Summary
In C. elegans, mRNA production is initially repressed in the embryonic germline by a protein unique to C. elegans germ cells, PIE-1. PIE-1 is degraded upon the birth of the germ cell precursors, Z2 and Z3. We have identified a chromatin-based mechanism that succeeds PIE-1 repression in these cells. A subset of nucleosomal histone modifications, methylated lysine 4 on histone H3 (H3meK4) and acetylated lysine 8 on histone H4 (H4acetylK8), are globally lost and the DNA appears more condensed. This coincides with PIE-1 degradation and requires that germline identity is not disrupted. Drosophila pole cell chromatin also lacks H3meK4, indicating that a unique chromatin architecture is a conserved feature of embryonic germ cells. Regulation of the germline-specific chromatin architecture requires functional nanos activity in both organisms. These results indicate that genome-wide repression via a nanos-regulated, germ cell-specific chromatin organization is a conserved feature of germline maintenance during embryogenesis.
Introduction
The germline is the generational reservoir of totipotent stem cells in most animals. How this lineage is established and maintained is of great interest to stem cell researchers, since mechanisms employed by germ cells likely overlap with those operating in somatic stem cells. During embryonic development, the germline in both C. elegans and Drosophila is maintained by repressive mechanisms, including transcriptional repression, that prevent the activation of somatic pathways in this tissue (Pirrotta, 2002; Seydoux and Schedl, 2001; Wylie, 1999). Transcriptional quiescence is also observed in the putative embryonic germline in Ascidians, indicating that this mode of repression is a widespread characteristic of the germ lineage (Tomioka et al., 2002). Disruption of transcriptional repression is detrimental to germ cells in flies and worms: the integrity of germline silencing mechanisms is thus a conserved imperative for propagation and survival of different species. Though both C. elegans and Drosophila employ transcriptional repression as one means to maintain germ cell identity, some aspects of this repression differ between these organisms. These differences, however, may reflect the different mechanisms of germ lineage separation in each species. In C. elegans, the early germline blastomeres (P blastomeres) each divide asymmetrically to give rise to both a somatic and a germline daughter through four cell divisions (Sulston et al., 1983; reviewed in Seydoux and Strome, 1999). The germline daughters from each division differ from their somatic counterparts in that they remain quiescent for RNA polymerase II (RNAPII) transcription while their somatic sisters become transcriptionally engaged (Seydoux and Fire, 1994; Seydoux et al., 1996). It is not until the symmetric division of the fourth P blastomere (P4) into Z2 and Z3 (Z2/Z3) that germline restriction occurs; i.e., the germ lineage separates completely from somatic lineages. PIE-1, a CCCH zinc finger protein that asymmetrically segregates into the P blastomeres in the early cell divisions, is required for transcriptional repression in the P blastomeres, possibly acting by inhibiting transcription elongation (Mello et al., 1996; Seydoux et al., 1996; Zhang et al., 2003). In pie-1 mutants, P2 prematurely activates transcription, and its descendents duplicate the somatic lineages of P2’s somatic sister, EMS. This illustrates that (1) transcriptional repression is necessary to retain germline fate, and (2) the default state in the absence of repression is activation of preprimed somatic pathways (Mello et al., 1992). PIE-1-dependent repression of germ cells lasts until after gastrulation and division of the P4 blastomere, at which time PIE-1 is degraded (Mello et al., 1996). Coincident with PIE-1 degradation, an RNAPII C-terminal domain (RNAPII CTD) phospho-epitope that correlates with transcription elongation, phospho-Ser2, appears in Z2/Z3 nuclei (Seydoux and Dunn, 1997). Despite the appearance of this epitope, however, few mRNAs are known to be produced (e.g., pgl-1 and nos-2 mRNAs; Kawasaki et al., 1998; Subramaniam and Seydoux, 1999), and proliferation ceases in these cells for the rest of embryogenesis.
In contrast to the C. elegans P lineage, the pole cells of Drosophila are restricted to a germ cell fate prior to their cellularization (Technau and Campos-Ortega, 1986; Underwood et al., 1980). Like C. elegans germ cells, the pole cells of Drosophila are also initially transcriptionally quiescent (Seydoux and Dunn, 1997; Zalokar, 1976). Two of the genes that have been shown to be required for the maintenance of transcriptional quiescence in the early pole cells are nanos (nos) and pumilio (pum). Mutations in nos and pum not only disrupt posterior patterning (the phenotype that resulted in their original identification) but also result in the premature activation of transcription in germ cells (Asaoka-Taguchi et al., 1999; Deshpande et al., 1999). In addition to nos and pum, mutations in germ cell-less (gcl) also cause premature transcriptional activation in pole cells, in this case at the time when pole buds are first formed (Leatherman et al., 2002).
Some aspects of these Drosophila mechanisms are conserved in C. elegans (for review, see Leatherman and Jongens, 2003). For example, knockdown of two C. elegans nanos-like genes, nos-1 and nos-2,causes phenotypes in the germ cell precursors Z2/Z3 similar to those observed in nos− pole cells, including premature proliferation and migration defects (Subramaniam and Seydoux, 1999). C. elegans pumilio-like puf genes are also required for germ cell function (Crittenden et al., 2002; Subramaniam and Seydoux, 1999, 2003). As with their Drosophila counterparts, the roles of the C. elegans nos and puf genes appear to be posttranscriptional. A global silencing mechanism that acts at the level of transcription and is conserved in both flies and worms has thus remained to be identified.
Global regulation of gene expression involves the regulation of higher-order chromatin assembly. Nucleosomal core histones have N-terminal tails, which can be modified in various ways including phosphorylation, ubiquination, acetylation, and methylation. It is hypothesized that these modifications may create a combinatorial “histone code” that directs transcriptional activity or repression (Strahl and Allis, 2000). Modifications that correlate with transcriptionally competent chromatin include acetylation of both histones H3 and H4 and methylation of H3 at lysine 4, lysine 36, and lysine 79 (H3meK4, H3meK36, and H3meK79, respectively). H3meK4 in particular is a highly conserved modification that, in virtually all cases, is found at high levels in euchromatin and at low or undetectable levels in heterochromatin (Kouzarides, 2002; Lachner and Jenuwein, 2002). H3meK4 also strongly correlates with conserved modes of global transcriptional regulation. For example, H3meK4 is excluded from the inactivated X chromosome in both mammalian dosage compensation and C. elegans spermatogenesis (Boggs et al., 2002; Kelly et al., 2002). Conversely, H3meK4 globally accumulates during whole genome activation in Tetrahymena (Strahl et al., 1999). In contrast to histone acetylation, for which deacetylases have been described, histone methylation is currently thought to be effectively irreversible, and its loss in chromatin requires either proteolytic cleavage of the N-terminal tails or histone replacement within the nucleosome (e.g., Kouzarides, 2002).
The mechanism of PIE-1-dependent transcriptional repression is not understood. We were interested to determine if PIE-1-mediated repression in C. elegans involves specific modes of chromatin organization and whether such mechanisms persist after the degradation of PIE-1. Surprisingly, we observed no obvious difference between the histone modification patterns in the P lineage and somatic blastomeres when the PIE-1 mode of transcriptional repression is engaged. In contrast, we observed a genome-wide disappearance of specific histone modifications (H3meK4, H4acetylK8) and a unique DNA compaction, specifically in Z2/Z3 after PIE-1 disappears. This remodeling event does not occur in a mutant in which PIE-1 degradation is defective. Drosophila pole cells similarly lack H3meK4, and nanos function is required for maintenance of the unique chromatin status in both species. These results indicate that chromatin-based mechanisms and their regulation are conserved guardians of germ cell totipotency among highly diverged species.
