is highest in G1. As with UNG2, MUTYH has a replication-associated function removing adenine misincorporated opposite 8-oxoG; it too is up-regulated during S phase and co-localizes with PCNA, thus appropriately attached to the replication apparatus.
Expression of the bifunctional NEIL1 glycosylase increases during S phase and it binds to a number of replication proteins, including PCNA, RPA, FEN-1 and WRN. It has recently been shown to be associated with the replication fork, presumably to act as a cow catcher before the fork to remove the oxidized DNA lesions it recognizes [63]. NEIL2 prefers lesions in single-stranded DNA and bubble structures; it associates with RNA polymerase II and other proteins involved with transcription, and has been implicated in transcription-coupled BER [64]. NEIL3 is found only in proliferating cells and cancer cells and appears to be important during embryo-genesis. Its cellular function remains to be elucidated (for a review see [62]). The expression of NEIL2 does not change during the cell cycle; however, NEIL3 appears to be upregulated in S phase and maintains its levels through G2/M. There are conflicting reports on the cell cycle expression of NTH1 and OGG1, although NTH1 seems to be expressed at higher levels than OGG1. The expression of nuclear and mitochondrial forms of OGG1 exhibit some cell cycle dependence.
Posttranslational modifications are often found in the disordered regions of the BER proteins and have been catalogued for most of them (for reviews see [25,26,65]). With respect to the DNA glycosylases, phosphorylation of OGG1, MUTYH, UNG2 and MBD4 have been observed to increase their catalytic activity. TDG, AGG/MPG, NEIL2 and OGG1 are acetylated, and TDG is SUMO-lated apparently altering its subcellular localization. Acetylation of NEIL2 inactivates it, while acetylation of OGG1, which increases in response to oxidative stress, increases its activity. Again, much remains to be learned about the roles that posttranslational modifications play in glycosylase function. APE1 can be phosphorylated by several different kinases at several different sites which have been shown to modulate APE1 activity, in some cases promoting inactivation. APE1 also undergoes a number of posttranslational modifications, including acetylations that affect its binding to Y-box binding protein 1 (YB-1); the Y box is a sequence motif found in certain promoters and enhancers (for a review see [66]). Interestingly, levels of acetylation of APE1 are increased in response to cellular stress.
There are a few studies on particular ubiquitylation sites on BER proteins which regulate their stability either positively or negatively (for a review see [67]). The E3 ubiquitin ligase, MULE, monoubiquitylates BER proteins when they are present in excess [68] while CHIP polyubiquitylates them [69], marking them for degradation. In fact, it has been suggested that any Pol β or Lig III/XRCC1 that is not part of a BER complex is ubiqutylated by CHIP for degradation [69,70]. The E3 ubiquitin ligase UBR3 appears to polyubiquitylate APE1 for degradation [71]. Clearly the various posttranslation modifications and the signal transduction pathways that influence them are in need of further elucidation.
BER also occurs in mitochondria and most of the glycosylases and APE1 are found in both nuclei and mitochondria. Nuclear localization sequences have been identified for NTH1, NEIL1, NEIL3, MUTYH and OGG1. NEIL2 does not contain a classic nuclear localization signal; nevertheless, it primarily localizes to the nucleus. OGG1 has seven splice variants. Some of these localize to both mitochondria and the nucleus while OGG1 β is exclusively found in mitochondria. More than 15 transcripts of MUTYH, formed by alternative splicing, have been identified that collectively are predicted to give rise to nine different isoforms with one of two abundant isoforms going to the nucleus and the other to the mitochondria (for a review see [72]). UNG1 is the only uracil glycosylase found in the mitochondria. AAG/MPG has also been found to localize to the mitochondria. Both AAG/MPG and UNG1 interact with mitochondrial single-stranded binding protein, which prevents their removing lesions from single-stranded DNA thus inhibiting the formation of potentially lethal AP sites and SSBs [73,74]. APE2 is most often localized to the nucleus but is also found in mitochondria [75]. Nuclear-encoded micro RNAs have also been found in the mitochondria and have been postulated to play a role in ROS signaling and damage response pathways (for a review see [76]).
Localization of BER glycosylases and downstream enzymes have been studied at damage-specific sites in the nucleus primarily using laser systems to create areas of damage. NTH1, OGG1, NEIL1 and NEIL2 as well as downstream BER enzymes were shown to accumulate at sites of laser irradiation. Similar studies with OGG1 showed it to be localized to specific regions following UVA radiation. The potential formation of “BER foci” is another area in need of experimental probing.
6. Base excision repair subpathways and crosspathways
Thus far this perspective has principally focused on classic BER initiated by a DNA glycosylase on a damaged base. In this section, repair of AP sites and SSBs as well as the relationship of BER to nucleotide incision repair (NIR), nucleotide excision repair (NER), ribonucleotide excision repair (RER) and crosslink repair will be discussed.
On a daily basis, both spontaneous depurinations and ROS-induced AP sites form between 10,000 – 20,000 sites of base loss per cell per day [77]. In addition, AP sites are intermediates in the BER pathway initiated by the monofunctional DNA glycosylases. It is currently assumed that recognition and initiation into the BER pathway at AP sites occurs by the action of APE1. However, there has been little investigation of the damage-specific search by this enzyme.
Single-strand breaks are one of the most common DNA damages being produced at levels of tens of thousands per cell per day (for reviews see [26,78–80]). They are formed by ROS, as intermediates of DNA repair pathways, and by topoisomerase 1 (TOP1), which acts to remove superhelical stress associated with replication and transcription. ROS produce 3′-phosphoglycolate or 3′-phosphoglycoaldehyde sugar residues on the 3′ end of the strand break, and these can be recognized and removed by APE1. Persistent or covalently-bound trapped TOP1 complexes are harmful since they can block both DNA and RNA polymerases. Tyrosyl-DNA phosphodiesterase 1 (TDP1) is capable of removing TOP1 crosslinked to a 3′ phosphate at a strand break generated by an abortive TOP1 reaction. The resulting 3′ phosphate group from the action of TDP1 is removed by PNK, which also removes the 3′P generated by the lyase reactions of NEIL1 and NEIL2. Abortive DNA ligations can leave 5′ AMP groups that can block replication and transcription. These residues are removed by aprataxin (APTX). Mutations in TDP1, APTX and PNK are genetically linked to specific neurological disorders (see Section 11).
PARP proteins, PARP1 and PARP2, are also involved in single-strand break repair (SSBR) and act as single-strand break sensors. Both PARP1 and PARP2 transfer the ADP ribose moiety from NAD to a number of proteins including themselves; PARP2 appears to act as a backup for PARP1. PARP1 also recruits XRCC1 to sites of SSBR [81].PARP1 has been shown to stably interact with OGG1 and NEIL1 [82,83] suggesting a possible role for PARP in events upstream of an SSBR although there are conflicting data suggesting that this is not the case [84].
A number of years ago it was demonstrated that APE1 has the ability to cleave damaged DNA upstream from various oxidative damages leaving a 3′ hydroxyl and 5′ phosphate next to a damaged nucleotide (for reviews see [25,80,85]). This pathway was called nucleotide incision repair or NIR. In fact, it was shown in cell free extracts that NIR activity is the predominant activity on 5-hydroxycytosine. In this case, the 5′ DNA strand containing the damage is cleaved by the activity of FEN1 and subsequent repair is accomplished through long patch BER.
Nucleotide excision repair (NER) has also been linked to oxidative damage repair (for a review see [86]). It has been known for over a decade [87,88] and more recently clarified [89] that NER can repair the distorting oxidized base, thymine glycol. Interestingly, in the absence of CSB or XPC/XPA which are involved in NER, 8-oxoG damage levels in DNA increase. As well, XPC can stimulate the activity of OGG1. In fact it has been recently demonstrated that in human fibroblasts, 8-oxoG can be removed by transcription-coupled repair utilizing OGG1 together with CSB, XPA and UVSSA [90]. CSB can also stimulate the activity of a number of glycosylases [91] and APE1 [92] and yeast NER can repair AP sites [93].
Ribonucleotides are incorporated into DNA at a rate of about 1–2 per kilobase. Because a ribonucleotide is less stable than a deoxyribonucleotide, genome stability can be affected. Ribonucleotides can be removed from DNA by ribonucleotide excision repair (RER) which is initiated by ribonuclease H2 (RNase H2) which is active on a DNA/RNA hybrid. RER has been reconstituted in vitro [94] and the products of RNase H2 incision, a 3′ OH and 5′ sugar, are substrates for a long patch BER reaction that includes PCNA, a replicative polymerase, FEN1 or Exo1 and Ligase I. Mice nullizygous for RNase H2 are embryonic lethals and mutations in RNase H2 cause a neuroinflammatory disorder, Aicarde-Goutières Syndrome [95,96].
In the past several years there has been evidence that BER may play a role in interstrand crosslink repair (for reviews see [80,97,98]). Interstrand crosslinks can be formed endogenously by malondialdehyde, a product of lipid peroxidation, as well as by exogenous agents, and can be repaired by NER or homology-directed repair (HDR). Psoralens, natural products found in plants, can also be activated to produce monoadducts and interstrand crosslinks. AGG/MPG appears to be important for the resistance of mouse embryonic stem cells to psoralen interstrand crosslinks but not to monoadducts [99]. Other studies demonstrated that NEIL1 can excise psoralen-induced monoadducts in duplex DNA thereby initiating the BER response pathway. These same workers found that NEIL1 can excise an unhooked interstrand crosslink remnant within a synthetic three-stranded structure, again initiating a classic BER reaction.
More recently it was found that cells deficient in BER display a platin-resistant phenotype, which is accompanied by enhanced excision of platin interstrand crosslinks [100]. These workers further demonstrated that cytosine residues that flank interstrand crosslinks are subject to spontaneous deamination. The subsequent BER-initiated excision of the resulting uracil also renders the cells resistant to platin. It turns out that when interstrand crosslinks are induced in cells by psoralen, NEIL1 is immediately recruited to the crosslink and it remains bound [101]. Thus cells lacking NEIL1 exhibit a much greater repair of the psoralen-induced interstrand crosslinks because the crosslinks are accessible to the NER and HDR repair enzymes. Taken together the data suggest that rather than being directly involved in the repair of the interstrand crosslinks, BER might actually inhibit repair of these crosslinks. These data do not rule out, however, the role that BER glycosylases may play in recognizing monoadducts, despite the fact that they are distorting, or the unhooked three-stranded remnants of an interstrand crosslink.
