SUMMARY
Neurotropic viruses, including mammalian reovirus, must disseminate from an initial site of replication to the central nervous system (CNS), often binding multiple receptors to facilitate systemic spread. Reovirus engages junctional adhesion molecule-A (JAM-A) to disseminate hematogenously. However, JAM-A is dispensable for reovirus replication in the CNS. We demonstrate that reovirus binds Nogo receptor NgR1, a leucine-rich-repeat protein expressed in the CNS, to infect neurons. Expression of NgR1 confers reovirus binding and infection of non-susceptible cells. Incubating reovirus virions with soluble NgR1 neutralizes infectivity. Blocking NgR1 on transfected cells or primary cortical neurons abrogates reovirus infection. Concordantly, reovirus infection is ablated in primary cortical neurons derived from NgR1-null mice. Reovirus virions bind to soluble JAM-A and NgR1, while infectious disassembly intermediates (ISVPs) bind only to JAM-A. These results suggest that reovirus uses different capsid components to bind distinct cell-surface molecules, engaging independent receptors to facilitate spread and tropism.
INTRODUCTION
Engagement of cellular receptors by viruses is a major determinant of pathogenesis, often dictating host range, mediating spread, and influencing virulence. Neurotropic viruses must navigate diverse cellular and tissue environments to disseminate from an initial site of replication in the periphery to tissues within the CNS, often guided by different receptors along the way (Schneider-Schaulies, 2000; Schweighardt and Atwood, 2001). Spread may occur hematogenously, providing direct entry to the CNS through vascular endothelium, or along nerves. Expression and engagement of specific cell-surface molecules likely dictates these distinct pathways of viral spread and tropism.
Mammalian reoviruses are nonenveloped, double-stranded RNA viruses that use both hematogenous and neural pathways to spread from an initial site of replication in the intestine to the CNS (Boehme et al., 2013b). Most mammals, including humans, are hosts for reovirus infection. While infection seldom results in human disease, rare cases of reovirus encephalitis in young children have been documented (Ouattara et al., 2011). Following peroral inoculation of newborn mice, reovirus is taken up by intestinal M cells and undergoes primary replication in gut-associated lymphoid tissue before accessing a variety of organs including the heart, liver, and brain (Morrison et al., 1991; Virgin et al., 1997). Reovirus displays exquisite serotype-specific patterns of dissemination, tropism within the CNS, and disease outcome. Type 1 (T1) reoviruses spread hematogenously, infect ependymal cells, and cause hydrocephalus, whereas type 3 (T3) reoviruses spread by both hematogenous and neural routes, infect neurons, and cause lethal encephalitis (Antar et al., 2009; Tyler et al., 1986; Weiner et al., 1980). Viral replication and neural pathology colocalize, with a strong predilection for the cerebral cortex, hippocampus, and thalamus (Antar et al., 2009; Oberhaus et al., 1997). Cellular factors that regulate these patterns of systemic dissemination and tropism are unknown. However, the viral σ1 attachment protein is a primary determinant of spread to the CNS (Tyler et al., 1986; Weiner et al., 1980), indicating a key role for receptor recognition in dictating the outcome of infection.
Reovirus initially tethers to the cell surface by σ1 engagement of sialylated glycans with low affinity (Barton et al., 2001a; Chappell et al., 2000; Reiss et al., 2012), which is followed by higher affinity binding to proteinaceous receptors via an adhesion-strengthening mechanism (Barton et al., 2001a). Junctional adhesion molecule A (JAMA), an immunoglobulin superfamily protein expressed in tight junctions and on hematopoietic cells, is the only known proteinaceous receptor for reovirus (Barton et al., 2001b; Campbell et al., 2005). Engagement of endothelial JAM-A is required for establishment of viremia and bloodstream dissemination from the intestine to sites of secondary replication in the host (Antar et al., 2009). However, JAM-A is not required for the distinct patterns of reovirus tropism or replication in the CNS. Moreover, reovirus replicates and causes neurologic disease in JAM-A-deficient (JAM-A−/−) mice following intracranial inoculation (Antar et al., 2009), indicating the existence of alternative neural receptors for reovirus. However, the identity of such receptors is unknown.
We used a whole genome siRNA screen to identify Nogo receptor NgR1 as a reovirus entry mediator. NgR1 is a glycosylphosphatidylinositol (GPI)-anchored, leucine-rich-repeat (LRR) protein expressed on the surface of neurons (Barton et al., 2003; Fournier et al., 2001; He et al., 2003; Wang et al., 2002b). When ligated by one of several myelin-associated proteins, NgR1 elicits intracellular signaling via coreceptors to regulate axonal plasticity in the developing brain and inhibit axonal outgrowth in the adult CNS (Akbik et al., 2012; Hunt et al., 2002; McGee and Strittmatter, 2003). Blockade of NgR1 or its myelin-derived ligands promotes axonal regeneration (Fournier et al., 2002; GrandPre et al., 2002; Li et al., 2004; Wang et al., 2011). As such, the NgR1 pathway has been targeted pharmacologically to improve recovery following stroke and other forms of CNS injury (Lee and Zheng, 2012).
We demonstrate here that NgR1 serves as a reovirus receptor. Expression of NgR1 permits reovirus binding and infection of otherwise non-susceptible cells. Blocking cell-surface NgR1 abrogates infection, including that of primary cortical neurons, a main target of reovirus infection in vivo. Importantly, reovirus infection is ablated in cortical neurons derived from NgR1-null mice. Reovirus virions bind directly to soluble JAM-A and NgR1. However, distinct capsid components mediate these interactions. These results indicate that NgR1 functions as a neural receptor for reovirus, providing evidence that multiple receptors are used to facilitate reovirus systemic spread and CNS tropism.
RESULTS
Expression of NgR1 confers reovirus infection of non-susceptible cells
Binding to JAM-A is required for reovirus bloodstream dissemination (Antar et al., 2009). However, JAM-A is not required for the distinct patterns of reovirus tropism in the brain nor does it serve as a reovirus receptor on cultures of primary cortical neurons (Figure S1). These data indicate that alternate receptors for reovirus are expressed on neurons. To identify host molecules required for reovirus-induced cell toxicity, including new receptor candidates, we performed a genome-wide siRNA screen using HeLa cells. NgR1 was identified as a top cellular target that when inhibited by siRNA knockdown, protected T3 reovirus-infected cells from cell death (Figure S2). Likewise, JAM-A was identified in the screen, suggesting that in some contexts both NgR1 and JAM-A are required for productive infection (Dermody and Mainou, unpublished). While NgR1 is expressed on cervical epithelial cells, which are the origin of HeLa cells, NgR1 expression is most prominent on the surface of CNS neurons (Wang et al., 2002b), including neural cell populations that are targeted by T3 reovirus in vivo (Antar et al., 2009; Oberhaus et al., 1997). Therefore, we investigated NgR1 as a putative reovirus receptor.
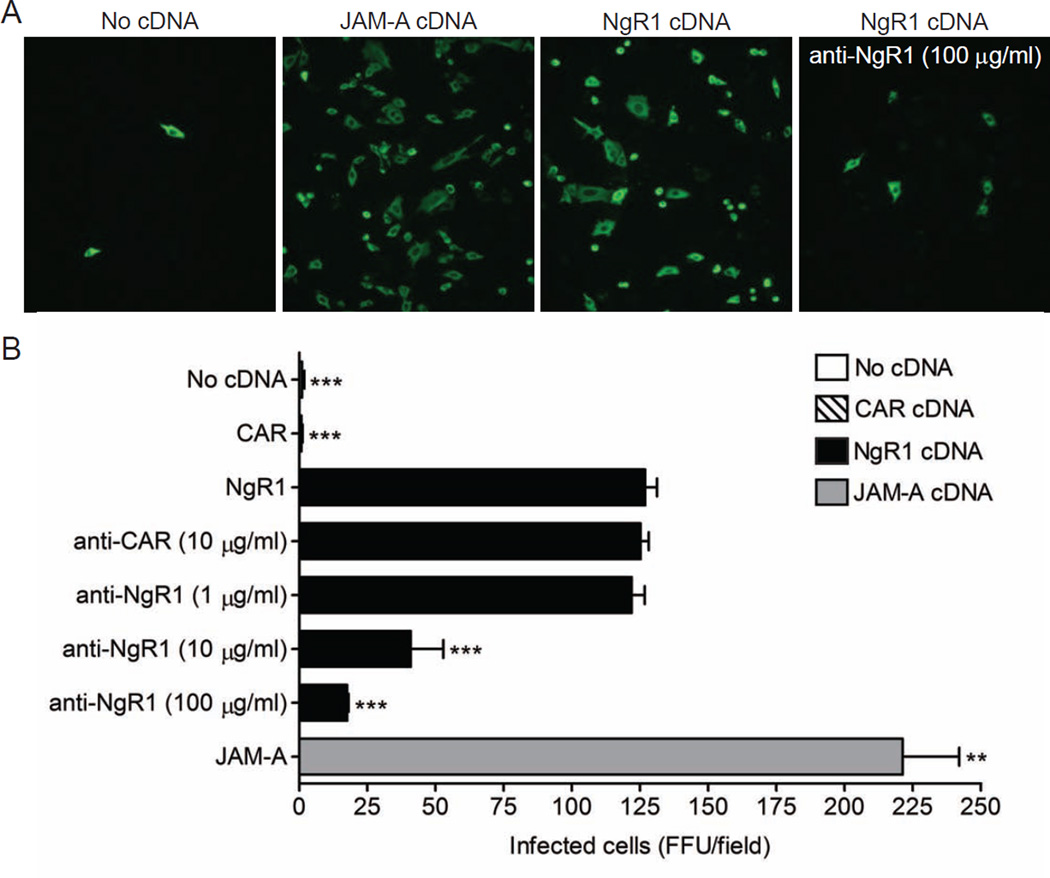
To test the hypothesis that NgR1 functions as a reovirus receptor, we determined whether expression of NgR1 allows infection of Chinese hamster ovary (CHO) cells, which are not susceptible to reovirus. CHO cells were transfected with plasmids encoding NgR1, JAM-A as a positive control, or the coxsackievirus and adenovirus receptor (CAR) as a negative control, and infected with strain T3SA-, which is neurotropic but incapable of binding sialic acid (SA) (Antar et al., 2009; Barton et al., 2001a). The use of T3SA- allows the role of NgR1 engagement to be isolated from that of SA in reovirus infectivity. As expected, JAM-A expression allowed reovirus infection of CHO cells, while CAR-transfected cells remained refractory (Figure 1). Expression of NgR1 also permitted reovirus infection of CHO cells, with a greater than 100-fold increase in the number of infected cells over that in mock- or CAR-transfected cells. Incubation of cells with NgR1-specific antibodies prior to infection abrogated NgR1-mediated infectivity in a dose-dependent manner, suggesting that NgR1 acts to promote reovirus infection at the viral attachment step (Figure 1). In contrast, treatment of transfected cells with neuraminidase (NA) had no effect on NgR1-mediated infectivity, indicating that sialylated glycans are not required for the observed infectivity enhancement (Figure S3). Thus, expression of NgR1 permits reovirus infection of normally non-susceptible cells.
