Abstract
Since the success of homologous recombination in altering mouse genome and the discovery of Cre-loxP system, the combination of these two breakthroughs has created important applications for studying the immune system in the mouse. Here, we briefly summarize the general principles of this technology and its applications in studying immune cell development and responses; such implications include conditional gene knockout and inducible and/or tissue-specific gene over-expression, as well as lineage fate mapping. We then discuss the pros and cons of a few commonly used Cre-expressing mouse lines for studying lymphocyte development and functions. We also raise several general issues, such as efficiency of gene deletion, leaky activity of Cre, and Cre toxicity, all of which may have profound impacts on data interpretation. Finally, we selectively list some useful links to the Web sites as valuable mouse resources.
Keywords: Cre, loxP, conditional knockout, transgenic, bacterial artificial chromosome (BAC), hematopoietic cells, fate mapping, inducible gene expression, lymphocyte cell development and differentiation
GENERAL INTRODUCTION TO THE Cre/LoxP SYSTEM
By altering the genome at specific gene loci through homologous recombination in mouse embryonic stem (ES) cells (reviewed in Capecchi, 1989a,b), Mario Capecchi and Oliver Smithies pioneered the generation of genetically modified mice in the late 1980s and early 1990s, which resulted in their sharing of the Nobel Prize in 2007 with Martin Evans, who was the first to culture mouse ES cells (Evans and Kaufman, 1981). Since then, hundreds of labs have used such technology to generate a variety of mice, each carrying a modified allele of a particular gene, often referred to as germ-line knockout mice; such knockout mice have provided valuable tools in examining the functions of a particular gene both in vitro and in vivo. However, in some cases, knocking out of a gene in the mouse germ line leads to embryonic lethality or a severe developmental defect in early progenitors, preventing the researcher from studying gene function in mature cells. Therefore, a better gene manipulation technology is required.
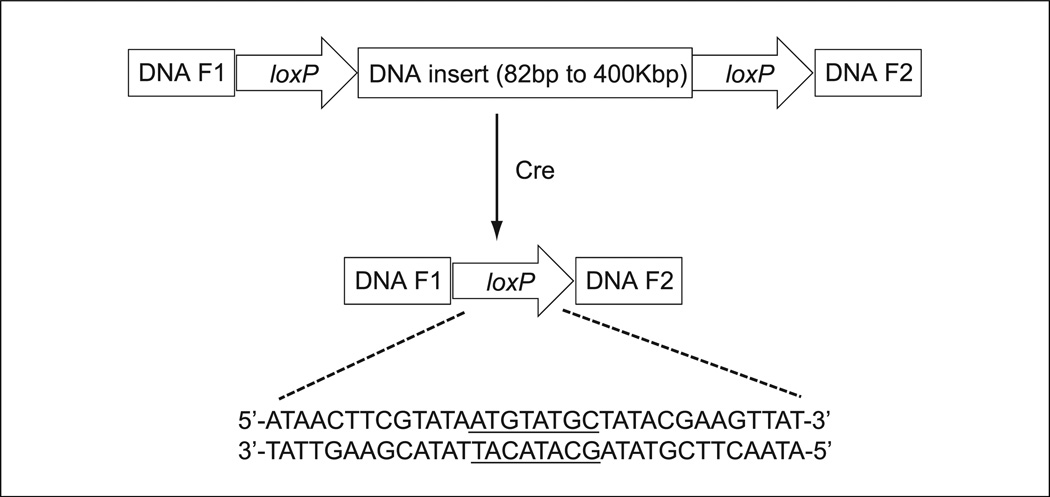
Cre (Causes recombination), a 38-kDa integrase encoded by bacteriophage P1, mediates site-specific recombination between 34-bp sequences referred to as loxP (locus of crossover (x) in P1 bacteriophage) sites (reviewed in Sauer, 1998; Nagy, 2000). A loxP site is composed of a nonpalindromic 8-bp sequence (GCATACAT or ATGTATGC) flanked on either side by 13-bp inverted repeats (ATAACTTCGTATA; Hamilton and Abremski, 1984; Hoess et al., 1982). Cre-based recombination between the two loxP sites leads to a reciprocal exchange of DNA strands (Fig. 10.34.1). Cre-mediated recombination requires a minimum of 82 bp between two loxP sites for efficient recombination, though there is no upper limit (Hoess and Abremski, 1985). Indeed, high deletion efficiency by Cre is still observed between two loxP sites that are 400 kb apart (Nagy, 2000). The two loxP sites can be in the same orientation on the same chromosome or a different chromosome, or in the opposite orientation, thus leading to deletion, inversion, duplication, or translocation of chromosomes (van der Weyden et al., 2002; Branda and Dymecki, 2004). Sauer and Henderson (1998) created a Cre-expressing mouse cell line and showed that Cre-mediated site-specific recombination occurred in vivo, indicating that the prokaryotic Cre-loxP system can function in mammalian cells.
Figure 10.34.1.
Principle of Cre-mediated deletion of DNA that is flanked by two loxP sites. DNA insert with its size from 82 bp up to 400 kbp between two loxP sites in a same direction is deleted by Cre-mediated DNA recombination. The full DNA sequence of a loxP site is shown.
Rajewsky’s group was the first to use the Cre-loxP technology in generating mouse models, including conditional inactivation of a target gene only in a selected cell population (Gu et al., 1993, 1994; Rajewsky et al., 1996). In a germ-line knockout strain, the target gene is inactivated in all cells throughout all developmental stages, whereas in conditional knockout, gene inactivation is either cell type specific or under temporal control. The specificity and timing of gene deletion are determined by the nature of Cre and its expression pattern. Up to now, there are more than 500 independent Cre mouse lines available (Nagy et al., 2009).
Not only Cre expression can be controlled by cell-type-specific regulatory elements or in an inducible way by tetracycline (or doxycycline; Gossen et al., 1995; Baron and Bujard, 2000; Bockamp et al., 2002) or by poly (I:C) through the production of endogenous interferon (note that this system may not be ideal for studying the immune system because of the involvement of type I interferon (Kuhn et al., 1995). Cre can also be engineered so that its activity is modulated by drugs (Metzger et al., 1995; Feil et al., 1996; Brocard et al., 1997). For example, the Cre protein can be fused to a mutant ligand-binding domain of the estrogen receptor that selectively binds to 17-β-estradiol analogs, e.g., tamoxifen, but not the endogenous estrogen (Feil et al., 1996; Brocard et al., 1997). The CreER fusion protein is normally present in the cytoplasm, but is translocated into the nucleus to induce gene excision upon the addition of the ligand. CreERT2, a newer version of CreER, which is 10-fold more sensitive to 4-hydroxytamoxifen (OHT) than CreER (Indra et al., 1999). Therefore, the use of CreERT2 is preferred.
