Abstract
Cyanobacteria are the only prokaryotes performing oxygenic photosynthesis. Non-diazotrophic strains such as the model Synechocystis sp. PCC 6803 depend on a balanced uptake and assimilation of inorganic carbon and nitrogen sources. The internal C/N ratio is sensed via the PII protein (GlnB). We analyzed metabolic changes of the ΔglnB mutant of Synechocystis sp. PCC 6803 under different CO2 availability. The identified metabolites provided a snapshot of the central C/N metabolism. Cells of the ΔglnB mutant shifted to carbon-limiting conditions, i.e. a decreased C/N ratio, showed changes in intermediates of the sugar storage and particularly of the tricarboxylic acid cycle, arginine, and glutamate metabolism. The changes of the metabolome support the notion that the PII protein is primarily regulating the N-metabolism whereas the changes in C-metabolism are probably secondary effects of the PII deletion.
Keywords: Inorganic carbon, PII-mutant, Primary metabolism, Metabolic fingerprinting, Synechocystis
1. Introduction
Cyanobacteria are the only oxygenic phototrophs among prokaryotes. During the photosynthetic carbon assimilation they convert inorganic carbon (Ci) into organic molecules [1]. CO2 is used as main carbon source and fixed via ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) in the Calvin-Benson cycle, whereas bicarbonate can additionally be incorporated into oxaloacetate via phosphoenolpyruvate (PEP) carboxylase. Not only Ci but also appropriate inorganic nitrogen sources are necessary for the production of cyanobacterial biomass. The variety of possible N-sources is much wider than that of C-sources. Many cyanobacterial strains are able to fix atmospheric N2. These diazotrophic strains are independent from external, combined nitrogen sources. However, even these strains prefer the utilization of NH4+ or NO3− as energetically cheaper inorganic N-sources over N2-fixation. Thus, NH4+ or NO3− can be used by almost all cyanobacterial strains to synthesize amino acids, mainly glutamate via the glutamine synthetase/glutamine 2-oxoglutarate (2OG) aminotransferase (GS/GOGAT) pathway. Many cyanobacterial strains also are able to use alternative organic N-compounds such as urea, arginine and some others [2].
In the natural environment, the amount and nature of C- as well as N-sources shows great variability. The acclimation toward these changes is well understood for unicellular, non-diazotrophic cyanobacterial model strains such as Synechocystis sp. PCC 6803 (hereafter Synechocystis). To cope with low and varying Ci amounts, Synechocystis as well as all other extent cyanobacteria evolved an efficient Ci concentrating mechanism (CCM) that combines active transporters for Ci and the compartmentalization of Rubisco together with carbonic anhydrase into the bacterial compartment carboxysome. This mechanism allows the accumulation of bicarbonate inside the cell and its conversion into CO2 near Rubisco inside the carboxysome [3,4]. Many genes coding for CCM components are up-regulated under Ci-limiting conditions [5]. Two transcriptional factors, CmpR [6] and NdhR (CcmR) [7,8], are known to be involved in this process. Recently, it has been shown that the promoter binding of these LysR-type transcriptional factors is modified by the association of specific metabolites such as 2-phosphoglycolate (2PG) and ribulose-1,5-bisphosphate in the case of the activator protein CmpR [9] or 2OG and NADP+ in the case of the repressor protein NdhR [10]. The altered gene expression results in an increased Ci affinity and a coordinated change in the metabolome [11,12]. The acclimation to low nitrogen availability also includes the activation of multiple transporters for combined N-sources as well as enzymes involved in specific routes of the N-assimilation. The transcriptional factor NtcA has been shown to be responsible for the coordinated regulation of corresponding low-N-responsive genes [2,13]. The promoter-binding activity of the activator protein NtcA also is stimulated by the association with the low molecular co-activator molecule 2OG [14,15,16].
However, the response toward different C- and N-levels must be coordinated to obtain a balance C/N ratio. The metabolite 2OG is believed to act as the central sensing molecule for the cellular C/N status, because 2OG is the direct precursor for the GS/GOGAT pathway, thus it connects C- and N-metabolism in cyanobacteria (see Figure 1). In addition to the 2OG-mediated regulation of the transcriptional factors NdhR and NtcA, 2OG also affects the structure and activity of another regulator protein, called PII [17]. PII proteins are found in all three kingdoms of life. In most cyanobacteria including Synechocystis it is encoded by the glnB gene and forms homotrimeric proteins, which beside 2OG can bind ADP or ATP. Furthermore, cyanobacterial PII proteins can be phosphorylated at a defined serine residue depending on the 2OG or ADP/ATP binding to the PII trimer [18,19]. Thus, it is believed that PII proteins monitor and regulate especially the N-metabolism depending on the C/N ratio and the energy status of the cell. This regulation is realized via the binding of PII to different effector proteins such as the key enzyme for arginine biosynthesis or the transporters for inorganic N-sources [17]. Of crucial importance is the control of PII over the transcriptional regulator NtcA that is realized via the PipX protein. The PipX protein is released from the PII-2OG complex formed under low N/high C conditions. This release causes the activation of NtcA due to the binding of NtcA to the co-activator molecules 2OG and PipX. This transcriptional active complex exists under N-limiting conditions and guarantees the activation of N-assimilatory genes. Under N-replete conditions, 2OG concentrations are low and PipX favors binding to PII rather than to NtcA. Thus, transcriptional activation by NtcA is abolished in the presence of sufficient N [20,21,22].
