Abstract
Proteins containing the zinc finger domain(s) are named zinc finger proteins (ZFPs), one of the largest classes of transcription factors in eukaryotic genomes. A large number of ZFPs have been studied and many of them were found to be involved in regulating normal growth and development of cells and tissues through diverse signal transduction pathways. Recent studies revealed that a small but increasing number of ZFPs could function as key transcriptional regulators involved in adipogenesis. Due to the prevalence of obesity and metabolic disorders, the investigation of molecular regulatory mechanisms of adipocyte development must be more completely understood in order to develop novel and long-term impact strategies for ameliorating obesity. In this review, we discuss recent work that has documented that ZFPs are important functional contributors to the regulation of adipogenesis. Taken together, these data lead to the conclusion that ZFPs may become promising targets to combat human obesity.
Keywords: Zinc finger, Adipose tissue, Adipocyte, Obesity, Zfp423, Adiposity, Adipogenesis
Introduction
As obesity and adiposity-associated diseases continue to be a huge worldwide health problem, various approaches have been proposed, studied, and/or tried to ameliorate this ever-increasing public health issue [1, 2]. Since to date energy balance and exercise-oriented approaches as well as pharmacological treatments have not been overly successful, academic researchers, clinical scientists, public health officials, as well as patients have been interested in effective strategies to lower the rate of obesity. In this regard, a more complete understanding of adipocyte biology has been of increasing interest to all for over 50 years [1, 2]. In the process of seeking understanding and solutions to obesity at organ, cellular, and molecular levels, significant biological processes involved in adipogenesis have been revealed, which inferred the process of precursor cells (arising from pluripotent cells) to form committed cells of the fat cell lineage and these preadipocytes then transform into lipid-containing mature adipocytes [3]. Although important signal transduction pathways and key transcriptional factors involved in adipogenesis have been brought to light within the complicated gene regulatory networks, these efforts have not yet resulted in robust and safe anti-obesity drugs. In multiple cases, anti-obesity drugs demonstrated limited efficacy accompanied by serious long-term side effects [2, 4]. During past investigations, zinc finger motif proteins emerged as key regulatory transcription factors during adipogenesis. This large class of proteins containing zinc finger motif(s) is more commonly referred to as the zinc finger proteins (ZFPs) and represents the largest transcription factor family in mammals [5]. By DNA/RNA binding, protein–protein interactions, transcription activation, and regulation of apoptosis, ZFP transcription factors play a crucial role in regulating diverse growth and development processes [6]. An increasing number of ZFPs involved in adipogenesis have been discovered; hence, this review focuses on recent advances in the most prominent ZFP members that are related to the regulation of adipogenesis. Advances in understanding of molecular regulatory factors during adipocyte growth and development may provide crucial new insights into possible solutions in the fight against obesity.
Adipose tissue development
Adipose tissue
Adipose tissue/fat was originally known by its function as an energy transit station in mammals via storing triglycerides and releasing fatty acids. Upon more recent research on cellular and molecular processes, however, adipose tissue was also found to secrete numerous adipokines, notably adiponectin and leptin, and currently is being considered as a major endocrine organ [7]. By influencing a variety of biological and physiological processes including thermoregulation, energy regulation, insulin sensitivity, tissue cross-talk, inflammatory reactions, and cardiovascular responses, fatty tissue plays an irreplaceable role in maintaining whole-body homeostasis.
Brown adipose tissue (BAT) and white adipose tissue (WAT) are the two types of adipose tissue present in mammals. BAT-derived adipocytes contain a large number of mitochondria, in which abundant uncoupling proteins 1(UCP1) are uniquely present in the inner mitochondrial membrane. These UCPs dissipate the matrix to inner-membrane space proton gradient thus uncoupling electron transport from ATP synthesis, which results in dissipation of energy as heat [8, 9]. Different from BAT, which plays a key role in thermogenesis, WAT stores a large amount of lipids and is the source of numerous adipokines, which are either involved in maintenance of whole-body energy balance, inflammation, and numerous other aspects of fat metabolism [10, 11]. From a whole-body distribution perspective, BAT is generally abundant in newborns and decreases with age, whereas WAT exists widely, especially in the intra-abdominal and subcutaneous tissues [12, 13]. In humans, the excessive accumulation of WAT is a main contributing factor to obesity, which also leads to adverse health effects, such as insulin resistance, heart disease, and metabolic disorders [14].
Adipose development and molecular regulation
Adipose tissue is a dynamic organ (as exhibited by circular lipogenic and lipolytic processes) that is controlled by systemic hormones and local paracrine and autocrine factors. Remodeling of fat tissue in response to fluctuations in nutrient availability, exercise, and medical care is essential for maintaining metabolic homeostasis. In vivo, both adipocyte hyperplasia (number increase) and hypertrophy (volume increase) contribute to adipose tissue expansion and/or compensation for adipocyte apoptosis [15] as outcomes of a series of molecular events and lipid synthesis. These processes are traditionally referred to as adipogenesis, which are characterized by initial precursor cell determinations of mesenchyme-derived cells programmed to differentiate into adipocyte lineage, followed by subsequent proliferation and terminal differentiation of preadipocytes to mature adipocytes resulting in cell expansion and finally cytoplasmic lipid accumulation. New ideas on the plasticity of adipocytes have led to a more flexible definition of adipogenesis [3], but understanding these mechanisms may allow us to identify therapeutic targets to prevent obesity and other metabolic disorders.
Adipocytes are primarily derived from multipotent mesenchymal stem cells (MSCs), which also have the capacity to develop into osteoblasts, chondrocytes, and myoblasts through various differentiation pathways, respectively [13, 16]. Studies of mesenchymal stem cell differentiation processes have revealed an array of “master genes” that are essential for progenitor cells into unique differentiation directions [17]. Through intricate adipogenic signaling pathways and epigenetic events, multipotent MSCs are progressively determined and then committed to the adipocyte lineage. The next stage of the process of adipogenesis is the re-entry of growth-arrested preadipocytes into the cell cycle and the completion of several rounds of clonal expansion [18]. Under adipogenic signals, the expression of C/EBPβ (CCAAT/enhancer binding protein β) is rapidly induced, consequently initiating adipocyte differentiation [19]. The subsequent expression of PPARγ (peroxisome proliferator-activated receptor γ) and C/EBPα (CCAAT/enhancer binding protein α) ends mitotic clonal expansion and is followed by a second growth arrest and terminal differentiation and maturation, which is characterized by lipid accumulation and the expression of aP2 (FABP4), leptin, and several other markers [19–21].
In the past decades, significant progress has been achieved in revealing the characteristics and molecular regulations of adipocytes by using several cell lines (such as 3T3-L1) as well as primary cell cultures. The prominent signaling cascades in preadipocyte determination, such as BMP (bone morphogenic protein), Wnt, and Hedgehog signaling pathways, and the master genes during adipocyte differentiation, notably PPARγ and C/EBPs (CCAAT/enhancer binding proteins), have broadened our knowledge about growth and development of adipocytes. However, the regulation of the early stage of adipose development, adipogenic commitment, is still largely unknown, especially regarding the adipocyte origin/determination and the underlying molecular mechanisms. It was recently discovered that zinc finger protein 423 (Zfp423) regulates PPARγ expression, in part through amplification of the BMP signaling pathway, playing a critical role in regulating the adipogenic potential/determination of progenitor cells [22]. In addition, an increasing number of ZFPs have been recently reported to regulate fat-cell determination and differentiation. Progressive understanding of ZFPs broadens our knowledge about adipogenesis and may allow us to identify therapeutic treatments against obesity.
An overview of ZFPs
The discovery of ZFPs
The first recognized zinc finger protein, general transcription factor IIIA (TFIIIA), was reported in 1983 from Xenopus immature oocytes [23]. This protein-activated transcription of Xenopus 5 S RNA synthesis by binding to its DNA control region while intrinsic zinc ion of TFIIIA was unexpectedly found to be required [23]. Amino acid sequence analysis revealed that TFIIIA contained several similar tandem units, each consisting of about 30 residues and two pairs of cysteine (Cys) and histidine (His) [24]. Proposed linear arrangement of these repeated domains showed that each domain might fold and center on a zinc ion independently [24]. By binding two pairs of Cys and His residues, each zinc iron draws the ends of adjacent units together, making the central residues to form a potential DNA-binding loop like raised “finger” [24]. This structure was later confirmed by nuclear magnetic resonance (NMR) spectroscopy [25] and is considered as the canonical zinc finger motif, accompanied by the discovery of a large number of other zinc-binding motifs over the past decades.
Basic structures of zinc fingers
Zinc finger proteins contain one or more zinc finger motif(s), which play a major role in its regulatory functions. Structurally, canonical zinc finger is a short polypeptide with a special secondary structure stabilized by a zinc ion bound to two conserved Cys and His residues within the motif. Based on the number and order of the Cys and the His residues, ZFPs are sorted into C2H2, C2HC, C2C2, C2HCC2C2, and C2C2C2C2 types (C represents Cys; H represents His) [26–28]. Among these, C2H2 (also known as CCHH/TFIIIA/Krüppel-like fingers) is the first found classical zinc finger, and represents one of the largest and most important families of DNA-binding proteins of eukaryotic transcription factors [29–31]. C2H2 is also described as CX2–4CX12HX2–6H (X represents variable amino acid residues) and contains two to three β strands in its N-terminal sequence and one α helix in the C-terminal half of the X12H [5, 32, 33]. The first recognized zinc finger protein TFIIIA belongs to the C2H2 category.
The specific affinity of zinc fingers for different ligands is associated with amino acid sequences, spatial structures, as well as finger numbers and their interactions. For example, a greater finger number always results in wider binding activities in the C2H2 type of ZFPs [32]. So far, in addition to the traditional way of binding DNA, some ZFPs can also bind to dsRNA/ssRNA, DNA, and RNA hybrid, or protein [26, 34, 35], and thereby function as a master regulator of a set of genes or work cooperatively with other DNA-binding proteins [36]. Diverse structures of zinc fingers enable the ZFP family to play critical roles in many cellular functions.
Roles of ZFPs in adipogenesis and adipocyte function
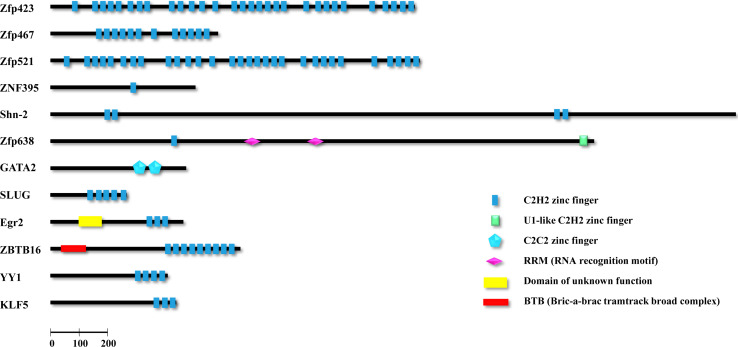
Zinc finger proteins are involved in different cellular responses, such as cell proliferation, growth, differentiation, metabolism, immunity, as well as the process of adipogenesis. Several members of the zinc-finger protein family, such as the GATA, KLF5, and Egr2 (Krox20), have been long known to play important roles in adipocyte development. As the increasing numbers of ZFP genes have currently been identified to be involved in adipose development, we believe that this prominent family of proteins and their proposed functions during adipocyte differentiation must be highlighted. In this review, we focus on recently discovered ZFPs occur in adipogenesis, which participate in adipocyte determination and differentiation, function as a master modulators or co-regulators via recruiting various co-activators and repressors (Table 1). In addition, schematic representations of these ZFPs are shown in Fig. 1. Since the ZFP superfamily has considerable members in mammals, some ZFP regulators like Sp1 and Sp3, which have been reported to participate in adipogenesis for a long time [37, 38], are not discussed here.
