Abstract
The eukaryotic stalk, which is responsible for the recruitment of translation factors, is a pentamer containing two P1–P2 dimers with unclear modes of action. In Saccharomyces cerevisiae, P1/P2 proteins (individual P1 and P2 proteins) are organized into two distinct dimers, P1A–P2B and P1B–P2A. To investigate the functional contribution of each dimer on the ribosome, RTA (ricin A chain), which binds to the stalk to depurinate the SRL (sarcin/ricin loop), was used as a molecular probe in yeast mutants in which the binding site for one or the other dimer on P0 was deleted. Ribosome depurination and toxicity of RTA were greatly reduced in mutants containing only P1A–P2B on the ribosome, whereas those with only P1B–P2A were reduced less in depurination and were unaffected in toxicity. Ribosomesn bearing P1B–P2A were depurinated by RTA at a similar level as wild-type, but ribosomes bearing P1A–P2B were depurinated at a much lower level in vitro. The latter ribosomes showed the lowest association and almost no dissociation with RTA by surface plasmon resonance. These results indicate that the P1B– P2A dimer is more critical for facilitating the access of RTA to the SRL, providing the first in vivo evidence for functional divergence between the two stalk dimers on the ribosome.
Keywords: individual P1 and P2 protein (P1/P2 protein), ribosomal stalk, ribosome, ribosome-inactivating protein, ricin A chain, sarcin/ricin loop
INTRODUCTION
The process of mRNA translation is facilitated by the interactions of ribosomes with a number of auxiliary protein factors called tGTPases (translational GTPases) [1]. The structure which is responsible for the recruitment of tGTPases and stimulation of GTP hydrolysis is a protein complex located on the large ribosomal subunit, termed the stalk [2–5]. The stalk architecture, which is composed of multiple copies of acidic proteins, represents a universally conserved entity across the three domains of life [6]. Despite sharing a common function, the stalk complexes are markedly different between the three domains of life [7]. In bacteria, the stalk structure can be found in two basic configurations, a pentamer, L10–(L12)4, in mesophiles and a heptamer, L10–(L12)6, in thermophiles [5,8]. In contrast, some cyanobacteria have eight copies of the L12 protein attached to the L10 protein, L10–(L12)8 [9]. In archaea, the ribosomal stalk is a heptamer, L10–(L12)6 [10], whereas in eukaryotes, it is a pentamer, P0–(P1–P2)2 [11]. Despite a wealth of crystallographic [5,12–19], NMR [20,21], cryo-EM [17,22,23] and SAXS [24,25] studies, the L12/P stalk represents the last structure on the ribosome for which the architecture and precise molecular function still remain poorly established.
In eukaryotes, the stalk is composed of P0, which constitutes the base of the stalk and anchors two copies of P1/P2 proteins (individual P1 and P2 proteins) [26], forming two P1–P2 heterodimers [27], organized together in a pentameric structure [11,28]. Lower eukaryotes, such as Saccharomyces cerevisiae, possess two P1/P2 protein forms, P1A, P1B, P2A and P2B [29], which preferentially form two heterodimers, P1A–P2B and P1B–P2A [30], bound to two separated specific contiguous sites on the P0 protein [31–33]. All stalk P-proteins have a highly conserved motif present at the CTD (C-terminal domain) containing a stretch of highly acidic and hydrophobic amino acids (EEEAKEESDDDMGFGLFD), regarded as their functional element directly involved in recruitment of external factors to the ribosome [34]. Recent crystallographic studies have reported only one CTD of the eukaryotic stalk counterpart, bacterial L12 dimer, interacting with the translation factors during the elongation cycle [18,19]. In eukaryotes, the C-terminal fragment of P0 is regarded as a minimal stalk element, conferring functionality of the stalk [35]. The function of the other ribosomal stalk P-proteins, which share the same conserved CTD, is not clear. Despite the high sequence similarity between P1/P2 proteins, their biophysical and functional properties are different [25,36]. Previous cross-linking experiments showed differences in the reactivity of P1 and P2 and suggested that the P1 protein, which displayed restricted reactivity, is internally located, whereas the P2 protein is more external and accessible to interact with the other cellular components [36]. Moreover, fluorescence correlation spectroscopy experiments suggested that the function of P1–P2 dimers is not equally distributed [36,37]. A recent study on silkworm ribosomes showed that both P1 and P2 contribute to ribosomal activity dependent on eEF-1a (eukaryotic elongation factor-1a) and eEF-2, and the relative contributions to protein synthesis seemed to be different between P1 and P2 [38]. In yeast cells, it was shown that the P1A–P2B dimer appears to be more relevant for cell fitness than P1B–P2A [39], which is considered to be a dynamic element of the stalk and undergo exchange with the cytoplasmic pool of free P-proteins [40]. However, the reason for the multiplication of P1–P2 dimers is not well understood and direct evidence for the distinct roles of the two stalk dimers is lacking.
Recent studies have shown the importance of the eukaryotic ribosomal stalk in facilitating the depurination activity of RIPs (ribosome-inactivating proteins) (reviewed in [41]), such as ricin [42], TCS (trichosanthin) [43–45], Stx1 (Shiga toxin 1) [46] and MOD (maize RIP) [47]. Ricin is a naturally occurring protein found in castor beans (Ricinus communis) comprising two subunits, a catalytic subunit, RTA (ricin A chain) and galactose/N-acetylgalactosamine-binding B subunit, RTB (ricin B chain), which are coupled together by a single disulfide bond [48]. Ricin depurinates a universally conserved adenine residue within the SRL (sarcin/ricin loop) of the 28S rRNA [49]. The SRL is an important component of the GAR (GTPase-associated region) [50]. Previous structural studies of the bacterial ribosome have shown that the SRL plays a key role in activation of the GTPase activity of EF-G (elongation factor-G), by opening the hydrophobic gate and placing the critical His87 into the proper position for GTP hydrolysis [50]. Depurination of the SRL prevents GTP hydrolysis by EF-G and subsequently causes an arrest of protein synthesis at the translocation step [51]. We showed that the ribosomal stalk is the docking site for RTA on the ribosome and this interaction is critical for ribosome depurination in yeast [42] and in human cells [52]. RTA interacts with the ribosomal stalk proteins through a highly conserved motif, present at the CTD of all stalk P-proteins [46]. Our recent results demonstrated that stalk binding stimulates depurination of the SRL by RTA on the ribosome [53].
In the present study, we use RTA as a molecular probe to investigate the contribution of the individual P1–P2 dimers to the stalk function. We show that the P1B–P2A dimer is more critical for binding RTA, depurination of the SRL by RTA and the toxicity of RTA than the P1A–P2B dimer on the ribosome. These results provide the first direct evidence for the differentiation in function between the two P1–P2 dimers on the ribosome.
MATERIALS AND METHODS
Yeast strains and plasmids
The two yeast strains carrying deleted forms of P0 protein were described previously [31]. In both strains, the gene encoding RPP0 (ribosomal P0 protein) had been modified to introduce deletions of fragments responsible for binding either the P1A–P2B or P1B–P2A dimer to generate P0Δ199–230 (P0ΔH1) and P0Δ230–258 (P0ΔH2) (Figure 1A). The wild-type strain BY4741 (MATa; his3Δ1; leu2 Δ0; met15Δ0; ura3Δ0) and the mutant strains were grown in either YPD medium [1 % (w/v) yeast extract/2% (w/v) peptone/2% (w/v) glucose] or minimal SD medium (synthetic defined medium) containing 2% glucose. The yeast strains were transformed with the vector containing the gene for pre-RTA (NT849) under the galactose-inducible GAL1 promoter and the LEU2 selectable marker [54]. The growth of yeast strains was performed in yeast minimal SD medium (Fischer Scientific) supplemented with 2% glucose with vigorous shaking at 30 °C. Cell growth was monitored at D600 using the SpectraMax®Plus384 reader (Molecular Devices). The doubling time was calculated using the online software Doubling Time (http://www.doubling-time.com/compute.php). Further information can be found in the Supplementary Online Materials and methods section (at http://www.biochemj.org/bj/460/bj4600059add.htm).