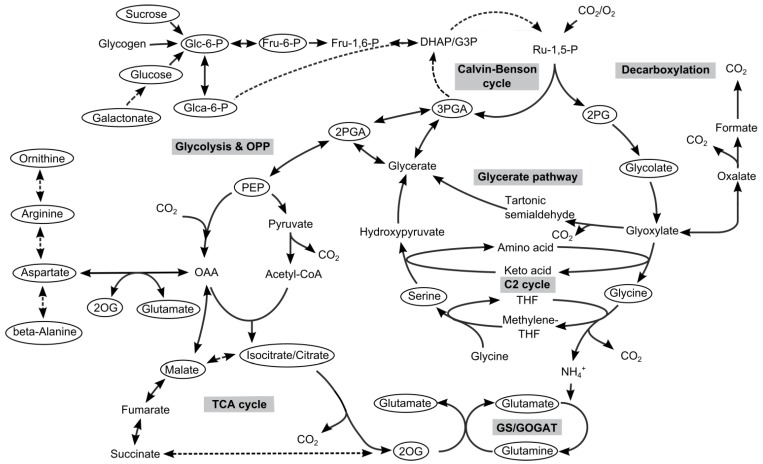
Figure 1.
Primary C- and N-metabolism of cyanobacteria. Metabolites highlighted by circles were determined in our metabolome profiling analysis.
Abbreviations: Glc-6-P, glucose-6-phosphate; Fru-6-P, fructose-6-phosphate; Glca-6-P, gluconate-6-phosphate; DHAP, dihydroxyacetone phosphate; G3P, glyceraldehyde-3-phosphate; 3PGA, 3-phosphoglycerate; 2PGA 2-phosphoglycerate; PEP, phosphoenolpyruvate, 2PG, 2-phosphoglycolate; 2OG, 2-oxoglutarate; OAA, oxaloacetate.
Previously, we showed that mutation of glnB in another model cyanobacterium Synechococcus elongatus PCC 7942 had only minor impact on the metabolome and mostly affected the down-regulation of N-assimilatory genes under Ci-limiting conditions (increase N/C ratio), whereas the up-regulation of Ci-specific genes was mostly non-affected [23]. Here we analyzed how the absence of PII affects the changes in the metabolome under varying Ci-levels in cells of Synechocystis.
2. Results and Discussion
2.1. Physiological Characterization
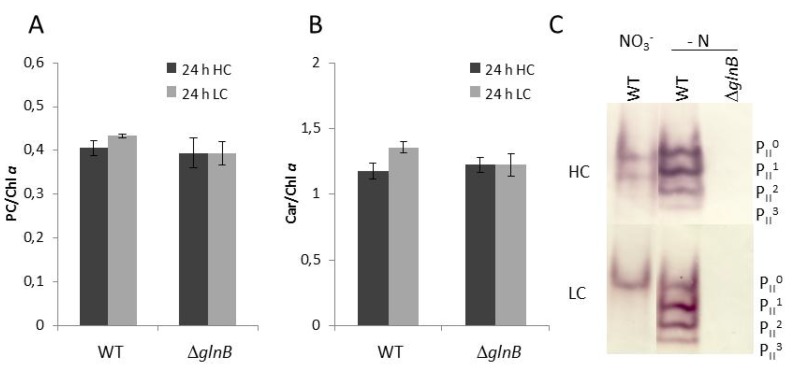
Under high CO2 conditions (HC, 5% CO2), the growth of the mutant ΔglnB was about two times slower than that of the wild type (WT) (data not shown). The transfer to low CO2 conditions (LC, 0.038% CO2) diminished the growth of the mutant and that of the WT to about the same extent. Our observation of decreased growth of the Synechocystis PII mutant at HC as well as LC corresponds to the previously published growth data of this mutant [24]. The PII mutant of Anabaena sp. PCC 7120 also showed only 50% of the growth rate of WT cells regardless if the cells were grown with nitrate or under N2-fixing conditions [25]. Moreover, both strains showed differences in color, i.e. the pigmentation of the Synechocystis PII mutant looked more yellowish under HC conditions. This different appearance is due to decreased phycocyanin content (phycocyanin in HC cells of WT 1.85 ± 0.08 and PII mutant 1.35 ± 0.07 µmol L−1 OD750−1 (n = 6)), whereas the pigment ratios showed only slight changes (Figure 2). This difference became smaller under LC conditions. Decreased chlorophyll a and especially phycocyanin contents also have been reported for PII mutant of S. elongatus PCC 7942 [26].
Figure 2.
Pigmentation and PII phosphorylation of cells of the wild type (WT) and the PII mutant ΔglnB of Synechocystis sp. PCC 6803 grown under high CO2 conditions (5%, HC) or low CO2 conditions (0.038%, LC). (A) Phycocyanin (PC) relative to chlorophyll a (Chla). (B) Carotenoids (Car) relative to chlorophyll a (Chla). (C) Detection of the different phosphorylated forms of the PII trimer in the WT and the ΔglnB in cells grown under HC or LC in standard BG11 medium (NO3−) or in BG11 medium without combined nitrogen source (- N). The different phosphorylation states of the PII trimer are visualized by immuno-blotting after separation of 10 µg of soluble proteins by native PAGE (PII0: dephosphorylated PII protein, PII3: complete phosphorylation of all three PII monomers).
We also checked how the different amounts of CO2 as well as nitrogen limitation affect the phosphorylation of the PII protein in WT cells. As shown previously [11,18], the PII phosphorylation state increased under N-limiting compared to nitrate-containing conditions in Synechocystis, whereas CO2-limiting conditions (LC) decreased the PII phosphorylation state. The PII protein became completely dephosphorylated under LC conditions in nitrate grown cells (Figure 2). The immuno-blot with protein extracts from the mutant ΔglnB showed no signals confirming the complete absence of PII protein in these cells.
2.2. Metabolome Analysis
Our metabolome data set comprised 693 mass features. As discussed recently [27], only a rather small fraction could be assigned to 79 defined metabolites (Supplemental Table 1). The identified metabolites allowed a good characterization of the primary carbon and associated nitrogen metabolism (Figure 1). The mutation of glnB had an important effect on the amounts and the pattern of identified metabolites. About 30% of these metabolites showed significant changes in cells of mutant ΔglnB compared to the WT.
2.2.1. Low Carbon Signature Metabolites
Previous investigations [11,12,23] on LC-shift-induced alterations in the metabolome of cyanobacteria revealed a general signature of metabolic changes. A major part of carbon is used for carbohydrate biosynthesis and glycogen storage under HC conditions. In contrast, LC conditions favor the export of carbon out of the Calvin-Benson cycle towards the glycolytic pathway, which leads to a LC-characteristic accumulation of 2-phosphoglycerate (2PGA) and PEP. In addition, the shift into CO2-limiting conditions activates at least transiently the photorespiratory 2-phosphoglycolate (2PG) metabolism leading to a transient accumulation of 2PG, glycolate, and glycine.