Table 1.
Direct roles of zinc finger proteins (ZFPs) in regulating white adipogenesis
| Main function stage | ZFP member | ZFP category | Cell type/model | Effect(s) on white adipogenesis in mammals | Reference |
|---|---|---|---|---|---|
| Determination | Zfp423 | C2H2 | Swiss 3T3; NIH 3T3; 3T3-L1; Zfp423−/−mice; Bovine IMF-SVs | Promotes adipocyte development by increasing PPARγ expression | [22, 43, 46, 47] |
| Zfp467 | C2H2 | Kusa 4b10; C57B1/6 mice; Mouse ADSCs | Promotes adipocyte development by increasing PPARγ expression; inhibits osteoblast development | [39, 48] | |
| Zfp521 | C2H2 | 3T3-L1; 3T3-F442A; C3H10T1/2; Primary MEFs; Zfp521+/− mice | Inhibits adipocyte development by negatively regulating Ebf1 and Zfp423; promotes bone development | [40] | |
| ZNF395 | C2H2 | hFIB; hBM-MSCs | Promotes adipocyte development by coordinating with PPARγ; may decrease osteoblast development | [41] | |
| Shn-2 | C2H2 | Shn-2−/− mice; Primary MEFs | Promotes adipogenesis via cooperating with Smad1/4 and C/EBPα to induce the expression of PPARγ | [42] | |
| Differentiation | Zfp638 | C2H2 | U2OS; 3T3-L1; C3H10T1/2 | Early positive regulator of preadipocyte differentiation by cooperating with C/EBPs and increasing PPARγ | [55] |
| GATA2 | C2C2 | 3T3-L1; 3T3-F442A; NIH3T3; COS-7; ob/ob mice; db/db mice; tub/tub mice; KKA Y yellow mice | Negatively control preadipocyte-to-adipocyte transition by suppressing PPARγ, C/EBPα, and C/EBPβ expression, and recruiting cofactors of FOGs, CTBPs, COUP-TFII, and SPI1 | [57, 59–62] | |
| GATA3 | C2C2 | 3T3-L1; 3T3-F442A; NIH3T3; COS-7; ob/ob mice; db/db mice; tub/tub mice; KKA Y yellow mice | Negatively control preadipocyte-to-adipocyte transition by suppressing PPARγ, C/EBPα, and C/EBPβ expression, and recruiting cofactors of FOGs and CTBPs | [57, 59, 60] | |
| SLUG | C2H2 | 3T3-L1; Primary MEFs; slug−/− mice; combi-Slug mice | Promotes preadipocyte differentiation | [64] | |
| Egr2 | C2H2 | 3T3-L1; NIH3T3 | Promotes preadipocyte differentiation partially through and in cooperation with C/EBPβ | [66, 67] | |
| Egr1 | C2H2 | 3T3-L1; db/db mice | Inhibits preadipocyte differentiation and decreases adipocyte insulin sensitivity (via PI3 K/Akt and Erk/MAPK signaling) | [67–69] | |
| ZBTB16 | C2H2 | 3T3-L1; Primary human SVs | Inhibits preadipocyte differentiation possibly by suppressing PPARγ and C/EBPα | [70] | |
| YY1 | C2H2 | 3T3-L1 | Promotes preadipocyte differentiation by suppressing CHOP-10 expression to release C/EBPβ | [74] | |
| KLF4 | C2H2 | 3T3-L1 | Promotes preadipocyte differentiation via activating C/EBPβ in cooperation with Egr2 | [89] | |
| KLF5 | C2H2 | 3T3-L1; NIH 3T3; MEFs; KLF5+/− mice | Promotes preadipocyte differentiation via activating the PPARγ in cooperation with C/EBPβ and C/EBPδ | [88] | |
| KLF6 | C2H2 | 3T3-L1; NIH 3T3 | Promotes preadipocyte differentiation via suppressing DLK1 and activating PPARγ, C/EBPα and C/EBPβ | [90] | |
| KLF8 | C2H2 | 3T3-L1; Primary mouse SVs | Promotes preadipocyte differentiation via activating the PPARγ and C/EBPα | [91] | |
| KLF9 | C2H2 | 3T3-L1; Primary rat SVs | Promotes adipocyte differentiation (middle stage) via activating the PPARγ in cooperation with C/EBPα | [86] | |
| KLF15 | C2H2 | 3T3-L1; NIH 3T3; C2C12; MEFs; KLF15+/− mice | Promotes adipocyte differentiation (middle stage) via activating the PPARγ in cooperation with C/EBPα; Positively regulates the expression of GLUT4 | [85, 87] | |
| KLF2 | C2H2 | 3T3-L1; MEFs; 3T3L1-KLF2 cell line; KLF2−/− mice | Negatively control preadipocyte-to-adipocyte transition by suppressing PPARγ and recovering DLK1 | [92, 93] | |
| KLF3 | C2H2 | 3T3-L1; MEFs; KLF3−/−mice | Inhibits adipogenesis via recruiting CTBP and suppressing C/EBPα promoter | [94] | |
| KLF7 | C2H2 | 3T3-L1; Human preadipocytes | Inhibits adipocyte differentiation via suppressing PPARγ, C/EBPα, aP2 and adipsin; contributes insulin resistance | [95] |
IMF-SVs intramuscular-derived stromal vascular cells, ADSCs adipose-derived stem cells, MEFs mouse embryonic fibroblasts, Ebf1 early B cell factor 1, hFIB human dermal fibroblasts, hBM-MSCs human bone-marrow derived mesenchymal stem cells, FOGs friend of GATA proteins, CTBPs C-terminal binding proteins, COUP-TFII COUP transcription factor II, SPI1 hematopoietic transcription factor PU.1, Combi-Slug slug overexpressing, SVs stromal vascular cells, CHOP-10 C/EBP homologous protein-10, DLK1 proto-oncogene delta-like 1 (also known as Pref-1), GLUT4 insulin-glucose transporter-4
Fig. 1.
Schematic representations of several ZFPs involved in white adipogenesis. GATA3 and Egr1, which have similar protein domains with GATA2 and Egr2, respectively, are not shown in this image. In addition, KLF5 is shown in this image representing KLF subfamily
Zinc finger proteins in white adipogenesis
Zinc finger proteins in adipogenic determination
Like other stem cell populations, adipogenic precursor cells require cooperation of multiple transcription factors to maintain their precursor state and/or regulate their differentiation directions [20]. Adipocytes derive from the same stem cell pools that also give rise to bone, cartilage, and muscle progenitors. Understanding the molecular switch between fat and other mesenchymal cell types appears to be of particular medical importance, with downstream implications for diseases like obesity and osteoporosis. Recent studies discovered that Zfp423, Zfp467, Zfp521, ZNF395, and Shn-2 members of the ZFP family have a pivotal role in adipocyte determination [22, 39–42].
Zfp423 promotes adipocyte commitment
Zinc finger protein 423 (Zfp423), which contains 30 Krüppel-like zinc fingers, was originally identified as a negative regulator of Ebf1 (early B cell factor 1), a basic transcription factor that participates in mesenchymal cell lineage determinations such as adipocyte and osteoblast differentiation [43–45]. It was recently discovered that Zfp423 directly participated in early adipose determination [22]. By comparing the differentially expressed genes in adipogenic and non-adipogenic fibroblast cell lines derived from 3T3 Swiss fibroblasts, Zfp423 as well as PPARγ, a dominant regulator of adipocyte differentiation, were found to be expressed abundantly in preadipose fibroblasts [22]. Under adipogenic signals, 3T3-L1 cell lines showed the greatest adipogenic potential as well as the highest mRNA and protein levels of Zfp423 [22]. In pro-differentiation cell culture conditions, ectopic expression of Zfp423 in non-adipogenic NIH 3T3 fibroblasts robustly activated expression of PPARγ and allowed cells to undergo adipogenic differentiation with accumulated lipids, while the knockdown of Zfp423 markedly reduced PPARγ expression and impaired the in vitro adipogenesis in 3T3-L1 preadipocytes [22]. Moreover, both brown and white adipocyte differentiations were significantly impaired in Zfp423-deficient mouse embryos [22]. Adipogenic potential was found to be related to Zfp423 status by Huang et al. [46] who selected several adipogenic clones from bovine stromal vascular (SV) cells possessing high and low adipogenic potential [46]. Increasing/decreasing Zfp423 in low/high adipogenic cells dramatically changed their adipogenic ability to a similar level of high/low adipogenic cells, respectively [46]. The lower adipogenic ability of adipogenic cells may result from the higher density of DNA methylation of the Zfp423 promoter [46].
Although Zfp423 expression increased significantly in preadipocytes compared with precursor cells, its expression did not change during the preadipocyte-to-adipocyte transition, suggesting Zfp423 function as a transcriptional regulator of preadipocyte commitment [47]. Interestingly, Zfp423 has a SMAD-binding domain that is required for bone morphogenic protein 4 (BMP4)-dependent adipogenesis. A Zfp423 mutant lacking this domain was still able to induce PPARγ and enhance adipocyte conversion in NIH 3T3 cell lines [22]. Zfp423 may promote adipogenesis in both a BMP-dependent and a BMP-independent manner. Future experiments will be required to uncover the whole pathways of how Zfp423 controls PPARγ expression, as well as the commitment of MSCs to adipogenic differentiation. In addition, regulating the DNA methylation of Zfp423 promoter to inhibit adipogenesis may be an alternative treatment to fight against obesity.
