Abstract
The high death rate of pancreatic cancer is attributed to the lack of reliable methods for early detection and underlying molecular mechanisms of its aggressive pathogenesis. Although MUC13, a newly identified transmembrane mucin, is known to be aberrantly expressed in ovarian and gastro-intestinal cancers, its role in pancreatic cancer is unknown. Herein, we investigated the expression profile and functions of MUC13 in pancreatic cancer progression. The expression profile of MUC13 in pancreatic cancer was investigated using a recently generated monoclonal antibody (clone PPZ0020) and pancreatic tissue microarrays. The expression of MUC13 was significantly (P < 0.005) higher in cancer samples compared with normal/nonneoplastic pancreatic tissues. For functional analyses, full-length MUC13 was expressed in MUC13 null pancreatic cancer cell lines, MiaPaca and Panc1. MUC13 overexpression caused a significant (P < 0.05) increase in cell motility, invasion, proliferation, and anchorage-dependent or -independent clonogenicity while decreasing cell–cell and cell-substratum adhesion. Exogenous MUC13 expression significantly (P < 0.05) enhanced pancreatic tumor growth and reduced animal survival in a xenograft mouse model. These tumorigenic characteristics correlated with the upregulation/phosphorylation of HER2, p21-activated kinase 1 (PAK1), extracellular signal-regulated kinase (ERK), Akt, and metastasin (S100A4), and the suppression of p53. Conversely, suppression of MUC13 in HPAFII pancreatic cancer cells by short hairpin RNA resulted in suppression of tumorigenic characteristics, repression of HER2, PAK1, ERK, and S100A4, and upregulation of p53. MUC13 suppression also significantly (P < 0.05) reduced tumor growth and increased animal survival. These results imply a role of MUC13 in pancreatic cancer and suggest its potential use as a diagnostic and therapeutic target.
Introduction
In 2011, pancreatic cancer is estimated to be detected in more than 44,000 people and to account for more than 37,000 deaths in the United States (1). With an overall 5-year survival rate of only 5%, pancreatic cancer is the fourth most lethal cancer, accounting for 6% of all cancer-related deaths in both men and women (1). Together, the aggressive nature of pancreatic cancer combined with vague symptoms and lack of screening mechanisms create a difficult disease to treat. The serum tumor marker CA19-9 may be helpful in diagnosing pancreatic cancer, but it lacks sensitivity and specificity to effectively screen asymptomatic patients (2). Therefore, the identification of sensitive and specific markers is needed for early detection and subsequent treatment of pancreatic cancer.
Mucins (MUC) have been identified as potential tumor markers and attractive therapeutic targets (3, 4). Mucins form a physical barrier which provides protection for epithelial cells under normal physiologic conditions. However, mucins may be involved in cancer development when expression, localization, or glycosylation patterns change. Such changes can lead to increased cell growth, transformation, and decreased immune surveillance (3, 4). Mucin 13 (MUC13) is a recently identified trans-membrane mucin which is normally expressed in the large intestine, trachea, kidney, small intestine, and gastric epithelium (5, 6). In recent studies, MUC13 has been shown to be aberrantly expressed in ovarian and gastrointestinal cancers (7–9). MUC13 has a large 151-amino acid tandem repeat domain, 3 epidermal growth factor (EGF)-like domains, and a sea urchin sperm protein enterokinase arginine (SEA) domain within the extracellular component, followed by a short 23-amino acid trans-membrane domain and a 69-amino acid cytoplasmic domain (5).
In this study, we show that MUC13 is overexpressed in pancreatic cancer and the exogenous expression of MUC13 augments tumorigenic features in pancreatic cancer cells, such as enhanced cell proliferation, cell motility, cell invasion, and in vivo tumor growth. Conversely, the suppression of MUC13 expression by short hairpin RNA (shRNA) in HPAFII cells shows the opposite effect. The expression of MUC13 correlates with the expression/activation of HER2, PAK1, ERK, Akt, and S100A4 and the decreased expression of p53. These results show, for the first time, the direct association of MUC13 with pancreatic cancer and its influence on pancreatic tumorigenesis.
Materials and Methods
Tissue specimens and immunohistochemistry
The tissue microarray slides (procured from AccuMax, ISU Abxis Co., Ltd and shown in Supplementary Data) and xenograft mouse tumor slides were stained using heat-induced antigen retrieval immunohistochemistry techniques with the Vector ABC kit (Vector Laboratories) with anti-MUC13 MAb (PPZ0020) and analyzed as previously described (9).
Cell culture, transfection procedure, and reagents
Human pancreatic cancer cells procured from American Type Cell Culture Collection were maintained at 37°C in recommended growth medium (MiaPaca:DMEM, HPAFII:DMEM/Ham’s F12) supplemented with 10% FBS and antibiotics (Hyclone Laboratories). To maintain authenticity of the cell lines, frozen stocks were prepared from initial stocks and every 6 months a new frozen stock was used for the experiments. The human MUC13 cloned in pcDNA3.1 or the empty vector was transfected into serum-starved MiaPaca and Panc1 pancreatic cancer cells using Lipofectamine (Invitrogen). Stable transfected cells were selected in medium containing 500 μg/mL G418 (Invitrogen). HPAFII cells were transduced with 5 different constructs of MUC13 specific shRNA lentiviral particles (Sigma) according to the manufacturer’s protocol. Stable cells were then selected and maintained in media containing 3 μg/mL puromycin (Sigma). As wild-type and vector control cells did not show any significant differences, results are shown for vector control cells in the majority of the assays to avoid redundancy.
RNA isolation, reverse transcription PCR, and real-time PCR
Total RNA, reverse transcription, and PCR analysis was done as previously described (9). Briefly, RNA was collected via RNeasy Mini Kit (Qiagen) and reverse transcription was done with the Superscript II RNase H-Reverse Transcriptase System (Invitrogen). The resulting cDNA samples were amplified using MUC13-specific and glyceraldehyde-3-phosphosphate dehydrogenase (GAPDH)-specific primers (9). Quantitative real-time PCR was done with the MUC13-specific TaqMan Gene Expression Assay (Applied Biosystems; ref. 9).
Immunofluorescence and confocal microscopy
Cells were grown at low density on chamber slides (Nalge Nunc Int.) for 24 hours and processed for immunofluorescence as described (9). Cells were incubated with anti-MUC13 MAb (clone PPZ020) or HER2 polyclonal rabbit antibody (Dako) followed with species specific Alexa Fluor 488 or 568 secondary antibodies (Invitrogen) and mounted in FluoroCare Anti-Fade mounting medium (BioCare Medical). Laser confocal microscopy was done with an Olympus Fluoview FV1000 confocal microscope (Olympus Corporation).
