Abstract
Lung cancer remains the most common cause of cancer deaths worldwide, yet there is currently a lack of diagnostic noninvasive biomarkers that could guide treatment decisions. Small molecules (<1500 Da) were measured in urine collected from 469 lung cancer patients and 536 population controls using unbiased liquid chromatography-mass spectrometry. Clinical putative diagnostic and prognostic biomarkers were validated by quantitation and normalized to creatinine levels at two different time points and further validated in an independent sample set, which comprises 80 cases and 78 population controls, with similar demographic and clinical characteristics when compared to the training set. Creatine riboside (IUPAC name: 2-{2-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)-oxolan-2-yl]-1-methylcarbamimidamido}acetic acid), a novel molecule identified in this study, and N-acetylneuraminic acid (NANA), were each significantly (P <0.00001) elevated in non–small cell lung cancer (NSCLC) and associated with worse prognosis (hazard ratio (HR) =1.81 [P =0.0002], and 1.54 [P =0.025], respectively). Creatine riboside was the strongest classifier of lung cancer status in all and stage I–II cases, important for early detection, and also associated with worse prognosis in stage I–II lung cancer (HR =1.71, P =0.048). All measurements were highly reproducible with intraclass correlation coefficients ranging from 0.82 – 0.99. Both metabolites were significantly (P <0.03) enriched in tumor tissue compared to adjacent non-tumor tissue (N =48), thus revealing their direct association with tumor metabolism. Creatine riboside and NANA may be robust urinary clinical metabolomic markers that are elevated in tumor tissue and associated with early lung cancer diagnosis and worse prognosis.
Keywords: lung cancer, metabolomics, urine, diagnosis, prognosis
Introduction
Lung cancer is the leading cause of cancer deaths in men and women in the United States (1, 2) and worldwide (3), and survival rates are dismal. When the disease is detected while it is still localized, the five-year survival rate is 53%, but that rate drops to 24% for regional disease and, even more significantly, to <5% for distant tumors (4). However, these survival rates could be improved substantially with the identification of biomarkers to support the accurate and reliable diagnosis and prognosis of lung cancer.
Current clinically accepted methods for detecting lung cancer include spiral CT scanning in smokers between the ages of 55 to 74 and a history of smoking 30 packs of cigarettes per year (5, 6). However, low-dose spiral CT (LDCT) scanning provides a high rate of false positives—96.4% overall, and 24% in combination with invasive testing (7). Moreover, spiral CT scanning may be of concern due to an increased lung cancer risk associated with radiation exposure (8). As a result, the medical community requires a concordant biomarker to better identify patients who should be screened or who should undergo invasive diagnostic work-ups. However, to date, no molecular biomarker for early stage lung cancer has been validated (9, 10).
Several biomarkers currently support the assessment of overall prognosis and guide therapy decisions. For example, the KRAS mutation in non–small cell lung cancer (NSCLC) confers a significantly shorter survival (HR =1.21) in stage IV disease (11), and the presence of an ALK or EGFR mutation indicates a responsive tumor to targeted therapies and longer survival (12–15). However, these biomarkers for lung cancer outcomes are based on tumor assays, an invasive approach that can be hindered by the limited availability of tissue.
Urine is now attracting increased attention as a biospecimen for detecting cancer biomarkers (16), not only because it is collected non-invasively but also because it is abundant and requires minimal preparation. For instance, one urinary cancer biomarker, PCA3, is currently applied clinically to detect prostate cancer (17). No clinically applied biomarkers exist yet for lung cancer. Nonetheless, promising urinary biomarkers include modified nucleosides (18–21), whose high levels indicate an increased RNA turnover and degradation and whose utility is being evaluated in clinical trials. However, modified nucleosides are elevated in many different tumor types, and therefore may not be cancer type specific (22).
Mass spectrometry–based metabolomic approaches are increasingly used for uncovering new biomarkers for diagnosis (23–28) and customized treatment (29), as well as for evaluating pathological characteristics of metastatic cells (30) and carcinogenic tobacco-smoke constituents (31, 32). The reliability and reproducibility of such approaches are robust (33) and the technologies are currently in place in clinical practice (34), making them strong candidates for uncovering potential biomarkers. Unfortunately, most studies suffer from limited sample sizes, poor quality control, and a lack of technical and biological validation.
To address these current limitations, we have taken a comprehensive approach utilizing state of the art methodology and a large sample size, and have uncovered robust and technically validated biomarkers that can aid diagnosis and guide therapeutic decisions in NSCLC. Initially, we measured small (<1500 Da) urinary molecules from 1,005 individuals with and without lung cancer (training set) to uncover metabolites that most strongly distinguished the two groups. We found that levels of four metabolites were elevated in lung cancer patients and best predicted their lung cancer status, independent of their gender, race, and self-reported smoking status: creatine riboside (a novel molecule identified in our study), N-acetylneuraminic acid (NANA), cortisol sulfate, and an as-yet-unidentified glucuronidated compound referred to as 561+. These results were confirmed in a validation set comprising 158 individuals, and abundances of significant metabolites were further validated through absolute quantitation and values normalized to urinary creatinine levels in order to control for kidney function. The applicability of these findings to lung cancer diagnosis in clinical practice is primarily focused on two of the urinary metabolites—creatine riboside and NANA—which were significantly more abundant in stage I tumors when compared to adjacent non-tumor lung tissues. This association in the tissue provides a direct link to altered tumor metabolism and importantly, elevated levels of these metabolites can be non-invasively detected in the urine. Notably, elevated levels of these metabolites are also associated with worse prognosis.
Materials and Methods
Study Subjects
Urine samples from 469 NSCLC patients prior to treatment and 536 population controls collected from 1998 to 2007 from the greater Baltimore, Maryland, area were employed as a training set (Table 1). Patients were recruited from pathology departments, pulmonary and thorasic clinics with the cooperation of attending physicians in seven hospitals: Baltimore Veterans Administration Medical Center, Bon Secours Hospital, MedStar Harbor Hospital, Sinai Hospital, Johns Hopkins Bayview Medical Center, The Johns Hopkins Hospital, and University of Maryland Medical Center. Population controls were identified from the Department of Motor Vehicles (DMV) lists and frequency-matched to cases by age, gender, and self reported race. Lung cancer patients were not diagnosed with other cancer types. Findings from the training set were replicated in an additional set of 80 recently diagnosed cases (years of diagnosis 2008–2010) and 78 population controls (recruited through the DMV), a sample set we refer to as a validation set (Table 1). These validation set samples have a similar distribution of demographic and clinical characteristics when compared to the training set. We also utilized 48 tumor and adjacent non-tumor stage I tissue pairs, of which 20 are a subset of the training set. Survival times were calculated as time of diagnosis to time of death or to follow-up (2010); death due to cancer was determined from the NDI extraction of the death certificates. This study was approved by the Institutional Review Boards of the seven institutions. Urine samples were collected at the time of interview when possible. If collected at a different time, a brief intake questionnaire was administered including recent smoking information. In each case, urine was collected in a plain, sterile 50 ml container and transported to the University of Maryland where it was split into 10 ml aliquots and stored at −80°C until used. Urines were thawed on wet ice at the time of use. Subjects were not required to fast or undergo any other preparatory procedure before urine collection. The time of interview and subsequent urine collection was recorded with the questionnaire data.
Table 1.
