Abstract
Circadian rhythms are essential to cardiovascular health and disease. Temporal coordination of cardiac structure and function has focused primarily at the physiological and gene expression levels, but these analyses are invariably incomplete, not the least because proteins underlie many biological processes. The purpose of this study was to reveal the diurnal cardiac proteome and important contributions to cardiac function. The 24-h day-night murine cardiac proteome was assessed by two-dimensional difference in gel electrophoresis (2D-DIGE) and liquid chromatography-mass spectrometry. Daily variation was considerable, as ∼7.8% (90/1,147) of spots exhibited statistical changes at paired times across the 24-h light- (L) dark (D) cycle. JTK_CYCLE was used to investigate underlying diurnal rhythms in corresponding mRNA. We next revealed that disruption of the L:D cycle altered protein profiles and diurnal variation in cardiac function in Langendorff-perfused hearts, relative to the L:D cycle. To investigate the role of the circadian clock mechanism, we used cardiomyocyte clock mutant (CCM) mice. CCM myofilaments exhibited a loss of time-of-day-dependent maximal calcium-dependent ATP consumption, and altered phosphorylation rhythms. Moreover, the cardiac proteome was significantly altered in CCM hearts, especially enzymes regulating vital metabolic pathways. Lastly, we used a model of pressure overload cardiac hypertrophy to demonstrate the temporal proteome during heart disease. Our studies demonstrate that time of day plays a direct role in cardiac protein abundance and indicate a novel mechanistic contribution of circadian biology to cardiovascular structure and function.
Keywords: cardiovascular, circadian, diurnal, proteomics, two-dimensional difference in gel electrophoresis
circadian rhythms are intrinsic 24-h rhythms that underlie behavior and physiology, as reviewed previously (52, 62, 65). They are crucial to many major processes in the cardiovascular system in humans and other mammals, including heart rate (51) and blood pressure (69), consistent with the sympathovagal balance of the autonomic nervous system (reviewed in Refs. 19, 22, 34, 40, and 74). Circadian rhythms are also associated with the timing of onset of adverse cardiac events [e.g., myocardial infarction (16, 47, 48, 61), ventricular tachyarrhythmia (79), and sudden cardiac death (88)], which peak early in the day. Rhythm disruptions occur in shift work and sleep disorders and are associated with increased prevalence of adverse cardiac events, exacerbation of heart disease, and metabolic disorders linked with heart disease, as evidenced by many clinical (5, 20, 21) and experimental studies (6, 39, 41, 56). Circadian rhythms are of central relevance to the healthy cardiovascular system, and their disruption contributes significantly to cardiovascular morbidity and mortality.
Recent studies demonstrate circadian variations not only in cardiac physiology, but also at the molecular level in the heart (95). Global mRNA profiling studies reveal that ∼13% of gene transcripts are rhythmic across 24 h light (L) and dark (D) diurnal (38, 41, 82) and circadian cycles (76); undoubtedly, the heart is transcriptionally a different organ in the day vs. the night. These rhythms are necessary to coordinate biological and biochemical processes crucial for maintaining the normal structure and function of the heart, and remodeling in cardiovascular disease (as reviewed in Refs. 7, 19, 40, 53, 74). However, our current understanding is incomplete. For example, the relative contribution of factors that are extrinsic (e.g., neurohormonal factors) vs. intrinsic (e.g., the cardiomyocyte circadian clock), which drive these rhythms are not fully known. Moreover, although many studies have investigated diurnal gene expression underlying time-of-day cardiovascular (and other organ) physiology, it is only recently that investigations have turned toward the proteins. Proteins are fundamentally important as they underlie many biological processes. Moreover, levels of mRNA and proteins do not always correlate with each other because of posttranscriptional mechanisms controlling translation rates, half-lives, and posttranslational modifications, for example (23, 50); thus, the proteome warrants independent investigation. This concept was demonstrated on a global scale by Reddy et al. (60), who reported that only ∼50% of fluctuations in the hepatic proteome could be accounted for by changes in mRNA levels. Notably, researchers are now turning their focus toward proteomics approaches applied to the circadian field, where large-scale quantitative protein abundance measurements will increase understanding of how protein rhythms underlie circadian physiology (24, 42, 46, 54, 60, 64, 83).
Proteomics has been established as a powerful tool for analyzing biological phenomena. Most recently, we and others have begun using large-scale proteomics approaches to demonstrate de novo time-of-day changes in the proteome (12, 14, 24, 42, 46, 60, 83); however, to date, no studies have investigated the global day-night cardiovascular proteome, despite its critical importance underlying cardiac structure and function. Here, we used two-dimensional difference in gel electrophoresis (2D-DIGE) and liquid chromatography mass spectrometry (LC/MS/MS) to discover that up to ∼7.8% (90/1,147 spots on 2D-DIGE) change in abundance in the normal heart over 24-h daily periods and that protein spots identified with LC/MS/MS and Western blot may have a corresponding cycling transcript by JTK_CYCLE gene expression analyses. In addition, we demonstrated that shortening the 24-h photoperiod to 20 h (to which the animals cannot entrain) alters the relationship between the L:D cycle and their endogenous rhythms, by using the Comprehensive Lab Animal Monitoring System (CLAMS), along with functional consequences in left ventricular developed pressure on Langendorff. Moreover, we also examined the cardiac proteome mechanistically using cardiomyocyte-specific clock mutant (CCM) mice. We detected changes in CCM vs. wild-type (WT) mice in myofilament ATPase activity, myofilament phosphorylation, and abundance of cardiac metabolic enzymes, revealing the importance of the intact molecular clock for the temporal cardiac proteome. Finally, we demonstrate important contributions of proteomic programming in a model of cardiovascular disease.
MATERIALS AND METHODS
Animals.
All animal work was conducted under the guidelines of the Canadian Council on Animal Care and were approved by the Animal Care Committee of the University of Guelph. The experimental approach is in Fig. 1, and the rationale for these approaches is in the Fig. 1 legend. For detection of the diurnal cardiac proteome (Fig. 1A), male C57BL/6 mice (8 wk old; Charles River Laboratories) were entrained to a 12:12-h light-dark cycle (12:12 L:D) for 2 wk at the University of Guelph. Animals were euthanized by isoflurane and cervical dislocation. Tissues were collected from animals killed every 4 h starting 1 h before lights on [Zeitgeber Time (ZT) 23] for 1 day/night cycle (n = 3/time). For the L:D-disrupted protocol (Fig. 1B), animals were housed under controlled conditions in a 12:12 L:D cycle for 2 wk, then on either their normal 12:12 L:D or an established diurnal disruption protocol that shortens the photoperiod (10:10 L:D) (32, 39). Animals were euthanized (n = 3/time point, 4 times; n = 12/group) for tissue collection. To investigate the role of the cardiomyocyte-specific clock (Fig. 1C), we used male CCM mice (FVB background, University of Alabama at Birmingham) and WT littermates. Animals were euthanized (n = 8/time point, 4 times; n = 32 per group), and ventricular tissues were collected. To investigate diurnal rhythms in heart disease (Fig. 1D), we used the transverse aortic constriction (TAC) model of cardiac hypertrophy in male C57BL/6 mice, as described previously (41). Briefly, cardiac hypertrophy was induced in mice anesthetized with isoflurane, and intubated and ventilated (0.3 l/min O2, 140 respirations/min; Harvard Apparatus 687). A thoracotomy was performed in the second left intercostal space, aorta distal to the subclavian artery cleared, and a silk suture (Ethicon 7–0) that was placed around a 27-gauge needle was used to constrict the arch. Sham-operated animals underwent the same surgical procedure, except the ligature was not tightened. Mice were administered buprenorphine (0.1 mg/kg) upon awakening and at 8 h and 24 h postoperation. At 1 wk postsurgery, animals were euthanized by isoflurane and cervical dislocation every 4 h starting 1 h before lights on (ZT23) for 1 diurnal cycle (n = 3/time point, 18 TAC, 18 sham), and serum and ventricular tissue was collected. Samples were frozen in liquid nitrogen and stored at −80°C until use.
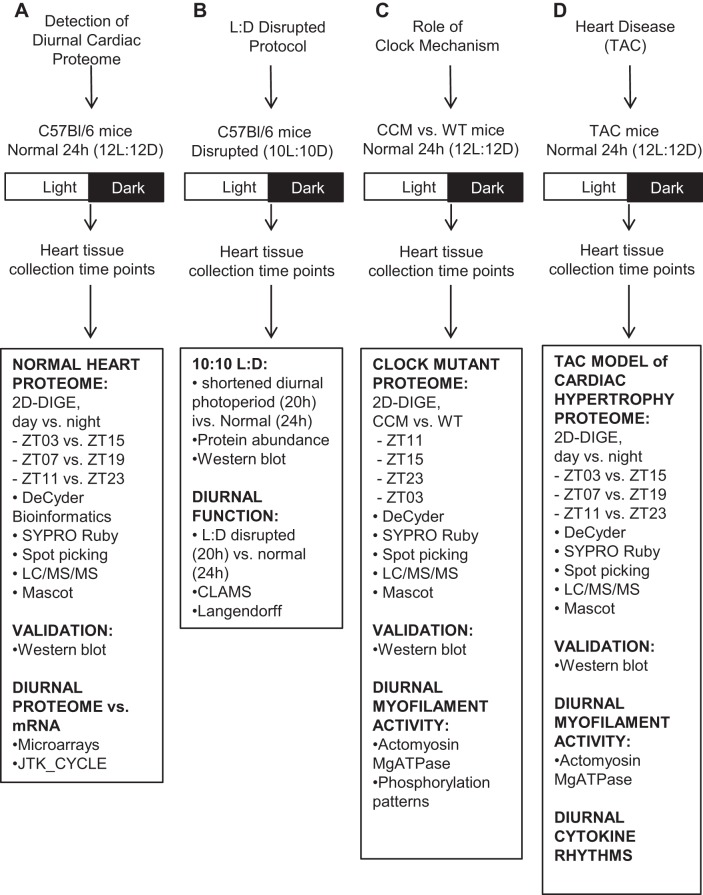
Fig. 1.
Experimental design. Four approaches were used to investigate circadian rhythms relevant to cardiovascular health and disease. A: model 1, detection of the diurnal cardiac proteome in male C57BL/6 mice housed in a normal 12:12-h light-dark (L:D) environment. This first classic approach investigates the cardiac proteome under conventional diurnal conditions. B: model 2, L:D disrupted protocol and cardiac protein abundance and heart function. Experimental and epidemiologic studies demonstrated that altered L:D increased risk of heart disease in humans and animals and exacerbated underlying heart disease, for example (21, 39, 41, 56). This second approach interrogates what happened when the L:D cycle was shortened. C: model 3, role of the cardiomyocyte-specific circadian clock (CCM) mechanism on the cardiac proteome and cardiac myofilament activity. Critical to circadian clock function is the transcription factor CLOCK (reviewed in Refs. 19, 40, 52, 62, 65, and many others). In this third model, CCM mice (17) were used to determine the importance of CLOCK (circadian clock mechanism) in the heart on the cardiac proteome. D: model 4, diurnal proteome in cardiovascular disease. Circadian rhythms play an important role in timing of onset of adverse cardiac events (for example, Refs. 45, 48, 49, 79, 89). In addition, blood pressure exhibits a 24-h cycling pattern; a major risk factor for cardiovascular disease occurs in hypertensive subjects that do not exhibit a dip during the night (87). Moreover, diurnal timing for cardiovascular therapies may benefit cardiac remodeling (43). Thus this fourth model begins to investigate what happens to the diurnal proteome in heart disease, using the pressure-overload induced cardiac hypertrophy (TAC) model in mice at 1 wk post TAC.
Protein purification.
Soluble heart proteins were collected following tissue homogenization in urea/CHAPs lysis buffer (10 mM Tris pH 8, 8 M urea, 4% wt/vol 3-[(3-cholamidopropyl) dimethylammonio]-1-propanesulfonate), with protease inhibitors (Roche; complete Mini EDTA-free). Tissue was homogenized on ice using Potter-Elvehjem grinder and centrifuged at 12,000 g, the supernatant was collected, and the protein concentration was measured by the Bradford assay (Bio-Rad).
2D-DIGE.
