Abstract
While gene flow between distantly related populations is increasingly recognized as a potentially important source of adaptive genetic variation for humans, fully characterized examples are rare. In addition, the role that natural selection for resistance to vivax malaria may have played in the extreme distribution of the protective Duffy-null allele, which is nearly completely fixed in mainland sub-Saharan Africa and absent elsewhere, is controversial. We address both these issues by investigating the evolution of the Duffy-null allele in the Malagasy, a recently admixed population with major ancestry components from both East Asia and mainland sub-Saharan Africa. We used genome-wide genetic data and extensive computer simulations to show that the high frequency of the Duffy-null allele in Madagascar can only be explained in the absence of positive natural selection under extreme demographic scenarios involving high genetic drift. However, the observed genomic single nucleotide polymorphism diversity in the Malagasy is incompatible with such extreme demographic scenarios, indicating that positive selection for the Duffy-null allele best explains the high frequency of the allele in Madagascar. We estimate the selection coefficient to be 0.066. Because vivax malaria is endemic to Madagascar, this result supports the hypothesis that malaria resistance drove fixation of the Duffy-null allele in mainland sub-Saharan Africa.
Keywords: Duffy blood group, vivax malaria, adaptive evolution, migration, Malagasy population genetics
1. Introduction
Scans for signatures of recent positive selection in human populations have now identified hundreds of candidate loci, suggesting that positive natural selection has been an important force shaping genetic diversity within and between human groups (e.g. [1,2]). For the vast majority of the candidate positive selection loci, we have no understanding of their biological consequences (phenotypes) or the environments under which the selective pressures were relevant. To date, most attention has been paid to identifying recently selected genomic regions within and between regional human groups. However, the role of natural selection in shaping the genetic diversity of admixed human populations has recently gained attention with the study of high-altitude-adapted alleles among admixed Tibetans [3] and the report that Eurasian populations may have acquired adaptive skin phenotype alleles through past admixture with Neandertals [4].
The Duffy blood group locus (the DARC gene), for which phenotypic consequences are known for a genetic variant [5] that is strongly differentiated between sub-Saharan African and non-African human populations [6], and for which a credible selection pressure hypothesis exists [7], presents an excellent opportunity to examine the role of natural selection in the Malagasy, a recently admixed population with major ancestry components from mainland sub-Saharan Africa and East Asia. The Duffy antigen is typically expressed on the surfaces of both white and red blood cells and is encoded by the Duffy antigen receptor for chemokines (DARC). There are two DARC protein polymorphisms, the FY*A and FY*B alleles, that differ by a single amino acid. A third polymorphism, FY*O or the Duffy-null allele, is a DARC promoter region single nucleotide polymorphism (SNP) that effects the loss of Duffy antigen protein expression on red blood cells [5].
The vivax malaria parasite, Plasmodium vivax, typically requires the presence of intact FY*A or FY*B antigens to invade red blood cells [7]. Individuals homozygous for the Duffy-null allele were once thought to be completely resistant to vivax malaria infection [7], though very rare cases of infections have been reported [8,9]. While mainland sub-Saharan Africans are nearly fixed for the Duffy-null allele, it is absent in all other human populations with no history of recent African admixture [6] except for Papua New Guineans who have acquired the allele through independent mutation and have it at very low frequency (approx. 2%) [10]. This extreme level of population differentiation is highly unusual across the genome [11]. Vivax malaria, in contrast, is currently, or was historically, found in all parts of the world with appropriate climates and Anopheles mosquito vectors except for sub-Saharan Africa [12].
The allele frequency pattern and genotype-disease association combination have been interpreted as evidence for a past history of natural selection for resistance to vivax malaria, thus favouring the Duffy-null allele, in mainland sub-Saharan Africans [13]. Indeed, relative to Europeans, Africans show reduced genetic diversity surrounding the SNP that causes loss of DARC expression, which is consistent with a past selective sweep [14,15]. However, questions have been raised about whether vivax malaria is a plausible agent of selection on the Duffy-null allele (e.g. [13,16], and see Discussion).
The Malagasy, the people of Madagascar, offer a unique opportunity to test hypotheses of recent selection for the Duffy-null allele in an environment with endemic vivax malaria. Madagascar was probably settled within the past 2300 years [17] by both Bantu-speaking sub-Saharan Africans and Austronesian speakers from southeast Asia [18]. A recent report suggests a possible human presence on Madagascar as early as 4000 years ago [19]; however, the context of the dated material is unclear owing to a complex stratigraphy. The strongest archaeological evidence points to an early human presence on Madagascar approximately 2300 years BP [17].
Genetic surveys focusing on the uniparentally inherited mitochondria and Y-chromosome have confirmed the dual southeast Asian/African origin of the Malagasy [20–22], with admixture proportions estimated to be near equal between the two ancestral groups [22], though with a difference between the more African coastal groups and the more Austronesian Central Highland groups [21,23]. A recent analysis of nuclear genomic SNP data from three southern coastal groups estimated the proportion of African ancestry to be 67% [24]. There was also some Near Eastern Islamic cultural influence on the Northwest coast of the island [25], though little evidence of a genetic legacy has yet been described [21,22].
While the oldest known settlement on Madagascar dates to 1300 BP [18], there are no clear cultural associations with the oldest sites, so it is not known from which source population they derive [25]. Thus, the sequence of arrivals on Madagascar is debated and uncertain [26,27]. By 1000 BP, clear archaeological connections to both mainland Africa and East Asia can be observed from pottery, domestic animal and domestic crop evidence [18,25,26,28]. The archaeological record suggests that Madagascar was relatively sparsely populated until about 1300 BP, followed by increases in the number and size of settlements [18,25], leading to the current census size of more than 23 million Malagasy [29].