Zfp467 promotes adipocyte commitment and suppresses osteoblast differentiation
Zinc finger protein 467 (Zfp467) is another recently identified potential co-regulator of preadipocyte commitment. Overexpression of Zfp467 in Kusa 4b10 cells (mouse marrow stromal cells) significantly decreased the rate of mineralization and increased adipocyte formation as shown by elevated levels of adipogenic markers (such as PPARγ) and reduced mRNA levels of osteoblast markers [39]. Conversely, knockdown of Zfp467 decreased expression of these adipocyte genes and impaired adipogenesis [39]. Similarly, a recent study indicated that Zfp467 played an important role in adipocyte/osteoblast differentiation of adipose-derived stem cells (ADSCs) [48]. Knockdown of Zfp467 in ADSCs inhibited adipocyte formation and stimulated osteoblast commitment with decreased expression of adipogenic markers and enhanced expression of osteogenic markers [48]. In addition, utilizing luciferase-reporter promoter function assay Quach et al. [39] showed Zfp467 significantly enhanced transactivation function of the PPARγ/RxRα heterodimer (PPAR-response element) thereby indicating a possible mechanism by which Zfp467 promotes adipocyte formation. Thus, like Zfp423, Zfp467 may also affect preadipocyte commitment on a transcriptional level to promote adipocyte differentiation and suppress osteoblast differentiation. Studies on Zfp467 may allow therapeutic strategies involving osteoporosis and adipogenesis, as Zfp467 regulates both adipocyte and osteoblast differentiation and si-Zfp467-based treatments are currently available [48].
Zfp521 inhibits adipocyte commitment and promotes bone development
Zinc finger protein 521 (Zfp521), also known as Evi3, contains 30 C2H2 zinc finger repeats [49]. Zfp521 is present in various tissues and cell types, especially in immature cells like MSCs and hematopoietic stem cells while these cells participate in diverse biological processes [50, 51]. A recent study showed that Zfp521 served as a key regulator of adipose cell commitment and differentiation by directly binding to Ebf1, which induces Zfp423 expression and is necessary for the generation of adipocyte progenitors by enabling the expression of PPARγ and C/EBPα [40]. Contrariwise through direct physical interaction with Ebf1, wild-type Zfp521 blocked Ebf1 stimulation for Zfp423, leading to the suppressed expression of Zfp423 [40]. In addition, Ebf1 bound to an intronic enhancer of the Zfp521 gene and repressed its expression, providing a negative feedback loop to regulate Zfp521 function [40]. Overexpression of Zfp521 in cells greatly inhibited adipogenic potential, whereas knockdown or genetic ablation of Zfp521 enhanced fat cell development [40]. Furthermore, Zfp521−/− embryos exhibited increased mass of interscapular BAT and subcutaneous white adipocytes [40]. Since Zfp521 has been known to promote bone development [52], Zfp521 appears to act as a critical switch in the commitment decision between adipogenic and osteogenic lineages.
ZNF395 promotes adipogenesis via coordinating with PPARγ
Except for the recently discovered dominant zinc finger genes, which play a key role in fat cell determination, the ZNF395 (zinc finger protein 395), was found to be a novel modulator in human mesenchymal stem cells (hMSCs) via functional coordination with PPARγ [41]. In hMSCs, ZNF395 was significantly induced by adipogenic agents [41]. The ablation of ZNF395 during hMSCs adipogenesis reduced adipocyte numbers, whereas co-transfection of ZNF395, together with PPARγ, significantly induced adipocyte population and white adipocyte markers from both hMSCs and human dermal fibroblasts compared to PPARγ transfection alone [41]. In addition, overexpression of ZNF395 alone failed to induce adipogenesis [41]. These observations suggest human ZNF395 may be an important co-modulator of (trans-) differentiation towards adipocytes. In a study involving obese and non-obese children, ZNF395 expression was strongly expressed in adipocytes of obese children when compared to non-obese children, which also suggests a role for ZNF395 as a strong modulator of adipogenesis in vivo [41]. Further experiments are required with ZNF395 to elucidate the molecular regulatory pathways and mechanisms underlying the noted enhanced adipogenesis.
Shn-2 promotes adipogenesis via coordinating with Smad1/4 and C/EBPα to induce PPARγ expression
Shn-2 (Schnurri-2) is a large zinc finger protein that has been demonstrated to regulate lymphogenesis [53] and bone development [54]. In addition, Shn-2 was recently reported to regulate adipogenesis via interacting with Smad1/4 and C/EBPα on the PPARγ promoter [42]. In an experiment to assess the function of Shn-2 in adipogenesis, Shn-2 knockout mice and its embryonic fibroblasts (MEFs) were used [42]. Compared to wild-type mice, Shn-2 null mice exhibited reduced WAT and increased insulin sensitivity, while BAT remains unchanged [42]. Moreover, Shn-2 null MEFs failed to efficiently differentiate into adipocytes in vitro. However, ectopic expression of PPARγ could compensate for the loss of Shn-2 [42]. Shn-2 entered the nucleus upon BMP-2 stimulation and, in cooperation with Smad1/4 and C/EBPα, induced the expression of PPARγ [42]. These results indicate that Shn-2-mediated BMP signaling has a critical role in adipogenesis.
Zinc finger proteins in preadipocyte differentiation
Zfp638 is the early positive regulator of preadipocyte differentiation
Zinc finger protein 638 (Zfp638), also known as ZNF638, is a novel regulator of adipogenesis [55]. During the differentiation of 3T3-L1 cells, Zfp638 was induced shortly after exposure to the induction mixture at both the protein and mRNA levels. In addition, the expression of Zfp638 peaked before PPARγ and decreased rapidly during later stages of differentiation, indicating a potential role of Zfp638 in the early stages of adipogenesis [55]. Immunostaining in 3T3-L1 cells undergoing adipocyte conversion revealed the nuclear localization of Zfp638 and identified the likely location where Zpf638 exerts its transcriptional regulation functions [55]. Ectopically, expression of Zfp638 in C3H10T1/2 mesenchymal stem cells increased the number of lipid-accumulating cells as well as the increased expression of specific markers of fat differentiation, including aP2 and PPARγ; Zpf638 knockdown inhibited differentiation and decreased expression of adipocyte-specific genes [55]. Moreover, Meruvu et al. [55] also showed that Zfp638 physically interacted and transcriptionally cooperated with C/EBPs, a process that leads to the expression of PPARγ. Zfp638 is a novel and early regulator of adipogenesis that works as a transcription cofactor of C/EBPs.
GATA2 and GATA3 are negative regulators of the preadipocyte-to-adipocyte transition
GATA2 and GATA3 belong to the C2C2-type zinc finger protein subfamily consisting of a highly conserved zinc finger DNA binding domain that recognizes the consensus DNA sequence (A/T)GATA(A/G) located in regulatory regions of GATA target genes [56]. In mammals, both GATA2 and GATA3 were highly expressed in white preadipocytes (adipocyte precursors) and decreased upon undergoing terminal differentiation, suggesting their potential roles in the regulation of adipocyte differentiation [57]. In an experiment to revert to a adipocyte phenotype, forced expression of GATA2 in mature adipocytes complemented PPARγ depletion and impaired adipocyte functionality with a more preadipocyte-like gene expression profile [58]. GATA2 and GATA3 control the preadipocyte-to-adipocyte transition and function as negative regulators of adipocyte differentiation [57]. Forced expression of GATA2 and GATA3 in 3T3-F442A preadipocytes inhibited adipocyte differentiation as determined by decreased accumulation of intracellular lipids as well as adipocyte differentiation markers (including the PPARγ, GLUT4, aP2, and adipsin) [57]. In addition, the expression of DLK1 (Pref-1) and AEBP-1 (preadipocyte markers and adipocyte differentiation suppressors), was maintained at levels comparable to those observed in undifferentiated preadipocytes, indicating the GATA-expressing cells were trapped at the preadipocyte stage [57]. This effect was mediated partially through direct binding to the PPARγ, C/EBPα, and C/EBPβ promoters and subsequently suppressing their basal transcriptional activities [57, 59]. Contrariwise, down-regulation of GATA2 and GATA3 allowed onset of adipogenesis and was associated with adiposity [57].
A series of experiments demonstrated that various cofactors cooperate with GATA to prevent adipogenesis. GATA2 and GATA3 regulate adipogenesis through recruiting cofactors of the friend of GATA family (FOGs), which, in turn, recruit co-regulators, including C-terminal binding proteins (CTBPs) [60]. A GATA2 mutant that was unable to bind FOG, displayed abnormal activity and caused enhanced proliferation and almost complete absence of the adipogenic program, suggesting the important function of this mutated region in adipocyte differentiation, or alternatively, a failure to exit the cell cycle [60]. Similarly, GATA2 failed to fully inhibit adipogenesis in the absence of COUP-TFII (a transcriptional factor that prevents adipogenesis) and showed additivity in repressing key adipocyte genes such as C/EBPα and GLUT4 together with COUP-TFII, indicating that GATA actions in adipogenesis require COUP-TFII cooperation [61]. Furthermore, GATA2 and SPI1 (hematopoietic transcription factor PU.1) have an additive inhibitory effect on C/EBP transactivation and adipogenesis [62]. These results implicate FOGs, CTBPs, COUP-TFII, SPI1, and possibly other factors as partners of GATA proteins in the control of adipocyte proliferation and differentiation. GATA2 and GATA3 are important regulators controlling preadipocyte-to-adipocyte conversion.
SLUG promotes preadipocyte differentiation
SLUG, also known as SNAI2, is one of the C2H2-type zinc finger transcriptional factors. Studies have indicated that Slug regulates epithelial–mesenchymal transitions (EMTs) and plays a key role in various physiological and pathological processes [63]. A recent study showed that SLUG is expressed in WAT tissue and serves as an important modulator for adipocyte differentiation and adipose development [64]. In both 3T3-L1 and mouse embryonic fibroblasts (MEFs), Slug expression was very high before adipogenic inducement and decreased during the differentiation, while PPARγ was apparent within 1 day and increased dramatically thereafter [64]. These results indicate that Slug is tightly controlled during preadipocyte differentiation. Slug-deficient mice had reduced WAT mass, while Slug-overexpressing mice (Combi-Slug) exhibited an increase in WAT size and this increase in the WAT tissue was restored by suppression of the Slug transgene [64]. WAT alterations induced by Slug were reversible [64]. In addition, Slug-deficient MEFs dramatically reduced adipogenic capacity in vitro, whereas Combi-Slug MEFs exhibited extensive lipid accumulation [64]. The analysis of adipogenic gene expression both in vivo and in vitro showed that PPARγ expression was altered, although Slug failed to directly regulate PPARγ promoter activity [64]. As Slug is a key regulator of adipocyte differentiation both in vivo and in vitro, and the loss of tight control of Slug expression can induce obesity and/or lipodystrophy in mice, further studies on the Slug regulatory mechanisms may allow the development of targeted drugs for the treatment of patients with obesity and lipodystrophy.