Soluble cardiac protein extracts (50 μg) were incubated in the dark with 200 pmol CyDye Fluors (GE Healthcare) for 30 min at 0°C, and the reaction quenched with 10 mM lysine, as described previously (26). For detection of the diurnal cardiac proteome (Fig. 1A), individual hearts collected 12 h apart were analyzed as 1) ZT23 (Cy5) vs. ZT11 (Cy3), or 2) ZT15 (Cy5) vs. ZT03 (Cy3), or 3) ZT07 (Cy5) vs. ZT19 (Cy3) (n = 3 gels/time point, 9 gels total). The rationale for pairing time points that were 12 h apart is in relation to mathematical modeling of circadian expression by cosine function, where peak and trough are ∼12 h apart (for example, Refs. 3, 28, 60, 77, 80). For studies investigating the clock mechanism (Fig. 1C), CCM samples were labeled with Cy3, and WT controls were labeled with Cy5. Gels were labeled as 1) ZT23, 2) ZT03, 3) ZT11, or 4) ZT15 (n = 3 gels/time, 12 gels total). For TAC heart disease (Fig. 1D), samples were labeled as ZT23 (Cy5) vs. ZT11 (Cy3), ZT15 (Cy5) vs. ZT03 (Cy3), or ZT07 (Cy3) vs. ZT19 (Cy5) (n = 3 gels/time, 9 gels total). For all studies, the internal standard was labeled with Cy2 and consisted of the pooled protein extracts used in each study, in accordance with field standards (1, 37, 81). For the first dimension, Cy-dye-labeled proteins were mixed with rehydration buffer (8 M urea, 1% CHAPS, 65 mM DTT, 0.5% vol/vol pharmalytes pH 3–10) and were added to 13-cm nonlinear immobilized pH gradient dry strips (pH 3–10) (GE Healthcare). Isoelectric focusing (IEF) was performed using a Protean cell (GE Healthcare). For the second dimension, IEF strips were equilibrated [75 mM Tris (pH 8.8), 6 M urea, 30% glycerol, 2% SDS, 1% wt/vol DTT, 2.5% wt/vol iodoacetamide], overlaid on 12% SDS-polyacrylamide gels, and electrophoresed using a DALT 6 apparatus (GE Healthcare). Gels were scanned with a Typhoon 9410 using the excitation/emission filters: 532 nm/580 nm for Cy3, 633 nm/670 nm for Cy5, and 488 nm/520 nm for Cy2. The photomultiplier tube was set at 550 and pixel size at 100 μm.
Bioinformatics.
2D-DIGE gels were analyzed with DeCyder 6.5 software (GE Healthcare). Difference in gel analysis was used to detect and match differentially labeled proteins within the gels. Protein abundance was calculated after background subtraction and normalization to the internal Cy2 control. Biological variance analysis was used to match multiple 2D-DIGE gels and calculate fold change (FC). Spots present in >75% of gels with volumes > 2 × 104 were included in the analyses. Normalized protein abundances were visualized in DeCyder where peak height and volume correlated with spot intensity. Normalized protein abundance and FC were visualized using graph view, and dashed lines represent individual FC, and the solid line is average FC.
In-gel digestion.
Spots were excised from 2D-DIGE SYPRO Ruby gels containing 300-μg pooled protein, using the Ettan Spot Picker (GE Healthcare). Trypsin digestion was carried out using in-gel digest kit (Thermo Scientific). Gel pieces were destained (25 mM ammonium bicarbonate, 50% vol/vol acetonitrile), reduced (50 mM Tris[2-carboxyethyl]phosphine in 25 mM ammonium bicarbonate), alkylated (100 mM iodoacetamide), then dried (acetonitrile), as per manufacturer's directions. Gel spots were rehydrated in 10 ng/μl trypsin digestion buffer and incubated at 30°C overnight. Solution containing tryptic peptide fragments was collected for LC/MS/MS.
LC/MS/MS analysis.
Peptide fragments were identified by nanoliquid chromatography (LC)-1D+ coupled to a hybrid linear ion trap/triple quadrupole mass spectrometer (QTRAP4000, ABSciex). Separation was performed using a linear binary gradient starting at 5% solvent B and ramping to 30% solvent B in 60 min; then, the gradient reached 100% B in 5 min and was held for 15 min before returning to the initial conditions. Solvent A contained 2% acetonitrile in water with 0.1% formic acid; solvent B contained 2% water in acetonitrile with 0.1% formic acid. Separation was performed at a flow-rate of 500 nl/min. The MS was set up in independent data acquisition mode where an enhanced mass scan triggered an enhanced product ion scan for ions between m/z 400 and m/z 1,000, with charge state 2 and 4, which exceeded 3,000 counts per second (cps). Mascot version 2.4, ProteinPilot v2.0.1, X!Tandem and Mouse Swiss Prot database were used for searching. Search parameters were trypsin, mouse, fixed modifications (carbamidomethyl), partial modifications [dehydration, deamidated (NQ), dioxidation (M), Gln→pyro-Glu (N-term Q), Gln→pyro-Glu (N-term E), oxidation (M)], and three missed cleavages. False positives were controlled by comparing molecular mass of the target spot on 2D-DIGE with the molecular weight (MW) of the identified protein and estimated to be <1%.
Western blot analysis.
Proteins identified by LC/MS/MS were independently validated at their 2D-DIGE identified time points (12-h paired time points) using pooled protein extracts (n = 3). In addition, time-of-day profiles (all 6 ZT time points) for proteins were independently determined from n = 3 individual samples taken at each time point across the diurnal cycle. Proteins were visualized by 12% SDS-PAGE [4–20% (Lonza) for PER2] and transferred to polyvinyl fluoride membrane (Bio-Rad). The following antisera were used: 1:10,000 mouse monoclonal STIP1 (Stressgen; SRA1500), 1:2,500 rabbit polyclonal IGF2 (Abcam; Ab9574), 1:100 rabbit polyclonal ALDH4A1 (Santa Cruz Biotechnology; sc130948), 1:1,000 rabbit polyclonal PER2 (Millipore, AB5428P), 1:1,000 goat polyclonal ALDH2 (Santa Cruz Biotechnology; sc48837), 1:2,000 rabbit polyclonal ECHS1 (ProteinTech; 11305–1-AP), 1:3,000 goat polyclonal LDHB (Abcam; ab2101), 1:1,500 rabbit polyclonal DLST (Aviva Systems Biology; OAAB01666), 1:1,500 rabbit polyclonal GOT2 (Aviva Systems Biology; ARP43518T100), 1:2,000 mouse monoclonal PDHE1a (Abcam; ab110330), 1:5,000 rabbit polyclonal CA2 (Santa Cruz Biotechnology; sc25596), 1:100 goat polyclonal FGB (Santa Cruz Biotechnology; sc18029), 1:200 rabbit polyclonal PSMA6 (Santa Cruz Biotechnology; sc67343), 1:200 goat polyclonal PCCA (Santa Cruz Biotechnology; sc68997), 1:1,000 rabbit polyclonal PDIA3 (Enzo; ADISPA585), and 1:1,000 rabbit polyclonal HSPB1 (Enzo; ADISPA801). Mouse monoclonal to β-actin at 1:25,000 (Millipore clone C4; MAB1501) was used as a loading control. Immunoreactive bands were visualized with ECLplus (GE Healthcare) and 1:5,000 HRP-conjugated secondary antibodies as applicable [goat anti-mouse (Sigma; A2304), rabbit anti-goat (Sigma; A5420), and goat anti-rabbit (Sigma; A6667).
mRNA expression and JTK analysis.
mRNA expression profiles were derived from our Affymetrix MOE430A GeneChip microarrays (GEO accession no. GSE36407) (41, 82). Briefly, a separate set of hearts were collected in the same diurnal manner as those used for protein studies, that is, every 4 h over 24 h starting at 1 h before lights on (ZT23) (ZT23, ZT03, ZT07, ZT11, ZT15, ZT19; n = 3 per time point, 18 samples total). Total RNA was isolated by TRIzol (Invitrogen) and assessed for high quality using a Nanodrop ND1000 (Thermo Scientific) and the Agilent Bioanalyzer 2100 (Agilent Technologies). Hybridization and scanning were in accordance with Affymetrix specifications, as described previously (82).The raw data set covered 22,690 transcripts (based on UniGene database build 107, June 2002) and were analyzed for diurnal rhythms in gene expression using the JTK_CYCLE nonparametric algorithm (28), as described on the CircaDB database (http://bioinf.itmat.upenn.edu/circa) (57).
Real-time PCR.
The microarray mRNA expression profiles corresponding to our proteins of interest were validated at peak and trough times by real-time PCR. The criteria for correlating the protein identifiers were as follows. Protein identifiers and corresponding tryptic peptide sequences provided by Mascot were used to retrieve protein identification data from SwissProt, including the protein full name, short name, identifier, references, and other cross-referencing material (see Supplemental Data S1). These were then used to retrieve the protein and mRNA sequences from the NCBI database. The NM_sequence gene reference identifier was used to search the Affymetrix microarray data.
For real-time PCR, total RNA from an independent set of normal C57BL/6 hearts collected every 4 h over 24 h was prepared using TRIzol (Invitrogen) as described previously (8, 82) and quality-assessed by Nanodrop (ThermoScientific). Amplification was performed on a VIIA7 real-time PCR system (Life Technologies) using the Power SYBR Green RNA-to-Ct one-step PCR kit (Life Technologies) under the following protocol: reverse transcription, 48°C for 30 min, 95°C for 10 min for 1 cycle, followed by amplification at 95°C for 15 s, 60°C for 1 min for 40 cycles. All primers were validated by twofold dilutions from 200 ng to 12.5 ng of mRNA and are listed in Supplemental Data S2. Real-time PCR samples were run in triplicate, and all values were normalized to histone using the delta delta CT method.
In silico circadian motif search.
The 2,000 base pair region upstream of the coding sequence for genes of interest (http://genome.ucsc.edu/cgi-bin/hgTables) were searched for putative circadian E/E′-box sequences (CANNTG, CACGTG, CACGTT), RORE motifs (AAAGTAGGTCA), and D-boxes ([A/G]T[G/T]A[T/C]GTAA[T/C]). We also searched the circadian mammalian promoter/enhancer database (PEDB, http://promoter.cdb.riken.jp/circadian.html) for E-boxes, D-boxes, and RREs that are predicted conserved in human vs. mouse, and searched circadian motifs identified in published reporter assays (35), and published ChIP and deep-sequencing analyses (63), and time-dependent patterns of circadian transcription (33).
CLAMS.
To investigate whole body behavioral and metabolic analyses, mice were housed in a CLAMS (Columbus Instruments). Recordings were first taken under a 12:12 L:D cycle (normal environment) for 5 days, after which the photoperiod was shortened to a 10:10 L:D for 5 days (6 L:D cycles). Daily patterns of respiratory exchange ratio (RER), energy expenditure, food intake, and physical activity were measured, as described previously(2).
Langendorff.
For ex vivo functional studies, hearts were excised, mounted, and perfused with Krebs-Henseleit buffer (118 mM NaCl, 25 mM NaHCO3, 1.2 mM KH2PO4, 4.7 mM KCl, 2.5 mM CaCl2, 1.2 mM MgSO4, 11 mM glucose, 0.2 mM EDTA, 0.5 mM Na pyruvate (95% O2-5% CO2, 37°C and 80-mmHg perfusion pressure). A balloon attached to a pressure transducer (AdInstruments) was inserted into the left ventricle and inflated to give end-diastolic pressures of 1–5 mmHg. Left ventricular developed pressure (LVDP) was determined after 30 min of stabilization (ADInstrument PowerLab, Chart Software v5.5.5, Colorado Creeks, USA). Function was assessed in hearts collected in the light period (murine sleep time) at 3 h after lights on, and in the dark period (murine wake time) at 3 h after lights off.
Myofilaments.
Cardiac myofilaments were isolated as described previously (90). Ventricular tissue was homogenized in ice-cold standard buffer (30 mM imidazole (pH 7.0), 60 mM KCl, 2 mM MgCl2, 0.2 mM PMSF, 0.01 mM leupeptin, 0.1 mM benzamidine). Tissue pellets were dissolved in skinning buffer (14.4 mM KCl, 60 mM imidazole (pH 7.0), 10 mM EGTA, 8.2 mM MgCl2, 5.5 mM ATP, 12 mM creatine phosphate, 10 U/ml bovine creatine phosphokinase, 0.2 mM PMSF, 0.01 mM leupeptin, 0.1 mM benzamidine, 1% Triton X100) for 45 min at 4°C. Resulting pellets were washed three times in ice-cold standard buffer, and protein concentrations were determined by the Bradford assay.
Actomyosin MgATPase.
We used an actomyosin Mg2+ ATPase assay to assess myofilament function. Myofilaments (50 μg) were incubated at 32°C for 10 min in mixtures of activating and relaxing buffer to create a range of free Ca2+. Activating buffer contained (in mM) 23.5 KCl, 20 imidazole (pH 7.0), 5 MgCl2, 3.2 ATP, 2 EGTA, 2.2 CaCl2, 0.2 PMSF, and 0.01 leupeptin; 0.1 benzamidine relaxing buffer consisted of 26 mM KCl, 5.1 mM MgCl2, 3.2 mM ATP, 2 mM EGTA, 20 mM imidazole (pH 7.0), 4.9 μM CaCl2. Free calcium was calculated using MaxChelator software (55). Reactions were quenched by adding an equal volume of ice-cold 10% trichloroacetic acid (TCA). Proteins were removed by centrifugation, and an equal volume of developing solution (0.5% FeSO4, 1% ammonium molybdate in 1 M H2SO4) was added to the supernatant. Absorbance was measured at 630 nm. ATPase values were plotted as the amount of phosphate released (nmol Pi·min−1·mg protein−1) vs. free Ca2+ (μM). Sigmoidal actomyosin Mg2+ ATPase activity-calcium relations were fit by a nonlinear procedure to a modified Hill equation: P = max·[Ca2+]H/([Ca2+]H + EC50H), where P is actomyosin MgATPase activity, max is the maximum value at saturating calcium, EC50 is the calcium concentration at which 50% of maximum is reached, and H is the slope of the relationship (Hill coefficient) (59).
Myofilament phosphorylation.