Sample characteristics of all sample sets presented in the study.
| Trainining Set | Validation Seta | Tissue Set | |||||
|---|---|---|---|---|---|---|---|
| All (N =1005) |
Cases (N =469) |
Population Controls (N =536) |
All (N =158) |
Cases (N =80) |
Population Controls (N =78) |
Tumor/Adjacent Normal Pairs (N =48) |
|
| Age | (mean = 66.4) | (mean = 66.2) | (mean = 66.6) | (mean = 66.7) | (mean = 64.2) | (mean = 68.7) | (mean = 68.9) |
| > mean | 519 | 240 | 279 | 82 | 35 | 47 | 27 |
| <=mean | 486 | 229 | 257 | 76 | 45 | 31 | 21 |
| Smoking Statusb | |||||||
| Ever | 10 | ||||||
| Current | 293 | 222 | 71 | 46 | 38 | 8 | 17 |
| Former | 463 | 214 | 249 | 73 | 31 | 42 | 17 |
| Never | 249 | 33 | 216 | 39 | 11 | 28 | 4 |
| Histology | |||||||
| ADC | 216 | 51 | 31 | ||||
| SCC | 122 | 14 | 16 | ||||
| NSCLC | 131 | 10 | 1 | ||||
| Gender | |||||||
| Female | 492 | 232 | 260 | 81 | 46 | 35 | 24 |
| Male | 513 | 237 | 276 | 77 | 34 | 43 | 24 |
| Raceb | |||||||
| African | |||||||
| American | 366 | 127 | 239 | 70 | 35 | 35 | 9 |
| Caucasian | 639 | 342 | 297 | 88 | 45 | 43 | 39 |
| Stagec | |||||||
| I–II | 213 | 31 | 48 | ||||
| III–IV | 103 | 41 | 0 | ||||
Five samples are missing histology, and eight samples are missing stage information.
Self-reported smoking status and race.
Only pathologically staged cases, according to the 7th edition of the Cancer Staging Manual of the American Joint Committee on Cancer, were utilized for stratified analyses.
Detailed clinical information derived from extensive questionnaires is available for each patient, including age, gender, self-reported race, self-reported smoking status (never smokers, having smoked less than 100 cigarettes in their lifetime; former smokers, having quit smoking at least six months prior to the interview date), pack years, histology, AJCC staging, and survival (Table 1). Lung cancer diagnosis was pathologically determined. Staging was performed by a pathologist using the seventh edition of the AJCC’s Cancer Staging Manual (35).
Study Design
All initial analyses were performed in a training set comprising 1,005 samples (Table 1). Results from Random Forest (36, 37) classifications and univariate Cox analysis were combined to identify 4 metabolites that were predictive of both lung cancer diagnosis and prognosis. Results were then confirmed in a quantitation set (N =198) comprising a subset of the training set samples, and a validation set of 158 urine samples independent of the training set samples (Table 1). Finally, the 4 metabolites of interest were measured in 48 matched tumor and adjacent non-tumor tissue pairs. The overall study design is depicted in Figure S1.
Untargeted Metabolite Profiling Using UPLC-ESI-QTOFMS
We analyzed urine samples using a quadrupole time-of-flight (QTOF) mass spectrometer (Premier, Waters), in positive (ESI+) and negative (ESI−) electrospray ionization modes, using a 50 × 2.1 mm Acquity 1.7 µm C18 column (Waters Corp, Milford, MA). Urine samples were diluted with an equal volume of 50% aqueous acetonitrile containing debrisoquine (ESI+ internal standard) and 4-nitrobenzoic acid (ESI− internal standard). Samples were centrifuged at 14,000 × g for 20 minutes at 4°C to precipitate proteins. Five µl was chromatographed on a 50 × 2.1 mm Acquity BEH 1.7 µm C18 column (Waters) using an Acquity UPLC system (Waters). The gradient mobile phase consisted of 0.1% formic acid (A) and acetonitrile containing 0.1% formic acid (B). A typical 10-min sample run (at 0.5ml/min) consisted of 0.5 min of 100% solvent A followed by a linear gradient to 80% A at 4 min, to 5% A at 8 min. After a 0.5 min wash step, the column was equilibrated to initial conditions for 1.5 min. The eluent was introduced by electrospray ionization into the QTOF mass spectrometer (Premier, Waters) operating in positive (ESI+) or negative (ESI−) ionization mode. The capillary and sampling cone voltages were set to 3,000 and 30 V, respectively. Source and desolvation temperatures were set to 120 °C and 350 °C, respectively, and the cone and desolvation gas flows were set to 50.0 and 650.0 L/h, respectively. To maintain mass accuracy, sulfadimethoxine at a concentration of 300 pg/µl in 50% aqueous acetonitrile was used as a lock mass and injected at a rate of 50 µl/min. For MS scanning, data were acquired in centroid mode from 50 to 850 m/z and for tandem MS the collision energy was ramped from 5 to 35 V.
To avoid artifacts based on sample injection order, the order was randomized. Four different quality control sets were included with the runs to assess machine sensitivity and sample carry over. First, 169 “pooled” samples, containing aliquots from 108 randomly selected urine samples were processed randomly throughout the run. Second, a standard cocktail containing theophylline, caffeine, hippuric acid, 4-nitrobenzoic acid, and nortriptyline (designated as MetMix) was injected every 100 samples. Third, 32 blanks were randomly injected to assess sample carryover. Fourth, 48 samples with 4 high-purity nicotine metabolite standards, including cotinine, nicotine-N’-oxide, anabasine, and trans-3’-hydroxycotinine (Sigma-Aldrich), were spiked into urine. Fifth, 10% of the samples were randomly selected and processed in duplicate at the end of the run to evaluate chromatogram consistency. Finally, debrisoquine and 4-nitrobenzoic acid were spiked into samples for runs in ESI+ and ESI− modes, respectively. Raw chromatograms and extracted and normalized ion counts can be accessed in the MetaboLights database with study identifier MTBLS28.
Metabolite Quantitation
Urine samples were processed with an equal volume of 50% aqueous acetonitrile containing chloropropamide and aminopimelic acid as internal standards and chromatographed on a 50 × 2.1 mm Acquity BEH 1.7 µm C18 column using an Acquity UPLC system (Waters). MRM transitions were monitored using a Xevo TQMS (Waters). In addition, samples were analyzed using hydrophilic interaction chromatography (HILIC) columns (Acquity UPLC BEH Amide 1.7 µm 50×2.1 mm) for the quantitation of creatine riboside and NANA. HILIC columns improve retention, separation, and detection of highly polar metabolites.
Tissue Metabolite Extraction and Quantitation
Tumor and matched adjacent non-tumor tissues were pulverized by cryogenic grinding (Cryomill®, Retsch GmbH, Haan, Germany) using a 5 mm stainless steel ball per sample. Average sample weight was 15mg (with a range between 3 and 30mg). A monophasic mixture of ice-cold chloroform:methanol:water (2:5:2, v:v:v) was used for extraction. Samples were centrifuged at 14,000 × g for 15 minutes at 4°C, dried down using vacuum evaporator (SpeedVac), reconstituted in 70% aqueous acetonitrile, of which 5uL was injected onto the Xevo TQMS system for analysis.
Statistical Analyses
Samples were classified as lung cancer or healthy controls using the R package randomForest (36, 37). For additional details regarding randomForest parameters used in data processing, please see Supplementary Materials and Methods.
Unconditional logistic regression was performed in STATA (Stata Statistical Software Release 11.2, College Station, TX), while controlling for race, gender, interview year, smoking status, pack years, and urine collection time. N-acetylneuraminic acid levels do show some diurnal variation (Figure S7), and therefore all analyses were also adjusted for the time of day urine was collected. Unconditional logistic regression analysis was performed on categorical variables calculated by dichotomizing metabolite abundances into high (>= 75th percentile) and low (< 75th percentile) based on the distribution of metabolite abundances in the population control subjects. Unconditional logistic regression models were used to estimate odds ratios (ORs) and 95% confidence intervals (CIs) for both univariate and multivariate models adjusted for race, gender, interview year, smoking status, pack years, and urine collection time. False discovery rates were calculated using the Benjamini and Hochberg method (38).