The mainland sub-Saharan African migrants brought the Duffy-null allele to Madagascar. Vivax malaria is currently endemic to Madagascar [12], probably brought to the island by Austronesians. A recent survey from across Madagascar found the frequency of the Duffy-null allele to be 0.83 [9]. If the sub-Saharan African migrants to Madagascar were fixed for the Duffy-null allele, then in the absence of further evolutionary processes the expected frequency of the Duffy-null allele in the Malagasy would equal the amount of sub-Saharan African ancestry, which has been estimated to be from 50 to 67%, depending on the population sample [21,22,24]. In this paper, we test the null hypothesis that genetic drift alone can explain the difference between the observed and expected frequencies of the Duffy-null allele in Madagascar, against an alternative hypothesis of a recent history of positive selection, using a series of computer simulations and genomic SNP data from Malagasy and comparative population samples.
2. Material and methods
We analysed genomic SNP data using the Affymetrix GeneChip Human 10 k 2.0 SNP Array for 19 Malagasy Merina individuals collected from the Central Highlands of Madagascar and a comparative dataset consisting of 16 Bantu speakers from South Africa, 20 Burunge from East Africa, 20 Mozambicans, 42 Europeans, 11 Mala and 11 Brahmin from India and 20 Chinese and Japanese individuals. We also genotyped the Duffy-positive/null causative SNP (rs2814778) in the Malagasy Merina samples, using Sanger Sequencing. Malagasy admixture was characterized using identity by state-multidimensional scaling (IBS-MDS) and the model-based clustering algorithm implemented in Admixture [30]. The expected change in Malagasy Duffy-null allele frequency in the absence of natural selection (genetic drift alone) for a variety of possible demographies was explored through forward evolution computer simulations using the program fastadsim.pl, written for this project. Fastadsim.pl was also used to estimate the expected amount of genomic allelic fixation given demographic scenarios compatible with the genetic drift hypothesis. Finally, we estimated the selection coefficient for the Duffy-null allele, assuming that selection could act only on the recessive phenotype. Detailed human research ethics, genotyping and analytical methods are provided in the electronic supplementary material, Materials and Methods.
3. Results
(a). Characterization of admixture in the Malagasy
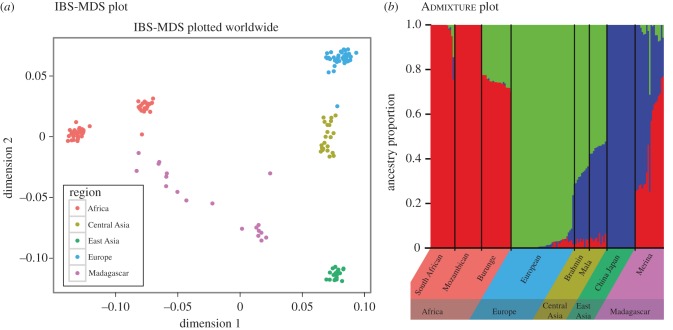
We genotyped 19 Malagasy individuals sampled from the Merina ethnic group located in the Central Highlands of Madagascar (including the capitol city, Antananarivo; the Merina are the largest Malagasy ethnic group), along with a comparative dataset. The final dataset consisted of 159 individuals genotyped for 11 552 SNPs. We used an IBS-MDS analysis to visualize the relationship of the Malagasy to the other worldwide populations from the SNP data (figure 1a). The first dimension largely separates mainland sub-Saharan Africans from the rest of the world, while the second dimension separates western and eastern Eurasians. The 19 Malagasy individuals are stretched along a gradient on the MDS plot from the sub-Saharan Africans to the East Asians, with one individual displaced in the direction of the Central Asians/Europeans. This pattern is not surprising given the known history of admixture in Madagascar [18].
Figure 1.
Malagasy population structure. (a) Multidimensional scaling analysis of 10 k SNP data for 19 Malagasy Merina individuals and a comparative dataset of 140 individuals from seven populations. (b) Admixture analysis of the SNP data, with estimated proportions of ancestry for each individual shown as a vertical line. The estimated proportion of mainland sub-Saharan African ancestry for the 19 Merina individuals ranges from 25 to 77%, with a mean of 48%. The ‘European’ ancestry in the Burunge of Tanzania probably reflects a back-to-Africa migration from the Middle East observed in many Eastern Africa populations [31–33]. (Online version in colour.)
To quantify admixture levels in the Malagasy individuals, we used the software Admixture [30]. The model with three ancestral populations, largely corresponding to sub-Saharan Africa, Europe and East Asia best explained the data (electronic supplementary material, figure S1). As expected, the Malagasy primarily have sub-Saharan African and East Asian ancestry, but are highly variable in their ancestry proportions (figure 1b). On average, our Malagasy population sample has an estimated 48% sub-Saharan African and 48% East Asian ancestry, in close agreement with estimates from mitochondrial and Y-chromosome DNA [22]. The Malagasy also have an average of 4% ‘European’ ancestry. While Arab traders had extensive contact with Madagascar over the past 1000 years [25], prior surveys of Y-chromosome and mtDNA have failed to identify a significant Middle Eastern genetic legacy in the Malagasy [21,22]. It is plausible that the ‘European/Central Asian’ ancestral component observed in our nuclear genomic analysis of the Malagasy can be attributed to a history of admixture from the Middle East, or possibly small-scale recent admixture during the period of French colonialism.
Our estimate of 48% average sub-Saharan African ancestry in the Highland Merina population is lower than the 67% average sub-Saharan African ancestry for three Madagascar southern coast populations that was recently reported by Pierron et al. [24], also using genome-wide SNP genotype data. This finding is broadly consistent with previously reported ancestry differences between highland and coastal populations based on mtDNA and Y-chromosome markers [21]. Moreover, we found the Merina to be much more variable in their ancestry proportions (s.d. = 20.2%) than the southern coastal populations (s.d. = 3.23%) [24]. We offer three non-mutually exclusive hypotheses that might explain this observed difference: (i) East Asian/sub-Saharan African admixture may have occurred more recently in the Merina, (ii) the Merina may have mated assortatively with respect to ancestry to a greater extent than the coastal populations, or (iii) some Malagasy from other ethnic groups with different ancestry proportions may have recently adopted the Merina ethnic affiliation, a plausible scenario given that the Merina are the dominant ethnic group and historic rulers of the island.
