Abstract
Citrullination, which is catalyzed by protein arginine deiminases (PADs 1–4 and 6), is a post-translational modification (PTM) that effectively neutralizes the positive charge of a guanidinium group by its replacement with a neutral urea. Given the sequence similarity of PAD2 across mammalian species and the genomic organization of the PAD2 gene, PAD2 is predicted to be the ancestral homologue of the PADs. Although PAD2 has long been known to play a role in myelination, it has only recently been linked to other cellular processes, including gene transcription and macrophage extracellular trap formation. For example, PAD2 deiminates histone H3 at R26, and this PTM leads to the increased transcription of more than 200 genes under the control of the estrogen receptor. Given that our understanding of PAD2 biology remains incomplete, we initiated mechanistic studies on this enzyme to aid the development of PAD2-specific inhibitors. Herein, we report that the substrate specificity and calcium dependence of PAD2 are similar to those of PADs 1, 3, and 4. However, unlike those isozymes, PAD2 appears to use a substrate-assisted mechanism of catalysis in which the positively charged substrate guanidinium depresses the pKa of the nucleophilic cysteine. By contrast, PADs 1, 3, and 4 use a reverse-protonation mechanism. These mechanistic differences will aid the development of isozyme-specific inhibitors.
Post-translational modifications (PTMs) are critical for life via their ability to regulate key cellular processes, including chromatin architecture, gene expression, enzyme activity, and protein stability.1 Among the more than 200 known PTMs (e.g., acetylation, phosphorylation, and methylation), citrullination involves the conversion of peptidyl-arginine into peptidyl-citrulline, which effectively neutralizes the positive charge of a guanidinium group by its replacement with a neutral urea.2 The enzymes that catalyze this modification, protein arginine deiminases (PADs 1–4 and 6), are principally found in mammals as well as a single bacterium, and their activity regulates a number of cellular processes, including gene expression, chromatin architecture, autophagy, and neutrophil extracellular trap (NET) formation.3−8
Importantly, chronic NET formation, or NETosis, is a hallmark of diseases including rheumatoid arthritis, lupus, and numerous cancers.9−14 For example, Demers and colleagues showed that in murine models of chronic myelogenous leukemia, breast, and lung cancer, neutrophil populations are sensitized by granulocyte colony stimulating factor (G-CSF) to undergo NETosis.15 As a result, excess DNA and decondensed chromatin prompts the occurrence of deep vein thrombosis, the second leading cause of cancer deaths.15 PAD4 activity is required for this process, as PADI4–/– mice do not form NETs and a pan-PAD inhibitor, Cl-amidine, blocks NET formation in vitro and in vivo.(16,17) The ability to inhibit NETosis provides a partial explanation for why Cl-amidine shows efficacy in several preclinical models, including RA, lupus, and cancer.14,18−21 Beyond NETosis, PAD2 also plays a role in tumorigenesis. For example, PAD2 is highly expressed in luminal breast cancers and helps define a HER2+ gene expression signature in primary invasive tumors.20 Additionally, PAD2 is the fifth most correlated gene with breast cancer recurrence and promotes HER2 expression via its recruitment to the HER2 proximal promoter, establishing a positive feedback loop as HER2 regulates PAD2 expression via PI3K signaling.20 Similar to its effect on ER target genes, the increased expression of HER2 likely reflects the ability of PAD2 to citrullinate histone H3. Further supporting a role for PAD2 in oncogenesis is the fact that Cl-amidine slows the growth of MCF10DCIS xenografts, a model of ductal carcinoma in situ in which PAD2 is highly expressed and promotes cellular growth.20 In total, these data suggest that the PADs, and PAD2 in particular, are interesting therapeutic targets for a range of diseases.
In humans, there are five PADs (PADs 1–4 and 6). These isozymes are highly conserved, with 50–55% overall sequence homology and close to 70% identity within the catalytic domain.2 Early crystallographic studies with human PAD4 showed that the PAD4 monomer consists of three distinct domains: the C-terminal catalytic domain and two immunoglobulin-like domains (IgG1 and IgG2) that are present within the N-terminal half of the protein.22 These structural studies also showed that PAD4 binds to five calcium ions: Ca1 and Ca2 are present in the catalytic domain, whereas the remaining three calciums (Ca3–5) bind in tandem within the IgG2 domain at the border of the catalytic domain.22 Although Ca1 and Ca2 bind within the catalytic domain, they do not participate directly in catalysis. Instead, all five calcium ions are important for triggering a series of conformational changes that move key active site residues into positions that are competent for catalysis.22−24 In previous work with PADs 1, 3, and 4, we showed that calcium increases PAD activity by >10 000-fold and that the enzymes require near millimolar levels of calcium for full activity.23−25 We also demonstrated that these isoforms possess similar, but nonidentical, substrate recognition motifs, and, on the basis of solvent isotope effects, pH rate profiles, and measurements of the pKa of the active site cysteine, we provided evidence that these isozymes use a reverse-protonation mechanism, wherein a fraction of the enzyme exists as the deprotonated thiolate and protonated imidazole and, upon substrate binding, catalysis proceeds via nucleophilic attack by the thiolate.24,25
Given its sequence similarity across mammalian species and the genomic organization of the PAD2 gene, PAD2 is predicted to be the ancestral homologue of the PADs.2 Although PAD2, the focus of this article, has long been known to play a role in myelination,26 it has only recently been linked to other cellular processes, including gene transcription and macrophage extracellular trap formation.7,20,27,28 For example, PAD2 deiminates histone H3 at R26, and this PTM leads to the increased transcription of more than 200 genes under the control of the estrogen receptor.7 Given that our understanding of PAD2 biology remains incomplete, we initiated mechanistic studies on this enzyme to aid the development of PAD2-specific inhibitors. Herein, we report that the substrate specificity and calcium dependence of PAD2 is similar to those of PADs 1, 3, and 4. However, unlike those isozymes, PAD2 appears to use a substrate-assisted mechanism of catalysis in which the positively charged substrate guanidinium promotes catalysis by depressing the pKa of the nucleophilic cysteine.
Experimental Procedures
Chemicals
Dithiothreitol (DTT) was acquired from Bioworld. 4-(2-Hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) was acquired from Research Products International. Tris(2-carboxyethyl) phosphine (TCEP) was acquired from AMRESCO. Ammonium iron(III) sulfate dodecahydrate, diacetyl monooxime (DAMO), and thiosemicarbazide were acquired from Sigma-Aldrich. Nα-Benzoyl-l-arginine methyl ester (BAME) was obtained from MP Biomedicals. Nα-Benzoyl-l-arginine ethyl ester (BAEE) was acquired from Sigma-Aldrich. Nα-Benzoyl-l-arginine amide (BAA) was obtained from Pfaltz & Bauer. Nα-Benzoyl-l-arginine (BA) was obtained from Acros. D2O and H218O (95%) were obtained from Cambridge Isotopes.