Results
Histone Modifications in P Blastomeres Are Consistent with an Open Chromatin Architecture
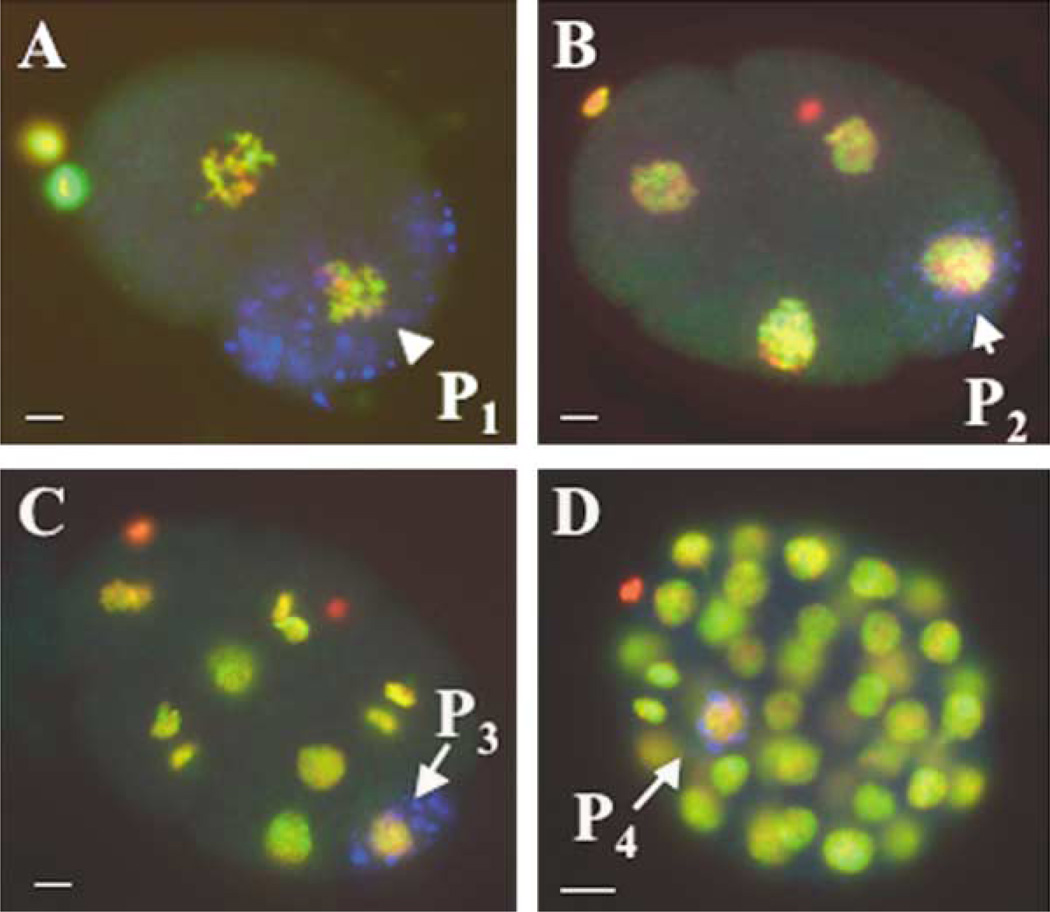
We tested whether PIE-1-mediated transcriptional repression impacts on chromatin assembly in the early germline (P) blastomeres. Early-stage embryos were fixed and stained with antibodies specific for individual modifications on histones H3 and H4, including dimethylated lysine 4 and acetylated lysines 9 and 14 of H3 (H3meK4, H3diacetyl) and acetylated lysines 5, 8, or 16 of H4 (H4acetylK5, H4acetylK8, H4acetylK16). We compared nuclear staining in the germ lineage to that in somatic lineages at different stages of embryogenesis. The germline blastomeres were identified by co-staining with an antibody (anti-PGL-1) that recognizes P granules, which are RNA/protein complexes specific to germ cell cytoplasm (for review, see Seydoux and Strome, 1999). The germ lineage is transcriptionally repressed at these stages, so we expected to see staining for markers of transcriptional competence only in the somatic lineages. Surprisingly, we saw no significant differences in staining between the somatic precursor cells and the P lineage using any of the modification-specific antibodies employed (P1–P4; Figures 1A–1D, and data not shown). DAPI staining also showed no significant differences in overall structure or condensation between somatic nuclei and the early P blastomeres, although some slight condensation was observed in later stages of P4 (e.g., Figure 3A below). The PIE-1 protein is active in the P cells at these stages (Figure 3; Seydoux et al., 1996; Mello et al., 1996), indicating that PIE-1-mediated global transcriptional repression does not involve a dramatic alteration of chromatin architecture. PIE-1 is therefore able to globally prevent transcription in spite of a permissive chromatin status.
Figure 1.
H3meK4 Is Present in All Nuclei of the Early Embryo
Fixed, whole-mount early-stage embryos were stained with anti-H3meK4 (green) and anti-PGL-1 (blue; marking the germ lineage) and counterstained with DAPI (red). H3meK4 is equally present in both AB and P1 (A; arrow) of 2-cell embryos, and persists in the germline blastomere chromatin through P2 (B), P3 (C), and P4 (D). Scale bar equals 5 µm.
Figure 3.
Loss of H3meK4 in Z2/Z3 Coincides with Loss of PIE-1
Fixed whole-mount embryos at various stages (columns A–C) were stained with DAPI (top) and costained with antibodies against H3dimethylK4 and PGL-1 (both in middle row) and PIE-1 (bottom).
Top row: DAPI staining of P4 blastomere in a 47-cell embryo (A, box); P4 dividing in a 90-cell embryo (B, box); and Z2/Z3 in a 200-cell embryo (C, box). Germline cells are enlarged in insets.
Middle row: Same embryos as in top panels costained with anti-H3meK4 (nuclear) and anti-PGL-1 (bright punctate staining around germ nuclei). H3meK4 staining is initially observed in P4, decreases as P4 divides, and is absent from Z2/Z3 (arrows). H3meK4 staining intensity is normally bright in condensed, mitotic DNA (arrowheads, middle and top panels of column B), but this is not readily evident in the dividing P4. Note that both primary antibodies in the middle row were raised in rabbits, so each epitope is only distinguishable by nuclear (H3meK4) versus perinuclear (PGL-1) when visualized with a common secondary antibody.
Bottom row: Same embryos as in top panels stained with anti-PIE-1. PIE-1 is also seen in P4 at 47 cells, decreases as P4 divides at ~90 cells, and is absent from Z2/Z3 at 200 cells. Scale bar in all panels equals 5 µm.
Specific Chromatin Alterations Occur at Germline Restriction in C. elegans
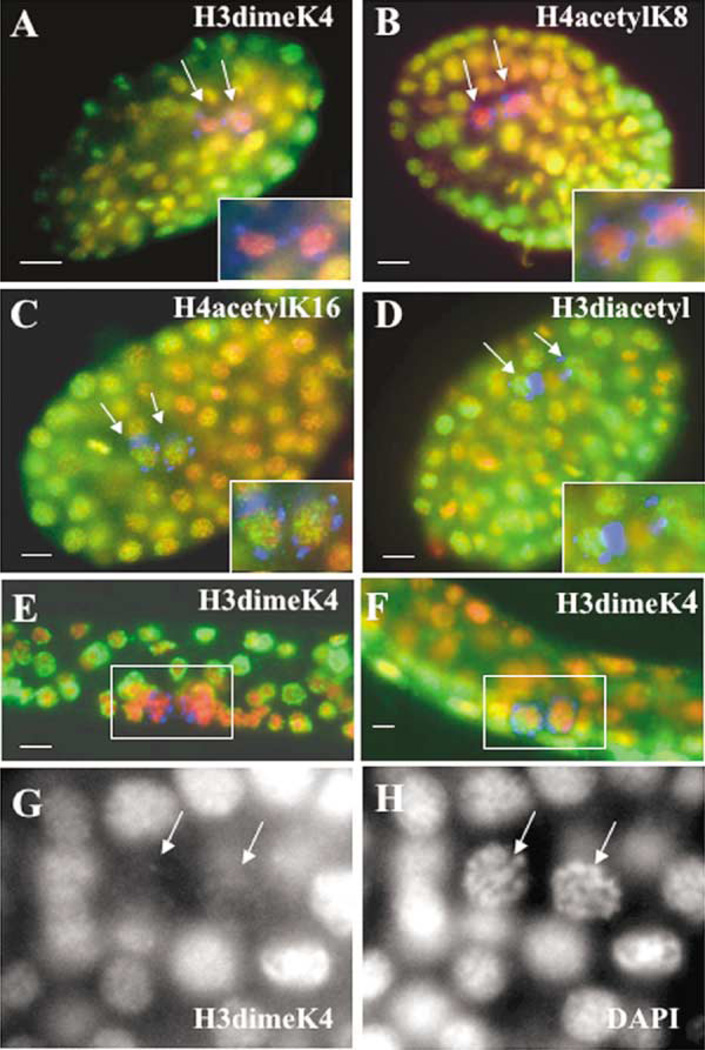
A difference in chromatin organization between the germ lineage and its somatic counterparts became strikingly apparent after the division of P4. H3meK4 was observed to specifically and globally disappear from the nuclei of Z2/Z3 (Figures 2A and 2G). Only one other modification, H4acetylK8, followed the same pattern as H3meK4 (Figure 2B). In contrast, H3diacetyl, H4acetylK16, and H4acetylK5 staining remained high in all nuclei in the embryo (Figures 2C and 2D, and data not shown). A very specific chromatin rearrangement therefore occurs in Z2/Z3 slightly before or after their birth.
Figure 2. Specific Chromatin Reorganization Occurs in the Primordial Germ Cells Z2/Z3.