Egr2 promotes adipocyte differentiation; Egr1 inhibits adipocyte differentiation
Egr2 (early growth response-2), also known as Krox20, belongs to the C2H2-type zinc finger protein family and has been involved in modulation of the cell cycle [65]. Recent studies indicated that Egr2 was abundantly expressed in adipose tissue in vivo and was transiently upregulated by serum stimulation in NIH3T3 fibroblasts [66]. However, the Egr2 expression was not readily detectable in mature adipocytes in vitro [66]. After exposing 3T3-L1 cells to the adipogenic induction cocktail, Egr2 was first induced after 15 min of exposure and peaked about 1 h post-induction [66]. Egr2 was quickly diminished after this and became undetectable at 6 h and thereafter [66]. The kinetics of Egr2 induction was similar to that of C/EBPβ and Egr2 was expressed earlier than C/EBPα and PPARγ, suggesting that it might play an early role in the process of adipogenesis [66]. Constitutive expression of Egr2 exhibited increased lipid accumulation as well as the expression of adipocyte markers in both 3T3-L1 cells and NIH3T3 cells, whereas knockdown of Egr2 significantly decreased lipid content and expression of adipocyte markers in 3T3-L1 cells [66]. Ectopic expression of Egr2 leaded to the induction of C/EBPβ expression and transactivation of C/EBPβ promoter [66]. However, Egr2 overexpression in C/EBPβ knockdown cell lines still exhibited enhanced adipogenesis compared with control cells [66]. These observations suggest that Egr2 exerts this adipogenic effect in C/EBPβ-dependent and -independent mechanisms. Furthermore, when Egr2 and C/EBPβ were co-expressed in NIH3T3 cells, cells exhibited greater adipogenesis than for expression of either gene alone further demonstrating that these two genes have a synergistic effect on adipogenesis [66]. Egr2 promotes expression of C/EBPβ and, in conjunction with C/EBPβ, facilitating terminal adipogenesis.
In addition to Egr2, Egr1 (Krox24) has also been identified as another early modulator of adipogenesis. Egr1 protein was rapidly induced after the addition of differentiation cocktail even before the expression of Egr2 [67]. In marked contrast to the effects of Egr2, differentiation was inhibited by ectopic expression of Egr1 and enhanced by knockdown of Egr1; these effects were particularly notable when IBMX (isobutylmethylxanthine) was omitted from the differentiation medium [67]. The pro-differentiation effects of IBMX involve suppression of the inhibitory influence of Egr1 [67]. However, Egr1 did not directly affect C/EBPβ protein expression as well as the activity of promoters of C/EBPα or PPARγ [67]. A recent study showed that Egr1 could tilt the signaling balance by blocking PI3 K/Akt signaling through PTEN and augmenting Erk/MAPK signaling through GGPPS (geranylgeranyl diphosphate synthase), resulting in insulin resistance in adipocytes [68, 69]. These data indicate that Egr1 and Egr2 exert opposing influences on adipocyte differentiation and that the precise regulation of both is required for maintaining the proper level of adipogenesis.
ZBTB16 inhibits adipocyte differentiation
By an integrated approach incorporating epigenomic profiling and motif enrichment analysis, ZBTB16 (zinc finger and BTB domain containing 16, also known as PLZF, Zfp145, and Zfp14), was selected as a candidate regulator of adipogenesis [70]. The expression of ZBTB16 was detected in both 3T3-L1 cells and human SV cells [70]. Gain- and loss-of-function assays found that overexpression of ZBTB16 in 3T3-L1 cells significantly repressed adipogenesis, as evidenced by reduced lipid accumulation and expression of adipogenic genes, such as PPARγ, C/EBPα, ADIPOQ, GLUT4, DGAT1, as well as FASN [70]. Conversely, knockdown of ZBTB16 enhanced 3T3-L1 adipogenesis [70]. ZBTB16 may function as a novel anti-adipogenic regulatory factor in vivo. However, further experiments are required to elucidate the underlying mechanisms.
YY1 promotes preadipocyte differentiation via suppressing CHOP-10 expression
YY1 (Yin Yang 1) is a ubiquitously expressed C2H2 zinc-finger transcription factor functioning as a transcriptional repressor or activator [71]. YY1 has been demonstrated to regulate normal cell proliferation and differentiation as well as cancer development via different signaling pathways [72]. Recently, a report has indicated that YY1, which could be induced by FBS or IGF-1 [73], contributed to the down-regulation of CHOP-10 (C/EBP homologous protein-10) in the early phase of the adipocyte differentiation program [74]. CHOP-10 inhibits adipocyte differentiation by sequestering C/EBPβ [75, 76]. YY1 bound to CHOP-10 promoter and suppressed its expression to stimulate adipocyte development [74]. Overexpression of YY1 decreased the transcription of CHOP-10, while knock-down of expression of YY1 increased CHOP-10 expression thereby inhibiting adipocyte differentiation [74]. YY1 may be a new adipocyte differentiation stimulator.
Members of KLFs enhance/inhibit adipocyte differentiation
KLFs (Krüppel-like factors) are a subfamily of zinc finger transcription factors that contains three highly conserved classical C2H2 zinc fingers [77]. The finger domain is located in carboxy-terminus and enables KLFs to specifically bind to GC-rich sequences and related GT or CACCC boxes in regulatory regions of target genes, while the non-DNA-binding region is highly variable and involved in gene activation/repression and interacts with other co-regulators [78–80]. Since the first mammalian KLF, named KLF1/EKLF (erythroid Krüppel-like factor), was first identified as a master regulator of erythropoiesis [81], the KLF subfamily has reportedly been involved in various cellular processes, including cell proliferation, differentiation, as well as apoptosis. Recently, a series of experiments demonstrated that many KLF members play vital roles in adipogenesis and adipose development (reviewed in [82, 83]), thus showing their potential therapeutic value in fighting obesity. A total of nine KLF members have been identified to be responsible for controlling white adipocyte development, of which KLF4, KLF5, KLF6, KLF8, KLF9, and KLF15 promote adipogenesis whereas KLF2, KLF3, and KLF7 inhibit adipogenesis.
KLF15 is the first KLF subfamily member that was reported to regulate adipocyte differentiation [84]. During the differentiation of 3T3-L1 preadipocytes, both KLF15 and KLF9 gene expression was dramatically increased in the middle stage of adipocyte differentiation [85, 86]. Gain- and loss-of-function approaches showed that overexpression of KLF15 in NIH 3T3 or C2C12 promoted adipogenesis while knockdown of KLF15/KLF9 blocked 3T3-L1 differentiation via directly suppressing the PPARγ expression [85–87]. Moreover, both KLF15 and KLF9 act synergistically with C/EBPα to increase the activity of the PPARγ gene promoter [85–87]. KLF4, KLF5, KLF6, and KLF8 expressions were induced at an early stage of differentiation [88–91]. Down-regulated expression of KLF5 inhibited adipocyte differentiation, whereas overexpression of KLF5 induced differentiation without hormonal stimulation [88]. In addition, neonatal KLF5+/− mice carried much less WAT mass than wild-type mice [88], suggesting that KLF5 plays a role in WAT development in vivo. KLF5 expression is induced by C/EBPβ and C/EBPδ, and KLF5, in turn, acts in concert with C/EBPβ and C/EBPδ to activate the PPARγ promoter [88]. KLF4 was expressed in 3T3-L1 cells within 30 min after exposure to a standard adipogenic cocktail [89]. Knockdown of KLF4 inhibited adipogenesis via directly decreasing C/EBPβ promoter activity [89]. Furthermore, KLF4 together with Egr2 cooperatively activated C/EBPβ expression, functioning as an early regulator of adipogenesis [89]. KLF6 was demonstrated as a positive regulator of adipogenesis since KLF6 repressed the expression of DLK1 [90], which is a master regulator of preadipocyte homeostasis and inhibits adipogenic differentiation. Forced expression of KLF6 strongly inhibited DLK1 expression in preadipocytes and NIH 3T3 cells, whereas down-regulation of KLF6 in 3T3-L1 cells prevented adipogenesis [90]. In addition, KLF6 could directly activate the expression of PPARγ, C/EBPα, and C/EBPβ, resulting in enhanced adipogenic differentiation [90]. KLF8 is a recently discovered adipogenesis-related member of KLFs. Expression knockdown of KLF8 decreased adipocyte differentiation, whereas overexpression of KLF8 resulted in enhanced adipogenesis [91]. Moreover, luciferase reporter assays indicated KLF8 was a new adipogenic regulator, which controlled terminal differentiation during adipogenesis via activating PPARγ and C/EBPα [91].
KLF2, KLF3, and KLF7 inhibit adipogenesis. KLF2 (LKLF) was expressed in preadipocytes and its role is diminished with the differentiation, functioning as a negative regulator of preadipocyte-to-adipocyte transition by directly inhibiting PPARγ promoter activity and recovering DLK1 function [92, 93]. Similarly, KLF3 (BKLF) was reported to inhibit adipocyte differentiation. KLF3 knockout mice had less WAT, with decreased size and number of adipose cells, while overexpression of KLF3 in 3T3-L1 inhibits adipocyte differentiation [94]. In vivo KLF3 recruits C-terminal binding protein (CtBP) co-repressors and suppresses C/EBPα promoter, thus negatively controlling adipogenesis and adipose development [94]. KLF7 (UKLF) has critical roles important to various tissue development processes. For example, KLF7 impaired insulin biosynthesis/secretion and adipocyte development (inhibit adipocyte differentiation and expression of PPARγ, C/EBPα, aP2 and adipsin), and suppressed hexokinase 2 gene expression in skeletal muscle, suggesting a new target for drug discovery in type 2 diabetes [95].
At this juncture, it needs to be mentioned that functional CACCC binding sites (one of the KLFs binding domains) were found in the control region of key adipogenic factors, such as C/EBPα and PPARγ [82], providing the structural basis for the adipogenic regulations of KLFs. Multiple KLFs are induced sequentially during adipocyte differentiation and they may work in concert to regulate adipocyte differentiation. In addition, interactions and reciprocal regulations may occur within the KLF family and among the KLFs and other adipogenic factors. For example, KLF2, KLF4, and KLF5, may activate KLF3, and KLF3 may then feedback to temper the activation [96]. Likewise, KLF4 activates C/EBPβ together with Egr2, while C/EBPβ reduces KLF4 expression via negative feedback [66, 89]. However, further studies are required to elucidate the relationship of these factors as well as the network of adipogenesis.
Summary of the roles of zinc finger protein members in white adipogenesis
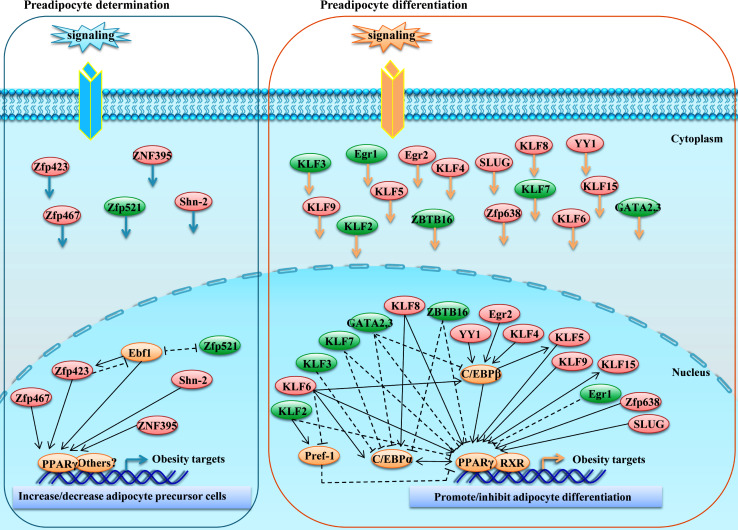
Adipogenesis occurs through expression of a complex and highly regulated transcriptional network. The development of fully differentiated mature adipocytes from mesenchymal precursor cells is an elegant progression involving the sequential activation of a battery of transcription factors. Under adipogenic signals, both the activation of pro-adipogenic regulators and the inhibition of anti-adipogenic regulators contribute to adipogenesis. Studies in the last decade have revealed an increasing number of ZFPs involved in inhibiting or promoting adipocyte development. As indicated in Fig. 2, ZFPs not only regulate fully differentiation of committed preadipocytes but also play a key role in fat cell determination, notably Zfp423, Zfp467, and Zfp521. Zinc finger proteins control adipogenesis by activating/inhibiting/recruiting PPARγ, C/EBPs, DLK1 (key modulators of adipogenesis) or other transcriptional factors. Studies of these novel modulators of fat cell determination and differentiation may provide an alternative strategy for treating obesity.