7. Base excision repair and telomeres
Telomeres are G-rich sequences at the ends of chromosomes that are progressively shortened as the cell differentiates (for reviews see [102,103]). Telomeres are maintained in proliferating stem cells and cancer cells by an RNA-dependent DNA polymerase called telomerase. These G-rich repeat regions consist of duplex DNA and, in addition, about 100–280 bases present in a single-stranded form at the end of chromosomes. It is important that the ends of chromosomes be protected in order to prevent recombination between chromosomes and thus chromosome aberrations and undesired DNA damage responses. This end protection is accomplished by a complex of proteins called shelterin (for reviews see [104,105]). The telomere shelterin protection complex includes telomeric repeat binding factors 1 and 2 (TRF1 and TRF2) as well as protection of telomere 1, POT1. TRF1 and TRF2 are double-stranded binding proteins while POT1 I is a telomeric single-stranded DNA binding protein. The G-rich telomere DNA is highly susceptible to oxidation and the resulting oxidation products may inhibit DNA replication and binding by the shelterin complex proteins resulting in telomere degradation. Also, telomere sequences have been shown to form G-quadruplex structures in vitro and along with POT1 are likely to play a role in telomere homeostasis in vivo [106–108]. G-quadruplex structures have also been proposed to inhibit DNA replication, transcription and translation [107].
Some years ago it was shown that TRF1, TRF2 and POT1 enhance the rate of individual steps of long patch BER [109]. TRF2 physically interacts with Pol β and induces telomere dysfunction [110] and as well interacts with other BER proteins [111]. More recently [112], using mouse tissues and MEFs from Nth nullizygous mice, telomeres were found to harbor a higher level of lesions recognized by NTH compared to non-telomeric sequences. As well, oxidative damage was repaired more slowly in telomeres than in non-telomeric DNA. Also, in adult bone marrow cells from Nth nullizygous mice, telomeres exhibited increased damage foci and increased telomere fragility when incubated in 20% oxygen. When subjected to replicative stress, these same cells suffered severe telomere loss. Interestingly, there were conflicting results obtained with Ogg1 nullizygous mice. In comparison to the wild type, telomere lengthening was observed in Ogg1−/− mouse tissues and primary embryonic fibroblasts (MEFs) cultivated in hypoxic conditions (3% oxygen). In contrast, telomere shortening was detected in Ogg1−/− mouse hematopoetic cells and primary MEFs cultivated in normoxic conditions (20% oxygen) or in the presence of an oxidant [113]. It should be noted that telomere shortening and lengthening phenotypes are difficult to compare between humans and mice, since mouse telomeres are about ten times longer than human telomeres. Recently the role of APE1 in telomere function has been examined using siRNA knockdown of APE1 in several cell culture lines [114]. Human APE1 consists of two regions, the N-terminus possesses a redox regulatory function and the C-terminus contains the AP endonuclease activity. Both functions are required to maintain telomere function. It turns out that loss of APE1 altered the binding of TRF2 to telomeres, which was in keeping with an earlier study showing that the presence of AP sites in telomeric DNA affected the binding of both TRF1 and TRF2 [115]. The resulting unpacking of the telomeric DNA led to telomeric DNA shortening, aberrant mitosis, and genomic instability.
Since G-quadruplex structures in telomeres may play important roles in maintaining telomere structure, either inhibiting or enhancing telomerase activity, understanding the potential role of BER proteins in processing lesions in quadruplex structures is important. It has recently been shown [116] that neither housekeeping enzyme, NTH1 or OGG1, can remove thymine glycol or 8-oxoG lesions, respectively, from telomeric quadruplex DNA at least from the particular sites examined. NEIL3, however, was able to remove both thymine glycol and the further oxidation products of 8-oxoG, Sp and Gh, from quadruplex structures, and the 8-oxoG oxidation products could also be removed by NEIL1. Further studies on the roles that BER proteins play in telomere maintenance and/or disruption could shed light on telomere homeostasis.
8. Base excision repair and transcriptional regulation mediated by oxidative DNA damage
Although it has been known for quite some time that reactive oxygen species are involved in a variety of cell signaling pathways, it is only recently that oxidative damage and its recognition by the BER enzymes were shown to be involved in transcriptional regulation of specific genes (for a review see [117]). Early on Ziel and co-workers observed that oxidative modifications were enriched in the hypoxic response element of the VEGF gene. When AP sites were introduced into this hypoxic region, reporter gene expression substantially increased compared to the wild type region [118]. Subsequently Perillo and co-workers showed that treatment of estrogen receptor positive MCF7 cells with estradiol induces the recruitment of the estrogen receptor to estrogen-responsive promoters. This recruitment was driven by receptor-targeted demethylation of H3-lysine 9 at both enhancer and promoter sites [119]. It turns out that the methylase that does this job, LSDI, launches a FAD-dependent oxidative reaction which results in the formation of hydrogen peroxide. This leads to oxidation of the nearby guanine bases to 8-oxoG. The subsequent recruitment of OGG1 and TOPOIIβ to the promoter results in transient nicks, which leads to conformational changes and subsequent estrogen receptor-induced transcriptional activation. This same group observed LSD1/BER-mediated transcriptional regulation of MYC--activated genes. The sequence required for MYC binding, CACGTC, is present in about 15% of the promoter regions in the human genome. In the MYC-responsive promoters of the Ncl and Cod genes, both OGG and APE1 were found to be engaged in this LSD1/BER-mediated transcriptional activation. Since MYC is one of the most common activators of genes involved in cellular proliferation, this is clearly relevant to cancer progression. G-quadruplex structures have also been shown to regulate c-MYC transcription [120]. Thus BER may play multiple roles in transcriptional activation induced by LSD1-mediated oxidative damage and/or quadruplex structures, roles that need to be further defined.
9. Base excision repair and active demethylation
In mammalian cells, about 3–4% of genomic cytosine is methylated to 5-methylcytosine in a CpG sequence. These sequences can be found in promoter regions and when methylated can influence transcription rates, usually by reducing the binding of essential transcription factors (for reviews see [121–123]). Recent studies have shown that BER may alter transcriptional patterns by actively demethylating DNA cytosine. Specifically, TET oxidases appear to first oxidize 5-methylcytosine to 5-hydroxymethylcytosine. Next, AID/APOBEC proteins can deaminate 5-methylcytosine to thymine, or 5-hydroxymethylcytosine to 5-hydroxymethyluracil. 5-Hydroxymethylcytosine can also be further oxidized by TET oxidases to 5-formylcytosine and 5-carboxylcytosine. These cytosine oxidation products are substrates for TDG, MBD4 and SMUG1, which can then initiate BER. TDG has been found to associate with a number of transcriptional co-regulators, including CBP/p300, which acetylate histone tails and purportedly make promoter regions more accessible for transcription factor binding. MDB4 is also involved in active DNA methylation. MDB4 not only contains a DNA glycosylase activity that removes thymines paired with G, but also contains a subunit that binds to CpG and can form a functional interaction with AID/APOBEC. Like TDG, MDB4 can also remove 5-hydroxymethyluracil paired with G, and recent studies show that phosphorylation of MDB4 may potentiate its activity directly on 5-methylcytosine paired with G. Both thymine and 5-hydroxymethyluracil (produced from 5-methylcytosine by deamination and deamination and oxidation, respectively) are substrates for TDG and MBD4, which both recognize T-G mispairs. Collectively these events can trigger transcription by demethylating promoter-containing cytosines to activate transcription, and also protect the functions of housekeeping genes by maintaining their associated CpG islands in an unmethylated state. Thus it appears that BER plays a critical role in transcriptional regulation during epigenetic reprogramming.
During embryogenesis there is a genome-wide epigenetic programming that first removes all methylation marks. It has been known for many years that specific methylation marks for tissue-specific programming are established by de novo DNA methylases and maintained during DNA replication by maintenance methylases. Recent studies have shed light on how genome-wide demethylation may occur, with the oxidized products of 5-methylcytosine playing an important role. TDG appears to be the glycosylase responsible for the genome-wide removal of these oxidized bases followed by BER processing that restores the original cytosine. Although other DNA glycosylases will recognize oxidized products of 5-methylcytosine, they do not appear to functionally overlap with TDG, since TDG is the only glycosylase which when knocked out in mice leads to a lethal phenotype. Knockouts of any of the downstream BER proteins are also lethal. Consistent with its key role in global demethylation, TDG is ubiquitously expressed during embryonic development and interacts with de novo DNA methylase, DNMT3a. This interaction prevents remethylation until genome wide demethylation is complete and reprogramming can occur. PARP1 also appears to be involved in this process. Much still needs to be learned about the role that BER plays in embryonic development.
10. Base excision repair functions in the immune system
Adaptive immunity ensues when a foreign antigen is recognized and germinal center B cells undergo affinity maturation by means of somatic hypermutation (SHM) and class switch recombination (CSR) which generate antibody diversity (for a review see [124]). Both of these processes are initiated by activation-induced deoxycytosine deaminase (AID) (for a review see [125]), which is upregulated upon antigen encounter and is recruited to transcribing Ig variable regions during somatic hypermutation. AID, a member of the APOBEC family of polynucleotide deaminases, is a processive enzyme that preferentially deaminates cytosine in a particular trinucleotide sequence context in single-stranded DNA leaving behind numerous uracil residues. When the replication fork passes these uracils, C → T transitions are fixed. Alternatively, uracil may be removed by UNG2 (which can process uracils in single-stranded DNA) in which case the resulting AP site can be bypassed by a translesion polymerase, leading to base substitutions at the AP site. Any uracils remaining in the newly synthesized duplex DNA are removed by BER. Error prone mismatch repair has also been shown to contribute to mutagenesis at sites removed from the original deaminated cytosine (for reviews see [121,125]. Class switch recombination switches the Ig isotype of the antibody by rearranging the constant region of the antibody heavy chain by breakage and joining of two switch regions. CSR is also initiated by AID, which is recruited to actively transcribing switch regions to deaminate cytosines. This is followed by uracil excision in either single-stranded or duplex DNA. The resulting closely opposed AP sites are cleaved by APE1, leaving a double-strand break substrate for class switch. Although APE2 was thought to play a role in this process [126,127], APE1 has been shown to be the essential nuclease [128].