Immunoblot analyses
Whole-cell lysates were prepared as described earlier (9–12). Proteins were analyzed by immunoblotting with anti-HER2, anti-phospho-HER2 (tyr1248), anti-p21-activated kinase 1 (PAK1), anti-phospho-PAK1, anti-p44/42 MAPK (ERK1/2), anti-phospho-p44/42 MAPK (ERK1/2), anti-AKT, anti-phospho-AKT (Thr308), anti-p53 antibodies (Cell Signaling Technology), and anti-β–actin (Sigma). Protein bands were detected using a chemiluminescence kit (Roche).
Aggregation assay
For aggregation assays, cells (5,000) were pipetted in a volume of 25 μL of growth media onto the inner surface of the lid of a Petri dish as described earlier (9, 13). After overnight incubation at 37°C, the drops of cell suspension were pipetted to disrupt the loose aggregates and photographed with a phase-contrast microscope.
Cell migration/cell invasion assays
Cellular motility was determined by an agarose bead-based cell motility assay as described earlier (9). Briefly, cells were mixed into a low melting point agarose solution and drops of suspension were placed onto plates. At 24 and 48 hours the plates were photographed using a phase-contrast microscope. Cell motility was also analyzed with a Boyden’s chamber assay as described earlier (9, 12). For cell invasion assays, BD Biocoat Matrigel Invasion Chambers (BD Biosciences) were used as per manufacturer’s suggestions. After 48 hours incubation, the invading cells were stained and counted in 10 fields of view.
Cell proliferation assay
Cells (2 × 104) were seeded in 6-well plates in 2 mL of growth medium. At 24, 48, 72, and 96 hours, the cells were counted in triplicate using an automated cell counter (Z1 Coulter Particle Counter, Beckman Coulter). Doubling time (Td) of the cells was calculated from the growth rate during the exponential growth phase (0–96 hours) using the formula, Td = 0.693t/ln(Nt/N0), wherein t is time in days, Nt is cell number at time t, and N0 is cell number at initial time as described earlier (9, 12).
Anchorage-dependent and -independent colony forming assays
For the anchorage-dependent colony forming assay, cells (1 × 103) were plated in 100 mm dishes, incubated for 10 days, washed with 1XPBS, fixed with 100% methanol and stained with hematoxylin. The colonies (>50 cells) were counted manually and plotted as described earlier (14). For the anchorage-independent colony forming assay, cells (2 × 104) were seeded in a 6-well plate in 1 mL of 0.3% complete media-agar over 2 mL of 0.6% complete media-agar. The cultures were maintained in a 37°C, 5% CO2 incubator for 15 days. The colonies were counted in 6 photomicrographs from each well.
Adhesion assays
100 μL of media containing 1.0 × 105 cells were seeded (in triplicate) into wells of a 96-well plate coated with collagen, fibronectin, laminin, and basement membrane complex proteins (Calbiochem) and incubated for 1 hour at 37°C as described earlier (15). After incubation, the wells were gently washed twice with 1XPBS. The adherent cells were then incubated with 100 μL of calcein-AM for 1 hour at 37°C. The fluorescence of the samples was measured using a fluorescence plate reader at excitation wavelength of 485 nm and emission wavelength of 515 nm.
In vivo tumor xenograft study
Six-week-old athymic Nu/nu nude mice were purchased from Charles River Laboratories International, Inc., and maintained in a pathogen-free environment. Each cell line was suspended in PBS and Matrigel (BD Bioscience) at a 1:1 ratio. Cell suspension (5 × 106 cells in 200 μL) was injected subcutaneously into the right flank of each mouse. Tumor volume (V) was estimated from the length (l), width (w), and height (h) of the tumor using the formula V = 0.52(l × w × h) as described previously (9). Mice were monitored until tumors reached 700 mm3 total volume, at which time mice were euthanized. Tumors were dissected and prepared for immunohistochemical analysis. All animal experiments were done using protocols approved by the Sanford Research/USD and The University of South Dakota Institutional Animal Care and Use Committee.
Statistical analyses
Student t test for independent analysis was applied to evaluate differences in in vitro tumorigenic properties. Linear regression analysis was done to estimate tumor volume as a function of time. Kaplan–Meier analysis was used to estimate animal survival, and differences in survival were analyzed by log-rank analysis. Significant differences were determined using a paired t test. Values of P < 0.05 were considered significant. SAS software version 9.1.3 (SAS Institute Inc.) was used for all analyses.
Results
MUC13 is overexpressed in human pancreatic cancers
Mucins have been reported to be overexpressed in pancreatic cancer (3, 16). However, the expression profile of MUC13 in pancreatic cancer has not been investigated. Therefore, we analyzed the expression pattern of MUC13 in normal/benign pancreas and pancreatic cancer tissue samples by immunohistochemistry using a MUC13 specific MAb (PPZ0020). MUC13 expression in normal, non-neoplastic pancreas samples was either undetectable or showed very faint staining. However, pancreatic cancer samples showed significantly (P < 0.05) higher MUC13 expression (Supplementary Fig. S1). MUC13 was predominately localized on the apical membrane in the majority of cases with cytoplasmic localization in some cases (Supplementary Fig. S1). To determine the correlation of MUC13 expression with cancer type, cancer samples were grouped into histologic types (well differentiated, moderately differentiated, and poorly differentiated). MUC13 expression was significantly higher (P < 0.05) in well and moderately differentiated types of pancreatic cancer compared with normal pancreatic tissue. A negative control antibody did not show any immunoreactivity.
MUC13 expression in pancreatic cancer cell lines
To select suitable cell lines for functional assays, we screened a panel of 5 pancreatic ductal adenocarcinoma cell lines (SW1990, Capan-1, HPAFII, MiaPaca and Panc-1) for MUC13 expression by reverse-transcription PCR (RT-PCR) and confocal microscopy. Of these cell lines, HPAFII and Capan-1 showed high MUC13 expression, while MiaPaca, Panc-1, and SW1990 did not show detectable MUC13 expression (Supplementary Fig. S2A and B). On the basis of this data, MiaPaca, Panc1, and HPAFII cell lines were selected for overexpression and knockdown studies, respectively, to determine functional roles of MUC13 in pancreatic cancer.
