Summary
Interstrand crosslinks (ICL) are one of the most hazardous types of DNA damage as they form a roadblock to all processes that involve strand separation. Repair of these lesions involves several different DNA repair pathways but the molecular mechanism is unclear. Here we describe a system that allows the examination of ICL repair, via a physiological mechanism, in vitro. This system, which uses Xenopus egg extracts in combination with a DNA template that contains a site-specific ICL, represents a unique tool to study the molecular mechanism of ICL repair.
Keywords: DNA repair, interstrand crosslink, Xenopus egg extract, DNA replication, crosslinked plasmid, cell-free system
1. Introduction
DNA interstrand crosslinks (ICLs) are toxic DNA lesions whose repair is poorly understood. ICL repair takes place primarily during S phase of the cell cycle, although there may be some repair activity in G1 phase as well. Genetic studies have identified several different DNA repair pathways that likely participate in ICL repair (1). However, the mechanistic details underlying this reaction remain unresolved. Progress has long been hampered by the lack of a system that recapitulates ICL repair in vitro. We recently developed a system that supports ICL repair in cell-free extracts, which allows the examination of various steps in the repair process (2). In this system, ICL repair is coupled to DNA replication, indicating it represents the primary S phase-specific repair pathway (2–4).
To recapitulate replication-coupled ICL repair, we make use of Xenopus laevis egg extracts that support efficient DNA replication of plasmid DNA templates (5). This is achieved by first incubating the DNA in a High-Speed Supernatant (HSS) of egg cytoplasm, which leads to pre-RC assembly via sequential recruitment of the origin recognition complex (ORC), Cdt1, Cdc6, and MCM2–7 to DNA. The subsequent addition of a highly concentrated NucleoPlasmic Extract (NPE) promotes initiation of a single round of DNA replication (6). For our studies of repair, a 5.6 kb plasmid DNA template that contains a sequence specific cisplatin ICL (pICL) is added to the extract system. Details on how to make such a crosslinked plasmid can be found in chapter xxx of this book.
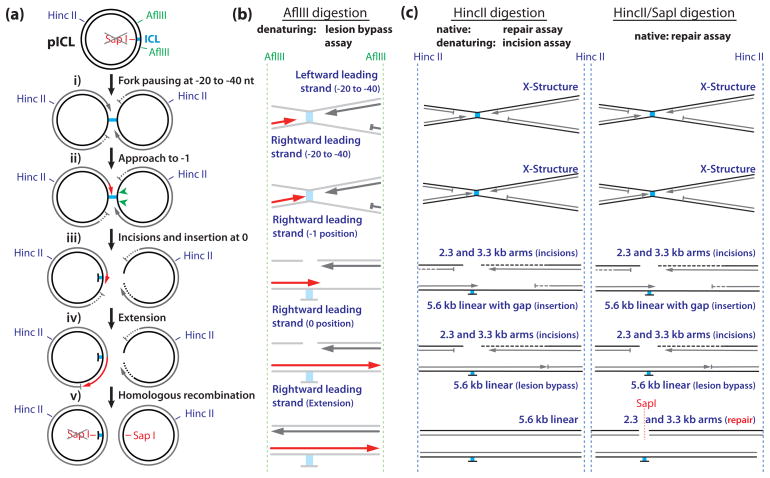
Incubation of pICL in HSS/NPE leads to the following series of events (2). First, two DNA replication forks converge on the lesion with their leading strands stalled between 20 and 40 nt from the lesion (Fig. 1a, i). Next, one of the forks resumes synthesis and pauses again 1 nucleotide before the crosslinked nucleotide (Fig. 1a, ii). Subsequently, endonucleolytic incisions on either side of the crosslink ‘unhook’ the ICL (Fig. 1a, iii), creating a DNA double strand break (DSB) in one sister and leaving a mono-adduct in the other. The lesion is bypassed by insertion of a nucleotide across from the adducted base (Fig. 1a, iii), followed by extension of the strand well beyond the ICL (Fig. 1a, iv). The final steps in repair involve homologous recombination to repair the dsDNA break in the broken sister chromatid (Fig 1a, v; David Long and J.C.W. in press). In cells, the mono-adduct in the other sister is likely removed via excision repair, but this reaction is inefficient in our extract system. Ultimately, 5 to 25 % of the replicated pICL is fully repaired, as measured by regeneration of a SapI restriction site that coincides with the crosslink in the parental plasmid. ICL repair in this system is fully dependent on, and directly coupled to, active DNA replication (2, 4).
Fig 1.
Schematic Overview of DNA repair intermediates and products generated by the various assays described in this chapter. (a) Cartoon of pICL showing the restriction sites and intermediates formed during repair. (b) DNA products analyzed on sequencing gels in the lesion bypass assay (Subheading 3.3). Dark grey and red strands are visible on the gel in Fig. 2. Lesion bypass of the rightward moving fork (red strands) can be followed at single nucleotide resolution (see Fig. 2). (c) Products analyzed under denaturing and native conditions in the incision and repair assays respectively (Subheadings 3.4 and 3.5).
This system is ideally suited to study the various steps in ICL repair and the roles of specific proteins in the repair process. Immunodepletion of specific factors from the extracts provides valuable insights into the function of each factor. Using this approach, we previously showed that the translesion DNA polymerase ζ is involved in the extension step during translesion synthesis(2). Furthermore, we demonstrated that the Fanconi anemia protein complex FANCD2-FANCI is required for ICL repair and that its depletion abrogates the incision and lesion bypass steps (4). In addition to immunodepletions, the extracts can also be easily manipulated by using specific inhibitors. For example, addition of a peptide derived from BRCA2 that inhibits Rad51 function showed that homologous recombination is a late step in ICL repair that acts downstream of the Fanconi anemia pathway (David Long and J.C.W. in press). Together with the ability to monitor specific ICL repair intermediates, these approaches make the system a powerful means to decipher the molecular mechanism of ICL repair.
This chapter describes several assays that examine specific steps in ICL repair. We first describe how to set up the ICL repair reaction using HSS/NPE and pICL. Detailed protocols on how to make HSS and NPE extracts can be found elsewhere (6). Then, we describe how to use the purified DNA repair intermediates to examine the lesion bypass reaction in detail, making use of high resolution sequencing gels. We explain how to analyze the dual incision step that unhooks the ICL. Finally, we outline an assay that allows the determination of the repair efficiency of the reaction.
2. Materials
2.1. Performing repair of pICL in egg extract
[α-P32]-dATP (3,000 Ci/mmol). Take the necessary radiation safety training and use standard precautions when working with this material.
1 M PC (Phosphocreatine disodium salt) (Sigma): dissolve in 10 mM sodium phosphate pH 7.0, store 50 μl aliquots at −20°C.
