Abstract
The dynamic process of cell fate specification is regulated by networks of regulatory genes. The architecture of the network defines the temporal order of specification events. To understand the dynamic control of the developmental process, the kinetics of mRNA and protein synthesis and the response of the cis-regulatory modules to transcription factor concentration must be considered. Here we review mathematical models for mRNA and protein synthesis kinetics which are based on experimental measurements of the rates of the relevant processes. The model comprises the response functions of cis-regulatory modules to their transcription factor inputs, by incorporating binding site occupancy and its dependence on biologically measurable quantities. We use this model to simulate gene expression, to distinguish between cis-regulatory execution of “AND” and “OR” logic functions, rationalize the oscillatory behavior of certain transcriptional auto-repressors and to show how linked subcircuits can be dealt with. Model simulations display the effects of mutation of binding sites, or perturbation of upstream gene expression. The model is a generally useful tool for understanding gene regulation and the dynamics of cell fate specification.
Keywords: Mathematical modeling, Gene regulation in development, Dynamics, Logic functions
Introduction
Throughout development the timing of gene activation is critical to the execution of the regulatory program. The topology of developmental gene regulatory networks (GRNs) specifies inputs into the regulatory system of each participating gene, and where this gene encodes a transcription factor, its outputs to target genes in the next tier of the hierarchical network. Thus any given domain of a GRN consists of prior, or upstream, and responding, or downstream, regulatory gene circuitry. In the operation of the GRN, time flows in the same direction as the causality determined in the GRN topology (except for feedbacks). Thus in terms of transcription dynamics, the measurable output of the GRN is a temporal sequence of cohorts of regulatory gene expressions. There is a one way logic relationship between overall GRN architecture and the temporal progression of transcription patterns: GRN topology predicts the kinetics of this progression, barring post-transcriptional modulations; however, it is almost impossible to infer network topology exclusively from dynamic expression data, except for linear cascades of such simplicity as are rarely seen in embryonic development.
The causal linkage between GRN topology and transcription kinetics produces many situations in experimental analysis of developmental regulatory systems where kinetic analysis provides great clarification. For example, for small GRN subcircuits, it is invariably illuminating to predict and mechanistically explain observed kinetic behavior. Appropriate kinetic analysis is required to determine how a regulatory system actually operates downstream of the GRN topology. Does it work as an irreversible, progressive developmental system in which the qualitative sequence of gene expressions is insensitive to exact levels of the prior transcription factors, and successive genes are activated long before any of the products attain steady state as in early development (Saulier-Le Drean et al., 1998; Nasiadka and Krause, 1999; Bolouri and Davidson, 2003) Or is it a system the qualitative outputs of which depend specifically on particular transcription factor levels, as for instance in the Dorsal gradient response genes of the Drosophila embryo (Stathopoulos and Levine, 2004; Levine and Davidson, 2005),or as in many postembryonic systems such as hematopoietic specification, e.g., Wallin et al. (1998) and Laslo et al. (2006). Another whole class of applications deals with the kinetic consequences of the types of logic operations cis-regulatory systems perform in integrating their various inputs (Yuh et al., 2001). Finally, kinetic models can explain the shape of quantitative accumulation time courses for mRNA or protein, and enable extraction of the degradation and synthesis rate constants for these molecules.
Incorporating the spatio-temporal expression pattern of the inputs and the response functions of cis-regulatory modules into a comprehensive mathematical model is a complicated task. The most natural approach is to build a model that simulates the dynamics of the biological system based on experimental study of the principal processes. Thorough studies of embryonic mRNA and protein synthesis, and identification of the rate limiting functions, were conducted in the 1960s, 70s, and early 80s (reviewed in chapter II, Davidson, 1986). Simple canonical mathematical equations that describe mRNA and protein kinetics were derived during these decades in many quarters. This set of equations has continued to be useful; for example it was recently applied to the extraction of turnover rates of maternal and zygotic mRNAs in sea urchin embryos (Howard-Ashby et al., 2006). Based on emerging experimental studies of cis-regulatory function in animal systems (Davidson, 2006), a model was recently developed to describe transcriptional gene regulation, incorporating the occupancy of binding sites by transcription factors (Bolouri and Davidson, 2003). This model integrates earlier work by many authors on the dependence of the occupancy on biologically measurable quantities such as transcription factor concentration, the total available DNA, and binding to specific versus nonspecific sites (see below for references).
Here we review the set of differential equations we and our colleagues find useful for modeling mRNA and protein synthesis, transcription factor interaction with cis-regulatory modules, and the resulting dynamics of gene expression. We demonstrate how AND and OR logic operations of the cis-regulatory modules can be included in the model. We show how the response functions of a cis-regulatory module can induce temporal variation of the output, even when the inputs are similar, and consider the dynamics produced by repressors that function only after their concentration crosses a threshold level. The quantitative approach in the following describes populations of molecules and cis-regulatory modules of given genes in all the cells expressing them. That is, the model variables are average concentrations, and the model parameters are the average rates for all the cells of a given territory. In developing animal systems the number of mRNA and protein molecules of each species is large, their turnover rates in general slow, and many cells contribute to any given territory or embryo or tissue, so that any stochastic transcriptional fluctuations at individual genes are inconsequential at the population level. The main feature of animal gene regulatory systems is that the regulation of cell fate specification depends essentially on the network architecture and the response functions of the constituent cis-regulatory modules, and not on hypersensitivity to small fluctuations. An average approximation model is thus directly useful for understanding and simulating the kinetics of gene regulation in animal development.
Transcriptional kinetics
The processes involved in mRNA synthesis
Two processes control the rate of primary transcript synthesis, viz., transcript initiation and RNA polymerase translocation (Fig. 1A). Transcripts are initiated when the RNA polymerase complex that binds to the promoter of the gene starts transcribing RNA. Transcription initiation rate, the number of initiations per minute, depends on the efficiency of the enhancer in activating transcription. But the maximal possible initiation rate depends on the RNA polymerase translocation rate, since the next polymerase cannot bind to the promoter before the currently transcribing polymerase has cleared about 100 bp of DNA (Davidson, 1986, p142–149).
Fig. 1.
Processes involved in transcription and translation. (A) For mRNA synthesis these are transcription initiation, RNA polymerase translocation, mRNA processing and mRNA export from the nucleus to the cytoplasm. The level of mRNA depends also on mRNA turnover rate. The processes that control protein level are translation and protein turnover rates. (B) The initiation rate controls the number of transcripts that are generated within a given time interval. The higher is the initiation rate the more mRNA copies are produced.
The translocation rate is largely sequence independent and temperature dependent. In sea urchin (S. purpuratus) embryos that are cultured at 15 °C, the translocation rate was measured to be 6–9 nucleotides per second (Aronson and Chen,1977; Davidson,1986). The translocation rate obeys the Q10 law, that is, for every 10 °C increase in temperature, there is about a 2–2.5 times increase in the translocation rate (Davidson, 1986, p144–145). Considering a translocation rate of 6–9 nucleotides per second, it takes the RNA polymerase about 11–17 s to transcribe 100 bp, and enable the next RNA polymerase to bind to the promoter. This means that the maximal initiation rate at 15 °C is about 5.5 initiations per minute, for one DNA copy of a gene. At higher temperature the initiation rate may be higher, according to the Q10 law for the translocation rate.
A typical eukaryote gene size is about 30 kb, including introns and exons. At 15 °C, at a translocation rate of 9 nucleotides per second, it takes about 56 min to complete the first primary transcript. This induces an inherent delay in the response to transcriptional activation that depends on gene size. That is, the first mRNA molecule will be generated only after the first RNA polymerase finished transcribing the entire gene. mRNA processing, that is, capping, splicing and polyadenylation, occurs while the primary RNA is being transcribed (Shuman, 1997), and therefore does not induce further delays. Once the first transcript is completed, the mRNA synthesis rate depends only on the initiation rate (Fig. 1B). mRNA export from the nucleus to the cytoplasm requires about 10–30 min depending on the mRNA (Schroder et al., 1989; Fuke and Ohno, 2008). This induces another delay in the response to transcriptional activation, since the mRNA can be translated into a protein only once it is exported from the nucleus and binds to the ribosomes.
Once in the cytoplasm the mRNA is targeted for degradation by various stochastically acting degradation mechanisms (Gorospe and Baglioni, 1994; Zubiaga et al., 1995; Wilusz et al., 2001; Moss, 2007). The common feature of the different degradation mechanisms is that the probability of degradation depends mostly on the 3′ untranslated region of the mRNA (3′ UTR sequence). The mRNA degradation process can include several steps, e.g., sequential poly-A tail cleavages, and therefore the time that individual mRNA molecules spend in the cytoplasm increases their probability of being degraded (Wilusz et al., 2001). However, when we consider a population of similar mRNA molecules we can still consider the average degradation rate as independent of the history of the molecules (Pedraza and Paulsson, 2008). These remarks refer to mRNAs degrading at the typical default rate for the given cell type, which appears to pertain to the majority of mRNA species. However, the sequence of some mRNAs confers on them great stability (Cabrera et al., 1984). In addition mRNAs that are microRNA targets in given cells may be destroyed at higher than average rates (Moss, 2007; Filipowicz et al., 2008). Nevertheless, in all cases, the degradation rate and the initiation rate are the parameters that determine the quantitative level of each mRNA, and these are the specific measurable variables quantitatively responsible for the expression levels of different genes.
