Abstract
Introduction
Dietary flaxseed (FS) displays antioxidant and anti-inflammatory properties in preclinical models of lung disease including radiation-induced pneumonopathy, however the mechanisms of lung radioprotection are incompletely understood. MicroRNAs (miRNAs) are short oligonucleotides that act as important posttranscriptional regulators of diverse networks including inflammatory response networks. Responses of miRNA profiles to diet and radiation exposure have been reported, but the potential contribution of miRNAs to diet-related radioprotection has never been tested.
Methods
In this exploratory pilot study, mice were fed 10% FS or a 0% FS isocaloric control diet and exposed to a single-fraction 13.5 Gy thoracic X-ray radiation treatment (XRT). Lung RNA was extracted 48 h post-XRT and small RNAs profiled by OpenArray.
Results
FS significantly modulated expression of multiple miRNAs, including 7 with P < 0.001. miR-150 was downregulated approximately 2.9-fold in the FS groups and is disproportionately integrated into immune response-related networks. Although few miRNAs were significantly changed by radiation, interaction between diet and radiation was observed. For example, miR-29c was greatly downregulated in the FS/Control group (10- to 50-fold) but slightly upregulated in the FS/radiation group. Compared with FS/control, the FS/radiation group experienced a 50% decrease of the p53-responsive miR-34a, which regulates senescence- and apoptosis-related factors.
Conclusions
FS induced significant changes in lung miRNA profile suggesting that modulation of small RNA by dietary supplements may represent a novel strategy to prevent adverse side-effects of thoracic radiotherapy. This pilot study provides insight into a potential mechanism of flaxseed’s radioprotection and provides a useful model-system to further explore and optimize such small RNA-based therapies.
Keywords: ROS, antioxidant, flaxseed, inflammation, lung fibrosis, lung injury, miR-150, miR-29c, miR-34a, miRNA, mouse model, radiation pneumonopathy
Introduction
The effectiveness of radiotherapy for thoracic malignancies is limited by the low tolerance of normal lung parenchyma to ionizing radiation.1,2 Clinically significant radiation-induced lung injury occurs in 30% of patients irradiated for lung cancer3 and 10–15% of other thoracic oncology patients.4 A greater proportion of patients have subclinical adverse effects from radiation of the lung, identifiable by imaging and/or physiological testing.5 Highly reactive oxygen species (ROS) and reactive nitrogen species (RNS) are induced in large quantities by radiation therapy (XRT) and have been implicated in this form of lung injury.6
There is currently no known effective pharmacologic therapy for the prevention of acute or chronic radiation pneumonopathy. To date, the only means to avoid life-threatening radiation pneumonopathy is to modify the irradiation technique to minimize the volume of normal lung that receives a significant radiation dose. A safe and effective biologic radioprotector would thus be extremely useful. Preclinical data suggest that antioxidant molecules and/or enzymes might offer protection of the lung.7-9 Our group has shown that systemic administration of polyethylene glycol conjugated antioxidant enzymes at the time of XRT alters several early biomarkers of lung injury, decreases apoptosis, and ameliorates late pulmonary fibrosis in a murine model.10 Although encouraging, this potential therapeutic is far from human clinical trials, and a safe, more accessible radioprotector is therefore needed.
Our group has identified dietary flaxseed (FS) as a potential radioprotector against radiation-induced lung injury in a murine model of thoracic XRT. FS is a non-toxic whole grain composed of high concentrations of omega-3 fatty acids and lignans. While omega-3 fatty acids reduce inflammation, lignans possess potent antioxidant properties. Specifically, secoisolariciresinol diglucoside (SDG) is the most abundant lignan present in FS and was shown in vitro to have direct hydroxyl radical scavenging properties and to inhibit lipid peroxidation11-13 as verified recently by our group in the context of oxidative lung injury.14,15 We confirmed the cell and tissue protective effects of the lignan precursor SDG on the abrogation of radiation-induced ROS generation in vitro. In addition, we provided evidence of radioprotection of FS and of dietary SDG formulations. Importantly, both FS and SDG formulations protected normal tissue damage from radiation and inflammation, while not decreasing lung tumor response to XRT using an orthotopic model.
In this pilot study, we investigated another potential mechanism of FS-induced radioprotection by examining modulation of microRNAs (miRNAs) in the lung in response to FS diet and radiation. miRNAs are short (approximately 22 nucleotides) RNA molecules which contribute to posttranscriptional gene regulation. As biomarkers, miRNAs hold tremendous clinical promise as diagnostics, prognostics, and predictors of response to therapy. The normal miRNA profile of a given cell or tissue type is often dysregulated in cancers and in response to inflammation. Beyond “bystander” effects, miRNAs may also be intimately involved in pathogenesis and thus represent targets of molecular therapies: expression enhancement strategies that replace a missing miRNA or antagomir approaches that block the actions of a specific miRNA. The latter has proven successful, for example, in a phase 2 trial of miraversen, an antisense inhibitor of the liver-enriched miR-122 that functions in the replication cycle of Hepatitis C virus. The ability to modulate endogenous miRNAs by dietary interventions would be a new and exciting therapeutic option.
Results
In this pilot study, we pre-fed mice 10% FS diet as compared with an isocaloric control for 3 wk prior to a single dose radiation challenge to the thorax16,17 and evaluated lung tissues 48 h post-exposure (Figure 1). The time post radiation exposure was based on previous studies where we determined that acute, damage-response genes are upregulated in lung tissues and abrogated by the FS diet.18 The dose of FS and the duration of pre-feeding was selected based on the kinetics of expression of antioxidant and cytoprotective genes15 which we determined to increase over control and reach a plateau by 2–3 wk on the diet. Comparisons were made between irradiated and non-irradiated lung tissues from flaxseed- or control diet-fed mice with respect to their miRNA profile.
Figure 1.
