Summary
The sizes of different neuronal populations within the central nervous system are precisely controlled, but whether neuronal number is coordinated between cell types is unknown. We examined the covariance structure of twelve different retinal cell types across thirty genetically distinct lines of mice, finding minimal co-variation when comparing synaptically-connected or developmentally-related cell types. Variation mapped to one or more genomic loci for each cell type, but rarely were these shared, indicating minimal genetic co-regulation of final number. Multiple genes, therefore, participate in the specification of the size of every population of retinal neuron, yet genetic variants work largely independent of one another during development to modulate those numbers, yielding substantial variability in the convergence ratios between pre- and post-synaptic populations. Density-dependent cellular interactions in the outer plexiform layer overcome this variability to ensure the formation of neuronal circuits that maintain constant retinal coverage and complete afferent sampling.
Keywords: recombinant inbred strain, quantitative trait locus, heritability, candidate gene, genetic variant
Introduction
Neuron number is determined by numerous developmental processes including proliferation, fate assignment, differentiation and apoptosis (Bassett and Wallace, 2012; Lui et al., 2011; Southwell et al., 2012). While the size of a neuronal population in different parts of the central nervous system (CNS) is known to be modulated genetically, the extent to which cell populations within a structure co-vary has been unexplored. The retina is an ideal structure for studying the precision of cellular demographics in the CNS, because most populations of neuron are confined to a single layer within a well-defined area, permitting accurate quantification. Variation in the number of different types of synaptically-connected neurons, furthermore, should have functional consequences since convergence and divergence ratios define the receptive field organization and response properties of postsynaptic neurons (Balasubramanian and Sterling, 2009; Xiong and Finlay, 1996). Because gene variants may bias fate choices within neuronal precursors, or modulate rates of apoptosis of interconnected cells (Whitney et al., 2009; 2011b), correlations (negative or positive) between developmentally and functionally related cell types are to be expected.
To gain insight into the covariance structure of multiple neuronal populations, we quantified twelve different populations of neurons in the retina (figure 1A, B) across a genetically diverse set of 30 lines of mice. These included two common inbred laboratory strains, the genomes of which having been fully sequenced (C57BL/6J and A/J), their isogenic F1 offspring, and 26 recombinant inbred (RI) strains (the AXB/BXA strain set). As each of these RI strains has been genotyped at high density using single nucleotide polymorphisms and microsatellite markers (Williams et al., 2001), we were able to map genomic sources of heritable variation in neuronal number to chromosomal positions (loci) and to examine the degree of joint or independent genetic control over the size of these twelve populations.
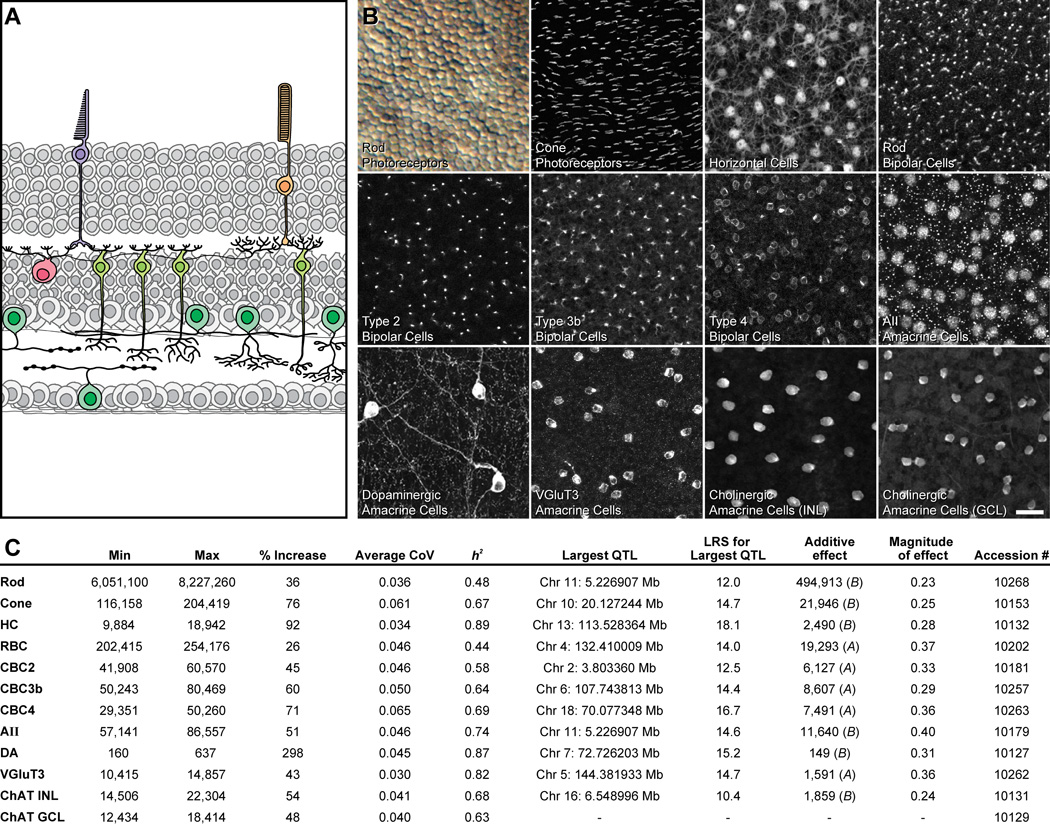
Figure 1. Conspicuous strain variation exists for all cell types, from which large-effect QTL can be mapped.
