Abstract
Objective
To investigate the prevalence and mechanisms of fluoroquinolone resistance in Shigella species isolated in Bangladesh and to compare with similar strains isolated in China.
Methods
A total of 3789 Shigella isolates collected from Clinical Microbiology Laboratory of icddr,b, during 2004–2010 were analyzed for antibiotic susceptibility. Analysis of plasmids, plasmid-mediated quinolone-resistance genes, PFGE, and sequencing of genes of the quinolone-resistance-determining regions (QRDR) were conducted in representative strains isolated in Bangladesh and compared with strains isolated in Zhengding, China. In addition, the role of efflux-pump was studied by using the efflux-pump inhibitor carbonyl cyanide-m-chlorophenylhydrazone (CCCP).
Results
Resistance to ciprofloxacin in Shigella species increased from 0% in 2004 to 44% in 2010 and S. flexneri was the predominant species. Of Shigella spp, ciprofloxacin resistant (CipR) strains were mostly found among S. flexneri (8.3%), followed by S. sonnei (1.5%). Within S. flexneri (n = 2181), 14.5% were resistance to ciprofloxacin of which serotype 2a was predominant (96%). MIC of ciprofloxacin, norfloxacin, and ofloxacin were 6–32 mg/L, 8–32 mg/L, and 8–24 mg/L, respectively in S. flexneri 2a isolates. Sequencing of QRDR genes of resistant isolates showed double mutations in gyrA gene (Ser83Leu, Asp87Asn/Gly) and single mutation in parC gene (Ser80Ile). A difference in amino acid substitution at position 87 was found between strains isolated in Bangladesh (Asp87Asn) and China (Asp87Gly) except for one. A novel mutation at position 211 (His→Tyr) in gyrA gene was detected only in the Bangladeshi strains. Susceptibility to ciprofloxacin was increased by the presence of CCCP indicating the involvement of energy dependent active efflux pumps. A single PFGE type was found in isolates from Bangladesh and China suggesting their genetic relatedness.
Conclusions
Emergence of fluoroquinolone resistance in Shigella undermines a major challenge in current treatment strategies which needs to be followed up by using empirical therapeutic strategies.
Introduction
Shigellosis caused by Shigella species is endemic throughout the world, and is one of the most important causes of global childhood mortality and morbidity. Globally, every year there are about 165 million cases of Shigella infection and 1.1 million Shigella-related deaths [1]. Shigella is transmitted efficiently in low-dose via fecal-oral route in areas, with poor hygienic conditions, and limited access to clean and potable water [2]. Of four Shigella species, shigellosis is predominantly caused by S. flexneri in the developing world especially in Asia, whereas S. sonnei is the predominant causative agent of this disease in developed as well as industrialized countries [1]. A recent multicenter study of the epidemiology and microbiology of shigellosis in Asia revealed that the incidence of this disease might even exceed previous estimations, as Shigella DNA could also be detected in up to one-third of culture-negative specimens [3].
Antimicrobial therapy is effective for the treatment of shigellosis. Increased resistance to commonly used antibiotics including ampicillin, streptomycin, sulfamethoxazole-trimethoprim, nalidixic acid and tetracycline among Shigella poses a major therapeutic challenge to control this disease [4]. One of the reasons for emergence of multi-drug resistant Shigella spp. is the unique capability of the pathogen to acquire resistance factors (transmissible genes) from the environment or from other bacteria [5]. Besides, indiscriminate use of antibiotics for the treatment of human infection and in animal husbandries in endemic areas triggers the increase of resistance to newer antibiotics [6]. Ciprofloxacin, a third generation fluoroquinolone, has been used successfully for the treatment of infectious diseases including shigellosis. Following a successful clinical trial, ciprofloxacin has been recommended for the treatment of both childhood and adult shigellosis caused by multiple antibiotic resistant Shigella spp. in Bangladesh [7]. However, this antibiotic is no longer effective for the treatment of shigellosis in south Asian region including Bangladesh because of the emergence of fluoroquinolone resistant S. dysenteriae type 1 and their (same clone) dissemination across the countries [8], [9].
Quinolone resistance emerges due to i) point mutations that result in amino acid substitution in chromosomal genes for DNA gyrase and topoisomerase IV, the targets of quinolone action and ii) changes in expression of efflux pumps and outer membrane permeability that control the accumulation of these agents inside the bacterial cell [10], [11]. In addition, a novel mechanism of plasmid-mediated quinolone resistance has recently been reported, and this involves DNA gyrase protection by a protein from the pentapeptide repeat family called Qnr [12], [13]. In Gram-negative organisms, the primary target of fluoroquinolones is the enzyme DNA gyrase, which is essential for DNA synthesis [14]. DNA gyrase consists of two A and two B subunits, encoded by the gyrA and gyrB genes, respectively. Most mutations have been shown to reside in a small region near the start of the gyrA gene, termed as quinolone resistance-determining region (QRDR), although mutations have also been reported in gyrB [15]. Genes encoding topoisomerase IV consists of two subunits parC and parE which have also been shown to be inhibited by fluoroquinolones. In Gram-negative bacteria, topoisomerase IV has been considered as the secondary target to flouroquinolones and alteration in parC is involved in the mechanism of resistance [16]. The level of MICs of fluoroquinolones has been shown to correlate with the type and number of amino acid substitution of the target genes within the QRDR [17]–[20]. In our previous study, possible mechanisms of fluoroquinolone resistance were analyzed in clinical strains of S. dysenteriae type 1 isolated from India, Nepal and Bangladesh [9].
