Abstract
In the renal collecting duct, binding of AVP to the V2 receptor triggers signaling changes that regulate osmotic water transport. Short-term regulation of water transport is dependent on vasopressin-induced phosphorylation of aquaporin-2 (AQP2) at Ser256. The protein kinase that phosphorylates this site is not known. We use Bayes' theorem to rank all 521 rat protein kinases with regard to the likelihood of a role in Ser256 phosphorylation on the basis of prior data and new experimental data. First, prior probabilities were estimated from previous transcriptomic and proteomic profiling data, kinase substrate specificity data, and evidence for kinase regulation by vasopressin. This ranking was updated using new experimental data describing the effects of several small-molecule kinase inhibitors with known inhibitory spectra (H-89, KN-62, KN-93, and GSK-650394) on AQP2 phosphorylation at Ser256 in inner medullary collecting duct suspensions. The top-ranked kinase was Ca2+/calmodulin-dependent protein kinase II (CAMK2), followed by protein kinase A (PKA) and protein kinase B (AKT). Liquid chromatography-tandem mass spectrometry (LC-MS/MS)-based in vitro phosphorylation studies compared the ability of three highly ranked kinases to phosphorylate AQP2 and other inner medullary collecting duct proteins, PKA, CAMK2, and serum/glucocorticoid-regulated kinase (SGK). All three proved capable of phosphorylating AQP2 at Ser256, although CAMK2 and PKA were more potent than SGK. The in vitro phosphorylation experiments also identified candidate protein kinases for several additional phosphoproteins with likely roles in collecting duct regulation, including Nedd4-2, Map4k4, and 3-phosphoinositide-dependent protein kinase 1. We conclude that Bayes' theorem is an effective means of integrating data from multiple data sets in physiology.
Keywords: Bayes' theorem, vasopressin, inner medullary collecting duct, aquaporin-2, urea channel UT-A
vasopressin-mediated control of water and urea transport in the inner medullary collecting duct (IMCD) is integral to the overall regulation of renal water excretion (48). In this collecting duct segment, the actions of vasopressin are mediated solely by the V2 receptor (14, 62), which is found in the basolateral plasma membrane. An increase in water and urea transport is seen within a few minutes of an imposed increase in vasopressin concentration, with a half-time of response of 8–9 min (66). Vasopressin also triggers long-term effects in the collecting duct via changes in gene transcription (16, 30). Prior studies demonstrated that vasopressin increases cAMP levels in native IMCD cells (55) and that addition of cAMP can mimic vasopressin by increasing the osmotic water and urea permeability of isolated perfused IMCDs (55). Vasopressin, working through the V2 receptor, also increases intracellular Ca2+ in the IMCD (5, 15, 55). The Ca2+ increase occurs in the form of aperiodic Ca2+ spikes that increase in frequency when vasopressin is added (43, 69). Inhibition of Ca2+ mobilization or inhibition of calmodulin markedly reduces the water permeability response in isolated perfused IMCD segments (8). Vasopressin has also been found to activate AKT and to reduce activity of several MAP kinases in the rat IMCD (23, 43).
A key event in the minute-by-minute regulation of water transport in the collecting duct is phosphorylation of the molecular water channel aquaporin-2 (AQP2) in the collecting duct. AQP2 is phosphorylated in four positions within the last 16 amino acids in its COOH-terminal tail: Ser256, Ser261, Ser264, and Ser269 (24). Studies with phosphospecific antibodies showed that all four sites are regulated by vasopressin (21). Among these vasopressin-regulated sites, phosphorylation at Ser256 appears to play a special role, such that it is necessary for trafficking of AQP2 to the apical plasma membrane (18, 29) and for phosphorylation of AQP2 at Ser264 and Ser269 (21).
It is widely held that vasopressin-induced phosphorylation of AQP2 at Ser256 is mediated by PKA in collecting duct cells (31, 37, 38, 46, 64). The evidence that PKA mediates AQP2 phosphorylation at Ser256 appears to be limited to three observations: 1) in mammals, the sequence surrounding Ser256 is consistent with a canonical basophilic PKA targeting motif (R-R-X-S, where X is any amino acid) (19); 2) H-89, a PKA-selective protein kinase inhibitor, reduces 32P incorporation at some site in the AQP2 protein (4, 63); and 3) recombinant PKA can phosphorylate the site corresponding to Ser256 of AQP2 when incubated in vitro with synthetic AQP2 peptides (21). However, various investigators have proposed that other protein kinases may be involved in AQP2 phosphorylation at Ser256. Specifically, there is evidence for phosphorylation at Ser256 by AKT (13), casein kinase purified from the Golgi apparatus (44), protein kinase G (PKG) (4), and protein kinase C-δ (PKCδ) (13). Thus the role of PKA (vs. other basophilic protein kinases) in phosphorylation at Ser256 and in vasopressin signaling in the renal collecting duct can be considered an open question.
In our previous phosphoproteomic studies, we identified several hundred proteins that showed increased phosphorylation in response to vasopressin in native rat IMCD cells (1, 23, 24) and cultured mouse mpkCCD cells (45). Analysis of the sequence surrounding the upregulated phosphorylation sites showed a dominant basophilic motif, indicating that vasopressin likely activates one or more protein kinases in the AGC protein kinase family [including PKA, PKG, PKC, AKT isoforms, and serum/glucocorticoid-regulated kinase (SGK) isoforms] or the Ca2+/calmodulin-dependent protein kinase (CAMK) protein kinase family (consisting of multiple calmodulin-activated kinases) (35). There are 134 kinases in these 2 families, and theoretically any of these could play a role in vasopressin-dependent signaling. To narrow the possibilities, we apply Bayesian analysis, i.e., repeated application of Bayes' rule (also known as Bayes' theorem), to rank the candidate protein kinases with regard to likelihood that they phosphorylate AQP2 at Ser256 in native IMCD cells. Initial Bayesian analysis using previously published transcriptomic data, proteomic profiling data, phosphorylation target motif data, and phosphoproteomic data narrowed the list to a few highly ranked candidates. Among these, CAMK IIδ (official gene symbol1 Camk2d), PKA catalytic subunits (official gene symbols Prkaca and Prkacb), and SGK (official gene symbol Sgk1) were identified as particularly promising (for simplicity, we refer to these as CAMK2, PKA, and SGK, except when necessary to designate the specific official gene symbol for clarity).
On the basis of prior probabilities for these three kinases, we conducted additional experimental studies to provide new data for extension of the Bayesian analysis. In the first set of studies, we used a set of kinase inhibitors with known selectivity spectra to further distinguish among candidate kinases with regard to their abilities to phosphorylate AQP2 at Ser256. In the second set of studies, we incubated dephosphorylated IMCD proteins with recombinant CAMK2, PKA, or SGK for proteome-wide identification of amino acid sites that each of these three kinases can phosphorylate in vitro. An important by-product of this study is extensive protein mass spectrometry (MS) data identifying proteins in the IMCD that can be phosphorylated by CAMK2, SGK, and/or PKA.2
METHODS
Use of Bayes' Theorem to Rank Candidate Protein Kinases
To rank protein kinases with respect to the likelihood that they phosphorylate AQP2 at Ser256, we used Bayes' rule to carry out large-scale data integration of multiple data sets (10): P(A|B) = P(B|A)P(A)/P(B), where P(A B) is the probability of A given B, P(B|A) is the probability of B given A, P(A) is the prior probability for A, and P(B) is the sum of probabilities of B over all A. We began with a list of all 521 protein kinases in the mouse genome extracted from UniProt/Swiss-Prot Protein Knowledgebase (http://www.uniprot.org/docs/pkinfam), giving each an equal prior probability P(A) of 1/521. We then sequentially calculated revised probabilities using the following data sets: 1) the IMCD transcriptome identified using Affymetrix expression arrays, as reported by Uawithya et al. (62) and downloaded from the URL http://helixweb.nih.gov/ESBL/Database/Transcriptomic/IMCDdatabase.html; 2) the rat IMCD proteome, concatenated from several studies using protein MS to identify proteins expressed in rat IMCD, downloaded from the URL http://helixweb.nih.gov/ESBL/Database/IMCD_Proteome/index.html; 3) classification of the protein kinases with regard to target specificity using kinase classes for all kinases enumerated at UniProt (pkinfam.txt; release 2013_10); and 4) evidence culled from the literature for each kinase with regard to regulation by vasopressin, cAMP, or Ca2+. The calculations (see Supplemental Data Sets 1 and 2 in Supplemental Material for this article, available online at the Journal website) provide a ranking of kinases with regard to potential roles in regulation of AQP2 phosphorylation with the following rationale. Data sets 1 and 2 address whether each kinase is expressed in the IMCD. Logically, kinases not expressed are unlikely to have a role in regulation of AQP2 phosphorylation or phosphorylation of other proteins in the IMCD. Data set 3 identifies the kinases in the kinome that are most likely to bind to and phosphorylate AQP2 at Ser256, given the predominance of basic amino acids upstream (sequence: VRRRQS*VELHS, where Ser256 is indicated by an asterisk). Thus basophilic kinases (i.e., kinases from the AGC or CAMK families) are considered more likely than kinases with other target specificities to phosphorylate AQP2 at Ser256. Given the glutamic acid (E) at position 258, casein kinases are also given a greater-than-even probability. Data set 4 is a list of kinases known to be regulated in response to vasopressin on the basis of published phosphoproteomic data. Data set 5 is a list of kinases known to be regulated in response to changes in intracellular cAMP or Ca2+. Since phosphorylation of AQP2 at Ser256 is regulated by vasopressin (which signals through changes in cAMP or Ca2+), kinases known to be regulated by vasopressin, cAMP, or Ca2+ are considered more likely than other kinases to be responsible for this phosphorylation event.
Probability parameters for the five levels of Bayesian integration using previously published data were assigned as follows: kinases with normalized values from microarray analysis >0.7 (P = 0.95), normalized values between 0.4 and 0.7 (P = 0.70), and normalized values <0.4 (P = 0.1) for data set 1; kinases identified in the proteome (P = 0.95) and all others (P = 0.5) for data set 2; basophilic kinases, i.e., members of the AGC or CAMK family (P = 0.8), CK1 family (P = 0.6), TK or RGC family (P = 0.1), and all other families (P = 0.3) for data set 3; kinases that are phosphorylated in response to vasopressin at a known activating site (P = 0.90), kinases that are phosphorylated in response to vasopressin with unknown effects on activity (P = 0.65), and all others (P = 0.5) for data set 4; and kinases known to be activated by cAMP (P = 0.9), kinases known to be activated by Ca2+ (P = 0.9), and all others (P = 0.5) for data set 5. In general, if there are no data for a given kinase in any of these data sets, we assign a value of 0.5 (coin toss), reflecting a lack of evidence for or against that kinase. Sensitivity analysis was carried out by perturbing each input probability. The resulting probabilities after all five stages of Bayesian data integration provide prior probabilities for interpretation of the experimental studies (see results).
Isolation of IMCDs From Rat Kidneys
To isolate native IMCD cells, rat kidney inner medullas were dissected, minced, and digested in enzyme isolation solution [3 mg/ml each of collagenase B and hyaluronidase in sucrose buffer (250 mM sucrose and 10 mM Tris, pH 7.4)] according to the method of Stokes et al. (57) with modifications (42). After digestion, the IMCD suspension was centrifuged at ∼60 g for 20 s. The supernatant, which contains non-IMCD segments, was discarded, and the pellet of IMCD cells was washed with sucrose buffer. The cells were spun and washed three additional times prior to use for experiments. All experiments were conducted in accordance with Animal Protocol H-0110R1, approved by the National Heart, Lung, and Blood Institute Animal Care and Use Committee.
IMCD Suspensions: Incubation With Inhibitors and Immunoblotting
IMCDs, isolated as described above, were resuspended in a physiological solution (in mM: 118 NaCl, 25 NaHCO3, 5 KCl, 4 Na2HPO4, 2 CaCl2, 1.2 MgSO4, and 5.5 glucose) for investigation of phosphorylation responses to protein kinase inhibitors, as described by Pisitkun et al. (43). The IMCD suspensions were preincubated for 25 min with one of the following protein kinase inhibitors (or their vehicles): H-89, a relatively nonselective inhibitor of basophilic kinases (7) (Calbiochem, San Diego, CA); KN-62, a CAMK2-selective inhibitor (58) (Calbiochem); KN-93, a CAMK2-selective inhibitor (58) (Calbiochem); or GSK-650394, a SGK-selective inhibitor (52) (Tocris Bioscience, Bristol, UK). All inhibitors were solubilized in 0.5% (final concentration) DMSO. A list of protein kinases inhibited (and not inhibited) by each agent was downloaded from the International Centre for Kinase Profiling (ICKP, Dundee, Scotland; http://www.kinase-screen.mrc.ac.uk/). After the preincubation period, desmopressin (dDAVP, 1 nM or vehicle) was added, and the cells were incubated for 15 min in the continued presence of the kinase inhibitors or vehicle. Samples were prepared for immunoblotting by pelleting via centrifugation followed by addition of 1× Laemmli solution with heating to 60°C, as described elsewhere (43). Total protein was measured by the bicinchoninic acid method, and initial gels were stained with Coomassie blue dye to ensure equal loading for subsequent immunoblotting.
Immunoblotting was performed as described elsewhere (43). Briefly, after solubilization in Laemmli buffer, IMCD protein samples (7 μg per lane) were resolved by SDS-PAGE on 4–15% polyacrylamide gels (Bio-Rad) and transferred electrophoretically onto nitrocellulose membranes. The membranes were then blocked with Odyssey blocking buffer (LI-COR, Lincoln, NE), rinsed, and probed with primary antibody overnight at room temperature. Primary antibodies were polyclonal antibodies directed to phosphorylated (Ser256) AQP2 and total AQP2 (23). Blots were incubated with the anti-rabbit fluorescently labeled secondary antibody Alexa Fluor 680 (Molecular Probes, Eugene, OR; 1:5,000 dilution) for detection of all primary antibodies. Data from the blots were integrated into the Bayesian analysis (see results).
