Abstract
Prostate-specific antigen (PSA) is recognized as an organ-specific marker with low specificity and sensitivity in discriminating prostate cancer (PCa) from other benign conditions, such as prostatic hyperplasia or chronic prostatitis. Thus, in the case of clinical suspicion, a PCa diagnosis cannot be made without a prostate biopsy. [-2]proPSA (p2PSA), a precursor of PSA, has been investigated as a new marker to accurately detect PCa. The aim of this systematic review was to discuss the available literature regarding the clinical validity and utility of p2PSA and its derivatives, p2PSA/fPSA (%p2PSA) and the Prostate Health Index (PHI). A systematic search of the PubMed and Scopus electronic databases was performed in accordance with the PRISMA statement (http://www.prisma-statement.org), considering the time period from January 1990 to January 2014 and using the following search terms: proprostate specific antigen, proenzyme PSA, proPSA, [-2]proPSA, p2PSA, Prostate Health Index, and PHI. To date, 115 studies have been published, but only 35 were considered for the qualitative analysis. These studies suggested that p2PSA is the most cancer-specific form of PSA, being preferentially expressed in PCa tissue and being significantly elevated in the serum of men with PCa. It is now evident that p2PSA, %p2PSA, and PHI measurements improve the specificity of the available tests (PSA and derivatives) in detecting PCa. Moreover, increasing PHI values seem to correlate with more aggressive disease. Some studies have compared p2PSA and its derivatives with other new biomarkers and found p2PSA to be significantly more accurate. Indeed, the implementation of these tests in clinical practice has the potential to significantly increase the physician's ability to detect PCa and avoid unnecessary biopsies, while also having an effective impact on costs. Further studies in large, multicenter, prospective trials are required to confirm these encouraging results on the clinical utility of these new biomarkers.
Keywords: [-2]proPSA, Diagnosis, Prostate cancer, Prostate health index
INTRODUCTION
Prostate-specific antigen (PSA) is widely known as a serum biomarker for the early detection of prostate cancer (PCa). Its introduction in clinical practice in the early 1990s changed PCa diagnosis and management. Currently in Western countries, PSA-based opportunistic or systematic screening has resulted in a stage migration to more organ-confined tumors at the time of diagnosis [1], with a consequently consistent reduction in PCa-related mortality observed over time [2]. However, PSA is recognized as an organ-specific marker but not a perfect PCa marker. Indeed, it has low specificity and sensitivity, especially in the total PSA (tPSA) range of 4 to 10 ng/mL (the so-called diagnostic gray zone) [3]. PSA levels may increase as a result of benign conditions such as benign prostatic hyperplasia (BPH) [4] and chronic prostatitis [5]. Moreover, PSA levels are also affected by biological variability, which may be related to differences in androgen levels, prostate manipulation, or ejaculation [6]. Finally, sample handling, laboratory processing, and assay standardization can all alter PSA measurements [7]. Owing to these factors, it is difficult to find an appropriate PSA cutoff for PCa diagnosis (which for many years was considered to be 4 ng/mL).
Thus, definitive diagnosis is still based on prostate biopsy. According to the European Association of Urology guidelines, the need for prostate biopsy should be determined by PSA level, suspicious digital rectal examination (DRE) result, patient's biological age, potential comorbidities, and therapeutic consequences. However, biopsies are positive in only approximately 30% of patients [8]. Consequently, prostate biopsy needs to be repeated if PSA rises or is persistently elevated, if the DRE result remains suspicious, or if there is a pathological diagnosis of atypical small acinar proliferation or extensive (multiple biopsy sites) prostatic intraepithelial neoplasia from a previous biopsy [9]. Finally, PCa is not rare among men with PSA levels of less than 4 ng/mL, with a risk ranging from 6.6% in men with PSA ≤0.5 ng/mL to 26.9% in men with PSA of 3.1 to 4.0 ng/mL [10]. It is also important to remember that clinically significant PCa (Gleason Score [GS] ≥7) may be diagnosed in 15% of patients with PSA levels of less than 4 ng/mL [11].
Considerable efforts have been made to find new markers to accurately detect but also discriminate between clinically significant and insignificant PCa. Accordingly, the introduction of several PSA derivatives (free PSA [fPSA], percentage of free PSA [%fPSA], PSA density, and PSA velocity) has improved the accuracy of tPSA in detecting PCa in clinical practice. Recently, fPSA was found to include several subforms, such as a precursor form of PSA (proPSA). PSA is an androgen-regulated chymotrypsin-like serine protease that is produced in high levels within the prostatic ductal and acinar epithelium. PSA has a 17-amino acid leader sequence (preproPSA) that is cleaved co-translationally to generate an inactive precursor protein (proPSA) with seven additional amino acids compared with mature PSA [12,13,14]. The partial removal of the leader sequence of the preproPSA leads to other truncated forms of proPSA. Thus, theoretically, seven isoforms of proPSA should exist, although only [-1], [-2], [-4], [-5], and [-7]proPSA have been found. There is still no evidence of [-3] or [-6]proPSA [15,16]. However, all forms of proPSA are enzymatically inactive [17]. It is possible to detect three truncated forms of proPSA in serum ([-2], [-4], and [-5/-7]proPSA), of which [-2]proPSA (p2PSA) is the most stable form [15,18].
Notably, p2PSA was found to be elevated in peripheral gland cancer tissue and to be specifically higher in serum from patients with PCa [18,19]. Thus, in the past decade, it has been under investigation as a potentially more accurate test for PCa detection in clinical practice. This systematic review focused on recently published studies investigating the clinical validity and utility of p2PSA and its derivatives, p2PSA/fPSA (%p2PSA) and the Prostate Health Index (PHI).
MATERIALS AND METHODS
1. Search strategy
A systematic search of the PubMed and Scopus electronic databases was performed by three investigators (A.A., G.L., M.L.) in accordance with the PRISMA statement (http://www.prisma-statement.org). Title, abstract, or keyword lists were searched, from January 1990 to January 2014, for combinations of the following free search terms: "proprostate specific antigen," "proenzyme PSA," "proPSA," "[-2]proPSA," "p2PSA," "prostate health index," and "PHI." The search was performed for each term alone or in combination with "prostate cancer" and "prostate biopsy."
2. Eligibility criteria
Titles and abstracts of each available study were reviewed, with a focus on the diagnostic and predictive characteristics of p2PSA, %p2PSA, and PHI compared with PSA and other available PCa biomarkers. Only scientific articles in English that reported original data were included. Priority was given to the most complete studies when the same population was reported and similar results were shown. Studies that failed to report a specific and detailed outcome or those not adding any novelty were excluded.
RESULTS
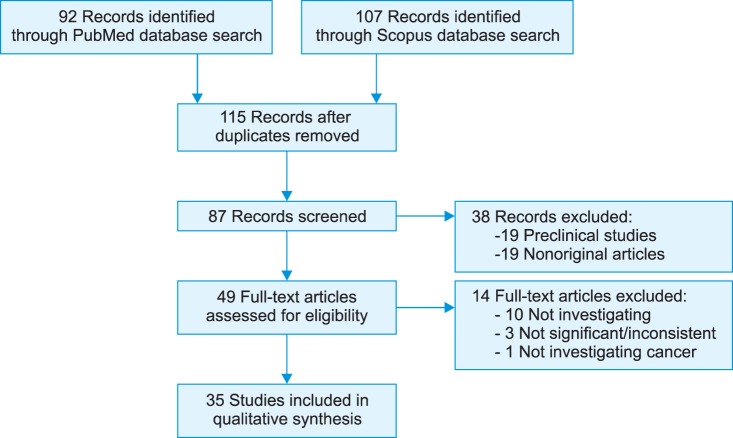
Of the more than 115 published papers, 35 were considered in this review (Fig. 1).