It has also been reported that fluoroquinolone resistance is often the result of a combination of target site mutations and enhanced expression of genes encoding efflux pumps in resistant bacteria [21]. In this study, we report the first isolation of fluoroquinolone resistant S. flexneri, S. boydii and S. sonnei in Bangladesh. In addition, the mechanism of chromosome mediated fluoroquinolone resistance in S. flexneri strains, which results from the combination of target site mutation and energy dependant active pumps involvement has been determined.
Materials and Methods
Bacterial strains
A total of 3,789 Shigella spp. were isolated from diarrheal patients attending the Dhaka treatment center of the International Centre for Diarrhoeal Disease Research, Bangladesh (icddr,b) between January 2004 and December 2010. These strains were isolated and identified as belonging to different species and serotypes by using standard microbiological, biochemical, and serological methods. Among the Shigella spp. 2,181 isolates were confirmed as S. flexneri, by using polyvalent commercial antisera (Denka Seiken, Tokyo, Japan). In this study 380 randomly selected ciprofloxacin resistant (CipR) Shigella spp. and 25 susceptible S. flexneri strains were used. Of these 380, 317 were S. flexneri, 3 were S. dysenteriae, 4 were S. boydii and 56 were S. sonnei. For comparison, 12 S. flexneri 2a strains isolated in Zhending, Hebei province, China in 2002 were included in the study. These isolates were obtained from the collection of the International Vaccine Institute, Seoul, Korea. Of these Chinese strains, 10 were completely resistant and 2 strains showed reduced susceptibility to ciprofloxacin [22]. Details of the representative (n = 25) S. flexneri 2a are presented in the Table 1. Escherichia coli (ATCC 25922) and Staphylococcus aureus (ATCC 25923) were also used as control strains for the antibiotic susceptibility tests.
Table 1. MIC and amino acid changes in gyrA and parC in representative S. flexneri 2a strains isolated in Bangladesh and China.
| Strain | Country | Resistance profile | Plasmidprofile | MIC (mg/L) | Substitutions in QRDRc | |||||||||
| Na | Cip | Cip+CCCP 4mg/L | Nor | Of | Azm | Cro | gyrA | parC | ||||||
| Ser-83a | Asp-87a | His-211a | Ser-80a | |||||||||||
| K105 | Bangladesh | Amp,Sxt,Na | P1 | 2 | 0.008 | 0.032 | 0.032 | 0.047 | 0.75 | 0.032 | -b | – | – | – |
| K304 | Bangladesh | Amp,Sxt | P1 | 2 | 0.008 | 0.032 | 0.032 | 0.047 | 0.75 | 0.032 | -b | – | – | – |
| K6224 | Bangladesh | Amp,Sxt,Na,Cip | P1 | >256 | 6 | 0.5 | 32 | 24 | 0.75 | 0.064 | Leu | Asn | Tyr | Ile |
| K6806 | Bangladesh | Amp,Sxt,Na,Cip | P1 | >256 | 6 | 0.5 | 8 | 16 | 0.75 | 0.032 | Leu | Asn | Tyr | Ile |
| K6813 | Bangladesh | Amp,Sxt,Na,Cip | P1 | >256 | 8 | 1 | 12 | 16 | 0.5 | 0.023 | Leu | Asn | Tyr | Ile |
| K6814 | Bangladesh | Amp,Sxt,Na,Cip | P1 | >256 | 6 | 0.5 | 16 | 12 | 0.5 | 0.023 | Leu | Asn | Tyr | Ile |
| K10224 | Bangladesh | Amp,Sxt,Na,Cip | P1 | >256 | 32 | 16 | 16 | 24 | 0.25 | 0.023 | Leu | Asn | Tyr | Ile |
| K6913 | Bangladesh | Amp,Sxt,Na,Cip | P1 | >256 | 8 | 1 | 12 | 16 | 0.5 | 0.023 | Leu | Asn | Tyr | Ile |
| K9482 | Bangladesh | Amp,Sxt,Na,Cip | P4 | >256 | 6 | 0.5 | 16 | 12 | 0.5 | 0.023 | Leu | Asn | Tyr | Ile |
| K6915 | Bangladesh | Amp,Sxt,Na,Cip | P1 | >256 | 6 | 0.5 | 16 | 24 | 0.25 | 0.023 | Leu | Asn | Tyr | Ile |
| K6916 | Bangladesh | Amp,Sxt,Na,Cip | P1 | >256 | 8 | 0.5 | 12 | 16 | 0.5 | 0.023 | Leu | Asn | Tyr | Ile |
| K9563 | Bangladesh | Amp,Sxt,Na,Cip | P4 | >256 | 6 | 0.5 | 16 | 12 | 0.5 | 0.023 | Leu | Asn | Tyr | Ile |
| K10347 | Bangladesh | Amp,Sxt,Na,Cip | P1 | >256 | 6 | 0.5 | 16 | 24 | 0.25 | 0.023 | Leu | Asn | Tyr | Ile |
| CH2535 | China | Amp,Sxt,Na,Cip | P3 | >256 | 6 | 0.5 | 10 | 12 | 1 | 0.032 | Leu | Gly | – | Ile |
| CH2765 | China | Sxt,Na,Cip | P1 | >256 | 6 | 0.5 | 16 | 8 | 0.5 | 0.023 | Leu | Gly | – | Ile |
| CH2970 | China | Sxt,Na,Cip | P1 | >256 | 6 | 2 | 16 | 12 | 1 | 0.023 | Leu | Gly | – | Ile |
| CH2971 | China | Sxt,Na,Cip | P2 | >256 | 6 | 2 | 24 | 24 | 1 | 0.023 | Leu | Gly | – | Ile |
| CH8603 | China | Amp,Sxt,Na,Cip | P2 | >256 | 6 | 0.5 | 16 | 32 | 1.5 | 0.047 | Leu | Asn | – | Ile |