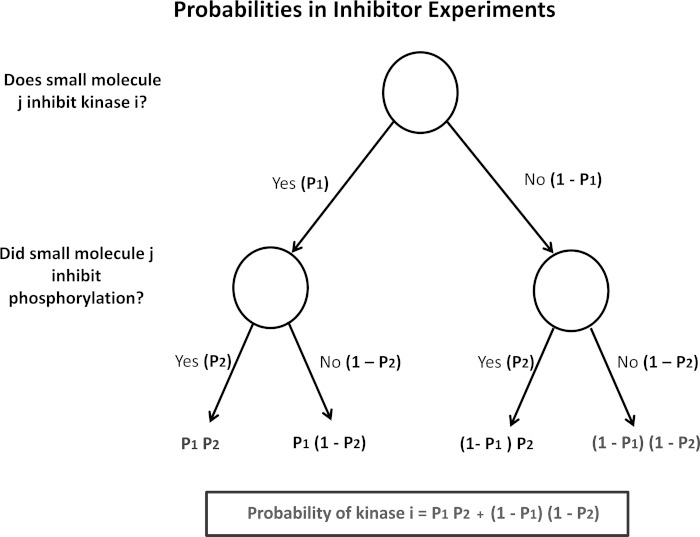
Probability parameters for Bayesian integration of data from inhibitor experiments were assigned according to the probability model shown in Fig. 1. The probabilities (P1) for in vitro inhibitor studies reported from the ICKP (see above) were assigned as follows: in the presence of inhibitors, kinases with residual activity <50% (P = 0.8), between 50% and 65% (P = 0.7), between 65% and 80% (P = 0.6), between 80% and 95% (P = 0.5), and >95% (P = 0.2). For in vivo phosphorylation experiments (immunoblotting data), the probabilities (P2) were assigned as follows: a statistically significant >20% decrease in phosphorylation of Ser256 (P = 0.9), a >20% decrease in phosphorylation that was not statistically significant, and <20% decrease in phosphorylation (P = 0.2). The resulting probabilities calculated from Fig. 1 for all four inhibitors and the result were integrated with the above-described Bayesian prior probabilities to obtain the final ranking of kinases.
Fig. 1.
Decision tree for calculation of input probabilities from inhibitor experiments. Calculated probabilities for each protein kinase (black box) take into account prior knowledge of the kinases that a given inhibitor can inhibit [downloaded from the International Centre for Kinase Profiling (ICKP; http://www.kinase-screen.mrc.ac.uk/)] and the results of immunoblotting to determine the effect of the inhibitor on aquaporin-2 (AQP2) phosphorylation at Ser256 (see methods for values of P1 and P2).
Identification of Substrates of PKA, CAMK2, and SGK Among IMCD Proteins
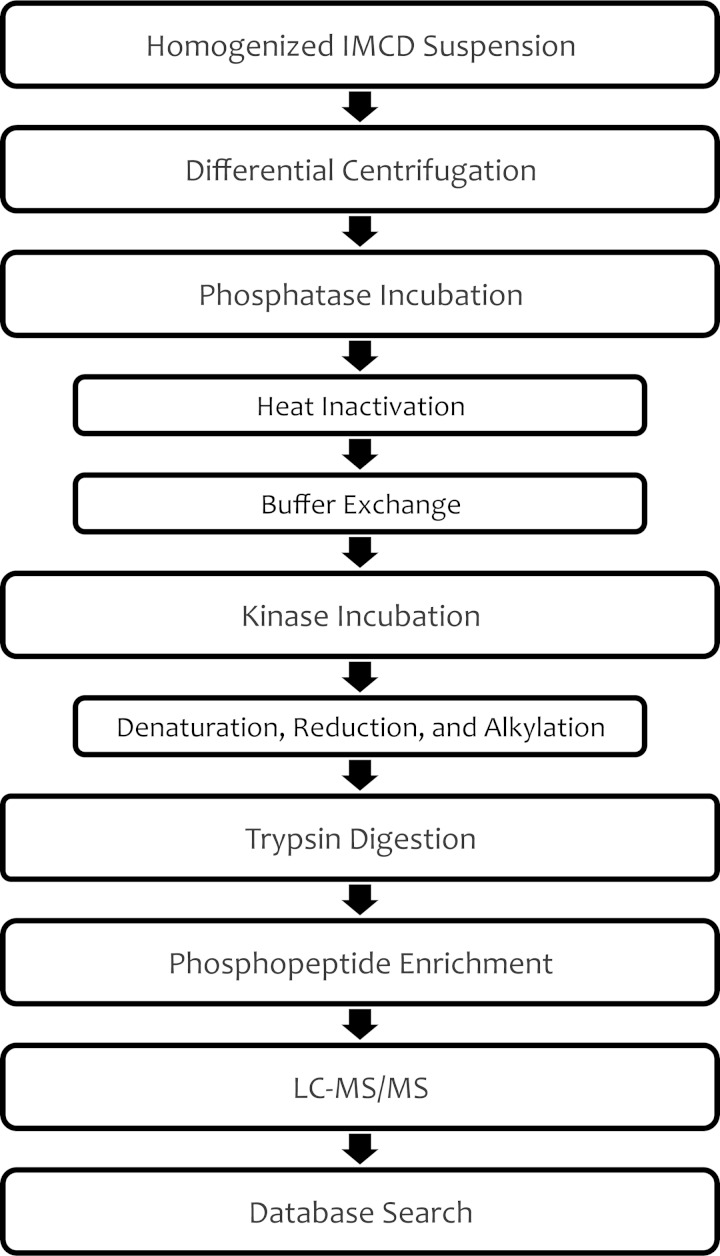
Liquid chromatography-tandem MS (LC-MS/MS) was used to identify potential substrates of PKA, CAMK2, and SGK in the proteome of native rat IMCD cells. Figure 2 outlines the experimental workflow. The basic idea is to use phosphoproteomic methodology to identify phosphorylation events (including phosphorylation of AQP2 at Ser256) resulting from in vitro incubation of cellular homogenates from native rat renal IMCDs with individual active, recombinant protein kinases. In general, we used the method described by Douglass et al. (13) with a few modifications to adapt the method to the present task. 1) We carried out differential centrifugation of the whole cell homogenates prior to incubation of each fraction with the recombinant kinases to reduce competition by high-abundance substrates, thus broadening the number of substrates that could be identified. 2) We added a second ion chromatography technique [beyond the immobilized metal affinity chromatography (IMAC) method used previously], metal oxide affinity chromatography, to increase the number of phosphopeptide identifications. 3) We adjusted the LC-MS/MS technique to increase phosphopeptide identification in the following ways: analysis of each sample twice and extension of the LC gradients from 1 h to 2 h. 4) To further increase phosphopeptide identification, we applied three independent spectral search algorithms (Sequest, InsPecT, and Mascot) to augment the efficiency of matching spectra to specific phosphopeptide sequences. 5) We used label-free quantification of phosphopeptide abundances to provide a quantitative basis for concluding that the kinase in question actually was responsible for a given phosphorylation event.
Fig. 2.
Phosphoproteomic workflow for identification of in vitro substrates of protein kinases. A suspension of homogenized inner medullary collecting ducts (IMCDs) from rat kidneys was separated into 5 fractions via differential centrifugation. Incubation with λ-phosphatase removed endogenous phosphorylation; then heat was applied to inactivate the phosphatase. After exchange of buffers, samples were incubated with one of the selected kinases or vehicle. Kinase reaction was halted by denaturation with urea; then samples were prepared for liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis.
Differential centrifugation.
Native rat IMCDs, prepared as described above, were homogenized with 5 repetitions of 15 slow strokes with a Potter-Elvehjem homogenizer and then centrifuged at 1,000 g for 10 min. The supernatant was removed and set aside for the subsequent higher-speed centrifugation. To increase the protein yield in the higher-speed fractions, the homogenization was repeated once with the Potter-Elvehjem homogenizer and a final time with an Omni International homogenizer (five 15-s pulses). The supernatants from the three homogenization rounds were pooled and fractionated via differential centrifugation, as described by Sachs et al. (47), yielding 1,000-, 4,000-, 17,000-, and 200,000-g pellets plus the 200,000-g supernatant. The protein concentration of each fraction was measured using the bicinchoninic acid method (Pierce Biotechnology, Rockford, IL).
Phosphatase incubation.
Fractions with a protein concentration less than the specified reaction concentration (∼6 μg/μl) were concentrated by spinning for 10 min at 14,000 g in 10-kDa-cutoff 0.5-ml centrifugal filters (Amicon Ultra, Millipore, Bedford, MA). For every 500 μg of proteins, 10 μl (4,000 units) of λ-phosphatase (New England BioLabs, Cambridge, MA) and an appropriate volume of phosphatase buffer for a reaction volume of 86.5 μl were added. To disrupt vesicles and allow greater exposure to the phosphatase, samples were sonicated (Misonix sonicator XL2020, power setting ∼6) for 1 min with 0.5-s pulses on ice. Samples were incubated for 20 h on a thermomixer set at 30°C and 600 rpm. The λ-phosphatase was inactivated and proteins were denatured by heating at 65°C for 1 h. To remove the MnCl2 required for phosphatase activity, a buffer exchange with 2.5× kinase reaction buffer [2.5× EDTA-free Halt protease inhibitor cocktail (Pierce Biotechnology) and 2.5× kinase buffer (125 mM Tris·HCl and 25 mM MgCl2; New England BioLabs)] was completed by spinning twice at 14,000 g for 30 min through 10-kDa-cutoff centrifugal filters (Millipore). A final protein concentration of 4.2 μg/μl was achieved by addition of 2.5× kinase reaction buffer to the retentate remaining after the buffer exchange.
Kinase incubation.
Forty microliters of 10 mM ATP (New England BioLabs), 1.5 μl of 100× EDTA-free Halt protease inhibitor (Pierce Biotechnology), and 60 μl of the phosphatase-treated sample (∼250-μg starting protein amount) were combined with PKA [catalytic subunit of cAMP-dependent protein kinase (New England BioLabs)], SGK [SGK I (CarnaBio, Natick, MA)], CAMK2 [CAMK IIδ (CarnaBio)], or vehicle, and water was added for a final reaction volume of 150 μl with a kinase concentration of 1.0 μM. For the CAMK2 samples, 0.5 mM CaCl2 and 0.03 μg/μl calmodulin (Sigma-Aldrich) were also added to the reaction mixture. Samples were incubated for 1 h at 30°C. Sonication with 0.5-s pulses for 1 min was performed before addition of the kinase and after 30 min of incubation. The kinase reaction was halted by addition of 450 μl of 8 M urea buffer [0.48 g of urea, 50 μl of 1 M Tris, pH 8, 15 μl of 5 M NaCl, 10 μl of EDTA-free 100× Halt protease and phosphatase inhibitor cocktail (Pierce Biotechnology), and 925 μl of H2O] to each 150-μl sample.
Trypsin digestion and phosphopeptide enrichment.
Samples were reduced with DTT, alkylated with iodoacetamide, and then digested with trypsin, as described by Hoffert et al. (24). The digested samples were enriched for phosphopeptides via IMAC using the Fe-NTA phosphopeptide enrichment kit (Pierce Biotechnology). To enhance phosphopeptide recovery, the flow-through and washes from the IMAC enrichment were pooled and further enriched with metal oxide affinity chromatography (TiO2 enrichment and clean-up kit, Pierce Biotechnology).
LC-MS/MS analysis.
All samples were analyzed first on a nanoflow LC system (Eksigent, Dublin, CA) and then on a mass spectrometer (LTQ Orbitrap Velos, Thermo Scientific, San Jose, CA). The samples from the IMAC elution were divided into two runs on the mass spectrometer (resulting in 2 technical replicates), while the samples from the TiO2 enrichment were analyzed in a single run. Loading of tryptic peptides onto a peptide trap cartridge (Agilent Technologies, Palo Alto, CA) occurred at a flow rate of 6 μl/min, and the subsequent reverse-phase separation on a PicoFrit column (New Objective, Woburn, MA) was performed at 300 nl/min using a linear gradient of 5–35% acetonitrile in 0.1% formic acid for a total gradient time of 2 h. Precursor masses (MS1 scans) and fragmented product masses (MS2 scans) were obtained in the Orbitrap and in the linear ion trap, respectively, using collision-induced dissociation.
Database searching.
Three search algorithms, Sequest (17), InsPecT (59), and Mascot (41), were employed for the identification of peptide ions from the mass spectra. The current version of the rat RefSeq database (National Center for Biotechnology Information) with concatenated forward and reverse sequences (allowing for target-decoy analysis) and also containing common MS contaminants (e.g., human keratin and porcine trypsin) was utilized for the searches. The precursor ion tolerance was set at 25 ppm, whereas the fragment ion tolerance was 0.8 Da. Three missed trypsin cleavage sites were allowed. Only the carbamidomethylation of cysteine (+57.021 Da) was set as a static modification. Variable modifications included oxidation of methionine (+15.995 Da), phosphorylation of serine, threonine, or tyrosine (+79.966 Da), and deamidation of asparagine or glutamine (+0.984 Da). Any known contaminant ions were excluded. Spectra were filtered to achieve a false discovery rate of 1% at the peptide level (estimated on the basis of target-decoy analysis). Phosphorylation sites were assigned using PhosSA (23) for Sequest results, phosphate localization score (PLS) for InsPecT results (PLS >7) (59), and PhosphoRS for Mascot results (pRS >0.99) (49).
Label-free quantification.
Quantification of peptide abundance (area under the curve of the reconstructed ion chromatogram elution profile) was achieved using QUOIL, an in-house software program designed for quantification of label-free peptides by LC-MS (67). Quantification consistency among three different elution windows (0.25, 0.4, and 0.55 min) was determined, and when quantification was inconsistent for a given peptide, elution profiles were inspected manually. Those that did not have satisfactory consistency and showed asymmetric or irregular profiles were excluded. Peptides not quantified in control samples result in an infinite fold change. These peptides were reported when the area under the elution profile curve for the kinase-treated sample was above the 95th percentile for all peptides (log2 ratio = ∞).
Bioinformatics for proteomics results.
Peptides were mapped to proteins using the in-house software ProMatch (60), available at http://helixweb.nih.gov/ESBL/ProMatch/. The consensus sequence logos of position-specific amino acid preferences were generated with the online tool PhosphoLogo (13), available at http://helixweb.nih.gov/PhosphoLogo/.