A: The twelve cell types examined shown in schematic cross-section of the retina. B: Images from wholemount preparations showing those sampled cell types. The calibration bar indicates 5 µm for the rod photoreceptors and 25 µm for the remaining cell types. C: Listed for each cell type is the range (min and max referring to the total number of cells present in the strain with the lowest and highest totals, respectively), the percentage increase from the lowest to the highest strain, and the average coefficient of variation across the strains; the estimated heritable component of that variation (h2); the genomic locus (chromosome and Mb, respectively) and magnitude of the largest QTL detected for each cell type, expressed as the likelihood ratio statistic (LRS) of that QTL, and in cell number (the “additive effect”; A or B indicating the haplotype correlating with an increase in trait values at each locus), and the magnitude of that effect as a proportion of the range observed across the RI strains. Finally, the accession numbers for the data in GeneNetwork (www.genenetwork.org) are also listed. Full details for each strain (including the mean, standard error, CoV and n) are provided in Table S1.
Results
Cell number is precisely specified within a strain
The populations of retinal neurons sampled included rod and cone photoreceptor cells, horizontal cells, four different types of bipolar cell, and five different types of amacrine cell (figure 1A, B). Neuronal number showed low variability within each strain, for every cell type examined (Table S1). The average coefficient of variation (CoV) across all strains for each cell type (figure 1C) ranged from 0.030 (for the VGluT3 amacrine cells) to 0.065 (for the Type 4 cone bipolar cells), and there was no correlation between average CoV and the size of the population across the twelve cell types (r = −0.26; p = 0.41). This modest and consistent level of within-strain variation demonstrates the sampling methods for these cells were reliable despite large variation in the proportion of retinal area sampled: the entire retina was sampled for the dopaminergic amacrine cells (being the sparsest of all cell types sampled), while less than a tenth of one percent of retinal area was sampled in the case of the rod photoreceptors (being the most numerous cell type). This accuracy by which the developing retina specifies neuron number is perhaps most remarkable when one considers the fact that the sizes of these populations differ by four orders of magnitude.
Large variation in cell number is present between strains
Such minimal variance in cell number within a strain is to be contrasted with the conspicuous variation between strains (Table S1). Rod photoreceptor number, for example, was lowest in the A/J strain, being 6,050,000 ± 169,000 cells (mean and s.e.m., and hereafter), climbing to 8,230,000 ± 51,000 cells in the BXA12 RI strain, a 36% increase (figure 3B). By contrast, VGluT3+ amacrine cell number was lowest in the BXA25 RI strain, being 10,400 ± 80 cells, reaching 14,900 ± 220 cells in BXA12, being a 43% increase (figure 3A). Variation across strains was significant and sizable for every cell type (figure 1C), in some cases approaching a two- or three-fold variation. Note as well that, for some cell types, the parental strains showed minimal difference in total number, yet substantial variation was present across the RI strain set (Table S1). We can estimate the magnitude of the genetic contribution controlling cell number by calculating the heritability (h2) for each cell type (Hegmann and Possidente, 1981), being the proportion of the total variance across all mice that can be ascribed to an effect of strain (figure 1C). For some cell types, nearly 90% of that variation among individual mice was due to an effect of genotype (e.g. horizontal cells, dopaminergic amacrine cells), whereas for others, heritability was only about half this value (e.g. rod photoreceptors, rod bipolar cells). Such variation in heritability is largely due to the relative magnitude of the strain differences for these cell types, rather than to any differences in average CoV between the cell types.
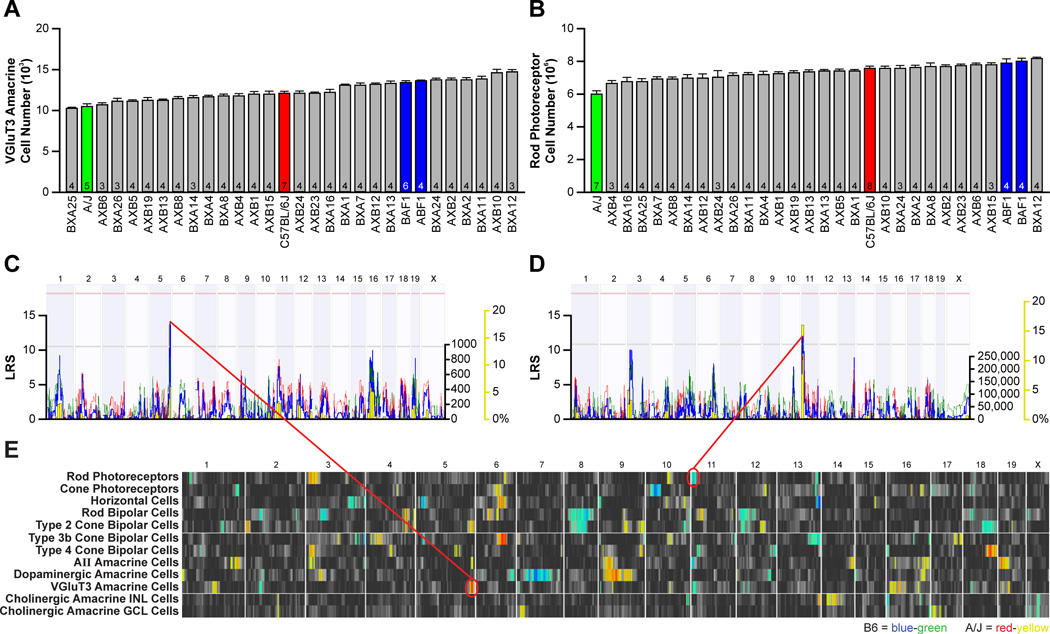
Figure 3. Strain variation for each cell type maps largely to independent genomic loci.