Gene manipulation may be achieved in a spatiotemporally regulated manner by tissue-specific expression of inducible CreERT2. Sometimes, Cre or CreERT2 can be introduced into the cells by retroviral or adenoviral transduction methods (Rohlmann et al., 1996; Wang et al., 1996; Stec et al., 1999; Zhu et al., 2004). A cell-membrane-permeable form of Cre protein, Tat-Cre, has also been used to delete conditional alleles in vitro (Wadia et al., 2004). The conditional knockout of a gene in a particular cell, tissue, or organ at a specific time point not only avoids the early lethality or severe developmental consequences, if any, but also helps study cell-intrinsic gene functions in particular cell types at certain developmental stages without the interference of a systemic effect, which could be potentially caused by a global deletion or over-expression.
Other similar systems such as FLP-FRT are being used in genome engineering in ES cells and transgenic mice (Dymecki et al., 1996a,b). FLP is a 423-amino acid monomeric peptide encoded within the 2-µm yeast plasmid of Saccharomyces cerevisiae that uses phosphotyrosine for energy, whereas FRT is composed of an 8-bp asymmetric spacer (TCTAGAAA or TTTCTAGA) surrounded by 13-bp repeats (GAAGTTCCTATTC). The asymmetric region dictates whether excision or inversion occurs after recombination. Both Cre and FLP recombinases belong to the tyrosine site–specific recombinase class; thus, they act similarly. However, FLP-mediated gene deletion is less inefficient. Therefore, an enhanced form of FLP, FLPe, has been developed, which makes the FLPe-FRT system an alternative to the Cre-loxP system (Rodriguez et al., 2000). Before the FLPe-FRT system is used, three loxP sites are usually employed for making a conditional knockout mouse strain (Gu et al., 1994). A pair of loxP sites flanks the DNA sequence encoding a selection marker, whereas another pair flanks the DNA fragment of interest (the two pairs share one loxP site). The selection of Cre-mediated partial excision, by EIIa-Cre (Lakso et al., 1996) for example, to remove only the DNA fragment encoding the selection marker, is somewhat difficult, resulting in a lengthy process for generating a conditional allele. Nowadays, the Cre-loxP and FLPe-FRT systems are often combined for preparing targeting constructs in such a way that FLPe-FRT system is responsible for removing the selection marker, whereas the Cre-loxP system takes care of the DNA fragment under study. The International Knockout Mouse Consortium (IKMC) often utilizes such a combined strategy for generating new mouse lines.
Besides the Cre-loxP and FLPe-FRT systems, Dre-rox, a related recombinase-DNA pair, has been reported to work in mice (Anastassiadis et al., 2009). Cre mutants and chimeric Cre/FLP may offer another alternative strategy (Hartung and Kisters-Woike, 1998; Shaikh and Sadowski, 2000). In addition, besides inducing recombination between two loxP sites, Cre can induce specific recombination between two lox511 sites; the lox511 site is an alternative recognition site for Cre that has a different spacer sequence compared to the loxP site (Soukharev et al., 1999). Since recombination between loxP and lox511 is very inefficient, the combination of loxP and lox511 pairs has been used for site-specific gene insertion. There are other integrases, such as phiC31 and phiBT1, that belong to the serine site-specific recombinase class and induce directional rather than reversible recombination. These have been reported to function in eukaryotic genome engineering, such as in yeast, Drosophila, and mammalian cells (Thyagarajan et al., 2001; Groth et al., 2004; Keravala and Calos, 2008; Xu et al., 2008). These alternatives may create more applications for sophisticated gene manipulation in the mouse in the future.
APPLICATIONS OF Cre-LoxP
Removal of the selection marker and generation of germ-line knockout
When a specific allele is targeted by an exogenous DNA fragment via homologous recombination, a selectable marker is required for efficiently obtaining the targeted allele. However, such a selectable marker sometimes affects the expression of the neighbor genes (Fiering et al., 1995), which may have related functions to the targeted gene. Therefore, in such cases, it is essential to remove the selectable marker by Cre-loxP or FLPe-FRT technology before proper investigation on the germ line knockout mouse line can be carried out. Furthermore, for generating a conditional knockout allele, the selection marker must be removed so that the allele flanked by two loxP sites (floxed) may function as a wild-type allele before its deletion. For this purpose, one may use a Cre or FLPe mouse line, in which Cre expression is under the control of CMV or β-actin promoters that are active in the germ line (Schwenk et al., 1995; Rodriguez et al., 2000). Such germ-line Cre mouse lines can also be used to generate the germ-line knockout allele from a conditional allele.
Conditional gene deletion
The Cre-loxP system has been widely used for conditional gene deletion in the mouse. The first step is to prepare a mouse model in which either the whole gene of interest or a critical gene segment is flanked by two loxP sites. The conditional mouse lines containing two loxP sites are usually designed to be wild-type mice. Because of such requirements, the selection of the sites to be inserted by loxP sequences is important. Usually, large introns with fewer regulatory elements are preferred; non-coding sequences that are conserved among species should be left intact. Occasionally, insertion of loxP sites into the gene locus may result in a hypomorphic allele.
Other conditional gene manipulations
A clever design of two loxP sites flanking a “STOP” cassette, which contains transcriptional and/or translational stop signals, has opened the door to generate many tools for studying the immune system. The general principle is that a loxP-STOP-loxP cassette is inserted in the front of a gene of interest so that this coded protein is only expressed when Cre deletes the loxP-STOP-loxP cassette. Depending on the nature of the gene following the STOP cassette, such technology has been utilized for different applications (Fig. 10.34.2). These applications include the following.
Figure 10.34.2.
Applications for the combination of Cre (or inducible Cre) with loxP-STOP-loxP cassette in front of a gene X. The expression of Cre or inducible Cre such as Cre-ERT2 is usually under the control of a tissue-specific promoter, whereas gene X is under another promoter (tissue-specific or not), but the expression of the gene X is blocked by the loxP-STOP-loxP cassette until the cassette is removed by Cre-mediated recombination. The system can be used for conditional gene induction if X encodes a general protein, for fate-mapping if X encodes a marker, or for selected cell ablation if X encodes a toxin.
Conditional transgenic gene expression
One can insert a gene to be overexpressed downstream of the loxP-STOP-loxP cassette and then prepare a mouse line. Such a mouse line will not express the transgene until Cre removes the STOP cassette in a tissue-specific or inducible manner. However, once Cre activates the transgene, it is irreversible.
Fate mapping
Because of the irreversible nature of the gene-segment deletion by Cre, the loxP-STOP-loxP has been used for cell-fate mapping (Jacob and Baltimore, 1999). The commonly used line for immunology research is the loxP-STOP-loxP conditional reporter line, including PLAP (Jacob and Baltimore, 1999), lacZ, GFP, and YFP, etc., integrated at the Rosa26 locus (Mao et al., 1999; Soriano, 1999; Novak et al., 2000; Srinivas et al., 2001). If a combination of different colors is needed (i.e., with an additional GFP as another marker), other mouse strains such as the ROSA26-STOP-tdTomato line (Madisen et al., 2010), expressing a red fluorescent protein upon Cremediated STOP removal, can be used for fate-mapping studies to minimize the compensation problems.
