Abstract
Nucleos(t)ide analog drugs profoundly suppress Hepatitis B Virus (HBV) replication but rarely cure the infection, so therapy is usually life-long. The nucleos(t)ide analogs inhibit the viral DNA polymerase and often push HBV to the brink of extinction, so it may be possible to eradicate HBV by suppressing HBV replication further. The HBV ribonuclease H (RNaseH) is a logical new drug target because it is the second of only two viral enzymes essential for viral replication. We recently developed a low throughput screening pipeline for inhibitors of the HBV RNaseH and viral replication. Here, we screened a series of twenty-three nitrogen-based polyoxygenated heterocycles including sixteen 2-hydroxyisoquinoline-1,3(2H,4H)-dione derivatives for anti-HBV RNaseH activity. Nine compounds inhibited the HBV RNaseH, but activity was marginal for eight of them. Compound #1 [2-hydroxyisoquinoline-1,3(2H,4H)-dione, HID] was the best hit with an IC50 of 28.1 µM and an EC50 of 4.2 µM. It preferentially suppressed accumulation of the viral plus-polarity DNA strand in replication inhibition assays, indicating that replication was blocked due to suppression of HBV RNaseH activity. It had a CC50 of 75 µM, yielding a therapeutic index of ~18. The EC50 value was 7-fold lower than the IC50, possibly due to cellular retention or metabolism of the compound, or higher affinity for the full-length enzyme than the recombinant form used for screening. These data indicate that the 2-hydroxyisoquinoline-1,3(2H,4H)-diones will have different structure-activity relationships for the HBV and HIV RNaseHs. Therefore, HID compounds may provide a foundation for development of more effective RNaseH inhibitors of HBV replication.
Keywords: Hepatitis B virus; ribonuclease H; 2-hydroxyisoquinoline-1,3(2H,4H)-diones; viral replication
Introduction
Hepatitis B virus (HBV) infection causes chronic hepatitis B (CHB), an inflammatory liver disease that affects up to 350 million people world-wide. CHB causes >600,000 deaths each year due to cirrhosis, liver failure, and hepatocellular carcinoma (Ganem and Prince, 2004; Gerlich, 2013; Sorrell et al., 2009).
CHB is primarily treated with nucleos(t)ide analogs (Kwon and Lok, 2011; Sorrell et al., 2009). These drugs can reduce viral titers by 4–5 log10, often to below the limit of clinical detection (Marcellin et al., 2008; van Bommel et al., 2010; Woo et al., 2010). However, only a few patients clear the virus as measured by disappearance of the HBV surface antigen (HBsAg) from serum and the rise of anti-HBsAg antibodies (Block et al., 2013; Woo et al., 2010; Wursthorn et al., 2010). Although the rate of seroconversion in patients on nucleos(t)ide analog therapy is higher than in untreated patients (Gish et al., 2010), the overwhelming majority remain chronically infected and dependent upon drug regimens costing up to $600 per month (Buti et al., 2009; Toy, 2013). Achieving higher rates of HBV clearance will thus necessitate development of new therapies that will probably be used in combination with the nucleos(t)ides to augment antiviral efficacy and drive the virus to extinction (Tavis et al., 2013b).
HBV replicates by reverse transcription catalyzed by the viral polymerase protein, which has two enzymatic activities in different domains. The DNA polymerase synthesizes the viral DNA and is the target of the nucleos(t)ide analogs (Ghany and Liang, 2007). The ribonuclease H (RNaseH) degrades the viral RNA after it has been copied into the minus-polarity DNA strand to permit synthesis of the viral plus-polarity DNA strand. The RNaseH is not targeted by existing drugs primarily due to difficulties in establishing screening assays. We recently developed a low-throughput screening pipeline for HBV RNaseH inhibitors (Hu et al., 2013; Tavis et al., 2013a), opening the door to anti-RNaseH drug discovery.
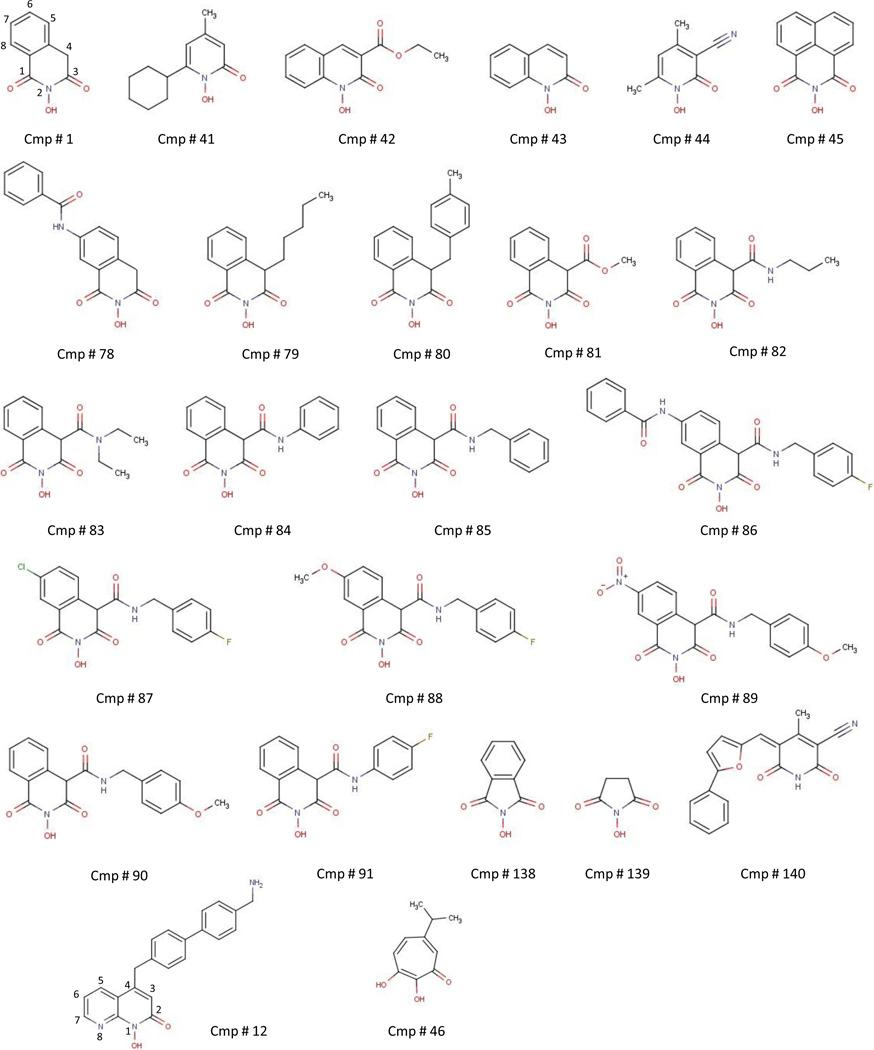
Many inhibitors of the HIV RNaseH have been identified (Ilina et al., 2012; Klumpp and Mirzadegan, 2006; Tramontano and Di Santo, 2010; Yu et al., 2008). We previously demonstrated that >20% of compounds selected for the ability to suppress the HIV RNaseH suppress the HBV enzyme, and found that a napthyridinone derivative (compound #12; structures are in Fig. 1) and β-thujaplicinol (compound #46) can suppress viral replication in cells by inhibiting the viral RNaseH (Hu et al., 2013; Tavis et al., 2013a). Here, we analyzed 2-hydroxyisoquinoline-1,3(2H,4H)-dione (HID) derivatives (Billamboz et al., 2008; Billamboz et al., 2011a; Billamboz et al., 2011b; Billamboz et al., 2013; Desimmie et al., 2013; Hang et al., 2004; Klumpp et al., 2003; Suchaud et al., 2014) and structurally related compounds for their ability to inhibit HBV RNaseH activity and viral replication based on the ability of this compound class to inhibit the HIV enzyme, with the goal of determining whether HID compounds may be suitable for development as anti-HBV drugs.
