Background
Tramadol is a mixed centrally acting opioid analgesic used to relieve moderate to severe pain. It is commonly used alone or in combination with nonopioid analgesics, for example, paracetamol (acetaminophen) in the treatment for postoperative, dental, cancer, neuropathic, and acute musculoskeletal pain [1–3]. It can also be used as an adjuvant to NSAID therapy in osteoarthritis patients [4]. Tramadol exerts its analgesic effect through at least two complementary and synergistic mechanisms: by activating the μ-opioid receptor and inhibiting the neurotransmitter reuptake. The opioid receptor-mediated analgesic effects are mainly attributed to the active metabolite M1 (O-desmethyltramadol), whereas the inhibition of the neurotransmitter reuptake is caused by the parent drug, thus enhancing the inhibitory effects on pain transmission in the spinal cord [1,5,6]. Therefore, both the parent drug and especially the M1 metabolite contribute toward the overall analgesic activity of tramadol.
Tramadol is administered as a racemic mixture of (+)-tramadol and (−)-tramadol enantiomers (also known as R, R and S, S tramadol, respectively). Tramadol is a synthetic opioid belonging to the class of weak opioid receptor agonists [step 2 analgesics according to the WHO ladder (http://www.who.int/cancer/palliative/painladder/en/)]. In comparison with typical opioid agonists such as morphine at equivalent doses, tramadol shows similar overall analgesic efficacy, and yet appears to have a lower potential for respiratory depression or physical dependence as well as a lower incidence of constipation [1,3,7–9]. However, cases of physical dependence and withdrawal syndrome have been reported, especially with long-term use of high tramadol doses [10–12]. The dose requirements for tramadol are variable and interactions with other drugs are common. Depending on the duration of treatment and settings, there might be certain variance in the equipotent use of tramadol and morphine. Tramadol has been shown to be approximately equipotent to morphine in the range of 4:1 to 5:1 ratio orally (tramadol:morphine) and 10:1 to 12:1 ratio parenterally [13–16]. Some patients experience inadequate pain relief or adverse effects with tramadol. The main reported adverse drug reactions (ADRs) for tramadol are nausea, vomiting, sweating, itching, constipation, headache, and central nervous system stimulation. Most of these reactions are dose dependent. Neurotoxicity to tramadol may manifest as seizures, which have been reported in patients receiving tramadol both at the recommended and the high dosage ranges in animal and human studies [17].
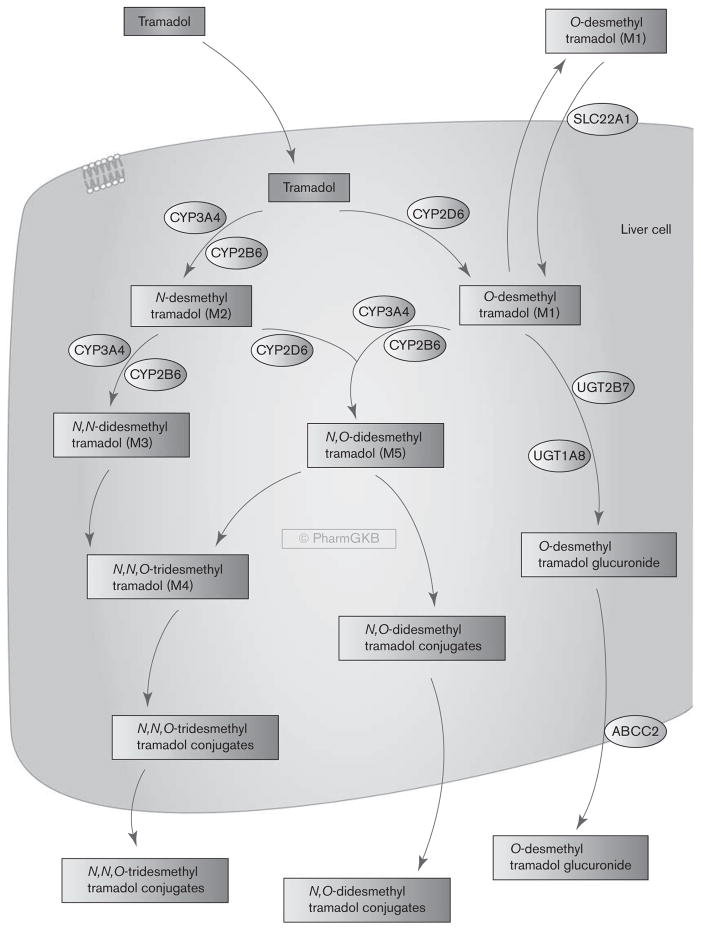
Tramadol metabolism to its major active metabolite M1 is predominantly mediated through CYP2D6 [18–20]; thus, CYP2D6 plays a significant role in tramadol pharmacokinetics and variability in drug responses [21,22]. The influence of the CYP2D6 genotype on plasma levels of tramadol and its metabolites as well as tramadol efficacy and ADR have been reported [18,21,23–25] (see the Pharmacogenomics section). This review briefly summarizes the metabolism and transport of tramadol (Fig. 1) and discusses genetic variations affecting the pharmacokinetics, efficacy, and toxicity of tramadol. Knowledge of genetic variants mediating the diverse pharmacological profile of tramadol may help optimize opioid therapy to achieve better control of pain and less adverse effects for the patients.
Fig. 1.
Stylized liver cell showing candidate genes involved in the metabolism and transport of tramadol. A fully interactive version is available online at http://www.pharmgkb.org/pathway/PA165946349.