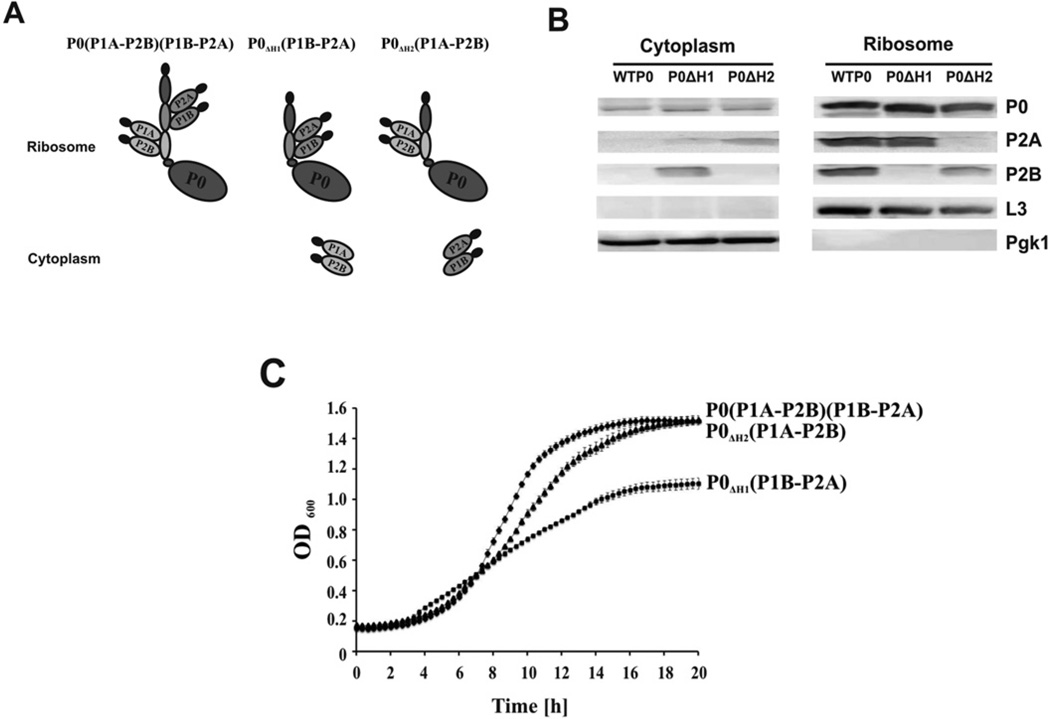
Figure 1. Analysis of the ribosomal stalk in the yeast mutants.
(A) Schematic representation of the yeast stalk mutants. (B) Immunoblot analysis of the yeast stalk mutants. Ribosomes (10 pmol) and the cytosolic fraction (40 µg of total protein) isolated from the P0ΔH1(P1B–P2A), and P0ΔH2(P1A–P2B) mutants and the isogenic wild-type were analysed by immunoblot analysis using monoclonal antibody against the C-termini of P-proteins (IB3B) to detect the P0 protein. Monoclonal antibodies against P2A (IBE3) and P2B (IAA9) were used to detect P2A and P2B respectively. Anti-L3 and anti-Pgk1 antibodies were used as loading controls for the ribosome and cytosol fractions respectively. The immunoblot analysis was repeated three times using different ribosome preparations. (C) Growth of yeast stalk mutants on minimal medium supplemented with 2 % glucose.
Preparation of ribosomes and immunoblot analysis
Exponentially growing yeast cells at a D600 value of 0.5 were collected and washed with ice-cold water, then re-suspended in buffer A (50 mM Tris/HCl, pH 7.5, 80 mM KCl and 15 mM MgCl2, containing 1 mg/ml heparin, 1 mM PMSF, 1 mM DTT and a protease inhibitor cocktail P8215 from Sigma; 1:1000 dilution). The cells were ground in liquid nitrogen, transferred to centrifuge tubes and centrifuged at 30000 g for 20 min in order to prepare a cell-free extract. Ribosomes were prepared by centrifugation of the cell-free extract at 200000 g in the above buffer supplemented with 1% Triton X-100. The resulting pellet was resuspended in 20 mM Hepes/KOH, pH 7.6, 20 mM magnesium acetate, 0.5 M KCl, 10% glycerol containing 1 mM GTP and 1 mM puromycin, and incubated at 30°C for 30 min. The mixture was then centrifuged at 10000 g for 15 min. The supernatant was applied on to a 5 ml solution of 20 mM Hepes/KOH, pH 7.6, 20 mM magnesium acetate and 0.5 M KCl, supplemented with 35% glycerol and then centrifuged at 200000 g for 5 h. The purified ribosomes were resuspended in 50 mM Hepes/KOH, pH 7.6, 12 mM magnesium acetate, 80 mM KCl, 0.1 mM PMSF, 1 mM DTT and 25% glycerol. The concentration of ribosomes was determined according to van der Zeist et al. [55]. All purification steps were performed at 4°C.
For immunoblot analysis the proteins were separated by SDS/PAGE (12% gel). Monoclonal antibodies specific against the conserved C-terminal peptide (3BH5) (a gift from Dr J.P. Ballesta, Centro de Biologia Molecular Severo Ochoa, Consejo Superior de Investigaciones Cientificas and Universidad Autonoma de Madrid, Madrid, Spain) were used for detection of the P0 protein. Monoclonal antibodies specific against P2A (IBE3) and P2B (IAA9) (gifts from Dr J.P. Ballesta) were used to detect the P2 proteins in the two dimers (P1A–P2B or P1B– P2A) present on the stalk [56]. The monoclonal antibodies against L3 (a gift from Dr J.R. Warner, Department of Cell Biology, Albert Einstein College of Medicine, NY, U.S.A.), Pgk1p (3-phosphoglycerate kinase; Invitrogen) were used as the loading controls for the ribosome and cytosol fractions respectively.
Yeast cell viability assay
Yeast cells harbouring pre-RTA vector (NT849) were grown in liquid SD medium with 2% glucose. The cells were collected by centrifugation and normalized to a D600 value of 0.1. Four serial dilutions (1:10) were made and 15 µl of each dilution were spotted on SD-Leu plates containing 2% glucose or 2% galactose and grown at 30°C for 2 days.
RTA expression and depurination in yeast
To analyse RTA expression in P0 mutants, the total protein extract was isolated from yeast as described by Zhang et al. [57]. Yeast cells (D600 of 5) were harvested at 0, 6 and 12 hpi (hours post-induction) and were first suspended in 0.5 ml of 2 M lithium acetate for 5 min on ice. Next, they were centrifuged at 6000 g and resuspended in 0.5 ml of 0.4 M NaOH for 5 min on ice. The cells were centrifuged again at 6000 g and then neutralized with 0.5 ml of 100 mM Tris/HCl, pH 6.8, and resuspended in 100 µl of 2 × SDS sample buffer and heated at 95 °C for 5 min. The extracts were centrifuged at 16000 g for 10 min, and the supernatants were collected. The samples were analysed using SDS/PAGE (12% gel). After being transferred on to nitrocellulose membranes, RTA was detected with monoclonal antibody PB10 (a gift from Dr N. Mantis, Division of Infectious Disease, Wadsworth Center, New York State Department of Health, NY, U.S.A.) [58]. The blot was stripped with 8 M guanidine hydrochloride and reprobed with an antibody against Dpm1 (dolichyl-phosphate mannosyltransferase 1) from Molecular Probes and developed using infrared imaging system (LI-COR, Odyssey). For depurination in vivo, total RNA was purified from D600 values of 0.3–0.8 of yeast cells using the Qiagen RNeasy Mini Kit with on-column DNase digestion. Total RNA (375 ng) was used for cDNA synthesis and to quantify the relative level of depurination. The extent of depurination was determined by comparison to yeast transformed with the empty vector using qRT-PCR (quantitative reverse transcription–PCR) by the ΔΔCT [comparative ΔCT (threshold cycle value)] method [59]. Two sets of primers were used in separate qRT-PCR reactions with the same sample. The first set of primers is highly specific for amplification only from rRNA where the SRL has been depurinated, whereas the second primer pair amplifies from rRNA regardless of the depurination state. The CT values for depurinated rRNA amplification are first normalized to CT values for total rRNA amplification to give the ΔCT. The normalized ΔCT values from yeast expressing RTA are then compared with those from yeast carrying a vector control plasmid to obtain the ΔΔCT. The fold difference in depurination between the two is determined using the formula 2−ΔΔCT as described previously [59].
Recombinant RTA purification
N-terminal 10× His-tagged recombinant RTA was purified using Ni-NTA (Ni2+-nitrilotriacetate) agarose from Qiagen. Protein showed a single band on SDS/PAGE by Coomassie Brilliant Blue R-250 staining and by immunoblot analysis and was active in in vitro translation inhibition and ribosome depurination assays [60].
Interaction of RTA with yeast ribosomes
The interactions were measured using a Biacore T200 system (GE Healthcare) with a CM3 chip. RTA was immobilized to Fc2 (flow cell 2) at 840 RU (resonance units) by amine coupling. Fc1 was activated and blocked as a control. The running buffer contained 10 mM Hepes, pH 7.6, 150 mM NaCl, 10 mM magnesium acetate, 50 µM EDTA and 0.005 % surfactant P20. Ribosomes were passed over both surfaces at 40 µl/min at different concentrations. The surface was regenerated by injection of 500 mM KCl in the running buffer for 20 s at a flow rate of 50 µl/min. The interactions were measured at 25 °C.