Late-stage (100–200 cells) and L1 larvae were stained with antibodies to specific histone modifications (green), anti-PGL-1 (blue), and counterstained with DAPI (red). The primordial germ cells Z2/Z3 (arrows) are enlarged in insets. Z2/Z3 are the only cells in the embryo that do not appreciably stain with anti-H3meK4 (A) or anti-H4acet-ylK8 (B), whereas anti-H4acetylK16 and anti-H3diacetyl (green) stain all cells (C and D, respectively). H3meK4 remains absent from Z2/ Z3 throughout embryogenesis and in starved L1 larvae (E), but returns to Z2/Z3 in fed L1 larvae (F). Note that there are still only two germ cells in (F, inset). The absence of H3meK4 in Z2/Z3 (G) correlates with a striking structural change visible by DAPI (H) when compared to surrounding somatic nuclei. Scale bar equals 5 µm.
Larval and adult germ cell chromatin, with the exception of the inactive X chromosome, is enriched in H3meK4 (Kelly et al., 2002; Reuben and Lin, 2002). We next examined the timing of the return of this modification in postembryonic germ cells. Z2/Z3 do not begin proliferation until after L1 larva begin feeding (Seydoux and Schedl, 2001); postembryonic cell divisions are inhibited in larvae hatched in the absence of food. We stained L1 larva, hatched in either the presence or absence of food, with the H3meK4 antibody. H3meK4 was not detected in Z2/Z3 in the starved hatchlings (Figure 2E) but was readily visible in Z2/Z3 of the fed larva prior to cell division (Figure 2F), indicating that the return of this modification coincides with a presumed need to globally commence transcription in advance of proliferation.
We next examined potential structural consequences of the loss of H3meK4 in Z2/Z3. Although DNA compaction is sensitive to fixation conditions, comparison with surrounding somatic nuclei in the same embryo consistently revealed a distinct change in overall structure specifically in Z2/Z3 (Figure 2H). This difference was more obvious using a low-concentration, short-exposure formaldehyde fixation procedure; standard methanol/acetone fixation caused a compaction of all nuclei. Increased compaction of Z2/Z3 DNA relative to DNA in surrounding nuclei, however, was still usually observed even with the more stringent fixation protocol (not shown). A comparable difference was not observed in the early P blastomeres (not shown), although some increased condensation was often apparent in P4 after gastrulation and prior to cell division (e.g., Figure 3A).
PIE-1 Degradation and Chromatin Remodeling Are Temporally Linked
The division of P4 to Z2/Z3 occurs after gastrulation around the 70- to 100-cell stage, which is coincident with the timing of PIE-1 depletion. To test if the degradation of PIE-1 is also coincident with loss of H3meK4, we costained embryos with antibodies for H3meK4 and PIE-1. H3meK4 and PIE-1 were both detected in the P lineage during the first four cell divisions (Figure 3, Column A, and data not shown). However, as P4 divides, a substantial decrease was observed in PIE-1 with some decreased staining for H3meK4 also observed (Figure 3, Column B). Once Z2/Z3 have formed, neither H3meK4 nor PIE-1 staining was observed (Figure 3, Column C). This demonstrates that chromatin remodeling in, and the loss of PIE-1 protein from, Z2/Z3 are temporally related.
During the course of the studies, we found that a maternal-effect lethal mutation, emb-4(hc60), exhibited defects in the normal degradation of PIE-1. Though it has been previously shown that PIE-1 levels in emb-4 mutant embryos are normal until at least the 28-cell stage (Tenenhaus et al., 1998), further analysis of emb-4 embryos revealed that PIE-1 is inappropriately stabilized in Z2/Z3 in later-stage embryos (Figure 4A). Since PIE-1 degradation and loss of H3meK4 are temporally related events in wild-type embryos, we wanted to determine if H3meK4 disappearance was similarly defective in emb-4 embryos. We therefore stained emb-4 embryos with antibodies to H3meK4 and found that this histone modification, like PIE-1, inappropriately persisted in Z2/ Z3 of older embryos (Figures 4C and 4D). A persistence of PIE-1 protein therefore correlates with a failure to activate the chromatin remodeling that occurs in Z2/Z3. Preliminary results suggest that H4acetylK8 loss in Z2/ Z3 occurs normally in emb-4 embryos, suggesting that some aspects of the specific remodeling events in Z2/ Z3 are unaffected by the retention of PIE-1 (P. Checchi, C.E.S., and W.G.K., unpublished results). The condensed DNA structure normally observed in Z2/Z3, however, was not apparent in these nuclei, indicating that loss of H3meK4 is more closely correlated with DNA condensation than is H4 acetylation (Figure 4E, compare to Figure 2H).
Figure 4.
Mutations Affecting Loss of H3meK4 Regulation in Z2/Z3
emb-4(A, C-E), pie-1 (F), or mex-1 (G) mutant embryos of 100 cells or more from homozygous mutant mothers were stained with either anti-PIE-1 (A, green) or anti-H3meK4 (B-D, F–I; green) and anti-PGL-1 (B–D, F–I; blue), and counterstained with DAPI (red in all panels). In emb-4 mutants, PIE-1 is abnormally detected in Z2/Z3 in later embryos, such as the ~155-cell embryo shown (A). H3meK4, normally absent at this stage (B), also persists in Z2/Z3 (C and D). The DNA compaction normally observed in these cells is not evident (E). Loss of H3meK4 does not occur in the somatically transformed somatic descendants of P2 in pie-1 mutant embryos (F) and mex-1 mutant embryos (G). H3meK4 is also observed in Z2/Z3 in nos-1(gv5) embryos exposed to nos-2 dsRNA (H and I). A 150-cell (H) and a late-stage embryo (2-fold; I) are shown. Insets in (H) show Z2/Z3 separately (arrowheads), as they were in different focal planes. Scale bar equals 5 µm.
Germ Cell Identity Is Required for Chromatin Remodeling at Germline Restriction
The emb-4 mutation illustrates a link between loss of PIE-1 activity and the loss of H3meK4 in Z2/Z3. We next analyzed mutations that disrupt PIE-1 activity in the earlier P blastomeres. pie-1 mutant embryos prematurely activate transcription in P2, causing the germline to be transformed into a somatic lineage identical to the descendants of P2’s sister, EMS (Mello et al., 1992). A similar, but later, transformation occurs in mex-1 mutants. MEX-1 is required for restricting PIE-1 to the germ lineage when P3 divides (Guedes and Priess, 1997). In mex-1 mutants, transcriptional repression is relieved and P3 gives rise to two somatic daughters (Mello et al., 1992; Schnabel et al., 1996). We stained embryos from pie-1 and mex-1 mutants with the H3meK4 antibody and found that neither lose this modification in the late descendants of P2 or P3, cells that would normally have become Z2/Z3 (Figures 4F and 4G). The loss of H3meK4 in the descendants of P2 and P3 therefore requires that germ cell identity be maintained in these cells and is not solely a temporal event. It is important to note, however, that a premature remodeling of germline chromatin in the P blastomeres lacking PIE-1 activity was not observed (not shown).
The MES and MEP-1 Complexes Are Not Required for Z2/Z3 Chromatin Remodeling
The relationship between the chromatin remodeling in Z2/Z3 and the loss of PIE-1 suggested PIE-1 may be directly or indirectly inhibiting the action of a germline-specific chromatin modifier. A variety of candidate genes were therefore tested for their potential roles in the regulation of this process.
MEP-1, identified in two-hybrid screens as a PIE-1-interacting protein, exists in a complex with two C. elegans orthologs of proteins that function in the mammalian NuRD nucleosome remodeling complex, HDA-1 and LET-418 (Unhavaithaya et al., 2002). The worm MEP-1 complex is required to prevent some aspects of germ cell differentiation in somatic cells, and its histone deacetylase activity can be inhibited in vitro by PIE-1 (Unhavaithaya et al., 2002). Its role in the germline, however, is unclear. Defects in any individual MEP-1 complex component result in similar phenotypes, suggesting the complex must be intact for full function (Unhavaithaya et al., 2002). We individually disrupted function of mep-1, hda-1, and let-418 by injecting dsRNA specific for each, and stained the embryos from injected mothers with either the H3meK4 or the H4acetylK8 antibodies. The extent of knockdown was determined by scoring injected brood siblings for the predicted phenotypes in parallel to those prepared for antibody analysis. No significant alterations in histone modification patterns were observed after depletion of any of the three genes, although siblings of the stained embryos exhibited high levels of phenotypic larval arrest (Table 1). The MEP-1 complex therefore either does not play a major role in regulating chromatin remodeling in Z2/Z3, or the degree of knockdown, while sufficient for phenotypic arrest, was insufficient to completely disable chromatin remodeling.
Table 1.