Myofilament isolates from flash-frozen isolates as were used for the actomyosin MgATPase assay (10 μg) were resolved using SDS-PAGE, and ProQ Diamond stain to evaluate protein phosphorylation. Load was determined using Coomassie blue stain. Gels were scanned with a Typhoon 9410.
Blood serum cytokines.
Serum cytokines were quantified using the murine Th1/Th2/Th17 cytometric bead array (BD BioSciences), according to the manufacturer's specifications.
Statistical analyses.
Values are expressed as means ± SE. Statistical comparisons were done using DeCyder values for normalized protein abundance (2D-DIGE) and either an unpaired Student's t-test or one-way ANOVA followed by the Tukey test for multiple groups in GraphPad Prism (GraphPad Software). Values of P < 0.05 were considered statistically significant.
RESULTS
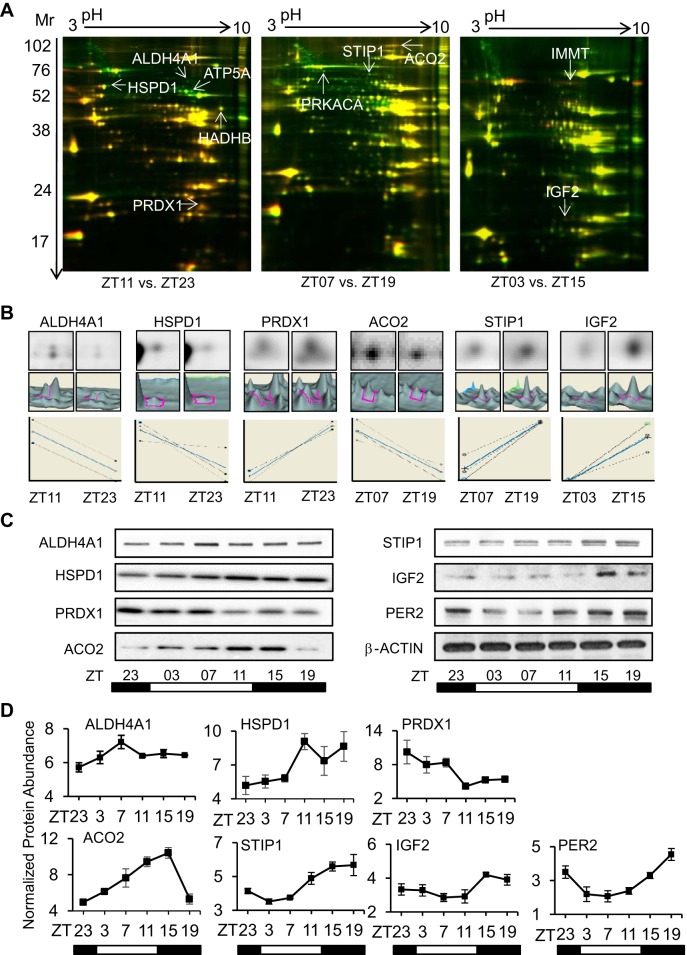
We first used a conventional diurnal environment to show that hearts from mice housed in the normal 24-h L:D cycle had evidence of a diurnal cardiac proteome, as indicated by the difference in spot abundance on 2D-DIGE (Fig. 2). The 2D-DIGE approach is a variant of two-dimensional (2D) gel electrophoresis that specifically addresses the variation between gels (which used to occur with 2D gels). Moreover, this technique has the advantage of running an internal standard as part of each separation (Cy2-labeled pools of all proteins run on the gel). It also allows for running up to two test samples simultaneously (thus, the paired time points) with more accurate and reliable results. Gels were run for each 12-h time point comparison (ZT11 vs ZT23, ZT07 vs. ZT19, and ZT03 vs. ZT15), using the soluble cardiac proteome from individual murine hearts (three individual murine hearts per time point, all gels run in triplicate), and a total of 1,147 protein spots were reliably detected on the gels (>75% of gels, spot volume >2 × 104) (Fig. 2A). Of these consistently detected spots, there were 382 uniquely identified as exhibiting altered abundance at paired time points, and 90 (7.8%) were significant (P < 0.05) by ANOVA (DeCyder software). Of these 90 spots, 38 exhibited statistically (P < 0.05) increased abundance in the light period (murine sleep time), and 52 were increased (P < 0.05) in the dark period (murine wake time).The DeCyder software illustration of abundance (top image), area under the peak (middle image), and relative expression (bottom graph) for six of these spots at paired time points is shown in Fig. 2B. Thus, overall, we estimated that about 7.8% of the soluble cardiac proteome varies in abundance over 24-h diurnal cycles.
Fig. 2.
Detection of diurnal cardiac proteome with two-dimensional difference in gel electrophoresis (2D-DIGE). A: cardiac protein samples collected 12 h apart were analyzed by 2D-DIGE, at L (light, ZT0–ZT12, murine sleep time) vs. D (dark, ZT12-ZT24, murine wake time) points. Left to right, ZT11 (Cy3) vs. ZT23 (Cy5), ZT07 (Cy5) vs. ZT19 (Cy3), and ZT03 (Cy3) vs. ZT15 (Cy5). Spots with FC ≥ 1.4 (P < 0.05) were identified by LC/MS/MS (arrows). B: representative DeCyder bioinformatics revealing single channel Cy3/Cy5 spot images (top) and 3D peaks (middle), and graphs (bottom) for proteins subsequently validated by Western blot. For all 2D-DIGE studies, an internal standard was labeled with Cy2, and consisted of the pooled protein extracts used in each study, in accordance with field standards. C) Independent validation by Western blot analysis, representative blots for ALDH4A1, HSPD1, PRDX1, ACO2, STIP1, and IGF2. PER2 is a positive control for rhythmic protein abundance, and β-actin is a loading control. D: densitometry of the proteins by triplicate Western blots (n = 3 samples per time point) for these proteins. See Supplemental Data S1 for protein data (names, replicate gel runs, molecular weight, Mascot search and query scores, other database information, peptides used for identification, measured/predicted peptide mass, specificity). Supplemental Data S3 provides DeCyder statistical analyses.
To independently demonstrate time-of-day changes and confirm that these spots correlated with proteins that vary in abundance in the heart over the 24-h diurnal cycle, we selected 10 high-stringency spots identified with 1.4-fold or greater difference in abundance at pairwise time points (ZT23 vs. ZT11; ZT03 vs. ZT15; ZT07 vs. ZT19) and significant (P < 0.05) by ANOVA (Fig. 2A arrows, Supplemental Data S3). The spots were identified by LC/MS/MS as δ-1-pyrrolinie-5-carboxylate dehydrogenase, mitochondrial (ALDH4A1) (2.4 ± 0.5 FC), cAMP-dependent protein kinase-α (PRKACA) (1.7 ± 0.4 FC), ATP synthase-α, mitochondrial (ATP5A1) (2.8 ± 0.3 FC), stress-induced phosphoprotein-1 (STIP1) (1.5 ± 0.1 FC), aconitate hydratase, mitochondrial (ACO2) (1.5 ± 0.1 FC), peroxiredoxin-1 (PRDX1) (1.95 ± 0.04 FC), trifunctional enzyme subunit-β, mitochondrial (HADHB) (1.6 ± 0.1 FC), insulin-like growth factor II (IGF2) (2.0 ± 0.3 FC), inner membrane protein, mitochondrial (IMMT) (1.5 ± 0.1 FC), and 60-kDa heat shock protein (HSPD1) (2.5 ± 0.5 FC). See Supplemental Data S1 for protein name, replicate gels, accession numbers, MW, Mascot search and query scores, other database information, number of peptides, tryptic sequences, measured/predicted peptide mass, and specificity. We independently validated proteins by Western blot analysis (Fig. 2C). Our choice of targets was determined by unique tryptic peptide matches per protein and the availability of commercial antisera. Moreover, we examined protein abundance over the 24-h diurnal cycle using samples from all six time points by Western blot analysis (Fig. 2C). Protein abundance (n = 3 hearts per time point) varied across the L:D cycle, as illustrated by densitometry analyses of the Western blots (Fig. 2D). The core circadian clock protein Period2 (PER2) was used as a positive control for rhythmic protein abundance and exhibited a 24-h cycle consistent with its known mRNA cycling pattern (38).
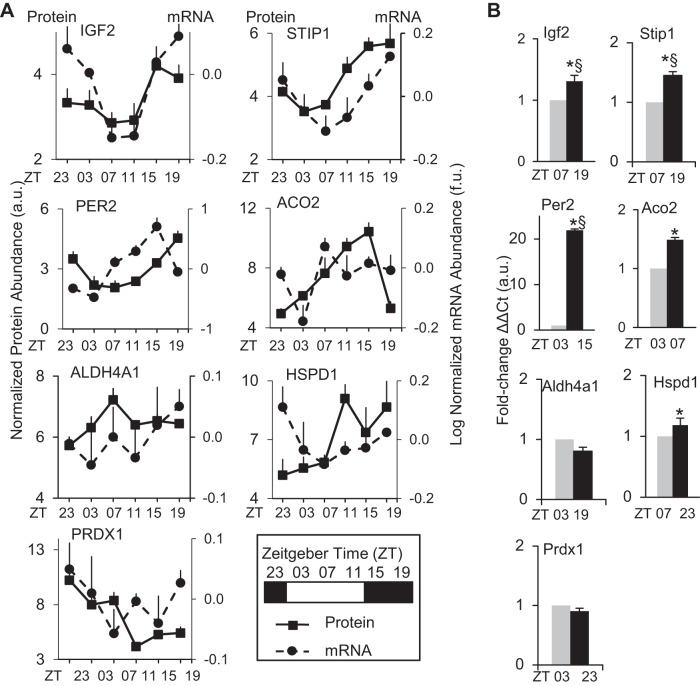
To determine whether proteins exhibiting diurnal profiles had an underlying rhythmic mRNA complement in the heart, we compared the protein profiles with our earlier diurnal (L:D) Affymetrix microarray gene expression data collected from independent sets of hearts over the 24-h day-night cycle (38, 41, 82). Diurnal mRNA rhythms were determined using the well-established JTK_CYCLE algorithm (28, 57). A total of 3,875 out of 22,690 (17%) rhythmic mRNA transcripts were detected using JTK (ADJ, P < 0.05), and, moreover, 599/22,690 (2.6%) were detected at highest stringency using JTK (BH, Q < 0.05). We found diurnal microarray data for all corresponding proteins of interest (Fig. 3A). The significant JTK P values were for Igf2 (P < 0.006) and Stip1 (P < 0.022), indicative of rhythmic expression and consistent with the positive control Per2 (P < 2.5e-05) (Supplemental Data S2). All the microarray data in Fig. 3A were further validated by using an independent set of hearts and examining the expression at the peak vs. trough time points and real-time PCR (Fig. 3B, Supplemental Data S2). Of the genes that significantly (P < 0.05) peaked in the dark phase, Igf2 exhibited a 1.3 ± 0.1 FC (ZT19 vs. ZT07), and Hspd1 had a 1.2 ± 0.1 FC (ZT23 vs. ZT07). For the genes that significantly (P < 0.05) peaked in the light phase, Aco2 showed a 1.5 ± 0.04 FC (ZT07 vs. ZT03). Aldh4a1, Stip1, and Prdx1 mRNA did not show a significant (P > 0.05) peak-trough change by real-time PCR. Per2 was used as a positive control for circadian cycling and exhibited a significant (P < 0.05) peak at ZT15 and a trough at ZT03, as anticipated. Moreover, the Per2 mRNA exhibited a delay between message and protein, as anticipated; however, interestingly, Igf2, Aldh4a1, and Aco2 were relatively coincident, and Stip1 appeared to exhibit a delay.
Fig. 3.
The diurnal cardiac proteome and transcriptome. A: diurnal profiles of microarray gene expression data (dashed line), compared with proteins identified by 2D-DIGE and LC/MS/MS, then validated by triplicate Western blot across all diurnal time points (solid line). Microarray gene expression values are provided as log-normalized mRNA abundance (f.u. = fluorescence units) using GeneSpring software. Rhythmic gene expression was assessed by JTK_CYCLE algorithm, and values are provided in Supplemental Data S2. For protein expression, values are provided as triplicate densitometry from Western blots (see Fig. 2D). For all graphs, the first (left) y-axis is normalized protein abundance (a.u., arbitrary units) and the second (right) y-axis is log-normalized mRNA abundance (f.u. = fluorescence units), and x-axis is time of day in zeitgebers as denoted in the legend key box inset. Values are expressed as means ± SE; n = 3/time point. B: quantitative real-time PCR was performed to validate the peak and trough mRNA expression time points from the microarrays. real-time PCR samples were run in triplicate, and all values were normalized to histone using the delta delta CT method. *P < 0.05, diurnal rhythm significant by JTK_CYCLE. §P < 0.05 peak:trough ratio validated by real-time PCR. See Supplemental Data S2 for mRNA analyses, including JTK_CYCLE, mRNA vs. protein ZT peaks, fold change, in silico data for the corresponding circadian promoter binding elements (canonical E box, E′box, D box, RORE), searches against published CLOCK/BMAL ChIP assays, and PCR primer sequences.