Cloning, Expression, and Purification of Wild-Type PAD2
The human PAD2 gene was cloned into the pET16B vector using NdeI/XhoI sites after PCR amplification using the following primers: forward 5′-AAAAAACATATGCTGCGCGAGCGG-3′ and reverse 5′-AAAAAACTCGAGTCAGGGCACCATGTGCCA-3′. The forward primer contains the NdeI restriction site (underlined) and 15 base pairs that correspond to the 5′-coding region of PAD2. The reverse primer contains the XhoI restriction site (underlined) followed by 15 base pairs that correspond to the coding region of the PAD2 gene. The resulting pET16b-PAD2 construct was sequenced to ensure that no mutations were incorporated. The PAD2 expression construct was then transformed into Escherichia coli BL21(DE3)pLysS cells (EMD Biosciences) for protein expression. The PAD2 purification procedure was adapted from previously described methods.29 Briefly, overnight cultures were used to inoculate 4 × 2 L of LB (20 g/L bacto-tryptone, 20 g/L bacto-yeast extract, and 10 g/L NaCl), and these cultures were grown at 37 °C and 250 rpm until the cultures reached an OD600 of 0.8. PAD2 expression was induced by the addition of IPTG (0.4 mM final) and allowed to proceed overnight at 16 °C and 200 rpm. Cells were harvested by centrifugation at 4800g for 10 min at 4 °C. The pellet was resuspended in 70 mL of lysis buffer (20 mM Tris-HCl, pH 8.0, 1% Triton X-100, 500 μM TCEP, 5 mM imidazole, and 500 mM NaCl) and lysed by eight cycles of sonication with an 8 s burst (duty cycle, 100%; output, 10). The lysate was clarified by centrifugation (15 min at 11 900g), and the supernatant was mixed with Ni-NTA–agarose resin (Invitrogen) pre-equilibrated with lysis buffer. Initial binding was carried out for 20 min at 4 °C with gentle stirring. The column was washed with wash buffer 1 (50 mL of 20 mM Tris-HCl, pH 8.0, 10% glycerol, 500 μM TCEP, 20 mM imidazole, and 500 mM NaCl) followed by wash buffer 2 (50 mL of 20 mM Tris-HCl, pH 8.0, 10% glycerol, 500 μM TCEP, 50 mM imidazole, and 500 mM NaCl). Elution buffer (20 mL of 20 mM Tris-HCl, pH 8.0, 10% glycerol, 500 μM TCEP, 250 mM imidazole, and 500 mM NaCl) was then allowed to equilibrate with the resin for 10 min, at which point the eluent was collected.
Fractions from the Ni-NTA column were analyzed by 12% SDS-PAGE and PAD activity assays (see below). The fraction containing pure full-length PAD2 was concentrated using a Centricon concentrator with a 10 kDa nominal molecular mass cutoff. Concentrated protein was then dialyzed (20 mM Tris-HCl, pH 7.6, 1 mM EDTA, 500 mM NaCl, 500 μM TCEP, and 10% glycerol), flash frozen in liquid nitrogen, and stored at −80 °C. PAD2 stored in this manner was stable for several months. Recombinant wild-type PAD2 was obtained in an overall yield of 4 mg/L at ≥95% purity.
H218O Incorporation Studies
The incorporation of 18O into BAEE was determined by performing a PAD2-catalyzed deimination reaction in either normal or 18O-labeled water. For these experiments, PAD2 (500 nM final) was incubated at 37 °C in reaction buffer (5 mM BAEE, 50 mM Tris-HCl, pH 7.6, 10 mM CaCl2, 2 mM DTT, and 50 mM NaCl) made with either normal or 18O-labeled water. For the 18O experiments, the mole percentage of 18O-labeled water was >70% final. After a 3 h incubation, the reaction mixture was analyzed by ESI mass spectrometry using an Agilent Technologies 1220 Infinity LC 6120 quadrupole LC/MS mass spectrometer in the positive ion mode. Formic acid (0.1%) was used as the organic modifier.
Substrate Specificity Studies
Kinetic assays were performed as described previously for PAD4.23 Briefly, this discontinuous colorimetric assay measures the formation of urea-containing compounds (e.g., citrulline, urea, methylurea, etc.). Steady-state kinetic parameters were determined with variable amounts of the substrate in reaction buffer (60 μL final volume). Peptide substrates were dissolved in 50 mM Tris-HCl, pH 7.6. Reaction mixtures were preincubated for 10 min at 37 °C, at which point PAD2 was added to a final concentration of 500 nM to initiate the reaction. Reactions were quenched by flash freezing in liquid nitrogen. For color development, 200 μL of freshly prepared COLDER solution (2.25 M H3PO4, 4.5 M H2SO4, 1.5 mM NH4Fe(SO4), 20 mM diacetyl monoxime, and 1.5 mM thiosemicarbazide) was added to the quenched reaction, and the mixture was vortexed to ensure complete mixing and then incubated for 30 min at 95 °C. The absorbance at 540 nm was then measured and compared to a citrulline standard curve to determine the concentration of citrulline produced during the reaction. All kinetic studies were performed in the linear range of PAD2 activity with respect to time and enzyme concentration. Assays were performed in duplicate. The initial rates were fit to eq 1
| 1 |
using the GraFit version 5.0.1.1 software package.30
Calcium Dependence Studies
Varying concentrations of calcium (0–10 mM) were incubated in calcium-free reaction buffer (50 mM NaCl, 2 mM DTT, 10 mM BAEE, and 100 mM Tris-HCl, pH 7.6). Reactions were preincubated at 37 °C for 10 min before the addition of PAD2 (500 nM final). The reactions were allowed to proceed for 10 min and then flash frozen in liquid nitrogen. Citrulline production was determined as described above in duplicate, and the data were fit to eq 2
| 2 |
where KD is the dissociation constant and n is the Hill coefficient. For these assays, PAD2 was dialyzed into EDTA-free long-term storage buffer (20 mM Tris-HCl, pH 7.6, 500 μM TCEP, 500 mM NaCl, and 10% glycerol).
pH Dependence Studies
pH profiles for PAD2 were generated by measuring the steady-state kinetic parameters for the deimination of BAEE between pH 6.5–9.8. Stock concentrations of BAEE were prepared in 50 mM buffer at the desired pH. Reaction mixtures containing 50 mM NaCl, 2 mM DTT, 100 mM buffer (Bis-tris, pH 6.5–7, or Tris-HCl, pH 7–8, and HEPES, 8–9.8), 10 mM CaCl2, and BAEE at various concentrations (0–10 mM in a final volume of 60 μL) were preincubated for 10 min prior to the addition of PAD2. These assays were performed in duplicate. The initial rates obtained from these experiments were fit to eq 1. The kcat and kcat/Km values obtained from this analysis were plotted as a function of pH and fit to eq 3
| 3 |
where ymax is the amount of activity at the pH optimum.
Solvent Isotope Effects
Solvent isotope effects (SIE) were measured in reaction buffer containing 100 mM Bis-Tris (pL 5.0–7.0), 100 mM Tris-HCl (pL 7.0–9.0), or 100 mM HEPES (pL 9.0–9.5) as well as 10 mM CaCl2, 2 mM DTT, 50 mM NaCl, and 500 nM PAD2 with various concentrations of BAEE (0–10 mM). The pL values were determined using the correction pL = pH + 0.4, and the final concentration of D2O was >96%. The kinetic parameters were determined using the methods described above.