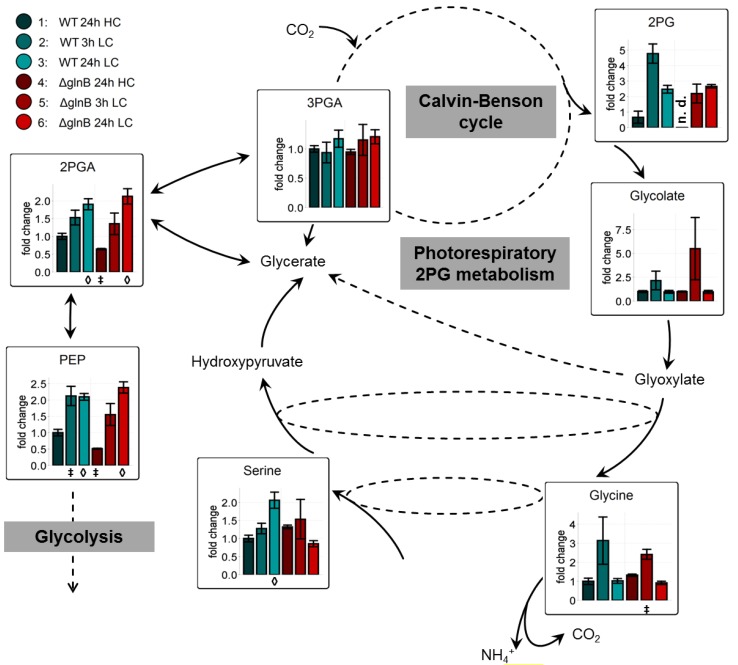
The mutation of glnB had a rather minor impact on this general changes. In our present data set, cells of the WT and the mutant ΔglnB showed almost the same relative increase in 2PGA, PEP, and 2PG in extracts from cells 24 h after the LC shift. Both strains also transiently accumulated increased amounts of the 2PG metabolism intermediates glycolate and glycine (Figure 3). Only the intermediate serine behaved differently in WT and mutant cells. Its amount increased in LC-shifted cells of the WT, whereas it showed a slightly lower level in LC compared to HC cells of the mutant ΔglnB. Collectively, these findings imply that PII is only little involved in the redirecting of carbon fluxes in Synechocystis under differing CO2 availability.
Figure 3.
Changes of the accumulation of low carbon signature metabolites in cells of the wild type (WT) and the PII mutant (ΔglnB) of Synechocystis sp. PCC 6803 after shifts from high CO2 (5%, HC) to low CO2 (0.038%, LC) conditions. Colored bars correspond to averaged values of three biological replicates measured by at least two technical replicates. Relative changes of metabolite pool sizes are represented by x-fold factors calculated relative to WT 24h HC. Error bars represent the standard errors (n.d. not detectable). Student’s T-test was applied to identify significant changes compared to WT 24 h HC.
‡: p-value ≤ 0.05, ◊: p-value ≤ 0.0). Abbreviations see Figure 1.
2.2.2. PII Deletion Increases Carbohydrate Consumption at Low CO2 Conditions
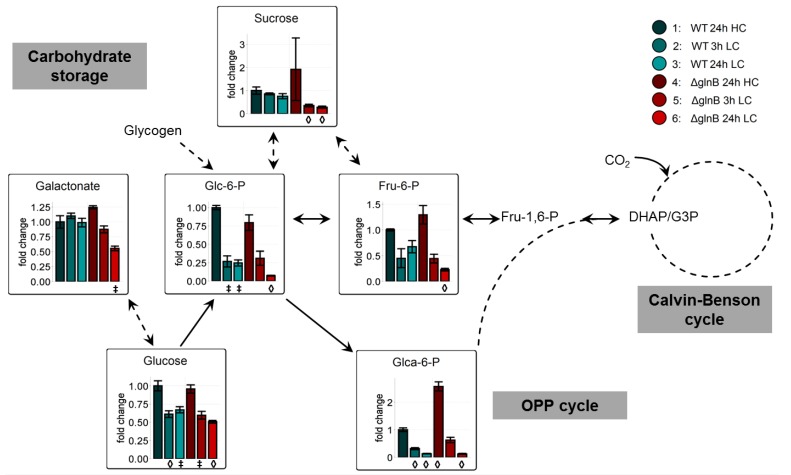
Another characteristic feature of LC-acclimated cyanobacterial cells is the decrease in the storage of glycogen and the pools of soluble carbohydrates [11,12]. This trend was confirmed under the experimental conditions of this study (Figure 4). Glucose and some phosphorylated sugars showed clearly lowered values in cells of the WT as well as the mutant ΔglnB when shifted from HC into LC conditions. In many cases, the decrease seemed to be more pronounced in cells of the mutant ΔglnB after 24 h growth at LC. For example, the amounts of fructose-6-phosphate (Fru-6-P) and glucose-6-phosphate (Glc-6-P) showed about half of the level of WT cells at this time point (Figure 4). Obviously, the cells of mutant ΔglnB used more of these Calvin-Benson cycle and Oxidative Pentose-Phosphate (OPP) cycle intermediates for biosynthetic purposes than WT cells. Alternatively, the HC grown cells of the PII mutant might have stored less glycogen than WT under HC, hence the storage is earlier consumed after LC shift leading to lowered final levels of this intermediates. Decreased glycogen accumulation has been shown for nitrate-grown cells of the S. elongatus PCC 7942 PII mutant [26].
Figure 4.