We explored mechanisms that could account for underlying mRNA patterns, to investigate whether they might be under clock mechanism control. To do this, we searched promoter sequences (retrieved from http://genome.ucsc.edu/cgi-bin/hgTables) for circadian elements (E/E′ box), and the (http://promoter.cdb.riken.jp/circadian.html) circadian mammalian promoter/enhancer database PEDB (35), and previously published ChIP assays with deep sequencing and reporter assays investigating circadian elements (33, 63). We detected elements in all mRNA that exhibited a diurnal rhythm, as well as those mRNA that were not rhythmic across the diurnal cycle by microarray and JTK_CYCLE or PCR analyses (Supplemental Data S2), suggesting that there are also other mechanisms underlying diurnal translational control.
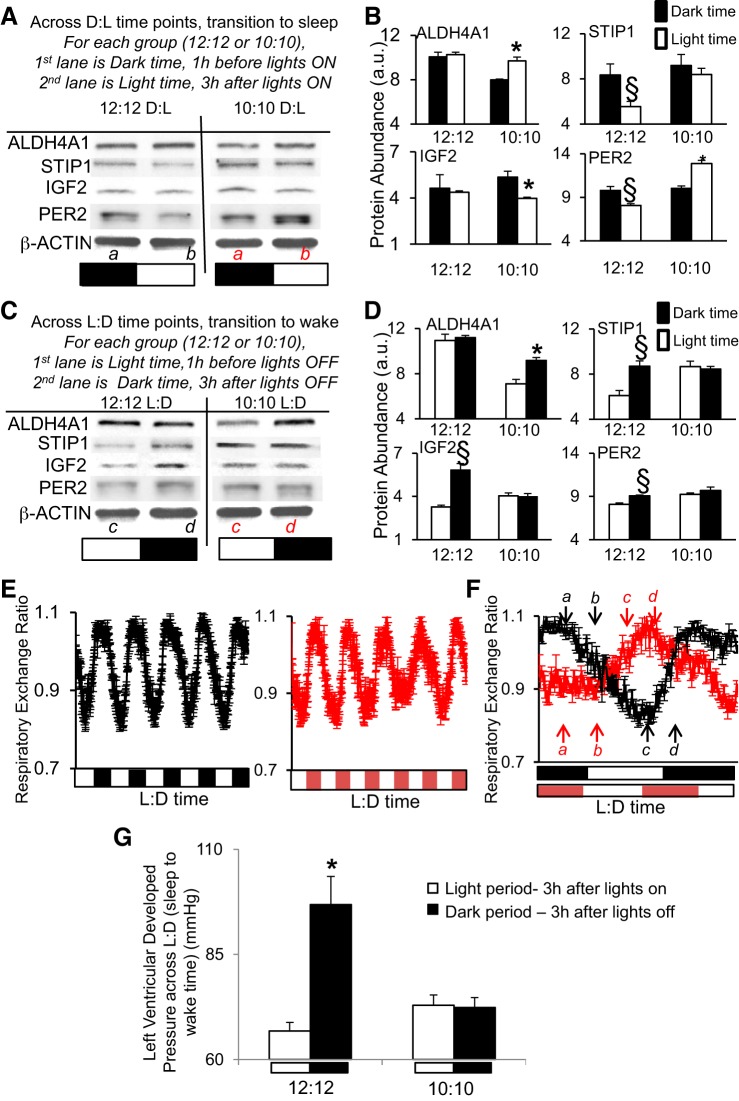
Next, we examined what happens when the diurnal cycle is altered to 10:10 L:D. The animals cannot entrain to this shortened photoperiod, and thus, it uncouples the L:D environment from their circadian rhythms (32). The rationale for altering L:D is that numerous experimental and epidemiologic studies have previously demonstrated that altered L:D cycles can affect cardiovascular health and disease in humans and rodents, for example (21, 39, 41, 56). Moreover, altering the 24-h L:D cycle affects molecular cardiac gene expression (40). Thus, we tested whether altering the L:D cycle also impacted protein abundance in the heart. Mice were subjected to an established L:D disruption protocol (10:10 L:D) (32, 41), and hearts were collected after 5 days. As a control, independent pooled sets of hearts were taken from mice under normal diurnal 12:12-h L:D, and they exhibited the same relative abundance of the sentinel proteins ALD4a1, STIP1, IGF2, and PER2 at D:L (transition to sleep time, Fig. 4, A and B) and L:D (transition to wake, Fig. 4, C and D) time points. However, profiles were significantly (P < 0.05) altered in hearts from diurnally disrupted mice (Fig. 4, A–D, Supplemental Data S4). To further investigate the effect of the shortened photoperiod on endogenous rhythms, we used a CLAMS. As shown in Fig. 4E (left, black lines), in the normal 12:12-h L:D environment, RER (a measure of substrate utilization, where lower RER in the light phase is indicative of increased fatty acid oxidation) exhibits a predictable diurnal profile that is lowest during the light phase (rodent sleep time) and highest in the dark (rodent wake time). The animals cannot entrain to the shortened 20-h photoperiod, and as shown in Fig. 4E (right, red lines), they continued to exhibit a clear endogenous rhythm in RER that did not follow the L:D cycle. The mice showed a similar pattern for the other CLAMS variables as well, including energy expenditure, food intake, and physical activity (Supplemental Data S4). These data were consistent with the findings of Karatsoreos et al. (32), which showed that the endogenous circadian rhythm in body temperature was maintained in the 10:10 L:D cycle. The times at which heart tissue was collected for protein analyses, in reference to both the L:D cycle and the endogenous rhythms, are illustrated in Fig. 4F. To pursue further the functional impact of L:D disruption, we assessed contractile function in Langendorff perfused hearts. Function was assessed in hearts collected in the light period at 3 h after lights on, and the dark period at 3 h after lights off. Consistent with LVDP as a measure of cardiac function ex vivo, hearts from animals maintained in the normal (12:12-h L:D) environment exhibited diurnal variation (P < 0.05) (light period, 66.81 ± 2.02 mmHg vs. dark, 96.85 ± 6.71 mmHg; Fig. 4G), revealing differences in cardiac function day vs. night, as anticipated (94). However, hearts from the 10:10 mice lacked this diurnal variation in LVDP relative to the L:D cycle (light: 72.91 ± 2.44 mmHg vs. dark: 72.37 ± 2.36 mmHg; Fig. 4G).
Fig. 4.
Diurnal disruption. Mice subjected to a shortened L:D environment (10:10 h) exhibited altered abundance of cardiac proteins ALDH4A1, STIP1, IGF2, PER2 relative to the L:D cycle, vs. mice housed in the normal 12:12-h environment. A: Western blot, showing dark (1 h before lights on) to light (3 h after lights on) transition times. B: densitometry relative protein abundance (in a.u.). C: Western blot, showing light (1 h before lights off) to dark (3 h after lights off) transition times. D: densitometry relative protein abundance (in a.u.). Images in A and C represent Western blots using pooled samples (n = 3 per time point). Graphs in B and D are from triplicate Western blots using individual samples (n = 3/time point) and scanned by densitometry (values are in Supplemental Data S4). Values are expressed as means ± SE; §P < 0.05 represents a statistical difference under normal 12:12-h L:D conditions, and *P < 0.05 represents a statistical difference in 10:10-h LD. PER2 is used as a positive cycling control. Bands are normalized to β-actin to control for loading. The lower case letters a,b,c,d in parts A and C denote the time points where samples were taken (red letters denote 10:10, while black letters denote 12:12 environment). These sampling times are relative to the L:D cycle, as illustrated in F. E: whole body metabolic substrate utilization rhythms (respiratory exchange ratio, RER) measured in a CLAMS over 12:12-h L:D cycle exhibited a diurnal rhythm that peaked in the dark (animals awake) and troughed in the light (sleep time) (left, black lines), and they continued to exhibit a clear endogenous rhythm in RER that did not follow the L:D cycle under the shortened 10:10-h L:D photoperiod (right, red lines). F: RER rhythms on the day of collection showing that sampling times are relative to the L:D cycle but differ in regards to the endogenous rhythm. n = 5/group, metabolic values are in Supplemental Data S4. The lower case letters a,b,c,d denote the times when samples were taken (red letters denote 10:10, while black letters denote 12:12 environment). G: there is normal diurnal variation in cardiac function on Langendorff (left ventricular developed pressure, LVDP) under 12:12-h L:D (left, lower during the light/animal sleep time, while higher during the dark phase/wake time); however, this difference is altered with respect to the L:D cycle in the 10:10 L:D diurnal disrupted group (right). Function was assessed in hearts collected in the light period (murine sleep time) at 3 h after lights on, and the dark period (murine wake time) at 3 h after lights off. Values are expressed as means ± SE. *P < 0.05 light vs. dark; n = 5 hearts/group.
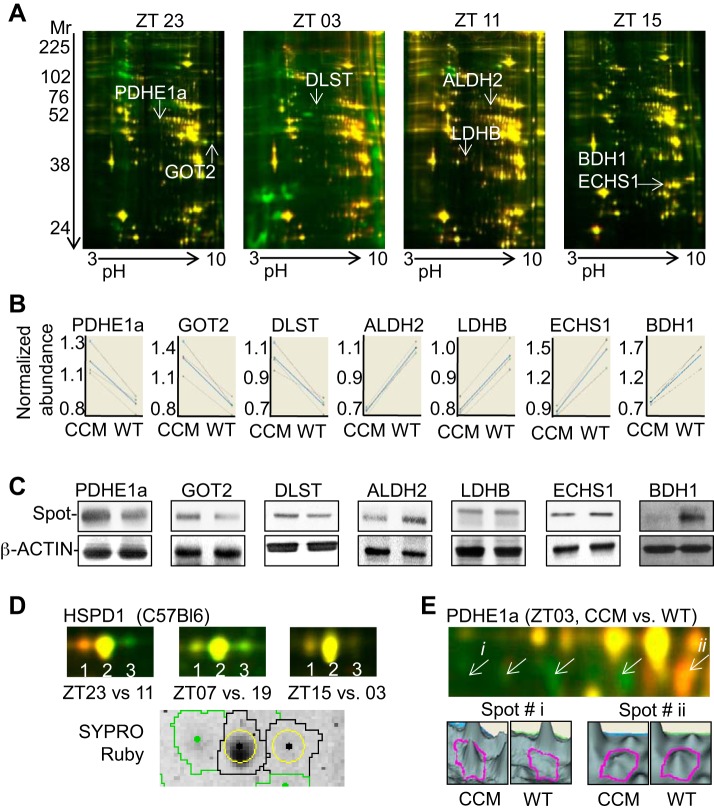
Critical to circadian clock function is the transcription factor CLOCK (reviewed in Refs. 19, 40, 52, 62, 65, and many others). To investigate the relative contribution of the cardiomyocyte clock on cardiac protein abundance, we next used CCM mice (17). We performed 2D-DIGE to investigate global changes in CCM vs. WT hearts across the diurnal cycle. Triplicate mouse hearts (n = 3) were examined at each time point. As shown in Fig. 5A, 2D-DIGE revealed that CCM hearts had 56 spots that differed (P < 0.05) in abundance (of 1,471 spots identified on these gels) vs. WT. A subset was identified by LC/MS/MS and Mascot (Fig. 5A, arrows), then independently validated by DeCyder (Fig. 5B), and Western blot analysis (Fig. 5C). Proteins with increased (P < 0.05) abundance in CCM hearts included pyruvate dehydrogenase E1a, mitochondrial (PDHE1a) (1.4 ± 0.1 FC), aspartate aminotransferase, mitochondrial (GOT2) (1.4 ± 0.02 FC), and dihydrolipoyllysine succinyltransferase of 2-oxoglutarate dehydrogenase, mitochondrial (DLST) (1.5 ± 0.1 FC), while proteins with decreased (P < 0.05) abundance were aldehyde dehydrogenase, mitochondrial (ALDH2) (1.4 ± 0.01 FC), l-lactate dehydrogenase B (LDHB) (1.3 ± 0.01 FC), enoyl-CoA hydratase, mitochondrial (ECHS1) (1.6 ± 0.1 FC), and d-β-hydroxybutyrate dehydrogenase, mitochondrial (BDH1) (2.1 ± 0.1 FC) (see Supplemental Data S1, S3 for details). Taken together, we estimated that 3.8% of soluble cardiac proteome exhibited differences in abundance in CCM, providing further support for a role for the circadian clock in regulating protein abundance in the heart.
Fig. 5.
The proteome in cardiomyocyte clock mutant (CCM) heart. Detection of de novo proteins with altered abundance in CCM (Cy3) vs. wild-type (WT; Cy5) hearts at four different time points (ZT23, ZT03, ZT11, ZT15) by 2D-DIGE (A) and DeCyder bioinformatics analyses (B). C: Western blots were performed to independently validate spots with FC ≥ 1.4 (P < 0.05) and selected for identification by LC/MS/MS. See Supplemental Data S1, S3 for tryptic peptide hits from mass spectrometry, protein identification, and DeCyder data. D: posttranslational modifications also occur at different times of day or night in the heart, with multiple rhythmic isoforms revealed for HSPD1 (upper, 2D-DIGE; lower, SYPRO ruby stain) in normal C57BL/6 hearts (D), and PDHE1a (upper, 2D-DIGE; lower, DeCyder peak views) in CCM hearts For all investigations, n = 3 per group.