Cell-type ablation
When the gene following the loxP-STOP-loxP cassette is a toxic protein, such as diphtheria toxin fragment A (DTA), the resulting mouse line can be used for specific cell–type ablation (Ivanova et al., 2005). Similarly, when diphtheria toxin receptor (DTR) is Cre-inducible and Cre expression is under the control of lineage-specific promoters, Cre-expressing cells can be efficiently eliminated by diphtheria toxin (DT) treatment (Buch et al., 2005). Alternatively, cell ablation can be achieved by inducing a cell surface marker, such as Thy1.1, which can be recognized by a depleting antibody (clone 19E12).
Applications combining the Cre-loxP system and TetR-inducible system
As discussed above, the Cre-loxP system is used for a conditional knockout or conditional transgenic so that gene expression is manipulated in a tissue-specific and/or temporally controlled manner. When Cre is fused to ERT2, its activity can be further regulated by tamoxifen. However, Cre-mediated gene deletion or activation is irreversible. Because of this feature, the Cre-loxP system is very useful for cell-fate mapping. However, it is not feasible to use this system to turn a particular gene on and off on demand.
The Escherichia coli tetracycline repressor (TetR)-based system (Gossen and Bujard, 1992; Gossen et al., 1995; Baron and Bujard, 2000), on the other hand, regulates gene expression in a reversible way (Felsher and Bishop, 1999). TetR proteins bind to antibiotic tetracycline or its derivatives such as doxycycline (Dox); Dox is much more cost effective, available, and efficient in regulating TetR proteins than tetracycline. TetR proteins also bind to the 19-bp operator sequences (tetO), which are built into the promoter of a transgene so that its expression is regulated by Dox and TetR proteins. There are “tet-off” and “tet-on” systems mediated by tetracycline-controlled transactivator (tTA) and reverse tTA (rtTA), respectively. When the tTA protein binds to Dox, it can no longer bind to DNA to activate gene expression (“tet-off”); on the other hand, rtTA only binds to the DNA and activates gene expression in the presence of Dox (“tet-on”).
Due to unique features of the Cre-loxP and the TetR-based systems, they can be combined for generating useful research tools (Belteki et al., 2005). When the loxP-STOP-loxP cassette is placed upstream of the DNA sequences encoding a TetR protein, the expression of tTA or rtTA will be regulated by a tissue-specific Cre. If the Cre is in an inducible form, such as CreERT2, the expression of TetR protein can then be limited to the specific tissue cells that are already developed during the period of tamoxifen treatment. Therefore, it may be useful for studying gene functions in fetal cells versus adult cells. If a TetR protein, whose expression is controlled by Cre, is designed to induce a transgene, and at the same time, the endogenous counterpart gene is flanked by two loxP sites, the combination of four modified alleles (tissue-specific Cre or inducible Cre; floxed endogenous gene; loxP-STOP-loxP restricted TetR; and TetR-inducible transgene) offers a perfect system to perform a temporal gene rescue experiment after gene deletion in a specific tissue. Since the expression of the transgene is titratable by Dox, this system may be best suited for studying dose effect of a particular gene in vivo.
Conversely, Cre expression can be under the control of the TetR system, and TetR expression may be designed to be tissue specific. This combination is identical to a tissue-specific CreERT2, but due to the titratable nature of the TetR system, the activity and toxicity of the Cre is better controlled.
TISSUE-SPECIFIC CRE TRANSGENIC MICE
Most of the existing Cre mouse lines can be found at the CREATE (Coordination of resources for conditional expression of mutated mouse alleles) consortium (http://creline.org/), which includes the Cre mouse database at Mouse Genome Informatics (MGI, http://loxP.creportal.org/). Some commonly used Cre mice for studying the immune system will be briefly discussed below and summarized in Table 10.34.1.
Table 10.34.1.
Commonly Used Cre Mouse Lines for Studying Lymphoid Cells
| Name | Tg/KI | Expression in cell types |
Note | Reference |
|---|---|---|---|---|
| ROSA26-CreERT2 | KI | Most cells except those in the brain | High deletion efficiency with tamoxifen treatment both in vitro and in vivo | Seibler et al. (2003) |
| Vav-Cre | Tg | All hematopoietic lineages, testis and ovaries | High deletion efficiency; may cause germ line deletion in some offspring | de Boer et al. (2003) |
| CD2-Cre | Tg | Common lymphoid progenitors (CLPs) | High deletion efficiency; some modified CD2-Cre lines may only delete genes in T cells but not B cells | Zhumabekok et al. (1995); de Boer et al. (2003) |
| Lck-Cre | Tg | Early DN stage in the thymus | Deletion efficiency varies | Lee et al. (2001) |
| CD4-Cre | Tg | Late DN to DP stage, deleting floxed genes in both CD4 and CD8 T cells | High deletion efficiency | Lee et al. (2001) |
| CD4-CreERT2 | Tg | Deleting floxed genes only CD4 but not CD8 T cells in the periphery | Inducible by tamoxifen; deletion efficiency up to 80% in vivo | Aghajani et al. (2012) |
| dLck-Cre (line 3779) | Tg | Late DP to SP stage | —70% deletion efficiency in CD4 T cells; higher efficiency (80% to 90%) in CD8 T cells; very low in Tregs | Wang et al. (2001) |
| OX40-Cre | KI | Tregs and activated CD4+ T cells | Endogenous OX40 gene is disrupted by Cre; very low efficiency in activated CD8 T cells | Yagi et al. (2010) |
| CD8a–Cre | Tg | Mature CD8+ but not CD4+ T cells | Also known as E8I–Cre; Cre expression driven by the core E8I enhancer and Cd8a promoter | Maekawa et al. (2008) |
| Granzyme-B-Cre | Tg | Activated CD4+ and CD8+ T cells | Cre driven by truncated granzyme B promoter | Jacob and Baltimore (1999) |
| Mb1-Cre | KI | Starting from Pre-Pro-B stage | Endogenous Mb1 gene encoding Igα signaling subunit of the BCR is disrupted by Cre; deletion efficiency is better than CD19-Cre | Hobeika et al. (2006) |
| CD19-Cre | KI | Starting Pro-B stage | Endogenous Cd19 gene is disrupted by Cre; deletion efficiency is 75% to 95% | Rickert et al. (1997) |
| CD19-CreERT2 | BAC Tg | Similar to CD19-Cre, but its activity requires tamoxifen treatment | Inducible by tamoxifen; deletion efficiency 25% to 60% | Boross et al. (2009) |
| Foxp3-YFPCre | KI | Only in Foxp3+ Tregs | YFP is dim; endogenous Foxp3 expression intact | Rubtsov et al. (2008) |
| Foxp3-GFPCreERT2 | KI | Only in Foxp3+ Tregs | Inducible but with low deletion efficiency (10% to 20%); endogenous Foxp3 expression intact | Rubtsov et al. (2010) |
| Id2-CreERT2 | KI | Id2-expressing cells: epithelial cells in the lung distal tips as well as progenitor of ILCs and T cells | Inducible but with low deletion efficiency; endogenous Id2 gene is disrupted by CreERT2 | Rawlins et al. (2009) |
Lymphocyte development and differentiation is an extensively studied area in the immunology field. T cells undergo multiple stages of development in the thymus, and these cells then further differentiate into effector cells in response to antigens in the periphery. Thus, T cell development and differentiation provide a perfect model for investigating fundamental immunological questions.