Figure 1. Expression of NgR1 confers reovirus infection of non-susceptible cells.
CHO cells were either mock transfected or transiently transfected with plasmid encoding NgR1, JAM-A, or CAR. After 48 h, cells were pretreated with PBS, a CAR-specific antibody, or increasing concentrations of NgR1-specific antibody prior to being adsorbed with reovirus T3SA- at an MOI of 10 PFU/cell. Cells were fixed at 20 h and scored for reovirus antigen (green) using indirect immunofluorescence. (A) Representative images are shown. (B) Results are expressed as the mean FFU/field for three fields of view in triplicate samples. Error bars indicate SD. **, P < 0.005; ***, P < 0.0005 (as determined by Student’s t test in comparison to NgR1-transfected cells). See also Figures S2, S3, and S6.
Expression of NgR1 allows reovirus binding to non-susceptible cells
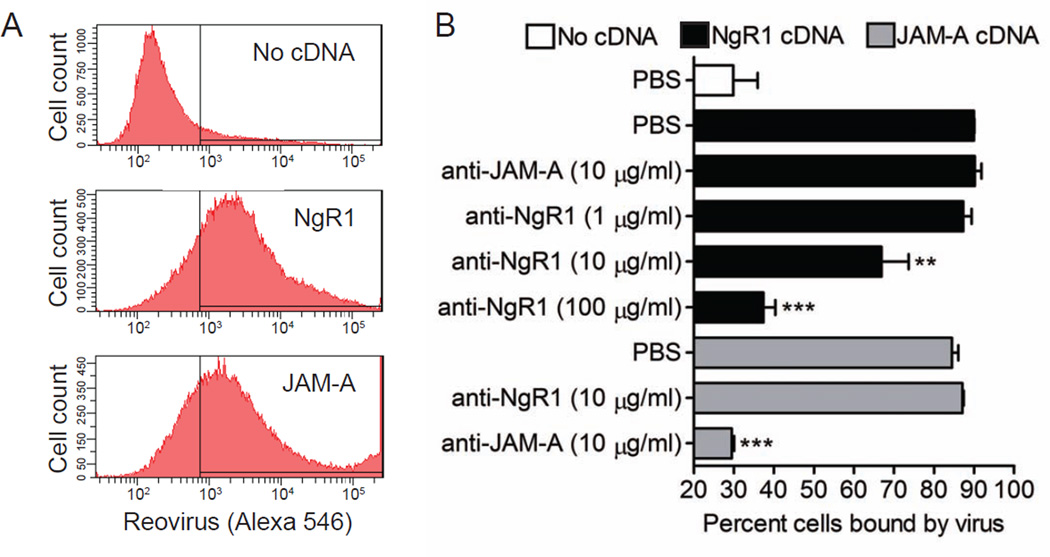
To determine whether NgR1 facilitates infection by enhancing viral attachment, we assessed the capacity of NgR1 expression to increase reovirus binding to the cell surface. CHO cells were mock transfected or transfected with plasmids encoding NgR1 or JAM-A, incubated with fluoresceinated reovirus T3SA-, and scored for reovirus binding using flow cytometry (Figure 2). Both JAM-A and NgR1 significantly increased the capacity of reovirus to bind CHO cells, with 85–90% of cells having bound viral particles compared with 29% of mock-transfected cells (Figure 2A and 2B). When cells were treated with antibodies directed against each receptor prior to infection, reovirus binding was inhibited in an antibody-specific and dose-dependent manner (Figure 2B). These data suggest that NgR1 confers reovirus infectivity by allowing virus binding to the cell surface.
Figure 2. Expression of NgR1 allows reovirus binding to non-susceptible cells.
CHO cells were either mock transfected or transiently transfected with plasmid encoding NgR1 or JAM-A. After 48 h, cells were treated with PBS or antibodies specific for JAM-A or NgR1 prior to being incubated with 105 particles/cell of Alexa Fluor 546-labeled reovirus T3SA- on ice for 1 h. The percentage of cells bound by virus was quantified by flow cytometry. (A) Representative flow cytometric profiles are shown. (B) Results are expressed as the percent cells bound by virus for triplicate samples. Error bars indicate SD. **, P < 0.005; ***, P < 0.0005 (as determined by Student’s t test in comparison to NgR1- or JAM-A-transfected cells).
Reovirus infectivity is abrogated when NgR1 is removed from the cell surface
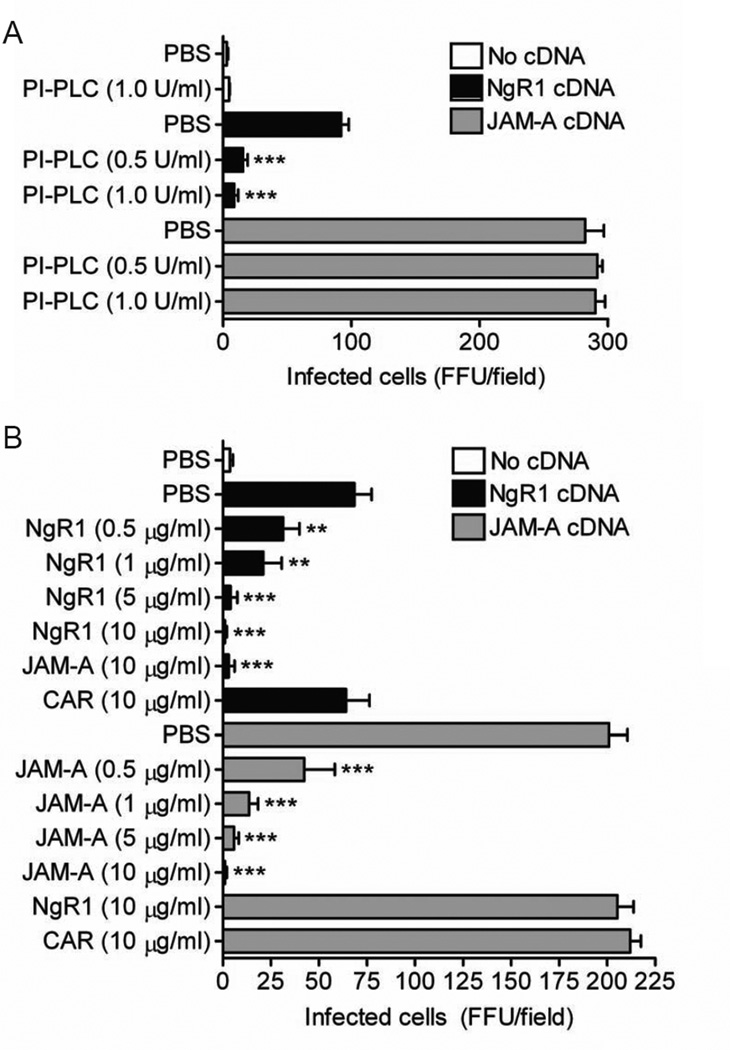
Phosphatidylinositol-specific phospholipase C (PI-PLC) cleaves phosphatidylinositol and serves as an efficient means to release most GPI-anchored proteins from the cell surface, including NgR1 (Fournier et al., 2001). As an additional approach to test whether surface-expressed NgR1 mediates reovirus infection, we used PI-PLC to remove the molecule from the cell surface prior to reovirus adsorption. CHO cells were mock transfected or transfected with plasmids encoding NgR1 or JAM-A, incubated with increasing concentrations of PI-PLC for 1 h prior to inoculation with reovirus T3SA-, and scored for reovirus antigen. Cleaved soluble NgR1 was detected in culture supernatants following PI-PLC treatment (Figure S4). NgR1 expression permitted reovirus infection of otherwise non-susceptible CHO cells as did JAM-A as a positive control (Figure 3A). PI-PLC treatment significantly diminished NgR1-mediated reovirus infectivity (Figure 3A), whereas PI-PLC treatment had no effect on infectivity mediated by JAM-A.
Figure 3. Reovirus infectivity is abrogated when NgR1 is removed from the cell surface or by treatment of virions with soluble NgR1 protein.
CHO cells were either mock transfected or transiently transfected with plasmid encoding NgR1 or JAM-A for 48 h. (A) Transfected cells were treated with PI-PLC, which cleaves GPI-anchored proteins, for 1 h prior to adsorption with reovirus T3SA- at an MOI of 10 PFU/cell. (B) Reovirus T3SA- was incubated with soluble Fc-tagged NgR1, JAM-A, or CAR at the indicated concentrations prior to inoculation of transfected cells at an MOI of 10 PFU/cell. (A and B) Cells were fixed at 20 h and scored for reovirus antigen using indirect immunofluorescence. Results are expressed as the mean FFU/field for three fields of view in triplicate samples. Error bars indicate SD. **, P < 0.005; ***, P < 0.0005 (as determined by Student’s t test in comparison to NgR1- or JAM-A-transfected cells). See also Figure S4.