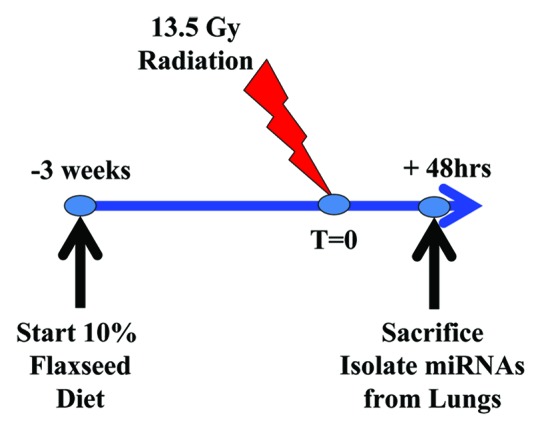
Thoracic radiation model and experimental plan. Mice are subjected to a single fraction thoracic radiation (13.5 Gy). Mice (n = 3 per group) were fed with 0% or 10% FS diet initiated prior (−3 wk) to single fraction X-ray radiation treatment (XRT). Mice were sacrificed at 48 h post-XRT and lung miRNA profile evaluated.
miRNA profiling: evaluation and quality control
miRNA profiling was conducted using the medium-throughput rodent miRNA “OpenArray” platform from Life Technologies. This platform includes TaqMan probes for 750 rodent miRNAs along with several small RNA control features. In contrast with the predecessor “TaqMan Low Density Array” (TLDA) platform, which requires the use of two 384-well PCR cards with 1 µL reaction vessels to profile one sample, OpenArray reactions are performed in 30 nL hydrophobic through-hole reaction vessels. Three thousand and seventy-four of these vessels are contained in each array, and three samples may be profiled per array. Four arrays may be processed simultaneously, allowing a 24-fold increase in throughput over the TLDA platform. At the same time, the ~30-fold decrease in reaction volume may result in lower sensitivity and greater variability, particularly at the low end of the abundance spectrum. Accordingly, we tested six samples in technical duplicate during different instrument runs to assess reproducibility. With the exception of several minor loading problems that were occasionally observed from sample to sample—a variable and small percentage of wells that were not filled and thus unreliable—the majority of features that were consistently detected in each duplicate sample were separated by 0.5 cycles or less. Only features that exceeded a minimum “amplification score” (assigned by the manufacturer’s software) were included in subsequent analysis. Most detected miRNAs amplified before 28 PCR cycles.
The number of features—miRNA or other small RNA—detected in each lung sample ranged from 294 to 326. Animals on a flaxseed diet and no radiation exposure had slightly fewer detected features (294 to 309) than were found in samples from other experimental groups (321 to 326). 274 miRNAs were consistently detected in all samples, along with all structural small RNA controls, while a negative control feature (a plant miRNA) was not detected. Unsupervised hierarchical clustering roughly recapitulated the experimental groupings (Fig. 2).
Figure 2.

Unsupervised hierarchical clustering of miRNAs reveals relationship of groups. Experimental groups were largely recapitulated by unsupervised hierarchical clustering of all results from OpenArray miRNA profiling. Clustering was performed by Pearson correlation with average linkage. CC, control diet, no radiation; CR, control diet, radiation; FC, flaxseed, no radiation; FR, flaxseed, radiation.
miRNA profile in lung tissues of flaxseed-fed mice
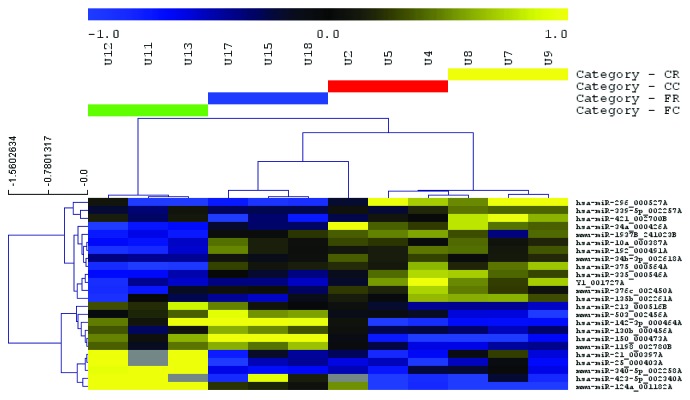
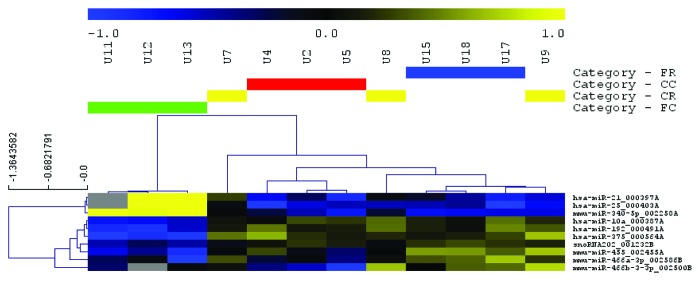
miRNAs and other features with expression differences between experimental conditions (as determined by two-way analysis of variance with a P value cutoff of 0.05) were used to hierarchically cluster samples. More than 20 features appeared to distinguish FS-fed mice from those on a control diet (Fig. 3). In contrast, few features were differentially expressed between irradiated and unexposed mice (Fig. 4). Several of these features were also detected in fewer than 100% of samples (Fig. 4), emphasizing a lack of reliable differences.
Figure 3.
Heatmap of miRNA profile in response to dietary flaxseed administration in mice Significantly differentially expressed, diet-associated miRNAs as identified by two-way ANOVA (P < 0.05) were used to cluster samples and features (Euclidean distance, average linkage). The expression heatmap legend (top) indicates relative abundance of median-centered expression values on a log2 scale. Yellow indicates reduced expression and blue indicates increased expression of the selected miRNAs for the given diets. In the heatmap, gray indicates failed detection. CC, control diet, no radiation; CR, control diet, radiation; FC, flaxseed, no radiation; FR, flaxseed, radiation.
Figure 4.