To apply conditional gene modification technology in T cell research, multiple Cre lines have been prepared to serve different purposes. The Lck-cre mouse line (Lee et al., 2001) is a commonly used tool for studying gene functions during early T cell development. The disadvantage of this Cre model is that the deletion efficiency is low and varies from mouse to mouse.
CD4-cre line (Lee et al., 2001) is frequently used for general purposes due to its high deletion efficiency. However, since CD4-Cre is turned on from late CD4/CD8 double negative (DN) stage and fully expressed at CD4/CD8 double positive (DP) stages, it is not an ideal model for studying early T cell development. Even for T cell development from DP to CD4 or CD8 single positive (SP) mature T cells, caution should be taken since the half-life of a particular mRNA and its protein product may be long enough to obscure the results. On the other hand, deletion of a particular gene by CD4-Cre may result in developmental defects or a complete development block. In such cases, other Cre lines are needed for studying gene functions in mature T cells.
Cre expression driven by the distal promoter of Lck (dLck-cre) becomes an important model to avoid thymic development problems. The dLck-Cre (line 3779) starts to be expressed at the late DP stage and reaches high activity at the SP stage (Wang et al., 2001). Therefore, one may avoid T cell developmental blockage and obtain na¨ıve mature CD4 T cells. For example, Gata3fl/fl-CD4-Cre mice display severe blockage from DP to SP CD4 T cells (Zhu et al., 2004); residual CD4 T cells developed in these mice show a highly activated phenotype. However, Gata3fl/fl-dLck-Cre mice show normal T cell development; up to 70% to 80% of na¨ıve CD4 T cells harvested from these mice have gene deletion (Yagi et al., 2010); deletion efficiency of dLck-Cre in mature CD8 T cells is somewhat higher, but dLck-Cre does not seem to be active in regulatory T cells. For studying gene functions specifically in regulatory T cells, the Foxp3-YFPCre mouse line is the best choice (Rubtsov et al., 2008).
An alternative choice is to use OX40-cre for studying peripheral T cells (Zhu et al., 2004; Klinger et al., 2009). However, OX40 is up-regulated by TCR-mediated signaling, and its maximum expression is on day 2 to day 3 after T cell activation. Thus, OX40-cre has limitations because it is not useful for studying early events during T cell differentiation. In addition, OX40-Cre is active in regulatory T cells, even in the thymus. Although OX40 is induced by TCR in CD8 T cells, the deletion efficiency in these cells is relatively lower compared to that in activated CD4 T cells. For studying gene function in activated CD8 T cells, granzyme-B-Cre (Jacob and Baltimore, 1999) can be used, although this Cre also deletes floxed genes in activated CD4 T cells.
CD4-Cre deletes floxed sequences in both CD4 and CD8 T cells. To delete floxed genes only in peripheral CD8 but not CD4 T cells, CD8a–Cre (Maekawa et al., 2008), also known as E8I–Cre, may be used. There are several Cre lines, such as Mb1-Cre (Hobeika et al., 2006; expressed at pre-pro B stage) and CD19-Cre (Rickert et al., 1997; expressed at pro-B stage), designed for studying gene functions in B cell development as well as in effector B cells. CD2-Cre (Zhumabekov et al., 1995) mediates DNA excision at the common lymphoid progenitor (CLP) stage, and thus may be used for studying gene function in both T and B cells (de Boer et al., 2003). Other Cre lines have been used for studying gene regulation in innate lymphoid cells (ILCs; Rawlins et al., 2009), macrophages (Clausen et al., 1999), and dendritic cells (DCs; Caton et al., 2007). All the hematopoietic cells are developed from hematopoietic stem cells (HSCs). To study gene functions in hematopoietic lineages, one may consider Vav-Cre (de Boer et al., 2003).
As mentioned above, when Cre is fused to a mutant estrogen receptor (ERT2), its activity can be modulated by addition of the estrogen receptor ligand tamoxifen. A mouse line carrying CreERT2 under the control of the Rosa26 locus has been prepared (Seibler et al., 2003). Upon tamoxifen treatment, CreERT2 will delete the floxed gene with high efficiency in most tissues except in the brain. Since the deletion is temporally controlled, this method is useful for studying gene function after cells have developed, and for following the cell fate of certain cells that are present at the time of tamoxifen treatment. However, since CreERT2 expression is rather ubiquitous, there is a certain limitation to this model. Recently, many groups have generated tissue-specific CreERT2 mouse strains, such as CD4-CreERT2 and CD19-CreERT2, which can be used for inducible deletion of floxed genes specifically in T cells and B cells, respectively (Boross et al., 2009; Aghajani et al., 2012). However, the deletion efficiency is somewhat compromised. Rudensky’s lab has developed a Foxp3-GFP-CreERT2 mouse line for the purpose of inducible inactivation of floxed gene in Tregs (Rubtsov et al., 2010). It is a Cre, ERT2, and GFP triple fusion protein. Although it has the advantage of being able to show Cre-expressing cells by GFP, this fusion may further reduce the deletion efficiency. Using the 2A peptide technology (Ryan et al., 1991; Donnelly et al., 2001; Szymczak et al., 2004; Szymczak and Vignali, 2005), our lab has recently developed a Tbx21-ZsGreen-T2A–CreERT2 mouse line in which an improved version of GFP (ZsGreen) and CreERT2 is connected by a 2A peptide; thus, ZsGreen marks T-bet-expressing cells and CreERT2 can efficiently delete floxed genes in these cells upon tamoxifen treatment (unpublished data). There are many more tissue-specific CreERT2 mouse lines. A database dealing with CreERT2 resource created by the Institut Clinique de la Souris (ICS; Illkirch-Strasbourg, France) can be found at http://www.ics-mci.fr/mousecre/.