Chicken B cell lines lacking UNG2 and UNG-deficient mice both show significantly perturbed SHM and CSR. In humans, recessive mutations in the UNG gene are associated with the hyper-IgM syndrome caused by a deficiency in CSR. There are a number of issues that require further examination. There have been reports that the catalytic activity of UNG2 is dispensable for efficient CSR. Also, there is residual SHM and CSR in Ung2 nullizygous mice. In fact, SMUG1 can partially restore SHM and CSR in Ung2−/− Msh2−/− mouse cells. However, as SMUG1 is downregulated upon B cell activation, this observation may not be biologically relevant. Moreover, mice nullizygous for MBD4 show no effect on either SHM or CSR, and the role of TDG remains to be elucidated. Interestingly, there has been a recent report [129] describing a mouse model heterozygous for a targeted disruption in POLB, where the variant exhibits very slow Pol β DNA repair polymerization. These mice show greatly increased somatic hypermutation, presumably because in the wild type mice, a substantial fraction of the uracils introduced by AID are actually repaired by BER (for a review see [130]). In addition, mice expressing the Pol β variant exhibit faulty V(D)J recombination. Of import, these mice develop lupus, suggesting that autoimmune disease results from an imbalance among the processes responsible for generating antibody diversity.
Since AID is upregulated in B cells undergoing SHM, it is important to understand how the proteins involved in generating SHM are targeted to the particular transcription complex undergoing SHM, since there are many fewer mutations in other actively transcribed lesions. Moreover, it remains to be determined what governs which uracils will be repaired accurately by BER and which will be converted to mutations, either by normal replication past uracil or translesion synthesis past an abasic site. It is interesting that SHM relies on the fact that AID is specific for single-stranded DNA and that UNG2 (and APE1 [131]) can process single-stranded DNA while the downstream members of the BER pathway require duplex DNA. Understanding these interrelationships is critical not only for our comprehension of immune disorders and autoimmunity but for cancer, since imbalances can lead to B cell malignancies (for a review see [132]).
Another member of the APOBEC family, APOBEC3G, is a cyto-sine deaminase that is expressed in T cells and catalyzes cytosine deaminations on reverse transcribed retroviral cDNA, when it is in its single-stranded form (for reviews see [121,125,133]). It has been known for several decades that HIV encodes the Vif protein, which is essential for virus infectivity and targets APOBEC3G for polyubiquitylation and subsequent degradation by the proteosme. HIV-Δ vif1 incorporates APOBEC3G into the viral capsid and, upon infecting a naive mouse T cell, the HIV DNA accumulates mutations, resulting in loss of infectivity. The target for APOBEC3G is a triplet cytosine sequence context in single-stranded DNA such that subsequent synthesis of the complementary strand results in C→T mutations. Additionally uracils may be removed from the single-stranded DNA by UNG2, leaving abasic sites that significantly impair the complementary DNA strand synthesis. Alternatively APE1 can cleave the DNA at UNG2-generated AP sites, leaving single-strand breaks. Despite what we know about HIV–Δ Avif1 infections, there is much to be learned about how APOBEC3G and BER may help suppress HIV and retroviral infectivity. APOBEC3G has also been implicated in cancer metastasis, although this process is not well understood.
11. Base excision repair and neurodegenerative diseases
Because brain cells have a high metabolic rate, they produce substantial amounts of ROS and there is a wealth of evidence linking oxidative damage and its lack of repair to the generation of neurodegenerative disorders (for reviews see [134–139]). for example, Cockayne Syndrome, a genetic disease, is defective in transcription-coupled NER. However, unlike NER-deficient xeroderma pigmentosum patients, Cockayne syndrome (CS) patients are not cancer prone but exhibit a number of neurological abnormalities and a drastic reduction in life span. Cells from CS patients of both CSA and CSB complementation groups are hypersensitive to oxidative damage and accumulate oxidative lesions in genomic DNA. Moreover, physical interactions between PARP1 and CSB as well as between CSB and NEIL1, OGG1 and APE1, have been observed. Since CS patients are not cancer prone, it is thought that the ineffective damage processing in these cells leads to cell death or senescence.
In a mouse model, focal cerebral ischemic injury was associated with impaired intracellular trafficking of Ogg1 to the mitochondria. Neurons from Ogg deficient mice are sensitive to oxidative stress and reduced OGG1 levels have also been associated with Alzheimer’s disease. The BER enzymes are often differentially expressed in the brain compared to other organs. For example, NTH1 expression in the brain is similar to other organs but OGG1 is lower in the brain compared to other tissues. PNK, APE1, XRCC1 and LigIIIα are generally highly expressed as is Pol β. Both XRCC1 and PNK have been linked to autosomal recessive neurological disorders. Mutations in the SSBR pathway proteins lead to the progressive neurodegenerative phenotypes, spinocerebellar ataxia with axonal neuropathy in the case of TOP1, and progressive ataxia with ocular motor apraxia and peripheral neuropathy in the case of APTX [139]. Since neurons do not replicate, the numbers of replication/repair proteins tend to be low and thus short patch BER appears to be the predominant mode of repair. Interestingly Ung, Ogg1 and Neil1 deficient mice exhibit increases in infarct size after middle cerebral artery occlusion and reperfusion while Neil3 appears to be involved in the repair of proliferating brain cells.
DNA repair, specifically BER of oxidative damage, has been implicated in trinucleotide repeat expansion (for a review see [140]), which has been associated with over 40 neurodegenerative diseases. OGG1 is involved in CAG trinucleotide repeat expansion in Huntington’s disease, as loss of Ogg1 in a mouse model suppresses trinucleotide expansion. Neil1 may also contribute to trinucleotide expansion. Apparently DNA glycosylase initiation of BER at oxidized bases may induce expansion in intermediates through strand slippage then BER processing would keep this expanded region in the DNA.
A number of laboratories have implicated oxidatively damaged RNA as an important factor in neurodegeneration (for reviews see [135,141]). Indeed, RNA is much more susceptible to oxidation than DNA and oxidatively damaged RNA can lead to abnormal expression of proteins and micro RNAs. Although the human AlkB homolog has been shown to be able to reverse alkylation damage on RNA bases, there is no evidence for repair mechanisms for oxidized RNA. However, a role for degrading damaged RNA has been suggested for APE1 since it has recently been shown to be able to cleave abasic sites in single-stranded message RNA as well as in a RNA/DNA hybrid [142]. This idea has been supported by experiments which showed that upon silencing of APE1 expression, global protein synthesis is significantly decreased. APE1 also interacts with proteins involved with ribosome assembly and RNA maturation in the cytoplasm. A recent review [143] describes the interaction between APE1 and nucleophosmin and how the nucleolus may serve as a hub for repair of oxidatively damaged RNA.
RNA oxidation has been implicated in the pathogenesis of Alzheimer's and Parkinson’s diseases as well as in Amyotrophic Lateral Sclerosis (ALS). Redox active metals accumulate in the brains of Alzheimer’s patients and are the source of hydroxyl radicals that generate oxidative damage. 50% of the mRNAs synthesized in the frontal cortex in Alzheimer’s patients contained oxidative damage compared to 2% in the age matched controls. RNA oxidation has also been shown to occur during the early stages of Parkinson’s disease. Interestingly, in familial ALS patients, about 20% have a mutation in SOD and thus are less able to quench ROS. These patients also exhibit increases in RNA oxidation. There is increasing evidence that RNA oxidation is a feature of neurons in the aging brain as well as in vulnerable neurons in early stage age-associated neurogenerative disorders suggesting that RNA oxidation contributes to the development of these disorders. We know very little about how RNA damage is handled in cells and how BER enzymes might be involved.
12. Base excision repair and cancer
Since BER removes approximately 40,000 endogenous lesions per human cell per day and cancer is for the most part a result of genomic damage and instability there is no doubt that BER plays an important role in cancer prevention. A number of reviews have been recently written on this topic [144–149]. A great deal has been learned about human disease phenotypes by observing mice that are nullizygous for particular genes. This has been more difficult with BER and cancer since the first step in the repair pathway is catalyzed by the DNA glycosylases that have redundant substrate specificities. Hence there is often not a clear phenotype in a nullizygous mouse for a single DNA glycosylase. Another complication is that all of the downstream enzymes in the pathway are absolutely required for development; therefore, nullizygous mice in these enzymes all exhibit embryonic lethality.
Mouse models for all of the uracil/thymine processing DNA glycosylases have been constructed and Ung deficient mice exhibit a large increase in uracil-containing DNA but a smaller increase in spontaneous mutation frequency compared to wild type, probably due to the presence of a second glycosylase, SMUG1. Ung deficient mice also display an increased incidence of spontaneous B cell lymphomas in their old age. Mice with a targeted allele replacement of MBD4 glycosylase show an increase in C→T transitions at CpG sites, in keeping with that enzyme’s substrate specificity. When crossed with a cancer prone APC heterozygous mouse, the MBD4−/− progeny showed accelerated tumor production associated with C→T transitions at CpG sites. SMUG1 knockout mice are phenotypically normal with a normal life span but they lack nearly all 5-hydroxymethyluracil activity. Ung−/− SMUG−/− double knockout mice were also viable; however, when crossed with an MSH2 knockout mouse, the double knockout mice had a greatly reduced life span and died from lymphomas. Interestingly, MBD4 is mutated in a large fraction of human colorectal cancers which have microsatellite instability. Nullizygous Tdg mice also exhibit embryonic lethality, as discussed in Section 9, in keeping with the putative role of this protein in active genome-wide embryonic DNA demethylation.
Chronic inflammation as a cause of cancer was suggested in 1863 by Virchow and there is substantial evidence to support this hypothesis (for a review see [150]). Reactive oxygen and nitrogen species are formed at the sites of inflammation and can initiate cell signaling pathways. In addition to apoptosis these can influence damage accumulation thus mutation accumulation resulting in tumor initiation and/or progression. Mice defective in Agg/Mpg are also viable and develop normally; however, the cells exhibit a moderate increase in sensitivity to alkylating agents. When the Agg−/− mice were treated with agents to induce alkylation damage that results in inflammation, they also exhibited a higher frequency of colon cancer. Agg−/− mice also show more severe gastric lesions than wild type when infected with Helicobacter pylori, implicating AAG and BER in the suppression of gastric cancers.