In addition to modulation of total protein levels, the cardiomyocyte clock may influence the proteome through posttranslational modifications. Our 2D-DIGE revealed multiple isoforms of proteins in the heart; it has been noted by others that isoforms carry different charges and, thus, are segregated by isoelectric focusing (60). For example, three distinct spots were present for 60-kDa heat shock protein, mitochondrial (HSPD1) in normal C57BL/6 hearts (Fig. 5D). Moreover, we found six distinct PDHE1a spots, which predominated in CCM vs. WTs at different times of day or night (Fig. 5E). PDHE1a protein at spot i exhibits a 1.88 increase at ZT03 (P < 0.05) in CCM vs. WT (1.70 ± 0.26 au vs. 0.90 ± 0.04 au) heart, and at spot ii a 1.57 decrease (P < 0.005) in CCM vs. WT (0.67 ± 0.03 au vs. 1.05 ± 0.01 au) heart. One likely posttranslational modification that expresses in a circadian manner is phosphorylation, as described by Reddy et al. (60); other possibilities may also exist.
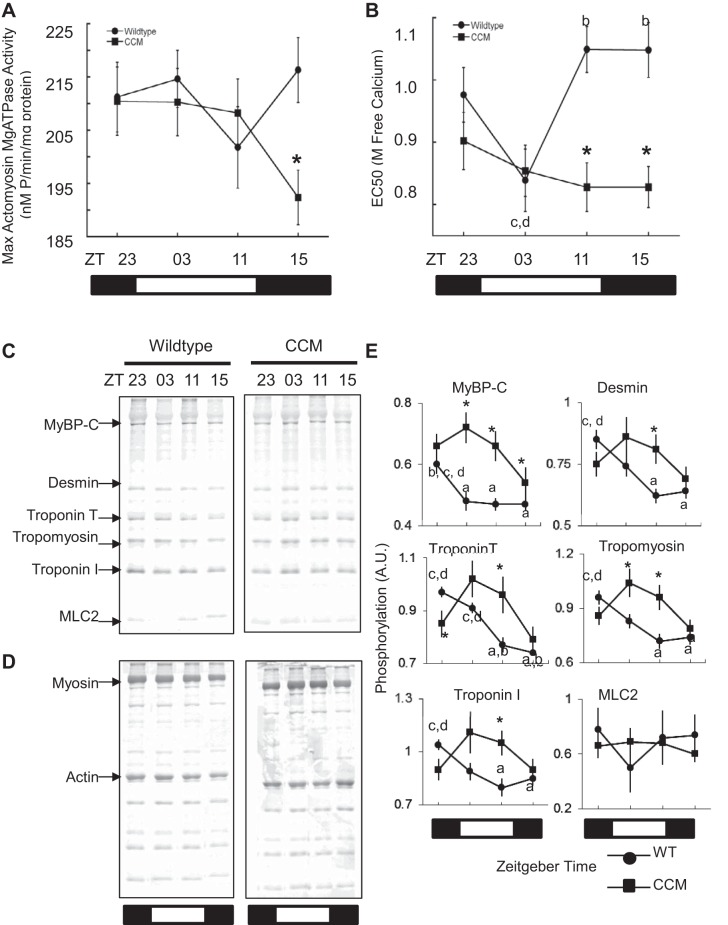
To examine whether the cardiomyocyte clock influenced diurnal myofilament function, we next purified cardiac myofilaments at different times across the diurnal cycle for WT and CCM hearts, and measured actomyosin MgATPase activity. The subcellular fraction was enriched for myofilaments and actinomyosin ATPase evaluated using established protocols (55, 59, 90). Compared with hearts from WT littermates, the CCM hearts lacked rhythmic myofilament calcium sensitivity (Fig. 6, A and B, Supplemental Data S5). For example, myofilament calcium sensitivity as measured by EC50 lacked a rhythm in CCM hearts, but was variable across the diurnal cycle in WT hearts (Fig. 6B). We examined the expression of myosin in the myofilament preparations and found no evidence of isoform shifts contributing to this effect. To further investigate diurnal changes in cardiac myofilaments, we examined phosphorylation patterns by SDS-PAGE and Pro-Q Diamond stain (Fig. 6, C and D). The myofilament phosphorylation samples were taken from the same flash-frozen tubes as the ATPase assay and run simultaneously. Consistent with the functional measurements across the diurnal cycle, CCM hearts exhibited altered (P < 0.05) myofilament protein phosphorylation at times across the diurnal cycle for cardiac myosin binding protein-C, desmin, troponins T and I, and tropomyosin compared with WT (Fig. 6E, Supplemental Data S5). These findings illustrate time-dependent posttranslational modifications of cardiac myofilament proteins, which are dependent on the cardiomyocyte circadian clock.
Fig. 6.
Diurnal variation in myofilament function and phosphorylation. To examine whether temporal posttranslational modifications were associated with myofilament function, cardiac myofilaments were purified at different times of day or night. Compared with normal WT hearts, the CCM hearts lacked diurnal variation in myofilament activity as measured by actomyosin MgATPase (A), and EC50 (B). Time-of-day myofilament phosphorylation patterns across the diurnal cycle revealed by SDS-PAGE and Pro-Q Diamond stain (C), and normalization by Coomassie blue stain (D), and densitometry values plotted (E). Myofilament ATPase and phosphorylation data are in Supplemental Data S5. Values are expressed as means ± SE. Letters a,b,c,d denote statistical significance, P < 0.05; for time points, see Supplemental Data S5. *P < 0.05; n ≥ 8 per time point.
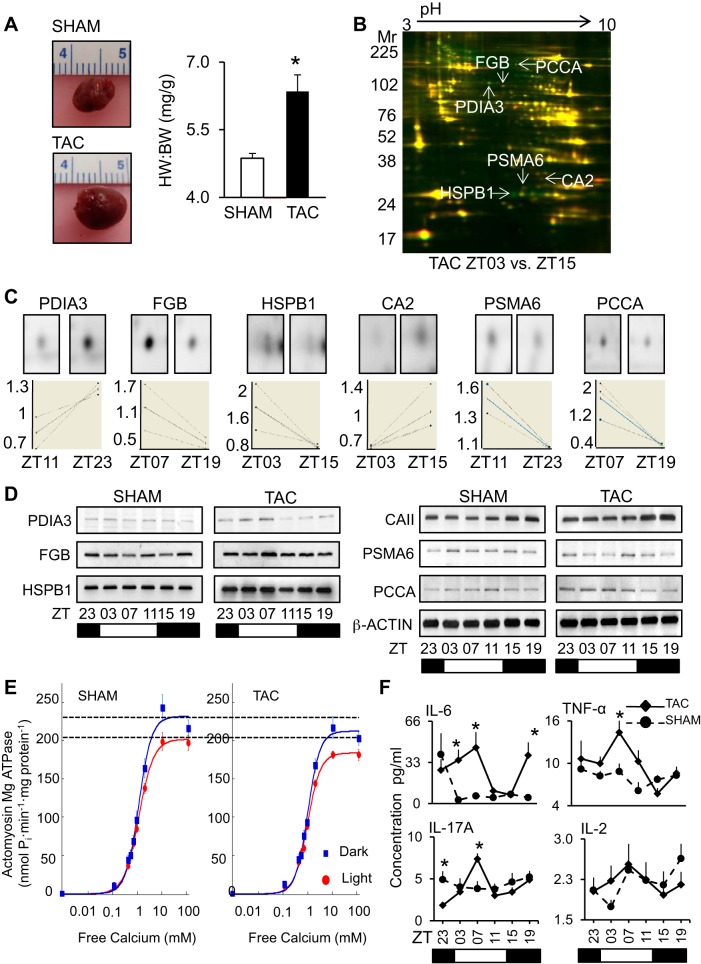
Finally, we examined what happens in heart disease. Circadian rhythms play an important role in timing of onset of adverse cardiovascular events, for example (45, 48, 49, 79, 89). In addition, blood pressure exhibits a 24-h cycling pattern; a major risk factor for cardiovascular disease occurs in hypertensive subjects that do not exhibit a dip during the night (87). Moreover, diurnal timing for cardiovascular therapies may benefit cardiac remodeling (43). Also, diurnal gene rhythms can be useful biomarkers of heart disease (82). Thus in this fourth model, we began to investigate what happened to the diurnal proteome in heart disease, using an established murine model of pressure overload-induced cardiac hypertrophy (TAC). As anticipated, increased (P < 0.01) heart weight-to-body weight ratio was observed in sham vs. TAC (4.9 ± 0.1 mg/g vs. 6.4 ± 0.4 mg/g) mice at 1 wk postsurgery (Fig. 7A, also Supplemental Data S6 for pathophysiology). Moreover, analysis by 2D-DIGE revealed that TAC hearts had 70 spots that differed (P < 0.05) in abundance across the diurnal cycle on 2D-DIGE (of the 997 identified). We selected six (arrows) with differences at pairwise times (ZT23 vs. ZT11, ZT03 vs. ZT15, and ZT07 vs. ZT19) for identification by LC/MS/MS (see Supplemental Data S1 and S3). One representative gel is shown in Fig. 7B, with the identified spots noted. Differences in abundance were also illustrated by DeCyder single channel (Fig. 7C, upper), graphs (lower), and validated by Western blot analyses across the diurnal time points (Fig. 7D). Proteins with increased (P < 0.05) abundance in TAC hearts in the light phase were fibrinogen beta chain (FGB) (ZT07 vs. ZT19, 1.6 ± 0.2 FC), heat shock protein b1 (HSPB1) (ZT03 vs. ZT15, 2.0 ± 0.4 FC), proteasome subunit 6a (PSMA6) (ZT11 vs. ZT23, 1.4 ± 0.1 FC), and propionyl-CoA carboxylase-α (PCCA) (ZT07 vs. ZT19, 3.7 ± 0.9 FC), and protein disulfide isomerase A3 (PDIA3) (ZT11 vs. ZT23, 1.5 ± 0.3 FC). Carbonic anhydrase 2 (CA2) (ZT03 vs. ZT15, 1.5 ± 0.2 FC) was increased (P < 0.05) during the dark. Moreover, we purified cardiac myofilaments and measured actomyosin MgATPase activity (Fig. 7E, Supplemental Data S7), revealing that TAC hearts exhibited a diurnal pattern of myofilament sensitivity (dark, 217.06 ± 14.24 vs. light, 181.31 ± 4.93 nmol Pi·min−1·mg−1); however, levels were reduced (P < 0.05) compared with sham hearts (dark, 242.69 ± 17.18 vs. light, 198.70 ± 11.82 nmol Pi·min−1·mg−1). We also investigated the peripheral cytokine proteome revealing that TAC alters (P < 0.05) diurnal cycling of innate serum cytokines IL-6, TNF-α, and IL-17α, but not Th1/Th2 cytokines implicated in other types of responses (Fig. 7F, Supplemental Data S8). Taken together, these studies confirm that coordination of cardiac function in health and disease is more complex than previously anticipated and that circadian regulation occurring across multiple levels, including transcriptional, translational, and posttranslational, can play a role in the disease process.
Fig. 7.
Altered day/night protein profiles in TAC heart. To demonstrate a time-of-day proteome underlying heart disease, we used a model of compensated heart disease (transverse aortic constriction; TAC). A: representative hearts from mice at 1 wk post-TAC were enlarged (left, top) vs. sham (left, bottom), and characterized by cardiac hypertrophy (right). TAC, black, sham, white. *P < 0.05; n = 36 (18 TAC, 18 sham). HW, heart weight, mg; BW, body weight, g. See Supplemental Data S6 for pathophysiology data (morphometrics, echocardiography, and hemodynamics). B: proteins with altered abundance in hearts at 1 wk post-TAC at different time points across the diurnal cycle were identified by 2D-DIGE. and C: DeCyder bioinformatics (top, single-channel spot images; bottom, graph views). D: independent validation of DeCyder bioinformatics by Western blot, using β-ACTIN as a loading control. One-week TAC profiles were further compared with the sham profiles on Western blots. See Supplemental Data S1 and S3 for tryptic peptide hits from LC/MS/MS, protein identification, and DeCyder data. E: hearts at 1 wk post-TAC were maintained with diurnal variation in myofilament activity, as measured by actomyosin MgATPase, but the diurnal calcium profiles were attenuated; also see Supplemental Data S7. F: compared with shams, the 1 wk post-TAC mice had altered diurnal profiles in serum cytokines important for early remodeling, as measured by multiplex ELISA (also see Supplemental Data S8). Values are expressed as means ± SE. *P < 0.05; n = 6 samples per time point, 6 time points total.
DISCUSSION
In this study, global proteomic approaches were used to test the hypothesis that the cardiac proteome differs in abundance over the 24-h day/night period, that temporal changes in proteins can be dependent on the cardiomyocyte circadian clock mechanism and that maintaining these rhythms is important for cardiac function. We found differences in abundance in ∼7.8% of the normal soluble cardiac proteome as analyzed by 2D-DIGE and mass spectrometry at 12-h paired time points across the diurnal cycle. This estimate is conservatively within the range in other murine tissues and other organisms (14, 29, 46, 60, 75, 83), albeit slightly lower than murine liver (up to 20%) (60). Daily oscillations in cellular function are due to a combination of fluctuations in the neurohormonal milieu (i.e., extrinsic factors) and cell autonomous rhythms (driven by circadian clocks). Manipulation of the L:D cycle affects both extrinsic and intrinsic factors, and, accordingly, disrupts rhythms in both the cardiac proteome and contractility relative to the diurnal cycle. Selective genetic disruption of the cardiomyocyte circadian clock attenuates rhythms in the subset of the cardiac proteome. There is also a temporal cardiac proteome underlying heart disease.