0.2 M ATP (Adenosine 5′-triphosphate disodium salt hydrate)(Sigma): dissolve in sterile water, adjust the pH to 7.0 with 10 M NaOH using pH indicator strips. Store 50 μl aliquots at −20°C.
5 mg/ml CPK (Creatine phosphokinase)(Sigma): dissolve in 50 mM NaCl, 50% glycerol and 10 mM HEPES-KOH, pH 7.5, store 250 μl aliquots at −20°C. These aliquots are stable for 2–6 months.
ATP regeneration mix: Combine 10 μl 1 M PC (see item 2), 5 μl 0.2 M ATP (see item 3), and 0.5 μl 5 mg/ml CPK (see item 4) immediately before use. Store on ice.
1 M DTT (Dithiotreitol)(Bio-rad): dissolve in sterile water, store 20 μl aliquots at −20°C.
10 × ELB-salts: 25 mM MgCl2, 500 mM KCl, 100 mM HEPES-KOH pH 7.7. Filter sterilize, and store at 4°C.
1 M sucrose: dissolve in water, filter sterilize and keep at 4°C. Discard this stock solution after a 1–2 months or when visibly contaminated.
ELB-sucrose: 1 × ELB salts (from a 10 × stock, see item 7), 0.25 M sucrose (from a 1 M stock, see item 8). Make fresh once a week.
0.5 mg/ml Nocodazole (Sigma): dissolve in DMSO, store 50 μl aliquots at −20°C.
75 ng/μl pICL in TE buffer. A detailed protocol for pICL preparation is described in chapter xx of this book.
75 ng/μl pControl in TE buffer (plasmid with the same sequence as pICL but not containing a crosslink).
75 ng/μl pQuant in TE buffer (see Note 1).
High Speed Supernatant (HSS). Preparation of HSS is described in detail in (6). Extract needs to be quality tested before use.
Nucleoplasmic extract (NPE). Preparation of NPE is described in detail in (6). Extract needs to be quality tested before use.
0.5 ml Safelock tubes (Eppendorf).
Stop solution I: 8 mM EDTA, 0.13% phosphoric acid, 10% ficoll, 5% SDS, 0.1% bromophenol blue in 80 mM Tris, pH 8.0.
Stop solution II: 0.5 % SDS, 25 mM EDTA pH 8.0 in 50 mM Tris pH 7.5.
20 mg/ml Proteinase K (Roche): dissolve in milliQ water, store 50 μl aliquots at −20°C.
2 mg/ml RNase, heat inactivated to remove background DNase activity.
2.2. Purification of pICL repair intermediates
10 mM Tris pH 8.0.
Phenol/Chloroform 1:1: Mix equal volumes of water-saturated phenol, pH 8.0 (USB)(include some of the water layer) and Chloroform (Fisher). Wait until the water and phenol/chloroform layers have completely separated. Store the solution protected from light at 4 °C for up to 2 months. Do not keep on ice as the solution becomes turbid.
0.5 ml Safelock tubes.
3 M Sodium acetate, pH 5.5. Dissolve in milliQ water and adjust pH with glacial acetic acid.
20 mg/ml muscle glycogen (Roche). Store in 20 μl aliquots at −20 °C
Ice cold 100 % Ethanol. Store at −20 °C.
Ice cold 70 % Ethanol. Store at −20 °C.
2.3. Detailed analysis of lesion bypass products on sequencing gels
AlflIII restriction enzyme, 500 U/ml (NEB). BSA solution and buffer 3 supplied with the enzyme.
Formamide loading buffer: Gel loading buffer II for denaturing PAGE (Ambion): 95% Formamide, 18 mM EDTA, and 0.025 % SDS, 0.025% Xylene Cyanol, and 0.025% Bromophenol Blue.
10 μM Primer for sequencing ladder (see Note 2).
[γ-P32]-ATP (3,000 Ci/mmol).
T4 polynucleotide kinase (PNK), 10,000 units/ml (NEB).
10 × PNK buffer (NEB)
Thermo Sequenase Cycle Sequencing Kit (USB/Affimetrix).
pControl (250 ng/μl).
Glass plates, large and small (see Note 3).
Spacers: sequencing gel spaces, 0.4 mm thick (GibcoBRL/Labrepco).
Comb: shark tooth comb, 0.4 mm thick (GibcoBRL/Labrepco).
Clips: large binder clips (Staples)
Windex window cleaner
Silanizing reagent: Safety Coat Silanizing Reagent, 250 mL (VWR)
Gel sealing tape (GibcoBRL/Labrepco).
40 % acrylamide: RapidGel-XL™ 40% Liquid Acrylamide Stock Solution (USB)
GTG buffer: 20 × Glycerol Tolerant Gel (GTG) Buffer (USB).
Running buffer: 0.8 × GTG buffer in water.
Urea (Fisher Scientific)
10 % Ammonium persulfate (APS) in water, store at 4 °C.
TEMED: N,N,N′,N′-tetramethylethylenediamine (Bio-Rad).
Needles, 18 gauge.
S2 sequencing apparatus (GibcoBRL/Labrepco)
High voltage power supply.
Gel loading tips, flat.
DEAE (Diethylaminoethyl) paper: DE81 ion exchange paper (Whatman).
Whatman paper: Whatman 3 MM chromatography paper (Whatman).
2.4. Timing of dual incisions
HincII restriction enzyme (NEB). BSA solution and enzyme.
3 M Sodium acetate, pH 5.5. Dissolve in milliQ water and adjust pH with glacial acetic acid. Keep at room temperature.
20 mg/ml glycogen from mussels (Roche). Store in 20 μl aliquots at −20 °C.
Ice cold 100 % Ethanol. Store at −20 °C.
Ice cold 70 % Ethanol. Store at −20 °C.
Alkaline loading buffer: 50 mM NaOH, 1 mM EDTA pH 8.0, 2.5 % Ficoll-400, 0.05 % Bromocresol green.
Alkaline running buffer: 50 mM NaOH, 1 mM EDTA pH 8.0.
0.25 M HCl: dilute concentrated HCl to 0.25 M in water.
NaOH/NaCl: 0.5M NaOH and 1.5M NaCl in water
Whatman paper: Whatman 3 MM chromatography paper (Whatman).
Membrane: Neutral nylon membrane (Perkin Elmer)
10 × SSC: 3M NaCl, 0.3 M NaCitrate, pH 7.0
0.5 NaCl and 0.5M Tris pH 7.5 in water
UV crosslinker: UV Stratalinker (stratagene, model 2400).
Random priming labeling kit (GE, Megaprime DNA Labeling System).
0.5M EDTA pH 8.0
G-50 columns (Illustra MicroSpin G-50 Columns, GE)
Hybridization mix: Ultrahyb (Ambion)
Hybridization oven
2 × SSC/0.1 % SDS and 0.1 × SCC/0.1 % SDS. Made from a 20 × SSC stock and a 10 % SDS stock in water.