| CH9608 | China | Amp,Sxt,Na | P3 | >256 | 0.5 | 0.5 | 3 | 2 | 1.5 | 0.032 | Leu | – | – | Ile |
| CH9681 | China | Amp,Sxt,Na | P1 | >256 | 0.5 | 0.5 | 2 | 2 | 1 | 0.032 | Leu | – | – | Ile |
| CH10365 | China | Amp,Sxt,Na,Cip | P2 | >256 | 6 | 0.5 | 16 | 16 | 0.75 | 0.023 | Leu | Gly | – | Ile |
| CH10416 | China | Amp,Sxt,Na,Cip | P2 | >256 | 6 | 2 | 24 | 24 | 0.75 | 0.023 | Leu | Gly | – | Ile |
| CH10430 | China | Amp,Sxt,Na,Cip | P2 | >256 | 6 | 2 | 24 | 16 | 1 | 0.032 | Leu | Gly | – | Ile |
| CH11108 | China | Amp,Sxt,Na,Cip | P2 | >256 | 6 | 2 | 16 | 24 | 1.5 | 0.047 | Leu | Gly | – | Ile |
| CH12806 | China | Amp,Sxt,Na,Cip | P2 | >256 | 6 | 0.5 | 16 | 16 | 0.75 | 0.023 | Leu | Gly | – | Ile |
Abbreviations: Na, nalidixic acid; Cip, ciprofloxacin; Nor, norfloxacin; Of, ofloxacin; Azm, azithromycin; Cro, ceftriaxone, CCCP, carbonyl cyanide-m-chlorophenylhydrazone aWild type, P1∶140, 4, 2.7, 2.1 MDa; P2∶140, 2.7, 2.1 MDa; P3∶140, 8, 2.7, 2.1MDa; P4∶140, 36–62, 2.7, 2.1MDa.
Serotyping
Serotype was confirmed using a commercially available antiserum kit (Denka Seiken, Tokyo, Japan) specific for all group factor antigens of S. dysenteriae, S. boydii and S. sonnei and in case of S. flexneri (i) all type and group-factor antigens and (ii) monoclonal antibody reagents specific for all S. flexneri type-and group-factor antigens (Reagensia AB, Stockholm, Sweden). Serotyping was performed by the slide agglutination test following a procedure described previously [23].
Susceptibility testing
Antibiotic susceptibility test was done by disc diffusion method following the guidelines of Clinical and Laboratory Standards Institute (CLSI) using commercially available antibiotic disc (Oxoid, Basingstoke, United Kingdom). The antibiotic discs used in this study were ampicillin (Amp; 10 µg), sulphamethoxazole-trimethoprim (Sxt; 25 µg), mecillinam (Mel; 25 µg), nalidixic acid (Na; 30 µg), ciprofloxacin (Cip; 5 µg), norfloxacin (Nor; 10 µg), ofloxacin (Of; 5 µg), azithromycin (Azm; 15 µg), and ceftriaxone (Cro; 30 µg) [24]. The minimum inhibitory concentrations (MICs) of nalidixic acid, ciprofloxacin, norfloxacin, ofloxacin, azithromycin, and ceftriaxone were determined by the E-test (AB Biodisk, Solna, Sweden).
Plasmid profile analysis
Plasmid DNA was prepared by the alkaline lysis method of Kado and Liu, with some modifications [25]. The molecular weight of the unknown plasmid DNA was assessed by comparing the mobility of the plasmids of known molecular mass as described previously [26].
Determination of resistance factor
A conjugation experiment between the multidrug-resistant (AmpR SxtR CipR) donors, S. flexneri serotype 2a K9482and K9563 strains, and the recipient, E. coli K-12 (Lac− F−), was carried out by a procedure described elsewhere [27]. Transconjugant colonies were selected on MacConkey agar plates containing Amp (50 mg/L). Plasmid analysis and antimicrobial susceptibility testing of the transconjugants were carried out to determine the transfer of plasmids with antibiotic resistance.
DNA sequence analysis
Chromosomal DNA from selected representative strains was prepared and purified by procedures described previously [28]. Polymerase chain reaction (PCR) for gyrA, gyrB, parC and pare were performed according to the procedures described earlier [29]–[32]. PCR amplicons were purified with the GFX™ PCR DNA and gel band purification kit (Amersham Pharmacia, USA). Sequencing of the amplicons was performed using the dideoxy-nucleotide chain termination method with the ABI PRISM® BigDye Terminator Cycle Sequencing Reaction kit (Perkin-Elmer Applied Biosystems, Foster City, California) on an automated sequencer (ABI PRISM™ 310). The chromatogram sequencing files were inspected using Chromas 2.23 (Technelysium, Queensland, Australia), and contigs were prepared using SeqMan II (DNASTAR, Madison, WI). Nucleotide and protein sequence similarity searches were performed using the National Center for Biotechnology Information (NCBI, National Institutes of Health, Bethesda, MD) BLAST (Basic Local Alignment Search Tool) server on GenBank database, release 138.0 [33]. Multiple sequence alignments were developed using CLUSTALX 1.81 [34]. Sequences were manually edited in the GeneDoc version 2.6.002 alignment editor.