FIG. 1.
Flow of information through the different phases of the systematic review. PSA, prostate-specific antigen; p2PSA, [-2]proPSA.
1. Clinical validity of proPSA isoforms in improving PSA specificity
Four important studies investigated the clinical validity of proPSA, which was defined as the sum of the [-2], [-4], and [-5/-7] forms. The first study by Sokoll et al. [20] involved archival serum obtained before biopsy from 119 men (31 PCa, 88 noncancer) with PSA of 2.5 to 4.0 ng/mL. The serum levels of tPSA, fPSA, and proPSA and the proPSA/fPSA ratio (%proPSA) were analyzed: PSA and %fPSA values were similar between the noncancer and PCa groups, whereas %proPSA was relatively higher in PCa patients (50.1% ± 4.4%) than in the noncancer group (35.5% ± 6.7%, p=0.07). The areas under the curve (AUCs) for %proPSA and %fPSA were 0.688 and 0.567, respectively. At fixed sensitivity (75%), the specificity was significantly greater for %proPSA (59%) than for %fPSA (33%, p<0.0001). These results were then confirmed in a follow-up study of the same group [21]. In multivariate logistic regression analyses, at fixed sensitivity (90%), the combination of proPSA with tPSA and %fPSA showed significantly higher specificity (44%) for early PCa detection than did the individual variables (13%, 23%, and 33%, respectively).
Catalona et al. [22] confirmed these results in a later study in which they analyzed serum specimens from 1,091 patients (635 noncancer, 456 PCa) who underwent prostate biopsies. In men with PSA of 2 to 4 ng/mL, %proPSA (at a threshold of 1.8) detected 90% of cancers, including all (16/16) extracapsular tumors and 96.6% (28/29) of cancers with a GS≥7.
In 2004, Mikolajczyk et al. [23] retrospectively evaluated the serum samples of 380 men (238 PCa, 142 noncancer) with tPSA of 4 to 10 ng/mL. Accordingly, %proPSA had a higher AUC than %p2PSA, fPSA, and complexed PSA (AUC: 0.69, 0.64, 0.63, and 0.57, respectively). In men with %fPSA>25, %p2PSA had the highest accuracy (AUC, 0.77). At a threshold of 2.5, %p2PSA had a sensitivity of 90% and would have resulted in 36% of prostate biopsies being avoided. However, in patients with %fPSA<15, at 90% sensitivity, %proPSA had a higher accuracy (AUC, 0.703; specificity, 36%) than did %p2PSA (AUC, 0.669; specificity, 21%).
2. Clinical validity of p2PSA and %p2PSA
As shown above, p2PSA is a more cancer-specific PSA isoform. Table 1 shows studies investigating p2PSA validity and their main results. Sokoll et al. [24] evaluated the relationship between p2PSA and PCa by using serum samples of 123 men (51% PCa, 49% noncancer) enrolled in the Early Detection Research Network study. Overall, the %fPSA was significantly lower, whereas p2PSA and %p2PSA were higher, in PCa patients. Additionally, in the PSA range of 2 to 10 ng/mL, p2PSA and %p2PSA continued to be significantly associated with PCa: the AUC for %p2PSA was 0.73 compared with 0.53 for %fPSA. Sokoll et al. [25] investigated the potential correlation between p2PSA and PCa aggressiveness and found that %p2PSA performed significantly better than %fPSA at lower (2-4 ng/mL) PSA levels.
TABLE 1.
Studies investigating the accuracy of p2PSA and %p2PSA in detecting PCa
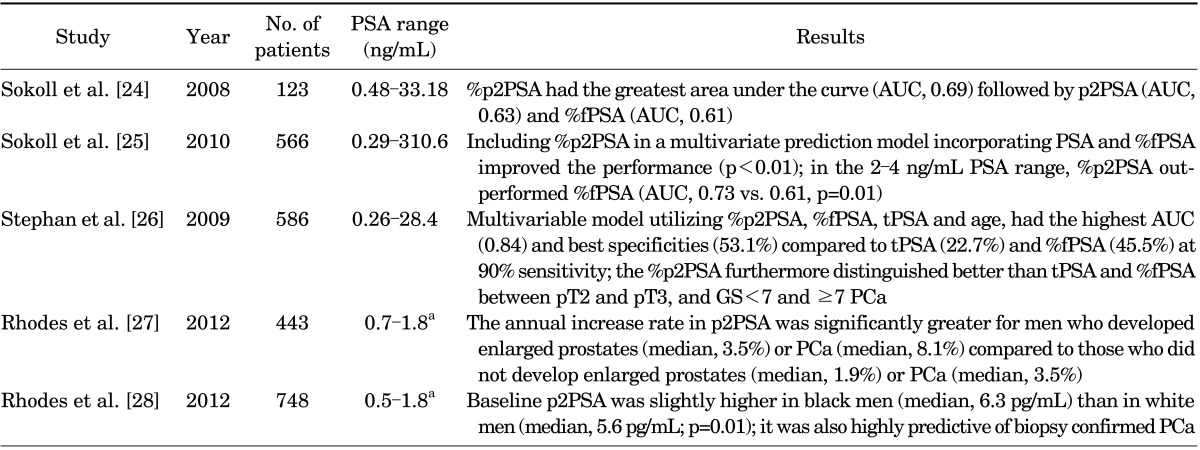
PSA, prostate-specific antigen; p2PSA, [-2]proPSA; %p2PSA, [-2]proPSA/fPSA; PCa, prostate cancer; tPSA, total PSA; fPSA, free PSA; %fPSA, fPSA/tPSA; GS, Gleason score; AUC, area under the curve.
a:25th-75th Percentiles.
However, multivariate regression models incorporating clinical information and p2PSA were shown to perform better than PSA forms individually. Stephan et al. [26] included 475 patients (264 PCa, 211 noncancer) with tPSA of 2 to 10 ng/mL and showed that the multivariable model including %p2PSA, %fPSA, tPSA, and age (but not prostate volume) reached the highest AUC (0.84) and specificity (53.1%) compared with tPSA (22.7%), %fPSA (45.5%), and %p2PSA (41.7%) alone at fixed sensitivity (90%).
More recently, p2PSA level changes over time were suggested as a potential predictor of PCa development. In 2012, Rhodes et al. [27] reported that p2PSA increased with advancing age and prostate volume. However, the greatest p2PSA level changes were seen in men who subsequently developed PCa (+8.1%/y) compared with those who did not (+3.5%/y) after a median follow-up of 7 years.