(b). Frequency of the Duffy-null allele in Madagascar
Menard et al. [9] sampled 661 Malagasy individuals from eight localities from across Madagascar and reported an overall Duffy-null allele frequency of 0.83. Their Highland population sample consisting of the Merina and Bezanozano ethnic groups (n = 69) had a Duffy-null allele frequency of 0.78 (108 of 138 chromosomes), while their southern coastal population sample from Farafangana consisting of the Antaisaka, Antaifasy and Zafisoro ethnic groups (n = 86) had a Duffy-null allele frequency of 0.92 (158 of 172 chromosomes).
We checked whether the frequency of the Duffy-null allele in our Merina Highland sample was consistent with the Highland population and overall Malagasy sample estimates of Menard et al. [9] with a PCR and Sanger sequencing genotyping method. We successfully genotyped 18 of 19 samples and observed 26 of 36 chromosomes with the Duffy-null allele (estimated allele frequency = 0.72), which is not significantly different from the Menard et al.'s [9] Highland population sample (Fisher's exact test; p = 0.505) or island-wide result (p = 0.112).
In our following analyses and simulations, we primarily compare our Merina genome-wide SNP genotype data and average sub-Saharan African ancestry estimates to the Menard et al.'s [9] Duffy-null allele frequency estimate of 0.78 for their Highland Madagascar sample, because this Duffy-null allele frequency estimate was based on a larger sample with lower associated sampling error (approx. 3.5%). However, we additionally evaluated our results in the context of the more conservative 0.72 frequency estimate obtained from our own Duffy-null allele genotyping experiments. Finally, we also considered the southern coastal population sample estimates of average sub-Saharan African ancestry = 0.67 from Pierron et al. [24] and Duffy-null allele frequency = 0.92 from Menard et al. [9].
(c). Computer simulations to test evolutionary hypotheses for the Duffy-null allele
Because mainland sub-Saharan Africans are nearly fixed for the Duffy-null allele, while non-admixed populations from other continents are effectively fixed for Duffy-positive alleles, in the absence of natural selection, the expected frequency of the Duffy-null allele in the Malagasy is equal to their proportion of sub-Saharan African ancestry, with differences from this proportion owing to genetic drift. The expected amount of genetic drift is a function of the effective population size and the number of generations of drift [34], which are unknown in the Malagasy. We used computer simulations to test whether the observed Duffy-null allele frequency is better explained by a null hypothesis of genetic drift or an alternative hypothesis of positive natural selection in the relatively short time since humans first arrived in Madagascar, under a range of demographic scenarios. Specifically, we wrote a forward-evolution computer simulation program, fastadsim.pl, which tracks allele frequency change in an admixed population, given: (i) allele frequencies in parent populations, (ii) admixture proportion, (iii) starting effective population size, (iv) migration rate from each parent population, (v) rate of population growth, and (vi) number of generations since admixture.
We sought to be conservative in our choice of simulation parameters. For all simulations, we used Duffy-null allele frequencies of 1.0 and 0.0 for the mainland sub-Saharan African and Eurasian parent populations, respectively. We chose starting effective population sizes (at the time of admixture) of 100, 200 and 500, and population growth parameters of 2, 5 and 10%, all of which are probably lower than those that reflect the true demographic history of Madagascar (hence, overestimating the amount of drift, and conservative with respect to our alternative hypothesis).
Finally, we simulated 33, 43 and 77 generations of drift. Genealogical and demographic estimates from a variety of societies suggest that human generations average approximately 30 years [33]. Given 30 year generation times, these choices correspond to hypothetical admixture beginning at approximately 1000, 1300 and 2300 years ago, respectively. It is not yet known when admixture actually began in Madagascar, but clear evidence of a mainland African culture on the island is absent until approximately 1000 years ago [25], making the simulations with 43 generations of drift (corresponding to the time of the earliest human settlements [25]) conservative with respect to the alternative hypothesis and the simulation of 77 generations of drift (corresponding to the best evidence for the earliest human presence on Madagascar [17]) probably unrealistic but in any case even more conservative. We performed 10 000 simulations for each possible combination of parameters.
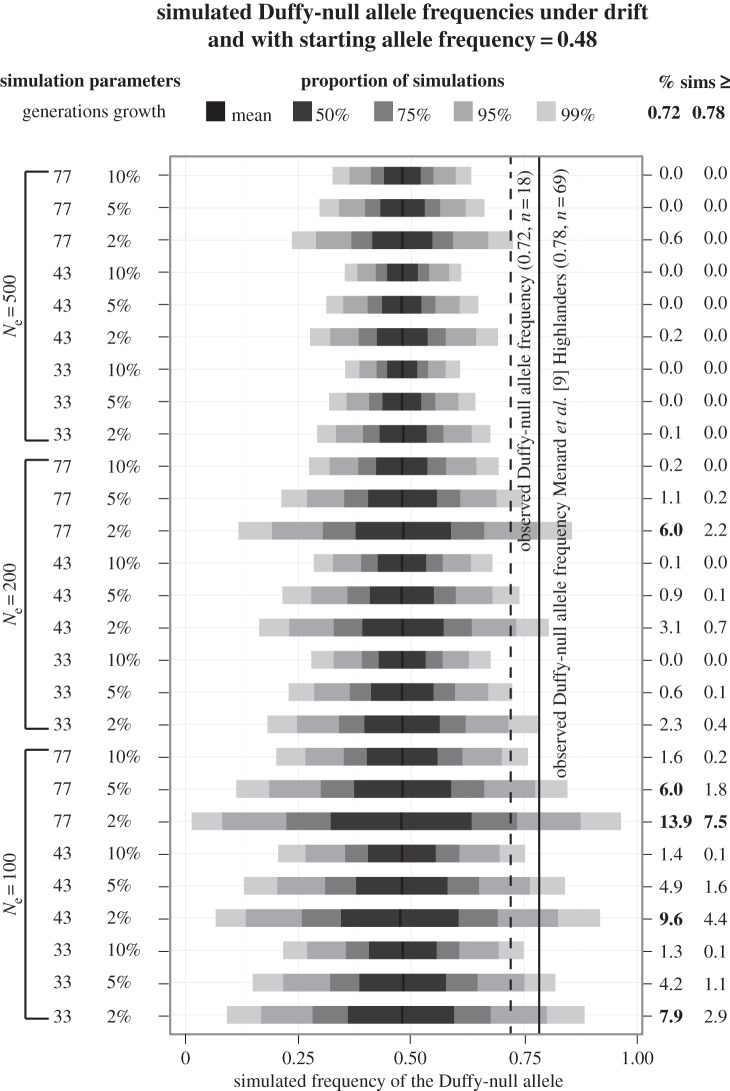
