Abstract
To obtain efficient non-viral vectors, a series of Gemini cationic lipids with carbamate linkers between headgroups and hydrophobic tails were synthesized. They have the hydrocarbon chains of 12, 14, 16 and 18 carbon atoms as tails, designated as G12, G14, G16 and G18, respectively. These Gemini cationic lipids were prepared into cationic liposomes for the study of the physicochemical properties and gene delivery. The DNA-bonding ability of these Gemini cationic liposomes was much better than their mono-head counterparts (designated as M12, M14, M16 and M18, respectively). In the same series of liposomes, bonding ability declined with an increase in tail length. They were tested for their gene-transferring capabilities in Hep-2 and A549 cells. They showed higher transfection efficiency than their mono-head counterparts and were comparable or superior in transfection efficiency and cytotoxicity to the commercial liposomes, DOTAP and Lipofectamine 2000. Our results convincingly demonstrate that the gene-transferring capabilities of these cationic lipids depended on hydrocarbon chain length. Gene transfection efficiency was maximal at a chain length of 14, as G14 can silence about 80 % of luciferase in A549 cells. Cell uptake results indicate that Gemini lipid delivery systems could be internalised by cells very efficiently. Thus, the Gemini cationic lipids could be used as synthetic non-viral gene delivery carriers for further study.
1 Introduction
Gene therapy requires safe and efficient carriers to transfer expressible genetic materials or silencing nucleic acids to target tissues. The most extensively used delivery tools can be divided into two general categories: viral and non-viral vectors. Development of non-viral vectors has been one of the primary areas of research because of several significant advantages, including greater carrier capacity, safety, ease of large-scale production, stability, potential to incorporate targeting ligands, and unlimited size. However, compared to viruses, which have evolved to overcome cellular barriers and immune defence mechanisms, non-viral gene carriers consistently exhibit significantly lower transfection efficiency.1–2 Among non-viral vectors, cationic liposomes- and cationic polymer-based vectors are the most heavily-investigated means to improve transfection efficiency. Although cationic polymers can condense DNA efficiently, exhibit better stability than cationic lipids, and the properties of polyplexes are more easily controlled than those of lipoplexes, cationic polymers usually display higher cytotoxicity and immunogenicity due to the “templating” properties of the polymeric backbone.3 To reduce toxicity, cationic amphiphiles have been utilised as an alternative to cationic polymers. In 1987, Felgner and colleagues first reported the design of cationic lipids for gene delivery.4 Since then, interest in the synthesis of efficient cationic lipids has surged globally. A number of strategies have been reported for the synthesis of versatile cationic lipids for gene delivery.5–9
The structure of cationic lipids generally comprises a hydrophilic cationic head group attached to hydrophobic tails via a linker. The polar head group provides association between positively charged liposomes and negatively charged nucleic acids by electrostatic complexation. The electrostatic properties of the cationic head group can affect the binding affinity of the liposomes for DNA, as well as the transfection efficiency. Generally, the toxicity of cationic lipids is due to the positive electrical charge of the head group.10 The hydrophobic tails in cationic lipids are usually composed of two long aliphatic chains. The two chains, generally 12–18 carbon atoms in length, can be either saturated or unsaturated (e.g., oleyl) and are not necessarily symmetrical.11 Many studies have shown that the length and type of aliphatic chain affect the transfection efficiency of a given lipid.
Most of linkers in the synthetic lipids are ether, ester or amide bonds. These are either too stable to be biodegraded, thus causing cytotoxicity (e.g., ethers), or liable to decompose in the circulatory system (e.g., esters). It is well known to chemists that carbamate bonds are stable under neutral conditions but liable to acid-catalysed hydrolysis. The pH inside lysosomes or endosomes is 1–2 units lower than in the cytoplasm. This can be used for the design of carbamate-linked lipids, as they will remain stable in plasma but decompose and release DNA because of the decrease in pH.12
Of the various forms of lipid, Gemini lipids are quite interesting, as they possess unusual aggregation and surface properties.13 Gemini lipids are a class of cationic lipids in which two lipid molecules are joined together by a spacer. In 1997, Engberts and colleagues first reported the novel Gemini surfactants for efficient, nontoxic in vitro gene delivery.14 In recent years, Gemini surfactants have been used industrially as emulsifiers and dispersants in detergents, cosmetics, personal hygiene products, coatings, and paint formulations. However, they have recently received more attention as gene transfection agents, due to their superior surface activity and DNA binding capabilities.15–19
The design of an efficient cationic lipid requires strategies to address a multitude of cellular barriers. Over the past decade, our laboratory has investigated the potential of cationic lipids as gene delivery vectors.1,20–24 During this process, we combined carbamates with Gemini lipids to give a novel type of cationic lipids. In the present work, we built two types of novel Gemini cationic lipids (termed mono-head quaternary ammonium and Gemini quaternary ammonium), with hydrophobic chains of various lengths (C12, C14, C16 and C18). Their usefulness for gene delivery was then evaluated.
