Abstract
Retinoic acid (RA) has diverse biological effects. The liver stores vitamin A, generates RA, and expresses receptors for RA. The current study examines the hepatic binding profile of two RA receptor isoforms, RARA (RARα) and RARB (RARβ), in response to RA treatment in mouse livers. Our data uncovered 35,521, and 14,968 genomic bindings for RARA and RARB, respectively. Each expressed unique and common bindings, implying their redundant and specific roles. RARB has higher RA responsiveness than RARB. RA treatment generated 18,821 novel RARB bindings but only 14,798 of RARA bindings, compared with the control group. RAR frequently bound the consensus hormone response element [HRE; (A/G)G(G/T)TCA], which often contained the motifs assigned to SP1, GABPA, and FOXA2, suggesting potential interactions between those transcriptional factors. Functional annotation coupled with principle component analysis revealed that the function of RAR target genes were motif dependent. Taken together, the cistrome of RARA and RARB revealed their extensive biological roles in the mouse liver. RAR target genes are enriched in various biological processes. The hepatic RAR genome-wide binding data can help us understand the global molecular mechanisms underlying RAR and RA-mediated gene and pathway regulation.
Keywords: ChIP-Seq, cistrome, liver, motif, retinoic acid
retinoic acid receptors (RAR) mediate the pleiotropic effects of retinoic acid (RA) including differentiation and anticancer. To investigate the differentiation effect of RA, the genome-wide RAR occupancy has been studied in multiple in vitro models. With the use of pan-anti-RAR antibody, the profile of RAR binding was examined in RA-induced differentiation of embryonic stem cells into spinal motor neurons (25). About 500 binding sites are constitutively bound by RAR in embryonic stem cells and RA treatment induced 1,417 new binding sites. Moreover, many of the RAR binding sites overlapped with ESR1 (estrogen receptor alpha), ESRRB (estrogen-related receptor beta), and NR5A2 (LRH-1) binding sites (25). RAR occupancy is cell type specific. Among 462 RAR binding sites in mouse stem cells, only 58 of them are identified in mouse embryonic fibroblasts (11).
In cell lines, RAR interact with other transcription factors to regulate gene expression. In acute promyelocytic cells, genome-wide binding analysis revealed that recruitment of histone deacetylase is important for RA-induced hematopoietic differentiation (26). In the breast cancer cell line MCF-7, both RARα (RARA) and γ (RARG) exerted antiproliferative and apoptotic effects and exhibited extensive genomic colocalization with ESR1 bindings. The genes commonly bound by RAR and ESR1 are antagonistically regulated by estrogen and RA (17). In addition to ESR1, RAR binding coincided with the binding of transcription factors FOXA1 and GATA3 (17, 33). In fact, FOXA1 is required for RAR recruitment (17). RA not only regulates ESR1 target genes, it also regulates the expression of androgen receptor (AR) target genes including TGM4, CDCA7L, and CDK6 (32). RARG plays a major role in the regulation of the TGM4 gene, whose expression is dependent on the presence of AR. RA and androgen antagonistically regulate CDCA7L and CDK6, suggesting the opposing effects of RAR and AR in the prostate. Collectively, the aforementioned data indicate the extensive interaction between RAR and other transcription factors in cell lines.
While some characterization of RAR genomic binding has been carried out recently, it remains unknown how RAR isoform binding targets are selected in the liver. The liver stores vitamin A (RA precursor) and produces RA binding proteins. It also expresses receptors for RA (5). Among RAR, RARA has the highest expression level in the liver. Although the basal level of RARβ (RARB) is low in the liver, RARB is inducible by RA. The inducibility of RARB can be used to predict the prognosis of cancer (2). When the RARB gene was cloned, it was cloned as a hepatitis B viral DNA integration site in the liver (10). In liver cancer cell lines, RARB expression is inhibited; in contrast, overexpression of RARB increases the susceptibility of liver cancer cells to the antiproliferative effect of RA (23, 36). The role of RAR in metabolism has also been studied in animal model. The RARA-dominant negative transgenic mice developed spontaneous steatosis and focal necrosis. Feeding the mice with RA reversed histological and biochemical abnormalities and inhibited the occurrence of liver tumors (42). Consistently, published literature demonstrated the effect of RA in regulating hepatic lipid homeostasis in vivo (1, 16). These studies indicated a significant role of RA and its receptors in liver carcinogenesis and metabolism.
To provide insight into the molecular mechanism underlying RA-mediated gene regulation through RAR in the liver, the current study analyzed the global differential binding profile of RARA and RARB in the liver in response to RA treatment in vivo for the first time. RARG was not included because the liver has a low expression level of this isoform (15). Since retinoid x receptor alpha (RXRA) is a key partner of RAR, RXRA bindings in the liver were included in data analysis. Our data revealed the differential role of RARA and RARB in response to RA in mouse liver. In addition, the data showed that the functions of RAR target genes are motif dependent.
MATERIALS AND METHODS
Animals and treatments.
C57BL/6 male mice (10 wk old; Jackson Laboratory) were fed with RA (Sigma-Aldrich, St. Louis, MO) via dietary supplementation (150 mg/kg diet) for 1 day. Control mice were fed with normal chow, which was not RA deficient. Livers were collected and frozen in liquid nitrogen immediately and stored in −80°C freezer until usage. Animal protocols were approved by the Institutional Animal Care and Use Committee of the University of Kansas Medical Center and the University of California, Davis.
Materials.
Antibodies for IgG (sc-2027), RARA (sc-551x), RARB (sc-552x), and RXRA (sc-553x) used in chromatin immunoprecipitation (ChIP) experiments were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-Pol II (05–623) was purchased from Millipore (Billerica, MA). The DNA purification kit (MinElute 28006) was purchased from Qiagen (Valencia, CA). Dynabeads used to pull down antibodies were purchased from Life Technologies (Gaithersburg, MD). Other ChIP-related chemicals were purchased from Sigma-Aldrich.
ChIP followed by sequencing.
ChIP was performed according to our published study (44). After fixation, chromatin was extracted from mouse liver tissue followed by sonication. The chromatin fragments were immunoprecipitated with anti-RARA, -RARB, and -RXRA antibodies. The target DNA fragments were obtained by reverse cross-linking and purification. Antibodies against IgG and RNA Pol II were used as negative and positive controls, respectively. DNA fragments obtained from ChIP were ligated with specified adaptors using the End-It DNA End Repair Kit (Illumina, Madison, WI), amplified, and size selected (175–225 bp) on an agarose gel followed by 35-bp single end read sequencing (High-Seq 2000; Illumina, Madison, WI).
ChIP-Seq data processing.
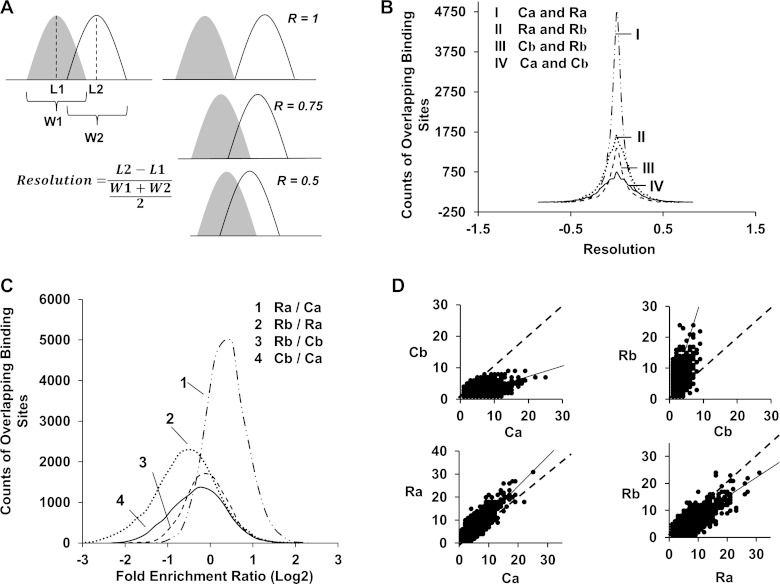
The sequences pulled down by anti-RARA, -RARB, and -RXRA antibodies were aligned to the mouse genome (mm10; http://hgdownload.cse.ucsc.edu/goldenPath/mm10/bigZips/) by Bowtie 0.12.7 (22) followed by peak-calling using MACS 1.4.1 (45) with default parameters in both programs. The called peaks were annotated against mouse genome database (GRCm38/mm10). Colocalization is defined as having at least 25% overlap in their peak widths. Resolution was introduced to evaluate the relative location of two potential overlapping peaks (see Fig. 2A for definition).