For simulations starting with a Duffy-null allele frequency of 0.48, a value based on our empirical observations of sub-Saharan African ancestry in the Merina, only the most extreme demographic scenario examined (initial Ne = 100, 2% growth, 77 generations of drift) had more than 5% of simulations finishing with a Duffy-null allele frequency greater than or equal to 0.78 (7.5% of simulations; figure 2). The Duffy-null allele frequency finished equal to or above 0.78 in only approximately 0.6% of all simulations run for 33 generations (564 out of 90 000), the admixture timing scenario most compatible with the archaeological record [25], and in zero simulations with an initial population size of 500 individuals. Also, the Duffy-null allele frequency finished equal to or greater than 0.72 in more than 5% of simulations in only five demographic scenarios, all with an initial Ne = 100, 77 generations of drift, or both, and thus probably more extreme than the actual demographic history of Madagascar (figure 2). The amount of genetic drift decreased when incorporating continuous migration from either the sub-Saharan African (electronic supplementary material, figure S3), or Austronesian (electronic supplementary material, figure S4) parent population. Consequently, we used the more conservative (with respect to the alternative hypothesis of natural selection) simultaneous admixture model in all subsequent analyses.
Figure 2.
Simulated frequencies of the Duffy-null allele under genetic drift. For each demographic scenario—composed of variable initial effective population sizes (Ne = 100, 200 or 500), number of generations (33, 43 and 77; equivalent to approx. 1000, 1300 or 2300 years, respectively) and levels of population growth (2, 5 or 10%)—forward-evolution simulations were performed from a starting allele frequency of 0.48 based on our estimate of African ancestry in the Malagasy. The final allele frequency was recorded for each of 10 000 simulations run for each demographic scenario. The bars show the 50, 75, 95 and 99% distributions of the final simulated allele frequencies for each demographic scenario. The solid vertical line indicates the observed frequency of 0.78 for the Duffy-null allele from a Highland Malagasy sample (n = 69) [9], while the dashed line indicates the observed frequency of 0.72 for the Duffy-null allele in our sample of 18 Merina. The percentage of simulations with an allele frequency greater than or equal to 0.72 and 0.78 are shown in the right-hand column (more than or equal to 5% shown in bold).
We obtained similar results from simulations based on the sub-Saharan ancestry proportion (67%) and Duffy-null allele frequency (0.92) that have been estimated for southern coastal populations [9,24]. Only three demographic scenarios produced more than 5% of simulations finishing with Duffy-null allele frequencies of at least 0.92, the least extreme of which had an initial Ne of 100 individuals, 2% population growth and 33 generations of drift (electronic supplementary material, figure S5). In aggregate, our simulation results suggest that genetic drift is only a viable (yet still unlikely) hypothesis for the observed frequency of the Duffy-null allele in the Malagasy if their demographic history was characterized by an extremely small initial effective population size (e.g. less than 200) at the onset of Austronesian and sub-Saharan African admixture.
(d). Genomic expectations for allelic fixation under the Duffy-null genetic drift hypothesis
Our simulation results suggest that under reasonable parameters for the demographic history of Malagasy admixture, genetic drift is an unlikely explanation for the high frequency of the Duffy-null allele in Madagascar. In other words, the influence of positive selection was probably necessary to achieve the observed high frequency of this allele. However, we did identify certain combinations of relatively extreme initial population size, population growth and time since admixture parameters under which genetic drift alone was sufficient to achieve an allele frequency of at least 0.78 or 0.72 in a small percentage of simulations with a starting allele frequency of 0.48. Yet, such strong genetic drift would simultaneously be expected to have measurable effects on genetic diversity across the entire genome, not just at the Duffy locus. Specifically, a substantial proportion of SNPs with non-intermediate allele frequencies at the onset of admixture would have become fixed. Therefore, we evaluated whether the observed levels of allelic fixation among Malagasy Merina SNPs with different initial estimated allele frequencies are consistent with the amount of genetic drift represented in those extreme demographic scenarios, to further assess the likelihood that positive selection at least partly explains the observed high frequency of the Duffy-null allele.
To do so, we used fastadsim.pl to estimate the expected level of allelic fixation owing to genetic drift across the genome, given two demographic scenarios that were most compatible with the Duffy-null allele genetic drift hypothesis under the different admixture and allele frequency estimate combinations. We then compared these results to the observed levels of SNP allelic fixation in the Malagasy Merina sample. Specifically, we estimated the initial allele frequency (at the onset of admixture) for each SNP in the Merina, using the Mozambique and East Asian samples as proxies for the mainland sub-Saharan African and Austronesian ancestral populations, respectively. We then binned SNPs according to their estimated initial minor allele frequencies with bin sizes of 0.025 and recorded the proportion of invariable SNPs in each bin. For the observed Malagasy data there was an average of 477 SNPs per bin, with no bin having fewer than 93 SNPs. The simulated data were based on 5000 SNPs per bin.