Fig. 1. Structures of the compounds used or discussed in this study.
2. Materials and methods
2.1. Compound acquisition and synthesis
Compounds (Table 1 and Fig. 1) were acquired commercially or were synthesized according to previously reported methods [#78 (Billamboz et al., 2008); #79–80 (Billamboz et al., 2011a); #81 (Billamboz et al., 2011b); #82–85 and #90–91 (Billamboz et al., 2013); #86–89 (Suchaud et al., 2014)]. The synthetic methods and compound characterizations are described in the supporting information. All compounds were dissolved in dimethyl sulfoxide (DMSO) and stored at −80°C.
Table 1.
Biochemical screening results
| Compound Number |
Namea | Genotype Db | Genotype Cb | Human RNAseH1b |
Genotype D IC50 (µM) |
Genotype C IC50 (µM) |
|---|---|---|---|---|---|---|
| 1 | TRC 939800 | +++ | +++ | ++ | 28.1 ± 9.7 | 30.4 ± 12.9 |
| 41 | Ciclopirox | − | − | + | ||
| 42 | Labotest 72543251 | − | − | + | ||
| 43 | Sigma PH008969 | − | + | − | >100 | |
| 44 | Labotest 12243782 | − | + | − | >100 | |
| 45 | TCI America H1040 | − | + | − | >100 | |
| 78 | MB1 | − | − | + | ||
| 79 | MB2 | − | − | − | ||
| 80 | MB3 | − | − | − | ||
| 81 | MB4 | − | − | +++ | ||
| 82 | MB71 | − | + | +++ | ||
| 83 | MB88 | + | − | +++ | ||
| 84 | MB105 | + | − | +++ | ||
| 85 | MB103 | + | − | +++ | ||
| 86 | VS42 | + | − | +++ | ||
| 87 | VS55 | − | − | +++ | >100 | |
| 88 | VS45 | − | − | +++ | ||
| 89 | VS51 | − | − | ++ | ||
| 90 | MB104 | − | − | ++ | ||
| 91 | MB106 | − | − | ++ | ||
| 138 | Sigma H53704 | − | − | − | ||
| 139 | Sigma 130672 | − | − | − | ||
| 140 | Chembridge 6325462 | − | − | ++ |
Common name or manufacturer's product number.
+, Detectable inhibition at 60 µM; ++, Inhibition at 20 µM; +++, Inhibition at 10 µM; −, No inhibition.
2.2. RNaseH expression and purification
HBV RNaseH and human RNaseH1 were expressed in E. coli, partially purified by nickel-affinity chromatography, and stored in liquid nitrogen as described (Tavis et al., 2013a).
2.3. Oligonucleotide-directed RNA cleavage assay
Olignonucleotide-directed RNA cleavage assays were conducted as previously described (Hu et al., 2013; Tavis et al., 2013a). Briefly, a 32P-labeled RNA was combined with a complementary DNA oligonucleotide or a non-complementary oligonucleotide. This substrate was incubated with RNaseH and test compounds in 50 mM Tris pH 8.0, 190 mM NaCl, 5 mM MgCl2, 3.5 mM DTT, 0.05% NP40, 6% glycerol, and 1% DMSO at 42°C for 90 minutes. The products were resolved by electrophoresis, detected by audioradiography, and quantified using ImageJ. IC50 values were calculated with GraphPad Prism.
2.4. HBV replication assay
HepDES19 cells contain a genotype D HBV genome under control of a tetracycline-repressible promoter (Guo et al., 2007). Cells (0.6 × 106) were seeded into 6-well plates and incubated in DMEM/F12, 10% fetal bovine serum (FBS), 1% penicillin/streptomycin (P/S) with 1 µl/ml tetracycline. Tetracycline was withdrawn after 24 hours. Compound #1 was applied to duplicate wells 48 hours later in culture medium containing a final DMSO concentration of 1%. Medium containing the compound was refreshed daily for the following two days. Cells were harvested and non-encapsidated nucleic acids were digested with micrococcal nuclease (New England Biolabs) as described (Hu et al., 2013). HBV DNA was purified from capsids using QIAamp Cador Pathogen Mini Kit according to the manufacturer’s instructions (Qiagen) with proteinase K incubation overnight at 37°C. TaqMan PCR was performed for 40 cycles at an annealing temperature of 60 °C. Primers and probe (IDT Inc.) for the plus-polarity strand were: 5’CATGAACAAGAGATGATTAGGCAGAG3’; 5’GGAGGCTGTAGGCATAAATTGG3’; 5’/56-FAM/CTGCGCACC/ZEN/AGCACCATGCA/3IABkFQ. Primers and probe for the minus-polarity strand were: 5’GCAGATGAGAAGGCACAGA3’; 5’CTTCTCCGTCTGCCGTT3’; 5’/56-FAM/AGTCCGCGT/ZEN/AAAGAGAGGTGCG/3IABkFQ.
2.5. Cytotoxicity assays
Ninety-six well plates were seeded with 1.0 × 104 HepDES19 cells per well and incubated in DMEM with 10% FBS plus 1% P/S, 1% non-essential amino acids, and 1% glutamine. Compound #1 was diluted in medium and applied to cells 24 hours after plating, with each concentration tested in triplicate. Medium containing the compound was refreshed daily for the next two days. Thiazolyl blue tetrazolium bromide (MTT, Sigma) was added to 0.25 mg/ml, the cultures were incubated for 60 minutes, metabolites were solubilized in acidic isopropanol, and absorbance was read at 570 nM. Cytotoxicity was also measured under the same culture conditions using the CytoTox-Glo assay (Promega) as recommended by the manufacturer.