Strains Scored for H3meK4 in Z2/Z3
| Genotype | Phenotypic Analysis | % Embryos with H3meK4 Staining in Z2/Z3 |
|---|---|---|
| N2 | N/A | 7 (3/46) |
| mes-2(bn11) | N/A | 7 (2/30) |
| mes-3(bn35) | N/A | 0 (0/26) |
| mes-4(bn67) | N/A | 8 (2/25) |
| mes-6(bn66) | N/A | 10 (2/20) |
| mep-1(RNAi)a | 0/366 lived to adult | 5 (1/22) |
| nos-1(gv5) | N/A | 0 (0/22) |
| nos-1(gv5); nos-2(RNAi) | 1215/2060 sterile (59%) | 62 (38/61) |
Mutant and RNAi analyses for defects in Z2/Z3 chromatin arrangement. Embryos from either homozygous mothers (mes mutants) or mothers injected with dsRNA targeting-specific genes (nos-2, mep-1, hda-1, and let-418) were fixed and costained with antibodies against H3meK4 and PGL-1. Late-stage embryos exhibiting two cells staining for PGL-1 (Z2/Z3) were analyzed by optical serial sectioning for detectable levels of costaining with H3meK4 in these cells. Parallel broods from dsRNA-injected mothers were scored for penetrance of phenotypes previously reported for RNAi of each gene.
Similar results seen with both hda-1(RNAi) and let-418(RNAi).
MES-2 and MES-6 are members of the Polycomb group of transcriptional repressors that, along with their interacting protein MES-3, are involved in germline chromatin assembly (Fong et al., 2002). MES-4, like MES-2, contains a SET domain, which is a conserved motif found in histone methyltransferases (Jenuwein, 2001; Xu et al., 2001). We therefore stained mes mutant embryos for defects in normal chromatin regulation in the embryonic germline: no defects were observed in any of the mes mutants (Table 1).
NANOS Activity Is Required to Maintain the Unique Chromatin Architecture in the Germline
The C. elegans nanos homologs nos-1 and nos-2 play important overlapping roles in worm primordial germ cell development (Subramaniam and Seydoux, 1999). Knockdown of both NOS-1 and NOS-2 activities results in sterile animals, a population of which exhibit germ cells that begin proliferation prematurely (Subramaniam and Seydoux, 1999). Loss of NOS-2 alone can also result in ectopic localization of the primordial germ cells (Subramaniam and Seydoux, 1999). We therefore examined H3meK4 levels in Z2/Z3 chromatin of embryos from nos-1(gv5);nos-2(RNAi) hermaphrodites. Animals homozygous for the nos-1(gv5) are viable and fertile; RNAi of nos-2 in these animals results in germ cell defects (Subramaniam and Seydoux, 1999). nos-1 mutant animals were therefore injected with nos-2 dsRNA, and parallel broods were assessed for sterility as adults or stained for H3meK4 in Z2/Z3. In our hands, 59% of the offspring of the injected animals grew up to be completely sterile adults (Table 1). An almost identical percentage of parallel brood embryos exhibited inappropriate staining for H3meK4 in one or both of the primordial germ cells, Z2/Z3 (Figures 4H and 4I). The C. elegans NANOS homologs, NOS-1 and NOS-2, therefore play important roles in either establishing and/or maintaining the chromatin-based mode of transcriptional repression in Z2/Z3.
A Similar Chromatin Architecture Is Conserved in Drosophila Pole Cells
While the progenitors of the soma and germline are also separated from each other during early embryogenesis in Drosophila, the processes that give rise to the physical segregation of these two distinct cell types in flies are quite different from those in worms. In light of these differences, it was of interest to determine if distinct chromatin states are also established in the somatic and germline nuclei of fly embryos. Staining of early Drosophila embryos with the H3meK4 antibody revealed that there is little detectable H3meK4 in any nuclei during the rapid synchronous nuclear divisions in the center of the embryo. Similarly, we did not detect H3meK4 in the nuclei of newly formed pole cells, or in somatic nuclei when they first migrate to the periphery of the embryo (not shown). H3meK4 was first detected in somatic nuclei between nuclear division cycles 12 and 13, after the nuclei around the periphery of the embryo have already undergone several rounds of nuclear division (Figure 5A). This change in the methylation status of histone H3 is coincident with, or slightly precedes, the time when transcription is broadly upregulated in the somatic nuclei of syncytial blastoderm embryos (Anderson and Lengyel, 1979; Edgar and Schubiger, 1986). High levels of H3meK4 are maintained through the cellular blastoderm stage, with all somatic nuclei staining with the H3meK4 antibody.
Figure 5.
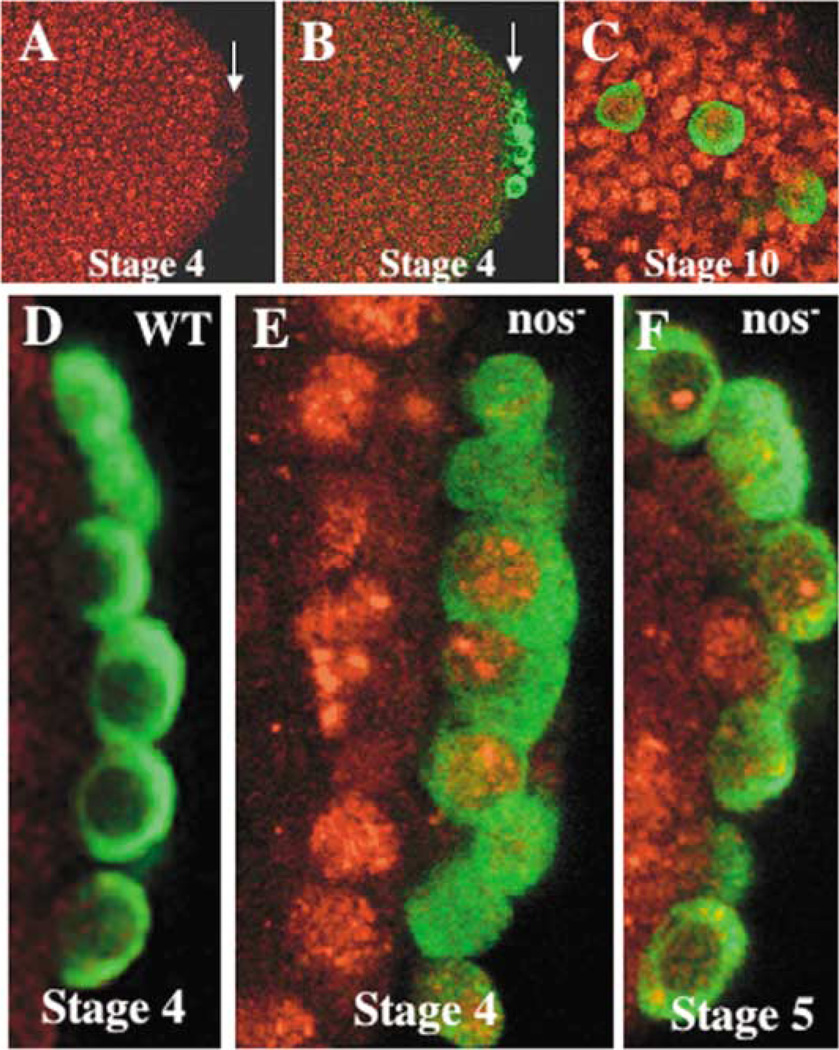
H3meK4 Is Absent in Early Wild-Type Drosophila Pole Cells but Present in nos Mutants
Wild-type embryos (0–8 hr) and nos− embryos (0–6 hr) were fixed and coimmunostained with anti-H3meK4 (red) and anti-Vasa (green) antibodies.
(A) Syncitial blastoderm embryo probed with anti-H3meK4. Arrow points to pole cell nuclei.
(B) Same embryo as in (A) showing anti-H3meK4 and anti-Vasa labeling.
(C) Stage 10 embryo showing the presence of H3meK4 signal in the germ cells labeled with anti-Vasa.
(D) Wild-type control pole cells in a stage 4, cellular blastoderm embryo at higher magnification stained with anti-H3meK4 and anti-Vasa.
(E) nos− pole cells at stage 4 showing H3meK4 and Vasa.
(F) nos− pole cells at stage 5 showing H3meK4 and Vasa.
In contrast to the somatic nuclei, little if any H3meK4 was detected in the pole cells (marked with anti-Vasa-specific antibody) of either syncytial blastoderm (see Figures 5A and 5B) or cellular blastoderm (not shown) embryos. Staining of embryos at later stages revealed that H3meK4 could still not be detected in germ cells in early gastrulation stage through midgut invagination (not shown). However, once the germ cells traversed the midgut wall and began migrating away from the hindgut toward the somatic gonadal precursor cells, H3meK4-specific signal became readily evident (Figure 5C). Therefore, from the time of their formation until stage 9 of embryogenesis, there is little if any H3meK4 in the germ cells’ chromatin. H3meK4 becomes readily detectable in the germ cells, however, coincident with the onset of transcription at stage 9/10 (Zalokar, 1976; Van Doren et al., 1998). The accumulation of H3meK4 in the germ cells, as in the soma, correlates temporally with transcriptional activation in these cells.