Survival analyses were performed on categorical variables of dichotomized metabolite abundances in SAS Enterprise Guide, version 4.2 (SAS Institute Inc.), and all reported P values are two-sided. Cox models with left truncation were performed to account for the lag time between diagnosis and urine collection dates (up to two years). Multivariate Cox models were adjusted for urine collection time, histology, stage, race, gender, interview year, pack years, smoking status, chemotherapy/radiation and surgery status. The proportional hazards assumption (39) was tested, and if it was not met, the hazard ratio function was calculated separately before and after a given time point. This cut-off was determined by the time at which the survival curves started to diverge/converge and by ensuring that the β coefficients of the signal-time term before and after were no longer significant.
Receiver operating characteristics (ROC) were conducted in STATA 11.2 to assess the predictive value of identified metabolites in lung cancer diagnosis using roctab and roccomp functions. Models were built using logistic regression on the continuous abundances of each metabolite individually, and on the combination of the four metabolites. For the comparison of ROC curves, rocreg function in STATA 11.2 was employed.
Non-parametric Wilcoxon test in STATA 11.2 was utilized to assess abundance differences of four metabolites, as detected in the urine of lung cancer patients when compared to population controls, for three sets (training, validation and quantitation sets).
Paired Student’s t-test in STATA 11.2 was used to assess abundance differences between forty eight tumor and forty eight adjacent non-tumor tissue samples. All reported P values are double sided.
Results
Quality Control Assessment of the Metabolomics Data
Initially, abundances of possible small (<1500 Da) urinary molecules in a training set comprising 1,005 urine and 521 quality control samples (Figure S1, Table 1) were measured using ultra-performance liquid chromatography-electrospray-ionization-quadrupole time of flight (UPLC-ESI-QTOF) mass spectrometry. After signal filtering (see Supplementary Materials and Methods for additional detail), a total of 1,807 signals were detected in the positive and 1,359 in the negative ionization mode, which represents a comprehensive pool of small urinary molecules. Signals here refer to unique m/z and retention time pairs and not unique metabolites. It is possible that a metabolite could be represented by multiple signals due to adduct formation and/or fragmentation occurring in the mass spectrometer.
The quality and robustness of our measurements were assessed using a variety of internal controls. First, the expected clustering of quality control samples (blanks, metmix, pools, nicotine standards) apart from the lung cancer and population control urine samples were observed in the multidimensional scaling analysis (see Materials and Methods for additional detail) (Figure S2A). Second, measurement reproducibility within a run was assessed by processing 169 (~15%) randomly selected, duplicate samples, and a strong correlation was observed with Pearson’s correlation coefficients >0.85 for the large majority of samples (Figure S2B). Third, the distribution of coefficients of variation (CVs) was assessed to ensure a small variation in quality control measurements. As expected, CVs were considerably smaller for the quality control samples compared to the study subject samples (P <0.00001, Figure S2C).
Predictions of Smoking Status
As a proof of principle, we aimed to classify individuals by their smoking status (smokers versus non-smokers of self-reported smoking status) to ensure that known metabolites related to tobacco smoke were detectable and strongly predictive of the self-reported smoking status. Random Forests (36, 37) was applied to the training set comprising 469 lung cancer cases and 536 population controls and 87% correct classification by smoking status was obtained (Figure S3A). The three most highly associated metabolites, ranked according to the importance score given by Random Forests, were well-known nicotine metabolites: cotinine, nicotine-N’-oxide, and trans-3’-hydroxycotinine. When stratified by smoking status, it became evident there was a global increase of these nicotine metabolites in current smokers compared to those who had formerly or never smoked (Figure S3B). This finding established the quality of measurements and the utility of our classification approach in identifying diagnostic metabolites of lung cancer.
Predictions of Lung Cancer Status
Classification of our training set samples using Random Forests resulted in 78.1% accuracy (True Positive Rate [TPR] = 76.5%, False Positive Rate [FPR] = 18.4%), by employing top predictive signals (Table S1, see Supplementary Materials and Methods for details regarding analysis). To account for possible differences in smoking habits between different genders and race, additional classifications of cases and controls were performed on samples stratified by self-reported race and gender. Using top predictive signals, we accurately categorized the following proportion of samples as lung cancer cases or controls: 77.7% for Caucasian males, 78.6% for Caucasian females, 84.9% for African-American males, and 82.3% for African-American females. TPRs and FPRs ranged from 70.0 to 81.7 and from 9.5 to 23.3, respectively (Table S1). Four metabolites contributed strongly to the classifications, independent of race and gender (Figure S4): NANA; cortisol sulfate; creatine riboside, novel metabolite identified in this study; and 561+, an unidentified metabolite with a mass/charge ratio of 561.3432+ that was confirmed to be a glucuronidated compound. We have conducted extensive validation methods to confirm the identity of novel creatine riboside, including ultraperformance liquid chromatography coupled to tandem mass spectrometry (UPLC-MS/MS) and 2D NMR (Figure S5, S6).
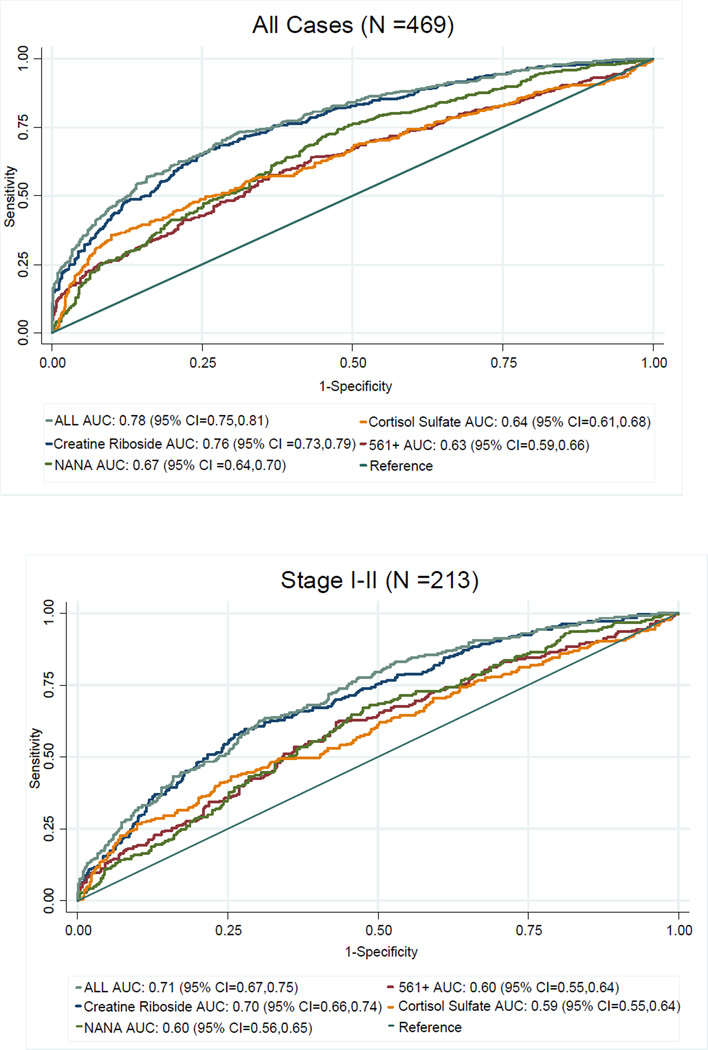
This study utilized a case control rather than a cohort setting and, as a result, could not be used for risk assessment. However, we took into account possible confounding factors of lung cancer classification, performing logistic regression in all cases and in stage I–II cases (Table 2), adjusting for race, gender, interview year, smoking status, pack years, and urine collection time (accounting for diurnal effects, Figure S7). Metabolite levels were dichotomized into high and low categorical variables based on the 75th percentile of population control abundances. As expected, associations with diagnosis were confirmed after adjusting for these potential confounders. ROC analysis resulted in areas under the curve (AUC) ranging from 0.63 to 0.76 for all cases, and 0.59 to 0.70 for stage I–II cases (Figure 1), using individual metabolites. Models using creatine riboside or all four biomarkers in all cases and in stage I–II cases were significantly more predictive (P <0.00001) than models using the other three metabolites individually, and these associations were independent of histology. Of note, lung cancer cases presented in this study were staged according to the latest 7th edition of the American Joint Committee on Cancer (AJCC) (35); however, 153 of 469 cases could not be restaged due to missing pathology reports, as reflected in the numbers of staged cases in Table 1.