2 Experimental procedures
Materials
DMEM, RPMI1640, fetal bovine serum (FBS), and Lipofectamine 2000 were purchased from Invitrogen Life Technologies (USA). DOPE and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) were purchased from Fluka (Switzerland). Anti-luciferase siRNA, missense siRNA, FAM labelled dsDNA (sense: 5’-CAA GGG ACT GGA AGG CTG GG), Texas Red labelled dsDNA (sense: 5’-CAA GGG ACT GGA AGG CTG GG) and Hoechst 33342 were purchased from Sigma-Aldrich (USA). NBD-PE and DOTAP were purchased from Avanti Polar Lipids, Inc. (USA). Hep-2 and A549 cells were obtained from the Institute of Biochemistry and Cell Biology (China). N,N’-carbonyldiimidazole (CDI), lauryl alcohol, tetradecyl alcohol, hexadecyl alcohol and octadecyl alcohol were purchased from Shanghai Chemical Industry Co. Ltd. (Shanghai, China). Diethylenetriamine (DETA) and methyl iodide (CH3I) were purchased from Shenyang Chemical Reagent Co. Ltd. (Shenyang, China). Agarose was purchased from Gene Tech (Shanghai, China). Green fluorescent protein (pGFP-N2) and luciferase (pGL3) plasmid vectors were purchased from Sangon Biotech Co. Ltd. (China), and extracted in our lab. All other chemicals were of reagent grade. All materials were used as received. All water used was purified using a Milli-Q Plus 185 water purification system (Millipore, USA), giving a resistivity greater than 18 MΩ cm.
Synthesis of cationic lipids
Gemini cationic lipids (G series) were readily obtained following the three-step process depicted in Scheme 1. Mono-head cationic lipids (M series) in Scheme 2 were synthesized through a similar route. The synthesis of these lipids was performed as follows, using G14 as an example: carbonyldiimidazole (40 mmol) was added to tetradecyl alcohol (40 mmol) in dichloromethane and kept at reflux for 3 h. Triethylenetetraamine (20 mmol in 30 mL tetrahydrofuran) was added. After another 3 h, the solvent was evaporated. The crude product was recrystallised from the mixture solvent of ethanol/water (v:v, 3:1) to give intermediate B as a white powder. For the construction of G14, intermediate B (4 mmol), a catalytic amount of potassium hydroxide and methyl iodide (6 mmol) were added to a 100 mL autoclave, then the mixture was allowed to react for 40 h at 160 °C. After the reaction was complete, excess methyl iodide was removed by heating. The resulting sticky paste was recrystallised from the mixture solvent of ethanol/water (v:v, 3:1) 3 times to obtain white lamellar crystals of G14.
Scheme 1.
Synthesis of Gemini cationic lipids with carbamate linkers (G12-G18). Reaction Conditions: (A) CH2Cl2, 40 °C, 3 h; (B) THF, 40 °C, 3 h; (C) 120–160 °C, 40 h.
Scheme 2.
The chemical structure of mono-head lipids. For their synthesis, triethylenetetraamine in Scheme 1 was replaced with diethylenetriamine.
Chemical analysis
All synthetic compounds were analysed by NMR, ESI-MS, IR and HPLC before being submitted to biological screening. 1H and 13C NMR spectra were measured at 400 and 100 MHz, respectively, on a Varian MercuryPlus 400 spectrometer. High resolution mass spectroscopy was performed using a Q-Tof Micro (Micromass Inc., Manchester, UK) with a resolving power of 5000, flow rate 5–20 µL min−1, capillary voltage 2.5–3.5 KV and ion source temperature 100 °C. IR spectra were recorded using a NICOLET 370 (Thermo, USA). HPLC was performed on an ELSD180 (Tianmei, China) with a C18 column (4.6 mm×150 mm, 5 µm), methanol/water mobile phase (85:15), column temperature 30 °C and flow rate 0.6 mL min−1. Characterisation data for cationic lipids are listed in the SI.
Liposomes preparation and characterisation
The protocol for preparation of liposomes based on M12-M18 and G12-G18 lipids was optimised to obtain high-quality delivery systems. The lipids were formulated in combination with the neutral lipid DOPE in a 1:1 molar ratio. To prepare liposomes, a suitable amount of lipid and DOPE were dissolved in 1 mL of chloroform in a 5 mL glass vial. The solvent was removed under a stream of nitrogen gas, followed by high-vacuum desiccation. The dry lipid film was resuspended in 1 mL distilled water to give liposomes in a concentration of approximately 1 mg mL−1. Liposomes solutions were subjected to several cycles of sonication in a bath sonicator and vigorous vortex mixing to form small vesicles. For the measurement of particle size and Zeta potential, 20 µL of the liposomes were diluted with distilled water (1 mL). Particle size and zeta potential were then measured three times using a Malvern ZetaSizer Nano series (Westborough, MA).
DNA-binding assay
Formation of lipoplexes between cationic liposomes and pDNA was confirmed by gel retardation assay. Cationic liposomes were miexed with pDNA in DMEM at N/P ratios from 0:1 to 8:1 and incubated for 20 min at room temperature. Each mixture was loaded onto a 1.2 % agarose gel containing 0.5 mg mL−1 ethidium bromide. The samples were subjected to electrophoresis at 100 V for 30 min, and DNA bands were visualised in a gel documentation unit (Syngene, Britain).
In vitro pDNA transfection
To prepare cationic liposomes/DNA complexes, plasmid DNA (pGFP-N2) and liposomes were each diluted in 100 µL Dulbecco’s modified Eagle’s medium (DMEM); then, the plasmid DNA solution was added to the liposomes solution. This mixture was incubated at room temperature for 20 min before use to allow the formation of DNA complexes. Tumour cells (5.0 × 104 cells well−1) were seeded in 24-well plates, and incubated at 37 °C under 5 % CO2 until approximately 80 % confluence was attained. The medium was removed and replaced with 100 µL serum-free DMEM per well. Cationic liposomes/DNA complexes were then added to the plates and kept at 37 °C under 5 % CO2 for 4 h. The medium was then replaced with DMEM containing 10 % FBS and 1 % antibiotics, and the cultures were maintained at 37 °C under 5 % CO2 for 48 h.