We also stained embryos for epitopes corresponding to H4acetylK5, H4acetylK8, H4acetylK12, H3diacetyl, and H3dimeK36. Only the antibodies specific for H3diacetyl and H4acetylK5 exhibited nuclear staining above background levels in early embryonic stages, and the appearance of these epitopes was temporally identical to that observed for H3meK4. In contrast to the H3meK4 modification, however, no differences were observed in the levels of these modifications in pole cell nuclei compared to somatic nuclei (data not shown). This indicates that a global lack of H3meK4, in combination with the presence of other modifications that correlate with transcriptional competence, is a conserved feature of chromatin structure in transcriptionally inert germ cell nuclei.
nos Is Required to Prevent Activation of Histone H3 K4 Methylation
Previous studies have demonstrated that nos function is required for either the establishment or maintenance of transcriptional quiescence in newly formed Drosophila pole cells (Deshpande et al., 1999). In light of our results showing a premature appearance of H3meK4 in C. elegans germ cells in the absence of nos activity, an obvious question is whether nos is also required to block this methylation in the early fly germline. To answer this question, fly embryos produced by nos mutant mothers were probed with H3meK4- and Vasa-specific antibodies. As can be seen in Figure 5E, H3meK4 can be detected in the pole cells of stage 4 nos− embryos at nuclear division cycle 14. In contrast, H3meK4 is completely absent from pole cell chromatin in wild-type embryos at this stage (Figure 5D). Moreover, H3meK4-specific signal was also detected in pole cells in stage 5 embryos that have just initiated gastrulation (Figure 5F). It is interesting to note that not all nos pole cells (27/ 42) display H3meK4-specific staining. This is consistent with previous findings that transcription is not activated in every pole cell of nos− blastoderm stage embryos and supports the suggestion that additional factors contribute to the establishment/maintenance of transcriptional quiescence in fly pole cells (Deshpande et al., 1999; Leatherman and Jongens, 2003).
Inactive Drosophila Pole Cells Are Enriched in Histone H3 Lysine 9 Methylation
While methylation of lysine 4 of histone H3 is a conserved mark of transcriptionally competent or active chromatin, methylation of lysine 9 of histone H3 (H3meK9) is a highly conserved modification that is enriched in silenced genomic regions, such as either facultative or constitutive heterochromatin (Lachner and Jenuwein, 2002). Consequently, it was of interest to determine if the transcriptionally quiescent pole cell nuclei are enriched in H3meK9 as compared to the transcriptionally active somatic nuclei. Drosophila embryos were probed with antibodies against H3meK9 (Figure 6, red) and Vasa to mark the pole cells (Figures 6B and 6C, green). In contrast to H3meK4, cleavage stage Drosophila embryonic nuclei exhibited readily detectable H3meK9 even prior to their migration. The modification was also detected in somatic nuclei after nuclear migration, where it appeared enriched in nuclear regions adjacent to the periphery of the embryo that correspond to telomeric and centromeric heterochromatin in the Rabl configuration (Csink and Henikoff, 1998). These regions, conversely, were observed to lack H3meK4 (not shown). H3meK9 is also detected in pole cells as soon as they are formed, with the level of H3meK9 antibody staining in the pole cell nuclei being considerably higher than in the somatic nuclei (Figure 6B). The observed enrichment of H3meK9 in the pole cell nuclei coincided with loss of the mitosis-specific modification, H3 phospho-Ser10, suggesting that a specific remodeling of the pole cell chromatin accompanies their exit from the cell cycle (data not shown).
Figure 6.
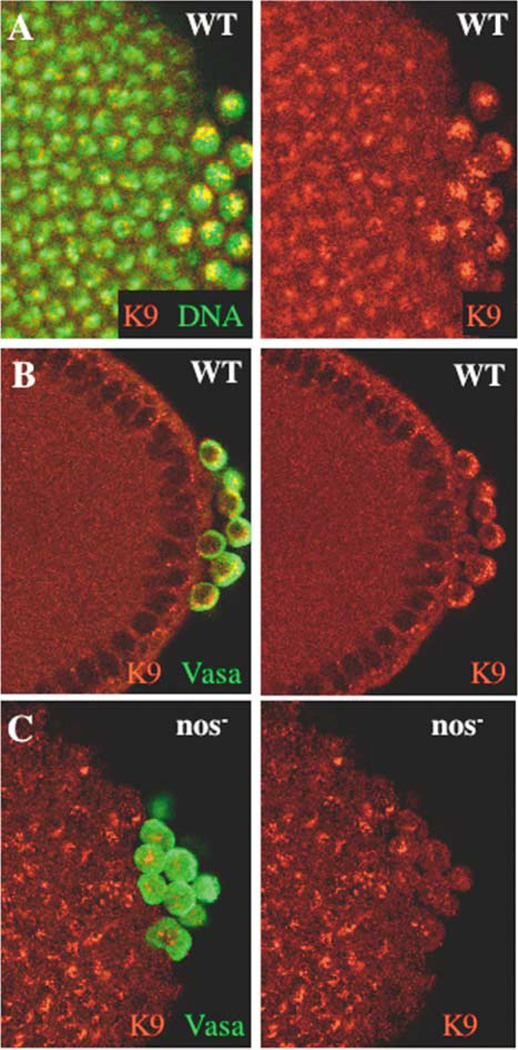
H3meK9 Is Enriched in Wild-Type Pole Cells
Wild-type and nos− embryos (0- to 6-hours-old) were fixed and costained with nuclear dye Hoechst (green) and anti-H3meK9 (red; A) or coimmunostained with anti-H3meK9 (red) and anti-Vasa (green) antibodies (B and C).
(A) Wild-type, cycle 13 embryo colabeled with Hoechst (green) and anti-H3meK9 (red). The left panel shows both labels; the right panel is H3meK9 alone.
(B) Wild-type, cycle 13 embryo costained for H3meK9 (red) and Vasa (green; left panel) and H3meK9 alone (right panel).
(C) nosBN, cycle 13 embryo costained for H3meK9 (red) and Vasa (green; left panel) and H3meK9 alone (right panel).
NANOS Is Required to Promote Methylation of Histone H3 Lysine 9
Since depletion of maternal nos activity resulted in a premature increased accumulation of H3meK4 in early pole cells, we investigated whether nos− pole cells exhibit a parallel reduction in H3meK9. Indeed, nos− pole cell nuclei do not exhibit the characteristic enrichment for H3meK9-specific signal seen in wild-type embryos. Furthermore, more than 50% of the pole cells examined show considerably reduced staining specific for nuclear H3meK9 compared to the neighboring somatic nuclei (Figure 6C). The known dichotomous roles for H3meK4 and H3meK9 in the regulation of transcriptional competence are therefore conserved in pole cells, and their relative abundances in pole cell chromatin are responsive to nos activity.
Discussion
Conserved Modes of Germline Silencing
Identifying the mechanisms that guide the separation of somatic and germ lineages is one of the oldest pursuits in developmental biology. How the totipotent germline is maintained during development has become increasingly relevant to modern science in the search for conserved mechanisms guarding stem cell identity. Our data provides new evidence for at least two modes of germline-specific repression that guard the germline during C. elegans embryogenesis, one of which is conserved in Drosophila. In the earliest phase, maternal PIE-1 activity in the germline P blastomeres prevents mRNA production through a mechanism that does not involve substantial, germline-specific alterations in chromatin architecture. After the degradation of PIE-1, however, a second mode involving a dramatic and specific remodeling of chromatin arises in the germ cell precursors, Z2/Z3. Whereas the former mode appears to be unique to C. elegans, our results illustrate that the latter mode, which bears the hallmarks of direct transcriptional repression provided by a specific mode of chromatin organization, is a conserved feature of germ cell maintenance.
In both worms and flies, lineage restriction to germ cell fate is marked by a global absence of H3meK4 and a more condensed chromatin structure. In the case of C. elegans, the absence of H3meK4 is the result of a specific depletion; in Drosophila the absence arises from preventing H3meK4 accumulation. This absence is maintained in both organisms until zygotic activation of the genome in germ cells. Premature activation, marked by premature accumulation of H3meK4, results in the loss of the germ cell lineage in both worms and flies. In both species, the lack of H3meK4 occurs in the presence of high levels of other histone modifications that often correlate with an open chromatin configuration, indicating a “dominance” of H3meK4 as an indicator of transcriptional activity.
The conserved correlation between H3meK4 presence and absence and global activation and silencing, respectively, is striking. The inactive X chromosome in mammals is globally depleted of H3meK4, as are silenced regions of the genome in fission yeast (reviewed in Lachner and Jenuwein, 2002). In Tetrahymena, the germinal micronucleus is transcriptionally silent and H3meK4 is missing. Sexual conjugation causes transformation of the micronucleus in to a transcriptionally active somatic macronucleus, which becomes enriched in H3meK4 (Strahl et al., 1999). Thus, even in protozoans, genome-wide diminishment of H3meK4 is a property of the “germline” and its accumulation accompanies “somatic” activation.