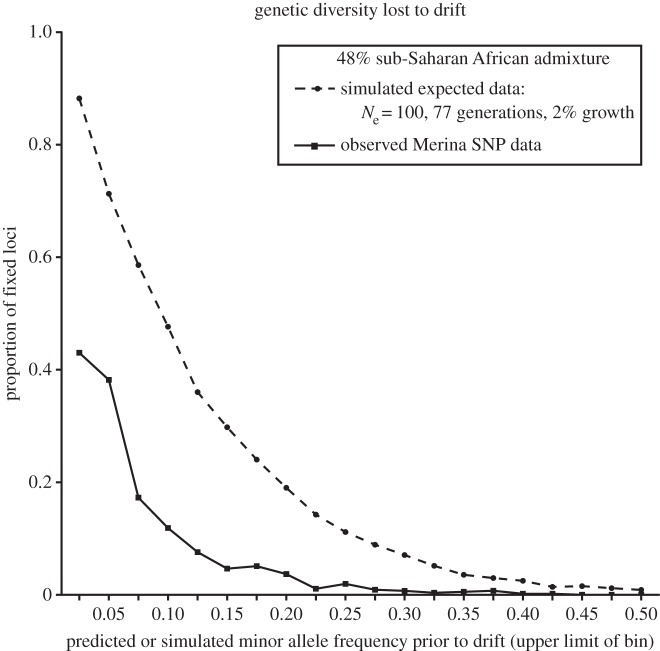
The observed level of allelic fixation in the Merina was much lower than expected under the extreme demographic scenarios that would be required for the Duffy-null genetic drift hypothesis and a Duffy-null allele frequency of 0.78 (figure 3). For example, given 48% admixture, initial Ne = 100 and 77 generations of drift, it is expected that 58.6% of SNPs with predicted starting allele frequencies of 0.050–0.075 would be observed to be fixed. However, we observe only 17.3% of such SNPs to be fixed in the Malagasy. The observed level of allelic fixation remains much lower than expected even under the least extreme demographic scenario compatible with drift and a Duffy-null allele frequency of 0.72 (electronic supplementary material, figure S6). The amount of genetic diversity observed in the Malagasy suggests a demographic history involving either a larger population size at the onset of admixture or a smaller number of generations than allowed by the demographic scenarios compatible with a genetic drift-only hypothesis to explain the high frequency of the Duffy-null allele in Madagascar.
Figure 3.
Theoretical versus observed levels of allelic fixation in the Malagasy. Proportions of fixed SNP loci in the Malagasy from simulated (dashed line) and observed (solid line) data, binned by predicted allele frequencies at the initial time of admixture based on 48% sub-Saharan African ancestry, and observed allele frequencies in present-day sub-Saharan African and East Asian population samples. The simulated results are based on a demographic scenario that was most compatible (but still unlikely) with an increase of the Duffy-null allele frequency from the admixture proportion of 0.48 to the observed Duffy-null frequency of 0.78 [9] owing to genetic drift alone (figure 2). The observed data are based on the Affymetrix 10 k SNP data from our population sample of 19 Merina Malagasy individuals.
In turn, we can also use this framework to identify a demographic scenario expected to closely result in the observed level of genome-wide allelic fixation in the Malagasy Merina, followed by genetic drift simulations with that demographic scenario to determine the likelihood of an allele frequency increase equal to or greater than that observed for the Duffy-null allele. Specifically, we found that the simulated loss of genetic diversity under a demographic history of initial Ne = 500, 2% population growth and 33 generations of drift was similar to that observed in our Merina genome-wide SNP data (electronic supplementary material, figure S7a). Under such a demographic history, genetic drift alone resulted in an allele frequency shift from 0.48 to 0.72 in only seven out of 10 000 simulations (0.07%), and never resulted in a final Duffy-null allele frequency increase to 0.78 or higher (electronic supplemental material, figure S7b). Thus, a demographic history compatible with patterns of Merina genetic diversity is highly unlikely to achieve the observed Duffy-null allele frequency in the same population in the absence of natural selection.
(e). Coefficient of selection and time to fixation
Because our analyses indicate that the observed difference between African ancestry proportions and Duffy-null allele frequency among the Malagasy is unlikely to have occurred in the absence of natural selection, we estimated the coefficients of selection, s, necessary to produce the observed frequency changes given various possible Duffy-null allele initial and current allele frequencies and numbers of generations of selection (electronic supplemental material, table S1). Given the best estimate of the initial Duffy-null allele frequency in the Merina (0.48), the best estimate of the current frequency (0.78) and 33 generations of selection (corresponding to the time when there is an archaeological presence from both Africa and Eurasia in Madagascar), we estimate s to be 0.066. For the southern coastal samples, given the best estimates of the initial Duffy-null allele frequency (0.67) and the current frequency (0.92), and 33 generations of selection, s = 0.065. Given a selection coefficient of 0.066, we thereby estimate that it would take 1646 generations (49 380 years given 30 year generations) for the Duffy-null allele to increase in frequency from 0.01 to 0.99.
4. Discussion
(a). Natural selection for the Duffy-null allele and the role of vivax malaria
Our empirical data and simulation results suggest that genetic drift alone is unlikely to explain the high frequency of the Duffy-null allele in Madagascar. Genetic drift alone is only a likely explanation for the large shift in Duffy-null allele frequency under a Malagasy demographic history characterized by an extremely small population size at the initial time of admixture and many generations of drift. Yet, such a scenario would be expected to simultaneously cause loss of allelic diversity across the genome at a level that is not compatible with observed Malagasy genomic diversity. We therefore conclude that positive natural selection has favoured the Duffy-null allele in Madagascar in the relatively short time since the allele was introduced to the island by migrants from mainland sub-Saharan Africa.
While it has been hypothesized that the Duffy-null allele was favoured in sub-Saharan Africa because it was protective against vivax malaria [35], this account has been questioned [13,16], because: (i) P. vivax was until recently thought to have been transferred to humans from macaques in Asia, rather than Africa [16], (ii) vivax malaria is rarely lethal and the strength of selection for the Duffy-null allele would be expected to have decreased as it increased in frequency and P. vivax became correspondingly more rare [13], and (iii) there was no evidence that vivax malaria was ever common in sub-Saharan Africa [13]. Livingstone [13], in particular, argued that some other selective pressure, related to resistance against an unknown pathogen, resulted in the fixation of the Duffy-null allele in sub-Saharan Africa, and that P. vivax was thereby excluded from invading the region because the people were not susceptible.