Heatmap of miRNA profile in irradiated mouse lungs. Significantly differentially expressed, diet-associated miRNAs as identified by two-way ANOVA (P < 0.05) were used to cluster samples and features (Euclidean distance, average linkage). The expression heatmap legend (top) indicates relative abundance of median-centered expression values on a log2 scale. Yellow indicates reduced expression and blue indicates increased expression of the selected miRNAs for the given diets. In the heatmap, gray indicates failed detection. There was comparatively little response of lung miRNA expression to radiation when apparent radiation-associated changes were identified. CC, control diet, no radiation; CR, control diet, radiation; FC, flaxseed, no radiation; FR, flaxseed, radiation.
qPCR validation of miRNA profile in lung tissues of irradiated flaxseed-fed mice
Selected miRNAs were assayed using individual qRT-PCR hydrolysis probe-based tests. These reactions differ from those performed with the OpenArray platform in that reverse transcription is conducted for only one to five miRNAs at a time, in contrast with the hundreds of miRNAs in the OpenArray system. Furthermore, there is no pre-amplification step. Because individual assays are less expensive, technical repeats and larger numbers of controls can be performed. Individual miRNA assays largely validated the profiling results. As indicated by the arrays, there was little modulation of the examined miRNAs in response to radiation, but significant differences were associated with FS diet.
Specifically, individual qPCR assays confirmed the response to diet of miRs-34a, -142-3p, and -150. miRs-142-3p and -150, which were significantly downmodulated by 2- to 4-fold in animals with FS diet, irrespective of radiation exposure (Fig. 5). In contrast, miR-34a was significantly upregulated by approximately 2-fold. One miRNA, miR-143, had a nominally significant P value for a slightfold-downregulation (Fig. 5). While this finding was of potential interest, the magnitude of downregulation and the small number of repeats reduced confidence in the result.
Figure 5.
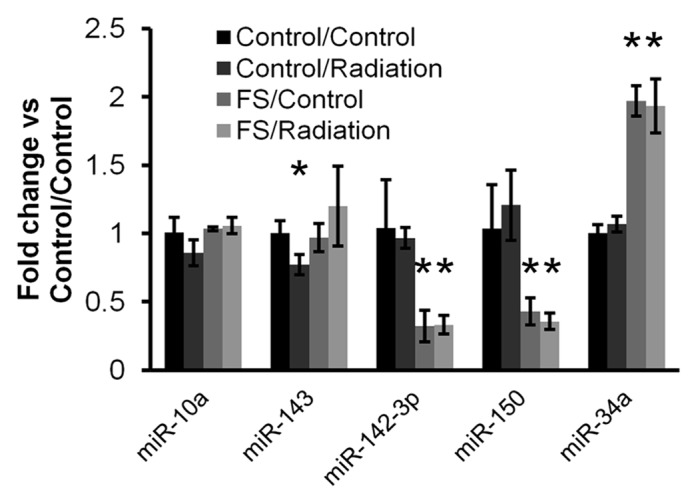
qRT-PCR confirmation: flaxseed alters the “set point” of several miRNAs. Mature miRNAs and two structural small RNA controls (U6 and sno-135) were quantitated by qRT-PCR for samples from the four indicated groups. Data were processed by relative Cq method, with normalization to the geometric mean of all assayed RNAs. Kruskal–Wallis tests were performed with the Dunn post-hoc test. Statistical tests were done with relative Cq-level data before transformation to fold change relative to the control/control condition (control diet and no radiation). Shown are the resulting fold change values for five mature miRNAs. Indicated comparisons (*) were significant at α of 0.05.
Comparison/correlation with previous gene expression results18
To determine whether flaxseed-associated miR-34a upregulation might contribute to downregulation of genes in murine lung tissue, we re-examined our previously published gene profiling data of murine lung tissue fed 0 or 10% FS18 to find genes in common between these data and a list of miR-34a targets generated by the stringent TargetScan algorithm.19 We note several inverse relationships between the upregulated miR-34a and downregulated genes, such as Pdgfra (downregulated −1.70 fold), Ubp1 (−1.57), Fgfr1 (−1.79), Bcl2 (−1.73), and Fbxo10 (−1.83).18 Downregulation of Fgfr1 has been confirmed by qPCR.18 These observations provide insight into miRNA-specific effects on lung gene expression after feeding a 10% flaxseed diet. Further investigation of the direct or indirect nature of miR-34a mediated regulation of these and other genes is warranted.
Discussion
miRNAs play significant roles in maintaining cellular function and tissue homeostasis, however, few reports have investigated diet-induced changes of the miRNA profile in normal tissue. Although just a pilot study, our approach integrates multiple innovations, beginning with novel uses of a mouse model of radiosensitization/radioprotection to investigate newly discovered, small RNA-mediated mechanisms of the effects of flaxseed diet. OpenArray is a state-of-the-art medium-throughput system that has a 24-fold improvement in throughput over the previous TaqMan low density array platform.20 Our findings are the first to report on miRNA changes in normal lung tissue of FS-fed mice in the presence or absence of radiation. Robust decreases in miR-142-3p and miR-150 and increases in miR-34a levels were detected by array and confirmed by qPCR. Previous studies have reported miRNA profile alterations of lung cancer cells exposed to ionizing radiation (for example,21). Our results show at 48 h post-radiation exposure few changes in irradiated, non-malignant lung as compared with non-irradiated lung. Examination of earlier or later time points or even different radiation doses might identify a different miRNA profile. An exciting finding of the study was the change of the miRNA profile by FS feeding which changed the “set point” of expression of several miRNAs in the lung, with or without radiation exposure. Importantly, this is supported by previous work with the model, showing differential regulation of several target genes of the upregulated miR-34a.18
Involvement of miRNAs in pathogenesis of lung disease
The identities and direction of regulation of FS-associated miRNAs are consistent with contributions to the previously reported effects of FS. The lung is highly susceptible to changes in gene expression that result from environmental exposures.22 miRNAs fulfill a modulatory role in lung pathogenesis.23 Significant loss of miRNAs associates with increased pulmonary inflammation.24 Noxious challenges, such as radiation exposure, sepsis, and environmental pollutants alter the expression of certain miRNAs and are correlated with specific lung disease processes.25 Modulating miRNA levels prior to exposure, as we have done here through a dietary intervention, might well moderate the damage of environmental insults.