Since guanine has the lowest redox potential of the four bases, the most frequently produced oxidative lesions result from oxidation of guanine. Organisms from bacteria to humans have evolved multiple mechanisms to deal with this problem (for reviews see [1,3]). Both 8-oxoG and FapyG opposite C are the preferred substrates for OGG. However if the replication fork bypasses the 8-oxoG or FapyG before they are removed, an A is often inserted opposite by the replicative polymerase and subsequent replications fix the G → T transversion mutations. To counteract this, MUTYH is able to remove adenine when it is incorporated opposite 8-oxoG and in concert with pol λ and long patch BER proteins, generate an 8-oxoG-C pair for OGG-initiated BER (for a review see [151]). Mice deficient in Ogg are viable and fertile, although they do accumulate 8-oxoG lesions and have an increased spontaneous mutation rate. MutY nullizygous mice are also viable and healthy but exhibit an increase in spontaneous mutation frequency. However, mice nullizygous for both Ogg and MutY exhibit a significantly increased tumor incidence, especially lymphomas, lung and ovarian tumors.
A direct relationship between MUTYH and colorectal cancers in humans has been established. A decade ago, a British family was diagnosed with multiple colorectal adenomas and carcinomas but family members lacked the standard inherited adenomatous polyposis coli (APC) defect. However, when the tumors of these families were examined, there was a very high proportion of G → T transversion mutations in the APC gene which are signatures of defects in MUTYH. It turns out that these patients have biallelic mutations in MUTYH and are predisposed to MUTYH-associated polyposis (MAP). Although colorectal cancers predominate in MAP patients, ovarian, bladder and skin cancers are also observed. There are numerous truncating and missense mutations observed in MAP patients. One of the most common mutations is in the glycosylase search wedge residue, mentioned in Section 3, which is used by Nth, Fpg and Nei, and presumably MUTYH, to locate lesions in DNA.
Nullizygous mice have also been generated for Nth1, Neil1 and Neil3, and all are viable and fertile, but show an increase in base lesions in genomic DNA of particular organs. Interestingly, a substrain of the Neil1 knockout mice, primarily males, develop metabolic syndrome, possibly due to the accumulation of unrepaired mitochondrial oxidative DNA damages. However, Nth−/−− Neil1−/−− double knockout mice, which have substantially reduced activity on oxidized pyrimidines and formamidopyrimidines, exhibit lung and liver tumors.
Although mice nullizygous for Ape1 are embryonic lethals, heterozygous mice for Ape1 are viable with no apparent abnormality. POLB null mice are similarly inviable but, unlike Ape1 heterozygotes, POLB heterozygotes exhibit higher levels of single-strand breaks and chromosomal aberrations, and hypersensitivity to alkylating agents. Pol β appears to play a very important role in suppressing carcinogenesis since 30% of human tumors studied to date carry POLB mutations that are not found in the germ line. Many of these tumor-associated variants induce genome instability and cellular transformation. Ligase III, ligase I and XRCC1 nullizygous mice are also embryonic lethals.
There are numerous genome wide studies attempting to associate germ line single nucleotide polymorphisms (SNPs) with predispositions to various types of cancer. Usually the most common variants in the population are studied because better statistical correlation can be obtained from epidemiological studies. Often these more common variants do not have significant alterations in their biochemical function and do not exhibit a convincing cancer phenotype. There is, however, increasing emphasis on correlating function or lack thereof with potential outcomes. For example, a recent paper [152] reported that a germ line variant of NTH1, present as a heterozygote in about 6% of the population, has a mutation in an active site residue, and as a result, is enzymatically inactive. This variant causes cellular transformation and genomic instability in a human breast epithelial line when present in a heterozygous state and is thus likely to predispose an individual possessing this variant to cancer. Functional and biological studies have also been done on Pol β and NEIL1 cancer variants [153–159]. Sometime in the not too distant future, when one’s exon sequences, determined by next generation sequencing, are part of every medical record, single SNPs, pathway SNPs and particular combinations of SNPs will be surely linked to cancer risk and outcomes.
13. Base excision repair and aging
It has been known for many decades, that DNA damage accumulates in cells over time and that this accumulation is linked to neurodegenerative disorders and cancer, both diseases of aging (see Sections 11 and 12). Implicit in this idea is that accumulation of damage and thus mutations are associated with the aging process (for a review see [160]) and that when speaking of endogenous damages, the BER system is incapable of keeping up.
Accumulation of oxidative damage in mitochondria and neuronal cells that have high metabolic rates have been associated with accumulation of damage and neurodegenerative disorders related to aging (Section 11). Endogenous oxidative damage is efficiently removed from mitochondrial DNA (for reviews see [161,162]) by short patch BER although long patch repair is now known to occur because FEN1 has been located in the mitochondria. Moreover, calorie restricted mice that have been shown to have a longer life span have also been shown to generate ROS at lower rates. Other evidence points to the role that deficiencies in mitochondrial BER, especially in postmitotic tissues, may play in cell function and survival. Mice deficient in Pol γ, which serves as both the replicative and repair polymerase in mitochondria, have been associated with the accumulation of point mutations and large mitochondrial DNA deletions, which are apparently caused by aberrant DNA repair. However, it is not clear if this is related to the aging process. Although increased DNA damage and lack of DNA repair is associated with aging, most of the data on possible links between these processes are correlative or circumstantial.
14. Summary and looking ahead
Base excision repair has certainly come a long way since Tomas Lindahl discovered the first DNA glycosylase in 1974 [163] and since we identified endonuclease III (Nth) the following year [164]. Since BER is evolutionarily conserved, the selective pressures over time tell us that the major job for BER is to repair endogenous DNA damage and maintain genome stability. In humans, this prevents outcomes such as cancer and in the case of oxidative damage, neurodegenerative diseases.
During the last several decades we have made major advances in understanding how the BER enzymes function under in vitro ensemble conditions using simple model substrates such as oligonucleotides containing a specific damage. We also have at our disposal the crystal structures of most BER proteins but not the structures of the BER proteins with their interacting partners. Single molecule approaches with lambda DNA tightropes, have increased our understanding of how glycosylases belonging to two structural families search for damage. Now we need to know if glycosylases in other families search in the same way. And, what about the search for lesions in nucleosomes and in higher order chromatin? We are in the early stages of our understanding about BER of damage in nucleosome structures and we know virtually nothing about BER and higher order chromatin either in terms of single BER enzymes or putative BERosome complexes. There is also much to work out on the role that BER plays in telomere homeostasis. Studies on cellular localization, cell cycle control, involvement of signaling pathways, post translational modification, targeted degradation and the involvement of micro RNAs in BER processing need to employ the latest technical advances and appropriate model systems. The extant data for the most part are a compilation of observations in multiple cell types and tissues, and it is far from providing us with a coherent story. BER subpath-ways and crosstalk among pathways, as well as involvement or lack thereof of BER in RNA damage processing also need clarification, and we know almost nothing about the relationships between BER and ROS-mediated signaling pathways. Importantly, further insights into the role of targeted oxidation at promoter sites are definitely called for. How widespread is this mechanism of gene activation? Further studies are warranted on the role of particular BER enzymes in embryonic genome-wide demethylation as are studies on the regulation of BER enzymes when they are functioning in adaptive immunity. And we know virtually nothing about the role of BER in aging? We are making major headways into understanding how and where BER functions and fortunately for all of us, there is still a great deal to do.
Acknowledgments
Work done in the Author’s laboratory was supported by NIH P01 CA098993 National Cancer Institute. The Author is grateful to Paul Wallace and Scott Kathe for designing Fig. 1 and to colleagues Sylvie Doublié, Joann Sweasy and especially to David Pederson for critically reading the manuscript.
Footnotes
Conflict of interest statement
There is no conflict of interest.