A, B: Variation in VGluT3 amacrine cell number (A) and rod photoreceptor number (B) across the 30 strains. Data represented as mean ± s.e.m. and n = number of retinas analyzed. C, D: The variation in VGluT3 amacrine cell number mapped to a locus on Chr 5 (C), where A alleles correlated with an increase in cell number. Variation in rod photoreceptor number mapped to a locus on Chr 11 (D), where B alleles correlated with an increase in rod number. E: QTL heat-maps derived from LRS scores using marker regression showed multiple QTL for many of the cell types. Blue-green regions indicate loci where the presence of B alleles correlate with an increase in trait values, while red-yellow regions indicate loci where A alleles correlate with an increase in trait values. Large-effect QTL (summarized in Figure 1C) are mapped for all but one cell type, and multiple QTL are often mapped for some cell types, but variation between cell types rarely maps to the same genomic locus.
Minimal co-variation exists between cell types
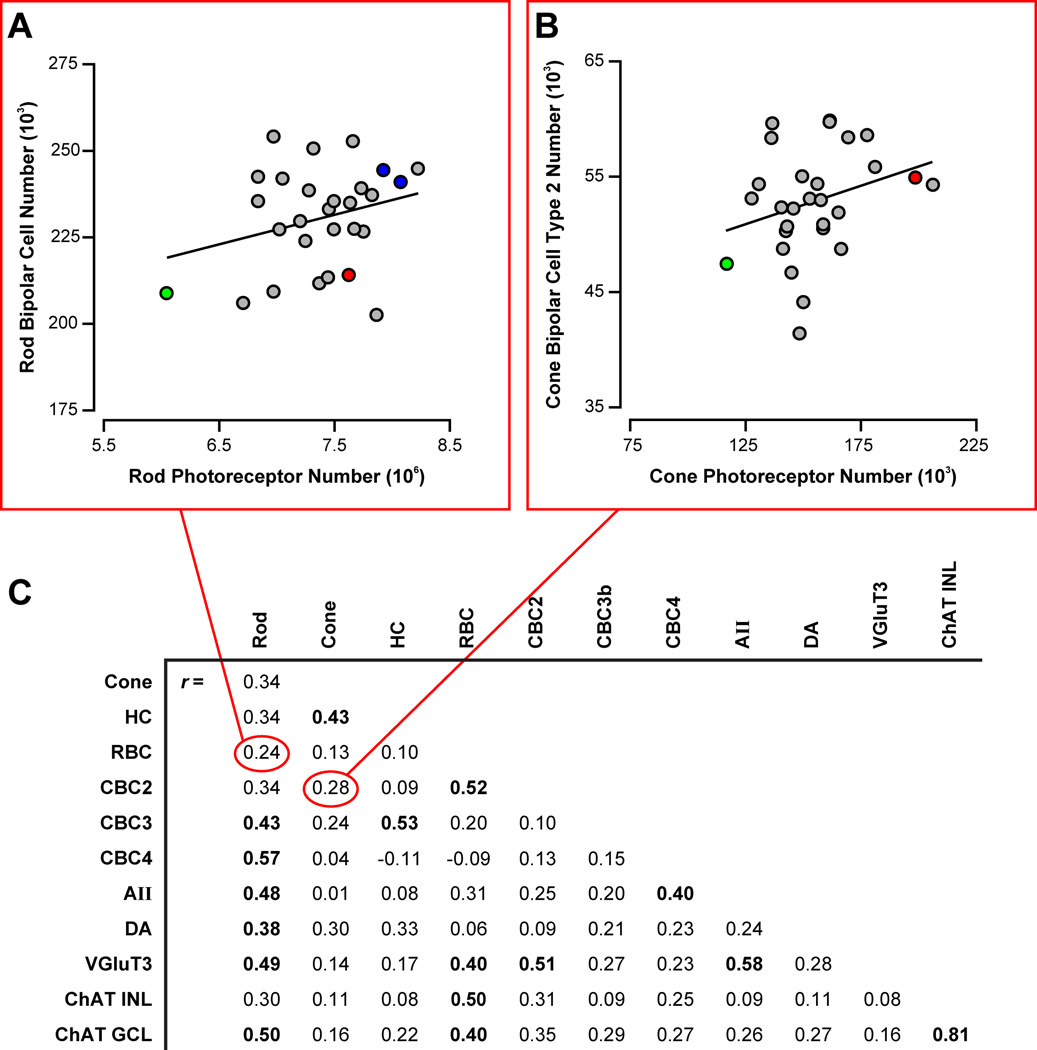
Because of the conspicuous variation in cell number across the strains, we searched for correlations between the different cell types; surprisingly, little co-variation was found between most cell types (figure 2). We looked first at developmentally-related cell types (i.e. sub-types of the major divisions of retinal nerve cell classes), failing to find significant correlations between the two major classes of photoreceptor, between the three types of cone bipolar cell, or between most comparisons of different amacrine cell subtypes. Likewise, synaptically connected cell types defining the radial pathways through the retina showed no significant correlations, between rods and rod bipolar cells (e.g. figure 2A), between rod bipolar cells and AII amacrine cells, or between cones and any of the three cone bipolar cells (e.g. figure 2B). A few cell types did show modest positive correlations (figure 2C), with coefficients around 0.5, for example, between the rod bipolar cells and the Type 2 cone bipolar cells (r = 0.52), or between the rods and the Type 4 cone bipolar cells (r = 0.57). A single exception was found for the two types of cholinergic amacrine cell in the retina, positioned on opposite sides of the inner synaptic layer within the retina and subserving distinct processing of ON versus OFF pathways. These two cell types are not synaptically connected, but are intimately related developmentally, and their numbers are highly correlated across the strains (r = 0.81). Their uniquely high correlation would suggest that none of the other cell types are as tightly related developmentally; indeed, the differentiation of these two cell types is thought to arise secondarily from within the same precursor population during early development (Kim et al., 2000), each being directed to settle within one of the two nuclear layers, rather than being stipulated cell-intrinsically through distinct differentiation pathways, as for instance, is assumed for the different types of cone bipolar cell (Strettoi et al., 2010; Xiang, 2013). They confirm the sufficiency of a dataset of 30 strains for detecting strong correlations, and while greater numbers of strains might elevate some of the lesser correlation coefficients to significance, the fact remains that no other comparison comes close to this single strong positive correlation, and not a one of them suggests a negative correlation.