Treatment of virions with soluble NgR1 neutralizes reovirus infectivity
If reovirus can bind cell-surface NgR1 to infect cells, incubation of virions with soluble NgR1 protein should neutralize NgR1-mediated infectivity. To test this hypothesis, we incubated T3SA- virions with soluble Fc-tagged versions of NgR1, JAM-A, or CAR prior to inoculation of CHO cells transfected with these receptor molecules. Virus treated with PBS or soluble CAR remained infectious and efficiently replicated in CHO cells expressing JAM-A or NgR1 (Figure 3B). As expected, treatment of virions with soluble JAM-A neutralized infection of JAM-A-expressing cells. Likewise, treatment of virions with soluble NgR1 effectively abolished infectivity of NgR1-expressing cells, suggesting that virions and soluble NgR1 interact to prevent the virus from binding cell-associated NgR1. Interestingly, while treatment of virions with soluble JAM-A cross-neutralized infection of NgR1-expressing cells, virions pretreated with soluble NgR1 remained capable of infecting JAM-A-expressing cells (Figure 3B). These results suggest that NgR1 and JAM-A bind distinct components of reovirus virions to mediate infection.
Blocking access to cell-surface NgR1 diminishes reovirus infection of neurons
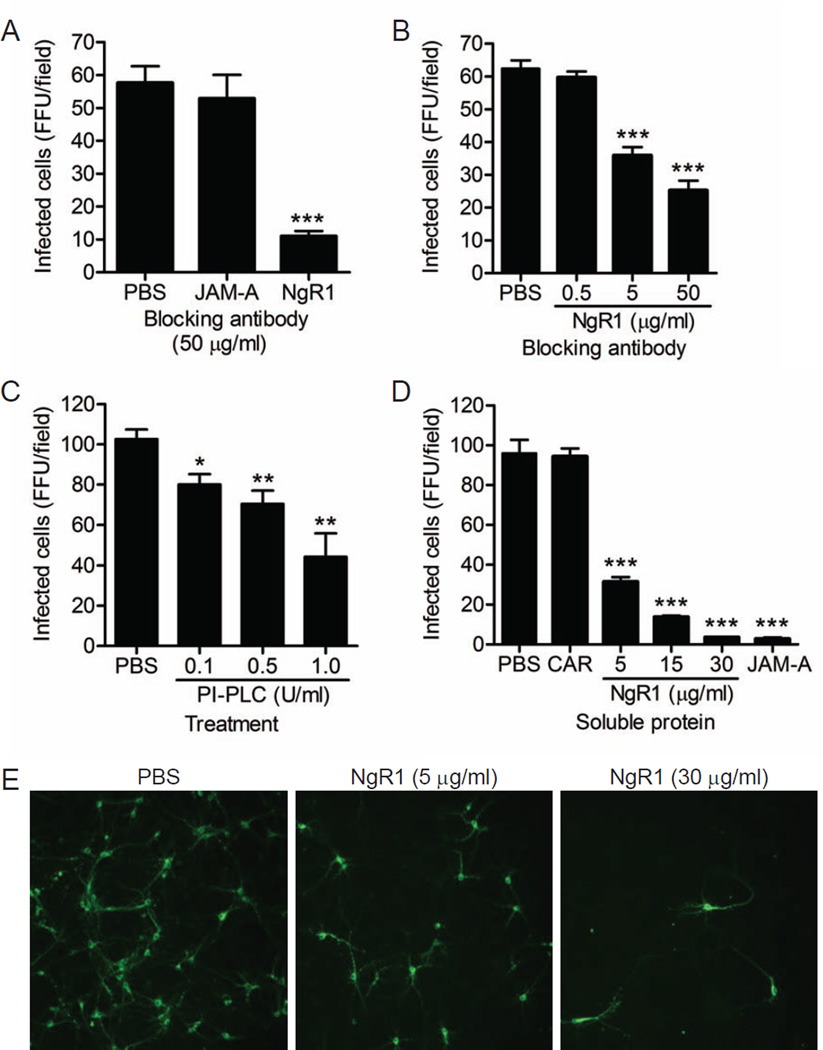
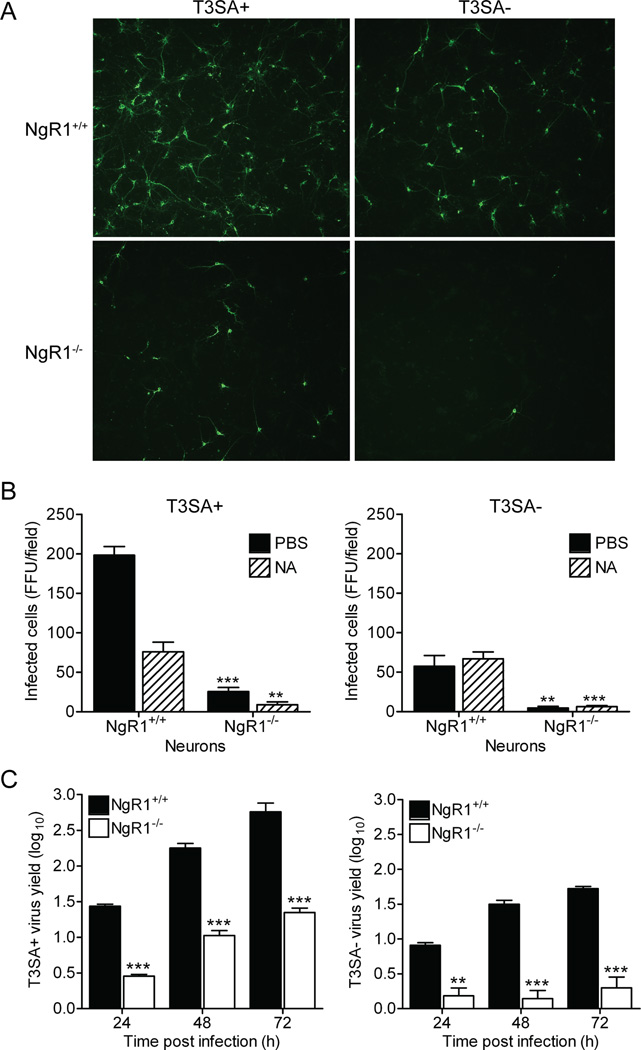
To determine whether NgR1 serves as a reovirus receptor on neurons, we established primary cultures of murine cortical neurons, which are a primary target of T3 reovirus infection in vivo (Antar et al., 2009). Cultured cortical neurons are susceptible to T3 reovirus infection but not T1 reovirus, mimicking the same patterns of serotype-specific tropism observed in vivo (Antar et al., 2009; Dichter and Weiner, 1984). While JAM-A does not serve as a reovirus receptor on cortical neurons (Figure S1), NgR1 is expressed on murine cortical neurons both in culture (Figure S5) and in vivo (Wang et al., 2002b). Cortical neurons prepared from wildtype (Figure 4A) or JAM-A−/− (Figure 4B) embryos were pretreated with either PBS or antibodies specific for JAM-A or NgR1 prior to inoculation with SA-binding reovirus strain T3SA+. While engagement of SA is not required for infection of cortical neurons, reovirus strains that bind SA infect these cells more efficiently than those that do not, allowing for a higher baseline level of infectivity (Antar et al., 2009; Frierson et al., 2012). Treatment of neurons with NgR1-specific antibodies prior to inoculation blocked T3SA+ infection by 60–80% and was dose-dependent, whereas JAM-A-specific antibodies had no effect on infection (Figure 4A and 4B). These results suggest that NgR1 facilitates reovirus infection of cortical neurons. Concordantly, treatment of cortical neurons with PI-PLC diminished T3SA+ infectivity (Figure 4C), providing evidence that GPI-anchored cell-surface molecules such as NgR1 facilitate reovirus infection of neurons.
Figure 4. Blocking access to cell-surface NgR1 diminishes reovirus infection of neurons.
Primary murine cortical neuron cultures prepared from (A) wildtype JAM-A+/+ or (B) isogenic JAM-A−/− embryos were pretreated with either PBS or antibodies specific for JAM-A or NgR1 prior to adsorption with reovirus T3SA+ at an MOI of 500 PFU/cell. (C) Neuron cultures prepared from wildtype cortices were treated with PI-PLC at increasing concentrations for 1 h prior to adsorption with reovirus T3SA+ at an MOI of 500 PFU/cell. (D) Reovirus T3SA+ was preincubated with increasing amounts of soluble Fc-tagged NgR1, JAM-A (30 µg/ml), or CAR (30 µg/ml) prior to inoculation of wildtype neuron cultures at an MOI of 500 PFU/cell. (A – D) Cells were fixed at 20 h and scored for reovirus antigen (green) using indirect immunofluorescence. Results are expressed as the mean FFU/field for three fields of view in triplicate samples. Representative images of an experiment in (D) are shown in (E). Error bars indicate SD. *, P < 0.05; **, P < 0.005; ***, P < 0.0005 (as determined by Student’s t test in comparison to PBS-treated infected cells). See also Figures S1 and S5.
To determine whether binding to soluble NgR1 neutralizes reovirus neural infection, we incubated T3SA+ with increasing concentrations of NgR1 prior to inoculation of cortical neuron cultures (Figure 4D and 4E). NgR1 treatment effectively abolished T3SA+ infection of cortical neurons, with near complete neutralization at the highest concentration of NgR1 protein tested (30 µg/ml). Importantly, incubation of T3SA+ with soluble CAR had no effect on infection. However, soluble JAM-A was capable of neutralizing T3SA+ neural infection (Figure 4D), even though JAM-A is not used as a receptor on these cells. These results indicate that either soluble JAM-A or NgR1 can neutralize the capacity of virions to infect NgR1-expressing cells. However, the determinants of virus binding and mechanisms of neutralization likely differ for the two receptor molecules.