Fig. 2.
Roles of ZFPs in white adipogenesis. The differentiation of multipotent mesenchymal precursors to mature adipocytes occurs in two stages. The first step of adipogenesis is the embryonic stem cell-derived MSCs transition to committed white preadipocytes. Then, upon hormone cocktail stimulation, committed white preadipocytes can become mature white adipocytes. During adipogenesis, various members of the ZFP family positively/negatively contribute to transcriptional control of preadipocyte differentiation as well as mesenchymal precursor cell linage determination. The solid arrow indicates possible direct transcriptional activation while the dashed arrow represents possible transcriptional suppression (red oval Pro-adipogenic ZFP; green oval Anti-adipogenic ZFP; and orange oval other adipogenic regulators)
Zinc finger proteins in brown adipocyte development
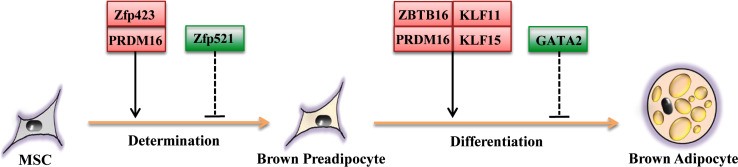
Among the above ZFP family members that participate in white adipogenesis, several proteins, such as Zfp423, Zfp521, GATA2, and KLF15 have been reported to be involved in brown adipose development [22, 40, 97, 98]. Moreover, another zinc finger transcriptional factor, ZBTB16, has also recently been suggested to play a role in brown adipocyte bioenergetics [99]. Conversely, zinc finger protein KLF11 and PRDM16 have been demonstrated to regulate brown adipogenesis without affecting white adipogenesis [97, 100–102]. The function of ZFPs in brown adipogenesis is shown in Fig. 3.
Fig. 3.
Roles of ZFPs in brown adipogenesis. Similar to white adipocyte development, the differentiation of multipotent mesenchymal precursors to brown adipocytes also occurs in two stages: brown preadipocyte determination and differentiation. Various members of the ZFP family positively/negatively contribute to transcriptional control of brown adipogenesis. The solid arrow indicates possible direct transcriptional activation while the dashed arrow represents possible transcriptional suppression (red square Pro-adipogenic ZFP; green square Anti-adipogenic ZFP)
PRDM16, Zfp423, and Zfp521 play roles in brown adipocyte determination
Like white adipocyte differentiation, brown adipocyte differentiation also requires regulation by PPARγ and C/EBPs [20, 103, 104]. For example, a zinc finger transcriptional regulator PRDM16 (PR domain containing 16) has recently been shown to have crucial roles in the control of brown fat determination and differentiation via interacting with C/EBPβ and PPARγ [102]. PRDM16 formed a complex with C/EBPβ to initiate the brown adipocyte cell formation, followed by the expression of adipogenic markers by recruiting PPARγ to a PRDM16 complex [100–102]. In addition, PRDM16 also forms a repressive complex with C terminal binding protein 1 (CTBP1) and CTBP2 to repress the expression of white adipocyte-specific genes [101]. Zfp423 was highly expressed in both WAT and BAT [22]. Knockdown of Zfp423 in brown adipocyte precursor cells showed dramatically decreased expression of PPARγ (a dominant and essential regulator of both white and brown adipocyte differentiation) as well as PRDM16 (a brown adipocyte determination factor) [22]. In ZFP423-deficient mouse embryos, both brown and white adipocyte differentiation was clearly impaired with a reduced mass of BAT and subcutaneous white adipocyte precursor cells [22]. The opposite results about the BAT development were found in Zfp521−/− embryos, which displayed significantly enlarged BAT depots as well as increased subcutaneous white adipocytes [40]. Both Zfp423 and Zfp521 are essential determination factors for brown adipocytes as well as white fat cells.
GATA2, KLF11, KLF15, and ZBTB16 participate in brown adipocyte differentiation/bioenergetics
Unlike WAT, which expressed both GATA2 and GATA3, BAT expressed only GATA2 and not GATA3 [98]. Overexpression of GATA2 strongly suppressed the expression of BAT-specific genes in brown adipocytes, such as UCP1, PGC1α (PPARγ-coactivator 1), and COX IV (cytochrome c oxidase IV), whereas disruption of a GATA2 allele in brown adipocytes resulted in significantly elevated differentiation as well as the expression of UCP1 and PGC1α [98]. GATA2 functions to suppress brown adipogenesis, and reduced expression of GATA2 in precursor cells potentiates brown adipocyte differentiation [98]. KLF15 and KLF11 belong to KLF subfamily and are highly expressed in BAT [97]. During brown adipocyte differentiation of muBM3.1 (mesenchymal stem cell line), KLF11 and KLF15 were dramatically induced [97]. Overexpression of KLF11 clearly enhanced the UCP1 expression level, whereas overexpression of KLF15 did not affect UCP1 but displayed an additivity effect on UCP1 expression when co-transfected with KLF11 [97]. This study also indicated KLF11 and KLF15 function to regulate brown adipogenesis via directly interacting with UCP1 promoter by GC- and GT-boxes, respectively [97]. Conversely, UCP1 expression was completely suppressed by KLF11 siRNA, whereas KLF15 knockdown partially suppressed UCP1 expression [97]. These results indicate that KLF11 is essential for brown adipogenesis and cooperation of KLF11 and KLF15 enhances brown adipocyte differentiation. A recent study identified that ZBTB16 is induced in both the BAT and skeletal muscle during acute adaptive thermogenesis [99]. ZBTB16 overexpression in brown adipocytes enhanced the thermogenic program, including genes involved in fatty acid oxidation, glycolysis, and mitochondrial function [99]. Mitochondrial biogenesis, as well as respiratory capacity and uncoupling, were also increased, accompanied by decreased triglyceride content and increased carbohydrate utilization in brown adipocytes [99]. In addition, cells overexpressing ZBTB16 upregulates brown fat-enriched markers such as UCP1, PGC1α, PPARα, and PRDM16 [99]. Notably, ZBTB16 expression is correlated with body weight, fat mass, and diabetes in vivo [99]. ZBTB16 may be a novel determinator of substrate utilization in brown adipocytes and adiposity in vivo. Since brown fat can increase energy expenditure in the form of heat through a specialized program of uncoupled respiration and thus physiological protect against cold and obesity [100, 102], studies in these areas may uncover new treatments for obesity and other metabolic disorders.
Clinical significance of fighting obesity
Despite the impressive increase in the understanding of various aspects of lipid metabolism, definitive treatments to combat obesity and its co-morbidities have not emerged, or produce adverse side effects [2]. Successfully targeting the adipocyte to prevent adipose accumulation requires completely understanding of adipose tissue development and expansion [1, 2, 4]. In mammals, a set of complex transcriptional networks are responsible for the changes in cell morphology and gene expression associated with adipocytes as well as adiposity. In the complex regulation of adipogenesis, ZFPs play an essential role, although their new putative functions and relationships in the regulatory networks still need to be described in detail in both murine and human cells. Zfp423, Zfp467, ZNF395, and Shn-2 positively regulate white preadipocyte determination, whereas Zfp521 inhibits preadipocyte commitment. ZFP638, SLUG, Egr2, YY1, KLF4, KLF5, KLF6, KLF8, KLF9, and KLF15 promote white preadipocyte differentiation mainly through activating PPARγ and C/EBPs. Conversely, GATA2, GATA3, Egr1, ZBTB16, KLF2, KLF3, and KLF7 are negative regulators during white adipocyte differentiation via suppressing PPARγ and C/EBPs or restoring DLK1. Among the above white adipogenesis regulators, Zfp423, Zfp521, KLF15, GATA2, and ZBTB16 also play a similar role in brown adipogenesis promotion/inhibition. In addition, PRDM16 and KLF11 positively regulate brown fat determination and differentiation respectively, without affecting white adipocyte development. ZFPs function as important contributors to intricate regulation network of adipogenesis, and may become promising targets to combat human obesity.
A better understanding of the various cascades of events in which ZFPs participate together within its superfamily or with other transcription factors and finally epigenetic factors to regulate adipose development will allow us to continue with relevant experiments on how commitment of cells is regulated to become depot fat or an obesity epidemic. Additionally, possible pharmaceutical compounds might be developed to specifically activate/suppress ZFP genes to regulate master regulators (such as PPARγ) of adipocyte differentiation. Alternatively, decreasing cells fated to be adipocytes may provide a novel way to inhibit excessive fat accumulation, as several ZFPs function to control the critical switch in the commitment decision of the adipogenic lineage. Understanding the fundamental mechanisms that regulate adipose progenitor cell proliferation/recruitment should provide additional targets that may be useful in prevention, rather than treatment, of obesity. Another possible way for the development of possible pharmacological approaches against obesity might involve the brown adipogenesis, as some ZFPs (such as ZBTB16) can increase energy expenditure of brown fat to physiological protect against obesity.
Potential therapeutic strategies for treating obesity through ZFPs might include several approaches. The first point is through the change of the spatial configuration of intrinsic ZFPs to alter the ZFP specificity and/or binding affinity as well as biological functions. Fundamentally, zinc finger motif(s), which play a major role in ZFP biological functions, are strongly stabilized by zinc ions. Moreover, amino acid residues, finger numbers, and finger–finger interactions contribute to the ZFP structure and function [32]. The disruption of such a stable structure of ZFPs represents a new way to regulate critical ZFPs involved in diseases. A case in point is the azodicarbonamide (ADA), an anti-HIV (human immunodeficiency virus) zinc finger inhibitor, which specifically targets the nucleocapsid protein 7 (NCp7, a zinc finger protein that is required for HIV replication) by attacking its cysteine residues and cause a covalent conformation change, which results in an ejection of the zinc from the zinc finger domain [105, 106]. Secondly, gene editing/modification of targeted zinc finger genes by engineered ZFPs (artificial ZFPs that contain specific binding domain and effector domain could effectively be used in gene modification) as well as other methods provide the possibility to regulate adipogenesis. For example, regulating the methylation level of ZFPs may be useful as methylation status of several ZFPs (such as Zfp423) is strongly correlated with adipogenic ability [46]. Additionally, overexpression/suppression/inactivation of targeted ZFPs through gene therapy is possible to treat adiposity, as gene therapy possesses highly targeting property and currently has been available in LPLD (lipoprotein lipase deficiency) disease treatment [107]. Developing novel inhibitors/activators/regulators targeting specific pro/anti-adipogenic genes/proteins have great potential to combat adiposity. Among the possible targets, ZFPs are promising for the development of therapeutic strategies to fight obesity.