Figure 2. Minimal co-variation between developmentally- or functionally-related cell types is present.
A, B: No significant correlation was detected between the number of rod (A) or cone (B) photoreceptors and the number of rod or type 2 cone bipolar cells, respectively. F1 strain data were not collected for cone photoreceptors (as indicated in Table S1), and so the F1 strains are excluded in B). C: The correlation matrix for cell number across the twelve cell types reveals only a single strong correlation, between cholinergic amacrine cells in the INL and in the GCL. Those with significant Pearson correlation coefficients (p < 0.05) are emboldened.
Large-effect QTL are identified for nearly every cell type
Variation in cell number mapped to one or more independent loci for nearly every cell type (figure 1C). For example, a quantitative trait locus (QTL) for rod photoreceptor number was identified proximally on Chr 11 at 11.07 Mb (near rs3023249), with two B alleles contributing 495,000 cells, being nearly one-quarter of the range across all strains (figure 3D). Variation in VGluT3-positive amacrine cells, in comparison, mapped a QTL to the distal end of Chr 5 at 145.20 Mb (rs3696754), although in this case, the presence of A alleles at this locus correlated with an increase in cell number, contributing 1,590 cells, or 36% of the difference across the strains (figure 3C). For all but one cell type examined, one or more loci were identified, and the single largest locus for each type accounted for between 20% and 40% of the range observed across the strains (figure 1C).
Variation between the cell types maps to independent genomic loci
Just as very few of the individual cell types co-varied with one another, so too was there minimal evidence for genetic co-regulation of cell number. Each of the QTLs identified appears to modulate primarily that cell type, with little evidence of variation in two or more cell types mapping to the same locus (figure 3E). Some of the modest correlations detected (figure 2C) occasionally mapped to coincident (and lesser) loci, suggesting some limited genetic co-regulation between these cell types (for instance, between rod bipolar cells and Type 2 cone bipolar cells, on Chrs. 4 and 12). Clearly, the variation in cell number identified across cell types cannot be explained by a master variant that modulates proliferation. For example, while retinal area showed slight and heritable variation across the strains that mapped a QTL on Chr 18, this locus did not coincide with any of the loci identified for cell type. A few cell types showed modest correlations with retinal area, the largest being rod photoreceptors (r = 0.53), but the variation in rod number was more substantially dependent upon rod density (r = 0.71). Overall, the variation in retinal cell number arises from genetic variants modulating cell types independent of each other, and independent of retinal area.