Downregulation of miRs-142-3p and -150 by FS
miR-142-3p appears to have important functions in lung development, inflammation, and carcinogenesis. Contributions to lung development and functioning are supported by high levels of expression in the human adult lung and significant differential expression in lung of neonate and adult mice.26,27 miR-142-3p has also been implicated in inflammatory responses,28,29 with significant upregulation at 3 and 6 h post-exposure in an lipopolysaccharide (LPS)-induced model.29 Bronchoalveolar lavage (BAL) fluid revealed an increase in polymorphonuclear (PMN) cell content. In light of previous reports that FS reduced BAL fluid white blood cell count and PMN content in a mouse model of radiation pneumonitis,17,30 our findings suggest a beneficial role of FS in reducing lung inflammation by reducing miR-142-3p. Of additional interest are associations of both of FS-downregulated miRNAs—miRs-142-3p and -150—and cancer: a positive correlation of miR-142-3p and lung carcinogenesis,31,32 as well as a role of increased miR-150 through deregulation of cell cycle control.33
FS increases miR-34a in lung; role in cell-cycle control and apoptosis
miR-34a, increased 2-fold by FS in this study, is significantly downregulated or undetectable in many cancers, including human lung cancer, and microarray analyses have identified a role for miR-34 family members in response to radiation damage.34,35 miR-34a exerts its effects through modulation of cell cycle control and apoptosis.36,37 Transactivated by p53, miR-34a declines with p53 dysregulation. Targets of miR-34a are disproportionately involved in cell cycle control.37,38 Interestingly, a recent report also implicates miR-150 as a suppressive agent in the p53/miR-34a regulatory network, with lower miR-150 activity associated with increased levels of miR-34a,39 similar to what we report here. The effects of miR-34a on the p53 network have made this miRNA an attractive therapeutic target in cancers.40,41 FS-enhanced lung levels of miR-34a may boost tissue defenses in response to radiation damage, cell cycle arrest and apoptotic cell death as shown in our earlier work.17
Flaxseed and other botanicals alter microRNA profile
Modulation of miRNAs by other botanicals and polyphenols has been the subject of notable investigations. Izzotti and coworkers were the first to report on miRNA regulation by chemopreventive agents in cigarette-exposed rat lung tissue.42 miRNAs can be modulated by botanicals in lung cancer.43-46 The effects of curcumin on a human retinal pigment epithelial cell line, ARPE-19, included downregulation of 20 and upregulation of 9 miRNAs.47 Effects of resveratrol on miRNA have also been studied.48 miR-34a was modulated by resveratrol in A549 human non-small cell lung cancer (NSCLC) cells.49 This treatment reduced the expression of oncogenic miRNAs and induced pro-apoptotic miRNAs in colon cancer cells.50,51 Two flavonoids, rhamnetin and cirsiliol, upregulated p53-dependent miR-34a expression in NSCLC cells, resulting in radiosensitization through increased apoptosis.52 Our study adds to the accumulating evidence of natural product-mediated miRNA control.
Downstream effects
Additional work is needed to identify the various targets of the miRNAs we found in this study, but several promising candidates of the upregulated miR-34a should be mentioned. A common theme in the miR-34a literature has been the reliable, multiply-verified targeting of Bcl2 by miR-34a,53-55 both directly and indirectly. Bcl2 was also downregulated in our model.18 Additionally, we found that miR-34a expression is negatively associated with FGFR1 following FS feeding. As a cellular receptor implicated in multiple cell signaling pathways, FGFR1 represents another potential target for inhibition in the treatment of malignancies that result via its cell signaling pathway.56
Taken together, our results as well as numerous reports in the literature suggest that FS-modulated miRNAs, miRs-34a, 142-3p, and -150, may play different but complementary, roles in tumor suppression and cancer radiosensitization in addition to contributing to salutary effects of dietary FS in protecting normal tissue. The therapeutic potential of miR-34a in particular is illustrated by MRX34, an injectable, liposome-formulated miR-34a mimic currently being evaluated in a phase I clinical trial in patients with liver cancer and liver metastases (trial NCT01829971).41 As results from our study and others show, modulation of miRNA levels by natural products such as FS may prove to be an additional avenue to explore in a variety of diseases linked to cell proliferation and apoptosis. We encourage and look forward to additional work in this area.
Materials and Methods
Animals
Our studies used female C57/BL6 mice, a strain well characterized in the field of pulmonary radioprotection.10,16,17,30,57 Mice were obtained from Charles River and irradiated at 6–8 wk of age under animal protocols approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Pennsylvania. Animals were housed in conventional cages under standardized conditions with controlled temperature and humidity and a 12:12-h day–night light cycle. Animals had free access to water and formulated study diets. For this study we used n = 3 mice for each irradiated (XRT, 13.5 Gy) or control group on test diets (0% FS, 10% FS).
Diet composition and dietary treatments
Two diets were used for this study, all based on a semi-purified AIN-93G diet which was modified to contain the test ingredient as previously described.16 Importantly, control (no test ingredient added) and experimental diets were isocaloric, isonitrogenous, and contained equal amounts of dietary lipid and carbohydrate. Mice were maintained on control (0% FS) or 10% FS-supplemented diet given ad libitum for three weeks prior to XRT and were maintained on the respective diets until conclusion of the study and tissue harvest.
Radiation procedure
The Small Animal Radiation Research Platform (SARRP), (Xstrahl) was used to irradiate animals with a custom-made beam collimator. This system uses a Varian model NDI-225–22 kV X-ray tube mounted on a gantry that rotates between 0 and 120 degrees. The custom collimator creates a 12.5 cm circular field with well-defined borders and with animals arranged in a circular, “head in” arrangement using a single central shield which provides uniform irradiation to the thoracic portion of multiple mice simultaneously. This set-up consists of a single, anterior 225 kV, 15 mA X-ray beam with a 0.15 mm Cu at an SSD of 35 cm that is designed to accurately reproduce the internal radiation dose distribution in mice that were used in previous studies.16-18,30,57 The dosimetry and shielding of this system has been tested extensively.28 The dose of radiation is a single fraction delivered via single AP (anterior-posterior) approach. The dose used is 13.5 Gy (roughly corresponding to LD50) as described in our previous work.16,30,57 For quality assurance, thermoluminescent dosimeters are placed over selected mice, to verify correct dose administration.