Pharmacodynamics
Tramadol consists of two enantiomers [(+)-tramadol and (−)-tramadol], both of which, along with metabolite M1, contribute toward overall analgesic activity by distinct but complementary mechanisms [1,6]. In vitro and clinical studies showed that the parent drug is only a weak μ-opioid receptor agonist, whereas the metabolite M1 is significantly more potent than tramadol μ-opioid receptor binding and in producing analgesia [22,26,27]. (+)-M1 has a significantly higher affinity for the μ-opioid receptor (encoded by gene OPRM1) (Ki = 0.0034 μmol/l) than the parent drug (+/−)-tramadol (Ki = 2.4 μmol/l) as well as (+/−)-M5 (Ki = 0.1 μmol/l) and (−)-M1 (Ki = 0.24 μmol/l) [26], suggesting that it is the compound primarily responsible for opioid receptor-mediated analgesia. The (+/−)-tramadol also contribute toward analgesia by inhibiting reuptake of the neurotransmitters serotonin [by binding to transporter hSERT (encoded by gene SLC6A4)] and norepinephrine [by binding to transporter hNET (encoded by gene SLC6A2)]. The racemic tramadol binds to hNET and hSERT with Ki values at 14.6 and 1.19 μmol/l, respectively (reviewed by Raffa et al. [28]). Studies with the enantiomers showed that (−)-tramadol is more potent in inhibiting norepinephrine uptake (Ki = 1.08 μmol/l) and stimulating α-adrenergic receptors. In contrast, the (+)-tramadol is more potent than (−)-tramadol for inhibiting serotonin uptake (Ki = 0.87 μmol/l) and enhancing serotonin release [28,29]. Both enantiomers act synergistically to enhance tramadol efficacy. These properties of tramadol may also contribute toward an increased risk of serotonin toxicity with concomitant use of serotoninergic drugs such as serotonin reuptake inhibitors (SSRIs) (e.g. fluoxetine, paroxetine, sertraline). SSRIs can not only inhibit tramadol metabolism by inhibiting CYP2D6 but also increase serotonin levels in the central nervous system, an effect also mediated by tramadol. Therefore, concomitant use of an SSRI and tramadol can lead to serotonin syndrome, and should be avoided [1,13,14,30]. The FDA-approved tramadol drug label now states: ‘In vitro drug interaction studies in human liver microsomes indicates that inhibitors of CYP2D6 such as fluoxetine and its metabolite norfluoxetine, amitriptyline, and quinidine inhibit the metabolism of tramadol to various degrees. The full pharmacological impact of these alterations in terms of either efficacy or safety is unknown. Concomitant use of SEROTONIN re-uptake INHIBITORS and MAO INHIBITORS may enhance the risk of adverse events, including seizures (see WARNINGS) and serotonin syndrome’ (from TRAMADOL HCL – tramadol hydrochloride tablet Package Insert, http://dailymed.nlm.nih.gov/dailymed/lookup.cfm?setid=ae7c54b1-b440-4cca-97e8-e5b825413d32).
Pharmacokinetics
Metabolism
After oral administration, tramadol is rapidly and almost completely absorbed [6]. The mean bioavailability of tramadol after a single oral dose is around 68% and increased to above 90% after multiple doses [3,31]. In healthy adult volunteers administered a 100 mg oral single dose of tramadol, the maximum plasma concentration (Cmax) was 308 μg/l and was reached in 1.6 h (tmax). The renal clearance was at 6.6 l/h [3]. Tramadol is extensively metabolized in the liver by O- and N-desmethylation and by conjugation reactions to form glucuronides and sulfate metabolites. The O-desmethylation of tramadol to its main active metabolite, O-desmethyltramadol (M1), is catalyzed by cytochrome P450 (CYP) 2D6 [19,20,32] (Fig. 1). Enantioselective analysis of tramadol and M1 showed (−)-M1 levels being generally higher than (+)-M1 levels [33,34]. The N-desmethylation to N-desmethyltramadol (M2, not pharmacologically active) is catalyzed by CYP2B6 and CYP3A4 [19]. M1 and M2 may then be further degraded to secondary metabolites, N,N-didesmethyltramadol (M3) and N, O-didesmethyltramadol (M5), and then to N,N, O-tridesmethyltramadol (M4) [19]. Among all its metabolites, only O-desmethyltramadol (M1) and to a lesser extent N,O-didesmethyltramadol (M5), are pharmacologically active. Following bioactivation in the liver, M1 is released into the blood, enters the central nervous system, and activates μ-opioid receptors. In the phase II metabolism, M1 is inactivated by glucuronidation in the liver, mostly through UGT2B7 and UGT1A8 [35]. The major route of excretion for tramadol and its metabolites is through the kidneys, with about 30% of the dose excreted in the urine as the parent drug and the rest excreted as metabolites. Capillary electrophoresis assay showed that in healthy volunteers administered 150 mg oral tramadol, about 11.4% of the dose was excreted as (−)-tramadol, 16.4% as (+)-tramadol, and 23.7% as O-desmethyltramadol glucuronide in the urine [36]. This study also observed a four-fold higher excretion of (−)-O-desmethyltramadol glucuronide than the (+)-O-desmethyltramadol glucuronide.
As tramadol metabolism is primarily mediated through CYP2D6, CYP3A4, and CYP2B6, drugs that inhibit CYP2D6 or induce or inhibit CYP3A4 and CYP2B6 function should be used with caution in patients taking tramadol because of a potential risk of drug interaction [37,38]. For example, carbamazepine, a potent inducer of CYP3A4, may increase tramadol metabolism through CYP3A4 to inactive metabolites and therefore reduce its analgesic effect [39] (TRAMADOL HCL tablet Package Insert, http://dailymed.nlm.nih.gov/dailymed/lookup.cfm?setid=ae7c54b1-b440-4cca-97e8-e5b825413d32). Paroxetine, a potent CYP2D6 inhibitor, has been shown to significantly inhibit the metabolism of tramadol to its active metabolite M1 and reduces, but does not abolish the analgesic effect of tramadol [40]. Escitalopram, a weak inhibitor of CYP2D6, inhibited O-desmethylation of (+)-tramadol, but did not reduce the hypoalgesic effect in experimental pain [41].