The expression of green fluorescent protein was measured using an inverted fluorescence microscope (Olympus IX71, Japan). Relative luciferase activity was assessed using a Bright-Glo™ Luciferase Assay System (Promega) and Synergy™ 2 Multi-Detection Microplate Reader (BioTek, USA). Briefly, the growth medium was removed from each well, and luciferase activity was measured after lysis buffer was added. Protein concentration in the cell lysate was determined using BCA protein assay reagent, allowing transfection efficiency to be determined from relative luciferase activity. The data are expressed as relative light units (RLU) per milligram of protein.
In vitro siRNA transfection
A549 cells expressing firefly luciferase were seeded in 12-well plates (1.0 × 105 per well) approximately 24 h before experiments. Cells were treated with different lipid formulations at the anti-luciferase siRNA concentration of 40 nM in Opti-MEM at 37 °C for 4 h. Cells were washed with DPBS, followed by incubation with lysis buffer at room temperature for 20 min. Fluorescence intensity in cell lysate was determined using a Perkin-Elmer LS 50B luminescence spectrometer (Norwalk, CT) (λex, 494 nm; λem, 519 nm). Cell lysate (5 µg per well) was dissolved in BCA protein assay reagent, and total protein concentrations determined at 570 nm. The silencing rate is expressed as relative light units (RLU) per microgram of total protein.
Cytotoxicity of the cationic liposomes
The cytotoxicity of the lipids M12-M18 and G12-G18 was assessed by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) reduction assay. The assay was performed in 96-well plates by maintaining the N/P weight ratios as used in the transfection experiments. Hep-2 cells were seeded in 96-well plates and incubated at 37 °C under 5 % CO2 for 24 h to get a confluence of about 80 %. After the replacement of medium, the liposomes/DNA complexes were added to the cells for further culture at 37 °C under 5 % CO2 for 24 h, and then MTT (20 µL, 5 mg mL−1, pH 7.4) was added and kept for 4.5 h. The absorbance at 570 nm was monitored by using the enzyme mark instrument (Sunrise Tecan, Australia).
Cellular uptake
A549 cells were seeded on 3.5 cm plates (Mat Tek) with glass bottom and incubated at 37 °C in humidified 5 % CO2. At the confluency of about 80 %, the culture medium was removed and washed by DPBS. Lipoplexes were obtained by adding liposomes with post-inserted NBD-PE (λex, 460 nm; λem, 535 nm) into a Texas Red (λex, 589 nm; λem, 615 nm) labeled dsDNA (mimic of siRNA). Lipoplexes were then added to the cells at a final dsDNA concentration of 40 nM. Following 4 hour culture at 37 °C in humidified 5 % CO2, cells were washed with DPBS extensively. Cells were further incubated 15 min with Hoeschst 33342 (λex, 350 nm; λem, 461 nm) to visualize nucleus of cells. Confocal laser scanning microscopy was carried out by using an Olympus FV-1000 MPE laser scanning microscope operated with FLUOVIEW software (Olympus, Tokyo, Japan).
For cytometry analysis, cells were seeded in a 12 well plate. At the cell confluency of about 80 %, lipoplexes obtained by adding liposomes into a FAM (λex, 492; λem, 517 nm) labeled dsDNA (mimic of siRNA) solution were then added to the cells at a final dsDNA concentration of 40 nM. Following 4 hour culture at 37 °C in humidified 5 % CO2, cells were washed with DPBS, trypsinized, and resuspended in DPBS at a concentration of 1×106 cells mL−1. Cells were detected and quantified by flow cytometry (Becton-Dickinson, Heidelberg, Germany). Results were processed using the Cellquest software (Becton-Dickinson).
3 Results and discussion
Design and synthesis of cationic lipids
When incorporating a carbamate group into the linker, it may be assumed that a decrease in pH will act as a trigger after entering endosomes, disconnecting the hydrophobic and hydrophilic portions of the lipoplex and releasing DNA.12 In a previous study, we synthesised several carbamate-linked cationic lipids, which showed some capacity for gene delivery. In this article, we report the design and synthesis of a series of novel Gemini cationic lipids (Scheme 1) with mono-head cationic lipids (Scheme 2) as controls (the synthetic scheme can be found in the SI).
We thus developed an environmentally friendly synthesis of carbamate-linked cationic lipids, using CDI to mediate the linkage of fatty alcohols and amine (Scheme 1). The advantages of this method include mild reaction conditions (which minimise side-reactions), regioselectivity reaction with primary amino groups in step B, the lack of amine hydrochloride formation (as observed when using acid chlorides), and simple purification. Imidazole, formed as a by-product when CDI reacts with hydroxyl groups and carboxylic acids or amines, can be easily removed by an acidic wash.25
This is the first reported use of our method for the synthesis of the novel lipids M12-M18 and G12-G18. They are composed of three biocompatible molecular moieties: bis-quaternary ammonium or mono-head quaternary ammonium (the cationic headgroup), carbamate (the linker), and two hydrocarbon chains (hydrophobic tail) (Scheme 1). For G14, intermediate B was obtained in a yield of approximately 60 % under the conditions given in scheme 1. Nevertheless, reactivity fell as chain length increased, so it was necessary to increase the reaction temperature for the intermediates of G16 and G18. Finally, CH3I treatment of intermediate B gave the final products in a yield of over 70 % after 3 recrystallisations from ethanol/water. The yield was influenced by reaction temperature, and alkali accelerated the quaternisation greatly. The structures and purity of mono-head and Gemini cationic lipids were characterised by electrospray ionisation-mass spectrometry (ESI-MS), proton nuclear magnetic resonance spectroscopy (1H NMR, 13C NMR), infrared (IR) spectroscopy, and high pressure liquid chromatography (HPLC). Taking G14 as an example, the 1H NMR peaks of the hydrocarbon chain were detected at 0.89, 1.26, 1.68 and 4.04 ppm; 1H NMR peaks for the carbamate NH, the methylenes between the two quaternary ammonium groups, the methylenes between carbamate and quaternary ammonium, and N-methyl in the head were at 6.20, 3.83, 4.69, 3.64 ppm, respectively. Molecular ions of these compounds were detected as [M-I]+ and [M-I]2+/2. For G14, these were 811.5563 (calculated: 811.5537) and 342.3250 (calculated: 342.3246), respectively. In the IR spectrum 3299.08 cm−1 and 1681.57 cm−1 represented νNH and νC=O, respectively, while 1548.85 cm−1 corresponded to δNH. The purity of all lipids obtained was over 95 % based on HPLC analysis. Analytical data of these compounds are listed in the SI.