RESULTS
Ranking of Protein Kinase Candidates for Phosphorylation of AQP2 at Ser256
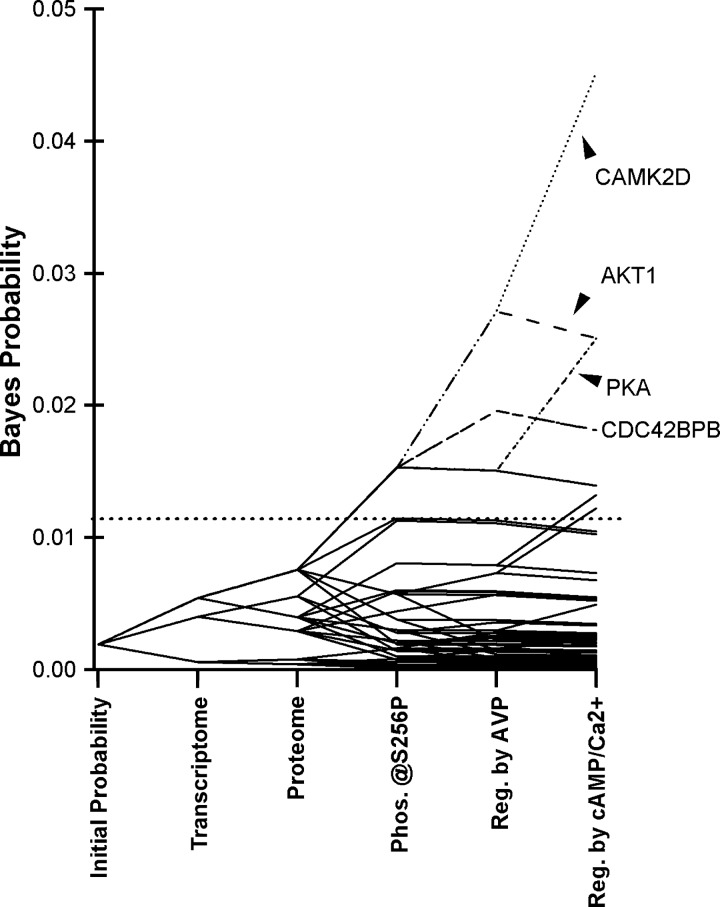
Prior to carrying out new experiments, we integrated previously reported data from several sources to provide a preliminary ranking of kinases that could play a role in phosphorylation of AQP2 at Ser256. Bayes' theorem was applied to rank all known kinases in the rat genome according to an aggregate probability score that takes into account the likelihood that the transcript is expressed in IMCD cells, the likelihood that the protein is expressed, the likelihood that the target specificity matches that of Ser256 of AQP2, and the likelihood that the kinase is regulated by vasopressin in the collecting duct. Evidence for regulation by vasopressin could be evidence for activating phosphorylation events in response to vasopressin (Table 1) or evidence for regulation by cAMP or elevation of intracellular Ca2+. Details of the calculations are provided in Supplemental Data Set 1. Figure 3 shows the results for all 521 protein kinases in the rat genome. Bayesian integration of multiple existing data sets resulted in the separation of a relatively small number of kinases from the entire population of 521. Table 2 lists the 20 top-ranked kinases. All are in the AGC or CAMK family, both of which have basophilic specificities. By a wide margin, Camk2d had the highest prior probability score based on the fact that it is strongly expressed in the IMCD at a transcript level (62), readily detectable at a protein level by MS (60), has the appropriate substrate target specificity for Ser256 of AQP2, and is regulated by vasopressin in two different ways: phosphorylation (23) and calmodulin activation resulting from increased intracellular Ca2+ (9). Next behind Camk2d on the list are Akt1 and the PKA gene products Prkaca and Prkacb.
Table 1.
Protein kinases whose activities are regulated by vasopressin based on previous phosphoproteomics studies
| Kinase (Gene Symbol) | Kinase Class | Phosphorylation Site(s) | Phosphorylation Change | Effect on Kinase Activity | Experimental Preparation | Reference No. |
|---|---|---|---|---|---|---|
| Akt1 | AGC | S473 | Up | Up | Rat IMCD | 43 |
| Akt1 | AGC | T308 | Up | Up | Rat IMCD | 43 |
| Camk2b | CAMK | T287 | Up | Up | Mouse mpkCCD | 45 |
| Camk2d | CAMK | T287 | Up | Up | Mouse mpkCCD | 45 |
| Gsk3a | CMGC | Y279 | Down | Down | Rat IMCD | 23 |
| Mapk1 | CMGC | T183 and Y185 | Down | Down | Rat IMCD | 43 |
| Mapk1 | CMGC | T183 and Y185 | Down | Down | Mouse mpkCCD | 45 |
| Mapk14 | CMGC | Y182 | Down | Down | Rat IMCD | 23 |
| Mapk3 | CMGC | T203 and Y205 | Down | Down | Rat IMCD | 43 |
| Mapk3 | CMGC | T203 and Y205 | Down | Down | Mouse mpkCCD | 45 |
| Mapk9 | CMGC | T183 and Y185 | Down | Down | Rat IMCD | 23 |
| Map2k1 | STE | S218 and S222 | Down | Down | Rat IMCD | 43 |
| Map2k2 | STE | S222 and 226 | Down | Down | Rat IMCD | 43 |
| Map2k3 | STE | S218 | Down | Down | Rat IMCD | 23 |
| Map2k5 | STE | S311 | Down | Down | Rat IMCD | 23 |
| Map2k6 | STE | S207 | Down | Down | Rat IMCD | 23 |
| Hck | TK | Y409 | Down | Down | Rat IMCD | 23 |
| Araf | TKL | S255 | Down | Down | Rat IMCD | 23 |
| Raf1 | TKL | S259 | Up | Down | Rat IMCD | 43 |
| Raf1 | TKL | S29 | Up | Down | Rat IMCD | 23 |
IMCD, inner medullary collecting duct; mpkCCD, cortical collecting duct cell line.
Fig. 3.
Bayes' analysis of rat IMCD kinases. All 521 kinases in the rat genome are assumed to be equally probable at the start of calculations. Probabilities were calculated using Bayes' rule from sequential integration of information from several data sets. Resulting values derived from preexisting information provide “prior probabilities” for assessment of new experimental data presented here. Horizontal dotted line indicates cutoff values for the top 20 kinases. CAMK2D, Ca2+/calmodulin kinase 2δ.
Table 2.
Calculated prior probabilities for the top-20 kinases with regard to likelihood of a role in AQP2 phosphorylation at Ser256
| Expression |
Regulated by AVP |
||||||
|---|---|---|---|---|---|---|---|
| Rank | Kinase Gene Symbol | Bayes' Probability | Transcript | Protein | Kinase Family | PTM | Ca2+/cAMP |
| 1 | Camk2d | 0.0452 | Yes | Yes | CAMK | Yes and activated | Yes |
| 2 | Akt1 | 0.0251 | Yes | Yes | AGC | Yes and activated | No |
| 2 | Prkaca | 0.0251 | Yes | Yes | AGC | Unknown | Yes |
| 2 | Prkacb | 0.0251 | Yes | Yes | AGC | Unknown | Yes |
| 5 | Cdc42bpb | 0.0181 | Yes | Yes | AGC | Yes | Yes |
| 5 | Prkcd | 0.0181 | Yes | Yes | AGC | Yes | Yes |
| 7 | Cask | 0.0139 | Yes | Yes | CAMK | Unknown | No |
| 7 | Dapk3 | 0.0139 | Yes | Yes | CAMK | Unknown | No |
| 7 | Mark2 | 0.0139 | Yes | Yes | CAMK | Unknown | No |
| 7 | Mark3 | 0.0139 | Yes | Yes | CAMK | Unknown | No |
| 7 | Nuak2 | 0.0139 | Yes | Yes | CAMK | Unknown | No |
| 7 | Pdpk1 | 0.0139 | Yes | Yes | AGC | Unknown | No |
| 7 | Pkn1 | 0.0139 | Yes | Yes | AGC | Unknown | No |
| 7 | Pkn2 | 0.0139 | Yes | Yes | AGC | Unknown | No |
| 7 | Prkci | 0.0139 | Yes | Yes | AGC | Unknown | No |
| 7 | Rock2 | 0.0139 | Yes | Yes | AGC | Unknown | No |
| 7 | Rps6ka1 | 0.0139 | Yes | Yes | AGC | Unknown | No |
| 18 | Camk1 | 0.0132 | Yes | No | CAMK | Unknown | No |
| 18 | Phkg2 | 0.0132 | Yes | No | CAMK | Unknown | No |
| 20 | Camkk2 | 0.0122 | Yes | Yes | Other | Yes | No |
AQP2, aquaporin-2; PTM, posttranslational modification.
Ability of Selective Protein Kinase Inhibitors to Decrease AQP2 Phosphorylation at Ser256
Next, we asked whether addition of new data using selective protein kinase inhibitors could refine the Bayesian ranking. Using suspensions of native IMCDs isolated from rat kidneys, we tested the ability of several small-molecule protein kinase inhibitors to reduce phosphorylation of AQP2 at Ser256 (Figs. 4–6). To aid in interpretation of the data, we downloaded the lists of protein kinases known to be inhibited and those known not to be inhibited by each inhibitor compound from the ICKP (www.kinase-screen.mrc.ac.uk/). These data are summarized in Table 3.
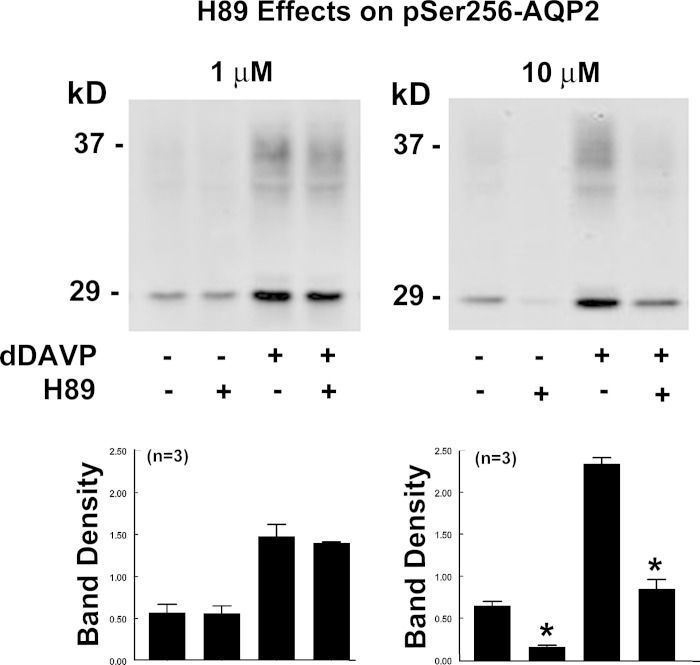
Fig. 4.
Effects of 1 and 10 μM H-89, a selective inhibitor of basophilic kinases, on AQP2 phosphorylation at Ser256 in rat IMCD suspensions. After 25 min of preincubation with inhibitor or vehicle, desmopressin (dDAVP, 1 nM or vehicle) was added, and the samples were incubated for 15 min in the continued presence of the kinase inhibitor or vehicle. Vehicle was 0.5% (final concentration) DMSO. *P < 0.01.
Fig. 6.
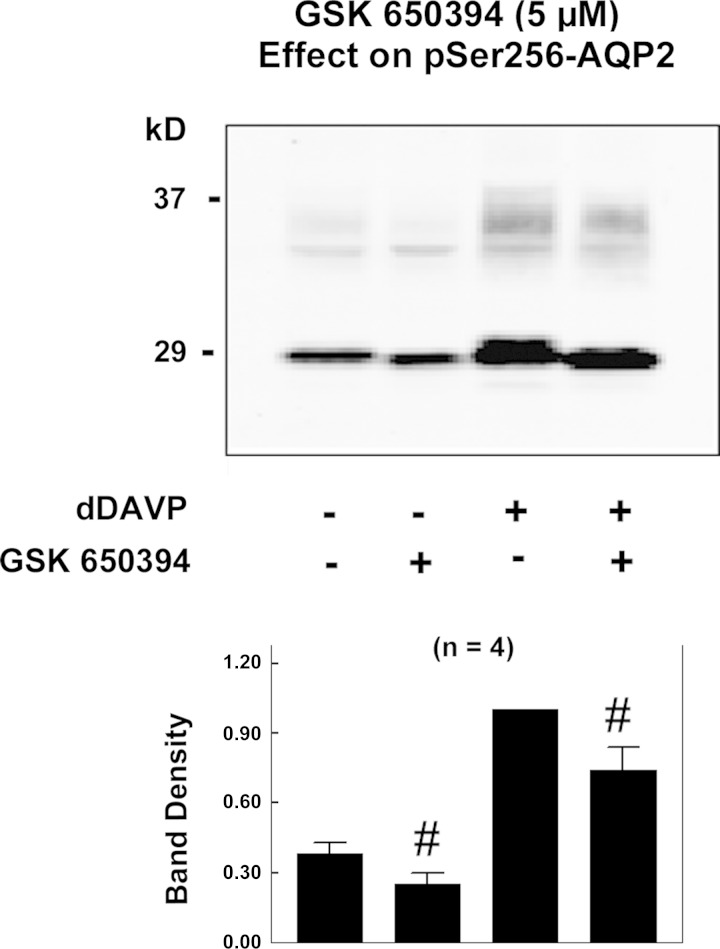
Effects of 5 μM GSK-650394, a selective inhibitor of serum/glucocorticoid-regulated kinase (SGK), on AQP2 phosphorylation at Ser256 in rat IMCD suspensions. After 25 min of incubation with inhibitor or vehicle, dDAVP (1 nM or vehicle) was added, and the samples were incubated for 15 min in the continued presence of the kinase inhibitor or vehicle. Vehicle was 0.5% (final concentration) DMSO. #P < 0.1.
Table 3.