The processes involved in protein synthesis
Once the mRNA enters the cytoplasm the ribosomes bind to it sequentially, and the message is translated. In a typical polysome the ribosomes are closely packed, with a center to center distance of about 135 bases (Martin and Miller, 1983). In sea urchin embryos at 15 °C the translation rate, i.e., the rate at which peptide is produced as the ribosomes progress along the message, was measured to be 1.8 amino acids/s=5.4 b/s (Goustin and Wilt, 1981). Therefore it takes the ribosome about 25 s to translate 135 bases, so the next ribosome can bind to the mRNA. That means the translation initiation rate is about 2 initiations per minute per mRNA molecule. Therefore, in a fully loaded polysome operating at steady state, this is also the rate of production of the completed protein, which is released as the ribosome leaves the mRNA. The average size of mRNA molecules in sea urchin embryos, as in many animal systems, is about 2.5 kb, so it takes about 8 min on average for each protein molecule to be translated as its ribosome traverses the mRNA.
Proteins are degraded with a probability that depends on their structure (Hershko and Ciechanover, 1998; Naujokat and Saric, 2007). Even though some of the degradation mechanisms involve multiple steps, and therefore the individual protein senesces through time, the average degradation rate for a population of protein can again be considered as independent of the history of the molecules (Pedraza and Paulsson, 2008). The translation rate is largely independent of the coding sequence, and is similar for all proteins in a given cell type at given conditions. Hence the parameters that control the level of a given protein are mainly its mRNA level and the protein degradation rate. MicroRNAs can bind to mRNA and prevent its translation (Moss, 2007; Filipowicz et al., 2008), and for mRNA regulated by microRNAs, the parameter that controls the protein level is the quantity of free mRNA remaining that can form polysomes.
Mathematical model for transcription kinetics
We review here the most generally used mathematical model for transcription kinetics. This approach is simple and intuitive, and is based on experimental observation of the rate limiting processes described above. Average numbers of mRNA and protein molecules are treated as continuous functions specified in a set of ordinary differential equations. This apparatus can be used, for example, to model the dynamic accumulation of either specific mRNA and protein or total populations of these molecules in the developing embryo.
The change of the number of mRNA molecules in a time interval equals the rate of flow of newly synthesized mRNA into the cytoplasm from the nucleus, minus the amount of mRNA that is degraded in the cytoplasm during this time (see Davidson, 1986, p 548–551). If processing is 100% efficient, i.e., every newly synthesized pre-mRNA is converted into a mature message, then the rate of flow into the cytoplasm is the same as the rate of transcriptional initiation of the pre-mRNA, Is. This is in fact almost always the case in sea urchin embryos (Cabrera et al., 1984; note, however, that incompletely efficient processing may be a general property of growing oocytes, as reviewed by Davidson, 1986, p.69, 359). But the equivalence of cytoplasmic mRNA entry flow rates and nuclear transcriptional initiation rates is of course only true if we consider these rates in molar terms, i.e., in terms of numbers of molecules, and not in mass terms, since pre-mRNAs may be 10–20× as large as mature mRNAs. Here we assume the molar transcription initiation rate, Is, is also the rate of flow of mRNA into the cytoplasm, again either for a particular species or the whole population:
| (1) |
Here mRNA(t) is in units of number of molecules of mRNA at time t, and the units of Is are number of mRNA molecules synthesized/time interval (usually minutes or seconds). kdm is the mRNA turnover rate in units of time−1 (e.g., per min) As mentioned above, in considering a population of mRNA molecules, we can assume that the degradation dynamics can be described by an average turnover rate. The turnover rate is the probability of mRNA degradation in a given time interval, expressed as the fraction of the population that will be degraded (“turned over”) in that interval. Therefore the number of mRNA molecules degraded in a given time interval equals the amount of mRNA times the turnover rate constant, as in Eq. (1); both terms on the right side of this equation represent mRNA/time.
Similarly, the change of the number of protein molecules in a time interval equals the number of protein molecules translated during that interval minus the number degraded in this time:
| (2) |
Here kdP is the protein turnover rate constant, and kt is the translation rate constant, and the units of both constants are time−1. We assume that for a population of protein molecules, the degradation dynamics can be described by an average turnover rate that is the probability of degradation in the given time interval, just as for the mRNA. The degradation of the protein is therefore equal to the product of the number of protein molecules that exist at any given time, and the turnover rate constant.
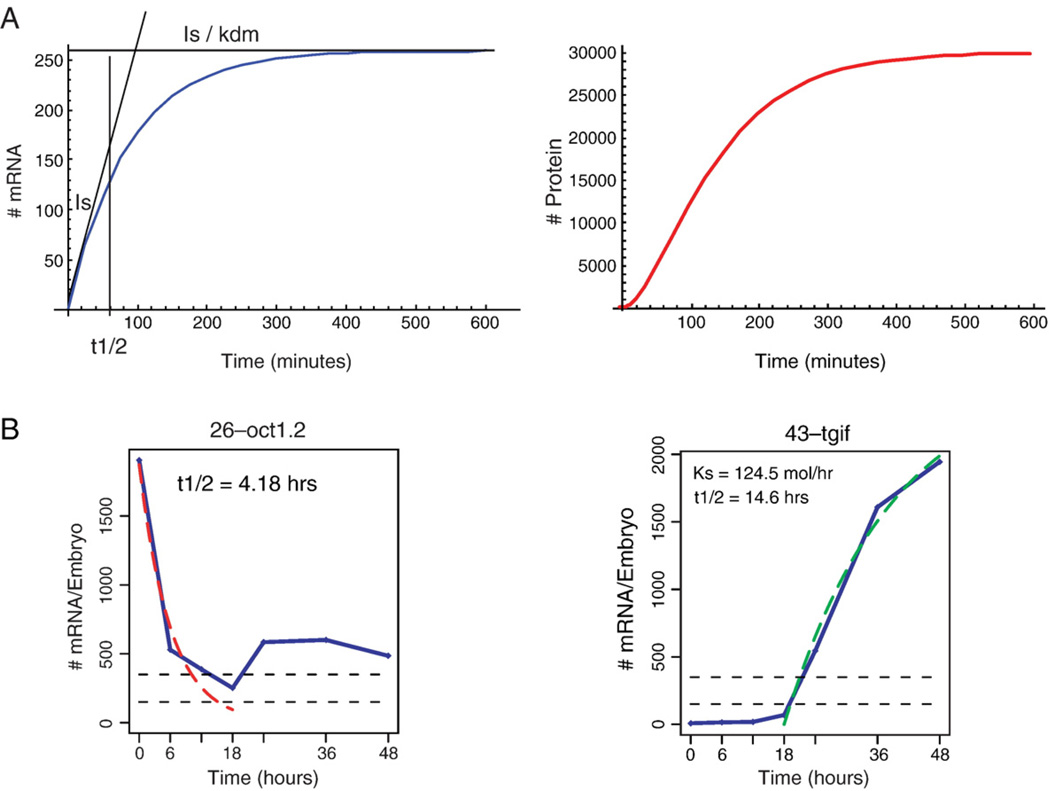
In Fig. 2A we present an illustration of mRNA and protein accumulation curves per cell that was obtained by assigning typical values to the kinetic constants, and solving Eqs. (1) and (2). It is important to note that even for the moderate initiation rate and the relatively rapid decay rates chosen in this simulation, for both the mRNA and the protein, (Is = 3 initiations/minute, kt = 2 protein molecules synthesized/(mRNA-minute), kdm=1.2% of the population decaying per minute, and kdp about the same), after 1 h the number of proteins per cell reaches about 5000 molecules. On the gene the actual frequency of initiation varies around the average. If we assume that the variation is according to the Poisson distribution, the standard deviation is the square root of the average, which is very low and has small effect on the rate of accumulation of protein.
Fig. 2.
mRNA and protein accumulation functions. (A) The mRNA and protein accumulation curves were obtained by substituting the following kinetic parameters in eqs. (1) and (2): Is=3 initiations/minute, kt=2 protein/(mRNA×minute), kdm=Ln2/60=0.012 min−1, and kdp=Ln2/40=0.014 min−1. The initiation rate, Is, is the initial linear slope of the mRNA accumulation curve. The maximal level of mRNA is the steady state level, Is/kdm. The half-life, t1/2=ln2/kdm, is the time when the mRNA accumulation function reaches half maximum. The simulation was done using Mathematica 5.2. (B) Use of the model to fit experimental measurements of mRNA levels in sea urchin embryos (Howard-Ashby et al., 2006). Left, for maternal transcripts, i.e., genes that their mRNA is present in the egg, the initiation rate is zero at early times and the mRNA level decays exponentially as e−kdmt, Eq. (1). A fit to the measured mRNA time course for the maternal phase of the gene oct1.2, results in half-lifetime of 4.18 h. (oct1.2 has a zygotic phase, i.e., transcription that starts after fertilization, initiated at 18 hpf, that was not fitted with the model). Right, Eq. (3) was used to fit the mRNA accumulation curve for the zygotic gene, tgif. The zygotic expression of this gene starts at about 18 hpf, so 18 hpf is the t=0 in this simulation. The result is an initiation rate of 124 molecules/hour and half-life of 14.6 h. Reprinted from Howard-Ashby et al., 2006. Dev. Biol. 300, 74–89; copyright Elsevier, Inc.
At early times when the mRNA level is still very low, the second term in the right hand side of Eq. (1) can be neglected and the equation becomes, dmRNA(t)/dt=Is. At this stage the mRNA increase is linear with time; i.e., it increases as Is · t (Fig. 2A left). At later times the system reaches a steady state where the time derivative is zero in Eq. (1), and therefore mRNASteadyState = Is/kdm (Fig. 2A). The general solution of Eq. (1), plotted in Fig. 2A, is:
| (3) |
The half-life or rise-time of a molecule is the time in which its level reaches half of the maximum. Since the maximal mRNA level is the steady state level, Is/kdm, at the half-life, t1/2, the mRNA level is Is/2kdm (Fig. 2A). Evaluating Eq. (3) when mRNA(t1/2)=Is/2kdm, we see that:
| (4) |
This is the relation between the half-life and the mRNA turnover rate. This relation enables the direct extraction of the turnover rate from the measurement of mRNA accumulation over time.
