Abstract
Background
Human Immunodeficiency Virus Type 1 (HIV-1) viral load testing at regular intervals is an integral component of disease management in Acquired Immunodeficiency Syndrome (AIDS) patients. The need in countries like India is therefore an assay that is not only economical but efficient and highly specific for HIV-1 sub type C virus. This study reports a SYBR Green-based HIV-1 real time PCR assay for viral load testing and is designed for enhanced specificity towards HIV-1 sub type C viruses prevalent in India.
Results
Linear regression of the observed and reference concentration of standards used in this study generated a correlation coefficient of 0.998 (p < 0.001). Lower limit of detection of the test protocol was 50 copies/ml of plasma. The assay demonstrated 100% specificity when tested with negative control sera. The Spearman coefficient of the reported assay with an US-FDA approved, Taqman probe-based commercial kit was found to be 0.997. No significant difference in viral load was detected when the SYBR Green based assay was used to test infected plasma stored at -20°C and room temperature for 7 days respectively (Wilcoxon signed rank test, p = 0.105). In a comparative study on 90 pretested HIV-1 positive samples with viral loads ranging from 5,000–25,000 HIV-1 RNA copies/ml and between two commercial assays it was found that the later failed to amplify in 13.33% and 10% samples respectively while in 7.77% and 4.44% samples the copy number values were reduced by >0.5 log value, a figure that is considered clinically significant by physicians.
Conclusion
The HIV-1 viral load assay reported in this study was found to be robust, reliable, economical and effective in resource limited settings such as those existing in India. PCR probes specially designed from HIV-1 Subtype C-specific nucleotide sequences originating from India imparted specificity towards such isolates and demonstrated superior results when compared to two similar commercial assays widely used in India.
Keywords: HIV-1, Gag, SYBR Green, RT PCR
Background
At present estimating HIV-1 RNA plasma concentration is the standard of care in monitoring the progression of HIV/AIDS and is believed to have clinically useful prognostic value [1, 2]. Three different commercial assays [NucliSens HIV-1 QT RNA Assay (bioMérieux, France), Quantiplex HIV-1 RNA 3.0 Assay (Bayer Diagnostics Division, USA) and COBAS TaqMan HIV-1 48 test (Roche Molecular Systems, USA)] presently dominate the HIV-1 viral load testing market but are prohibitively expensive [3].
Real time PCR technology is currently considered the gold standard for HIV-1 viral load detection because of its high assay specificity, sensitivity and wide linear range of detection [4]. TaqMan chemistry is widely popular and accepted method but in order to develop this on real time PCR platform for quantification of HIV-1 viral RNA, three conserved regions within the viral genome are required all in close proximity which is often difficult due to the heterogenic genomic structure of HIV-1. The problem of sub-optimal performance by TaqMan chemistry based commercial assay is mostly encountered in countries where non B subtype of HIV-1 is prevalent because these assays are widely validated in USA and Europe where HIV-1 subtype B dominates [3, 5]. In addition to the TaqMan chemistry, SYBR Green chemistry based real time PCR is also widely used for HIV-1 viral RNA quantification assays.
In this study, we report a real time RT-PCR assay specifically optimized for HIV-1 C subtype that uses SYBR Green chemistry for quantitative detection of HIV-1 viral RNA in human plasma. The assay is designed to target a conserved region within the gag gene of the viral genome and is less expensive compared to those which employ dual-labeled TaqMan probes. The protocol gave consistent results across a wide range of clinical samples.
Results
Linear range, detection limit and inter-assay precision
The linearity of the assay was determined using reference standards constructed specifically for this assay by using a recombinant plasmid harboring the amplified gag gene fragment as insert. Linear regression of the observed and reference concentrations yielded a correlation coefficient of 0.998 (p < 0.001). A standard curve was generated by plotting the threshold cycles of reference standards versus their log concentrations (Figure 1). The dissociation curve of all reference standards showed a melting temperature of 85.5°C (Figure 2).In order to compute the detection limit samples with known viral load in the range of 10 to 500 HIV-1 RNA copies/ml were analyzed. The results showed detectable amplification in 5 (25%) of the replicates for standard having 10 HIV-1 RNA copies/ml, in 10 (50%) of the replicates for those having 30 HIV-1 RNA copies/ml, and 20 (100%) having 50, 100 and 500 copies/ml of HIV-1 RNA respectively. The data is represented as a bar diagram where the log concentrations are compared with their corresponding threshold cycles (Figure 3). From this analysis, the lower limit of the assay was fixed at 50 copies/ml of HIV-1 RNA.
