Abstract
The Panicum mosaic virus-like translation enhancer (PTE) functions as a cap-independent translation enhancer (3’CITE) in members of several Tombusviridae genera including 7/19 carmoviruses. For nearly all PTE, a kissing-loop connects the element with a hairpin found in several conserved locations in the genomic RNA (5’ terminal hairpin or ~100 nt from the 5’end) and small subgenomic RNA (~63 nt from the 5’end). Moving the interaction closer to the 5’end in reporter mRNAs using Saguaro cactus virus (SCV) sequences had either a minimal or substantial negative effect on translation. Movement of the kissing loop from position 104 to the SCV 5’ terminal hairpin also reduced translation by 4-fold. These results suggest that relocating the PTE kissing loop closer to the 5’end reduces PTE efficiency, in contrast to results for the Barley yellow dwarf BTE and Tomato bushy stunt virus Y-shaped 3’CITEs , suggesting that different 3’CITEs have different bridging requirements.
Keywords: Cap-independent translation, RNA virus translation, long-distance RNA:RNA interactions, Carmovirus, Saguaro cactus virus
Introduction
Viruses are obligate parasites that rely on their hosts’ translational machinery for production of encoded proteins. In the host, recruitment of the 43S ribosomal subunit complex to the 5’end of most cellular mRNAs requires a 5’ 7mG cap and a 3’ poly-A tail. The 5’ cap is recognized by eukaryotic translation initiation factor eIF4E, which is a part of the eIF4F complex that contains additional translation initiation factors including the core scaffold protein eIF4G. eIF4G interacts with eIF3 to recruit the 43S ribosomal subunit complex to the 5’end. The 43S complex then scans in the 3’ direction until encountering an initiation codon in an optimal context. After 60S subunit joining, translation elongation commences (Aitken and Lorsch, 2012; Pestova et al., 2007). 5’ bound eIF4G also interacts with poly-A binding protein, which when bound to the 3’ poly-A tail circularizes the mRNA, which is thought to synergistically enhance translational efficiency by placing terminating ribosomal subunits near the 5’end for recycling (Wells et al., 1998).
Many plant and animal viruses with RNA genomes lack a 5’ cap and/or a 3’ poly-A tail and have evolved non-canonical mechanisms for both attracting ribosomes and circularizing their genomes to allow for ribosome recycling (Firth and Brierley, 2012; Walsh et al., 2013; Walsh and Mohr, 2011). Some viruses snatch caps from cellular mRNAs (Garcin et al., 1995) while other viruses replace the cap with a viral-encoded protein that becomes covalently attached at the 5’end and directly recruits translation initiation factors such as eIF4E and eIF3 (Daughenbaugh et al., 2003; Goodfellow et al., 2005). Many animal RNA viruses conduct cap-independent translation by attracting ribosomes to extensive, highly structured cis-acting RNA elements known as internal ribosome entry sites (IRES). Viral IRES are located near , or span the initiation codon and recruit required initiation factors or ribosomal subunits directly (Hertz and Thompson, 2011; Martinez-Salas et al., 2012). Plant viruses can also contain 5’ IRES, which for Tobacco etch potyvirus recruits eIF4G within the eIF4F complex as well as the 40S ribosomal subunit for initiation of polyprotein translation (Basso et al., 1994; Gallie, 2001). In the carmovirus Pelargonium flower break virus (PFBV), an internal IRES is proposed to direct translation of the coat protein (CP) from the genomic (g)RNA (Fernández-Miragall and Hernández, 2011). This element may allow for early translation of the CP, which serves as a suppressor of the plant RNA silencing defense system, prior to synthesis of the subgenomic RNA (sgRNA) that serves as the mRNA for the majority of CP production.
Positive-sense plant viruses with no 5’ cap more frequently have critical translation enhancers near or within the 3’UTR of their genomes (Guo et al., 2001; Miller et al., 2007; Simon and Miller, 2013). These cis-acting, cap-independent translation elements (CITEs) have predominantly been found in viruses belonging to the Tombusvirus and Luteovirus families. To date, 7 classes of 3’CITEs have been described in 6 genera of plant viruses, with some classes found in multiple genera (Simon and Miller, 2013). Although 3’CITEs have diverse secondary structures, most recruit translation initiation factors or ribosomes in a structure and/or sequence dependent manner. These bound translation components are then repositioned at the 5’end via long-distance RNA:RNA kissing loop interactions involving hairpins in the 5’UTR or nearby coding region and a hairpin associated with the 3’CITE. Placement of either 3’ bound ribosomes (Gao et al., 2013; Gao et al., 2012) or initiation factors near the 5’end results in ribosome entry at or near the 5’end of the genome, with experimental evidence of ribosome scanning in the 3’ direction to the initiation codon (Guo et al., 2001; Miller et al., 2007; Nicholson et al., 2010; Simon and Miller, 2013).
The “PTE” class of 3’CITEs was originally identified in the panicovirus Panicum mosaic virus (PMV) (Batten et al., 2006) and later in the umbravirus Pea enation mosaic virus (PEMV) (Wang et al., 2009) and seven carmoviruses, including Saguaro cactus virus (SCV) (Chattopadhyay et al., 2011). In PMV and PEMV, the PTE lies fully within the 3’UTR whereas in SCV and several other carmoviruses, the PTE spans the junction between the CP ORF and the 3’UTR (Fig. 1B). Characteristic PTE structural components include a lower supporting stem followed by a guanosine-rich asymmetric loop (G-domain) and a three-way branched structure with two hairpins (H1 and H2; Fig. 1B). In PEMV, a pseudoknot formed between sequences in the G-domain and in the region linking the two hairpins is the binding site for eIF4E; this pseudoknot has been proposed to be a basic feature of PTE (Wang et al., 2011; Wang et al., 2009).
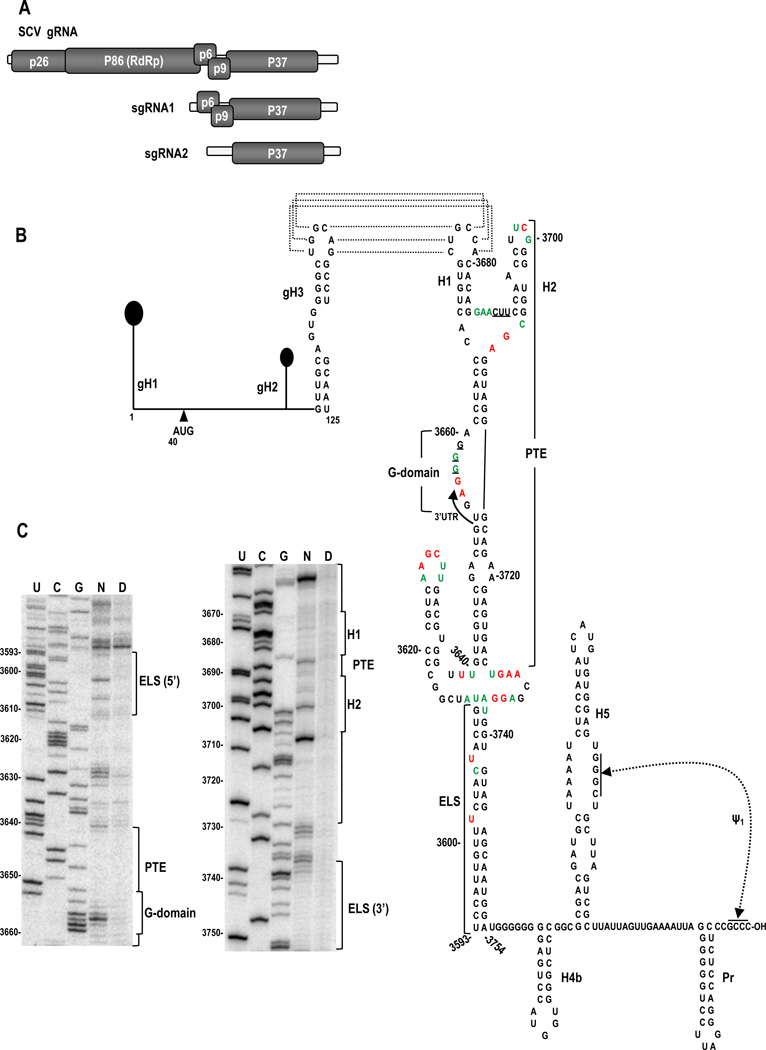
Fig. 1.