In a subsequent study [28], the same group reported that the baseline p2PSA levels in black men were slightly higher than those in white men (median, 6.3 pg/mL vs. 5.6 pg/mL, respectively). Furthermore, more interestingly, white men (from the Olmsted County Study of Urinary Symptoms and Health Status among Men cohort) with higher baseline p2PSA had an almost eight-fold higher risk of subsequent PCa diagnosis (hazard ratio, 7.8; 95% confidence interval, 2.2-27.8). Thus, baseline p2PSA and p2PSA changes over time might be useful predictors of PCa development, and this warrants further investigation.
These clinical studies investigating p2PSA showed very promising results. However, they were all retrospective, involving serum specimens collected in different preanalytical settings and stored for up to 16 years [28].
3. PHI: clinical validity and utility
Because p2PSA appears to have the highest predictive ability when associated with other variables, Beckman Coulter Inc. developed the PHI, a mathematical algorithm that is defined as follows: (p2PSA/fPSA) · √tPSA. Fifteen studies have investigated the utility of p2PSA and PHI, and the main results of these studies are reported in Table 2.
TABLE 2.
Studies investigating the accuracy of the PHI in detecting PCa
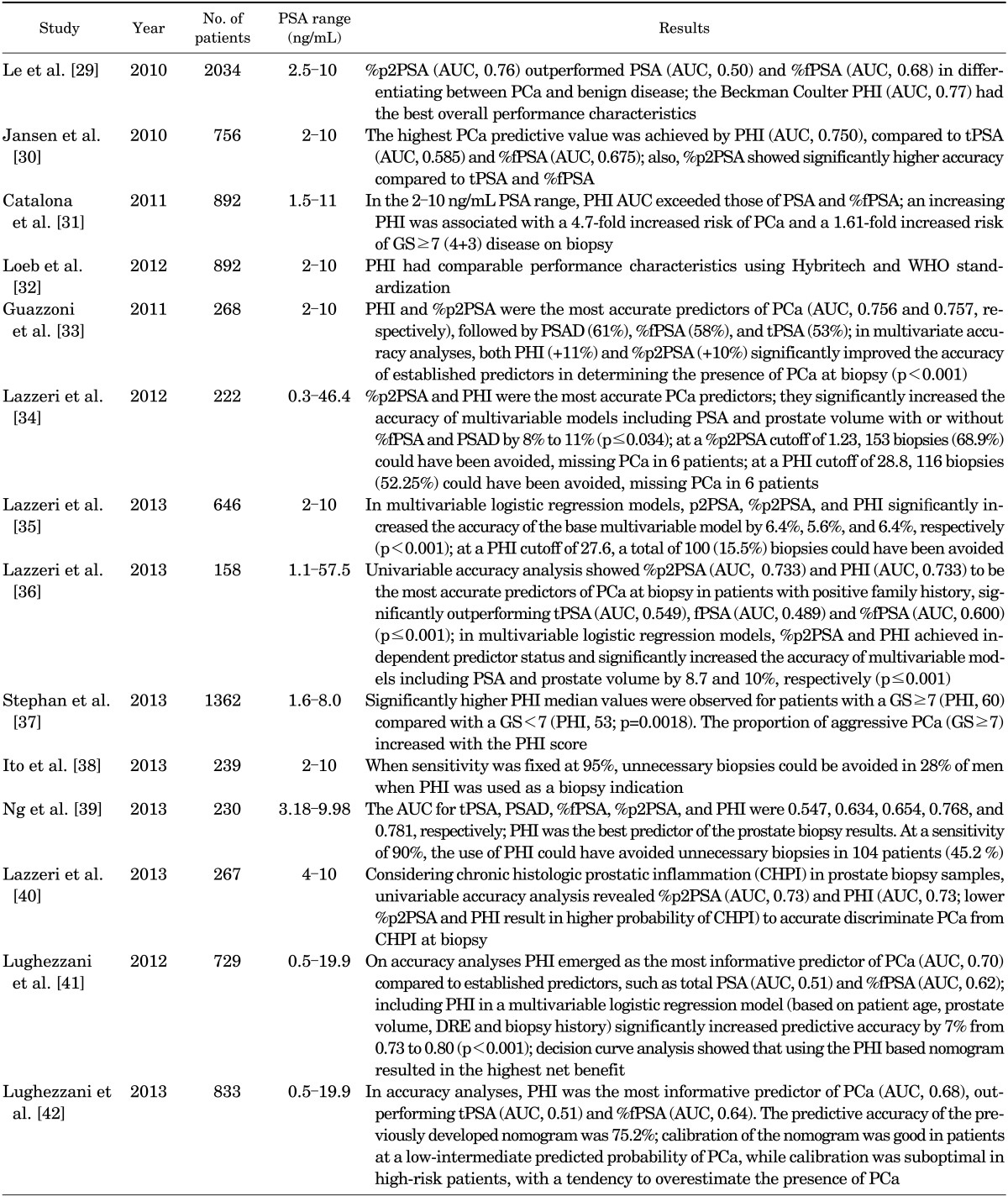
PSA, prostate-specific antigen; tPSA, total PSA; fPSA, free PSA; %fPSA, fPSA/tPSA; PSAD, PSA density; p2PSA, [-2]proPSA; %p2PSA, [-2]proPSA/fPSA; PHI, Prostate Health Index; PCa, prostate cancer; GS, Gleason score; WHO, World Health Organization; DRE, digital rectal examination; AUC, area under the receiver-operating characteristic curve.
Le et al. [29] were the first to evaluate the predictive ability of p2PSA and PHI in a prospective PCa screening setting. Their study involved 2,034 men undergoing PCa screening: 322 patients were advised to undergo prostate biopsy for an elevated PSA level (>2.5 ng/mL) and/or suspicious DRE. Eventually, only 74 patients underwent prostate biopsy; 63 of them had a tPSA level of 4 to 10 ng/mL and a normal DRE result. ROC analysis showed that PHI had the highest predictive ability (AUC, 0.77), followed by %p2PSA (AUC, 0.76) and %fPSA (AUC, 0.68). tPSA alone lacked sensitivity and specificity in the range of 2.5 to 10 ng/mL (AUC, 0.50). At a sensitivity of 88.5%, PHI and %p2PSA outperformed %fPSA or tPSA (specificity: 64.9% and 48.6% vs. 40.5% and 24.3%, respectively).
In 2010 Jansen et al. [30] retrospectively evaluated serum samples of 405 patients enrolled in the Rotterdam arm of the ERSPC study and 351 samples from Innbruck Medical University to investigate the use of p2PSA, PHI, and benign prostatic hyperplasia-associated PSA. The authors found significantly higher PCa predictive value and specificity for PHI and %p2PSA. However, p2PSA had limited additional value in identifying aggressive PCa (GS≥ 7). At 90% sensitivity, PHI and %p2PSA had the highest specificity (31%-35%) compared with tPSA (only 10%-16%).