Ribosome depurination in vitro
The reaction mixture contained 40 nM yeast ribosomes, reaction buffer (10 mM Tris/HCl, pH 7.4, 60 mM KCl and 10 mM MgCl2), with different concentrations of RTA or PAP (pokeweed antiviral protein) in a total reaction volume of 100 µl. The reaction was started by adding RTA or PAP to the reaction mixture and incubating at 30 °C for 10 min. The reaction was stopped by adding 100 µl of 2× RNA extraction buffer (50 mM Tris/HCl, pH 8.8, 240 mM NaCl, 20 mM EDTA and 2% SDS). The RNA was extracted with phenol and then phenol/chloroform, and precipitated with sodium acetate and ethanol overnight at −20 °C. The extent of depurination was determined using qRT-PCR as described previously [59].
RESULTS
Characterization of the yeast stalk mutants carrying individual P1A–P2B or P1B–P2A dimers on the ribosome
To determine the functional contribution of the individual P1– P2 stalk dimers to the interaction of ribosomes with RTA, we used yeast mutants where the gene encoding the RPP0 had been modified to introduce deletions of the helices responsible for binding either P1A–P2B or P1B–P2A dimer [31]. Composition of the stalk complexes on the ribosome is shown in Figure 1(A). Wild-type strain carries all five stalk P-proteins organized in a pentameric configuration: P0–(P1A–P2B)-(P1B–P2A). The P0 deletion mutant, P0ΔH1 carries deletion of helix 1 (amino acid positions 199–230) responsible for binding P1A–P2B and P0ΔH2 carries deletion of helix 2 (amino acid positions 230–258) responsible for binding P1B–P2A, resulting in trimeric configurations of the stalk, P0ΔH1(P1B–P2A) and P0ΔH2(P1A–P2B) respectively. Immunoblot analysis was used to examine the stalk composition in the mutants (Figure 1B). Monoclonal antibodies specific for the conserved C-termini of all P-proteins showed that P0ΔH1(P1B–P2A) and P0ΔH2(P1A–P2B) contained truncated forms of P0 protein on the ribosome. Immunoblot analysis using monoclonal antibodies specific for yeast P2A or P2B proteins showed presence of P2A protein on the P0ΔH1(P1B–P2A) ribosomes, but not on the P0ΔH2(P1A– P2B) ribosomes. In contrast, P2B protein was detected on the P0ΔH2(P1A–P2B) ribosomes, but not on the P0ΔH2(P1B–P2A) ribosomes (Figure 1B), confirming the proper composition of the stalk trimers. These results were in agreement with previous characterization of the stalk composition in these mutants by isoelectric focusing [31].
The growth rate of yeast strains was monitored on minimal medium (Figure 1C). The absence of P1A–P2B dimer on the ribosome caused a severe reduction in the growth rate of P0ΔH1(P1B–P2A), whereas the absence of P1B–P2A on the ribosome did not cause a substantial reduction in the growth rate of P0ΔH2(P1A–P2B). The P0ΔH1(P1B–P2A) strain had a doubling time close to 8 h (7.76 ± 0.6 h) compared with wild-type, which had a doubling time of approximately 3 h (2.92 ± 0.2 h) and P0ΔH2(P1A–P2B), which had a doubling time of 3.5 h (3.51 ± 0.2 h). These results are consistent with previously reported studies which demonstrated that removal of the P1A– P2B–binding site exerted the strongest effect on the growth of the mutant strains, indicating that the P1A–P2B dimer is more important for the metabolic fitness of yeast [31].
The yeast mutant lacking the P1B–P2A dimer on the ribosome is resistant to the cytotoxicity of RTA
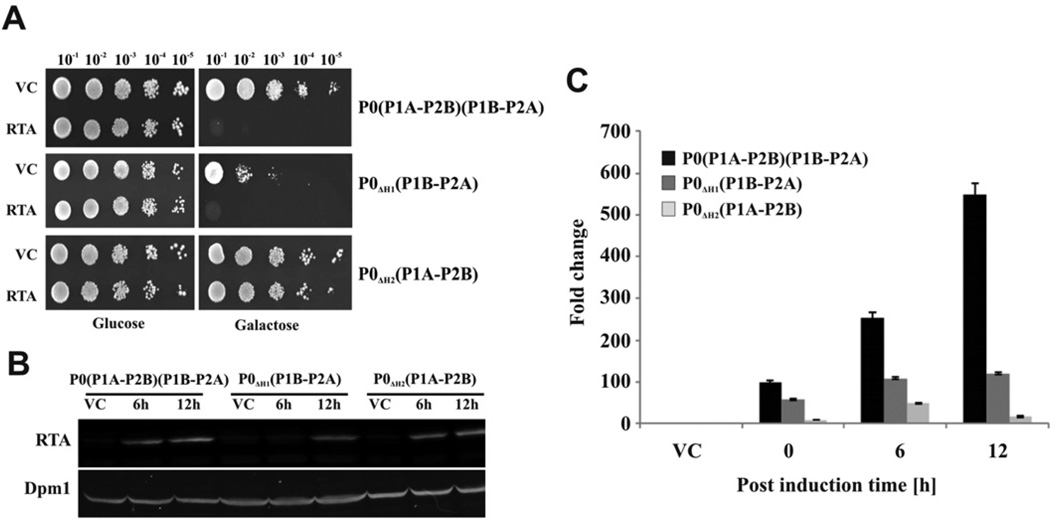
To examine the sensitivity of the yeast mutants with different stalk configurations to RTA, each mutant and the isogenic wild-type were transformed with a plasmid carrying the gene encoding pre-RTA under the GAL1 promoter. Cells were grown in liquid medium containing glucose, serially diluted, and placed on to agar plates containing glucose or galactose and grown at 30 °C for 3 days (Figure 2A). RTA clearly reduced the viability of the wild-type yeast and P0ΔH1(P1B–P2A) mutant carrying only the P1B–P2A dimer on the ribosome, but not P0ΔH2(P1A– P2B) mutant carrying only the P1A–P2B dimer on the ribosome (Figure 2A). A viability assay was also performed by examining cell survival after RTA expression. The cells grown on liquid medium containing glucose were transferred to the medium containing galactose to induce pre-RTA expression and an equal number of cells were spotted on a plate containing glucose at 0, 6 and 12 hpi of galactose (Supplementary Figure S1 at http://www.biochemj.org/bj/460/bj4600059add.htm). Expression of RTA greatly reduced the survival rate of the wild-type and P0ΔH1(P1B–P2A) mutant. In contrast, viability of the P0ΔH2(P1A– P2B) mutant was not affected by RTA. These results indicate that the P1B–P2A dimer on the ribosome is more important for the toxicity of RTA than the P1A–P2B dimer.
Figure 2. Analysis of viability, expression and ribosome depurination by RTA in the yeast stalk mutants.
(A) Viability of yeast stalk mutants expressing RTA was measured by plating serial dilutions on galactose plates. Yeast cells transformed with a plasmid carrying gene encoding pre-RTA under the GAL1 promoter were first grown SD medium supplemented with 2% glucose and then serially diluted and plated on to SD medium supplemented with 2% galactose and grown at 30°C for 3 days. Cells carrying the VC (empty vector) were used as controls. (B) Immunoblot analysis of RTA expression in yeast cells at 6 and 12 hpi. Total protein extracted from equal amount of cells (D600 of 0.8) was analysed by immunoblot analysis using monoclonal antibodies (PB10) against RTA and IR-labelled secondary antibodies. The blot was visualized using a LI-COR IR imaging system. Monoclonal antibodies against Dpm1 were used as the loading control. (C) Depurination activity of RTA in the yeast stalk mutants determined by qRT-PCR analysis. Total RNA was purified from D600 values of 0.3–0.8 of yeast cells using Qiagen RNeasy Mini Kit with on-column DNase digestion. Total RNA (375 ng) was used for cDNA synthesis. The relative level of depurination in cells expressing RTA was measured by qRT-PCR in comparison with cells containing the VC by the ΔΔCT method [59]. The fold increase in depurination in yeast expressing RTA compared with yeast harbouring the VC is shown. The experiment was repeated three times, the error bars show S.D.