Changes of carbohydrate metabolism in cells of the wild type (WT) compared to the PII mutant (ΔglnB) of Synechocystis sp. PCC 6803 after shift from high CO2 (5%, HC) to low CO2 (0.038%, LC) conditions. Colored bars correspond to averaged x-fold values of three biological replicates measured by at least two technical replicates. Factors are calculated relative to WT 24h HC. Error bars represent standard error (n.d. = not detectable). Student’s T-test was applied to identify significant changes compared to WT 24 h HC.
‡: p-value ≤ 0.05, ◊: p-value ≤ 0.01). Abbreviations see Figure 1.
2.2.3. PII Deletion Strongly Decreases Ammonia Assimilation via GS/GOGAT
Compared to the missing or rather slight changes in the values of metabolites involved in Calvin-Benson cycle, glycolysis, photorespiration, OPP cycle, and carbohydrate metabolism, we found dramatic changes in metabolites connected to GS/GOGAT and N-detoxification between WT and ΔglnB cells. The known decrease in glutamate and glutamine levels after LC shifts in WT cells was correlated with a decreased expression of the corresponding genes in LC-shifted cells [5]. Similarly, glnA expression also became decreased in WT cells of S. elongatus PCC 7942 [23]. However, the decrease in glutamate and glutamine levels was much more pronounced in cells of the mutant ΔglnB. Already at HC, the contents of these amino acids were only 30% compared to WT, but under LC conditions the pools of these amino acids approximated the detection limit (Figure 5). Their decrease might be at least partly related to a reduced activity of GS/GOGAT. Decreased GS activity has been shown in the PII mutant of Anabaena sp. PCC 7120 [25]. The decrease of glnA expression under LC conditions was about two-times higher in the ΔglnB mutant of S. elongatus PCC 7949 compared to WT [23]. However, for the Synechocystis PII mutant rather increased GS activities and glnA mRNA levels have been reported, whereas the nitrite reductase was somewhat lower compared to WT [28]. Thus, beside regulatory effects on GS amount and activity, this decrease is probably more directly related to the strong decrease in the 2OG level in the glnB of Synechocystis. WT cells showed a stable 2OG pool at 3 h LC and a slight decrease at longer times at LC. In contrast, 2OG was below the detection threshold in extracts from the mutant ΔglnB regardless of the growth conditions (Figure 5). This finding contrasts the behavior of LC-shifted S. elongatus PCC 7942 WT and ΔglnB cells, which showed rather an increase in 2OG than a decrease under LC conditions [23]. It has been shown that the ΔglnB mutants of S. elongatus PCC 7942 contained a point-mutated, inactive gene encoding for the PipX protein [29]. In contrast, the Synechocystis ΔglnB mutant used in the current study shows no alterations in the DNA sequence of the pipX gene (ssl0105) including 250 bp of the up- and downstream regions (personal communication of the Forchhammer group, University Tübingen, Germany). Thus, the different behavior of the S. elongatus PCC 7942 and Synechocystis ΔglnB mutants could be based on this genotypic difference. The lowered 2OG level in cells of the Synechocystis PII mutant is unlikely to be explained by a limited flux of carbon through glycolysis or into the TCA cycle, because glycolytic intermediates such as 2PGA and PEP did not show significant differences between cells of the WT and the ΔglnB mutant (see Figure 3).
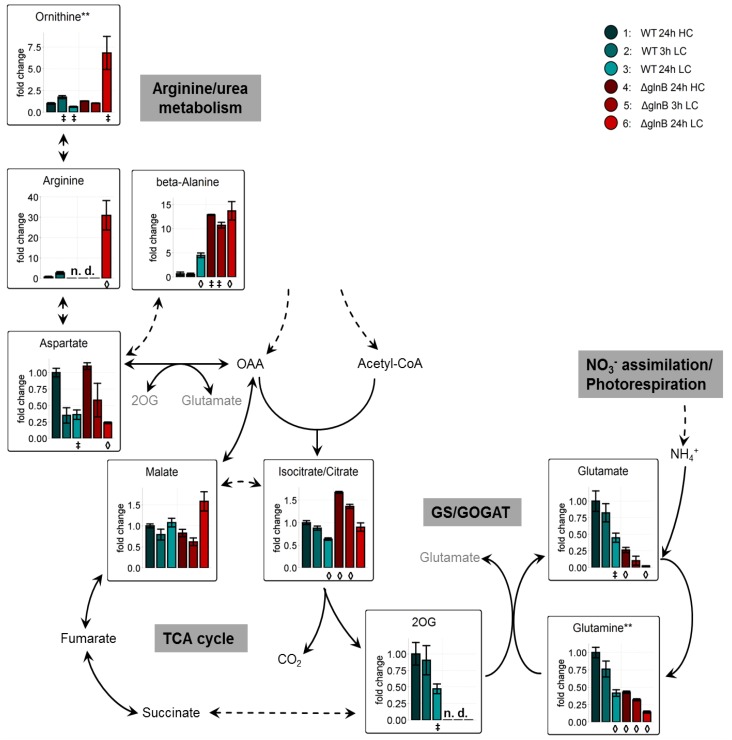
Figure 5.
Changes of primary N-metabolism and its precursors in cells of the wild type (WT) compared to the PII mutant (ΔglnB) of Synechocystis sp. PCC 6803 after shift from high CO2 (5%, HC) to low CO2 (0.038%, LC) conditions. Colored bars correspond to averaged x-fold values of three biological replicates measured by at least two technical replicates. Factors are calculated relative to WT 24h HC. Error bars represent standard error (n.d. = not detectable). Student’s T-test was applied to identify significant changes compared to WT 24 h HC.
‡: p-value ≤ 0.05, ◊: p-value ≤ 0.01. Abbreviations see Figure 1. ** Due to the chemical derivatization required for the GC-MS based profiling of primary metabolites the glutamine measurements represent the sum of endogenous pyroglutamate, glutamine, and minor contributions of glutamate. The ornithine measurements represent endogenous ornithine, citrulline, and arginine. The arginine measurement is taken from a specific chemical derivative.