2.5. Determining ICL repair efficiency
HincII and SapI restriction enzymes (NEB). BSA solution and buffer 4 supplied with the enzymes.
10 × DNA loading buffer: 50 % glycerol, 2 × TBE buffer, 25 mM EDTA and 0.1 % Bromophenol Blue).
DEAE (Diethylaminoethyl) paper: DE81 ion exchange paper (Whatman).
Whatman paper: Whatman 3 MM chromatography paper (Whatman).
Phosphorimager
Quantification software: we use Quantity One software from Bio-Rad, the basic version can be downloaded for free from the Bio-Rad website
3. Methods
3.1. Performing replication/repair reaction of pICL in HSS/NPE
Preparing an ICL repair reaction is very similar to a DNA replication reaction (described in detail in (6)), the main differences being that repair involves use of pICL as a template and requires longer incubation times. Starting from a master replication reaction, samples are taken over a time course of up to 4 hours. The following protocol is designed to yield enough repair intermediates to analyze 10 time points using the assays described in this chapter. It is freely scalable, allowing further analyses of the isolated repair intermediates (e.g. by native 2D electrophoresis, electron microscopy, or ChIP). As ICL repair in this system is dependent on efficient DNA replication, it is important to take samples to check the replication efficiency. In addition, samples can be taken from the reaction to analyze post-translational protein modifications during repair using Western blotting (see Note 4).
Unless otherwise stated, all incubations are performed at 20 to 22°C.
Prepare the following solutions just before you start the reaction: ATP regeneration mix, 0.5 M DTT, ELB-sucrose (see Note 5).
Thaw a 33 μl HSS aliquot, keep on ice after thawing. Add 1 μl of the ATP regeneration mix and 0.2 μl of 0.5 mg/ml nocodazole (see Note 6). Mix by pipetting up and down a few times.
Transfer 30 μl of HSS-mix to a fresh tube and add pICL and pQuant to final concentrations of 6 ng/μl and 0.3 ng/μl, respectively. This will result in a pICL concentration of 2 ng/μl, and pQuant of 0.1 ng/μl in the HSS/NPE mixture (see Notes 1 and 7). Avoid diluting the extract more than 20 %. Mix by gently pipetting up and down 2 times (see Note 8). Transfer the HSS-mix to 20–22 °C for 20 min prior to addition of NPE (see Note 9).
Thaw 40 μl of NPE and keep it on ice after thawing. Add 2.4 μl ATP regeneration mix, 1.0 μl 0.5 M DTT and 2 μl of [α-P32]-dATP (see Notes 10 and 11). Add 32 μl ELB-sucrose to dilute the NPE (see Note 12). Mix by gently pipetting 2–3 times. Incubate the NPE mix at 20–22 °C for 15 minutes prior to addition to HSS.
Add 72 μl of NPE mix to the 36 μl of HSS mix to start the replication/repair reaction.
At each time point take 9 μl from the reaction mix and add 90 μl stop solution II in a 0.5 ml safelock tube (see Note 13). From this sample, DNA repair intermediates will be extracted for further analysis. At the same time take an additional 1 μl from the reaction mix and add it to 5 μl stop solution I. This sample will be run on a native agarose gel to assess replication efficiency.
When all samples are collected, add 7 μl RNase to the samples that will be extracted and incubate at 37°C for 30 minutes. Then, add 3 μl Proteinase K and incubate for at least 1 hour at 37 °C, or overnight at room temperature. These samples are now ready to be extracted.
To the samples that will be used to determine the replication efficiency, add 1 μl proteinase K. Incubate for at least 1 hour at 37 °C, or overnight at room temperature and proceed as described in (6).
3.2. Purification of pICL repair intermediates
Briefly spin samples from subheading 3.1 step 7 to collect possible condensation on the lid.
Dilute each sample with 40 μl of 10 mM Tris pH 8.0 (see Note 14).
Add 150 μl Phenol/Chloroform 1:1. Close the tubes firmly and mix by carefully inverting the tubes 10–15 times (do not vortex).
Centrifuge samples at 16,000 × g for 4 minutes at room temperature. During this step, prepare a new series of Safelock tubes.
Extract the DNA, which is in the upper aqueous phase, and add it to a new tube. Be conservative to prevent contamination with the phenol/chloroform layer. It is important to get equal amounts from each sample. We usually take ~140 μl of the upper layer.
To the extracted DNA, add an equal volume of chloroform and mix by inverting the tubes 10–15 times. Centrifuge at 16,000 × g for 4 minutes at room temperature.
Prepare a new series of tubes. In this case we do not use the Safelock tubes as they are made of a thick layer of plastic that prevents clear vision of the pellet after precipitation.
To each tube add: 12 μl (1/10th of the volume) 3 M Sodium acetate pH 5.5 and 1 μl glycogen.
Extract the aqueous phase from the chloroform extraction tubes. We usually remove ~120 μl, and add it to the tubes containing sodium acetate and glycogen.
Add 280 μl ice cold 100 % ethanol, close tubes firmly, and mix by carefully inverting 15 times. Incubate on ice for 15 minutes.
Centrifuge samples at 18,000 × g for 30 minutes at 4 °C.
Aspirate the supernatant carefully (see Note 15) and add 250 μl ice cold 70 % ethanol to wash the pellet.
Centrifuge at 18,000 × g for another 5 minutes at 4 °C and aspirate the supernatant (see Note 15). Try to get all the fluid off.
Air dry the pellets for no longer than 10 minutes (see Note 16).
Resolubilize the pellet in 10 μl 10 mM Tris pH 8.0. Incubate at room temperature for at least half an hour and mix by tapping the tube. Do not vortex or pipet up and down. Store at 4 °C until further use (see Note 17).