Particle size and ζ-potential
After the preparation of liposomes as depicted in the experimental procedures, they were subjected to the measurement of particle size and zeta potential by using dynamic light scattering analysis (DLS) method. The average particle sizes of these liposomes were from 100 nm to 200 nm, similar to those of DOTAP and Lipofectamine 2000 (Fig. 1A). It has been proved that this particle size range was suitable for gene delivery.26 The zeta potential of liposomes is an indirect measure of the surface charge; it can be used to evaluate the extent of interaction of the liposomal cationic surface charges with the anionic charges of DNA.27 In this study, the zeta potentials of the liposomes were between 38–55 mV (Fig. 1B), which should maintain the stability of liposomes for at least 3 months at 4 °C. This has been proved by the data of particle size listed in the SI.
Fig. 1.
Particle size and zeta potential of liposomes from the M and G series. Liposomes (20 µL) were diluted in 1 mL distilled water. Particle size (A) and zeta potential (B) were measured using a Malvern ZetaSizer. The liposomes PDI of M12, M14, M16, M18, G12, G14, G16, G18, DOTAP and Lipofectamine were 0.203, 0.275, 0.353, 0.352, 0.207, 0.204, 0.180, 0.136, 0.230 and 0.277, respectively.
After the addition of DNA to liposomes, lipoplexes formed due to electrostatic forces. The range of zeta potentials may also ensure the formation of lipoplexes. Zeta potential is a function of lipid:DNA ratios in lipoplexes, as shown in Fig. 2A and SI. As increasing the N/P charge ratios, zeta potential tended to increase from negative (−21.6 mV at an N/P ratio of 1) to positive values (around +40 mV for higher N/P ratios). The structure of lipoplexes with a positive zeta potential is different from those with a negative potential.28 Lipoplexes with positive zeta potential may have an aggregated multilamellar structure (LCα).29 Negative zeta potential leads to free plasmids or protruding DNA strings. The average particle size reaches a maximum (about 230 nm) at an N/P ratio of 3:1 or 4:1, with a bigger PDI (Fig. 2B and SI). A DNA complex size of approximately 200 nm and a positively charged surface are desirable for cellular uptake.30
Fig. 2.
Zeta potential and particle size of G14 lipoplexes at different N/P weight ratios. Lipoplexes (20 µL) were diluted in 1 mL distilled water. Zeta potential (A) and particle size (B) were measured using a Malvern ZetaSizer. The PDI at an N/P ratio of 1, 2, 3, 4, 6, and 8 was 0.192, 0.173, 0.242, 0.302, 0.169 and 0.171, respectively.
This increase in both size and PDI was caused by adsorption of DNA molecules to the outside of the cationic liposomes, resulting in different lipoplex structures. In some cases, an excess of cationic liposomes to DNA leads to entrapment of DNA molecules between the lamellae in clusters of aggregated multilamellar structures.31 In other studies, the lamellar structure,32 a highly ordered tubular structure, the multilamellar structure LCα, and inverted hexagonal structures HCII and HI have also been found.29 In this study, when N/P ratios were over 6, DNA was highly condensed, giving smaller particle sizes and PDI (Fig. 2B). During lipoplex formation, it has been determined that a single plasmid is surrounded by sufficient cationic liposomes to completely neutralise the negative charge of DNA, providing the complex with a net positive charge that is attracted to the negatively charged cell surface. This may be associated with effective transfection.4 However, after the entry of lipoplexes into cells, the release of DNA from the matrix is required for gene expression.
Lipoplex formation confirmed by gel electrophoresis
Liposomes can facilitate pDNA entry into cells and also protect pDNA from degradation by DNase. DNA degradation is a limiting factor in gene therapy.33 To identify the appropriate ratio of liposomes to DNA for condensation, we evaluated their ability to promote DNA condensation and to inhibit its migration in agarose gel electrophoresis. The results show that the pDNA formed two major bands, corresponding to supercoiled pDNAs and open circular pDNAs. After the addition of liposomes, pDNA was completely retarded at different N/P weight ratios for different liposomes-M12, 6:1; M14, 6:1; M16, 8:1; M18 over 8:1; G12, 2:1; G14, 3:1; G16, 3:1 and G18, 6:1. As controls DOTAP and Lipofectamine 2000 retarded pDNA at N/P ratios of 6:1 and 3:1, respectively. These results demonstrate that cationic liposomes can form DNA-liposomes complexes. The data of gel electrophoresis are listed in the SI. The DNA-bonding ability of cationic liposomes containing Gemini lipids was much better than those containing mono-head lipids, as Gemini lipids bear more charges than their mono-head counterparts. In the same series of liposomes, bonding ability declined with an increase in tail length. It was also observed that the increasing trend in DNA-binding ability was obvious with the increase in lipid-to-DNA ratios. The lower N/P ratios of Gemini liposomes for retardation compared with the control DOTAP and mono-head liposomes may reduce the dose of Gemini cationic liposomes required to obtain maximal transfection efficiency. That is one of the advantages for Gemini cationic liposomes.