Both Is and t1/2 can be therefore acquired directly from measurement of mRNA accumulation time courses, as for example in the recent QPCR measurements of Howard-Ashby et al. (2006) reproduced in Fig. 2B. The measurement gives the total number of mRNA molecules of a certain species for the whole embryo, and therefore the measured initiation rate is the total initiation rate for all the cells in which the gene is expressed. This and other studies showed that the mRNA half life of maternal and zygotic mRNA in sea urchin embryos varies from a few hours to immeasurably long (>48 h) (Cabrera et al., 1984; Lee et al., 1992; Howard-Ashby et al., 2006). The initiation rate, Is, varies from zero, for genes not detectably expressed, to about 10 molecules per minute.
Cis-regulatory target site occupancy and the parameters that control it
The basic problem in modeling GRN kinetics is how to compute the activity (i.e., the kinetics of expression) of a gene in terms of the cis-regulatory inputs produced by the upstream genes. The method must also be capable of building in the mode of operation of the downstream cis-regulatory system, and of dealing with repression as well as activation. There are many approaches to modeling gene expression (Smolen et al., 2000; de Jong, 2002; Giurumescu et al., 2006; Tomlin and Axelrod, 2007; Zhu et al., 2007). Here we review and further develop the model presented by Bolouri and Davidson (2003), using kinetic values typical of sea urchin embryos. This approach utilized in turn several earlier treatments, the most important of which were those of Ackers et al. (1982) and Emerson et al. (1985). The initial objective must be computation of cis-regulatory target site binding in terms of the concentrations and the qualitative properties of the transcription factors that recognize and bind these sites. This is the essential first step because this is the relationship which causally links upstream to downstream genes in the GRN topology.
Binding site occupancy
Transcription factor–DNA interactions have long been treated as classical thermodynamic equilibrium problems. More recently statistical mechanics models have been derived that deal with the actual microscopic sequence of events when the protein approaches and binds the DNA (McGhee and von Hippel, 1974; Bintu et al., 2005; Lipniacki et al., 2006; Murugan, 2006; Ribeiro, 2007; Zhu et al., 2007). However our problem is not this, but rather the average probability that the target site will be occupied as a function of two parameters: the overall concentration of the factor and the stability of the DNA–protein complex once formed. This probability, a readout of the thermodynamic equilibrium treatment, is the extremely useful parameter “occupancy.”
If Ds is the molar concentration of non-occupied specific sites in a given genome, and PDs is the molar concentration of transcription factor–DNA complexes, the occupancy, YP, is defined:
| (5) |
That is, Yp is the ratio between the occupied sites, PDs, and the entire number of specific binding sites in the genome, occupied and unoccupied. Measurement of PDs in a living embryo cell is experimentally demanding. However, numerous studies indicate that equilibrium kinetic studies carried out in vitro provide quantitative parameters that can be referred to reactions of the same proteins in vivo, once certain obvious differences such as the concentrations of the reactants and of the salt in the medium are accounted for (Emerson et al., 1985; Calzone et al.,1988; Calzone et al., 1991; Hoog et al., 1991; Walsh and Carroll, 2007).
Considering the processes of formation of a transcription factor– DNA complex, and its decay, the rate of change in the amount of the factor–DNA complex can be seen to depend on complex formation and dissociation rates, kaS and kdS, respectively (Fig. 3A):
| (6) |
Fig. 3.
Occupancy and transcription. (A) The occupancy of a binding site depends on the ratio of its association and dissociation rate constants, kaS and kdS, respectively, and on the transcription factor concentration. (B) Initiation rate dependence on occupancy for different activation strengths, kb (Eq. (12)). Red curve, kb=5, green curve, kb=20 and blue curve kb=50. At low occupancy the initiation rate increases linearly with the occupancy with a slope of kb. In this example we consider Imax=11 initiations per minute, as calculated in text for 2 gene copies at 15 °C. The simulation was done using Mathematica 5.2. (C) Cooperative binding to the DNA increases the stability of the factor–DNA complex. The cooperativity constant, Kq, indicates how much the two factor–DNA complex is stabilized compared to independent binding of the two factors. Free energy contributions for DNA–protein and protein–protein interactions are indicated by green and yellow arrows respectively.
Here the association rate, kaS, is in terms of per mol/L×min, or (mol/L)−1×min−1. The dissociation rate, kdS, is in min−1 and the free protein concentration, P, is in mol/L. When the left side of Eq. (6) is zero, the binding reaction is said to be in equilibrium, and thus the ratio of complex to free potential reactants defines the equilibrium constant, Ks:
| (7) |
Ks depends on the chemistry of the factor–DNA interaction and thus basically on the primary sequence of the transcription factor: it is an intrinsic character of the protein that reflects the “affinity” of the transcription factor for the specific site to which it binds. More correctly, Ks indicates the stability of the site-specific DNA–protein complex, and thus in comparing diverse interactions that display widely different values of Ks, the differences are seen to depend almost entirely on the different values of the dissociation rate, kdS (Riggs et al., 1970; Calzone et al., 1988; Okahata et al., 1998; Wen et al., 2000; Cranz et al., 2004).The association rate, kaS is similar between different transcription factors, as it depends directly on how fast they diffuse, and plus or minus a factor of around two, most transcription factors are more or less similar in size.
But transcription factors do not just execute all or nothing binding interactions. They contain basic domains, and all react non-specifically but measurably with the acid phosphate bridge of the genomic DNA backbone. As a general rule of thumb, the ratio of the stability of specific to non-specific complexes is 4 to 6 orders of magnitude. Since nonspecific sites are presented by every nucleotide phosphate, the number of these sites is the number of non-occluded base pairs, and in the enormous genomes of animal cells the factors are thus either hopping from one nonspecific site on the DNA to another, or are (relatively) stably bound to one of the specific sites. Essentially, non-specifically bound factor is concentrated in the vicinity of the DNA by its weak affinity for any DNA base pair. It was pointed out three decades ago that were it not for this, most factors would never find their target sites at their concentrations in animal cell nuclei (Lin and Riggs, 1975).
The non-specific DNA–protein interactions can be described similarly to the specific interactions (Emerson et al. 1985), except that unlike Ks, Kn, the non-specific equilibrium constant, is almost the same for every type of factor:
| (8) |
Here PDn is the protein-nonspecific DNA complex and Dn is nonspecific unoccupied sites, both in molar concentration (Fig. 3A). The ratio between the specific and the non-specific equilibrium constants, Ks /Kn=Kr, can be measured experimentally by means we shortly take up, and, as indicated above, Kr is usually 104−106 (Calzone et al., 1988). Kr, the relative equilibrium constant, is a quantitative measure of specific site binding in the presence of the sea of non-specific sites, the actual case in the nucleus.
Since transcription factors in the nucleus are essentially all either non-specifically or specifically bound to the DNA (Emerson et al., 1985; Elf et al., 2007), the total transcription factor concentration, P0, is the sum of specific and non-specific protein–DNA complexes: P0=PDN+PDS. We can now use Kr, P0 and Eqs. (5)–(8) to derive two extremely useful expressions. The first of these provides the means of measuring all the parameters:
| (9) |
The size of the genome is several orders of magnitude larger than the number of proteins of any given transcription factor species, and thus Dn, the number of unoccupied nonspecific sites, can be approximated from the amount of open chromatin (non-occluded), which is about 90% of the total genome (Felsenfeld and Groudine, 2003). Now the other two parameters on the right side of Eq. (9), Kr and P0, can both be determined in vitro by means of gel shift experiments in which the amount of the transcription factor–DNA complex, PDs, is measured as the amount of Ds is experimentally increased (Calzone et al., 1988). As is evident from its form, P0 is the saturation plateau at high Ds, and Kr can be inferred from the initial slope of the function in Eq. (9) at low Ds, where (Emerson et al., 1985; Calzone et al.,1988; Calzone et al., 1991; Hoog et al., 1991; Walsh and Carroll, 2007).
The second relationship provides a new definition of occupancy in terms of the relative equilibrium constants that pertain to conditions inside the cell (Bolouri and Davidson, 2003):
| (10) |
Because of the huge size of the genomes of animal cells, even though Kn ≪ Ks, almost all the protein is bound to non-specific sites, and so P0 ~ PDn, and Eq. (10) can be written:
| (11) |
We see that at low transcription factor concentrations the occupancy increases linearly with the transcription factor concentration, with a slope of Kr/Dn. That is, the higher is the relative equilibrium constant the steeper is the increase of the occupancy for a given factor concentration. The maximal occupancy probability is 1, which means that the binding sites are all always occupied. In the following section we relate the occupancy to the transcription initiation rate and generate equations that describe gene regulatory circuits.