Genome organization and structure of the 5’ and 3’ terminal regions of SCV. (A) SCV genome organization. See text for details. (B) Secondary structure of the 5’ and 3’ ends of the SCV genomic RNA. The 5’ 125 nt harbors three predicted hairpins. The initiation codon for p26 is indicated. The 3’ region of SCV contains the PTE, which starts in the CP ORF and terminates in the 3’UTR (arrow). Residues potentially involved in a pseudoknot necessary for eIF4E binding to the PEMV PTE are underlined. 3’UTR elements H4b, H5, Pr and Ψ1 are conserved in most other carmoviruses. The long distance kissing-loop interaction between gH3 and PTE H1 is shown with dotted lines. The 3’ structure shown is one of the mfold-predicted structures for the PTE and surrounding region that was in agreement with the SHAPE-derived flexibility profile. Full length SCV gRNA was modified with NMIA and sites of modified bases were revealed by primer extension of 32P-labelled oligonucleotides following denaturing polyacrylamide gel electrophoresis. Residues in the structure corresponding to high and low reactivity to NMIA are denoted by red and green colors, respectively. ELS, elongated lower stem. (C) Denaturing acrylamide gel showing primer extension products of the PTE and its surrounding region. N, NMIA treated RNA; D, DMSO treated RNA; U, G and C, sequencing ladders generated by dideoxy sequencing of the RNA with the same primer used in modification lane. Positions of the 5’ and 3’ sides of the ELS, and conserved features in PTE are indicated to the right of each gel.
In general, PTE hairpin H1 contains a 4 to 6 base-pair (bp) stem and a 5- to 8-nt apical loop, whereas the 3’ hairpin is heterogeneous in length and structure (Chattopadhyay et al., 2011). With the exception of the PEMV PTE, the apical loops of all PTE H1 have known or putative kissing-loop interacting partners in apical loops of hairpins positioned in several distinctive locations within the genome. At the 5’end of the gRNA, the partner sequence is found in one of two locations: in PMV and three carmoviruses (Carnation mottle virus [CarMV]; Galinsoga mosaic virus [GaMV]; and Hibiscus chlorotic ringspot virus [HCRSV]), the proposed PTE-pairing partner is the apical loop of a 5’ terminal hairpin (gH1), which is a general component of most carmoviruses whether or not they contain a 3’PTE. In contrast, for carmoviruses SCV, PFBV, and Honeysuckle ringspot virus (HnRSV), the PTE-pairing partner is in the apical loop of a hairpin located in very similar positions within the 5’ proximal ORF (Simon and Miller, 2013). The presence of pairing partners for carmoviral PTE at one of two distinctive locations near the 5’end of the gRNA suggests that proper positioning of the long-distance interaction might be needed for appropriate function. The sequences involved in the kissing-loop interactions are also highly conserved (UGCCA/UGGCA), and either sequence can be located in the 5’ or 3’ position (Simon and Miller, 2013).
The PTE-containing carmovirus SCV is a single stranded, positive-sense RNA virus lacking both a 5’ cap and 3’ poly-A tail. The compact 3859 nt SCV gRNA has a 39 nt 5’UTR followed by 4 overlapping ORFs in addition to the 3’ proximal CP ORF (Fig. 1A). The 5’ proximal ORF encodes replication-required protein p26 and ribosomal-readthrough product p86, which is the viral RNA-dependent RNA polymerase. sgRNA1 encodes p6 and p9, which are presumptive movement proteins based on sequence similarities with other carmovirus movement proteins (Weng and Xiong, 1997). The 224 nt 3’UTR terminates with a series of hairpins and one pseudoknot (H4b, H5, Pr and Ψ1) (Fig. 1B, right), with several of these elements (H5, Pr, Ψ1) conserved in all carmoviruses with the exception of GaMV. The SCV PTE, which substantially enhances translation of luciferase reporter constructs that contain its 5’ interacting sequence, is located upstream of these conserved elements, spanning the CP termination codon (Chattopadhay et al 2011). Mutational analyses of a reporter construct assayed in protoplasts and in-line probing RNA structure assays indicated that the apical loop of PTE hairpin H1 participates in an essential, long-distance RNA:RNA interaction with the apical loop of hairpin gH3 located in the p26 ORF (Fig. 1B), and hairpin sgH1 in the 5’UTR of sgRNA2 (see Fig. 5A).
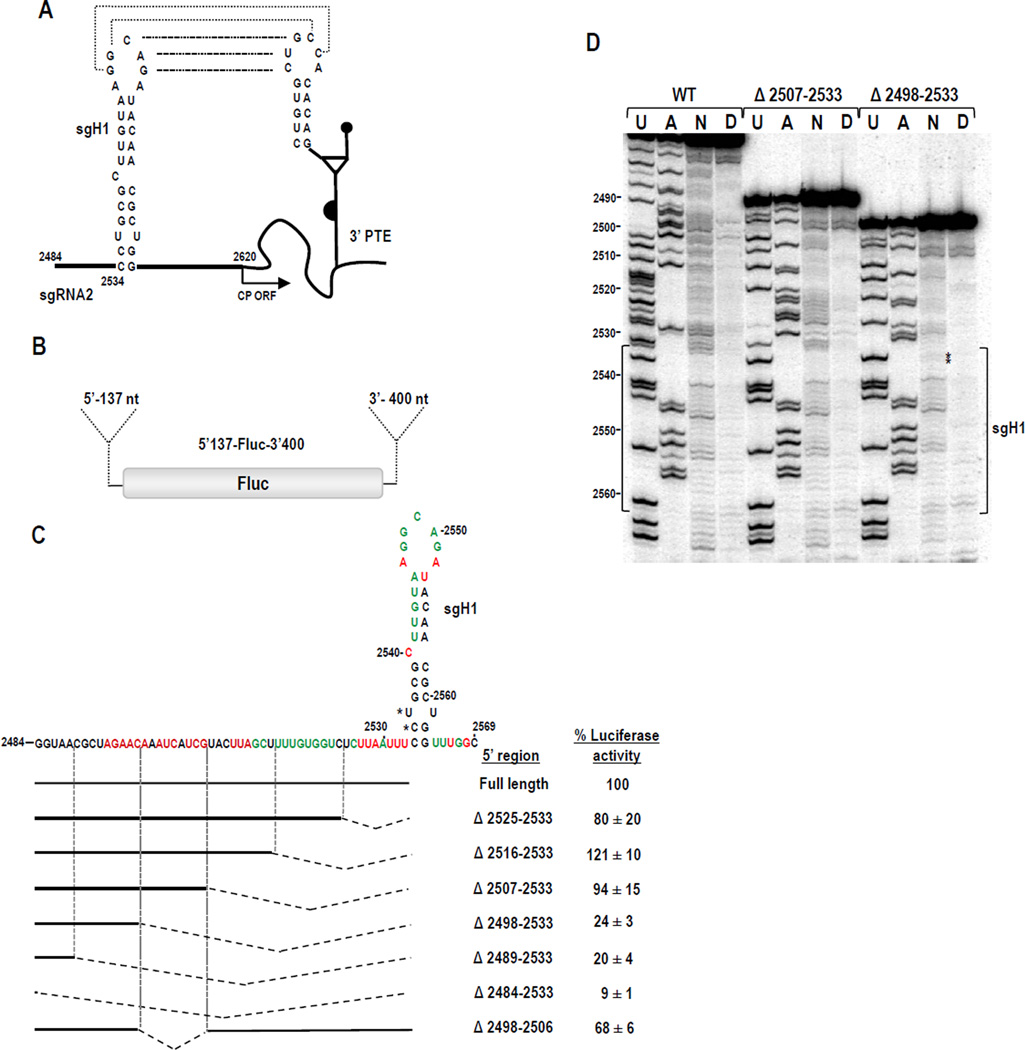
Fig. 5.