Interestingly isocitrate, the precursor of 2OG synthesis, showed almost a twofold increase in cells of the mutant ΔglnB compared to WT under all growth conditions. One possible explanation could be a decreased activity of the isocitrate dehydrogenase (IDH) activity in the mutant leading to an accumulation of isocitrate. It has been found that the expression of the icd gene coding for IDH is about twofold decreased in LC-shifted Synechocystis cells [5]. Correspondingly, an increased expression of icd was reported for N-limited cyanobacteria including Synechocystis [30]. Similarly decreased icd mRNA levels were found in LC-treated cells of S. elongatus PCC 7942, but the expression was not further reduced in the corresponding mutant ΔglnB [23]. The transcriptional regulation fits to the main role of IDH to synthesize the precursor 2OG for glutamate synthesis, which is decreasingly needed under lowered N/C conditions. In addition to the expression level, the IDH activity is known to be regulated on biochemical level in cyanobacterial cells. For example, the purified IDH from Synechocystis showed inhibition by 2OG and NADPH2 [31]. Whereas the 2OG inhibition is not relevant to explain the increased isocitrate levels in cells of the mutant ΔglnB, higher amounts of NADPH2 can be assumed in mutant cells due to the lowered N-assimilation, one major sink for redox in cyanobacterial cells. Whether or not IDH activity also is directly redox-regulated is a matter of discussion. IDH of S. elongatus PCC 7942 showed no sign of redox regulation [32], whereas IDH from Anabaena cylindrica could be activated by spinach thioredoxin [33]. Recently, the IDH was found to be among the redox-regulated proteins upon nitrogen starvation in Prochlorococcus sp. SS120 [34]. Unfortunately, direct redox-effects were not investigated with the purified Synechocystis IDH [31]. However, it is likely that the redox state in cells of the mutant ΔglnB is changed leading to decreased IDH activity and thus contributing to the decreased isocitrate consumption. A lack of carbon flux in the direction to amino acids such as glutamate via 2OG could also explain the increase in malic acid in long-term LC cultivated cells of the mutant ΔglnB (Figure 5). Like glutamate, the amount of aspartate drops to low levels in cells of the mutant ΔglnB 24 h after the shift to LC conditions. Possibly, then the oxaloacetate is used to a lesser extent for aspartate biosynthesis leading to an enhanced C-flux toward malate.
Dramatic changes also occurred in the levels of arginine and ornithine. Our analysis indicated that these two N-containing metabolites increased 30fold (arginine) and 7fold (ornithine) in cells of the mutant ΔglnB after 24 h in LC. The increase in arginine could be related to the diminished cyanophycin accumulation in the Synechocystis PII mutant [35]. It has been reported that when ammonia was added to N-limited Synechocystis WT cells high amounts of cyanophycin were accumulated, whereas the PII mutant did not show this typical elevated cyanophycin levels. However, the addition of arginine also induced the cyanophycin accumulation in the PII mutant albeit to only 30% of the WT level [35]. These differences were attributed to the changed activity of the N-acetyl-L-glutamate kinase (NAGK), the key enzyme for arginine biosynthesis, which is known to be directly controlled by the PII protein [17]. This enzyme is less active in the PII mutant [35], since the PII-induced reduction of the feedback inhibition of NAGK by arginine is abolished in the mutant. Thus, the de novo synthesis of arginine should be lowered in the mutant. Probably, the accumulation of arginine as well as ornithine is rather a sign of a deregulated N assimilation. PII is known to down-regulate nitrate uptake and assimilation under conditions of excess nitrate in the medium [17], i.e., LC conditions leading to a decreased C/N ratio. Gene expression analysis with a S. elongatus 7942 ΔglnB mutant indicated that PII protein was important to allow NtcA the nitrogen-deprivation regulated activation (such as glnA) or repression (such as rbcS) of genes, since these responses were almost completely abolished in in a glnB mutant as well as in the ntcA defective strain [36]. Accordingly, the Synechocystis mutant ΔglnB has been shown to excrete nitrite due to an uncontrolled uptake of excess nitrate [37]. Clearly increased nitrate reductase activity also was found in nitrate-grown cells of the glnB mutant of Anabaena sp. PCC 7120 and the excess of combined nitrogen compounds was excreted here as ammonia instead of nitrite from the mutant cells [25].
Strikingly, the second precursor of cyanophycin, aspartate shows an inverse behavior to arginine in the Synechocystis mutant ΔglnB (Figure 5). The aspartate response resembles the response-pattern of carbohydrates, i.e., carbohydrates are decreased under LC conditions and deplete further in cells of the mutant ΔglnB. Additional to cyanophycin and protein synthesis, aspartate can be consumed by the reaction of aspartate decarboxylase especially under conditions of deregulated N assimilation. This reaction generates beta-alanine. Most probably, the decreased aspartate pool is connected to the more than 10fold accumulation of beta-alanine under LC conditions (Figure 5). This pathway could be used to detoxify excess nitrogen in cells of the Synechocystis glnB mutant. In the presence of an intact PII protein, N assimilation is down-regulated under LC conditions. Accordingly, WT cells that were exposed for 24 h to LC conditions decrease expression of many N-regulated genes [5,23] and accumulate much lower amounts of beta-alanine. Together our observations support the hypothesis that beta-alanine generated via aspartate may represent an indicator of an N/C imbalance.
3. Experimental Section
3.1. Strains and Culture Conditions
Axenic cells of the Synechocystis sp. PCC 6803 wild type and the mutant defective in the PII protein ΔglnB::Sp [24] were routinely grown in medium BG11 [38] at pH 8, 29 °C and continuous light of 120 µmol photons m−2·s−1. Mutant cells were cultivated in BG11 medium supplemented with 20 µg mL−1 spectinomycin (Sp). Cells were pre-cultured under high CO2 (HC) conditions by sparking the cultures with air enriched with 5% CO2. Low carbon (LC) conditions were induced by transferring cells into BG11 medium pH 7 and sparking with ambient air (0.038% CO2). To test PII phosphorylation under N-limiting conditions, cells were grown for 16 h in nitrate-free BG11 medium under HC or LC conditions. Contamination by heterotrophic bacteria was checked by spreading of 0.2 ml culture on LB-plates and incubating them for 48 h at 30 °C.