3. Results
3.1. Biochemical screening against the HBV RNaseH
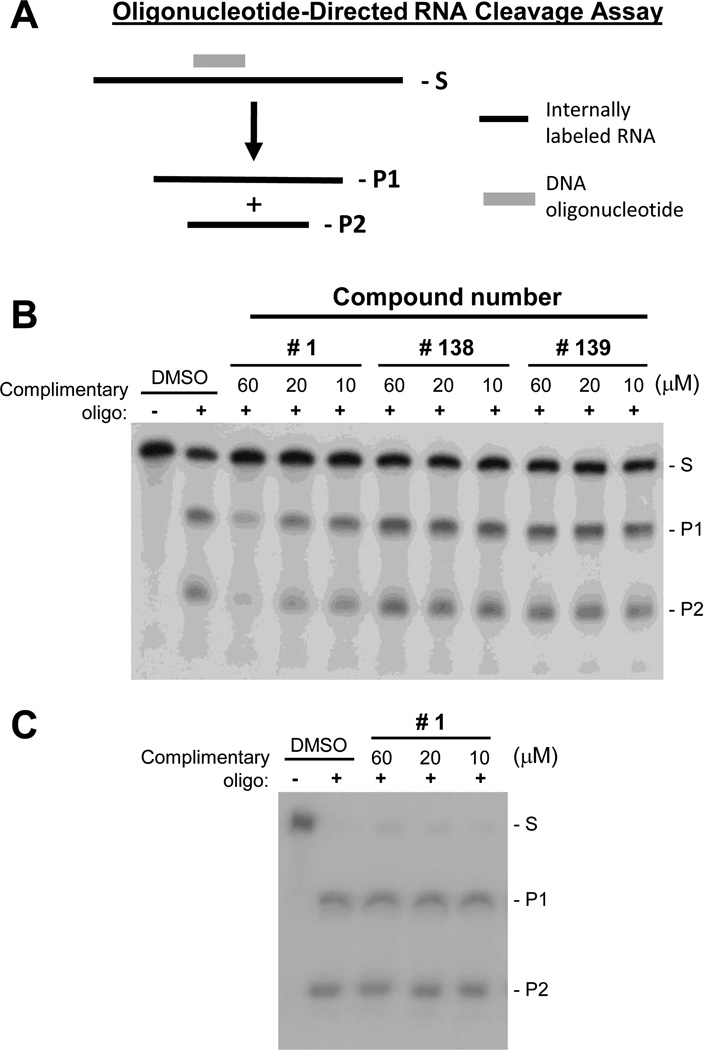
Twenty-three HID or related compounds (Table 1 and Fig. 1) were screened for anti-HBV RNaseH activity. First, a qualitative primary screening assay was performed against recombinant genotype D HBV RNaseH, then a qualitative assay was conducted against a genotype C isolate, and finally IC50 values were determined for select compounds against the genotype D and/or C enzymes. Screening employed an oligonucleotide-directed RNA cleavage assay (Fig. 2A) in which a 264 nucleotide long internally 32P-labeled RNA is annealed to a 20 nucleotide complementary DNA oligonucleotide or a non-complementary oligonucleotide as a specificity control. This heteroduplex substrate is incubated with the RNaseH along with the test compounds, and the cleavage products are resolved by electrophoresis (Hu et al., 2013; Tavis et al., 2013a).
Fig. 2. Representative primary biochemical screening assays for the HBV RNaseH.
A. The oligonucleotide-directed RNaseH assay. Internally radiolabeled RNA is bound to a complementary DNA oligonucleotide, and the RNaseH cleaves the RNA within the DNA:RNA heteroduplexes. RNA, black line; DNA, grey line; S, RNA substrate; P1 and P2, RNaseH cleavage products. B. Representative primary screening assay employing HBV genotype D RNaseH. DMSO, vehicle control. C. E. coli RNaseH activity was not affected by compound #1 in the oligonucleotide-directed cleavage assay under identical conditions.
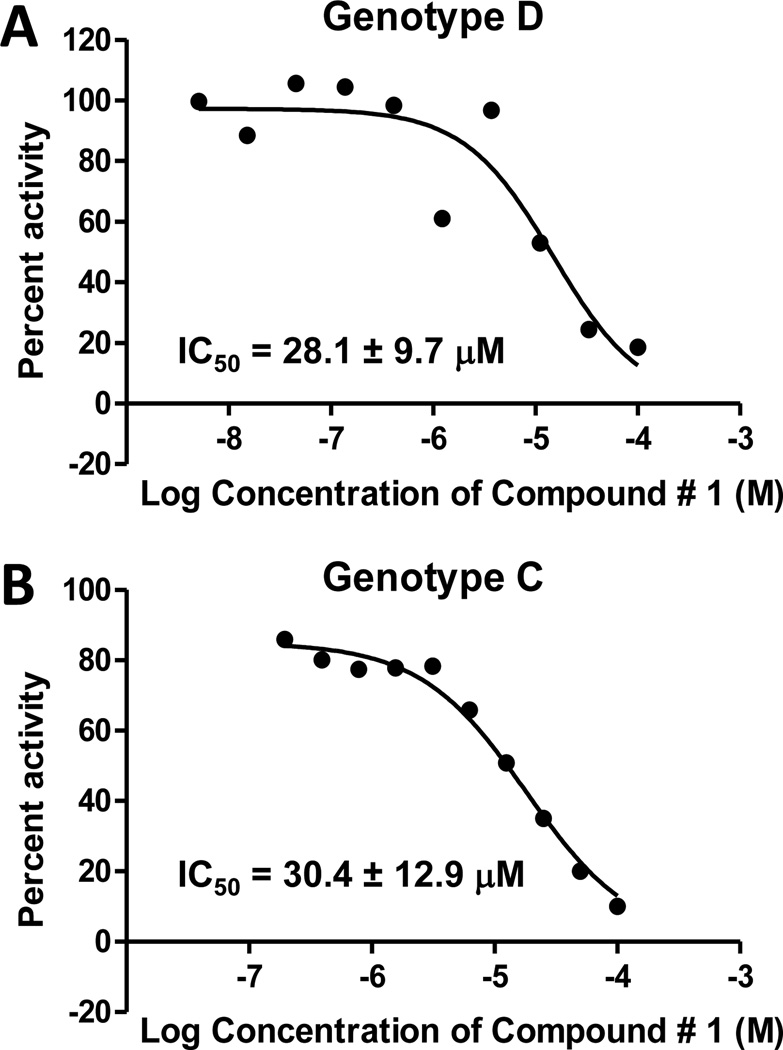
The compounds were employed at 60, 20, and 10 µM in the qualitative screens. Nine compounds had activity at ≤60 µM against either the genotype D or C enzymes, but activity was very weak for eight of them. The only compound with substantial activity was #1 [2-hydroxyisoquinoline-1,3(2H,4H)-dione, HID], which inhibited both of these versions of the HBV enzyme at 10 µM (Table 1 and Fig. 2B). The potential for these results to be due to contamination of our enzyme extracts with E. coli RNaseH was excluded because compound #1 was inactive against the bacterial RNaseH (Fig. 2C). IC50 values were determined for compound #1 against both genotype D and C enzymes, and also against compounds #43, 44, 45, and 87 for comparison. Compound #1 had IC50 values of 28.1 ± 9.7 and 30.4 ± 12.9 µM against the genotype D and C enzymes, respectively, whereas IC50 values were >100 µM for the other compounds (Fig. 3 and Table 1).