RNA isolation
RNA was isolated and purified from mouse lung tissue 48 h post-radiation exposure (including non-irradiated controls) using a modified Trizol protocol with column clean-up. Twenty to thirty milligrams of frozen tissue was placed into 1 mL Trizol (Invitrogen,) in 2 mL screw-cap tubes, and Lysing matrix D (MP Biomedicals) was added. Tissue was disrupted using a desktop bead beater for 2 times 30 s, with 5 min on ice interspersed. Tubes were spun to collect beads and homogenate, and 200 µL chloroform was added. Tubes were firmly capped and shaken vigorously for 1 min, then centrifuged at 12 000 × g in an Eppendorf C2415 centrifuge for 15 min at 4 °C. The aqueous phase was transferred to a new tube, with care taken not to disturb the interphase. 100% ethanol was added in a volume at least 1.25 times that of the recovered aqueous phase for each tube. Sample was then applied to columns from a mirVana miRNA isolation kit (Ambion) to obtain total RNA. Washing and elution was done per the manufacturer’s protocol. RNA concentration and purity was analyzed by NanoDrop. RNA was stored at −80 °C.
QuantStudio OpenArray profiling
Profiling was conducted as described previously.20 One hundred nanograms of RNA per sample was reverse transcribed using the RT primers of rodent-specific primer pools A and B (Life Technologies). cDNA was pre-amplified for 12 cycles using Rodent Pre-Amp primer pools A and B, also per manufacturer’s protocol. Pre-amplified product was diluted 1:40 with 0.1× TE, pH 8.0. After mixing 1:1 with TaqMan OpenArray master mix, sample was loaded onto OpenArray plates by robot and amplified with the QuantStudio system. For six samples, technical duplicates were performed.
OpenArray data processing and analysis
Threshold cycles were assigned to each amplification reaction using ExpressionSuite software (Life Technologies). This software uses a proprietary algorithm to calculate a “relative threshold cycle” or Crt, similar to the Cycle of quantitation (Cq) or threshold cycle (Ct) assigned by other platforms but taking amplification efficiency into account for each individual reaction.. Additionally, the software assigns an “amplification score” to each reaction for rapid assessment of data reliability. Data were filtered to exclude reactions with a minimum amplification score and maximum Crt of 1.24 and 32, respectively (almost all features had Crt < 28). Several features with abnormally early Crt values were discarded after amplification curve inspection; these included mmu-miRs-1894-3p, -1896, and -1942, and hsa-miRs-188-3p and -5p. Data were exported to Microsoft Excel. Features detected in fewer than 14 of 18 runs (12 samples, 6 replicates) were excluded from further consideration, with the exception of miR-122, which was detected in all flaxseed-fed samples but in only two others. Following application of this detection filter, replicate values were averaged.
Data normalization by quantiles, as well as analysis, was performed with R/Bioconductor tools including affy/preprocesscore; the MultiExperiment Viewer (MeV; http://www.tm4.org/mev.html); and Microsoft Excel as described previously.58,59 Mean-centered normalized data were used to cluster samples hierarchically (Pearson distance, average linkage). Differences between treatment groups were assessed by two-way ANOVA, and features displaying differential regulation (P < 0.005) as assessed by diet or radiation exposure comparisons were used for clustering analysis. Interaction of diet and radiation was also assessed.
Validation by individual stem-loop qPCR assays
Individual stem-loop primer reverse transcription/hydrolysis probe qPCR assays (Life Technologies) were used to quantitate 12 mature miRNAs (miRs-10a, -16, -21, -25, -26a, -34a, 142-3p, -143, -150, -181c, -340, and -375) and two structural small RNAs, snoRNA-135 and snRNA U6, following manufacturer’s recommendations and with minor modifications as described previously.60,61 Data were normalized to the geometric mean of all measured RNAs using the delta-delta Cq method. Differences between groups were assessed by the Kruskal-Wallis test followed by the Dunn post-test for multiple comparisons.
miRNA target prediction
TargetScan19 was used to predict targets of the significantly upregulated miR-34a.
Data availability
Raw and normalized data have been deposited with the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE57123.
Disclosure of Potential Conflicts of Interest
The authors declare that they have no competing interests.
Acknowledgments and Funding
The authors wish to thank Dr. Cameron Koch for helping with the irradiation procedure and the designing of a custom collimator for the SARRP, as well as Roxann Ashworth and the DNA Analysis Facility of The Johns Hopkins University. This work was funded in part by: NIH-R01 CA133470 (M.C.S.), NIH-RC1AI081251 (M.C.S.), NIH-R21AI102659 (K.W.W.), and the University of Pennsylvania Research Foundation (M.C.S.).
Author Contributions
M.C.S. designed the study and individual experiments, analyzed data, wrote the manuscript, and supervised project and lab personnel; R.P. performed animal experiments, biochemical assays and conducted data analysis; E.A. conducted animal experiments and tissue analyses; M.A.M. performed RNA purification and qPCR assays and analysis; K.W.W. performed miRNA analysis, qPCR confirmation, and data analyses, and contributed in writing of the manuscript.
