Abstract
We have employed upstream open reading frames (uORFs) to systematically tune the translation levels of recombinant proteins. We present the design principles that guided the development of this technology and provide information that may help others in implementing synthetic uORFs for their own applications. We also report on recent applications to our own research projects, including the coupling of uORF and translation initiation site (TIS) engineering with small molecule-inducible post-translational control. Finally, we discuss opportunities to investigate and potentially engineer gene-specific translational responses to cellular stress.
Keywords: upstream open reading frames, uORF, translation initiation, ribosome scanning, cellular stress, post-transcriptional regulation
Preface
A focus of our lab is the tuning of biomolecular systems in human cells. We believe that living systems and the levels of their many molecular components have been tuned and optimized during the course of evolution. Furthermore, diseases could result from the aberrant optimization of gene expression or molecular signaling pathways. Thus, in dissecting these systems, whether one is taking a traditional genetics approach (e.g., genetic complementation or restoration of a deficiency) or engineering a system from a set of defined biomolecular components, we would argue that it is prudent, and perhaps most informative, to evaluate recombinant gene expression at physiologically relevant levels. We realized that if we were to quantitatively study and engineer biomolecular systems, we would need to develop new methods to precisely control expression levels. Common expression vectors typically utilize strong transcriptional promoters and generate high levels of recombinant gene expression. Yet when screening proto-oncogene candidates, we found that the strong viral promoters generated such high levels of ectopic expression that expression could have actually suppressed proliferation or been toxic. When we attempted to control levels using a small-molecule inducible transcription system, the tetracycline transactivator,1 we found that in expressing the Ras oncogene, the basal, “leaky” level of transcription generated in the expression “off” state was already sufficient to stimulate cell proliferation. In some cases, inducing Ras oncogene expression above this basal level actually caused the proliferation rate to decrease. To reduce toxic side effects from overexpression, we previously have engineered a library of promoters to produce lower levels of transcription.2,3 Although we were successful, ensuing work revealed two potential problems. First, promoters sometimes exhibited different expression levels that varied with cell type. In hindsight, this was not surprising, since in nature transcription is often controlled by promoter elements that are specific for different cells or tissues. Second, our observations suggested that weak promoters generating low levels of ectopic expression could be more prone to epigenetic silencing than strong promoters. Altogether, to address these challenges we pursued an expression tuning method that (1) could generate a broad range of expression levels, particularly those on the low and very low end of the expression spectrum, (2) worked predictably in all cell types, and (3) was not controlled at the transcriptional level.
Sequence-Dependent Control of Translation in Eukaryotes