Transport
Drug transporters play important roles in drug absorption, distribution, and excretion and may affect the analgesic and tolerability profile of tramadol. The bidirectional transport of racemic tramadol and its metabolites have been investigated in a few in vitro and clinical studies [5,42,43]. Tramadol and its metabolites are not substrates of the P-glycoprotein (P-gp) (ABCB1) in vitro [5]. More recently, Tzvetkov et al. [43] reported that the hepatic reuptake of M1, but not of tramadol, is mediated by SLC22A1 (OCT1). The claim is supported both by in-vitro and by clinical data. SLC22A1 (OCT1) is a polyspecific organic cation transporter that is strongly expressed in the sinusoidal membranes of the human liver. The data of Tzvetkov and colleagues suggest that after M1 is produced and excreted from the liver, it may be taken back up by OCT1 (Fig. 1). Therefore, OCT1 may affect the plasma concentrations of M1 and thus affect its opioidergic efficacy. The authors also found that tramadol is an inhibitor of OCT1. However, the inhibition potency was rather low and clinically relevant drug–drug interactions on the basis of inhibition of OCT1 by tramadol are not very probable.
Pharmacogenomics
There is considerable variability in the pharmacokinetic and pharmacodynamic of tramadol depending on the genetic background [2,44]. This has been partly ascribed to the CYP2D6 polymorphisms as CYP2D6 plays a critical role in generating the M1 metabolite that contributes to the major opioid analgesic effect. Genetic variations of CYP2D6 have been shown to affect not only the pharmacokinetics of tramadol and M1 but also the analgesic efficacy in volunteer and patient studies as well as pharmacodynamic responses [18,21,23–25,45]. In addition to CYP2D6, a few other studies have explored the role of drug transporters and pharmacological targets in tramadol efficacy or toxicity [42,43,46].
Metabolizing enzyme variants
Tramadol is metabolized primarily by CYP2D6, a phase I metabolizing enzyme responsible for the activation or clearance of about 25% of all marked drugs. CYP2D6 is highly polymorphic, with more than 100 alleles defined by the Cytochrome P450 Nomenclature Committee (http://www.cypalleles.ki.se/cyp2d6.htm). CYP2D6 activity varies considerably within a population, which leads to distinct phenotypes. The CYP2D6 phenotype can be classified according to the metabolizer status into ultra-rapid metabolizers (UMs), extensive metabolizers (EMs), intermediate metabolizers (IMs), and poor metabolizers (PMs). The EMs carry two active, the IMs one inactive and one reduced activity, and the PMs two inactive CYP2D6 alleles. The UMs carry at least three active CYP2D6 alleles because of gene duplication/multiplication [47–49]. The following CYP2D6 alleles are considered active: *1, *2, *27, *33, *35, *45,*46, *39, *48, *53. The alleles *3, *4, *5, *6, *7, *8, *11,*12, *13, *14, *15, *16, *18, *19, *20, *21, *31, *36, *38, *40, *42, *44, *47, *51, *56,*57, *62 are considered inactive or nonfunctional. The alleles *9, *10, *17, *29, *41, *49, *50, *54, *55, *59, *69, *72 are considered to have reduced function or decreased activity [50]. CYP2D6 plays a pivotal role in the pharmacokinetics and analgesic efficacy of tramadol. Several reduced or none functional CYP2D6 alleles as well as alleles with multiple gene copies have significant impacts on clinical outcome in patients under tramadol medication [22,24,27,33,45,51–55]. Pharmacokinetic studies have shown that the impact of CYP2D6 phenotypes on tramadol pharmacokinetics was similar after single oral, multiple oral, or intravenous administrations [33,52,55]. CYP2D6 PMs had increased exposure to tramadol, but markedly reduced M1 formation when compared with individuals with normal CYP2D6 activity (EMs) [22,27,33,52–55]. PMs tend to experience significantly reduced analgesic effects: in a prospective study enrolling 300 postoperative patients, the percentage of nonresponders was significantly higher in the PM group (46.7%) compared with the EM group (21.6%; P =0.005) [25]. Tramadol loading doses were also higher in PMs and more patients required rescue medication for pain relief after surgery. In another study including 174 patients receiving intravenous tramadol for postoperative analgesia, comparable findings were reported [33]: the nonresponse rates to tramadol treatment increased four-fold compared with the other genotypes (P <0.001) and the concentration of the (+)-M1 metabolite was markedly reduced in PM versus EM or UM patients. In healthy Chinese individuals, the reduced function allele CYP2D6*10 (a very common allele in Asians with allele frequency of 52.4%) was associated with altered pharmacokinetics of tramadol, but CYP2D6*2 had no effect [56]. Chinese volunteers carrying the CYP2D6*10/*10 allele had significantly reduced tramadol clearance as compared with patients carrying the CYP2D6*1/*1, CYP2D6*2/*2 alleles, and CYP2D6*2/*10 (P <0.01). Another study with Chinese gastric cancer patients recovering from gastrectomy showed that patients with a homozygous CYP2D6*10 allele had significantly higher tramadol total consumption than patients without the CYP2D6*10 or heterozygous for CYP2D6*10 [57]. In contrast to PM patients who have reduced CYP2D6 function and decreased tramadol efficacy, individuals who are CYP2D6 UMs tend to have increased sensitivity to tramadol and also an increased risk for adverse events. Maximum plasma concentrations of the active metabolite M1 were significantly higher in the UM group compared with the EM group in a pharmacokinetic study in healthy male volunteers [24]. Pain thresholds and pain tolerance were increased and a more pronounced meiosis was reported in UMs under tramadol medication as compared with EMs. However, UMs also seem to be specifically prone to opioid/tramadol-related side effects when compared with those with normal CYP2D6 activity, probably because of the overall higher (+)-M1 concentrations. Almost 50% of the UM group experienced nausea compared with only 9% of the EM group [24]. Severe life-threatening adverse events were reported in two case reports. One patient with renal impairment carrying the CYP2D6 gene duplication experienced respiratory depression [45] and another UM developed a near-fatal cardiotoxicity, in both cases because of tramadol use [51].