Glossary
Abbreviations:
- FS
flaxseed
- ROS
reactive oxygen species
- qRT-PCR
quantitative reverse transcriptase-polymerase chain reaction
- SARRP
small animal radiation research platform
- SEM
standard error means
- TLDA
TaqMan Low Density Array
- XRT
X-ray treatment
- miRNA
microRNA
REFERENCES
- 1.Machtay M. Pulmonary complications of anticancer treatment.: Churchill Livingston, 2003. [Google Scholar]
- 2.Wang JY, Chen KY, Wang JT, Chen JH, Lin JW, Wang HC, Lee LN, Yang PC. Outcome and prognostic factors for patients with non-small-cell lung cancer and severe radiation pneumonitis. Int J Radiat Oncol Biol Phys. 2002;54:735–41. doi: 10.1016/S0360-3016(02)02994-2. [DOI] [PubMed] [Google Scholar]
- 3.Robnett TJ, Machtay M, Vines EF, McKenna MG, Algazy KM, McKenna WG. Factors predicting severe radiation pneumonitis in patients receiving definitive chemoradiation for lung cancer. Int J Radiat Oncol Biol Phys. 2000;48:89–94. doi: 10.1016/S0360-3016(00)00648-9. [DOI] [PubMed] [Google Scholar]
- 4.Hughes-Davies L, Tarbell NJ, Coleman CN, Silver B, Shulman LN, Linggood R, Canellos GP, Mauch PM. Stage IA-IIB Hodgkin’s disease: management and outcome of extensive thoracic involvement. Int J Radiat Oncol Biol Phys. 1997;39:361–9. doi: 10.1016/S0360-3016(97)00085-0. [DOI] [PubMed] [Google Scholar]
- 5.Marks LB, Fan M, Clough R, Munley M, Bentel G, Coleman RE, Jaszczak R, Hollis D, Anscher M. Radiation-induced pulmonary injury: symptomatic versus subclinical endpoints. Int J Radiat Biol. 2000;76:469–75. doi: 10.1080/095530000138466. [DOI] [PubMed] [Google Scholar]
- 6.Riley PA. Free radicals in biology: oxidative stress and the effects of ionizing radiation. Int J Radiat Biol. 1994;65:27–33. doi: 10.1080/09553009414550041. [DOI] [PubMed] [Google Scholar]
- 7.Molteni A, Ward WF, Ts’ao CH, Solliday NH, Dunne M. Monocrotaline-induced pulmonary fibrosis in rats: amelioration by captopril and penicillamine. Proc Soc Exp Biol Med. 1985;180:112–20. doi: 10.3181/00379727-180-42151. [DOI] [PubMed] [Google Scholar]
- 8.Epperly M, Bray J, Kraeger S, Zwacka R, Engelhardt J, Travis E, Greenberger J. Prevention of late effects of irradiation lung damage by manganese superoxide dismutase gene therapy. Gene Ther. 1998;5:196–208. doi: 10.1038/sj.gt.3300580. [DOI] [PubMed] [Google Scholar]
- 9.Vujaskovic Z, Feng QF, Rabbani ZN, Samulski TV, Anscher MS, Brizel DM. Assessment of the protective effect of amifostine on radiation-induced pulmonary toxicity. Exp Lung Res. 2002;28:577–90. doi: 10.1080/01902140290096791. [DOI] [PubMed] [Google Scholar]
- 10.Machtay M, Scherpereel A, Santiago J, Lee J, McDonough J, Kinniry P, Arguiri E, Shuvaev VV, Sun J, Cengel K, et al. Systemic polyethylene glycol-modified (PEGylated) superoxide dismutase and catalase mixture attenuates radiation pulmonary fibrosis in the C57/bl6 mouse. Radiother Oncol. 2006;81:196–205. doi: 10.1016/j.radonc.2006.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prasad K. Hydroxyl radical-scavenging property of secoisolariciresinol diglucoside (SDG) isolated from flax-seed. Mol Cell Biochem. 1997;168:117–23. doi: 10.1023/A:1006847310741. [DOI] [PubMed] [Google Scholar]
- 12.Prasad K. Antioxidant Activity of Secoisolariciresinol Diglucoside-derived Metabolites, Secoisolariciresinol, Enterodiol, and Enterolactone. Int J Angiol. 2000;9:220–5. doi: 10.1007/BF01623898. [DOI] [PubMed] [Google Scholar]
- 13.Kitts DD, Yuan YV, Wijewickreme AN, Thompson LU. Antioxidant activity of the flaxseed lignan secoisolariciresinol diglycoside and its mammalian lignan metabolites enterodiol and enterolactone. Mol Cell Biochem. 1999;202:91–100. doi: 10.1023/A:1007022329660. [DOI] [PubMed] [Google Scholar]
- 14.Kinniry P, Amrani Y, Vachani A, Solomides CC, Arguiri E, Workman A, Carter J, Christofidou-Solomidou M. Dietary flaxseed supplementation ameliorates inflammation and oxidative tissue damage in experimental models of acute lung injury in mice. J Nutr. 2006;136:1545–51. doi: 10.1093/jn/136.6.1545. [DOI] [PubMed] [Google Scholar]
- 15.Lee JC, Bhora F, Sun J, Cheng G, Arguiri E, Solomides CC, Chatterjee S, Christofidou-Solomidou M. Dietary flaxseed enhances antioxidant defenses and is protective in a mouse model of lung ischemia-reperfusion injury. Am J Physiol Lung Cell Mol Physiol. 2008;294:L255–65. doi: 10.1152/ajplung.00138.2007. [DOI] [PubMed] [Google Scholar]
- 16.Christofidou-Solomidou M, Tyagi S, Pietrofesa R, Dukes F, Arguiri E, Turowski J, Grieshaber PA, Solomides CC, Cengel KA. Radioprotective role in lung of the flaxseed lignan complex enriched in the phenolic secoisolariciresinol diglucoside (SDG) Radiat Res. 2012;178:568–80. doi: 10.1667/RR2980.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee JC, Krochak R, Blouin A, Kanterakis S, Chatterjee S, Arguiri E, Vachani A, Solomides CC, Cengel KA, Christofidou-Solomidou M. Dietary flaxseed prevents radiation-induced oxidative lung damage, inflammation and fibrosis in a mouse model of thoracic radiation injury. Cancer Biol Ther. 2009;8:47–53. doi: 10.4161/cbt.8.1.7092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dukes F, Kanterakis S, Lee J, Pietrofesa R, Andersen ES, Arguiri E, Tyagi S, Showe L, Amrani Y, Christofidou-Solomidou M. Gene expression profiling of flaxseed in mouse lung tissues-modulation of toxicologically relevant genes. BMC Complement Altern Med. 2012;12:47. doi: 10.1186/1472-6882-12-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu IW, Espinoza DA, McAlexander MA, Witwer KW. OpenArray profiling reveals no differential modulation of miRNA by positive and negative CD4+ T-cell immunoselection. Exp Hematol. 2014;42:11–3. doi: 10.1016/j.exphem.2013.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shin S, Cha HJ, Lee EM, Lee SJ, Seo SK, Jin HO, Park IC, Jin YW, An S. Alteration of miRNA profiles by ionizing radiation in A549 human non-small cell lung cancer cells. Int J Oncol. 2009;35:81–6. [PubMed] [Google Scholar]