Nucleotide sequences accession number
The nucleotide sequences reported in this paper were submitted in GenBank using the National Center for Biotechnology Information (NCBI, Bethesda, MD) Sequin, version 5.26 under accession numbers- DQ681123-30 for gyrA, DQ681105-13 for gyrB, DQ681114-22 for parC and GQ452292 for parE.
Amplification of qnrA, qnrB, qnrS and aac(6¢)-Ib-cr
Multiplex PCR of some representative strains was performed to detect the qnrA, qnrB, qnrS and aac(6¢)-Ib-cr according to the procedure described earlier [21]. The primer sequences are 5′-AGAGGATTTCTCACGCCAGGA-3′ and 5′-GGCTGGCCGATTATGATTGGT- 3′for qnrA, 5′-GGCTGGCCGATTATGATTGGT-3′ and 5′-CGCGTGCGATGAGATAACC-3′ for qnrB, 5′-TGCCACTTTGATGTCGCAGAT-3′ and 5′-CGCACGGAACTCTATACCGTAG-3′ for qnrS, and 5′-ATCTCATATCGTCGAGTGGTGG-3′ and 5′-CGCTTTCTCGTAGCATCGGAT-3′ for aac(6¢)-Ib-cr.
Synergy tests
Synergy tests were performed according to the procedure described elsewhere using ciprofloxacin and the efflux pump inhibitor carbonyl cyanide-m-chlorophenylhydrazone (CCCP) [35], [36]. Concentration of CCCP was evaluated and optimized to avoid the cytotoxic effect of CCCP on bacterial growth [37]. CCCP was added to the Mueller–Hinton agar at a concentration of 4 mg/L. Susceptibility testing for ciprofloxacin resistant (CipR) strains was performed by agar dilution plates, with and without CCCP (Sigma Aldrich, USA).
Pulsed-field gel electrophoresis (PFGE)
Genomic DNA of S. flexneri was prepared in agarose blocks and digested with the restriction enzyme XbaI (New England Biolabs) according to the PulseNet program descried elsewhere [38]. DNA fragments were separated by pulsed-field gel electrophoresis on a CHEF-MAPPER apparatus (Bio-Rad). Analysis of the TIFF images was carried out by the BioNumerics software (Applied Maths, Belgium) using the dice coefficient and unweighted-pair group method using average linkages to generate dendrogram with 1.0% tolerance values.
Results and Discussion
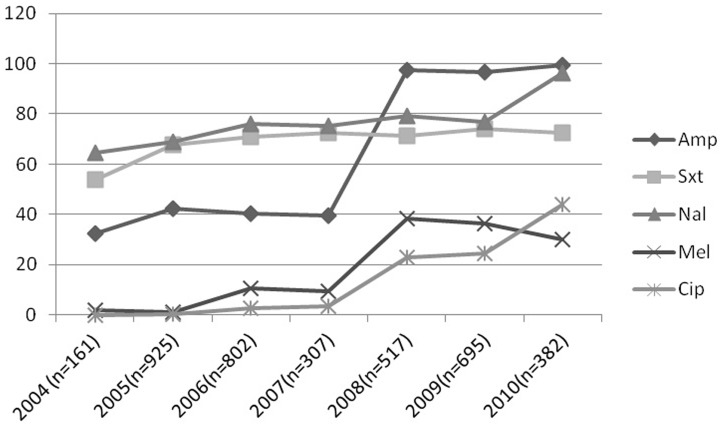
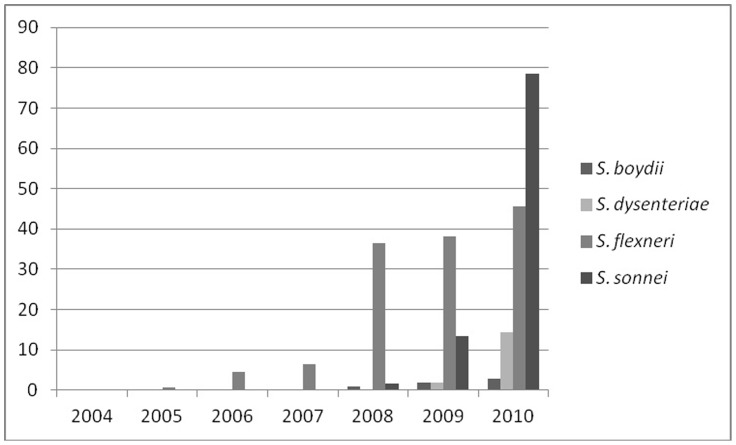
Of 3,789 Shigella spp. isolated between January 2004 and December 2010 at icddr,b, S. flexneri was the predominant serotype (n = 2181, 57.5%), followed by S. boydii (n = 753, 20%), S. sonnei (n = 574, 15%) and S. dysenteriae (n = 281, 7.4%). In S. flexneri strains, serotype 2a was the most abundant (40%), followed by serotype 3a (19%), 1b (11%), type 6 (10%), 2b (9%), 1c (5%), type 4 (3%), 4X (2%) and Y variant (1%). Of S. boydii, predominant serotype was S. boydii 12 (28%), followed by serotype 1 (10%), 4 (9%), 18 (9%), 5 (7%), and 14 (7%). In case of S. dysenteriae, predominant serotype was serotype 2 (40%) followed by novel S. dysenteriae (17%), serotype 3 (15%), and serotype 4 (13%). Resistance to Amp, Sxt, Na, Mel and Cip in Shigella spp. increased from 32%, 54%, 65%, 2% and 0% in 2004 to 99.5%, 73%, 96%, 30% and 44% respectively in 2010 (Figure 1). During the study period, CipR strains were mostly found among S. flexneri (8.3%), followed by S. sonnei (1.5%). Among the S. flexneri (n = 2181) strains, frequency of resistance to ciprofloxacin (n = 317, 14.5%) increased from 0.7% (n = 3) in 2005 to 45.5% (n = 87) in 2010. Of CipR S. flexneri, serotype 2a was the predominant (96%), followed by type 6 (1.6%), serotype 3a (1.3%), serotype Y variant (0.6%) and serotype 1c (0.3%). Resistance to ciprofloxacin among S. sonnei, S. boydii and S. dysenteriae also started and increased substantially between 2008 and 2010 (Figure 2). It is interesting to note that S. flexneri 2a and S. sonnei isolated in 2010, 89% (83/93) and 72% (77/107) were resistant to ciprofloxacin respectively.
Figure 1. Antibiotic resistance pattern of Shigella spp. isolated between 2004 and 2010 in Dhaka, Bangladesh.
Figure 2. Prevalence of CipR Shigella spp. isolated between 2004 and 2010 in Dhaka, Bangladesh.