On the basis of the data described above, the US FDA-approved drug label now contains information on metabolism of tramadol by CYP2D6: ‘CYP2D6 poor metabolizers may have higher tramadol concentrations, and concomitant use of CYP2D6 or CYP3A4 inhibitors such as fluoxetine, paroxetine and quinidine could result in significant drug interactions’. The Royal Dutch Pharmacists Association – Pharmacogenetics Working Group (DPWG) has also evaluated therapeutic dose recommendations for tramadol on the basis of CYP2D6 genotypes [58]. For CYP2D6 PM and IM genotypes, they recommend selecting an alternative drug (not oxycodone or codeine) and/or being extra alert to symptoms of insufficient pain relief. For UM genotypes, they recommend using a 30% decreased dose and being alert to an increased risk of ADRs (e.g. nausea, vomiting, constipation, respiratory depression, confusion, urinary retention) or select an alternative drug [e.g. paracetamol (acetaminophen), NSAID, morphine, but not oxycodone or codeine, which also require CYP2D6 for metabolism] [58].
Transporter variants
P-gp (encoded by the ABCB1 gene) is an important drug efflux transporter in the intestine, kidney, liver, and at the blood–brain barrier. P-gp affects the bioavailability, eliminations, and penetration in the brain of many drugs including some opioids. In a study with healthy volunteers, a functional variant in ABCB1, rs1045642 (3435C > T), has been associated with tramadol pharmacokinetics [42]. Individuals carrying the 3455TT geno-type had increased Cmax values of tramadol, but this variant neither affected the tramadol metabolic ratio nor the parent compound’s clearance. However, a later in vitro study using Caco-2 cells showed that ABCB1 did not transport tramadol or its metabolites [5]. A more recent study showed that the hepatic reuptake of the active metabolite M1, but not the parent drug tramadol, is mediated by SLC22A1 (OCT1) [43]. SLC22A1 is highly polymorphic. A number of SLC22A1 polymorphisms have been associated with reduced protein activity as well as with an impact on drug disposition, response, and toxicity [59,60]. Because of the presence of five common loss-of-function polymorphisms, about 9% of Whites lack an active OCT1 allele and a further 41% have only one active allele [60,61]. Tzvetkov and colleagues showed that individuals carrying none or only one active OCT1 allele had significantly higher plasma concentrations of M1 metabolite and longer lasting meiosis, suggesting stronger opioidergic effects. In vitro investigations also supported this association showing that OCT1 overexpression increased M1 uptake 2.4-fold. This increase in uptake could be reversed by OCT1 inhibitors and was absent when the common loss-of-function OCT1 variants were overexpressed [43]. It should be noted that these loss-of-function polymorphisms are much rarer in Africans and are almost missing in Asians [60].
Pharmacodynamic gene variants
Tramadol is a synthetic opioid that acts at the μ-opioid receptor (encoded by gene OPRM1) [6,62]. Opioid receptors belong to the group of G protein-coupled receptors. They bind to endogenous opioids (e.g. dynorphins, enkephalins, endorphins, endomorphins, and nociceptin) as well as to opioid drugs. They are expressed widely in the brain, spinal cord, and digestive tracts. By targeting the μ-opioid receptor, tramadol exerts its analgesic as well as its addictive properties. One of the most widely studied variant in OPRM1 is the A118G base exchange (rs1799971) in exon 1 [63]. The A to G substitution confers an asparagine to aspartate exchange and results in reduced OPRM1 mRNA and protein levels [63]. This variant is considerably more common in Asians than in Whites, with a minor allele frequency of 24–45% in Asians and around 10–17% in Whites [64,65]. This variant has been linked to response to many opioids, including morphine, fentanyl, alfentanil as well as tramadol (reviewed by Stamer and colleagues [64,66]). However, the results have been controversial, with both positive and negative associations reported for this SNP on various opioid phenotypes, for example, dose requirement, analgesic efficacy, and side effects [65]. In a study of 160 Korean patients treated with tramadol, the A118G variant has been associated with ADRs [46]. Korean patients carrying the GG genotype were less likely to develop nausea/vomiting (6.3-fold lower odds) when treated with tramadol as compared with patients with the AA genotype, suggesting that a high risk of nausea/vomiting may be linked to high levels of μ-opioid receptor expression. However, this effect has not been replicated and a recent meta-analysis including 14 studies and 3446 patients on opioid therapies showed very small clinical effects for the A118G variant [65]. Although the metastudy involves many opioids (not tramadol specific), the overall results suggest that the clinical relevance of the OPRM1 A118G variant as a solitary genetic marker is limited, but this variant may be considered a part of the complex genotypes along with other genetic and non-genetic factors underlying pain and analgesia [65,67].