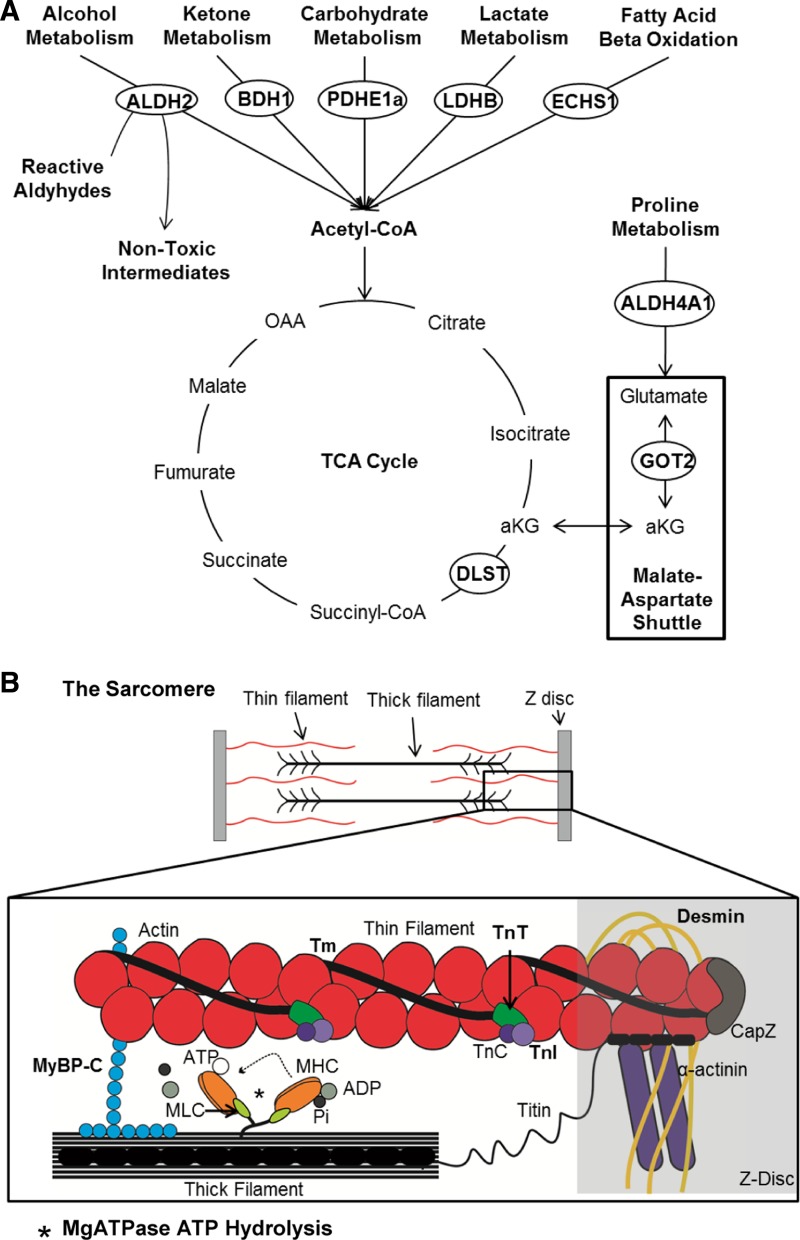
In our first study in the normal heart, we identified several mitochondrial metabolic proteins involved in substrate generation for the citric acid cycle [HADHB (84), ALDH4a1(85)], the citric acid cycle itself (ACO2), and ATP production [ATP5A1(31)]. HADHB, a component of the trifunctional protein complex responsible for fatty acid β-oxidation, was most abundant during the sleep phase, which corresponds with times of greater reliance on fatty acids by the heart (92). Conversely, ALDH4a1 was most abundant during the L:D transition times, making it tempting to speculate on alternative substrates for energy production during this transitional metabolic period. Moreover, ACO2, which catalyzes the conversion of citrate to isocitrate in the citric acid cycle was least abundant during ALD4a1 peak abundance, thus providing further support for time-of-day activity. Decreased isocitrate production may be compensated for by increased α-ketogluturate production, allowing the TCA cycle to continue. Since our studies focused on soluble proteins, many of the diurnal proteins identified included enzymes predominantly associated with vital cardiac metabolic pathways. Indeed, it is tempting to speculate that mitochondrial content, in general, exhibits a circadian pattern. This would be consistent with the notion of diurnal metabolism, and moreover, it was recently demonstrated that mitochondrial proteins involved in metabolic pathways are influenced by circadian clock-driven acetylation (44). A more detailed look at the day-night cardiac mitochondrial proteome is likely worthy of further investigation, although beyond the scope of the current study.
We also investigated the underlying diurnal mRNA expression by JTK_CYCLE, which revealed statistically significant rhythms for up to 17% of the cardiac transcriptome, which is consistent with reports by others (38, 41, 76, 82). These included several of our genes of interest, supporting the notion that transcription can underlie rhythmic protein abundance. Moreover, the Per2 mRNA exhibited a delay between message and protein as anticipated, and interestingly, Igf2, Aldh4a1, and Aco2 were relatively coincident and Stip1 appeared to exhibit a delay. Our in silico analyses revealed the presence of E-boxes in many additional genes, suggesting that translational control through promoter activity of known circadian elements alone is not sufficient to explain these corresponding mRNA profiles. Additional mechanisms likely are also involved in regulating diurnal protein abundance in the heart. These findings are consistent with reports in other tissues such as liver (60). Collectively, these studies promote new understanding of cardiac processes, and diurnal regulation provides a means for promoting protein abundance and flexibility over a 24-h period.
Next, we examined what happens to the cardiac proteome when L:D rhythms were altered. We and others have previously demonstrated that altered L:D adversely affects the expression of diurnal gene rhythms in the heart and cardiac function (18, 39–41, 56, 93); however, effects on the cardiac proteome have not been similarly investigated. This study revealed altered cardiac protein profiles with altered L:D. A CLAMS was used to demonstrate that the mice do not synchronize to the shortened L:D cycle, consistent with previous studies (32). Therefore, heart proteins were sampled in reference to the L:D cycle, but there was still a clear endogenous rhythm in body physiology that likely contributed to altered protein abundance. Moreover, there was loss of diurnal variation in LVDP on Langendorff analyses in the hearts of diurnally disrupted mice, relative to the L:D cycle. Thus, the altered diurnal environment can impact cardiac protein abundance and heart function relative to the L:D cycle. These findings have implications for cardiovascular health and disease of individuals subjected to rhythm disruptions, such as shift workers, patients with sleep disorders (including sleep apnea), nondipping hypertensives, as well as others associated with the 24/7 demands of society.
Our findings also suggest that protein abundance in the heart is regulated, in part, by the cardiomyocyte clock. For example, we found significantly altered abundance of GOT2 and BDH1 in CCM hearts. Similarly, previous microarray studies revealed altered got2 and bdh1 gene expression in CCM vs. WT hearts (7), consistent with our findings. Also, regulation of PDHE1a and LDHB by the circadian clock may be at the translational level contributing to the observed rhythms in glucose and lactate metabolism and are supported by previous observations that rhythmic cardiac glucose and lactate metabolism are disrupted in CCM mice (15). Lastly, it is tempting to speculate that the posttranslational modification of PDH observed in CCM hearts is potentially phosphorylation. PDH, when phosphorylated, is inactivated. Decreased PDH activity, plus decreased LDHB, ECHS1, and BDH1 could limit acetyl-CoA production in CCM hearts. A mechanistic illustration of the metabolic proteins with altered abundance in CCM hearts is in Fig. 8A.
Fig. 8.
Circadian control of critical cardiac processes. A: protein abundance differs in CCM vs. WT heart at different times of day or night, including rate-limiting enzymes important for vital metabolic pathways that provide substrate for the trichloroacetic acid (TCA) cycle. Identified proteins are circled. B: schematic diagram of the sarcomere, the functional unit of the cardiomyocyte. Contraction is achieved through complex interactions between the thin and thick filaments and the Z-disc. The thin filaments comprise proteins, including actin subunits, α-helix tropomyosin (Tm), and the troponin complex [troponin T (TnT), troponin I (TnT), and troponin C (TnC)]. Thick filaments are composed of a number of proteins, including myosin binding protein C (MyBP-C), and myosin heads consisting of myosin heavy chain (MHC) and myosin light chain (MLC) and magnesium-dependent ATPase (MgATPase). Z-discs consist of demsin, α-actinin, titin, and actin-capping protein (capZ) and other proteins to anchor filaments. Calcium binding to the troponin complex allows results in thin-filament conformational changes, permitting the myosin head of the thick filament to bind actin, leading to sarcomere contraction. When the myosin head is released, ATP is used by the Mg-dependent ATPase to hydrolyze ATP to ADP + Pi, and return the myosin head back to its starting conformation.
The heart responds to diurnal variations in drive, altering metabolic expression, oxidative metabolism, and energy handling. While significant research has been undertaken to understand the circadian variances in these areas, relatively little is known about how myofilaments—the central contractile element and largest consumer of energy in cardiac myocytes—are affected by circadian rhythms. Recently, investigators identified a link between cardiac myofilaments and the circadian clock when they reported the movement of CLOCK protein between cardiac Z-discs and nuclei of cells under stress (4); however, subsequent work failed to repeat these findings(86). Interestingly, recent work by Collins et al. (11) reports novel time-of-day variation in myocardial contractility that could not be fully explained by differences in calcium handling. Moreover, a comprehensive cardiac myofilament subproteome has been described with potential for studying dynamic changes important for myofilament contraction (91). Roles for myofilaments have been hypothesized; however, no previous study has examined diurnal variations in myofilament function or regulation. Thus, despite these and other studies suggesting that cardiac myofilaments are targeted by the circadian clock, to our knowledge, the current study is the first investigation of circadian variations in cardiac myofilament function.
A novel finding of our experiments is that myofilament function exhibits a temporal oscillation in the heart. A mechanistic illustration of these myofilament proteins is provided (Fig. 8B).The observed rhythms may be due in part to daily variations in cAMP levels (58), a molecule known to regulate bioavailability of the cAMP-dependent protein kinase A, which is crucial for phosphorylating myofilament proteins(72). Consistent with this notion is that we found rhythms in PRKACA, the major catalytic subunit of cAMP protein-dependent kinase (PKA) responsible for regulation of cardiac structure and function via posttranslational modification of numerous intracellular targets. PRKACA was most abundant in normal hearts during the early waking hours, which may allow for increased PKA activity and modulation of cardiac function via myofilament phosphorylation. Moreover, we found altered ATPase activity in CCM hearts, further indicating a role for the clock mechanism in myofilament structure and function. These diurnal differences in myofilament function were associated with observable alterations in phosphorylation of several key myofilament proteins important for sarcomere function (72, 73). Consistent with these findings of a role for the circadian mechanism in sarcomeric structure/function is that GSK3b exhibits diurnal activity(30) and is mechanistically tied to the cardiac circadian clock mechanism (16) and has been shown recently to phosphorylate cMyBP-C (36). A critical implication of these findings relates to cardiac hypertrophy (TAC mice), in which we observed a reduction in myofilament sensitivity to Ca2+. Moreover, these findings are consistent with recent reports of circadian oscillations in calcineurin activity and protein phosphorylation in normal vs. TAC hypertrophy and failing hearts (67). There are also diurnal variations in myocellular excitation-contraction coupling that relate to calcium homeostasis (10). Other avenues could also be worthy of future study. For example, we enriched for myofilaments consistent with our established protocols (55, 59, 90); however, future studies could use 2D-DIGE to examine preparations enriched for sarcomeric proteins, including alpha-myosin and beta-myosin heavy-chain changes in disease, and circadian changes in phosphorylation of sarcomeric proteins identified by isoelectric focusing. In summary, these findings indicate novel time-dependent posttranslational mechanisms within cardiac sarcomeres that influence myofilament contractility and relaxation over the course of the day.
Perspectives and Significance
Clinical and experimental studies have revealed that circadian rhythms play an important role in the pathophysiology of heart disease [e.g., timing of onset of myocardial infarction (MI) (9, 27, 48), severity of MI (16, 61, 78), time of day of ventricular tachyarrhythmias (79), temporal nocturnal hypertensive profiles (25, 70), and response to angiotensin-converting enzyme inhibitors (43, 71)]. Furthermore, a flurry of recent studies has associated cardiovascular structure and function with an underlying rhythmic genome by microarray and bioinformatics analyses (38, 66, 76). There are altered diurnal genomic expression profiles in TAC heart disease (41, 82). Moreover, diurnal gene expression profiles in the heart are regulated by both the diurnal environment (40, 41) and the cardiomyocyte clock mechanism (17, 19). However, the diurnal proteome has not been similarly investigated. Although genes encode proteins, it is the proteins that actually perform many important cellular functions. Here, we use proteomic approaches to reveal a temporal cardiac proteome, regulation by both the environment and the cardiac clock, and altered diurnal protein abundance in heart disease. These findings support the notion that temporal regulation of substrate utilization optimizes metabolic efficiency and cardiac function in a normal diurnal environment, and elucidates an important yet relatively unrecognized role in heart disease. The implications are that de novo studies, such as these, investigating the diurnal cardiac metabolic, sarcomeric, and myocellular proteome, can lead to new approaches to understand and treat heart disease.