Ribosome depurination by RTA is reduced more in the yeast mutant lacking P1B – P2A dimer on the ribosome
Since the yeast mutant lacking P1B–P2A dimer on the ribosome was resistant to RTA, we examined the level of RTA expression in the yeast mutants by immunoblot analysis using monoclonal antibodies (PB10) against RTA and IR-labelled secondary antibodies. The blot was visualized using a LI-COR IR imaging system (Figure 2B). The level of expression of RTA was similar in the wild-type strain and in the RTA-resistant P0ΔH2(P1A–P2B) strain at 6 and 12 hpi. The expression of RTA was lower and clearly detected only at 12 hpi in P0ΔH1(P1B–P2A). We confirmed these results by transforming each mutant with a C-terminal EGFP fusion of pre-RTA (pre-RTA-EGFP), which has been shown to be enzymatically active and toxic in S. cerevisiae [61]. The pre-RTA-EGFP expression was detected in the P0ΔH2(P1A–P2B) mutant after 6 and 12 hpi as in the wild-type. In P0ΔH1(P1B–P2A) mutant, the RTA-EGFP fluorescence signal was lower, but detectable at 12 hpi, suggesting delayed protein expression, possibly due to the growth defect observed in this strain (Supplementary Figure S2 at http://www.biochemj.org/bj/460/bj4600059add.htm). These results show that the resistance to RTA observed in P0ΔH2(P1A–P2B) is not due to the lack of RTA expression.
The depurination of the SRL by RTA was examined in the yeast mutants by qRT-PCR analysis as described previously [59,62]. Total RNA was isolated from the transformed yeast strains and the level of depurination was examined at 0, 6 and 12 hpi (Figure 2C). Ribosome depurination by RTA was reduced 5.4-fold at 6 hpi and 32-fold at 12 hpi in P0ΔH2(P1A–P2B) mutant compared with the wild-type. The level of depurination in the P0ΔH1(P1B– P2A) strain was reduced 2.4-fold at 6 hpi and 5-fold at 12 hpi compared with wild-type. However, despite the low expression of RTA in P0ΔH1(P1B–P2A), the level of depurination in this strain was higher than in P0ΔH2(P1A–P2B), consistent with the lower viability of P0ΔH1(P1B–P2A) expressing RTA (Figure 2A and Supplementary Figure S1). The lowest level of ribosome depurination observed in P0ΔH2(P1A–P2B) correlated well with the increased viability of this strain (Figure 2A), demonstrating that the presence of the P1B–P2A dimer on the ribosome is more critical for ribosome depurination by RTA in vivo.
RTA interacts differently with ribosomes carrying either the P1A–P2B or P1B–P2A dimer
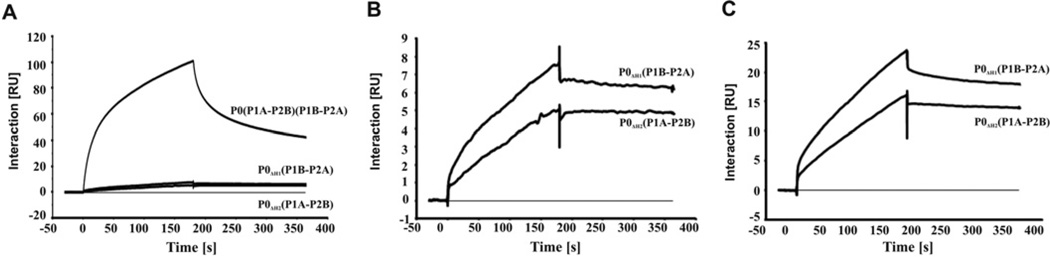
We examined the interaction of RTA with ribosomes isolated from the yeast mutants using SPR (surface plasmon resonance) with a Biacore T200 system [60,63]. The wild-type ribosomes bound RTA with an initial rapid association and dissociation phase followed by a slower association and dissociation phase (Figure 3A), which did not fit the 1:1 interaction model [60]. The absence of either P1A–P2B or P1B–P2A dimmer on the ribosome greatly reduced the interaction with RTA (Figure 3A). When the interaction of RTA with the mutant ribosomes was analysed, ribosomes carrying P1B–P2A showed faster association and dissociation with RTA than ribosomes carrying P1A–P2B (Figure 3B and 3C). The interaction pattern of ribosomes carrying P1B–P2A with RTA resembled the interaction pattern of wild-type ribosomes with RTA, showing rapid initial association and dissociation followed by slower association and dissociation. In contrast, ribosomes carrying P1A–P2B showed slower association and almost no dissociation. These results indicate that the dynamic interactions of RTA with ribosomes bearing the individual P1–P2 dimers are different. The absence of either dimer diminishes binding of RTA to the ribosome. However, the absence of P1B–P2A on the ribosome causes a greater decrease in the interaction with RTA than the absence of P1A–P2B.
Figure 3. Interaction of RTA with ribosomes from the stalk mutants.
(A) Interaction of RTA with ribosomes isolated from wild-type and stalk mutants was analysed with a Biacore T200 system at a 1 nM ribosome concentration. (B) Interaction of RTA with the stalk mutant ribosomes at a 1 nM concentration. (C) Interaction of RTA with the stalk mutant ribosomes at a 5 nM concentration. The interactions were measured using a Biacore T200 system with a CM3 chip. RTA was immobilized to Fc2 at 840 RU by amine coupling. Fc1 was activated and blocked as a control. Ribosomes from wild-type yeast or the stalk mutants were passed over both surfaces at 40 µl/min. Running buffer consisted of 10 mM Hepes, pH 7.6, 150 mM NaCl, 10 mM magnesium acetate, 5 µM EDTA and 0.005% surfactant P20.
In vitro depurination of SRL is greatly reduced in ribosomes lacking P1B–P2A proteins
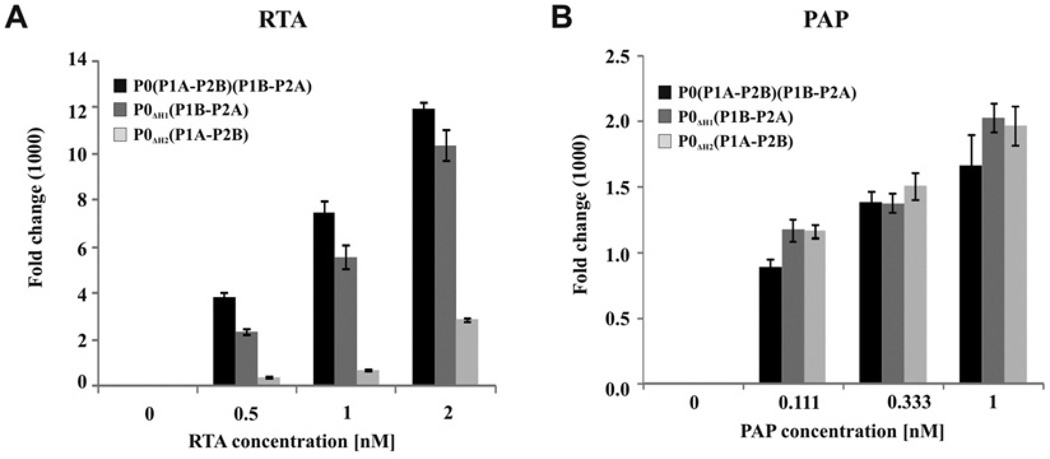
To determine whether the differences observed in the interaction of stalk mutant ribosomes with RTA affect depurination, we examined the depurination level of SRL using ribosomes isolated from the stalk mutants. As shown in Figure 4(A), RTA depurinated wild-type ribosomes in a concentration-dependent manner. Depurination of ribosomes carrying P1A–P2B was greatly reduced compared with wild-type ribosomes. In contrast, RTA depurinated ribosomes carrying P1B–P2A almost as well as wild-type ribosomes in vitro. To determine whether the differences in depurination may be due to differences in the accessibility of SRL, we performed a control experiment with PAP from Phytolacca americana. Previous results showed that PAP binds to ribosomal protein L3 to gain access to the SRL [64], and does not require the ribosomal stalk for ribosome depurination [42,44]. PAP depurinated ribosomes from the stalk mutants at a similar level (Figure 4B), indicating that the differences observed in depurination of the stalk mutants is specific to the depurination activity of RTA and has no effect on the depurination activity of PAP. These results demonstrate that the P1B–P2A dimer on the ribosome is more critical for depurination of the SRL by RTA than the P1A–P2B dimer.
Figure 4. Depurination of ribosomes from the stalk mutants.
Depurination of ribosomes isolated from the stalk mutants by (A) RTA or (B) PAP. Different concentrations of RTA or PAP were used in a reaction mixture containing 40 nM yeast ribosomes and reaction buffer (10 mM Tris/HCl, pH 7.4,60 mM KCl and 10 mM MgCl2) in a total reaction of 100 µl for 10 min at 30 °C. The extent of depurination was determined by qRT-PCR in comparison with no toxin control used by the ΔΔCT as described previously [59]. The experiment was repeated three times, the error bars show S.D.