Ciprofloxacin-resistance was first observed in S. dysenteriae type 1 isolated in Bangladesh in 2003 and at the same time isolation rate of S. dysenteriae type 1 was abruptly reduced [8]. Since 2004 there is no report of isolation of S. dysenteriae type 1 in Bangladesh. Since S. flexneri is the most predominant species of Shigella and serotype 2a is the most predominant subserotype, S. flexneri 2a strains were selected for further detail study in order to understand the ciprofloxacin resistance mechanisms of this pathogen. The range of MIC values of ciprofloxacin, norfloxacin, and ofloxacin for S. flexneri 2a, were 6–32 mg/L, 8–32 mg/L, and 8–24 mg/L, respectively (Table 1). These CipR strains were also resistant to Amp, Sxt, and Na, but were susceptible to Mel, Azm and Cro. The MIC of Cip for 12 strains isolated in China was 6 mg/L and 0.5 mg/L for resistant and reduced susceptible strains, respectively (Table 1).
According to a previous study, at least 4 mutation points at positions 81 (Gly81→Ser), 83 (Ser83 → Leu), 87 (Asp87 → Asn or Gly), and 92 (Met92→ Lys) have been identified in the gyrA gene associated with quinolone and/or fluoroquinolone resistance of Shigella spp. [39]. In the present study, all fluoroquinolone resistant S. flexneri 2a strains from Bangladesh and China had double mutations in gyrA (Ser83 → Leu, Asp87 → Asn or Gly) and a single mutation in parC (Ser80 → Ile). A similar type of findings was observed in S. dysenteriae type 1 strains [8]. In case of Chinese strains, a single mutation at position 83 in gyrA (Ser → Leu) and at position 80 of parC (Ser→ Ile) was associated with decreased susceptibility to fluoroquinolone but not fully resistance. In addition, difference between CipR strains isolated in Bangladesh and China was found in amino acid substitution at position 87, which was Asp87 → Asn in case of Bangladeshi strains whereas it was Asp87 → Gly in case of Chinese strains. However, one of these Chinese strains (CH8603) had the same amino acid substitution at position 87 (Table 1) similar as Bangladeshi strains. This is in accordance with the previous study where double mutations in gyrA (Ser83 → Leu, Asp87 → Gly) were found to be associated with fluoroquinolone resistance in S. flexneri 2a strains [40]. It appears that like E. coli, amino acid substitution in gyrA at positions 83 and 87 is common for fluoroquinolone resistance in Shigella species. In E. coli, mutation in other positions of gyrA gene outside the QRDR region of fluoroquinolone resistant strains has been reported, which has yet been reported in Shigella spp. [41]. In this study, we had found a new mutation point outside the QRDR region in gyrA gene at position 211 (His211→Tyr) in fluoroquinolone resistant S. flexneri 2a strains that was absent in fluoroquinolone susceptible S. flexneri 2a strains (Table 1). The novel substitution within gyrA is however of interest. Further studies such as site directed mutagenesis analysis are suggested to confirm the association between fluoroquinolone resistance and the amino acid substitution at position 211 in S. flexneri. No mutations in gyrB and parE genes were found in any of the S. flexneri 2a strains in the study, which corroborated with our previous findings [9] and the finding was also compared with the E. coli control strain (ATCC 25922) to avoid the biasness (data not shown). In addition to substitution in structural genes (gyrA/B, parC/E), plasmid mediated quinolone resistance has also been reported that can be determined by the presence of the Qnr gene [12]. Several surveys, based on molecular approaches consisting of PCR and sequencing, indicated a higher rate of the association between Qnr-positive and ESBL-positive isolates [42]. In this study none of the strains was found to carry the Qnr gene that excluded the possibility of plasmid mediated quinolone resistance in S. flexneri isolated in Bangladesh and these result corroborated with our previous observations also [9].