Summary
The genetic factors underlying the variation in tramadol response are complex, and likely involve multiple genes involved in the metabolism, transport, and response to the drug, as well as receptors that are implicated in patients’ pain perception and modulation. CYP2D6 plays a critical role in the formation of active metabolites from tramadol. The associations between genetic polymorphisms of CYP2D6 and therapeutic failure or adverse events to tramadol have been reported in multiple case reports and clinical studies. However, the clinical utility of CYP2D6 testing for tramadol dosing remains to be established. Genetic variations in the drug transporters have also been implicated in tramadol efficacy and toxicity. Loss-of-function genetic polymorphisms in the liver uptake transporter OCT1 were recently suggested to affect the hepatic uptake of M1, the active metabolite of tramadol. Although the effects were found both in vitro and in vivo, independent validations of the human findings are required to consider OCT1 polymorphisms in the treatment guidelines. Most of the existing pharmacogenetic studies of tramadol response enrolled a limited number of patients and reported a limited number of adverse events. There is a need for larger, well-designed studies that consider both pharmacokinetic and pharmacodynamic aspects and investigate possible candidate genes to improve the robustness of the reported associations and to identify novel genetic markers. Prospective trials that show improvement in clinical outcomes will be required to establish the value of genetic testing in pain therapy.
Acknowledgments
The authors thank Fen Liu for assistance with the graphics. PharmGKB is supported by the NIH/NIGMS (U01GM61374).
Footnotes
Conflicts of interest
There are no conflicts of interest.
References
- 1.Reeves RR, Burke RS. Tramadol: basic pharmacology and emerging concepts. Drugs Today (Barc) 2008;44:827–836. doi: 10.1358/dot.2008.44.11.1289441. [DOI] [PubMed] [Google Scholar]
- 2.Leppert W. Tramadol as an analgesic for mild to moderate cancer pain. Pharmacol Rep. 2009;61:978–992. doi: 10.1016/s1734-1140(09)70159-8. [DOI] [PubMed] [Google Scholar]
- 3.Scott LJ, Perry CM. Tramadol: a review of its use in perioperative pain. Drugs. 2000;60:139–176. doi: 10.2165/00003495-200060010-00008. [DOI] [PubMed] [Google Scholar]
- 4.Beaulieu AD, Peloso PM, Haraoui B, Bensen W, Thomson G, Wade J, et al. Once-daily, controlled-release tramadol and sustained-release diclofenac relieve chronic pain due to osteoarthritis: a randomized controlled trial. Pain Res Manag. 2008;13:103–110. doi: 10.1155/2008/903784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kanaan M, Daali Y, Dayer P, Desmeules J. Uptake/efflux transport of tramadol enantiomers and O-desmethyltramadol: focus on P-glycoprotein. Basic Clin Pharmacol Toxicol. 2009;105:199–206. doi: 10.1111/j.1742-7843.2009.00428.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grond S, Sablotzki A. Clinical pharmacology of tramadol. Clin Pharmacokinet. 2004;43:879–923. doi: 10.2165/00003088-200443130-00004. [DOI] [PubMed] [Google Scholar]
- 7.Houmes RJ, Voets MA, Verkaaik A, Erdmann W, Lachmann B. Efficacy and safety of tramadol versus morphine for moderate and severe postoperative pain with special regard to respiratory depression. Anesth Analg. 1992;74:510–514. doi: 10.1213/00000539-199204000-00007. [DOI] [PubMed] [Google Scholar]
- 8.Preston KL, Jasinski DR, Testa M. Abuse potential and pharmacological comparison of tramadol and morphine. Drug Alcohol Depend. 1991;27:7–17. doi: 10.1016/0376-8716(91)90081-9. [DOI] [PubMed] [Google Scholar]
- 9.Lanier RK, Lofwall MR, Mintzer MZ, Bigelow GE, Strain EC. Physical dependence potential of daily tramadol dosing in humans. Psychopharmacology (Berl) 2010;211:457–466. doi: 10.1007/s00213-010-1919-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tjäderborn M, Jönsson AK, Ahlner J, Hägg S. Tramadol dependence: a survey of spontaneously reported cases in Sweden. Pharmacoepidemiol Drug Saf. 2009;18:1192–1198. doi: 10.1002/pds.1838. [DOI] [PubMed] [Google Scholar]
- 11.Rodriguez Villamañan JC, Albaladejo Blanco C, Sanchez Sanchez A, Carvajal A, Martin Arias L, Garcia del Pozo J. Withdrawal syndrome after long-term treatment with tramadol. Br J Gen Pract. 2000;50:406. [PMC free article] [PubMed] [Google Scholar]
- 12.Radbruch L, Glaeske G, Grond S, Münchberg F, Scherbaum N, Storz E, et al. Topical review on the abuse and misuse potential of tramadol and tilidine in Germany. Subst Abus. 2013;34:313–320. doi: 10.1080/08897077.2012.735216. [DOI] [PubMed] [Google Scholar]
