Abstract
Human dental pulp stem cells (DPSCs) isolated from adult dental pulp are multipotent mesenchymal stem cells that can be directed to differentiate into osteogenic/odontogenic cells and also trans-differentiate into neuronal cells. The utility of DPSC has been explored in odontogenic differentiation for tooth regeneration. Alcohol abuse appears to lead to periodontal disease, tooth decay and mouth sores that are potentially precancerous. Persons who abuse alcohol are at high risk of having seriously deteriorated teeth, gums and compromised oral health in general. It is currently unknown if alcohol exposure has any impact on adult stem cell maintenance, stem cell fate determination and plasticity, and stem cell niche environment. Here we provide detailed experimental methods, analysis and information associated with our data deposited into Gene Expression Omnibus (GEO) under GSE57255. Our data provide transcriptomic changes that are occurring by EtOH treatment of DPSCs at 24-hour and 48-hour time point.
Keywords: Human dental pulp stem cells, Alcohol, Transcriptome
| Specifications | |
|---|---|
| Organism/cell line/tissue | Human dental pulp stem cells |
| Sex | N/A |
| Sequencer or array type | Affymetrix Human Genome Plus 2.0 |
| Data format | Raw and analyzed |
| Experimental factors | Normal human dental pulp stem cells treated with various concentrations of ethanol in vitro |
| Experimental features | Time and dose dependency exposure experiment to compare molecular effects of ethanol on normal human dental pulp stem cells |
| Consent | N/A |
| Sample source location | N/A |
Direct link to deposited data
Experimental design, materials and methods
Cell culture
Early passage DPSCs (P1–P2) isolated from deciduous teeth have been obtained from Dr. Songtao Shi (Univ. of Southern California) and cultured in alpha-MEM supplemented with 20% fetal bovine serum (v/v), 2 mM l-glutamine, 100 μM l-ascorbate-2-phosphate, 50 u/ml penicillin and 50 μg/ml streptomycin [1]. Exponentially growing DPSCs were treated with different concentrations of ethanol diluted from absolute ethanol (FW = 21.7 M). For acute exposure, cells were fed with media containing given concentrations of ethanol (0, 1, 5, 10, 20, 50 mM) for 24 or 48 h.
RNA isolation
Total RNA was isolated from DPSCs treated with ethanol (0, 1, 5, 10, 20, 50 mM) for 24 or 48 h. RNA was extracted using RNeasy purification kit, following the manufacturer's instruction (Qiagen, Valencia, CA). Isolated RNA was further purified by DNase treatment (Ambion/Life Technologies, Grand Island, NY). RNA purity and concentration were determined by NanoDrop, ND-1000 spectrophotometer (Thermo Scientific, Indianapolis, IN) and microfluidics-based platform 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). RNA concentration ranged from 56.3 ng/μl to 109 ng/μl. RNA concentration ≥ 50 ng/μl is recommended. 260/280 ratio ranged from 2.01 to 2.09. Ideal 260/280 ratio for pure RNA is 2.0.
Gene expression microarray analysis
Biological duplicate samples were hybridized to Affymetrix Human Genome Plus 2.0 (Cat.# 900469). We set the target intensity (TGT) at 500. The sensitivity of the system was measured by %P using the 3′ biased Affymetrix HG-U133A 2.0 arrays. %P ranged from 41.1 to 45.1% demonstrating the ability to detect a large number of transcripts across a wide range of abundance. All 24 arrays were assessed for recommended standard quality control metrics by Affymetrix including image quality, signal distribution and pair wise scatter plots and passed. mas5.CHP files were generated for each array by MAS 5.0 (Affymetrix, Santa Clara, CA) and combined to a final RESULTS.MAS5.TXT file.
Data analysis
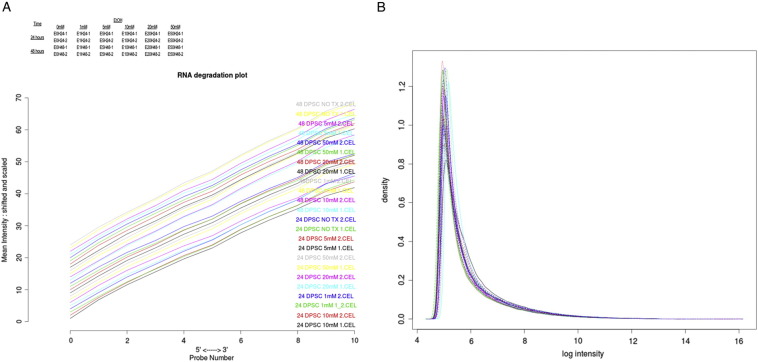
Degradation plot was prepared with each curve corresponding to a single chip and visualizing the chip-averaged dependency between probe intensity and probe position (Fig. 1A). Raw data was initially analyzed for the quality of microarray analysis by log density estimates of the data across all arrays (Fig. 1B).