Afterwards, Catalona et al. [31] conducted a multicenter, double-blind, case-control clinical trial to validate PHI in the PSA range of 2.0 to 10.0 ng/mL. Of 1,372 men enrolled in eight medical centers from October 2003 to June 2009, 892 patients met the eligibility criteria: age≥50 years, normal DRE result, and PSA of 1.5 to 11 ng/mL. PHI was found to have the greatest PCa predictive accuracy (AUC, 0.703) compared with %fPSA (AUC, 0.648), fPSA (AUC, 0.615), p2PSA (AUC, 0.557), and tPSA (AUC, 0.525), directly correlating with GS (p=0.013), with an AUC of 0.724 for GS≥ 4+3 disease. Moreover, men with a PHI>55.0 had a 52% likelihood of being diagnosed with PCa at biopsy compared with 26% of men with a PHI<25.0. In particular, compared with a PHI<25.0, the relative risk of PCa detection was 1.6-, 3.0-, and 4.7-fold higher at PHI values of 25.0-34.9, 35.0-54.9, and ≥55.0, respectively. At a PHI cutoff of 21.3, GS was ≥7 in 25% of missed cancers, resulting in the authors suggesting careful surveillance.
The same group recently published a prospective, multicenter study involving 892 men undergoing prostate biopsy [32]. The AUC for PHI (0.704) was significantly higher than for %fPSA (0.649, p=0.005) and tPSA (0.527, p<0.001) in men with a PSA of 1.6 to 7.8 ng/mL (World Health Organization [WHO] calibration [corresponding to 2-10 ng/mL Hybritech calibration]). Moreover, higher PHI values were associated with higher PCa risk and GS. The authors concluded that PHI had comparable performance characteristics by use of Hybritech and WHO standardization.
In 2011, Guazzoni et al. [33] conducted a prospective observational study of 268 consecutive men with PSA between 2 and 10 ng/mL and normal DRE results who underwent prostate biopsy. In this cohort, %p2PSA and PHI were the strongest predictors of positive prostatic biopsy outcome. PHI and %p2PSA improved the accuracy of a base multivariate model (including tPSA, fPSA, prostate volume, and age) by 11% and 10%, respectively (p<0.001). Similarly, in patients with tPSA of 4 to 10 ng/mL, the inclusion of PHI and %p2PSA significantly increased the multivariate predictive accuracy from 72% to 83% (+11%) in both models (p<0.001).
Furthermore, Lazzeri et al. [34] prospectively evaluated a clinical cohort of men with previous negative biopsies but persistent suspicion of PCa. Again, %p2PSA and PHI were the most accurate predictors of disease. In multivariable logistic regression models, %p2PSA and PHI achieved independent predictor status and significantly increased the accuracy of multivariable models by 8% to 11% (p≤0.034). At a PHI cutoff of 28.8, 116 biopsies (52.25%) could have been avoided and PCa would have been overlooked in 6 patients, but none with a GS≥7, demonstrating a real clinical utility.
Recently, Lazzeri et al. [35] confirmed previous results in an observational, prospective, multicenter European cohort (PROMEtheuS Project). This study involved 646 patients from five European urology centers with tPSA of 2 to 10 ng/mL who were subjected to initial prostate biopsy for suspected PCa. p2PSA, %p2PSA, and PHI significantly increased the accuracy of the base multivariable model by 6.4%, 5.6%, and 6.4%, respectively (p<0.001). At 90% sensitivity, the PHI cutoff of 27.6 could result in the avoidance of 100 biopsies (15.5%), with 26 cancers (9.8%) being overlooked (23 with GS 6, 3 with GS 3+4).
Interestingly, the same group [36] published a nested case-control study from the same PROMEtheuS database, evaluating 158 patients with a positive family history for PCa (at least one first-degree relative with PCa), in a PSA range of 1.1 to 57.5 ng/mL. %p2PSA and PHI were directly associated with GS and were more accurate than tPSA, fPSA, and %fPSA in predicting PCa. At 90% sensitivity, the thresholds for %p2PSA and PHI were 1.20 and 25.5, sparing a total of 39 (24.8%) and 27 biopsies (17.2%), respectively, and missing 2 cases (3.8%) of PCa, each with a GS of 7. Again %p2PSA and PHI significantly increased the accuracy of multivariable models by 8.7% and 10%, respectively (p≤0.001). Although the PROMEtheuS study had a well-planned observational design, the main limitation was that patients were included for their PSA and DRE-related risk of PCa and not through a p2PSA screening protocol.
Another European multicenter study was published by Stephan et al. [37]. This study involved 1,362 patients with tPSA between 1.6 and 8.0 g/L (668 PCa, 694 noncancer). Serum concentrations of tPSA and fPSA were both calibrated against a WHO reference material. %p2PSA and PHI were significantly higher in all PCa subcohorts (positive initial or repeat biopsy results or negative DRE result) compared with patients without PCa (p<0.0001). PHI had the largest AUC (0.74) and provided significantly better clinical performance for predicting PCa compared with %p2PSA (AUC, 0.72; p=0.018), p2PSA (AUC, 0.63; p<0.0001), %fPSA (AUC, 0.61), or tPSA (AUC, 0.56). Significantly higher PHI was observed for patients with GS ≥7 (PHI 60) compared with GS<7 (PHI 53, p=0.0018). The proportion of aggressive PCa (GS≥7) increased with PHI.
Two recent studies from Asia confirm previous results in another population setting. Ito et al. [38] reported data on 239 consecutive men with tPSA between 2.0 and 10.0 ng/mL who underwent prostate biopsy. When PHI was used as a biopsy indicator and sensitivity was fixed at 95%, unnecessary biopsies could be avoided in 28% of men. Accordingly, Ng et al. [39] retrospectively analyzed archived serum samples from 230 patients over 50 years of age who had undergone their first prostate biopsy with a PSA of 4 to 10 ng/mL and a negative DRE result. PHI was found to be the best predictor of the prostate biopsy results. At a sensitivity of 90%, the use of PHI could have resulted in the avoidance of unnecessary biopsies in 104 patients (45.2%).
Interestingly, Lazzeri et al. [40] showed that p2PSA, %p2PSA, and PHI values might specifically discriminate PCa from chronic histologic prostatic inflammation (CHPI) or BPH, but not CHPI from BPH, in men with tPSA of 4 to 10 ng/mL and normal DRE. Univariable accuracy analysis revealed %p2PSA (AUC, 0.73) and PHI (AUC, 0.73) to be the most accurate predictors of CHPI at biopsy (lower %p2PSA and PHI result in higher probability of CHPI), outperforming the other biomarkers. Again, multivariable models including p2PSA, %p2PSA, and PHI showed the highest net benefit in discriminating between patients with and without PCa in a probability of pathologic outcome range (threshold probability) between 25% and 90%.
Finally, Lughezzani et al. [41] developed and validated, on over 729 patients, a PHI-based nomogram to predict PCa at extended prostate biopsy. Including PHI in a multivariable logistic regression model based on patient age, prostate volume, DRE, and biopsy history significantly increased predictive accuracy by 7% from 0.73 to 0.80 (p<0.001). Decision curve analysis showed that using the PHI-based nomogram resulted in the highest net benefit. This nomogram was also externally validated in a recent multicenter European study [42].
Overall, studies to date suggest that %p2PSA and PHI are more accurate than standard reference tests in predicting prostate biopsy outcome and could result in the avoidance of unnecessary biopsies.
4. p2PSA and PHI as predictors of final histology in radical prostatectomy specimens
Further studies are necessary before definitively proving that PHI and p2PSA can predict PCa aggressiveness on prostate biopsies, as well as on final histology after radical prostatectomy (RP), potentially limiting overtreatment.