Our goal was to systematically control the protein levels encoded by genes of interest. Eukaryotes can control protein levels at the point of translation initiation. To translate a gene, first the 43S ribosomal preinitiation complex loads at the methylated 5′ cap of mRNAs4 (Fig. 1). The complex consists of the 40S subunit, initiator Met-tRNA, and eukaryotic initiation factors (eIFs) 1, 1A, 2, 3, and 5 (Fig. 2). Upon loading, it scans for a translation initiation site (TIS, Figs. 1 and 2), which consists of a start codon (usually, but not necessarily AUG) and its adjacent bases. If the complex recognizes a TIS (Fig. 2), eIFs 1, 2, 3, and 5 detach and eIF5B joins. This leads to recruitment of the 60S subunit and shedding of the remaining eIFs. Protein synthesis then proceeds.5 Yet initiation does not occur every time a ribosomal complex reaches a TIS sequence. The complex recognizes the TIS at a certain frequency governed by the sequence. By controlling this frequency, the cells can control the level of translation.6 The TIS sequence is thought to be defined by the bases from the ‒6 to +4 (or +5) positions of open reading frames (ORFs), where +1 is the first base of the start codon.7-9 Kozak reported GCCACCAUGG (start codon underlined) as an optimal TIS10,11 for high translation levels and as a result, this TIS sequence is often employed for the expression of recombinant proteins. In addition, Kozak found that translation was generally high when a purine7 (A or G, abbreviated as R) was found at position −3. Although less important than the −3 purine, Kozak also reported that a G at +4 could bolster translation, particularly when the −3 position was a pyrimidine (C or U).7,9
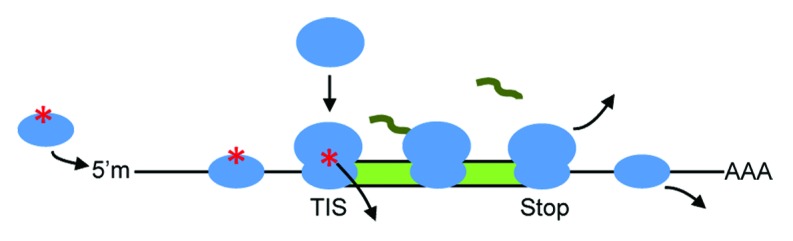
Figure 1. Eukaryotic translation of mRNA. Methylated 5′ cap, 5′m; translation initiation site, TIS (a motif that also includes the start codon); stop codon; ribosomal subunits, ovals; protein-coding ORF, green rectangle; protein, squiggle; polyA, AAA. The asterisk denotes factors that complete the ribosomal preinitiation complex.
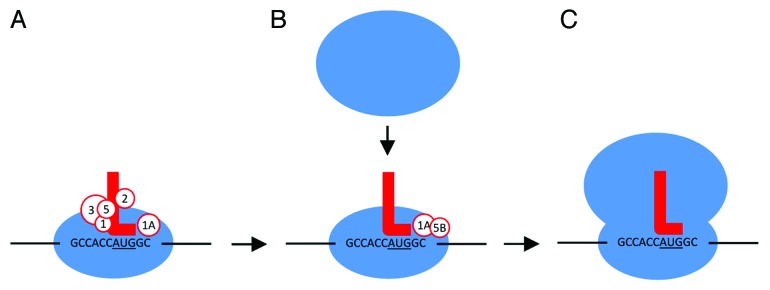
Figure 2. The TIS sequence and eIFs mediate initiation of translation. (A) 43S preinitiation complex (40S, eIFs 1, 1A, 2, 3, 5, and initiator Met-tRNA) recognizes a TIS sequence (in this example, GCCACCAUGGC with start codon underlined). (B) eIF5B is loaded while others detach. This allows the joining of the 60S ribosomal subunit to form the 80S complex. After shedding the remaining eIFs, (C) protein elongation begins. Initiator Met-tRNA, red L; eIFs, white circles; bases represent the TIS sequence.
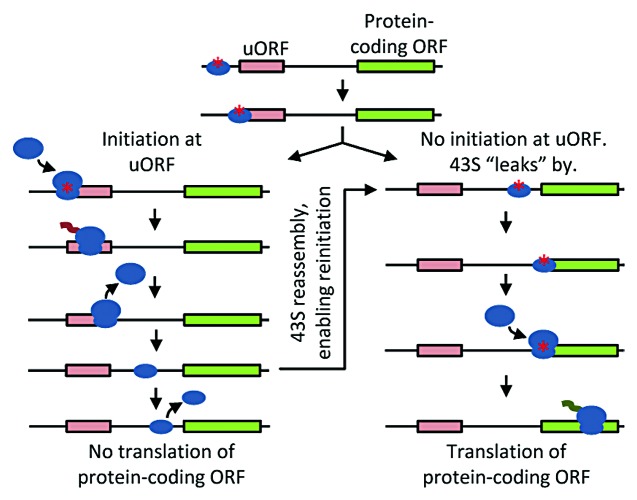
Eukaryotes can also control translation levels with ORFs upstream of the protein-coding ORF (uORF, Fig. 3). Originally, it was believed that for every one mRNA transcript only one ORF was translated. The ribosome scans until it reaches the first TIS and then translates the ORF. After reaching the stop codon, it disassembles and exits (Fig. 3, left). Because the ribosome exits, even if a second downstream ORF were present, that ORF would not be translated. Yet exceptions to this one-message-one-ORF rule have always been known. Two independent studies have reported that 6%12 and 13%13 of yeast genes utilize at least one uORF. Kochetov et al. reported that 20% of plant genes utilize uORFs.14 It still came as something of a surprise when genome sequence analysis revealed that half of all human protein-coding genes also encode one or more uORFs.15 While most of these human genes contain one to four uORFs, there are thousands that have more than four. In fact, 658 human genes have ten or more uORFs (RefSeq release 55, September 2012). These uORFs are often short, with a geometric mean of 13 codons (here the number includes the stop codon). While they are translated, they typically do not encode proteins with biological activity. Although we do not fully understand the roles of uORFs, given their abundance, they likely play a major role in regulating and tuning the translation of protein-coding ORFs. By introducing ORFs upstream of a preproinsulin reporter gene, Kozak was one of the first to demonstrate that protein-coding ORFs are still translated when a transcript contains a uORF.16 Although these protein-coding ORFs can be translated in the presence of a uORF, the level of translation is typically decreased.16