Fig. 2.
Analysis of DNA regions colocalized in RARA and RARB bindings in control and in response to RA. A: calculation of resolution (R), which is used to represent the relative location between 2 colocalized binding sites. Two bindings with R values of 0.5, 0.75, and 1 are shown as examples. B: distribution of resolutions for colocalized binding sites between the 2 indicated groups. “C” and “R” represent control and RA treatment while “a” and “b” represent RARA and RARB. C: distribution of fold enrichment ratios (log2) for colocalized bindings between the 2 indicated groups. D: number of binding sites per overlapped target gene. Each point represents 1 overlapped gene. Dashed lines represent the theoretical trend if the 2 indicated nuclear receptors have comparable binding density and solid lines represent the actual trend.
Biological functional annotation.
Official gene symbols of target genes were annotated by the DAVID program (Database for Annotation, Visualization and Integrated Discovery, v6.7; http://david.abcc.ncifcrf.gov/) (18). Annotations were performed against one of the databases of Gene Ontology Biological Processes (GOBP) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. Biological processes or pathways with assigned P ≤ 0.01 were selected for further analysis.
Motif analysis.
MEME-ChIP program was used to discover potential motifs in target bindings (24). The FIMO program was used to screen all studied binding sequences against discovered motifs by MEME-ChIP (12). The number of binding sites with specific motifs was plotted against the relative location of motifs from peak centers, showing the motif distribution profile in bindings of a nuclear receptor.
Repeats of hormone response element (HRE) with a sequence of (A/G)G(G/T)TCA were studied in RARA and RARB bindings. To reduce the noise, the FIMO-identified HRE motifs with P ≤ 0.0001 were selected to study the presence of direct repeat (DR), inverted repeat (IR), and everted repeat (ER). For each type of repeat, numbers of identified repeats were plotted against the number of nucleotides between the two half sites. The baselines were adjusted by using loess alogrithm in R program (31). HRE repeat profiles in RARA and RARB bindings of control and RA-treated mouse livers were visualized as histograms using SPSS (V20; IBM).
Hepatic gene expression profiling.
Microarray was performed using Affymetrix 430 A_2 Chips (Santa Clara, CA) to study the differential expression of hepatic genes in control and RA-treated mice (n = 3 per group) based on our published method (16). Data were annotated using the Affymetrix Expression Console (MAS5).
Statistical analysis.
Heat maps were generated by heatmap.2 function of gplots package 2.12.1 in R program (41). SPSS (V20; IBM) dimension reduction function with principle component analysis (PCA) algorithm was used to describe the global change in gene expression and functional profile of a set of genes. The orthogonal partial least square discriminant analysis (OPLSDA) function in SIMCA-P program (Umetrics, San Jose, CA) was used to differentiate the global profiles of two sets of genes and identify markers contributing to this differentiation.
Data access.
ChIP-Seq data of RXRA, RARA, and RARB were deposited in the Gene Expression Omnibus (GEO) at the National Center for Biotechnology Information (NCBI) with the accession number GSE53736. ChIP-Seq data were also submitted to the NCBI Sequence Read Archive (SRA) database via NCBI GEO with the accession numbers SRX403582 (RARA in control group), SRX403583 (RARB in control group), SRX403584 (RXRA in control group), SRX403585 (RARA in RA treatment group), SRX403586 (RARB in RA treatment group), and SRX403587 (RXRA in RA treatment group).
Mouse hepatic transcriptome (detected by cDNA microarray) data before and after RA-treatment were deposited in the NCBI GEO database with the accession number GSE50028.
RESULTS
Number of RARA and RARB binding sites and target genes in control and RA-treated mouse livers.
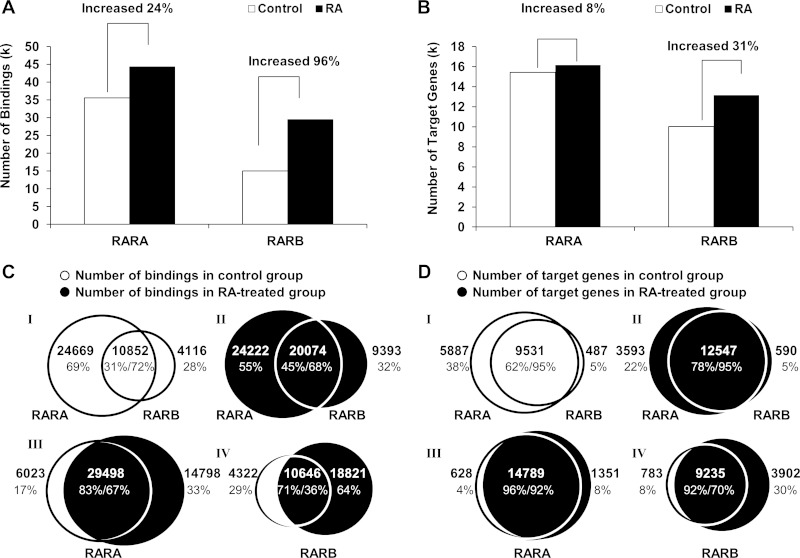
MACS called 35,521 peaks from RARA bindings, which was substantially greater than those of RARB bindings (14,968) in the control mouse livers (Fig. 1A). After RA treatment, the number of bindings for RARA and RARB increased. In the control mouse liver, there were 24,669 and 4,116 biding sites that were uniquely bound by RARA and RARB, respectively (Fig. 1C, I). In the RA-treated group, the number of RARA-unique binding sites remained similar to the control group (24,222), while the number of RARB-unique binding doubled (9,393) (Fig. 1C, II). RA treatment removed 6,023 RARA binding sites presented in the control group and induced 14,798 new RARA binding sites (Fig. 1C, III). For RARB, RA eliminated 4,322 binding sites and generated 18,821 novel bindings (Fig. 1C, IV).
Fig. 1.
Hepatic genome-wide binding of retinoic acid receptors (RARA and RARB) in control and RA-treated mouse livers. A and B: total number of bindings and target genes, respectively, for RARA and RARB in response to RA. RARB has higher RA responsiveness than RARA. C and D: showed the number of overlapping and unique binding sites and target genes between 2 indicated groups. I: comparison between RARA and RARB in control. II: comparison between RARA and RARB after RA treatment. III: comparison between control and RA treatment in RARA bindings and target genes. IV: comparison between control and RA treatment in RARB bindings and target genes. The right and left percentage numbers displayed in the overlapped area represent the percentages of the overlapped peaks or target genes in right and left circles, respectively.
The number of RARA and RARB target genes was also studied. RA only induced 723 new RARA target genes while induced 3,119 new RARB target genes (Fig. 1B). Although RA treatment generated 14,798 novel RARA bindings (Fig. 1C, III), these bindings were only associated with 1,351 RARA target genes (Fig. 1D, III). Thus there were about 10 bindings per target gene and the density of binding was high. On the other hand, 3,902 novel RARB target genes were identified in the RA-treated group (Fig. 1D, IV). Taken together, based on the number of bindings and target genes, RARB was more responsive than RARA to RA treatment.
Analysis of the overlapping bindings and common target genes of RARA and RARB.