Conjugation experiment was carried out for some selected strains of this serotype 2a and the outcome indicated that resistance to ciprofloxacin is not co transferred with that of ampicillin resistance and not that resistance to ciprofloxacin is plasmid mediated (Table 2). Mechanisms like role of AcrAB-TolC efflux pump and MAR (multiple antimicrobial resistances) phenotype in fluoroquinolone resistance, decreased expression of outer membrane porins, or over-expression of multidrug efflux pumps are also important for detail analysis of fluroquinolone resistance. To elucidate the involvement of efflux pumps in fluoroquinolone resistance in Shigella isolates, synergy experiments were performed by using the efflux pump inhibitor CCCP and ciprofloxacin. A degree of synergy between CCCP and ciprofloxacin was observed using the agar dilution method. The concentration of CCCP was evaluated and optimized to control the cytotoxic effect of CCCP on bacterial growth [37]. The MICs of ciprofloxacin resistance were decreased up to 16 fold in an agar plate containing CCCP when compared to those observed in CCCP-free medium (Table 1). These results indicate that efflux pumps may be involved in the fluoroquinolone resistance of S. flexneri because there were changes in MICs in the presence or absence of CCCP. It will be interesting to find out the specific mechanisms of efflux pumps mediated fluoroquinolone resistance in S. flexneri.
Table 2. Transfer of resistance plasmid to E. coli K-12 by conjugation.
| Strain | Parent Strain | Transconjugant | ||
| Resistance pattern | Plasmid profile (MDa) | Resistance pattern | Plasmid profile (MDa) | |
| K9482 | AmpR, SxtR, NaR, CipR | 140, 62, 2.7, 2.1 | AmpR, SxtR, | 62 |
| K9563 | AmpR, SxtR, NaR, CipR | 140, 36, 2.7, 2.1 | AmpR | 36 |
Plasmid profile and PFGE analysis have long been used as molecular fingerprinting tools for bacterial strains. All CipR strains from Bangladesh were clustered in two plasmid pattern (P1 and P4), while Chinese strains were divided into three patterns (P1, P2 and P3). Plasmids of P2 pattern (140, 2.7 and 2.1 MDa) were commonly present in all strains irrespective of their resistance pattern and origin of isolation.
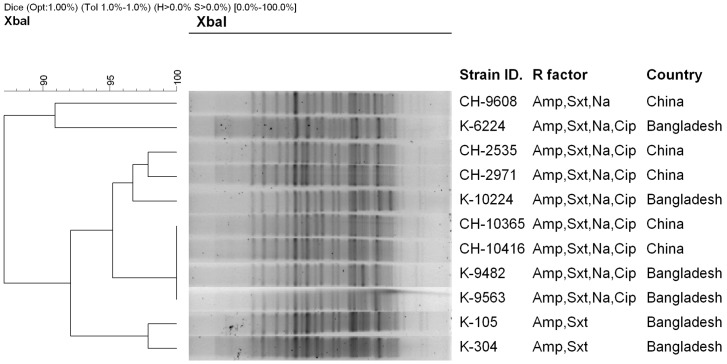
PFGE analysis of representative S. flexneri 2a strains isolated from Bangladesh and China, either resistant or susceptible to flouroquinolones revealed a closely related banding pattern with 90% similarity indices. Four S. flexneri 2a strains, two from Bangladesh (K9482 and K9563) and 2 from China (CH10365 and CH10416) had identical banding pattern and were member of single clone (Figure. 3) indicating dissemination of a genetically related S. flexneri 2a in these countries, which is alarming.
Figure 3. Dendrogram generated by BioNumeric software, showing distances calculated by the dice similarity index of PFGE XbaI profiles for S. flexneri 2a isolated from Bangladesh and compared with strains isolated in China.
The degree of similarity (%) is shown on the scale.
Emergence of fluoroquinolone resistance in S. flexneri 2a has undermined the current treatment strategies of shigellosis as serotype 2a has been identified as the predominant causative agent of endemic shigellosis. Currently, pivmecillinam has been recommended as an alternative drug of fluoroquinolones for the treatment of shigellosis. However, how long can it be used as an effective alternative- is a burning question? In this report, we have shown for the first time the emergence of multi-drug resistance (MDR), especially the fluoroquinolone resistant S. flexneri in Bangladesh with their comparative genetic relatedness of the strains from China [43]. This study suggests the need for continuous monitoring of the MDR Shigella spp. as well as the elucidation of their molecular mechanisms of drug resistance. This knowledge will help in the development of new antimicrobial strategies, to stop or reduce the emergence and spread of these MDR pathogens.
Acknowledgments
We gratefully acknowledge Dr. Lorenz von Seidlein, International Vaccine Institute, Seoul, Korea, for supplying S. flexneri strains. The authors would like to thank the members of the Enteric and Food Microbiology Laboratory, CFWD, icddr,b for their sincere cooperation throughout the study.