Effect of sgH1 location on translation of reporter constructs and sgH1 structure. (A) The long distance kissing-loop interaction between sgH1 and the PTE is shown with dotted lines. Start of the CP ORF is indicated. (B) sgRNA parental reporter construct 5’137-Fluc-3’400 contains Fluc flanked by the 5’UTR (137 nt) of sgRNA2 and the 3’ 400 nt. (C) Effect of deletions on translation of sgH1. Residues of sgH1 with strong or moderate reactivity to NMIA within 5’137-Fluc-3’400 transcripts are denoted by red and green colors, respectively. The position of sgH1 was altered by deleting upstream sequence. Deletions are denoted by hatched lines. Experimental constructs were assayed in Arabidopsis protoplasts co-inoculated with a Renilla luciferase reporter control construct. Levels of luciferase activity are presented as a percentage of the activity of the parental construct. Results are from three independent experiments with standard deviations given. (D) SHAPE autoradiogram comparing sgH1 structure in parental WT transcripts, and transcripts containing deletions Δ2507-2533 or Δ2489-2533. U and A, ladder lanes; N, NMIA; D, DMSO. Position of sgH1 is bracketed. Asterisks denote residues with reproducible, slightly increased flexibility.
In this report, we continue our analysis of the SCV PTE to examine the hypothesis that conserved positioning of PTE-interacting hairpins is important for efficient PTE function as a translation enhancer. Using reporter constructs assayed in vivo, we determined that: 1) relocating 5’ partner hairpins at least 27-nt closer to the 5’end does not substantially reduce function, suggesting that an alternative explanation is responsible for the strict conservation of the kissing loop location; 2) gH1, found at the 5’terminus of most carmoviruses, impacts translation mediated by SCV 5’ sequences; 3) transferring the gH3 loop to gH1 allows for translation but at a significantly reduced level; and 4) movement of sgRNA2 hairpin sgH1 36-nt closer to the 5’end can significantly impact translation despite only minor apparent changes to the structure of the hairpin. These results add to our understanding of the functioning of PTE-type translational enhancers and requirements for 3’CITE kissing-loop interactions.
Results
The SCV PTE is supported by an extended base stem that is conserved in PTE-containing carmoviruses
The structure of the SCV PTE was previously determined using in-line structure probing of a 343-nt fragment that was co-terminal with the 3’end of the gRNA (Chattopadhay et al 2011). This fragment, when located downstream of luciferase in a reporter construct containing the 5’ 125 nt of SCV, was able to enhance translation by 75-fold compared with control constructs. For the current study, we employed SHAPE (selective 2’ hydroxyl acylation analyzed by primer extension) to examine the structure of the PTE within the full-length SCV gRNA. SHAPE interrogates the local backbone flexibility of the RNA by modifying 2’OH groups of unconstrained bases using the acylating agent, NMIA (Wilkinson et al., 2006). The sites of modification are resolved by reverse transcriptase-mediated primer extension, which is impeded by 2’ acylated nucleotides. An RNA structural map is generated in which flexibility of the nucleoside correlates with the level of SHAPE modification.
The SHAPE-derived structure of the PTE in the full-length gRNA (Fig. 1B and C) was consistent with the structure previously defined by in-line probing of the 343 nt fragment (Chattopadhay et al 2011). All residues in the lower stem, including the A-A and G-A non-Watson-Crick pairs, were inflexible using both assays, denoting that a stable, paired lower stem also exists in the PTE within the full-length gRNA. The 5’ side purines in the linker sequence between the two hairpins were flexible in both assays, whereas the adjacent pyrimidines were inflexible. Residues on the 3’ side of the 3-way junction region were also flexible by both procedures. The different techniques and/or different sized templates also revealed differences in residue flexibility in the PTE and surrounding sequences. For example, residues 3655-3658 in the 8-nt G domain were sensitive to NMIA in the full-length gRNA whereas residues 3653-3657 were susceptible to in-line cleavage within the shorter fragment. Furthermore, in the apical loop of PTE hairpin H2, different sets of residues were flexible in the two assays. The most striking difference was in the apical loop of PTE hairpin H1, which participates in the kissing-loop interaction. Five of six loop residues were flexible in the 343-nt fragment, whereas all loop residues were inflexible in gRNA that contains the 5’ partner sequences located in gRNA hairpin gH3 and sgRNA hairpin sgH1 (Fig. 1B and C). This latter result suggests that the PTE is paired with one of its interacting sequences in the full-length gRNA.
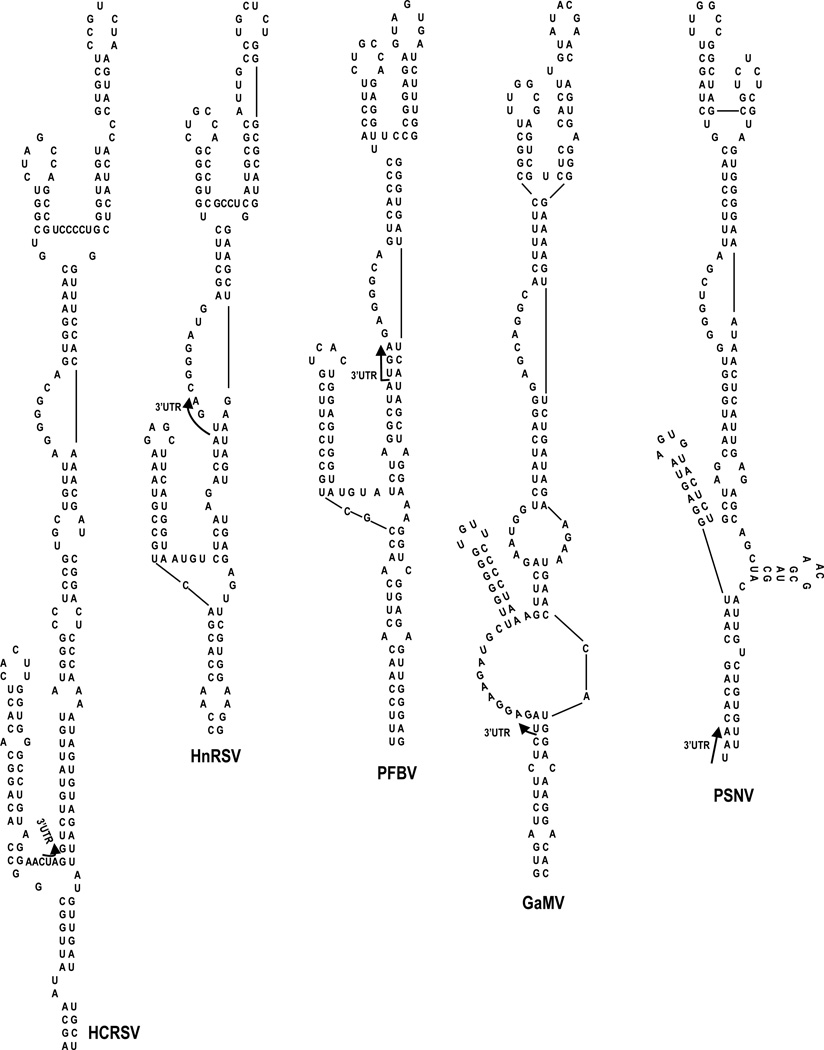
Previous mfold computational predictions (Zuker, 2003) combined with in-line RNA structure probing suggested that the PTE was upstream of a small hairpin and long single-stranded region, followed by conserved carmovirus hairpin H4b (Chattopadhay et al 2011). SHAPE probing of SCV gRNA did not support the predicted structure upstream of H4b. Rather, the SHAPE flexibility profile was consistent with the PTE branching off of a 3-way junction, which also contains a 5’side hairpin and an elongated base stem (ELS) (Fig. 1B and C). Examination of the other six PTE-containing carmoviruses using mfold (Zuker, 2003) revealed that a similar structural configuration was one of the predicted structures for all of these viruses (Fig. 2 and data not shown). It is currently unknown if any of the presumptive conserved elements adjacent to the PTE contribute to translation or other viral function.