- 13.Wilder-Smith CH, Schimke J, Osterwalder B, Senn HJ. Oral tramadol, a mu-opioid agonist and monoamine reuptake-blocker, and morphine for strong cancer-related pain. Ann Oncol. 1994;5:141–146. doi: 10.1093/oxfordjournals.annonc.a058765. [DOI] [PubMed] [Google Scholar]
- 14.Grond S, Radbruch L, Meuser T, Loick G, Sabatowski R, Lehmann KA. High-dose tramadol in comparison to low-dose morphine for cancer pain relief. J Pain Symptom Manage. 1999;18:174–179. doi: 10.1016/s0885-3924(99)00060-3. [DOI] [PubMed] [Google Scholar]
- 15.Vickers MD, Paravicini D. Comparison of tramadol with morphine for postoperative pain following abdominal surgery. Eur J Anaesthesiol. 1995;12:265–271. [PubMed] [Google Scholar]
- 16.Stamer UM, Maier C, Grond S, Veh-Schmidt B, Klaschik E, Lehmann KA. Tramadol in the management of post-operative pain: a double-blind, placebo- and active drug-controlled study. Eur J Anaesthesiol. 1997;14:646–654. doi: 10.1046/j.1365-2346.1994.00214.x. [DOI] [PubMed] [Google Scholar]
- 17.Bekjarovski N, Chaparoska D, Radulovikj-Bekjarovska S. Seizures after use and abuse of tramadol. Prilozi. 2012;33:313–318. [PubMed] [Google Scholar]
- 18.Gan SH, Ismail R, Wan Adnan WA, Zulmi W. Impact of CYP2D6 genetic polymorphism on tramadol pharmacokinetics and pharmacodynamics. Mol Diagn Ther. 2007;11:171–181. doi: 10.1007/BF03256239. [DOI] [PubMed] [Google Scholar]
- 19.Subrahmanyam V, Renwick AB, Walters DG, Young PJ, Price RJ, Tonelli AP, Lake BG. Identification of cytochrome P-450 isoforms responsible for cistramadol metabolism in human liver microsomes. Drug Metab Dispos. 2001;29:1146–1155. [PubMed] [Google Scholar]
- 20.Paar WD, Poche S, Gerloff J, Dengler HJ. Polymorphic CYP2D6 mediates O-demethylation of the opioid analgesic tramadol. Eur J Clin Pharmacol. 1997;53:235–239. doi: 10.1007/s002280050368. [DOI] [PubMed] [Google Scholar]
- 21.Stamer UM, Stüber F. Codeine and tramadol analgesic efficacy and respiratory effects are influenced by CYP2D6 genotype. Anaesthesia. 2007;62:1294–1295. doi: 10.1111/j.1365-2044.2007.05360_1.x. author reply 1295–1296. [DOI] [PubMed] [Google Scholar]
- 22.Enggaard TP, Poulsen L, Arendt-Nielsen L, Brosen K, Ossig J, Sindrup SH. The analgesic effect of tramadol after intravenous injection in healthy volunteers in relation to CYP2D6. Anesth Analg. 2006;102:146–150. doi: 10.1213/01.ane.0000189613.61910.32. [DOI] [PubMed] [Google Scholar]
- 23.Halling J, Weihe P, Brosen K. CYP2D6 polymorphism in relation to tramadol metabolism: a study of Faroese patients. Ther Drug Monit. 2008;30:271–275. doi: 10.1097/FTD.0b013e3181666b2f. [DOI] [PubMed] [Google Scholar]
- 24.Kirchheiner J, Keulen JT, Bauer S, Roots I, Brockmöller J. Effects of the CYP2D6 gene duplication on the pharmacokinetics and pharmacodynamics of tramadol. J Clin Psychopharmacol. 2008;28:78–83. doi: 10.1097/JCP.0b013e318160f827. [DOI] [PubMed] [Google Scholar]
- 25.Stamer UM, Lehnen K, Höthker F, Bayerer B, Wolf S, Hoeft A, Stuber F. Impact of CYP2D6 genotype on postoperative tramadol analgesia. Pain. 2003;105:231–238. doi: 10.1016/s0304-3959(03)00212-4. [DOI] [PubMed] [Google Scholar]
- 26.Gillen C, Haurand M, Kobelt DJ, Wnendt S. Affinity, potency and efficacy of tramadol and its metabolites at the cloned human mu-opioid receptor. Naunyn Schmiedebergs Arch Pharmacol. 2000;362:116–121. doi: 10.1007/s002100000266. [DOI] [PubMed] [Google Scholar]
- 27.Poulsen L, Arendt-Nielsen L, Brøsen K, Sindrup SH. The hypoalgesic effect of tramadol in relation to CYP2D6. Clin Pharmacol Ther. 1996;60:636–644. doi: 10.1016/S0009-9236(96)90211-8. [DOI] [PubMed] [Google Scholar]
- 28.Raffa RB, Buschmann H, Christoph T, Eichenbaum G, Englberger W, Flores CM, et al. Mechanistic and functional differentiation of tapentadol and tramadol. Expert Opin Pharmacother. 2012;13:1437–1449. doi: 10.1517/14656566.2012.696097. [DOI] [PubMed] [Google Scholar]
- 29.Raffa RB, Friderichs E, Reimann W, Shank RP, Codd EE, Vaught JL, et al. Complementary and synergistic antinociceptive interaction between the enantiomers of tramadol. J Pharmacol Exp Ther. 1993;267:331–340. [PubMed] [Google Scholar]
- 30.Fox MA, Jensen CL, Murphy DL. Tramadol and another atypical opioid meperidine have exaggerated serotonin syndrome behavioural effects, but decreased analgesic effects, in genetically deficient serotonin transporter (SERT) mice. Int J Neuropsychopharmacol. 2009;12:1055–1065. doi: 10.1017/S146114570900011X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lintz W, Barth H, Osterloh G, Schmidt-Böthelt E. Bioavailability of enteral tramadol formulations. 1st communication: capsules. Arzneimittelforschung. 1986;36:1278–1283. [PubMed] [Google Scholar]
- 32.Paar WD, Frankus P, Dengler HJ. The metabolism of tramadol by human liver microsomes. Clin Investig. 1992;70:708–710. doi: 10.1007/BF00180294. [DOI] [PubMed] [Google Scholar]