Conclusions
Obesity is a growing world epidemic and is reported to be involved in many human diseases. Fat accumulation in vivo is mediated by the cascade of events regulated by a large number of transcription factors that are involved in the processes controlling fat cell determination and differentiation of preadipocytes into mature fat cells, resulting in increased numbers as well as expanded volumes of adipocytes. Extensive and detailed research in fat cell biology has been conducted for a long time; these studies were accompanied with considerable progress of transcription factors in fat cell regulation networks. To date, we all know that both adipocyte hypertrophy and hyperplasia contribute to overweight and PPARγ and C/EBPs are the main regulators of preadipocyte differentiation. Such extensive research notwithstanding, there are still, however, no efficient drugs available for patients suffering from adiposity. Development of successful therapeutic anti-obesity drugs is difficult and fat cell regulatory networks are much more complicated than we initially thought. Zinc finger proteins are a class of regulatory proteins that participate in a variety of cellular activities, such as development, differentiation, and tumor suppression, as well as adipogenesis, as discussed. Recently emerging ZFPs during adipogenesis show an important role in regulating this process, which may provide opportunities to further understand adipocyte development, especially in fat fate determination. Studies on ZFPs would open the door to study fat cell origin and determination and may set up another milestone in understanding fat biology. Summarizing and highlighting ZFPs functions in regulating adipogenesis can well serve as a theoretical basis for developing novel and efficient anti-obesity drugs. In summary, here we reviewed the recent studies relating principal ZFPs actions on adipose development, especially the principal ZFP regulators in white adipogenesis (determination/differentiation). ZFPs regulatory networks with other adipogenic factors (such as PPARγ and C/EBPs) highlight the ZFPs as potential target of obesity and its associated diseases treatment strategies.
Acknowledgments
The research was supported by the China National ‘863’ Program (#2011AA100307-02, #2013AA102505), the China National Natural Science Foundation (#31272411, #31000997), the China National Twelfth ‘5 Year’ Science and Technology Support Project (#2011BAD28B04-03), the China GMO New Varieties Major Project (#2011ZX08007-002), the China National Beef and Yak Industrial Technology System (CARS-38), the Program for Changjiang Scholars and Innovative Research Team of China (IRT0940), the Science and Technology Coordination and Innovation Project of Shaanxi Province (2011KTCL02-07), as well as the National Institutes of Health, USA (R01HD067449).
Conflict of interest
The authors have declared that no conflicts of interest exist.
Footnotes
S. Wei and L. Zhang contributed equally to this work.
Contributor Information
Linsen Zan, Phone: +86-29-87091923, FAX: +86-29-87091148, Email: zanlinsen@163.com.
Michael V. Dodson, Phone: +1-509-3359644, FAX: +1-509-3351082, Email: dodson@wsu.edu
References
- 1.Dodson MV, Mir PS, Hausman GJ, Guan LL, Du M, Jiang Z, Fernyhough ME, Bergen WG. Obesity, metabolic syndrome, and adipocytes. J Lipids. 2011;2011:721686. doi: 10.1155/2011/721686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dodson MV, Boudina S, Albrecht E, Bucci L, Fernyhough-Culver M, Wei S, Bergen WG, Amaral AJ, Moustaid-Moussa N, Poulos S, Hausman GJ. A long journey to effective obesity treatments: is there light at the end of the tunnel? Exp Biol Med. 2013 doi: 10.1177/1535370213477603. [DOI] [PubMed] [Google Scholar]
- 3.Fernyhough ME, Helterline DI, Vierck JL, Hausman GJ, Hill RA, Dodson MV. Dedifferentiation of mature adipocytes to form adipofibroblasts: more than just a possibility. Adipocytes. 2005;1:17–24. [Google Scholar]
- 4.Fernyhough ME, Bucci LR, Hausman GJ, Antonio J, Vierck JL, Dodson MV. Gaining a solid grip on adipogenesis. Tissue Cell. 2005;37:335–338. doi: 10.1016/j.tice.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 5.Ganss B, Jheon A. Zinc finger transcription factors in skeletal development. Crit Rev Oral Biol Med. 2004;15:282–297. doi: 10.1177/154411130401500504. [DOI] [PubMed] [Google Scholar]
- 6.Leon O, Roth M. Zinc fingers: DNA binding and protein–protein interactions. Biol Res. 2000;33:21–30. doi: 10.4067/s0716-97602000000100009. [DOI] [PubMed] [Google Scholar]
- 7.Havel PJ. Update on adipocyte hormones: regulation of energy balance and carbohydrate/lipid metabolism. Diabetes. 2004;53(Suppl 1):S143–S151. doi: 10.2337/diabetes.53.2007.s143. [DOI] [PubMed] [Google Scholar]
- 8.Rousset S, Alves-Guerra MC, Mozo J, Miroux B, Cassard-Doulcier AM, Bouillaud F, Ricquier D. The biology of mitochondrial uncoupling proteins. Diabetes. 2004;53(Suppl 1):S130–S135. doi: 10.2337/diabetes.53.2007.s130. [DOI] [PubMed] [Google Scholar]
- 9.Townsend K, Tseng Y. Brown adipose tissue: recent insights into development, metabolic function and therapeutic potential. Adipocyte. 2012;1:13–24. doi: 10.4161/adip.18951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ashrafi K (2007) Obesity and the regulation of fat metabolism. In: WormBook (ed) The C. elegans Research Community, Wormbook, United Kingdom [DOI] [PMC free article] [PubMed]
- 11.Ntaios G, Gatselis NK, Makaritsis K, Dalekos GN. Adipokines as mediators of endothelial function and atherosclerosis. Atherosclerosis. 2013;227:216–221. doi: 10.1016/j.atherosclerosis.2012.12.029. [DOI] [PubMed] [Google Scholar]
- 12.Billon N, Monteiro MC, Dani C. Developmental origin of adipocytes: new insights into a pending question. Biol Cell. 2008;100:563–575. doi: 10.1042/BC20080011. [DOI] [PubMed] [Google Scholar]
- 13.Wei S, Zan L, Hausman GJ, Rasmussen TP, Bergen WG, Dodson MV (2013) Dedifferentiated adipocyte-derived progeny cells (DFAT cells): potential stem cells of adipose tissue. Adipocyte. 10.4161/adip.23784 [DOI] [PMC free article] [PubMed]
- 14.German AJ, Ryan VH, German AC, Wood IS, Trayhurn P. Obesity, its associated disorders and the role of inflammatory adipokines in companion animals. Vet J. 2010;185:4–9. doi: 10.1016/j.tvjl.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 15.Jo J, Gavrilova O, Pack S, Jou W, Mullen S, Sumner AE, Cushman SW, Periwal V. Hypertrophy and/or hyperplasia: dynamics of adipose tissue growth. PLoS Comput Biol. 2009;5:e1000324. doi: 10.1371/journal.pcbi.1000324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chamberlain G, Fox J, Ashton B, Middleton J. Concise review: mesenchymal stem cells: their phenotype, differentiation capacity, immunological features, and potential for homing. Stem Cells. 2007;25:2739–2749. doi: 10.1634/stemcells.2007-0197. [DOI] [PubMed] [Google Scholar]
- 17.Kolf CM, Cho E, Tuan RS. Mesenchymal stromal cells. Biology of adult mesenchymal stem cells: regulation of niche, self-renewal and differentiation. Arthr Res Ther. 2007;9:204. doi: 10.1186/ar2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Camp HS, Ren D, Leff T. Adipogenesis and fat-cell function in obesity and diabetes. Trends Mol Med. 2002;8:442–447. doi: 10.1016/s1471-4914(02)02396-1. [DOI] [PubMed] [Google Scholar]
- 19.Zamani N, Brown CW. Emerging roles for the transforming growth factor-{beta} superfamily in regulating adiposity and energy expenditure. Endocr Rev. 2011;32:387–403. doi: 10.1210/er.2010-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cristancho AG, Lazar MA. Forming functional fat: a growing understanding of adipocyte differentiation. Nat Rev Mol Cell Biol. 2011;12:722–734. doi: 10.1038/nrm3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gregoire FM, Smas CM, Sul HS. Understanding adipocyte differentiation. Physiol Rev. 1998;78:783–809. doi: 10.1152/physrev.1998.78.3.783. [DOI] [PubMed] [Google Scholar]
- 22.Gupta RK, Arany Z, Seale P, Mepani RJ, Ye L, Conroe HM, Roby YA, Kulaga H, Reed RR, Spiegelman BM. Transcriptional control of preadipocyte determination by Zfp423. Nature. 2010;464:619–623. doi: 10.1038/nature08816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanas JS, Hazuda DJ, Bogenhagen DF, Wu FY, Wu CW. Xenopus transcription factor A requires zinc for binding to the 5 S RNA gene. J Biol Chem. 1983;258:14120–14125. [PubMed] [Google Scholar]
- 24.Miller J, McLachlan AD, Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985;4:1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee MS, Gippert GP, Soman KV, Case DA, Wright PE. Three-dimensional solution structure of a single zinc finger DNA-binding domain. Science. 1989;245:635–637. doi: 10.1126/science.2503871. [DOI] [PubMed] [Google Scholar]
- 26.Mackay JP, Crossley M. Zinc fingers are sticking together. Trends Biochem Sci. 1998;23:1–4. doi: 10.1016/s0968-0004(97)01168-7. [DOI] [PubMed] [Google Scholar]
- 27.Sanchez-Garcia I, Rabbitts TH. The LIM domain: a new structural motif found in zinc-finger-like proteins. Trends Genet. 1994;10:315–320. doi: 10.1016/0168-9525(94)90034-5. [DOI] [PubMed] [Google Scholar]
- 28.Klug A, Schwabe JW. Protein motifs 5 Zinc fingers. FASEB J. 1995;9:597–604. [PubMed] [Google Scholar]
- 29.Tupler R, Perini G, Green MR. Expressing the human genome. Nature. 2001;409:832–833. doi: 10.1038/35057011. [DOI] [PubMed] [Google Scholar]
- 30.Laity JH, Lee BM, Wright PE. Zinc finger proteins: new insights into structural and functional diversity. Curr Opin Struct Biol. 2001;11:39–46. doi: 10.1016/s0959-440x(00)00167-6. [DOI] [PubMed] [Google Scholar]