Inactivation Studies
Inactivation reactions containing 10 mM CaCl2 and 100 mM of buffer (pH 6.5–9.0) were incubated with 5.0 μM PAD2 at 37 °C for 10 min before adding either iodoacetamide or 2-chloroacetamidine (dissolved in 50 mM buffer) to initiate the reaction (60 μL final volume). At various time points (0–30 min), an aliquot (6 μL) was removed and added to assay buffer, which was preincubated for 10 min at 37 °C to measure residual PAD2 activity (60 μL total volume). Reactions were allowed to proceed for 15 min before being flash frozen in liquid nitrogen. Citrulline production was measured according to the methodology described above, and the residual activity data were fit to eq 4.
| 4 |
where v is the velocity, v0 is the initial velocity, k is the pseudo-first-order rate constant for inactivation, and t is time. In the absence of inactivator saturation in the v versus [I] plot, the second-order rate constants of enzyme inactivation, i.e., kinact/KI, were determined by fitting the data to eq 5
| 5 |
where kinact is the maximal rate of inactivation, KI is the concentration of inactivator that yields half-maximal inactivation, and [I] is the concentration of inactivator. When inactivator saturation was observed, the data were fit to eq 6
| 6 |
The kinact/KI values thus obtained were then plotted versus pH and subsequently fit to eq 7
| 7 |
where ymin is the minimum rate and ymax is the maximum rate of inactivation.
Results
Calcium Dependence
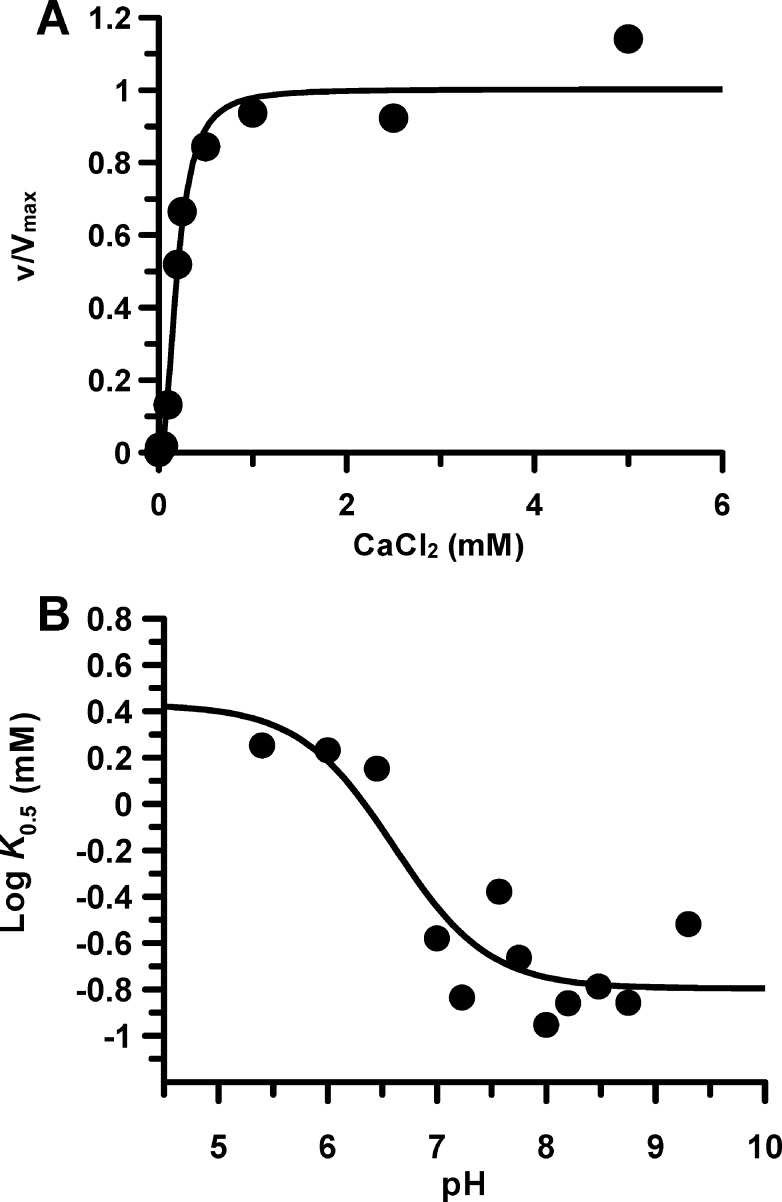
To gain insights into the calcium dependence of PAD2, we determined the concentration of calcium that is required for half-maximal activity, i.e., K0.5. At the pH optimum (pH 7.6), the K0.5 is 200 ± 18.9 μM with a Hill coefficient of 2.2 ± 0.5 (Figure 1A). These values are similar to those obtained for PAD4, and the fact that the Hill coefficient is >2 indicates that at least three calcium ions are required to activate the enzyme. To further explore the calcium dependence of PAD2, we measured the K0.5 over a range of pH values (pH 5.5–9.5). The results of these studies indicate that there is a monotonic decrease in K0.5 as the pH increases (pKa = 6.6 ± 0.3), which is likely caused by deprotonation of one or more acidic residues (Asp and Glu) that are required for calcium binding (Figure 1B). This result differs substantially from PAD4, where the K0.5 increases at pH values > 7.6. The reason for this difference is unclear but may reflect differences in the catalytic mechanism or the mechanism of enzyme activation.
Figure 1.
Calcium dependence of PAD2 catalysis. (A) Calcium dependence of PAD2 (500 nM final) measured at the pH optimum using reaction buffer with varying concentrations of calcium (0–5 mM). (B) K0.5, the concentration of calcium that yields half-maximal activity, plotted on a logarithmic scale against pH.
Substrate Specificity Studies
To initially characterize the substrate specificity of PAD2, we evaluated a series of small molecule substrate mimetics (e.g., BAEE, BAA, BAME, and BA) as well as the free amino acid, l-arginine, and agmatine. The latter two compounds were tested because they are processed by other members of the guanindinum-modifying superfamily of enzymes.31,32 Consistent with previous results for PADs 1, 3 and 4, the deimination rates for l-arginine and agmatine were nearly undetectable and close to background levels (Table 1). By contrast, the benzoylated arginine series was efficiently processed, indicating that an N-terminal amide is critical and sufficient for substrate recognition. Among this series, PAD2 selectively deiminates BAEE with a kcat/Km value of 11 700 M–1 s–1. By comparison, BAME, BAA, and BA were significantly poorer substrates; the kcat/Km values are 1800, 680, and 390 M–1 s–1, respectively. Although the Km values for these substrates are generally similar in value (BAEE, 270 ± 60; BAME, 240 ± 40; BAA, 480 ± 70; BA, 1600 ± 100 μM), the kcat values are reduced 5–10-fold (Table 1). In the case of BAEE, this may suggest that the guanidinium is better positioned for nucleophilic attack by Cys647, the active site nucleophile. The greater than 10-fold preference for BAEE over BAA also likely explains why Cl-amidine, whose structure is based on the BAA scaffold, preferentially (∼10-fold) inhibits PAD4 over PAD2.