In vitro pDNA and siRNA transfection
In vitro transfection using liposomes from lipids M12-M18 and G12-G18 was evaluated using pGFP-N2 plasmid DNA and pGL3 plasmid DNA against Hep-2 cells. Expression of pGFP-N2 is illustrated in Fig. 3A and the SI, which indicates that G series cationic lipids were much better than M series in terms of transfection efficiency. Among M and G series, lipids with carbon chain length of 14 showed higher transfection efficiency than other chain lengths. G14 could maintain high transfection at the weight ratios from 1:1 to 4:1. Compared to Lipofectamine 2000 and DOTAP, G14 showed similar or higher transfection efficiency at an N/P weight ratio of 3:1. This was also confirmed by delivering pGL3 plasmid as shown in Fig. 3B and Fig. 4.
Fig. 3.
Gene expression in Hep-2 cells using G14 liposome at different N/P ratios. As controls, DOTAP and Lipofectamine 2000 were used at the N/P ratio of 3:1. (DOTAP showed nearly the same transfection efficiency for the N/P ratios of 3:1 and 6:1 as shown in the SI, so we used 3:1 to keep the consistency.) Images were taken 48 h after transfection. (A: Fluorescence microscopic images (20 × 10) of GFP. B: Luciferase expression of pGL3.)
Fig. 4.
Luciferase expression in Hep-2 cells in the presence of mono-head cationic lipids and Gemini cationic lipids, compared to DOTAP and Lipofectamine 2000 at a N/P weight ratio of 3:1. (DOTAP showed nearly the same transfection efficiency for the N/P ratios of 3:1 and 6:1 as shown in the SI, so we used 3:1 to keep the consistency.)
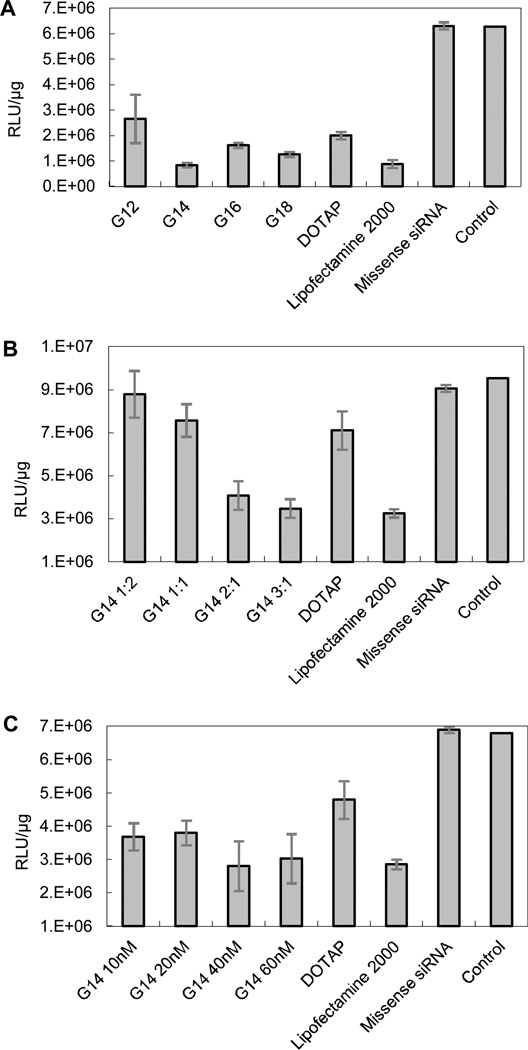
As Gemini cationic lipids were more effective than mono-head cationic lipids in terms of pDNA delivery, G series lipids were screened for the ability to deliver siRNA to A549 lung carcinoma cells, which can stably express firefly luciferase (Fig. 5). Efficacy of siRNA delivery by these Gemini cationic lipids was determined by treating cells with siRNA-Gemini complexes, prepared using a firefly luciferase targeting siRNA (siLuc).
Fig. 5.
In vitro screening of Gemini cationic lipids for siRNA delivery. A549 cells expressing firefly luciferase were treated with firefly luciferase targeting siRNA-Gemini complexes. Firefly luciferase activity (RLU µg−1) after treatment with siRNA-Gemini lipid complexes in triplicate is shown at a 3:1 (wt/wt) ratio for different Gemini cationic lipids (A); at N/P ratios of 1:2, 1:1, 2:1 and 3:1 for G14 (B) and at siRNA concentrations of 10, 2, 40 and 60 nM for G14 (C). For DOTAP and Lipofectamine 2000, the N/P ratio was 3:1 and the siRNA concentration was 40 nM. Missense siRNA was delivered by using G14 at the N/P ratio of 3:1.