Accordingly, Guazzoni et al. [43] conducted an observational, prospective study of 350 consecutive men diagnosed with clinically localized PCa who underwent RP. Preoperative %p2PSA and PHI were significantly higher in patients with pT3 disease, pathological GS≥7 and those with GS upgrading (p<0.001). These measures might therefore be useful in the preoperative counseling of patients with newly diagnosed, clinically localized PCa.
Interestingly, in 2013 Heidegger et al. [44] found that p2PSA values were highly differentiated (p<0.001) between GS≥8 and GS≤7 as early as 3 years before diagnosis and that preoperative p2PSA values were significantly higher in men with pT3a or higher compared with pT2c or lower PCa up to 4 years before diagnosis (p<0.01). p2PSA was shown to have a high positive predictive value concerning GS≥8 and GS≤7 and also extraprostatic extension.
Although these two studies were well designed and conducted, and the results were strong, they do not completely resemble the general population and need to be confirmed in larger multicenter studies.
5. p2PSA and PHI in active surveillance regimens
PSA screening has resulted in an increasing number of patients being diagnosed with potentially low-risk, clinically insignificant cancers. To reduce overtreatment, active surveillance (AS) has been proposed as an alternative strategy for these patients [9]. An effective program should include regular periodic DREs, PSA testing, and repeated prostate biopsies.
Makarov et al. [45] assessed the association of proPSA with outcomes among men with PCa in AS. The authors found that the p2PSA/%fPSA ratio in serum was significantly higher at diagnosis in men with unfavorable biopsy results (0.87±0.44) than in those with favorable biopsy results (0.65±0.36, p=0.02). Moreover, p2PSA/%fPSA (hazard ratio, 2.53; p=0.02) was significantly associated with an unfavorable biopsy result in Kaplan-Meier and Cox analyses.
In 2011, the same group analyzed the role of the PHI in this same cohort of patients [46]. The PHI was significantly greater in men who ultimately had unfavorable biopsy findings (37.23±15.76 vs. 30.60±12.28, p=0.03). Moreover, PHI (p=0.003) and p2PSA/%fPSA (p=0.004) were significant predictors of unfavorable biopsy conversion in a Cox regression analysis.
Tosoian et al. [47] reported data from 167 men scheduled in a single-institution AS program. Risk of biopsy reclassification was significantly associated with lower %fPSA (p=0.002) and higher %p2PSA (p<0.0001) and PHI (p<0.0001) at baseline.
Recently, Hirama et al. [48] evaluated the predictive impact of baseline p2PSA and related indexes on the pathological reclassification at 1 year in 67 patients enrolled over 134 candidates for AS. %p2PSA and PHI at baseline were significantly different between the reclassification and nonreclassification groups (2.44 vs. 1.88 [p=0.003] and 60.3 vs. 47.8 [p=0.01], respectively). Multivariate logistic regression analysis revealed baseline %p2PSA and PHI (both p=0.008) to be the only independent predictive factors for pathological upgrade at the 1-year mark during AS.
Therefore, baseline p2PSA and derivative values seem to help to identify those men at risk of future unfavorable reclassification during AS, but further studies are needed to define the role of these variables in selecting men who would most benefit from AS.
6. p2PSA and other molecular markers
Recently, four studies compared the accuracy of p2PSA in detecting PCa with that of other interesting biomarkers, in particular, urinary prostate cancer antigen 3 (PCA3). Ferro et al. [49] were the first to report that %p2PSA, PHI, and PCA3 are comparably good indicators of malignancy (AUC: 0.73, 0.77, and 0.71, respectively). PHI had the highest AUC, but it was not statistically different from PCA3 (p=0.368).
In a subsequent study by the same group [50], PHI (AUC, 0.82; p<0.001), PCA3 (AUC, 0.77; p=0.015), and their combination (AUC, 0.83; p<0.001) improved the diagnostic accuracy (AUC, 0.72) of the base multivariable model (including age, PSA, %fPSA, DRE, and prostate volume). However, the AUC of the multivariable model did not improve over both PHI and PCA3 alone (p>0.05).
Scattoni et al. [51] showed that PHI accuracy was higher than that of PCA3 at both the initial prostate biopsy (AUC: 0.69 vs. 0.57) and the repeat biopsy (AUC: 0.72 vs. 0.63), although accuracy in these two settings (initial or repeat biopsy) was not statistically different. Including PCA3 in the base multivariable model (PSA, %fPSA, prostate volume) did not increase predictive accuracy in either setting (AUC: 0.79 vs. 0.80 and 0.75 vs. 0.76, respectively). Conversely, PHI improved the predictive accuracy of the base model by 5% (AUC: 0.79 to 0.84) and 6% (AUC: 0.75 to 0.81) in initial and repeat settings, respectively.
Moreover, Stephan et al. [52] compared PHI, PCA3, and the transmembrane protease, serine 2 (TMPRSS2):v-ets erythroblastosis virus E26 oncogene homolog (avian) (ERG) gene fusion (TMPRSS2:ERG). These markers showed the highest accuracy (AUC: 0.68, 0.74, 0.63, respectively). PCA3 had the largest AUC, although it was not statistically different from that of PHI. The combination of both markers modestly enhanced diagnostic power (AUC gain: 0.01-0.04). Although PCA3 had the highest AUC also in the repeat-biopsy cohort, the highest AUC for PHI was observed in DRE-negative patients with PSA of 2 to 10 ng/mL.
If these results are confirmed in larger multicenter studies, PHI seems to be the best compromise between diagnostic accuracy and ease of sampling and analysis.
7. Cost-effectiveness of p2PSA and PHI
Owing to its high accuracy in predicting PCa, PHI could result in the avoidance of a considerable number of negative prostate biopsies, thus reducing direct costs. In two subsequent studies, Nichol et al. [53,54] evaluated the cost-effectiveness of PHI.
In the first study [53], the authors constructed two budget impact models by using PSA cutoff values of ≥2 ng/mL (model #1) and ≥4 ng/mL (model #2) for recommending a prostate biopsy in a hypothetical health plan with 100,000 male members aged 50 to 75 years old. The budgetary impact on the 1-year expected total costs for PCa detection was calculated. The addition of PHI to the current PSA screening strategies (using tPSA and %fPSA) increased the total cost of blood tests by $51,524 (model #1) and $13,611 (model #2), but produced higher cost savings in model #1 ($356,647) than in model #2 ($94,219) with a small short-term reduction in the number of positive tests.
In the second study [54], the same group evaluated the cost-effectiveness of early PCa detection with PHI associated with a PSA test compared with the PSA test alone from a United States of America societal perspective. Over 25 annual screening cycles, the strategy of PSA plus PHI was estimated to save $1,199 or $443 with an expected gain of 0.08 or 0.03 quality-adjusted life years per person for PSA thresholds of ≥2 and ≥4 ng/mL, respectively. Because the strategy of PSA plus PHI is expected to increase the number of true-positive tests while reducing false-positives in men aged 50 to 75 years, the authors suggested that the increased total costs of the laboratory assay (PSA+fPSA+p2PSA) could be offset by reducing unnecessary prostate biopsies.