DISCUSSION
The eukaryotic stalk has a pentameric organization, where the two copies of P1–P2 heterodimers are bound to P0 protein, which anchors the whole structure to the ribosome. The stalk proteins are the only ribosomal proteins present in multiple copies on the ribosome [65]. The function of the multiplication of the stalk proteins, which share a conserved peptide at their C-termini, is not well-understood. It has been suggested that very early in evolution, probably only one dimer was present on the ancestral ribosome [9] and multiplication of stalk proteins was probably coupled with the evolution of translation factors and the diversification of life forms. The first indication about the differences in the function and structure of the individual stalk proteins was provided by cross-linking experiments in bacteria, where it was shown that one of the L12 stalk dimers has a bent conformation with the CTD interacting with protein L11 at the stalk base, whereas the second one has an extended conformation that is able to react with proteins far from the stalk [66]. This model has been further confirmed by an NMR study, which showed that among the four CTDs of the L12 proteins, two are mobile and two are interacting tightly with the ribosome [67], and recent crystallographic studies showed that only one CTD is interacting with the translation factors [18,19]. Results obtained for eukaryotic P1/P2 proteins also suggested that their function is not equally distributed [36–38]. However, despite numerous studies, which focused on the structure and function of the stalk, there is no direct evidence indicating that multiple stalk dimers differ in their function on the ribosome.
To understand the role of individual P1–P2 stalk dimers independently of their position on the ribosome, we used S. cerevisiae mutants, in which the binding site for one or the other dimer was deleted. Mutants containing deletions in the specific binding sites for P1A–P2B or P1B–P2A were used, instead of mutants lacking the individual P1/P2 proteins, since deletion of an individual P-protein may cause an aberrant stalk structure and protein composition [68]. Deletion of the binding sites for either the P1A–P2B or P1B–P2A dimer on P0 resulted in ribosomes with well-defined trimeric stalk configurations, P0ΔH1(P1B–P2A) or P0ΔH2(P1A–P2B), where each dimer is the same distance away from the stalk base. Therefore we were able to evaluate the function of each dimer independently of their position relative to the stalk base. First, the two trimeric stalk configurations had a differential effect on the growth of yeast cells (Figure 1C). The lack of P1A–P2B dimer on the ribosome caused a significant reduction in growth rate. In contrast, deletion of P1B–P2A did not have a substantial effect on the metabolic fitness of yeast cells. Our results are consistent with previously reported studies, which demonstrated that removal of the individual binding sites for P1– P2 proteins on P0 had a differential impact on culture growth and thus on the metabolic fitness of yeast [31]. They are also in agreement with the observation that the P1A–P2B dimer plays a key role in the mechanism of the stalk assembly, whereas the P1B–P2A dimer plays a subsidiary function and may represent a dynamic regulatory element(s) of the stalk [28]. In addition, the co-operative nature of the stalk dimers was demonstrated by our previously reported analysis showing that RTA interacts more efficiently with the stalk pentamer than with either trimer [63]. More recently, it was demonstrated that the stalk acts as a collaborative entity in polypeptide synthesis [38]. These results indicate that pentameric organization of the stalk constitutes an orchestrated entity where all elements contribute independently to the overall activity of the stalk structure.
In the present study, we show the divergence between the two pairs of P1–P2 dimers on the ribosome with respect to the toxicity and the depurination activity of RTA. The yeast mutant containing only P1A–P2B dimer on the ribosome was resistant to RTA (Figure 2A). In contrast, yeast with only the P1B–P2A dimer on the ribosome was sensitive to RTA. These results indicate that the P1B–P2A dimer on the ribosome contributes significantly to RTA toxicity. Ribosome depurination by RTA was reduced relative to the wild-type when either dimer was missing from the ribosome. However, the mutant with only the P1B–P2A on the ribosome was more susceptible to RTA-mediated depurination than the mutant with only the P1A–P2B (Figure 2C). Since RTA expression in the mutant with only P1B–P2A was much lower, the in vivo depurination data were consistent with the viability analysis and suggested that the P1B–P2A dimer mainly contributes to the overall activity of the pentameric stalk structure towards RTA. The mutant with only P1A–P2B, which grew faster and expressed a higher level of RTA, showed lower level of depurination by RTA (Figure 2C), indicating that the differences in the level of depurination in the stalk mutants in vivo were not due to the differences in the level of RTA expression. We currently do not have an explanation for the lower expression level of RTA in the P0AH1(P1B–P2A) mutant. However, ribosomes isolated from this mutant were depurinated by RTA at a similar level as wild-type ribosomes in vitro (Figure 4A). These results demonstrate that P1B–P2A dimer has higher capacity to interact with external factors, such as RTA than the P1A–P2B dimer, and that the P1A–P2B dimer alone is not sufficient to target RTA to the SRL for depurination. Since the CTD of P1/P2, which is critical for RTA interaction, is identical in all P-proteins, we can conclude that the overall architecture of the dimers and their mobility on the ribosome must play an important role in their activity towards external factors, like RTA. These results provide direct evidence that P-protein dimers differ with respect to their function on the ribosome in vivo.
The functional differentiation of P1–P2 dimers was further verified by interaction studies between RTA and ribosomes with modified stalk structures using SPR. We observed very dynamic interaction between RTA and wild-type ribosomes. As shown previously, this was not a simple 1:1 interaction [60]. It was characterized by an initial rapid association and dissociation, followed by a slower association and dissociation, consistent with our previous data [60]. In contrast, ribosomes isolated from the mutant strains with altered stalk composition showed markedly reduced binding to RTA, but with clear differences in the association and dissociation profiles (Figure 3). Although ribosomes carrying only the P1B–P2A dimer bound RTA considerably less than wild-type ribosomes, the association and dissociation profile of these ribosomes was similar to wild-type ribosomes with an initial fast association and dissociation followed by slower association and dissociation. In contrast, ribosomes with only the P1A–P2B dimer showed slower association and almost no dissociation from RTA making it impossible to determine the association (ka) or dissociation (kd) rate constants. Moreover, the complexity of the interactions between RTA and wild-type and mutant ribosomes, which showed either very slow dissociation [P0ΔH1 (P1B–P2A)] or almost no dissociation [P0ΔH2(P1A–P2B)], made comparison of the equilibrium dissociation constants (Kd) meaningless. The direct comparison of the interaction of RTA with the isolated P1A–P2B and P1B–P2A stalk dimers could not be performed due to the differences in the physical stability of these complexes. Previous results showed that only P1A and P2B proteins interact with high affinity to form a stable P1A–P2B complex in solution. In contrast, although P1B and P2A have the ability to interact with each other, the P1B–P2A heterodimer is not fully stable in solution and may need P0 for full stability [30,68,69]. In our previous study, we compared the interaction of RTA with the native yeast stalk pentamer and the trimeric stalk complexes, containing a C-terminal fragment of P0 and P1–P2 proteins [63]. We did not see a difference in the pattern of interaction of RTA with either trimer. Since the dimers and the entire stalk are attached in a flexible manner to the large subunit of the ribosome, the mobility of the dimers may be different when they are free in solution compared with when they are attached to the ribosome. The stalk exists as a pentamer on the ribosome, and the co-operative cross-talk among ribosome components contributes significantly to stalk function. Therefore the interactions reported in the present study between RTA and intact ribosomes containing one or the other dimer representa more complete viewof the ribosome interactions of RTA.
The differential role of the individual stalk dimers on the ribosome was further verified by the depurination analysis in vitro. Depurination of ribosomes with the P1A–P2B dimer alone was substantially reduced compared with wild-type ribosomes, whereas depurination of ribosomes with only the P1B–P2A dimer was reduced less, indicating that under these conditions the P1B– P2A dimer can support RTA action. Control experiments showed that the deletion of the binding sites for the P1–P2 dimers on P0 had no effect on depurination of the mutant ribosomes by PAP, which does not require the stalk to access the SRL [64,44]. Although the binding of RTA to ribosomes carrying P1B–P2A was considerably lower than binding of RTA to wild-type ribosomes, the level of depurination of ribosomes carrying P1B–P2A was reduced less. This may be because the Biacore assay measures binding kinetics in real time, whereas the qRT-PCR assay used to measure depurination is an end point assay. Alternatively, efficient depurination may require rapid association and dissociation of RTA from the ribosome. The major difference in the interaction of RTA with ribosomes carrying one or the other dimer was in the pattern of dissociation. A high dissociation rate indicates that RTA will spend a short time in each ribosome complex, whereas a low rate indicates that it will spend more time in the complex. If RTA cannot dissociate from ribosomes, it will not be able to depurinate ribosomes catalytically. These results demonstrate that the differences in the stalk composition on the ribosome are the source of the difference in the depurination activity of RTA. The stalk complex with P1B–P2A dimer alone promotes RTA action, which is dependent on fast association and fast dissociation. In contrast, ribosomes bearing the P1A–P2B dimer alone, which show the lowest association and almost no dissociation, are depurinated less by RTA. Therefore the architecture of the P1B– P2A dimer, rather than the CTD alone plays a critical role in the depurination of the SRL by RTA. We showed recently that the ribosome-binding surface of RTA is on the opposite side of the surface which contains the active site and suggested a model whereby the interaction of RTA with the stalk stimulates depurination of the SRL by orienting the active site of RTA towards the SRL [53]. On the basis of the data demonstrated in the present study, together with our recently published results [53], we propose that both dimers co-operate to recruit RTA to the ribosome, but the P1B–P2A dimer has a leading role in optimal depurination of the SRL by RTA.