Kinase inhibitor sensitivity profiles for top-20 kinases in Table 2
| Kinase (Gene Symbol) | H-89 (1 μM) | H-89 (10 μM) | KN-93 (10 μM) | KN-62 (10 μM) | GSK-650394A (5 μM) |
|---|---|---|---|---|---|
| Camk2d* | <25 | 50–100 | 50–100 | 50–100 | <25 |
| Akt1 | 50–100 | 50–100 | 50–100 | <25 | 25–50 |
| Prkaca | 50–100 | 50–100 | 50–100 | <25 | <25 |
| Prkacb | 50–100 | 50–100 | 50–100 | <25 | <25 |
| Cdc42bpb* | 50–100 | 50–100 | 50–100 | <25 | 25–50 |
| Prkcd* | 25–50 | 25–50 | 25–50 | <25 | 25–50 |
| Cask* | <25 | 50–100 | 50–100 | 50–100 | <25 |
| Dapk3* | <25 | 50–100 | <25 | <25 | ND |
| Mark2 | <25 | 50–100 | <25 | <25 | 25–50 |
| Mark3 | <25 | 50–100 | <25 | <25 | 25–50 |
| Nuak2* | 50–100 | 50–100 | <25 | ND | 50–100 |
| Pdpk1 | <25 | <25 | <25 | <25 | <25 |
| Pkn1* | 50–100 | 50–100 | 50–100 | <25 | 50–100 |
| Pkn2 | 50–100 | 50–100 | 50–100 | <25 | 50–100 |
| Prkci* | <25 | 50–100 | <25 | <25 | <25 |
| Rock2 | 50–100 | 50–100 | 50–100 | <25 | 25–50 |
| Rps6ka1 | 50–100 | 50–100 | 50–100 | <25 | <25 |
| Camk1 | <25 | 50–100 | 50–100 | 50–100 | <25 |
| Phkg2* | 25–50 | 50–100 | <25 | <25 | 50–100 |
| Camkk2 | <25 | <25 | 25–50 | <25 | 50–100 |
Values represent percent inhibition. Data are from the International Centre for Kinase Profiling (Dundee, Scotland).
Effect was obtained from the nearest homolog: Camk1 for Camk2d, Rock2 for Cdc42bpb, Prkca for Prkcd, Camk1 for Cask, Dapk1 for Dapk3, Nuak1 for Nuak2, Pkn2 for Pkn1, Prkcz for Prkci, and Phkg1 for Phkg2.
ND, not determined.
Figure 4 shows effects of H-89, which is widely viewed as a PKA inhibitor but also inhibits a substantial number of other basophilic protein kinases (Table 3). H-89 at 1 μM had no effect on AQP2 phosphorylation at Ser256 in the absence or presence of the V2 receptor-selective vasopressin analog dDAVP. However, 10 μM H-89 significantly decreased AQP2 phosphorylation at Ser256 in the absence and presence of dDAVP.
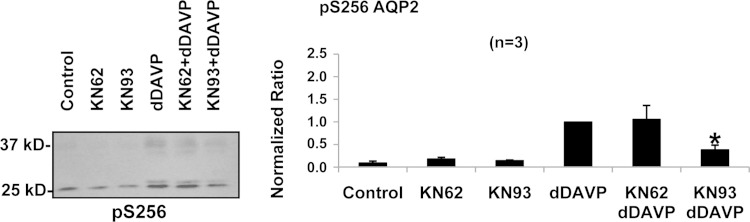
Figure 5 shows the effects of two additional small-molecule inhibitors, KN-62 (10 μM) and KN-93 (5 μM) (58). These agents are widely regarded as selective inhibitors of calmodulin-regulated kinases. They have somewhat different profiles with regard to the kinases they inhibit (Table 3). As described above, these experiments were carried out in isolated IMCD suspensions from rats. Neither agent had an effect on AQP2 phosphorylation in the absence of dDAVP. However, KN-93 reduced dDAVP-dependent phosphorylation at Ser256. In contrast, KN-62 did not significantly alter dDAVP-dependent Ser256 phosphorylation.
Fig. 5.
Effects of selective inhibitors of calmodulin-regulated kinases, KN-62 (10 μM) and KN-93 (5 μM), on AQP2 phosphorylation at Ser256 in rat IMCD suspensions. After 25 min of preincubation with inhibitor or vehicle, dDAVP (1 nM or vehicle) was added, and the samples were incubated for 15 min in the continued presence of the kinase inhibitors or vehicle. Vehicle was 0.5% (final concentration) DMSO. *P < 0.01.
Figure 6 shows effects of a selective inhibitor of SGK, GSK-650394 (5 μM), on Ser256 phosphorylation. GSK-650394 is not specific for Sgk1, but it inhibits a number of other basophilic kinases (Table 3). This inhibitor had only a small effect on Ser256 phosphorylation (26% inhibition, P = 0.06, based on densitometry of both bands combined). However, there was a marked qualitative effect on the morphology of the 29-kDa band corresponding to a loss of the upper portion, thought to be associated with high mannose glycosylation in the endoplasmic reticulum (20). The 37-kDa mature glycosylation band was consistently decreased in all four experiments.
Using the prior probabilities calculated above and the inhibitor data, we extended the Bayesian analysis of the protein kinases likely to phosphorylate AQP2 at Ser256. For this analysis, we used the decision tree shown in Fig. 1 to estimate the relative probability of a role for a given kinase in phosphorylation of AQP2 at Ser256 based on inhibition of Ser256 phosphorylation and the kinase target spectrum of each protein kinase. These probabilities were used to update overall probabilities using Bayes' rule (Table 4). The updated Bayesian ranking showed that, among the top-20 kinases likely to phosphorylate AQP2 at Ser256, Camk2 is the highest-ranked candidate, followed by Prkaca and Prkacb and then Akt1.
Table 4.
Bayesian ranking of the top-20 kinases before and after integration of data from inhibitor experiments
| From Preexisting Data |
After-Inhibitor Data |
||||
|---|---|---|---|---|---|
| Rank | Kinase | Bayes' Probability | Rank | Kinase | Bayes' Probability |
| 1 | Camk2d | 0.0452 | 1 | Camk2d | 0.2065 |
| 2 | Akt1 | 0.0251 | 2 | Prkaca | 0.1275 |
| 2 | Prkaca | 0.0251 | 2 | Prkacb | 0.1275 |
| 2 | Prkacb | 0.0251 | 4 | Akt1 | 0.0937 |
| 5 | Cdc42bpb | 0.0181 | 5 | Cdc42bpb | 0.0462 |
| 5 | Prkcd | 0.0181 | 6 | Mark3 | 0.0424 |
| 7 | Cask | 0.0139 | 7 | Prkcd | 0.0406 |
| 7 | Dapk3 | 0.0139 | 8 | Rps6ka1 | 0.0394 |
| 7 | Mark2 | 0.0139 | 8 | Dapk3 | 0.0305 |
| 7 | Mark3 | 0.0139 | 10 | Mark2 | 0.0305 |
| 7 | Nuak2 | 0.0139 | 11 | Prkci | 0.0294 |
| 7 | Pdpk1 | 0.0139 | 12 | Rock2 | 0.0289 |
| 7 | Pkn1 | 0.0139 | 13 | Pdpk1 | 0.0281 |
| 7 | Pkn2 | 0.0139 | 13 | Pkn1 | 0.0255 |
| 7 | Prkci | 0.0139 | 15 | Pkn2 | 0.0255 |
| 7 | Rock2 | 0.0139 | 16 | Cask | 0.0206 |
| 7 | Rps6ka1 | 0.0139 | 17 | Nuak2 | 0.0196 |
| 18 | Camk1 | 0.0132 | 18 | Camkk2 | 0.0140 |
| 18 | Phkg2 | 0.0132 | 19 | Camk1 | 0.0124 |
| 20 | Camkk2 | 0.0122 | 20 | Phkg2 | 0.0112 |
Ability of PKA, CAMK2, and SGK to Phosphorylate AQP2 In Vitro
Among the protein kinases listed in Table 4 are isoforms of CAMK, PKA, and AKT. SGK, an abundant kinase in the IMCD (39), fell slightly out of the top 20. This analysis provided the motivation for further study of these kinases to determine which, if any, are capable of phosphorylating AQP2 (as well as other proteins at vasopressin-regulated phosphorylation sites). Because AKT and SGK are closely related and are likely to have the same or similar target specificities, we elected to use SGK for the profiling experiments. [Previous studies demonstrated that Akt1 is capable of phosphorylating AQP2 at Ser256 (13).] We used protein MS to investigate the ability of PKA, CAMK2, and SGK to phosphorylate AQP2 at Ser256 and other proteins in vitro. We incubated rat IMCD homogenates with active recombinant kinases and then performed phosphoproteomic analysis of the product. This approach used published methodology (13) with modifications described in methods (Fig. 2).
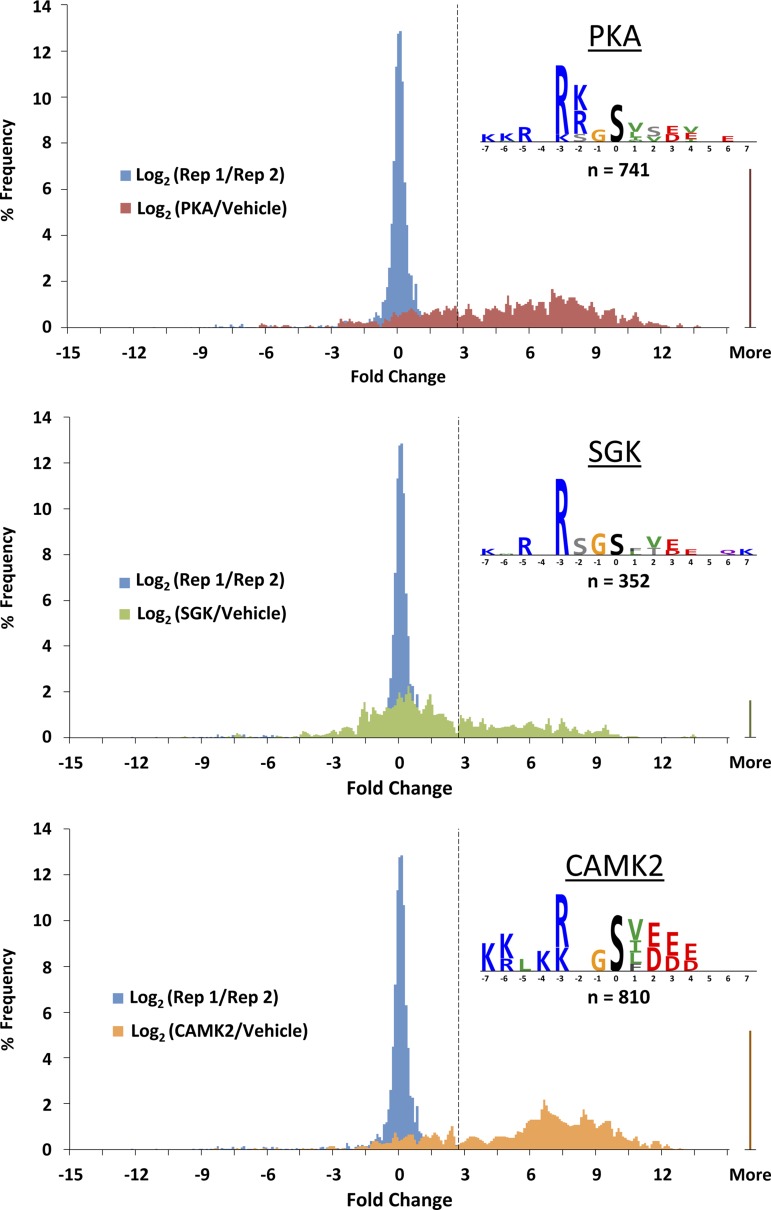
Figure 7 shows histograms representing general features of the in vitro phosphorylation results. The procedure employed in vitro dephosphorylation of IMCD homogenates followed by incubation with PKA, SGK, or CAMK2. In general, although extensive dephosphorylation prior to kinase incubation is attainable, a low-level residual phosphorylation is seen for many sites (13). To address whether the kinase in question catalyzes increased phosphorylation at these sites, in Fig. 7 we report the base 2 logarithm of the ratio of MS signals (area under the reconstructed ion chromatogram peak) with and without the kinase incubation and compare this value with the intrinsic variability of the method (dashed vertical line) calculated from replicate MS runs with the same sample. Thus all values to the right of the dashed line are considered to have been phosphorylated in vitro by the designated recombinant kinase. The sites labeled “More” are those in which phosphorylation was not detectable without the kinase incubation. The number of sites that were phosphorylated by each kinase were as follows: 741 for PKA, 352 for SGK, and 810 for CAMK2. Several sites, including Ser256 of AQP2, were found to be phosphorylated by more than one kinase (see below). Consequently, the total number of sites found among the three kinases was less than the sum of sites for the three protein kinases (total = 1,468 sites).
Fig. 7.
Histograms and logos from MS quantification. For each kinase, 2 histograms are overlaid: blue histograms repeating in each panel represent inherent variability of the method as measured by the base 2 logarithm of the ratio of the MS signal for a peptide in the 1st replicate (Rep 1) to the signal for the same peptide in the 2nd replicate (Rep 2); the other colored histograms are of the base 2 logarithms of the ratios of the MS signals with and without the kinase incubation. Dashed vertical line delineates 2 SDs above zero based on the replicate comparison (blue histograms). Logos of the phosphorylation consensus sequences based on the unique phosphorylation sites identified in the study for each kinase were created in PhosphoLogo and are shown for PKA, SGK, and CAMK2.
From the sequences obtained, we calculated phosphorylation sequence preference logos for each of the three protein kinases, reporting the amino acids that occur in a given position more frequently than would occur by random sampling of the 20 amino acids (Fig. 7, insets). Position 0 represents the phosphorylated S or T. In this display of amino acid preferences from PhosphoLogo software (13), the height of an individual character is used to indicate relative preference for the depicted amino acid. Two observations stand out: 1) these sequence preferences confirmed the identification of all three as “basophilic protein kinases,” with basic amino acids (R and K) overrepresented upstream from the phosphorylated residue, and 2) these sequence preferences closely resembled previously reported kinase-specific logos from MS-based (13) and peptide array-based (25) profiling.