Variation in pre- and post-synaptic cell number modulates dendritic differentiation
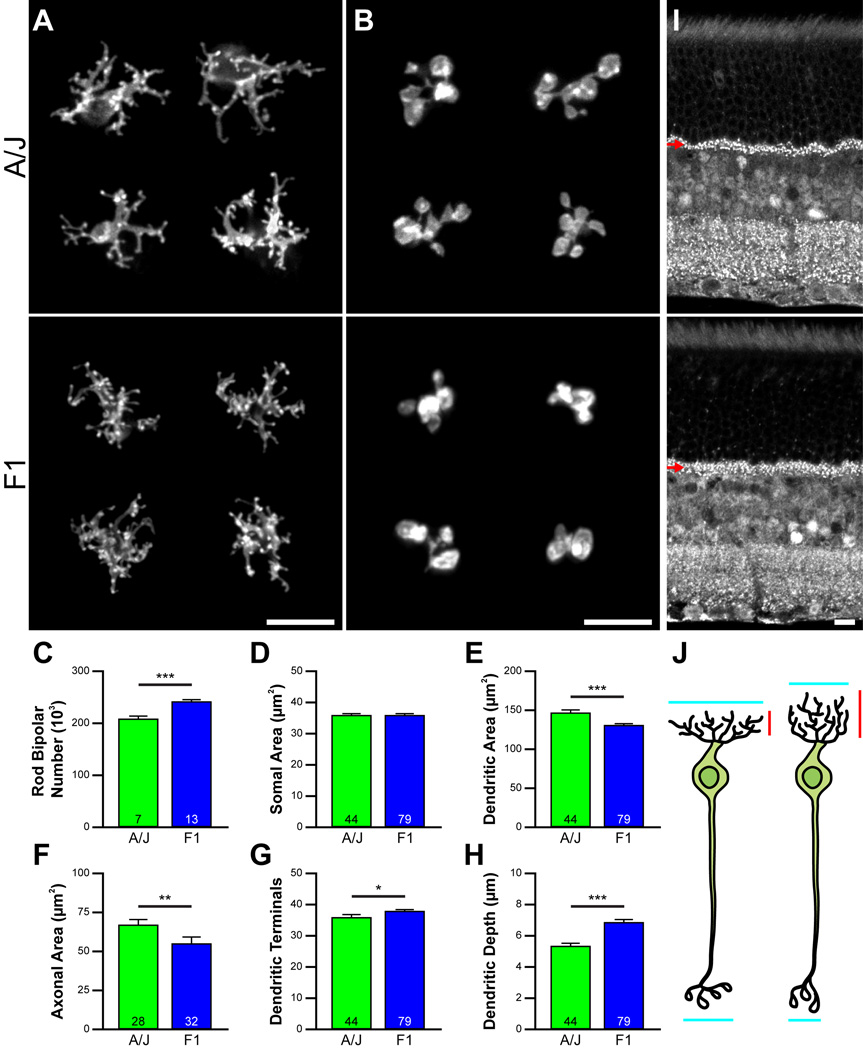
Such variation between the sizes of pre- and post-synaptic populations (e.g. figure 2A) may affect their differentiation. We examined the morphology of the rod bipolar cells in two of the strains of mice, the A/J and F1 strains (figure 4A, B), the latter containing 16% more rod bipolar cells (figure 4C). While soma size was not different between them (figure 4D), the dendritic field areas of the F1 strain were significantly smaller than those in the A/J strain (figure 4E), as were the areas of the axonal arbors (figure 4F). Curiously, the total number of dendritic terminal endings within these narrower F1 dendritic fields was slightly but significantly larger (figure 4G), suggesting the presence of a larger number of rod photoreceptors afferent to these rod bipolar cells, borne out by the estimates of total rod photoreceptor number in these two strains (figure 3B), and consistent with the presence of a thicker outer plexiform layer (OPL) containing a distribution of labeled synaptic ribbons that spanned a greater depth (figure 4I, arrows). The larger numbers of dendritic terminals formed by those rod bipolar cell dendrites in the F1 strain (figure 4G) were likewise distributed across a greater depth of the OPL (figure 4H). Rod bipolar cells, consequently, have their dendritic and axonal arbor growth constrained by the local density of homotypic neighbors, while their dendritic depth and number of terminal endings are driven by the density of afferents within their dendritic field (figure 4J, indicated by the blue and red segments, respectively).
Figure 4. Variation in pre- and post-synaptic cell number modulates dendritic differentiation.
A, B: Rod bipolar cell dendritic and axonal arbors differ in area in A/J and F1 strains (A), the latter strain having 16% more rod bipolar cells than the former strain (C). D-F: Soma areas do not differ (D), but dendritic field area (E) and axonal arbor area (F) are each larger in the A/J strain. G: Despite their smaller dendritic areas, the rod bipolar cells in the F1 strain have significantly more dendritic terminals. H, I: The F1 strain has a substantially larger rod photoreceptor population (figure 3B), distributing their pre-synaptic terminals (labeled with an antibody to CtBP2) across a greater depth of the OPL (I, arrows). The dendritic arbors of their rod bipolar cells are themselves distributed across a greater depth of the OPL (H). J: Local homotypic cell density, therefore, constrains dendritic field extent (blue), while local afferent density drives terminal formation and distribution within that field area (red). Calibration bars indicate 10 µm. Data represented as mean ± s.e.m. and n = number of retinas analyzed for C, number of cells analyzed for D–H. *, **, and *** indicate p < 0.05, 0.01, and 0.001, respectively.
Discussion
We have demonstrated that the number of neurons within a retinal population is precisely specified. This level of specification is comparable across populations showing a thousand-fold variation in their number, regardless of whether they are packed side-by-side, form sparse yet regular mosaics, or approach a random distribution. Cell number is, however, a complex trait controlled by multiple genetic variants, made clear by the graded yet substantial within-class variation in total number across the different strains of mice, for every cell type.
This large variation in cell number present across the RI strains was mapped to genomic loci for all but one of the cell types investigated, but rarely did that variation map to the same chromosomal loci. Variation in cell number, therefore, reflects the actions of multiple variants affecting the final numbers of each cell type largely independent of one another. This lack of genetic co-regulation is manifest in the largely uncorrelated variation between different pairs of neuron types. Hence, while certain cell types are known to share transcriptional regulatory control during their development (Bassett and Wallace, 2012; Ohsawa and Kageyama, 2008), variants in these genes appear not to be major co-determinants of the variation in cell number in maturity.
These results do not, of course, prove the negative, that genomic co-regulation does not contribute to the determination of retinal cell number. First, it remains possible that allelic variants in genes contributing small-to-modest effect size for multiple cell types have gone undetected due to an insufficient fractioning of the genome with only 26 RI strains. And second, there may be allelic variants in genes contributing large and common effects in various cell classes, but these may not differ between the parental genomes examined in the present study. Our data simply indicate that the genetic variation discriminating these two parental strains has a far greater influence on the variation in the numbers of individual cell types, rather than on co-variation between the cell types.