In summary, the results shown in the present study provide the first experimental evidence for the functional divergence between the P1–P2 dimers in their interaction with an RIP independently of their position on the ribosome. We showed that the P1B–P2A dimer is likely to represent a more dynamic element, which can interact with the external factors such as RTA better than the P1A–P2B dimer. The results have broad implications for our understanding of the mode of operation of the ribosomal stalk, which seems to have dual function, first, strictly related to its action as a component of the GTPase centre, involved in binding tGTPases and stimulation of GTP hydrolysis and secondly, in the regulation of the stalk [4,38,63]. Therefore we propose that the stalk acts as a collaborative entity where each one of the two P1–P2 dimers appears to have defined rolesin the recruitment, delivery and temporary stabilization of the external factors recruited to the ribosome.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr N. Mantis for providing the PB10 monoclonal RTA antibody; Dr J.P. Ballesta for providing the monoclonal antibody against the C-terminal part of P-proteins (IB3B) and monoclonal antibodies against P2A (IBE3) and P2B (IAA9); Dr J.R. Warner for providing the monoclonal antibody against L3; and Dr J. Irvin (Department of Chemistry, Southwest Texas State University, TX, U.S.A.) for providing purified PAP. The authors thank Jennifer Nielsen-Kahn for critically reading the paper before submission.
FUNDING
This work was supported by the National Institutes of Health [grant number AI072425 (to N.E.T.)] and the National Institutes of Health Fogarty International Research Collaboration Award (FIRCA) [grant number TW008418 (to N.E.T. and M.T.)].
Abbreviations
- ΔCT
threshold cycle value
- ΔΔCT
comparative ΔCT
- CTD
C-terminal domain
- Dpm1
dolichyl-phosphate mannosyltransferase 1
- eEF
eukaryotic elongation factor
- EF-G
elongation factor-G
- Fc
flow cell
- hpi
hours post-induction
- P1/P2
individual P1 and P2 protein
- PAP
pokeweed antiviral protein
- qRT-PCR
quantitative reverse transcription-PCR
- RIP
ribosome-inactivating protein
- RPP0
ribosomal P0 protein
- RTA
ricin A chain
- RU
resonance unit
- SD medium
synthetic defined medium
- SPR
surface plasmon resonance
- SRL
sarcin/ricin loop
- tGTPase
translational GTPase
Footnotes
AUTHOR CONTRIBUTION
Przemysław Grela and Xiao-Ping Li performed the experiments. Nilgun Tumer, Przemysław Grela and Xiao-Ping Li conceived the experiments and analysed the data. Marek Tchórzewski contributed reagents. Nilgun Tumer, Przemysław Grela and Marek Tchórzewski wrote the paper. This work was carried out in Nilgun Tumer’s laboratory.
REFERENCES
- 1.Liljas A, Ehrenberg M. Structural Aspects of Protein Synthesis. 2nd Edition. Singapore: World Scientific Publishing; 2012. pp. 1–492. [Google Scholar]
- 2.Helgstrand M, Mandava CS, Mulder FA, Liljas A, Sanyal S, Akke M. The ribosomal stalk binds to translation factors IF2, EF-Tu, EF-G and RF3 via a conserved region of the L12 C-terminal domain. J. Mol. Biol. 2007;365:468–479. doi: 10.1016/j.jmb.2006.10.025. [DOI] [PubMed] [Google Scholar]
- 3.Mandava CS, Peisker K, Ederth J, Kumar R, Ge X, Szaflarski W, Sanyal S. Bacterial ribosome requires multiple L12 dimers for efficient initiation and elongation of protein synthesis involving IF2 and EF-G. Nucleic Acids Res. 2012;40:2054–2064. doi: 10.1093/nar/gkr1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nomura N, Honda T, Baba K, Naganuma T, Tanzawa T, Arisaka F, Noda M, Uchiyama S, Tanaka I, Yao M, et al. Archaeal ribosomal stalk protein interacts with translation factors in a nucleotide-independent manner via its conserved C terminus. Proc. Natl. Acad. Sci. U.S.A. 2012;109:3748–3753. doi: 10.1073/pnas.1112934109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diaconu M, Kothe U, Schlunzen F, Fischer N, Harms JM, Tonevitsky AG, Stark H, Rodnina MV, Wahl MC. Structural basis for the function of the ribosomal L7/12 stalk in factor binding and GTPase activation. Cell. 2005;121:991–1004. doi: 10.1016/j.cell.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 6.Gonzalo P, Reboud JP. The puzzling lateral flexible stalk of the ribosome. Biol. Cell. 2003;95:179–193. doi: 10.1016/s0248-4900(03)00034-0. [DOI] [PubMed] [Google Scholar]
- 7.Grela P, Bernado P, Svergun D, Kwiatowski J, Abramczyk D, Grankowski N, Tchorzewski M. Structural relationships among the ribosomal stalk proteins from the three domains of life. J. Mol. Evol. 2008;67:154–167. doi: 10.1007/s00239-008-9132-2. [DOI] [PubMed] [Google Scholar]
- 8.Ilag LL, Videler H, McKay AR, Sobott F, Fucini P, Nierhaus KH, Robinson CV. Heptameric (L12)6/L10 rather than canonical pentameric complexes are found by tandem MS of intact ribosomes from thermophilic bacteria. Proc. Natl. Acad. Sci. U.S.A. 2005;102:8192–8197. doi: 10.1073/pnas.0502193102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davydov II, Wohlgemuth I, Artamonova II, Urlaub H, Tonevitsky AG, Rodnina MV. Evolution of the protein stoichiometry in the L12 stalk of bacterial and organellar ribosomes. Nat. Commun. 2013;4:1387. doi: 10.1038/ncomms2373. [DOI] [PubMed] [Google Scholar]
- 10.Maki Y, Hashimoto T, Zhou M, Naganuma T, Ohta J, Nomura T, Robinson CV, Uchiumi T. Three binding sites for stalk protein dimers are generally present in ribosomes from archaeal organism. J. Biol. Chem. 2007;282:32827–32833. doi: 10.1074/jbc.M705412200. [DOI] [PubMed] [Google Scholar]
- 11.Guarinos E, Santos C, Sanchez A, Qiu DY, Remacha M, Ballesta JP. Tag-mediated fractionation of yeast ribosome populations proves the monomeric organization of the eukaryotic ribosomal stalk structure. Mol. Microbiol. 2003;50:703–712. doi: 10.1046/j.1365-2958.2003.03733.x. [DOI] [PubMed] [Google Scholar]
- 12.Leijonmarck M, Liljas A. Structure of the C-terminal domain of the ribosomal protein L7/L12 from Escherichia coli at 1.7 Å. J. Mol. Biol. 1987;195:555–579. doi: 10.1016/0022-2836(87)90183-5. [DOI] [PubMed] [Google Scholar]
- 13.Kravchenko O, Mitroshin I, Nikonov S, Piendl W, Garber M. Structure of a two-domain N-terminal fragment of ribosomal protein L10 from Methanococcus jannaschii reveals a specific piece of the archaeal ribosomal stalk. J. Mol. Biol. 2010;399:214–220. doi: 10.1016/j.jmb.2010.04.017. [DOI] [PubMed] [Google Scholar]
- 14.Kavran JM, Steitz TA. Structure of the base of the L7/L12 stalk of the Haloarcula marismortui large ribosomal subunit: analysis of L11 movements. J. Mol. Biol. 2007;371:1047–1059. doi: 10.1016/j.jmb.2007.05.091. [DOI] [PubMed] [Google Scholar]
- 15.Naganuma T, Nomura N, Yao M, Mochizuki M, Uchiumi T, Tanaka I. Structural basis for translation factor recruitment to the eukaryotic/archaeal ribosomes. J. Biol. Chem. 2010;285:4747–4756. doi: 10.1074/jbc.M109.068098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wahl MC, Bourenkov GP, Bartunik HD, Huber R. Flexibility, conformational diversity and two dimerization modes in complexes of ribosomal protein L12. EMBO J. 2000;19:174–186. doi: 10.1093/emboj/19.2.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao YG, Selmer M, Dunham CM, Weixlbaumer A, Kelley AC, Ramakrishnan V. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science. 2009;326:694–699. doi: 10.1126/science.1179709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tourigny DS, Fernandez IS, Kelley AC, Ramakrishnan V. Elongation factor G bound to the ribosome in an intermediate state of translocation. Science. 2013;340:1235490. doi: 10.1126/science.1235490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Y, Feng S, Kumar V, Ero R, Gao YG. Structure of EF-G-ribosome complex in a pretranslocation state. Nat. Struct. Mol. Biol. 2013;20:1077–1084. doi: 10.1038/nsmb.2645. [DOI] [PubMed] [Google Scholar]