Fig. 3. Dose-response curves with compound #1 in the oligonucleotide-directed RNA cleavage assay against the HBV RNaseH.
A. Genotype D RNaseH. B. Genotype C RNaseH. The curves are from representative assays. The numerical values are the average ± one standard deviation from three or four independent assays.
3.2. Counter-screening against human RNaseH1
The compounds were counter-screened against recombinant human RNaseH1 in an initial effort to identify inhibitors with the least probability of being toxic. Thirteen compounds were active against the human enzyme at 10 or 20 µM, and two had activity at 60 µM (Table 1). The inhibition patterns differed between the HBV and human RNaseHs, with greater activity against the human enzyme usually being observed. Therefore, many HID compounds inhibited human RNaseH1, but the inhibition patterns against the HBV and human and RNaseHs were distinct.
3.3. HBV replication inhibition
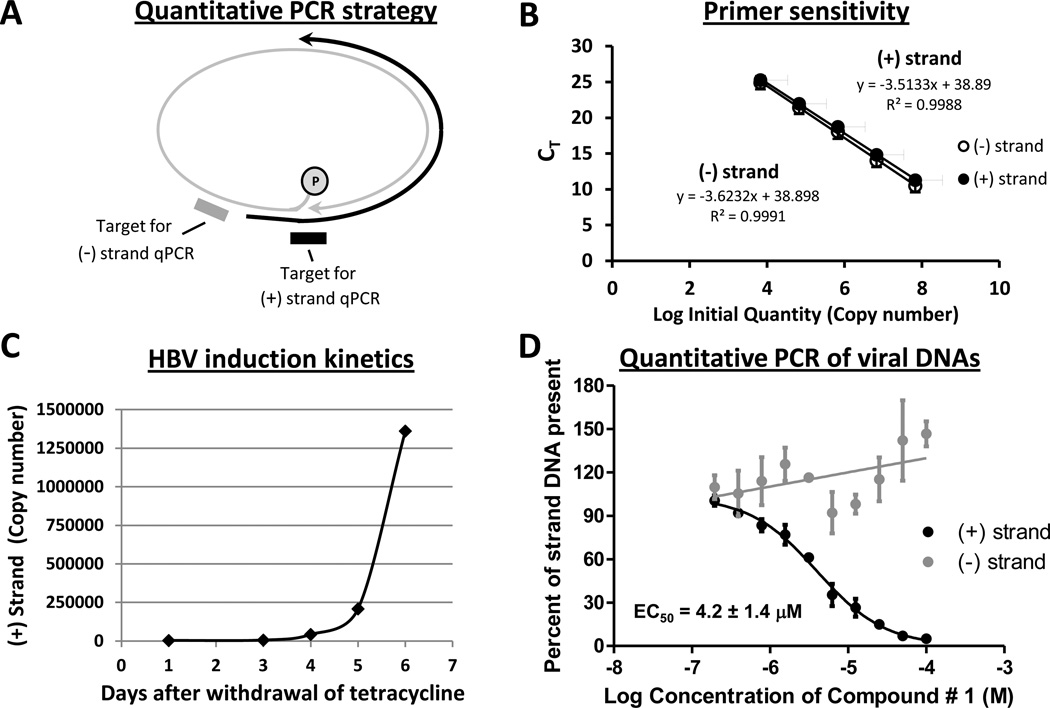
Compound #1 was tested against HBV replication because it inhibited genotypes D and C RNaseHs well but had only moderate activity against human RNaseH1. Inhibiting the HBV RNaseH during viral replication suppresses production of the viral positive-polarity DNA strand and causes truncation of many minus-polarity DNA strands (Chen et al., 1994; Hu et al., 2013; Tavis et al., 2013a). Therefore, we employed strand-preferential quantitative PCR to measure accumulation of each viral DNA strand in the presence of compound #1 (Fig. 4A). In this assay, plus-polarity DNA preferential PCR depends upon amplification of the viral DNA across the gap in the minus-polarity DNA strand. Minus-polarity DNA preferential PCR depends on placing the amplification primers just upstream of the start site for the plus-polarity strand; this detects few plus-polarity strands because most plus-strand DNAs are shorter than full length (Tavis and Badtke, 2009). Fig. 4B demonstrates that the plus- and minus-polarity preferential PCR primers detected double-stranded HBV DNA with equal efficiency.
Fig. 4. Effect of compound #1 on HBV replication.
A. Basis for the strand-preferential PCR reactions. The minus-polarity preferential primers/probe are upstream of the start site for the plus-polarity DNA, and the plus-polarity preferential primers/probe cross the gap in the minus-polarity DNA. Grey, minus-polarity DNA strand; black, plus-polarity strand; oval, the covalently-attached viral polymerase protein; arrows, 3’ ends of the DNAs. B. Relative sensitivity of the plus- and minus-polarity preferential primers against a double-stranded HBV DNA template. A linear HBV DNA was serially diluted and used as a template for quantitative PCR employing the strand-preferential primers. Filled circles, plus-polarity preferential primers; open circles, minus-polarity preferential primers. Error bars are ± one standard deviation from three independent assays. C. HBV induction kinetics in HepDES19 cells. Tetracycline was withdrawn from the medium, cytoplasmic HBV capsid particles were purified 1 to 6 days later, and plus-polarity HBV DNA derived from the capsid particles was measured by quantitative PCR. D. Inhibition of HBV plus-polarity synthesis by compound #1. Viral nucleic acids were purified from cytoplasmic capsid particles from cells replicating HBV in the presence of varying concentrations of compound #1. Quantitative PCR preferential for plus- and minus-polarity strands of HBV DNA was performed to measure the effect of compound # 1 on accumulation of plus- (black) and minus- (grey) strand HBV DNA. The results were normalized to the DMSO vehicle control. Data are the mean ± one standard error of the mean from three independent experiments each performed in duplicate.
HBV replication was measured in HepDES19 cells (Guo et al., 2007), which are HepG2 cells stably transfected with a HBV genotype D genome under control of a tetracycline-repressible promoter. HBV induction kinetics were established by withdrawing tetracycline from the medium and harvesting intracellular capsid particles over the next 6 days. The cytoplasmic extracts containing the capsid particles were treated with micrococcal nuclease to destroy contaminating chromosomal DNA, the viral DNAs were released from the capsid particles by proteinase K digestion, and then the amount of plus-polarity HBV DNA recovered within the capsids was measured by qualitative PCR (Fig. 4C). Control experiments in which the PCR reactions were done without releasing DNA from the capsid particles failed to detect HBV DNA, confirming that the DNAs detected were derived from the HBV capsids (data not shown). Newly synthesized HBV DNA became detectable 3 days after withdrawal of tetracycline from the medium, and sufficient viral DNA to support reliable quantification of HBV replication accumulated by 5 days after withdrawal of tetracycline.