The lack of H3meK4 in fly pole cells is mirrored by enrichment for H3meK9. This is not observed in Z2/Z3 in C. elegans. The reason for this difference is not understood but may reflect a more substantial role for this modification in genome regulation in Drosophila, which has a more highly repetitive genome than that of C. elegans, and consequently more abundant classically defined heterochromatin. Indeed, in contrast to Drosophila, H3meK9 is only cytologically enriched at telomeres in C. elegans embryos and on the highly condensed, unpaired X chromosome during male meiosis (Kelly et al., 2002; Reuben and Lin, 2002). In both examples, the enrichment for H3meK9 is always mirrored by the absence of H3meK4.
A Conserved Role for nanos
Posttranscriptional regulation plays a major role in maintaining the embryonic germline in both worms and flies. The conserved classes of proteins involved include the pumilio (Pufs) and nanos families (Leatherman and Jongens, 2003). nanos activities are similarly required for proper migration of the primordial germ cells, maintenance of their mitotic quiescence, proper proliferation of the germline after hatching, and, as we show in this report, the regulation of chromatin organization in Z2/ Z3 in both species (Subramaniam and Seydoux, 1999). In addition, nanos has also been shown by several labs to be required to maintain transcriptional quiescence in pole cells (Asaoka et al., 1998; Asaoka-Taguchi, et al., 1999; Deshpande et al., 1999). Similar nanos functions may also be required in mammals, where two of three nanos homologs are required for fertility (Tsuda et al., 2003). The phenotype of nanos-3 null mice is consistent with specification of primordial germ cells occurring normally, followed by an inability to maintain germ cell identity during migration. This phenotype is strikingly similar to those observed in nanos(−) worms and flies.
nanos function is essential to maintain embryonic germline repression prior to normal activation of proliferation, presumably through its characterized roles in posttranscriptional regulation. Only a few direct targets of nanos regulation in germ cells have been identified, including maternal cyclin mRNAs (Asaoka-Taguchi et al., 1999). Targets that are the effectors of H3meK4 addition have yet to be identified, but conceivably include an H3 lysine 4-specific methyltransferase. The phenotypes observed upon removal of nanos are also only partially penetrant in both organisms. It is thus likely that other conserved, partially redundant systems exist. A clear candidate is germ cell-less (gcl), which is required in Drosophila to maintain transcriptional quiescence (Leatherman et al., 2002). Preliminary experiments suggest that H3meK4 regulation is also disrupted in gcl mutants (G.D. and P.D.S., unpublished results). A predicted C. elegans gene with substantial homology to gcl has been identified, but its role in these processes has not as yet been assessed (W.G.K., unpublished data).
It appears clear in flies that nos is required to maintain the repressive state of the germline and that part of this involves preventing accumulation of H3meK4. The pole cells begin life lacking this modification, and NANOS function is required to prevent its addition to pole cell chromatin. It is not as clear-cut in worms, since H3meK4 is initially present and then removed at germline restriction. Our detection of H3meK4 in Z2/Z3 of nos-depleted embryos could therefore conceivably reflect a role for nos in either (presumably indirectly) promoting the removal of H3meK4 after PIE-1 and/or preventing the re-addition of H3meK4 before hatching. Given the conservation of most other phenotypes caused by the loss of nos activity in multiple organisms, we favor the latter role for C. elegans NOS proteins.
Specific Chromatin Remodeling in Z2/Z3
At approximately the 100-cell stage in C. elegans, PIE-1 levels decrease and this is concurrent with a global loss of a distinct subset of histone modifications, H3meK4 and H4acetylK8, in Z2/Z3. The presence of other histone tail modifications in Z2/Z3 indicates that the loss of H3meK4 and H4acetylK8 epitopes are not due to a general cleavage of histone tails, but rather a specific chromatin-remodeling event. While removal of H4acetylK8 is likely to be performed by a histone deacetylase (HDAC), an activity that demethylates lysines has yet to be identified in any organism (Lachner et al., 2003). The loss of H3meK4 could therefore represent either a replication-independent or a replication-coupled replacement of histone H3, perhaps by a germline-specific H3 variant. We have frequently observed a diminishment of H3meK4 in P4 after gastrulation and prior to its entry into mitosis, which could indicate an S phase-related event (Figure 4, and data not shown).
We cannot formally rule out that the histone H3 and H4 epitopes recognized by these antibodies could theoretically be masked by the addition of other modifications, but we consider this to be unlikely. In the case of H3meK4, increased methylation of lysine 4 could potentially decrease binding of the dimethylK4 antibody used, but antibodies against H3trimethylK4 yielded the same pattern as those recognizing H3dimethylK4 (not shown). In addition, other kinds of modifications that can block recognition of either H3meK4 or H4acetylK8 by the antibodies used are, to our knowledge, currently unprecedented.
The loss of PIE-1 protein, H3meK4, and H4acetylK8 occur near or at the same time, suggesting that these events are linked. The perdurance of H3meK4 observed in emb-4(hc60) and the parallel perdurance of PIE-1 in this mutant suggests a requirement for PIE-1 to be degraded for the remodeling to occur, but we cannot rule out that these are separable events caused by the emb-4 mutation (the emb-4 locus has not yet been identified). If PIE-1’s degradation is required, an early product of initial transcriptional activation allowed by PIE-1 depletion and/or the loss of a direct PIE-1 inhibition could activate a chromatin-remodeling complex to continue repression of the genome. For example, PIE-1 has been shown to be a direct inhibitor of the MEP-1/HDAC complex’s deacetylase activity (Unhavaithaya et al., 2002). However, we were unable to detect a role for this complex in the chromatin remodeling we observe in Z2/ Z3, and PIE-1 perdurance in emb-4 mutants does not prevent loss of H4acetylK8 (not shown), so other remodeling complexes may be involved.
A Unique Requirement for PIE-1
Given the substantial conservation of modes of germline repression that exist, it is initially unclear why C. elegans has evolved a unique mode of repression in the early P blastomeres that requires PIE-1. In contrast to nos, the requirement for a PIE-1 mode of repression does not appear to be conserved in flies. PIE-1 is a CCCH protein for which no ortholog has been identified in any organism. How PIE-1 achieves transcriptional repression in the P lineage is not fully understood. Recent results support a model in which PIE-1 blocks transcription elongation through an internal peptide motif that mimics the repeated motif found in the C-terminal domain (CTD) of RNA polymerase II (RNAPII) (Zhang et al., 2003). According to this model, the PIE-1 motif decoys and competes with the recognition of the RNAPII CTD by the PTEFb kinase complex, thereby inhibiting phosphorylation of the CTD, and subsequent release of the polymerase complex, for transcriptional elongation (Zhang et al., 2003). Our data is consistent with this model. In the P blastomeres, the chromatin organization of the genome appears to be very “permissive,” with global patterns of nucleosomal histone modifications consistent with a generally open chromatin structure. This suggests that access to promoters is not likely to be generally blocked by an inhibitory chromatin architecture and that the repression of transcription mediated by PIE-1 probably occurs at a subsequent step. Indeed, absent PIE-1, the default state of the early germline is somatic activation driven by maternal transcription factors, and there is no evidence for other intrinsic mechanisms operating to inhibit somatic differentiation. For example, maternal transcription factors that are used to specify early somatic lineages are normally present, but not functional, in the P lineage. In the pie-1 mutant, these factors are immediately functional and readily recognize and drive transcription from their target promoters, indicative of a genome primed for transcriptional activity (reviewed in Seydoux and Strome, 1999). A mechanism involving facultative heterochromatin assembly throughout the genome, as occurs in pole cells, is therefore unlikely to be either involved or, as discussed below, desired.
Why has such a system evolved in C. elegans? We propose that the answer lies in the early divisions of the P blastomeres, which do not have counterparts in Drosophila embryogenesis. These early asymmetric divisions represent a germline that is not fully restricted— somatic lineages are produced from the P blastomeres. Given the rapidity, the strong maternal component, and the constrained geometric restrictions that are all required for faithful induction and completion of early cleavage divisions, all blastomeres may require an open chromatin status at the time of their birth for correct temporal coordination. Drosophila pole cells, by virtue of their precommitted status, can apparently afford to establish and maintain a chromatin-based repressive mechanism to be remodeled after gastrulation.
In contrast, the C. elegans P blastomeres are not precommitted. Establishing a chromatin-based mechanism that would have to be remodeled for zygotic activation in their somatic daughters may result in a temporal delay the somatic lineages could ill-afford. A PIE-1 mode of repression that acts analogous to the starting gate at a horse race therefore makes sense: it restrains the polymerase at the promoter in the dually committed P blastomeres, and the removal of PIE-1 in somatic daughters after cell division allows immediate release, after which it’s “off to the races” with somatic differentiation. Given the nonnuclear activities described for PIE-1 (Tenenhaus et al., 2001), its role in transcriptional repression may be an evolved adaptation, and there may be numerous other nonhomologous proteins that have been adapted in other systems to provide similar roles.
