Abstract

Caveolin-3 (Cav3) is an unconventional membrane protein that serves as a critical scaffolding hub in caveolae and is genetically linked to various muscle disorders. In this work, we report the expression, purification, and characterization of full-length human Cav3. To mimic the palmitoylation of endogenous Cav3, we developed a generally applicable approach to covalently attached thioalkyl chains at natively modified cysteine residues. Nuclear magnetic resonance measurements indicate that lipidation exerts only a modest and local effect on the Cav3 structure, with little impact on the structures of the N-terminal domain, the scaffolding domain, and the extreme C-terminus.
Caveolins are integral membrane proteins that constitute the major scaffolding component of caveolae, which are omega-shaped membrane invaginations enriched in cholesterol, glycosphingolipids, and sphingomyelin.1,2 Mutations in caveolins are known to cause a variety of disorders ranging from cancer to muscular diseases,3−6 and structural studies are needed to gain insight into the molecular etiology of such diseases. There are three proteins in the caveolin family.7−9 The most extensively characterized caveolin is caveolin-1 (Cav1), which features a kinked reentrant integral membrane segment that is flanked by cytoplasmic N- and C-terminal regions.10−17 Significantly less is known about caveolin-3 (Cav3). However, Cav1 and Cav3 have 61% similar sequences and likely adopt a similar topology (Figure 1A, top left). Furthermore, both proteins feature multiple palmitoylation sites18 and form higher-order oligomers under certain conditions.19 In this work, we present the first structural characterization of the monomeric form of full-length human Cav3 and assess the impact of lipid modifications on its structural properties.
Figure 1.

Topology and in vitro lipidation of Cav3. (A) Cartoon depicting the predicted topology of monomeric Cav3 at the membrane, with the positions of the natively palmitoylated cysteine residues indicated (top). Cysteine thiols were first modified by the addition of excess 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB, Ellman’s reagent) (middle). 3-Thio-6-nitrobenzoate groups were then disulfide exchanged with thioalkyl chains in the presence of excess octanethiol (bottom). (B) Representative sodium dodecyl sulfate–polyacrylamide gel electrophoresis gel (Coomassie stain) of purified lipidated Cav3.
Cav3 was expressed in Escherichia coli and purified as described in the Supporting Information, with consistent yields of 2–3 mg of >90% pure protein/L of culture. To identify optimal conditions for solution nuclear magnetic resonance (NMR) studies of Cav3, we assessed the 1H–15N TROSY NMR spectrum of Cav3 in the presence of 10 different detergents commonly used for structural studies of integral membrane proteins. A visible precipitate formed during the preparation of Cav3 in β-n-decyl maltoside (DM) and lauryldimethylamine oxide (data not shown), indicating these detergents are not suitable for structural studies. The 1H–15N TROSY spectra of Cav3 in the other eight detergents varied significantly in quality (Figure S1 of the Supporting Information), confirming that the structure, dynamics, and oligomeric state of Cav3 are sensitive to the properties of the membrane environment. The indole NH side chain peaks of the four tryptophan residues in Cav3 could be resolved only in micelles of lyso-palmitoylphosphatidylcholine (LPPC) or lyso-palmitoylphosphatidylglycerol (LPPG), suggesting that the structural order of Cav3 is maximized and that the protein is monomeric under these conditions. Furthermore, the largest number of amide peaks could be observed in LPPG micelles (106 of the 153 expected peaks), a number slightly greater than the number in LPPC (95 peaks). Therefore, of the conditions evaluated, LPPG micelles provided the most favorable membrane mimetic for Cav3. The optimal pH (6.5) and temperature (318 K) for solution NMR measurements were also identified by 1H–15N TROSY screening.