Kinetic models for transcriptional control of gene expression
Modeling transcriptional activation
The transcription initiation rate, Is [see Eq. (1)] depends on the binding site occupancy just defined, and on the efficiency of activation by the transcription factor. Many mechanisms are used by transcription factors to activate transcription, including interactions with various co-factors, interactions with chromatin remodeling enzymes, and direct interactions with the transcription apparatus. We require a mathematical model that relates the activation efficiency and the occupancy to the initiation rate, irrespective of the precise mechanism used. This expression should simply reflect the phenomenological behavior of the system. As the factor concentration increases, the occupancy of the binding site increases, and so does the initiation rate. For single site occupancy, we assume that the increase of the initiation rate is linear with the occupancy. However the initiation rate induced by strong activators at high occupancy might approach the maximal rate that translocation rate for the polymerase permits. When this limit is approached the rate of increase in transcription with occupancy will slow down; activation cannot be effected if the preceding RNA polymerase has not yet moved out of the way. Therefore, if Imax is the maximal initiation frequency that the translocation rate allows, one way of modeling the initiation rate dependence on the occupancy is:
| (12) |
Here YP is occupancy as above and kb represents the efficiency with which a given degree of occupancy causes a given amount of transcription initiation and is a measure of the activator strength (number/minute; Bolouri and Davidson, 2003). An illustration of initiation rate dependence on occupancy for different activation strengths, according to this model, is depicted in Fig. 3B. At low occupancy the initiation rate increases linearly with occupancy, with a slope of . For low activation strength (red curve) the initiation rate is in the linear region even at maximal occupancy, YP=1. For strong activation (blue curve), at high occupancy the initiation rate approaches the maximal initiation rate possible by the RNA polymerase translocation rate, as discussed above.
This relationship can be used to analyze the dynamic expression of regulatory genes that are functionally interconnected in a GRN (Bolouri and Davidson, 2003). The expressions for mRNA and protein synthesis, Eqs. (1)–(4) are written in terms of the number of molecules, while the protein concentrations in the occupancy expressions, Eqs. (5)–(11) are in molar terms. Thus the number of molecules of transcription factor has to be converted into molar units aswell, dividing by Avogadro's number and the nuclear volume (about 4×10−15 L per nucleus for sea urchin embryos). In considering the kinetics with which an upstream regulatory gene causes the appearance of transcripts of its downstream target gene, an additional factor is the inherent delay in the response due to the time it takes the RNA polymerase to transcribe the gene. The average delay is the product of the gene size and the RNA polymerase translocation rate. The first mRNA molecule is generated only after the entire gene is transcribed. After this time the mRNA generation depends only on the initiation rate. Taking transcription time into consideration, and introducing for Is in Eq. (1) the kinetic relation between occupancy and transcription rate in Eq. (12), we have:
| (13) |
Here Tm is the transcriptional delay in minutes. The relation describing the kinetics of appearance of the protein, Eq. (2), is unchanged. Using average rate constants for the sea urchin embryo, Bolouri and Davidson (2003) showed that the typical time interval in this system between activation of an upstream gene and of its target genes is 2–3 h, a result consistent with many sequential GRN time course measurements, e.g., Oliveri et al. (2008). Furthermore the downstream target genes are activated long before the upstream activator ever achieves steady state.
Modeling cis-regulatory logic
Usually genes are controlled by multiple regulatory inputs (Davidson, 2006). The function that the cis-regulatory modules execute upon these inputs can be reduced to basic AND, OR and NOT logic functions (Yuh et al., 1998; Yuh et al., 2001; Buchler et al., 2003; Istrail and Davidson, 2005; Istrail et al., 2007). When a cis-regulatory module is activated when either of its two inputs is present, then it acts as an “Additive OR” gate. In that case each factor contributes to the total initiation rate, which can be expressed as the sum of the initiation rates generated by the two inputs, A and B:
| (14) |
Here the indices A and B stand for the kinetic functions and parameters of the two activators, A and B. The maximal initiation rate is divided by the number of terms since the maximal initiation rate of the transcriptional system is always limited by the RNA polymerase translocation rate. The contribution of each factor to the initiation rate depends on its efficiency, expressed by kb. The general equation for N binding sites of transcription factors that behave as additive OR logic is a sum of all contributions multiplied by the maximal initiation rate divided by the number of terms.
When a cis-regulatory module is active only when both of two inputs are present it acts as an “AND” gate (experimental examples reviewed in Istrail and Davidson 2005). This is a very potent cis-regulatory information processing device often encountered in GRNs controlling spatial specification processes in development. That is, the downstream factor is activated only in the embryonic domain where its two inputs spatially overlap (Davidson, 2006). For AND gates, the single occupancy term Yp in Eqs. (12) and (13) is replaced with the product of the occupancies of each factor, YA(t)×YB(t). That is, when either one factor is absent, the initiation rate is zero. In general, when there are N binding sites of transcription factors that act as an AND gate, the single occupancy term Yp in Eqs. (12) and (13) is replaced with the product of the occupancies of each of the factors.
In many cases the DNA binding of two transcription factors is cooperative (Driever and Nusslein-Volhard, 1989; Garrity et al., 1994; Sugawara et al., 1995; Thanos and Maniatis, 1995; Ma et al., 1996; Burz et al., 1998; Sigvardsson et al., 2002; Walsh and Carroll, 2007). That means that the factors form a more stable complex once together on the DNA than when each of them occupies the DNA alone (Fig. 3C).We incorporate this effect in the equations by adding a cooperativity factor, Kq, to the double occupancy expression (Ackers et al., 1982; Bolouri and Davidson, 2003):
| (15) |
Here A(t) and B(t) are the molar concentrations of the two transcription factors (protein levels). The cooperativity constant, Kq, indicates how much the two factor–DNA complex is stabilized compared to independent binding of the two factors (i.e., Kq measures the free energy contributed to the complex by interaction between the bound proteins; Ackers et al., 1982). When Kq=1, the binding of the factors is not cooperative and the expression for the double occupancy is simply the product of the two single occupancies. When Kq>1 there is cooperative binding, and the double occupancy increase is steeper when the two factors are present. This equation can also be used to describe the cooperative binding of homodimers, where A(t)=B(t).
The expression for the kinetics of mRNA appearance controlled by a cis-regulatory AND gate is thus:
| (16) |
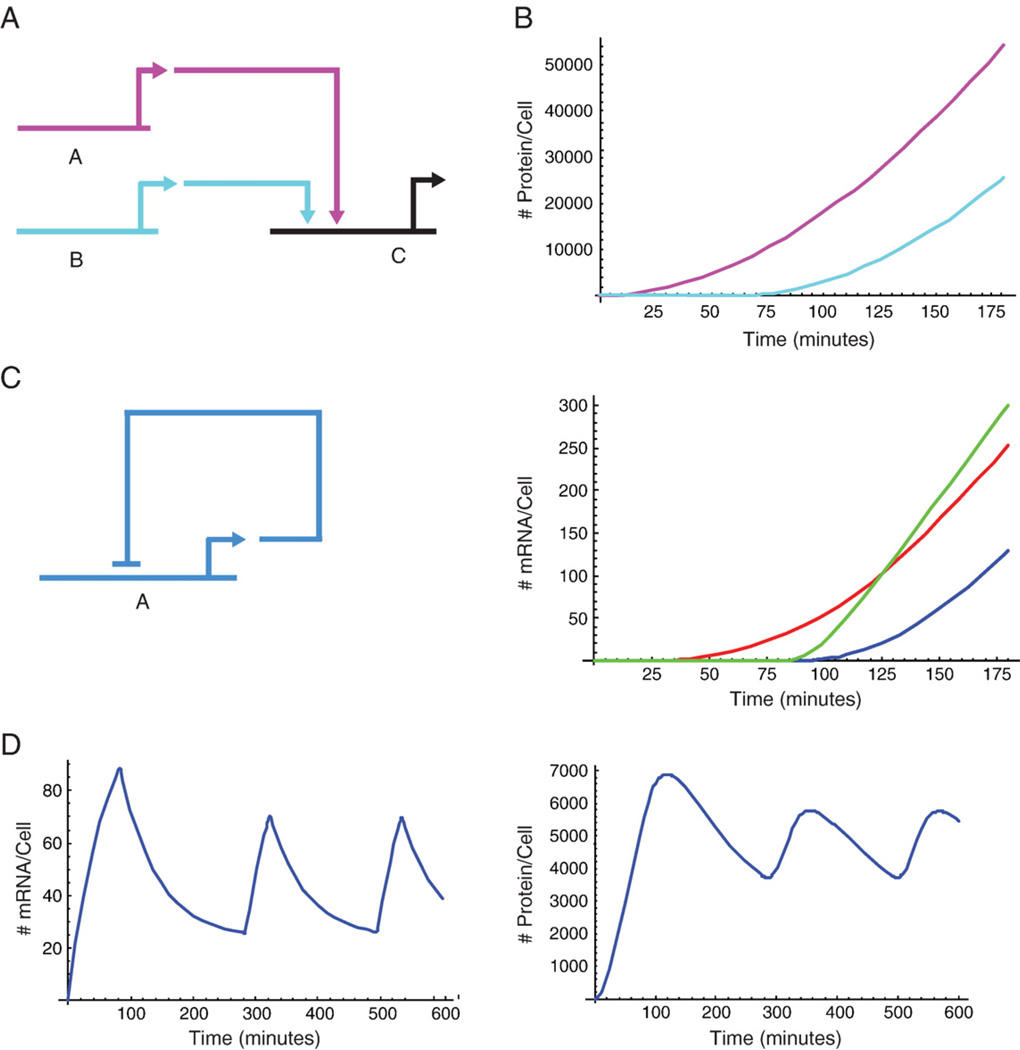
The kinetics of “Additive OR”, “AND” and “cooperative-AND” gates can be distinguished, as illustrated in Figs. 4A, B. In this example gene c is activated by transcription factors a and b, (Fig. 4A). Gene a is turned on at t=0, and gene b is turned on at t=60 min. a and b protein levels are plotted on the upper panel of Fig. 4B. The resulting expression levels of c mRNA for these 3 kinds of gate are plotted in the lower panel of Fig. 4B. When c is regulated by “a Additive OR b”, its level starts increasing immediately after a appears, and the mRNA accumulation slope increases once b is present (red curve). When c is regulated by “a AND b”, its level starts increasing only after both a and b are present in the system (green and blue curves.). The mRNA level of the “cooperative-AND” gate (green curve) increases more rapidly than of the “AND” gate (blue curve), since the occupancy is higher for the same input concentrations when the binding is cooperative. A cis-regulatory module that is activated by an “a AND b” gate, is shut off when the site where either factor binds is mutated. A cis-regulatory module that is activated by an “a Additive OR b” gate is still active after a single mutation of either a or b sites. The observed change is only in the decrease of the mRNA accumulation slope and in the time of activation, which will be delayed if the early input site is mutated. Additive kinetic behavior of inputs is often observed experimentally, as e.g., by Nam et al. (2007).