Fig. 2.
mfold-predicted secondary structures of the PTE region in other carmoviruses. Phylogenetic analysis revealed that the PTEs of PFBV, HnRSV and GaMV have structural configurations similar to the PTE of SCV. All four PTEs protrude from a 3-way junction containing an elongated lower stem and a short 5’ side hairpin.
Efficient translation of PTE-containing reporter constructs does not require that the 5’ gRNA interacting sequence be located at a specific position
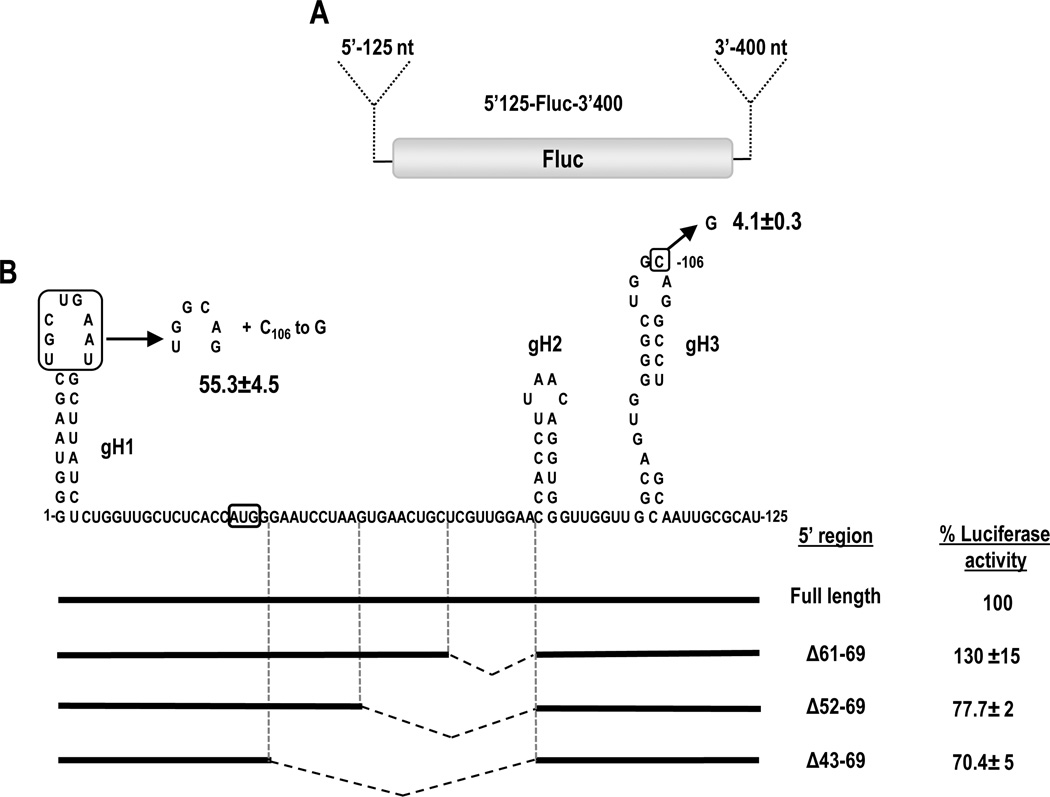
In SCV, PFBV and HnRSV, the sequences at the 5’end of the gRNA that engage in the kissing-loop interaction with the PTE are similarly positioned 100 to 104 nt from the 5’end, which for these viruses lies within their first ORF (Table 1). Pea stem necrosis virus (PSNV), which has an extended 5’UTR compared to other carmoviruses, contains a putative PTE-partner sequence 107 nt from the 5’end within its 5’UTR. To determine if efficient translation enhancement requires specific positioning of the SCV PTE-interacting sequence relative to the 5’end of the gRNA, the location of gH3 was altered in luciferase reporter constructs containing the 5’ 125 nt and 3’ 400-nt of SCV (5’125-Fluc-3’400; Fig. 3A). The 5’ 125 nt of SCV gRNA contains the 39-nt UTR along with the beginning of the p26 coding region, which includes gH3 (Chattopadhyay et al. 2011). Wild-type (WT) parental and mutant constructs were co-inoculated with a control Renilla luciferase (RLuc) construct into Arabidopsis protoplasts, and luciferase activity was measured 18-hr later. Deletion of 9 nt (positions 61-69) upstream of a small hairpin (gH2) adjacent to gH3 resulted in enhanced luciferase activity (130% of WT). Deletions of 18 and 27 nt resulted in only moderate reductions in luciferase activity (to 78% and 70% of WT, respectively) (Fig. 3B). This moderate loss of luciferase activity could be accounted for by repositioning gH3 closer to the 5’ end or could be due to specific deletion of the 27 bases upstream of gH2. These results, however, suggest that gH3 can be repositioned at least 27 nt closer to the 5’end in 5’125-Fluc-3’400 without a substantial loss in luciferase activity.
Table 1.
Location of the PTE 5’ kissing loop sequences in Carmoviruses
| Virus | PTE kissing-loop sequence |
Position of kissing-loop sequence in gRNA 5’ region |
Position of kissing-loop sequence in sgRNA2 relative to 5’ end |
|---|---|---|---|
| SCV | CUGCCA | UGGCAG 104 nt, in ORF |
AGGCAG 64 nt |
| PFBV | CUGCCA | UGGCAG 100 nt, in ORF |
UGGCAG 63 nt |
| HnRSV | CUGCCA | UGGCAG 104 nt, in ORF |
UGGCAG 62 nt* |
| PSNV | UGGC | GCCA 107 nt, in 5’UTR |
GGCCAAC 65 nt |
| GaMV | UUGGCG | CGCCAA 5’ Terminal HP |
CGCCAA ND** |
| CarMV | CUGCCA | UGGCGG 5’ Terminal HP |
GGCAG 64 nt |
| HCRSV | UAGCCA | UGGC 5’ Terminal HP |
GGCUG 64 nt* |
Estimated based on phylogenetic conservation of sgRNA2 start sites shown in Table 2
Start site for sgRNA2 could not be estimated
Fig. 3.
Effect of gH3 location on translation of reporter constructs. (A) Parental reporter construct 5’125-Fluc-3’400 contains firefly luciferase (Fluc) flanked by the 5’ 125 nt and 3’ 400 nt of SCV gRNA. (B) The position of gH3 was altered by deletions as well as replacement of gH1 loop sequence with that of gH3. The later alteration was combined with a single mutation in the gH3 loop (C106 to G) that disrupts the natural kissing-loop interaction. Deletions are denoted by hatched lines. Experimental constructs were assayed in Arabidopsis protoplasts co-inoculated with a control Renilla luciferase reporter construct. Levels of luciferase activity are presented as a percentage of the activity of the parental construct. Results are from three independent experiments with standard deviations given. The initiation codon of the p26 ORF is boxed.