Cav3 is known to be natively palmitoylated at three positions within its C-terminal domain (C106, C116, and C129), modifications that likely impact Cav3 lipid–membrane interactions, oligomerization, and function (reviewed in ref (2)). To mimic the effects of these modifications, we devised a thiol-mediated approach to artificially lipidate the cysteine residues in Cav3 in vitro. However, to restrict lipidation to the three natively palmitoylated residues (the fifth, sixth, and eighth Cys sites from the N-terminus), it was first necessary to mutate the six natively unmodified cysteine residues in Cav3. We generated a series of conservative mutations at the unmodified cysteine residues. Those predicted to fall within the membrane domain were mutated to alanine (C72A, C92A, and C98A), while cysteines predicted to fall within the soluble domains were mutated to serine (C19S, C124S, and C140S). This mutant is termed 568C-Cav3. To assess the impact of the substitutions, we compared the 1H–15N TROSY spectra of wild-type (WT) Cav3 and 568C-Cav3 in LPPG micelles. Only modest differences in peak positions were apparent in the spectrum of 568C-Cav3 (Figure 2A), which most likely correspond to residues proximal to the mutation sites. Indeed, the majority of the peaks in the two spectra overlap. Furthermore, the far-UV CD spectra of these proteins are quite similar (Figure S2 of the Supporting Information). Together, these data suggest that these substitutions do not perturb the overall fold of the protein under these conditions.
Figure 2.
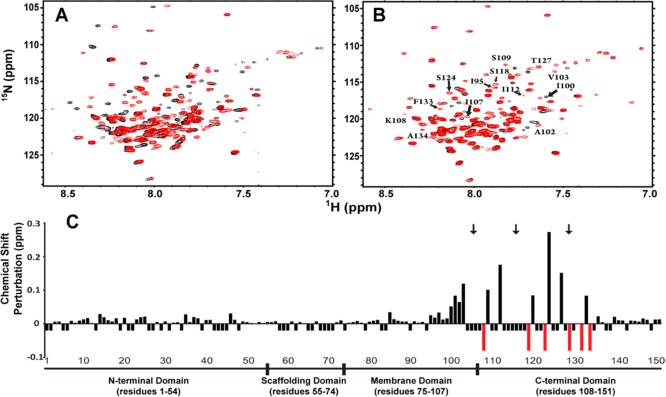
Effects of mutation and lipidation on the structural properties of Cav3. (A) 1H–15N TROSY spectra of wild-type (WT) Cav3 (black) and 568C-Cav3 (red) in 100 mM imidazole (pH 6.5), 1 mM EDTA, 1 mM TCEP, 1.5 mM DSS, and 66 mM (WT) or 112 mM (568C-Cav3) LPPG were recorded on a 900 MHz NMR spectrometer. (B) The 1H–15N TROSY spectrum of lipidated 568C-Cav3 (red) in 100 mM imidazole (pH 6.5), 1 mM EDTA, 1 mM TCEP, 1.5 mM DSS, and 110 mM LPPG was recorded on a 900 MHz NMR spectrometer. Residues influenced by lipidation are indicated. The 1H–15N TROSY spectrum of nonlipidated 568C-Cav3 from panel A is plotted for reference (black). (C) The magnitude of the change in the chemical shift for each assigned amide resonance in the 1H–15N TROSY spectrum of lipidated Cav3 relative to 568C-Cav3 is indicated by the height of the bar. Negative bars indicate the positions for which the change in chemical shift could not be determined because of spectral overlap or missing assignments. Red bars indicate peaks seen only in the spectrum of lipidated 568C-Cav3. Black arrows indicate lipidation sites.
We next introduced alkyl chains into 568C-Cav3 at native palmitoylation sites according to the scheme shown in Figure 1A (details in the Supporting Information). The 1H–15N HSQC spectra of octylated 568C-Cav3 and tetradecylated 568C-Cav3 are nearly indistinguishable (Figure S3 of the Supporting Information), which suggests the length of the acyl chain modification is not a critical factor governing the structural properties of the protein under these conditions. Therefore, we chose to proceed with structural characterization of octylated 568C-Cav3 (hereafter termed lipidated Cav3) because of the relatively high yield of these modifications [>90, 88, and 76% for Cys 129, 106, and 116, respectively (Figure S4 of the Supporting Information)]. While lipidation did not reach 100%, the spectrum predominantly reflects the fully lipidated species because the minor population of incompletely modified protein is dispersed into subpopulations with minor signals (Figure S4 of the Supporting Information). Lipidated Cav3 migrates as a single sodium dodecyl sulfate–polyacrylamide gel electrophoresis band (Figure 1B). Lipidation of 568C-Cav3 resulted in the perturbation of several chemical shifts as well as an improvement in the quality of the NMR spectra with regard to the number of amide peaks (Figure 2B); 118 amide peaks were identified in the spectrum of lipidated protein versus 112 for unmodified 568C-Cav3. These differences were slowly reversed upon reduction of the disulfide bonds using tris(2-carboxyethyl)phosphine (TCEP) (Figure S4 of the Supporting Information), which confirms that the spectral perturbations arise as a direct result of lipidation.