To measure the effect of compound #1 on HBV replication, HepDES19 cells were incubated for two days without tetracycline to initiate HBV replication. The cultures were then treated with compound #1 for three additional days, with provision of fresh medium plus compound every 24 hours. HBV DNAs were purified from viral capsids and viral DNA levels were measured by quantitative PCR. Compound #1 suppressed HBV replication in qualitative replication inhibition assays at 60, 20, and 10 µM (Table 2). Quantitative assays revealed an EC50 of 4.2 ± 1.4 µM against plus-polarity HBV DNA (Fig. 4D). The minus-polarity DNA strand in the same samples was not suppressed by treatment with compound #1; rather, its accumulation was somewhat enhanced. Therefore, synthesis of mature HBV genomes was suppressed by compound #1 via inhibition of the viral RNaseH activity.
Table 2.
Inhibition of HBV replication by compound #1
| Replication inhibitiona | Cytotxicityb | ||
|---|---|---|---|
| Qualitative | +++ | MTT | 74.7 ± 24 |
| Quantitative | 4.2 ± 1.4 | Cell rupture | 87 |
+++, inhibition at ≤10 µM or CC50 (µM)
CC50 (µM)
3.4. Toxicity
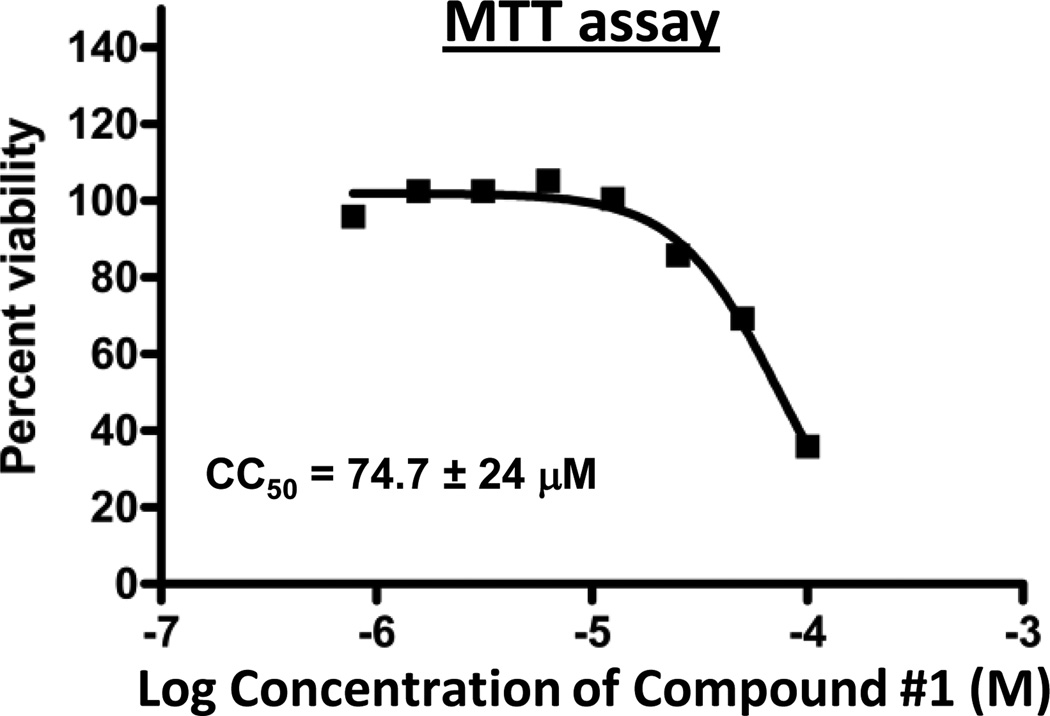
Cytotoxicity of compound #1 was assessed in HepDES19 cells using an MTT assay under culture conditions analogous to those used for the replication inhibition assays (Fig. 5) or by measuring cell rupture (Table 2). CC50 values were 74.7 ± 24 and 87 µM by the MTT and cell-rupture assays, respectively. These values were much higher than the EC50 for compound #1, leading to a therapeutic index (TI) of 17.8.
Fig. 5. MTT toxicity assay for compound #1.
An MTT assay was conducted in the presence of varying concentrations of compound #1 under culture conditions analogous to the replication assay in Fig. 4D. The curve is from a representative assay. The CC50 value is the average ± one standard deviation from three independent assays.
4. Discussion
Screening compound classes known to suppress HIV RNaseH activity previously led us to identify a napthyridinone derivative [compound #12 (Tavis et al., 2013a)] and β-thujaplicinol [compound #46 (Hu et al., 2013)] as inhibitors of HBV RNaseH activity and HBV genomic replication. Here, 15 HID compounds and 8 polyoxygenated nitrogen-containing heterocycles were screened against the HBV RNaseH. Eight very weak and one robust inhibitor were identified (Table 1).
The compounds were screened against HBV RNaseH enzymes from both genotypes C and D to maximize chances of finding hits with efficacy against genetically divergent HBV isolates. There was discord in efficacy of the compounds between the two genotypes for the weak inhibitors, but this was not surprising given that their activity was at the detection limit of our assays. However, compound #1 was equally effective against both genotype C and D enzymes (Table 1 and Fig. 3). Therefore, it is possible to identify HID compounds that can inhibit RNaseH enzymes from multiple HBV genotypes.
The compounds were counter-screened against human RNaseH1 as a first step toward eliminating compounds with unacceptable toxicity. Fifteen compounds inhibited the human RNaseH1, and many had robust activity at 10 µM (Table 1). Compound #1 was modestly active against the human RNaseH1, with detectable inhibition as low as 20 µM. Therefore, development of HIDs for use in humans will require care to exploit structure-activity relationship (SAR) differences between the human and HBV RNaseH enzymes.
A previous study of the HID scaffold with substitutions at the 7-carbon position revealed widely varying degrees of inhibition of the HIV RNaseH (Billamboz et al., 2008). Inhibition of the HIV RNaseH did not predict inhibition of the HBV enzyme. For example, compound #1 has an IC50 of 5.9 µM against the HIV RNaseH (Billamboz et al., 2008; Billamboz et al., 2011b), which is ~5-fold lower than its efficacy against the HBV enzyme, while compound #81 strongly inhibits the HIV RNaseH (IC50 = 61 nM) (Billamboz et al., 2011b) but had negligible activity against the HBV enzyme (Table 1).