- 33.Stamer UM, Musshoff F, Kobilay M, Madea B, Hoeft A, Stuber F. Concentrations of tramadol and O-desmethyltramadol enantiomers in different CYP2D6 genotypes. Clin Pharmacol Ther. 2007;82:41–47. doi: 10.1038/sj.clpt.6100152. [DOI] [PubMed] [Google Scholar]
- 34.Payne KA, Roelofse JA, Shipton EA. Pharmacokinetics of oral tramadol drops for postoperative pain relief in children aged 4 to 7 years – a pilot study. Anesth Prog. 2002;49:109–112. [PMC free article] [PubMed] [Google Scholar]
- 35.Lehtonen P, Sten T, Aitio O, Kurkela M, Vuorensola K, Finel M, Kostiainen R. Glucuronidation of racemic O-desmethyltramadol, the active metabolite of tramadol. Eur J Pharm Sci. 2010;41:523–530. doi: 10.1016/j.ejps.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 36.Soetebeer UB, Schierenberg MO, Schulz H, Andresen P, Blaschke G. Direct chiral assay of tramadol and detection of the phase II metabolite O-demethyl tramadol glucuronide in human urine using capillary electrophoresis with laser-induced native fluorescence detection. J Chromatogr B Biomed Sci Appl. 2001;765:3–13. doi: 10.1016/s0378-4347(01)00366-8. [DOI] [PubMed] [Google Scholar]
- 37.Nelson EM, Philbrick AM. Avoiding serotonin syndrome: the nature of the interaction between tramadol and selective serotonin reuptake inhibitors. Ann Pharmacother. 2012;46:1712–1716. doi: 10.1345/aph.1Q748. [DOI] [PubMed] [Google Scholar]
- 38.Reus VI, Rawitscher L. Possible interaction of tramadol and antidepressants. Am J Psychiatry. 2000;157:839. doi: 10.1176/appi.ajp.157.5.839. [DOI] [PubMed] [Google Scholar]
- 39.Klotz U. Tramadol – the impact of its pharmacokinetic and pharmacodynamic properties on the clinical management of pain. Arzneimittelforschung. 2003;53:681–687. doi: 10.1055/s-0031-1299812. [DOI] [PubMed] [Google Scholar]
- 40.Laugesen S, Enggaard TP, Pedersen RS, Sindrup SH, Brøsen K. Paroxetine, a cytochrome P450 2D6 inhibitor, diminishes the stereoselective O-demethylation and reduces the hypoalgesic effect of tramadol. Clin Pharmacol Ther. 2005;77:312–323. doi: 10.1016/j.clpt.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 41.Noehr-Jensen L, Zwisler ST, Larsen F, Sindrup SH, Damkier P, Brosen K. Escitalopram is a weak inhibitor of the CYP2D6-catalyzed O-demethylation of (+)-tramadol but does not reduce the hypoalgesic effect in experimental pain. Clin Pharmacol Ther. 2009;86:626–633. doi: 10.1038/clpt.2009.154. [DOI] [PubMed] [Google Scholar]
- 42.Slanar O, Nobilis M, Kvétina J, Matousková O, Idle JR, Perlik F. Pharmacokinetics of tramadol is affected by MDR1 polymorphism C3435T. Eur J Clin Pharmacol. 2007;63:419–421. doi: 10.1007/s00228-006-0255-3. [DOI] [PubMed] [Google Scholar]
- 43.Tzvetkov MV, Saadatmand AR, Lötsch J, Tegeder I, Stingl JC, Brockmöller J. Genetically polymorphic OCT1: another piece in the puzzle of the variable pharmacokinetics and pharmacodynamics of the opioidergic drug tramadol. Clin Pharmacol Ther. 2011;90:143–150. doi: 10.1038/clpt.2011.56. [DOI] [PubMed] [Google Scholar]
- 44.Slanar O, Dupal P, Matouskova O, Vondrackova H, Pafko P, Perlik F. Tramadol efficacy in patients with postoperative pain in relation to CYP2D6 and MDR1 polymorphisms. Bratisl Lek Listy. 2012;113:152–155. doi: 10.4149/bll_2012_036. [DOI] [PubMed] [Google Scholar]
- 45.Stamer UM, Stüber F, Muders T, Musshoff F. Respiratory depression with tramadol in a patient with renal impairment and CYP2D6 gene duplication. Anesth Analg. 2008;107:926–929. doi: 10.1213/ane.0b013e31817b796e. [DOI] [PubMed] [Google Scholar]
- 46.Kim E, Choi CB, Kang C, Bae SC Ultracet Study Group. Adverse events in analgesic treatment with tramadol associated with CYP2D6 extensive-metaboliser and OPRM1 high-expression variants. Ann Rheum Dis. 2010;69:1889–1890. doi: 10.1136/ard.2009.124347. [DOI] [PubMed] [Google Scholar]
- 47.Zanger UM, Raimundo S, Eichelbaum M. Cytochrome P450 2D6: overview and update on pharmacology, genetics, biochemistry. Naunyn Schmiedebergs Arch Pharmacol. 2004;369:23–37. doi: 10.1007/s00210-003-0832-2. [DOI] [PubMed] [Google Scholar]
- 48.Gaedigk A, Simon SD, Pearce RE, Bradford LD, Kennedy MJ, Leeder JS. The CYP2D6 activity score: translating genotype information into a qualitative measure of phenotype. Clin Pharmacol Ther. 2008;83:234–242. doi: 10.1038/sj.clpt.6100406. [DOI] [PubMed] [Google Scholar]