Fig. 1.
A. Degradation plot: Each curve corresponds to a single chip and visualizes the chip-averaged dependency between probe intensity and probe position. B. Log density estimates (histograms) of the data across arrays.
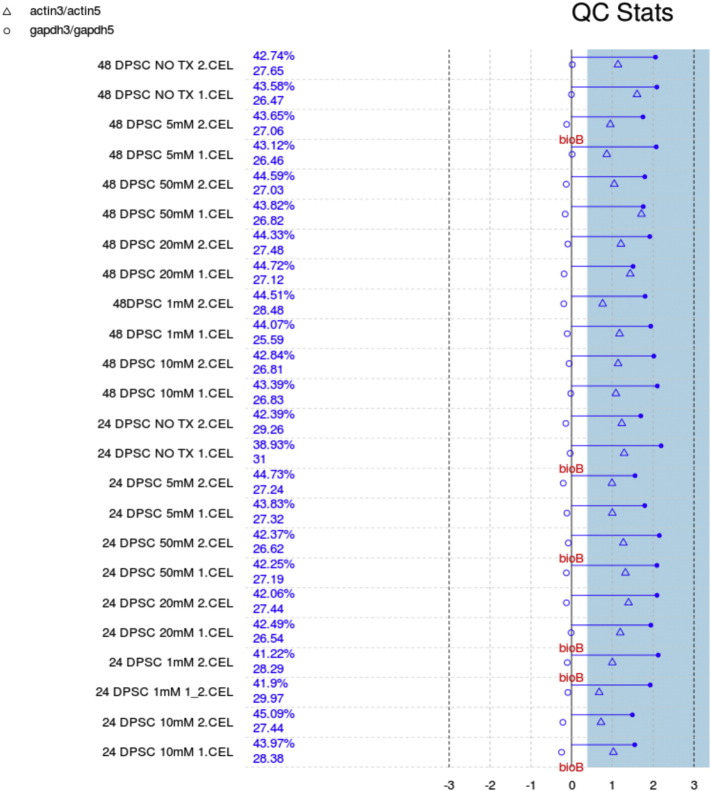
We performed background correction (Fig. 2), quantile normalization and log transformation with Robust Multi-array Average (RMA) approach on Affymetrix gene expression data using “Affy” R package (Fig. 3) [2].
Fig. 2.
Quality control stats. Each array is presented by a separated line. The blue bar represents the region where all scale factors fall within 3 fold of the mean scale factor for all chips. The chips passed all the QC metrics except that less than 30% absence of BioB is pointed to flag, indicating adequate quality data with acceptable inefficiency in hybridization step.
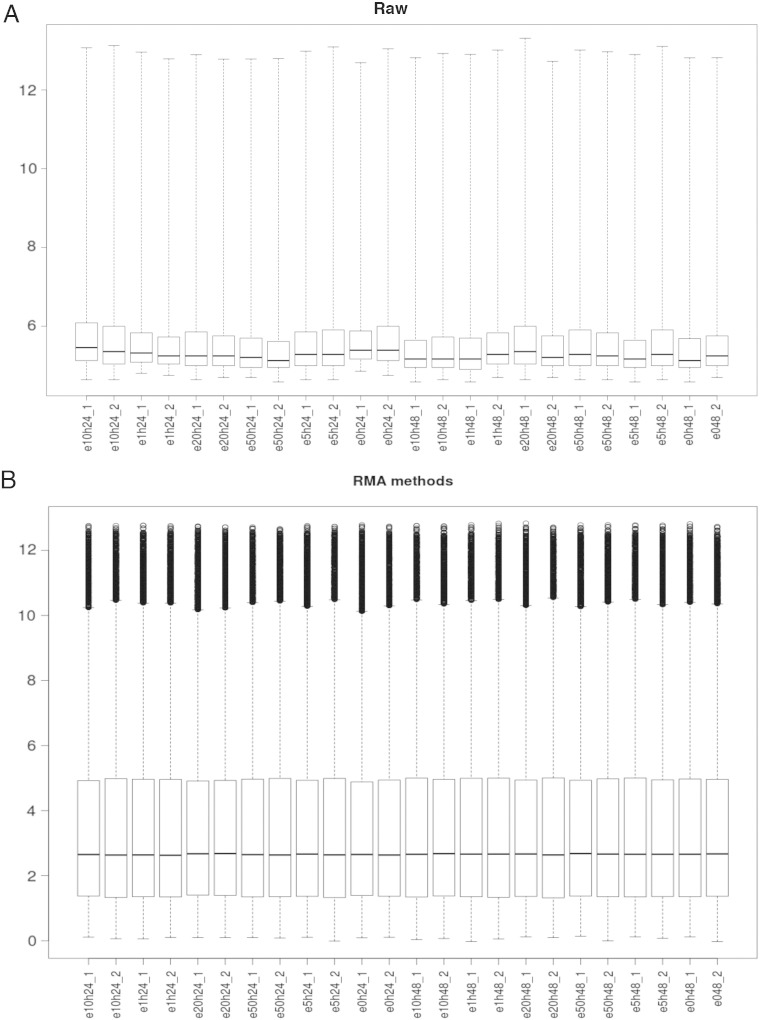
Fig. 3.