The colocalization of RARA and RARB bindings in the mouse genome was evaluated by resolution (R) as presented in Fig. 2A. A majority of the overlapping binding sites fell within R values of ±0.5, which confirmed that the identified overlapping bindings are localized in the same region (Fig. 2B). Differences between RARA and RARB on their overlapping binding sites and common target genes were analyzed based on fold enrichment (FE) ratios and binding densities. FE represents the specific binding frequency of a nuclear receptor in a DNA region. Before RA treatment, RARA had a higher binding frequency than RARB on their overlapping binding sites as RARB-to-RARA FE ratios (log2) distributed surround −0.25 (Fig. 2C, 4). RA treatment magnified the difference between the binding frequency of RARA and RARB (Fig. 2C, 2). RA treatment had the opposite effects on the binding frequency for RARA and RARB because the FE ratio for RARA shifted to the right while RARB shifted to the left (Fig. 2C, 1 and 3). Binding sites densities on common target genes are represented by the values of BPG (numbers of binding sites per overlapped gene). Correlation analysis showed that the BPG values of RARA and RARB had a positive correlation with the actual trend line skewed towards RARA (Fig. 2D, Cb vs. Ca), indicating that RARA had a greater binding site density than RARB in the control group. The binding densities of both RARA and RARB increased following RA treatment, particularly in RARB. Thus RA treatment shifted the trend line away from RARA toward RARB, minimizing the difference of binding densities between them (Fig. 2D, Rb vs. Ra).
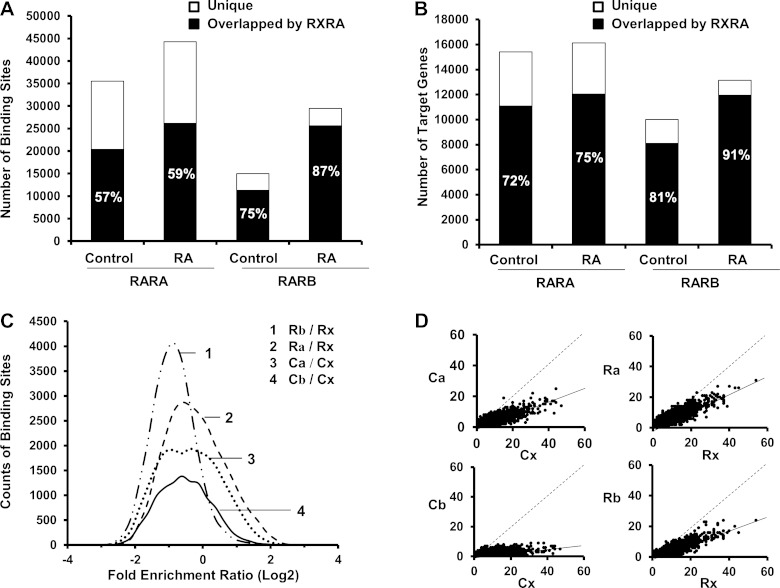
Interaction between RAR and RXRA.
As RXRA is a partner of RAR, their overlapping binding and cotarget genes were analyzed. RXRA bound to ∼75% of RARB-bound regions but bound to <60% of RARA-bound regions in control and RA groups (Fig. 3A). In terms of the number of target genes, the majority of hepatic RARA (72%) and RARB (81%) targets were also RXRA target genes in the control mouse liver (Fig. 3B). RA treatment only had a modest effect in changing these percentages. Compared with RARA, binding sites and target genes of RARB overlapped with RXRA at higher percentages, implying the essentialness of RXRA for RARB. Distribution of FE ratios of RAR to RXRA were plotted in Fig. 3C. All curves had peak centers locating in the negative area on the x-axis, indicating that RXRA had higher binding frequencies than RAR in the target DNA regions. RA treatment did not change the distribution profile of RARA-to-RXRA FE ratios (Fig. 3C, 2 vs. 3). However, it shifted the distribution profile of RARB-to-RXRA FE ratios further to the left (Fig. 3C, 1 vs. 4). Since RA increased the FE of RARA (Fig. 2C, 1), the unchanged RARA-to-RXRA FE ratios suggested that RA induced the FE of RXRA as well (Fig. 3C, 1 vs. 4). RXRA had a higher binding site density than RARA and RARB in the control liver (Fig. 3D, Ca vs. Cx, and Cb vs. Cx), causing the actual trend line to be closer to RXRA. RA treatment increased the binding site density of RARA and RARB. However, their binding site densities were still lower than the RA-induced RXRA binding site density (Fig. 3D, Ra vs. Rx, and Rb vs. Rx).
Fig. 3.
Comparison of hepatic bindings between RAR and RXRA in control and RA-treated mice. A: occupancy of RXRA in RAR bindings. B: number of genes uniquely targeted by each RAR or cotargeted by RAR-RXR. Percentages of RAR-RXR cotarget bindings and genes are shown in each column. C: distribution of fold enrichment ratios (log2) for colocalized bindings between RAR and RXRA (1-4). D: number of binding sites per overlapped target gene. Each point represents 1 overlapped gene between the 2 indicated nuclear receptors. Dashed lines represent the theoretical trend if the 2 indicated nuclear receptors have comparable binding density, and solid lines represent the actual trend. “C” and “R” represent “Control” and “RA treatment”; “a,” “b,” and “x” represent “RARA,” “RARB,” and “RXRA,” respectively.
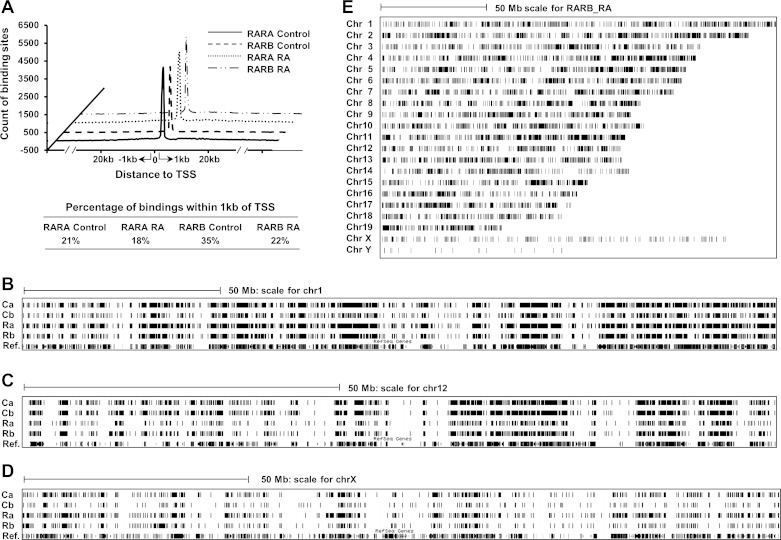
Location of RARA and RARB bindings in chromosomes and relative to transcriptional start sites of target genes.
Distances to the transcriptional start sites (TSS) of target genes were plotted against the number of binding sites at the corresponding location (Fig. 4A). In the control group, only 21% of total RARA bindings and 35% of total RARB bindings were located within 1 kb from the TSS. After RA treatment, the bindings in the same region were reduced to 18 and 22% for RARA and RARB, respectively. However, as indicated by the peak area under the curves, this reduction in percentages of both RARA and RARB bindings in the regions near the TSS was not visibly apparent in Fig. 4A. Since RA increased the numbers of RARA and RARB bindings (Fig. 1A), most RA-induced RARA and RARB bindings should be located beyond 1 kb from the TSS. To describe the relative locations between bindings and target genes globally, RAR binding sites and the mouse reference genes were aligned to each chromosome. Global bindings on chromosome 1, 12, and X were randomly picked up and presented in Fig. 4, B–D. In each chromosome, RAR binding densities closely correlated with the reference gene densities, indicating that these bindings were gene dependent and not random. This correlation made it possible to predict genes that are regulated by RAR. RARA and RARB binding profiles in 21 mouse chromosomes were visualized. Figure 4E is an example of the chromosomal distribution of RARB bindings in the RA-treated group. RARB bound to chromosomes 1 to 19 with similar binding density without showing significant binding bias. However, RARB binding density was lower on the sex chromosomes. RA treatment did not change the binding profile of RARB (data not shown). RARA binding profiles were similar to those of RARB in control and RA-treated groups (data not shown).
Fig. 4.
Location of RARA and RARB binding sites in mouse genome. A: binding locations relative to the transcriptional start sites (TSS) of RAR target genes. B, C, and D: locations of RARA and RARB binding sites in control and RA-treated livers relative to the locations of the reference (ref.) genes included in Ref-Seq database. Chromosomes 1 (B), 12 (C), and X (D) were randomly selected as examples. Ca, Cb, Ra, and Rb represent the same definitions as indicated in the legend of Fig. 3. E: chromosomal distribution of RARB binding sites in RA-treated mouse liver. Each bar represents 1 binding site.