- 49.Zhou SF. Polymorphism of human cytochrome P450 2D6 and its clinical significance: part I. Clin Pharmacokinet. 2009;48:689–723. doi: 10.2165/11318030-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 50.Crews KR, Gaedigk A, Dunnenberger HM, Klein TE, Shen DD, Callaghan JT, et al. Clinical Pharmacogenetics Implementation Consortium. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for codeine therapy in the context of cytochrome P450 2D6 (CYP2D6) genotype. Clin Pharmacol Ther. 2012;91:321–326. doi: 10.1038/clpt.2011.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Elkalioubie A, Allorge D, Robriquet L, Wiart JF, Garat A, Broly F, Fourrier F. Near-fatal tramadol cardiotoxicity in a CYP2D6 ultrarapid metabolizer. Eur J Clin Pharmacol. 2011;67:855–858. doi: 10.1007/s00228-011-1080-x. [DOI] [PubMed] [Google Scholar]
- 52.Pedersen RS, Damkier P, Brøsen K. Enantioselective pharmacokinetics of tramadol in CYP2D6 extensive and poor metabolizers. Eur J Clin Pharmacol. 2006;62:513–521. doi: 10.1007/s00228-006-0135-x. [DOI] [PubMed] [Google Scholar]
- 53.Levo A, Koski A, Ojanperä I, Vuori E, Sajantila A. Post-mortem SNP analysis of CYP2D6 gene reveals correlation between genotype and opioid drug (tramadol) metabolite ratios in blood. Forensic Sci Int. 2003;135:9–15. doi: 10.1016/s0379-0738(03)00159-2. [DOI] [PubMed] [Google Scholar]
- 54.Slanar O, Nobilis M, Kvetina J, Idle JR, Perlik F. CYP2D6 polymorphism, tramadol pharmacokinetics and pupillary response. Eur J Clin Pharmacol. 2006;62:75–76. doi: 10.1007/s00228-005-0039-1. author reply 77–78. [DOI] [PubMed] [Google Scholar]
- 55.García-Quetglas E, Azanza JR, Sádaba B, Muñoz MJ, Gil I, Campanero MA. Pharmacokinetics of tramadol enantiomers and their respective phase I metabolites in relation to CYP2D6 phenotype. Pharmacol Res. 2007;55:122–130. doi: 10.1016/j.phrs.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 56.Li Q, Wang R, Guo Y, Wen S, Xu L, Wang S. Relationship of CYP2D6 genetic polymorphisms and the pharmacokinetics of tramadol in Chinese volunteers. J Clin Pharm Ther. 2010;35:239–247. doi: 10.1111/j.1365-2710.2009.01102.x. [DOI] [PubMed] [Google Scholar]
- 57.Wang G, Zhang H, He F, Fang X. Effect of the CYP2D6*10 C188T polymorphism on postoperative tramadol analgesia in a Chinese population. Eur J Clin Pharmacol. 2006;62:927–931. doi: 10.1007/s00228-006-0191-2. [DOI] [PubMed] [Google Scholar]
- 58.Swen JJ, Nijenhuis M, de Boer A, Grandia L, Maitland-van der Zee AH, Mulder H, et al. Pharmacogenetics: from bench to byte–an update of guidelines. Clin Pharmacol Ther. 2011;89:662–673. doi: 10.1038/clpt.2011.34. [DOI] [PubMed] [Google Scholar]
- 59.Kerb R, Brinkmann U, Chatskaia N, Gorbunov D, Gorboulev V, Mornhinweg E, et al. Identification of genetic variations of the human organic cation transporter hOCT1 and their functional consequences. Pharmacogenetics. 2002;12:591–595. doi: 10.1097/00008571-200211000-00002. [DOI] [PubMed] [Google Scholar]
- 60.Shu Y, Leabman MK, Feng B, Mangravite LM, Huang CC, Stryke D, et al. Pharmacogenetics Of Membrane Transporters Investigators. Evolutionary conservation predicts function of variants of the human organic cation transporter, OCT1. Proc Natl Acad Sci USA. 2003;100:5902–5907. doi: 10.1073/pnas.0730858100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tzvetkov MV, Saadatmand AR, Bokelmann K, Meineke I, Kaiser R, Brockmöller J. Effects of OCT1 polymorphisms on the cellular uptake, plasma concentrations and efficacy of the 5-HT(3) antagonists tropisetron and ondansetron. Pharmacogenomics J. 2012;12:22–29. doi: 10.1038/tpj.2010.75. [DOI] [PubMed] [Google Scholar]
- 62.Yanarates O, Dogrul A, Yildirim V, Sahin A, Sizlan A, Seyrek M, et al. Spinal 5-HT7 receptors play an important role in the antinociceptive and antihyperalgesic effects of tramadol and its metabolite, O-desmethyltramadol, via activation of descending serotonergic pathways. Anesthesiology. 2010;112:696–710. doi: 10.1097/ALN.0b013e3181cd7920. [DOI] [PubMed] [Google Scholar]
- 63.Zhang Y, Wang D, Johnson AD, Papp AC, Sadée W. Allelic expression imbalance of human mu opioid receptor (OPRM1) caused by variant A118G. J Biol Chem. 2005;280:32618–32624. doi: 10.1074/jbc.M504942200. [DOI] [PubMed] [Google Scholar]
- 64.Stamer UM, Zhang L, Stüber F. Personalized therapy in pain management: where do we stand? Pharmacogenomics. 2010;11:843–864. doi: 10.2217/pgs.10.47. [DOI] [PubMed] [Google Scholar]
- 65.Walter C, Doehring A, Oertel BG, Lötsch J. μ-opioid receptor gene variant OPRM1 118A > G: a summary of its molecular and clinical consequences for pain. Pharmacogenomics. 2013;14:1915–1925. doi: 10.2217/pgs.13.187. [DOI] [PubMed] [Google Scholar]
- 66.Sadhasivam S, Chidambaran V. Pharmacogenomics of opioids and perioperative pain management. Pharmacogenomics. 2012;13:1719–1740. doi: 10.2217/pgs.12.152. [DOI] [PubMed] [Google Scholar]
- 67.Walter C, Lötsch J. Meta-analysis of the relevance of the OPRM1 118A > G genetic variant for pain treatment. Pain. 2009;146:270–275. doi: 10.1016/j.pain.2009.07.013. [DOI] [PubMed] [Google Scholar]