Figure 3. Schematic of leaky scanning and reinitiation. 43S complex loaded with tRNA plus initiation factors and primed for initiation, small blue oval with red asterisk; 40S subunit not competent for initiation, small blue oval without asterisk; 60S subunit, large blue oval; ORFs, rectangles; squiggle represents ongoing translation.
There are two mechanisms by which uORFs allow and regulate translation of a downstream, protein-coding ORF (Fig. 3).5,11,17 First, downstream translation can occur through a leaky scanning mechanism (Fig. 3, directly to right decision fork), where ribosomes fail to recognize the uORF and instead initiate at the downstream, protein-coding ORF. Second, downstream translation can occur through reinitiation (Fig. 3). In this case, the ribosome does recognize and initiate translation of the uORF. Upon reaching the stop codon, the ribosomal subunits will typically disassemble and release from the mRNA. Yet if certain initiation factors are associated with the ribosome when the subunits separate, the 40S subunit can remain bound to the mRNA and resume scanning. Factors eIF3 and eIF4F are believed to remain bound to the ribosome during the translation of short ORFs, while other eIFs, including eIF2, can potentially bind de novo to the 40S subunit once it resumes scanning.18 Furthermore, the presence of eIF4A and eIF4B enhances scanning in the 3′ direction and can promote scanning over greater distances.19 If the scanning 40S subunit continues on the mRNA for a sufficient period, it can then completely reload with initiation factors and an initiator Met-tRNA to reassemble a functional 43S preinitiation complex. The preinitiation complex can then “reinitiate” translation at the next ORF.
Tuning Recombinant Gene Expression
To tune the expression of recombinant proteins, we sought to control the efficiency of translation, i.e., the amount of protein synthesized per mRNA transcript. To accomplish this goal we varied the TIS sequence and introduced uORFs. The uORFs affected the flux of ribosomes reaching the protein-coding ORF, and the TIS sequence affected the probability of a ribosome, once it reaches the protein-coding ORF, of initiating translation (Fig. 4). From an engineering and design standpoint, we set the following objectives: (1) utilize a short RNA leader sequence that can be easily added to a protein-coding ORF by PCR, (2) minimize changes to the amino acid sequence of the protein of interest, and (3) leverage the leaky scanning mechanism and minimize reinitiation (so that the two mechanisms cannot potentially work against each other). To achieve these objectives, we varied only the −3, −2, and −1 TIS positions adjacent to the start codon. Although the +4 position has been shown to significantly affect translation levels, by not varying the +4 position, we minimized changes to the amino acid sequence of the protein of interest. In designing a short synthetic uORF, we reasoned that in order to reproduce the behavior found in nature, the uORF needed to encode a peptide of at least two amino acids. In this way, the A, P, and E sites of translating ribosomes would be properly utilized and we would preserve any effects due to initiation factors and tRNA interactions or peptide bond formation. To minimize reinitiation, we tried to limit the window of opportunity for the 40S subunit to reload with initiation factors. Because Kozak previously detected minimal reinitiation when the separation between the uORF and protein-coding ORF was relatively short (8 bases between the uORF stop codon and protein-coding ORF start codon), we only evaluated similarly short distances.