- 31.Ciftci-Yilmaz S, Mittler R. The zinc finger network of plants. Cell Mol Life Sci. 2008;65:1150–1160. doi: 10.1007/s00018-007-7473-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iuchi S. Three classes of C2H2 zinc finger proteins. Cell Mol Life Sci. 2001;58:625–635. doi: 10.1007/PL00000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pavletich NP, Pabo CO. Crystal structure of a five-finger GLI-DNA complex: new perspectives on zinc fingers. Science. 1993;261:1701–1707. doi: 10.1126/science.8378770. [DOI] [PubMed] [Google Scholar]
- 34.Shi Y, Berg JM. Specific DNA-RNA hybrid binding by zinc finger proteins. Science. 1995;268:282–284. doi: 10.1126/science.7536342. [DOI] [PubMed] [Google Scholar]
- 35.Brown RS. Zinc finger proteins: getting a grip on RNA. Curr Opin Struct Biol. 2005;15:94–98. doi: 10.1016/j.sbi.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 36.Wolfe SA, Nekludova L, Pabo CO. DNA recognition by Cys2His2 zinc finger proteins. Annu Rev Biophys Biomol Struct. 2000;29:183–212. doi: 10.1146/annurev.biophys.29.1.183. [DOI] [PubMed] [Google Scholar]
- 37.Tang QQ, Jiang MS, Lane MD. Repressive effect of Sp1 on the C/EBPalpha gene promoter: role in adipocyte differentiation. Mol Cell Biol. 1999;19:4855–4865. doi: 10.1128/mcb.19.7.4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang MS, Lane MD. Sequential repression and activation of the CCAAT enhancer-binding protein-alpha (C/EBPalpha) gene during adipogenesis. Proc Natl Acad Sci USA. 2000;97:12519–12523. doi: 10.1073/pnas.220426097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quach JM, Walker EC, Allan E, Solano M, Yokoyama A, Kato S, Sims NA, Gillespie MT, Martin TJ. Zinc finger protein 467 is a novel regulator of osteoblast and adipocyte commitment. J Biol Chem. 2011;286:4186–4198. doi: 10.1074/jbc.M110.178251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kang S, Akerblad P, Kiviranta R, Gupta RK, Kajimura S, Griffin MJ, Min J, Baron R, Rosen ED. Regulation of early adipose commitment by Zfp521. PLoS Biol. 2012;10:e1001433. doi: 10.1371/journal.pbio.1001433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hasegawa R, Tomaru Y, de Hoon M, Suzuki H, Hayashizaki Y, Shin JW. Identification of ZNF395 as a novel modulator of adipogenesis. Exp Cell Res. 2013;319:68–76. doi: 10.1016/j.yexcr.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 42.Jin W, Takagi T, Kanesashi SN, Kurahashi T, Nomura T, Harada J, Ishii S. Schnurri-2 controls BMP-dependent adipogenesis via interaction with Smad proteins. Dev Cell. 2006;10:461–471. doi: 10.1016/j.devcel.2006.02.016. [DOI] [PubMed] [Google Scholar]
- 43.Schupp M, Lazar MA. Fingered for a fat fate. Cell Metab. 2010;11:244–245. doi: 10.1016/j.cmet.2010.02.014. [DOI] [PubMed] [Google Scholar]
- 44.Akerblad P, Lind U, Liberg D, Bamberg K, Sigvardsson M. Early B-cell factor (O/E-1) is a promoter of adipogenesis and involved in control of genes important for terminal adipocyte differentiation. Mol Cell Biol. 2002;22:8015–8025. doi: 10.1128/MCB.22.22.8015-8025.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hesslein DG, Fretz JA, Xi Y, Nelson T, Zhou S, Lorenzo JA, Schatz DG, Horowitz MC. Ebf1-dependent control of the osteoblast and adipocyte lineages. Bone. 2009;44:537–546. doi: 10.1016/j.bone.2008.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang Y, Das AK, Yang QY, Zhu MJ, Du M. Zfp423 promotes adipogenic differentiation of bovine stromal vascular cells. PLoS One. 2012;7:e47496. doi: 10.1371/journal.pone.0047496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Janesick A, Blumberg B. Endocrine disrupting chemicals and the developmental programming of adipogenesis and obesity. Birth Defects Res C Embryo Today. 2011;93:34–50. doi: 10.1002/bdrc.20197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.You L, Pan L, Chen L, Chen JY, Zhang X, Lv Z, Fu D. Suppression of zinc finger protein 467 alleviates osteoporosis through promoting differentiation of adipose derived stem cells to osteoblasts. J Transl Med. 2012;10:11. doi: 10.1186/1479-5876-10-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Warming S, Liu P, Suzuki T, Akagi K, Lindtner S, Pavlakis GN, Jenkins NA, Copeland NG. Evi3, a common retroviral integration site in murine B-cell lymphoma, encodes an EBFAZ-related Krüppel-like zinc finger protein. Blood. 2003;101:1934–1940. doi: 10.1182/blood-2002-08-2652. [DOI] [PubMed] [Google Scholar]
- 50.Wu M, Hesse E, Morvan F, Zhang JP, Correa D, Rowe GC, Kiviranta R, Neff L, Philbrick WM, Horne WC, Baron R. Zfp521 antagonizes Runx2, delays osteoblast differentiation in vitro, and promotes bone formation in vivo. Bone. 2009;44:528–536. doi: 10.1016/j.bone.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shen S, Pu J, Lang B, McCaig CD. A zinc finger protein Zfp521 directs neural differentiation and beyond. Stem Cell Res Ther. 2011;2:20. doi: 10.1186/scrt61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stephens JM. The fat controller: adipocyte development. PLoS Biol. 2012;10:e1001436. doi: 10.1371/journal.pbio.1001436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takagi T, Harada J, Ishii S. Murine Schnurri-2 is required for positive selection of thymocytes. Nat Immunol. 2001;2:1048–1053. doi: 10.1038/ni728. [DOI] [PubMed] [Google Scholar]
- 54.Saita Y, Takagi T, Kitahara K, Usui M, Miyazono K, Ezura Y, Nakashima K, Kurosawa H, Ishii S, Noda M. Lack of Schnurri-2 expression associates with reduced bone remodeling and osteopenia. J Biol Chem. 2007;282:12907–12915. doi: 10.1074/jbc.M611203200. [DOI] [PubMed] [Google Scholar]
- 55.Meruvu S, Hugendubler L, Mueller E. Regulation of adipocyte differentiation by the zinc finger protein ZNF638. J Biol Chem. 2011;286:26516–26523. doi: 10.1074/jbc.M110.212506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Patient RK, McGhee JD. The GATA family (vertebrates and invertebrates) Curr Opin Genet Dev. 2002;12:416–422. doi: 10.1016/s0959-437x(02)00319-2. [DOI] [PubMed] [Google Scholar]
- 57.Tong Q, Dalgin G, Xu H, Ting CN, Leiden JM, Hotamisligil GS. Function of GATA transcription factors in preadipocyte-adipocyte transition. Science. 2000;290:134–138. doi: 10.1126/science.290.5489.134. [DOI] [PubMed] [Google Scholar]
- 58.Schupp M, Cristancho AG, Lefterova MI, Hanniman EA, Briggs ER, Steger DJ, Qatanani M, Curtin JC, Schug J, Ochsner SA, McKenna NJ, Lazar MA. Re-expression of GATA2 cooperates with peroxisome proliferator-activated receptor-gamma depletion to revert the adipocyte phenotype. J Biol Chem. 2009;284:9458–9464. doi: 10.1074/jbc.M809498200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tong Q, Tsai J, Tan G, Dalgin G, Hotamisligil GS. Interaction between GATA and the C/EBP family of transcription factors is critical in GATA-mediated suppression of adipocyte differentiation. Mol Cell Biol. 2005;25:706–715. doi: 10.1128/MCB.25.2.706-715.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jack BH, Crossley M. GATA proteins work together with friend of GATA (FOG) and C-terminal binding protein (CTBP) co-regulators to control adipogenesis. J Biol Chem. 2010;285:32405–32414. doi: 10.1074/jbc.M110.141317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu Z, Yu S, Hsu CH, Eguchi J, Rosen ED. The orphan nuclear receptor chicken ovalbumin upstream promoter-transcription factor II is a critical regulator of adipogenesis. Proc Natl Acad Sci USA. 2008;105:2421–2426. doi: 10.1073/pnas.0707082105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang F, Tong Q. Transcription factor PU.1 is expressed in white adipose and inhibits adipocyte differentiation. Am J Physiol Cell Physiol. 2008;295:C213–C220. doi: 10.1152/ajpcell.00422.2007. [DOI] [PubMed] [Google Scholar]
- 63.Mathsyaraja H, Ostrowski MC. Setting Snail2′s pace during EMT. Nat Cell Biol. 2012;14:1122–1123. doi: 10.1038/ncb2616. [DOI] [PubMed] [Google Scholar]
- 64.Perez-Mancera PA, Bermejo-Rodriguez C, Gonzalez-Herrero I, Herranz M, Flores T, Jimenez R, Sanchez-Garcia I. Adipose tissue mass is modulated by SLUG (SNAI2) Hum Mol Genet. 2007;16:2972–2986. doi: 10.1093/hmg/ddm278. [DOI] [PubMed] [Google Scholar]
- 65.Chavrier P, Zerial M, Lemaire P, Almendral J, Bravo R, Charnay P. A gene encoding a protein with zinc fingers is activated during G0/G1 transition in cultured cells. EMBO J. 1988;7:29–35. doi: 10.1002/j.1460-2075.1988.tb02780.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen Z, Torrens JI, Anand A, Spiegelman BM, Friedman JM. Krox20 stimulates adipogenesis via C/EBPbeta-dependent and -independent mechanisms. Cell Metab. 2005;1:93–106. doi: 10.1016/j.cmet.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 67.Boyle KB, Hadaschik D, Virtue S, Cawthorn WP, Ridley SH, O’Rahilly S, Siddle K. The transcription factors Egr1 and Egr2 have opposing influences on adipocyte differentiation. Cell Death Differ. 2009;16:782–789. doi: 10.1038/cdd.2009.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yu X, Shen N, Zhang ML, Pan FY, Wang C, Jia WP, Liu C, Gao Q, Gao X, Xue B, Li CJ. Egr-1 decreases adipocyte insulin sensitivity by tilting PI3 K/Akt and MAPK signal balance in mice. EMBO J. 2011;30:3754–3765. doi: 10.1038/emboj.2011.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shen N, Yu X, Pan FY, Gao X, Xue B, Li CJ. An early response transcription factor, Egr-1, enhances insulin resistance in type 2 diabetes with chronic hyperinsulinism. J Biol Chem. 2011;286:14508–14515. doi: 10.1074/jbc.M110.190165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mikkelsen TS, Xu Z, Zhang X, Wang L, Gimble JM, Lander ES, Rosen ED. Comparative epigenomic analysis of murine and human adipogenesis. Cell. 2010;143:156–169. doi: 10.1016/j.cell.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li J, Song JS, Bell RJ, Tran TN, Haq R, Liu H, Love KT, Langer R, Anderson DG, Larue L, Fisher DE. YY1 regulates melanocyte development and function by cooperating with MITF. PLoS Genet. 2012;8:e1002688. doi: 10.1371/journal.pgen.1002688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Deng Z, Cao P, Wan MM, Sui G. Yin Yang 1: a multifaceted protein beyond a transcription factor. Transcription. 2010;1:81–84. doi: 10.4161/trns.1.2.12375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Flanagan JR. Autologous stimulation of YY1 transcription factor expression: role of an insulin-like growth factor. Cell Growth Differ. 1995;6:185–190. [PubMed] [Google Scholar]