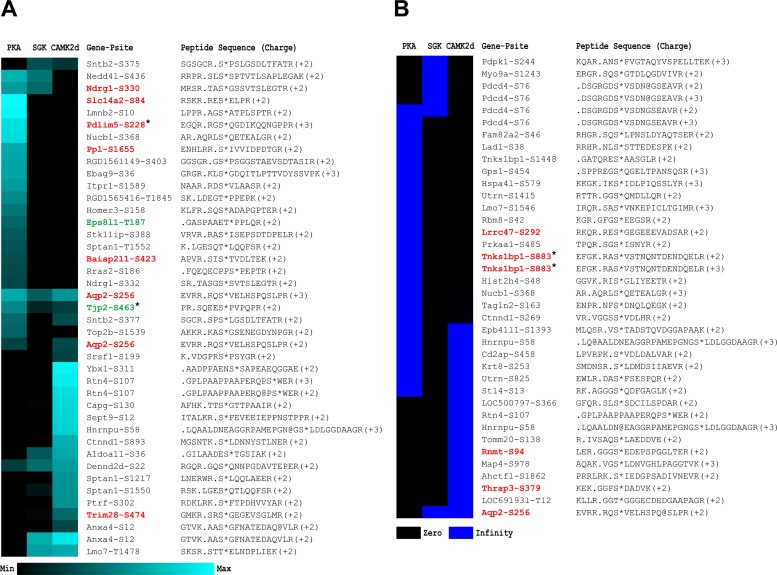
Figure 8 lists the sites phosphorylated by one or more of the kinases of this study that were also identified and quantified in intact IMCDs in previous studies (23) by Bansal et al. (1). Those shown in red were reported to be upregulated by vasopressin, and those shown in green were reported to be downregulated. Ser256 of AQP2 was found multiple times. The triply and doubly charged peptides in Fig. 8A were phosphorylated about equally by PKA and CAMK2 and somewhat less by SGK (see heat map representation of quantification). We also found a third peptide with a 2+ charge that was phosphorylated by CAMK2 and SGK (Fig. 8B). Thus all three kinases proved to be capable of phosphorylating AQP2 at Ser256.
Fig. 8.
Vasopressin-regulated sites with demonstrated phosphorylation by PKA, SGK, and/or CAMK2. Phosphorylation sites identified in this study were cross-referenced with 2 phosphoproteomic studies that revealed phosphorylation sites physiologically regulated by AVP: asterisk indicates sites from Bansal et al. (1), while all other sites were quantified by Hoffert et al. (23). Physiologically regulated sites are juxtaposed with a heat map of the corresponding phosphorylation responses from the 3 kinases. Sites shown in red were, on average, significantly upregulated by AVP in their respective study; sites shown in green were, on average, significantly downregulated. A: peptides with defined log2 ratios. B: peptides with phosphorylation that was not detectable without the kinase incubation and, thus, with a log2 (kinase/vehicle) of infinity. Peptide sequences have been extended (identified sequences are to the right of the period) to reveal the 6 amino acids upstream from the phosphorylation site.
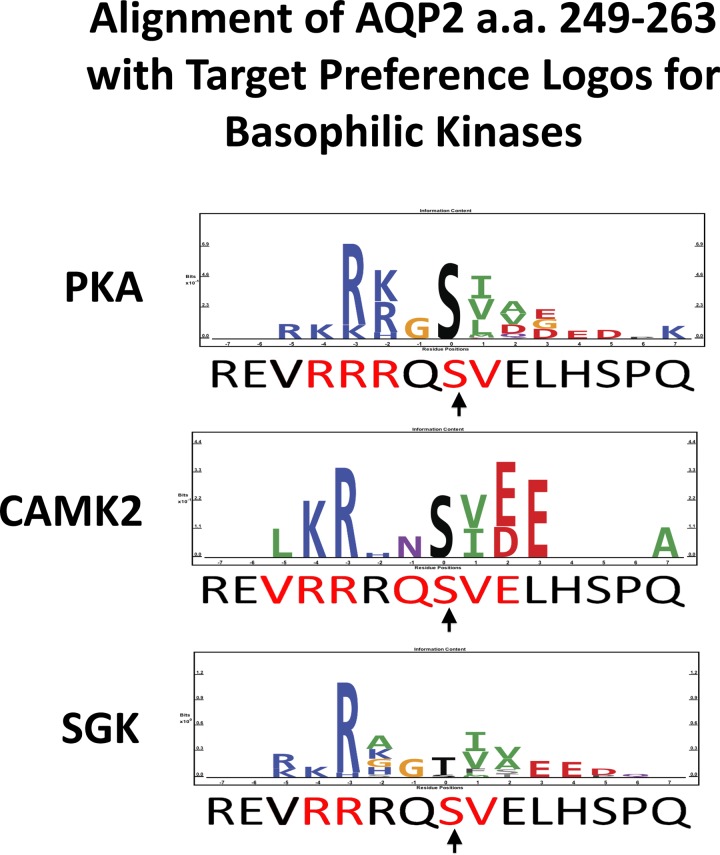
Figure 9 shows the AQP2 sequence from amino acid 249 to 263 [left to right, centered on Ser256 (arrow)] compared with previously reported protein kinase target preferences for PKA, CAMK2, and SGK (13). As described above, the height of an individual character is used to indicate relative preference for the depicted amino acid. The amino acids in the AQP2 sequence matching the respective target preference sequences are shown in red. “Matching” means that the identical amino acid is seen or that there is a conservative substitution based on charge and polarity of the side chain. There are five amino acid matches for PKA, seven for CAMK2, and only four for SGK. Thus, solely on the basis of this criterion, CAMK2 appears to be a somewhat better candidate than PKA or SGK for a role in Ser256 phosphorylation. This is consistent with the rank order of candidate kinases from the Bayesian analysis (Table 4).
Fig. 9.
Rat AQP2 sequence from amino acid 249 to 263 [left to right, centered on Ser256 (arrow)] compared with previously reported protein kinase target preferences for PKA, CAMK2, and SGK. Number of matched amino acids (red amino acids in AQP2 sequence) is as follows: 5 for PKA, 7 for CAMK2, and 4 for SGK.
General Results of Phosphoproteomic Analysis
A by-product of the MS method used in this study is a large number of proteins, in addition to Ser256 of AQP2, that were identified as potential substrates for the three kinases. All sites quantified, regardless of whether they were induced by a kinase, are reported on the online database https://helixweb.nih.gov/ESBL/Database/iKinSub/. These sites are specific to the rat proteome, although we have mapped many of them to other species, including human (click on “Download” at the URL mentioned above).
Most of the phosphorylation sites in Fig. 8 were found in the present study to be phosphorylated by only one of the three basophilic protein kinases tested. In addition, many sites, including Ser256 of AQP2, were found to be capable of being phosphorylated by more than one kinase. This finding is consistent with the idea that the basophilic kinases have overlapping specificities, as suggested by the phosphorylation logos shown in Fig. 9.
Since a major goal of our work is to understand how transport function is regulated by vasopressin in the renal collecting duct, we have listed in Table 5 all solute transporter and channel proteins found to be phosphorylated by PKA, SGK, or CAMK2 in this study. Phosphorylation sites were found in aquaporin-4 (Aqp4), two components of the catalytic portion of the vacuolar H+-ATPase (Atp6v1g3 and Atp6v1g1), the α1-subunit of the Na+-K+-ATPase (ATP1a1), the α-subunit of the H+-K+-ATPase (gastric form, Atp4a), a CFTR-related ATP binding cassette transporter (Abcc6), an inositol triphosphate-regulated Ca2+ release channel (Itpr1), an intracellular Mg2+ channel (Mmgt1), a bumetanide-sensitive Na+-K+-2Cl− cotransporter (Slc12a1), a Cl−/HCO3− exchanger [Slc4a2, or “anion exchanger-2” (AE-2)], an Na+/H+ exchanger (Slc9a1), and several mitochondrial ion transporters. Among these, the only site found to be phosphorylated solely by SGK was Ser240 of Slc4a2, also known as AE-2.
Table 5.
Transport proteins phosphorylated by PKA, SGK, or CAMK2
| Accession No. | Gene Symbol | Peptide Sequence(Charge) | Site | Phosphorylated by |
|---|---|---|---|---|
| NP_036957 | Aqp4 | GKDS*SGEVLSSV(+2) | S315 | PKA |
| NP_036957 | Aqp4 | KGKDS*SGEVLSSV(+2) | S315 | PKA |
| NP_037041 | Aqp2 | RQS*VELHSPQSLPR(+2) | S256 | PKA CAMK2 |
| NP_037041 | Aqp2 | RQS*VELHSPQSLPR(+3) | S256 | PKA SGK CAMK2 |
| NP_037041 | Aqp2 | RQS*VELHSPQ@SLPR(+2) | S256 | SGK CAMK2 |
| NP_037041 | Aqp2 | RQS*VELHSPQS(+2) | S256 | PKA |
| NP_037041 | Aqp2 | RRQS*VELHSPQSLPR(+3) | S256 | PKA SGK CAMK2 |
| NP_001099461 | Atp6v1 g3 | IMGSQS*HLSDELEEQTLEK(+3) | S67 | PKA CAMK2 |
| NP_001100130 | Atp6v1 g1 | EAAALGSHGS*CSSEVEKETQEK(+3) | S68 | PKA |
| NP_036636 | Atp1a1 | RNS*VFQQGMK(+2) | S943 | PKA |
| NP_036636 | Atp1a1 | IISANGCKVDN@SS*LTGESEPQTR(+3) | S217 | CAMK2 |
| NP_036636 | Atp1a1 | MS*INAEDVVVGDLVEVK(+2) | S179 | PKA CAMK2 |
| NP_036636 | Atp1a1 | ILDRCSS*ILLHGK(+3) | S520 | PKA |
| NP_036636 | Atp1a1 | NGEKMS*INAEDVVVGDLVEVK(+3) | S179 | PKA |
| NP_036636 | Atp1a1 | AVAGDAS*ESALLK(+2) | S452 | CAMK2 |
| NP_036636 | Atp1a1 | IISANGCKVDNSS*LTGESEPQTR(+3) | S217 | CAMK2 |
| NP_036641 | Atp4a | VDNSS*LTGESEPQTR(+2) | S227 | CAMK2 |
| NP_112275 | Abcc6 | SSLT*WGLLR(+2) | T1308 | SGK CAMK2 |
| NP_001007236 | Itpr1 | AMS*LVSSDSEGEQNELR(+2) | S2681 | PKA |
| NP_001007236 | Itpr1 | RDS*VLAASR(+2) | S1589 | PKA |
| NP_001100440 | Mmgt1 | VLFRPS*DATNSSNLDALSSNTSLK(+3) | S104 | PKA |
| NP_001100440 | Mmgt1 | VLFRPSDAT*NSSNLDALSSNTSLK(+3) | T107 | PKA |
| NP_062007 | Slc12a1 | VNRPS*LQEIHEQLAK(+2) | S126 | PKA |
| NP_062007 | Slc12a1 | VNRPS*LQEIHEQLAK(+3) | S126 | PKA |
| NP_808877 | Slc14a2 | RES*ELPR(+2) | S84 | PKA |
| NP_808877 | Slc14a2 | RRES*ELPR(+2) | S84 | PKA |
| XP_002726232 | Slc25a12 | GTGS*VVGELMYK(+2) | S357 | PKA |
| NP_001121016 | Slc25a24 | QLLAGGVAGAVS*R(+2) | S208 | CAMK2 |
| NP_620800 | Slc25a3 | EKGS*TASQVLQR(+2) | S291 | PKA |
| NP_058744 | Slc4a2 | IGS*MTGVEQALLPR(+2) | S240 | SGK |
| NP_036784 | Slc9a1 | IGS*DPLAYEPK(+2) | S707 | PKA SGK CAMK2 |
| NP_112644 | Vdac2 | YQLDPTAS*ISAK(+2) | S244 | CAMK2 |
SGK, serum/glucocorticoid-regulated kinase; CAMK, Ca2+/calmodulin-dependent kinase.
Table 6 shows sites phosphorylated by one or more of the three protein kinases in protein phosphatases, other protein kinases, and ubiquitin ligases. Interestingly, one protein on this list, 3-phosphoinositide-dependent protein kinase 1 (also known as PDK1), the kinase responsible for phosphorylation and activation of AKT and SGK isoforms, was phosphorylated at several sites by SGK. Among these sites, Ser244 is recognized as an amino acid that must be phosphorylated for PDK1 to have protein kinase activity. Thus PDK1 appears to be part of a positive-feedback loop in which it phosphorylates and activates SGK (and AKT), which phosphorylates and activates PDK1. Another important kinase on this list is Mark2 (also known as Par-1), a CAMK family kinase involved in cell polarity determination and Wnt signaling in epithelia. Tables 5 and 6 sustain the view that a substantial fraction of putative target sites can be phosphorylated by more than one basophilic protein kinase.
Table 6.