Fig. 4.
Simple GRN subcircuits and kinetic outputs. (A) Subcircuit in which regulatory genes a and b produce factors that activate the expression of gene c. (B) Time courses for expression of a, b and c, assuming different logic gates. Upper panel: Time courses for protein output of a (magenta) and b (cyan). b is activated 60 min after the activation of a and both factors are activated at constant initiation rate of Is=2. Bottom panel: Time course for c mRNA under different cis-regulatory gates processing inputs from a and b genes. Red curve: c is regulated by a Additive OR b inputs, Eq. (14). Blue curve: c is regulated by a AND b inputs, Eq. (16), Kq=1. Green curve: c is regulated by a AND b inputs and the binding of a and b is cooperative, Eq. (16), Kq=20. The parameters used in this simulation are: Relative equilibrium constant, Kr=105, activation strength, kb=5, mRNA turnover rate kdm=0.001 min−1, protein turnover kdp=0.002 min−1, translation rate, kt=2 protein/(mRNA×minute), mRNA transcription delay, Tm=20 min. The number of non-specific sites, Dn, was estimated as 90% of the total sea urchin genome, which is 8×108, so Dn=7.2×108. The initial levels of all genes, a, b and c was assumed to be zero at time zero. (C) Auto-repression sub-circuit. (D) Time courses of mRNA (left) and protein (right) for an auto-repressor operating according to the threshold model (Eq. 18). The kinetic parameters used in this simulation are: kt=2, Is=2, Kr=105, kdm=kdp=0.017 min−1, Y0=0.36, B0=0.2, Dn=7.2×108 and Tm=20 min. Simulations were done using Mathematica 5.2.
Modeling transcriptional repression
Repression is commonly used in development to exclude ectopic expression of regulatory genes, and to set boundaries of spatial regulatory states (Arnosti et al., 1996; Fujioka et al., 1999; Oliveri et al., 2006; Oliveri and Davidson, 2007). Various mechanisms induce transcriptional repression (Emerson et al., 1987; Levine and Manley, 1989; Gray et al., 1994; Gray and Levine, 1996b; Barolo and Levine, 1997; Nibu et al., 2003; Janssens et al., 2006). Most repressors or corepressors recruit chromatin remodeling proteins that locally modify the histone acetylation or methylation status and thereby silence gene expression (Lee et al., 2001; Nuthall et al., 2002; Nuthall et al., 2004; Di Caro et al., 2007; Tai et al., 2007). Some repressors can interact directly with the transcription complex to block transcription initiation (Ptashne, 2007). More rare mechanisms of repression are competition of repressor and activator for binding to the same site (Kamachi and Kondoh, 1993; Sekido et al., 1997) and repressor binding next to an activator and interfering with the activator interaction with the transcription complex (Gray and Levine, 1996a; Janssens et al., 2006).
Many, perhaps most repression processes are multistep: the initial sequence-specific repressor recruits other proteins which progressively install silencing (Pikaart et al., 1998; Mutskov and Felsenfeld, 2004; Stirzaker et al., 2004; Santoro and Grummt, 2005; Dodd et al., 2007). The kinetics of the process will depend on the mechanism and the nature of the secondary processes. The thermodynamic approach that we use here to describe activation is therefore not suitable to describe most repression processes, in particular not the irreversible ones, where the gene remains silenced even after the repressor is not present. Irreversible repression and silencing have been modeled by others (Dodd et al., 2007; Sedighi and Sengupta, 2007), and lies beyond the scope of this review.
One particularly interesting aspect of transcriptional repression often encountered in developmental GRNs is autorepression of regulatory gene expression. This can result in occurrence of a temporal peak of expression which is ultimately extinguished as the factor achieves repressive occupancy of its own cis-regulatory system. Or, under particular circumstances it can produce an oscillation (e.g., Hirata et al., 2002; Nuthall et al., 2002; Bernard et al., 2006; Rateitschak and Wolkenhauer, 2007; Bessho and Kageyama, 2003). That is, the regulatory gene produces a factor which at high concentrations binds to its own cis-regulatory system and then turns itself off (Fig. 4C). The mRNA and factor concentration then decay, the bound repressor leaves, and the gene turns on again. A model for such a system is shown in Eqs. 17 and 18. To represent the transition between activation and repression, we use a step function to generate the initiation rate. In this model, the initiation rate can have two values, depending on the occupancy of the repressor binding site. When the repressor site occupancy is below the critical threshold level, the initiation rate is unaffected and equals the maximal rate enabled by the enhancer. When the repressor site occupancy is above the critical level repression occurs and remains until the repressor decays. YR(t) is the occupancy of the repressor binding site at time t, Y0 is the threshold level, and the step function is defined as:
| (17) |
Here B0 is set so Is×B0 is the basal expression level, and Is is the maximal initiation rate that the enhancer generates. The rate of change in the mRNA output of a downstream gene is then:
| (18) |
The equation for protein accumulation is unchanged and is described by Eq. (2). To illustrate the oscillatory kinetics that result from the threshold behavior built into Eqs. (17) and (18) their solution is plotted in Fig. 4D. Here a steady state balance between repression and activation is replaced by oscillation because the initiation rate can only have two values, matching “on” and “off” levels of activity, and therefore it cannot be tuned to achieve equal rates of generation and turnover. The period of the oscillations depends directly on the turnover rate of the mRNA and the protein. The lower the turnover rate, the longer the period of the oscillations. For long lived mRNA and protein the expression profile has a single peak followed by a slow decay.
Modeling compound circuits
This model can be used to simulate GRN circuits. The equations should respond to the circuit topology and the logic the cis-regulatory modules of the circuit genes apply on their inputs. We demonstrate the use of the model to simulate a network subcircuit that is based on common features we observe in the sea urchin endomesoderm specification GRN, Fig. 5A (Davidson, 2006; Ben-Tabou de-Leon and Davidson, 2007; Oliveri et al., 2008). Gene A activates gene B. Gene B has a positive feedback into its own cis-regulatory module. The expression of gene A is transient and decays with time, but due to the positive feedback, B keeps itself on even after A is off. This is a common lock down mechanism used by GRNs to maintain a specification state (Davidson, 2006; Istrail et al., 2007). Gene B activates gene C, and genes B and C together are required to activate gene D in a coherent feed forward structure (Mangan and Alon, 2003). The following equations describe the accumulation of the mRNA of the different factors:
| (19) |
| (20) |
| (21) |
| (22) |
Fig. 5.
Compound GRN circuit. (A) Schematic diagram of the circuit. Gene A activates gene B. Gene B has a positive feedback into its own cis-regulatory module. Gene B activates gene C, and genes B and C together activate gene D. The cis-regulatory module of B executes Additive OR logic on A and B, and the cis-regulatory module of D executes AND logic on B and C. (B) Time courses of the mRNA expression levels of genes A (magenta), B (cyan), C (dark blue) and D (green). (C) Time course of the protein expression levels of genes A, B, C and D, color code similar to (B). The parameters used in this simulation are: Relative equilibrium constant, Kr=105, activation strength for all the equations, kb=2, mRNA turnover rates: kdmA=0.001 min−1, kdmB=kdmC=kdmD= 0.005 min−1, protein turnover rates: kdp=0.01 min−1, kdpB=kdpC=kdpD=0.008 min−1, translation rate, kt=2 protein/(mRNA×minute), mRNA transcription delay, Tm=40 min, cooperativity factor Kq=1. The number of non-specific sites, Dn, was estimated as 90% of the total sea urchin genome, which is 8×108, so Dn=7.2×108. The initial levels of the protein A, and the mRNA and protein of B, C and D were assumed to be zero at time zero. The initial mRNA level of gene A, mA(0)=500 molecules per cell. Simulations were done using Mathematica 5.2.
Here mA(t), mB(t), mC(t) and mD(t) are the number of mRNA molecules per cell of the genes A, B, C and D respectively. The indices A, B and C stand for the kinetic functions and parameters of the activators, A, B and C. Since either the presence of transcription factor A or the presence of transcription factor B is sufficient to drive gene B expression we use the expression of “A Additive OR B” to represent the function of B cis-regulatory module on its inputs. “B AND C” expression is used to represent the function of D cis-regulatory module. The equation for the protein accumulation for all factors is similar to Eq. (2).
In Figs. 5B, C we present the solution for the set of the coupled Eqs. (19)–(22) with the initial conditions of zero concentration at time zero for all mRNA and protein, except for gene A which its mRNA level is assumed to be 500 molecules at time zero. The mRNA of gene A decays exponentially, while its protein level increases due to translation of mRNA, and eventually decays. Gene B is activated once A protein is present, and it keeps itself on even after A is off, due to its positive feedback on itself. The transcription factor B then activates C, and together they activate gene D, in a timely manner. The use of coherent feed forward structure as a timing device is quiet common in the GRN of the sea urchin skeletonic lineage (Amore and Davidson, 2006; Oliveri et al. 2008).