Table 2.
Association of top four metabolites with lung cancer diagnosis (unconditional logistic regression) in the training set in all cases (top) and cases of stages I–II (bottom).
| All Cases (N= 469) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Metabolitec | Univariate | Multivariatea | ||||||
| Controls (%)d | Cases (%)d | OR (95% CI) | P | FDRb | OR (95% CI) | P | FDRb | |
| Creatine Riboside | 134 (25.0) | 304 (64.8) | 5.50 (4.21, 7.26) | 8.35E-35 | 2.64E-31 | 5.05 (3.57, 7.14) | 4.93E-20 | 1.56E-16 |
| Cortisol Sulfate | 134 (25.0) | 227 (48.4) | 2.84 (2.17, 3.71) | 1.69E-14 | 2.68E-11 | 2.56 (1.83, 3.58) | 3.52E-08 | 2.79E-05 |
| N -acetylneuraminc acid | 134 (25.0) | 213 (34.8) | 2.50 (1.91, 3.26) | 1.87E-11 | 5.38E-09 | 2.13 (1.52, 2.98) | 1.11E-05 | 1.25E-03 |
| 561+ | 134 (25.0) | 201 (34.1) | 2.25 (1.72, 2.94) | 2.90E-09 | 4.37E-07 | 1.89 (1.34, 2.67) | 3.17E-04 | 0.01 |
| Stage I – II Cases (N= 213) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Metabolitec | Univariate | Multivariatea | ||||||
| Controls (%)d | Cases (%)d | OR (95% CI) | P | FDRb | OR (95% CI) | P | FDRb | |
| Creatine Riboside | 134 (25.0) | 116 (54.5) | 3.59 (2.57, 5.01) | 5.59E-14 | 1.77E-10 | 3.34 (2.07, 5.39) | 7.85E-07 | 0.002 |
| Cortisol Sulfate | 134 (25.0) | 88 (41.3) | 2.11 (1.51, 2.95) | 1.26E-05 | 0.003 | 1.84 (1.14, 2.98) | 0.013 | 0.295 |
| N -acetylneuraminc acid | 134 (25.0) | 74 (34.7) | 1.60 (1.13, 2.25) | 0.007 | 0.076 | 1.72 (1.05, 2.81) | 0.030 | 0.347 |
| 561+ | 134 (25. 0) | 7 6 (35.7) | 1.66 (1.18, 2.34) | 0.003 | 0.046 | 1.30 (0.80, 2.12) | 0.296 | 0.728 |
Adjusted for race, gender, interview year, smoking status, pack years and urine collection time
False discovery rate (FDR) based on Benjamini and Hochberg
Levels dichotomized to high and low based on the 75th percentile of population control abundances (low = referent)
Numbers of controls and cases with high levels of the corresponding metabolite
Figure 1.
Receiver Operating Characteristic (ROC) analysis of individual metabolites and their combination in the training set in all cases (top), and in stage I–II (bottom) cases.
Association with Tobacco Smoke Exposure
To investigate whether the urinary metabolomic markers are correlated to tobacco smoke exposure, metabolite levels stratified by cigarettes per day (cpd) were investigated. We observed that the number of cigarettes per day was not associated with urinary levels of creatine riboside and NANA, nor was it associated with cortisol sulfate and 561+ (Figure S8). A correlation between abundances of each metabolite and cotinine (accepted indicator of exposure to tobacco smoke) was also investigated and no correlation was observed (data not shown). Additionally, logistic regression classification was stratified by smoking status: all four metabolites are also significantly associated with lung cancer status in never smokers (data not shown), further confirming that these metabolites are not associated with smoking.
Association with Prognosis
We next investigated whether the four metabolites found to be most robust in predicting lung cancer status are associated with prognosis, and whether they, therefore, may have utility in predicting patient outcome. Metabolite levels were dichotomized into high and low categorical variables based on the 75th percentile of the population control abundances. After adjusting for gender, race, stage, histology, smoking status, pack years, interview year, urine collection time, chemotherapy and/or radiation and surgery status, we found that high levels of NANA (HR =1.54 [P =0.025] in the first 15 months), cortisol sulfate (HR =1.63 [P =0.0001], creatine riboside (HR =1.81 [P =0.0002] in the first 45 months), and 561+ (HR =1.95 [P =0.0001] in the first 20 months) were associated with worse survival rates (Table 3, Figure 2A). In stage I–II cases, creatine riboside (HR =1.71 [P =0.048]) and 561+ (HR =8.63 [P =0.001]) were also associated with worse survival, independent of putative clinical cofactors (Table 3, Figure S9A). The time cut-offs presented here are chosen to meet the proportional hazards assumption test (39), details of which can be found in the Materials and Methods.
Table 3.
Association of top four metabolites with lung cancer survival (Cox proportional hazards regression) in the training set in all cases (top) and cases of stages I–II (bottom).
| All Cases (N= 469) | ||||||
|---|---|---|---|---|---|---|
| Metabolitec | Univariate | Multivariatea | ||||
| HR (95% CI) | P | FDRb | HR (95% CI) | P | FDRb | |
| N -acetylneuraminic acid | ||||||
| <= 15 months | 1.74 (1.22 – 2.48) | 0.002 | 0.06 | 1.54 (1.06 – 2.25) | 0.025 | 0.09 |
| > 15 months | 1.14 (0.82 – 1.57) | 0.44 | 1.27 (0.90 – 1.80) | 0.17 | ||
| Cortisol sulfate | 1.53 (1.21 – 1.94) | 0.0004 | 0.01 | 1.63 (1.27 – 2.08) | 0.0001 | 0.02 |
| Creatine riboside | ||||||
| <= 45 months | 2.05 (1.54 – 2.71) | < 0.0001 | 0.0005 | 1.81 (1.33 – 2.45) | 0.0002 | 0.002 |
| > 45 months | 0.86 (0.38 – 1.95) | 0.72 | 0.78 (0.34 – 1.83) | 0.57 | ||
| 561+ | ||||||
| <= 20 months | 2.32 (1.70 – 3.15) | < 0.0001 | 0.001 | 1.95 (1.39 – 2.74) | 0.0001 | 0.009 |
| > 20 months | 1.05 (0.70 – 1.55) | 0.83 | 0.86 (0.56 – 1.32) | 0.48 | ||
| Stage I–II Cases (N= 213) | ||||||
|---|---|---|---|---|---|---|
| Metabolitec | Univariate | Multivariatea | ||||
| HR (95% CI) | P | FDRb | HR (95% CI) | P | FDRb | |
| N -acetylneuraminic acid | 0.70 (0.41 – 1.19) | 0.18 | 0.89 | 0.56 (0.32 – 1.00) | 0.052 | 0.80 |
| Cortisol sulfate | 1.45 (0.90 – 2.32) | 0.12 | 0.89 | 1.39 (0.84 – 2.29) | 0.20 | 0.84 |
| Creatine riboside | 1.78 (1.08 – 2.93) | 0.02 | 0.81 | 1.71 (1.01 – 2.92) | 0.048 | 0.67 |
| 561+ | ||||||
| <= 15 months | 7.83 (2.23 – 27.51) | 0.001 | 0.60 | 8.63 (2.40 – 31.05) | 0.001 | 0.27 |
| > 15 months | 0.83 (0.45 – 1.52) | 0.54 | 0.84 (0.43 – 1.67) | 0.63 | ||
Adjusted for gender, race, stage (unless stratified), histology, smoking status, pack years, interview year, urine collection time, chemotherapy and/or radiation status, and surgery status
False discovery rate (FDR) based on Benjamini and Hochberg
Levels dichotomized into high and low based on the 75th percentile of population control abundances (low = referent)
Figure 2.