Experimental Procedures
General Methods and Strains
Worms
The techniques described by Brenner (1974) were used for animal maintenance and handling. All animals were grown at 16°C or 20°C unless otherwise noted. The C. elegans N2 (Bristol) strain was used as wild-type in this study. The other C. elegans strains used are mes-2(bn11) unc-4(e120)/mnC1 dpy-10(e128) unc-52(e444)II, mes-3(bn35) dpy-5(e61)I; sDp2(I;f), dpy-11(e224) mes-4(bn67) V/nT1[unc-?(n754) let-? qIs50](IV;V), and mes-6(bn66) dpy-20(e1282)IV/ nT1[unc-?(n754) let-?](IV;V), all generously gifted to us by Dr. Susan Strome, Indiana University. Dr. Geraldine Seydoux, Johns Hopkins Medical School, kindly donated the strain dpy-18(e364) pie-1(zu127)/qC1. The Caenorhabditis Genetics Center provided the strains nos-1(gv5)II, emb-4(hc60)V, and rol-1(e91) mex-1(zu121)/ mnC1 dpy-10(e128) unc-52(e444)II. The m−z− homozygous mes embryos used in immunocytochemistry were the F1 progeny of fertile m+z− homozygous mes mothers identified by linked recessive mutations. The temperature-sensitive emb-4 mutant embryos used in immunocytochemistry were the progeny of L4 larvae raised to 25°C overnight.
Flies
0- to 6-hour-old embryos were collected from nos− females mated either with wild-type or nos− males. Homozygous females carrying either nos BN or nos RC mutation were used for embryo collections. Similar results were obtained with both the alleles. As a wild-type control, embryos from either Oregon r or w1 females were used.
Antibodies
The following primary antibodies were used at the indicated dilutions: rabbit antiacetylated histone H4 (acetyl-K8, -K16,1:1000,1:3000) (Serotec), and rabbit antiacetylated histone H3 (acetyl-K9, -K14; 1:500) (Upstate Biotechnology). Rabbit antihistone H3 dimethyl K4 (1:500) and antihistone H3 dimethylK9 (1:250) were gifts from Dr. C. David Allis, Rockefeller U. Epitopes recognized by these antibodies are referred to as H3meK4 and H3meK9, respectively, throughout the paper. The anti-PGL-1 antibody (undiluted OIC1D4) and the anti-PIE-1 antibody (1:100) were gifts from Drs. S. Strome (Indiana U.) and G. Seydoux (Johns Hopkins School of Medicine), respectively. Dr. P. Lasko (MacGill U.) donated an anti-VASA antibody used in preliminary studies. Secondary antibodies purchased from Molecular Probes were used at the indicated dilutions: Alexafluor; 594 goat anti-rabbit IgG (1:500), Alexafluor; 488 goat anti-mouse IgG (1:500). Either DAPI (Sigma, 1 µg/ml) or Hoechst 33342 (Molecular Probes, 1:2000 dilution) were used to counterstain DNA.
Immunocytochemistry
A methanol/acetone fixation procedure (Strome and Wood, 1983) was used for whole-mount fixation and antibody staining of embryos. In some cases, where indicated, embryos were fixed in 2.5% formaldehyde for 2 min, followed by a 2 min post-fix in −20°C (95%) ethanol prior to washing and antibody addition. L1 larvae were collected for antibody staining by washing N2 hermaphrodites twice with M9 buffer and dissolving hatched animals in sodium hypochlorite solution (1 M NaOH/10% bleach). After two additional M9 washes, the embryos were collected and hatched overnight in 200 µl of M9 in a humidity chamber at 20°C. The next day, half of the starved L1 larvae were fixed for antibody staining as described above. The remaining L1 larvae were fed E. coli (OP50 strain) for 6 hr before fixation and antibody staining.
Embryos were staged by counting DAPI-stained nuclei in optical Z-sections of each embryo using a Leica DMRA microscope outfitted with a Cooke Sensicam. Images were collected and analyzed with Volume Scan (Vaytek) and Image-Pro Plus (Media Cybernetics) software. Whole-mount antibody staining of Drosophila embryos was carried out as previously described (Deshpande et al., 1995).
RNA-Mediated Interference (RNAi) Analysis
Sense and antisense transcripts corresponding to the full-length coding region of nos-2, exons 1–3 of mep-1, exon 2 of hda-1, and exon 3 of let-418 were generated using the Ribomax Large Scale RNA Production kit (Promega) and annealed to produce dsRNA. nos-2 dsRNA was injected into adult nos-1(gv5) hermaphrodites. mep-1, hda-1,and let-418 dsRNAs were injected separately into adult N2 hermaphrodites. Injected hermaphrodites were recovered overnight and dissected the following day, and their embryos were fixed for antibody staining as described above. Parallel broods were assessed for sterility as adults for nos-2 RNA injections, embryonic lethality for hda-1 RNA injections, and larval arrest for mep-1 and let-418 injections.
Acknowledgments
The authors wish to thank Drs. B. Yedvobnick, V. Finnerty, and K. Bhat and members of their labs for their expert guidance and assistance in many of the initial Drosophila experiments performed during the course of these studies. We also thank Drs. G. Seydoux, T. Schedl, E. Smith, and S. L’Hernault for helpful discussions and provision of some reagents and strains. This work was supported by an Emory University Research Council Grant (W.G.K.) and grants from the NIH to W.G.K. (GM63102), C.E.S. (T32GM08367), and P.S. (other NIH grants). Some of the C. elegans strains used in this study were provided by the Caenorhabditis Genetics Center, and we thank T. Stiernagle for her continuing assistance with strain requests.
References
- Anderson KV, Lengyel JA. Rates of synthesis of major classes of RNA in Drosophila embryos. Dev. Biol. 1979;70:217–231. doi: 10.1016/0012-1606(79)90018-6. [DOI] [PubMed] [Google Scholar]
- Asaoka M, Sano H, Obara Y, Kobayashi S. Maternal Nanos regulates zygotic gene expression in germline progenitors of Drosophila melanogaster. Mech. Dev. 1998;78:153–158. doi: 10.1016/s0925-4773(98)00164-6. [DOI] [PubMed] [Google Scholar]
- Asaoka-Taguchi M, Yamada M, Nakamura A, Hanyu K, Kobayashi S. Maternal Pumilio acts together with Nanos in germline development in Drosophila embryos. Nat. Cell Biol. 1999;1:431–437. doi: 10.1038/15666. [DOI] [PubMed] [Google Scholar]
- Boggs BA, Cheung P, Heard E, Spector DL, Chinault AC, Allis CD. Differentially methylated forms of histone H3 show unique association patterns with inactive human X chromosomes. Nat. Genet. 2002;30:73–76. doi: 10.1038/ng787. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crittenden SL, Bernstein DS, Bachorik JL, Thompson BE, Gallegos M, Petcherski AG, Moulder G, Barstead R, Wickens M, Kimble J. A conserved RNA-binding protein controls germline stem cells in Caenorhabditis elegans. Nature. 2002;417:660–663. doi: 10.1038/nature754. [DOI] [PubMed] [Google Scholar]
- Csink AK, Henikoff S. Large-scale chromosomal movements during interphase progression in Drosophila. J.Cell Biol. 1998;143:13–22. doi: 10.1083/jcb.143.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande G, Stukey J, Schedl P. scute (sis-b) function in Drosophila sex determination. Mol. Cell. Biol. 1995;15:4430–4440. doi: 10.1128/mcb.15.8.4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande G, Calhoun G, Yanowitz JL, Schedl PD. Novel functions of nanos in downregulating mitosis and transcription during the development of the Drosophila germline. Cell. 1999;99:271–281. doi: 10.1016/s0092-8674(00)81658-x. [DOI] [PubMed] [Google Scholar]
- Edgar BA, Schubiger G. Parameters controlling transcriptional activation during early Drosophila development. Cell. 1986;44:871–877. doi: 10.1016/0092-8674(86)90009-7. [DOI] [PubMed] [Google Scholar]
- Fong Y, Bender L, Wang W, Strome S. Regulation of the different chromatin states of autosomes and X chromosomes in the germ line of C. elegans. Science. 2002;296:2235–2238. doi: 10.1126/science.1070790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guedes S, Priess JR. The C. elegans MEX-1 protein is present in germline blastomeres and is a P granule component. Development. 1997;124:731–739. doi: 10.1242/dev.124.3.731. [DOI] [PubMed] [Google Scholar]