References
- 1.Duclos S, Doublie S, Wallace SS. Consequences and repair of oxidative DNA damage. In: Greim H, Albertini JJ, editors. Issues in Toxicology: The Cellular Response to the Genotoxic Insult: The Question of Threshold for Genotoxic Carconogens. London: The Royal Society of Chemistry; 2012. pp. 109–152. [Google Scholar]
- 2.Krokan HE, Bjoras M. Base excision repair, Cold Spring Harbor Perspect. Biol. 2013;5:a012583. doi: 10.1101/cshperspect.a012583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.David SS, O’Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prakash A, Doublie S, Wallace SS. The Fpg/Nei family of DNA glycosylases:substrates, structures, and search for damage, and search for damage. Prog. Mol. Biol. Transl. Sci. 2012;110:71–91. doi: 10.1016/B978-0-12-387665-2.00004-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu M, Bandaru V, Bond JP, Jaruga P, Zhao X, Christov PP, Burrows CJ, Rizzo CJ, Dizdaroglu M, Wallace SS. The mouse ortholog of NEIL3 is a functional DNA glycosylase in vitro and in vivo. Proc. Nat. Acad. Sci. U.S.A. 2010;107:4925–4930. doi: 10.1073/pnas.0908307107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burrows CJ, Muller JG, Kornyushyna O, Luo W, Duarte V, Leipold MD, David SS. Structure and potential mutagenicity of new hydantoin products from guanosine and 8-oxo-7,8-dihydroguanine oxidation by transition metals. Environ. Health Perspect. 2002;110(Suppl. 5):713–717. doi: 10.1289/ehp.02110s5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li M, Wilson 3rd DM. Human apurinic/apyrimidinic endonuclease 1. Antioxid. Redox Signal. 2014;20:678–707. doi: 10.1089/ars.2013.5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilson SH, Kunkel TA. Passing the baton in base excision repair. Nat. Struct. Biol. 2000;7:176–178. doi: 10.1038/73260. [DOI] [PubMed] [Google Scholar]
- 9.Prasad R, Beard WA, Batra VK, Liu Y, Shock DD, Wilson SH. A review of recent experiments on step-to-step “hand-off of the DNA intermediates in mammalian base excision repair pathways. Mol. Biol. 2011;45:586–600. [PMC free article] [PubMed] [Google Scholar]
- 10.Hanssen-Bauer A, Solvang-Garten K, Sundheim O, Pena-Diaz J, Andersen S, Slupphaug G, Krokan HE, Wilson 3rd DM, Akbari M, Otterlei M. XRCC1 coordinates disparate responses and multiprotein repair complexes depending on the nature and context of the DNA damage. Environ. Mol. Mutagen. 2011;52:623–635. doi: 10.1002/em.20663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parlanti E, Locatelli G, Maga G, Dogliotti E. Human base excision repair complex is physically associated to DNA replication and cell cycle regulatory proteins. Nucleic Acids Res. 2007;35:1569–1577. doi: 10.1093/nar/gkl1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim YJ, Wilson 3rd DM. Overview of base excision repair biochemistry. Curr. Mol. Pharmacol. 2012;5:3–13. doi: 10.2174/1874467211205010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huffman JL, Sundheim O, Tainer JA. DNA base damage recognition and removal: new twists and grooves. Mutat. Res. 2005;577:55–76. doi: 10.1016/j.mrfmmm.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 14.Hitomi K, Iwai S, Tainer JA. The intricate structural chemistry of base exci-sion repair machinery: implications for DNA damage recognition, removal,and repair. DNA Repair. 2007;6:410–428. doi: 10.1016/j.dnarep.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 15.Yang W. Structure and mechanism for DNA lesion recognition. Cell Res. 2008;18:184–197. doi: 10.1038/cr.2007.116. [DOI] [PubMed] [Google Scholar]
- 16.Wallace SS. DNA glycosylases search for and remove oxidized DNA bases. Environ. Mol. Mutagen. 2013;54:691–704. doi: 10.1002/em.21820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duclos S, Aller P, Jaruga P, Dizdaroglu M, Wallace SS, Doublie S. Structural and biochemical studies of a plant formamidopyrimidine-DNA glycosylase reveal why eukaryotic Fpg glycosylases do not excise 8-oxoguanine. DNA Repair. 2012;11:714–725. doi: 10.1016/j.dnarep.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beard WA, Wilson SH. Structural design of a eukaryotic DNA repair polymerase: DNA polymerase beta. Mutat. Res. 2000;460:231–244. doi: 10.1016/s0921-8777(00)00029-x. [DOI] [PubMed] [Google Scholar]
- 19.Idriss HT, Al-Assar O, Wilson SH. DNA polymerase beta. Int. J. Biochem. Cell Biol. 2002;34:321–324. doi: 10.1016/s1357-2725(01)00131-5. [DOI] [PubMed] [Google Scholar]
- 20.Pascal JM. DNA and RNA ligases: structural variations and shared mechanisms. Curr. Opin. Struct. Biol. 2008;18:96–105. doi: 10.1016/j.sbi.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 21.Howes TR, Tomkinson AE. DNA ligase I, the replicative DNA ligase. Sub-cell. Biochem. 2012;62:327–341. doi: 10.1007/978-94-007-4572-8_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tomkinson AE, Sallmyr A. Structure and function of the DNA ligases encoded by the mammalian LIG3 gene. Gene. 2013;531:150–157. doi: 10.1016/j.gene.2013.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Imamura K, Wallace SS, Doublie S. Structural characterization of a viral NEIL1 ortholog unliganded and bound to abasic site-containing DNA. J. Biol. Chem. 2009;284:26174–26183. doi: 10.1074/jbc.M109.021907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Imamura K, Averill A, Wallace SS, Doublie S. Structural characterization of viral ortholog of human DNA glycosylase NEIL1 bound to thymine glycol or 5-hydroxyuracil-containing DNA. J. Biol. Chem. 2012;287:4288–4298. doi: 10.1074/jbc.M111.315309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Almeida KH, Sobol RW. A unified view of base excision repair: lesion-dependent protein complexes regulated by post-translational modification. DNA Repair. 2007;6:695–711. doi: 10.1016/j.dnarep.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hegde ML, Izumi T, Mitra S. Oxidized base damage and single-strand break repair in mammalian genomes: role of disordered regions and posttransla-tional modifications in early enzymes. Prog. Mol. Biol. Transl. Sci. 2012;110:123–153. doi: 10.1016/B978-0-12-387665-2.00006-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hegde ML, Tsutakawa SE, Hegde PM, Holthauzen LM, Li J, Oezguen N, Hilser VJ, Tainer JA, Mitra S. The disordered C-terminal domain of human DNA glycosylase NEIL1 contributes to its stability via intramolecular interactions. J. Mol. Biol. 2013;425:2359–2371. doi: 10.1016/j.jmb.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Francis AW, David SS. Escherichia coli MutY and Fpg utilize a processive mechanism fortarget location. Biochemistry. 2003;42:801–810. doi: 10.1021/bi026375+. [DOI] [PubMed] [Google Scholar]
- 29.Zharkov DO, Grollman AP. The DNA trackwalkers: principles of lesion search and recognition by DNA glycosylases. Mutat. Res. 2005;577:24–54. doi: 10.1016/j.mrfmmm.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 30.Sidorenko VS, Zharkov DO. Correlated cleavage of damaged DNA by bacterial and human 8-oxoguanine-DNA glycosylases. Biochemistry. 2008;47:8970–8976. doi: 10.1021/bi800569e. [DOI] [PubMed] [Google Scholar]
- 31.Friedman JI, Stivers JT. Detection of damaged DNA bases by DNA glycosylase enzymes. Biochemistry. 2010;49:4957–4967. doi: 10.1021/bi100593a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zharkov DO, Mechetin GV, Nevinsky GA. Uracil-DNA glycosylase: struc-tural, thermodynamic and kinetic aspects of lesion search and recognition. Mutat. Res. 2010;685:11–20. doi: 10.1016/j.mrfmmm.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee AJ, Warshaw DM, Wallace SS. Insights into the glycosylase search for damage from single molecule fluorescence microscopy. DNA Repair. 2014 doi: 10.1016/j.dnarep.2014.01.007. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bennett SE, Sanderson RJ, Mosbaugh DW. Processivity of Escherichia coli and rat liver mitochondrial uracil–DNA glycosylase is affected by NaCl concentration. Biochemistry. 1995;34:6109–6119. doi: 10.1021/bi00018a014. [DOI] [PubMed] [Google Scholar]
- 35.Porecha RH, Stivers JT. Uracil DNA glycosylase uses DNA hopping and short-range sliding to trap extrahelical uracils. Proc. Nat. Acad. Sci. U.S.A. 2008;105:10791–10796. doi: 10.1073/pnas.0801612105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hedglin M, O’Brien PJ. Hopping enables a DNA repair glycosylase to search both strands and bypass a bound protein. ACS Chem. Biol. 2010;5:427–436. doi: 10.1021/cb1000185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blainey PC, van Oijen AM, Banerjee A, Verdine GL, Xie XS. A base-excision DNA-repair protein finds intrahelical lesion bases by fast sliding in contact with DNA. Proc. Nat. Acad. Sci. U.S.A. 2006;103:5752–5757. doi: 10.1073/pnas.0509723103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blainey PC, Luo G, Kou SC, Mangel WF, Verdine GL, Bagchi B, Xie XS. Nonspecifically bound proteins spin while diffusing along DNA. Nat. Struct. Mol. Biol. 2009;16:1224–1229. doi: 10.1038/nsmb.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dunn AR, Kad NM, Nelson SR, Warshaw DM, Wallace SS. Single Qdot-labeled glycosylase molecules use a wedge amino acid to probe for lesions while scanning along DNA. Nucleic Acids Res. 2011;39:7487–7498. doi: 10.1093/nar/gkr459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nelson SR, Dunn AR, Kathe SD, Warshaw DM, Wallace SS. Two glycosylase families diffusively scan DNA using a wedge residue to probe for and identify oxidatively damaged bases. Proc. Nat. Acad. Sci. U.S.A. 2014 doi: 10.1073/pnas.1400386111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Odell ID, Wallace SS, Pederson DS. Rules of engagement for base excision repair in chromatin. J. Cell. Physiol. 