Kaplan-Meier survival estimates in the training set are depicted for the top four predictive metabolites in A) all lung cancer patients. The P values reported in the Kaplan-Meier plots reflect the maximum likelihood estimates generated using a univariate Cox model, taking into account left truncation (the lag time between diagnosis and time of urine collection). B) The combination of the top four predictive metabolites is shown for all cases. Only metabolites that showed statistically significant associations with survival, independent of clinical cofactors (see Materials and Methods), were combined. Metabolite levels were dichotomized into high and low based on the 75th percentile of population controls abundances.
Significantly, the combination of these metabolites and their associations with survival demonstrates an independent and additive effect (Figure 2B, Figure S9B, Table S2), suggesting that in combination, these four markers may be of value in therapy decisions, therefore improving patient outcomes. Although this study was limited in the representation of African-Americans, stratification by self-reported race highlighted cortisol sulfate as most strongly associated with survival in African-Americans (Table S3).
Validation in Independent Sample Sets and Assessment of Metabolite Stability
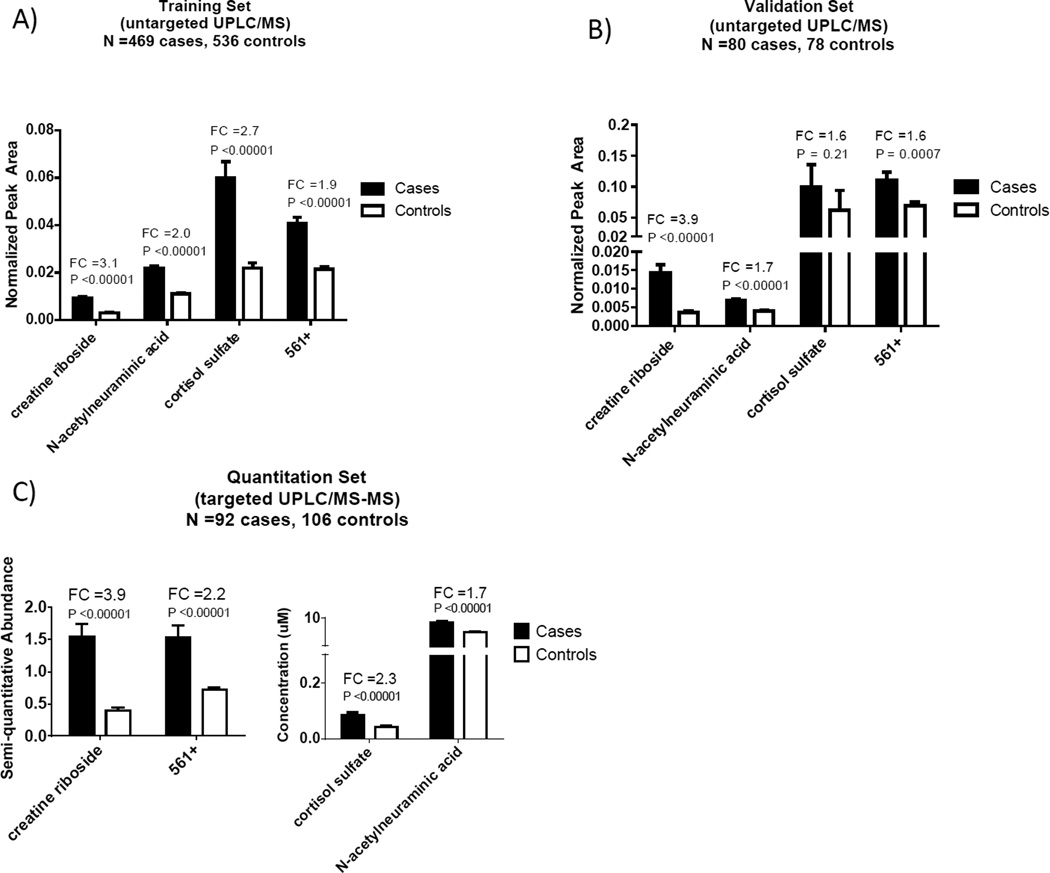
When compared to the training set, creatine riboside, NANA, and 561+ were confirmed to be elevated in the urine of lung cancer patients in an independent validation set comprising 158 more recently diagnosed cases (P <0.0007) (Figure 3A, 3B). Although cortisol sulfate was not found to be significantly elevated in cases, possibly due to insufficient power, the expected trend of the levels being higher in lung cancer patients was observed. Measurements of these metabolites were technically validated on a quantitative Xevo triple quadrupole mass spectrometer in a subset (N =198) of the training set, representing similar distributions of age, gender, and racial composition to the training cohort (P <0.00001, Figure 3C). Conscious of the importance of measurement reproducibility, especially in clinical laboratory practice, the stability of metabolites in storage over time and after a freeze-thaw cycle was studied. The reproducibility of metabolite measurements obtained by a second quantitation carried out two years later on the same samples resulted in intraclass correlation coefficients (ICC) from 0.82 to 0.99 (Table S4). These high ICCs strongly suggest that these metabolites are sufficiently stable and reproducible and may be used as biomarkers of lung cancer diagnosis in clinical practice.
Figure 3.
Abundance and validation of metabolites that were top contributors in the classification of patients as lung cancer or healthy controls. Untargeted and MSTUS normalized UPLC-MS abundances (mean and standard error of the mean (SEM)) are depicted for A) the training set containing 469 lung cancer cases and 536 controls, B) the validation set comprising 80 cases and 78 controls. Quantitated UPLC-MS/MS abundances (mean and SEM) in C) a subset of the training set containing 92 cases and 106 controls. FC=fold change
Link to Tumor Metabolome
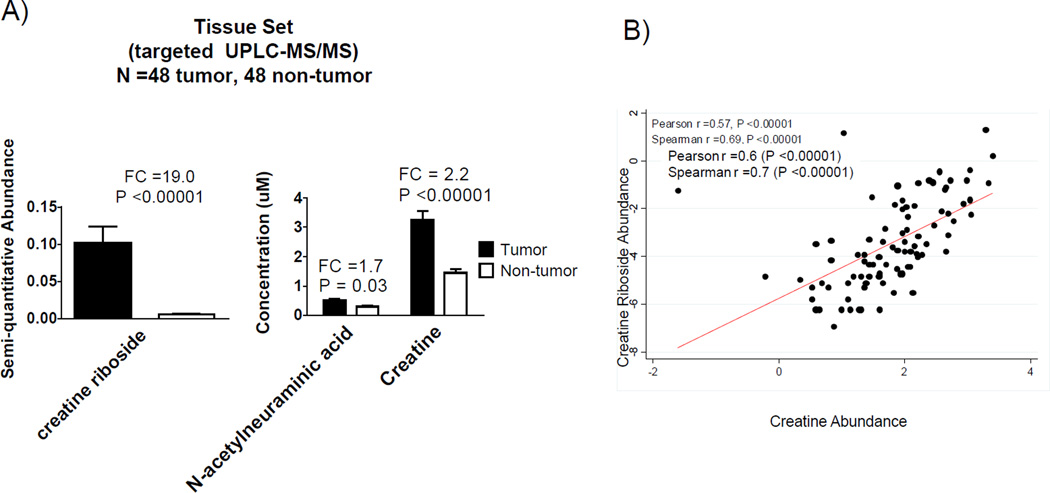
We next assessed the presence of creatine riboside, NANA, cortisol sulfate and metabolite 561+ in 48 tumor tissues resected from stage I adeno- and squamous cell-carcinoma patients. Their detection in tissue would indicate a direct relationship to lung tumor metabolism. Creatine riboside and NANA were significantly more abundant in tumor compared to adjacent non-tumor tissue. Creatine was also elevated in the tumor compared to non-tumor tissue (Figure 4A) and correlates with creatine riboside (Figure 4B), further confirming the formation of creatine riboside from creatine. These important findings suggest that creatine riboside and NANA are products of altered lung tumor metabolism that can be detected in non-invasively obtained urine.