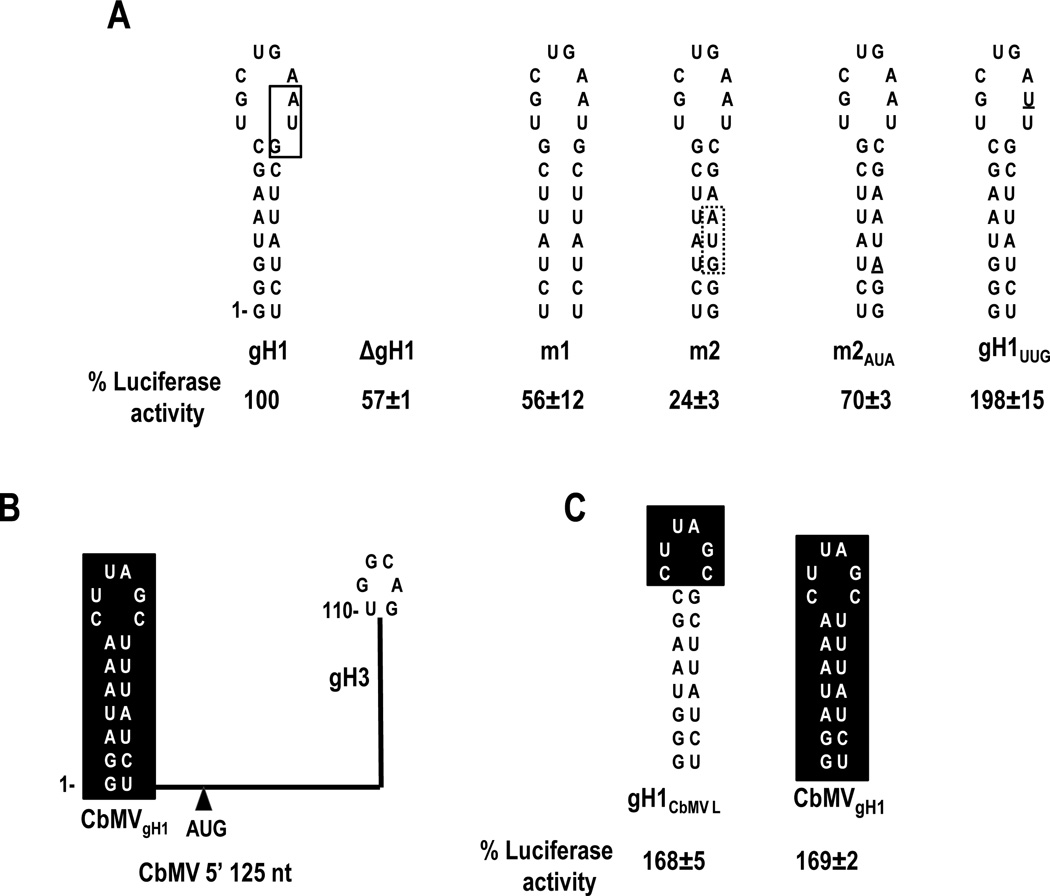
Most carmovirus gRNAs, including SCV, contain 5’ terminal sequences that can fold into a stable hairpin (gH1). In GaMV, HCRSV and Carnation mottle virus (CarMV), the PTE-complementary sequence is located in the apical loop of gH1. To investigate the impact on translation when gH1 of SCV is altered to contain the PTE-interacting sequence, the apical loop of gH1 was replaced with the apical loop of gH3 (Fig. 3B). In addition, a single point mutation (C106 to G) was engineered in the loop of gH3 to disrupt the natural long distance RNA:RNA interaction. This single mutation by itself reduced luciferase activity in protoplasts to 4.1% of WT (Fig. 3B). When the construct also contained gH1 with the apical loop of gH3, luciferase activity improved 13-fold to 55% of WT. This suggests that moving the interacting sequence to the 5’ terminus can support the required RNA:RNA interaction. However translation was reduced by nearly 50% from WT levels, suggesting either that a preference exists for the WT location of the interacting sequence, or that SCV hairpin gH1 plays a role in translation that is disrupted when its apical loop is altered.
gH1 is required for efficient translation of the SCV reporter construct
To determine if gH1 is important for SCV translation, the entire hairpin was deleted, producing construct ΔgH1. Luciferase activity of ΔgH1 was 57% of WT (Fig. 4A), suggesting that the hairpin, or 24-nt in this location, improves translation. To determine the importance of the gH1 stem, the stem was disrupted by replacing the 5’ side sequence with the 3’ side sequence (construct m1). m1 luciferase activity (56% of WT) was similar to that of ΔgH1, suggesting that the hairpin is not acting as a simple spacer element. When compensatory mutations were included in the 3’ side of gH1 to restore the stem (construct m2), luciferase activity decreased to 24% of WT (Fig. 4A). One possibility for why the restored gH1 stem in construct m2 did not re-establish efficient translation was the inadvertent introduction of an out-of-frame AUG codon into the gH1 stem (Fig. 4A). To determine if this upstream AUG reduced translation initiation from the p26 initiation codon, the AUG in the m2 gH1stem was mutated to AUA generating construct m2AUA. Eliminating the AUG in m2 increased luciferase activity to 72% of WT, suggesting that the upstream out-of-frame AUG is interfering with luciferase translation. This prompted an examination of a naturally occurring out-of-frame AUG located in WT gH1 for its effect on translation (Fig. 4A, solid box). When the SCV gH1 AUG was altered to UUG (construct gH1UUG), luciferase activity increased to 198% of WT, suggesting that the gH1 AUG was negatively impacting translation. However, a second possibility was that the alteration in the loop of gH1 affected a function associated with gH1 that was independent of the presence of an initiation codon. To test for this possibility, the apical loop or entire gH1 sequence was replaced with gH1 sequences from carmovirus Calibrachoa mottle virus (CbMV), generating constructs gH1CbMVAL and CbMVgH1, respectively. CbMV is predicted to contain a TED-like 3'CITE that engages in a long-distance kissing-loop interaction with the apical loop of a coding region hairpin located in a similar ORF position as the SCV PTE-interacting hairpin (110 nt from the 5’end; Simon and Miller, 2013). As with SCV, CbMV also contains a 5' terminal gH1 hairpin but without an AUG (Fig. 4B). Our reasoning was that if gH1 or its apical loop are important for translation, then the CbMV gH1 or CbMV gH1 apical loop replacements might provide effective substitutions. gH1CbMVL and CbMVgH1 increased translation to 168% and 169% of WT, respectively (Fig. 4C). These results suggest that CbMV gH1 can functionally replace SCV gH1 in our translation assays, and enhanced translation of constructs containing gH1UUG, gH1CBMVL and CBMVgH1 is due to the absence of an upstream AUG.
Fig. 4.
Mutational analysis of gH1. (A) Alterations were generated in 5’125-Fluc-3’400. A natural AUG located upstream and out-of-frame with the initiator AUG is boxed in a solid line in SCV gH1 (left). An out-of-frame AUG that was introduced into the stem in construct m2 when the 3’ side sequence was replaced with the 5’ side sequence is boxed with a dotted line. This AUG was altered to AUA in construct m2AUA. gH1UUG contains a single base change (underlined) that eliminates the AUG in SCV gH1. Luciferase activity values were determined from three independent experiments and are presented as a percentage of the activity of the parental construct. (B) Schematic representation showing the organization of predicted hairpins in the 5’125 nt of CbMV. CbMV contains a putative I-shaped 3’CITE capable of forming a kissing-loop interaction with its gH3 that is very similar to the SCV kissing-loop interaction (Simon and Miller, 2013). CbMVgH1 does not contain an AUG. (C) Hairpins that were substituted for SCV gH1 in 5’125-Fluc-3’400. Results are from three independent experiments with standard deviations given.
Since deletion of gH1 (construct ΔgH1) or compensatory mutations in the gH1 stem (construct m2AUA) also removed any upstream out-of-frame initiation codons, the level of luciferase activity produced by these constructs should be more accurately compared with that of gH1UUG. Thus ΔgH1 reduced translation to 28% of gH1UUG and m2AUA only restored translation to 36% of gH1UUG. In addition, since transferring the PTE-interacting sequence to gH1 (Fig. 3) also eliminated the gH1 AUG, the level of luciferase acidity obtained by this construct should also be compared with that of gH1UUG. Thus, transferring the RNA:RNA interaction site to the 5’ terminal hairpin produced only 25% of the luciferase activity of gH1UUG. These results together suggest that (1) SCV gH1 plays a role in translation; (2) the gH1 AUG impacts translation from the in-frame AUG in 5’125-Fluc-3’400; and (3) CbMV gH1, which shares structural but no sequence similarity with gH1 of SCV, can functionally replace SCV gH1.
Translation directed by the sgRNA2 5’UTR can be affected by altering the location of the PTE interacting sequence
Carmoviral PTE also engage in a long-distance kissing-loop interaction with hairpin sgH1 located in the central region of their sgRNA2 5’UTR (Chattopadhyay et al. 2011; Fig. 5A). All sgH1 interacting sequences in PTE-containing carmoviruses are found in the apical loops of diverse hairpins located in the center of their sgRNA2 5’UTR, with the interacting sequences occupying nearly identical positions relative to the 5’end (62 to 65 nt downstream) (Table 1). Although the sgRNA2 transcription start sites have not been mapped for two carmoviruses (HnRSV and HCRSV), there is strong phylogenetic conservation of sequences just upstream of carmovirus sgRNA2 start sites (Table 2). In addition, the 5’ terminal sequences of carmovirus sgRNA2 contain a carmovirus consensus sequence (G2-3 A/U3) similar or identical to the sequence found at the 5’end of their gRNA (Guan et al., 2000).