THINGS TO BE CONSIDERED
Deletion efficiency
An important issue to be considered for conditional knockout using the Cre-loxP system is the deletion efficiency. The efficiency is determined by several factors, including Cre activity, the nature of floxed genes, and the function of the floxed genes in cell proliferation and/or survival. The expression levels of a tissue-specific Cre are determined by the activity of similar regulatory elements that also control an endogenous lineage-specific gene. If the Cre expression level is not high enough to mediate recombination, it is possible to modify these regulatory elements during vector construction (such as to remove a silencer); however, such modifications may also result in the loss of cell specificity in Cre expression. A humanized Cre (hCre or iCre), which exhibits higher activity than regular Cre in mammalian cells, has been utilized to improve deletion efficiency (Shimshek et al., 2002). In the case of BAC transgenic–driven Cre expression, there is an opportunity to select for high copy numbers within the transgenic founders. However, the higher the Cre expression, the higher the cell toxicity that may be noticed, while the deletion efficiency is improved.
Gene deletion efficiency is also determined by the nature of the floxed gene to be deleted. Even using a same Cre line, the deletion efficiency may vary from gene to gene. Although it has not been carefully assessed, several possibilities may explain such variation: e.g., epigenetic modifications and DNA looping at the floxed genes may affect Cre-mediated recombination; or if loxP sites are inserted near a transcription factor binding element, the accessibility of Cre protein to the loxP sequences may be altered. Therefore, it is difficult to predict the exact deletion efficiency for a particular Cre-loxP combination.
If the floxed gene is crucial for the development of a specific cell type and this cell type has capacity to undergo tremendous cell expansion before its maturation, it will appear that the deletion efficiency is extremely low due to a counter selection. Similarly, if the gene to be deleted plays a critical role in cell proliferation and/or survival in mature cells, cells that escape deletion may preferentially expand. For example, GATA3 is important for the expansion of Th2 cells. Retroviral expression of Cre resulted in deletion of the floxed Gata3. However, when such cells were maintained in culture for a substantial length of time, i.e., 2 to 3 weeks, cells that had not deleted the Gata3 dominated the culture.
Therefore, deletion efficiency is a major concern in using conditional knockout mice. It always a good idea to check deletion efficiency at different stages of development either by genotyping or by single-cell analysis, such as flow cytometry, to make sure that the phenotype observed is consistent with the genotype. In some cases, the alteration in phenotype may be underestimated because the cells that have not undergone gene deletion are also included in the analysis.
Leaky deletion in the germ line
Conditional deletion of a floxed gene segment is dependent on tissue specificity of the Cre expression. Sometimes, Cre expression is leaky, thus resulting in widespread recombination. Therefore, it is not uncommon that a “lineage-specific” Cre line may occasionally delete floxed genes in germ line, and such a knockout allele may then heritably remain in the offspring. This occurs possibly because many cell type–specific genes are transiently expressed during early embryogenesis. Therefore, one should exclude the possibility of germ-line deletion when studying conditional knockout mice. Several lines have been reported to delete genes in the germ line despite the fact that they are designed to be cell-type specific. For example, OX40-Cre, if carried by male mice, may delete floxed gene in the germ line of some offspring. Thus, it is recommended that OX40-Cre should be carried by the female parent. In other cases, the Cre gene may need to be carried by the male parent. It has been noticed that Vav-Cre often induces gene deletion in the germ line possibly due to its expression in the testis and ovaries (de Boer et al., 2003). Therefore, when a new Cre line is established or used, it is better to test whether the female or male mouse is better suited to carry the Cre for breeding purposes. Furthermore, each parent should be screened for germ-line deletion when they are set up for further breeding.
Copy number matters
In some cases, DNA containing loxP sites is introduced into the mouse by transgenic technology, which will generate founders that carry an insertion of the DNA fragments in multiple copies. However, to assess the effect of Cre-mediated gene excision between two loxP sites, selection of a single copy is required. Otherwise, the observed effect may be resulted from the deletion of excessive copies rather than the deletion of flanked sequence. In addition, some BAC vectors contain a loxP site. If the vector sequence is not completely removed before microinjection, the resulting BAC transgenic lines may carry several loxP sequences, depending on the copy numbers. Upon Cre activation, the transgene, which is expressed with multiple copies, may no longer be expressed when it becomes a single copy since the remaining copy could represent a partial integration.
Cre toxicity
Cre has cell toxicity when it is expressed at high levels (Loonstra et al., 2001). Pseudo-loxP sites are present in the mouse genome, and Cre-mediated recombination between these pseudo-loxP sites may occur with low efficiency (Thyagarajan et al., 2000). However, when a codon-optimized Cre with a nuclear localization signal is highly expressed, cell toxicity becomes a major concern. Therefore, it is crucial to include proper controls, i.e., Cre+ mice without floxed genes, when the Cre-loxP system is utilized to study cell survival and proliferation. To minimize the Cre toxicity, it is better to control the Cre expression or activity in an inducible manner. Alternatively, a self-inactivating Cre, i.e., Cre gene flanked by two loxP sites, may be considered.
USEFUL RESOURCES AND DATABASES RELATED TO MOUSE LINES
The International Knockout Mouse Consortium (IKMC, http://www.knockoutmouse.org/) provides a large collection of mice that carry mutant protein-coding genes. The mouse models have been and/or are being prepared through gene trapping and/or gene targeting in ES cells derived from the C57BL/6 mouse strain. A substantial number of the genes in the mouse genome have been targeted, many of which have conditional knockout potential. The IKMC includes the following four programs: Knockout Mouse Project (KOMP) in the U.S. (http://www.nih.gov/science/models/mouse/knockout/); European Conditional Mouse Mutagenesis Program (EUCOMM) in Europe (http://www.knockoutmouse.org/about/eucomm); North American Conditional Mouse Mutagenesis Project (NorCOMM) in Canada (http://www.norcomm.org/index.htm); and Texas A&M Institute for Genomic Medicine (TIGM) in the US (http://www.tigm.org/).
International Mouse Strain Resource (IMSR, http://www.findmice.org/index) and Mouse Genome Informatics (MGI, http://www.informatics.jax.org/) are two valuable resources for finding mutant mice that were either published or deposited into public repositories.
The CREATE (Coordination of resources for conditional expression of mutated mouse alleles) consortium (http://creline.org/) provides a resource for Cre mouse strains. It includes four international databases: CRE mouse database at MGI (http://www.creportal.org/); CRE-X-Mice (http://nagy.mshri.on.ca/cre/); Crezoo at Fleming (http://bioit.fleming.gr/crezoo/); and MouseCre (http://www.ics-mci.fr/mousecre/).
CONCLUSIONS
The conditional gene modification technology, especially the inducible Cre-loxP system, has created tremendous opportunities and tools for immunologists to study the principles of the immune systems in the mouse. Now, we have many choices to inactivate or overexpress a specific genes in a tissue-specific and/or temporally controlled manner. Furthermore, we often apply this technology to study lineage relationship and cell-fate mapping. With the advance of new technologies in gene targeting, such as TALEN technology (reviewed in Carlson et al., 2012), it becomes much easier and faster to generate genetically modified alleles. Research in the immunology field will benefit greatly from the development of new tools and technologies.