Table 1. Kinetic Parameters of Potential PAD2 Substrates.
| substrate | Km (μM) | kcat (s–1) | kcat/Km (M–1 s–1) |
|---|---|---|---|
| l-arginine | NDb | NDb | <9a |
| agmatine | NDb | NDb | <9a |
| BAEE | 270 ± 60 | 3.2 ± 0.1 | 11 700 |
| BAME | 240 ± 40 | 0.43 ± 0.02 | 1800 |
| BAA | 480 ± 70 | 0.32 ± 0.01 | 680 |
| BA | 1600 ± 100 | 0.63 ± 0.02 | 390 |
| histone H3 | NDb | NDb | 1200a |
| histone H4 | NDb | NDb | 2400a |
| AcH4–5 | 850 ± 300 | 0.22 ± 0.05 | 290 |
| AcH4–15 | 1000 ± 70 | 1.4 ± 0.1 | 1400 |
| AcH4–15R3MMA | NDb | NDb | <2a |
| AcH4–21 | 710 ± 100 | 0.72 ± 0.08 | 1000 |
Estimated using the equation v = kcat/Km([Et][S]).
ND, not determined.
To probe more physiologically relevant substrates, we next tested histones H3 and H4 and showed that both proteins were deiminated with comparable efficiency to that of BAME. Because histone H4 was a slightly better substrate, we also evaluated a small series of histone tail analogues (the sequences of these peptides are provided in Table S1). Notably, these peptides were generally deiminated with comparable efficiency to that of histone H4 (Table 1). The one exception is the AcH4–5 peptide, which showed a 3-fold decrease in kcat/Km, which suggests that longer-range interactions may be important for substrate recognition. To determine whether PAD2 could catalyze the hydrolysis of a methylated arginine residue, monomethylated arginine was incorporated into the AcH4–15 peptide in place of arginine 3. The fact that the kcat/Km is decreased by ≥500-fold indicates that PAD2 does not catalyze the so-called “demethylimination” reaction, consistent with previous studies on PAD2 and other PAD isozymes.8,23,25,29 In total, these studies indicate that like PADs 1, 3, and 4, PAD2 shows strong substrate promiscuity.23,25
Proposed Mechanism of Catalysis
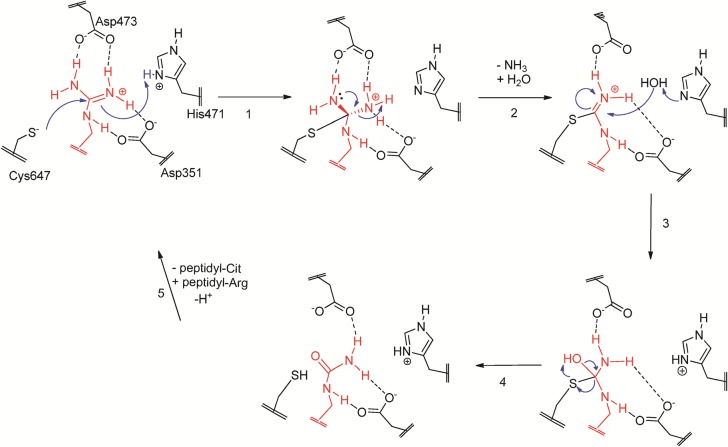
The PAD active site contains a Cys–His catalytic dyad (Cys647 and His471 in PAD2), which is reminiscent of the structures of cysteine proteases such as papain.33 In addition, the PADs contain two aspartyl groups (Asp345 and Asp374) that position the guanidinium for nucleophilic attack by the active site cysteine. In the first step, Cys647, the active site thiolate, attacks the guanidinum carbon of arginine. In this step, His471 acts as a general acid, protonating the guanidinium group with concomitant electrostatic stabilization by Asp351 and Asp473. The newly formed S-alkyl tetrahedral intermediate collapses as ammonia leaves. Within the third step, water is activated for nucleophilic attack by His471, resulting in the formation of a second tetrahedral intermediate, which ultimately collapses to form the citrullinated product (Figure 2). Consistent with a hydrolysis mechanism, 18O was incorporated into the product (BCEE) when the reaction was performed in 18O-labeled water (Table S2).
Figure 2.
Proposed mechanism of PAD2 catalysis. In the first step, the active site thiolate, Cys647, attacks the guanidinum carbon of arginine. Within this step, His471 acts as a general acid, protonating the guanidinium group with concomitant electrostatic stabilization by Asp351 and Asp473. The newly formed S-alkyl tetrahedral intermediate collapses as ammonia leaves. Within the third step, water is activated for nucleophilic attack by His471, which now acts as a general base. Attack of the water molecule results in the formation of the second tetrahedral intermediate, which ultimately collapses to form the citrullinated product.