Figure 4.
Linking urinary metabolites to lung cancer tissue metabolome. A) Levels of creatine riboside, N-acetylneuraminic acid and creatine in a paired tumor/adjacent non-tumor tissue set containing 48 stage I adenocarcinoma and squamous cell carcinoma tumors and 48 adjacent non-tumor samples. B) Correlation between creatine riboside and creatine quantitated in tumor tissue samples.
Discussion
A paucity of noninvasive biomarkers for detection and prognostic assessment plagues the lung cancer field, and most pre-clinical studies aimed to identify putative biomarkers suffer from limited sample sizes (10). Our assessment of 469 cases and 536 population controls revealed two urinary biomarkers for the detection and prognosis of NSCLC: creatine riboside and NANA. Although we also identified cortisol sulfate and 561+ as robust putative biomarkers predictive of lung cancer status—independent of race and gender—creatine riboside and NANA were also elevated in tumor compared to adjacent non-tumor tissue, thereby providing a direct link with metabolic changes in the tumor, and allowing for non-invasive detection of these tumor-specific metabolites in easily obtainable urine. This finding may eventually be able to guide therapeutic decisions in improving lung cancer patient outcomes. However, the utility of these metabolites has not been evaluated in other cancers, and their potential to aid early diagnosis of lung cancer remains to be further evaluated. Although there are currently accepted technologies for early detection of lung cancer, such as LDCT, a complementary biomarker is needed; while LDCT has a very high sensitivity and almost no lung lesion goes undetected, it performs poorly in distinguishing benign from malignant nodules. We speculate that creatine riboside and NANA may aid in the early detection of lung cancer, possibly as an adjunct to LDCT, and may perhaps decrease its high false positive rate of 96.4% (7). Of note, creatine riboside was the strongest classifier of lung cancer status in all cases but also in stage I–II lung cancer. Pending future studies addressing the mechanism of creatine riboside generation and potential causal relationship to lung cancer, this novel metabolite may eventually serve as a therapeutic target in clinical practice.
Therapeutic decisions, including surgery for earlier stages of cancer, adjuvant chemotherapy, and/or radiation therapy, are based on tumor size, molecular biomarkers, morphological features, and gross tumor characteristics (40). However, the assessment of high risk requires refinement, especially for completely resected stage I NSCLC, where no trial has shown any significant survival benefit in stage IB (41, 42) and where there is a possibly detrimental effect of adjuvant chemotherapy for stage IA patients (43). We propose that these metabolites could be useful in guiding such therapy decisions. In particular, the association of creatine riboside with worse prognosis in stage I–II lung cancer patients and its elevated levels in tumors makes creatine riboside a candidate for aiding in therapeutic decisions. Furthermore, the combination of all metabolites should be explored, as the combination of all four metabolites was most strongly associated with prognosis in all stages, while the combination of creatine riboside and 561+ was most strongly associated with prognosis in stage I–II NSCLC patients.
Creatine riboside is also of special interest, as it has not been previously reported. Markedly higher serum levels of the creatine kinase isoenzyme BB, an enzyme responsible for the conversion of creatine into a phosphocreatine, an important energy reserve, have been observed in lung cancer patients (44, 45). Additionally, cancer cells have a higher energy requirement compared to quiescent normal cells (46); as a result, creatine riboside may be a product of both high creatine within the tumor, as reported in our study, and high phosphate flux. While creatine riboside as a compound has not been described until now, increased mutagenicity of creatine and ribose pyrolysis products in cooked foods has been reported (47), suggesting a functional role of creatine riboside in tumorigenesis. Since creatine riboside is the strongest predictor of lung cancer diagnosis in our study, including stage I–II cases, its abundance may be a useful complement to LDCT in further distinguishing malignant from benign nodules detected at screening and preventing unnecessary and invasive diagnostic work-ups.
NANA and cortisol sulfate have been previously reported in the context of cancer. N-acetylneuraminic acid is one of the two most common forms of sialic acid and plays a role in cell signaling, binding and transportation of positively charged molecules, attraction and repulsion of cells and molecules, and immunity (48). In cancer, these sialylated conjugates protect malignant cells from cellular defense systems. Elevated levels of NANA have been found in various cancer types, including lung cancer (49). Sialic acid as a blood biomarker for prognosis has been assessed with mixed results, although, to our knowledge, not in lung cancer. Due to the role that NANA plays on the cell surface of mammalian cells, this marker may not be lung cancer specific, allowing for a possibility of its utility in other cancers. Regarding cortisol sulfate, high urinary levels were reported in breast cancer (50), and deregulated cortisol metabolism was reported in critical illness (51), which may in part be due to the induction of pro-inflammatory cytokines, activators of cortisol production (52, 53).
This study and the conclusion that these metabolites may have clinical applications for the diagnosis and prognosis of lung cancer are notable for several reasons. First, urine is abundant, allows for noninvasive sampling, and does not require extensive processing (54). Second, mass spectrometry–based approaches are cost-effective on a per-sample basis and allow for fast screening with minimal processing, making it suitable for clinical settings. Third, measurements of the metabolites reported here are highly reproducible, indicating their stability in urine over time, despite freeze-thaw cycles (ICCs >0.82). And finally, the robustness of these biomarkers against age, gender, and race points to their universal applicability.
The current study, however, is not without its limitations. Because metabolism can vary due to dietary and drug intake (55, 56), we were unable to adjust for these factors. In addition, we were unable to rule out selection, type of controls, and participation rates biases. An evaluation of these putative biomarkers in a prospective setting and their utility for risk assessment also remains to be carried out. The majority of the patients (323) had urine specimens collected prior to the administration of chemotherapy and/or radiation. We have determined that there are no differences in metabolite levels between those patients who had received treatment and those who had not (Figure S10A). Furthermore, only 37 out of 469 patients had undergone surgery before urine collection, with no significant differences in metabolite levels between the two groups (Figure S10B). The Cox regression survival analysis was controlled for treatment and surgery status, to ensure no confounding by the aforementioned variables. Furthermore, normalization to urinary creatinine levels is expected to eliminate the potential of altered kidney function to affect metabolite levels.
Overall, our findings indicate that creatine riboside and NANA may be useful in the diagnosis and prognosis of NSCLC, as they showed strong associations with these outcomes and were deregulated in tumor tissue. Undoubtedly, measurement of these metabolites in urine using mass spectrometry provides great potential for the detection of lung cancer in the clinic and may lead to the identification of novel therapeutic strategies and targets. Additionally, the results of this study lay the groundwork for assessing the direct impact of these metabolites in lung tumorigenesis (and possibly other cancers).