With the expansion of our knowledge, our questions become more and more sophisticated and precise. For example, we have now realized that some particular cell types can only be defined by a combination of two markers. Thus, it is challenging to manipulate gene expression in these cells in a lineage-specific manner. A plausible solution is to construct the locus of one marker to control FLP expression and the locus of another marker to control FRT-STOP-FRT-Cre expression. Therefore, only in cells simultaneously expressing both markers will the conditionally modified gene locus flanked by two loxP sites or the transgene following a loxP-STOP-loxP cassette undergo Cre-mediated recombination. With utilization of other, similar recombination systems in combination with doxycycline-inducible systems, gene manipulation specifically in cell subsets co-expressing three or more markers is also possible. Finally, many biological processes are regulated by a combination of several factors in a quantitative way. Thus, combining gene knockout with inducible but titratable gene rescue will be an important future application of conditional gene manipulations in the mouse.
ACKNOWLEDGMENTS
S.S. and J.Z. are supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health, U.S.A, no. Z0I-A1001169.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
LITERATURE CITED
- Aghajani K, Keerthivasan S, Yu Y, Gounari F. Generation of CD4CreER(T(2)) transgenic mice to study development of peripheral CD4-T-cells. Genesis. 2012;50:908–913. doi: 10.1002/dvg.22052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastassiadis K, Fu J, Patsch C, Hu S, Weidlich S, Duerschke K, Buchholz F, Edenhofer F, Stewart AF. Dre recombinase, like Cre, is a highly efficient site-specific recombinase in E. coli, mammalian cells and mice. Dis. Model Mech. 2009;2:508–515. doi: 10.1242/dmm.003087. [DOI] [PubMed] [Google Scholar]
- Baron U, Bujard H. Tet repressor-based system for regulated gene expression in eukaryotic cells: Principles and advances. Methods Enzymol. 2000;327:401–421. doi: 10.1016/s0076-6879(00)27292-3. [DOI] [PubMed] [Google Scholar]
- Belteki G, Haigh J, Kabacs N, Haigh K, Sison K, Costantini F, Whitsett J, Quaggin SE, Nagy A. Conditional and inducible transgene expression in mice through the combinatorial use of Cre-mediated recombination and tetracycline induction. Nucleic Acids Res. 2005;33:e51. doi: 10.1093/nar/gni051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bockamp E, Maringer M, Spangenberg C, Fees S, Fraser S, Eshkind L, Oesch F, Zabel B. Of mice and models: Improved animal models for biomedical research. Physiol. Genomics. 2002;11:115–132. doi: 10.1152/physiolgenomics.00067.2002. [DOI] [PubMed] [Google Scholar]
- Boross P, Breukel C, van Loo PF, van der Kaa J, Claassens JW, Bujard H, Schönig K, Verbeek JS. Highly B lymphocyte-specific tamoxifen inducible transgene expression of CreER T2 by using the LC-1 locus BAC vector. Genesis. 2009;47:729–735. doi: 10.1002/dvg.20549. [DOI] [PubMed] [Google Scholar]
- Branda CS, Dymecki SM. Talking about a revolution: The impact of site-specific recombinases on genetic analyses in mice. Dev. Cell. 2004;6:7–28. doi: 10.1016/s1534-5807(03)00399-x. [DOI] [PubMed] [Google Scholar]
- Brocard J, Warot X, Wendling O, Messaddeq N, Vonesch JL, Chambon P, Metzger D. Spatio-temporally controlled site-specific somatic mutagenesis in the mouse. Proc. Natl. Acad. Sci. U.S.A. 1997;94:14559–14563. doi: 10.1073/pnas.94.26.14559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buch T, Heppner FL, Tertilt C, Heinen TJ, Kremer M, Wunderlich FT, Jung S, Waisman A. A Cre-inducible diphtheria toxin receptor mediates cell lineage ablation after toxin administration. Nat. Methods. 2005;2:419–426. doi: 10.1038/nmeth762. [DOI] [PubMed] [Google Scholar]
- Capecchi MR. Altering the genome by homologous recombination. Science. 1989a;244:1288–1292. doi: 10.1126/science.2660260. [DOI] [PubMed] [Google Scholar]
- Capecchi MR. The new mouse genetics: Altering the genome by gene targeting. Trends Genet. 1989b;5:70–76. doi: 10.1016/0168-9525(89)90029-2. [DOI] [PubMed] [Google Scholar]
- Carlson DF, Fahrenkrug SC, Hackett PB. Targeting DNA with fingers and TALENs. Mol. Ther. Nucleic Acids. 2012;1:e3. doi: 10.1038/mtna.2011.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caton ML, Smith-Raska MR, Reizis B. Notch-RBP-J signaling controls the homeostasis of CD8-dendritic cells in the spleen. J. Exp. Med. 2007;204:1653–1664. doi: 10.1084/jem.20062648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen BE, Burkhardt C, Reith W, Renkawitz R, Forster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999;8:265–277. doi: 10.1023/a:1008942828960. [DOI] [PubMed] [Google Scholar]
- de Boer J, Williams A, Skavdis G, Harker N, Coles M, Tolaini M, Norton T, Williams K, Roderick K, Potocnik AJ, Kioussis D. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur. J. Immunol. 2003;33:314–325. doi: 10.1002/immu.200310005. [DOI] [PubMed] [Google Scholar]
- Donnelly ML, Luke G, Mehrotra A, Li X, Hughes LE, Gani D, Ryan MD. Analysis of the aphthovirus 2A/2B polyprotein ‘cleavage’ mechanism indicates not a proteolytic reaction, but a novel translational effect: A putative ribosomal ‘skip’. J. Gen. Virol. 2001;82:1013–1025. doi: 10.1099/0022-1317-82-5-1013. [DOI] [PubMed] [Google Scholar]
- Dymecki SM. A modular set of Flp, FRT and lacZ fusion vectors for manipulating genes by site-specific recombination. Gene. 1996a;171:197–201. doi: 10.1016/0378-1119(96)00035-2. [DOI] [PubMed] [Google Scholar]
- Dymecki SM. Flp recombinase promotes site-specific DNA recombination in embryonic stem cells and transgenic mice. Proc. Natl. Acad. Sci. U.S.A. 1996b;93:6191–6196. doi: 10.1073/pnas.93.12.6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- Feil R, Brocard J, Mascrez B, LeMeur M, Metzger D, Chambon P. Ligand-activated site-specific recombination in mice. Proc. Natl. Acad. Sci. U.S.A. 1996;93:10887–10890. doi: 10.1073/pnas.93.20.10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsher DW, Bishop JM. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol. Cell. 1999;4:199–207. doi: 10.1016/s1097-2765(00)80367-6. [DOI] [PubMed] [Google Scholar]