pH Profiles
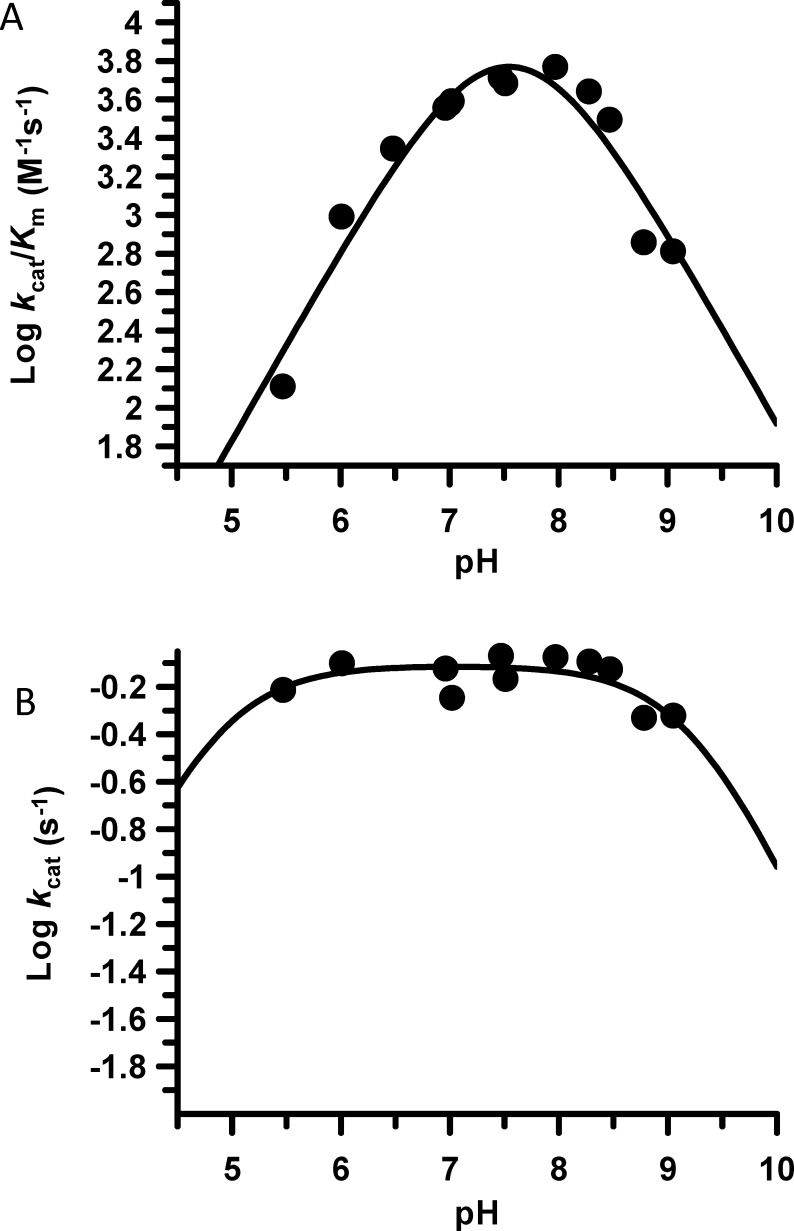
To provide insight into the PAD2 catalytic mechanism, we determined the steady-state kinetic parameters for BAEE over a range of pH values (5.5–9). The plots of kcat/Km versus pH are bell-shaped, yielding apparent pKa values of 7.5 and 7.6 for the ascending and descending limbs, respectively (Figure 3A). The pKa values are apparent because the narrowness of the profile precludes an accurate determination of the individual pKa values. Nevertheless, this data is consistent with other PAD enzymes, including PAD4, and on the basis of the simple assumption that the reaction rates will rise as the concentration of the more reactive thiolate species increases with increasing pH, the ascending limb most likely corresponds to the pKa of Cys647.24 By contrast, the descending limb likely corresponds to His471, as a loss of activity is expected following deprotonation of the imidazolium form of His471. In contrast to the narrow kcat/Km versus pH profile, the kcat profile is relatively flat over the entire pH range, with pKa values ≤ 4.9 and ≥ 9.2, indicating that the rate-limiting step is relatively pH-insensitive (Figure 3B).
Figure 3.
pH dependence of PAD2 catalysis. (A) pH rate profiles constructed using PAD2 (0.5 μM final) in reaction buffer with varying concentrations of BAEE (0–10 mM) at various pH values. The second-order rate constant, kcat/Km, is plotted on a logarithmic scale versus pH. (B) kcat, which measures the rate-limiting step, is plotted on a logarithmic scale versus pH.
Solvent Isotope Effects
To provide deeper insights into the catalytic mechanism, we repeated the above experiments but used D2O as the solvent in place of H2O (Figure 4). Overall, the plots of both kcat/Km and kcat versus pH are similar to those obtained in H2O and are, in fact, nearly overlapping. The solvent isotope effects (SIE) on kcat and kcat/Km are only 1.2 ± 0.06 and 0.8 ± 0.14-fold, respectively, at the pH optimum. The relatively small SIE observed on kcat suggests that proton transfer contributes minimally to the rate-limiting step of the reaction. Most notable, however, is the relatively small inverse SIE on kcat/Km. This result is in stark contrast to the large and inverse SIE observed for PAD4 (SIE = 0.43 ± 0.07). A similarly large and inverse SIE was obtained for PAD1. These results suggest that the PAD2 mechanism differs from the one employed by these isozymes.
Figure 4.
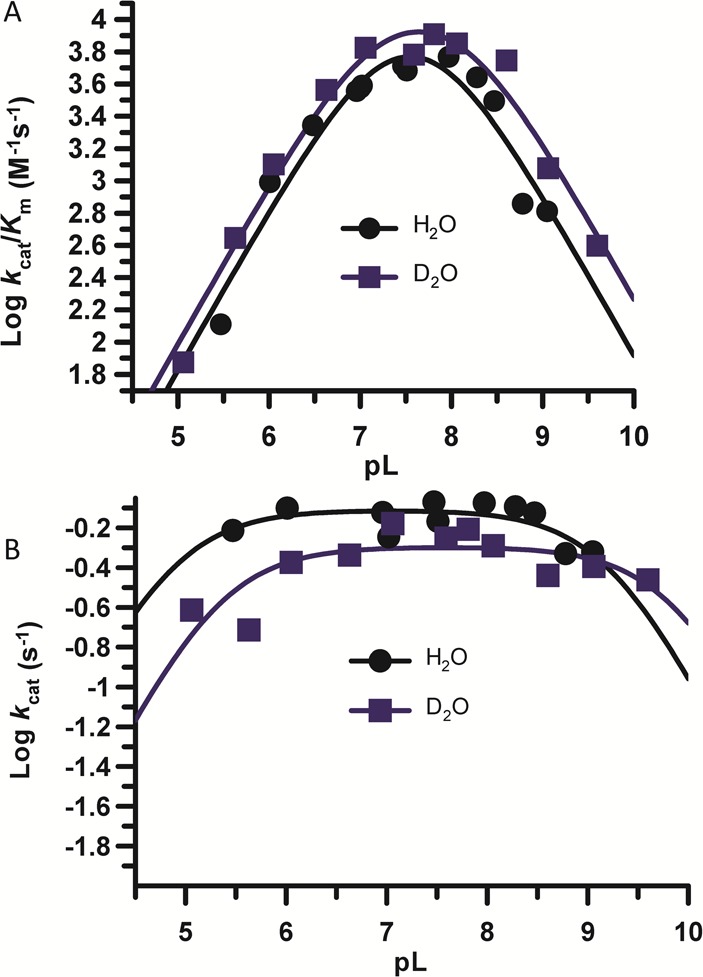
pH dependence of PAD2 catalysis in H2O and D2O. (A) pH rate profiles constructed using PAD2 (500 nM final) in reaction buffer with varying concentrations of BAEE (0–10 mM) in H2O or ≥95% D2O. The second-order rate constant, kcat/Km, is plotted on a logarithmic scale versus pL for both H2O and D2O. (B) kcat is plotted on a logarithmic scale versus pL for both H2O and D2O.