3.3. Detailed analysis of lesion bypass products on sequencing gels
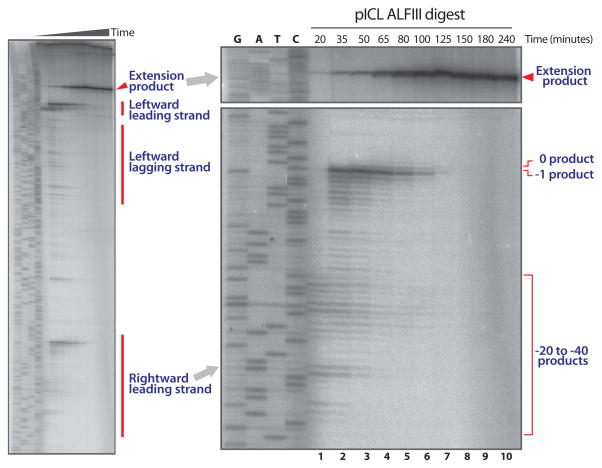
Lesion bypass in ICL repair is a stepwise process (Fig 1a, and (2)) that can be followed over time using high-resolution sequencing gels. We make use of two AflIII sites on pICL situated 149 nt to the left and 540 nt to the right of the ICL (Fig 1a). Digestion of repair intermediates with AflIII generates products assignable to leading and lagging strand products (see Note 18)(Fig 1b and 2). This way it is possible to show the pausing of the leading strand at the −20 to −40 position (Fig 1b and Fig 2, lane 1), the approach to the −1 position (Fig 1b and Fig 2, lanes 2–4), the insertion of a nucleotide across from the damaged base (Fig 1b and Fig 2, lanes 4 and 5), and the extension of the leading strand beyond the lesion (Fig 1b and Fig 2, lane 5–10)(see Note 19). This is a powerful assay to monitor various steps of lesion bypass under specific conditions (e.g. addition of a specific inhibitor or immunodepletion of a specific protein). The procedure is described in four steps: sample preparation (3.3.1.), generating the sequencing ladder (3.3.2.), pouring the sequencing gel (3.3.3.), and running the sequencing gel (3.3.4.).
Fig. 2.
Analysis of lesion bypass during ICL repair (Subheading 3.3). AflIII digested repair intermediates are separated on a sequencing gel. Leading and lagging strands of the leftward moving fork, leading strands of the rightward moving fork, and lesion bypass product (‘Extension product’) are indicated on the left panel. Right panel shows an enlargement of the rightward moving leading strand on which −20 to −40, −1, 0, and extension products are indicated.
3.3.1. Sample preparation
Digest 3 μl of the purified repair intermediates (subheading 3.2 step 15) with 0.4 μl AflIII (2 units) in a maximal volume of 5 μl (see Note 20).
Incubate the reaction at 37 °C for 3 hours.
Briefly spin the samples and add 2.5 μl formamide loading buffer to each sample. The samples are ready for use or can be stored at −80 °C.
3.3.2. Preparing the sequencing ladder
Prepare a 10 μM solution of a suitable primer (see Note 2).
Setup an end-labeling reaction by mixing 1 μl of this primer with 3.5 μl [γ-P32]-ATP, 0.5 μl 10 × PNK buffer, and 0.2 μl T4 polynucleotide kinase.
Incubate at 37 °C for 10 minutes.
Incubate at 95 °C for 2 minutes to inactivate the PNK (see Note 21).
During these incubations, prepare the sequencing reactions. We use the Thermo Sequenase Cycle Sequencing Kit. Take 4 PCR tubes and add 4 μl of the ddGTP, ddATP, ddTTP, and ddCTP termination mixes provided by the kit to each tube.
Make a master stock of the reaction mix by adding 2 μl 10 × reaction buffer provided by the kit, 1 μl of the end-labeled primer from step 4, and 0.7 μl pControl (at 250 ng/μl) to 11.8 μl milliQ water. Mix and add 2 μl of sequenase provided by the kit just prior to starting the reaction.
Add 4 μl of the reaction mix to each tube of termination mixes, mix and run the following protocol in a PCR machine: 1) 95 °C for 30 seconds, 2) 60 °C for 30 seconds, 3) 72 °C for 90 seconds, 4) GoTo step 2 for 50 times, 6) Hold at 4 °C.
When the reaction is finished, add 4 μl formamide loading buffer to each sample and store at −80 °C until use.
3.3.3. Pouring the sequencing gel
Thoroughly clean the inside of the inner and outer glass plates with Windex window cleaner, rinse with water and dry. Rinse the plates with 100 % ethanol and air dry. Mark the outside of the plate and treat the inside with care (see Note 22).
Silanize the smaller glass plate by applying a few milliliters of silanizing reagent on a tissue and spreading it out on the glass plate. Rub thoroughly over the entire surface. Leave the plate to dry for about half an hour. Repeat step 1 (see Notes 23 and 24).
Assemble the gel sandwich with the spacers. Make sure the plates are well aligned and the inside of the plates is on the inside of the sandwich. Add a clip to each side to fix the sandwich.
Tape the bottom of the gel sandwich and the bottom half of the sides with sequencing gel tape (see Notes 25 and 26). Take care not to wrinkle the tape as this will cause leaking. Add 4 additional clips to each side of the gel sandwich.
Prepare the gel mix by adding 14 ml 40 % acrylamide, 3.2 ml GTG buffer, and 38.4 g Urea to 34.8 ml milliQ water (see Notes 27 and 28).
Filter the gel mix through a 22 μM filter using a vacuum filter setup.
Degas the mix by leaving it under vacuum for about 10 minutes.
Add 480 μl 10 % APS and 40 μl TEMED, and fill a 50 ml syringe with the gel mix. Add a needle to the syringe and immediately start injecting the mix into the gap between the gel plates. Do this by keeping the gel sandwich vertically and slightly tilted to one side. Inject gel mix from one side (see Note 29).
When the space between the plates is almost filled, lay down the gel horizontally. If enough of the gel mix was added, the entire gel space will now fill up and excess gel mix will come out at the top. Immediately insert the flat side of the comb in between the glass plates, 4 to 5 millimeters deep. Add one or two clips to the top of the sandwich.
Leave the gel for at least 2 hours to complete polymerization (see Note 30).
3.3.4. Running the sequencing gel
Remove the clips and the tape from the gel sandwich.
Rinse the sides of the gel sandwich to remove precipitated urea.
Mount the gel into the running apparatus.
Add running buffer to the upper compartment and verify that the gel properly seals this compartment.
Carefully remove the comb and thoroughly rinse the slot with running buffer using a syringe with a needle (see Note 31).
Insert the comb back between the glass plates, this time with the teeth pointing down. At this stage, do not let the comb touch the gel surface but leave about 1 millimeter in between (see Note 32).
Add running buffer to the lower buffer compartment and prerun the gel at 60 Watts for 40 minutes to 2 hours. The gel should warm up to approximately 50 °C during this period.
Just prior to running, prepare the samples and the sequencing ladder by incubating them at 75 °C for 2 minutes. Snap cool them on ice/water and keep them on ice until use.
After prerunning, the comb can be pushed into the gel until the teeth are about 1 millimeter deep to create the wells for the samples. Choose an area of the gel with high quality wells (a flat bottom surface) and carefully rinse the wells with running buffer using a syringe and needle.
Using ultrathin flat gel loading tips, load 1 to 4 μl of each sample (see Note 33). It is important to load the samples as a tight band at the bottom of each well. This will improve the resolution of the bands (see Note 34).
Run the gel at 60 Watts for about 2 hours and 20 minutes until the Xylene Cyanol dye has run 30 cm from the top of the gel (see Note 35).