- 74.Huang HY, Li X, Liu M, Song TJ, He Q, Ma CG, Tang QQ. Transcription factor YY1 promotes adipogenesis via inhibiting CHOP-10 expression. Biochem Biophys Res Commun. 2008;375:496–500. doi: 10.1016/j.bbrc.2008.07.151. [DOI] [PubMed] [Google Scholar]
- 75.Tang QQ, Lane MD. Role of C/EBP homologous protein (CHOP-10) in the programmed activation of CCAAT/enhancer-binding protein-beta during adipogenesis. Proc Natl Acad Sci USA. 2000;97:12446–12450. doi: 10.1073/pnas.220425597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huang H, Lane MD, Tang QQ. Effect of serum on the down-regulation of CHOP-10 during differentiation of 3T3-L1 preadipocytes. Biochem Biophys Res Commun. 2005;338:1185–1188. doi: 10.1016/j.bbrc.2005.10.057. [DOI] [PubMed] [Google Scholar]
- 77.Suske G, Bruford E, Philipsen S. Mammalian SP/KLF transcription factors: bring in the family. Genomics. 2005;85:551–556. doi: 10.1016/j.ygeno.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 78.Bieker JJ. Krüppel-like factors: three fingers in many pies. J Biol Chem. 2001;276:34355–34358. doi: 10.1074/jbc.R100043200. [DOI] [PubMed] [Google Scholar]
- 79.Kaczynski J, Cook T, Urrutia R. Sp1- and Krüppel-like transcription factors. Genome Biol. 2003;4:206. doi: 10.1186/gb-2003-4-2-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pearson R, Fleetwood J, Eaton S, Crossley M, Bao S. Krüppel-like transcription factors: a functional family. Int J Biochem Cell Biol. 2008;40:1996–2001. doi: 10.1016/j.biocel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 81.Miller IJ, Bieker JJ. A novel, erythroid cell-specific murine transcription factor that binds to the CACCC element and is related to the Krüppel family of nuclear proteins. Mol Cell Biol. 1993;13:2776–2786. doi: 10.1128/mcb.13.5.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Brey CW, Nelder MP, Hailemariam T, Gaugler R, Hashmi S. Krüppel-like family of transcription factors: an emerging new frontier in fat biology. Int J Biol Sci. 2009;5:622–636. doi: 10.7150/ijbs.5.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wu Z, Wang S. Role of Krüppel-like transcription factors in adipogenesis. Dev Biol. 2013;373:235–243. doi: 10.1016/j.ydbio.2012.10.031. [DOI] [PubMed] [Google Scholar]
- 84.Uchida S, Tanaka Y, Ito H, Saitoh-Ohara F, Inazawa J, Yokoyama KK, Sasaki S, Marumo F. Transcriptional regulation of the CLC-K1 promoter by myc-associated zinc finger protein and kidney-enriched Krüppel-like factor, a novel zinc finger repressor. Mol Cell Biol. 2000;20:7319–7331. doi: 10.1128/mcb.20.19.7319-7331.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mori T, Sakaue H, Iguchi H, Gomi H, Okada Y, Takashima Y, Nakamura K, Nakamura T, Yamauchi T, Kubota N, Kadowaki T, Matsuki Y, Ogawa W, Hiramatsu R, Kasuga M. Role of Krüppel-like factor 15 (KLF15) in transcriptional regulation of adipogenesis. J Biol Chem. 2005;280:12867–12875. doi: 10.1074/jbc.M410515200. [DOI] [PubMed] [Google Scholar]
- 86.Pei H, Yao Y, Yang Y, Liao K, Wu JR. Krüppel-like factor KLF9 regulates PPARgamma transactivation at the middle stage of adipogenesis. Cell Death Differ. 2011;18:315–327. doi: 10.1038/cdd.2010.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gray S, Feinberg MW, Hull S, Kuo CT, Watanabe M, Sen-Banerjee S, DePina A, Haspel R, Jain MK. The Krüppel-like factor KLF15 regulates the insulin-sensitive glucose transporter GLUT4. J Biol Chem. 2002;277:34322–34328. doi: 10.1074/jbc.M201304200. [DOI] [PubMed] [Google Scholar]
- 88.Oishi Y, Manabe I, Tobe K, Tsushima K, Shindo T, Fujiu K, Nishimura G, Maemura K, Yamauchi T, Kubota N, Suzuki R, Kitamura T, Akira S, Kadowaki T, Nagai R. Krüppel-like transcription factor KLF5 is a key regulator of adipocyte differentiation. Cell Metab. 2005;1:27–39. doi: 10.1016/j.cmet.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 89.Birsoy K, Chen Z, Friedman J. Transcriptional regulation of adipogenesis by KLF4. Cell Metab. 2008;7:339–347. doi: 10.1016/j.cmet.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li D, Yea S, Li S, Chen Z, Narla G, Banck M, Laborda J, Tan S, Friedman JM, Friedman SL, Walsh MJ. Krüppel-like factor-6 promotes preadipocyte differentiation through histone deacetylase 3-dependent repression of DLK1. J Biol Chem. 2005;280:26941–26952. doi: 10.1074/jbc.M500463200. [DOI] [PubMed] [Google Scholar]
- 91.Lee H, Kim HJ, Lee YJ, Lee MY, Choi H, Lee H, Kim JW. Krüppel-like factor KLF8 plays a critical role in adipocyte differentiation. PLoS One. 2012;7:e52474. doi: 10.1371/journal.pone.0052474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Banerjee SS, Feinberg MW, Watanabe M, Gray S, Haspel RL, Denkinger DJ, Kawahara R, Hauner H, Jain MK. The Krüppel-like factor KLF2 inhibits peroxisome proliferator-activated receptor-gamma expression and adipogenesis. J Biol Chem. 2003;278:2581–2584. doi: 10.1074/jbc.M210859200. [DOI] [PubMed] [Google Scholar]
- 93.Wu J, Srinivasan SV, Neumann JC, Lingrel JB. The KLF2 transcription factor does not affect the formation of preadipocytes but inhibits their differentiation into adipocytes. Biochemistry. 2005;44:11098–11105. doi: 10.1021/bi050166i. [DOI] [PubMed] [Google Scholar]
- 94.Sue N, Jack BH, Eaton SA, Pearson RC, Funnell AP, Turner J, Czolij R, Denyer G, Bao S, Molero-Navajas JC, Perkins A, Fujiwara Y, Orkin SH, Bell-Anderson K, Crossley M. Targeted disruption of the basic Krüppel-like factor gene (Klf3) reveals a role in adipogenesis. Mol Cell Biol. 2008;28:3967–3978. doi: 10.1128/MCB.01942-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kawamura Y, Tanaka Y, Kawamori R, Maeda S. Overexpression of Krüppel-like factor 7 regulates adipocytokine gene expressions in human adipocytes and inhibits glucose-induced insulin secretion in pancreatic beta-cell line. Mol Endocrinol. 2006;20:844–856. doi: 10.1210/me.2005-0138. [DOI] [PubMed] [Google Scholar]
- 96.Eaton SA, Funnell AP, Sue N, Nicholas H, Pearson RC, Crossley M. A network of Krüppel-like Factors (Klfs). Klf8 is repressed by Klf3 and activated by Klf1 in vivo. J Biol Chem. 2008;283:26937–26947. doi: 10.1074/jbc.M804831200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yamamoto K, Sakaguchi M, Medina RJ, Niida A, Sakaguchi Y, Miyazaki M, Kataoka K, Huh NH. Transcriptional regulation of a brown adipocyte-specific gene, UCP1, by KLF11 and KLF15. Biochem Biophys Res Commun. 2010;400:175–180. doi: 10.1016/j.bbrc.2010.08.039. [DOI] [PubMed] [Google Scholar]
- 98.Tsai J, Tong Q, Tan G, Chang AN, Orkin SH, Hotamisligil GS. The transcription factor GATA2 regulates differentiation of brown adipocytes. EMBO Rep. 2005;6:879–884. doi: 10.1038/sj.embor.7400490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Plaisier CL, Bennett BJ, He A, Guan B, Lusis AJ, Reue K, Vergnes L. Zbtb16 has a role in brown adipocyte bioenergetics. Nutr Diabetes. 2012;2:e46. doi: 10.1038/nutd.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kajimura S, Seale P, Kubota K, Lunsford E, Frangioni JV, Gygi SP, Spiegelman BM. Initiation of myoblast to brown fat switch by a PRDM16-C/EBP-beta transcriptional complex. Nature. 2009;460:1154–1158. doi: 10.1038/nature08262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kajimura S, Seale P, Tomaru T, Erdjument-Bromage H, Cooper MP, Ruas JL, Chin S, Tempst P, Lazar MA, Spiegelman BM. Regulation of the brown and white fat gene programs through a PRDM16/CtBP transcriptional complex. Genes Dev. 2008;22:1397–1409. doi: 10.1101/gad.1666108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, Scime A, Devarakonda S, Conroe HM, Erdjument-Bromage H, Tempst P, Rudnicki MA, Beier DR, Spiegelman BM. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tanaka T, Yoshida N, Kishimoto T, Akira S. Defective adipocyte differentiation in mice lacking the C/EBPbeta and/or C/EBPdelta gene. EMBO J. 1997;16:7432–7443. doi: 10.1093/emboj/16.24.7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nedergaard J, Petrovic N, Lindgren EM, Jacobsson A, Cannon B. PPARgamma in the control of brown adipocyte differentiation. Biochim Biophys Acta. 2005;1740:293–304. doi: 10.1016/j.bbadis.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 105.Turpin JA, Terpening SJ, Schaeffer CA, Yu G, Glover CJ, Felsted RL, Sausville EA, Rice WG. Inhibitors of human immunodeficiency virus type 1 zinc fingers prevent normal processing of gag precursors and result in the release of noninfectious virus particles. J Virol. 1996;70:6180–6189. doi: 10.1128/jvi.70.9.6180-6189.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rice WG, Turpin JA, Huang M, Clanton D, Buckheit RW, Jr, Covell DG, Wallqvist A, McDonnell NB, DeGuzman RN, Summers MF, Zalkow L, Bader JP, Haugwitz RD, Sausville EA. Azodicarbonamide inhibits HIV-1 replication by targeting the nucleocapsid protein. Nat Med. 1997;3:341–345. doi: 10.1038/nm0397-341. [DOI] [PubMed] [Google Scholar]
- 107.Ylä-Herttuala S. Endgame: Glybera finally recommended for approval as the first gene therapy drug in the European Union. Mol Ther. 2012;20:1831–1832. doi: 10.1038/mt.2012.194. [DOI] [PMC free article] [PubMed] [Google Scholar]