- 22.Reddy SP. The antioxidant response element and oxidative stress modifiers in airway diseases. Curr Mol Med. 2008;8:376–83. doi: 10.2174/156652408785160925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Angulo M, Lecuona E, Sznajder JI. Role of MicroRNAs in lung disease. Arch Bronconeumol. 2012;48:325–30. doi: 10.1016/j.arbr.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Flora S, Balansky R, D’Agostini F, Cartiglia C, Longobardi M, Steele VE, Izzotti A. Smoke-induced microRNA and related proteome alterations. Modulation by chemopreventive agents. Int J Cancer. 2012;131:2763–73. doi: 10.1002/ijc.27814. [DOI] [PubMed] [Google Scholar]
- 25.Sessa R, Hata A. Role of microRNAs in lung development and pulmonary diseases. Pulm Circ. 2013;3:315–28. doi: 10.4103/2045-8932.114758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harris KS, Zhang Z, McManus MT, Harfe BD, Sun X. Dicer function is essential for lung epithelium morphogenesis. Proc Natl Acad Sci U S A. 2006;103:2208–13. doi: 10.1073/pnas.0510839103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Williams AE, Moschos SA, Perry MM, Barnes PJ, Lindsay MA. Maternally imprinted microRNAs are differentially expressed during mouse and human lung development. Dev Dyn. 2007;236:572–80. doi: 10.1002/dvdy.21047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chaudhuri AD, Yelamanchili SV, Marcondes MC, Fox HS. Up-regulation of microRNA-142 in simian immunodeficiency virus encephalitis leads to repression of sirtuin1. FASEB J. 2013;27:3720–9. doi: 10.1096/fj.13-232678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moschos SA, Williams AE, Perry MM, Birrell MA, Belvisi MG, Lindsay MA. Expression profiling in vivo demonstrates rapid changes in lung microRNA levels following lipopolysaccharide-induced inflammation but not in the anti-inflammatory action of glucocorticoids. BMC Genomics. 2007;8:240. doi: 10.1186/1471-2164-8-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Christofidou-Solomidou M, Tyagi S, Tan KS, Hagan S, Pietrofesa R, Dukes F, Arguiri E, Heitjan DF, Solomides CC, Cengel KA. Dietary flaxseed administered post thoracic radiation treatment improves survival and mitigates radiation-induced pneumonopathy in mice. BMC Cancer. 2011;11:269. doi: 10.1186/1471-2407-11-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qian S, Ding JY, Xie R, An JH, Ao XJ, Zhao ZG, Sun JG, Duan YZ, Chen ZT, Zhu B. MicroRNA expression profile of bronchioalveolar stem cells from mouse lung. Biochem Biophys Res Commun. 2008;377:668–73. doi: 10.1016/j.bbrc.2008.10.052. [DOI] [PubMed] [Google Scholar]
- 32.Kaduthanam S, Gade S, Meister M, Brase JC, Johannes M, Dienemann H, Warth A, Schnabel PA, Herth FJ, Sültmann H, et al. Serum miR-142-3p is associated with early relapse in operable lung adenocarcinoma patients. Lung Cancer. 2013;80:223–7. doi: 10.1016/j.lungcan.2013.01.013. [DOI] [PubMed] [Google Scholar]
- 33.Zhang N, Wei X, Xu L. miR-150 promotes the proliferation of lung cancer cells by targeting P53. FEBS Lett. 2013;587:2346–51. doi: 10.1016/j.febslet.2013.05.059. [DOI] [PubMed] [Google Scholar]
- 34.Hermeking H. p53 enters the microRNA world. Cancer Cell. 2007;12:414–8. doi: 10.1016/j.ccr.2007.10.028. [DOI] [PubMed] [Google Scholar]
- 35.Buscaglia LE, Li Y. Apoptosis and the target genes of microRNA-21. Chin J Cancer. 2011;30:371–80. doi: 10.5732/cjc.011.10132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamakuchi M, Ferlito M, Lowenstein CJ. miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci U S A. 2008;105:13421–6. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–52. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tarasov V, Jung P, Verdoodt B, Lodygin D, Epanchintsev A, Menssen A, Meister G, Hermeking H. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle. 2007;6:1586–93. doi: 10.4161/cc.6.13.4436. [DOI] [PubMed] [Google Scholar]
- 39.Wang DT, Ma ZL, Li YL, Wang YQ, Zhao BT, Wei JL, Qi X, Zhao XT, Jin YX. miR-150, p53 protein and relevant miRNAs consist of a regulatory network in NSCLC tumorigenesis. Oncol Rep. 2013;30:492–8. doi: 10.3892/or.2013.2453. [DOI] [PubMed] [Google Scholar]
- 40.Di Martino MT, Leone E, Amodio N, Foresta U, Lionetti M, Pitari MR, Cantafio ME, Gullà A, Conforti F, Morelli E, et al. Synthetic miR-34a mimics as a novel therapeutic agent for multiple myeloma: in vitro and in vivo evidence. Clin Cancer Res. 2012;18:6260–70. doi: 10.1158/1078-0432.CCR-12-1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.A Multicenter Phase I Study of MRX34, MicroRNA miR-RX34 Liposome Injectable Suspension. Mina Therapeutics I; 2013 Apr 8 [updated 2013 Oct 28]. Available from: http://www.clinicaltrials.gov/show/NCT01829971