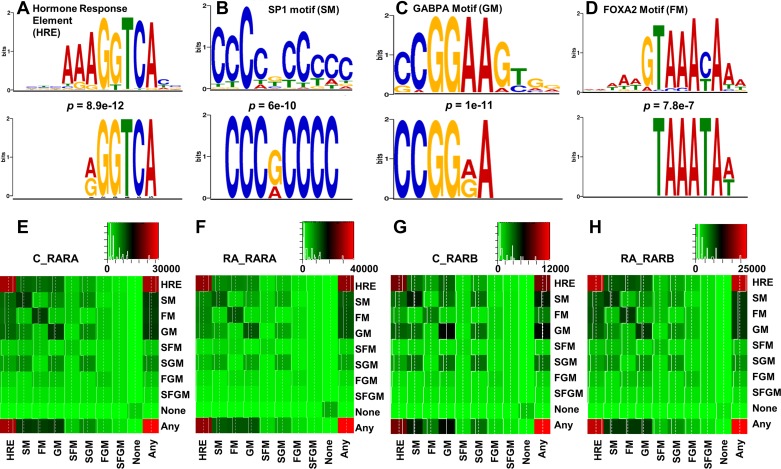
Identification of RAR binding motifs.
Four RARA and RARB binding motifs [(A/G)G(G/T)TCA, CCC(G/A)CCCC, CCGG(A/G)A, and TAAATA(A/T)] were discovered using the MEME-ChIP program (Fig. 5, A–D). The TOMTOM program identified transcription factors SP1, GABPA, and FOXA2 as the most commonly associated binding proteins for CCC(G/A)CCCC, CCGG(G/A)A, and TAAATA(A/T), respectively. (A/G)G(G/T)TCA is the consensus HRE. Since ChIP was performed using antibodies specific for RARA and RARB, the identified HRE should represent direct RAR bindings. The FIMO program extracted further information regarding the bindings that contained the specified motifs discovered by MEME-ChIP. Results showed that HRE occurred in the majority of RARA and RARB bindings followed by the motifs of GABPA, SP1, and FOXA2 in control and RA-treated groups. To understand the potential coregulation of genes by these transcription factors, the co-occurrence of these motifs within an RAR binding was analyzed (Fig. 5, E–H). Most RAR bindings contained at least one of the four motifs. In the control liver, the HRE motif was present in 26,164 RARA bindings, followed by GABPA (12,708), SP1 (12,429), and FOXA2 (12,167) motifs (Fig. 5E). Coexistence of HRE and any one of the SP1, GABPA, or FOXA2 motifs was observed in >10,000 RARA bindings. Coexistence of multiple motifs within an RAR binding was not limited to HRE. For example, both SP1 and GABPA motifs were found in 7,448 RARA bindings, which was more than half of the total bindings containing either SP1 or GABPA motifs. However, FOXA2 was rarely found to coexist with SP1 or GABPA. Moreover, coexistence of all four motifs was also rare. These motif profiles were similar between RARA and RARB as well as between control and RA-treated groups (Fig. 5, E–H).
Fig. 5.
Motifs identified from RARA and RARB bindings in control and RA-treated mice. Based on P values given by the MEME-ChIP program, (A/G)G(G/T)TCA (A), CCC(G/A)CCCC (B), CCGG(A/G)A (C), and TAAAA(A/T) (D) are the 4 most common motifs in all RAR bindings (bottom). TOMTOM program identified the corresponding transcription factors to motifs shown in A, B, C, and D as nuclear hormone receptors, SP1, GABPA, and FOXA2, respectively (top). Heat maps showing co-occurrence of motifs for hormone response element (HRE), SP1 (SM), GABPA (GM), and FOXA2 (FM) in control RARA (E), RA-treated RARA (F), control RARB (G), and RA-treated RARB bindings (H). “SFM,” “SGM,” and “FGM” represent the binding with motifs of both SP1 and FOXA2, SP1, and GABPA, as well as FOXA2 and GABPA. “SFGM” represents bindings with 3 motifs (SP1, FOXA2, and GABPA) simultaneously. “Any” represents bindings with any 1 of the 4 motifs, while “None” represents bindings without any motifs. The number of bindings in each co-occurrence is color coded to the scale on the top right corner of each heat map.
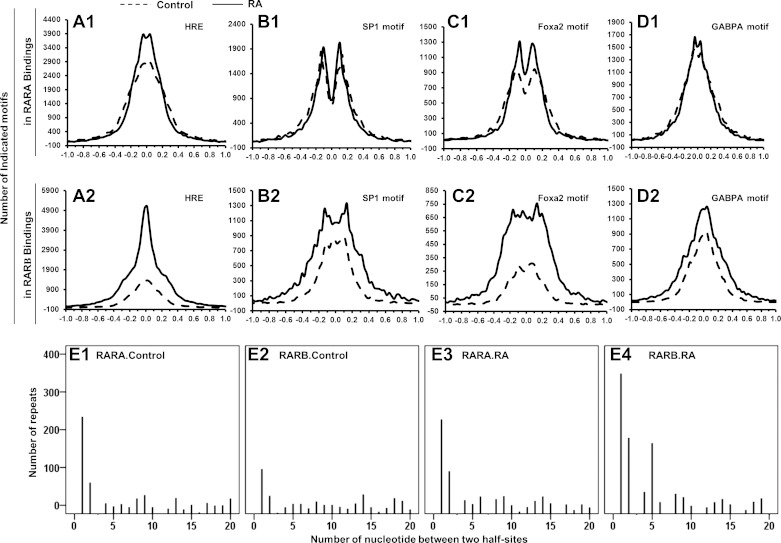
Distribution of motif relative to the binding summits.
The consistent colocalization of transcription factor bindings within the RAR bindings implies the potential interaction between them. To confirm that these observations were not merely coincidental, we analyzed the location of these binding motifs relative to the RAR binding summit. Figure 6, A1 and A2, shows that the locations of these four motifs were not randomly distributed in the RAR bindings. The HRE and GABPA motifs were located at the peak summits, while the SP1 and FOXA2 motifs were not. However, the SP1 and FOXA2 motifs gathered in a specific region within the RAR binding sites. The close proximity of these motifs to the peak summit suggested potential genome-wide interactions among these transcription factors. RA treatment increased the motif numbers of HRE, SP1, GABPA, or FOXA2 in RARB bindings (Fig. 6, A2, B2, C2, and D2). In contrast, RA did not significantly affect the motif numbers of those transcription factors in RARA bindings (Fig. 6, A1, B1, C1, and D1).
Fig. 6.
Motif distribution and HRE repeat in RARA and RARB bindings. A1 and A2, B1 and B2, C1 and C2, and D1 and D2: distribution of HRE, SP1, GABPA, and FOXA2, respectively, in RARA and RARB bindings. E1–E4: representation of direct repeat profile in RAR bindings.
HRE repeats were also analyzed. In the control mouse liver, DR1 was the predominant HRE repeat in RARA bindings (Fig. 6, E1). DR1 in RARB bindings of control mouse liver was not as frequent as in RARA bindings; however, DR1 was still the most frequent HRE repeat in RARB bindings (Fig. 6, E2). After RA treatment, the number of DR2 increased for RARA binding (Fig. 6, E3). The RA-induced change in RARB bindings was much greater than in RARA bindings. Three DRs (DR1, 2, and 5) were observed in RARB bindings in the RA-treated mouse liver (Fig. 6, E4). There was no ER and IR observed for RARA and RARB bindings. Thus, in the mouse liver, DR1 was the predominant form for RARA and RARB binding. DR2 was a RA-dependent HRE for both RARA and RARB binding, while DR5 was a RA-dependent HRE for RARB binding.
Motif-associated functional annotation and pathway analysis based on RARA and RARB bindings.