Figure 1.
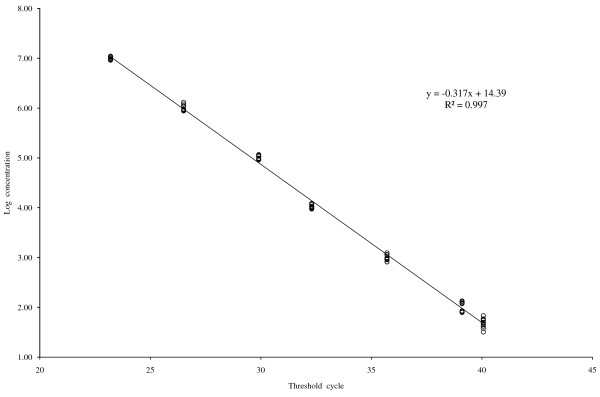
Standard curve for quantification of HIV-1 viral load. Log concentration of serial diluted reference standards plotted against their corresponding Threshold Cycles.
Figure 2.
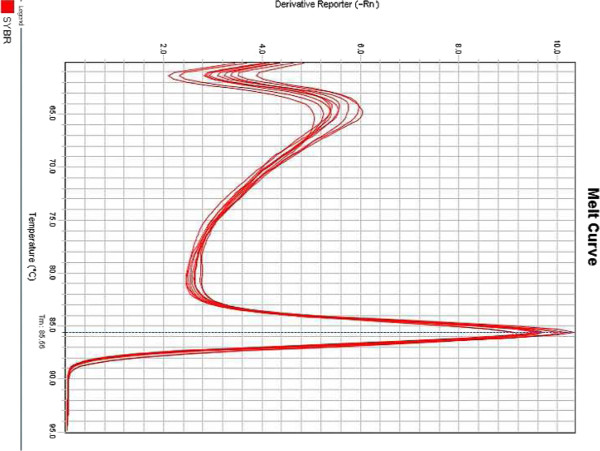
Dissociation curve of the HIV-1 reference standards. The negative first derivative of fluorescent signal (Y-axis) was plotted against Temperature (X-axis).
Figure 3.
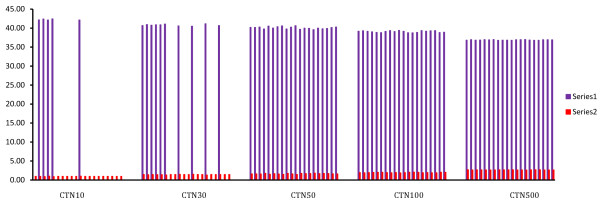
Bar diagram used for finding the detection limit of the assay. Log concentration of the dilution series plotted with their corresponding Threshold Cycles. Series 1: Threshold Cycles; Series 2: Log concentration of viral load; CTN10: 10 HIV-1 RNA copies/ml; CTN30: 30 HIV-1 RNA copies/ml; CTN50: 50 HIV-1 RNA copies/ml; CTN100: 100 HIV-1 RNA copies/ml; CTN500: 500 HIV-1 RNA copies/ml.
In order to evaluate the inter-assay precision of the test protocol the reference panel comprising of 25 samples with known HIV-1 viral load were tested in triplicates through 10 independent experiments. Statistical analysis of the data for standard deviation (SD) and coefficient of variation are shown in Table 1. The coefficient of variation was found to increase as the reference viral load approached detection limit of the assay.
Table 1.