Start preheating the gel dryer at 80 °C 10 minutes before the electrophoresis is finished. Cut a piece of DEAE paper and a piece of Whatman paper at the size of the gel.
Remove the gel sandwich from the apparatus and rinse the bottom with water to reduce radioactive contamination.
Lay down the gel sandwich on a piece of paper and carefully lift the upper (small) glass plate by wedging a piece of plastic/metal in between the plates (see Note 36). As the small glass plate is silanized, the gel should stick to the large plate. Try to remove the small glass plate in one movement, if it retouches the gel, the gel will likely stick to the plate and break.
Carefully lay down the DEAE paper (see Note 37) on the gel starting on one side and moving to the other by rolling a plastic pipet over it. Try to prevent wrinkles and bubbles. Put the Whatman paper on top of the DEAE paper.
Lift up both papers starting from one corner of the gel and peel off the entire gel. It should stick well to the paper.
Immediately cover the gel with a piece of saran wrap, try to avoid wrinkles, and dry the gel for at least 45 minutes in a gel dryer at 80 °C.
Expose the dried gel to a phosphor screen for 16 to 72 hours and scan the screen using a phosphorimager (for an example see Fig. 2).
3.4. Timing of dual incisions
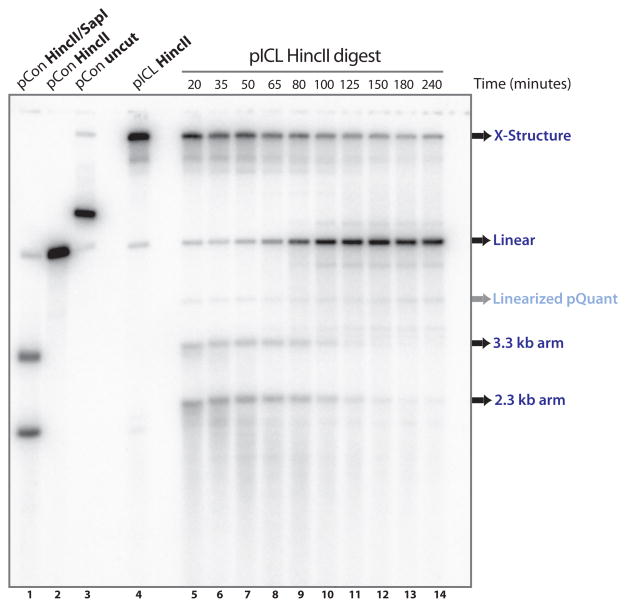
During ICL repair, the crosslink is released from one of the DNA strands by dual incisions on either side of the lesion. To analyze these incisions we use denaturing gel electrophoresis of HincII-digested repair intermediates (see Fig 1c, left panel)(4). Before any incisions have taken place, the parental DNA migrates in gels as a large X-shaped molecule because the two strands are connected by the ICL (Fig. 3, lane 4). Dual incisions result in the collapse of this large X-shaped molecule into a linear molecule and 2 arm fragments (Fig 3, lane 8–14, for size markers lane 1 and 2). The arm fragments disappear over time due to resection and homologous recombination (Fig 3, lanes 10–14,(4)). As the parental strands are not radioactively labeled, we use Southern blotting for their detection. The 2.3 and 3.3 kb arm fragments visible at early time points (Fig 3, lane 5–7) are leading and lagging strand products of replication. The procedure is described in three steps: preparing the samples (3.4.1.), running and transferring the alkaline gel (3.4.2.), and hybridization and detection (3.4.3).
Fig. 3.
Examination of dual incisions during ICL repair. HincII digested repair intermediates are separated on a denaturing gel and visualized using Southern blotting (land 5–14) (Subheading 3.4). Undigested and HincII and/or SapI digested pControl and pICL are shown in lane 1–4 and serve as size markers. X-structures, linears, and 2.3/3.3 arm fragments are indicated. pQuant is visible due to some background reactivity with the probe.
3.4.1. Preparing the samples
Digest 1 μl of the purified repair intermediates (subheading 3.2 step 15) with 0.4 μl HincII (4 units) in a volume of 20 μl. Incubate samples at 37 °C for 3 hours (see Notes 38 and 39).
Precipitate the DNA by adding 2 μl 3 M Sodium acetate pH 5.5, 0.2 μl 0.5 M EDTA pH 8.0, and 55 μl ice cold 100 % ethanol. Mix by carefully inverting 1015 times and incubate on ice for 15 minutes.
Centrifuge samples at 18,000 × g for 30 minutes at 4 °C.
Aspirate the supernatant carefully (see Note 40) and add 100 μl ice cold 70 % ethanol to the pellet.
Centrifuge at 18,000 × g for another 5 minutes at 4 °C and aspirate the supernatant (see Note 40).
Air dry the pellets for about 10 minutes at room temperature.
Resolubilize the pellet in 10 μl alkaline loading buffer.
3.4.2. Running and transferring the alkaline gel
Poor a large (20 × 30 cm) 0.9% agarose gel in milliQ water. Use a comb with thin (1 mm) slots (see Note 41). Let it solidify completely. Incubate the gel in 2.5 L alkaline running buffer half an hour before starting the run.
Load the samples and start the run at 0.8 V/cm (cm referring to the distance between the electrodes). Run until the samples have just entered the gel (this will take around 40 minutes).
Put a glass plate on top of the gel covering at least the area from the wells to halfway the gel. Run for another 18 hours at 0.8 V/cm (until the dye front is halfway the gel).
Cut the gel just above the dye front and incubate the top half for 7 minutes in 1 L 0.25 M HCl while slowly shaking.
Rinse gel with water and incubate for 30 minutes in 1 L NaOH/NaCl.
-
Transfer the DNA to the membrane by making the following stack on top of a tray:
1 large piece of Whatman paper (prewet in 10 × SCC)
gel
membrane (prewet in milliQ followed by 10 × SCC)
6 gel-sized whatman papers (prewet in 10 × SCC)
a big stack of paper towels
a glass plate or tray
-
a weight (1–2 kg)
The stack is placed in a larger tray that contains 10 × SSC to a level just below the tray the stack is assembled on. The large piece of Whatman paper hangs over the sides of the inner tray into the buffer. After covering the membrane with the Whatman papers air bubbles are removed by rolling with a pipet.
Transfer for 18 hours.
Disassemble the stack and incubate the membrane in 250 ml 0.5M Tris pH 7.5, 0.5M NaCl for 15 minutes.
Air dry the membrane.
Crosslink the DNA to the membrane using the ‘Autocrosslink’ settings of the UV crosslinker (output is 120,000 microloules/cm2).
3.4.3. Hybridization and detection
The probe is generated using the random prime labeling kit (GE). pControl, linearized by digestion with HincII, is used as a template. For this, 50 ng of linearized pControl in 21 μl milliQ water is added to 5 μl of the random primers provided by the kit.