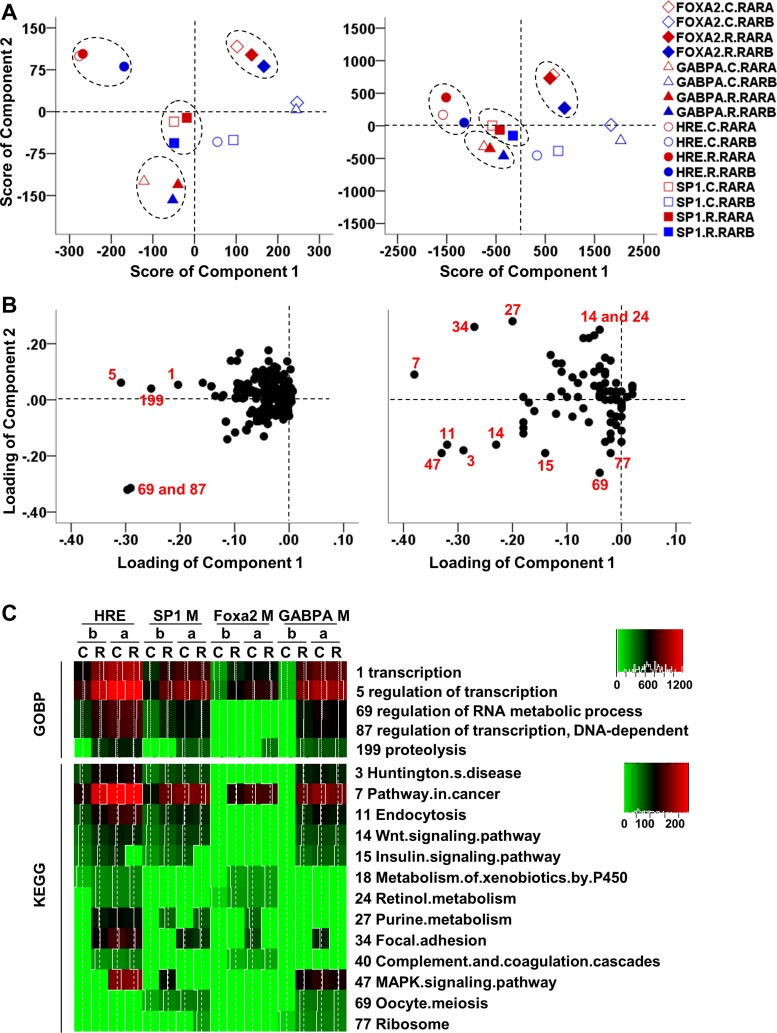
GOBP analysis showed that HRE, SP1, FOXA2, or GABPA motif-associated RARA or RARB target genes were enriched in hundreds biological processes (P < 0.01; Supplemental Dataset S1; Supplemental Material for this article is available online at the Am J Physiol Gastrointest Liver Physiol website). PCA differentiated the functional profiles of these genes. Spatial orientation of the representative points on the PCA score plot defined the similarity and difference in the functional profiles of the different motif-associated gene sets (Fig. 7A, left). Among the four motif-associated target gene sets, only RARB target genes in the control were not grouped with the others due to their distant locations. The data showed that RA had a minimal effect on the functional profile of HRE-associated RARA target genes in contrast to a greater RA-induced change on RARB target genes. Similarly, an RA effect was also apparent in RARB target genes associated with SP1, GABPA, and FOXA2 motifs. Indicated by the loading plot, a majority of the loading values fell below zero in component 1 (Fig. 7B, left). Thus FOXA2-associated genes were not enriched as much as other motif-associated genes in those biological processes. Based on the loading values on component 1 (Fig. 7B, left), five biological processes (labeled by number) were identified as the major contributors to differentiate the four motif-associated groups identified by the score plot (Fig. 7A, left). These processes include the regulation of transcription (process 1, 5, and 87), RNA metabolic process (process 69), and proteolysis (process 199). In these five biological processes, HRE-associated genes were enriched the most compared with SP1, GABPA, and FOXA2-associated genes (Fig. 7C, top). In addition to GOBP, the KEGG pathway database was also used to functionally annotate the motifs-associated genes (Supplemental Dataset S2). In the PCA score plot using KEGG database (Fig. 7A, right), the motif-associated gene sets were distributed in a similar pattern as that using GOBP database. Identified pathways with a higher contribution toward the differentiation among the four groups were labeled in the loading plots (Fig. 7B, right). The numbers of gene involved in these pathways were visualized in a heat map (Fig. 7C, bottom).
Fig. 7.
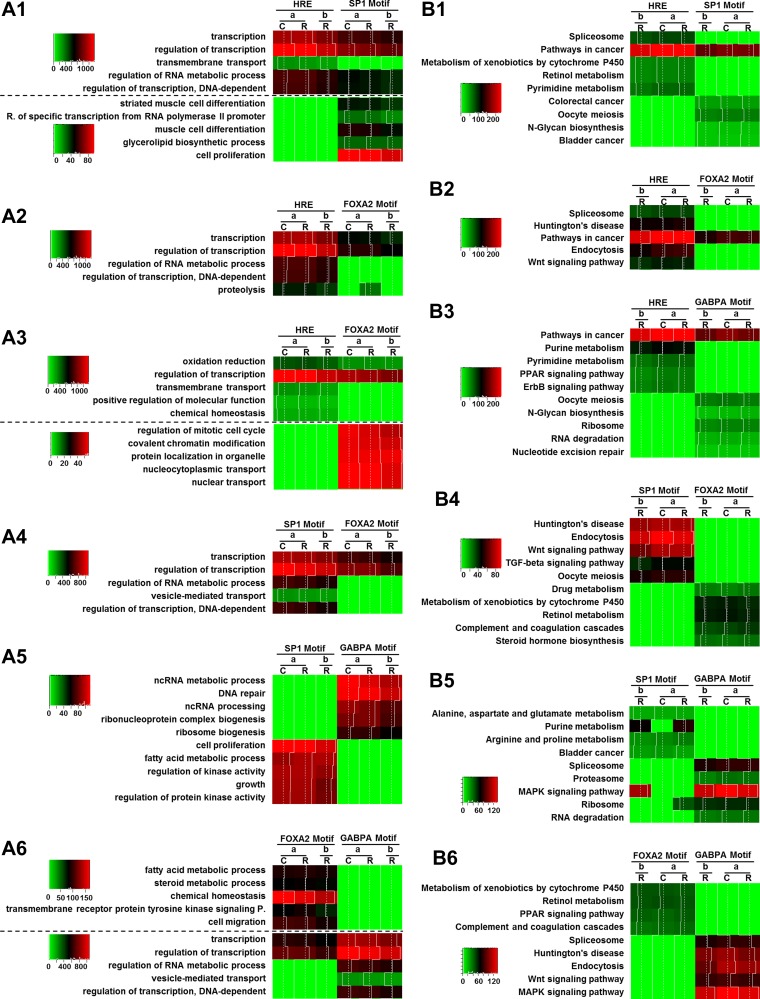
Profile differentiation for motif-associated RAR target genes at the biological process and pathway level by principle component analysis (PCA). Genes associated with a specific motif were grouped as a unit while the number of genes involved in various gene ontology biological processes (GOBP) or Kyoto Encyclopedia of Genes and Genome (KEGG) were treated as variables. A: PCA score plots based on data annotated with GOBP (left) and KEGG (right) database. Each shape and color represents a unit with a different combination of nuclear receptors, treatment groups (control as C and RA treatment as R), and motifs. Units with similar gene function profiles are grouped together within an ellipse. B: PCA loading plots based on data annotated with GOBP (left) and KEGG (right) database. Each point represents a biological process or KEGG pathway. C: identified GOBP (top) and KEGG pathways (bottom) that contribute significantly in differentiating the groups established in score plots. The numbers of genes involved in the various biological processes or pathways are color coded by the scale on the top right corner of each heat map. “C” and “R” represent control and RA treatment, while “a” and “b” represents RARA and RARB, respectively. “HRE,” “SP1M,” “GABPAM,” and “FOXA2M” represent HRE, SP1 motif, GABPA motif, and FOXA2 motif, respectively.