2013;228:258–266. doi: 10.1002/jcp.24134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nilsen H, Lindahl T, Verreault A. DNA base excision repair of uracil residues in reconstituted nucleosome core particles. EMBO J. 2002;21:5943–5952. doi: 10.1093/emboj/cdf581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beard BC, Wilson SH, Smerdon MJ. Suppressed catalytic activity of base excision repair enzymes on rotationally positioned uracil in nucleosomes. Proc. Nat. Acad. Sci. U.S.A. 2003;100:7465–7470. doi: 10.1073/pnas.1330328100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prasad A, Wallace SS, Pederson DS. Initiation of base excision repair ofoxidative lesions in nucleosomes by the human, bifunctional DNA glycosylase NTH1. Mol. Cell. Biol. 2007;27:8442–8453. doi: 10.1128/MCB.00791-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Odell ID, Newick K, Heintz NH, Wallace SS, Pederson DS. Non-specificDNA binding interferes with the efficient excision of oxidative lesions fromchromatin by the human DNA glycosylase, NEIL1. DNA Repair. 2010;9:134–143. doi: 10.1016/j.dnarep.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hinz JM, Rodriguez Y, Smerdon MJ. Rotational dynamics of DNA on the nucleosome surface markedly impact accessibility to a DNA repair enzyme. Proc. Nat. Acad. Sci. U.S.A. 2010;107:4646–4651. doi: 10.1073/pnas.0914443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Odell ID, Barbour JE, Murphy DL, Della-Maria JA, Sweasy JB, Tomkinson AE, Wallace SS, Pederson DS. Nucleosome disruption by DNA ligase III-XRCC1 promotes efficient base excision repair. Mol. Cell. Biol. 2011;31:4623–4632. doi: 10.1128/MCB.05715-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rodriguez Y, Smerdon MJ. The structural location of DNA lesions in nucleosome core particles determines accessibility by base excision repair enzymes. J. Biol. Chem. 2013;288:13863–13875. doi: 10.1074/jbc.M112.441444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maher RL, Prasad A, Rizvanova O, Wallace SS, Pederson DS. Contribution of DNA unwrapping from histone octamers to the repair of oxidatively damaged DNA in nucleosomes. DNA Repair. 2013;12:964–971. doi: 10.1016/j.dnarep.2013.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Menoni H, Gasparutto D, Hamiche A, Cadet J, Dimitrov S, Bouvet P, Angelov D. ATP-dependent chromatin remodeling is required for base excision repair in conventional but not in variant H2A. Bbd nucleosomes. Mol. Cell. Biol. 2007;27:5949–5956. doi: 10.1128/MCB.00376-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakanishi S, Prasad R, Wilson SH, Smerdon M. Different structural states in oligonucleosomes are required for early versus late steps of base excision repair. Nucleic Acids Res. 2007;35:4313–4321. doi: 10.1093/nar/gkm436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Finkel T. Signal transduction by reactive oxygen species. J. Cell. Biol. 2011;194:7–15. doi: 10.1083/jcb.201102095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Finkel T. Signal transduction by mitochondrial oxidants. J. Biol. Chem. 2012;287:4434–4440. doi: 10.1074/jbc.R111.271999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hajas G, Bacsi A, Aguilera-Aguirre L, Hegde ML, Tapas KH, Sur S, Radak Z, Ba X, Boldogh I. 8-Oxoguanine DNA glycosylase-1 links DNA repair to cellular signaling via the activation of the small GTPase Rac1. Free Radical Biol. Med. 2013;61C:384–394. doi: 10.1016/j.freeradbiomed.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.German P, Szaniszlo P, Hajas G, Radak Z, Bacsi A, Hazra TK, Hegde ML, Ba X, Boldogh I. Activation of cellular signaling by 8-oxoguanine DNA glycosylase-1-initiated DNA base excision repair. DNA Repair. 2013;12:856–863. doi: 10.1016/j.dnarep.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boldogh I, Hajas G, Aguilera-Aguirre L, Hegde ML, Radak Z, Bacsi A, Sur S, Hazra TK, Mitra S. Activation of ras signaling pathway by 8-oxoguanine DNAglycosylase bound to its excision product, 8-oxoguanine. J. Biol. Chem. 2012;287:20769–20773. doi: 10.1074/jbc.C112.364620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Willis J, Patel Y, Lentz BL, Yan S. APE2 is required for ATR-Chk1 checkpoint activation in response to oxidative stress. Proc. Nat. Acad. Sci. U.S.A. 2013;110:10592–10597. doi: 10.1073/pnas.1301445110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sampath H, McCullough AK, Lloyd RS. Regulation of DNA glycosylases and their role in limiting disease. Free Radical Res. 2012;46:460–478. doi: 10.3109/10715762.2012.655730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saha T, Rih JK, Roy R, Ballal R, Rosen EM. Transcriptional regulation of the base excision repair pathway by BRCA1. J. Biol. Chem. 2010;285:19092–19105. doi: 10.1074/jbc.M110.104430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saha T, Smulson M, Rosen EM. BRCA1 regulation of base excision repair pathway. Cell Cycle. 2010;9:2471–2472. doi: 10.4161/cc.9.13.12084. [DOI] [PubMed] [Google Scholar]
- 61.Inoue M, Shen GP, Chaudhry MA, Galick H, Blaisdell JO, Wallace SS. Expression of the oxidative base excision repair enzymes is not induced in TK6 human lymphoblastoid cells after low doses of ionizing radiation. Radiat. Res. 2004;161:409–417. doi: 10.1667/3163. [DOI] [PubMed] [Google Scholar]
- 62.Liu M, Doublie S, Wallace SS. Neil3, the final frontier for the DNA glycosylases that recognize oxidative damage. Mutat. Res. 2013;743–744:4–11. doi: 10.1016/j.mrfmmm.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hegde ML, Hegde PM, Bellot LJ, Mandal SM, Hazra TK, Li GM, Boldogh I, Tomkinson AE, Mitra S. Prereplicative repair of oxidized bases in the human genome is mediated by NEIL1 DNA glycosylase together with replication proteins. Proc. Nat. Acad. Sci. U.S.A. 2013;110:E3090–E3099. doi: 10.1073/pnas.1304231110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Banerjee D, Mandal SM, Das A, Hegde ML, Das S, Bhakat KK, Boldogh I, Sarkar PS, Mitra S, Hazra TK. Preferential repair of oxidized base damage in the transcribed genes of mammalian cells. J. Biol. Chem. 2011;286:6006–6016. doi: 10.1074/jbc.M110.198796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dianov GL, Hubscher U. Mammalian base excision repair: the forgotten archangel. Nucleic Acids Res. 2013;41:3483–3490. doi: 10.1093/nar/gkt076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dolfini D. R. Mantovani. Targeting the Y/CCAAT box in cancer: YB-1 (YBX1) or NF-Y? Cell Death Differ. 2013;20:676–685. doi: 10.1038/cdd.2013.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Khoronenkova SV, Dianov GL. The emerging role of Mule and ARF in the regulation of base excision repair. FEBS Lett. 2011;585:2831–2835. doi: 10.1016/j.febslet.2011.06.015. [DOI] [PubMed] [Google Scholar]
- 68.Parsons JL, Tait PS, Finch D, Dianova II, Edelmann MJ, Khoronenkova SV, Kessler BM, Sharma RA, McKenna WG, Dianov GL. Ubiquitin ligase ARF-BP1/Mule modulates base excision repair. EMBO J. 2009;28:3207–3215. doi: 10.1038/emboj.2009.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Parsons JL, Tait PS, Finch D, Dianova II, Allinson SL, Dianov GL. CHIP-mediated degradation and DNA damage-dependent stabilization regulate base excision repair proteins. Mol. Cell. 2008;29:477–487. doi: 10.1016/j.molcel.2007.12.027. [DOI] [PubMed] [Google Scholar]
- 70.Sobol RW. CHIPping away at base excision repair. Mol. Cell. 2008;29:413–415. doi: 10.1016/j.molcel.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 71.Meisenberg C, Tait PS, Dianova II, Wright K, Edelmann MJ, Ternette N, Tasaki T, Kessler BM, Parsons JL, Kwon YT, Dianov GL. Ubiquitin ligase UBR3 regulates cellular levels of the essential DNA repair protein APE1 and is required for genome stability. Nucleic Acids Res. 2012;40:701–711. doi: 10.1093/nar/gkr744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Oka S, Nakabeppu Y. DNA glycosylase encoded by MUTYH functions as a molecular switch for programmed cell death under oxidative stress to suppress tumorigenesis. Cancer Sci. 2011;102:677–682. doi: 10.1111/j.1349-7006.2011.01869.x. [DOI] [PubMed] [Google Scholar]
- 73.Wollen Steen K, Doseth B, Akbari PWMM, Kang D, Falkenberg M, Slupphaug G. mtSSB may sequester UNG1 at mitochondrial ssDNA and delay uracil processing until the dsDNA conformation is restored. DNA Repair. 2012;11:82–91. doi: 10.1016/j.dnarep.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 74.van Loon B, Samson LD. Alkyladenine DNA glycosylase (AAG) localizes to mitochondria and interacts with mitochondrial single-stranded binding protein (mtSSB) DNA Repair. 2013;12:177–187. doi: 10.1016/j.dnarep.2012.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tsuchimoto D, Sakai Y, Sakumi K, Nishioka K, Sasaki M, Fujiwara T, Nakabeppu Y. Human APE2 protein is mostly localized in the nuclei andto some extent in the mitochondria, while nuclear APE2 is partly associated with proliferating cell nuclear antigen. Nucleic Acids Res. 2001;29:2349–2360. doi: 10.1093/nar/29.11.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wan G, Liu Y, Han C, Zhang X, Lu X. Noncoding RNAs in DNA Repair and Genome Integrity. Antioxid. Redox Signal. 2014;20:655–677. doi: 10.1089/ars.2013.5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lindahl T, Barnes DE. Repair of endogenous DNA damage. Cold Spring Harbor Symp. Quant. Biol. 2000;65:127–133. doi: 10.1101/sqb.2000.65.127. [DOI] [PubMed] [Google Scholar]
- 78.Caldecott KW. Single-strand break repair and genetic disease. Nat. Rev. Genet. 2008;9:619–631. doi: 10.1038/nrg2380. [DOI] [PubMed] [Google Scholar]
- 79.Svilar D, Goellner EM, Almeida KH, Sobol RW. Base excision repair and lesion-dependent subpathways for repair of oxidative DNA damage. Antioxid. Redox Signal. 2011;14:2491–2507. doi: 10.1089/ars.2010.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Berquist BR, Wilson DM., 3rd Pathways for repairing and tolerating the spectrum of oxidative DNA lesions. Cancer Lett. 2012;327:61–72. doi: 10.1016/j.canlet.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Campalans A, Kortulewski T, Amouroux R, Menoni H, Vermeulen W, Radicella JP. Distinct spatiotemporal patterns and PARP dependence of XRCC1 recruitment to single-strand break and base excision repair. Nucleic Acids Res. 2013;41:3115–3129. doi: 10.1093/nar/gkt025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Noren Hooten N, Kompaniez K, Barnes J, Lohani A, Evans MK. Poly(ADP-ribose) polymerase 1 (PARP-1) binds to 8-oxoguanine-DNA glycosylase (OGG1) J. Biol. Chem. 2011;286:44679–44690. doi: 10.1074/jbc.M111.255869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Noren Hooten N, Fitzpatrick M, Kompaniez K, Jacob KD, Moore BR, Nagle J, Barnes J, Lohani A, Evans MK. Coordination of DNA repair by NEIL1 and PARP-1: a possible link to aging. Aging. 2012;4:674–685. doi: 10.18632/aging.100492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Strom CE, Johansson F, Uhlen M, Szigyarto CA, Erixon K, Helleday T. Poly(ADP-ribose) polymerase (PARP) is not involved in base excision repair but PARP inhibition traps a single-strand intermediate. Nucleic Acids Res. 2011;39:3166–3175. doi: 10.1093/nar/gkq1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ischenko AA, Saparbaev MK. Alternative nucleotide incision repair pathway for oxidative DNA damage. Nature. 2002;415:183–187. doi: 10.1038/415183a. [DOI] [PubMed] [Google Scholar]