Exogenous MUC13 expression enhances tumorigenesis in pancreatic cancer cells
To investigate MUC13 functions in pancreatic tumorigenesis, full-length MUC13 (MUC13F-pcDNA3.1) was overexpressed in MUC13-null cell lines, MiaPaca and Panc1, and several pools of clones were screened for MUC13 expression. Two pools, MP2 and MP4, were selected for further analysis due to their different expression levels of MUC13 (MP2: low; MP4: high) as determined by RT-PCR (Fig. 1A, left). These cells also showed homogenous expression of MUC13 in more than 90% of the cells as shown by confocal microscopy (Fig. 1A, right). Immunoblotting was not done for MUC13 expression analysis due to the lack of anti-MUC13 antibodies appropriate for immunoblotting. High (MP4) and low (MP2) MUC13 expressing cells were selected to determine the effect of variable MUC13 expression in pancreatic cancer cells. The MiaPaca pcDNA3.1 (MPPC) vector-only transfected cells exhibited cell characteristics indistinguishable from the parental, MUC13 negative cell line (Fig. 1A, a and b). In a similar fashion, MUC13 expressing and MUC13 null stable cell lines were generated from Panc1 cells (Supplementary Fig. S4A).
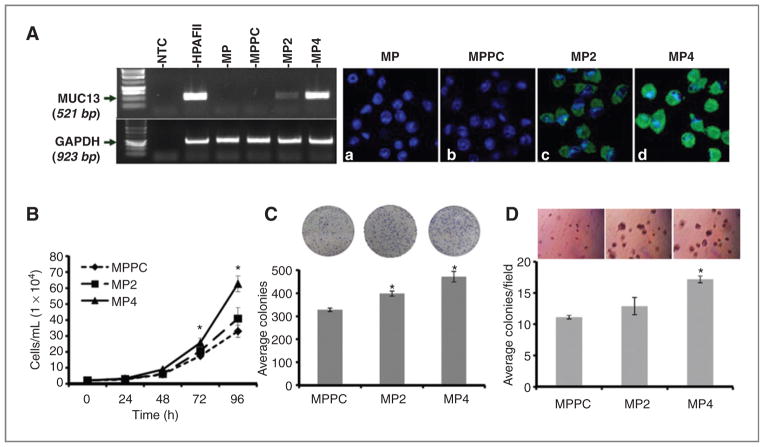
Figure 1.
MUC13-alters cell pro-liferative characteristics in MiaPaca cells. A, RT-PCR and confocal microscopy analysis. Left, RT-PCR analysis of MUC13 expression in HPAFII and MUC13 transfected MiaPaca cells (MP2 and MP4). GAPDH was used as an internal control. Right. confocal microscopy images of MiaPaca-derived cells after immunofluorescence staining with anti-MUC13 MAb (green). MUC13 expression is evident only in MUC13-transfected cells (c, d) but not in wild-type (MP) or vector control cells (MPPC; a and b). 4′,6-diamidino-2-phenylindole (blue) was used as a nuclear stain. Original magnification, ×400. B, cell proliferation assay. The number of MiaPaca-derived cells was determined after 24, 48, 72, and 96 hours of culture. C, anchorage-dependent colony forming assay. The MiaPaca-derived colonies were counted 10 days after plating. D, anchorage-independent soft agar colony forming assays. The MiaPaca-derived colonies were counted 15 days after plating in media-agar. For B, C, and D, mean ± SE; n = 3; *, P < 0.05.
Multiple in vitro assays were done to determine the effects of MUC13 expression on pancreatic cancer cell growth. High MUC13-expressing cells (MP4) showed significantly (P < 0.05) enhanced cell proliferation compared with vector control cells (Fig. 1B). Both MP2 and MP4 cells displayed lower cell doubling times of 17.6 and 16.5 hours, respectively, compared with 19.3 hours of MPPC control cells (Supplementary Fig. S3A). In anchorage-dependent and independent colony formation assays, high MUC13-expressing cells (MP4) formed significantly (P < 0.05) higher number of colonies than MUC13-null cells (Fig. 1C and D). Similarly, MUC13 transfected Panc1 stable cells (Panc-1-M13D) have also shown a significant increase in cell proliferation and colony formation compared with respective vector control cells (Supplementary Fig. S4A–C).
MUC13 knockdown suppresses growth of pancreatic cancer cells
To further confirm the role of MUC13 in pancreatic tumorigenesis, we knocked down MUC13 expression in HPAFII cells, which express high levels of MUC13, by using transduction with MUC13-specific shRNA lentiviral particles. A scrambled shRNA (sh-V-GFP) HPAFII stable cell line was also generated by transduction with GFP-shRNA lentiviral particles. Knockdown of MUC13 was confirmed by RT-PCR (Fig. 2A, left panel) and confocal microscopy (Fig. 2A, right panel). In vector control HPAFII sh-V-GFP cells, MUC13 expression was similar (Fig. 2A, b) compared with wild-type HPAFII cells (Fig. 2A, a). However, MUC13 expression in the knockdown cells was much lower than wild-type and vector cells, with shMUC13D having relatively greater knockdown of MUC13 compared with shMUC13A (Fig. 2A, c and d). Two pools of cells showing downregulation of MUC13 were selected to determine the effect of MUC13 expression in pancreatic cancer cells. Interestingly, MUC13 knockdown cells (shMUC13D) showed significant (P < 0.05) inhibition in cell proliferation compared with vector control cells (Fig. 2B) and displayed relatively higher cell doubling times (29.6 hours) as compared with vector control HPAFII (26.3 hours) cells (Supplementary Fig. S3B). In addition, in anchorage-dependent and -independent colony forming assays, MUC13 knockdown cells (shMUC13D) formed significantly (P < 0.05) less colonies than vector control (sh-V-GFP; Fig. 2C and D).
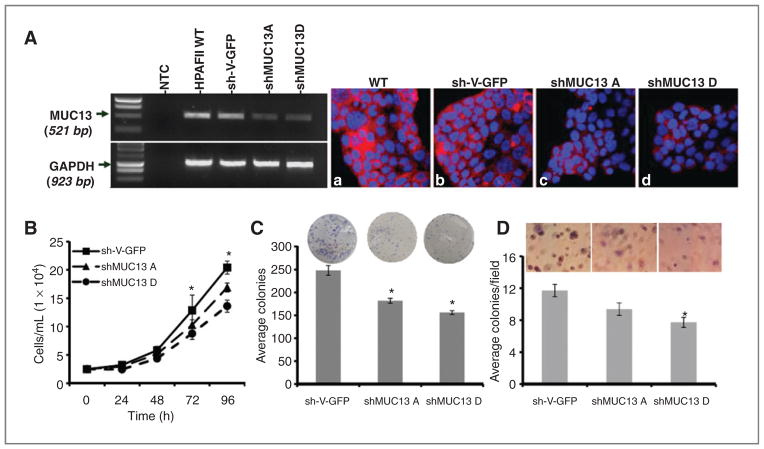
Figure 2.