- 20.Bocharov EV, Sobol AG, Pavlov KV, Korzhnev DM, Jaravine VA, Gudkov AT, Arseniev AS. From structure and dynamics of protein L7/L12 to molecular switching in ribosome. J. Biol. Chem. 2004;279:17697–17706. doi: 10.1074/jbc.M313384200. [DOI] [PubMed] [Google Scholar]
- 21.Lee KM, Yusa K, Chu LO, Wing-Heng Yu C, Oono M, Miyoshi T, Ito K, Shaw PC, Wong KB, Uchiumi T. Solution structure of human P1*P2 heterodimer provides insights into the role of eukaryotic stalk in recruiting the ribosome-inactivating protein trichosanthin to the ribosome. Nucleic Acids Res. 2013;41:8776–8787. doi: 10.1093/nar/gkt636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Datta PP, Sharma MR, Qi L, Frank J, Agrawal RK. Interaction of the G’ domain of elongation factor G and the C-terminal domain of ribosomal protein L7/L12 during translocation as revealed by cryo-EM. Mol. Cell. 2005;20:723–731. doi: 10.1016/j.molcel.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 23.Anger AM, Armache JP, Berninghausen O, Habeck M, Subklewe M, Wilson DN, Beckmann R. Structures of the human and Drosophila 80S ribosome. Nature. 2013;497:80–85. doi: 10.1038/nature12104. [DOI] [PubMed] [Google Scholar]
- 24.Grela P, Gajda MJ, Armache JP, Beckmann R, Krokowski D, Svergun DI, Grankowski N, Tchorzewski M. Solution structure of the natively assembled yeast ribosomal stalk determined by small-angle X-ray scattering. Biochem. J. 2012;444:205–209. doi: 10.1042/BJ20120115. [DOI] [PubMed] [Google Scholar]
- 25.Grela P, Helgstrand M, Krokowski D, Boguszewska A, Svergun D, Liljas A, Bernado P, Grankowski N, Akke M, Tchorzewski M. Structural characterization of the ribosomal P1A–P2B protein dimer by small-angle X-ray scattering and NMR spectroscopy. Biochemistry. 2007;46:1988–1998. doi: 10.1021/bi0616450. [DOI] [PubMed] [Google Scholar]
- 26.Wool IG, Chan YL, Gluck A, Suzuki K. The primary structure of rat ribosomal proteins P0, P1, and P2 and a proposal for a uniform nomenclature for mammalian and yeast ribosomal proteins. Biochimie. 1991;73:861–870. doi: 10.1016/0300-9084(91)90127-m. [DOI] [PubMed] [Google Scholar]
- 27.Shimizu T, Nakagaki M, Nishi Y, Kobayashi Y, Hachimori A, Uchiumi T. Interaction among silkworm ribosomal proteins P1, P2 and P0 required for functional protein binding to the GTPase-associated domain of 28S rRNA. Nucleic Acids Res. 2002;30:2620–2627. doi: 10.1093/nar/gkf379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grela P, Krokowski D, Gordiyenko Y, Krowarsch D, Robinson CV, Otlewski J, Grankowski N, Tchorzewski M. Biophysical properties of the eukaryotic ribosomal stalk. Biochemistry. 2010;49:924–933. doi: 10.1021/bi901811s. [DOI] [PubMed] [Google Scholar]
- 29.Planta RJ, Mager WH. The list of cytoplasmic ribosomal proteins of Saccharomyces cerevisiae . Yeast. 1998;14:471–477. doi: 10.1002/(SICI)1097-0061(19980330)14:5<471::AID-YEA241>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 30.Tchorzewski M, Boguszewska A, Dukowski P, Grankowski N. Oligomerization properties of the acidic ribosomal P proteins from Saccharomyces cerevisiae: effect of P1A protein phosphorylation on the formation of the P1A–P2B hetero-complex. Biochim. Biophys. Acta. 2000;1499:63–73. doi: 10.1016/s0167-4889(00)00108-7. [DOI] [PubMed] [Google Scholar]
- 31.Krokowski D, Boguszewska A, Abramczyk D, Liljas A, Tchorzewski M, Grankowski N. Yeast ribosomal P0 protein has two separate binding sites for P1/P2 proteins. Mol. Microbiol. 2006;60:386–400. doi: 10.1111/j.1365-2958.2006.05117.x. [DOI] [PubMed] [Google Scholar]
- 32.Hagiya A, Naganuma T, Maki Y, Ohta J, Tohkairin Y, Shimizu T, Nomura T, Hachimori A, Uchiumi T. A mode of assembly of P0, P1, and P2 proteins at the GTPase-associated center in animal ribosome: in vitro analyses with P0 truncation mutants. J. Biol. Chem. 2005;280:39193–39199. doi: 10.1074/jbc.M506050200. [DOI] [PubMed] [Google Scholar]
- 33.Lalioti VS, Perez-Fernandez J, Remacha M, Ballesta JP. Characterization of interaction sites in the Saccharomyces cerevisiae ribosomal stalk components. Mol. Microbiol. 2002;46:719–729. doi: 10.1046/j.1365-2958.2002.03179.x. [DOI] [PubMed] [Google Scholar]
- 34.Tchorzewski M. The acidic ribosomal P proteins. Int. J. Biochem. Cell Biol. 2002;34:911–915. doi: 10.1016/s1357-2725(02)00012-2. [DOI] [PubMed] [Google Scholar]
- 35.Remacha M, Jimenez-Diaz A, Bermejo B, Rodriguez-Gabriel MA, Guarinos E, Ballesta JP. Ribosomal acidic phosphoproteins P1 and P2 are not required for cell viability but regulate the pattern of protein expression in Saccharomyces cerevisiae . Mol. Cell. Biol. 1995;15:4754–4762. doi: 10.1128/mcb.15.9.4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qiu D, Parada P, Marcos AG, Cardenas D, Remacha M, Ballesta JP. Different roles of P1 and P2 Saccharomyces cerevisiae ribosomal stalk proteins revealed by cross-linking. Mol. Microbiol. 2006;62:1191–1202. doi: 10.1111/j.1365-2958.2006.05445.x. [DOI] [PubMed] [Google Scholar]
- 37.Garcia-Marcos A, Sanchez SA, Parada P, Eid J, Jameson DM, Remacha M, Gratton E, Ballesta JP. Yeast ribosomal stalk heterogeneity in vivo shown by two-photon FCS and molecular brightness analysis. Biophys. J. 2008;94:2884–2890. doi: 10.1529/biophysj.107.121822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baba K, Tumuraya K, Tanaka I, Yao M, Uchiumi T. Molecular dissection of the silkworm ribosomal stalk complex: the role of multiple copies of the stalk proteins. Nucleic Acids Res. 2013;41:3635–3643. doi: 10.1093/nar/gkt044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Remacha M, Santos C, Bermejo B, Naranda T, Ballesta JP. Stable binding of the eukaryotic acidic phosphoproteins to the ribosome is not an absolute requirement for in vivo protein synthesis. J. Biol. Chem. 1992;267:12061–12067. [PubMed] [Google Scholar]
- 40.Tsurugi K, Ogata K. Evidence for the exchangeability of acidic ribosomal proteins on cytoplasmic ribosomes in regenerating rat liver. J. Biochem. 1985;98:1427–1431. doi: 10.1093/oxfordjournals.jbchem.a135410. [DOI] [PubMed] [Google Scholar]
- 41.Tumer NE, Li XP. Interaction of ricin and Shiga toxins with ribosomes. Curr. Top. Microbiol. Immunol. 2012;357:1–18. doi: 10.1007/82_2011_174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chiou JC, Li XP, Remacha M, Ballesta JP, Tumer NE. The ribosomal stalk is required for ribosome binding, depurination of the rRNA and cytotoxicity of ricin A chain in Saccharomyces cerevisiae . Mol. Microbiol. 2008;70:1441–1452. doi: 10.1111/j.1365-2958.2008.06492.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chan DS, Chu LO, Lee KM, Too PH, Ma KW, Sze KH, Zhu G, Shaw PC, Wong KB. Interaction between trichosanthin, a ribosome-inactivating protein, and the ribosomal stalk protein P2 by chemical shift perturbation and mutagenesis analyses. Nucleic Acids Res. 2007;35:1660–1672. doi: 10.1093/nar/gkm065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Juri Ayub M, Smulski CR, Ma KW, Lewin MJ, Shaw PC, Wong KB. The C-terminal end of P proteins mediates ribosome inactivation by trichosanthin but does not affect the pokeweed antiviral protein activity. Biochem. Biophys. Res. Commun. 2008;369:314–319. doi: 10.1016/j.bbrc.2008.01.170. [DOI] [PubMed] [Google Scholar]