Overall, all four Gemini lipids showed gene silence compared with the control. Three of them were better than DOTAP, and G14 was comparable to Lipofectamine 2000, silencing nearly 80% of luciferase in A549 cells. The ratio of delivery material to nucleic acids is known to affect the delivery potential of lipoplexes. In this experiment, several N/P weight ratios were investigated to determine the optimal ratio for delivery of siRNA. All ratios were below 3:1 to prevent cytotoxicity. The results in Fig. 5B show that with an increase in N/P ratio silencing of firefly luciferase was enhanced, but the difference in rate of silencing between ratios of 2:1 and 3:1 was only approximately 6 %. Therefore, the ratio of 3:1 should be suitable for the delivery of siRNA using G14. The effects of four concentrations of G14 (10 nM, 20 nM, 40 nM and 60 nM) on the silencing results are shown in Fig. 5C. This indicates that 40 nM was a suitable concentration for in vitro siRNA delivery to lung carcinoma cells. Much work has been carried out on modifying cationic lipids for use in gene transfection, to determine if there is a ‘best’ chain length.34–35
However, solid conclusions are rarely obtained, for the conclusions of these studies are frequently contradictory. Most studies have found that cationic lipids with shorter saturated chains are generally important for acquiring high transfection efficiency. It is possible that shorter hydrocarbon chains increase the fluidity of bilayers, thus favouring a higher rate of intermembrane transfer of lipid monomers. Lipid membrane mixing may result in disruption of the endosome, and consequent siRNA escape due to endosomal degradation. In this study, G14 showed superior siRNA delivery. It may maintain an appropriate balance between fluidity and rigidity for entering into cells, and have the ability to release siRNA in cells due to the carbamate linker. We can also see that missense RNA could not silence luciferase gene, though it was delivered by G14. This ensures the specificity of siRNA.
Cellular uptake
To study whether liposomes could enhance the delivery of siRNA using luciferase stably expressed in A549 cells, cell uptake was measured by flow cytometry at a constant N/P ratio of 3:1 with FAB labelled dsRNA (a model of siRNA) at 40 nM. Cells were cultured for 4 h after treatment with lipoplexes, then cells were trypsinised and resuspended for FITC signal analysis by flow cytometer. Through the selection of live cells to give P1 cells and by comparison with control cells, P2 cells were designated as the type exhibiting uptake. Histograms of cell number versus fluorescence intensity are shown in Fig. 6A for G14, control, DOTAP and Lipofectamine 2000. The uptake rate from P2/P1 is shown in Fig. 6B, indicating superior uptake of G series liposomes than of the commercial products. G14 attained 65 % uptake, 20 % higher than Lipofectamine 2000. This could partially explain why G14 exhibited better transfection efficiency than other liposomes, on the hypothesis they had nearly the same rate of release from endosomes.
Fig. 6.
Flow cytometry analysis of A549 cells after 4 hours of treatment with lipoplexes of liposomes and FAB labelled dsDNA at an N/P ratio of 3:1. Cells were trypsinised and resuspended; the FITC signal was analysed using a flow cytometer. (A) Cell counts based on the uptake of lipoplexes using P2 for control, G14, DOTAP and Lipofectamine 2000 (ordinate indicates the number of cells, and abscissa indicates fluorescence intensity); (B) Uptake rate based on P2, accounting for selected cells (P1) for different lipoplexes. “Oligo” indicates cells were treated with dsDNA without liposomes.
The internalisation of G14 lipoplexes was also monitored by confocal microscopy. In this experiment, FAM-labelled dsDNA, a model for siRNA, was complexed with NBD-PE-labelled G14 liposomes, and incubated with A549 cells for 4 h to study the sub-cellular distribution of the delivered dsDNA. Fig. 7 shows that G14 liposomes promoted internalisation of dsDNA by cells. Green fluorescence was mainly observed in the cytosol around the nuclei; red fluorescence was distributed in both the cytosol and the nuclei. Most complexes were evenly spread throughout the cytoplasm, though some punctate distribution was also observed. The confocal images are similar to those observed in a recent study, although those researchers used lipid-modified cationic polymers as the delivery system.36–37
Fig. 7.
Confocal microscopy of transfected cells, with G14 liposomes and dsDNA (40 nM) labelled with NBD-PE and Texas red, respectively. After removal of the transfection solution, cells were washed with DPBS and dyed with Hoechst 33342, diluted 1000-fold in medium for 15 min. Confocal laser scanning microscopy was carried out using an Olympus FV-1000 MPE laser scanning microscope operated with FLUOVIEW software and magnification of × 60 in oil (Olympus, Tokyo, Japan). Panels: (A) Nuclei; (B) Liposomes labelled with NBD-PE; (C) dsDNA labelled with Texas red; (D) Cells with bright field; (E) Superimposition of A, B, C and D.
The results indicate that after 4 h transfection most dsDNA was released from the lipoplexes, though some was still combined with G14 liposomes. Overall, the cell uptake studies clearly demonstrate that G14 can effectively deliver dsDNA into cells. After entry into cells, the liposomes formed from G14 may readily release nucleic acids due to the carbamate linker in G14 structure with the decrease in pH. It was already confirmed by Charlie in 2012.38
Cytotoxicity of cationic liposomes
As an important factor in biocompatibility, in vitro cytotoxicity of gene delivery materials is usually measured by the MTT method.39–40 MTT-based cell viabilities after exposure to lipids M12-M18 and G12-G18 were evaluated in Hep-2 cells across the entire range of lipid/DNA charge ratios used in the transfection experiments. Cells tolerated all the lipids at relatively low N/P ratios (under 4:1), equal to or higher than the commercial liposomes Lipofectamine 2000 and DOTAP (Fig. 8). When N/P ratios were increased to 6:1 and 8:1, cytotoxicity was found to rise dramatically, with 70 % cell viability or lower. Cationic lipids can exert cytotoxic effects by interacting with critical enzymes such as PKC. Some research shows cationic amphiphiles containing a steroid backbone are more potent inhibitors of PKC than their straight-chain analogues; thus, they also have higher toxicity.41 Compared with mono-head cationic lipids, Gemini cationic lipids did not show any greater cytotoxicity, despite their more charge per molecule. However, Gemini cationic lipids were more efficient for in vitro transfection than their counterparts at relatively low N/P ratios. Therefore, these novel Gemini cationic lipids display potential for application in gene delivery and deserve to be studied further.