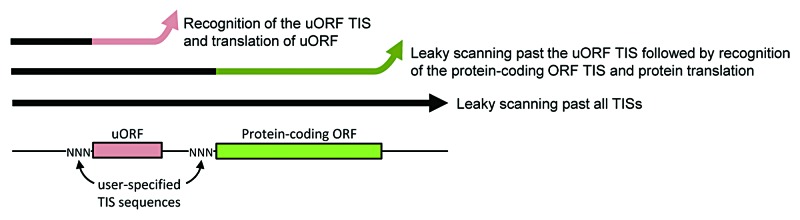
Figure 4. Schematic of protein translation tuning achieved by specifying TIS sequences of upstream and protein-coding ORFs. Because of the possibility of leaky scanning, ribosomes can take three possible paths (thick-lined arrows). By varying the three TIS bases (NNN) preceding the start codon of uORFs or protein-coding ORFs, the distribution of ribosomes over these paths is controlled. In doing so, one controls the flux of ribosomes that both reach and translate the protein of interest. Additional control can also be achieved by using multiple uORFs in series and non-AUG start codons (not shown).
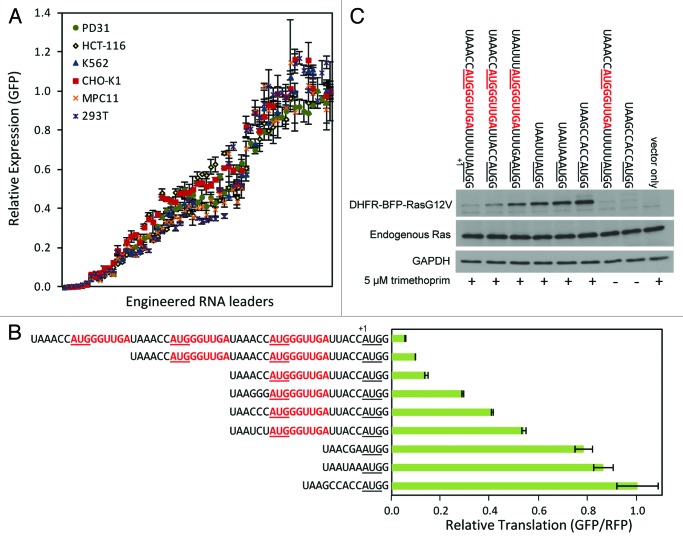
In varying TIS sequences to control protein expression (without using uORFs), we found that variation of the bases preceding an AUG start codon could achieve a range of high expression levels. In line with the findings of Kozak, the TIS ACCAUGG generated one of the highest levels of translation. However, by utilizing TIS with different bases at the −3, −2, and −1 positions (maintaining a +4G and AUG start codon), we were never able to generate an expression level less than approximately half that of the level produced by the TIS ACCAUGG. When we inserted synthetic uORFs, we now were able to achieve lower expression levels. By combining different strategies that included insertion of one or more uORFs, variation of TIS sequences at both uORFs and protein-coding ORFs, and also use of non-AUG start codons, we were able to achieve expression of green fluorescent protein (GFP) over three orders of magnitude (Fig. 5A). These levels were also generally reproducible across different cell types, vectors, and genes of interest. Mathematical modeling suggested that the uORF regulation of protein translation levels could be predicted solely by the leaky scanning model when RNA leaders utilized only one uORF. By employing more efficient TIS sequences to initiate translation of uORFs, fewer ribosomes were available to translate the downstream protein-coding ORF. Conversely, by employing less efficient TIS sequences to initiate translation of uORFs, more ribosomes were available to translate the downstream protein-coding ORF. Therefore, we were able to control the flux of ribosomes that reach and translate a protein-coding ORF of interest. For the longer leaders that utilized two and three uORFs in series, where now the distance between the first uORF and protein-coding ORF was also longer, our modeling indicated that reinitiation may be contributing to translation of the protein-coding ORF.