In addition to such lack of co-variation among developmentally-related cell types, synaptically-connected cell types also failed to show significant co-variation in their numbers, consistent with other studies showing a lack of spatial correlation between them (Rockhill et al., 2000). Differentiation programs, however, while directing cell type-specific dendritic patterning and outgrowth, may also equip neurons to modulate their morphogenesis in response to varying homotypic and afferent densities. Such developmental plasticity enables two populations of synaptically-connected neurons, the numbers of which vary widely and largely independent of one another, to conserve a fundamental design feature: a uniform dendritic coverage by the post-synaptic cell to sample the entirety of the population of their afferents. Curiously, this plasticity would appear to be the rule in the OPL (Lee et al., 2011; Reese et al., 2005), whereas within the inner plexiform layer, there are multiple exceptions to it (Farajian et al., 2004; Keeley and Reese, 2010a; Lin et al., 2004).
Independent genetic control between populations has now been shown to be present at three different levels of organization within the central nervous system: first, between interconnected local circuit neurons within a structure (shown here); second, between long-range interconnected populations of projection neurons (Seecharan et al., 2003); and third, between interconnected brain regions (Hager et al., 2012). Collectively, these three levels of independent control argue against strong developmental constraints upon neuron number or brain structure size within species. Natural selection occurs at the behavioral level, and behavior is remarkably well buffered from such enormous variation in cell number through the actions of afferent- and homotypic-dependent plasticity instructing the formation of neuronal circuits (Reese et al., 2011).
Experimental Procedures
Mice
Parental C57BL/6J and A/J strain mice, their reciprocal F1 offspring (B6AF1/J and AB6F1/J), and recombinant inbred (RI) mice of the AXB/BXA strain-set were obtained from the Jackson Laboratory or bred in-house at the Animal Resource Center at UCSB. All RI strain mice were females for the analysis of eleven of the cell types; the cone photoreceptor analysis was the exception, including equal numbers of male and female mice in each strain. All of the parental and F1 mice were of both sex. Adult mice ~1–3 months of age were heavily anesthetized and then perfused intracardially with 0.9% saline followed by 4% paraformaldehyde in 0.1 M sodium phosphate buffer (pH 7.2 at 20°C). Retinas were dissected and prepared as wholemounts as previously described (Whitney et al., 2009), extreme care being taken to maintain the entirety of the retina intact. All procedures were conducted under authorization by the Institutional Animal Care and Use Committee at UCSB, and conform to the AVMA Guidelines on Euthanasia.
Labeling of specific cell types
Whole retinas were labeled using standard immunofluorescence procedures, as previously described (Whitney et al., 2009; 2011b). The two retinas from each mouse were each double-labeled with different primary antibodies raised in distinct species, enabling multiple cell types to be counted from each mouse. The primary antibodies used included: 1) a cocktail of rabbit polyclonal antibodies to red/green and blue cone opsin to label all cone outer segments (each at 1:1,000; Millipore; AB5405 and AB5407); 2) a mouse monoclonal antibody to calbindin D-28 to label horizontal cells (1:10,000; Sigma; C9848); 3) a rabbit polyclonal antibody to Protein Kinase C to label rod bipolar cell axons (1:10,000; Cambio; CA-1042); 4) a mouse monoclonal antibody to synaptotagmin2 to label the Type 2 cone bipolar cell axons (1:100; Zebrafish International Resource Center; ZDB-ATB-081002-25); 5) a mouse monoclonal antibody to Protein Kinase A regulatory subunit IIβ to label Type 3b cone bipolar cell dendritic stalks (1:3,000; BD Biosciences; 610626); 6) a mouse monoclonal antibody to Calsenilin to label Type 4 cone bipolar cells (1:1,000; Millipore; 05–756); 7) an affinity-purified rabbit polyclonal antibody to Prox1 to label AII amacrine cells (1:1,000; Covance; PRB-238C); 8) a mouse monoclonal antibody to tyrosine hydroxylase to label dopaminergic amacrine cells (1:10,000; Sigma; T1299); 9) a guinea pig polyclonal antibody to the vesicular glutamate transporter 3 (VGluT3), to label the population of VGluT3 amacrine cells (1:3,000; Millipore; AB5421); and 10) an affinity-purified goat polyclonal antibody to choline acetyltransferase (ChAT) to label cholinergic amacrine cells in the inner nuclear layer (INL) and in the ganglion cell layer (GCL) (1:50; Millipore; AB144P). Primary antibodies were subsequently detected using donkey host secondary antibodies to rabbit, mouse, guinea pig or goat IgGs conjugated to Cy2 or Cy3 (1:200; Jackson ImmunoResearch Labs), or to Alexa Fluor 488 or Alexa Fluor 546 (1:200; Life Technologies). Examples of labeled specimens are provided in figure 1B.
In order to sample the rod photoreceptor population, the above protocol was modified by further fixation of the flattened retina in glutaraldehyde in order to enhance Nomarski contrast of individual inner segments for subsequent viewing. Retinas were then rinsed and incubated overnight in peanut agglutinin conjugated to Alexa Fluor 488 (PNA-488) (1:200; Life Technologies; L21409) to label the sheaths of the cone outer and inner segments to discriminate them from the rods.