Fig. 8.
Cytotoxicity of the mono-head cationic liposomes (A) and Gemini cationic liposomes (B) prepared with DOPE at a molar ratio of 1:1 against Hep-2 cells, 48 h after the addition of lipoplexes formulated from liposomes and GFP plasmids. The cell viability of negative control cells was designated as 1. The positive controls were Lipofectamine 2000 and DOTAP, which were formulated with GFP plasmids to form lipoplexes at a ratio of 3:1, as indicated in the protocols.
4 Conclusions
Although hundreds of cationic lipids have been studied,42 there is still a need to develop new gene transfer systems. Our objective in this study was to develop cationic lipids with a high gene- transfer efficiency that could be readily and inexpensively synthesised. We have synthesised several novel Gemini cationic lipids with hydrocarbon chains of varied length and tested for their gene-transferring capabilities in Hep-2 and A549 cells.
Compared with their mono-head counterparts, they have shown superior pDNA binding ability and more efficient in vitro transfection of both pDNA and siRNA. Of these Gemini cationic lipids, G14 showed more potential for gene delivery. G14 was as efficient as the commercial transfection reagents Lipofectamine 2000 and superior to DOTAP. What’s more, liposomes formed from G14 displayed low cytotoxicity, even though it bears more charge. The appropriate balance between fluidity and rigidity may partially explain it’s superior properties. Despite these promising results, further research is clearly needed to clarify the complex mechanisms involved in gene transfer mediated by Gemini cationic lipids. The improvement of these structures may yield new compounds worthy of exploration. We also expect further developments toward the use of this type of cationic lipid in successful in vivo gene transfer.
Supplementary Material
Acknowledgements
This research is financially supported by the National Natural Science Foundation of China (20876027 and 21176046), Roche, Program for New Century Excellent Talents in University (NCET-08-0654), the Fundamental Research Funds for the Central Universities (DC12010104) and NIH grants (CA129835, CA129421, CA149363).
Footnotes
Electronic Supplementary Information (ESI) available: The synthesis route for mono-head quaternary ammoniums; IR, NMR, MS and HPLC characterisation of all lipids synthesized by the article; In vitro transfection images. See DOI: 10.1039/b000000x/
Notes and references
- 1.Zhang SB, Zhao YN, Zhao BD, Wang B. Bioconjug. Chem. 2010;21:1003. doi: 10.1021/bc900261c. [DOI] [PubMed] [Google Scholar]
- 2.Simoes S, Filipe A, Femeca H, Mano M, Penacho N, Duzgunes N. Expert Opin. on Drug Deliv. 2005;11:375. doi: 10.1517/17425247.2.2.237. [DOI] [PubMed] [Google Scholar]
- 3.Wasungu I, Hoekstra D. J. Control. Release. 2006;116:255. doi: 10.1016/j.jconrel.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 4.Felgner P, Gadek T, Holm M. Proc. Natl. Acad. Sci. 1987;84:7413. doi: 10.1073/pnas.84.21.7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gall TL, Loizeau D, Picquet E, Carmoy N, Yaouanc JJ, Burel-Deschamps L. J. Med. Chem. 2010;53:1496. doi: 10.1021/jm900897a. [DOI] [PubMed] [Google Scholar]
- 6.Shan YB, Luo T, Peng C, Sheng RL, Cao A, Cao XY. Biomaterials. 2012;33:3025. doi: 10.1016/j.biomaterials.2011.12.045. [DOI] [PubMed] [Google Scholar]
- 7.Qu W, Qin SY, Kuang Y, Zhuo RX, Zhang XZ. J. Materials Chem. B. 2013;1:2147. doi: 10.1039/c3tb00226h. [DOI] [PubMed] [Google Scholar]
- 8.Karmali PP, Chaudhuri A. Med. Res. Rev. 2007;27:696. doi: 10.1002/med.20090. [DOI] [PubMed] [Google Scholar]
- 9.Bhattacharya S, Bajaj A. Chem. Commun. 2009;35:4632. doi: 10.1039/b900666b. [DOI] [PubMed] [Google Scholar]
- 10.Sen J, Chaudhuri A. J. Med. Chem. 2005;48:812. doi: 10.1021/jm049417w. [DOI] [PubMed] [Google Scholar]
- 11.Lv HT, Zhang SB, Wang B, Cui SH, Yan D. J. Control. Release. 2006;114:100. doi: 10.1016/j.jconrel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 12.Liu DL, Hu JJ, Qiao WH, Li ZS, Zhang SB, Cheng LB. Bioorganic & Medicinal Chemistry Lett. 2005;15:3147. doi: 10.1016/j.bmcl.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 13.Bhattacharya S, Bajaj A. J. Phys. Chem. B. 2007;111:2463. doi: 10.1021/jp068383w. [DOI] [PubMed] [Google Scholar]