Figure 5. Control of protein translation levels. (A) Varying GFP expression levels achieved in different cell lines using MoMLV retroviral vectors equipped with engineered RNA leaders. Leaders utilizing different TIS sequences and uORFs were used to generate lower expression levels. Leaders with varying TIS sequences and no uORFs were used to generate higher expression levels. (B) Lentiviral expression of GFP in HCT-116 cells normalized to an internal RFP reference. (C) Immunoblot of retroviral expression of DHFR-BFP-RasG12V in NIH-3T3 cells with (+) and without (-) trimethoprim, the small molecule inducer of DHFR fusion protein stability. Sequences of the RNA leaders are given with uORFs in red bold type (including stop codons); start codons are underlined; the first base of the start codon encoding a protein of interest is indicated by the +1 base position. Error bars represent standard deviations from triplicate experiments.
For researchers that would like to utilize our RNA leaders to control translation levels in mammalian cells, we recommend that researchers start with the leader sequences in Figure 5B. These sequences can simply be added as a 5′ extension on oligonucleotide primers and used to PCR-amplify a protein-coding ORF of interest; one then just conventionally inserts the PCR product into a vector of choice. Alternatively, researchers may use our vectors that already contain the engineered RNA leaders (available through Addgene). To date, our lab has utilized our RNA leader sequences to tune the expression levels of GFP, mCherry, mTagBFP, mKate, c-Myc, p53, p21, H-Ras,20 FKBP, and DHFR. For applications where the protein of interest can be toxic or applications where conditional or dynamic control of expression is desired, we have found it desirable to specify the magnitude of expression by controlling translation and to conditionally turn on expression using small-molecule inducible control of protein localization (estrogen-receptor fusion with nuclear localization induced by addition of 4-hydroxytamoxifen) and stability (DHFR-fusion stabilized by trimethoprim, Fig. 5C).
While our previous results have suggested that unintended secondary structures do not significantly affect translation levels, those that choose to employ our leader sequences should still be aware of possible gene-specific effects. Strong secondary structures could still affect translation levels. Additionally, it is important to be aware of internal, in-frame AUG sequences that could lead to translation of a truncated, aberrantly active or dominant-negative protein due to either leaky scanning or reinitiation. In a few cases, we have observed a larger deviation across cell types when specifying the TIS sequence and not utilizing uORFs. Thus when we have the option, we generally choose leader sequences that regulate translation using uORFs and initiate translation of the protein of interest using an efficient TIS (e.g., ACCAUGG); this should minimize the occurrence of unintentionally truncated protein products.
In principle, the tuning of ectopic gene expression by employing uORFs and manipulating translation initiation efficiency should work in all eukaryotes. Use of synthetic uORFs to regulate ribosome flux will not work in bacteria or archaea in a manner that abides by the leaky scanning model; instead of the eukaryotic mechanism of 5′ cap loading followed by scanning, these non-eukaryotes load ribosomes at internal Shine-Dalgarno RBS sequences immediately 5′ of ORFs. Kozak has previously shown that the efficiency of TIS sequences is similar in vertebrates,10,21 and we believe that the relative translation levels generated by our RNA leader sequences likely will be reproducible in all vertebrate cells. Furthermore, our ongoing analysis of TIS sequences (unpublished) suggests that high efficiency TIS sequences perhaps are more conserved between all eukaryotes than previously thought. Thus we predict that TIS and uORF programming of translation levels can be extended to animals including lower metazoans, plants, and yeast, though some of the sequences may need to be tailored.