Temporal and spatial models
From the point of view of modeling, the unique feature of animal development is spatial specification of transcriptional regulatory state. Other biological processes, for example those of bacteria and yeast, or physiological processes, share with development temporal progressions in transcriptional expression, temporal aspects of gene interactions, and temporal modulation of expression due to external factors. But the parameters of the types of model we discuss in this paper are not sufficient to explain the processes that in animal development cause adjacent cells at species-specific locations to express the sets of regulatory genes that causally determine localized fate and function. To model the crucial spatial aspects of development will require an entirely different set of approaches that include spatial parameters and that explicitly display the transformations in spatial output executed by GRN subcircuits. These will depend on the structure of the subcircuit and on the combinatorial logic operations performed by the cis-regulatory modules which determine regulatory gene expression in the spatial compartments of the embryo, according to their hardwired genomic design. There are innumerable models purporting to describe spatial distribution of gradients of factors affecting gene expression, but at the end of all such roads are the cis-regulatory apparatuses that read and transduce the input they see into regulatory gene expression. Besides, gradients account for only a minor fraction of spatial gene expression changes in development. Much signaling in spatial specification operates by short range, cell-bound signal reception, as in Notch signaling; and a huge variety of mechanisms not involving signaling that direct spatial patterns of gene expression exist, such as double negative gates (Davidson and Levine, in press; Oliveri et al., 2008), repression cascades (Liberman and Stathopolous, in press), localization in eggs (Davidson, 2001), etc. etc. Ultimately all spatial specification in species-specific pattern formation processes, no matter what the form of the input, depends causally on cis-regulatory input processing functions. Kinetic models such as those we consider here have many and various uses, such as those touched on in Introduction, and without them we could never satisfactorily deal with quantitative phenomena of gene expression. But we must not confuse ourselves by thinking that models which illuminate how things operate once the genomic apparatus has spoken, so to speak, explain the logic behind the speech. That lies in the genomic sequence and in the organization of GRNs.
Concluding remark
A thought provoking implication of the kinetic analysis summarized in this paper is that the dynamics of gene regulatory circuits follow simply from the network topology and the function of cis-regulatory modules on their inputs, given the basic rates of the biological processes of transcription, and mRNA and protein synthesis and turnover. The kinetic parameters are of course temperature dependant, but for a given system they are approximate constants which, according to the gene regulatory network structure, control the overall dynamics of regulatory life. Therefore, it is not necessary to invoke a special clock mechanism or to imagine the existence of more complicated computational apparatus to explain GRN kinetics. The levels and identity of the transcription factors in a given cell identifies the regulatory state of the cell at every point in development. Transcriptional response to regulatory states, in the activation of downstream regulatory genes, leads to the onset of the next regulatory state, and so on until specification and differentiation are achieved. Thus, it is just the temporal change of transcription factor levels that functions as the underlying clock of specification, the chain of events that replaces the central synchronizing clock used in many manmade computational machines (Istrail et al., 2007; Smith et al., 2007; Smith et al., 2008).
Acknowledgments
The authors thank Jongmin Nam and Joel Smith for critical review of the manuscript and very insightful comments. Research was supported by NIH grant GM61005. Smadar Ben-Tabou de-Leon was supported by the Human Frontiers Science Program Organization.
Contributor Information
Smadar Ben-Tabou de-Leon, Email: smadar@caltech.edu.
Eric H. Davidson, Email: davidson@caltech.edu, jrigg@caltech.edu.
References
- Ackers GK, Johnson AD, Shea MA. Quantitative model for gene regulation by lambda phage repressor. Proc. Natl. Acad. Sci. U.S.A. 1982;79:1129–1133. doi: 10.1073/pnas.79.4.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amore G, Davidson EH. cis-Regulatory control of cyclophilin, a member of the ETS-DRI skeletogenic gene battery in the sea urchin embryo. Dev. Biol. 2006;293:555–564. doi: 10.1016/j.ydbio.2006.02.024. [DOI] [PubMed] [Google Scholar]
- Arnosti DN, Barolo S, Levine M, Small S. The eve stripe 2 enhancer employs multiple modes of transcriptional synergy. Development. 1996;122:205–214. doi: 10.1242/dev.122.1.205. [DOI] [PubMed] [Google Scholar]
- Aronson AI, Chen K. Rates of RNA chain growth in developing sea urchin embryos. Dev. Biol. 1977;59:39–48. doi: 10.1016/0012-1606(77)90238-x. [DOI] [PubMed] [Google Scholar]
- Barolo S, Levine M. hairy mediates dominant repression in the Drosophila embryo. EMBO J. 1997;16:2883–2891. doi: 10.1093/emboj/16.10.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Tabou de-Leon S, Davidson EH. Gene regulation: gene control network in development. Annu. Rev. Biophys. Biomol. Struct. 2007;36:191–212. doi: 10.1146/annurev.biophys.35.040405.102002. [DOI] [PubMed] [Google Scholar]
- Bernard S, Cajavec B, Pujo-Menjouet L, Mackey MC, Herzel H. Modelling transcriptional feedback loops: the role of Gro/TLE1 in Hes1 oscillations. Philos. Transact A Math. Phys. Eng. Sci. 2006;364:1155–1170. doi: 10.1098/rsta.2006.1761. [DOI] [PubMed] [Google Scholar]
- Bessho Y, Kageyama R. Oscillations, clocks and segmentation. Curr. Opin. Genet. Dev. 2003;13:379–384. doi: 10.1016/s0959-437x(03)00083-2. [DOI] [PubMed] [Google Scholar]
- Bintu L, Buchler NE, Garcia HG, Gerland U, Hwa T, Kondev J, Phillips R. Transcriptional regulation by the numbers: models. Curr. Opin. Genet. Dev. 2005;15:116–124. doi: 10.1016/j.gde.2005.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolouri H, Davidson EH. Transcriptional regulatory cascades in development: initial rates, not steady state, determine network kinetics. Proc. Natl. Acad. Sci. USA. 2003;100:9371–9376. doi: 10.1073/pnas.1533293100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchler NE, Gerland U, Hwa T. On schemes of combinatorial transcription logic. Proc. Natl. Acad. Sci. USA. 2003;100:5136–5141. doi: 10.1073/pnas.0930314100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burz DS, Rivera-Pomar R, Jackle H, Hanes SD. Cooperative DNA-binding by Bicoid provides a mechanism for threshold-dependent gene activation in the Drosophila embryo. EMBO J. 1998;17:5998–6009. doi: 10.1093/emboj/17.20.5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera CV, Lee JJ, Ellison JW, Britten RJ, Davidson EH. Regulation of cytoplasmic mRNA prevalence in sea urchin embryos. Rates of appearance and turnover for specific sequences. J. Mol. Biol. 1984;174:85–111. doi: 10.1016/0022-2836(84)90366-8. [DOI] [PubMed] [Google Scholar]
- Calzone FJ, Theze N, Thiebaud P, Hill RL, Britten RJ, Davidson EH. Developmental appearance of factors that bind specifically to cis-regulatory sequences of a gene expressed in the sea urchin embryo. Genes Dev. 1988;2:1074–1088. doi: 10.1101/gad.2.9.1074. [DOI] [PubMed] [Google Scholar]
- Calzone FJ, Hoog C, Teplow DB, Cutting AE, Zeller RW, Britten RJ, Davidson EH. Gene regulatory factors of the sea urchin embryo. I. Purification by affinity chromatography and cloning of P3A2, a novel DNA-binding protein. Development. 1991;112:335–350. doi: 10.1242/dev.112.1.335. [DOI] [PubMed] [Google Scholar]
- Cranz S, Berger C, Baici A, Jelesarov I, Bosshard HR. Monomeric and dimeric bZIP transcription factor GCN4 bind at the same rate to their target DNA site. Biochemistry. 2004;43:718–727. doi: 10.1021/bi0355793. [DOI] [PubMed] [Google Scholar]
- Davidson EH. Gene Activity in Early Development. Orlando: Academic press; 1986. [Google Scholar]
- Davidson EH. Genomic Regulatory Systems: Development and Evolution. San-Diego: Academic Press; 2001. [Google Scholar]
- Davidson EH. Gene Regulatory Networks in Development and Evolution. San-Diego: Academic Press; 2006. The Regulatory Genome. [Google Scholar]
- Davidson EH, Levine M. Properties of developmental gene regulatory networks. Proc. Natl. Acad. Sci. USA. doi: 10.1073/pnas.0806007105. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong H. Modeling and simulation of genetic regulatory systems: a literature review. J Comput. Biol. 2002;9:67–103. doi: 10.1089/10665270252833208. [DOI] [PubMed] [Google Scholar]