3.2. Metabolome Analysis
Cells grown in liquid media under standard HC or LC conditions were harvested by fast filtration in the light and immediately frozen in liquid nitrogen as described previously [23]. All experiments were repeated using three independent cell cultures. From each culture at least two replicates were analyzed. Metabolites were determined using a metabolite-targeted profiling method, and a quantitatively standardized and calibrated variant of the previously established gas chromatography electron ionization time-of-flight MS (GC-EI-TOF-MS) profiling analysis [11,39]. Relative amounts of metabolites were estimated by adding a defined amount of an internal standard, D-13C6-sorbitol (CAS 121067-66-1) and L-2,3,3,3-d4-alanine- (CAS 18806-29-6) (Sigma-Aldrich Chemie GmbH, Munich, Germany), to the cell material. The amounts were normalized to biomass using the optical density at 750 nm (OD750) of each sample [11,12]. Guidelines for manually supervised metabolite identification were the presence of at least 3 specific mass fragments per compound and a retention index deviation < 1.0% [40]. Metabolites were routinely assessed by relative changes expressed as response ratios, i.e. x-fold factors of LC-shifted cells in comparison to the condition WT 24 h HC.
3.3. Statistical Analyses
Statistical testing, e.g. Student’s T-test was performed using log10-transformed response ratios. Statistical assessments were performed using the Microsoft-Excel 2010 program.
3.4. Physiological Characterization
The growth of the cells was monitored by measurements of OD750. Pigment concentrations were calculated after evaluation of in vivo absorption spectra as described by reference [23]. To monitor the PII protein phosphorylation status, soluble proteins were isolated from 50 mL of cells. Equal amounts of proteins (10 µg) were separated in 15% acrylamide gels under native conditions and subsequently blotted onto nylon membranes. The PII protein bands were detected by a PII-specific antibody (received from K. Forchhammer, University of Tübingen, Tübingen, Germany). The immune-blotting method is described in more detail by reference [11].
4. Conclusions
The comparison of metabolic changes in LC-shifted cells of the Synechocystis WT and the ΔglnB mutant supported the notion that the PII protein represents an important regulator of C/N homoeostasis. As shown previously for S. elongatus PCC 7942 [23], the absence of PII resulted in marked changes in the levels of many metabolites. However, metabolites of the primary C metabolism, which are known to be regulated by LC-shift conditions [11,12], were not so much affected in cells of ΔglnB. The lesser effects of C-specific metabolites correlated well with the almost unchanged expression of genes for the primary C metabolism as well as CCM components in LC-shifted cells of the S. elongatus PCC 7942 PII mutant [23]. Thus, we assume that another regulator is responsible for C-specific changes instead of PII.
However, the levels and pattern of metabolites associated with N assimilation were drastically affected in the Synechocystis PII mutant. Usually, shifts to LC induce a coordinated down-regulation of the N assimilation. Our results clearly indicate that this coordinated down-regulation of the N assimilation under LC conditions is not functioning in the PII mutant at the metabolic level. Many metabolites of the N metabolism such as ornithine, arginine, beta-alanine, and aspartate showed a pattern that gives a clear indication for N/C imbalance. Significantly decreased levels of 2OG and increased levels of citrate and isocitrate but not of arginine were also reported for the mutant defective the transcriptional regulator cyAbrB [41]. These authors proposed that the metabolic changes in this mutant might be due to a low expression of glnB in the ΔcyabrB2 mutant, which is a direct target of cyAbrB. It is interesting to note that a deregulation of the LC-induced genes for CCM components also have been reported to occur in cells of the ΔcyabrB2 mutant [42]. Future studies will reveal if this regulatory protein is somehow connected with PII to regulate specifically the C assimilation as it was shown for the regulator NtcA responsible for the PII-dependent specific regulation of N-assimilatory genes.
Acknowledgements
Our work on cyanobacterial metabolomics is supported by the DFG (Deutsche Forschungsgemeinschaft, German Research Foundation). Karl Forchhammer (Department of Microbiology, Eberhard Karls University of Tübingen, Tübingen, Germany) is acknowledged for providing the PII mutant, the PII-specific antibody, and unpublished information regarding the genotype of the PII mutant.
Supplementary Files
Supplementary XLSX (XLSX, 240 KB)
Author Contributions
The study was mostly designed by Martin Hagemann and Joachim Kopka. Doreen Schwarz and Isabel Orf collected and analyzed the data. The manuscript was written by Martin Hagemann and Joachim Kopka with valuable contributions from Doreen Schwarz and Isabel Orf.