To determine which domains of the protein are perturbed by lipidation, we utilized the amide resonance assignments of the Cav3 spectrum deposited by our lab in the BioMagResBank (BMRB, entry 19903). Figure 2C reveals that the largest chemical shift perturbations caused by lipidation occur near the modified cysteine sites, including the proximal membrane domain. Additionally, the lipidated residue C129 represents one of the six emergent peaks in the spectrum of lipidated 568C-Cav3 (but too broad to detect in nonlipidated samples), suggesting that modification may suppress local conformational dynamics on the microsecond-to-millisecond time scale. Furthermore, residues near the modification sites also exhibited increased transverse 15N relaxation rates (R2) and hetero-NOE values (Figure S5 of the Supporting Information), consistent with locally dampened motions. Together, these findings suggest these modifications likely alter the local conformation of the chain by anchoring the segments of the protein near the modification sites to the micelle environment.
It has previously been suggested that palmitoylation may alter the properties of nearby transmembrane segments, including their tilt angle relative to the membrane.20−22 However, lipidation does not appear to significantly influence the secondary structure (Figure S2 of the Supporting Information) or overall topology of Cav3. Indeed, the structure and dynamics of the entire N-terminal domain, the scaffold domain, and the extreme C-terminus appear to be insensitive to the effects of lipidation (Figure 2C and Figure S5 of the Supporting Information). This suggests that biological functions performed by these regions are unlikely to be conformationally modulated by lipidation, at least not in the absence of other factors associated with native membrane conditions. It is noteworthy that palmitoylation of caveolins has been suggested to enhance oligomerization of Cav1 in native membranes.23 However, lipidation of Cav3 does not induce its oligomerization under the micellar conditions utilized in this work or impact the conformation of the N-terminal segment (residues 34–74), which is known to mediate oligomerization under native conditions.12 Questions for future work include whether these results will be different in actual lipid bilayers and/or in the presence of cholesterol. The possibility that disulfide-linked alkyl chains are imperfect mimics for thioester-linked chains also cannot be ruled out.
The structural impact of Cav3 lipidation appears to be only local and modest. Thus, our results suggest that any effects of lipidation on Cav3 function are unlikely to occur as a result of a large-scale conformational change. This work represents the first structural characterization of a monomeric full-length human caveolin. The expression, purification, and NMR strategies detailed herein will allow a multitude of new structural studies of Cav3, complementing previous work.19 Furthermore, these preliminary solution NMR measurements represent an important first step toward determining the three-dimensional structure of Cav3. Finally, considering that lipid modifications of proteins are of general importance for cellular signaling,24−28 the method reported herein for surrogate lipidation in place of native cysteine palmitoylation should have broad applicability to other lipidated proteins.
Acknowledgments
We thank Prof. Anne Kenworthy for her comments on a preliminary version of the manuscript.
Supporting Information Available
Materials and methods, supporting references, and Figures S1–S5. This material is available free of charge via the Internet at http://pubs.acs.org.