Engineering Translational Regulation in Response to Cellular Stress
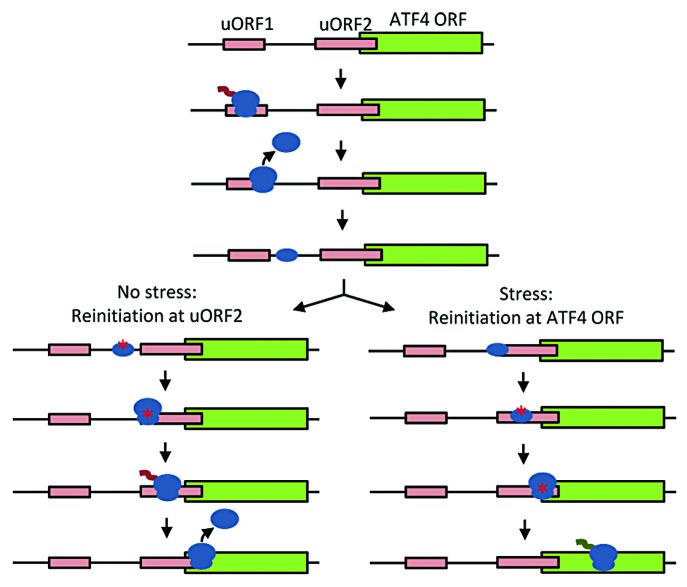
With our invention, we controlled ribosomal leaky scanning and minimized reinitiation in order to precisely specify protein expression levels. Next, we envision that both leaky scanning and reinitiation can be programmed simultaneously to generate RNA leader sequences that regulate expression in response to cellular stress, e.g., protein folding or endoplasmic reticulum stress, oxidative stress, hypoxia, or nutrient deprivation. In nature, cells regulate translation both globally and in a sequence-specific manner.22-24 As a general response to stress, translation is decreased in a non-sequence dependent manner, helping cells divert more energy and resources toward surviving and alleviating the stress. Because the response occurs at the translational level and does not experience lags involved with transcriptional responses, cells can respond to stress rapidly. First, stress leads to activation of the eIF2α kinase family; for example, PERK is activated by endoplasmic reticulum stress and hypoxia.22,24 PERK phosphorylates the essential initiation factor eIF2α. Phosphorylation inhibits the formation of eIF2-GTP and formation of the 43S preinitiation complex. This ultimately suppresses translation globally.24 Yet for certain critical genes, stress induces an increase in translation and uORFs are known to play a regulatory role. There are two well-studied examples of stress response genes regulated by uORFs: GCN4 in yeast25-27 and ATF4 in mammalian cells.28-30 Both utilize a similar strategy that leverages reinitiation (Fig. 6). Under non-stressed conditions, ribosomes translate uORF1 of ATF4. A fraction of 40S subunits continue along the mRNA and after some amount of time they reload with initiation factors and initiator Met-tRNA to regenerate a functional 43S preinitiation complex. The complex then reinitiates translation at uORF2. Because uORF2 is out of frame with the ATF4 ORF and terminates after the start of ATF4, ATF4 is not translated. Under stress, uORF1 is again translated, but because stress has led to a decrease in the supply of eIF2-GTP, the 40S subunit does not reload in time to initiate at uORF2. Yet, because of the additional distance before reaching ATF4, the 40S subunit does have enough time to reload and reinitiate at ATF4. Thus under stress, ATF4 translation increases.
Figure 6. Schematic of stress-induced translation of ATF4. After translating uORF1, under non-stressed conditions, the 40S subunit (small oval) quickly reloads with eIF2-GTP, Met-tRNA, and other eIFs (red asterisk) and reinitiates translation at uORF2. Under stress, the 40S subunit is not reloaded until later, reinitiating translation at ATF4.