Incubate at 98 °C for 5 minutes. Quickly spin down and add: 5 μl 10 × buffer (kit), 4 μl dCTP, dGTP and dTTP each (kit), 5 μl [α-P32]-dATP, and 2 μl Klenow (kit).
Incubate at 37 °C for 1 hour and stop the reaction by adding 2 μl 0.5M EDTA.
Remove the unincorporated [α-P32]-dATP by spinning through a G-50 column according to manufacturer’s protocol.
Preheat the hybridization mix at 68 °C until the precipitation has dissolved.
Prehybridize the blot for at least 30 minutes at 42 °C in 22 ml hybridization mix.
Incubate the probe at 98 °C for 3 minutes and snap cool on water/ice slurry for 2 minutes. Add the probe to the hybridization mix and incubate the blot overnight at 42 °C in a hybridization oven.
Wash the membrane twice with prewarmed (42 °C) 2 × SSC/0.1% SDS for 5 minutes. Then, wash the blot twice with prewarmed 0.1 × SSC/0.1% SDS for 15 minutes. Air dry the membrane and expose to a phosphoimager screen for 4 to 24 hours. Scan the screen using a phosphorimager.
3.5. Determining ICL repair efficiency
The recognition site of the restriction endonuclease SapI overlaps with the crosslink in pICL (Fig 1a). Because the crosslink prevents cleavage by SapI (2), repair of pICL can be monitored by the regeneration of the SapI site. Regeneration of the SapI site is determined by dual digestion with both SapI and HincII (HincII cuts only once in pICL)(Fig 1c.) and quantification of the 2.3 and 3.3 kb dual digestion products. A complication is that even in unrepaired molecules, any incisions near the ICL (during the repair process, (2)) would result in 2.3 and 3.3 kb fragments after HincII digestion (Fig 1a and c). To identify the background level of these products, we digest an equal volume of the repair intermediates with HincII only and subtract these products from the HincII/SapI digestion products (a detailed description of how this is done is given below). This assay determines the percentage of the input pICL that is repaired ‘error free’ (no errors at the SapI site). We first describe how to run the repair gel (3.5.1.) followed by an explanation how to determine the repair efficiency from this gel (3.5.2.).
3.5.1. Running the pICL repair gel
Digest 1 μl of the purified repair intermediates (subheading 3.2 step 15) with 0.4 μl HincII (4 units), and 1 μl with 0.4 μl HincII and 0.6 μl SapI (1.2 units), in a volume of 20 μl. Incubate samples at 37 °C for 3 hours (see Notes 42 and 43).
Briefly spin down the samples. Add 5 μl DNA loading buffer to each sample and load the entire sample on a large (20 × 30 cm) 0.8 % agarose gel in 1 × TBE. Run the gel ~3 hours at 4.5 V/cm (cm referring to the distance between the electrodes), until the dye front is half way the gel (see Note 44).
Cut the gel just above the dye front. Discard the bottom half of the gel containing non-incorporated nucleotides in an appropriate radioactive waste container.
Dry the gel in a stack containing: saran wrap, approximately 20 paper towels, a piece of Whatman paper, a piece of DEAE paper, the gel, and again DEAE paper, Whatman paper, paper towels and saran wrap. Dry the gel by applying a weight of about 1 kg on top of the stack for approximately 1 h. Dispose of the paper towels, taking care to check whether they are radioactive.
Finish drying by placing the gel surrounded by the DEAE and Whatman paper in a gel dryer. Cover with saran wrap and vacuum dry at 80°C for 30 to 60 minutes.
Wrap the dried gel in saran wrap and expose it to a phosphor screen for 2 to 18 hours. Scan the screen using a phosphorimager.
3.5.2. Interpreting the repair gel and determining repair efficiency
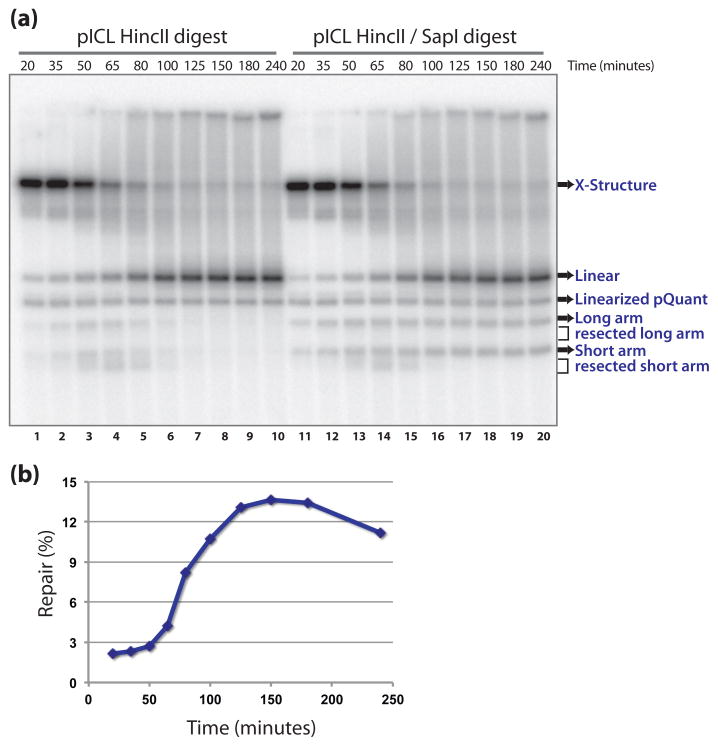
An example of a repair gel is given in Figure 4a. After HincII digestion (Fig. 4a, lane 1–10), the major species at early time points is the large X-shaped structure that is formed when replication forks converge at the crosslink (Fig. 1a and c, (2)). At later time points, dual incisions take place that unhook the crosslink from one strand, resulting in the formation of a linear molecule and two arm fragments (Fig. 1c and Fig. 4a lane 3–6). The arm fragments are degraded over time (2). The HincII/SapI digested samples (Fig 4a, lane 11–20) show the same species but in addition show a buildup of 2.3 and 3.3 kb arm fragments at later time points which represent the repair products (Fig 1c and Fig. 4a lanes 15–20). All the lanes on the repair gel show the linearized pQuant, which will be used for normalization (see Note 45).
First, using quantification software, determine the signal present in the linearized pQuant bands of each lane (Fig 4a). Subtract the background counts from a representative area of the same volume (see Note 46).
Determine the amount of signal present in the 2.3 and 3.3 kb arm fragments in each lane (draw a box around both). Include the signal in the degradation products just under the arm fragments. Subtract background signal obtained from a representative background area of the same volume.
Normalize all pQuant bands to the one in the first lane and multiply the signal in the arm fragments with the normalization factor. This will adjust for unequal recovery of products upon extraction and/or unequal loading.