HID compounds inhibit the HIV RNaseH by chelating the active site divalent cations via interactions with the three oxygen atoms of the N-hydroxyimide function of the isoquinoline ring (Billamboz et al., 2011b; Klumpp et al., 2003). Our results are consistent with this mechanism also applying to the HBV RNaseH. Furthermore, the data establish four constraints on the SAR for HID compounds against the HBV enzyme. First, all three oxygens in the isoquinoline heterocycle appear to be needed for robust inhibition of the HBV enzyme. Compounds #41–44 lacking one keto function were poorly inactive against the HBV RNaseH. Second, the inactivity of compounds #138 and 139 against the HBV enzyme indicates that the nitrogen-containing six-member ring cannot be contracted to five atoms despite the apparent conservation of the metal-binding N-hydroxyimide function. This is similar to previous observations with influenza endonuclease inhibitors (Parkes et al., 2003). Third, introducing extended side chains at position 4 of the scaffold is detrimental (compounds #79–91), although compounds #83–86 showed weak inhibition of HBV genotype D at 60 µM. This is in contrast to the HIV RNaseH, in which some compounds displayed significant inhibition. For example, compound #79 had an IC50 of 13.2 µM (Billamboz et al., 2011a), and compounds #86 and 89 induced 41.2 % and 67.2% inhibition at 10 µM, respectively (Suchaud et al., 2014). The discrepancy between HIV and HBV inhibition was even greater with the addition of an electron-withdrawing group such as a methoxycarbonyl moiety at position 4, which greatly bolstered anti-RNaseH activity of compound #81 against the HIV enzyme (Billamboz et al., 2011b). Finally, comparing compounds #1 and #45 reveals that joining positions 4 and 5 of the scaffold with a phenyl ring is not permissible. This supports a metal chelating mechanism for HBV RNase H inhibition similar to the HIV RNaseH and integrase mechanism. Indeed, we previously showed that enolization of the keto function at position 3 is required for optimal magnesium chelation (Billamboz et al., 2011b), and this process is not possible for compound #45.
Only compound #1 met the criteria of our screening pipeline for assessment against HBV replication. It had an EC50 of 4.2 ± 1.4 µM against replication of a genotype D HBV genome, compared to ~5 µM for β-thujaplicinol [compound #46, (Hu et al., 2013)] and <10 µM for the napthyridinone derivative [compound #12, (Tavis et al., 2013a)]. Suppressing HBV RNaseH activity blocks accumulation of the viral plus-polarity DNA strand (Chen et al., 1994; Hu et al., 2013; Tavis et al., 2013a). As predicted from the biochemical analyses, inhibition of HBV replication by compound #1 occurred by blocking the viral RNaseH activity because accumulation of the plus-polarity HBV DNA strand was strongly inhibited, while the minus-polarity DNA strand was not suppressed (Fig 4D). Moderate cytotoxicity was observed, with CC50 values of about 75 and 87 µM by the MTT and cell rupture assays, respectively (Table 2 and Fig. 5). This led to a TI of 17.8.
Compound #1 inhibited HBV replication at a ~7-fold lower concentration than it suppressed the HBV RNaseH in the biochemical assays (EC50 = 4.2 µM vs. IC50 = 28.1 µM for genotype D). This was unexpected given that the EC50 values for the other compounds that we have tested were similar or higher than their IC50 values [(Hu et al., 2013; Tavis et al., 2013a) and unpublished]. This could be due to cellular retention or metabolism of the compound, or to higher affinity for the full-length enzyme than the truncated recombinant form used for screening.
One HID compound, 2-hydroxy-4-methoxycarbonylisoquinoline-1,3(2H,4H)-dione [compound #81 in this paper = compound # 3 in (Billamboz et al., 2011b)], was previously found to inhibit HIV replication in MT-4 cells with an EC50 of 13.4 µM, presumably by blocking the HIV RNaseH. However, several other HIDs did not clearly suppress HIV replication, in part due to toxicity. Thus, the EC50 values for most of these compounds against HIV, including compound #1, exceeded their CC50 values (Billamboz et al., 2008; Billamboz et al., 2011a; Billamboz et al., 2011b). This has precluded their development as anti-HIV agents.
Compound #1 shares some similarities to a napthyridinone compound that we previously reported to inhibit HBV replication (compound #12, Fig. 1) (Tavis et al., 2013a). They both display a bicyclic framework and a triad of metal chelating heteroatoms, but there are also major differences. First, the metal-chelating moiety is embedded into the bicyclic framework in the napthyridinone derivative since the N-8 nitrogen atom participates in metal chelation with the hydroxyl and keto functions at positions 1 and 2 (Fig. 1). In contrast, the three-oxygen metal-chelating moiety is located on a single ring in compound #1 (Fig. 1). Second, the orientation of the metal-chelating moieties relative to the aromatic ring is different. Third, there is a long side aminomethyldiphenylmethyl chain extending from the position 4 on the 1-hydroxy-1,8-naphthyridin-2(1H)-one ring. Thus, comparing compounds #1 and 12 may help guide development of an effective HBV RNaseH inhibitor. A crystal structure of the HBV RNaseH catalytic core would be very helpful for the design of catalytic inhibitors, but a structure is not available.
This series of HIDs showed that substitution of the scaffold at position 4 by alkyl groups and by ester and amide functions did not favor HBV RNaseH inhibition. The only potent compound was the unsubstituted compound #1, so other modifications of this hit, particularly on the aromatic ring, will be necessary to gain further insight into the SARs of the HID scaffold. Positioning a long side chain on the aromatic ring analogous to the naphthyridinone structure is an interesting possibility for pharmacomodulation.
As the design of clinically useful anti-RNaseH compounds becomes possible, so does the possibility of creating novel combination drug therapies for CHB that could suppress viral replication further than is currently possible in most patients. Combination therapy would be predicted to reduce the incidence of drug resistance seen with the inexpensive, older nucleos(t)ide such as lamivudine (Shaw et al., 2006; Zoulim and Locarnini, 2009). Therefore, development of RNaseH inhibitors could extend the time that the NAs remain effective. This could reduce the cost of treatment, which would be particularly important in the developing world where the bulk of CHB patients reside. However, the optimal outcome from development of anti-RNaseH drugs would be to devise a combination therapy that suppresses viral replication far enough to eradicate CHB. This would alleviate the immense health burdens imposed by HBV on the millions of afflicted people in the world.
Highlights.
An HID compound inhibits HBV RNaseH activity with a low micromolar IC50.
The compound inhibits HBV replication in culture by blocking the viral RNaseH.
HBV replication is inhibited with an EC50 of 4.2 µM and a TI of about 18.
HID compounds may provide a promising scaffold for anti-HBV drug development.
Acknowledgments
This work was supported by Saint Louis University seed funds from the Molecular Microbiology Department, the School of Medicine, and the Cancer Center. It was also supported in part by a grant from the Friends of the Saint Louis University Liver Center and the NIH/National Center for Advancing Translational Sciences (NCATS) grant UL1 TR000448. The chemical part of this work was financially supported by grants from le Ministère de l’Enseignement Supérieur et de la Recherche Française and l’Agence Nationale de la Recherche contre le Sida (ANRS). The funding sources had no role in the study design, data interpretation, or publication of the results.