Funding Statement
This research study was funded by the icddr,b’s core donors and Government of Bangladesh through IHP-HNPRP. icddr,b acknowledges with gratitude the commitment of Government of Bangladesh through IHP-HNPRP to its research efforts. icddr,b also gratefully acknowledges the following donors which provide unrestricted support: Australian Agency for International Development (AusAID), Government of the People’s Republic of Bangladesh, Canadian International Development Agency (CIDA), Swedish International Development Cooperation Agency (Sida), and the Department for International Development, UK (DFID). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Kotloff KL, Winickoff JP, Ivanoff B, Clemens JD, Swerdlow DL, et al. (1999) Global burden of Shigella infections: implications for vaccine development and implementation of control strategies. Bull World Health Organ 77: 651–666. [PMC free article] [PubMed] [Google Scholar]
- 2. Weissman JB, Gangorosa EJ, Schmerler A, Marier RL, Lewis JN (1975) Shigellosis in day-care centres. Lancet 1: 88–90. [DOI] [PubMed] [Google Scholar]
- 3. von Seidlein L, Kim DR, Ali M, Lee H, Wang X, et al. (2006) A multicentre study of Shigella diarrhoea in six Asian countries: disease burden, clinical manifestations and microbiology. PLoS Med 3: e353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dutta S, Rajendran K, Roy S, Chatterjee A, Dutta P, et al. (2002) Shifting serotypes, plasmid profile analysis and antimicrobial resistance pattern of Shigellae strains isolated from Kolkata, India during 1995–2000. Epidemiol Infect 129: 235–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sack RB, Rahman M, Yunus M, Khan EH (1997) Antimicrobial resistance in organisms causing diarrheal disease. Clin Infect Dis 24: S102–05. [DOI] [PubMed] [Google Scholar]
- 6. Barbosa TM, Levy SB (2000) The impact of antibiotic use on resistance development and persistence. Drug Resist Updat 3(5): 303–311. [DOI] [PubMed] [Google Scholar]
- 7. Salam MA, Dhar U, Khan WA, Bennish ML (1998) Randomised comparison of ciprofloxacin suspension and pivmecillinam for childhood shigellosis. Lancet 15: 522–27. [DOI] [PubMed] [Google Scholar]
- 8. Talukder KA, Khajanchi BK, Islam MA, Dutta DK, Islam Z, et al. (2004) Genetic relatedness of ciprofloxacin resistant Shigella dysenteriae type 1 strains isolated in south Asia. J Antimicrob Chemother 54: 730–34. [DOI] [PubMed] [Google Scholar]
- 9. Talukder KA, Khajanchi BK, Islam MA, Islam Z, Dutta DK, et al. (2006) Fluoroquinolone resistance linked to both gyrA and parC mutations in the quinolone resistance-determining region of Shigella dysenteriae type 1. Curr Microbiol 52: 108–11. [DOI] [PubMed] [Google Scholar]
- 10. Drlica K, Zhao X (1997) DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol Mol Biol Rev 61: 377–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Poole K (2000) Efflux-mediated resistance to fluoroquinolones in gram-negative bacteria. Antimicrob Agents Chemother 44: 2233–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang M, Sahm DF, Jacoby GA, Hooper DC (2004) Emerging plasmid-mediated quinolone resistance associated with the qnr gene in Klebsiella pneumoniae clinical isolates in the United States. Antimicrob Agents Chemother 48: 1295–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tran JH, Jacoby GA, Hooper DC (2005) Interaction of the plasmid-encoded quinolone resistance protein Qnr with Escherichia coli DNA gyrase. Antimicrob Agents Chemother. 49(1): 118–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gellert M, Mizuuchi K, O’Dea MH, Itoh T, Tomizawa JI (1977) Nalidixic acid resistance: a second genetic character involved in DNA gyrase activity. Proc Natl Acad Sci U S A 74: 4772–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Piddock LJ (1995) Mechanisms of resistance to fluoroquinolones: state-of-the-art 1992–1994. Drugs 49: 29–35. [DOI] [PubMed] [Google Scholar]
- 16. Everett MJ, Jin YF, Ricci V, Piddock LJ (1996) Contributions of individual mechanisms to fluoroquinolone resistance in 36 Escherichia coli strains isolated from humans and animals. Antimicrob Agents Chemother 40: 2380–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Truong QC, Nguyen Van JC, Shlaes D, Gutmann L, Moreau NJ (1997) A novel, double mutation in DNA gyrase A of Escherichia coli conferring resistance to quinolone antibiotics. Antimicrob Agents Chemother 41: 85–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Komp Lindgren P, Karlsson A, Hughes D (2003) Mutation rate and evolution of fluoroquinolone resistance in Escherichia coli isolates from patients with urinary tract infections. Antimicrob Agents Chemother 47(10): 3222–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Turner AK, Nair S, Wain J (2006) The acquisition of full fluoroquinolone resistance in Salmonella Typhi by accumulation of point mutations in the topoisomerase targets. J Antimicrob Chemother 58(4): 733–40. [DOI] [PubMed] [Google Scholar]
- 20. Morgan-Linnell SK, Zechiedrich L (2007) Contributions of the combined effects of topoisomerase mutations toward fluoroquinolone resistance in Escherichia coli. Antimicrob Agents Chemother 51(11): 4205–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kim JY, Kim SH, Jeon SM, Park MS, Rhie HG, et al. (2008) Resistance to fluoroquinolones by the combination of target site mutations and enhanced expression of genes for efflux pumps in Shigella flexneri and Shigella sonnei strains isolated in Korea. Clin Microbiol Infect 14: 760–65. [DOI] [PubMed] [Google Scholar]
- 22. Wang XY, Du L, Von Seidlein L, Xu ZY, Zhang YL, et al. (2005) Occurrence of shigellosis in the young and elderly in rural China: results of a 12-month population-based surveillance study. Am J Trop Med Hyg 7: 416–22. [PubMed] [Google Scholar]
- 23. Talukder KA, Dutta DK, Safa A, Ansaruzzaman M, Hassan F, et al. (2001) Altering trends in the dominance of Shigella flexneri serotypes and emergence of serologically atypical Shigella flexneri strains in Dhaka, Bangladesh. J Clin Microbiol 39: 3757–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Disk Susceptibility Tests- Tenth Edition (2009) Approved Standard M02-A10. CLSI, Wayne, PA, USA.