NgR1 is required for efficient reovirus infection of cortical neurons
To directly test whether NgR1 is required for reovirus infection of neurons, we infected primary cortical neuron cultures prepared from embryonic wildtype and NgR1-null (NgR1−/−) mice (Kim et al., 2004) with T3 reovirus. While reovirus T3SA+ and T3SA- efficiently infected wildtype neurons, infection of NgR1−/− neurons by these strains was markedly diminished (Figure 5A and 5B). Residual T3SA+ infectivity was further ablated following treatment of NgR1−/− neurons with neuraminidase (NA) prior to infection (Figure 5B). Additionally, yields of reovirus T3SA+ and T3SA- were significantly diminished in NgR1−/− neurons (Figure 5C). These results provide direct evidence that NgR1 is required for efficient infection of cortical neurons, with glycan engagement enhancing NgR1-mediated infectivity.
Figure 5. NgR1 is required for efficient reovirus infection of primary cortical neurons.
Primary murine cortical neuron cultures prepared from wildtype NgR1+/+ or isogenic NgR1−/− embryos were pretreated with either PBS or NA prior to adsorption with reovirus T3SA+ or T3SA- at an MOI of 500 PFU/cell. Cells were fixed at 20 h and scored for reovirus antigen (green) using indirect immunofluorescence. (A) Representative images are shown. (B) Results are expressed as the mean FFU/field for three fields of view in triplicate samples. (C) Primary murine cortical neuron cultures prepared from wildtype NgR1+/+ or isogenic NgR1−/− embryos were adsorbed with reovirus T3SA+ or T3SA- at an MOI of 10 PFU/cell. Titers of virus in cell lysates at the indicated intervals were determined by plaque. Results are expressed as mean viral yields for triplicate samples. Error bars indicate SD. **, P < 0.005; ***, P < 0.0005 (as determined by Student’s t test in comparison to NgR1+/+ neurons). See also Figure S7.
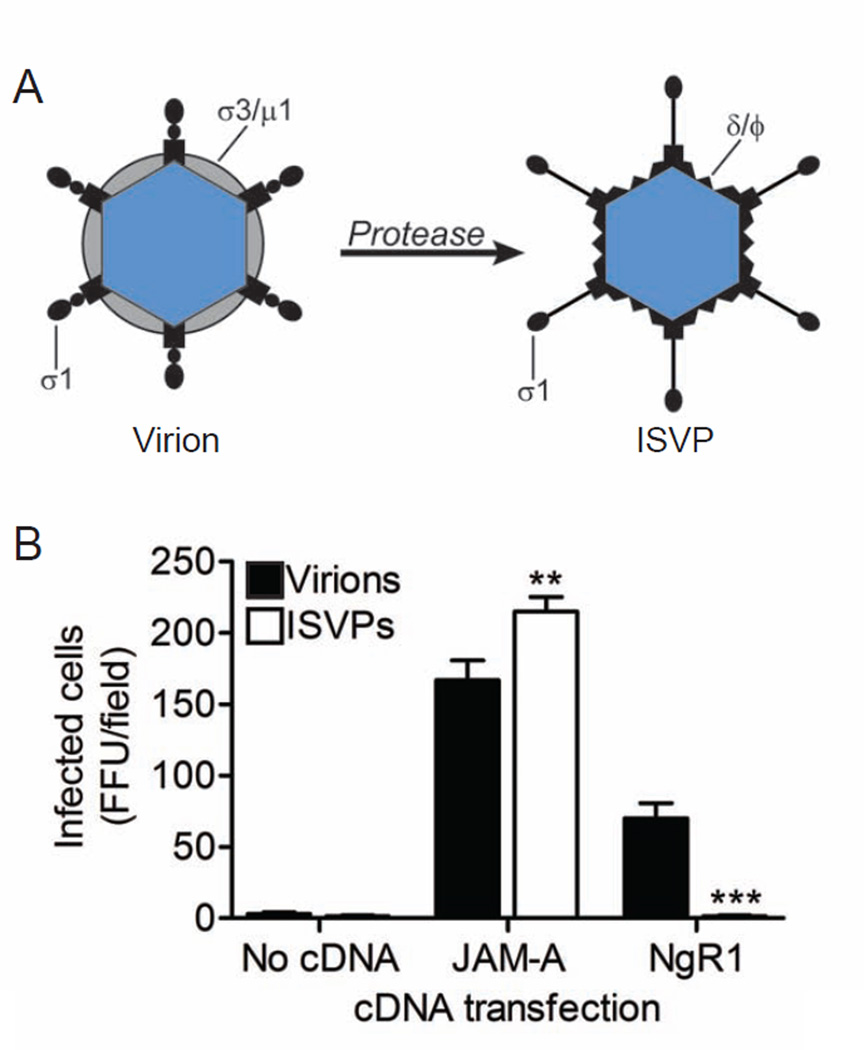
NgR1 mediates infection by reovirus virions but not ISVPs
Exposure of reovirus virions to proteases in the intestinal lumen or in endocytic vesicles leads to formation of ISVPs (Ebert et al., 2002; Johnson et al., 2009; Silverstein et al., 1972), which are obligate intermediates in reovirus disassembly (Sturzenbecker et al., 1987). ISVPs are characterized by loss of outer-capsid protein σ3, an extended conformer of σ1, and cleavage of outer-capsid protein µ1 to form particle-associated fragments δ and Φ (Figure 6A). While ISVPs can bind sialylated glycans and JAM-A to enter cells (Barton et al., 2001b), they do not require endocytic proteolysis to initiate infection. To determine whether cell-surface NgR1 can mediate infection by ISVPs, we transfected CHO cells with NgR1 or JAM-A and compared infectivity using equal particle numbers of T3SA- virions or ISVPs. Both JAM-A and NgR1 allowed infection by reovirus virions (Figure 6B). However, ISVPs failed to infect NgR1-expressing cells in contrast to JAM-A-expressing cells (Figure 6B). These findings indicate that reovirus uses different viral components to engage JAM-A and NgR1. Furthermore, our data suggest that reovirus engages NgR1 using either a virion-associated conformer of σ1 or outer-capsid protein σ3. Since σ3 occludes µ1 on virions, it is unlikely that µ1 contributes to NgR1 binding.
Figure 6. NgR1 mediates infection of reovirus virions but not ISVPs.
(A) Reovirus virions undergo proteolytic disassembly leading to formation of ISVPs. ISVPs are characterized by loss of outer-capsid protein σ3, an extended conformer of σ1, and cleavage of outer-capsid protein µ1 to form δ and Φ. (B) CHO cells were transiently transfected with plasm ids encoding NgR1 or JAM-A and adsorbed with equal particle numbers of reovirus T3SA- virions or ISVPs. Cells were fixed at 20 h scored for reovirus antigen using indirect immunofluorescence. Results are expressed as the mean FFU/field for three fields of view in triplicate samples. Error bars indicate SD. **, P < 0.005; ***, P < 0.0005 (as determined by Student’s t test in comparison to cells infected with virus).
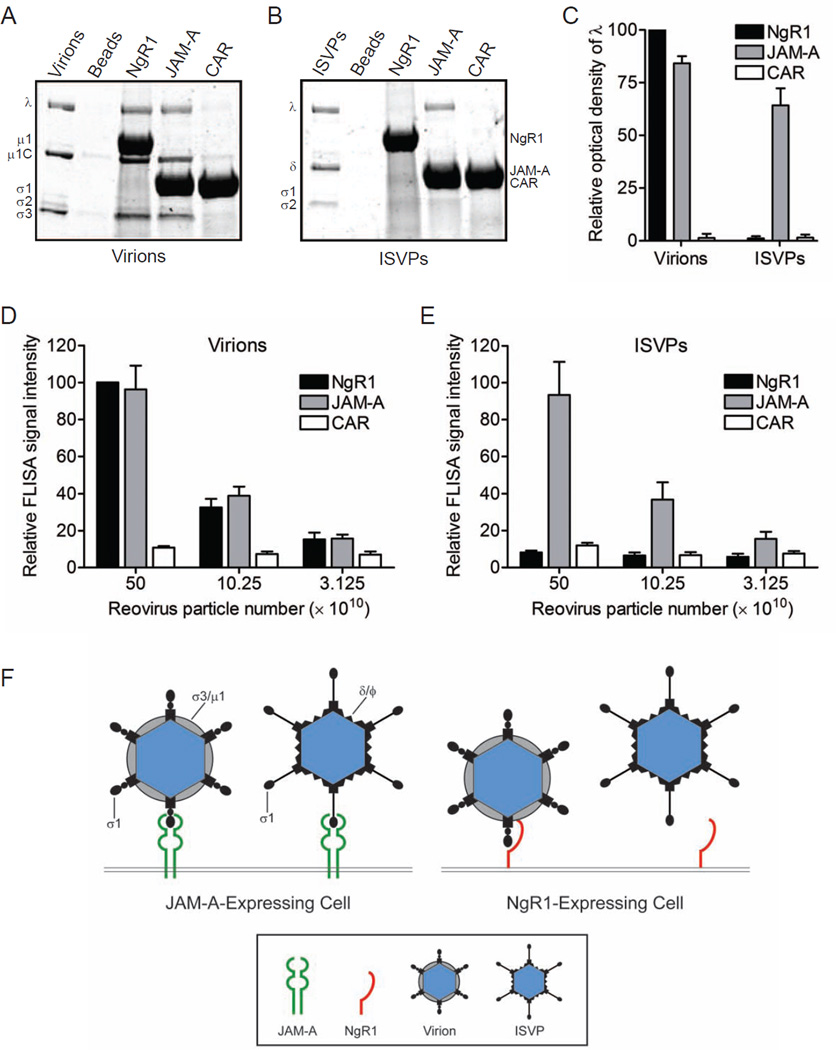
Reovirus virions but not ISVPs bind to soluble NgR1
To determine whether reovirus particles bind directly to NgR1, we used soluble Fc-tagged receptor proteins to assess viral binding in independent assays. Fc-tagged versions of NgR1, JAM-A, or CAR were immobilized onto protein G beads and incubated with 1011 particles of either reovirus T3SA+ virions (Figure 7A) or ISVPs (Figure 7B) in a receptor-binding-bead pulldown assay. Beads were washed extensively and resuspended in protein-dissociation buffer. Precipitated material was resolved by SDS-polyacrylamide gel electrophoresis and visualized by colloidal blue staining. The intensity of bands corresponding to precipitated reovirus λ proteins was determined to quantify virus-receptor interactions. In contrast to unconjugated control or CAR-conjugated beads, reovirus virions were precipitated by JAM-A and NgR1 (Figure 7A and 7C). Concordant with the infectivity data, reovirus ISVPs were precipitated by JAM-A but not NgR1 (Figure 7B and 7C). These results indicate that reovirus virions bind directly to NgR1.