Boxplot of intensity for each sample on (A) raw data and (B) after normalization and log transformation using Robust Multi-array Average (RMA) method.
We removed probes with expression lower than the overall sample median; 27,327 out of 54,676 probes were kept for further analysis (Fig. 3). Given that the sensitivity of array platforms is generally considered lower than deep sequencing (with enough sequence depth), clearly a significant percentage of genes are either not expressed or beyond the detection limit in a typical array experiment. Filtering out this group of genes would potentially be beneficial to DEG (differentially expressed gene) detection.
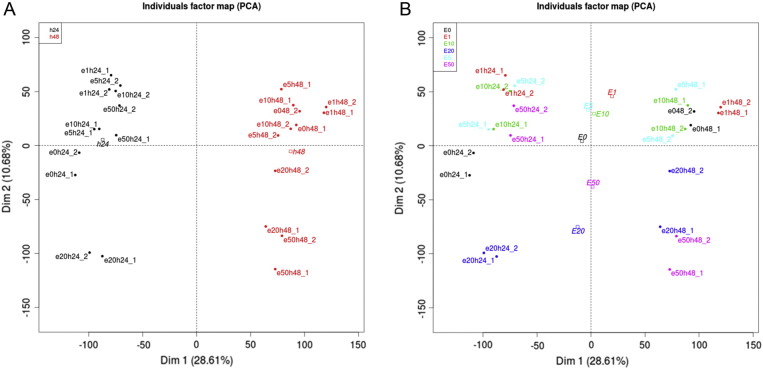
Principal Component Analysis was performed to detect expression data separation by EtOH treatment time (Fig. 4A) and by doses (Fig. 4B). Treatment time shows quite consistent grouping irrespective of doses, but doses in combination of treatment time show high degree of variations in gene expression.
Fig. 4.
Principal Component Analysis (PCA). PCA was performed on RMA dataset after filtering 50% probes with low expression and reducing original 54,676 probes to 27,327 probes. A. Separation by treatment time and B. separation by EtOH doses.
We constructed linear regression models to evaluate the effect of dose and time on gene expression (Fig. 5). For each gene we fit a separate model in terms of dose, time, and the dose by time interaction effect. In these models dose was treated as a continuous variable. In addition to the linear model, we performed a regression spline analysis with moderated F-test to detect the probes differentially expressed over time with increasing dose, with the assumption that expression changes smoothly along with the increasing of the dose rather than making discrete jumps from one dose to another. “Limma” R packages were used for this analysis. Q-values were generated to control the false discovery rate using “Qvalue” R package (http://www.bioconductor.org/packages/release/bioc/html/qvalue.html). A set of significant probes with cut-off of p < 0.05 for interaction effects was selected from each model for further functional analysis. All statistical analysis was performed using R-3.0.2 (http://cran.r-project.org/bin/windows/base/old/3.0.2/).
Fig. 5.
Interaction from linear model ranked by Q values.
Acknowledgment
This work was supported by UCLA Faculty Seed Grant and NIH/NIAAA R01 grant (R01AA21301) to Y.K.
References
- 1.Gronthos S., Mankani M., Brahim J., Robey P.G., Shi S. Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo. Proc. Natl. Acad. Sci. U. S. A. 2000;97:13625–13630. doi: 10.1073/pnas.240309797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gautier L., Cope L., Bolstad B.M., Irizarry R.A. Affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]