- 86.Pascucci B, D’Errico M, Parlanti E, Giovannini S, Dogliotti E. Role of nucleotide excision repair proteins in oxidative DNA damage repair: an updating. Biochemistry. 2011;76:4–15. doi: 10.1134/s0006297911010032. [DOI] [PubMed] [Google Scholar]
- 87.Lin JJ, Sancar A. A new mechanism for repairing oxidative damage to DNA: (A)BC excinuclease removes AP sites and thymine glycols from DNA. Biochemistry. 1989;28:7979–7984. doi: 10.1021/bi00446a002. [DOI] [PubMed] [Google Scholar]
- 88.Kow YW, Wallace SS, Van Houten B. UvrABC nuclease complex repairsthymine glycol, an oxidative DNA base damage. Mutat. Res. 1990;235:147–156. doi: 10.1016/0921-8777(90)90068-g. [DOI] [PubMed] [Google Scholar]
- 89.Brown KL, Roginskaya M, Zou Y, Altamirano A, Basu AK, Stone MP. Bind-ing of the human nucleotide excision repair proteins XPA and XPC/HR23B tothe 5R-thymine glycol lesion and structure of the cis-(5R, 6S) thymine glycol epimer in the 5′-GTgG-3′ sequence: destabilization of two base pairs at the lesion site. Nucleic Acids Res. 2010;38:428–440. doi: 10.1093/nar/gkp844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Guo J, Hanawalt PC, Spivak G. Comet-FISH with strand-specific probes reveals transcription-coupled repair of 8-oxoGuanine in human cells. Nucleic Acids Res. 2013;41:7700–7712. doi: 10.1093/nar/gkt524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Muftuoglu M, de Souza-Pinto NC, Dogan A, Aamann M, Stevnsner T, Rybanska I, Kirkali G, Dizdaroglu M, Bohr VA. Cockayne syndrome group B protein stimulates repair of formamidopyrimidines by NEIL1 DNA glycosylase. J. Biol. Chem. 2009;284:9270–9279. doi: 10.1074/jbc.M807006200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wong HK, Muftuoglu M, Beck G, Imam SZ, Bohr VA, Wilson DM., 3rd Cockayne syndrome B protein stimulates apurinic endonuclease 1 activity and protects against agents that introduce base excision repair intermediates. Nucleic Acids Res. 2007;35:4103–4113. doi: 10.1093/nar/gkm404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Torres-Ramos CA, Johnson RE, Prakash L, Prakash S. Evidence for the involvement of nucleotide excision repair in the removal of abasic sites in yeast. Mol. Cell. Biol. 2000;20:3522–3528. doi: 10.1128/mcb.20.10.3522-3528.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sparks JL, Chon H, Cerritelli SM, Kunkel TA, Johansson E, Crouch RJ, Burgers PM. RNase H2-initiated ribonucleotide excision repair. Mol. Cell. 2012;47:980–986. doi: 10.1016/j.molcel.2012.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Reijns MA, Rabe B, Rigby RE, Mill P, Astell KR, Lettice LA, Boyle S, Leitch A, Keighren M, Kilanowski F, Devenney PS, Sexton D, Grimes G, Holt IJ, Hill RE, Taylor MS, Lawson KA, Dorin JR, Jackson AP. Enzymatic removal of ribonucleotides from DNA is essential for mammalian genome integrity and development. Cell. 2012;149:1008–1022. doi: 10.1016/j.cell.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rabe B. Aicardi-Goutieres syndrome: clues from the RNase H2 knock-out mouse. J. Mol. Med. 2013;91:1235–1240. doi: 10.1007/s00109-013-1061-x. [DOI] [PubMed] [Google Scholar]
- 97.Wilson DM, 3rd, Seidman MM. A novel link to base excision repair? Trends Biochem. Sci. 2010;35:247–252. doi: 10.1016/j.tibs.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mace-Aime G, Couve S, Khassenov B, Rosselli F, Saparbaev MK. The Fanconi anemia pathway promotes DNA glycosylase-dependent excision of inter-strand DNA crosslinks. Environ. Mol. Mutagen. 2010;51:508–519. doi: 10.1002/em.20548. [DOI] [PubMed] [Google Scholar]
- 99.Maor-Shoshani A, Meira LB, Yang X, Samson LD. 3-Methyladenine DNA glycosylase is important for cellular resistance to psoralen interstrand crosslinks. DNA Repair. 2008;7:1399–1406. doi: 10.1016/j.dnarep.2008.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kothandapani A, Dangeti VS, Brown AR, Banze LA, Wang XH, Sobol RW, Patrick SM. Novel role of base excision repair in mediating cisplatin cytotox-icity. J. Biol. Chem. 2011;286:14564–14574. doi: 10.1074/jbc.M111.225375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.McNeill DR, Paramasivam M, Baldwin J, Huang J, Vyjayanti VN, Seidman MM, Wilson 3rd DM. NEIL1 responds and binds to psoralen-induced DNA interstrand crosslinks. J. Biol. Chem. 2013;288:12426–12436. doi: 10.1074/jbc.M113.456087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Murnane JP. Telomere dysfunction and chromosome instability. Mutat. Res. 2012;730:28–36. doi: 10.1016/j.mrfmmm.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Webb CJ, Wu Y, Zakian VA. DNA repair at telomeres: keeping the ends intact. Cold Spring Harbor Perspect. Biol. 2013;5 doi: 10.1101/cshperspect.a012666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.de Lange T. How telomeres solve the end-protection problem. Science. 2009;326:948–952. doi: 10.1126/science.1170633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pinto AR, Li H, Nicholls C, Liu JP. Telomere protein complexes and interactions with telomerase in telomere maintenance. Front. Biosci. 2011;16:187–207. doi: 10.2741/3683. [DOI] [PubMed] [Google Scholar]
- 106.Yang Q, Xiang J, Yang S, Zhou Q, Li Q, Tang Y, Xu G. Verification of specific G-quadruplex structure by using a novel cyanine dye supramolecular assembly: I. recognizing mixed G-quadruplex in humantelomeres. Chem. Commun. 2009:1103–1105. doi: 10.1039/b820101c. [DOI] [PubMed] [Google Scholar]
- 107.Lipps HJ, Rhodes D. G-quadruplex structures: in vivo evidence and function. Trends Cell Biol. 2009;19:414–422. doi: 10.1016/j.tcb.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 108.Wang H, Nora GJ, Ghodke H, Opresko PL. Single molecule studies of physiologically relevant telomeric tails reveal POT1 mechanism for promoting G-quadruplex unfolding. J. Biol. Chem. 2011;286:7479–7489. doi: 10.1074/jbc.M110.205641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Miller AS, Balakrishnan L, Buncher NA, Opresko PL, Bambara RA. Telomere proteins POT1, TRF1 and TRF2 augment long-patch base excision repair in vitro. Cell Cycle. 2012;11:998–1007. doi: 10.4161/cc.11.5.19483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fotiadou P, Henegariu O, Sweasy JB. DNA polymerase beta interacts with TRF2 and induces telomere dysfunction in a murine mammary cell line. Cancer Res. 2004;64:3830–3837. doi: 10.1158/0008-5472.CAN-04-0136. [DOI] [PubMed] [Google Scholar]
- 111.Muftuoglu M, Wong HK, Imam SZ, Wilson 3rd DM, Bohr VA, Opresko PL. Telomere repeat binding factor 2 interacts with base excision repair proteins and stimulates DNA synthesis by DNA polymerase beta. Cancer Res. 2006;66:113–124. doi: 10.1158/0008-5472.CAN-05-2742. [DOI] [PubMed] [Google Scholar]
- 112.Vallabhaneni H, O’Callaghan N, Sidorova J, Liu Y. Defective repair of oxidative base lesions by the DNA glycosylase Nth1 associates with multiple telomere defects. PLoS Genet. 2013;9:e1003639. doi: 10.1371/journal.pgen.1003639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wang Z, Rhee DB, Lu J, Bohr CT, Zhou F, Vallabhaneni H, de Souza-Pinto NC, Liu Y. Characterization of oxidative guanine damage and repair in mammalian telomeres. PLoS Genet. 2010;6:e1000951. doi: 10.1371/journal.pgen.1000951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Madlener S, Strobel T, Vose S, Saydam O, Price BD, Demple B, Say-dam N. Essential role for mammalian apurinic/apyrimidinic (AP) endonuclease Ape1/Ref-1 in telomere maintenance. Proc. Nat. Acad. Sci. U.S.A. 2013;110:17844–17849. doi: 10.1073/pnas.1304784110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Opresko PL, Fan J, Danzy S, Wilson 3rd DM, Bohr VA. Oxidative damage in telomeric DNA disrupts recognition by TRF1 and TRF2. Nucleic Acids Res. 2005;33:1230–1239. doi: 10.1093/nar/gki273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhou J, Liu M, Fleming AM, Burrows CJ, Wallace SS. Neil3 and NEIL1 DNA glycosylases remove oxidative damages from quadruplex DNA and exhibit preferences for lesions in the telomeric sequence context. J. Biol. Chem. 2013;288:27263–27272. doi: 10.1074/jbc.M113.479055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Li J, Braganza A, Sobol RW. Base excision repair facilitates a functional relationship between guanine oxidation and histone demethylation. Antioxid. Redox Signal. 2013;18:2429–2443. doi: 10.1089/ars.2012.5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ziel KA, Grishko V, Campbell CC, Breit JF, Wilson GL, Gillespie MN. Oxidants in signal transduction: impact on DNA integrity and gene expression. FASEB J.: Off. Publ. Fed. Am. Soc. Exp. Biol. 2005;19:387–394. doi: 10.1096/fj.04-2805com. [DOI] [PubMed] [Google Scholar]
- 119.Perillo B, Ombra MN, Bertoni A, Cuozzo C, Sacchetti S, Sasso A, Chiariotti L, Malorni A, Abbondanza C, Avvedimento EV. DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science. 2008;319:202–206. doi: 10.1126/science.1147674. [DOI] [PubMed] [Google Scholar]