Figure 7. Reovirus virions but not ISVPs bind to soluble NgR1.
Soluble Fc-tagged NgR1, JAM-A, or CAR was immobilized onto protein G beads. Receptor-conjugated beads were incubated with 1011 particles of either reovirus T3SA+ (A) virions or (B) ISVPs, washed extensively, resuspended and boiled in protein-dissociation buffer to release bound material. Precipitated proteins were resolved by SDS-polyacrylamide gel electrophoresis and visualized by colloidal blue staining. (C) Reovirus λ band intensity was quantified by the Odyssey imaging system for three independent experiments. Results are expressed as the mean optical density relative to that of NgR1-precipated λ bands. Error bars indicate SD. (D, E) Plates were coated with soluble Fc-tagged NgR1, JAM-A, or CAR and adsorbed with serial dilutions of reovirus T3SA+ virions (D) or ISVPs (E) in a FLISA binding assay. Wells were washed and incubated with reovirus-specific antibodies and fluorescent secondary antibodies to detect virions or ISVPs bound to receptor protein. Signal intensity was quantified using the Odyssey imaging system. Results are shown as the mean relative FLISA signal for three independent experiments. Error bars indicate SD. (F) Both reovirus virions and ISVPs efficiently bind JAM-A. However, ISVPs are incapable of engaging NgR1, suggesting that the reovirus ligand for NgR1 is either σ3 or the virion-associated conformer of σ1.
In a complementary FLISA binding assay, ELISA plates were coated with soluble Fc-tagged NgR1, JAM-A, or CAR and adsorbed with serial dilutions of either T3SA+ virions (Figure 7D) or ISVPs (Figure 7E). After extensive washing, plates were incubated with reovirus-specific antibodies and fluorescent secondary antibodies to detect virions or ISVPs bound to immobilized receptor. Consistent with the bead-binding assay, reovirus virions bound both JAM-A and NgR1, but ISVPs were capable of binding only JAM-A. Together, our results demonstrate that NgR1 directly interacts with virions, supporting the hypothesis that NgR1 mediates reovirus infection at the cell attachment step.
DISCUSSION
Reovirus spreads systemically by both hematogenous and neural routes to target specific cell populations in the CNS. JAM-A, the only previously known proteinaceous receptor for reovirus, is required for bloodstream spread (Antar et al., 2009). However, JAM-A is dispensable for reovirus neural dissemination and tropism, indicating the existence of alternative receptors for reovirus in the nervous system. Results presented in this report demonstrate that NgR1 serves as a neural entry mediator for reovirus, satisfying criteria of a functional viral receptor. Expression of NgR1 by otherwise non-susceptible cells permits reovirus attachment and infection. Antibodies specific for NgR1 effectively block reovirus binding to and infectivity of NgR1-expressing cells, including cultured primary cortical neurons. T3 reovirus fails to infect cortical neurons derived from NgR1−/− mice, providing evidence that NgR1 is required for neural infection by reovirus. Incubation of virions with soluble NgR1 neutralizes reovirus infection of susceptible cells, suggesting specific binding of virions by the receptor molecule. Finally and importantly, soluble NgR1 engages reovirus virions using two independent binding assays, supporting a direct interaction between reovirus and NgR1.
NgR1 is a GPI-anchored neural receptor belonging to the LRR family of proteins. Expressed on axons of maturing and adult neurons, NgR1 engages ligands associated with myelin on the surface of oligodendrocytes. NgR1 ligands are structurally diverse and include members of the reticulin (Nogo-A) (Fournier et al., 2001), immunoglobulin (myelin-associated glycoprotein) (Domeniconi et al., 2002; Liu et al., 2002), and LRR-containing (oligodendrocyte myelin glycoprotein) (Wang et al., 2002a) protein families. Eight central LRR domains flanked by LRR N- and C-terminal subdomains are responsible for binding ligands within the concave surface of NgR1 (Barton et al., 2003; Lauren et al., 2007). Following engagement of myelin-associated proteins, NgR1 regulates neuronal remodeling in the maturing brain, guiding axonal outgrowth during postnatal refinement of neural circuitry (Hu and Strittmatter, 2004; Hunt et al., 2002). Axonal expression of NgR1 increases with age, as does myelination and the corresponding expression of myelin-associated ligands on oligodendrocytes (Wang et al., 2002b). As such, high concentrations of receptor and ligand interact in the adult brain to effectively restrict neural outgrowth and block axonal repair.
T3 reovirus infects neurons and causes encephalitis in newborn mice. However, adult mice are protected from reovirus CNS disease, becoming resistant to infection within the first few weeks of life. In the newborn CNS, NgR1 is expressed at lower levels on neurons (Wang et al., 2002b). However, the majority of the newborn CNS is unmyelinated. Therefore, the corresponding low concentrations of myelin-associated NgR1 ligands early in life may leave NgR1 free to engage reovirus in the newborn CNS. In fact, reovirus preferentially infects unmyelinated nerve terminals in vivo (Mann et al., 2002), where competition for NgR1 by myelin-associated ligands would presumably be absent. As the CNS matures and myelination becomes more evident, a richer environment of cellular ligands may outcompete reovirus for binding to NgR1, possibly explaining why myelinated neurons are not targeted for reovirus invasion and potentially providing a mechanism for the age restriction of reovirus encephalitis (Tardieu et al., 1983).
If NgR1 serves as a reovirus receptor, expression of the molecule should correspond to areas of reovirus tropism in the brain. Remarkably, sites of NgR1 expression parallel those targeted by T3 reovirus, including the thalamus, the middle and outer layers of the cerebral cortex, the CA3 region of the hippocampus, and cerebellar Purkinjie cells (Antar et al., 2009; Wang et al., 2002b). Factors that facilitate the precise targeting of reovirus to these specific brain regions are not understood. However, host receptor expression is hypothesized as a probable mechanism (Weiner et al., 1980). Importantly, JAM-A does not contribute to reovirus neurotropism, as T3 reovirus infects similar regions in the brain of wildtype and JAM-A−/− mice (Antar et al., 2009). The finding that NgR1 is required for infection of primary cortical neurons suggests that NgR1 may serve as a reovirus receptor in at least a subset of CNS neurons.
Reovirus serotypes display striking differences in CNS tropism, with T1 reovirus infecting ependymal cells causing hydrocephalus and T3 reovirus infecting neurons causing encephalitis. These differences are genetically linked to the σ1 protein (Weiner et al., 1980), which supports the idea that receptors for T1 reovirus are expressed on ependymal cells and those for T3 reovirus are expressed on neurons. As is the case for JAM-A (Barton et al., 2001b; Campbell et al., 2005), NgR1 is capable of allowing infection by both T1 and T3 reovirus, at least in transfected CHO cells (Figure S6). However, cultured primary cortical neurons, which express NgR1, are susceptible only to T3 reovirus and remain refractory to T1 strains (Figure S7; Antar et al., 2009; Dichter and Weiner, 1994). While NgR1 may be capable of binding both serotypes when ectopically expressed in cultured, non-neuronal cells, it is possible that an independent cofactor is required for efficient T1 reovirus binding, entry, or replication that is not expressed by neurons. Alternatively, additional receptors for reovirus may act in concert with NgR1 to promote infection in the CNS. In this regard, it is possible that differences in the cell-surface carbohydrates bound by T1 and T3 reovirus, including the GM2 and GM3 gangliosides, respectively, influence CNS tropism (Reiss et al., 2012). Glycan expression may route the virus to different regions of the brain, where proteinaceous receptors are available for binding and entry into neurons or ependymal cells. In support of this idea, glycan binding enhances reovirus infection of neurons in culture (Figure 5B) and permits efficient neural spread in vivo (Frierson et al., 2012). Studies using NgR1-null mice, which are viable and produce fertile offspring (Kim et al., 2004), will better define the role of NgR1 in reovirus infection in vivo.