- 14.Woude IVD, Wagenaar A, Meekel AAP, Beest MBA, Ruiters MHJ, Engberts JBFN. Proc. Natl. Acad.. Sci.. USA. 1997;94:1160. doi: 10.1073/pnas.94.4.1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cashion MP, Li XL, Geng Y. Langmuir. 2010;26:678. doi: 10.1021/la902287b. [DOI] [PubMed] [Google Scholar]
- 16.Perrone S, Usai M, Lazzari P, Tucker SJ, Wallace HM, Zanda M. Bioconjugate Chem. 2013;24:176. doi: 10.1021/bc3004292. [DOI] [PubMed] [Google Scholar]
- 17.Koeda S, Umezaki K, Noji T, Lkeda A, Kawakami K, Kondo M. Langmuir. 2013;37:11667. doi: 10.1021/la402167v. [DOI] [PubMed] [Google Scholar]
- 18.Misra SK, Biswas J, Kondaiah P, Bhattacharya S. PLoS ONE. 2013;7:68305. doi: 10.1371/journal.pone.0068305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muñoz-Úbeda M, Misra SK, Barrán-Berdón AL, Datta S, Aicart-Ramos C, Castro-Hartmann P. Biomacromolecules. 2012;12:3926. doi: 10.1021/bm301066w. [DOI] [PubMed] [Google Scholar]
- 20.Zhang SB, Xu YM, Wang B, Qiao WH, Liu DL, Li ZS. J. Control. Release. 2004;100:165. doi: 10.1016/j.jconrel.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 21.Zhang SB, Zhao BD, Jiang HM, Wang B, Ma BC. J. Control. Release. 2007;123:1. doi: 10.1016/j.jconrel.2007.07.016. [DOI] [PubMed] [Google Scholar]
- 22.Ma BC, Zhang SB, Jiang HM, Zhao BD, Lv HT. J. Control. Release. 2007;123:184. doi: 10.1016/j.jconrel.2007.08.022. [DOI] [PubMed] [Google Scholar]
- 23.Zhao YN, Zhang SB, Cui SH, Wang B, Zhang SF. Expert Opin. Drug Deliv. 2012;1:127. doi: 10.1517/17425247.2011.630387. [DOI] [PubMed] [Google Scholar]
- 24.Wang B, Zhang SB, Cui SH, Yang BL, Zhao YN, Hao XM. Biotechnology Letters. 2012;1:19. doi: 10.1007/s10529-011-0748-8. [DOI] [PubMed] [Google Scholar]
- 25.Zhi DF, Zhang SB, Qureshi F, Zhao YN, Cui SH, Wang B. Bioorganic & Medicinal chemistry letters. 2012;22:3837. doi: 10.1016/j.bmcl.2012.01.097. [DOI] [PubMed] [Google Scholar]
- 26.Chan CL, Majzoub RN, Shirazi RS, Ewert KK, Chen YJ, Liang KS. Biomaterials. 2012;33:4928. doi: 10.1016/j.biomaterials.2012.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perrie Y, Gregoriadis G. Biochim. Biophys. Acta. 2000;1475:125. doi: 10.1016/s0304-4165(00)00055-6. [DOI] [PubMed] [Google Scholar]
- 28.Jiang M, Gan L, Zhu CL, Dong Y, Liu JP, Gan Y. Biomaterials. 2012;7:1. doi: 10.1016/j.biomaterials.2012.06.079. [DOI] [PubMed] [Google Scholar]
- 29.Ewert K, Evans HM, Ahmad A, Slack NL, Lin AJ, Martin-Herranz A. Adv. Genet. 2005;53:119. doi: 10.1016/S0065-2660(05)53005-0. [DOI] [PubMed] [Google Scholar]
- 30.Khan M, Ang CY, Wiradharma N, Yong LK, Liu SQ, Liu LH. Biomaterials. 2012;33:4673. doi: 10.1016/j.biomaterials.2012.02.067. [DOI] [PubMed] [Google Scholar]
- 31.ustafsson J, rvidson G, Karlsson G, Almgren M. Biochim. Biophys. Acta. 1995;1235:305. doi: 10.1016/0005-2736(95)80018-b. [DOI] [PubMed] [Google Scholar]
- 32.Hulst R, Muizebelt I, Oosting P, Pol C, Wagenaar A, Šmisterová J. Eur. J. Org. Chem. 2004;4:835. [Google Scholar]
- 33.Tripathi SK, Singh VP, Gupta KC, Kumar P. J. Materials Chem. B. 2013;1:2515. doi: 10.1039/c3tb00481c. [DOI] [PubMed] [Google Scholar]
- 34.McGregor C, Perrin C, Monck M, Camilleri P, Kirkby AJ. J. Am. Chem. Soc. 2001;123:6215. doi: 10.1021/ja005681c. [DOI] [PubMed] [Google Scholar]
- 35.Sen J, Chaudhuri A. J. Med. Chem. 2005;48:812. doi: 10.1021/jm049417w. [DOI] [PubMed] [Google Scholar]
- 36.Charlie M, Hsu HU. Biomaterials. 2012;33:7834. doi: 10.1016/j.biomaterials.2012.06.093. [DOI] [PubMed] [Google Scholar]
- 37.Li JH, Wang CL, Wang Q, Cader MZ, Lu J, Evans DG, Duan X, O'Hara D. J. Materials Chem. B. 2013;1:61. doi: 10.1039/c2tb00081d. [DOI] [PubMed] [Google Scholar]
- 38.Charlie YMH, Hasan U. Biomaterials. 2012;31:7834. [Google Scholar]
- 39.Zhou YF, Yang B, Ren XY, Liu ZZ, Deng Z, Chen L. Biomaterials. 2012;33:4731. doi: 10.1016/j.biomaterials.2012.03.014. [DOI] [PubMed] [Google Scholar]
- 40.Xiu M, Zhao NN, Yang WT, Xu FJ. Acta Biomaterialia. 2013;9:7439. doi: 10.1016/j.actbio.2013.04.010. [DOI] [PubMed] [Google Scholar]
- 41.Bottega R, Epand RM. Biochemistry. 1992;31:9025. doi: 10.1021/bi00152a045. [DOI] [PubMed] [Google Scholar]
- 42.Liu D, Ren T, Gao X. Curr. Med. Chem. 2003;10:1307. doi: 10.2174/0929867033457386. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.