Table 2.
Estimating sgRNA2 start sites for HnRSV and HCRSV
| Virus | Known or putative (*) sgRNA 5’ start site | gRNA 5’ end |
|---|---|---|
| SCV | GGGUAA | |
| CarMV | GGGUAA | |
| PFBV | GGGAUA | |
| PSNV | GGGGAU | |
| TCV | GGUAAU | |
| HnRSV | GGGGUUUU | |
| HCRSV | GGGAAA |
To determine whether the location of sgH1 affects translation directed by the sgRNA2 5’UTR, sgH1 was moved closer to the 5’end by sequentially deleting upstream sequences in a parental luciferase construct containing the complete 5’UTR (137 nt) and 3’ 400 nt (5’137-Fluc-3’400; Fig. 5B). Deletions of 9, 18 or 27 nt (positions 2525-2533, 2516-2533 and 2507-2533) resulted in 80%, 121% and 94% of the WT parental luciferase activity, suggesting that relocating the interacting sequences at least 27 nt closer to the 5’end was not detrimental to translation of the reporter construct in vivo. In contrast, deletion of an additional 9 or 18 nt (positions 2489-2533 and 2498-2533) reduced luciferase activity by 4 to 5-fold. Deleting all upstream sequences (thus placing the kissing-loop hairpin at the 5’ terminus) resulted in a further decrease in translation to 9% of WT luciferase levels (Fig. 5C).
The nearly 4-fold reduction in translation that occurred when constructs contained a 36-nt deletion compared with a 27-nt deletion could reflect either a positional requirement for sgH1 or the loss of a required element or both. To help determine if the decrease in luciferase activity was caused by disruption of a required element, a construct was generated that contained only the 9-nt deletion between positions 2498 and 2506 (Fig. 5C). This 9-nt deletion caused a 32% loss in translation activity. Randomizing the entire sequence between the 5’end and sgH1 resulted in a similar loss of luciferase activity (33%). These results suggest that the 4-fold reduction in translation due to deletion of positions 2498-2533 could be due to a combination of effects: loss of an element that contributes to translation as well as an additional effect caused by the 36-nt deletion.
To determine if the 36-nt deletion caused an alteration in the structure of sgH1 (i.e., the deletion resulted in the loss of exposure of the sgH1 apical loop), RNA transcripts derived from parental construct 5’137-Fluc-3’400, and deletion constructs 5’137Δ27-Fluc-3’400 (deletion of positions 2507-2533) and 5’137Δ36-Fluc-3’400 (deletion of positions 2498-2533) were subjected to SHAPE analysis in the vicinity of sgH1. The pattern of residues susceptible to NMIA in sgH1in the WT luciferase construct (Fig. 5D, left) was mainly consistent with the hairpin structure previously proposed using in-line probing (Chattopadhyay et al., 2011). Residues in the apical loop participating in the kissing-loop interaction were more weakly modified by NMIA than the adjacent loop residues, suggesting that the long-distance interaction was present in a portion of the transcripts. The bulged cytidylate at position 2540 was flexible according to SHAPE, whereas the U-U non-Watson Crick pair near the base of the stem was inflexible, suggesting that this nucleotide pair is forming non-canonical hydrogen bonds. The residues in the upper portion of the 5’ side stem were also weakly modified.
The susceptibility to NMIA of sgH1 residues in RNA transcripts of parental construct 5’137-Fluc-3’400 and 5’137Δ27-Fluc-3’400 were indistinguishable (Fig. 5D). Residue flexibility of sgH1 in 5’137Δ36-Fluc-3’400 was also very similar with two exceptions. Two residues, the bulged uridylate at position 2536 and the adjacent cytidylate at position 2535, showed a reproducible low level of modification by NMIA (Fig. 5D, asterisks). The lack of discernible difference in residue flexibility in the sgH1 apical loop suggests that the kissing-loop interaction with the PTE would still be available. Thus an alternative explanation, such as a positional effect, likely accounts for the reduced translational activity of 5’137Δ36-Fluc-3’400.
Discussion
At least four classes of 3’CITEs (PTE, TSS, ISS, TED-like) have been found associated with viruses in the carmovirus genus, with the PTE being the most common (Simon and Miller, 2013). The best studied PTE is the element associated with umbravirus PEMV (Wang et al., 2011; Wang et al., 2009). However, this PTE is exceptional in not being directly involved in a long-distance interaction. Thus, to better understand how PTE function in general, analyses of more canonical PTE are required. To this end, we examined the conformation of the SCV PTE and surrounding sequences within a full-length gRNA transcript and the consequences of altering the conserved placement of 5’ sequences that interact with the PTE and bridge the 5’ and 3’ ends, thereby circularizing the translated RNAs.
The SHAPE-derived structure of the SCV PTE within full-length gRNA (Fig. 1B) was consistent with the 3-way branched structure previously predicted by in-line structure probing of a shorter 3’ terminal 343-nt fragment (Chattopadhyay et al., 2011) and by SHAPE probing of a short fragment centered on the PTE (Wang et al., 2011). The major difference between the current and previous structures was the inflexibility of the 3H1 apical loop in the full-length structure. This finding strongly suggests that these residues in full-length transcripts are inflexible due to their base-pairing with one of the pairing partners (gH1 and sgH1). In the full-length fragment, the SHAPE flexibility profile of the surrounding sequences was most consistent with the PTE jutting out of a 3-way junction, which also contains a lower stem and short 5’ side hairpin. The 10 lowest free energy mfold -generated structures of the 3’ 800 nt of SCV also contained a similar conformation, although the PTE portion of the structure was not correctly folded (data not shown). Similar structures in this region were predicted for all PTE-containing carmoviruses, suggesting that this RNA conformation is phylogenetically conserved. However, it is not likely that this overall conformation is required for PTE function as the PTE region in panicovirus PMV is not predicted to adopt a similar structure (data not shown). In addition, the PEMV PTE is known to be just downstream of a second 3’CITE, the kl-TSS ribosome-binding translation enhancer, and thus this region in PEMV also does not adopt a similar conformation. However, the PEMV PTE differs from carmovirus PTE in that it does not participate in a long distance interaction, a function associated with the adjacent kl-TSS (Gao et al. 2011).
For several carmovirus gRNAs, the PTE-interacting sequences are found either in the loop of 5’ terminal hairpin gH1 or 100 to 107 nt from the 5’end also in the apical loop of a hairpin. In sgRNA2, the location of the interacting sequence is even more precise: 62 to 65 nt from the 5’end of sgRNA2 in the apical loop of a hairpin (sgH1 in SCV). The PMV 5’ sequence that is complementary with its PTE is also located in loop of a 5’ terminal hairpin (Batten et al., 2006) as are the sequences that form kissing-loops with a number of other 3’CITEs in the Luteoviridae and Tombusviridae (Guo et al., 2001; Miller and White, 2006; Shen and Miller, 2004). Based on the strict positional conservation of 5’ sequences that partner with PTE in the critical long-distance kissing-loop interaction, our original hypothesis was that the position of the 5’ partner sequences relative to the 5’ end of the translated RNA was associated with PTE function as a translation enhancer. However, moving SCV gH3 or sgH1 27 nt closer to the 5’end of their respective reporter transcripts produced either only a modest 30% reduction in luciferase activity (gH3; Fig. 3B), or had a negligible effect (sgH1; Fig. 5C). This suggests that the strict conservation of PTE-interacting sequences may be either (1) an evolutionary remnant; (2) necessary for a function not directly connected with PTE-mediated translation; or (3) necessary for translation of the viral RNA and not the reporter construct. Interestingly, extending the 27-nt deletion to 36 nt by deleting positions 2498 to 2506 reduced translation mediated by the sgRNA2 5’UTR by 4-fold. Deletion of position 2498 to 2506 alone also reduced translation, but by only 32%, suggesting that the 4-fold reduction in translation resulting from the 36-nt deletion was not due simply to alteration of an important element in the region. The 36-nt deletion only slightly affected the structure near the base of sgH1 and did not alter the weak flexibility of the apical loop residues, indicating that the kissing-loop interaction involving these residues was likely still present. The 4-fold decrease in translation that occurred using the construct containing the 36-nt deletion thus may be, at least in part, a consequence of the altered position of sgH1 relative to the 5’end.