Experiments using reovirus virions and ISVPs demonstrate that ISVPs are incapable of engaging NgR1, while both virions and ISVPs efficiently bind JAM-A. Key structural differences between these particle forms may offer insight into the identity of the reovirus ligand for NgR1. During the transition from virions to ISVPs, outer-capsid protein σ3 is shed and σ1 undergoes conformational rearrangement from a more compact form found on virions to an extended, flexible fiber on ISVPs (Dryden et al., 1993; Furlong et al., 1988). Based on these observations, the reovirus ligand for NgR1 is either σ3 or the compact, virion-associated conformer of σ1 (Figure 7F). Cellular receptors have not been reported for σ3, as all available data for mammalian orthoreovirus are consistent with σ1 serving as the viral attachment protein. However, fiberless reovirus strains exist in nature. For example, baboon reovirus (BRV) is a fusogenic reovirus that lacks an adhesion fiber but does express σB, which is a σ3 homolog (Yan et al., 2011). BRV causes meningoencephalomyelitis in juvenile baboons, demonstrating that a fiber is not required for reovirus neural spread or disease (Kumar et al., 2013; Leland et al., 2000). While there are no reports of cellular receptors for BRV, the NgR1-encoding gene is conserved in primates. Alternatively, conformational changes in σ1 associated with the transition of virions to ISVPs have been hypothesized to alter the capacity of σ1 to interact with cell-surface receptors (Bokiej et al., 2012; Dryden et al., 1993).
It is possible that different cellular environments encountered by reovirus in vivo may influence what form of the virus is exposed to host cells. ISVPs are formed by protease digestion of virions in the intestinal lumen. Therefore, functional reovirus receptors at that site should allow engagement by ISVPs. In contrast, intact virions may be the infectious form of the virus that disseminates via neural routes to deliver reovirus to the brain. As such, the capacity of reovirus to use different viral components to engage multiple, structurally distinct receptor molecules may facilitate systemic spread in vivo.
Much remains to be learned about how NgR1 mediates reovirus infection. It is possible that the kinetics of reovirus binding or entry differ depending on whether NgR1 or JAM-A is engaged. Additionally, virus may be routed by these receptors to different intracellular compartments. Of note, following binding to JAM-A, β1 integrin mediates internalization of reovirus into the endocytic pathway (Maginnis et al, 2006). Cellular mediators that facilitate reovirus entry following NgR1 engagement are unknown. However, integrins are implicated in signaling events following binding of cellular ligands to NgR1 (Hu and Strittmatter, 2008). The NF-κB pathway is activated following reovirus entry and serves a critical function in reovirus neuropathology by precipitating apoptosis (O’Donnell et al., 2005). NgR1 lacks a transmembrane domain and instead relies on coreceptors to induce intracellular signaling, primarily through RhoA-dependent pathways (Nikolic, 2002). Defining the downstream signaling events that are initiated by reovirus binding to NgR1, and the coreceptors responsible, may lead to a better understanding of neural cell fate following reovirus infection.
The engagement of specific cellular receptors has major consequences for viral tropism, pathogenesis, and virulence. Viruses that spread systemically, like reovirus, may rely on the coordinated engagement of distinct receptors to navigate diverse host environments in vivo. Indeed, measles virus engages specific receptors to facilitate spread from the periphery to the CNS (Delpeut et al., 2012). By virtue of the multiple tissue types encountered in the infected host, reovirus is an ideal system to investigate stepwise mechanisms of spread and tropism used by neurotropic viruses. Identifying tissue-specific receptors for reovirus, such as NgR1, will facilitate a better understanding of reovirus pathogenesis. Importantly, viral encephalitis remains a significant cause of morbidity and mortality worldwide (Griffin, 2011). Understanding cellular factors that mediate viral neuroinvasion and neurotropism is paramount to developing strategies directed against agents of viral encephalitis. Additionally, reovirus is currently being tested as an anticancer therapeutic in Phase I-III clinical trials (Maitra et al., 2012). New knowledge about cellular factors that influence reovirus infection of specific host cells may enable improved oncolytic vector design by facilitating more precise targeting of tumor cells. Our data also highlight an additional role for NgR1 as a pathogen receptor. Since pathogen-sensing is a well-known property of other LRR proteins including the Toll-like receptors and variable lymphocyte receptors, it is possible that neural LRR proteins in addition to NgR1 function as pathogen recognition factors.
EXPERIMENTAL PROCEDURES
Cells, viruses, and antibodies
CHO cells were grown in Ham’s F12 medium (Gibco) supplemented to contain 10% fetal bovine serum (Invitrogen), 100 U/ml penicillin, 100 µg/ml streptomycin, and 0.25 mg/ml amphotericin B. Primary murine cortical neurons were prepared from murine brain tissue at embryonic day 15 as described (Antar et al., 2009; Boehme et al., 2013a). L929 cells were maintained as described (Barton et al., 2001a).
Reovirus strains T3SA- and T3SA+ consist of nine gene segments from reovirus T1L and the S1 gene segment from either non-SA-binding strain T3C44 (T3SA-) or SA-binding strain T3C44-MA (T3SA+) (Barton et al., 2001a). Purified virions were prepared by cesium chloride gradient centrifugation from infected L929 cells (Furlong et al., 1988). Viral titers were determined by plaque assay using L929 cells (Virgin et al., 1988). ISVPs were generated by treating virions with α-chymotrypsin (Sigma) (Baer and Dermody, 1997). Reovirus virions were labeled with succinimidyl ester Alexa Fluor 546 (A546; Invitrogen) to generate fluoresceinated particles (Mainou and Dermody, 2012).
Purified immunoglobulin G (IgG) fractions of rabbit antisera raised against reovirus (Barton et al., 2001a; Wetzel et al., 1997) were used to detect reovirus antigen by indirect immunofluorescence, along with Alexa Fluor-conjugated secondary antibodies (Invitrogen). JAM-A-specific monoclonal antibody J10.4 (provided by Charles Parkos, Emory University) was used to block human JAM-A on transfected cells. JAM-A-specific monoclonal antibody BV20 (provided by Elisabetta Dejana, FIRC Institute of Molecular Oncology, University of Milan) was used to block murine JAM-A on cortical neurons. NgR1-specific polyclonal antibody AF1208 (R&D Systems) was used to block NgR1 on CHO cells and cortical neurons. Antibodies against cell-surface proteins were preincubated with cells at various concentrations in PBS at 37°C for 1 h prior to adsorption of virus.
Plasmids, transient transfection, and quantitation of reovirus infectivity
Plasmids encoding human JAM-A and human CAR have been described (Barton et al., 2001b). Plasmid encoding human NgR1 was obtained from OriGene Technologies. CHO cells were transfected with 0.5 mg of plasmid using FuGENE 6 transfection reagent (Promega). After 48 h, transfected cells were adsorbed with reovirus virions or ISVPs at an MOI of 10 PFU/cell. Cells were fixed after 20 h, and reovirus antigen was visualized by indirect immunofluorescence (Barton et al., 2001a).
Assessment of reovirus binding by flow cytometry
Monolayers of transfected CHO cells were washed with PBS, detached with Cellstripper (Cellgro), quenched with PBS containing 2% FBS, pelleted, and incubated with 105 particles of A546-labeled reovirus T3SA- in PBS on ice for 1 h (Mainou and Dermody, 2012). Cells were washed and fixed in PBS containing 1% paraformaldehyde. For some experiments, anti-receptor antibodies were incubated with detached cells at room temperature for 1 h prior to adsorption with virus. Cells were analyzed using an LSRII flow cytometer (BD Bioscience) and quantified using FlowJo software.
Virus replication
Primary cortical neuron cultures were adsorbed with reovirus at an MOI of 10 PFU/cell for 1 h, washed with PBS, and incubated for various intervals. Cells were frozen and thawed twice prior to determination of viral titers by plaque assay using L929 cells (Virgin et al., 1988). Viral yields were calculated according to the following formula: log10 yieldtx = log10 (pfu/ml)tx - log10 (pfu/ml)t0, where tx is the time postinfection.
PI-PLC treatment
GPI-anchored proteins were removed from the cell surface by incubating cells with various concentrations of PI-PLC (Molecular Probes) at 37°C for 1 h. Cells were washed with serum-free media prior to adsorption with reovirus, fixed at 20 h, and scored for reovirus antigen.
Neutralization of reovirus with soluble receptor protein
Purification of soluble Fc-tagged human JAM-A and CAR proteins has been described (Barton et al., 2001b). Fc-tagged human NgR1 protein was obtained from R&D Systems. Reovirus T3SA- was incubated with soluble Fc-tagged NgR1, JAM-A, or CAR at various concentrations in PBS at 4°C overnight. Mock-treated virus was incubated in PBS at 4°C overnight. Cells were inoculated at an MOI of 10 PFU/cell, fixed at 20 h, and scored for reovirus antigen.
Neuraminidase treatment
Terminal SA residues were removed from cell-surface glycans by incubation with 40 mU/mL A. ureafaciens neuraminidase at 37°C for 1 h. Cells were washed prior to absorption with reovirus. After 20 h incubation, cells were fixed and scored for reovirus antigen.
Receptor-binding bead pull-down assay
Soluble Fc-tagged NgR1, JAM-A, or CAR was immobilized onto protein G beads (Dynabeads®, Life Technologies) by incubating 25 µg protein per 100 µl beads in PBS at 4°C for 4 h. Receptor-conjugated beads were washed with PBS and incubated with 1011 particles of reovirus T3SA+ virions or ISVPs at 4°C overnight. Beads were washed extensively, resuspended in protein-dissociation buffer, and boiled to release bound material. Precipitated proteins were resolved by SDS-polyacrylamide gel electrophoresis and visualized by colloidal blue staining (Invitrogen). Intensity of the reovirus λ bands was quantified using the Odyssey Imaging System (LI-COR Biosciences). Results are expressed as the optical density relative to that of NgR1-precipated λ bands.