Supplementary Material
Acknowledgments
We thank Dr. Raymond Jones, John Cottrell, and Audrey Salabes at the University of Maryland and Baltimore Veterans Administration Medical Center for tissue and data collection, and Mr. Leoni Leondaridis of Advance Medical Systems Consultants for the coordination of data from the NDI. We also thank the Proteomics and Metabolomics Shared Resource at the Georgetown Lombardi Comprehensive Cancer Center, part of Georgetown University Medical Center and MedStar Georgetown University Hospital—specifically, Mr. Marc Bourbeau and Dr. Amrita Cheema. We utilized the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health, Bethesda, MD.
Grant Support:
The work presented in this manuscript has been partially funded by the NIH grant # ES022186.
Footnotes
The authors disclose no potential conflicts of interest.
References
- 1.Jemal A, Simard EP, Dorell C, Noone AM, Markowitz LE, Kohler B, et al. Annual Report to the Nation on the Status of Cancer, 1975–2009, Featuring the Burden and Trends in Human Papillomavirus (HPV)-Associated Cancers and HPV Vaccination Coverage Levels. J Natl Cancer Inst. 2013 doi: 10.1093/jnci/djs491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Boyle PLB, editor. The World Cancer Report 2008. Lyon, France: IARC; 2008. [Google Scholar]
- 4.Horner M, Ries LAG, Krapcho M, et al. SEER Cancer Statistics Review, 1975–2006. Bethesda, MD: National Cancer Institute; 2009. [Google Scholar]
- 5.Jaklitsch MT, Jacobson FL, Austin JH, Field JK, Jett JR, Keshavjee S, et al. The American Association for Thoracic Surgery guidelines for lung cancer screening using low-dose computed tomography scans for lung cancer survivors and other high-risk groups. J Thorac Cardiovasc Surg. 2012;144(1):33–38. doi: 10.1016/j.jtcvs.2012.05.060. [DOI] [PubMed] [Google Scholar]
- 6.Kang JX. Identification of metabolic biomarkers for personalized nutrition. Journal of nutrigenetics and nutrigenomics. 2012;5(2):I–II. doi: 10.1159/000342702. [DOI] [PubMed] [Google Scholar]
- 7.Aberle DR, Adams AM, Berg CD, Black WC, Clapp JD, Fagerstrom RM, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med. 2011;365(5):395–409. doi: 10.1056/NEJMoa1102873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brenner DJ. Radiation risks potentially associated with low-dose CT screening of adult smokers for lung cancer. Radiology. 2004;231(2):440–445. doi: 10.1148/radiol.2312030880. [DOI] [PubMed] [Google Scholar]
- 9.Vansteenkiste J, Dooms C, Mascaux C, Nackaerts K. Screening and early detection of lung cancer. Ann Oncol. 2012;23(Suppl 10):x320–x327. doi: 10.1093/annonc/mds303. [DOI] [PubMed] [Google Scholar]
- 10.Hassanein M, Callison JC, Callaway-Lane C, Aldrich MC, Grogan EL, Massion PP. The state of molecular biomarkers for the early detection of lung cancer. Cancer Prev Res (Phila) 2012;5(8):992–1006. doi: 10.1158/1940-6207.CAPR-11-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson ML, Sima CS, Chaft J, Paik PK, Pao W, Kris MG, et al. Association of KRAS and EGFR mutations with survival in patients with advanced lung adenocarcinomas. Cancer. 2013;119(2):356–362. doi: 10.1002/cncr.27730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350(21):2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 13.Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, et al. EGF receptor gene mutations are common in lung cancers from "never smokers" and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101(36):13306–13311. doi: 10.1073/pnas.0405220101. PMCID: 516528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361(10):947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 15.Antoniu SA. Crizotinib for EML4-ALK positive lung adenocarcinoma: a hope for the advanced disease? Evaluation of Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363(18):1693–1703. doi: 10.1517/14728222.2011.550880. [DOI] [PubMed] [Google Scholar]; Expert Opin Ther Targets. 2011;15(3):351–353. doi: 10.1517/14728222.2011.550880. [DOI] [PubMed] [Google Scholar]
- 16.Schmidt C. Urine biomarkers may someday detect even distant tumors. J Natl Cancer Inst. 2009;101(1):8–10. doi: 10.1093/jnci/djn482. [DOI] [PubMed] [Google Scholar]
- 17.Groskopf J, Aubin SM, Deras IL, Blase A, Bodrug S, Clark C, et al. APTIMA PCA3 molecular urine test: development of a method to aid in the diagnosis of prostate cancer. Clin Chem. 2006;52(6):1089–1095. doi: 10.1373/clinchem.2005.063289. [DOI] [PubMed] [Google Scholar]
- 18.Henneges C, Bullinger D, Fux R, Friese N, Seeger H, Neubauer H, et al. Prediction of breast cancer by profiling of urinary RNA metabolites using Support Vector Machine-based feature selection. BMC Cancer. 2009;9:104. doi: 10.1186/1471-2407-9-104. PMCID: 2680413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsu WY, Chen WT, Lin WD, Tsai FJ, Tsai Y, Lin CT, et al. Analysis of urinary nucleosides as potential tumor markers in human colorectal cancer by high performance liquid chromatography/electrospray ionization tandem mass spectrometry. Clin Chim Acta. 2009;402(1–2):31–37. doi: 10.1016/j.cca.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 20.Jeng LB, Lo WY, Hsu WY, Lin WD, Lin CT, Lai CC, et al. Analysis of urinary nucleosides as helper tumor markers in hepatocellular carcinoma diagnosis. Rapid Commun Mass Spectrom. 2009;23(11):1543–1549. doi: 10.1002/rcm.4034. [DOI] [PubMed] [Google Scholar]
- 21.Xu G, Schmid HR, Lu X, Liebich HM, Lu P. Excretion pattern investigation of urinary normal and modified nucleosides of breast cancer patients by RP-HPLC and factor analysis method. Biomed Chromatogr. 2000;14(7):459–463. doi: 10.1002/1099-0801(200011)14:7<459::AID-BMC7>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 22.Borek E, Baliga BS, Gehrke CW, Kuo CW, Belman S, Troll W, et al. High turnover rate of transfer RNA in tumor tissue. Cancer research. 1977;37(9):3362–3366. [PubMed] [Google Scholar]
- 23.Kim YS, Maruvada P, Milner JA. Metabolomics in biomarker discovery: future uses for cancer prevention. Future Oncol. 2008;4(1):93–102. doi: 10.2217/14796694.4.1.93. [DOI] [PubMed] [Google Scholar]
- 24.Kind T, Tolstikov V, Fiehn O, Weiss RH. A comprehensive urinary metabolomic approach for identifying kidney cancerr. Anal Biochem. 2007;363(2):185–195. doi: 10.1016/j.ab.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 25.Matsumura K, Opiekun M, Oka H, Vachani A, Albelda SM, Yamazaki K, et al. Urinary volatile compounds as biomarkers for lung cancer: a proof of principle study using odor signatures in mouse models of lung cancer. PLoS One. 2010;5(1):e8819. doi: 10.1371/journal.pone.0008819. PMCID: 2811722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, Yu J, et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457(7231):910–914. doi: 10.1038/nature07762. PMCID: 2724746. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 27.Yang Q, Shi X, Wang Y, Wang W, He H, Lu X, et al. Urinary metabonomic study of lung cancer by a fully automatic hyphenated hydrophilic interaction/RPLC-MS system. J Sep Sci. 2010;33(10):1495–1503. doi: 10.1002/jssc.200900798. [DOI] [PubMed] [Google Scholar]