Knockdown of MUC13 reduces in vitro tumorigenesis in HPAFII cells. A, RT-PCR and confocal microscopy analysis. Left, RT-PCR analysis of MUC13 expression in HPAFII-MUC13 knockdown stable cell lines. GAPDH was used as an internal control. Right, confocal microscopy images of HPAFII-MUC13 knockdown stable cell lines after immunofluorescence staining with anti-MUC13 MAb (red). DAPI (blue) was used as a nuclear stain. Original magnification ×400. B, cell proliferation assay. The number of HPAFII-derived cells was determined after 24, 48, 72, and 96 hours of culture. C, anchorage-dependent colony forming assay. HPAFII-derived colonies were counted 10 days after plating. D, anchorage-independent soft agar colony forming assays. HPAFII-derived colonies were counted 15 days after plating in media-agar. For B, C, and D, mean ± SE; n = 3; *, P < 0.05.
MUC13 expression correlates with motility, invasion, and adhesion of pancreatic cancer cells
To elucidate the role of MUC13 in invasion and metastasis, we compared cell motility, invasion and adhesion in MUC13 overexpressing and MUC13 knockdown cells. In agarose-bead, Boyden’s chamber, and Matrigel invasion assays, MUC13-expressing clones showed higher cellular motility and invasion compared with vector control cells (Fig. 3A–C). Because an antiadhesive property would make cancer cells more motile and invasive, cells were assayed for their adhesion to extracellular matrix components and MUC13-expressing cells were less adhesive to fibronectin, basement membrane complex (BMC), and laminin than MUC13-nonexpressing cells (Fig. 3D). In addition, an aggregation assay showed that high MUC13-expressing cells (MP4) formed smaller and looser cell aggregates than MUC13-nonexpressing cells (MPPC; data not shown). Accordingly, MUC13 knockdown cells (shMUC13D) showed a significant reduction in cellular motility and invasion compared with the vector control (Fig. 4A and B). In adhesion assays, MUC13 knockdown cells (shMUC13D) were more adhesive to collagen IV, laminin, BMC, and fibronectin compared with control cells (Fig. 4C). Moreover, MUC13 knockdown cells (shMUC13D) formed larger and tighter cell aggregates whereas the vector control formed smaller and looser cells aggregates (Fig. 4D).
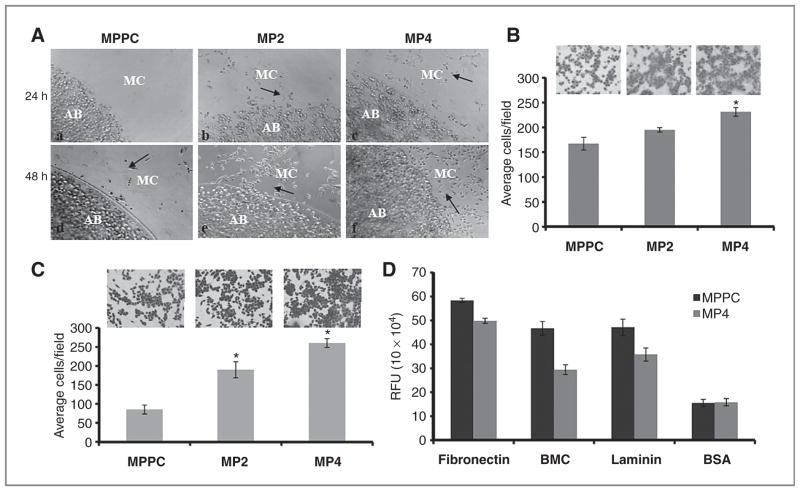
Figure 3.
MUC13 expression increases cellular migration and invasion. A, cellular motility assay. MiaPaca-derived clones were mixed into agarose solution and dropped onto fibronectin/bovine serum albumin (BSA)-coated plates. MUC13-expressing cells showed an increase in cell migration (b, c and e, f) compared with MUC13-null cells (a, d) at 24 and 48 hours. Black arrows indicate representative areas with migratory cells. AB, agarose bead; MC, migratory cells. Original magnification, ×100. B, quantitative Boyden’s chamber cell migration assays. Migratory cells were counted following 48 hours incubation in the presence of a serum gradient. Mean number of cells in 10 fields of view. Mean ± SE; n = 3; *, P < 0.05. C, Matrigel cell invasion assay. Invasive cells were counted following 48 hours incubation in BD Biocoat Matrigel invasion chambers. Mean number of cells in 10 fields of view. Mean ± SE; n = 3; *, P < 0.05. D, extracellular matrix cell adhesion assays. The number of adhesive cells was determined after 1 hour incubation in wells coated with fibronectin, laminin, and BMC. BSA was used as a negative control. Mean ± SE; n = 6; *, P < 0.05.
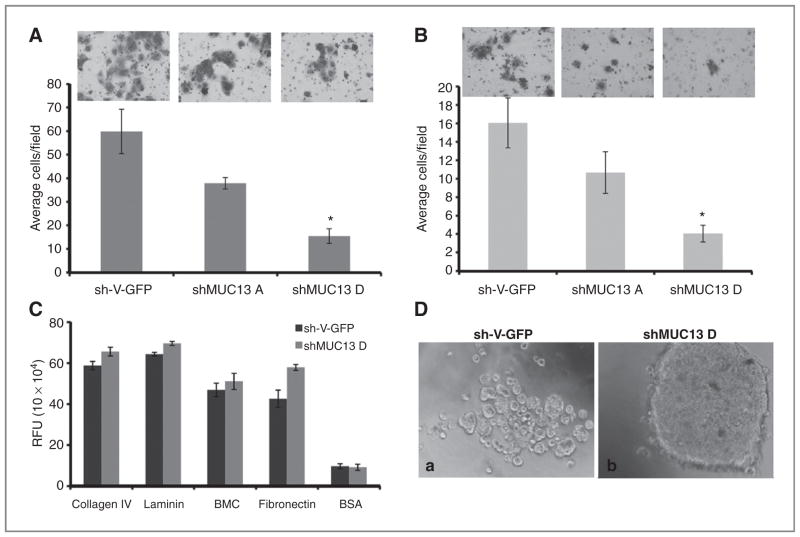
Figure 4.