Receptor-binding fluorescence-linked immunosorbent assay (FLISA)
High-binding 96-well Immulon assay plates (Fisher Scientific) were coated with 20 µg/ml soluble Fc-tagged NgR1, JAM-A, or CAR in carbonate-bicarbonate buffer at 4°C overnight. Wells were incubated with LI-COR blocking buffer (LI-COR Biosciences) at 4°C for 4 h and washed with PBS containing 0.05% Tween 20. Reovirus T3SA+ virions or ISVPs serially diluted in TBS containing 0.1% BSA and 0.05% Tween were added to the wells and incubated at 4°C overnight. Wells were washed prior to adding reovirus-specific monoclonal antibody 9BG5 (Burstin et al., 1982) at room temperature for 2 h. Wells were washed and incubated with fluorescent secondary antibodies (LI-COR) at room temperature for 2 h (Bokiej et al., 2012). Following washing, signal intensity was quantified using the Odyssey Imaging System. Results are expressed as the FLISA signal relative to that of NgR1-coated wells.
Statistical analysis
Means of results from triplicate samples were compared using an unpaired Student’s t test. Statistical analyses were performed using Prism software (GraphPad Software, Inc.). P values of < 0.05 were considered to be statistically significant.
Supplementary Material
HIGHLIGHTS.
NgR1 is a neural entry mediator for mammalian reovirus.
Expression of NgR1 permits infection of non-susceptible cells.
NgR1 is required for efficient infection of primary cortical neurons.
Distinct capsid proteins bind NgR1 and JAM-A, likely with unique disease functions.
ACKNOWLEDGEMENTS
We are grateful to members of the Dermody laboratory for many useful discussions. We thank Karl Boehme, Carolyn Coyne, Julie Pfeiffer, and Thilo Stehle for critical review of the manuscript. NgR1 was identified by siRNA screening conducted with assistance from Borden Lacey’s laboratory and the Vanderbilt High-Throughput Screening Facility. Soluble Fc-tagged JAM-A and CAR were produced and purified by the Vanderbilt Antibody and Protein Resource. We are grateful to Jeffrey Bergelson for providing Fc-CAR plasmid and CAR-specific antiserum. We thank Charles Parkos and Elisabetta Dejana for providing JAM-A-specific antibodies.
This research was supported by Public Health Service awards T32 CA009385 (J.L.K.), F32 AI081486 (J.L.K.), F32 AI080108 (B.A.M.), and R37 AI038296 (T.S.D.), an RNAi Discovery Grant from Thermo Fisher Scientific, and the Elizabeth B. Lamb Center for Pediatric Research. This work also was supported by grants to S.M.S. from the NIH and the Falk Medical Research Trust. Additional support was provided by the Vanderbilt Flow Cytometry Shared Resource (P30 DK058404), the Vanderbilt Institute of Chemical Biology, and the Vanderbilt Ingram Cancer Center (P30 CA68485).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
J.L.K. designed and performed the experiments, analyzed the results, and wrote the manuscript. B.A.M. and D.M.S. designed and performed the experiments and analyzed the results. Y.S. and S.M.S. provided critical reagents and experimental advice. T.S.D. designed the experiments, analyzed the results, and wrote the manuscript.
REFERENCES
- Akbik F, Cafferty WB, Strittmatter SM. Myelin associated inhibitors: a link between injury-induced and experience-dependent plasticity. Exp. Neurol. 2012;235:43–52. doi: 10.1016/j.expneurol.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antar AAR, Konopka JL, Campbell JA, Henry RA, Perdigoto AL, Carter BD, Pozzi A, Abel TW, Dermody TS. Junctional adhesion molecule-A is required for hematogenous dissemination of reovirus. Cell Host Microbe. 2009;5:59–71. doi: 10.1016/j.chom.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer GS, Dermody TS. Mutations in reovirus outer-capsid protein σ3 selected during persistent infections of L cells confer resistance to protease inhibitor E64. J. Virol. 1997;71:4921–4928. doi: 10.1128/jvi.71.7.4921-4928.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton ES, Connolly JL, Forrest JC, Chappell JD, Dermody TS. Utilization of sialic acid as a coreceptor enhances reovirus attachment by multistep adhesion strengthening. J. Biol. Chem. 2001a;276:2200–2211. doi: 10.1074/jbc.M004680200. [DOI] [PubMed] [Google Scholar]
- Barton ES, Forrest JC, Connolly JL, Chappell JD, Liu Y, Schnell F, Nusrat A, Parkos CA, Dermody TS. Junction adhesion molecule is a receptor for reovirus. Cell. 2001b;104:441–451. doi: 10.1016/s0092-8674(01)00231-8. [DOI] [PubMed] [Google Scholar]
- Barton WA, Liu BP, Tzvetkova D, Jeffrey PD, Fournier AE, Sah D, Cate R, Strittmatter SM, Nikolov DB. Structure and axon outgrowth inhibitor binding of the Nogo-66 receptor and related proteins. EMBO J. 2003;22:3291–3302. doi: 10.1093/emboj/cdg325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehme KW, Hammer K, Tollefson WC, Konopka-Anstadt JL, Kobayashi T, Dermody TS. Nonstructural Protein sigma1s Mediates Reovirus-Induced Cell Cycle Arrest and Apoptosis. J. Virol. 2013a;87:12967–12979. doi: 10.1128/JVI.02080-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehme KW, Lai CM, Dermody TS. Mechanisms of reovirus bloodstream dissemination. Adv. Virus Res. 2013b;87:1–35. doi: 10.1016/B978-0-12-407698-3.00001-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokiej M, Ogden KM, Ikizler M, Reiter DM, Stehle T, Dermody TS. Optimum length and flexibility of reovirus attachment protein σ1 are required for efficient viral infection. J. Virol. 2012;86:10270–10280. doi: 10.1128/JVI.01338-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burstin SJ, Spriggs DR, Fields BN. Evidence for functional domains on the reovirus type 3 hemagglutinin. Virology. 1982;117:146–155. doi: 10.1016/0042-6822(82)90514-1. [DOI] [PubMed] [Google Scholar]
- Campbell JA, Shelling P, Wetzel JD, Johnson EM, Wilson GAR, Forrest JC, Aurrand-Lions M, Imhof B, Stehle T, Dermody TS. Junctional adhesion molecule-A serves as a receptor for prototype and field-isolate strains of mammalian reovirus. J. Virol. 2005;79:7967–7978. doi: 10.1128/JVI.79.13.7967-7978.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell JD, Duong JL, Wright BW, Dermody TS. Identification of carbohydrate-binding domains in the attachment proteins of type 1 and type 3 reoviruses. J. Virol. 2000;74:8472–8479. doi: 10.1128/jvi.74.18.8472-8479.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delpeut S, Noyce RS, Siu RW, Richardson CD. Host factors and measles virus replication. Curr. Opin. Virol. 2012;2:773–783. doi: 10.1016/j.coviro.2012.10.008. [DOI] [PubMed] [Google Scholar]
- Dichter MA, Weiner HL. Infection of neuronal cell cultures with reovirus mimics in vitro patterns of neurotropism. Ann. Neurol. 1984;16:603–610. doi: 10.1002/ana.410160512. [DOI] [PubMed] [Google Scholar]
- Domeniconi M, Cao Z, Spencer T, Sivasankaran R, Wang K, Nikulina E, Kimura N, Cai H, Deng K, Gao Y, et al. Myelin-associated glycoprotein interacts with the Nogo66 receptor to inhibit neurite outgrowth. Neuron. 2002;35:283–290. doi: 10.1016/s0896-6273(02)00770-5. [DOI] [PubMed] [Google Scholar]
- Dryden KA, Wang G, Yeager M, Nibert ML, Coombs KM, Furlong DB, Fields BN, Baker TS. Early steps in reovirus infection are associated with dramatic changes in supramolecular structure and protein conformation: analysis of virions and subviral particles by cryoelectron microscopy and image reconstruction. J. Cell Biol. 1993;122:1023–1041. doi: 10.1083/jcb.122.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert DH, Deussing J, Peters C, Dermody TS. Cathepsin L and cathepsin B mediate reovirus disassembly in murine fibroblast cells. J. Biol. Chem. 2002;277:24609–24617. doi: 10.1074/jbc.M201107200. [DOI] [PubMed] [Google Scholar]
- Fournier AE, Gould GC, Liu BP, Strittmatter SM. Truncated soluble Nogo receptor binds Nogo-66 and blocks inhibition of axon growth by myelin. J. Neurosci. 2002;22:8876–8883. doi: 10.1523/JNEUROSCI.22-20-08876.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier AE, GrandPre T, Strittmatter SM. Identification of a receptor mediating Nogo-66 inhibition of axonal regeneration. Nature. 2001;409:341–346. doi: 10.1038/35053072. [DOI] [PubMed] [Google Scholar]
- Frierson JM, Pruijssers AJ, Konopka JL, Reiter DM, Abel TW, Stehle T, Dermody TS. Utilization of sialylated glycans as coreceptors enhances the neurovirulence of serotype 3 reovirus. J. Virol. 2012;86:13164–13173. doi: 10.1128/JVI.01822-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlong DB, Nibert ML, Fields BN. Sigma 1 protein of mammalian reoviruses extends from the surfaces of viral particles. J. Virol. 1988;62:246–256. doi: 10.1128/jvi.62.1.246-256.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GrandPre T, Li S, Strittmatter SM. Nogo-66 receptor antagonist peptide promotes axonal regeneration. Nature. 2002;417:547–551. doi: 10.1038/417547a. [DOI] [PubMed] [Google Scholar]
- Griffin DE. Viral encephalomyelitis. PLoS Pathog. 2011;7:e1002004. doi: 10.1371/journal.ppat.1002004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He XL, Bazan JF, McDermott G, Park JB, Wang K, Tessier-Lavigne M, He Z, Garcia KC. Structure of the Nogo receptor ectodomain: a recognition module implicated in myelin inhibition. Neuron. 2003;38:177–185. doi: 10.1016/s0896-6273(03)00232-0. [DOI] [PubMed] [Google Scholar]