- Jenuwein T. Re-SET-ting heterochromatin by histone methyltransferases. Trends Cell Biol. 2001;11:266–273. doi: 10.1016/s0962-8924(01)02001-3. [DOI] [PubMed] [Google Scholar]
- Kawasaki I, Shim YH, Kirchner J, Kaminker J, Wood WB, Strome S. PGL-1, a predicted RNA-binding component of germ granules, is essential for fertility in C. elegans. Cell. 1998;94:635–645. doi: 10.1016/s0092-8674(00)81605-0. [DOI] [PubMed] [Google Scholar]
- Kelly WG, Schaner CE, Dernburg AF, Lee MH, Kim SK, Villeneuve AM, Reinke V. X-chromosome silencing in the germline of C. elegans. Development. 2002;129:479–492. doi: 10.1242/dev.129.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Histone methylation in transcriptional control. Curr. Opin. Genet. Dev. 2002;12:198–209. doi: 10.1016/s0959-437x(02)00287-3. [DOI] [PubMed] [Google Scholar]
- Lachner M, Jenuwein T. The many faces of histone lysine methylation. Curr. Opin. Cell Biol. 2002;14:286–298. doi: 10.1016/s0955-0674(02)00335-6. [DOI] [PubMed] [Google Scholar]
- Lachner M, O’Sullivan RJ, Jenuwein T. An epigenetic road map for histone lysine methylation. J. Cell Sci. 2003;116:2117–2124. doi: 10.1242/jcs.00493. [DOI] [PubMed] [Google Scholar]
- Leatherman JL, Jongens TA. Transcriptional silencing and translational control: key features of early germline development. Bioessays. 2003;25:326–335. doi: 10.1002/bies.10247. [DOI] [PubMed] [Google Scholar]
- Leatherman JL, Levin L, Boero J, Jongens TA. germ cell-less acts to repress transcription during the establishment of the Drosophila germ cell lineage. Curr. Biol. 2002;12:1681–1685. doi: 10.1016/s0960-9822(02)01182-x. [DOI] [PubMed] [Google Scholar]
- Mello CC, Draper BW, Krause M, Weintraub H, Priess JR. The pie-1 and mex-1 genes and maternal control of blastomere identity in early C. elegans embryos. Cell. 1992;70:163–176. doi: 10.1016/0092-8674(92)90542-k. [DOI] [PubMed] [Google Scholar]
- Mello CC, Schubert C, Draper B, Zhang W, Lobel R, Priess JR. The PIE-1 protein and germline specification in C. elegans embryos. Nature. 1996;382:710–712. doi: 10.1038/382710a0. [DOI] [PubMed] [Google Scholar]
- Pirrotta V. Silence in the germ. Cell. 2002;110:661–664. doi: 10.1016/s0092-8674(02)00967-4. [DOI] [PubMed] [Google Scholar]
- Reuben M, Lin R. Germline X chromosomes exhibit contrasting patterns of histone H3 methylation in C. elegans. Dev. Biol. 2002;245:71–82. doi: 10.1006/dbio.2002.0634. [DOI] [PubMed] [Google Scholar]
- Schnabel R, Weigner C, Hutter H, Feichtinger R, Schnabel H. mex-1 and the general partitioning of cell fate in the early C. elegans embryo. Mech. Dev. 1996;54:133–147. doi: 10.1016/0925-4773(95)00466-1. [DOI] [PubMed] [Google Scholar]
- Seydoux G, Dunn MA. Transcriptionally repressed germ cells lack a subpopulation of phosphorylated RNA polymerase II in early embryos of Caenorhabditis elegans and Drosophila melanogaster. Development. 1997;124:2191–2201. doi: 10.1242/dev.124.11.2191. [DOI] [PubMed] [Google Scholar]
- Seydoux G, Fire A. Soma-germline asymmetry in the distributions of embryonic RNAs in Caenorhabditis elegans. Development. 1994;124:2823–2834. doi: 10.1242/dev.120.10.2823. [DOI] [PubMed] [Google Scholar]
- Seydoux G, Schedl T. The germline in C. elegans: origins, proliferation, and silencing. Int. Rev. Cytol. 2001;203:139–185. doi: 10.1016/s0074-7696(01)03006-6. [DOI] [PubMed] [Google Scholar]
- Seydoux G, Strome S. Launching the germline in Caenorhabditis elegans: regulation of gene expression in early germ cells. Development. 1999;126:3275–3283. doi: 10.1242/dev.126.15.3275. [DOI] [PubMed] [Google Scholar]
- Seydoux G, Mello CC, Pettitt J, Wood WB, Priess JR, Fire A. Repression of gene expression in the embryonic germ lineage of C. elegans. Nature. 1996;382:713–716. doi: 10.1038/382713a0. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Ohba R, Cook RG, Allis CD. Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in Tetrahymena. Proc. Natl. Acad. Sci. USA. 1999;96:14967–14972. doi: 10.1073/pnas.96.26.14967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome S, Wood WB. Generation of asymmetry and segregation of germ-line granules in early C. elegans embryos . Cell. 1983;35:15–25. doi: 10.1016/0092-8674(83)90203-9. [DOI] [PubMed] [Google Scholar]
- Subramaniam K, Seydoux G. nos-1 and nos-2, two genes related to Drosophila nanos, regulate primordial germ cell development and survival in Caenorhabditis elegans. Development. 1999;126:4861–4871. doi: 10.1242/dev.126.21.4861. [DOI] [PubMed] [Google Scholar]
- Subramaniam K, Seydoux G. Dedifferentiation of primary spermatocytes into germ cell tumors in C. elegans lacking the Pumilio-like protein PUF-8. Curr. Biol. 2003;13:134–139. doi: 10.1016/s0960-9822(03)00005-8. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Technau G, Campos-Ortega J. Lineage analysis of transplanted individual cells in embryos of Drosophila melanogaster III. Commitment and proliferative capabilities of pole cells and midgut progenitors. Rouxs Arch. Dev. Biol. 1986;195:489–498. doi: 10.1007/BF00375889. [DOI] [PubMed] [Google Scholar]
- Tenenhaus C, Schubert C, Seydoux G. Genetic requirements for PIE-1 localization and inhibition of gene expression in the embryonic germ lineage of Caenorhabditis elegans. Dev. Biol. 1998;200:212–224. doi: 10.1006/dbio.1998.8940. [DOI] [PubMed] [Google Scholar]
- Tenenhaus C, Subramaniam K, Dunn MA, Seydoux G. PIE-1 is a bifunctional protein that regulates maternal and zygotic gene expression in the embryonic germ line of Caenorhabditis elegans. Genes Dev. 2001;15:1031–1040. doi: 10.1101/gad.876201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomioka M, Miya T, Nishida H. Repression of zygotic gene expression in the putative germline cells in Ascidian embryos. Zoolog. Sci. 2002;19:49–55. doi: 10.2108/zsj.19.49. [DOI] [PubMed] [Google Scholar]
- Tsuda M, Sasaoka Y, Kiso M, Abe K, Haraguchi S, Kobayashi S, Saga Y. Conserved role of nanos proteins in germ cell development. Science. 2003;301:1239–1241. doi: 10.1126/science.1085222. [DOI] [PubMed] [Google Scholar]
- Underwood EM, Caulton JH, Allis CD, Mahowald AP. Developmental fate of pole cells in Drosophila melanogaster. Dev. Biol. 1980;77:303–314. doi: 10.1016/0012-1606(80)90476-5. [DOI] [PubMed] [Google Scholar]
- Unhavaithaya Y, Shin TH, Miliaras N, Lee J, Oyama T, Mello CC. MEP-1 and a homolog of the NURD complex component Mi-2 act together to maintain germline-soma distinctions in C. elegans. Cell. 2002;111:991–1002. doi: 10.1016/s0092-8674(02)01202-3. [DOI] [PubMed] [Google Scholar]
- Van Doren M, Williamson AL, Lehmann R. Regulation of zygotic gene expression inDrosophila primordial germ cells. Curr. Biol. 1998;8:243–246. doi: 10.1016/s0960-9822(98)70091-0. [DOI] [PubMed] [Google Scholar]
- Wylie C. Germ cells. Cell. 1999;96:165–174. doi: 10.1016/s0092-8674(00)80557-7. [DOI] [PubMed] [Google Scholar]
- Xu L, Fong Y, Strome S. The Caenorhabditis elegans maternal-effect sterile proteins, MES-2, MES-3, and MES-6, are associated in a complex in embryos. Proc. Natl. Acad. Sci. USA. 2001;98:5061–5066. doi: 10.1073/pnas.081016198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalokar M. Autoradiographic study of protein and RNA formation during early development of Drosophila eggs. Dev. Biol. 1976;49:425–437. doi: 10.1016/0012-1606(76)90185-8. [DOI] [PubMed] [Google Scholar]
- Zhang F, Barboric M, Blackwell TK, Peterlin BM. A model of repression: CTD analogs and PIE-1 inhibit transcriptional elongation by P-TEFb. Genes Dev. 2003;17:748–758. doi: 10.1101/gad.1068203. [DOI] [PMC free article] [PubMed] [Google Scholar]