Inhibition of MUC13 suppresses invasive phenotype of HPAFII cells. A, Boyden’s chamber cell migration assays. Migratory cells were counted following 48 hours incubation in the presence of a serum gradient. Mean number of cells in 10 fields of view. Mean ± SE; n = 3; *, P < 0.05. B, Matrigel cell invasion assay. Invasive cells were counted following 48 hours incubation in BD Biocoat Matrigel invasion chambers. Mean number of cells in 10 fields of view. Mean ± SE; n = 3; *, P < 0.05. C, extracellular matrix cell adhesion assays. The number of adhesive cells was determined after 1 hour incubation in wells coated with collagen, fibronectin, laminin, and BMC. BSA was used as a negative control. Mean ± SE; n = 6; *, P < 0.05. D, aggregation assays. Representative phase contrast images of aggregation assays after overnight incubation. Original magnification, ×200.
MUC13 expression influences tumorigenic cell signaling pathways
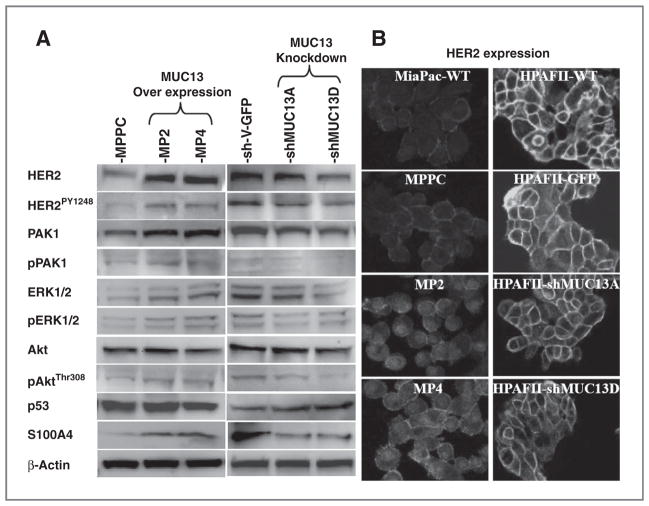
Recently, in pancreatic cancer cells, the transmembrane mucin MUC4 was shown to interact with and stabilize HER2 at the cell membrane (17). As a transmembrane mucin with 3 EGF-like domains, MUC13 may also increase HER2 stability and signaling. To investigate this possibility, we examined HER2 expression and its phosphorylation status in MUC13-overexpressing and MUC13-knockdown cells. MUC13 overexpressing cells have increased HER2 and MUC13 knockdown cells have decreased HER2 expression (Fig. 5A and B). Accordingly, the tyrosine phosphorylation of HER2 (pY1248-HER2) increased in MUC13-overexpressing cells and decreased in MUC13-knockdown cells (Fig. 5A and B). We also examined downstream targets of HER2, such as Akt, PAK1, and ERK, which are parts of the phosphoinositide-3-kinase/Akt and mitogen-activated protein kinase (MAPK) signal transduction pathways and play crucial roles in cancer progression (4, 18–24). As expected these targets also show altered expression/activation levels in MUC13 overexpressing and knockdown pancreatic cancer cells (Fig. 5A). Total and phosphorylated PAK1 and ERK were upregulated with MUC13-overexpression and downregulated with MUC13 knockdown (Fig. 5A). Even though total Akt levels remained the same, phosphorylated Akt was upregulated with MUC13-overexpression and was downregulated with MUC13 knockdown (Fig. 5A).
Figure 5.
MUC13 affects tumorigenic signaling pathways. A, immunoblot assay. Whole cell lysates from MiaPaca MUC13 overexpressing and HPAFII MUC13 knockdown cells were immunoblotted with the indicated antibodies. β-Actin was used as an internal control. Each experiment was repeated a minimum of 3 times and a representative blot is shown. B, confocal microscopy. Confocal images of MUC13 overexpressing MiaPaca (MP2 and MP4) and MUC13 knockdown HPAFII (shMUC13A and shMUC13D) derived cells after immunofluorescence for HER2. Original magnification, ×600 for MiaPaca and ×400 for HPAFII cells. HER2 expression is increased upon overexpression of MUC13 in MiaPaca cells and decreased upon knockdown of MUC13 in HPAFII cells.
Given the importance of aberrant p53 in cancer development, we also examined the effect of MUC13 on p53 expression. Our data suggest downregulation of p53 at both RNA (data not shown) and protein levels in high MUC13-overexpressing cells compared with control cells (Fig. 5A). Conversely, p53 expression was upregulated upon knocking down MUC13 in HPAFII cells compared with control cells (Fig. 5A). These results imply a novel regulatory mechanism of p53 by MUC13 overexpression.
To investigate how MUC13 expression influences invasion of pancreatic cancer cells, we determined the expression of S100A4 in MUC13-overexpressing and MUC13-knockdown cells. A member of the S100 calcium-binding proteins, S100A4 is linked to cancer invasion and metastasis (25, 26). MUC13-overexpressing cells had higher RNA (data not shown) and protein levels of S100A4 compared with control cells (Fig. 5A). However, MUC13-knockdown cells (shMUC13D) showed lower RNA (data not shown) and protein levels of S100A4 compared with control cells (Fig. 5A).