- Di Caro V, Cavalieri V, Melfi R, Spinelli G. Constitutive promoter occupancy by the MBF-1 activator and chromatin modification of the developmental regulated sea urchin alpha-H2A histone gene. J Mol. Biol. 2007;365:1285–1297. doi: 10.1016/j.jmb.2006.10.098. [DOI] [PubMed] [Google Scholar]
- Dodd IB, Micheelsen MA, Sneppen K, Thon G. Theoretical analysis of epigenetic cell memory by nucleosome modification. Cell. 2007;129:813–822. doi: 10.1016/j.cell.2007.02.053. [DOI] [PubMed] [Google Scholar]
- Driever W, Nusslein-Volhard C. The bicoid protein is a positive regulator of hunchback transcription in the early Drosophila embryo. Nature. 1989;337:138–143. doi: 10.1038/337138a0. [DOI] [PubMed] [Google Scholar]
- Elf J, Li GW, Xie XS. Probing transcription factor dynamics at the single-molecule level in a living cell. Science. 2007;316:1191–1194. doi: 10.1126/science.1141967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson BM, Lewis CD, Felsenfeld G. Interaction of specific nuclear factors with the nuclease-hypersensitive region of the chicken adult beta-globin gene: nature of the binding domain. Cell. 1985;41:21–30. doi: 10.1016/0092-8674(85)90057-1. [DOI] [PubMed] [Google Scholar]
- Emerson BM, Nickol JM, Jackson PD, Felsenfeld G. Analysis of the tissue-specific enhancer at the 3′ end of the chicken adult beta-globin gene. Proc. Natl. Acad. Sci. USA. 1987;84:4786–4790. doi: 10.1073/pnas.84.14.4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenfeld G, Groudine M. Controlling the double helix. Nature. 2003;421:448–453. doi: 10.1038/nature01411. [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of posttranscriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Fujioka M, Emi-Sarker Y, Yusibova GL, Goto T, Jaynes JB. Analysis of an even-skipped rescue transgene reveals both composite and discrete neuronal and early blastoderm enhancers, and multi-stripe positioning by gap gene repressor gradients. Development. 1999;126:2527–2538. doi: 10.1242/dev.126.11.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuke H, Ohno M. Role of poly (A) tail as an identity element for mRNA nuclear export. Nucleic Acids Res. 2008;36:1037–1049. doi: 10.1093/nar/gkm1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrity PA, Chen D, Rothenberg EV, Wold BJ. Interleukin-2 transcription is regulated in vivo at the level of coordinated binding of both constitutive and regulated factors. Mol. Cell. Biol. 1994;14:2159–2169. doi: 10.1128/mcb.14.3.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giurumescu CA, Sternberg PW, Asthagiri AR. Intercellular coupling amplifies fate segregation during Caenorhabditis elegans vulval development. Proc. Natl. Acad. Sci. USA. 2006;103:1331–1336. doi: 10.1073/pnas.0506476103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorospe M, Baglioni C. Degradation of unstable interleukin-1 alpha mRNA in a rabbit reticulocyte cell-free system. Localization of an instability determinant to a cluster of AUUUA motifs. J Biol. Chem. 1994;269:11845–11851. [PubMed] [Google Scholar]
- Goustin AS, Wilt FH. Protein synthesis, polyribosomes, and peptide elongation in early development of Strongylocentrotus purpuratus . Dev. Biol. 1981;82:32–40. doi: 10.1016/0012-1606(81)90426-7. [DOI] [PubMed] [Google Scholar]
- Gray S, Levine M. Short-range transcriptional repressors mediate both quenching and direct repression within complex loci in Drosophila . Genes Dev. 1996a;10:700–710. doi: 10.1101/gad.10.6.700. [DOI] [PubMed] [Google Scholar]
- Gray S, Levine M. Transcriptional repression in development. Curr. Opin. Cell Biol. 1996b;8:358–364. doi: 10.1016/s0955-0674(96)80010-x. [DOI] [PubMed] [Google Scholar]
- Gray S, Szymanski P, Levine M. Short-range repression permits multiple enhancers to function autonomously within a complex promoter. Genes Dev. 1994;8:1829–1838. doi: 10.1101/gad.8.15.1829. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Hirata H, Yoshiura S, Ohtsuka T, Bessho Y, Harada T, Yoshikawa K, Kageyama R. Oscillatory expression of the bHLH factor Hes1 regulated by a negative feedback loop. Science. 2002;298:840–843. doi: 10.1126/science.1074560. [DOI] [PubMed] [Google Scholar]
- Hoog C, Calzone FJ, Cutting AE, Britten RJ, Davidson EH. Gene regulatory factors of the sea urchin embryo. II. Two dissimilar proteins, P3A1 and P3A2, bind to the same target sites that are required for early territorial gene expression. Development. 1991;112:351–364. doi: 10.1242/dev.112.1.351. [DOI] [PubMed] [Google Scholar]
- Howard-Ashby M, Materna SC, Brown CT, Chen L, Cameron RA, Davidson EH. Identification and characterization of homeobox transcription factor genes in Strongylocentrotus purpuratus, and their expression in embryonic development. Dev. Biol. 2006;300:74–89. doi: 10.1016/j.ydbio.2006.08.039. [DOI] [PubMed] [Google Scholar]
- Istrail S, Davidson EH. Logic functions of the genomic cis-regulatory code. Proc. Natl. Acad. Sci. USA. 2005;102:4954–4959. doi: 10.1073/pnas.0409624102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Istrail S, Ben-Tabou De-Leon S, Davidson EH. The regulatory genome and the computer. Dev. Biol. 2007;310:187–195. doi: 10.1016/j.ydbio.2007.08.009. [DOI] [PubMed] [Google Scholar]
- Janssens H, Hou S, Jaeger J, Kim AR, Myasnikova E, Sharp D, Reinitz J. Quantitative and predictive model of transcriptional control of the Drosophila melanogaster even skipped gene. Nat. Genet. 2006;38:1159–1165. doi: 10.1038/ng1886. [DOI] [PubMed] [Google Scholar]
- Kamachi Y, Kondoh H. Overlapping positive and negative regulatory elements determine lens-specific activity of the delta 1-crystallin enhancer. Mol. Cell. Biol. 1993;13:5206–5215. doi: 10.1128/mcb.13.9.5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laslo P, Spooner CJ, Warmflash A, Lancki DW, Lee HJ, Sciammas R, Gantner BN, Dinner AR, Singh H. Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell. 2006;126:755–766. doi: 10.1016/j.cell.2006.06.052. [DOI] [PubMed] [Google Scholar]
- Lee JJ, Calzone FJ, Davidson EH. Modulation of sea urchin actin mRNA prevalence during embryogenesis: nuclear synthesis and decay rate measurements of transcripts from five different genes. Dev. Biol. 1992;149:415–431. doi: 10.1016/0012-1606(92)90296-s. [DOI] [PubMed] [Google Scholar]
- Lee MS, Son MY, Park JI, Park C, Lee YC, Son CB, Kim YS, Paik SG, Yoon WH, Park SK, Hwang BD, Lim K. Modification of octamer binding transcriptional factor is related to H2B histone gene repression during dimethyl sulfoxide-dependent differentiation of HL-60 cells. Cancer Lett. 2001;172:165–170. doi: 10.1016/s0304-3835(01)00654-1. [DOI] [PubMed] [Google Scholar]
- Levine M, Davidson EH. Gene regulatory networks for development. Proc. Natl. Acad. Sci. USA. 2005;102:4936–4942. doi: 10.1073/pnas.0408031102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M, Manley JL. Transcriptional repression of eukaryotic promoters. Cell. 1989;59:405–408. doi: 10.1016/0092-8674(89)90024-x. [DOI] [PubMed] [Google Scholar]
- Liberman LM, Stathopolous AM. Design flexibility in cis-regulatory control of gene expression: synthetic and comparative evidence. Dev. Biol. doi: 10.1016/j.ydbio.2008.12.020. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Riggs AD. The general affinity of lac repressor for E. coli DNA: implications for gene regulation in procaryotes and eucaryotes. Cell. 1975;4:107–111. doi: 10.1016/0092-8674(75)90116-6. [DOI] [PubMed] [Google Scholar]
- Lipniacki T, Paszek P, Marciniak-Czochra A, Brasier AR, Kimmel M. Transcriptional stochasticity in gene expression. J Theor. Biol. 2006;238:348–367. doi: 10.1016/j.jtbi.2005.05.032. [DOI] [PubMed] [Google Scholar]
- Ma X, Yuan D, Diepold K, Scarborough T, Ma J. The Drosophila morphogenetic protein Bicoid binds DNA cooperatively. Development. 1996;122:1195–1206. doi: 10.1242/dev.122.4.1195. [DOI] [PubMed] [Google Scholar]
- Mangan S, Alon U. Structure and function of the feed-forward loop network motif. Proc. Natl. Acad. Sci. USA. 2003;100:11980–11985. doi: 10.1073/pnas.2133841100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin KA, Miller OL., Jr Polysome structure in sea urchin eggs and embryos: an electron microscopic analysis. Dev. Biol. 1983;98:338–348. doi: 10.1016/0012-1606(83)90364-0. [DOI] [PubMed] [Google Scholar]
- McGhee JD, von Hippel PH. Theoretical aspects of DNA–protein interactions: co-operative and non-co-operative binding of large ligands to a one-dimensional homogeneous lattice. J Mol. Biol. 1974;86:469–489. doi: 10.1016/0022-2836(74)90031-x. [DOI] [PubMed] [Google Scholar]
- Moss EG. Heterochronic genes and the nature of developmental time. Curr. Biol. 2007;17:R425–R434. doi: 10.1016/j.cub.2007.03.043. [DOI] [PubMed] [Google Scholar]
- Murugan R. Stochastic transcription initiation: time dependent transcription rates. Biophys. Chemist. 2006;121:51–56. doi: 10.1016/j.bpc.2005.12.010. [DOI] [PubMed] [Google Scholar]