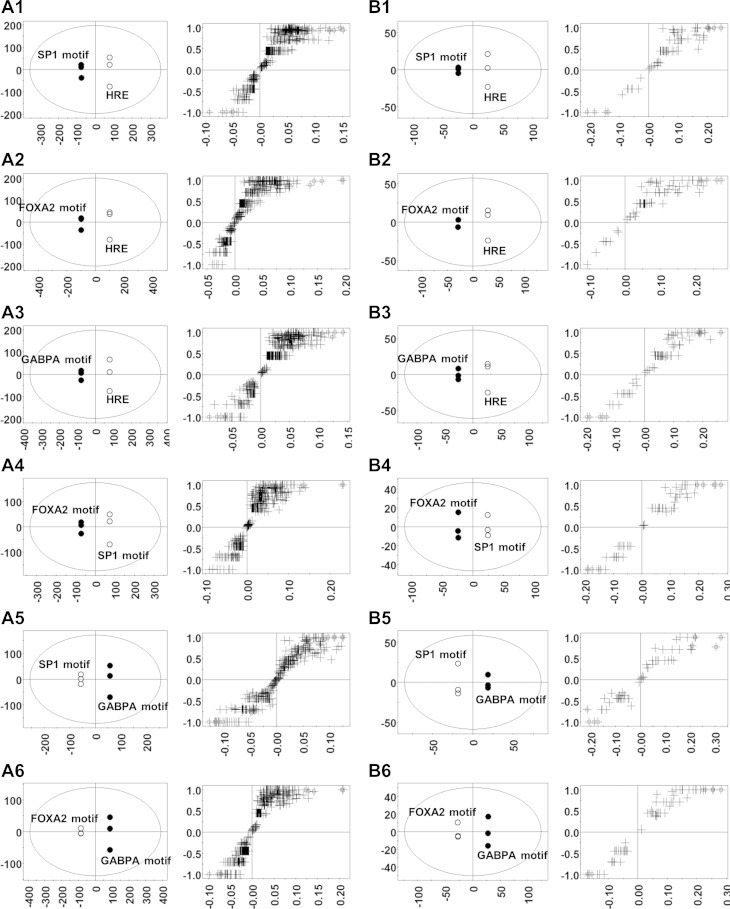
Further OPLSDA were conducted to identify biological processes and pathways that can differentiate any two out of the four motif-associated groups shown in Fig. 7A. OPLSDA score plots differentiated all six comparisons significantly (Fig. 8), which is consistent with the PCA score plots. According to the S plots, corresponding biological processes and pathways with greater values of both reliability (indicated by the y-axis of S plots) and contribution (indicated by the x-axis of S plots) were identified. The numbers of genes involved in these identified biological processes and pathways were visualized as heat maps (Fig. 9). In general, OPLSDA extracted additional information based on output of PCA. For example, PCA did not identify pathways that were predominantly enriched by FOXA2 motif-associated genes. OPLSDA showed that FOXA2 motif-associated genes were enriched more than GABPA motif-related genes in biological processes including fatty acid metabolism, steroid metabolism, and chemical homeostasis (Fig. 9, A6), as well as pathways including P450-mediated xenobiotic metabolism, retinol metabolism, and peroxisome proliferator-activated receptor (PPAR) signaling pathways (Fig. 9, B6).
Fig. 8.
Identification of biological processes and pathways by orthogonal partial least square discriminant analysis (OPLSDA) to differentiate the biological function profile of 2 group of genes. Target genes of a specific nuclear receptor with the same motif were grouped as a unit while the number of genes involved in various biological processes or pathways were treated as variables. OPLSDA based on GOBP and KEGG database is shown in A1-A6 and B1-B6, respectively. A1-A6 and B1-B6, left: score plots. A1-A6 and B1-B6, right: S plots. The x-axis and y-axis of the score plots represent the scores of components 1 and 2. Based on the contribution (x-axis) and reliability (y-axis) shown in the S plots, up to 5 biological processes were identified to differentiate each 2 of the 4 groups shown in score plots of Fig. 7.
Fig. 9.
Visualization of the number of genes in biological processes and pathways identified from OPLSDA. The numbers of motif-associated genes involved in identified GOBP (A1-A6) and KEGG pathways (B1-B6) are color coded to the scale on the left of each heat map. “C” and “R” represent control and RA-treated mice, while “a” and “b” represent RARA and RARB.
Correlation between RAR bindings and hepatic gene expressions in response to RA.
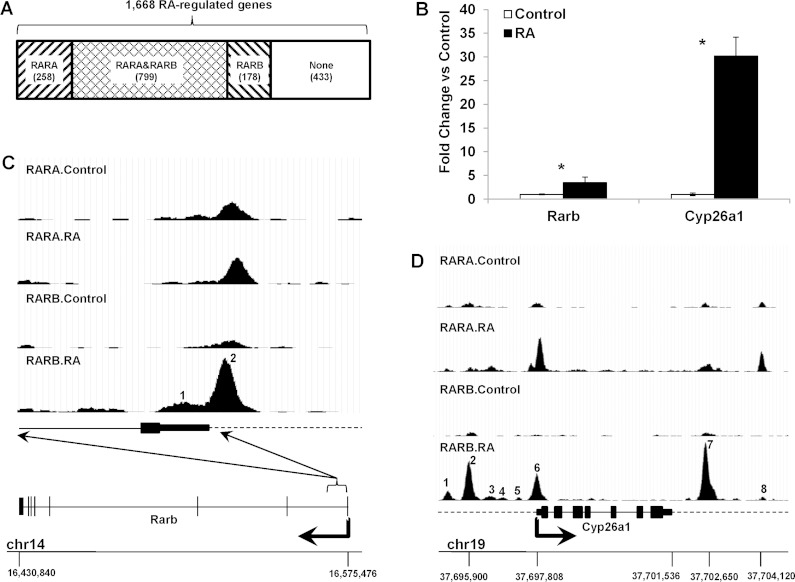
In addition to DNA bindings, the expression of hepatic genes was studied. After RA treatment, 1,668 hepatic genes were significantly regulated, and bindings associated with these RA-regulated genes were classified as RA dependent or independent based on their response to RA. All RA-generated, -removed, -enhanced, and -repressed (>1.5 times in fold enrichment values) bindings were considered as RA-dependent bindings. Among the 1,668 RA-regulated hepatic genes, the binding of RARA and/or RARB to 1,235 genes was RA dependent (Fig. 10A). Two RA target genes, Rarb and Cyp26a1, were selected to show the correlated RA effect on RAR bindings and gene expressions. RA significantly induced Rarb and Cyp26a1 mRNA levels by more than 3 and 40 times, respectively (Fig. 10B). Accordingly, RA induced RARB bindings in the Rarb promoter region (Fig. 10C). Peak 2 in Fig. 10C was consistent with the reported RAR binding (37), while a small RA-induced peak (peak 1) was also observed near peak 2. Regarding the Cyp26a1, RA induced RARA and RARB bindings on multiple places (Fig. 10D). Among those bindings, peaks 2 and 6 shown in Fig. 10D have already been reported (46), while others are novel findings of the present study. Among these binding sites, except for peak 1, all other peaks were RA dependent in RARA and RARB bindings. Peak 1 was RA dependent in RARB, but not RARA, binding.
Fig. 10.
Correlation between RA-dependent RAR bindings and hepatic gene expressions. A: number of RA-regulated hepatic genes that showed RA dependence in RARA and/or RARB binding. B: RA significantly induced the mRNA levels of Cyp26a1 and Rarb. C: RA induced RARB bindings in promoter region of Rarb gene (peaks 1 and 2). D: RA induced RARA and RARB bindings in the Cyp26a1 gene (peaks 1-8) *P < 0.01.
DISCUSSION
This article is the first to characterize the mouse hepatic genome-wide binding profiles of RARA and RARB at the physiological level and in response to RA treatment. Genomic binding profiles of different nuclear receptors have been studied in the liver. The numbers of binding sites range from 16,005 for PPARA, to 7,794 for FXR, to ∼3,500 for LXR and PXR (4, 9, 39). Therefore, the identification of 35,521 genome bindings for RARA and 14,968 binding sites for RARB suggested that RARA and RARB still have many unidentified roles in the liver. Moreover, because RARA and RARB have common and unique bindings before and after RA treatment, these two RAR isoforms must have redundant and specific roles in the liver. Genome-wide RAR binding studies have been done in several cell lines, which reported different numbers of binding sites. (11, 25, 29). In breast cancer cells, 7,346 RARA and 3,916 RARG binding sites were detected by ChIP-Seq (17). Conversely, with the use of pan-anti-RAR antibody, only <2,000 RAR binding sites were found in embryonic stem cells (25). The numbers of nuclear receptor binding sites found in different models indicate the relative biological significance of nuclear receptors in a specific cell type or organ.