DISCUSSION
This review has summarized current knowledge about the early diagnosis of PCa with p2PSA, %p2PSA, and PHI and has presented indications that %p2PSA and PHI may discriminate men with or without PCa with higher accuracy than the reference standard tests. Furthermore, the results of observational prospective international studies support the association between these new biomarkers and cancer aggressiveness [35,37]. Several authors have shown that PHI correlates with the GS and might result in the avoidance of unnecessary biopsies without missing significant PCa [31,34,35,37]. PHI was also shown to be a useful clinical marker in patients with a positive family history of PCa [36]. Thus, the results reported above suggest that the new diagnostic tests may be particularly useful in patients with a tPSA range of 2/4 to 10 ng/ml. Furthermore, a strong correlation between %p2PSA and PHI and the pathological characteristics in whole gland samples was found after RP. Finally, we also reported the results of studies comparing p2PSA and derivatives with other available biomarkers, in particular PCA3. These studies showed a slightly higher accuracy for PHI than for PCA3 but an improvement in accuracy with their combination. Moreover, it is notable that other biomarkers (TMPRSS2:ERG, 4Ks, miRNAs, circulating tumor cells) are emerging and these will need to be taken into account in future studies. To our knowledge, only one study recently compared TMPRSS2: ERG to p2PSA [52], and this study was reported in this review.
The present systematic review had a number of limitations that must be taken into account. Most of the studies were retrospective and different biopsy protocols were used. Even though the gold standard for biopsy was used, some groups limited the number of biopsy cores to 12, whereas others extended the core number to 18 to 24, possibly causing significant heterogeneity. Further heterogeneity was found regarding study design (retrospective, prospective, screening), race (most of the studies included Caucasian men), and preanalytic and analytic phases. Another limitation was the potential duplication of results in related publications that could bias the conclusions. In fact, there may have been some overlap between subsequent studies, but being that this article was not a meta-analysis, we decided to include all the studies because of their different aims. Finally, the issue of costs remains unsolved.
CONCLUSIONS
p2PSA and PHI are more accurate than the currently used tests (PSA and derivatives) in predicting the presence of PCa at biopsy. Their implementation in clinical practice has the potential to significantly increase physicians' ability to detect PCa and avoid unnecessary biopsies. Further work is needed to confirm and generalize these conclusions to wider populations.
Footnotes
The authors have nothing to disclose.
References
- 1.Hoffman RM, Stone SN, Espey D, Potosky AL. Differences between men with screening-detected versus clinically diagnosed prostate cancers in the USA. BMC Cancer. 2005;5:27. doi: 10.1186/1471-2407-5-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 3.Catalona WJ, Partin AW, Slawin KM, Brawer MK, Flanigan RC, Patel A, et al. Use of the percentage of free prostate-specific antigen to enhance differentiation of prostate cancer from benign prostatic disease: a prospective multicenter clinical trial. JAMA. 1998;279:1542–1547. doi: 10.1001/jama.279.19.1542. [DOI] [PubMed] [Google Scholar]
- 4.Hedelin H, Johansson N, Stroberg P. Relationship between benign prostatic hyperplasia and lower urinary tract symptoms and correlation between prostate volume and serum prostate-specific antigen in clinical routine. Scand J Urol Nephrol. 2005;39:154–159. doi: 10.1080/00365590510007685. [DOI] [PubMed] [Google Scholar]
- 5.Simardi LH, Tobias-MacHado M, Kappaz GT, Taschner Goldenstein P, Potts JM, Wroclawski ER. Influence of asymptomatic histologic prostatitis on serum prostate-specific antigen: a prospective study. Urology. 2004;64:1098–1101. doi: 10.1016/j.urology.2004.08.060. [DOI] [PubMed] [Google Scholar]
- 6.Roehrborn CG, Pickens GJ, Carmody T., 3rd Variability of repeated serum prostate-specific antigen (PSA) measurements within less than 90 days in a well-defined patient population. Urology. 1996;47:59–66. doi: 10.1016/s0090-4295(99)80383-5. [DOI] [PubMed] [Google Scholar]
- 7.Link RE, Shariat SF, Nguyen CV, Farr A, Weinberg AD, Morton RA, et al. Variation in prostate specific antigen results from 2 different assay platforms: clinical impact on 2304 patients undergoing prostate cancer screening. J Urol. 2004;171(6 Pt 1):2234–2238. doi: 10.1097/01.ju.0000127736.86597.e7. [DOI] [PubMed] [Google Scholar]
- 8.Vickers AJ, Cronin AM, Roobol MJ, Hugosson J, Jones JS, Kattan MW, et al. The relationship between prostate-specific antigen and prostate cancer risk: the Prostate Biopsy Collaborative Group. Clin Cancer Res. 2010;16:4374–4381. doi: 10.1158/1078-0432.CCR-10-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heidenreich A, Bastian PJ, Bellmunt J, Bolla M, Joniau S, van der Kwast T, et al. EAU guidelines on prostate cancer. part 1: screening, diagnosis, and local treatment with curative intent-update 2013. Eur Urol. 2014;65:124–137. doi: 10.1016/j.eururo.2013.09.046. [DOI] [PubMed] [Google Scholar]
- 10.Thompson IM, Pauler DK, Goodman PJ, Tangen CM, Lucia MS, Parnes HL, et al. Prevalence of prostate cancer among men with a prostate-specific antigen level < or =4.0 ng per milliliter. N Engl J Med. 2004;350:2239–2246. doi: 10.1056/NEJMoa031918. [DOI] [PubMed] [Google Scholar]
- 11.Stamey TA, Yang N, Hay AR, McNeal JE, Freiha FS, Redwine E. Prostate-specific antigen as a serum marker for adenocarcinoma of the prostate. N Engl J Med. 1987;317:909–916. doi: 10.1056/NEJM198710083171501. [DOI] [PubMed] [Google Scholar]
- 12.Kumar A, Mikolajczyk SD, Goel AS, Millar LS, Saedi MS. Expression of pro form of prostate-specific antigen by mammalian cells and its conversion to mature, active form by human kallikrein 2. Cancer Res. 1997;57:3111–3114. [PubMed] [Google Scholar]
- 13.Lovgren J, Rajakoski K, Karp M, Lundwalla, Lilja H. Activation of the zymogen form of prostate-specific antigen by human glandular kallikrein 2. Biochem Biophys Res Commun. 1997;238:549–555. doi: 10.1006/bbrc.1997.7333. [DOI] [PubMed] [Google Scholar]
- 14.Takayama TK, Fujikawa K, Davie EW. Characterization of the precursor of prostate-specific antigen. Activation by trypsin and by human glandular kallikrein. J Biol Chem. 1997;272:21582–21588. doi: 10.1074/jbc.272.34.21582. [DOI] [PubMed] [Google Scholar]