GRANTS
This work was supported by grants from the Canadian Institutes of Health Research (MOP119518 to T. A. Martino) and the National Heart, Lung, and Blood Institute (HL-074259, M. Young).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
P.P., W.G.P., S.A., F.J.A., E.V.T., W.F.R., A.M., J.A.S., D.C.W., M.E.Y., and T.A.M. performed experiments; P.P., W.G.P., S.A., F.J.A., E.V.T., A.M., J.A.S., D.C.W., and T.A.M. analyzed data; P.P., W.G.P., F.J.A., E.V.T., and T.A.M. prepared figures; P.P., W.G.P., S.A., F.J.A., E.V.T., W.F.R., A.M., J.A.S., G.K., D.C.W., M.E.Y., and T.A.M. approved final version of manuscript; W.G.P., J.A.S., G.K., M.E.Y., and T.A.M. conception and design of research; P.P., W.G.P., S.A., F.J.A., E.V.T., D.C.W., and T.A.M. interpreted results of experiments; W.G.P., M.E.Y., and T.A.M. edited and revised manuscript; T.A.M. drafted manuscript.
Supplementary Material
ACKNOWLEDGMENTS
The authors sincerely thank Dr. John Hogenesch at the Institute for Biomedical Informatics, Institute for Translation Medicine and Therapeutics, University of Pennsylvania Perelman School of Medicine, Philadelphia PA, for analysis of the microarray data using JTK_CYCLE, and for his thoughtful comments and helpful advice.
REFERENCES
- 1.Alban A, David SO, Bjorkesten L, Andersson C, Sloge E, Lewis S, Currie I. A novel experimental design for comparative two-dimensional gel analysis: two-dimensional difference gel electrophoresis incorporating a pooled internal standard. Proteomics 3: 36–44, 2003. [DOI] [PubMed] [Google Scholar]
- 2.Alibhai FJ, Tsimakouridze EV, Chinnappareddy N, Wright DC, Billia F, O'Sullivan L, Pyle WG, Sole MJ, Martino TA. Short-term disruption of diurnal rhythms following murine myocardial infarction adversely affects long term myocardial structure and function. Circ Res 114: 1713–1722, 2014. [DOI] [PubMed] [Google Scholar]
- 3.Bingham C, Arbogast B, Guillaume GC, Lee JK, Halberg F. Inferential statistical methods for estimating and comparing cosinor parameters. Chronobiologia 9: 397–439, 1982. [PubMed] [Google Scholar]
- 4.Boateng SY, Goldspink PH. Assembly and maintenance of the sarcomere night and day. Cardiovasc Res 77: 667–675, 2008. [DOI] [PubMed] [Google Scholar]
- 5.Bradley TD, Floras JS. Sleep apnea and heart failure: Part II: central sleep apnea. Circulation 107: 1822–1826, 2003. [DOI] [PubMed] [Google Scholar]
- 6.Bray MS, Ratcliffe WF, Grenett MH, Brewer RA, Gamble KL, Young ME. Quantitative analysis of light-phase restricted feeding reveals metabolic dyssynchrony in mice. Int J Obes 37: 843–852, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bray MS, Shaw CA, Moore MW, Garcia RA, Zanquetta MM, Durgan DJ, Jeong WJ, Tsai JY, Bugger H, Zhang D, Rohrwasser A, Rennison JH, Dyck JR, Litwin SE, Hardin PE, Chow CW, Chandler MP, Abel ED, Young ME. Disruption of the circadian clock within the cardiomyocyte influences myocardial contractile function, metabolism, and gene expression. Am J Physiol Heart Circ Physiol 294: H1036–H1047, 2008. [DOI] [PubMed] [Google Scholar]
- 8.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162: 156–159, 1987. [DOI] [PubMed] [Google Scholar]
- 9.Cohen MC, Rohtla KM, Lavery CE, Muller JE, Mittleman MA. Meta-analysis of the morning excess of acute myocardial infarction and sudden cardiac death. Am J Cardiol 79: 1512–1516, 1997. [DOI] [PubMed] [Google Scholar]
- 10.Collins HE, Rodrigo GC. Inotropic response of cardiac ventricular myocytes to beta-adrenergic stimulation with isoproterenol exhibits diurnal variation: involvement of nitric oxide. Circ Res 106: 1244–1252, 2010. [DOI] [PubMed] [Google Scholar]
- 11.Collins HE, Turrell HE, Samani NJ, Rodrigo GC. Diurnal variation in excitation-contraction coupling is lost in the adult spontaneously hypertensive rat heart. J Hypertens 31: 1214–1223, 2013. [DOI] [PubMed] [Google Scholar]
- 12.Dallmann R, Viola AU, Tarokh L, Cajochen C, Brown SA. The human circadian metabolome. Proc Natl Acad Sci USA 109: 2625–2629, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics 20: 1453–1454, 2004. [DOI] [PubMed] [Google Scholar]
- 14.Deery MJ, Maywood ES, Chesham JE, Sladek M, Karp NA, Green EW, Charles PD, Reddy AB, Kyriacou CP, Lilley KS, Hastings MH. Proteomic analysis reveals the role of synaptic vesicle cycling in sustaining the suprachiasmatic circadian clock. Curr Biol: 19: 2031–2036, 2009. [DOI] [PubMed] [Google Scholar]
- 15.Durgan DJ, Pat BM, Laczy B, Bradley JA, Tsai JY, Grenett MH, Ratcliffe WF, Brewer RA, Nagendran J, Villegas-Montoya C, Zou C, Zou L, Johnson RL, Jr, Dyck JR, Bray MS, Gamble KL, Chatham JC, Young ME. O-GlcNAcylation, novel post-translational modification linking myocardial metabolism and cardiomyocyte circadian clock. J Biol Chem 286: 44606–44619, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Durgan DJ, Pulinilkunnil T, Villegas-Montoya C, Garvey ME, Frangogiannis NG, Michael LH, Chow CW, Dyck JR, Young ME. Short communication: ischemia/reperfusion tolerance is time-of-day-dependent: mediation by the cardiomyocyte circadian clock. Circ Res 106: 546–550, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Durgan DJ, Trexler NA, Egbejimi O, McElfresh TA, Suk HY, Petterson LE, Shaw CA, Hardin PE, Bray MS, Chandler MP, Chow CW, Young ME. The circadian clock within the cardiomyocyte is essential for responsiveness of the heart to fatty acids. J Biol Chem 281: 24254–24269, 2006. [DOI] [PubMed] [Google Scholar]
- 18.Durgan DJ, Tsai JY, Grenett MH, Pat BM, Ratcliffe WF, Villegas-Montoya C, Garvey ME, Nagendran J, Dyck JR, Bray MS, Gamble KL, Gimble JM, Young ME. Evidence suggesting that the cardiomyocyte circadian clock modulates responsiveness of the heart to hypertrophic stimuli in mice. Chronobiol Int 28: 187–203, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Durgan DJ, Young ME. The cardiomyocyte circadian clock: emerging roles in health and disease. Circ Res 106: 647–658, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Floras JS. Sleep apnea in heart failure: implications of sympathetic nervous system activation for disease progression and treatment. Curr Heart Fail Rep 2: 212–217, 2005. [DOI] [PubMed] [Google Scholar]
- 21.Furlan R, Barbic F, Piazza S, Tinelli M, Seghizzi P, Malliani A. Modifications of cardiac autonomic profile associated with a shift schedule of work. Circulation 102: 1912–1916, 2000. [DOI] [PubMed] [Google Scholar]
- 22.Guo YF, Stein PK. Circadian rhythm in the cardiovascular system: chronocardiology. Am Heart J 145: 779–786, 2003. [DOI] [PubMed] [Google Scholar]
- 23.Hanash S. Integrated global profiling of cancer. Nat Rev Cancer 4: 638–644, 2004. [DOI] [PubMed] [Google Scholar]
- 24.Hatcher NG, Atkins N, Jr, Annangudi SP, Forbes AJ, Kelleher NL, Gillette MU, Sweedler JV. Mass spectrometry-based discovery of circadian peptides. Proc Natl Acad Sci USA 105: 12527–12532, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hermida RC, Ayala DE, Mojon A, Fernandez JR. Influence of circadian time of hypertension treatment on cardiovascular risk: results of the MAPEC study. Chronobiol Int 27: 1629–1651, 2010. [DOI] [PubMed] [Google Scholar]
- 26.Hobson DJ, Rupa P, Diaz GJ, Zhang H, Yang M, Mine Y, Turner PV, Kirby GM. Proteomic analysis of ovomucoid hypersensitivity in mice by two-dimensional difference gel electrophoresis (2D-DIGE). Food Chem Toxicol 45: 2372–2380, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu K, Ivanov P, Hilton MF, Chen Z, Ayers RT, Stanley HE, Shea SA. Endogenous circadian rhythm in an index of cardiac vulnerability independent of changes in behavior. Proc Natl Acad Sci USA 101: 18223–18227, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hughes ME, Hogenesch JB, Kornacker K. JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J Biol Rhythms 25: 372–380, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hwang H, Cho MH, Hahn BS, Lim H, Kwon YK, Hahn TR, Bhoo SH. Proteomic identification of rhythmic proteins in rice seedlings. Biochim Biophys Acta 1814: 470–479, 2011. [DOI] [PubMed] [Google Scholar]
- 30.Iitaka C, Miyazaki K, Akaike T, Ishida N. A role for glycogen synthase kinase-3β in the mammalian circadian clock. J Biol Chem 280: 29397–29402, 2005. [DOI] [PubMed] [Google Scholar]
- 31.Kagawa Y, Sone N, Hirata H, Yoshida M. Structure and function of H+-ATPase. J Bioenerg Biomembr 11: 39–78, 1979. [DOI] [PubMed] [Google Scholar]
- 32.Karatsoreos IN, Bhagat S, Bloss EB, Morrison JH, McEwen BS. Disruption of circadian clocks has ramifications for metabolism, brain, and behavior. Proc Natl Acad Sci USA 108: 1657–1662, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, Takahashi JS. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 338: 349–354, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krauchi K. How is the circadian rhythm of core body temperature regulated? Clin Auton Res 12: 147–149, 2002. [DOI] [PubMed] [Google Scholar]
- 35.Kumaki Y, Ukai-Tadenuma M, Uno KD, Nishio J, Masumoto KH, Nagano M, Komori T, Shigeyoshi Y, Hogenesch JB, Ueda HR. Analysis and synthesis of high-amplitude Cis-elements in the mammalian circadian clock. Proc Natl Acad Sci USA 105: 14946–14951, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuster DW, Sequeira V, Najafi A, Boontje NM, Wijnker PJ, Witjas-Paalberends ER, Marston SB, Dos Remedios CG, Carrier L, Demmers JA, Redwood C, Sadayappan S, van der Velden J. GSK3β phosphorylates newly identified site in the proline-alanine-rich region of cardiac myosin-binding protein C and alters cross-bridge cycling kinetics in human. Circ. Res. 112: 633–639, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lilley KS, Friedman DB. All about DIGE: quantification technology for differential-display 2D-gel proteomics. Expert Rev Proteomics 1: 401–409, 2004. [DOI] [PubMed] [Google Scholar]
- 38.Martino T, Arab S, Straume M, Belsham DD, Tata N, Cai F, Liu P, Trivieri M, Ralph M, Sole MJ. Day/night rhythms in gene expression of the normal murine heart. J Mol Med 82: 256–264, 2004. [DOI] [PubMed] [Google Scholar]
- 39.Martino TA, Oudit GY, Herzenberg AM, Tata N, Koletar MM, Kabir GM, Belsham DD, Backx PH, Ralph MR, Sole MJ. Circadian rhythm disorganization produces profound cardiovascular and renal disease in hamsters. Am J Physiol Regul Integr Comp Physiol 294: R1675–R1683, 2008. [DOI] [PubMed] [Google Scholar]
- 40.Martino TA, Sole MJ. Molecular time: an often overlooked dimension to cardiovascular disease. Circ Res 105: 1047–1061, 2009. [DOI] [PubMed] [Google Scholar]
- 41.Martino TA, Tata N, Belsham DD, Chalmers J, Straume M, Lee P, Pribiag H, Khaper N, Liu PP, Dawood F, Backx PH, Ralph MR, Sole MJ. Disturbed diurnal rhythm alters gene expression and exacerbates cardiovascular disease with rescue by resynchronization. Hypertension 49: 1104–1113, 2007. [DOI] [PubMed] [Google Scholar]
- 42.Martino TA, Tata N, Bjarnason GA, Straume M, Sole MJ. Diurnal protein expression in blood revealed by high throughput mass spectrometry proteomics and implications for translational medicine and body time of day. Am J Physiol Regul Integr Comp Physiol 293: R1430–R1437, 2007. [DOI] [PubMed] [Google Scholar]
- 43.Martino TA, Tata N, Simpson JA, Vanderlaan R, Dawood F, Kabir MG, Khaper N, Cifelli C, Podobed P, Liu PP, Husain M, Heximer S, Backx PH, Sole MJ. The primary benefits of angiotensin-converting enzyme inhibition on cardiac remodeling occur during sleep time in murine pressure overload hypertrophy. J Am Coll Cardiol 57: 2020–2028, 2011. [DOI] [PubMed] [Google Scholar]