Quantification of cell number
The methodologies and results for the quantification of cone photoreceptors, horizontal cells, and dopaminergic amacrine cells have previously been published (Whitney et al., 2009; 2011a; 2011b). Fields containing rod photoreceptor inner segments were imaged using a 60× oil immersion objective on a Nikon FXA fluorescence photomicroscope and an Olympus DP11 digital camera, each field being imaged twice: first, using differential interference contrast optics to discriminate individual inner segments; and second, under fluorescence to detect PNA-positive profiles (cones). The latter counts were then subtracted from the former to determine rod number per field. Each retina was sampled multiple times in a 1 mm square grid (average of 14 samples per retina), sampling a field size of 225 µm2 at each location.
For all other cell types, each retinal quadrant was sampled, either at a mid-eccentric location, or at both a peripheral and central location. Retinas were sampled using an Olympus microscope equipped with a Sony video camera and linked to a computer running Bioquant Nova Prime (R&M Biometrics), sampling field sizes of approximately 16,000 µm2 for rod bipolar cells and Type 4 bipolar cells; 32,000 µm2 for Type 2 bipolar cells, Type 3b bipolar cells, and AII amacrine cells; 60,000 µm2 for the cholinergic amacrine cells; and 175,000 µm2 for VGluT3 amacrine cells. For the cholinergic amacrine cells, images were collected at two depths in the retina, sampling from the INL and GCL independently.
The outline of each retina was subsequently measured using Bioquant Nova Prime. Retinal area was multiplied by average cell density in order to estimate the total number of cells per retina. Three to four retinas were routinely counted in every RI and F1 strain for each cell type, with three exceptions where we could only obtain an n of two mice at the time of sampling a particular cell type. Parental strains were frequently sampled in larger numbers, averaging around seven retinas for each cell type. For a few cell types, one or two strains were unavailable at the time of sampling for those types, and so the analysis for those cell types was conducted with only 25 or 24 RI strains.
All counting was done by eye (i.e. without automated image-analysis to extract labeled profiles, etc), marking individual profiles on the screen to ensure no double-counting within a field. As a further assurance against bias in the process of counting, particularly for the more densely packed cell types (i.e. the rod photoreceptor population, all four bipolar cell populations, and the AII amacrine cell population), as well as for the VGluT3 population, counting for these cell types was conducted “blind” to strain, with mice of different strains being randomly intermingled.
Calculating heritability
Heritability was estimated by computing the ratio: (0.5 * X) / ((0.5 * X) + Y), where X = Variance(Strain) and Y = Variance(Error). Only the RI strains have been included in the calculation of heritability (Hegmann and Possidente, 1981).
QTL mapping
Quantitative trait locus (QTL) mapping was performed using the mapping module available at GeneNetwork (www.GeneNetwork.org). Only the RI strain data was included in this mapping. Every RI strain has a unique mix of A and B haplotypes across every chromosome (with the exceptions of the Y chromosome and the mitochondrial genome) due to the nearly random recombination events occurring in the course of inbreeding different pairs of F2 siblings (Williams et al., 2001). QTL mapping estimates the genetic control of a trait with the variation of A versus B alleles throughout the genome. GeneNetwork implements simple and composite interval mapping for each trait, and estimates the genome-wide p value of a type I error by permutation. It also generates “heat-maps” for each trait that enable easy comparisons between multiple traits, though in this case using marker regression rather than interval mapping. Those heat-maps identify any locus with a likelihood ratio statistic (LRS) score greater than 0.5 of the suggestive LRS threshold, climbing to a maximum (for each cell type) set by the significant LRS threshold for that cell type (indicated by blue-green or red-orange in figure 3E). This liberal criterion maximizes the likelihood of detecting lesser QTLs that individually might not garner attention, but if reinforced across multiple cell types, should draw further scrutiny. The suggestive (p = 0.67) and significant (p = 0.05) LRS thresholds are computed for each cell type individually, based on 1000 random permutations of the strain data for that cell type (indicated by the horizontal grey and pink lines in figures 3C and D). The suggestive threshold was used as the minimal criterion for defining a mapped QTL in figure 1C. The primary cell type data (means and s.e.m.) have been permanently deposited in GeneNetwork, with their phenotype accession identifiers for the mouse AXB/BXA Published Phenotypes database being indicated in figure 1C.
Morphometric analysis of dendritic morphology
Individual rod bipolar cells in the A/J and F1 strains were injected with DiI to study their morphology. All procedures for labeling and analysis have recently been described (Keeley and Reese, 2010b). Comparisons of rod bipolar cell morphology between these two strains were tested for statistical significance using Student’s t tests.
Supplementary Material
Highlights.
Mouse strains exhibit large variations in retinal cell number
Minimal co-variation in cell number exists between different retinal cell types
Variation between the cell types maps to independent genomic loci
Variation in pre- and post-synaptic cell number modulates dendritic differentiation
Acknowledgements
This research was supported by the National Institutes of Health (EY-019968; RR-022585)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Keeley et al. find that the numbers of different types of retinal neurons vary independently of one another. Strong determinants of variation map to independent loci. Thus, retinal neuron cell-type allocation is not tightly correlated or coregulated. Rather, density-dependent cellular interactions overcome variability to maintain coverage and complete afferent sampling.