When the entire sequence upstream of sgH1 was randomized, translation was reduced by a similar level as with the deletion of position 2498 to 2506, suggesting that an element contributing to translation may reside in that location. The 9-nt sequence contains a portion of a CA-rich sequence (AACAAAUCA) and similar CA-rich motifs are found in 5′ UTRs of both gRNAs and sgRNAs of TCV and tombusvirus Tomato bushy stunt virus (TBSV) and in the 5′ UTRs of CarMV and Cardamine chlorotic fleck virus (CCFV) sgRNA2 (Qu and Morris, 2000). This single-stranded region has been postulated to provide a suitable landing site for ribosome docking with the viral mRNA (Qu and Morris, 2000). SHAPE structure probing of 5’137-Fluc-3’400 transcripts also indicated that this sequence is single-stranded (Fig. 5B).
All carmoviruses, with the exception of the two viruses with ribosome-binding 3’CITEs (TCV and CCFV), have a predicted hairpin (gH1) at the 5’end of their gRNAs. A subset of the PTE-containing carmoviruses have the kissing-loop sequence within the apical loop of this hairpin, as does TGP-carmo isolate 1, which is predicted to have a TED-like 3’CITE (Simon and Miller, 2013). Transposition of the SCV PTE-interacting sequence from its distal location to the loop of gH1 resulted in significantly reduced translation, despite removal of an upstream out-of-frame AUG residing partially within the gH1 loop that reduced translation of the parental reporter construct by ~50% (Fig. 3B). This result suggests that gH1 in at least some carmoviruses may have a function in translation other than simply as a potential scaffold for the long-distance interaction. A separate role for SCV gH1 in translation is suggested by the inefficient translation that resulted when the stem of gH1 was disrupted, which was only partially restored by compensatory mutations and only when an inadvertent potential initiation codon within the stem was mutated. Although this result suggests a sequence-specific requirement for proper gH1 function, CbMV gH1 was able to effectively substitute for SCV gH1, despite having only limited sequence similarity. These results for the PTE differ from results obtained using similar reporter constructs and UTR sequences from Barley yellow dwarf luteovirus (BYDV). The BYDV BTE 3’CITE also engages in a kissing-loop interaction with 5’ sequences, which for the gRNA are located 104 nt from the 5’ end and for sgRNA1, are located in a 5’terminal hairpin. In this system, relocating the gRNA 5’ interacting sequences to a 5’terminal hairpin had only a limited 26% decrease in translation efficiency (Rakotondrafara et al., 2006). A similar study in TBSV, which contains a Y-shaped 3’CITE, also indicated that repositioning the 5’ interacting sequence relative to either the 5’ terminus or the initiation codon in an experimental mRNA did not substantially affect translation efficiency (Fabian and White, 2006).
It has been proposed that the kissing-loop interactions that connect 3’CITEs to the 5’ ends of their translated mRNAs function more efficiently when the 5’ sequences are located proximal to the 5’ end. This proximity is proposed to assist ribosomes in using the translation factor-bound 3’CITEs to dock with the template and finding the 5’ terminus of the mRNA prior to scanning to the initiation codon (Rakotondrafara et al., 2006). This hypothesis was based partially on finding that increasing the distance of the kissing-loop interacting sequence relative to the 5’ end in BYDV reduced translation efficiency. Our results for the SCV PTE kissing-loop interaction do not currently support this hypothesis. With one exception (9-nt deletion in the gRNA), all deletions that repositioned the kissing-loop sequence closer to the 5’ end had either a limited effect on translation or a strong negative effect on translation. In addition, sgRNA2 in PTE-containing carmoviruses must express the CP at high levels despite having the interacting sequence at least 63-nt distal to the 5’end. Movement of sgRNA2 sgH1 to the 5’end by deleting intervening sequences (which when randomized only reduced translation by 33%) reduced translation by over 10-fold (Fig. 5C). Furthermore carmoviruses that have the gRNA kissing-loop sequence at the 5’ end in gH1 or at least 100-nt distal to the 5’ end in a downstream hairpin likely require synthesis of comparable levels of their replication proteins. It seems likely that additional factors also dictate the evolutionary placement of the 5’ kissing-loop interacting sequences for 3’CITEs.
Material and methods
Generation of constructs
Previously described single luciferase reporter constructs 5’125-Fluc-3’400 and 5’137-Fluc-3’400 were the parental constructs used for the alterations generated (Chattopadhyay et al. 2011). The 5’ end of the reporter constructs contain either the 5’ 125 nt of the SCV gRNA or the 5’ UTR of SCV sgRNA2 (137 nt) upstream of the firefly luciferase (Fluc) gene. The 3’ end contains the 3’ 400 nt of SCV. Oligonucleotide-mediated site-directed mutagenesis was used to generate specific mutations in the 5’ 125 nt and the 5’ 137 nt regions (oligonucleotide sequences available upon request). All mutations were verified by sequencing. Plasmids were linearized with SspI and used as templates for in vitro transcription using T7 RNA polymerase.
In vivo translation in protoplasts
Protoplasts were generated from callus cultures of Arabidopsis thaliana ecotype col-0, using cellulase and pectinase to remove the cell wall as previously described (Zhang et al., 2006). Uncapped, in vitro transcribed RNA (30 µg) was inoculated into 5×106 protoplasts using 50% poly-ethylene glycol (PEG) along with 10 µg of uncapped in vitro transcripts containing a Renilla luciferase (RLuc) reporter as an internal control. Protoplasts were harvested at 18 hpi, cells were lysed using 1x passive lysis buffer (Promega) and luciferase activity was measured using a Modulus™ Microplate Multimode Reader (Turner Biosystem).
SHAPE structure probing
Full-length SCV gRNA transcripts were used as templates to assay RNA structure in the PTE and surrounding region. Transcripts synthesized from 5’137-Fluc-3’400, 5’137Δ27-Fluc-3’400 and 5’137Δ36-Fluc-3’400 were used to assay the structure of sgH1. Six pmoles of in vitro transcribed RNAs were heated at 65°C for 5 min and snap-cooled on ice for 2 min. Transcripts were folded in SHAPE Folding Buffer-2 (80 mM Tris-HCl pH 8, 11 mM Mg(CH3COO)2, 160 mM NH4Cl) for 20 min at 37°C. Three pmoles of the folded RNA was combined with either N-methylisatoic anhydride (NMIA) or DMSO at a final concentration of 15 mM. RNA reaction mixtures were incubated for 35 min at 37°C (5 half-lives of NMIA) followed by phenol extraction and ethanol precipitation. Oligonucleotides were 5’end radiolabeled using γ-[32P]-ATP and polynucleotide kinase and primer extension reactions performed using Superscript III reverse transcriptase (Invitrogen) as previously described (Wilkinson et al., 2006). For SCV gRNAs, oligonucletides used were complementary to positions 3826-3851 and 3715-3738. For 5’137-Fluc-3’400 and derivatives, the oligonucleotide (5’-GTTTTTGGCGTCTTCCATGAGC-3’) was complementary to sequence in the Fluc coding region. RNA products of the reverse transcription reactions were re-suspended in 8 µl of 0.5x TE buffer and subjected to electrophoresis through 8% denaturing polyacrylamide gels and radioactive bands visualized using a phosphorimager. Sequencing ladders were generated by Sanger sequencing methods using unmodified RNA (2 pmoles), Superscript III reverse transcriptase, and the same 5’-end labeled oligonucleotides as used for the modified RNA. RNA secondary structural maps were generated from structure probing results and best fitted mfold predictions.
Research highlights.
Proximity to the 5’end of the kissing-loop interaction can negatively impact translation of SCV
Phylogenetically conserved 5’ terminal hairpin in carmoviruses is involved in translation.
Natural upstream out-of-frame AUG in SCV genomic RNA significantly reduces translation of a reporter mRNA.