Regulatory proteins phosphorylated by PKA, SGK, or CAMK2
| Accession No. | Gene Symbol | Peptide Sequence(Charge) | Site | Phosphorylated by |
|---|---|---|---|---|
| NP_001093976 | Cdk18 | RAS*LSDIGFGK(+2) | S109 | PKA |
| NP_001099225 | Pkn2 | AS*SLGEIDDSSELR(+2) | S581 | PKA SGK CAMK2 |
| NP_001099344 | Mylk | SS*LTPVLGTESDATVK(+2) | S1252 | PKA |
| NP_001099344 | Mylk | SSLT*PVLGTESDATVK(+2) | T1254 | PKA |
| NP_001100374 | Map4k4 | KGS*VVNVNPTNTR(+2) | S839 | PKA |
| NP_062015 | Prkaa1 | SGS*ISNYR(+2) | S485 | PKA |
| NP_062222 | Slk | INS*TATPDQDREK(+2) | S1062 | PKA |
| NP_062222 | Slk | KKT*LEEEFAR(+2) | T1156 | PKA |
| NP_067731 | Mark2 | RSS*DQAVPAIPTSNSYSK(+2) | S409 | PKA |
| NP_112343 | Pdpk1 | ANS*FVGTAQYVSPELLTEK(+2) | S244 | SGK |
| NP_112343 | Pdpk1 | IGS*FDETCTR(+2) | S179 | SGK |
| NP_112343 | Pdpk1 | KIGS*FDETCTR(+2) | S179 | SGK |
| NP_112343 | Pdpk1 | ANS*FVGTAQYVSPELLTEK(+3) | S244 | SGK |
| NP_112343 | Pdpk1 | ANSFVGTAQY*VSPELLTEK(+3) | Y251 | SGK |
| NP_112343 | Pdpk1 | ILGEGS*FSTVVLAR(+2) | S95 | SGK CAMK2 |
| NP_112343 | Pdpk1 | LLVLDAT*KR(+2) | T325 | SGK |
| NP_001101361 | Ppp2r5a | SQGS*QAELHPLPQLK(+2) | S49 | PKA |
| NP_446342 | Ppp1r12a | RIS*EMEEELK(+2) | S941 | PKA CAMK2 |
| NP_476481 | Ppp2r1a | KLS*TIALALGVER(+2) | S36 | PKA |
| NP_543183 | Ppp4r1 | AS*TLDAHDEAGGAEQR(+2) | S534 | CAMK2 |
| NP_001008301 | Nedd4l | SLSS*PTVTLSAPLEGAK(+2) | S437 | PKA |
| NP_001008301 | Nedd4l | SLS*SPTVTLSAPLEGAK(+2) | S436 | PKA SGK |
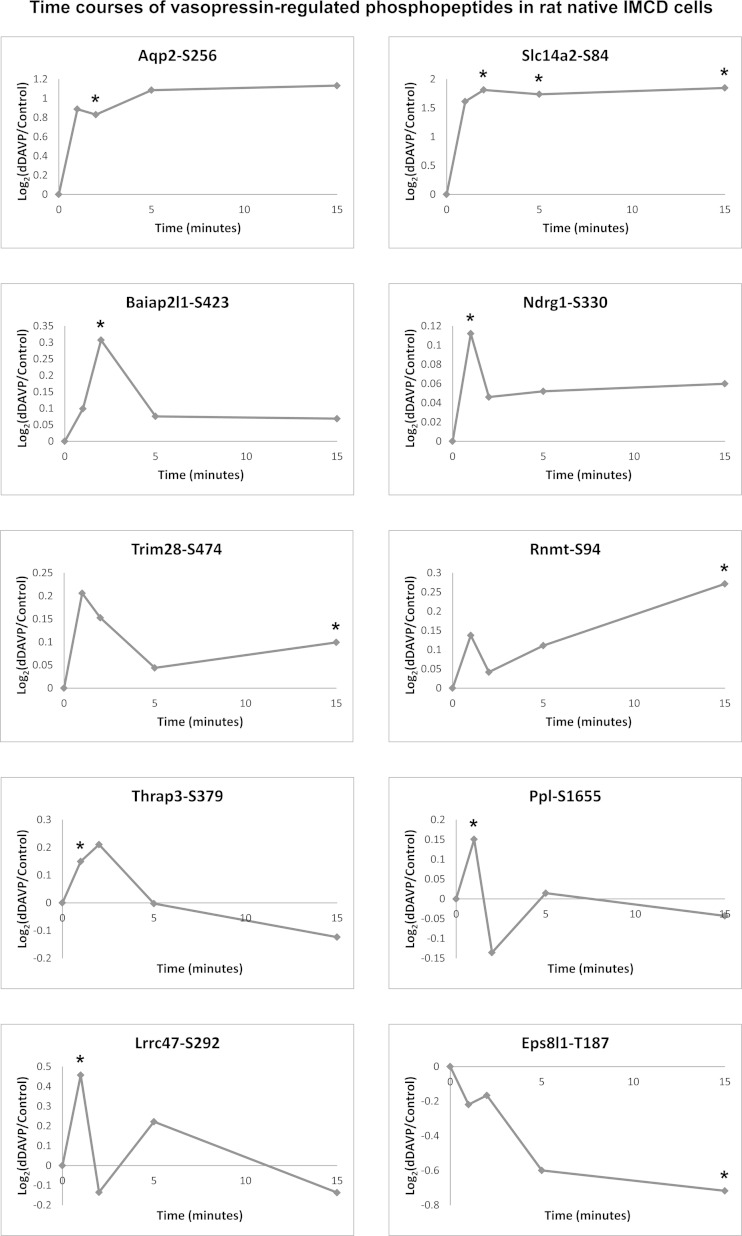
Figure 8 identifies the phosphorylation sites in IMCD identified in this study that were also quantified in previous studies (1, 23). Those previously found to be regulated by vasopressin are shown in red (increased) and green (decreased). To aid in the interpretation of Fig. 8, we constructed Fig. 10, showing the time course of phosphorylation changes in response to the vasopressin analog dDAVP in suspensions of living rat IMCD. These data were gleaned from the supplemental material of a previous publication (23). Two of these sites [Ser256 of AQP2 and Ser84 of the urea channel Slc14a2 (UT-A1/3)] showed a rapid increase in phosphorylation within 1 min of dDAVP addition and a sustained high level of phosphorylation. As seen in Fig. 8, unlike Ser256 of AQP2, Ser84 of Slc14a2 was phosphorylated only by PKA. Most of the other vasopressin targets in Fig. 10 showed early transient increases in phosphorylation, suggesting that they are dynamic, intermediate nodes in the vasopressin signaling network. An exception is Thr187 of a putative scaffold protein called epidermal growth factor receptor kinase substrate 8-like protein 1 (Eps8l1), which showed a sustained decrease in phosphorylation in the intact cell studies (Fig. 10) similar to the time course of phosphorylation of AQP2 at Ser261 in response to vasopressin (22).
Fig. 10.
Time course of phosphorylation changes in response to dDAVP in isolated rat IMCD suspensions. Values are from iTRAQ quantification of peptides using LC-MS/MS, as presented by Hoffert et al. (23).
Our analysis of protein kinase substrates (summarized in Fig. 8) utilized subcellular fractionation of native IMCDs as a technical procedure to decrease competition among substrates for recognition by the individual exogenous kinases in in vitro incubations (see methods). In addition, the fractions in which the target proteins were found may provide information relevant to the potential physiological roles of the phosphorylation events. The fractions that contained the upregulated (red-highlighted) phosphoproteins of Fig. 8 are therefore presented in Table 7. A strong correlation was found between the subcellular fractions containing the phosphoproteins and the kinases that phosphorylated them. In particular, Table 7 shows that the phosphoproteins found in the 17,000-g pellet (enriched in plasma membrane, endoplasmic reticulum, Golgi, and endosomal proteins) were phosphorylated predominantly by PKA (compare with Fig. 8). In contrast, the upregulated phosphoproteins not found in the 17,000-g pellet (Rnmt, Trim28, and Thrap3) were found to be phosphorylated in vitro predominantly by CAMK2. These results therefore raise the possibility that, in the native IMCD, PKA and CAMK2 reside in and are active in different cellular compartments. Consistent with this view, previous proteomics studies of subcellular fractions in mpkCCD cells showed that the PKA catalytic subunit Prkacb is localized to the cytoplasmic, but not the nuclear extract, fraction, whereas Camk2d is found in the nuclear extract and the cytoplasmic fraction (50). Thus PKA and CAMK2 could be involved in phosphorylation at Ser256 of AQP2 in different compartments.
Table 7.
Upregulated phosphoproteins: subcellular fractions
| Differential Centrifugation Fraction* |
|||||||
|---|---|---|---|---|---|---|---|
| Accession No. | Gene Symbol | Protein Name | 1K | 4K | 17K | 200K | 200K Supernatant |
| NP_001008300 | Rnmt | mRNA cap guanine methyltransferase | Present | Present | |||
| NP_001009693 | Thrap3 | Thyroid hormone receptor-associated 3 | Present | Present | |||
| NP_001011991 | Ndrg1 | NDRG1 | Present | Present | Present | ||
| NP_001029312 | Baiap2l1 | Brain-specific angiogenesis inhibitor | Present | ||||
| NP_001100446 | Ppl | Periplakin | Present | Present | Present | Present | Present |
| NP_001129138 | Lrrc47 | Leucine-rich repeat-containing 47 | Present | Present | Present | ||
| NP_037041 | Aqp2 | AQP2 | Present | Present | Present | Present | |
| NP_062220 | Slc14a2 | Urea transporter 2 isoform 1 | Present | Present | Present | ||
| NP_446368 | Trim28 | Transcription intermediary factor 1β | Present | ||||
| NP_808877 | Slc14a2 | Urea transporter 2 isoform 2 | Present | Present | Present | Present | |
| XP_001069984 | Tnks1bp1 | Tankyrase 1-binding protein 1 | Present | Present | Present | Present | |
Bold font indicates proteins predominantly phosphorylated by PKA in vitro; italicized font indicates proteins most strongly phosphorylated by CAMK2 in vitro (based on Fig. 8). Ser256 of AQP2 was phosphorylated nearly equally by PKA and CAMK2.
Fractions were obtained by differential centrifugation as described in methods and are designated as follows: 1K, 1,000-g pellet; 4K, 4,000-g pellet; 17K, 17,000-g pellet; 200K, 200,000-g pellet; 200K Supernatant, supernatant from 200,000-g spin. Gene Ontology (GO) analysis of all proteins (phosphorylated and nonphosphorylated) revealed the fractions in which individual GO cellular component terms are maximal as follows (percentage of total in fraction): 1K, nucleus (35.1), intermediate filament (3.0), tight junction (2.0), myosin complex (2.0); 4K, mitochondrion (18.6), early endosome (1.8), cell cortex (1.5); 17K, plasma membrane (20.8), endoplasmic reticulum (11.0), Golgi apparatus (7.2), endosome (4.0); 200K, ribonucleoprotein complex (11.6), ribosome (7.6), proteasome complex (5.8), microtubule (4.4); 200K supernatant, cytosol (13.9), cytoskeleton (13.3), extracellular space (2.5), actin cytoskeleton (3.1).
DISCUSSION
Phosphorylation at Ser256 of AQP2
As reviewed in the introduction, phosphorylation at Ser256 of AQP2 is a key event in the causal chain that connects vasopressin binding to the V2 vasopressin receptor to the trafficking of AQP2 to the apical plasma membrane of collecting duct cells. This trafficking process is responsible for the ability of vasopressin to regulate water excretion in short (10–30 min) time frames. As also reviewed in the introduction, the protein kinase or kinases responsible for AQP2 phosphorylation at Ser256 remain an open question. We have used a combination of Bayesian analysis, inhibitor studies of AQP2 phosphorylation using immunoblotting, and kinase target substrate profiling to rank the 521 protein kinases in the rat genome with regard to the likelihood that they play a role in phosphorylation of AQP2 at Ser256 in the rat IMCD.
Bayesian analysis has not been extensively used in physiology, although it has been much more widely employed in the area of medicine. For example, the politically controversial conclusion that smoking causes cancer was reached in the 1950s using data integration from multiple sources via Bayesian statistics (11). More recently, the Bayesian approach has been employed to address age dependence of risks and benefits of screening mammography (2) and whether the prostate-specific antigen test should be used for routine screening for prostate cancer (65). The benefit for physiology research is that Bayesian analysis allows new physiological data to be interpreted in the context of prior information, instead of in isolation, as seen with purely frequentist statistical approaches, such as t-tests and ANOVA. A readable discussion of the advantages of Bayesian statistics over frequentist statistics has been presented by Davidoff (12). We used a Bayesian approach to address the following question: Which among the 521 protein kinases expressed in the rat genome are most likely to phosphorylate AQP2 at Ser256 in rat renal IMCD cells? The best candidates should be expressed in the IMCD at mRNA and protein levels, should have a target substrate specificity matching the amino acid sequence surrounding Ser256, and should be regulated by vasopressin. Data from prior sources addressing these criteria were integrated using Bayes' rule to rank all candidate kinases (Fig. 3). In these results, the four kinases that are our main focus ranked in the following order of prior probabilities: CAMK2 > PKA = Akt > SGK. The resulting relative probability values became Bayesian “prior” probabilities for interpretation of the new data describing the effects of various small-molecule agents to inhibit Ser256 phosphorylation in the presence of vasopressin. A critical factor of the inhibitor data analysis was published information from the ICKP reporting the protein kinases that are inhibited by each agent and the protein kinases that are not inhibited (http://www.kinase-screen.mrc.ac.uk/). The ICKP results for the agents used in the present study are summarized in Table 3. It is clear from these data that the agents H-89, KN-62 and KN-93, and GSK-650394, which nominally target PKA, CAMK2, and SGK, respectively, have broader kinase inhibitor spectra than has been commonly assumed in physiological studies. The kinase profiling data and the measured effects (or lack of effects) of each inhibitor can be integrated to obtain joint probability measures of the likelihood that a given kinase is necessary for Ser256 phosphorylation in the rat IMCD (Fig. 1). These values and the prior probabilities calculated from preexisting data (Table 2) could then be integrated using Bayes' rule to calculate posterior probabilities for each kinase with regard to its role in phosphorylation of AQP2 at Ser256 (Table 4). These posterior probabilities further support the following rank order: CAMK2 > PKA > SGK. To summarize the inhibitor data, H-89 inhibited Ser256 phosphorylation at the high dose, and not the low dose, strengthening the argument for CAMK2. KN-93, but not KN-62, inhibited Ser256 phosphorylation, strengthening the argument for PKA. GSK-650394 had only a marginal effect on vasopressin-dependent Ser256 phosphorylation, suggesting a minor role for SGK.
Next, we asked the following question: Among CAMK2, PKA, and SGK, which are capable of phosphorylating AQP2 at Ser256? We incubated dephosphorylated IMCD proteins with purified, recombinant CAMK2, PKA, or SGK and then, using a technique described by Douglass et al. (13), performed protein MS to identify amino acid sites phosphorylated by each kinase. A database of all phosphorylation sites found in these experiments (https://helixweb.nih.gov/ESBL/Database/iKinSub/) has been made available as a resource. From the general results, it was clear that many sites could be phosphorylated by more than one of the three kinases tested (Fig. 8). This observation is compatible with the view that many basophilic kinases have overlapping specificities (13, 34). Indeed, all three protein kinases proved capable of phosphorylating AQP2 at Ser256 (Fig. 8). In contrast, another vasopressin-regulated phosphorylation site, Ser84 of the urea channel UT-A1 (Slc14a2), was phosphorylated by PKA, but not by CAMK2 or SGK.
The previous findings that protein kinases in the same family tend to have overlapping substrate specificities (13, 34) were confirmed in the present study. For this reason, assignment of target specificity probabilities for the initial Bayesian analysis (Fig. 3) did not make specificity assumptions beyond those that could be inferred from the family classification for each kinase. For example, AGC and CAMK family members (all basophilic kinases) were assumed to have the same high probability of being capable of phosphorylating AQP2 at Ser256. This may be too conservative, and future studies may benefit from greater use of existing target specificity data for each kinase. An example is provided in Fig. 9, showing that the experimentally determined substrate target motif for CAMK2 is a closer fit than those of PKA and SGK to the sequence surrounding Ser256 of AQP2. Nevertheless, factors that cause kinases to associate with their targets may be of greater importance than the substrate target specificity in determining what substrates are phosphorylated by what kinases (33). For a given kinase to phosphorylate a given substrate, the two must come into physical contact and, therefore, must be localized in a common subcellular compartment. For some kinases, such as PKA, anchoring proteins, such as Akap proteins, may play an important role in compartmentation (32). Thus there is a need for comprehensive proteomics of subcellular fractions of collecting duct cells to determine the location of all candidate kinases with respect to their potential substrates.