Precision of the SYBR Green Real time PCR assay determined in this study
| Reference concentration (copies/ml) | Measured concentration (copies/ml) | Standard deviation (SD) | Coefficient of variation (CV) |
|---|---|---|---|
| 52 | 50 | 1.65 | 0.0325 |
| 124 | 120 | 3.06 | 0.0251 |
| 344 | 345 | 0.47 | 0.0014 |
| 871 | 863 | 5.42 | 0.0063 |
| 1,235 | 1,250 | 10.14 | 0.0082 |
| 16,383 | 16,358 | 17.68 | 0.0011 |
| 267,816 | 267,098 | 507.70 | 0.0019 |
| 31,293 | 31,299 | 4.48 | 0.0001 |
| 541 | 534 | 4.71 | 0.0088 |
| 1,476 | 1,466 | 6.60 | 0.0045 |
| 4,572 | 4,555 | 12.26 | 0.0027 |
| 2571,210 | 2569,050 | 1,526.88 | 0.0006 |
| 76 | 89 | 9.43 | 0.1145 |
| 43,110 | 43,135 | 18.15 | 0.0004 |
| 5,447 | 5,414 | 23.10 | 0.0043 |
| 21,312 | 21,393 | 57.28 | 0.0027 |
| 96 | 94 | 1.65 | 0.0174 |
| 635,469 | 635,592 | 86.97 | 0.0001 |
| 626 | 620 | 4.48 | 0.0072 |
| 54,886 | 54,853 | 23.81 | 0.0004 |
| 8,742 | 8,781 | 27.58 | 0.0031 |
| 917,838 | 917,648 | 133.88 | 0.0001 |
| 74,511 | 74,552 | 29.23 | 0.0004 |
| 944 | 965 | 15.32 | 0.0161 |
| 9,648 | 9,984 | 237.82 | 0.0242 |
The standard deviation and the coefficient of variation were determined using data from 10 independent experiments.
Sensitivity and specificity of the in-house HIV-I RT PCR assay
Clinical sensitivity of the assay was determined using samples from the reference panel. All 25 samples were found to be positive using the in-house SYBR Green RT PCR assay indicating 100% sensitivity. The specificity of the assay was also found to be 100% since none of the samples showed any detectable amplification from our reference panel of HIV-1-negative samples (Figure 4). This was further confirmed by analyzing the melting temperature profile of the amplicons using dissociation curve analysis. No significant fluorescent signals were detected from any non-specific amplification including primer dimers.
Figure 4.
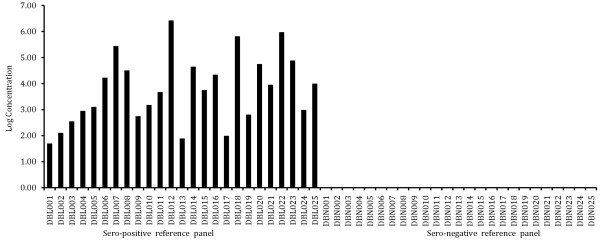
Bar diagram showing sensitivity and specificity of the assay. Sero-positive reference panel represented by DBL001 - DBLO25. Amplification of all samples in this panel indicates 100% sensitivity. Sero-negative reference panel represented by DBN001 – DBN025. No amplification in all samples in this panel indicates 100% specificity.
Comparison of viral load measurements using the in-house and the US-FDA approved artus HIV-1 RG RT-PCR kit
Viral load of 100 HIV-1 sero-positive samples was determined both by a commercially available US FDA-approved artus HIV-1 RG RT-PCR kit (Qiagen, Germany) as well as the SYBR Green RT PCR assay described in this study. The log concentration of HIV-1 RNA copies/ml of samples were compared by plotting results obtained using the present study protocol with those obtained using artus HIV-1 RG RT-PCR kit (Figure 5). The spearman coefficient of the in house assay with that of artus HIV-1 RG RT-PCR kit was found to be 0.997.
Figure 5.