Other than ATF4, researchers have investigated few other mammalian genes that utilize uORFs to regulate gene-specific expression in response to stress. Yet with the widespread frequency of uORFs in the human genome, it is possible that the expression of numerous genes is specifically tuned in response to stress. Many genes just may be tuned and regulated more subtly than ATF4. In order for cells to quickly tune proteome-wide expression in response to cellular stress, we hypothesize that cells broadly utilize a genetic code of RNA elements, including not only uORF and TIS elements, but also non-AUG start codons,31 stop codons and their adjacent sequences,32 inter-ORF spacer elements,16,33 and secondary structures.34 To investigate how these genetic elements are combined to regulate translation, one may take not only a classic molecular genetics approach, but also a synthetic approach. By building and combining synthetic elements, functional and operational rules can be assigned. Armed with a better understanding of how RNA elements regulate stress responses, researchers will be able to modulate the activity of known regulatory RNA leaders (e.g., that of ATF4) or engineer stress responsive sequences de novo.
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
Acknowledgments
This work was supported by an NSF CAREER Award (0846392), the NIH (5R21AG040360-02) and the Ellison Medical Foundation (AG-NS-0550-09).
References
- 1.Peacock RW, Sullivan KA, Wang CL. Tetracycline-regulated expression implemented through transcriptional activation combined with proximal and distal repression. ACS Synth Biol. 2012;1:156–62. doi: 10.1021/sb200029a. [DOI] [PubMed] [Google Scholar]
- 2.Ferreira JP, Peacock RW, Lawhorn IE, Wang CL. Modulating ectopic gene expression levels by using retroviral vectors equipped with synthetic promoters. Syst Synth Biol. 2011;5:131–8. doi: 10.1007/s11693-011-9089-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferreira JP, Lawhorn IE, Peacock RW, Wang CL. Quantitative assessment of Ras over-expression via shotgun deployment of vectors utilizing synthetic promoters. Integr Biol (Camb) 2012;4:108–14. doi: 10.1039/c1ib00082a. [DOI] [PubMed] [Google Scholar]
- 4.Myasnikov AG, Simonetti A, Marzi S, Klaholz BP. Structure-function insights into prokaryotic and eukaryotic translation initiation. Curr Opin Struct Biol. 2009;19:300–9. doi: 10.1016/j.sbi.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 5.Hinnebusch AG. Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiol Mol Biol Rev. 2011;75:434–67. doi: 10.1128/MMBR.00008-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferreira JP, Overton KW, Wang CL. Tuning gene expression with synthetic upstream open reading frames. Proc Natl Acad Sci U S A. 2013;110:11284–9. doi: 10.1073/pnas.1305590110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–92. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 8.Kozak M. At least six nucleotides preceding the AUG initiator codon enhance translation in mammalian cells. J Mol Biol. 1987;196:947–50. doi: 10.1016/0022-2836(87)90418-9. [DOI] [PubMed] [Google Scholar]
- 9.Kozak M. Recognition of AUG and alternative initiator codons is augmented by G in position +4 but is not generally affected by the nucleotides in positions +5 and +6. EMBO J. 1997;16:2482–92. doi: 10.1093/emboj/16.9.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–48. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kozak M. Emerging links between initiation of translation and human diseases. Mamm Genome. 2002;13:401–10. doi: 10.1007/s00335-002-4002-5. [DOI] [PubMed] [Google Scholar]
- 12.Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–9. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lawless C, Pearson RD, Selley JN, Smirnova JB, Grant CM, Ashe MP, Pavitt GD, Hubbard SJ. Upstream sequence elements direct post-transcriptional regulation of gene expression under stress conditions in yeast. BMC Genomics. 2009;10:7. doi: 10.1186/1471-2164-10-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kochetov AV, Syrnik OA, Rogozin IB, Glazko GV, Komarova ML, Shumnyĭ VK. [Context organization of mRNA 5′-untranslated regions of higher plants] Mol Biol (Mosk) 2002;36:649–56. [PubMed] [Google Scholar]