References
- Balasubramanian V, Sterling P. Receptive fields and functional architecture in the retina. J Physiol. 2009;587:2753–2767. doi: 10.1113/jphysiol.2009.170704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett EA, Wallace VA. Cell fate determination in the vertebrate retina. Trends in Neuroscience. 2012;35:565–573. doi: 10.1016/j.tins.2012.05.004. [DOI] [PubMed] [Google Scholar]
- Farajian R, Raven MA, Cusato K, Reese BE. Cellular positioning and dendritic field size of cholinergic amacrine cells are impervious to early ablation of neighboring cells in the mouse retina. Visual Neuroscience. 2004;21:13–22. doi: 10.1017/s0952523804041021. [DOI] [PubMed] [Google Scholar]
- Hager R, Lu L, Rosen GD, Williams RW. Genetic architecture supports mosaic brain evolution and independent brain-body size regulation. Nat Commun. 2012;3:1079. doi: 10.1038/ncomms2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegmann JP, Possidente B. Estimating genetic correlations from inbred strains. Behavior Genetics. 1981;11:103–114. doi: 10.1007/BF01065621. [DOI] [PubMed] [Google Scholar]
- Keeley PW, Reese BE. Morphology of dopaminergic amacrine cells in the mouse retina: Independence from homotypic interactions. Journal of Comparative Neurology. 2010a;518:1220–1231. doi: 10.1002/cne.22270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeley PW, Reese BE. Role of afferents in the differentiation of bipolar cells in the mouse retina. J Neurosci. 2010b;30:1677–1685. doi: 10.1523/JNEUROSCI.5153-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I-B, Lee E-J, Kim M-K, Park D-K, Chun M-H. Choline acetyltransferase-immunoreactive neurons in the developing rat retina. Journal of Comparative Neurology. 2000;427:604–616. doi: 10.1002/1096-9861(20001127)427:4<604::aid-cne8>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- Lee SC, Cowgill EJ, Al-Nabulsi A, Quinn EJ, Evans SM, Reese BE. Homotypic regulation of neuronal morphology and connectivity in the mouse retina. J Neurosci. 2011;31:14126–14133. doi: 10.1523/JNEUROSCI.2844-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin B, Wang SW, Masland RH. Retinal ganglion cell type, size, and spacing can be specified independent of homotypic dendritic contacts. Neuron. 2004;43:475–485. doi: 10.1016/j.neuron.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Lui JH, Hansen DV, Kriegstein AR. Development and evolution of the human neocortex. Cell. 2011;146:18–36. doi: 10.1016/j.cell.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsawa R, Kageyama R. Regulation of retinal cell fate specification by multiple transcription factors. Brain Research. 2008;1192:90–98. doi: 10.1016/j.brainres.2007.04.014. [DOI] [PubMed] [Google Scholar]
- Reese BE, Keeley PW, Lee SC, Whitney IE. Developmental plasticity of dendritic morphology and the establishment of coverage and connectivity in the outer retina. Developmental Neurobiology. 2011;71:1273–1285. doi: 10.1002/dneu.20903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese BE, Raven MA, Stagg SB. Afferents and homotypic neighbors regulate horizontal cell morphology, connectivity and retinal coverage. J Neurosci. 2005;25:2167–2175. doi: 10.1523/JNEUROSCI.4876-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockhill RL, Euler T, Masland RH. Spatial order within but not between types of retinal neurons. PNAS. 2000;97:2303–2307. doi: 10.1073/pnas.030413497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seecharan DJ, Kulkarni AL, Lu L, Rosen GD, Williams RW. Genetic control of interconnected neuronal populations in the mouse primary visual system. J Neurosci. 2003;23:11178–11188. doi: 10.1523/JNEUROSCI.23-35-11178.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southwell DG, Paredes MF, Galvao RP, Jones DL, Froemke RC, Sebe JY, Alfaro-Cervello C, Tang Y, Garcia-Verdugo JM, Rubenstein JL, et al. Intrinsically determined cell death of developing cortical interneurons. Nature. 2012;491:109–113. doi: 10.1038/nature11523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strettoi E, Novelli E, Mazzoni F, Barone I, Damiani D. Complexity of retinal cone bipolar cells. Progress in Retinal and Eye Research. 2010;29:272–283. doi: 10.1016/j.preteyeres.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney IE, Raven MA, Ciobanu DC, Poché RA, Ding Q, Elshatory Y, Gan L, Williams RW, Reese BE. Genetic modulation of horizontal cell number in the mouse retina. Proc Nat'l Acad Sci U S A. 2011b;108:9697–9702. doi: 10.1073/pnas.1103253108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney IE, Raven MA, Ciobanu DC, Williams RW, Reese BE. Multiple genes on chromosome 7 regulate dopaminergic amacrine cell number in the mouse retina. Investigative Ophthalmology and Visual Science. 2009;50:1996–2003. doi: 10.1167/iovs.08-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney IE, Raven MA, Lu L, Williams RW, Reese BE. A QTL on chromosome 10 modulates cone photoreceptor number in the mouse retina. Investigative Ophthalmology and Visual Science. 2011a;52:3228–3236. doi: 10.1167/iovs.10-6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RW, Gu J, Qi S, Lu L. The genetic structure of recombinant inbred mice: high-resolution consensus maps for complex trait analysis. Genome Biology. 2001;2 doi: 10.1186/gb-2001-2-11-research0046. research0046.0041-0046.0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang M. Intrinsic control of mammalian retinogenesis. Cell and Molecular Life Sciences. 2013;70:2519–2532. doi: 10.1007/s00018-012-1183-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong M, Finlay BL. What do developmental mapping rules optimize? Progress in Brain Research. 1996;112:351–361. doi: 10.1016/s0079-6123(08)63341-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.