We thank Drs. Duane Grandgenett and Mark Buller for helpful advice. Potential applications of these compounds against HBV are covered by U.S. Patent Application 13/072201 (Pending).
Abbreviations
- HBV
Hepatitis B virus
- CHB
chronic hepatitis B
- NA
nucleos(t)ide analogs
- RNaseH
ribonuclease H
- HIV
human immunodeficiency virus
- HID
2-hydroxyisoquinoline-1,3(2H,4H)-dione compounds
- DMSO
dimethyl sulfoxide
- IC50
50% inhibitory concentration
- MTT
thiazolyl blue tetrazolium bromide
- EC50
50% effective concentration
- CC50
50% cytotoxic concentration
- TI
therapeutic index
- SAR
structure-activity relationship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Catherine W. Cai, Email: ccai@slu.edu.
Elena Lomonosova, Email: lomonoe@slu.edu.
Eileen A. Moran, Email: eileenmoran@slu.edu.
Xiaohong Cheng, Email: xcheng1@slu.edu.
Kunjan B. Patel, Email: kpatel65@slu.edu.
Fabrice Bailly, Email: Fabrice.Bailly@univ-lille1.fr.
Philipe Cotelle, Email: Philippe.Cotelle@univ-lille1.fr.
Marvin J. Meyers, Email: mmeyers8@slu.edu.
References
- Billamboz M, Bailly F, Barreca ML, De LL, Mouscadet JF, Calmels C, Andreola ML, Witvrouw M, Christ F, Debyser Z, Cotelle P. Design, synthesis, and biological evaluation of a series of 2-hydroxyisoquinoline-1,3(2H,4H)-diones as dual inhibitors of human immunodeficiency virus type 1 integrase and the reverse transcriptase RNase H domain. J.Med.Chem. 2008;51:7717–7730. doi: 10.1021/jm8007085. [DOI] [PubMed] [Google Scholar]
- Billamboz M, Bailly F, Lion C, Calmels C, Andreola ML, Witvrouw M, Christ F, Debyser Z, De Luca L, Chimirri A, Cotelle P. 2-hydroxyisoquinoline-1,3(2H,4H)-diones as inhibitors of HIV-1 integrase and reverse transcriptase RNase H domain: influence of the alkylation of position 4. European journal of medicinal chemistry. 2011a;46:535–546. doi: 10.1016/j.ejmech.2010.11.033. [DOI] [PubMed] [Google Scholar]
- Billamboz M, Bailly F, Lion C, Touati N, Vezin H, Calmels C, Andreola ML, Christ F, Debyser Z, Cotelle P. Magnesium chelating 2-hydroxyisoquinoline-1,3(2H,4H)-diones, as inhibitors of HIV-1 integrase and/or the HIV-1 reverse transcriptase ribonuclease H domain: discovery of a novel selective inhibitor of the ribonuclease H function. J.Med.Chem. 2011b;54:1812–1824. doi: 10.1021/jm1014692. [DOI] [PubMed] [Google Scholar]
- Billamboz M, Suchaud V, Bailly F, Lion C, Demeulemeester J, Calmels C, Andreola ML, Christ F, Debyser Z, Cotelle P. 4-Substituted 2-hydroxyisoquinoline-1,3(2H,4H)-diones, as a novel class of HIV-1 integrase inhibitors. ACS. Med. Chem. Lett. 2013;4:606–611. doi: 10.1021/ml400009t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block TM, Gish R, Guo H, Mehta A, Cuconati A, Thomas London W, Guo JT. Chronic hepatitis B: What should be the goal for new therapies? Antiviral Res. 2013;98:27–34. doi: 10.1016/j.antiviral.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buti M, Brosa M, Casado MA, Rueda M, Esteban R. Modeling the cost-effectiveness of different oral antiviral therapies in patients with chronic hepatitis B. J.Hepatol. 2009;51:640–646. doi: 10.1016/j.jhep.2009.04.013. [DOI] [PubMed] [Google Scholar]
- Chen Y, Robinson WS, Marion PL. Selected Mutations of the Duck Hepatitis B Virus P Gene RNase H Domain Affect both RNA Packaging and Priming of Minus-Strand DNA Synthesis. J.Virol. 1994;68:5232–5238. doi: 10.1128/jvi.68.8.5232-5238.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desimmie BA, Demeulemeester J, Suchaud V, Taltynov O, Billamboz M, Lion C, Bailly F, Strelkov SV, Debyser Z, Cotelle P, Christ F. 2-Hydroxyisoquinoline-1,3(2H,4H)-diones (HIDs), novel inhibitors of HIV integrase with a high barrier to resistance. ACS chemical biology. 2013;8:1187–1194. doi: 10.1021/cb4000426. [DOI] [PubMed] [Google Scholar]
- Ganem D, Prince AM. Hepatitis B virus infection--natural history and clinical consequences. N.Engl.J Med. 2004;350:1118–1129. doi: 10.1056/NEJMra031087. [DOI] [PubMed] [Google Scholar]
- Gerlich WH. Medical virology of hepatitis B: how it began and where we are now. Virology journal. 2013;10:239. doi: 10.1186/1743-422X-10-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghany M, Liang TJ. Drug targets and molecular mechanisms of drug resistance in chronic hepatitis B. Gastroenterology. 2007;132:1574–1585. doi: 10.1053/j.gastro.2007.02.039. [DOI] [PubMed] [Google Scholar]
- Gish RG, Chang TT, Lai CL, de Man R, Gadano A, Poordad F, Yang J, Brett-Smith H, Tamez R. Loss of HBsAg antigen during treatment with entecavir or lamivudine in nucleoside-naive HBeAg-positive patients with chronic hepatitis B. J Viral Hepat. 2010;17:16–22. doi: 10.1111/j.1365-2893.2009.01146.x. [DOI] [PubMed] [Google Scholar]
- Guo H, Jiang D, Zhou T, Cuconati A, Block TM, Guo JT. Characterization of the intracellular deproteinized relaxed circular DNA of hepatitis B virus: an intermediate of covalently closed circular DNA formation. J Virol. 2007;81:12472–12484. doi: 10.1128/JVI.01123-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hang JQ, Rajendran S, Yang Y, Li Y, In PW, Overton H, Parkes KE, Cammack N, Martin JA, Klumpp K. Activity of the isolated HIV RNase H domain and specific inhibition by N-hydroxyimides. Biochem Biophys Res Commun. 2004;317:321–329. doi: 10.1016/j.bbrc.2004.03.061. [DOI] [PubMed] [Google Scholar]
- Hu Y, Cheng X, Cao F, Huang A, Tavis JE. beta-Thujaplicinol inhibits hepatitis B virus replication by blocking the viral ribonuclease H activity. Antiviral Res. 2013;99:221–229. doi: 10.1016/j.antiviral.2013.06.007. [DOI] [PubMed] [Google Scholar]