- 120.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Nat. Acad. Sci. U.S.A. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jacobs AL, Schar P. DNA glycosylases: in DNA repair and beyond. Chromosoma. 2012;121:1–20. doi: 10.1007/s00412-011-0347-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dalton SR, Bellacosa A. DNA demethylation by TDG. Epigenomics. 2012;4:459–467. doi: 10.2217/epi.12.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sjolund AB, Senejani AG, Sweasy JB. MBD4 and TDG: multifaceted DNA glycosylases with ever expanding biological roles. Mutat. Res. 2013;743–744:12–25. doi: 10.1016/j.mrfmmm.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu. Rev. Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- 125.Jaszczur M, Bertram JG, Pham P, Scharff MD, Goodman MF. AID and Apobec3G haphazard deamination and mutational diversity. Cell. Mol. Life Sci.: CMLS. 2013;70:3089–3108. doi: 10.1007/s00018-012-1212-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Schrader CE, Guikema JE, Wu X, Stavnezer J. The roles of APE1, APE2, DNApolymerase beta and mismatch repair in creating S region DNA breaks duringantibody class switch. Philos. Trans. R. Soc. London, Ser. B: Biol. Sci. 2009;364:645–652. doi: 10.1098/rstb.2008.0200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Guikema JE, Gerstein RM, Linehan EK, Cloherty EK, Evan-Browning E, Tsuchimoto D, Nakabeppu Y, Schrader CE. Apurinic/apyrimidinic endonuclease 2 is necessary for normal B cell development and recovery of lymphoid progenitors after chemotherapeutic challenge. J. Immunol. 2011;186:1943–1950. doi: 10.4049/jimmunol.1002422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Masani S, Han L, Yu K. Apurinic/apyrimidinic endonuclease 1 is the essential nuclease during immunoglobulin class switch recombination. Mol. Cell. Biol. 2013;33:1468–1473. doi: 10.1128/MCB.00026-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Senejani AG, Liu Y, Kidane D, Maher SE, Zeiss CJ, Park HJ, Kashgarian M, McNiff JM, Zelterman D, Bothwell AL, Sweasy JB. Mutation of POLB causes lupus in mice. Cell Rep. 2014;6:1–8. doi: 10.1016/j.celrep.2013.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Saribasak H, Gearhart PJ. Does DNA repair occur during somatic hypermutation? Semin. Immunol. 2012;24:287–292. doi: 10.1016/j.smim.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Marenstein DR, Wilson 3rd DM, Teebor GW. Human AP endonuclease (APE1) demonstrates endonucleolytic activity against AP sites in single-stranded DNA. DNA Repair. 2004;3:527–533. doi: 10.1016/j.dnarep.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 132.Alt FW, Zhang Y, Meng FL, Guo C, Schwer B. Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell. 2013;152:417–429. doi: 10.1016/j.cell.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Holmes RK, Malim MH, Bishop KN. APOBEC-mediated viral restriction: not simply editing? Trends Biochem. Sci. 2007;32:118–128. doi: 10.1016/j.tibs.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 134.Hegde ML, Mantha AK, Hazra TK, Bhakat KK, Mitra S, Szczesny B. Oxidative genome damage and its repair: implications in aging and neurodegenerative diseases. Mech. Ageing Develop. 2012;133:157–168. doi: 10.1016/j.mad.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Mantha AK, Sarkar B, Tell G. A short review on the implications of base excision repair pathway for neurons: relevance to neurodegenerative diseases. Mitochondrion. 2013 doi: 10.1016/j.mito.2013.10.007. [DOI] [PubMed] [Google Scholar]
- 136.Iyama T, Wilson 3rd DM. DNA repair mechanisms in dividing and non-dividing cells. DNA Repair. 2013;12:620–636. doi: 10.1016/j.dnarep.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bucholtz N, Demuth I. DNA-repair in mild cognitive impairment and Alzheimer’s disease. DNA Repair. 2013;12:811–816. doi: 10.1016/j.dnarep.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 138.Canugovi C, Misiak M, Ferrarelli LK, Croteau DL, Bohr VA. The role of DNA repair in brain related disease pathology. DNA Repair. 2013;12:578–587. doi: 10.1016/j.dnarep.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Rulten SL, Caldecott KW. DNA strand break repair and neurodegeneration. DNA Repair. 2013;12:558–567. doi: 10.1016/j.dnarep.2013.04.008. [DOI] [PubMed] [Google Scholar]
- 140.Liu Y, Wilson SH. DNA base excision repair: a mechanism of trinucleotide repeat expansion. Trends Biochem. Sci. 2012;37:162–172. doi: 10.1016/j.tibs.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kong Q, Lin CL. Oxidative damage to RNA: mechanisms, consequences, anddiseases. Cell. Mol. Life Sci.: CMLS. 2010;67:1817–1829. doi: 10.1007/s00018-010-0277-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Berquist BR, McNeill DR, Wilson DM., 3rd Characterization of abasicendonuclease activity of human Ape1 on alternative substrates, as well aseffects of ATP and sequence context on AP site incision. J. Mol. Biol. 2008;379:17–27. doi: 10.1016/j.jmb.2008.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Antoniali G, L Lirussi, Poletto M, Tell G. Emerging roles of the nucleolus in regulating the DNA damage response: the noncanonical DNA repair enzyme APE1/Ref-1 as a paradigmatical example. Antioxid. Redox Signal. 2014;20:621–639. doi: 10.1089/ars.2013.5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Chan KK, Zhang QM, Dianov GL. Base excision repair fidelity in normal and cancer cells. Mutagenesis. 2006;21:173–178. doi: 10.1093/mutage/gel020. [DOI] [PubMed] [Google Scholar]
- 145.Sweasy JB, Lang T, DiMaio D. Is base excision repair a tumor suppressor mechanism? Cell Cycle. 2006;5:250–259. doi: 10.4161/cc.5.3.2414. [DOI] [PubMed] [Google Scholar]
- 146.Paz-Elizur T, Sevilya Z, Leitner-Dagan Y, Elinger D, Roisman LC, Livneh Z. DNA repair of oxidative DNA damage in human carcinogenesis: potential application forcancerriskassessment and prevention. Cancer Lett. 2008;266:60–72. doi: 10.1016/j.canlet.2008.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Nemec AA, Wallace SS, Sweasy JB. Variant base excision repair proteins: contributors to genomic instability. Semin. Cancer Biol. 2010;20:320–328. doi: 10.1016/j.semcancer.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wilson 3rd DM, Kim D, Berquist BR, Sigurdson AJ. Variation in base excision repair capacity. Mutat. Res. 2011;711:100–112. doi: 10.1016/j.mrfmmm.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wallace SS, Murphy DL, Sweasy JB. Base excision repair and cancer. Cancer Lett. 2012;327:73–89. doi: 10.1016/j.canlet.2011.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kidane D, Chae WJ, Czochor J, Eckert KA, Glazer PM, Bothwell AL, Sweasy JB. Interplay between DNA repair and inflammation, and the link to cancer. Crit. Rev. Biochem. Mol. Biol. 2014 doi: 10.3109/10409238.2013.875514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.van Loon B, Markkanen E, Hubscher U. Oxygen as a friend and enemy: how to combat the mutational potential of 8-oxo-guanine. DNA Repair. 2010;9:604–616. doi: 10.1016/j.dnarep.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 152.Galick HA, Kathe S, Liu M, Robey-Bond S, Kidane D, Wallace SS, Sweasy JB. Germ-line variant of human NTH1 DNA glycosylase induces genomic instability and cellular transformation. Proc. Nat. Acad. Sci. U.S.A. 2013;110:14314–14319. doi: 10.1073/pnas.1306752110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Prakash A, Carroll BL, Sweasy JB, Wallace SS, Doublie S. Genome and can-cer single nucleotide polymorphisms of the human NEIL1 DNA glycosylase:activity, structure, and the effect of editing. DNA Repair. 2014;14:17–26. doi: 10.1016/j.dnarep.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Donigan KA, Hile SE, Eckert KA, Sweasy JB. The human gastric cancer-associated DNA polymerase beta variant D160N is a mutator that induces cellular transformation. DNA Repair. 2012;11:381–390. doi: 10.1016/j.dnarep.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Nemec AA, Donigan KA, Murphy DL, Jaeger J, Sweasy JB. Colon cancer-associated DNA polymerase beta variant induces genomic instability and cellular transformation. J. Biol. Chem. 2012;287:23840–23849. doi: 10.1074/jbc.M112.362111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Murphy DL, Donigan KA, Jaeger J, Sweasy JB. The E288K colon tumor variant of DNA polymerase beta is a sequence specific mutator. Biochemistry. 2012;51:5269–5275. doi: 10.1021/bi3003583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Donigan KA, Tuck D, Schulz V, Sweasy JB. DNA polymerase beta variant Ile260Met generates global gene expression changes related to cellular transformation. Mutagenesis. 2012;27:683–691. doi: 10.1093/mutage/ges034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Yamtich J, Nemec AA, Keh A, Sweasy JB. Agermline polymorphism of DNA polymerase beta induces genomic instability and cellular transformation. PLoS Genet. 2012;8:e1003052. doi: 10.1371/journal.pgen.1003052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Eckenroth BE, Towle-Weicksel JB, Sweasy JB, Doublie S. The E295K cancer variant of human polymerase beta favors the mismatch conformational pathway during nucleotide selection. J. Biol. Chem. 2013;288:34850–34860. doi: 10.1074/jbc.M113.510891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Vijg J, Suh Y. Genome instability and aging. Annu. Rev. Physiol. 2013;75:645–668. doi: 10.1146/annurev-physiol-030212-183715. [DOI] [PubMed] [Google Scholar]
- 161.Gredilla R, Bohr VA, Stevnsner T. Mitochondrial DNA repair and association with aging—an update. Exp. Gerontol. 2010;45:478–488. doi: 10.1016/j.exger.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Alexeyev M, Shokolenko I, Wilson G, LeDoux S. The maintenance of mitochondrial DNA integrity—critical analysis and update. Cold Spring Harbor Perspect. Biol. 2013;5:a012641. doi: 10.1101/cshperspect.a012641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Lindahl T. An N-glycosidase from Escherichia coli that releases free uracil from DNA containing deaminated cytosine residues. Proc. Nat. Acad. Sci. U.S.A. 1974;71:3649–3653. doi: 10.1073/pnas.71.9.3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Strniste GF, Wallace SS. Endonucleolytic incision of x-irradiated deoxyribonucleic acid by extracts of Escherichia coli. Proc. Nat. Acad. Sci. U.S.A. 1975;72:1997–2001. doi: 10.1073/pnas.72.6.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]