Acknowledgements
This work was supported by NSF (MCB 1157906) to AES. MC and MK were supported by NIH Institutional Training Grant 2T32AI051967-06A1. We are grateful to HHMI for supporting KK and undergraduate researchers at the University of Maryland.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aitken CE, Lorsch JR. A mechanistic overview of translation initiation in eukaryotes. Nat Struct Mol Biol. 2012;19:568–576. doi: 10.1038/nsmb.2303. [DOI] [PubMed] [Google Scholar]
- Basso J, Dallaire P, Charest PJ, Devantier Y, Laliberte JF. Evidence for an internal ribosome entry site within the 5' non-translated region of Turnip mosaic potyvirus RNA. J Gen Virol. 1994;75:3157–3165. doi: 10.1099/0022-1317-75-11-3157. [DOI] [PubMed] [Google Scholar]
- Batten JS, Desvoyes B, Yamamura Y, Scholthof KBG. A translational enhancer element on the 3'-proximal end of the Panicum mosaic virus genome. FEBS Lett. 2006;580:2591–2597. doi: 10.1016/j.febslet.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay M, Shi K, Yuan X, Simon AE. Long-distance kissing loop interactions between a 3' proximal Y-shaped structure and apical loops of 5' hairpins enhance translation of Saguaro cactus virus. Virology. 2011;417:113–125. doi: 10.1016/j.virol.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daughenbaugh KF, Fraser CS, Hershey JWB, Hardy ME. The genome-linked protein VPg of the Norwalk virus binds eIF3, suggesting its role in translation initiation complex recruitment. EMBO J. 2003;22:2852–2859. doi: 10.1093/emboj/cdg251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR, White KA. Analysis of a 3'-translation enhancer in a tombusvirus: a dynamic model for RNA-RNA interactions of mRNA termini. RNA. 2006;12:1304–1314. doi: 10.1261/rna.69506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Miragall O, Hernández C. An internal ribosome entry site directs translation of the 3'-gene from Pelargonium flower break virus genomic RNA: implications for infectivity. PLoS One. 2011;6:e22617. doi: 10.1371/journal.pone.0022617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth AE, Brierley I. Non-canonical translation in RNA viruses. J Gen Virol. 2012;93:1385–1409. doi: 10.1099/vir.0.042499-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie DR. Cap-independent translation conferred by the 5 ' leader of tobacco etch virus is eukaryotic initiation factor 4G dependent. J Virol. 2001;75:12141–12152. doi: 10.1128/JVI.75.24.12141-12152.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Gulay SP, Kasprzak W, Dinman JD, Shapiro BA, Simon AE. The Kissing-Loop T-Shaped Structure Translational Enhancer of Pea Enation Mosaic Virus Can Bind Simultaneously to Ribosomes and a 5 ' Proximal Hairpin. J Virol. 2013;87:11987–12002. doi: 10.1128/JVI.02005-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Kasprzak W, Stupina VA, Shapiro BA, Simon AE. A ribosome-binding, 3' translational enhancer has a T-shaped structure and engages in a long-distance RNA-RNA interaction. J Virol. 2012;86:9828–9842. doi: 10.1128/JVI.00677-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcin D, Lezzi M, Dobbs M, Elliott RM, Schmaljohn C, Kang CY, Kolakofsky D. The 5'ends of Hantaan virus (Bunyaviridae) RNAs suggest a primer-and-realign mechanism for the intiation of RNA-synthesis. J Virol. 1995;69:5754–5762. doi: 10.1128/jvi.69.9.5754-5762.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodfellow I, Chaudhry Y, Gioldasi I, Gerondopoulos A, Natoni A, Labrie L, Laliberte JF, Roberts L. Calicivirus translation initiation requires an interaction between VPg and eIF4E. EMBO Rep. 2005;6:968–972. doi: 10.1038/sj.embor.7400510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan HC, Carpenter CD, Simon AE. Analysis of cis-acting sequences involved in plus-strand synthesis of a turnip crinkle virus-associated satellite RNA identifies a new carmovirus replication element. Virology. 2000;268:345–354. doi: 10.1006/viro.1999.0153. [DOI] [PubMed] [Google Scholar]
- Guo L, Allen EM, Miller WA. Base-pairing between untranslated regions facilitates translation of uncapped, nonpolyadenylated viral RNA. Mol Cell. 2001;7:1103–1109. doi: 10.1016/s1097-2765(01)00252-0. [DOI] [PubMed] [Google Scholar]
- Hertz MI, Thompson SR. Mechanism of translation initiation by Dicistroviridae IGR IRESs. Virology. 2011;411:355–361. doi: 10.1016/j.virol.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Salas E, Pineiro D, Fernandez N. Alternative Mechanisms to Initiate Translation in Eukaryotic mRNAs. Comp. Funct. Genomics. 2012;2012 doi: 10.1155/2012/391546. Article ID 391546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller WA, Wang Z, Treder K. The amazing diversity of cap-independent translation elements in the 3'-untranslated regions of plant viral RNAs. Biochem Soc Trans. 2007;35:1629–1633. doi: 10.1042/BST0351629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller WA, White KA. Long-distance RNA-RNA interactions in plant virus gene expression and replication. Ann Rev Phytopath. 2006;44:447–467. doi: 10.1146/annurev.phyto.44.070505.143353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson BL, Wu B, Chevtchenko I, White KA. Tombusvirus recruitment of host translational machinery via the 3' UTR. RNA. 2010;16:1402–1419. doi: 10.1261/rna.2135210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Lorsch JR, Hellen CUT. The mechanism of translation initiation in eukaryotes. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2007. pp. 87–128. [Google Scholar]
- Qu F, Morris TJ. Cap-independent translational enhancement of turnip crinkle virus genomic and subgenomic RNAs. J. Virol. 2000;74:1085–1093. doi: 10.1128/jvi.74.3.1085-1093.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakotondrafara AM, Polacek C, Harris E, Miller WA. Oscillating kissing stem-loop interactions mediate 5' scanning-dependent translation by a viral 3'-cap-independent translation element. RNA. 2006;12:1893–1906. doi: 10.1261/rna.115606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen RZ, Miller WA. The 3' untranslated region of tobacco necrosis virus RNA contains a barley yellow dwarf virus-like cap-independent translation element. J Virol. 2004;78:4655–4664. doi: 10.1128/JVI.78.9.4655-4664.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon AE, Miller WA. 3' cap-independent translation enhancers of plant viruses. Annu Rev Microbiol. 2013;67:21–42. doi: 10.1146/annurev-micro-092412-155609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh D, Mathews MB, Mohr I. Tinkering with Translation: Protein Synthesis in Virus-Infected Cells. Cold Spring Harb Perspect Biology. 2013;5:a012351. doi: 10.1101/cshperspect.a012351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh D, Mohr I. Viral subversion of the host protein synthesis machinery. Nat Rev Microbiol. 2011;9:860–875. doi: 10.1038/nrmicro2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Parisien M, Scheets K, Miller WA. The cap-binding translation initiation factor, eIF4E, binds a pseudoknot in a viral cap-independent translation element. Structure. 2011;19:868–880. doi: 10.1016/j.str.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Treder K, Miller WA. Structure of a viral cap-independent translation element that functions via high affinity binding to the eIF4E subunit of eIF4F. J Biol Chem. 2009;284:14189–14202. doi: 10.1074/jbc.M808841200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells SE, Hillner PE, Vale RD, Sachs AB. Circularization of mRNA by eukaryotic translation initiation factors. Mol Cell. 1998;2:135–140. doi: 10.1016/s1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]
- Weng ZM, Xiong ZG. Genome organization and gene expression of saguaro cactus carmovirus. J Gen Virol. 1997;78:525–534. doi: 10.1099/0022-1317-78-3-525. [DOI] [PubMed] [Google Scholar]
- Wilkinson KA, Merino EJ, Weeks KM. Selective 2'-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat Protoc. 2006;1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhang G, McCormack JC, Simon AE. Evolution of virus-derived sequences for high-level replication of a subviral RNA. Virology. 2006;351:476–488. doi: 10.1016/j.virol.2006.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucl. Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]