Our results contribute to this debate. Specifically, given that the Duffy-null allele is protective against vivax malaria and that vivax malaria is endemic to Madagascar, resistance to P. vivax infection may have been the selective force that has driven the frequency increase of the Duffy-null allele among the Malagasy. While it is not possible at this time to completely exclude key roles for non-malarial selective agents in this evolutionary process, our finding is consistent with the hypothesis that selection for vivax malaria resistance drove the fixation of the Duffy-null allele in mainland sub-Saharan Africa. Moreover, African great apes were recently discovered to be infected with P. vivax [36], and phylogenetic analysis of these and human isolates suggest that all human P. vivax descends from a single clade of the African great ape P. vivax strains, indicating an African origin for human P. vivax [37]. This finding supports the long-term presence of P. vivax in Africa, perhaps allowing for the approximately 49 000 years that we estimate would have been required for selection to fix the Duffy-null allele in Africa.
Given these findings, we propose the following hypothesis for the evolution of the Duffy-null allele in Africa. First, P. vivax initially infected humans in Africa via transmission from sympatric apes [37] and become endemic across Africa prior to the expansion of modern humans out of Africa 50 000–100 000 years ago [38]. The dispersal of modern humans out of Africa may then have led to the spread of P. vivax around the world, resulting in endemicity wherever suitable habitat exists [12]. The Duffy-null allele may be relatively old, as suggested by the occurrence of the allele on both the FY*A and FY*B haplotype backgrounds [10,15,39]. The Duffy-null allele may have risen to appreciable frequency either through drift or selection in West Africa where P. vivax was probably first transmitted to humans [37], but may have still been at low enough frequency in East Africa at the time of the out-of-Africa migration that it was lost during the concomitant bottleneck. This time frame would allow for the long time required for the Duffy-null allele to reach fixation in sub-Saharan Africa given selection for the recessive phenotype and would be consistent with previous estimates indicating that the FY*A and FY*B alleles had not been fully replaced by the Duffy-null allele by approximately 15 000 years ago [39]. In this scenario, much of the selection for the Duffy-null allele would have occurred prior to adoption of agriculture, when population densities would have been relatively low. Consistent with this notion, compared with Plasmodium falciparum malaria, P. vivax malaria endemicity may be possible with lower population densities owing to its relatively long incubation period, relatively lower virulence and recurring nature [13].
(b). Natural selection in admixed populations
The role of recent natural selection in admixed human populations is increasingly recognized as a potentially important force shaping genomic diversity. Current interest in this topic has been fuelled in part by recent reports of admixture between anatomically modern humans and Neandertals [40] and Denisovans [41]. Comparison of a large sample of Eurasian genomes to the Neandertal genome suggests that while purifying selection may have disproportionately removed Neandertal genomic segments containing testes-expressed genes from the Eurasian gene pool, several Neandertal genomic segments containing genes involved in skin pigmentation appear to have increased in frequency at a rate consistent with adaptive introgression [4].
Additionally, studies of modern human populations have scanned the genome for regions with over- or under-represented ancestry relative to the average amount of admixture in African American [42,43], Latino [44–46] and South African Khoe–San hunter–gatherer [47] populations, which could reflect recent positive selection for previously population-specific alleles. The two studies of African Americans identified several such candidate regions, though none overlapped between the studies and none have been associated with a phenotype [42,43]. A scan in Puerto Ricans identified three candidate loci, including the immune system genes of the major histocompatibility complex (MHC) [44]. While this is an intriguing result, the MHC region might be particularly prone to false positive identifications of adaptive introgression in admixed populations because it is typically under strong balancing selection [48]; thus, deeply diverged lineages associated with balancing selection at this locus may have originated prior to the initial population split and been retained, rather than introduced through recent admixture. The Khoe–San study highlighted a region of potential adaptive introgression from Bantu-speaking agriculturalists nearby a gene known to be involved in skin pigmentation [47], although the specific effect of the purported adaptive allele is unknown.
A recent study presented a strong case for selection related to high-altitude adaptation in Tibetan populations, whose genomes reflect past admixture between ancestral populations related to both the lowland Han Chinese and the highland Nepalese Sherpa [3]. The admixed Tibetans have significantly higher proportions of ancestry attributed to the high-altitude ancestral population nearby the EGLN1 and EPAS1 genes than that observed across the remainder of the genome [3]. EGLN1 and EPAS1 variants are thought to play a major role in the elevated haemoglobin phenotype seen in Tibetans and confer a selective advantage in the high-altitude environment [49–51].
Our report of a probable history of positive selection for the Duffy-null allele in the admixed Malagasy is also a strong example of recent selection in an admixed human population, especially considering the clear DARC antigen phenotype associated with the Duffy-null allele and the known associated benefit of high resistance to P. vivax infection. These results highlight the important role that natural selection may play in shaping the genetic diversity of recently admixed populations and the roles of migration and gene flow in providing a potential source of adaptive genetic variants.
Supplementary Material
Acknowledgements
We thank all of the participants in our study, and G. Campbell, T. de Ravel, P. Willem, A. Rakouth and S. Quanbeck for their assistance with sample collection in Madagascar. J.A.H. thanks Austin Burt and James Rosindell for helpful discussions about the analysis.
The Malagasy and South African samples were collected with approval from the Human Research Ethics Committee of the University of Witwatersrand.
Data accessibility
The 10k SNP genotype data generated for this study and the version of the forward simulation program, fastadsim.pl, used for analyses in this paper, have been deposited in the dryad database (doi:10.5061/dryad.r1d6g).
Funding statement
Funding for this work was provided by the College of the Liberal Arts, Penn State University (to G.H.P.), grant no. PTDC/BIA-BDE/68999/2006 (to J.R.) from Fundação para a Ciência e a Tecnologia (FCT, Portugal) and by projects ‘Variabilidade Biológica Humana em Moçaambique’ and ‘STEPS’ at the Pedagogic University and the University Eduardo Mondlane of Mozambique, respectively. H.S. thanks the National Health Laboratory Service, South African Medical Research Council, National Research Council and University of the Witwatersrand for jointly supporting her research.