- 15.Mikolajczyk SD, Grauer LS, Millar LS, Hill TM, Kumar A, Rittenhouse HG, et al. A precursor form of PSA (pPSA) is a component of the free PSA in prostate cancer serum. Urology. 1997;50:710–714. doi: 10.1016/S0090-4295(97)00449-4. [DOI] [PubMed] [Google Scholar]
- 16.Peter J, Unverzagt C, Krogh TN, Vorm O, Hoesel W. Identification of precursor forms of free prostate-specific antigen in serum of prostate cancer patients by immunosorption and mass spectrometry. Cancer Res. 2001;61:957–962. [PubMed] [Google Scholar]
- 17.Jansen FH, Roobol M, Jenster G, Schroder FH, Bangma CH. Screening for prostate cancer in 2008 II: the importance of molecular subforms of prostate-specific antigen and tissue kallikreins. Eur Urol. 2009;55:563–574. doi: 10.1016/j.eururo.2008.11.040. [DOI] [PubMed] [Google Scholar]
- 18.Mikolajczyk SD, Millar LS, Wang TJ, Rittenhouse HG, Marks LS, Song W, et al. A precursor form of prostate-specific antigen is more highly elevated in prostate cancer compared with benign transition zone prostate tissue. Cancer Res. 2000;60:756–759. [PubMed] [Google Scholar]
- 19.Mikolajczyk SD, Marker KM, Millar LS, Kumar A, Saedi MS, Payne JK, et al. A truncated precursor form of prostate-specific antigen is a more specific serum marker of prostate cancer. Cancer Res. 2001;61:6958–6963. [PubMed] [Google Scholar]
- 20.Sokoll LJ, Chan DW, Mikolajczyk SD, Rittenhouse HG, Evans CL, Linton HJ, et al. Proenzyme psa for the early detection of prostate cancer in the 2.5-4.0 ng/ml total psa range: preliminary analysisProenzyme psa for the early detection of prostate cancer in the 2 ng/ml total psa range: preliminary analysis. Urology. 2003;61:274–276. doi: 10.1016/s0090-4295(02)02398-1. [DOI] [PubMed] [Google Scholar]
- 21.Khan MA, Partin AW, Rittenhouse HG, Mikolajczyk SD, Sokoll LJ, Chan DW, et al. Evaluation of proprostate specific antigen for early detection of prostate cancer in men with a total prostate specific antigen range of 4.0 to 10.0 ng/ml. J Urol. 2003;170:723–726. doi: 10.1097/01.ju.0000086940.10392.93. [DOI] [PubMed] [Google Scholar]
- 22.Catalona WJ, Bartsch G, Rittenhouse HG, Evans CL, Linton HJ, Horninger W, et al. Serum pro-prostate specific antigen preferentially detects aggressive prostate cancers in men with 2 to 4 ng/ml prostate specific antigen. J Urol. 2004;171(6 Pt 1):2239–2244. doi: 10.1097/01.ju.0000127737.94221.3e. [DOI] [PubMed] [Google Scholar]
- 23.Mikolajczyk SD, Catalona WJ, Evans CL, Linton HJ, Millar LS, Marker KM, et al. Proenzyme forms of prostate-specific antigen in serum improve the detection of prostate cancer. Clin Chem. 2004;50:1017–1025. doi: 10.1373/clinchem.2003.026823. [DOI] [PubMed] [Google Scholar]
- 24.Sokoll LJ, Wang Y, Feng Z, Kagan J, Partin AW, Sanda MG, et al. [-2]proenzyme prostate specific antigen for prostate cancer detection: a national cancer institute early detection research network validation study. J Urol. 2008;180:539–543. doi: 10.1016/j.juro.2008.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sokoll LJ, Sanda MG, Feng Z, Kagan J, Mizrahi IA, Broyles DL, et al. A prospective, multicenter, National Cancer Institute Early Detection Research Network study of [-2]proPSA: improving prostate cancer detection and correlating with cancer aggressiveness. Cancer Epidemiol Biomarkers Prev. 2010;19:1193–1200. doi: 10.1158/1055-9965.EPI-10-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stephan C, Kahrs AM, Cammann H, Lein M, Schrader M, Deger S, et al. A [-2]proPSA-based artificial neural network significantly improves differentiation between prostate cancer and benign prostatic diseases. Prostate. 2009;69:198–207. doi: 10.1002/pros.20872. [DOI] [PubMed] [Google Scholar]
- 27.Rhodes T, Jacobson DJ, McGree ME, St Sauver JL, Girman CJ, Lieber MM, et al. Longitudinal changes of benign prostate-specific antigen and [-2]proprostate-specific antigen in seven years in a community-based sample of men. Urology. 2012;79:655–661. doi: 10.1016/j.urology.2011.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rhodes T, Jacobson DJ, McGree ME, St Sauver JL, Sarma AV, Girman CJ, et al. Distribution and associations of [-2]proenzyme-prostate specific antigen in community dwelling black and white men. J Urol. 2012;187:92–96. doi: 10.1016/j.juro.2011.09.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Le BV, Griffin CR, Loeb S, Carvalhal GF, Kan D, Baumann NA, et al. [-2]Proenzyme prostate specific antigen is more accurate than total and free prostate specific antigen in differentiating prostate cancer from benign disease in a prospective prostate cancer screening study. J Urol. 2010;183:1355–1359. doi: 10.1016/j.juro.2009.12.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jansen FH, van Schaik RH, Kurstjens J, Horninger W, Klocker H, Bektic J, et al. Prostate-specific antigen (PSA) isoform p2PSA in combination with total PSA and free PSA improves diagnostic accuracy in prostate cancer detection. Eur Urol. 2010;57:921–927. doi: 10.1016/j.eururo.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 31.Catalona WJ, Partin AW, Sanda MG, Wei JT, Klee GG, Bangma CH, et al. A multicenter study of [-2]pro-prostate specific antigen combined with prostate specific antigen and free prostate specific antigen for prostate cancer detection in the 2.0 to 10.0 ng/ml prostate specific antigen range. J Urol. 2011;185:1650–1655. doi: 10.1016/j.juro.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loeb S, Sokoll LJ, Broyles DL, Bangma CH, van Schaik RH, Klee GG, et al. Prospective multicenter evaluation of the Beckman Coulter Prostate Health Index using WHO calibration. J Urol. 2013;189:1702–1706. doi: 10.1016/j.juro.2012.11.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guazzoni G, Nava L, Lazzeri M, Scattoni V, Lughezzani G, Maccagnano C, et al. Prostate-specific antigen (PSA) isoform p2PSA significantly improves the prediction of prostate cancer at initial extended prostate biopsies in patients with total PSA between 2.0 and 10 ng/ml: results of a prospective study in a clinical setting. Eur Urol. 2011;60:214–222. doi: 10.1016/j.eururo.2011.03.052. [DOI] [PubMed] [Google Scholar]
- 34.Lazzeri M, Briganti A, Scattoni V, Lughezzani G, Larcher A, Gadda GM, et al. Serum index test %[-2]proPSA and Prostate Health Index are more accurate than prostate specific antigen and %fPSA in predicting a positive repeat prostate biopsy. J Urol. 2012;188:1137–1143. doi: 10.1016/j.juro.2012.06.017. [DOI] [PubMed] [Google Scholar]