pKa Determination
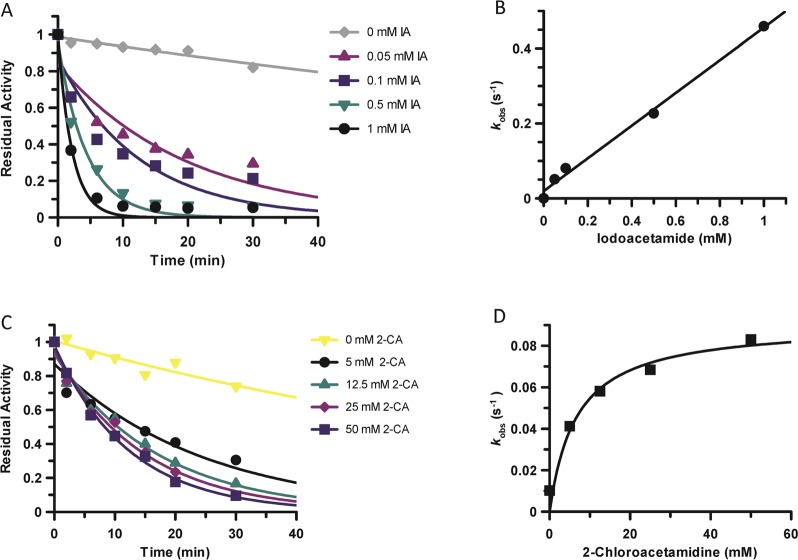
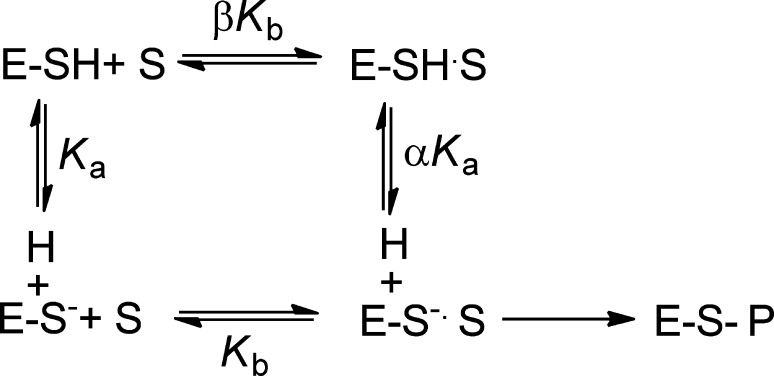
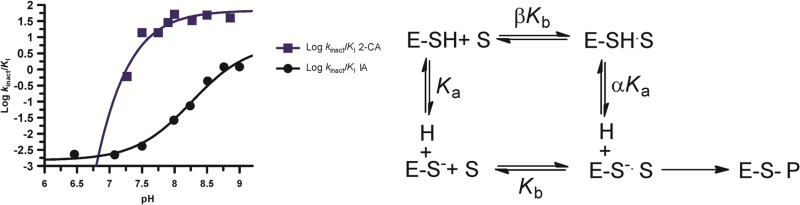
In light of the previous data, we measured the pKa value of Cys647, the active site nucleophile, by measuring the rates of inactivation afforded by both iodoacetamide and 2-chloroacetamidine as a function of pH. Both compounds are well-studied nonspecific affinity labels that preferentially modify the active site cysteine in the PADs and related enzymes.31,34,35 The key difference between the two compounds is their overall charge: iodoacetamide is neutral, whereas 2-chloroacetamidine, a guanidinium mimetic, is positively charged. For these experiments, residual PAD2 activity was measured after incubation with different concentrations of the two compounds to obtain values for kobs, the pseudo-first-order rate constant of inactivation. These experiments were repeated over a range of pH values (6.5–9 for iodoacetamide and 7.25–9 for 2-chloroacetamidine). The kobs values were then plotted against inactivator concentration to obtain values for kinact/KI (Figure 5). Note that the kobs versus inactivation plots were fit to eq 5 or 6 to obtain kinact/KI values from either the slope of the line or from the ratio of kinact and KI, respectively. Plots of kinact/KI versus pH were then generated and fit to eq 7 to obtain pKa values for the active site cysteine. On the basis of the iodoacetamide inactivation experiments, the pKa of the active site cysteine is 8.2. This result is consistent with the values obtained for the corresponding cysteines in PADs 1 and 4, where, unlike other cysteine hydrolases (e.g., papain), the architecture of the PAD active site does not promote the formation of a highly reactive low pKa thiolate in appreciable quantities. Instead, these enzymes use a reverse-protonation mechanism, wherein a fraction of the enzyme exists as the deprotonated thiol and protonated imidazole. Although the concentration of the thiolate form of the enzyme is low, catalysis still proceeds via nucleophilic attack by the thiolate form.24,25 For PAD2, however, when using 2-chloroacetamidine as the inactivator, the pKa value is reduced to 7.2, a full log unit (Figure 6). This result stands in stark contrast to that obtained for PADs 1 and 4, where the pKa values obtained with iodoacetamide and 2-chloroacetamidine are >8 and similar in value. Because kinact/KI reports on all steps up to and including the first irreversible step of the reaction, these data suggest that, upon formation of the initial encounter complex, the positively charged nature of 2-chloroacetamidine depresses the pKa of Cys647, likely via electrostatic stabilization, which is predicted for a substrate-assisted mechanism of enzyme catalysis (Scheme 1).
Figure 5.
Kinetics of inactivation by iodoacetamide and 2-chloroacetamidine. (A) Representative inactivation experiments when using iodoacetamide (IA) (0–1 mM) and measuring the residual activity of PAD2 for time points from 0 to 30 min. (B) Observed rates of inactivation, kobs, from panel A are plotted against inhibitor concentration for subsequent calculation of the second-order rate constant, kinact/KI. (C) Representative inactivation experiments when using 2-chloroacetamidine (2-CA). (D) Concentration of 2-CA versus kobs from panel C.
Figure 6.
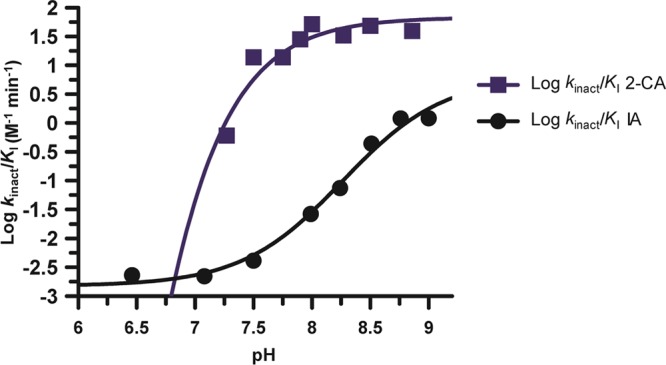
pKa value of the active site thiolate, Cys647, in PAD2. The log of the second-order rate constants, kinact/KI, obtained from both the iodoacetamide and 2-chloroacetamidine inactivation experiments is plotted against pH (6.5–9 for IA 7.25–9 for 2-CA).
Scheme 1. Substrate-Assisted versus Reverse-Protonation Mechanisms of Catalysis.
Discussion
Guanidinium-modifying enzymes have been suggested to use a variety of mechanisms to promote catalysis at the guanidinium center. For example, our group proposed that PAD4 used a reverse-protonation mechanism, whereas the Fast group suggested that the positively charged nature of the substrate guanidinium facilitates catalysis by depressing the pKa of the active site cysteine in dimethylarginine dimethylaminohydrolase (DDAH), a related enzyme.36 In this substrate-assisted mechanism, one would predict that upon forming an initial encounter complex with a positively charged inactivator, the pKa of Cys647 would be similarly depressed (Scheme 1). Our pKa studies with 2-chloroacetamidine, a positively charged inactivator, show this predicted effect. This data suggests that PAD2, like DDAH, uses a substrate-assisted mechanism rather than a reverse-protonation mechanism. By contrast, no such pKa shift was observed for PAD4 with 2-chloroacetamidine, which yielded a pKa value of 7.9 ± 0.2, nearly identical to the value obtained with iodoacetamide (8.2 ± 0.1).