References
- 1.Sabeti PC, et al. 2007. Genome-wide detection and characterization of positive selection in human populations. Nature 449, 913–918. ( 10.1038/nature06250) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pickrell JK, et al. 2009. Signals of recent positive selection in a worldwide sample of human populations. Genome Res. 19, 826–837. ( 10.1101/gr.087577.108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jeong C, Alkorta-Aranburu G, Basnyat B, Neupane M, Witonsky DB, Pritchard JK, Beall CM, Di Rienzo A. 2014. Admixture facilitates genetic adaptations to high altitude in Tibet. Nat. Commun. 5, 3281 ( 10.1038/ncomms4281) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vernot B, Akey JM. 2014. Resurrecting surviving Neandertal lineages from modern human genomes. Science 343, 1017–1021. ( 10.1126/science.1245938) [DOI] [PubMed] [Google Scholar]
- 5.Tournamille C, Colin Y, Cartron JP, Le Van Kim C. 1995. Disruption of a GATA motif in the Duffy gene promoter abolishes erythroid gene expression in Duffy-negative individuals. Nat. Genet. 10, 224–228. ( 10.1038/ng0695-224) [DOI] [PubMed] [Google Scholar]
- 6.Howes RE, et al. 2011. The global distribution of the Duffy blood group. Nat. Commun. 2, 266 ( 10.1038/ncomms1265) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller LH, Mason SJ, Clyde DF, McGinniss MH. 1976. The resistance factor to Plasmodium vivax in blacks: the Duffy-blood-group genotype, FyFy. N. Engl. J. Med. 295, 302–304. ( 10.1056/NEJM197608052950602) [DOI] [PubMed] [Google Scholar]
- 8.Cavasini CE, et al. 2007. Plasmodium vivax infection among Duffy antigen-negative individuals from the Brazilian Amazon region: an exception? Trans. R. Soc. Trop. Med. Hyg. 101, 1042–1044. ( 10.1016/j.trstmh.2007.04.011) [DOI] [PubMed] [Google Scholar]
- 9.Menard D, et al. 2010. Plasmodium vivax clinical malaria is commonly observed in Duffy-negative Malagasy people. Proc. Natl Acad. Sci. USA 107, 5967–5971. ( 10.1073/pnas.0912496107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zimmerman PA, et al. 1999. Emergence of FY*A(null) in a Plasmodium vivax-endemic region of Papua New Guinea. Proc. Natl Acad. Sci. USA 96, 13 973–13 977. ( 10.1073/pnas.96.24.13973) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Durbin RM, et al. 2010. A map of human genome variation from population-scale sequencing. Nature 467, 1061–1073. ( 10.1038/nature09534) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guerra CA, et al. 2010. The international limits and population at risk of Plasmodium vivax transmission in 2009. PLoS Negl. Trop. Dis. 4, e774 ( 10.1371/journal.pntd.0000774) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Livingstone FB. 1984. The Duffy blood groups, vivax malaria, and malaria selection in human populations: a review. Hum. Biol. 56, 413–425. [PubMed] [Google Scholar]
- 14.Hamblin MT, Thompson EE, Di Rienzo A. 2002. Complex signatures of natural selection at the Duffy blood group locus. Am. J. Hum. Genet. 70, 369–383. ( 10.1086/338628) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamblin MT, Di Rienzo A. 2000. Detection of the signature of natural selection in humans: evidence from the Duffy blood group locus. Am. J. Hum. Genet. 66, 1669–1679. ( 10.1086/302879) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Escalante AA, Cornejo OE, Freeland DE, Poe AC, Durrego E, Collins WE, Lal AA. 2005. A monkey's tale: the origin of Plasmodium vivax as a human malaria parasite. Proc. Natl Acad. Sci. USA 102, 1980–1985. ( 10.1073/pnas.0409652102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perez VR, Godfrey LR, Nowak-Kemp M, Burney DA, Ratsimbazafy J, Vasey N. 2005. Evidence of early butchery of giant lemurs in Madagascar. J. Hum. Evol. 49, 722–742. ( 10.1016/j.jhevol.2005.08.004) [DOI] [PubMed] [Google Scholar]
- 18.Dewar RE, Richard AF. 2012. Madagascar: a history of arrivals, what happened, and will happen next*. Annu. Rev. Anthropol. 41, 495–517. ( 10.1146/annurev-anthro-092611-145758) [DOI] [Google Scholar]
- 19.Dewar RE, Radimilahy C, Wright HT, Jacobs Z, Kelly GO, Berna F. 2013. Stone tools and foraging in northern Madagascar challenge Holocene extinction models. Proc. Natl Acad. Sci. USA 110, 12 583–12 588. ( 10.1073/pnas.1306100110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cox MP, Nelson MG, Tumonggor MK, Ricaut FX, Sudoyo H. 2012. A small cohort of Island southeast Asian women founded Madagascar. Proc. R. Soc. B 279, 2761–2768. ( 10.1098/rspb.2012.0012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tofanelli S, Bertoncini S, Castri L, Luiselli D, Calafell F, Donati G, Paoli G. 2009. On the origins and admixture of Malagasy: new evidence from high-resolution analyses of paternal and maternal lineages. Mol. Biol. Evol. 26, 2109–2124. ( 10.1093/molbev/msp120) [DOI] [PubMed] [Google Scholar]
- 22.Hurles ME, Sykes BC, Jobling MA, Forster P. 2005. The dual origin of the Malagasy in Island southeast Asia and East Africa: evidence from maternal and paternal lineages. Am. J. Hum. Genet. 76, 894–901. ( 10.1086/430051) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tofanelli S, Bertoncini S. 2010. Origin and evolutionary history of the Malagasy. In Encyclopedia of Life Sciences. Chichester, UK: John Wiley & Sons Ltd. [Google Scholar]
- 24.Pierron D, et al. 2014. Genome-wide evidence of Austronesian-Bantu admixture and cultural reversion in a hunter-gatherer group of Madagascar. Proc. Natl Acad. Sci. USA 111, 936–941. ( 10.1073/pnas.1321860111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dewar RE, Wright HT. 1993. The culture history of Madagascar. J. World Prehistory 7, 417–466. ( 10.1007/BF00997802) [DOI] [Google Scholar]
- 26.Beaujard P. 2011. The first migrants to Madagascar and their introduction of plants: linguistic and ethnological evidence. Azania Archaeol. Res. Afr. 46, 169–189. ( 10.1080/0067270X.2011.580142) [DOI] [Google Scholar]
- 27.Blench R. 2007. New palaeozoogeographical evidence for the settlement of Madagascar. Azania Archaeol. Res. Afr. 42, 69–82. ( 10.1080/00672700709480451) [DOI] [Google Scholar]
- 28.Pearson MP, Godden K. 2010. Pastoralists, warriors and colonists: the archaeology of southern Madagascar. Oxford, UK: Archaeopress. [Google Scholar]
- 29.Central Intelligence Agency 2014. Madagascar. In The world factbook. See http://www.cia.gov/library/publications/the-world-factbook/.