For every time point, subtract the arm fragments generated by incisions in the repair process (after HincII-only digestion, Fig 4a, lane 1–10) from the arm fragments generated by both the incisions and the repair products (after HincII/SapI digestion, Fig 4a, lane 11–20). This is now the signal of the repair products only.
To determine the percentage of the replicated plasmid that resulted in repaired products we first calculate the total amount of replicated pICL by multiplying the intensity of the pQuant band with the dilution factor (how much more pICL was added compared to pQuant). Then, determine the percentage the repair products represent from this total.
This way, calculate the percentage repair for each time point and plot against time (Fig 4b) (see Note 47).
Fig. 4.
Determination of ICL repair efficiency. (a) Analysis of HincII (lane 1–10) and HincII/SapI (lane 11–20) digested repair intermediates on a native agarose gel (Subheading 3.5). (b) Calculated repair efficiency plotted against time.
4. Notes
Materials
A low amount of pQuant is added to the repair reaction to determine repair efficiency (see subheading 3.5.2). Generally, the concentration of pQuant should be similar to the expected concentration of repair products. Therefore, if the concentration of pICL is 2 ng/μl, and 10 % repair is expected, 0.2 ng/μl pQuant should be added. pQuant can be any 3–4 kb plasmid with little homology to pICL (to prevent adding a potential donor sequence for homologous recombination). It should be cut once with HincII and not with SapI. We use pCDF-DUET (Novagen) that we modified to contain only one HincII restriction site. For accurate quantification it is important that linearized pQuant can be separated from any fragments of pICL on native agarose gels (section 3.5 and Figure 4). Digestion of pQuant with AflIII should not generate any short fragments (<700 bp) that might obscure bands in the lesion bypass assay (section 3.3 and Figure 2).
Choose a sequencing primer that starts at the digestion site used in section 3.3.1. For the example described here, samples are digested with AflIII and the gel is run under conditions that allow nucleotide resolution of fragments between ~80 and ~160 nucleotides. One of the AflIII sites is located at a distance of ~150 nucleotides from the ICL and the primer starts exactly at this restriction site. This way, the sequence and size of the products present in the sequencing ladder correspond to those of the newly synthesized (and therefore radioactively labeled) leading strands of the repair intermediates.
The size of the glass plates depends on the sequencing apparatus. We use a model S2 sequencing apparatus (GibcoBRL/Labrepco). The large plate is 42 × 33 cm and the small plate 39 × 33 cm.
It can also be useful to take samples for western blot analysis at this stage. These samples can be used to examine the activation of the Fanconi anemia pathway by monitoring the ubiquitylation of FANCD2 and FANCI (2, 7). Another useful marker to study is the phosphorylated form of Chk1 (2), which is used as a measure of activation of the DNA damage checkpoint kinase ATR (see Note 10).
Use ELB-sucrose for dilution of stock solutions (DTT, plasmids etc.).
In principle, a low speed supernatant (LSS) extract can replace HSS in this procedure. This, however, only works if plasmid DNA is used as a template. If chromatin DNA is used as a template, nuclei formation and DNA replication initiates before NPE addition and synchronization is lost.
The plasmid concentration in a replication/repair reaction generally should not be lower than ~0.3 ng/μl since this will not support efficient DNA replication (8). Optimal concentration is between 0.5 and 10 ng/μl. For a repair reaction we use a concentration of pICL between 1 and 3 ng/μl.
When using pICL, do not vortex and try to avoid extensive pipetting, preferably mix by stirring with the pipet tip in the solution (if pipetting up and down is needed, do it carefully) to make sure no additional damage is added to the potentially fragile crosslinked template.
This deviates from the DNA replication protocol as described in (6) where a 30 minute incubation is described. In a repair reaction this time is reduced to avoid potential processing of the ICL in HSS.
[α-P32]-dATP can be added to either HSS or NPE reaction mixes. Since incorporation of nucleotides only starts a few minutes after adding NPE to HSS, [α-P32]-dATP can also be added immediately after combining HSS and NPE. This is useful for taking non-radioactive samples of the reaction for analysis by Western blotting.
The amount of [α-P32]-dATP that is added to a repair reaction depends on several variables including what assays will be performed with the samples. A sequencing gel requires more [α-P32]-dATP incorporation than a repair assay. Second, the efficiency of replication. In case of low anticipated replication efficiency (for example after extensive depletion) addition of more [α-P32]-dATP may be necessary to increase the radioactivity of the samples. Finally it depends on how fresh the batch of [α-P32]-dATP is. The amount of fresh [α-P32]-dATP we use in this example is enough to perform all assays described in this chapter using samples from a single repair reaction.
NPE often requires dilution with ELB-sucrose to allow rapid and complete replication (6). The optimal degree of dilution has to first be established for every extract.
It is important that the repair reaction samples are collected in Safelock tubes. This prevents radioactive contamination during subsequent phenol/chloroform extraction.
We found that if samples are not diluted at this step a crystalline white pellet forms at the ethanol precipitation step that inhibits digestion of the samples by restriction endonucleases.
The pellet after ethanol precipitation and wash steps is very small and often does not stick well to the bottom of the tube. Take extreme care during aspiration, as it is very easy to lose the pellet. We usually process no more then 4 samples at a time and leave the rest spinning.
Do not overdry the pellets and do not use a vacuum dryer. This will cause problems in dissolving the pellet, likely due to the fact that repair intermediates contain complex structures.
These purified repair intermediate samples are used for the assays described below. We have used these samples up to 10 days after generating them but we recommend using the samples as soon as possible for maximal signal intensity (T1/2 for 32P is 14.3 days).
The method we used to distinguish between leading and lagging strand products is described in supplementary figure 8 of (2).
Note that the glycerol and salt concentrations as well as the sample volume loaded on the gel can affect the migration of DNA molecules on denaturing polyacrylamide gels. Therefore we recommend loading equal volumes of samples and sequencing ladder. In case the migration of the DNA intermediates differs from the expected, the sequence ladder samples can be precipitated and taken up in the same solution as the repair intermediate samples.
It is important to keep the volume of the digestion reaction small to be able to load sufficient repair intermediates for detection. We use AflIII to digest our products because this results in fragments of suitable sizes for the analysis on sequencing gels. In case a plasmid with a different sequence is used, choose an appropriate restriction enzyme.
At this stage, unincorporated nucleotides can be removed by spinning the sample through a G-50 column. This will reduce radioactive contamination during the following steps.
Only touch the silanized surface of the glass plate with soft tissues.
If the gel sticks to both plates upon separation causing it to rupture, it sometimes helps to silanize the large glass plate as well as the small one.
Silanization of the glass plates does not need to be done before every experiment. The plates can be used up to 10 times before resilanization is required.