MUC13 expression influences in vivo tumorigenicity
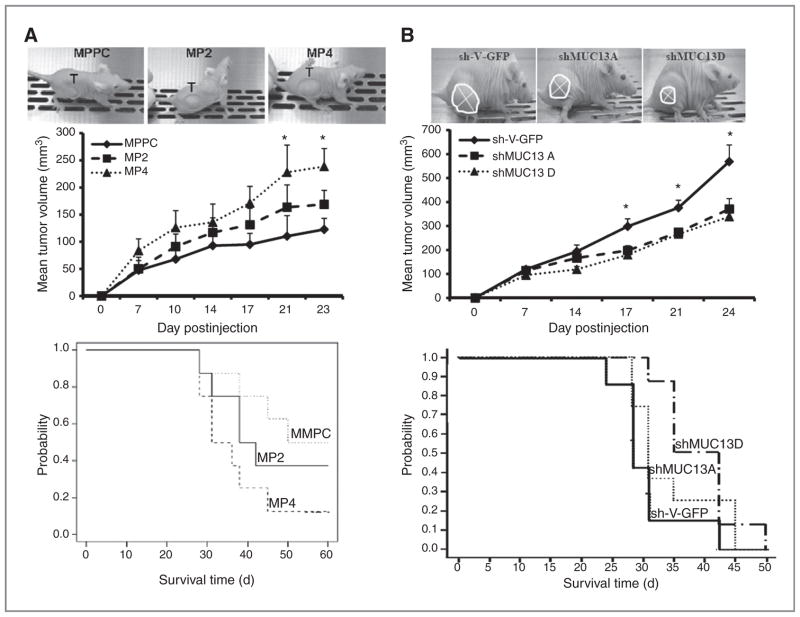
To determine the tumorigenic potential of MUC13 in vivo, we examined the tumor growth pattern of MUC13-expressing cells in a pancreatic cancer xenograft mouse model. MiaPaca cells expressing high MUC13 (MP4) formed significantly larger (P < 0.05) tumors and had reduced survival compared with MUC13 null (MPPC) cells (Fig. 6A). To confirm the association of MUC13 expression with tumor growth, we used MUC13 knockdown HPAFII cells for in vivo tumorigenesis and survival studies. Tumors from the MUC13 knockdown cells grew slower, were significantly smaller (P < 0.01), and mice had significantly improved survival compared with the mice injected with control cells (Fig. 6B). Analysis of xenograft tumors by immunohistochemistry showed a change in HER2 and p53 expression in cells overexpressing MUC13 (MP4) or in cells with suppressed MUC13 expression (shMUC13D; Supplementary Fig. S5A and B). Similar to the in vitro analysis, in xenograft tumors, MUC13 over-expressing cells have increased HER2 and decreased p53, while MUC13 knockdown cells have decreased HER2 and increased p53.
Figure 6.
MUC13 expression alters in vivo tumorigenesis and survival in nude mice xenograft model. A, top, photographs of mice with tumors from MiaPaca-derived MUC13 expressing (MP2 and MP4) and nonexpressing vector control (MPPC) cells. Tumors are circled and the graph shows the average tumor volume on days postinjections. Mean of at least 8 mice/group. Mean ± SE; *, P < 0.05. Bottom. Kaplan–Meier plot showing survival of mice injected with MiaPaca-derived cells. B, top, photographs of tumors established with MUC13 knockdown HPAFII (shMUC13A and shMUC13D) and scrambled sh-vector control (sh-V-GFP). Tumors are circled and the graph shows the average tumor volume on days post injection. Mean of at least 8 mice/group. Mean ± SE; *, P < 0.05. Bottom, Kaplan–Meier plot showing survival of mice injected with HPAFII-derived cells.
Discussion
The high mortality rate from pancreatic cancer is primarily attributed to the inability to detect pancreatic cancer at an early stage, limited knowledge regarding the etiology of the aggressiveness of the disease, and lack of an effective treatment modality (2). These facts accentuate the need to identify novel molecular markers for early diagnosis of pancreatic cancer and to reveal underlying molecular mechanisms of the aggressiveness of the disease. In this study, for the first time, we have investigated the expression profile of a newly identified transmembrane mucin, MUC13, and its functional role in pancreatic cancer progression. Our data show that MUC13 is highly overexpressed in pancreatic tumors compared with undetectable or faint expression in normal pancreas. MUC13 expression correlated with the differentiation status of the tumor and was highest in well-differentiated tumor samples and lowest in poorly differentiated tumor samples (Supplementary Fig. S1). Our findings suggest that MUC13 is a pancreatic tumor-associated molecule and may be useful for pancreatic cancer diagnosis. Given that biochemical analysis suggests that MUC13 is cleaved within the SEA domain (5, 6), our extracellular luminal staining results suggest that MUC13 may be shed from pancreatic cancer cells and released into the blood. In addition, we have seen a progressive increase in expression and aberrant localization of MUC13 in pancreatic intraepithelial (PanIN) lesions (data not shown). For these reasons, there is a strong possibility that a MUC13-based serum immunoassay can be developed as a screening method for pancreatic cancer. Moreover, the intense membrane staining (Supplementary Fig. S1) also indicates that MUC13 may be an excellent target for antibody-guided therapy of pancreatic cancer.
To elucidate the functional roles and signaling pathways influenced by MUC13 in pancreatic cancer, we exogenously expressed MUC13 in MiaPaca and Panc1 cells which resulted in increased cell proliferation and colony forming efficiency of these cells. Conversely, MUC13 knockdown in HPAFII caused a decrease in cell proliferation and colony forming efficiency. The cellular response to altered MUC13 expression was more pronounced in cells showing higher expression or greater knockdown of MUC13, indicating a variable effect depending upon the level of MUC13 expression. In our xenograft mouse studies, MUC13 expression significantly augmented pancreatic tumor growth and reduced mice survival (Fig. 6A). In contrast, suppression of MUC13 expression markedly reduced tumor growth and improved animal survival (Fig. 6B). Because of its 3 EGF domains, MUC13 may influence expression, stabilization, recycling, and activation of EGF receptors, such as HER2, which in turn may modulate the EGF receptor cascade and its downstream signaling. Our data show that MUC13 modulates HER2 expression and its phosphorylation at tyrosine 1248, which in turn, induces oncogenic downstream targets of HER2, Akt and ERK (Fig. 5). Activation of PI3K/Akt and MAPK signaling pathways by HER2 have been implicated in the regulation of tumorigenesis (24, 27, 28).
Mutations in p53 have been well documented in a variety of cancers, including pancreatic cancer, and such mutations often create an oncogenic environment favoring tumorigenesis. Currently, our data suggest that MUC13 expression decreases the expression of p53. At this time, it is unknown if MUC13 regulates the expression of wild-type and/or mutant p53 as both forms can be detected by the immunologic methods (IHC and WB) used in our study (the cell lines used have mutated p53; refs. 29, 30). It has been reported that suppression of mutant p53 can lead to a decrease in MiaPaca cell proliferation through enhanced Id2 expression (31). Our results may seem contradictory to the previous study as MUC13 overexpression was associated with decreased p53 and increased cell proliferation. However, our data also show that MUC13 expression activates growth signaling pathways (HER2, PAK1, ERK, and Akt). Therefore, upon MUC13 overexpression, any inhibition in cell growth due to the reduction of mutant p53 would be countered by increased signaling through potent growth signaling pathways. The link between MUC13 and p53 expression may be mediated by intermediates (e.g., through activation of HER2) or could be mediated directly by the cytoplasmic tail of MUC13 (MUC13-CT). Another member of the transmembrane mucin family, MUC1, has been shown to inhibit transcription of p53 through binding of the MUC1 cytoplasmic tail (MUC1-CT) with Kruppel-like factor 4 (KLF4) which enhances binding to PE21 (a p53 repressor; ref. 32). A similar interaction is possible with the cytoplasmic tail of MUC13 although future experiments are needed to resolve this issue.