We limited the Bayesian analysis in this study to one phosphorylation target, Ser256 of AQP2. Future studies are needed to extend the Bayesian approach to allow prediction of the protein kinases most likely to be responsible for all vasopressin-regulated phosphorylation sites in collecting duct cells. These predictions, coupled with similar predictions for phosphatases, would provide a stepping stone to resolution of a complete vasopressin signaling network in collecting duct cells.
Phosphorylation of Other Collecting Duct Proteins
An ancillary objective of this study is to identify additional phosphorylation sites in other collecting duct proteins that could play roles in AQP2 regulation. The phosphorylation sites that were previously found to be increased by vasopressin and were phosphorylated by one or more of the basophilic kinases investigated in this study are shown in red in Fig. 8. Among these, four, Trim28, periplakin, Ndrg1, and tankyrase 1-binding protein (Tnks1bp1), appear to be well annotated and have plausible roles in the regulation of AQP2. Collecting duct-specific annotations of these proteins are presented in the appendix.
Three additional phosphoproteins reported in Table 6, Nedd4-2 (gene symbol Nedd4l), mitogen-activated protein kinase kinase kinase kinase 4 (Map4k4), and 3-phosphoinositide-dependent protein kinase I or PDK1 (gene symbol Pdpk1), also have potential roles in vasopressin signaling.
Nedd4-2 is an E3 ubiquitin ligase that has been shown to decrease the abundance of the epithelial Na+ channel (ENaC) in the plasma membrane. Inhibition of Nedd4l by SGK- or PKA-mediated Nedd4l phosphorylation, therefore, increases ENaC activity and may mediate vasopressin's action to increase ENaC-mediated Na+ transport (54). As summarized by Fig. 8A, PKA and SGK phosphorylate Nedd4l at one of its putative regulatory sites, namely, Ser436. Snyder et al. (53) demonstrated Nedd4l phosphorylation by SGK and PKA at three additional Nedd4l sites (Ser327, Ser221, and Thr246). Phosphorylation at a different site, Thr439, was found to be upregulated by vasopressin in the IMCD (23). Since ENaC is not strongly expressed in the IMCD (62), we speculate that Nedd4l may ubiquitylate other transport proteins there, perhaps AQP2 (28).
Another potentially important regulatory protein in Table 6 is Map4k4, an STE20 family protein kinase that is upstream from c-jun NH2-terminal kinase (Jnk), a proline-directed kinase that is strongly downregulated in the IMCD in response to vasopressin (23). Map4k4 phosphorylates c-raf and related proteins. Phosphorylation of Map4k4 at Ser839 by PKA (Table 6) provides a possible pathway that could bridge the canonical vasopressin signaling pathway to the mitogen-activated protein kinase signaling pathway.
A third important regulatory protein in Table 6 is PDK1 (gene symbol Pdpk1). PDK1 is an AGC family kinase that is activated in endosomes and the plasma membrane by phosphatidylinositol 3-phosphate. PDK1 is believed to activate SGK (as well as the related kinase AKT) through phosphorylation of its activation loop (40). Since Table 6 reveals several sites on PDK1 that can be phosphorylated by its substrate SGK, the results point to a feedback mechanism that could modulate the pathway.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
D.B., J.L.W., C.-L.C., J.D.H., M.A.K., and T.P. are responsible for conception and design of the research; D.B. and J.L.W. performed the experiments; D.B., V.R., J.L.W., C.-L.C., M.A.K., and T.P. analyzed the data; D.B., V.R., J.L.W., C.-L.C., J.D.H., M.A.K., and T.P. interpreted the results of the experiments; D.B., V.R., J.L.W., C.-L.C., M.A.K., and T.P. prepared the figures; D.B. drafted the manuscript; D.B., V.R., J.L.W., C.-L.C., J.D.H., M.A.K., and T.P. edited and revised the manuscript; D.B., V.R., J.L.W., C.-L.C., J.D.H., M.A.K., and T.P. approved the final version of the manuscript.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Guanghui Wang (National Heart, Lung, and Blood Institute Proteomics Core Facility) for assistance with the mass spectrometry.
The study was carried out in the intramural program of the National Heart, Lung, and Blood Institute (Project ZO1-HL-001285, M. A. Knepper).
Present address of T. Pisitkun: Faculty of Medicine, Chulalongkorn University, Bangkok, Thailand. T. Pisitkun is currently supported by CU Research Cluster: 2014 Ratchadapisek Sompoch Endowment Fund, Chulalongkorn University.
Appendix
Here we present collecting duct-specific annotations of four vasopressin-regulated proteins that are of particular interest with regard to regulation of collecting duct function.
Trim28.
Trim28 (also called KAP1) is a well-studied multidomain protein that functions, together with KRAB domain-containing zinc-finger transcription factors, to carry out repression of specific genes (26). It does this in part by providing a scaffold for a variety of histone modifiers. Trim28's transcript is abundant in the rat IMCD, expressed at a level approximately fivefold over the median of all transcripts (62). The Trim28 protein was found by protein MS in nuclear and cytoplasmic fractions in IMCD cells (60) and in mpkCCD cells (50). In isolated IMCD suspensions, phosphorylation at Ser474 was markedly increased by vasopressin addition at 30 min, but not at earlier time points (23). The site was found to be phosphorylated by CAMK2 (Fig. 8A). This phosphorylation site has been previously identified in transcriptional regulation in other cell types through its ability to modulate binding of heterochromatin-binding protein 1 (HP1, gene symbol Cbx3) (6), raising the possibility that it could be involved in regulation of AQP2 gene expression in the IMCD.
Periplakin.
Periplakin (gene symbol Ppl) is a cytoskeletal protein that binds to actin and intermediate filaments and plays a role in cell adhesion (Swiss-Prot Q9R269). It has been demonstrated to bind to the COOH-terminal tail of a number of G protein-coupled receptors, including the vasopressin V2 receptor, and this interaction is believed to modulate signaling (68). The periplakin transcript is extremely abundant in the rat IMCD and is expressed at a level ∼13-fold over the median of all transcripts (62). It has been readily identified in cytoplasmic and nuclear fractions of IMCD, indicating that, like many junctional and cytoskeletal proteins, periplakin can move into the nucleus (60). Indeed, periplakin has been found to increase in its abundance in the nuclei of cultured mpkCCD cells in response to vasopressin, indicative of vasopressin-induced nuclear translocation (50). In isolated IMCD suspensions, phosphorylation at Ser1655 was significantly increased by vasopressin addition within 30 s (23). Ser1655 was found to be preferentially phosphorylated by PKA in this study (Fig. 8A).
N-myc downregulated gene 1.
The protein coded by N-myc downregulated gene 1 (NDRG1, also called CAP43) is a multifunctional protein involved in membrane recycling (27) and regulation of apoptosis (56). It has been identified as an effector for Rab4a in early endosomes and is involved in the regulation of cadherin-1 recycling and, thus, in the maintenance of adherens junctions (27). NDRG1 is highly expressed in the rat renal IMCD, with a transcript abundance ∼19 times the median transcript abundance (62). It is also readily detectable by MS of IMCD proteins (47). In cultured mpkCCD cells, it has been detected in cytoplasmic and nuclear fractions (http://helixweb.nih.gov/ESBL/Database/mNPD/). Ser330 (Fig. 8A) is situated in a polyphosphorylated region near the COOH terminus of NDRG1 just upstream from a tandem repeat of a hydrophilic sequence (GTRSRSHTSE) of unknown function (Swiss-Prot Q29597). In isolated IMCD suspensions, Ser330 phosphorylation was significantly increased by vasopressin (23). In this study, we identify this site as a potential substrate for SGK and PKA (Fig. 8A). Phosphorylation by SGK at this site has been previously documented and has been proposed to be a priming event for subsequent phosphorylation by GSK 3-β (36). Our results are also consistent with results from another study showing that Ser330 of NDRG1 may also be phosphorylated by PKA (61). Vasopressin strongly inhibits apoptosis in the IMCD (23), and it appears possible that NDRG1 phosphorylation plays a role in this process.
Immunoprecipitation and protein MS were used in a recent proteomics study to address whether a multiprotein complex is associated with NDRG1 (61). Among other proteins, NDRG1 appears to bind to β-catenin and E-cadherin, so it fits with proposed roles for β-catenin in vasopressin-regulated transcription (3, 50). In addition, it binds to the α1-subunit of the Na+-K+-ATPase (ATP1a1), the major Na+ pump isoform in the IMCD (62). We found in mpkCCD cells that NDRG1 is significantly upregulated in total protein and mRNA abundance in response to long-term treatment with vasopressin (30).
Tankyrase 1-binding protein.
Tankyrase 1-binding protein (Tnks1bp1, Swiss-Prot P58871) is a polyphosphorylated 182-kDa protein identified as a binding partner and potential modulator of tankyrase-1 (51). The latter is a poly-ADP-ribosyltransferase that plays a key role in Wnt signaling by poly-ADP-ribosylating Axin1 and Axin2, key components of the β-catenin destruction complex. Tnks1bp1 is moderately expressed in the rat IMCD, with a transcript abundance 1.75 times the median value (62). The protein is also readily detectable in rat IMCD by MS (http://dir.nhlbi.nih.gov/papers/lkem/imcd/). The phosphorylation site in question, Ser883, was significantly increased (by 67%) in response to vasopressin in isolated IMCD suspensions (1). In the present study, this site was phosphorylated by PKA, but not by CAMK2 or SGK (Fig. 8B). Given our observation that β-catenin abundance in the nucleus increases in response to vasopressin (3, 50) and the well-established role for β-catenin as a transcriptional coregulator, we speculate that PKA-mediated phosphorylation of Tnks1bp1 could play a role in vasopressin-dependent gene regulation in the collecting duct.
Footnotes
Official gene symbols are designated by italics.
This article is the topic of an Editorial Focus by Rudi Prihandoko and Andrew B. Tobin (43a).
REFERENCES
- 1.Bansal AD, Hoffert JD, Pisitkun T, Hwang S, Chou CL, Boja ES, Wang G, Knepper MA. Phosphoproteomic profiling reveals vasopressin-regulated phosphorylation sites in collecting duct. J Am Soc Nephrol 21: 303–315, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berry DA. Breast cancer screening: controversy of impact. Breast 22 Suppl 2: S73–S76, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolger SJ, Gonzales Hurtado PA, Hoffert JD, Saeed F, Pisitkun T, Knepper MA. Quantitative phosphoproteomics in nuclei of vasopressin-sensitive renal collecting duct cells. Am J Physiol Cell Physiol 303: C1006–C1020, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouley R, Breton S, Sun T, McLaughlin M, Nsumu NN, Lin HY, Ausiello DA, Brown D. Nitric oxide and atrial natriuretic factor stimulate cGMP-dependent membrane insertion of aquaporin 2 in renal epithelial cells. J Clin Invest 106: 1115–1126, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Champigneulle A, Siga E, Vassent G, Imbert-Teboul M. V2-like vasopressin receptor mobilizes intracellular Ca2+ in medullary collecting tubules. Am J Physiol Renal Fluid Electrolyte Physiol 265: F35–F45, 1993. [DOI] [PubMed] [Google Scholar]
- 6.Chang CW, Chou HY, Lin YS, Huang KH, Chang CJ, Hsu TC, Lee SC. Phosphorylation at Ser473 regulates heterochromatin protein 1 binding and corepressor function of TIF1β/KAP1. BMC Mol Biol 9: 61, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chijiwa T, Mishima A, Hagiwara M, Sano M, Hayashi K, Inoue T, Naito K, Toshioka T, Hidaka H. Inhibition of forskolin-induced neurite outgrowth and protein phosphorylation by a newly synthesized selective inhibitor of cyclic AMP-dependent protein kinase, N-[2-(p-bromocinnamylamino)ethyl]-5-isoquinolinesulfonamide (H-89), of PC12D pheochromocytoma cells. J Biol Chem 265: 5267–5272, 1990. [PubMed] [Google Scholar]
- 8.Chou CL, Yip KP, Knepper MA. Role of Ca/calmodulin in vasopressin-stimulated aquaporin-2 trafficking in rat collecting duct (Abstract). J Am Soc Nephrol 10: 13A, 1999.9890304 [Google Scholar]
- 9.Chou CL, Yip KP, Michea L, Kador K, Ferraris J, Wade JB, Knepper MA. Regulation of aquaporin-2 trafficking by vasopressin in renal collecting duct: roles of ryanodine-sensitive Ca2+ stores and calmodulin. J Biol Chem 275: 36839–36846, 2000. [DOI] [PubMed] [Google Scholar]
- 10.Congdon P. Bayesian Statistical Modeling. Chichester, UK: Wiley, 2006. [Google Scholar]
- 11.Cornfield J, Haenszel W, Hammond EC, Lilienfeld AM, Shimkin MB, Wynder EL. Smoking and lung cancer: recent evidence and a discussion of some questions. J Natl Cancer Inst 22: 173–203, 1959. [PubMed] [Google Scholar]
- 12.Davidoff F. Standing statistics right side up. Ann Intern Med 130: 1019–1021, 1999. [DOI] [PubMed] [Google Scholar]
- 13.Douglass J, Gunaratne R, Bradford D, Saeed F, Hoffert JD, Steinbach PJ, Knepper MA, Pisitkun T. Identifying protein kinase target preferences using mass spectrometry. Am J Physiol Cell Physiol 303: C715–C727, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ecelbarger CA, Chou CL, Lolait SJ, Knepper MA, DiGiovanni SR. Evidence for dual signaling pathways for V2 vasopressin receptor in rat inner medullary collecting duct. Am J Physiol Renal Fluid Electrolyte Physiol 270: F623–F633, 1996. [DOI] [PubMed] [Google Scholar]