- Hu F, Strittmatter SM. Regulating axon growth within the postnatal central nervous system. Semin. Perinatol. 2004;28:371–378. doi: 10.1053/j.semperi.2004.10.001. [DOI] [PubMed] [Google Scholar]
- Hu F, Strittmatter SM. The N-terminal domain of Nogo-A inhibits cell adhesion and axonal outgrowth by an integrin-specific mechanism. J. Neurosci. 2008;28:1262–1269. doi: 10.1523/JNEUROSCI.1068-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt D, Coffin RS, Anderson PN. The Nogo receptor, its ligands and axonal regeneration in the spinal cord; a review. J. Neurocytol. 2002;31:93–120. doi: 10.1023/a:1023941421781. [DOI] [PubMed] [Google Scholar]
- Johnson EM, Doyle JD, Wetzel JD, McClung RP, Katunuma N, Chappell JD, Washington MK, Dermody TS. Genetic and pharmacologic alteration of cathepsin expression influences reovirus pathogenesis. J. Virol. 2009;83:9630–9640. doi: 10.1128/JVI.01095-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JE, Liu BP, Park JH, Strittmatter SM. Nogo-66 receptor prevents raphespinal and rubrospinal axon regeneration and limits functional recovery from spinal cord injury. Neuron. 2004;44:439–451. doi: 10.1016/j.neuron.2004.10.015. [DOI] [PubMed] [Google Scholar]
- Kumar S, Dick EJ, Jr., Reddy BY, Yang A, Mubiru J, Hubbard GB, Owston MA. Reovirus-associated meningoencephalomyelitis in baboons. Vet. Pathol. 2013 doi: 10.1177/0300985813497487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauren J, Hu F, Chin J, Liao J, Airaksinen MS, Strittmatter SM. Characterization of myelin ligand complexes with neuronal Nogo-66 receptor family members. J. Biol. Chem. 2007;282:5715–5725. doi: 10.1074/jbc.M609797200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JK, Zheng B. Role of myelin-associated inhibitors in axonal repair after spinal cord injury. Exp. Neurol. 2012;235:33–42. doi: 10.1016/j.expneurol.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leland MM, Hubbard GB, Sentmore HT, Soike KF, 3rd, Hilliard JK. Outbreak of Orthoreovirus-induced meningoencephalomyelitis in baboons. Comp. Med. 2000;50:199–205. [PubMed] [Google Scholar]
- Li S, Liu BP, Budel S, Li M, Ji B, Walus L, Li W, Jirik A, Rabacchi S, Choi E, et al. Blockade of Nogo-66, myelin-associated glycoprotein, and oligodendrocyte myelin glycoprotein by soluble Nogo-66 receptor promotes axonal sprouting and recovery after spinal injury. J. Neurosci. 2004;24:10511–10520. doi: 10.1523/JNEUROSCI.2828-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu BP, Fournier A, GrandPre T, Strittmatter SM. Myelin-associated glycoprotein as a functional ligand for the Nogo-66 receptor. Science. 2002;297:1190–1193. doi: 10.1126/science.1073031. [DOI] [PubMed] [Google Scholar]
- Maginnis MS, Forrest JC, Kopecky-Bromberg SA, Dickeson SK, Santoro SA, Zutter MM, Nemerow GR, Bergelson JM, Dermody TS. β1 integrin mediates internalization of mammalian reovirus. J. Virol. 2006;80:2760–2770. doi: 10.1128/JVI.80.6.2760-2770.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mainou BA, Dermody TS. Transport to late endosomes is required for efficient reovirus infection. J. Virol. 2012;86:8346–8358. doi: 10.1128/JVI.00100-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitra R, Ghalib MH, Goel S. Reovirus: a targeted therapeutic-progress and potential. Mol. Cancer Res. 2012;10:1514–1525. doi: 10.1158/1541-7786.MCR-12-0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann MA, Knipe DM, Fischbach GD, Fields BN. Type 3 reovirus neuroinvasion after intramuscular inoculation: direct invasion of nerve terminals and age-dependent pathogenesis. Virology. 2002;303:222–231. doi: 10.1006/viro.2002.1699. [DOI] [PubMed] [Google Scholar]
- McGee AW, Strittmatter SM. The Nogo-66 receptor: focusing myelin inhibition of axon regeneration. Trends Neurosci. 2003;26:193–198. doi: 10.1016/S0166-2236(03)00062-6. [DOI] [PubMed] [Google Scholar]
- Morrison LA, Sidman RL, Fields BN. Direct spread of reovirus from the intestinal lumen to the central nervous system through vagal autonomic nerve fibers. Proc. Natl. Acad. Sci. USA. 1991;88:3852–3856. doi: 10.1073/pnas.88.9.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolic M. The role of Rho GTPases and associated kinases in regulating neurite outgrowth. Int. J. Biochem. Cell Biol. 2002;34:731–745. doi: 10.1016/s1357-2725(01)00167-4. [DOI] [PubMed] [Google Scholar]
- O’Donnell SM, Hansberger MW, Connolly JL, Chappell JD, Watson MJ, Pierce JM, Wetzel JD, Han W, Barton ES, Forrest JC, et al. Organ-specific roles for transcription factor NF-kB in reovirus-induced apoptosis and disease. J. Clin. Invest. 2005;115:2341–2350. doi: 10.1172/JCI22428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberhaus SM, Smith RL, Clayton GH, Dermody TS, Tyler KL. Reovirus infection and tissue injury in the mouse central nervous system are associated with apoptosis. J. Virol. 1997;71:2100–2106. doi: 10.1128/jvi.71.3.2100-2106.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouattara LA, Barin F, Barthez MA, Bonnaud B, Roingeard P, Goudeau A, Castelnau P, Vernet G, Paranhos-Baccala G, Komurian-Pradel F. Novel human reovirus isolated from children with acute necrotizing encephalopathy. Emerging Infect. Dis. 2011;17:1436–1444. doi: 10.3201/eid1708.101528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss K, Stencel JE, Liu Y, Blaum BS, Reiter DM, Feizi T, Dermody TS, Stehle T. The GM2 glycan serves as a functional co-receptor for serotype 1 reovirus. PLoS Path. 2012;8:e1003078. doi: 10.1371/journal.ppat.1003078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider-Schaulies J. Cellular receptors for viruses: links to tropism and pathogenesis. J. Gen. Virol. 2000;81:1413–1429. doi: 10.1099/0022-1317-81-6-1413. [DOI] [PubMed] [Google Scholar]
- Schweighardt B, Atwood WJ. Virus receptors in the human central nervous system. J. Neurovirol. 2001;7:187–195. doi: 10.1080/13550280152403236. [DOI] [PubMed] [Google Scholar]
- Silverstein SC, Astell C, Levin DH, Schonberg M, Acs G. The mechanism of reovirus uncoating and gene activation in vivo. Virology. 1972;47:797–806. doi: 10.1016/0042-6822(72)90571-5. [DOI] [PubMed] [Google Scholar]
- Sturzenbecker LJ, Nibert ML, Furlong DB, Fields BN. Intracellular digestion of reovirus particles requires a low pH and is an essential step in the viral infectious cycle. J. Virol. 1987;61:2351–2361. doi: 10.1128/jvi.61.8.2351-2361.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardieu M, Powers ML, Weiner HL. Age-dependent susceptibility to reovirus type 3 encephalitis: role of viral and host factors. Ann. Neurol. 1983;13:602–607. doi: 10.1002/ana.410130604. [DOI] [PubMed] [Google Scholar]
- Tyler KL, McPhee DA, Fields BN. Distinct pathways of viral spread in the host determined by reovirus S1 gene segment. Science. 1986;233:770–774. doi: 10.1126/science.3016895. [DOI] [PubMed] [Google Scholar]
- Virgin HW, IV, Bassel-Duby R, Fields BN, Tyler KL. Antibody protects against lethal infection with the neurally spreading reovirus type 3 (Dearing) J. Virol. 1988;62:4594–4604. doi: 10.1128/jvi.62.12.4594-4604.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgin HW, Tyler KL, Dermody TS. In: Reovirus. In Viral Pathogenesis. Nathanson N, editor. New York: Lippincott-Raven; 1997. pp. 669–699. [Google Scholar]
- Wang KC, Koprivica V, Kim JA, Sivasankaran R, Guo Y, Neve RL, He Z. Oligodendrocyte-myelin glycoprotein is a Nogo receptor ligand that inhibits neurite outgrowth. Nature. 2002a;417:941–944. doi: 10.1038/nature00867. [DOI] [PubMed] [Google Scholar]
- Wang X, Chun SJ, Treloar H, Vartanian T, Greer CA, Strittmatter SM. Localization of Nogo-A and Nogo-66 receptor proteins at sites of axon-myelin and synaptic contact. J. Neurosci. 2002b;22:5505–5515. doi: 10.1523/JNEUROSCI.22-13-05505.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Duffy P, McGee AW, Hasan O, Gould G, Tu N, Harel NY, Huang Y, Carson RE, Weinzimmer D, et al. Recovery from chronic spinal cord contusion after Nogo receptor intervention. Ann. Neurol. 2011;70:805–821. doi: 10.1002/ana.22527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner HL, Powers ML, Fields BN. Absolute linkage of virulence and central nervous system tropism of reoviruses to viral hemagglutinin. J. Infect. Dis. 1980;141:609–616. doi: 10.1093/infdis/141.5.609. [DOI] [PubMed] [Google Scholar]
- Wetzel JD, Chappell JD, Fogo AB, Dermody TS. Efficiency of viral entry determines the capacity of murine erythroleukemia cells to support persistent infections by mammalian reoviruses. J. Virol. 1997;71:299–306. doi: 10.1128/jvi.71.1.299-306.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Parent KN, Goodman RP, Tang J, Shou J, Nibert ML, Duncan R, Baker TS. Virion structure of baboon reovirus, a fusogenic orthoreovirus that lacks an adhesion fiber. J. Virol. 2011;85:7483–7495. doi: 10.1128/JVI.00729-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.