MUC13-overexpresion in MiaPaca cells increased motility and invasion, while suppression of MUC13 decreased cellular motility and invasion in HPAFII cells. Overexpression of PAK1 enhances migration of breast, hepatocellular, and colorectal cancer cells and regulates the actin cytoskeleton during cell motility (33, 34). In addition, S100A4 is associated with metastasis and invasion (25, 26). Cancers expressing high levels of S100A4 have a worse prognosis compared with S100A4 negative cancers (35). Interestingly, PAK1 and S100A4 increased with MUC13 expression and decreased with MUC13 suppression (Fig. 5). In this study, we provide evidence to suggest that MUC13 plays a role in the invasiveness of human pancreatic cancer through the expression and/or activation of PAK1 and S100A4.
MUC13 has a heavily glycosylated extracellular tandem repeat domain (5) which may contribute to the antiadhesiveness of cells (16). Herein, we provide functional evidence that the overexpression of MUC13 in MiaPaca cells resulted in reduced cell–cell and cell-matrix adhesion. On the contrary, MUC13 knockdown in HPAFII cells resulted in increased cell–cell and cell-matrix adhesion (Figs. 3 and 4). Reducedcell–cell adhesion due to altered expression or function of cadherins may allow cancer cells to disseminate from sites of localized cancer and invade surrounding tissues (36, 37). Therefore, MUC13 induced changes in cell adhesion characteristics may potentiate invasion, metastasis, and aggressiveness of pancreatic cancer.
In conclusion, our data provide novel evidence for the aberrant expression of MUC13 in pancreatic tumors and indicates the role of MUC13 in pancreatic tumorigenesis and progression. We propose that MUC13 is overexpressed in pancreatic cancer and induces cellular motility, proliferation, and invasion through modulation of HER2, PAK1, Akt, S100A4, and p53 expression/activation. Further investigations are needed to understand the comprehensive molecular mechanisms of MUC13 and HER2 interplay in pancreatic cancer. This work is the first demonstration of the direct association of MUC13 with pancreatic cancer.
Supplementary Material
Acknowledgments
The authors thank Cathy Christopherson for editorial assistance. The authors also thank Dr. Keith Miskimins for his constructive suggestions. The authors also thank Sanford Research/USD Core facilities, which are supported by P20 RR17662 and P20RR024219 COBRE grants.
Grant Support
This work was supported by Department of Defense (DoD) PC073887, Governor’s 2010, and NIH R01 (CA142736) Grants awarded to S.C. Chauhan and DoD PC073643 awarded to M. Jaggi.
Footnotes
Note: Supplementary data for this article are available at Molecular Cancer Therapeutics Online (http://mct.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011;61:212–36. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- 2.Freelove R, Walling AD. Pancreatic cancer: diagnosis and management. Am Fam Physician. 2006;73:485–92. [PubMed] [Google Scholar]
- 3.Kufe DW. Mucins in cancer: function, prognosis and therapy. Nat Rev Cancer. 2009;9:874–85. doi: 10.1038/nrc2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chauhan SC, Kumar D, Jaggi M. Mucins in ovarian cancer diagnosis and therapy. J Ovarian Res. 2009;2:21. doi: 10.1186/1757-2215-2-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williams SJ, Wreschner DH, Tran M, Eyre HJ, Sutherland GR, McGuckin MA. Muc13, a novel human cell surface mucin expressed by epithelial and hemopoietic cells. J Biol Chem. 2001;276:18327–36. doi: 10.1074/jbc.M008850200. [DOI] [PubMed] [Google Scholar]
- 6.Maher DM, Gupta BK, Nagata S, Jaggi M, Chauhan SC. Mucin 13: structure, function, and potential roles in cancer pathogenesis. Mol Cancer Res. 2011;9:531–7. doi: 10.1158/1541-7786.MCR-10-0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shimamura T, Ito H, Shibahara J, Watanabe A, Hippo Y, Taniguchi H, et al. Overexpression of MUC13 is associated with intestinal-type gastric cancer. Cancer Sci. 2005;96:265–73. doi: 10.1111/j.1349-7006.2005.00043.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walsh MD, Young JP, Leggett BA, Williams SH, Jass JR, McGuckin MA. The MUC13 cell surface mucin is highly expressed by human colorectal carcinomas. Hum Pathol. 2007;38:883–92. doi: 10.1016/j.humpath.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 9.Chauhan SC, Vannatta K, Ebeling MC, Vinayek N, Watanabe A, Pandey KK, et al. Expression and functions of transmembrane mucin MUC13 in ovarian cancer. Cancer Res. 2009;69:765–74. doi: 10.1158/0008-5472.CAN-08-0587. [DOI] [PubMed] [Google Scholar]
- 10.Singh AP, Chauhan SC, Andrianifahanana M, Moniaux N, Meza JL, Copin MC, et al. MUC4 expression is regulated by cystic fibrosis transmembrane conductance regulator in pancreatic adenocarcinoma cells via transcriptional and post-translational mechanisms. Oncogene. 2007;26:30–41. doi: 10.1038/sj.onc.1209764. [DOI] [PubMed] [Google Scholar]
- 11.Jaggi M, Rao PS, Smith DJ, Wheelock MJ, Johnson KR, Hemstreet GP, et al. E-cadherin phosphorylation by protein kinase D1/protein kinase C{mu} is associated with altered cellular aggregation and motility in prostate cancer. Cancer Res. 2005;65:483–92. [PubMed] [Google Scholar]
- 12.Singh AP, Moniaux N, Chauhan SC, Meza JL, Batra SK. Inhibition of MUC4 expression suppresses pancreatic tumor cell growth and metastasis. Cancer Res. 2004;64:622–30. doi: 10.1158/0008-5472.can-03-2636. [DOI] [PubMed] [Google Scholar]