Many nuclear receptors have isoforms, and the different biological function of those isoforms is always of a great interest. For instance, RARB is a tumor suppressor gene and its expression level can be used to predict the prognosis of RA treatment in cancer (3, 21), whereas RARA is not. Our data illustrate for the first time the apparent differences between RARA and RARB in response to RA at the hepatic genome-wide level. Although the number of bindings for RARB is substantially less than that of RARA in the control group, there was a greater dynamic shift in RARB upon RA treatment. To elucidate this difference, we studied the relationship between binding and the absolute gene expression level of RARA and RARB using unpublished data derived from RNA-sequencing generated in our laboratory. Based on the absolute expression level of transcripts, hepatic Rara was expressed about three times higher than Rarb in the control liver. After RA treatment, Rarb transcript levels doubled and approached the level of Rara. In contrast, RA had no effect on the level of Rara transcript in our experimental model. Thus the absolute amount of transcripts may have something to do with preferentially binding in response to RA treatment. Regarding biological functions, the functions of RARA and RARB target genes were found to be more diverse compared with other RXRA partners, which are mainly involved in metabolism (8).
The location of binding is frequently used to predict whether the binding leads to gene transcription. It also demonstrates the mechanism of gene regulation. Although many RAR binding sites are located beyond 1 kb from TSS, those bindings can be functionally significant because the folding and looping of chromosomes can bring two distant regions closer to each other (30, 40). By visualizing the binding locations of RAR on 21 mouse chromosomes and observing RARA and RARB binding to regions surrounding the reference genes, our data strongly suggest that these bindings are associated with gene expression and regulation. However, we did notice that a few RAR bindings occurred in regions that lack coding genes. These bindings may potentially regulate noncoding RNA, which can be found in the gene desert regions (27).
RXRA, abundantly expressed in the liver, is a major partner of RAR (5, 16, 44). However, RAR also responds to RA in an RXR-independent manner (34). Consistently, our data showed that ∼50% of RARA and 25% of RARB bindings do not overlap with the bindings of RXRA. Conversely, RXRA also has its unique bindings (data not shown). In RXRA and RAR common target genes, the binding densities of RXRA are higher than its RAR counterparts. These RXRA-unique bindings are potentially co-occupied with other nuclear receptors such as PPARA, FXR, LXR, and PXR (8).
The binding of a transcription factor to DNA identified from ChIP-Seq can be either direct or indirect (19). Our data showed that RAR frequently binds to the SP1 motif. SP1 binds to the GC-rich region and regulates embryogenesis (20). It has been shown that RARA/RXRA works with SP1 to regulate TGFB1 in the absence of an RAR response element (35), suggesting the available of indirect interaction between RAR and DNA. Furthermore, SP1 and RAR physically interact with each other to regulate the expression of PLAU (38) and FOLR2 (14). In the present study, we further demonstrated the potential interaction between RAR and SP1 in the hepatic genome; ∼50% of both RARA and RARB bindings contained the SP1 motif. Among these SP1 motif-positive RAR bindings, half of them contained the SP1 motif exclusively, while the other half contained both the SP1 and HRE motifs. Thus RAR may either directly bind to DNA or indirectly bind to DNA via interaction with SP1.
In addition to SP1, FOXA2 and GABPA may also have cross talk with RAR. FOXA2 is known to interact with RAR in regulating the sonic hedgehog expression (7). The identification of FOXA2 motif in many RARA and RARB binding sites provided evidence that RAR participates in FOXA2-mediated gene regulation. Regarding GABPA, there is currently no experimental data showing the direct interaction between GABPA and RAR. However, GABPA is required for myeloid differentiation (43), which is also regulated by RARA (13), indicating a potential collaboration between GABPA and RAR. Our data might be the first to show the potential global interaction between RAR and GABPA. It is important to note that like RAR, GABPA also physically interacts with SP1 (13), suggesting the cross talk among multiple transcription factors. By analyzing the ChIP-Seq data of SP1, FOXA2, and GABPA deposited in the GEO database (6, 28), we also found the presence of HRE in the binding sites of these transcription factors. Taken together, these findings clearly indicate the complex role of those transcription factors in the hepatic genome.
Taken together, the cistrome of RARA and RARB revealed their extensive biological roles in the mouse liver. RARB bindings showed higher RA responsiveness than RARA. RAR binding sites frequently contain the SP1, FOXA2, and GABPA binding motifs, suggesting the potential interactions among them. RAR target genes were enriched in many biological processes and pathways, which showed a motif-dependent style. Moreover, most RA-regulated genes also showed RAR-dependent binding in response to RA. The hepatic RAR genome-wide binding data presented in this article help us understand the global molecular mechanisms underlying RAR and RA-mediated gene and pathway regulation.
GRANTS
This work was supported by National Institutes of Health Grants DK-092100 and CA-53596 (to Y.-J. Wan).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: Y.H. and Y.-J.Y.W. conception and design of research; Y.H. performed experiments; Y.H. and J.T. analyzed data; Y.H. and Y.-J.Y.W. interpreted results of experiments; Y.H. and J.T. prepared figures; Y.H. drafted manuscript; Y.H., J.T., and Y.-J.Y.W. edited and revised manuscript; Y.H., J.T., and Y.-J.Y.W. approved final version of manuscript.
Supplementary Material
ACKNOWLEDGMENTS
We thank Lisa Teixeira for assistance in preparing the manuscript.