- 15.Calvo SE, Pagliarini DJ, Mootha VK. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc Natl Acad Sci U S A. 2009;106:7507–12. doi: 10.1073/pnas.0810916106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kozak M. Effects of intercistronic length on the efficiency of reinitiation by eucaryotic ribosomes. Mol Cell Biol. 1987;7:3438–45. doi: 10.1128/mcb.7.10.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Merrick WC. Eukaryotic protein synthesis: still a mystery. J Biol Chem. 2010;285:21197–201. doi: 10.1074/jbc.R110.111476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackson RJ, Hellen CU, Pestova TV. Termination and post-termination events in eukaryotic translation. Adv Protein Chem Struct Biol. 2012;86:45–93. doi: 10.1016/B978-0-12-386497-0.00002-5. [DOI] [PubMed] [Google Scholar]
- 19.Skabkin MA, Skabkina OV, Hellen CU, Pestova TV. Reinitiation and other unconventional posttermination events during eukaryotic translation. Mol Cell. 2013;51:249–64. doi: 10.1016/j.molcel.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferreira JP, Wang CL. Optimization of oncogene expression through intra-population competition. Biotechnol J. 2013;8:1476–84. doi: 10.1002/biot.201300037. [DOI] [PubMed] [Google Scholar]
- 21.Kozak M. Determinants of translational fidelity and efficiency in vertebrate mRNAs. Biochimie. 1994;76:815–21. doi: 10.1016/0300-9084(94)90182-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamasaki S, Anderson P. Reprogramming mRNA translation during stress. Curr Opin Cell Biol. 2008;20:222–6. doi: 10.1016/j.ceb.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sheikh MS, Fornace AJ., Jr. Regulation of translation initiation following stress. Oncogene. 1999;18:6121–8. doi: 10.1038/sj.onc.1203131. [DOI] [PubMed] [Google Scholar]
- 24.Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol. 2012;13:89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- 25.Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol. 2005;59:407–50. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- 26.Hinnebusch AG, Jackson BM, Mueller PP. Evidence for regulation of reinitiation in translational control of GCN4 mRNA. Proc Natl Acad Sci U S A. 1988;85:7279–83. doi: 10.1073/pnas.85.19.7279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mueller PP, Hinnebusch AG. Multiple upstream AUG codons mediate translational control of GCN4. Cell. 1986;45:201–7. doi: 10.1016/0092-8674(86)90384-3. [DOI] [PubMed] [Google Scholar]
- 28.Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6:1099–108. doi: 10.1016/S1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 29.Lu PD, Harding HP, Ron D. Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J Cell Biol. 2004;167:27–33. doi: 10.1083/jcb.200408003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vattem KM, Wek RC. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc Natl Acad Sci U S A. 2004;101:11269–74. doi: 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hann SR, Sloan-Brown K, Spotts GD. Translational activation of the non-AUG-initiated c-myc 1 protein at high cell densities due to methionine deprivation. Genes Dev. 1992;6:1229–40. doi: 10.1101/gad.6.7.1229. [DOI] [PubMed] [Google Scholar]
- 32.Grant CM, Hinnebusch AG. Effect of sequence context at stop codons on efficiency of reinitiation in GCN4 translational control. Mol Cell Biol. 1994;14:606–18. doi: 10.1128/mcb.14.1.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grant CM, Miller PF, Hinnebusch AG. Requirements for intercistronic distance and level of eukaryotic initiation factor 2 activity in reinitiation on GCN4 mRNA vary with the downstream cistron. Mol Cell Biol. 1994;14:2616–28. doi: 10.1128/MCB.14.4.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kozak M. Downstream secondary structure facilitates recognition of initiator codons by eukaryotic ribosomes. Proc Natl Acad Sci U S A. 1990;87:8301–5. doi: 10.1073/pnas.87.21.8301. [DOI] [PMC free article] [PubMed] [Google Scholar]