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Tabita F.R. The biochemistry and molecular regulation of carbon dioxide metabolism in cyanobacteria. In: Bryant D.A., editor. The Molecular Biology of Cyanobacteria. Kluwer Academic Publishers; Dordrecht, The Netherlands: 1994. pp. 437–467. [Google Scholar]
- 2.Flores E., Herrero A. Nitrogen assimilation and nitrogen control in cyanobacteria. Biochem. Soc. Trans. 2005;33:164–167. doi: 10.1042/BST0330164. [DOI] [PubMed] [Google Scholar]
- 3.Kaplan A., Hagemann M., Bauwe H., Kahlon S., Ogawa T. Carbon acquisition by cyanobacteria: Mechanism, comparative genomics and evolution. In: Herrero A., Flores E., editors. The Cyanobacteria: Molecular Biology, Genomics and Evolution. Caister Academic Press; Norfolk, UK: 2008. pp. 305–334. [Google Scholar]
- 4.Rae B.D., Long B.M., Whitehead L.F., Förster B., Badger M.R., Price G.D. Cyanobacterial carboxysomes: Microcompartments that facilitate CO2 fixation. J. Mol. Microbiol. Biotechnol. 2013;23:300–307. doi: 10.1159/000351342. [DOI] [PubMed] [Google Scholar]
- 5.Eisenhut M., von Wobeser E.A., Jonas L., Schubert H., Ibelings B.W., Bauwe H., Matthijs H.C., Hagemann M. Long-term response toward carbon limitation in wild type and glycolate turnover mutants of the cyanobacterium Synechocystis sp. strain PCC 6803. Plant Physiol. 2007;144:1946–1959. doi: 10.1104/pp.107.103341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Omata T., Gohta S., Takahashi Y., Harano Y., Maeda S. Involvement of a CbbR homolog in low CO2-induced activation of the bicarbonate transporter operon in cyanobacteria. J. Bacteriol. 2001;183:1891–1898. doi: 10.1128/JB.183.6.1891-1898.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Figge R.M., Cassier-Chauvat C., Chauvat F., Cerff R. Characterization and analysis of an NAD(P)H dehydrogenase transcriptional regulator critical for the survival of cyanobacteria facing inorganic carbon starvation and osmotic stress. Mol. Microbiol. 2001;39:455–468. doi: 10.1046/j.1365-2958.2001.02239.x. [DOI] [PubMed] [Google Scholar]
- 8.Wang H.L., Postier B.L., Burnap R.L. Alterations in global patterns of gene expression in Synechocystis sp. PCC 6803 in response to inorganic carbon limitation and the inactivation of ndhR, a LysR family regulator. J. Biol. Chem. 2004;279:5739–5751. doi: 10.1074/jbc.M311336200. [DOI] [PubMed] [Google Scholar]
- 9.Nishimura T., Takahashi Y., Yamaguchi O., Suzuki H., Maeda S., Omata T. Mechanism of low CO2-induced activation of the cmp bicarbonate transporter operon by a LysR family protein in the cyanobacterium Synechococcus elongatus strain PCC 7942. Mol. Microbiol. 2008;68:98–109. doi: 10.1111/j.1365-2958.2008.06137.x. [DOI] [PubMed] [Google Scholar]
- 10.Daley S.M., Kappell A.D., Carrick M.J., Burnap R.L. Regulation of the cyanobacterial CO2-concentrating mechanism involves internal sensing of NADP+ and α-ketogutarate levels by transcription factor CcmR. PLoS One. 2012;7:e41286. doi: 10.1371/journal.pone.0041286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eisenhut M., Huege J., Schwarz D., Bauwe H., Kopka J., Hagemann M. Metabolome phenotyping of inorganic carbon limitation in cells of wild type and photorespiratory mutants of the cyanobacterium Synechocystis sp. strain PCC 6803. Plant. Physiol. 2008;148:2109–2120. doi: 10.1104/pp.108.129403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huege J., Goetze J., Schwarz D., Bauwe H., Hagemann M., Kopka J. Modulation of the major paths of carbon in photorespiratory mutants of Synechocystis. PLoS One. 2011;6:e16278. doi: 10.1371/journal.pone.0016278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohashi Y., Shi W., Takatani N., Aichi M., Maeda S., Watanabe S., Yoshikawa H., Omata T. Regulation of nitrate assimilation in cyanobacteria. J. Exp. Bot. 2011;62:1411–1424. doi: 10.1093/jxb/erq427. [DOI] [PubMed] [Google Scholar]
- 14.Muro-Pastor M.I., Reyes J.C., Florencio F.J. Cyanobacteria perceive nitrogen status by sensing intracellular 2-oxoglutarate levels. J. Biol. Chem. 2001;276:38320–38328. doi: 10.1074/jbc.M105297200. [DOI] [PubMed] [Google Scholar]
- 15.Tanigawa R., Shirokane M., Maeda S.S., Omata T., Tanaka K., Takahashi H. Transcriptional activation of NtcA-dependent promoters of Synechococcus sp. PCC 7942 by 2-oxoglutarate in vitro. Proc. Natl. Acad. Sci. USA. 2002;99:4251–4255. doi: 10.1073/pnas.072587199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao M.X., Jiang Y.L., He Y.X., Chen Y.F., Teng Y.B., Chen Y., Zhang C.C., Zhou C.Z. Structural basis for the allosteric control of the global transcription factor NtcA by the nitrogen starvation signal 2-oxoglutarate. Proc. Natl. Acad. Sci. USA. 2010;107:12487–12492. doi: 10.1073/pnas.1001556107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forchhammer K. PII signal transducers: novel functional and structural insights. Trends Microbiol. 2008;16:65–72. doi: 10.1016/j.tim.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 18.Forchhammer K., Tandeau de Marsac N. The PII protein in the cyanobacterium Synechococcus sp. strain PCC 7942 is modified by serine phosphorylation and signals the cellular N-status. J. Bacteriol. 1994;176:84–91. doi: 10.1128/jb.176.1.84-91.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fokina O., Herrmann C., Forchhammer K. Signal-transduction protein PII from Synechococcus elongatus PCC 7942 senses low adenylate energy charge in vitro. Biochem. J. 2011;440:147–156. doi: 10.1042/BJ20110536. [DOI] [PubMed] [Google Scholar]
- 20.Espinosa J., Forchhammer K., Burillo S., Contreras A. Interaction network in cyanobacterial nitrogen regulation: PipX, a protein that interacts in a 2-oxoglutarate dependent manner with PII and NtcA. Mol. Microbiol. 2006;61:457–469. doi: 10.1111/j.1365-2958.2006.05231.x. [DOI] [PubMed] [Google Scholar]
- 21.Osanai T., Tanaka K. Keeping in touch with PII: PII-interacting proteins in unicellular cyanobacteria. Plant Cell Physiol. 2007;48:908–914. doi: 10.1093/pcp/pcm072. [DOI] [PubMed] [Google Scholar]