- 15.Ecelbarger CA, Chou CL, Lolait SJ, Knepper MA, DiGiovanni SR. Evidence for dual signaling pathways for V2 vasopressin receptor in rat inner medullary collecting duct. Am J Physiol Renal Fluid Electrolyte Physiol 270: F623–F633, 1996. [DOI] [PubMed] [Google Scholar]
- 16.Ecelbarger CA, Nielsen S, Olson BR, Murase T, Baker EA, Knepper MA, Verbalis JG. Role of renal aquaporins in escape from vasopressin-induced antidiuresis in rat. J Clin Invest 99: 1852–1863, 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eng JK, McCormack AL, Yates JR., 3rd An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom 5: 976–989, 1994. [DOI] [PubMed] [Google Scholar]
- 18.Fushimi K, Sasaki S, Marumo F. Phosphorylation of serine 256 is required for cAMP-dependent regulatory exocytosis of the aquaporin-2 water channel. J Biol Chem 272: 14800–14804, 1997. [DOI] [PubMed] [Google Scholar]
- 19.Fushimi K, Uchida S, Hara Y, Hirata Y, Marumo F, Sasaki S. Cloning and expression of apical membrane water channel of rat kidney collecting tubule. Nature 361: 549–552, 1993. [DOI] [PubMed] [Google Scholar]
- 20.Hendriks G, Koudijs M, van Balkom BW, Oorschot V, Klumperman J, Deen PM, van der Sluijs P. Glycosylation is important for cell surface expression of the water channel aquaporin-2 but is not essential for tetramerization in the endoplasmic reticulum. J Biol Chem 279: 2975–2983, 2004. [DOI] [PubMed] [Google Scholar]
- 21.Hoffert JD, Fenton RA, Moeller HB, Simons B, Tchapyjnikov D, McDill BW, Yu MJ, Pisitkun T, Chen F, Knepper MA. Vasopressin-stimulated increase in phosphorylation at Ser269 potentiates plasma membrane retention of aquaporin-2. J Biol Chem 283: 24617–24627, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoffert JD, Nielsen J, Yu MJ, Pisitkun T, Schleicher SM, Nielsen S, Knepper MA. Dynamics of aquaporin-2 serine-261 phosphorylation in response to short-term vasopressin treatment in collecting duct. Am J Physiol Renal Physiol 292: F691–F700, 2007. [DOI] [PubMed] [Google Scholar]
- 23.Hoffert JD, Pisitkun T, Saeed F, Song JH, Chou CL, Knepper MA. Dynamics of the G protein-coupled vasopressin V2 receptor signaling network revealed by quantitative phosphoproteomics. Mol Cell Proteomics 11: M111, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoffert JD, Pisitkun T, Wang G, Shen RF, Knepper MA. Quantitative phosphoproteomics of vasopressin-sensitive renal cells: regulation of aquaporin-2 phosphorylation at two sites. Proc Natl Acad Sci USA 103: 7159–7164, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, Villen J, Haas W, Sowa ME, Gygi SP. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143: 1174–1189, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iyengar S, Farnham PJ. KAP1 protein: an enigmatic master regulator of the genome. J Biol Chem 286: 26267–26276, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kachhap SK, Faith D, Qian DZ, Shabbeer S, Galloway NL, Pili R, Denmeade SR, DeMarzo AM, Carducci MA. The N-Myc down regulated gene1 (NDRG1) is a Rab4a effector involved in vesicular recycling of E-cadherin. PLos One 2: e844, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kamsteeg EJ, Hendriks G, Boone M, Konings IB, Oorschot V, van der SP, Klumperman J, Deen PM. Short-chain ubiquitination mediates the regulated endocytosis of the aquaporin-2 water channel. Proc Natl Acad Sci USA 103: 18344–18349, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Katsura T, Gustafson CE, Ausiello DA, Brown D. Protein kinase A phosphorylation is involved in regulated exocytosis of aquaporin-2 in transfected LLC-PK1 cells. Am J Physiol Renal Physiol 272: F817–F822, 1997. [PubMed] [Google Scholar]
- 30.Khositseth S, Pisitkun T, Slentz DH, Wang G, Hoffert JD, Knepper MA, Yu MJ. Quantitative protein and mRNA profiling shows selective post-transcriptional control of protein expression by vasopressin in kidney cells. Mol Cell Proteomics 10: M110.–004036., 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klussmann E, Maric K, Rosenthal W. The mechanisms of aquaporin control in the renal collecting duct. Rev Physiol Biochem Pharmacol 141: 33–95, 2000. [DOI] [PubMed] [Google Scholar]
- 32.Klussmann E, Maric K, Wiesner B, Beyermann M, Rosenthal W. Protein kinase A anchoring proteins are required for vasopressin-mediated translocation of aquaporin-2 into cell membranes of renal principal cells. J Biol Chem 274: 4934–4938, 1999. [DOI] [PubMed] [Google Scholar]
- 33.Linding R, Jensen LJ, Ostheimer GJ, van Vugt MA, Jorgensen C, Miron IM, Diella F, Colwill K, Taylor L, Elder K, Metalnikov P, Nguyen V, Pasculescu A, Jin J, Park JG, Samson LD, Woodgett JR, Russell RB, Bork P, Yaffe MB, Pawson T. Systematic discovery of in vivo phosphorylation networks. Cell 129: 1415–1426, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Linding R, Jensen LJ, Pasculescu A, Olhovsky M, Colwill K, Bork P, Yaffe MB, Pawson T. NetworKIN: a resource for exploring cellular phosphorylation networks. Nucleic Acids Res 36: D695–D699, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science 298: 1912–1934, 2002. [DOI] [PubMed] [Google Scholar]
- 36.Murray JT, Campbell DG, Morrice N, Auld GC, Shpiro N, Marquez R, Peggie M, Bain J, Bloomberg GB, Grahammer F, Lang F, Wulff P, Kuhl D, Cohen P. Exploitation of KESTREL to identify NDRG family members as physiological substrates for SGK1 and GSK3. Biochem J 384: 477–488, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nielsen S, Frokiaer J, Marples D, Kwon TH, Agre P, Knepper MA. Aquaporins in the kidney: from molecules to medicine. Physiol Rev 82: 205–244, 2002. [DOI] [PubMed] [Google Scholar]
- 38.Noda Y, Sasaki S. Molecular mechanisms and drug development in aquaporin water channel diseases: molecular mechanism of water channel aquaporin-2 trafficking. J Pharm Sci 96: 249–254, 2004. [DOI] [PubMed] [Google Scholar]
- 39.Pearce D, Verrey F, Chen SY, Mastroberardino L, Meijer OC, Wang J, Bhargava A. Role of SGK in mineralocorticoid-regulated sodium transport. Kidney Int 57: 1283–1289, 2000. [DOI] [PubMed] [Google Scholar]
- 40.Pearce LR, Komander D, Alessi DR. The nuts and bolts of AGC protein kinases. Nat Rev Mol Cell Biol 11: 9–22, 2010. [DOI] [PubMed] [Google Scholar]
- 41.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20: 3551–3567, 1999. [DOI] [PubMed] [Google Scholar]
- 42.Pisitkun T, Bieniek J, Tchapyjnikov D, Wang G, Wu WW, Shen RF, Knepper MA. High-throughput identification of IMCD proteins using LC-MS/MS. Physiol Genomics 25: 262–275, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pisitkun T, Jacob V, Schleicher SM, Chou CL, Yu MJ, Knepper MA. Akt and ERK1/2 pathways are components of the vasopressin signaling network in rat native IMCD. Am J Physiol Renal Physiol 295: F1030–F1043, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43a.Prihandoko R, Tobin AB. Challenges of assigning protein kinases to in vivo phosphorylation events. Focus on “Use of LC-MS/MS and Bayes' theorem to identify protein kinases that phosphorylate aquaporin-2 at Ser256.” Am J Physiol Cell Physiol (May 14, 2014). 10.1152/ajpcell.00136.2014. [DOI] [PubMed] [Google Scholar]
- 44.Procino G, Carmosino M, Marin O, Brunati AM, Contri A, Pinna LA, Mannucci R, Nielsen S, Kwon TH, Svelto M, Valenti G. Ser-256 phosphorylation dynamics of aquaporin 2 during maturation from the ER to the vesicular compartment in renal cells. FASEB J 17: 1886–1888, 2003. [DOI] [PubMed] [Google Scholar]
- 45.Rinschen MM, Yu MJ, Wang G, Boja ES, Hoffert JD, Pisitkun T, Knepper MA. Quantitative phosphoproteomic analysis reveals vasopressin V2-receptor-dependent signaling pathways in renal collecting duct cells. Proc Natl Acad Sci USA 107: 3887, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robben JH, Knoers NV, Deen PM. Cell biological aspects of the vasopressin type-2 receptor and aquaporin 2 water channel in nephrogenic diabetes insipidus. Am J Physiol Renal Physiol 291: F257–F270, 2006. [DOI] [PubMed] [Google Scholar]
- 47.Sachs AN, Pisitkun T, Hoffert JD, Yu MJ, Knepper MA. LC-MS/MS analysis of differential centrifugation fractions from native inner medullary collecting duct of rat. Am J Physiol Renal Physiol 295: F1799–F1806, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sands JM, Layton HE. The physiology of urinary concentration: an update. Semin Nephrol 29: 178–195, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Savitski MM, Lemeer S, Boesche M, Lang M, Mathieson T, Bantscheff M, Kuster B. Confident phosphorylation site localization using the Mascot delta score. Mol Cell Proteomics 10: M110, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schenk LK, Bolger SJ, Luginbuhl K, Gonzales PA, Rinschen MM, Yu MJ, Hoffert JD, Pisitkun T, Knepper MA. Quantitative proteomics identifies vasopressin-responsive nuclear proteins in collecting duct cells. J Am Soc Nephrol 23: 1008–1018, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seimiya H, Smith S. The telomeric poly(ADP-ribose) polymerase, tankyrase 1, contains multiple binding sites for telomeric repeat binding factor 1 (TRF1) and a novel acceptor, 182-kDa tankyrase-binding protein (TAB182). J Biol Chem 277: 14116–14126, 2002. [DOI] [PubMed] [Google Scholar]
- 52.Sherk AB, Frigo DE, Schnackenberg CG, Bray JD, Laping NJ, Trizna W, Hammond M, Patterson JR, Thompson SK, Kazmin D, Norris JD, McDonnell DP. Development of a small-molecule serum- and glucocorticoid-regulated kinase-1 antagonist and its evaluation as a prostate cancer therapeutic. Cancer Res 68: 7475–7483, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Snyder PM, Olson DR, Kabra R, Zhou R, Steines JC. cAMP and serum and glucocorticoid-inducible kinase (SGK) regulate the epithelial Na+ channel through convergent phosphorylation of Nedd4-2. J Biol Chem 279: 45753–45758, 2004. [DOI] [PubMed] [Google Scholar]
- 54.Soundararajan R, Lu M, Pearce D. Organization of the ENaC-regulatory machinery. Crit Rev Biochem Mol Biol 47: 349–359, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Star RA, Nonoguchi H, Balaban R, Knepper MA. Calcium and cyclic adenosine monophosphate as second messengers for vasopressin in the rat inner medullary collecting duct. J Clin Invest 81: 1879–1888, 1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stein S, Thomas EK, Herzog B, Westfall MD, Rocheleau JV, Jackson RS, Wang M, Liang P. NDRG1 is necessary for p53-dependent apoptosis. J Biol Chem 279: 48930–48940, 2004. [DOI] [PubMed] [Google Scholar]
- 57.Stokes JB, Grupp C, Kinne RK. Purification of rat papillary collecting duct cells: functional and metabolic assessment. Am J Physiol Renal Fluid Electrolyte Physiol 253: F251–F262, 1987. [DOI] [PubMed] [Google Scholar]
- 58.Sumi M, Kiuchi K, Ishikawa T, Ishii A, Hagiwara M, Nagatsu T, Hidaka H. The newly synthesized selective Ca2+/calmodulin dependent protein kinase II inhibitor KN-93 reduces dopamine contents in PC12h cells. Biochem Biophys Res Commun 181: 968–975, 1991. [DOI] [PubMed] [Google Scholar]
- 59.Tanner S, Shu H, Frank A, Wang LC, Zandi E, Mumby M, Pevzner PA, Bafna V. InsPecT: identification of posttranslationally modified peptides from tandem mass spectra. Anal Chem 77: 4626–4639, 2005. [DOI] [PubMed] [Google Scholar]
- 60.Tchapyjnikov D, Li Y, Pisitkun T, Hoffert JD, Yu MJ, Knepper MA. Proteomic profiling of nuclei from native renal inner medullary collecting duct cells using LC-MS/MS. Physiol Genomics 40: 167–183, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tu LC, Yan X, Hood L, Lin B. Proteomics analysis of the interactome of N-myc downstream regulated gene 1 and its interactions with the androgen response program in prostate cancer cells. Mol Cell Proteomics 6: 575–588, 2007. [DOI] [PubMed] [Google Scholar]
- 62.Uawithya P, Pisitkun T, Ruttenberg BE, Knepper MA. Transcriptional profiling of native inner medullary collecting duct cells from rat kidney. Physiol Genomics 32: 229–253, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Valenti G, Procino G, Carmosino M, Frigeri A, Mannucci R, Nicoletti I, Svelto M. The phosphatase inhibitor okadaic acid induces AQP2 translocation independently from AQP2 phosphorylation in renal collecting duct cells. J Cell Sci 113: 1985–1992, 2000. [DOI] [PubMed] [Google Scholar]
- 64.Valenti G, Procino G, Tamma G, Carmosino M, Svelto M. Aquaporin 2 trafficking. Endocrinology 146: 5063–5070, 2005. [DOI] [PubMed] [Google Scholar]
- 65.Vollmer RT. Predictive probability of serum prostate-specific antigen for prostate cancer: an approach using Bayes rule. Am J Clin Pathol 125: 336–342, 2006. [PubMed] [Google Scholar]
- 66.Wall SM, Han JS, Chou CL, Knepper MA. Kinetics of urea and water permeability activation by vasopressin in rat terminal IMCD. Am J Physiol Renal Fluid Electrolyte Physiol 262: F989–F998, 1992. [DOI] [PubMed] [Google Scholar]
- 67.Wang G, Wu WW, Zeng W, Chou CL, Shen RF. Label-free protein quantification using LC-coupled ion trap or FT mass spectrometry: reproducibility, linearity, and application with complex proteomes. J Proteome Res 5: 1214–1223, 2006. [DOI] [PubMed] [Google Scholar]
- 68.Ward RJ, Jenkins L, Milligan G. Selectivity and functional consequences of interactions of family A G protein-coupled receptors with neurochondrin and periplakin. J Neurochem 109: 182–192, 2009. [DOI] [PubMed] [Google Scholar]
- 69.Yip KP. Coupling of vasopressin-induced intracellular Ca2+ mobilization and apical exocytosis in perfused rat kidney collecting duct. J Physiol 538: 891–899, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.