- 28.Yuan JM, Gao YT, Murphy SE, Carmella SG, Wang R, Zhong Y, et al. Urinary levels of cigarette smoke constituent metabolites are prospectively associated with lung cancer development in smokers. Cancer research. 2011;71(21):6749–6757. doi: 10.1158/0008-5472.CAN-11-0209. PMCID: 3392910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fan TW, Lane AN, Higashi RM. The promise of metabolomics in cancer molecular therapeutics. Curr Opin Mol Ther. 2004;6(6):584–592. [PubMed] [Google Scholar]
- 30.Denkert C, Budczies J, Kind T, Weichert W, Tablack P, Sehouli J, et al. Mass spectrometry-based metabolic profiling reveals different metabolite patterns in invasive ovarian carcinomas and ovarian borderline tumors. Cancer research. 2006;66(22):10795–10804. doi: 10.1158/0008-5472.CAN-06-0755. [DOI] [PubMed] [Google Scholar]
- 31.Church TR, Anderson KE, Caporaso NE, Geisser MS, Le CT, Zhang Y, et al. A prospectively measured serum biomarker for a tobacco-specific carcinogen and lung cancer in smokers. Cancer Epidemiol Biomarkers Prev. 2009;18(1):260–266. doi: 10.1158/1055-9965.EPI-08-0718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hecht SS, Murphy SE, Stepanov I, Nelson HH, Yuan JM. Tobacco smoke biomarkers and cancer risk among male smokers in the Shanghai Cohort Study. Cancer Lett. 2012 doi: 10.1016/j.canlet.2012.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hsu PC, Zhou B, Zhao Y, Ressom HW, Cheema AK, Pickworth W, et al. Feasibility of identifying the tobacco-related global metabolome in blood by UPLC-QTOF-MS. J Proteome Res. 2012 doi: 10.1021/pr3007705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strathmann FG, Hoofnagle AN. Current and future applications of mass spectrometry to the clinical laboratory. Am J Clin Pathol. 2011;136(4):609–616. doi: 10.1309/AJCPW0TA8OBBNGCK. [DOI] [PubMed] [Google Scholar]
- 35.Edge S, Byrd DR, Compton CC, Fritz AG, Greene FL, Trotti A. AJCC Cancer Staging Manual. 7th ed. Springer-Verlag; 2010. [Google Scholar]
- 36.Breiman L. Random Forests. Machine Learning. 2001;45:5–32. [Google Scholar]
- 37.Ho TK. Random Decision Forest. Proceedings of the 3rd International Conference on Document Analysis and Recognition; Montreal, QC. 1995. pp. 278–282. [Google Scholar]
- 38.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57(1):11. [Google Scholar]
- 39.Breslow NE. Analysis of Survival Data under the Proportional Hazards Model. International Statistical Review / Revue Internationale de Statistique. 1978;43:45–57. [Google Scholar]
- 40.Ettinger DS, Akerley W, Bepler G, Blum MG, Chang A, Cheney RT, et al. Non-small cell lung cancer. J Natl Compr Canc Netw. 2010;8(7):740–801. doi: 10.6004/jnccn.2010.0056. [DOI] [PubMed] [Google Scholar]
- 41.Winton T, Livingston R, Johnson D, Rigas J, Johnston M, Butts C, et al. Vinorelbine plus cisplatin vs. observation in resected non-small-cell lung cancer. N Engl J Med. 2005;352(25):2589–2597. doi: 10.1056/NEJMoa043623. [DOI] [PubMed] [Google Scholar]
- 42.Pisters KM, Evans WK, Azzoli CG, Kris MG, Smith CA, Desch CE, et al. Cancer Care Ontario and American Society of Clinical Oncology adjuvant chemotherapy and adjuvant radiation therapy for stages I–IIIA resectable non small-cell lung cancer guideline. J Clin Oncol. 2007;25(34):5506–5518. doi: 10.1200/JCO.2007.14.1226. [DOI] [PubMed] [Google Scholar]
- 43.Pignon JP, Tribodet H, Scagliotti GV, Douillard JY, Shepherd FA, Stephens RJ, et al. Lung adjuvant cisplatin evaluation: a pooled analysis by the LACE Collaborative Group. J Clin Oncol. 2008;26(21):3552–3559. doi: 10.1200/JCO.2007.13.9030. [DOI] [PubMed] [Google Scholar]
- 44.Neri B, Bartalucci S, Gemelli MT, Tommasi M, Bacalli S. Creatine kinase isoenzyme BB: a lung cancer associated marker. Int J Biol Markers. 1988;3(1):19–22. doi: 10.1177/172460088800300104. [DOI] [PubMed] [Google Scholar]
- 45.Gazdar AF, Zweig MH, Carney DN, Van Steirteghen AC, Baylin SB, Minna JD. Levels of creatine kinase and its BB isoenzyme in lung cancer specimens and cultures. Cancer research. 1981;41(7):2773–2777. [PubMed] [Google Scholar]
- 46.Warburg O. On the origin of cancer cells. Science. 1956;123(3191):309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 47.Iwaoka WT, Krone CA, Sullivan JJ, Johnson CA. Effect of pH and ammonium ions on mutagenic activity in cooked beef. Cancer Lett. 1981;12(4):335–341. doi: 10.1016/0304-3835(81)90176-2. [DOI] [PubMed] [Google Scholar]
- 48.Schauer R. Sialic acids: fascinating sugars in higher animals and man. Zoology (Jena) 2004;107(1):49–64. doi: 10.1016/j.zool.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 49.Krolikowski FJ, Reuter K, Waalkes TP, Sieber SM, Adamson RH. Serum sialic acid levels in lung cancer patients. Pharmacology. 1976;14(1):47–51. doi: 10.1159/000136578. [DOI] [PubMed] [Google Scholar]
- 50.Ghosh PC, Lockwood E, Pennington GW. Abnormal excretion of corticosteroid sulphates in patients with breast cancer. Br Med J. 1973;1(5849):328–330. doi: 10.1136/bmj.1.5849.328. PMCID: 1588192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boonen E, Vervenne H, Meersseman P, Andrew R, Mortier L, Declercq PE, et al. Reduced cortisol metabolism during critical illness. N Engl J Med. 2013;368(16):1477–1488. doi: 10.1056/NEJMoa1214969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bornstein SR, Chrousos GP. Clinical review 104: Adrenocorticotropin (ACTH)- and non-ACTH-mediated regulation of the adrenal cortex: neural and immune inputs. J Clin Endocrinol Metab. 1999;84(5):1729–1736. doi: 10.1210/jcem.84.5.5631. [DOI] [PubMed] [Google Scholar]
- 53.Bornstein SR, Engeland WC, Ehrhart-Bornstein M, Herman JP. Dissociation of ACTH and glucocorticoids. Trends Endocrinol Metab. 2008;19(5):175–180. doi: 10.1016/j.tem.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 54.Heavner DL, Richardson JD, Morgan WT, Ogden MW. Validation and application of a method for the determination of nicotine and five major metabolites in smokers' urine by solid-phase extraction and liquid chromatography-tandem mass spectrometry. Biomed Chromatogr. 2005;19(4):312–328. doi: 10.1002/bmc.463. [DOI] [PubMed] [Google Scholar]
- 55.Mellert W, Kapp M, Strauss V, Wiemer J, Kamp H, Walk T, et al. Nutritional impact on the plasma metabolome of rats. Toxicol Lett. 2011;207(2):173–181. doi: 10.1016/j.toxlet.2011.08.013. [DOI] [PubMed] [Google Scholar]
- 56.Johnson CH, Patterson AD, Idle JR, Gonzalez FJ. Xenobiotic metabolomics: major impact on the metabolome. Annual review of pharmacology and toxicology. 2012;52:37–56. doi: 10.1146/annurev-pharmtox-010611-134748. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.