- 25. Talukder KA, Islam MA, Dutta DK, Ansaruzzaman M, Hassan F, et al. (2002) Phenotypic and genotypic characterization of serologically atypical strains of Shigella flexneri type 4 isolated in Dhaka, Bangladesh. J Clin Microbiol 40: 2490–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Talukder KA, Islam MA, Khajanchi BK, Dutta DK, Islam Z, et al. (2003) Temporal shifts in the dominance of serotypes of Shigella dysenteriae from 1999 to 2002 in Dhaka, Bangladesh. J Clin Microbiol 41: 5053–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Munshi MH, Sack DA, Haider K, Ahmed ZU, Rahaman MM, et al. (1987) Plasmid-mediated resistance to nalidixic acid in Shigella dysenteriae type 1. Lancet ii: 419–21. [DOI] [PubMed] [Google Scholar]
- 28. Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, et al. (1995) Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol 33: 2233–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jacoby GA, Chow N, Waites KB (2003) Prevalence of plasmid-mediated quinolone resistance. Antimicrob Agents Chemother 47: 559–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rahman M, Mauff G, Levy J, Couturier M, Pulverer G, et al. (1994) Detection of 4-quinolone resistance mutation in gyrA gene of Shigella dysenteriae type 1 by PCR. Antimicrob Agents Chemother 38: 2488–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vila J, Ruiz J, Goñi P, De Anta MT (1996) Detection of mutations in parC in quinolone-resistant clinical isolates of Escherichia coli . Antimicrob Agents Chemother 40: 491–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang H, Dzink-Fox JL, Chen M, Levy SB (2001) Genetic characterization of highly fluoroquinolone-resistant clinical Escherichia coli strains from China: role of acrR mutations. Antimicrob Agents Chemother 45: 1515–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–10. [DOI] [PubMed] [Google Scholar]
- 34. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pournaras S, Maniati M, Spanakis N, Ikonomidis A, Tassios PT, et al. (2005) Spread of efflux pump-overexpression, non-metallo-β-lactamase-producting,metopenem-resistant but ceftazidime-susceptible Pseudomonas aeruginosa in a region with bla VIM endemicity. J Antimicrob Chemother 56: 761–64. [DOI] [PubMed] [Google Scholar]
- 36. Vito R, Piddock L (2003) Accumulation of garenoxacin by Bacteroides fragilis compare with that of five fluoroquinolones. J Antimicrob Chemother 52: 605–09. [DOI] [PubMed] [Google Scholar]
- 37. Cho H, Oh Y, Park S, Lee Y (2001) Concentration of CCCP should be optimized to detect the efflux system in quinolone-susceptible Escherichia coli. J Microbiol 39: 62–66. [Google Scholar]
- 38. Pichel M, Brengi SP, Cooper KL, Ribot EM, Al-Busaidy S, et al. (2012) Standardization and International Multicenter Validation of a PulseNet Pulsed-Field Gel Electrophoresis Protocol for Subtyping Shigella flexneri Isolates. Foodborne Pathog Dis 9: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ding J, Ma Y, Gong Z, Chen Y (1999) A study on the mechanism of the resistance of Shigellae to fluoroquinolones. Zhonghua Nei Ke Za Zhi 38: 550–53. [PubMed] [Google Scholar]
- 40. Oonaka K, Fukuyama M, Tanaka M, Sato K, Ohnishi K, et al. (1998) Mechanism of resistance of Shigella flexneri 2a resistant to new quinolone antibiotics. Kansenshogaku Zasshi 72: 365–70. [DOI] [PubMed] [Google Scholar]
- 41. Friedman SM, Lu T, Drlica K (2001) Mutation in the DNA gyrase A Gene of Escherichia coli that expands the quinolone resistance-determining region. Antimicrob Agents Chemother 45: 2378–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cattoir V, Poirel L, Rotimi V, Soussy CJ, Nordmann P (2007) Multiplex PCR for detection of plasmid-mediated quinolone resistance qnr genes in ESBL-producing enterobacterial isolates. J Antimicrob Chemother 60: 394–97. [DOI] [PubMed] [Google Scholar]
- 43. Pu XY, Pan JC, Wang HQ, Zhang W, Huang ZC, et al. (2009) Characterization of fluoroquinolone-resistant Shigella flexneri in Hangzhou area of China. J Antimicrob Chemother 63: 917–20. [DOI] [PubMed] [Google Scholar]