- 30.Alexander DH, Novembre J, Lange K. 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664. ( 10.1101/gr.094052.109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pagani L, et al. 2012. Ethiopian genetic diversity reveals linguistic stratification and complex influences on the Ethiopian gene pool. Am. J. Hum. Genet. 91, 83–96. ( 10.1016/j.ajhg.2012.05.015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pickrell JK, Patterson N, Loh PR, Lipson M, Berger B, Stoneking M, Pakendorf B, Reich D. 2014. Ancient west Eurasian ancestry in southern and eastern Africa. Proc. Natl Acad. Sci. USA 111, 2632–2637. ( 10.1073/pnas.1313787111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hodgson JA, Mulligan CJ, Al-Meeri A, Raaum RL. 2014. Early back-to-Africa migration into the Horn of Africa. PLoS Genet. 10, e1004393 ( 10.1371/journal.pgen.1004393) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hartl DL, Clark AG. 2007. Principles of population genetics, 4th edn Sunderland, MA: Sinauer. [Google Scholar]
- 35.Carter R. 2003. Speculations on the origins of Plasmodium vivax malaria. Trends Parasitol. 19, 214–219. ( 10.1016/S1471-4922(03)00070-9) [DOI] [PubMed] [Google Scholar]
- 36.Prugnolle F, et al. 2013. Diversity, host switching and evolution of Plasmodium vivax infecting African great apes. Proc. Natl Acad. Sci. USA 110, 8123–8128. ( 10.1073/pnas.1306004110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu W, et al. 2014. African origin of the malaria parasite Plasmodium vivax. Nat. Commun. 5, 3346 ( 10.1038/ncomms4346) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mellars P. 2006. A new radiocarbon revolution and the dispersal of modern humans in Eurasia. Nature 439, 931–935. ( 10.1038/nature04521) [DOI] [PubMed] [Google Scholar]
- 39.Seixas S, Ferrand N, Rocha J. 2002. Microsatellite variation and evolution of the human Duffy blood group polymorphism. Mol. Biol. Evol. 19, 1802–1806. ( 10.1093/oxfordjournals.molbev.a004003) [DOI] [PubMed] [Google Scholar]
- 40.Green RE, et al. 2010. A draft sequence of the Neandertal genome. Science 328, 710–722. ( 10.1126/science.1188021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reich D, et al. 2010. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature 468, 1053–1060. ( 10.1038/nature09710) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bryc K, et al. 2010. Genome-wide patterns of population structure and admixture in West Africans and African Americans. Proc. Natl Acad. Sci. USA 107, 786–791. ( 10.1073/pnas.0909559107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jin W, Xu S, Wang H, Yu Y, Shen Y, Wu B, Jin L. 2012. Genome-wide detection of natural selection in African Americans pre- and post-admixture. Genome Res. 22, 519–527. ( 10.1101/gr.124784.111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang H, Choudhry S, Mei R, Morgan M, Rodriguez-Cintron W, Burchard EG, Risch NJ. 2007. Recent genetic selection in the ancestral admixture of Puerto Ricans. Am. J. Hum. Genet. 81, 626–633. ( 10.1086/520769) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brisbin A, et al. 2012. PCAdmix: principal components-based assignment of ancestry along each chromosome in individuals with admixed ancestry from two or more populations. Hum. Biol. 84, 343–364. ( 10.3378/027.084.0401) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Basu A, Tang H, Zhu X, Gu CC, Hanis C, Boerwinkle E, Risch N. 2008. Genome-wide distribution of ancestry in Mexican Americans. Hum. Genet. 124, 207–214. ( 10.1007/s00439-008-0541-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schlebusch CM, et al. 2012. Genomic variation in seven Khoe–San groups reveals adaptation and complex African history. Science 338, 374–379. ( 10.1126/science.1227721) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hedrick PW. 1999. Balancing selection and MHC. Genetica 104, 207–214. ( 10.1023/A:1026494212540) [DOI] [PubMed] [Google Scholar]
- 49.Simonson TS, et al. 2010. Genetic evidence for high-altitude adaptation in Tibet. Science 329, 72–75. ( 10.1126/science.1189406) [DOI] [PubMed] [Google Scholar]
- 50.Beall CM, et al. 2010. Natural selection on EPAS1 (HIF2alpha) associated with low hemoglobin concentration in Tibetan highlanders. Proc. Natl Acad. Sci. USA 107, 11 459–11 464. ( 10.1073/pnas.1002443107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yi X, et al. 2010. Sequencing of 50 human exomes reveals adaptation to high altitude. Science 329, 75–78. ( 10.1126/science.1190371) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 10k SNP genotype data generated for this study and the version of the forward simulation program, fastadsim.pl, used for analyses in this paper, have been deposited in the dryad database (doi:10.5061/dryad.r1d6g).