We use proper gel sealing tape for this. We have tried many types of more general tape but none were adequate (for example the gel-mix dissolves the glue of the tape, or the tape is very difficult to remove from the glass plates after use).
A rubber casting-clamp is available for sealing the gel sandwich, which can be used instead of tape.
Heating of the acrylamide mix in a microwave oven for 15 seconds helps to dissolve urea.
The percentage of the gel can be varied depending on the size of the product generated by digestion. In this case we use a 7 % gel to get nucleotide resolution of fragments between ~80 and ~160 nucleotides. A 5 % acrylamide gel allows examination of products up to 550 nt at nucleotide resolution.
Occasionally, bubbles appear on the side of the gel during pouring. This is usually a sign that the glass plates need to be resilanized. If the samples are loaded at a distance of a centimeter or more from the bubbles, they will not affect the migration of the samples.
Alternatively it can be left overnight to polymerize, cover the top of the gel with wet tissues to prevent it from drying out.
Carefully inspect the surface of the gel made by the comb. Mark areas where the surface is not perfectly straight. Because this surface forms the bottom of the sample slots it should be as straight as possible to ensure high resolution.
If the teeth of the comb are inserted into the gel during the prerunning of the gel it is very difficult to seal the wells properly for sample loading.
Try to load the samples as quickly as possible after stopping the pre-run, the gel should not cool down too much. Load similar volumes of the repair samples and sequencing ladder samples (see Note 19).
We have found that the last sample of a series often shows less tight bands compared to the previous ones. We solve this by adding 2 or more sequencing ladder samples after the last sample.
Running time depends on the percentage of the gel and the size of the products. Here we examine a ~150 nt product by running it on a 7 % gel for 2.2 hours at 60 Watts. However, a 500 nt product can be examined at nucleotide resolution on a 5 % gel by running it for ~5.5 hours at 55 Watts.
If there are air bubbles on one side of the gel, open the gel from the other side as they can cause the gel to break.
It’s critical to use positively charged DEAE paper as it will help to retain small DNA molecules.
For the incision assay it is preferable to prepare samples without [α-P32]-dATP incorporation. This way, double detection of the signal from the nascent strands is prevented (once due to body labeling and once due to the hybridization with the radioactive probe in the Southern blot).
Suitable markers for the Southern blot can be generated by digestion of similar amounts of pICL and pControl with HincII, and pControl with HincII/SapI.
Because there is very little DNA in the samples and no glycogen was added, the pellet is difficult to see. The pellet does seem to be firmly stuck to the side of the tube as we have never lost a pellet at this stage (all tubes should be put into the centrifuge in a known orientation, so that one knows where to expect the pellet after centrifugation).
We found it is important to use a comb with thin (1 mm) slots, as it increases the resolution of the gel.
We sometimes observe problems with SapI digestion. To keep SapI active during the entire digestion period we add 0.3 μl per sample at the beginning and another 0.3 μl SapI after 2 hours.
In some cases, contaminants in the purified DNA samples inhibit proper digestion. From the gel it is not always obvious whether they have been properly digested as one will always find a certain degree of heterogeneous, large products regardless of the quality of digestion. Therefore it is best to have an independent digestion control. For this, set up a replication/repair reaction with pControl in parallel with pICL, which will result in radioactive products that generate the same 2.3 and 3.3 kb fragments after HincII/SapI digestion. Alternatively, it is possible to include a non-radioactive sample of pControl during the extraction procedure. Digestion of the extracted pControl with HincII/SapI and detection of the products on an agarose gel stained with Sybr gold allows verification of digestion.
Running a large gel is required to properly separate the repair products from linearized pQuant. In addition, it is important to stop the gel before unincorporated radioactive nucleotides migrate off the gel (when the dye front it halfway down the gel). This helps to decrease background radioactivity on the gel and reduces radioactive waste.
In both the HincII and the HincII/SapI digested samples, at later time points, a pronounced high molecular weight smear is often observed. This high molecular weight smear likely consists of partially degraded X-shape structures and repair intermediates formed by homologous recombination ((4) and David Long and J.C.W in press)
The pQuant band may overlap with considerable background smearing that is not equal in each lane of the gel. If this is the case, we choose the background signal for each lane separately in an area close to the pQuant band.
If there is a small fraction of non-crosslinked plasmid in the pICL prep, these product will show up as ‘repair products’ at the earliest time points (in the example in Fig. 1 this represents 2%). The actual repair products are formed only after about 1 hours and show a sharp increase at that time (2). To determine the true repair efficiency, former should be subtracted from the latter.
References
- 1.Legerski RJ. Repair of DNA interstrand cross-links during S phase of the mammalian cell cycle. Environ Mol Mutagen. 2010;51:540–551. doi: 10.1002/em.20566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Raschle M, Knipscheer P, Enoiu M, Angelov T, Sun J, Griffith JD, Ellenberger TE, Scharer OD, Walter JC. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell. 2008;134:969–980. doi: 10.1016/j.cell.2008.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akkari YM, Bateman RL, Reifsteck CA, Olson SB, Grompe M. DNA replication is required To elicit cellular responses to psoralen-induced DNA interstrand cross-links. Mol Cell Biol. 2000;20:8283–8289. doi: 10.1128/mcb.20.21.8283-8289.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knipscheer P, Raschle M, Smogorzewska A, Enoiu M, Ho TV, Scharer OD, Elledge SJ, Walter JC. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science. 2009;326:1698–1701. doi: 10.1126/science.1182372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walter J, Sun L, Newport J. Regulated chromosomal DNA replication in the absence of a nucleus. Mol Cell. 1998;1:519–529. doi: 10.1016/s1097-2765(00)80052-0. [DOI] [PubMed] [Google Scholar]
- 6.Lebofsky R, Takahashi T, Walter JC. DNA replication in nucleus-free Xenopus egg extracts. Methods Mol Biol. 2009;521:229–252. doi: 10.1007/978-1-60327-815-7_13. [DOI] [PubMed] [Google Scholar]
- 7.Sobeck A, Stone S, Costanzo V, de Graaf B, Reuter T, de Winter J, Wallisch M, Akkari Y, Olson S, Wang W, Joenje H, Christian JL, Lupardus PJ, Cimprich KA, Gautier J, Hoatlin ME. Fanconi anemia proteins are required to prevent accumulation of replication-associated DNA double-strand breaks. Mol Cell Biol. 2006;26:425–437. doi: 10.1128/MCB.26.2.425-437.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lebofsky R, van Oijen AM, Walter JC. DNA is a co-factor for its own replication in Xenopus egg extracts. Nucleic Acids Res. 2011;39:545–555. doi: 10.1093/nar/gkq739. [DOI] [PMC free article] [PubMed] [Google Scholar]