- Mutskov V, Felsenfeld G. Silencing of transgene transcription precedes methylation of promoter DNA and histone H3 lysine 9. EMBO J. 2004;23:138–149. doi: 10.1038/sj.emboj.7600013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam J, Su YH, Lee PY, Robertson AJ, Coffman JA, Davidson EH. Cis-regulatory control of the nodal gene, initiator of the sea urchin oral ectoderm gene network. Dev. Biol. 2007;306:860–869. doi: 10.1016/j.ydbio.2007.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasiadka A, Krause HM. Kinetic analysis of segmentation gene interactions in Drosophila embryos. Development. 1999;126:1515–1526. doi: 10.1242/dev.126.7.1515. [DOI] [PubMed] [Google Scholar]
- Naujokat C, Saric T. Concise review: role and function of the ubiquitin–proteasome system in mammalian stem and progenitor cells. Stem Cells. 2007;25:2408–2418. doi: 10.1634/stemcells.2007-0255. [DOI] [PubMed] [Google Scholar]
- Nibu Y, Senger K, Levine M. CtBP-independent repression in the Drosophila embryo. Mol. Cell. Biol. 2003;23:3990–3999. doi: 10.1128/MCB.23.11.3990-3999.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuthall HN, Husain J, McLarren KW, Stifani S. Role for Hes1-induced phosphorylation in Groucho-mediated transcriptional repression. Mol. Cell. Biol. 2002;22:389–399. doi: 10.1128/MCB.22.2.389-399.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuthall HN, Joachim K, Stifani S. Phosphorylation of serine 239 of Groucho/TLE1 by protein kinase CK2 is important for inhibition of neuronal differentiation. Mol. Cell. Biol. 2004;24:8395–8407. doi: 10.1128/MCB.24.19.8395-8407.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okahata Y, Niikura K, Sugiura Y, Sawada M, Morii T. Kinetic studies of sequence-specific binding of GCN4-bZIP peptides to DNA strands immobilized on a 27-MHz quartz-crystal microbalance. Biochemistry. 1998;37:5666–5672. doi: 10.1021/bi980037k. [DOI] [PubMed] [Google Scholar]
- Oliveri P, Davidson EH. Development. Built to run, not fail. Science. 2007;315:1510–1511. doi: 10.1126/science.1140979. [DOI] [PubMed] [Google Scholar]
- Oliveri P, Walton KD, Davidson EH, McClay DR. Repression of mesodermal fate by foxa, a key endoderm regulator of the sea urchin embryo. Development. 2006;133:4173–4181. doi: 10.1242/dev.02577. [DOI] [PubMed] [Google Scholar]
- Oliveri P, Tu Q, Davidson EH. Global regulatory logic for specification of an embryonic cell lineage. Proc. Natl. Acad. Sci. USA. 2008;105:5955–5962. doi: 10.1073/pnas.0711220105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedraza JM, Paulsson J. Effects of molecular memory and bursting on fluctuations in gene expression. Science. 2008;319:339–343. doi: 10.1126/science.1144331. [DOI] [PubMed] [Google Scholar]
- Pikaart MJ, Recillas-Targa F, Felsenfeld G. Loss of transcriptional activity of a transgene is accompanied by DNA methylation and histone deacetylation and is prevented by insulators. Genes Dev. 1998;12:2852–2862. doi: 10.1101/gad.12.18.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M. Repressors. Curr. Biol. 2007;17:R740–R741. doi: 10.1016/j.cub.2007.06.053. [DOI] [PubMed] [Google Scholar]
- Rateitschak K, Wolkenhauer O. Intracellular delay limits cyclic changes in gene expression. Math. Biosci. 2007;205:163–179. doi: 10.1016/j.mbs.2006.08.010. [DOI] [PubMed] [Google Scholar]
- Ribeiro AS. Dynamics of a two-dimensional model of cell tissues with coupled stochastic gene networks. Phys. Rev. E Stat. Nonlinear Soft Matter Phys. 2007;76:051915. doi: 10.1103/PhysRevE.76.051915. [DOI] [PubMed] [Google Scholar]
- Riggs AD, Bourgeois S, Cohn M. The lac repressor–operator interaction. 3. Kinetic studies. J. Mol. Biol. 1970;53:401–417. doi: 10.1016/0022-2836(70)90074-4. [DOI] [PubMed] [Google Scholar]
- Santoro R, Grummt I. Epigenetic mechanism of rRNA gene silencing: temporal order of NoRC-mediated histone modification, chromatin remodeling, and DNA methylation. Mol. Cell. Biol. 2005;25:2539–2546. doi: 10.1128/MCB.25.7.2539-2546.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saulier-Le Drean B, Nasiadka A, Dong J, Krause HM. Dynamic changes in the functions of Odd-skipped during early Drosophila embryogenesis. Development. 1998;125:4851–4861. doi: 10.1242/dev.125.23.4851. [DOI] [PubMed] [Google Scholar]
- Schroder HC, Friese U, Bachmann M, Zaubitzer T, Muller WE. Energy requirement and kinetics of transport of poly(A)-free histone mRNA compared to poly(A)-rich mRNA from isolated L-cell nuclei. Eur. J. Biochem. 1989;181:149–158. doi: 10.1111/j.1432-1033.1989.tb14706.x. [DOI] [PubMed] [Google Scholar]
- Sedighi M, Sengupta AM. Epigenetic chromatin silencing: bistability and front propagation. Phys. Biol. 2007;4:246–255. doi: 10.1088/1478-3975/4/4/002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekido R, Murai K, Kamachi Y, Kondoh H. Two mechanisms in the action of repressor deltaEF1: binding site competition with an activator and active repression. Genes Cells. 1997;2:771–783. doi: 10.1046/j.1365-2443.1997.1570355.x. [DOI] [PubMed] [Google Scholar]
- Shuman S. Origins of mRNA identity: capping enzymes bind to the phosphorylated C-terminal domain of RNA polymerase II. Proc. Natl. Acad. Sci. USA. 1997;94:12758–12760. doi: 10.1073/pnas.94.24.12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigvardsson M, Clark DR, Fitzsimmons D, Doyle M, Akerblad P, Breslin T, Bilke S, Li R, Yeamans C, Zhang G, Hagman J. Early B-cell factor, E2A, and Pax-5 cooperate to activate the early B cell-specific mb-1 promoter. Mol. Cell. Biol. 2002;22:8539–8551. doi: 10.1128/MCB.22.24.8539-8551.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J, Theodoris C, Davidson EH. A gene regulatory network subcircuit drives a dynamic pattern of gene expression. Science. 2007;318:794–797. doi: 10.1126/science.1146524. [DOI] [PubMed] [Google Scholar]
- Smith J, Kraemer E, Liu H, Theodoris C, Davidson E. A spatially dynamic cohort of regulatory genes in the endomesodermal gene network of the sea urchin embryo. Dev. Biol. 2008;313:863–875. doi: 10.1016/j.ydbio.2007.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolen P, Baxter DA, Byrne JH. Modeling transcriptional control in gene networks—methods, recent results, and future directions. Bull. Math. Biol. 2000;62:247–292. doi: 10.1006/bulm.1999.0155. [DOI] [PubMed] [Google Scholar]
- Stathopoulos A, Levine M. Whole-genome analysis of Drosophila gastrulation. Curr. Opin. Genet. Dev. 2004;14:477–484. doi: 10.1016/j.gde.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Stirzaker C, Song JZ, Davidson B, Clark SJ. Transcriptional gene silencing promotes DNA hypermethylation through a sequential change in chromatin modifications in cancer cells. Cancer Res. 2004;64:3871–3877. doi: 10.1158/0008-5472.CAN-03-3690. [DOI] [PubMed] [Google Scholar]
- Sugawara M, Scholl T, Mahanta SK, Ponath PD, Strominger JL. Cooperativity between the J and S elements of class II major histocompatibility complex genes as enhancers in normal and class II-negative patient and mutant B cell lines. J Exp. Med. 1995;182:175–184. doi: 10.1084/jem.182.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai KY, Shiah SG, Shieh YS, Kao YR, Chi CY, Huang E, Lee HS, Chang LC, Yang PC, Wu CW. DNA methylation and histone modification regulate silencing of epithelial cell adhesion molecule for tumor invasion and progression. Oncogene. 2007;26:3989–3997. doi: 10.1038/sj.onc.1210176. [DOI] [PubMed] [Google Scholar]
- Thanos D, Maniatis T. Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- Tomlin CJ, Axelrod JD. Biology by numbers: mathematical modelling in developmental biology. Nat. Rev. Genet. 2007;8:331–340. doi: 10.1038/nrg2098. [DOI] [PubMed] [Google Scholar]
- Wallin JJ, Gackstetter ER, Koshland ME. Dependence of BSAP repressor and activator functions on BSAP concentration. Science. 1998;279:1961–1964. doi: 10.1126/science.279.5358.1961. [DOI] [PubMed] [Google Scholar]
- Walsh CM, Carroll SB. Collaboration between Smads and a Hox protein in target gene repression. Development. 2007;134:3585–3592. doi: 10.1242/dev.009522. [DOI] [PubMed] [Google Scholar]
- Wen SC, Roder K, Hu KY, Rombel I, Gavva NR, Daftari P, Kuo YY, Wang C, Shen CK. Loading of DNA-binding factors to an erythroid enhancer. Mol. Cell. Biol. 2000;20:1993–2003. doi: 10.1128/mcb.20.6.1993-2003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz CJ, Wormington M, Peltz SW. The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell Biol. 2001;2:237–246. doi: 10.1038/35067025. [DOI] [PubMed] [Google Scholar]
- Yuh CH, Bolouri H, Davidson EH. Genomic cis-regulatory logic: experimental and computational analysis of a sea urchin gene. Science. 1998;279:1896–1902. doi: 10.1126/science.279.5358.1896. [DOI] [PubMed] [Google Scholar]
- Yuh CH, Bolouri H, Davidson EH. Cis-regulatory logic in the endo16 gene: switching from a specification to a differentiation mode of control. Development. 2001;128:617–629. doi: 10.1242/dev.128.5.617. [DOI] [PubMed] [Google Scholar]
- Zhu R, Ribeiro AS, Salahub D, Kauffman SA. Studying genetic regulatory networks at the molecular level: delayed reaction stochastic models. J Theor. Biol. 2007;246:725–745. doi: 10.1016/j.jtbi.2007.01.021. [DOI] [PubMed] [Google Scholar]
- Zubiaga AM, Belasco JG, Greenberg ME. The nonamer UUAUUUAUU is the key AU-rich sequence motif that mediates mRNA degradation. Mol. Cell. Biol. 1995;15:2219–2230. doi: 10.1128/mcb.15.4.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]