- 45.Too PH, Ma MK, Mak AN, Wong YT, Tung CK, Zhu G, Au SW, Wong KB, Shaw PC. The C-terminal fragment of the ribosomal P protein complexed to trichosanthin reveals the interaction between the ribosome-inactivating protein and the ribosome. Nucleic Acids Res. 2009;37:602–610. doi: 10.1093/nar/gkn922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCluskey AJ, Poon GM, Bolewska-Pedyczak E, Srikumar T, Jeram SM, Raught B, Gariepy J. The catalytic subunit of Shiga-like toxin 1 interacts with ribosomal stalk proteins and is inhibited by their conserved C-terminal domain. J. Mol. Biol. 2008;378:375–386. doi: 10.1016/j.jmb.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 47.Yang Y, Mak AN, Shaw PC, Sze KH. Solution structure of an active mutant of maize ribosome-inactivating protein (MOD) and its interaction with the ribosomal stalk protein P2. J. Mol. Biol. 2010;395:897–907. doi: 10.1016/j.jmb.2009.10.051. [DOI] [PubMed] [Google Scholar]
- 48.Olsnes S, Pihl A. Ricin: a potent inhibitor of protein synthesis. FEBS Lett. 1972;20:327–329. doi: 10.1016/0014-5793(72)80098-x. [DOI] [PubMed] [Google Scholar]
- 49.Endo Y, Tsurugi K. The RNA N-glycosidase activity of ricin A-chain. The characteristics of the enzymatic activity of ricin A-chain with ribosomes and with rRNA. J. Biol. Chem. 1988;263:8735–8739. [PubMed] [Google Scholar]
- 50.Voorhees RM, Schmeing TM, Kelley AC, Ramakrishnan V. The mechanism for activation of GTP hydrolysis on the ribosome. Science. 2010;330:835–838. doi: 10.1126/science.1194460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi X, Khade PK, Sanbonmatsu KY, Joseph S. Functional role of the sarcin-ricin loop of the 23S rRNA in the elongation cycle of protein synthesis. J. Mol. Biol. 2012;419:125–138. doi: 10.1016/j.jmb.2012.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.May KL, Li XP, Martinez-Azorin F, Ballesta JP, Grela P, Tchorzewski M, Tumer NE. The P1/P2 proteins of the human ribosomal stalk are required for ribosome binding and depurination by ricin in human cells. FEBS J. 2012;279:3925–3936. doi: 10.1111/j.1742-4658.2012.08752.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li XP, Kahn PC, Kahn JN, Grela P, Tumer NE. Arginine residues on the opposite side of the active site stimulate the catalysis of ribosome depurination by ricin A chain by interacting with the P-protein stalk. J. Biol. Chem. 2013;288:30270–30284. doi: 10.1074/jbc.M113.510966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li XP, Baricevic M, Saidasan H, Tumer NE. Ribosome depurination is not sufficient for ricin-mediated cell death in Saccharomyces cerevisiae. Infect. Immun. 2007;75:417–428. doi: 10.1128/IAI.01295-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van der Zeijst BA, Kool AJ, Bloemers HP. Isolation of active ribosomal subunits from yeast. Eur. J. Biochem. 1972;30:15–25. doi: 10.1111/j.1432-1033.1972.tb02066.x. [DOI] [PubMed] [Google Scholar]
- 56.Vilella MD, Remacha M, Ortiz BL, Mendez E, Ballesta JP. Characterization of the yeast acidic ribosomal phosphoproteins using monoclonal antibodies. Proteins L44/L45 and L44’ have different functional roles. Eur. J. Biochem. 1991;196:407–414. doi: 10.1111/j.1432-1033.1991.tb15831.x. [DOI] [PubMed] [Google Scholar]
- 57.Zhang T, Lei J, Yang H, Xu K, Wang R, Zhang Z. An improved method for whole protein extraction from yeast Saccharomyces cerevisiae . Yeast. 2011;28:795–798. doi: 10.1002/yea.1905. [DOI] [PubMed] [Google Scholar]
- 58.O’Hara JM, Neal LM, McCarthy EA, Kasten-Jolly JA, Brey RN, 3rd, Mantis NJ. Folding domains within the ricin toxin A subunit as targets of protective antibodies. Vaccine. 2010;28:7035–7046. doi: 10.1016/j.vaccine.2010.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pierce M, Kahn JN, Chiou J, Tumer NE. Development of a quantitative RT-PCR assay to examine the kinetics of ribosome depurination by ribosome inactivating proteins using Saccharomyces cerevisiae as a model. RNA. 2011;17:201–210. doi: 10.1261/rna.2375411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li XP, Chiou JC, Remacha M, Ballesta JP, Tumer NE. A two-step binding model proposed for the electrostatic interactions of ricin a chain with ribosomes. Biochemistry. 2009;48:3853–3863. doi: 10.1021/bi802371h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yan Q, Li XP, Tumer NE. N-glycosylation does not affect the catalytic activity of ricin A chain but stimulates cytotoxicity by promoting its transport out of the endoplasmic reticulum. Traffic. 2012;13:1508–1521. doi: 10.1111/j.1600-0854.2012.01404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Melchior WB, Jr, Tolleson WH. A functional quantitative polymerase chain reaction assay for ricin, Shiga toxin, and related ribosome-inactivating proteins. Anal. Biochem. 2010;396:204–211. doi: 10.1016/j.ab.2009.09.024. [DOI] [PubMed] [Google Scholar]
- 63.Li XP, Grela P, Krokowski D, Tchorzewski M, Tumer NE. Pentameric organization of the ribosomal stalk accelerates recruitment of ricin A chain to the ribosome for depurination. J. Biol. Chem. 2010;285:41463–41471. doi: 10.1074/jbc.M110.171793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hudak KA, Dinman JD, Tumer NE. Pokeweed antiviral protein accesses ribosomes by binding to L3. J. Biol. Chem. 1999;274:3859–3864. doi: 10.1074/jbc.274.6.3859. [DOI] [PubMed] [Google Scholar]
- 65.Subramanian AR. Copies of proteins L7 and L12 and heterogeneity of the large subunit of Escherichia coli ribosome. J. Mol. Biol. 1975;95:1–8. doi: 10.1016/0022-2836(75)90330-7. [DOI] [PubMed] [Google Scholar]
- 66.Traut RR, Dey D, Bochkariov DE, Oleinikov AV, Jokhadze GG, Hamman B, Jameson D. Location and domain structure of Escherichia coli ribosomal protein L7/L12: site specific cysteine crosslinking and attachment of fluorescent probes. Biochem. Cell Biol. 1995;73:949–958. doi: 10.1139/o95-102. [DOI] [PubMed] [Google Scholar]
- 67.Mulder FA, Bouakaz L, Lundell A, Venkataramana M, Liljas A, Akke M, Sanyal S. Conformation and dynamics of ribosomal stalk protein L12 in solution and on the ribosome. Biochemistry. 2004;43:5930–5936. doi: 10.1021/bi0495331. [DOI] [PubMed] [Google Scholar]
- 68.Krokowski D, Tchorzewski M, Boguszewska A, Grankowski N. Acquisition of a stable structure by yeast ribosomal P0 protein requires binding of P1A–P2B complex: in vitro formation of the stalk structure. Biochim. Biophys. Acta. 2005;1724:59–70. doi: 10.1016/j.bbagen.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 69.Tchorzewski M, Krokowski D, Boguszewska A, Liljas A, Grankowski N. Structural characterization of yeast acidic ribosomal P proteins forming the P1A–P2B heterocomplex. Biochemistry. 2003;42:3399–3408. doi: 10.1021/bi0206006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.