- 44.Masri S, Patel VR, Eckel-Mahan KL, Peleg S, Forne I, Ladurner AG, Baldi P, Imhof A, Sassone-Corsi P. Circadian acetylome reveals regulation of mitochondrial metabolic pathways. Proc Natl Acad Sci USA 110: 3339–3344, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mehta RH, Manfredini R, Hassan F, Sechtem U, Bossone E, Oh JK, Cooper JV, Smith DE, Portaluppi F, Penn M, Hutchison S, Nienaber CA, Isselbacher EM, Eagle KA, International Registry of Acute Aortic Dissection. I. Chronobiological patterns of acute aortic dissection. Circulation 106: 1110–1115, 2002. [DOI] [PubMed] [Google Scholar]
- 46.Moller M, Sparre T, Bache N, Roepstorff P, Vorum H. Proteomic analysis of day-night variations in protein levels in the rat pineal gland. Proteomics 7: 2009–2018, 2007. [DOI] [PubMed] [Google Scholar]
- 47.Mukamal KJ, Muller JE, Maclure M, Sherwood JB, Mittleman MA. Increased risk of congestive heart failure among infarctions with nighttime onset. Am Heart J 140: 438–442, 2000. [DOI] [PubMed] [Google Scholar]
- 48.Muller JE, Stone PH, Turi ZG, Rutherford JD, Czeisler CA, Parker C, Poole WK, Passamani E, Roberts R, Robertson T, Sobel BE, Willerson JT, Braunwald E, MILIS Study Group. Circadian variation in the frequency of onset of acute myocardial infarction New Engl J Med 313: 1315–1322, 1985. [DOI] [PubMed] [Google Scholar]
- 49.Muller JE, Tofler GH, Willich SN, Stone PH. Circadian variation of cardiovascular disease and sympathetic activity. J Cardiovasc Pharmacol 10 Suppl 2: S104–S109; discussion S110–S101, 1987. [PubMed] [Google Scholar]
- 50.Nickel W, Rabouille C. Mechanisms of regulated unconventional protein secretion. Nat Rev Mol Cell Biol 10: 148–155, 2009. [DOI] [PubMed] [Google Scholar]
- 51.Ohishi K, Nagasato K, Aoi W, Nakamura T, Ichinose K, Nishiura Y, Satoh A, Tsujihata M, Shibata Y, Nagataki S. Circadian rhythms of blood pressure and heart rate in patients with human T-lymphotropic virus type-I-associated myelopathy. The Tohoku J Exp Med 169: 67–75, 1993. [DOI] [PubMed] [Google Scholar]
- 52.Panda S, Hogenesch JB, Kay SA. Circadian rhythms from flies to human. Nature 417: 329–335, 2002. [DOI] [PubMed] [Google Scholar]
- 53.Paschos GK, FitzGerald GA. Circadian clocks and vascular function. Circ Res 106: 833–841, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Patel VR, Eckel-Mahan K, Sassone-Corsi P, Baldi P. CircadiOmics: integrating circadian genomics, transcriptomics, proteomics and metabolomics. Nat Methods 9: 772–773, 2012. [DOI] [PubMed] [Google Scholar]
- 55.Patton C, Thompson S, Epel D. Some precautions in using chelators to buffer metals in biological solutions. Cell Calcium 35: 427–431, 2004. [DOI] [PubMed] [Google Scholar]
- 56.Penev PD, Kolker DE, Zee PC, Turek FW. Chronic circadian desynchronization decreases the survival of animals with cardiomyopathic heart disease. Am J Physiol Heart Circ Physiol 275: H2334–H2337, 1998. [DOI] [PubMed] [Google Scholar]
- 57.Pizarro A, Hayer K, Lahens NF, Hogenesch JB. CircaDB: a database of mammalian circadian gene expression profiles. Nucleic Acids Res 41: D1009–D1013, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Prosser RA, Gillette MU. Cyclic changes in cAMP concentration and phosphodiesterase activity in a mammalian circadian clock studied in vitro. Brain Res 568: 185–192, 1991. [DOI] [PubMed] [Google Scholar]
- 59.Pyle WG, Sumandea MP, Solaro RJ, De Tombe PP. Troponin I serines 43/45 and regulation of cardiac myofilament function. Am J Physiol Heart Circ Physiol 283: H1215–H1224, 2002. [DOI] [PubMed] [Google Scholar]
- 60.Reddy AB, Karp NA, Maywood ES, Sage EA, Deery M, O'Neill JS, Wong GK, Chesham J, Odell M, Lilley KS, Kyriacou CP, Hastings MH. Circadian orchestration of the hepatic proteome. Curr Biol 16: 1107–1115, 2006. [DOI] [PubMed] [Google Scholar]
- 61.Reiter R, Swingen C, Moore L, Henry TD, Traverse JH. Circadian dependence of infarct size and left ventricular function after ST elevation myocardial infarction. Circ Res 110: 105–110, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature 418: 935–941, 2002. [DOI] [PubMed] [Google Scholar]
- 63.Rey G, Cesbron F, Rougemont J, Reinke H, Brunner M, Naef F. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol 9: e1000595, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Robles MS, Mann M. Proteomic approaches in circadian biology. Handbook Exp Pharmacol 389–407, 2013. [DOI] [PubMed] [Google Scholar]
- 65.Roenneberg T, Merrow M. Circadian clocks - the fall and rise of physiology. Nat Rev Mol Cell Biol 6: 965–971, 2005. [DOI] [PubMed] [Google Scholar]
- 66.Rudic RD, McNamara P, Reilly D, Grosser T, Curtis AM, Price TS, Panda S, Hogenesch JB, FitzGerald GA. Bioinformatic analysis of circadian gene oscillation in mouse aorta. Circulation 112: 2716–2724, 2005. [DOI] [PubMed] [Google Scholar]
- 67.Sachan N, Dey A, Rotter D, Grinsfelder DB, Battiprolu PK, Sikder D, Copeland V, Oh M, Bush E, Shelton JM, Bibb JA, Hill JA, Rothermel BA. Sustained hemodynamic stress disrupts normal circadian rhythms in calcineurin-dependent signaling and protein phosphorylation in the heart. Circ Res 108: 437–445, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Saldanha AJ. Java Treeview–extensible visualization of microarray data. Bioinformatics 20: 3246–3248, 2004. [DOI] [PubMed] [Google Scholar]
- 69.Shea SA, Hilton MF, Hu K, Scheer FA. Existence of an endogenous circadian blood pressure rhythm in humans that peaks in the evening. Circ Res 108: 980–984, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Smolensky MH, Haus E. Circadian rhythms and clinical medicine with applications to hypertension. Am J Hypertens 14: 280S–290S, 2001. [DOI] [PubMed] [Google Scholar]
- 71.Smolensky MH, Hermida RC, Ayala DE, Tiseo R, Portaluppi F. Administration-time-dependent effects of blood pressure-lowering medications: basis for the chronotherapy of hypertension. Blood Press Monit 15: 173–180, 2010. [DOI] [PubMed] [Google Scholar]
- 72.Solaro RJ, Henze M, Kobayashi T. Integration of troponin I phosphorylation with cardiac regulatory networks. Circ Res 112: 355–366, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Solaro RJ, Kobayashi T. Protein phosphorylation and signal transduction in cardiac thin filaments. J Biol Chem 286: 9935–9940, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sole MJ, Martino TA. Diurnal physiology: core principles with application to the pathogenesis, diagnosis, prevention, and treatment of myocardial hypertrophy and failure. J Appl Physiol 107: 1318–1327, 2009. [DOI] [PubMed] [Google Scholar]
- 75.Stockel J, Jacobs JM, Elvitigala TR, Liberton M, Welsh EA, Polpitiya AD, Gritsenko MA, Nicora CD, Koppenaal DW, Smith RD, Pakrasi HB. Diurnal rhythms result in significant changes in the cellular protein complement in the cyanobacterium Cyanothece 51142. PloS One 6: e16680, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ. Extensive and divergent circadian gene expression in liver and heart. Nature 417: 78–83, 2002. [DOI] [PubMed] [Google Scholar]
- 77.Straume M. DNA microarray time series analysis: automated statistical assessment of circadian rhythms in gene expression patterning. Methods Enzymol 383: 149–166, 2004. [DOI] [PubMed] [Google Scholar]
- 78.Suarez-Barrientos A, Lopez-Romero P, Vivas D, Castro-Ferreira F, Nunez-Gil I, Franco E, Ruiz-Mateos B, Garcia-Rubira JC, Fernandez-Ortiz A, Macaya C, Ibanez B. Circadian variations of infarct size in acute myocardial infarction. Heart 97: 970–976, 2011. [DOI] [PubMed] [Google Scholar]
- 79.Tofler GH, Gebara OC, Mittleman MA, Taylor P, Siegel W, Venditti FJ, Jr, Rasmussen CA, Muller JE, The CPI Investigators. Morning peak in ventricular tachyarrhythmias detected by time of implantable cardioverter/defibrillator therapy. Circulation 92: 1203–1208, 1995. [DOI] [PubMed] [Google Scholar]
- 80.Tong YL. Parameter estimation in studying circadian rhythms. Biometrics 32: 85–94, 1976. [PubMed] [Google Scholar]
- 81.Tonge R, Shaw J, Middleton B, Rowlinson R, Rayner S, Young J, Pognan F, Hawkins E, Currie I, Davison M. Validation and development of fluorescence two-dimensional differential gel electrophoresis proteomics technology. Proteomics 1: 377–396, 2001. [DOI] [PubMed] [Google Scholar]
- 82.Tsimakouridze EV, Straume M, Podobed PS, Chin H, LaMarre J, Johnson R, Antenos M, Kirby GM, Mackay A, Huether P, Simpson JA, Sole M, Gadal G, Martino TA. Chronomics of pressure overload-induced cardiac hypertrophy in mice reveals altered day/night gene expression and biomarkers of heart disease. Chronobiol Int 29: 810–821, 2012. [DOI] [PubMed] [Google Scholar]
- 83.Tsuji T, Hirota T, Takemori N, Komori N, Yoshitane H, Fukuda M, Matsumoto H, Fukada Y. Circadian proteomics of the mouse retina. Proteomics 7: 3500–3508, 2007. [DOI] [PubMed] [Google Scholar]
- 84.Uchida Y, Izai K, Orii T, Hashimoto T. Novel fatty acid β-oxidation enzymes in rat liver mitochondria. II. Purification and properties of enoyl-coenzyme A (CoA) hydratase/3-hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase trifunctional protein J Biol Chem 267: 1034–1041, 1992. [PubMed] [Google Scholar]
- 85.Valle D, Goodman SI, Harris SC, Phang JM. Genetic evidence for a common enzyme catalyzing the second step in the degradation of proline and hydroxyproline. J Clin Invest 64: 1365–1370, 1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang J, Dube DK, White J, Fan Y, Sanger JM, Sanger JW. Clock is not a component of Z-bands. Cytoskeleton 69: 1021–1031, 2012. [DOI] [PubMed] [Google Scholar]
- 87.White WB. Ambulatory blood pressure monitoring: dippers compared with non-dippers. Blood Press Monit 5 Suppl 1: S17–S23, 2000. [PubMed] [Google Scholar]
- 88.Willich SN, Goldberg RJ, Maclure M, Perriello L, Muller JE. Increased onset of sudden cardiac death in the first three hours after awakening. Am J Cardiol 70: 65–68, 1992. [DOI] [PubMed] [Google Scholar]
- 89.Willich SN, Levy D, Rocco MB, Tofler GH, Stone PH, Muller JE. Circadian variation in the incidence of sudden cardiac death in the Framingham Heart Study population. Am J Cardiol 60: 801–806, 1987. [DOI] [PubMed] [Google Scholar]
- 90.Yang F, Aiello DL, Pyle WG. Cardiac myofilament regulation by protein phosphatase type 1α and CapZ. Biochem Cell Biol 86: 70–78, 2008. [DOI] [PubMed] [Google Scholar]
- 91.Yin X, Cuello F, Mayr U, Hao Z, Hornshaw M, Ehler E, Avkiran M, Mayr M. Proteomics analysis of the cardiac myofilament subproteome reveals dynamic alterations in phosphatase subunit distribution. Mol Cell Proteom 9: 497–509, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Young ME. The circadian clock within the heart: potential influence on myocardial gene expression, metabolism, and function. Am J Physiol Heart Circ Physiol 290: H1–H16, 2006. [DOI] [PubMed] [Google Scholar]
- 93.Young ME, Bray MS. Potential role for peripheral circadian clock dyssynchrony in the pathogenesis of cardiovascular dysfunction. Sleep Med 8: 656–667, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Young ME, Razeghi P, Cedars AM, Guthrie PH, Taegtmeyer H. Intrinsic diurnal variations in cardiac metabolism and contractile function. Circ Res 89: 1199–1208, 2001. [DOI] [PubMed] [Google Scholar]
- 95.Young ME, Razeghi P, Taegtmeyer H. Clock genes in the heart: characterization and attenuation with hypertrophy. Circ Res 88: 1142–1150, 2001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.