- 42.Izzotti A, Calin GA, Steele VE, Cartiglia C, Longobardi M, Croce CM, De Flora S. Chemoprevention of cigarette smoke-induced alterations of MicroRNA expression in rat lungs. Cancer Prev Res (Phila) 2010;3:62–72. doi: 10.1158/1940-6207.CAPR-09-0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Banerjee N, Talcott S, Safe S, Mertens-Talcott SU. Cytotoxicity of pomegranate polyphenolics in breast cancer cells in vitro and vivo: potential role of miRNA-27a and miRNA-155 in cell survival and inflammation. Breast Cancer Res Treat. 2012;136:21–34. doi: 10.1007/s10549-012-2224-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang H, Bian S, Yang CS. Green tea polyphenol EGCG suppresses lung cancer cell growth through upregulating miR-210 expression caused by stabilizing HIF-1α. Carcinogenesis. 2011;32:1881–9. doi: 10.1093/carcin/bgr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang CH, Yue J, Sims M, Pfeffer LM. The curcumin analog EF24 targets NF-κB and miRNA-21, and has potent anticancer activity in vitro and in vivo. PLoS One. 2013;8:e71130. doi: 10.1371/journal.pone.0071130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang J, Du Y, Wu C, Ren X, Ti X, Shi J, Zhao F, Yin H. Curcumin promotes apoptosis in human lung adenocarcinoma cells through miR-186* signaling pathway. Oncol Rep. 2010;24:1217–23. doi: 10.3892/or_00000975. [DOI] [PubMed] [Google Scholar]
- 47.Howell JC, Chun E, Farrell AN, Hur EY, Caroti CM, Iuvone PM, Haque R. Global microRNA expression profiling: curcumin (diferuloylmethane) alters oxidative stress-responsive microRNAs in human ARPE-19 cells. Mol Vis. 2013;19:544–60. [PMC free article] [PubMed] [Google Scholar]
- 48.Lançon A, Kaminski J, Tili E, Michaille JJ, Latruffe N. Control of MicroRNA expression as a new way for resveratrol to deliver its beneficial effects. J Agric Food Chem. 2012;60:8783–9. doi: 10.1021/jf301479v. [DOI] [PubMed] [Google Scholar]
- 49.Bae S, Lee EM, Cha HJ, Kim K, Yoon Y, Lee H, Kim J, Kim YJ, Lee HG, Jeung HK, et al. Resveratrol alters microRNA expression profiles in A549 human non-small cell lung cancer cells. Mol Cells. 2011;32:243–9. doi: 10.1007/s10059-011-1037-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tili E, Michaille JJ, Adair B, Alder H, Limagne E, Taccioli C, Ferracin M, Delmas D, Latruffe N, Croce CM. Resveratrol decreases the levels of miR-155 by upregulating miR-663, a microRNA targeting JunB and JunD. Carcinogenesis. 2010;31:1561–6. doi: 10.1093/carcin/bgq143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tili E, Michaille JJ, Alder H, Volinia S, Delmas D, Latruffe N, Croce CM. Resveratrol modulates the levels of microRNAs targeting genes encoding tumor-suppressors and effectors of TGFβ signaling pathway in SW480 cells. Biochem Pharmacol. 2010;80:2057–65. doi: 10.1016/j.bcp.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kang J, Kim E, Kim W, Seong KM, Youn H, Kim JW, Kim J, Youn B. Rhamnetin and cirsiliol induce radiosensitization and inhibition of epithelial-mesenchymal transition (EMT) by miR-34a-mediated suppression of Notch-1 expression in non-small cell lung cancer cell lines. J Biol Chem. 2013;288:27343–57. doi: 10.1074/jbc.M113.490482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cole KA, Attiyeh EF, Mosse YP, Laquaglia MJ, Diskin SJ, Brodeur GM, Maris JM. A functional screen identifies miR-34a as a candidate neuroblastoma tumor suppressor gene. Mol Cancer Res. 2008;6:735–42. doi: 10.1158/1541-7786.MCR-07-2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tryndyak VP, Ross SA, Beland FA, Pogribny IP. Down-regulation of the microRNAs miR-34a, miR-127, and miR-200b in rat liver during hepatocarcinogenesis induced by a methyl-deficient diet. Mol Carcinog. 2009;48:479–87. doi: 10.1002/mc.20484. [DOI] [PubMed] [Google Scholar]
- 55.Kojima K, Fujita Y, Nozawa Y, Deguchi T, Ito M. MiR-34a attenuates paclitaxel-resistance of hormone-refractory prostate cancer PC3 cells through direct and indirect mechanisms. Prostate. 2010;70:1501–12. doi: 10.1002/pros.21185. [DOI] [PubMed] [Google Scholar]
- 56.Ho HK, Yeo AH, Kang TS, Chua BT. Current strategies for inhibiting FGFR activities in clinical applications: opportunities, challenges and toxicological considerations. Drug Discov Today. 2014;19:51–62. doi: 10.1016/j.drudis.2013.07.021. [DOI] [PubMed] [Google Scholar]
- 57.Pietrofesa R, Turowski J, Tyagi S, Dukes F, Arguiri E, Busch TM, Gallagher-Colombo SM, Solomides CC, Cengel KA, Christofidou-Solomidou M. Radiation mitigating properties of the lignan component in flaxseed. BMC Cancer. 2013;13:179. doi: 10.1186/1471-2407-13-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Witwer KW, Clements JE. Evidence for miRNA expression differences of HIV-1-positive, treatment-naive patients and elite suppressors: a re-analysis. Blood. 2012;119:6395–6. doi: 10.1182/blood-2012-02-412742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Witwer KW, Watson AK, Blankson JN, Clements JE. Relationships of PBMC microRNA expression, plasma viral load, and CD4+ T-cell count in HIV-1-infected elite suppressors and viremic patients. Retrovirology. 2012;9:5. doi: 10.1186/1742-4690-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McAlexander MA, Phillips MJ, Witwer KW. Comparison of Methods for miRNA Extraction from Plasma and Quantitative Recovery of RNA from Cerebrospinal Fluid. Front Genet. 2013;4:83. doi: 10.3389/fgene.2013.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Witwer KW, Sarbanes SL, Liu J, Clements JE. A plasma microRNA signature of acute lentiviral infection: biomarkers of CNS disease. AIDS. 2011;204:1104–14. doi: 10.1097/QAD.0b013e32834b95bf. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw and normalized data have been deposited with the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE57123.