- 22.Llácer J.L., Espinosa J., Castells M.A., Contreras A., Forchhammer K., Rubio V. Structural basis for the regulation of NtcA-dependent transcription by proteins PipX and PII. Proc. Natl. Acad. Sci. USA. 2010;107:15397–15402. doi: 10.1073/pnas.1007015107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwarz D., Nodop A., Hüge J., Purfürst S., Forchhammer K., Michel K.P., Bauwe H., Kopka J., Hagemann M. Metabolic and transcriptomic phenotyping of inorganic carbon acclimation in the cyanobacterium Synechococcus elongatus PCC 7942. Plant Physiol. 2011;155:1640–1655. doi: 10.1104/pp.110.170225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hisbergues M., Jeanjean R., Joset F., Tandeau de Marsac N., Bedu S. Protein PII regulates both inorganic carbon and nitrate uptake and is modified by a redox signal in Synechocystis PCC 6803. FEBS Lett. 1999;463:216–220. doi: 10.1016/S0014-5793(99)01624-5. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y., Pu H., Wang Q., Cheng S., Zhao W., Zhang Y., Zhao J. PII is important in regulation of nitrogen metabolism but not required for heterocyst formation in the cyanobacterium Anabaena sp. PCC 7120. J. Biol. Chem. 2007;282:33641–33648. doi: 10.1074/jbc.M706500200. [DOI] [PubMed] [Google Scholar]
- 26.Forchhammer K., Tandeau de Marsac N. Functional analysis of the phosphoprotein PII (glnB gene product) in the cyanobacterium Synechococcus sp. strain PCC 7942. J. Bacteriol. 1995;177:2033–2040. doi: 10.1128/jb.177.8.2033-2040.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwarz D., Orf I., Kopka J., Hagemann M. Recent applications of metabolomics toward cyanobacteria. Metabolites. 2013;3:72–100. doi: 10.3390/metabo3010072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takatani N., Omata T. Effects of PII deficiency on expression of the genes involved in ammonia utilization in the cyanobacterium Synechocystis sp. strain PCC 6803. Plant Cell Physiol. 2006;47:679–688. doi: 10.1093/pcp/pcj037. [DOI] [PubMed] [Google Scholar]
- 29.Espinosa J., Castells M.A., Laichoubi K.B., Contreras A. Mutations at pipX suppress lethality of PII-deficient mutants of Synechococcus elongatus PCC 7942. J. Bacteriol. 2009;191:4863–4869. doi: 10.1128/JB.00557-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muro-Pastor M.I., Reyes J.C., Florencio F.J. The NADP+-isocitrate dehydrogenase gene (icd) is nitrogen regulated in cyanobacteria. J. Bacteriol. 1996;178:4070–4076. doi: 10.1128/jb.178.14.4070-4076.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muro-Pastor M.I., Florencio F.J. Purification and properties of the NADP-isocitrate dehydrogenase from the unicellular cyanobacterium Synechocystis sp. PCC 6803. Eur. J. Biochem. 1992;203:99–105. doi: 10.1111/j.1432-1033.1992.tb19833.x. [DOI] [PubMed] [Google Scholar]
- 32.Friga G.M., Farkas G.L. Isolation and properties of an isocitrate dehydrogenase from Anacystis nidulans. Arch. Microbiol. 1981;129:331–334. doi: 10.1007/BF00406456. [DOI] [PubMed] [Google Scholar]
- 33.Papen H., Neuer G., Refaian M., Bothe H. The isocitrate dehydrogenase from cyanobacteria. Arch. Microbiol. 1983;134:73–79. doi: 10.1007/BF00429411. [DOI] [PubMed] [Google Scholar]
- 34.McDonagh B., Domínguez-Martín M.A., Gómez-Baena G., López-Lozano A., Diez J., Bárcena J.A., García Fernández J.M. Nitrogen starvation induces extensive changes in the redox proteome of Prochlorococcus sp. strain SS120. Environ. Microbiol. Rep. 2012;4:257–267. doi: 10.1111/j.1758-2229.2012.00329.x. [DOI] [PubMed] [Google Scholar]
- 35.Maheswaran M., Ziegler K., Lockau W., Hagemann M., Forchhammer K. PII-regulated arginine synthesis controls acccumulation of cyanophycin in Synechocystis sp. PCC 6803. J. Bacteriol. 2006;188:2730–2734. doi: 10.1128/JB.188.7.2730-2734.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fadi Aldehni M., Sauer J., Spielhaupter C., Schmid R., Forchhammer K. Signal transduction protein PII is required for NtcA-regulated gene expression during nitrogen deprivation in the cyanobacterium Synechococcus elongatus strain PCC 7942. J. Bacteriol. 2003;185:2582–2591. doi: 10.1128/JB.185.8.2582-2591.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kloft N., Forchhammer K. Signal transduction protein PII phosphatase PphA is required for light-dependent control of nitrate utilization in Synechocystis sp. strain PCC 6803. J. Bacteriol. 2005;187:6683–6690. doi: 10.1128/JB.187.19.6683-6690.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rippka R. Isolation and purification of cyanobacteria. Methods Enzymol. 1988;167:3–27. doi: 10.1016/0076-6879(88)67004-2. [DOI] [PubMed] [Google Scholar]
- 39.Wagner C., Sefkow M., Kopka J. Construction and application of a mass spectral and retention time index database generated from plant GC/EI-TOF-MS metabolite profiles. Phytochemistry. 2003;62:887–900. doi: 10.1016/S0031-9422(02)00703-3. [DOI] [PubMed] [Google Scholar]
- 40.Hummel J., Strehmel N., Selbig J., Walther D., Kopka J. Decision tree supported substructure prediction of metabolites from GC-MS profiles. Metabolomics. 2010;6:322–333. doi: 10.1007/s11306-010-0198-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaniya Y., Kizawa A., Miyagi A., Kawai-Yamada M., Uchimiya H., Kaneko Y., Nishiyama Y., Hihara Y. Deletion of the transcriptional regulator cyAbrB2 deregulates primary carbon metabolism in Synechocystis sp. PCC 6803. Plant Physiol. 2013;162:1153–1163. doi: 10.1104/pp.113.218784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lieman-Hurwitz J., Haimovich M., Shalev-Malul G., Ishii A., Hihara Y., Gaathon A., Lebendiker M., Kaplan A. A cyanobacterial AbrB-like protein affects the apparent photosynthetic affinity for CO2 by modulating low-CO2-induced gene expression. Environ. Microbiol. 2009;11:927–936. doi: 10.1111/j.1462-2920.2008.01818.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary XLSX (XLSX, 240 KB)