- Fiering S, Epner E, Robinson K, Zhuang Y, Telling A, Hu M, Martin DI, Enver T, Ley TJ, Groudine M. Targeted deletion of 5'HS2 of the murine beta-globin LCR reveals that it is not essential for proper regulation of the beta-globin locus. Genes Dev. 1995;9:2203–2213. doi: 10.1101/gad.9.18.2203. [DOI] [PubMed] [Google Scholar]
- Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. U.S.A. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen M, Freundlieb S, Bender G, Müller G, Hillen W, Bujard H. Transcriptional activation by tetracyclines in mammalian cells. Science. 1995;268:1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H, Zou YR, Rajewsky K. Independent control of immunoglobulin switch recombination at individual switch regions evidenced through Cre-loxP-mediated gene targeting. Cell. 1993;73:1155–1164. doi: 10.1016/0092-8674(93)90644-6. [DOI] [PubMed] [Google Scholar]
- Gu H, Marth JD, Orban PC, Mossmann H, Rajewsky K. Deletion of a DNA polymerase beta gene segment in T cells using cell type-specific gene targeting. Science. 1994;265:103–106. doi: 10.1126/science.8016642. [DOI] [PubMed] [Google Scholar]
- Hamilton DL, Abremski K. Site-specific recombination by the bacteriophage P1 lox-Cre system: Cre-mediated synapsis of two lox sites. J. Mol. Biol. 1984;178:481–486. doi: 10.1016/0022-2836(84)90154-2. [DOI] [PubMed] [Google Scholar]
- Hartung M, Kisters-Woike B. Cre mutants with altered DNA binding properties. J. Biol. Chem. 1998;273:22884–22891. doi: 10.1074/jbc.273.36.22884. [DOI] [PubMed] [Google Scholar]
- Hobeika E, Thiemann S, Storch B, Jumaa H, Nielsen PJ, Pelanda R, Reth M. Testing gene function early in the B cell lineage in mb1-cre mice. Proc. Natl. Acad. Sci. U.S.A. 2006;103:13789–13794. doi: 10.1073/pnas.0605944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoess RH, Abremski K. Mechanism of strand cleavage and exchange in the Cre-lox site-specific recombination system. J. Mol. Biol. 1985;181:351–362. doi: 10.1016/0022-2836(85)90224-4. [DOI] [PubMed] [Google Scholar]
- Hoess RH, Ziese M, Sternberg N. P1 site-specific recombination: Nucleotide sequence of the recombining sites. Proc. Natl. Acad. Sci. U.S.A. 1982;79:3398–3402. doi: 10.1073/pnas.79.11.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indra AK, Warot X, Brocard J, Bornert JM, Xiao JH, Chambon P, Metzger D. Temporally-controlled site-specific mutagenesis in the basal layer of the epidermis: Comparison of the recombinase activity of the tamoxifen-inducible Cre-ER(T) and Cre-ER(T2) recombinases. Nucleic Acids Res. 1999;27:4324–4327. doi: 10.1093/nar/27.22.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova A, Signore M, Caro N, Greene ND, Copp AJ, Martinez-Barbera JP. In vivo genetic ablation by Cre-mediated expression of diphtheria toxin fragment A. Genesis. 2005;43:129–135. doi: 10.1002/gene.20162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob J, Baltimore D. Modelling T-cell memory by genetic marking of memory T cells in vivo. Nature. 1999;399:593–597. doi: 10.1038/21208. [DOI] [PubMed] [Google Scholar]
- Keravala A, Calos MP. Site-specific chromosomal integration mediated by phiC31 integrase. Methods Mol. Biol. 2008;435:165–173. doi: 10.1007/978-1-59745-232-8_12. [DOI] [PubMed] [Google Scholar]
- Klinger M, Kim JK, Chmura SA, Barczak A, Erle DJ, Killeen N. Thymic OX40 expression discriminates cells undergoing strong responses to selection ligands. J. Immunol. 2009;182:4581–4589. doi: 10.4049/jimmunol.0900010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- Lakso M, Pichel JG, Gorman JR, Sauer B, Okamoto Y, Lee E, Alt FW, Westphal H. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc. Natl. Acad. Sci. U.S.A. 1996;93:5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PP, Fitzpatrick DR, Beard C, Jessup HK, Lehar S, Makar KW, Pérez-Melgosa M, Sweetser MT, Schlissel MS, Nguyen S, Cherry SR, Tsai JH, Tucker SM, Weaver WM, Kelso A, Jaenisch R, Wilson CB. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 2001;15:763–774. doi: 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- Loonstra A, Vooijs M, Beverloo HB, Allak BA, van Drunen E, Kanaar R, Berns A, Jonkers J. Growth inhibition and DNA damage induced by Cre recombinase in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 2001;98:9209–9214. doi: 10.1073/pnas.161269798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa Y, Minato Y, Ishifune C, Kurihara T, Kitamura A, Kojima H, Yagita H, Sakata-Yanagimoto M, Saito T, Taniuchi I, Chiba S, Sone S, Yasutomo K. Notch2 integrates signaling by the transcription factors RBP-J and CREB1 to promote T cell cytotoxicity. Nat. Immunol. 2008;9:1140–1147. doi: 10.1038/ni.1649. [DOI] [PubMed] [Google Scholar]
- Mao X, Fujiwara Y, Orkin SH. Improved reporter strain for monitoring Cre recombinase-mediated DNA excisions in mice. Proc. Natl. Acad. Sci. U.S.A. 1999;96:5037–5042. doi: 10.1073/pnas.96.9.5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger D, Clifford J, Chiba H, Chambon P. Conditional site-specific recombination in mammalian cells using a ligand-dependent chimeric Cre recombinase. Proc. Natl. Acad. Sci. U.S.A. 92:6991–6995. doi: 10.1073/pnas.92.15.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A. Cre recombinase: The universal reagent for genome tailoring. Genesis. 2000;26:99–109. [PubMed] [Google Scholar]
- Nagy A, Mar L, Watts G. Creation and use of a cre recombinase transgenic database. Methods Mol. Biol. 2009;530:365–378. doi: 10.1007/978-1-59745-471-1_19. [DOI] [PubMed] [Google Scholar]
- Novak A, Guo C, Yang W, Nagy A, Lobe CG. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis. 2000;28:147–155. [PubMed] [Google Scholar]
- Rajewsky K, Gu H, Kühn R, Betz UA, Müller W, Roes J, Schwenk F. Conditional gene targeting. J. Clin. Invest. 1996;98:600–603. doi: 10.1172/JCI118828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlins EL, Clark CP, Xue Y, Hogan BL. The Id2+ distal tip lung epithelium contains individual multipotent embryonic progenitor cells. Development. 2009;136:3741–3745. doi: 10.1242/dev.037317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickert RC, Roes J, Rajewsky KB. Lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res. 1997;25:1317–1318. doi: 10.1093/nar/25.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez CI, Buchholz F, Galloway J, Sequerra R, Kasper J, Ayala R, Stewart AF, Dymecki SM. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat. Genet. 2000;25:139–140. doi: 10.1038/75973. [DOI] [PubMed] [Google Scholar]