This work was supported by National Institutes of Health (NIH) Grants U54 GM094608 and by RO1 GM106672. The NMR instrumentation used in this work was supported by NIH Grant S10 RR025677-01 and National Science Foundation Grant DBI-0922862.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Brown D. A.; London E. (1998) Annu. Rev. Cell Dev. Biol. 14, 111–136. [DOI] [PubMed] [Google Scholar]
- Parat M.-O. (2009) Int. Rev. Cell Mol. Biol. 273, 117–162. [DOI] [PubMed] [Google Scholar]
- Galbiati F.; Razani B.; Lisanti M. P. (2001) Trends Mol. Med. 7, 435–441. [DOI] [PubMed] [Google Scholar]
- Gazzerro E.; Sotgia F.; Bruno C.; Lisanti M. P.; Minetti C. (2010) Eur. J. Hum. Genet. 18, 137–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams T. M.; Lisanti M. P. (2004) Ann. Med. 36, 584–595. [DOI] [PubMed] [Google Scholar]
- Woodman S. E.; Sotgia F.; Galbiati F.; Minetti C.; Lisanti M. P. (2004) Neurology 62, 538–543. [DOI] [PubMed] [Google Scholar]
- Rothberg K. G.; Heuser J. e.; Donzell W. C.; Ying Y. S.; Glenney J. R.; Anderson R. G. (1992) Cell 68, 673–682. [DOI] [PubMed] [Google Scholar]
- Scherer P. E.; Okamoto T.; Chun M.; Nishimoto I.; Lodish H. F.; Lisanti M. P. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 131–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z.; Scherer P. E.; Okamoto T.; Song K.; Chu C.; Kohtz D. S.; Nishimoto I.; Lodish H. F.; Lisanti M. P. (1996) J. Biol. Chem. 271, 2255–2261. [DOI] [PubMed] [Google Scholar]
- Dupree P.; Parton R. G.; Raposo G.; Kurzchalia T. V.; Simons K. (1993) EMBO J. 12, 1597–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monier S.; Parton R. G.; Vogel F.; Behlke J.; Henske A.; Kurzchalia T. V. (1995) Mol. Biol. Cell 6, 911–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargiacomo M.; Scherer P. E.; Tang Z.; Kubler E.; Song K. S.; Sanders M. C.; Lisanti M. P. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 9407–9411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.; Glover K. J. (2012) Biochim. Biophys. Acta 1818, 1158–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui H.; Root K. T.; Lee J.; Glover K. J.; Im W. (2014) Biophys. J. 106, 1371–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Lan C.; Gallay J.; Vincent M.; Neumann J. M.; de Foresta B.; Jamin N. (2010) Eur. Biophys. J. 39, 307–325. [DOI] [PubMed] [Google Scholar]
- Le Lan C.; Neumann J. M.; Jamin N. (2006) FEBS Lett. 580, 5301–5305. [DOI] [PubMed] [Google Scholar]
- Rieth M. D.; Lee J.; Glover K. J. (2012) Biochemistry 51, 3911–3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzen D. J.; Hastings W. R.; Lublin D. M. (1995) J. Biol. Chem. 270, 6838–6842. [DOI] [PubMed] [Google Scholar]
- Whiteley G.; Collins R. F.; Kitmitto A. (2012) J. Biol. Chem. 287, 40302–40316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrami L.; Kunz B.; Iacovache I.; van der Goot F. G. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 5384–5389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph M.; Nagaraj R. (1995) J. Biol. Chem. 270, 16749–16755. [DOI] [PubMed] [Google Scholar]
- Joseph M.; Nagaraj R. (1995) J. Biol. Chem. 270, 19439–19445. [DOI] [PubMed] [Google Scholar]
- Monier S.; Dietzen D. J.; Hastings W. R.; Lublin D. M.; Kurzchalia T. V. (1996) FEBS Lett. 388, 143–149. [DOI] [PubMed] [Google Scholar]
- Aicart-Ramos C.; Valero R. A.; Rodriguez-Crespo I. (2011) Biochim. Biophys. Acta 1808, 2981–2994. [DOI] [PubMed] [Google Scholar]
- Conibear E.; Davis N. G. (2010) J. Cell Sci. 123, 4007–4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder M.; Deschenes R. (2006) Methods 40, 125–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaskovic S.; Blanc M.; van der Groot F. G. (2013) FEBS J. 280, 2766–2774. [DOI] [PubMed] [Google Scholar]
- Salaun C.; Greaves J.; Chamberlain L. H. (2010) J. Cell Biol. 191, 1229–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.