- 35.Lazzeri M, Haese A, de la Taille A, Palou Redorta J, McNicholas T, Lughezzani G, et al. Serum isoform [-2]proPSA derivatives significantly improve prediction of prostate cancer at initial biopsy in a total PSA range of 2-10 ng/ml: a multicentric European study. Eur Urol. 2013;63:986–994. doi: 10.1016/j.eururo.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 36.Lazzeri M, Haese A, Abrate A, de la Taille A, Redorta JP, McNicholas T, et al. Clinical performance of serum prostate-specific antigen isoform [-2]proPSA (p2PSA) and its derivatives, %p2PSA and the prostate health index (PHI), in men with a family history of prostate cancer: results from a multicentre European study, the PROMEtheuS project. BJU Int. 2013;112:313–321. doi: 10.1111/bju.12217. [DOI] [PubMed] [Google Scholar]
- 37.Stephan C, Vincendeau S, Houlgatte A, Cammann H, Jung K, Semjonow A. Multicenter evaluation of [-2]proprostate-specific antigen and the prostate health index for detecting prostate cancer. Clin Chem. 2013;59:306–314. doi: 10.1373/clinchem.2012.195784. [DOI] [PubMed] [Google Scholar]
- 38.Ito K, Miyakubo M, Sekine Y, Koike H, Matsui H, Shibata Y, et al. Diagnostic significance of [-2]pro-PSA and prostate dimension-adjusted PSA-related indices in men with total PSA in the 20-100 ng/mL range. World J Urol. 2013;31:305–311. doi: 10.1007/s00345-012-0927-9. [DOI] [PubMed] [Google Scholar]
- 39.Ng CF, Chiu PK, Lam NY, Lam HC, Lee KW, Hou SS. The Prostate Health Index in predicting initial prostate biopsy outcomes in Asian men with prostate-specific antigen levels of 4-10 ng/mL. Int Urol Nephrol. 2014;46:711–717. doi: 10.1007/s11255-013-0582-0. [DOI] [PubMed] [Google Scholar]
- 40.Lazzeri M, Abrate A, Lughezzani G, Gadda GM, Freschi M, Mistretta F, et al. Relationship of chronic histologic prostatic inflammation in biopsy specimens with serum isoform [-2]proPSA(p2PSA), %p2PSA, and prostate health index in men with a total prostate-specific antigen of 4-10 ng/ml and normal digital rectal examination. Urology. 2014;83:606–612. doi: 10.1016/j.urology.2013.10.016. [DOI] [PubMed] [Google Scholar]
- 41.Lughezzani G, Lazzeri M, Larcher A, Lista G, Scattoni V, Cestari A, et al. Development and internal validation of a Prostate Health Index based nomogram for predicting prostate cancer at extended biopsy. J Urol. 2012;188:1144–1150. doi: 10.1016/j.juro.2012.06.025. [DOI] [PubMed] [Google Scholar]
- 42.Lughezzani G, Lazzeri M, Haese A, McNicholas T, de la Taille A, Buffi NM, et al. Multicenter European External Validation of a Prostate Health Index-based Nomogram for Predicting Prostate Cancer at Extended Biopsy. Eur Urol. 2013 Dec 16; doi: 10.1016/j.eururo.2013.12.005. [Epub] http://dx.doi.org/10.1016/j.eururo.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 43.Guazzoni G, Lazzeri M, Nava L, Lughezzani G, Larcher A, Scattoni V, et al. Preoperative prostate-specific antigen isoform p2PSA and its derivatives, %p2PSA and prostate health index, predict pathologic outcomes in patients undergoing radical prostatectomy for prostate cancer. Eur Urol. 2012;61:455–466. doi: 10.1016/j.eururo.2011.10.038. [DOI] [PubMed] [Google Scholar]
- 44.Heidegger I, Klocker H, Steiner E, Skradski V, Ladurner M, Pichler R, et al. [-2]proPSA is an early marker for prostate cancer aggressiveness. Prostate Cancer Prostatic Dis. 2014;17:70–74. doi: 10.1038/pcan.2013.50. [DOI] [PubMed] [Google Scholar]
- 45.Makarov DV, Isharwal S, Sokoll LJ, Landis P, Marlow C, Epstein JI, et al. Pro-prostate-specific antigen measurements in serum and tissue are associated with treatment necessity among men enrolled in expectant management for prostate cancer. Clin Cancer Res. 2009;15:7316–7321. doi: 10.1158/1078-0432.CCR-09-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Isharwal S, Makarov DV, Sokoll LJ, Landis P, Marlow C, Epstein JI, et al. ProPSA and diagnostic biopsy tissue DNA content combination improves accuracy to predict need for prostate cancer treatment among men enrolled in an active surveillance program. Urology. 2011;77:763.e1–763.e6. doi: 10.1016/j.urology.2010.07.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tosoian JJ, Loeb S, Feng Z, Isharwal S, Landis P, Elliot DJ, et al. Association of [-2]proPSA with biopsy reclassification during active surveillance for prostate cancer. J Urol. 2012;188:1131–1136. doi: 10.1016/j.juro.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hirama H, Sugimoto M, Ito K, Shiraishi T, Kakehi Y. The impact of baseline [-2]proPSA-related indices on the prediction of pathological reclassification at 1 year during active surveillance for low-risk prostate cancer: the Japanese multicenter study cohort. J Cancer Res Clin Oncol. 2014;140:257–263. doi: 10.1007/s00432-013-1566-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ferro M, Bruzzese D, Perdona S, Mazzarella C, Marino A, Sorrentino A, et al. Predicting prostate biopsy outcome: prostate health index (phi) and prostate cancer antigen 3 (PCA3) are useful biomarkers. Clin Chim Acta. 2012;413:1274–1278. doi: 10.1016/j.cca.2012.04.017. [DOI] [PubMed] [Google Scholar]
- 50.Ferro M, Bruzzese D, Perdona S, Marino A, Mazzarella C, Perruolo G, et al. Prostate Health Index (Phi) and Prostate Cancer Antigen 3 (PCA3) significantly improve prostate cancer detection at initial biopsy in a total PSA range of 2-10 ng/ml. PLoS One. 2013;8:e67687. doi: 10.1371/journal.pone.0067687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scattoni V, Lazzeri M, Lughezzani G, De Luca S, Passera R, Bollito E, et al. Head-to-head comparison of prostate health index and urinary PCA3 for predicting cancer at initial or repeat biopsy. J Urol. 2013;190:496–501. doi: 10.1016/j.juro.2013.02.3184. [DOI] [PubMed] [Google Scholar]
- 52.Stephan C, Jung K, Semjonow A, Schulze-Forster K, Cammann H, Hu X, et al. Comparative assessment of urinary prostate cancer antigen 3 and TMPRSS2:ERG gene fusion with the serum [-2]proprostate-specific antigen-based prostate health index for detection of prostate cancer. Clin Chem. 2013;59:280–288. doi: 10.1373/clinchem.2012.195560. [DOI] [PubMed] [Google Scholar]
- 53.Nichol MB, Wu J, An JJ, Huang J, Denham D, Frencher S, et al. Budget impact analysis of a new prostate cancer risk index for prostate cancer detection. Prostate Cancer Prostatic Dis. 2011;14:253–261. doi: 10.1038/pcan.2011.16. [DOI] [PubMed] [Google Scholar]
- 54.Nichol MB, Wu J, Huang J, Denham D, Frencher SK, Jacobsen SJ. Cost-effectiveness of Prostate Health Index for prostate cancer detection. BJU Int. 2012;110:353–362. doi: 10.1111/j.1464-410X.2011.10751.x. [DOI] [PubMed] [Google Scholar]