- Ilina T, Labarge K, Sarafianos SG, Ishima R, Parniak MA. Inhibitors of HIV-1 Reverse Transcriptase-Associated Ribonuclease H Activity. Biology. 2012;1:521–541. doi: 10.3390/biology1030521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumpp K, Hang JQ, Rajendran S, Yang Y, Derosier A, Wong KI, Overton H, Parkes KE, Cammack N, Martin JA. Two-metal ion mechanism of RNA cleavage by HIV RNase H and mechanism-based design of selective HIV RNase H inhibitors. Nucleic Acids Res. 2003;31:6852–6859. doi: 10.1093/nar/gkg881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumpp K, Mirzadegan T. Recent progress in the design of small molecule inhibitors of HIV RNase H. Curr.Pharm.Des. 2006;12:1909–1922. doi: 10.2174/138161206776873653. [DOI] [PubMed] [Google Scholar]
- Kwon H, Lok AS. Hepatitis B therapy. Nat.Rev.Gastroenterol.Hepatol. 2011;8:275–284. doi: 10.1038/nrgastro.2011.33. [DOI] [PubMed] [Google Scholar]
- Marcellin P, Heathcote EJ, Buti M, Gane E, De Man RA, Krastev Z, Germanidis G, Lee SS, Flisiak R, Kaita K, Manns M, Kotzev I, Tchernev K, Buggisch P, Weilert F, Kurdas OO, Shiffman ML, Trinh H, Washington MK, Sorbel J, Anderson J, Snow-Lampart A, Mondou E, Quinn J, Rousseau F. Tenofovir disoproxil fumarate versus adefovir dipivoxil for chronic hepatitis B. N.Engl.J.Med. 2008;359:2442–2455. doi: 10.1056/NEJMoa0802878. [DOI] [PubMed] [Google Scholar]
- Parkes KE, Ermert P, Fassler J, Ives J, Martin JA, Merrett JH, Obrecht D, Williams G, Klumpp K. Use of a pharmacophore model to discover a new class of influenza endonuclease inhibitors. Journal of medicinal chemistry. 2003;46:1153–1164. doi: 10.1021/jm020334u. [DOI] [PubMed] [Google Scholar]
- Shaw T, Bartholomeusz A, Locarnini S. HBV drug resistance: mechanisms, detection and interpretation. J.Hepatol. 2006;44:593–606. doi: 10.1016/j.jhep.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Sorrell MF, Belongia EA, Costa J, Gareen IF, Grem JL, Inadomi JM, Kern ER, McHugh JA, Petersen GM, Rein MF, Strader DB, Trotter HT. National Institutes of Health Consensus Development Conference Statement: management of hepatitis B. Ann.Intern.Med. 2009;150:104–110. doi: 10.7326/0003-4819-150-2-200901200-00100. [DOI] [PubMed] [Google Scholar]
- Suchaud V, Bailly F, Lion C, Calmels C, Andreola ML, Christ F, Debyser Z, Cotelle P. Investigation of a novel series of 2-hydroxyisoquinoline-1,3-(2H,4H)-diones (HIDs) as human immunodeficiency virus type 1 integrase inhibitors. J.Med.Chem. 2014 doi: 10.1021/jm500109z. In Press. [DOI] [PubMed] [Google Scholar]
- Tavis JE, Badtke MP. Hepadnaviral Genomic Replication. In: Cameron CE, G”tte M, Raney KD, editors. Viral Genome Replication. New York: Springer Science+Business Media, LLC; 2009. pp. 129–143. [Google Scholar]
- Tavis JE, Cheng X, Hu Y, Totten M, Cao F, Michailidis E, Aurora R, Meyers MJ, Jacobsen EJ, Parniak MA, Sarafianos SG. The hepatitis B virus ribonuclease h is sensitive to inhibitors of the human immunodeficiency virus ribonuclease h and integrase enzymes. PLoS pathogens. 2013a;9:e1003125. doi: 10.1371/journal.ppat.1003125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavis JE, Gehring AJ, Hu Y. How further suppression of virus replication could improve current HBV treatment. Expert review of anti-infective therapy. 2013b;11:755–757. doi: 10.1586/14787210.2013.814846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toy M. Cost-effectiveness of viral hepatitis B & C treatment. Best practice & research. Clinical gastroenterology. 2013;27:973–985. doi: 10.1016/j.bpg.2013.08.020. [DOI] [PubMed] [Google Scholar]
- Tramontano E, Di Santo R. HIV-1 RT-associated RNase H function inhibitors: Recent advances in drug development. Curr Med Chem. 2010;17:2837–2853. doi: 10.2174/092986710792065045. [DOI] [PubMed] [Google Scholar]
- van Bommel F, De Man RA, Wedemeyer H, Deterding K, Petersen J, Buggisch P, Erhardt A, Huppe D, Stein K, Trojan J, Sarrazin C, Bocher WO, Spengler U, Wasmuth HE, Reinders JG, Moller B, Rhode P, Feucht HH, Wiedenmann B, Berg T. Long-term efficacy of tenofovir monotherapy for hepatitis B virus-monoinfected patients after failure of nucleoside/nucleotide analogues. Hepatology. 2010;51:73–80. doi: 10.1002/hep.23246. [DOI] [PubMed] [Google Scholar]
- Woo G, Tomlinson G, Nishikawa Y, Kowgier M, Sherman M, Wong DK, Pham B, Ungar WJ, Einarson TR, Heathcote EJ, Krahn M. Tenofovir and entecavir are the most effective antiviral agents for chronic hepatitis B: a systematic review and Bayesian metaanalyses. Gastroenterology. 2010;139:1218–1229. doi: 10.1053/j.gastro.2010.06.042. [DOI] [PubMed] [Google Scholar]
- Wursthorn K, Jung M, Riva A, Goodman ZD, Lopez P, Bao W, Manns MP, Wedemeyer H, Naoumov NV. Kinetics of hepatitis B surface antigen decline during 3 years of telbivudine treatment in hepatitis B e antigen-positive patients. Hepatology. 2010;52:1611–1620. doi: 10.1002/hep.23905. [DOI] [PubMed] [Google Scholar]
- Yu F, Liu X, Zhan P, De Clercq E. Recent advances in the research of HIV-1 RNase H inhibitors. Mini reviews in medicinal chemistry. 2008;8:1243–1251. doi: 10.2174/138955708786141052. [DOI] [PubMed] [Google Scholar]
- Zoulim F, Locarnini S. Hepatitis B virus resistance to nucleos(t)ide analogues. Gastroenterology. 2009;137:1593–1608. doi: 10.1053/j.gastro.2009.08.063. [DOI] [PubMed] [Google Scholar]