Although the molecular basis for this mechanistic switch between PADs 2 and 4 is unknown, it likely relates to subtle differences in the active site architecture because the two enzymes adopt similar conformations in the catalytically competent calcium-bound state. For example, in the structure of the PAD4C645A–calcium complex, the active site cysteine and histidine, Cys645 and His471, appear to form a thiolate–imidazolium ion pair that is thought to position the active site thiolate for nucleophilic attack on the substrate guanidinium. PAD2 shows a similar active site arrangement when bound to calcium (Slade et al. unpublished data). The distance between the thiolate and imidazolium in PAD2 is quite long (4.7 Å), suggesting that the strength of the thiolate–imidazolium ion pair is correspondingly weak and may not provide sufficient energy to promote catalysis via a reverse-protonation mechanism, and as a consequence, PAD2 must rely on a substrate-assisted mechanism in which the positive charge of the substrate depresses the pKa of Cys647 and further activates the enzyme. Although the specific reasons for the divergent mechanisms are unclear, the requirement for substrate to bind and facilitate thiol deprotonation may afford greater protection against the nonspecific inactivation of PAD2 by reactive oxygen and/or nitrogen species. Consistent with this notion is the fact that PAD2 is highly expressed in macrophages, which are known to generate a highly oxidative environment upon activation.2
Overall, our kinetic and mechanistic studies of PAD2 suggest that, unlike the other members of the PAD family, this isozyme uses a substrate-assisted mechanism of catalysis. In addition to potentially affording greater protection against reactive oxygen and nitrogen species, these results suggest that PAD2-selective irreversible inhibitors can be identified by simply modifying the identity of the electrophilic warhead. As such, our studies bring us a step closer to identifying potent, selective, and bioavailable inhibitors for PAD2, an important therapeutic target for breast cancer.
Glossary
Abbreviations Used
- PAD
protein arginine deiminase
- PTM
post-translational modification
- BAEE
Nα-benzoyl-l-arginine ethyl ester
- BAME
Nα-benzoyl-l-arginine methyl ester
- BAA
Nα-benzoyl-l-arginine amide
- BA
Nα-benzoyl-l-arginine
- SIE
solvent isotope effect
- IA
iodoacetamide
- 2-CA
2-chloroacetamidine
Supporting Information Available
Sequences of histone H4 tail peptides; PAD2 catalysis of the hydrolytic deimination of BAEE. This material is available free of charge via the Internet at http://pubs.acs.org.
Financial support for this work was provided by NIH (grant GM079357 to P.R.T), TSRI, and a graduate fellowship from the Scheller Foundation (to C.J.D.).
The authors declare the following competing financial interest(s): P.R.T. is a cofounder of and consultant to Padlock Therapeutics.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Walsh C. (2006) Posttranslational Modification of Proteins: Expanding Nature’s Inventory; Roberts and Company Publishers, Englewood, CO. [Google Scholar]
- Vossenaar E. R.; Zendman A. J.; van Venrooij W. J.; Pruijn G. J. (2003) PAD, a growing family of citrullinating enzymes: genes, features and involvement in disease. Bioessays 25, 1106–1118. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Li M.; Stadler S.; Correll S.; Li P.; Wang D.; Hayama R.; Leonelli L.; Han H.; Grigoryev S. A.; Allis C. D.; Coonrod S. A. (2009) Histone hypercitrullination mediates chromatin decondensation and neutrophil extracellular trap formation. J. Cell Biol. 184, 205–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophorou M. A.; Castelo-Branco G.; Halley-Stott R. P.; Oliveira C. S.; Loos R.; Radzisheuskaya A.; Mowen K. A.; Bertone P.; Silva J. C.; Zernicka-Goetz M.; Nielsen M. L.; Gurdon J. B.; Kouzarides T. (2014) Citrullination regulates pluripotency and histone H1 binding to chromatin. Nature 507, 104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Li P.; Wang S.; Hu J.; Chen X. A.; Wu J.; Fisher M.; Oshaben K.; Zhao N.; Gu Y.; Wang D.; Chen G. (2012) Anticancer PAD inhibitors regulate the autophagy flux and the mammalian target of rapamycin complex 1 activity. J. Biol. Chem. 287, 25941–25953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. H.; Coonrod S. A.; Kraus W. L.; Jelinek M. A.; Stallcup M. R. (2005) Regulation of coactivator complex assembly and function by protein arginine methylation and demethylimination. Proc. Natl. Acad. Sci. U.S.A. 102, 3611–3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. S.; Bolt M.; Guertin M. J.; Chen W.; Zhang S.; Cherrington B. D.; Slade D. J.; Dreyton C. J.; Subramanian V.; Bicker K. L.; Thompson P. R.; Mancini M. A.; Lis J. T.; Coonrod S. A. (2012) Peptidylarginine deiminase 2-catalyzed histone H3 arginine 26 citrullination facilitates estrogen receptor alpha target gene activation. Proc. Natl. Acad. Sci. U.S.A. 109, 13331–13336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Wysocka J.; Sayegh J.; Lee Y. H.; Perlin J. R.; Leonelli L.; Sonbuchner L. S.; McDonald C. H.; Cook R. G.; Dou Y.; Roeder R. G.; Clarke S.; Stallcup M. R.; Allis C. D.; Coonrod S. A. (2004) Human PAD4 regulates histone arginine methylation levels via demethylimination. Science 306, 279–283. [DOI] [PubMed] [Google Scholar]
- Chang X.; Han J. (2006) Expression of peptidylarginine deiminase type 4 (PAD4) in various tumors. Mol. Carcinog. 45, 183–196. [DOI] [PubMed] [Google Scholar]
- Garcia-Romo G. S.; Caielli S.; Vega B.; Connolly J.; Allantaz F.; Xu Z.; Punaro M.; Baisch J.; Guiducci C.; Coffman R. L.; Barrat F. J.; Banchereau J.; Pascual V. (2011) Netting neutrophils are major inducers of type I IFN production in pediatric systemic lupus erythematosus. Sci. Transl. Med. 3, 73ra20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan M. J. (2011) Neutrophils in the pathogenesis and manifestations of SLE. Nat. Rev. Rheumatol. 7, 691–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande R.; Ganguly D.; Facchinetti V.; Frasca L.; Conrad C.; Gregorio J.; Meller S.; Chamilos G.; Sebasigari R.; Riccieri V.; Bassett R.; Amuro H.; Fukuhara S.; Ito T.; Liu Y. J.; Gilliet M. (2011) Neutrophils activate plasmacytoid dendritic cells by releasing self-DNA–peptide complexes in systemic lupus erythematosus. Sci. Transl. Med. 3, 73ra19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang X.; Yamada R.; Sawada T.; Suzuki A.; Kochi Y.; Yamamoto K. (2005) The inhibition of antithrombin by peptidylarginine deiminase 4 may contribute to pathogenesis of rheumatoid arthritis. Rheumatology 44, 293–298. [DOI] [PubMed] [Google Scholar]