- Rohlmann A, Gotthardt M, Willnow TE, Hammer RE, Herz J. Sustained somatic gene inactivation by viral transfer of Cre recombinase. Nat. Biotechnol. 1996;14:1562–1565. doi: 10.1038/nbt1196-1562. [DOI] [PubMed] [Google Scholar]
- Rubtsov YP, Rasmussen JP, Chi EY, Fontenot J, Castelli L, Ye X, Treuting P, Siewe L, Roers A, Henderson WR, Jr, Muller W, Rudensky AY. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity. 2008;28:546–558. doi: 10.1016/j.immuni.2008.02.017. [DOI] [PubMed] [Google Scholar]
- Rubtsov YP, Niec RE, Josefowicz S, Li L, Darce J, Mathis D, Benoist C, Rudensky AY. Stability of the regulatory T cell lineage in vivo. Science. 2010;329:1667–1671. doi: 10.1126/science.1191996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan MD, King AM, Thomas GP. Cleavage of foot-and-mouth disease virus polyprotein is mediated by residues located within a 19 amino acid sequence. J. Gen. Virol. 1991;72:2727–2732. doi: 10.1099/0022-1317-72-11-2727. [DOI] [PubMed] [Google Scholar]
- Sauer B. Inducible gene targeting in mice using the Cre/lox system. Methods. 1998;14:381–392. doi: 10.1006/meth.1998.0593. [DOI] [PubMed] [Google Scholar]
- Sauer B, Henderson N. Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc. Natl. Acad. Sci. U.S.A. 1998;85:5166–5170. doi: 10.1073/pnas.85.14.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenk F, Baron U, Rajewsky K. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 1995;23:5080–5081. doi: 10.1093/nar/23.24.5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibler J, Zevnik B, Küter-Luks B, Andreas S, Kern H, Hennek T, Rode A, Heimann C, Faust N, Kauselmann G, Schoor M, Jaenisch R, Rajewsky K, Kühn R, Schwenk F. Rapid generation of inducible mouse mutants. Nucleic Acids Res. 2003;31:e12. doi: 10.1093/nar/gng012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaikh AC, Sadowski PD. Chimeras of the Flp and Cre recombinases: Tests of the mode of cleavage by Flp and Cre. J. Mol. Biol. 2000;302:27–48. doi: 10.1006/jmbi.2000.3967. [DOI] [PubMed] [Google Scholar]
- Shimshek DR, Kim J, Hübner MR, Spergel DJ, Buchholz F, Casanova E, Stewart AF, Seeburg PH, Sprengel R. Codonimproved Cre recombinase (iCre) expression in the mouse. Genesis. 2002;32:19–26. doi: 10.1002/gene.10023. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Soukharev S, Miller JL, Sauer B. Segmental genomic replacement in embryonic stem cells by double lox targeting. Nucleic Acids Res. 1999;27:e21. doi: 10.1093/nar/27.18.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stec DE, Davisson RL, Haskell RE, Davidson BL, Sigmund CD. Efficient liver-specific deletion of a floxed human angiotensinogen transgene by adenoviral delivery of Cre recombinase in vivo. J. Biol. Chem. 1999;274:21285–21290. doi: 10.1074/jbc.274.30.21285. [DOI] [PubMed] [Google Scholar]
- Szymczak AL, Vignali DA. Development of 2A peptide-based strategies in the design of multicistronic vectors. Exp. Opin. Biol. Ther. 2005;5:627–638. doi: 10.1517/14712598.5.5.627. [DOI] [PubMed] [Google Scholar]
- Szymczak AL, Workman CJ, Wang Y, Vignali KM, Dilioglou S, Vanin EF, Vignali DA. Correction of multi-gene deficiency in vivo using a single ‘self-cleaving’ 2A peptide-based retroviral vector. Nat. Biotechnol. 2004;22:589–594. doi: 10.1038/nbt957. [DOI] [PubMed] [Google Scholar]
- Thyagarajan B, Guimaraes MJ, Groth AC, Calos MP. Mammalian genomes contain active recombinase recognition sites. Gene. 2000;244:47–54. doi: 10.1016/s0378-1119(00)00008-1. [DOI] [PubMed] [Google Scholar]
- Thyagarajan B, Olivares EC, Hollis RP, Ginsburg DS, Calos MP. Site-specific genomic integration in mammalian cells mediated by phage phiC31 integrase. Mol. Cell Biol. 2001;21:3926–3934. doi: 10.1128/MCB.21.12.3926-3934.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Weyden L, Adams DJ, Bradley A. Tools for targeted manipulation of the mouse genome. Physiol. Genomics. 2002;11:133–164. doi: 10.1152/physiolgenomics.00074.2002. [DOI] [PubMed] [Google Scholar]
- Wadia JS, Stan RV, Dowdy SF. Transducible TAT-HA fusogenic peptide enhances escape of TAT-fusion proteins after lipid raft macropinocytosis. Nat. Med. 2004;10:310–315. doi: 10.1038/nm996. [DOI] [PubMed] [Google Scholar]
- Wang Q, Strong J, Killeen N. Homeostatic competition among T cells revealed by conditional inactivation of the mouse Cd4 gene. J. Exp. Med. 2001;194:1721–1730. doi: 10.1084/jem.194.12.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Krushel LA, Edelman GM. Targeted DNA recombination in vivo using an adenovirus carrying the cre recombinase gene. Proc. Natl. Acad. Sci. U.S.A. 1996;93:3932–3936. doi: 10.1073/pnas.93.9.3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Lee NC, Dafhnis-Calas F, Malla S, Smith MC, Brown MR. Site-specific recombination in Schizosaccharomyces pombe and systematic assembly of a 400 kb transgene array in mammalian cells using the integrase of Streptomyces phage phiBT1. Nucleic Acids Res. 2008;36:e9. doi: 10.1093/nar/gkm1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi R, Junttlia IS, Wei G, Urbahn JF, Jr, Zhao K, Paul WE, Zhu J. The transcription factor GATA3 actively represses RUNX3 protein-regulated production of interferon-gamma. Immunity. 2010;32:507–517. doi: 10.1016/j.immuni.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Min B, Hu-Li J, Watson CJ, Grinberg A, Wang Q, Killeen N, Urban JF, Jr, Guo L, Paul WE. Conditional deletion of Gata3 shows its essential function in T(H)1-T(H)2 responses. Nat. Immunol. 2004;5:1157–1165. doi: 10.1038/ni1128. [DOI] [PubMed] [Google Scholar]
- Zhumabekov T, Corbella P, Tolaini M, Kioussis D. Improved version of a human CD2 minigene based vector for T cell-specific expression in transgenic mice. J. Immunol. Methods. 1995;185:133–140. doi: 10.1016/0022-1759(95)00124-s. [DOI] [PubMed] [Google Scholar]