REFERENCES
- 1.Amengual J, Ribot J, Bonet ML, Palou A. Retinoic acid treatment enhances lipid oxidation and inhibits lipid biosynthesis capacities in the liver of mice. Cell Physiol Biochem 25: 657–666, 2010. [DOI] [PubMed] [Google Scholar]
- 2.Balmer JE, Blomhoff R. Gene expression regulation by retinoic acid. J Lipid Res 43: 1773–1808, 2002. [DOI] [PubMed] [Google Scholar]
- 3.Berg WJ, Nanus DM, Leung A, Brown KT, Hutchinson B, Mazumdar M, Xu XC, Lotan R, Reuter VE, Motzer RJ. Up-regulation of retinoic acid receptor beta expression in renal cancers in vivo correlates with response to 13-cis-retinoic acid and interferon-alpha-2a. Clin Cancer Res 5: 1671–1675, 1999. [PubMed] [Google Scholar]
- 4.Boergesen M, Pedersen TA, Gross B, van Heeringen SJ, Hagenbeek D, Bindesboll C, Caron S, Lalloyer F, Steffensen KR, Nebb HI, Gustafsson JA, Stunnenberg HG, Staels B, Mandrup S. Genome-wide profiling of liver x receptor, retinoid x receptor, and peroxisome proliferator-activated receptor alpha in mouse liver reveals extensive sharing of binding sites. Mol Cell Biol 32: 852–867, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bushue N, Wan YJ. Retinoid pathway and cancer therapeutics. Adv Drug Deliv Rev 62: 1285–1298, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calabrese JM, Sun W, Song L, Mugford JW, Williams L, Yee D, Starmer J, Mieczkowski P, Crawford GE, Magnuson T. Site-specific silencing of regulatory elements as a mechanism of X inactivation. Cell 151: 951–963, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang BE, Blader P, Fischer N, Ingham PW, Strahle U. Axial (HNF3beta) and retinoic acid receptors are regulators of the zebrafish sonic hedgehog promoter. EMBO J 16: 3955–3964, 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chawla A, Repa JJ, Evans RM, Mangelsdorf DJ. Nuclear receptors and lipid physiology: opening the X-files. Science 294: 1866–1870, 2001. [DOI] [PubMed] [Google Scholar]
- 9.Cui JY, Gunewardena SS, Rockwell CE, Klaassen CD. ChIPing the cistrome of PXR in mouse liver. Nucleic Acids Res 38: 7943–7963, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dejean A, de Thé H. Hepatitis B virus as an insertional mutagene in a human hepatocellular carcinoma. Mol Biol Med 7: 213–222, 1990. [PubMed] [Google Scholar]
- 11.Delacroix L, Moutier E, Altobelli G, Legras S, Poch O, Choukrallah MA, Bertin I, Jost B, Davidson I. Cell-specific interaction of retinoic acid receptors with target genes in mouse embryonic fibroblasts and embryonic stem cells. Mol Cell Biol 30: 231–244, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics 27: 1017–1018, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grignani F, Ferrucci PF, Testa U, Talamo G, Fagioli M, Alcalay M, Mencarelli A, Grignani F, Peschle C, Nicoletti I, Pelicci PG. The acute promyelocytic leukemia-specific PML-RAR alpha fusion protein inhibits differentiation and promotes survival of myeloid precursor cells. Cell 74: 423–431, 1993. [DOI] [PubMed] [Google Scholar]
- 14.Hao H, Qi H, Ratnam M. Modulation of the folate receptor type beta gene by coordinate actions of retinoic acid receptors at activator Sp1/ets and repressor AP-1 sites. Blood 101: 4551–4560, 2003. [DOI] [PubMed] [Google Scholar]
- 15.Haq R, Pfahl M, Chytil F. Retinoic acid affects the expression of nuclear retinoic acid receptors in tissues of retinol-deficient rats. Proc Natl Acad Sci USA 88: 8272–8276, 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He Y, Gong L, Fang Y, Zhan Q, Liu HX, Lu Y, Guo GL, Lehman-McKeeman L, Fang J, Wan YJ. The role of retinoic acid in hepatic lipid homeostasis defined by genomic binding and transcriptome profiling. BMC Genomics 14: 575, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hua S, Kittler R, White KP. Genomic antagonism between retinoic acid and estrogen signaling in breast cancer. Cell 137: 1259–1271, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57, 2009. [DOI] [PubMed] [Google Scholar]
- 19.Jothi R, Cuddapah S, Barski A, Cui K, Zhao K. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Res 36: 5221–5231, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaczynski J, Cook T, Urrutia R. Sp1- and Kruppel-like transcription factors. Genome Biol 4: 206, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khuri FR, Lotan R, Kemp BL, Lippman SM, Wu H, Feng L, Lee JJ, Cooksley CS, Parr B, Chang E, Walsh GL, Lee JS, Hong WK, Xu XC. Retinoic acid receptor-beta as a prognostic indicator in stage I non-small-cell lung cancer. J Clin Oncol 18: 2798–2804, 2000. [DOI] [PubMed] [Google Scholar]
- 22.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li C, Wan YJ. Differentiation and antiproliferation effects of retinoic acid receptor beta in hepatoma cells. Cancer Lett 124: 205–211, 1998. [DOI] [PubMed] [Google Scholar]
- 24.Machanick P, Bailey TL. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics 27: 1696–1697, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mahony S, Mazzoni EO, McCuine S, Young RA, Wichterle H, Gifford DK. Ligand-dependent dynamics of retinoic acid receptor binding during early neurogenesis. Genome Biol 12: R2, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martens JH, Brinkman AB, Simmer F, Francoijs KJ, Nebbioso A, Ferrara F, Altucci L, Stunnenberg HG. PML-RARalpha/RXR alters the epigenetic landscape in acute promyelocytic leukemia. Cancer Cell 17: 173–185, 2010. [DOI] [PubMed] [Google Scholar]
- 27.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet 10: 155–159, 2009. [DOI] [PubMed] [Google Scholar]
- 28.Michaud J, Praz V, James Faresse N, Jnbaptiste CK, Tyagi S, Schutz F, Herr W. HCFC1 is a common component of active human CpG-island promoters and coincides with ZNF143, THAP11, YY1, and GABP transcription factor occupancy. Genome Res 23: 907–916, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moutier E, Ye T, Choukrallah MA, Urban S, Osz J, Chatagnon A, Delacroix L, Langer D, Rochel N, Moras D, Benoit G, Davidson I. Retinoic acid receptors recognize the mouse genome through binding elements with diverse spacing and topology. J Biol Chem 287: 26328–26341, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ptashne M, Gann A. Transcriptional activation by recruitment. Nature 386: 569–577, 1997. [DOI] [PubMed] [Google Scholar]
- 31.R Development Core Team. R: A Language and Environment for Statistical Computing. Wien, Austria: R Foundation for Statistical Computing, 2011. [Google Scholar]
- 32.Rivera-Gonzalez GC, Droop AP, Rippon HJ, Tiemann K, Pellacani D, Georgopoulos LJ, Maitland NJ. Retinoic acid and androgen receptors combine to achieve tissue specific control of human prostatic transglutaminase expression: a novel regulatory network with broader significance. Nucleic Acids Res 40: 4825–4840, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ross-Innes CS, Stark R, Holmes KA, Schmidt D, Spyrou C, Russell R, Massie CE, Vowler SL, Eldridge M, Carroll JS. Cooperative interaction between retinoic acid receptor-alpha and estrogen receptor in breast cancer. Genes Dev 24: 171–182, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schrader M, Wyss A, Sturzenbecker LJ, Grippo JF, LeMotte P, Carlberg C. RXR-dependent and RXR-independent transactivation by retinoic acid receptors. Nucleic Acids Res 21: 1231–1237, 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shimada J, Suzuki Y, Kim SJ, Wang PC, Matsumura M, Kojima S. Transactivation via RAR/RXR-Sp1 interaction: characterization of binding between Sp1 and GC box motif. Mol Endocrinol 15: 1677–1692, 2001. [DOI] [PubMed] [Google Scholar]
- 36.Soprano DR, Qin P, Soprano KJ. Retinoic acid receptors and cancers. Annu Rev Nutr 24: 201–221, 2004. [DOI] [PubMed] [Google Scholar]
- 37.Sucov HM, Murakami KK, Evans RM. Characterization of an autoregulated response element in the mouse retinoic acid receptor type beta gene. Proc Natl Acad Sci USA 87: 5392–5396, 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suzuki Y, Shimada J, Shudo K, Matsumura M, Crippa MP, Kojima S. Physical interaction between retinoic acid receptor and Sp1: mechanism for induction of urokinase by retinoic acid. Blood 93: 4264–4276, 1999. [PubMed] [Google Scholar]
- 39.Thomas AM, Hart SN, Kong B, Fang J, Zhong XB, Guo GL. Genome-wide tissue-specific farnesoid X receptor binding in mouse liver and intestine. Hepatology 51: 1410–1419, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang JC, Giaever GN. Action at a distance along a DNA. Science 240: 300–304, 1988. [DOI] [PubMed] [Google Scholar]
- 41.Warnes GR, Bolker B, Bonebakker L, Gentleman R, Huber W, Liaw A, Lumley T, Maechler M, Magnusson A, Moeller S. Gplots: various R programming tools for plotting data. In: R package version 2.12.1. 2013. http://CRAN.R-project.org/package=gplots. [Google Scholar]
- 42.Yanagitani A, Yamada S, Yasui S, Shimomura T, Murai R, Murawaki Y, Hashiguchi K, Kanbe T, Saeki T, Ichiba M, Tanabe Y, Yoshida Y, Morino S, Kurimasa A, Usuda N, Yamazaki H, Kunisada T, Ito H, Shiota G. Retinoic acid receptor alpha dominant negative form causes steatohepatitis and liver tumors in transgenic mice. Hepatology 40: 366–375, 2004. [DOI] [PubMed] [Google Scholar]
- 43.Yang ZF, Drumea K, Cormier J, Wang J, Zhu X, Rosmarin AG. GABP transcription factor is required for myeloid differentiation, in part, through its control of Gfi-1 expression. Blood 118: 2243–2253, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhan Q, Fang Y, He Y, Liu HX, Fang J, Wan YJ. Function annotation of hepatic retinoid x receptor a based on genome-wide DNA binding and transcriptome profiling. PLoS One 7: e50013, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y, Zolfaghari R, Ross AC. Multiple retinoic acid response elements cooperate to enhance the inducibility of CYP26A1 gene expression in liver. Gene 464: 32–43, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.