- Dwivedi N.; Upadhyay J.; Neeli I.; Khan S.; Pattanaik D.; Myers L.; Kirou K. A.; Hellmich B.; Knuckley B.; Thompson P. R.; Crow M. K.; Mikuls T. R.; Csernok E.; Radic M. (2012) Felty’s syndrome autoantibodies bind to deiminated histones and neutrophil extracellular chromatin traps. Arthritis Rheum. 64, 982–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demers M.; Krause D. S.; Schatzberg D.; Martinod K.; Voorhees J. R.; Fuchs T. A.; Scadden D. T.; Wagner D. D. (2012) Cancers predispose neutrophils to release extracellular DNA traps that contribute to cancer-associated thrombosis. Proc. Natl. Acad. Sci. U.S.A. 109, 13076–13081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmers S.; Teijaro J. R.; Arandjelovic S.; Mowen K. A. (2011) PAD4-mediated neutrophil extracellular trap formation is not required for immunity against influenza infection. PLoS One 6, e22043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P.; Li M.; Lindberg M. R.; Kennett M. J.; Xiong N.; Wang Y. (2010) PAD4 is essential for antibacterial innate immunity mediated by neutrophil extracellular traps. J. Exp. Med. 207, 1853–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis V. C.; Gizinski A. M.; Banda N. K.; Causey C. P.; Knuckley B.; Cordova K. N.; Luo Y.; Levitt B.; Glogowska M.; Chandra P.; Kulik L.; Robinson W. H.; Arend W. P.; Thompson P. R.; Holers V. M. (2011) N-α-Benzoyl-N5-(2-chloro-1-iminoethyl)-l-ornithine amide, a protein arginine deiminase inhibitor, reduces the severity of murine collagen-induced arthritis. J. Immunol. 186, 4396–4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange S.; Gogel S.; Leung K. Y.; Vernay B.; Nicholas A. P.; Causey C. P.; Thompson P. R.; Greene N. D.; Ferretti P. (2011) Protein deiminases: new players in the developmentally regulated loss of neural regenerative ability. Dev. Biol. 355, 205–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElwee J. L.; Mohanan S.; Griffith O. L.; Breuer H. C.; Anguish L. J.; Cherrington B. D.; Palmer A. M.; Howe L. R.; Subramanian V.; Causey C. P.; Thompson P. R.; Gray J. W.; Coonrod S. A. (2012) Identification of PADI2 as a potential breast cancer biomarker and therapeutic target. BMC Cancer 12, 500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chumanevich A. A.; Causey C. P.; Knuckley B. A.; Jones J. E.; Poudyal D.; Chumanevich A. P.; Davis T.; Matesic L. E.; Thompson P. R.; Hofseth L. J. (2011) Suppression of colitis in mice by Cl-amidine: a novel peptidylarginine deiminase (PAD) inhibitor. Am. J. Physiol.: Gastrointest. Liver Physiol. 300, G929–G938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arita K.; Hashimoto H.; Shimizu T.; Nakashima K.; Yamada M.; Sato M. (2004) Structural basis for Ca2+-induced activation of human PAD4. Nat. Struct. Mol. Biol. 11, 777–783. [DOI] [PubMed] [Google Scholar]
- Kearney P. L.; Bhatia M.; Jones N. G.; Luo Y.; Glascock M. C.; Catchings K. L.; Yamada M.; Thompson P. R. (2005) Kinetic characterization of protein arginine deiminase 4: a transcriptional corepressor implicated in the onset and progression of rheumatoid arthritis. Biochemistry 44, 10570–10582. [DOI] [PubMed] [Google Scholar]
- Knuckley B.; Bhatia M.; Thompson P. R. (2007) Protein arginine deiminase 4: evidence for a reverse protonation mechanism. Biochemistry 46, 6578–6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuckley B.; Causey C. P.; Jones J. E.; Bhatia M.; Dreyton C. J.; Osborne T. C.; Takahara H.; Thompson P. R. (2010) Substrate specificity and kinetic studies of PADs 1, 3, and 4 identify potent and selective inhibitors of protein arginine deiminase 3. Biochemistry 49, 4852–4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musse A. A.; Li Z.; Ackerley C. A.; Bienzle D.; Lei H.; Poma R.; Harauz G.; Moscarello M. A.; Mastronardi F. G. (2008) Peptidylarginine deiminase 2 (PAD2) overexpression in transgenic mice leads to myelin loss in the central nervous system. Dis. Models & Mech. 1, 229–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherrington B. D.; Morency E.; Struble A. M.; Coonrod S. A.; Wakshlag J. J. (2010) Potential role for peptidylarginine deiminase 2 (PAD2) in citrullination of canine mammary epithelial cell histones. PLoS One 5, e11768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanan S.; Horibata S.; McElwee J. L.; Dannenberg A. J.; Coonrod S. A. (2013) Identification of macrophage extracellular trap-like structures in mammary gland adipose tissue: a preliminary study. Front. Immunol. 4, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musse A. A.; Polverini E.; Raijmakers R.; Harauz G. (2008) Kinetics of human peptidylarginine deiminase 2 (hPAD2)—reduction of Ca2+ dependence by phospholipids and assessment of proposed inhibition by paclitaxel side chains. Biochem. Cell Biol. 86, 437–447. [DOI] [PubMed] [Google Scholar]
- Leatherbarrow R. J. (2004) GraFit, version 5.0, Erathicus Software, Staines, UK.
- Jones J. E.; Dreyton C. J.; Flick H.; Causey C. P.; Thompson P. R. (2010) Mechanistic studies of agmatine deiminase from multiple bacterial species. Biochemistry 49, 9413–9423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linsky T.; Fast W. (2010) Mechanistic similarity and diversity among the guanidine-modifying members of the pentein superfamily. Biochim. Biophys. Acta 1804, 1943–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers J. C.; Asgian J. L.; Ekici O. D.; James K. E. (2002) Irreversible inhibitors of serine, cysteine, and threonine proteases. Chem. Rev. 102, 4639–4750. [DOI] [PubMed] [Google Scholar]
- Stone E. M.; Schaller T. H.; Bianchi H.; Person M. D.; Fast W. (2005) Inactivation of two diverse enzymes in the amidinotransferase superfamily by 2-chloroacetamidine: dimethylargininase and peptidylarginine deiminase. Biochemistry 44, 13744–13752. [DOI] [PubMed] [Google Scholar]
- Jones J. E.; Causey C. P.; Lovelace L.; Knuckley B.; Flick H.; Lebioda L.; Thompson P. R. (2010) Characterization and inactivation of an agmatine deiminase from Helicobacter pylori. Bioorg Chem. 38, 62–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone E. M.; Costello A. L.; Tierney D. L.; Fast W. (2006) Substrate-assisted cysteine deprotonation in the mechanism of dimethylargininase (DDAH) from Pseudomonas aeruginosa. Biochemistry 45, 5618–5630. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.