- 13.Redfield A, Nieman MT, Knudsen KA. Cadherins promote skeletal muscle differentiation in three-dimensional cultures. J Cell Biol. 1997;138:1323–31. doi: 10.1083/jcb.138.6.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yallapu MM, Othman SF, Curtis ET, Gupta BK, Jaggi M, Chauhan SC. Multi-functional magnetic nanoparticles for magnetic resonance imaging and cancer therapy. Biomaterials. 2010;32:1890–905. doi: 10.1016/j.biomaterials.2010.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chaturvedi P, Singh AP, Moniaux N, Senapati S, Chakraborty S, Meza JL, et al. MUC4 mucin potentiates pancreatic tumor cell proliferation, survival, and invasive properties and interferes with its interaction to extracellular matrix proteins. Mol Cancer Res. 2007;5:309–20. doi: 10.1158/1541-7786.MCR-06-0353. [DOI] [PubMed] [Google Scholar]
- 16.Hollingsworth MA, Swanson BJ. Mucins in cancer: protection and control of the cell surface. Nat Rev Cancer. 2004;4:45–60. doi: 10.1038/nrc1251. [DOI] [PubMed] [Google Scholar]
- 17.Chaturvedi P, Singh AP, Chakraborty S, Chauhan SC, Bafna S, Meza JL, et al. MUC4 mucin interacts with and stabilizes the HER2 onco-protein in human pancreatic cancer cells. Cancer Res. 2008;68:2065–70. doi: 10.1158/0008-5472.CAN-07-6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wickenden JA, Watson CJ. Signalling downstream of PI3 kinase in mammary epithelium: a play in 3 Akts. Breast Cancer Res. 2010;12:202. doi: 10.1186/bcr2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adam L, Vadlamudi R, Kondapaka SB, Chernoff J, Mendelsohn J, Kumar R. Heregulin regulates cytoskeletal reorganization and cell migration through the p21-activated kinase-1 via phosphatidylinositol-3 kinase. J Biol Chem. 1998;273:28238–46. doi: 10.1074/jbc.273.43.28238. [DOI] [PubMed] [Google Scholar]
- 20.Vadlamudi RK, Barnes CJ, Rayala S, Li F, Balasenthil S, Marcus S, et al. p21-activated kinase 1 regulates microtubule dynamics by phosphorylating tubulin cofactor B. Mol Cell Biol. 2005;25:3726–36. doi: 10.1128/MCB.25.9.3726-3736.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vadlamudi RK, Li F, Adam L, Nguyen D, Ohta Y, Stossel TP, et al. Filamin is essential in actin cytoskeletal assembly mediated by p21-activated kinase 1. Nat Cell Biol. 2002;4:681–90. doi: 10.1038/ncb838. [DOI] [PubMed] [Google Scholar]
- 22.Grothey A, Hashizume R, Ji H, Tubb BE, Patrick CW, Jr, Yu D, et al. C-erbB-2/HER-2 upregulates fascin, an actin-bundling protein associated with cell motility, in human breast cancer cell lines. Oncogene. 2000;19:4864–75. doi: 10.1038/sj.onc.1203838. [DOI] [PubMed] [Google Scholar]
- 23.Xiong HQ. Molecular targeting therapy for pancreatic cancer. Cancer Chemother Pharmacol. 2004;54(Suppl 1):S69–77. doi: 10.1007/s00280-004-0890-2. [DOI] [PubMed] [Google Scholar]
- 24.Reddy KB, Nabha SM, Atanaskova N. Role of MAP kinase in tumor progression and invasion. Cancer Metastasis Rev. 2003;22:395–403. doi: 10.1023/a:1023781114568. [DOI] [PubMed] [Google Scholar]
- 25.Boye K, Maelandsmo GM. S100A4 and metastasis: a small actor playing many roles. Am J Pathol. 2010;176:528–35. doi: 10.2353/ajpath.2010.090526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sherbet GV, Lakshmi MS. S100A4 (MTS1) calcium binding protein in cancer growth, invasion and metastasis. Anticancer Res. 1998;18:2415–21. [PubMed] [Google Scholar]
- 27.Arias-Romero LE, Chernoff J. p21-activated kinases in Erbb2-positive breast cancer: a new therapeutic target? Small Gtpases. 2011;1:124–8. doi: 10.4161/sgtp.1.2.14109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moasser MM. The oncogene HER2: its signaling and transforming functions and its role in human cancer pathogenesis. Oncogene. 2007;26:6469–87. doi: 10.1038/sj.onc.1210477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruggeri B, Zhang SY, Caamano J, DiRado M, Flynn SD, Klein-Szanto AJ. Human pancreatic carcinomas and cell lines reveal frequent and multiple alterations in the p53 and Rb-1 tumor-suppressor genes. Oncogene. 1992;7:1503–11. [PubMed] [Google Scholar]
- 30.Sipos B, Moser S, Kalthoff H, Torok V, Lohr M, Kloppel G. A comprehensive characterization of pancreatic ductal carcinoma cell lines: towards the establishment of an in vitro research platform. Virchows Arch. 2003;442:444–52. doi: 10.1007/s00428-003-0784-4. [DOI] [PubMed] [Google Scholar]
- 31.Yan W, Liu G, Scoumanne A, Chen X. Suppression of inhibitor of differentiation 2, a target of mutant p53, is required for gain-of-function mutations. Cancer Res. 2008;68:6789–96. doi: 10.1158/0008-5472.CAN-08-0810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wei X, Xu H, Kufe D. Human mucin 1 oncoprotein represses transcription of the p53 tumor suppressor gene. Cancer Res. 2007;67:1853–8. doi: 10.1158/0008-5472.CAN-06-3063. [DOI] [PubMed] [Google Scholar]
- 33.Kumar R, Gururaj AE, Barnes CJ. p21-activated kinases in cancer. Nat Rev Cancer. 2006;6:459–71. doi: 10.1038/nrc1892. [DOI] [PubMed] [Google Scholar]
- 34.Coniglio SJ, Zavarella S, Symons MH. Pak1 and Pak2 mediate tumor cell invasion through distinct signaling mechanisms. Mol Cell Biol. 2008;28:4162–72. doi: 10.1128/MCB.01532-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Silva Rudland S, Martin L, Roshanlall C, Winstanley J, Leinster S, Platt-Higgins A, et al. Association of S100A4 and osteopontin with specific prognostic factors and survival of patients with minimally invasive breast cancer. Clin Cancer Res. 2006;12:1192–200. doi: 10.1158/1078-0432.CCR-05-1580. [DOI] [PubMed] [Google Scholar]
- 36.Wheelock MJ, Shintani Y, Maeda M, Fukumoto Y, Johnson KR. Cadherin switching. J Cell Sci. 2008;121:727–35. doi: 10.1242/jcs.000455. [DOI] [PubMed] [Google Scholar]
- 37.Hirohashi S. Inactivation of the E-cadherin-mediated cell adhesion system in human cancers. Am J Pathol. 1998;153:333–9. doi: 10.1016/S0002-9440(10)65575-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.