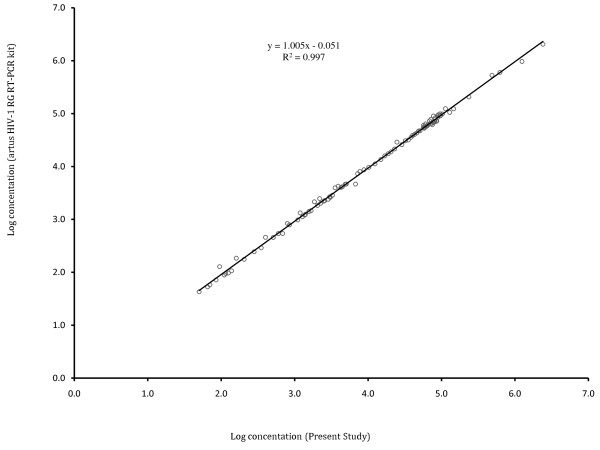
Correlation of viral loads as determined by artus HIV-1 RG RT-PCR kit and Present Study.
Comparison of sensitivity with two commercial HIV-1 RNA detection assays
The detection limit of HIV RNA Real-Quant (Sacace Biotechnologies S.r.l, Italy) and HIV-1 Real Time RT PCR kit (Life River, Shanghai ZJ Biotech, China) are 500 and 1,000 HIV-1 RNA copies/ml respectively. Out of the 90 samples whose viral load varied between 5000 - 25,000 HIV-1 RNA copies/ml as determined by artus HIV-1 RG RT-PCR kit (Qiagen, Germany), 12 (13.33%) and 9 (10.0%) samples fail to show any detectable amplification when tested using HIV RNA Real-TM Quant (Sacace Biotechnologies S.r.l, Italy) and HIV-1 Real Time RT PCR kit (Life River, Shanghai ZJ Biotech, China) respectively. While 7 (7.77%) and 4 (4.44%) samples showed underestimation of viral load at a value that is greater than 0.5 log when compared with reference panel results. All the above mentioned samples had comparable results when tested using the protocol described in this study as shown in Table 1.
Effect of ambient temperature on stability of HIV-1 RNA
Effect of storage of plasma at room temperature was studied by analyzing 10 plasma samples each stored in (1) -20°C freezer and (2) room temperature (25°C) for 7 days. These samples were subjected to viral load testing and each reaction was performed in triplicate. Results indicate a w- value of 11.5, mean difference of 1.01, sum of positive ranks equaling to 43.5 and sum of negative ranks equaling to 11.5 respectively. As the critical value of w for n = 10 (p ≤ 0.05) was 8, the result was not significant meaning that there was no significant difference in viral load detected in both the groups (Wilcoxon signed rank test, p = 0.105) (Figure 6). The box and whisker plot in Figure 6 show differences between medians (DBM), over all visible spread (OVS) and DBM as a percentage of OVS within the two groups equaling to 0.013, 1.114 and 1.17% respectively indicating no significant difference between the two data sets.
Figure 6.
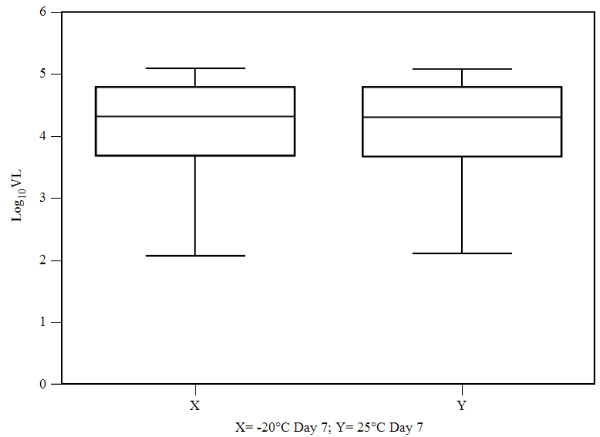
Comparison of viral load from 10 plasma samples after storage at (X) -20°C degree and (Y) room temperature (25°C) for 7 days prior to SYBR Green Real Time PCR assay presented in a box and whisker plot.
Discussion
At present SYBR Green chemistry based real time PCR is widely used for development of sensitive and economical quantitative PCR assays due to its competitive performance when compared to TaqMan chemistry based counterpart tests. In this study we described a SYBR Green chemistry based sensitive real time PCR assay for quantitative detection of HIV-1 viral load in human plasma samples.
Two assay panels were created for the study. The reference study panel comprised of 25 HIV-1 sero-positive samples with reference viral load assigned to them using artus HIV-1 RG RT-PCR kit (Qiagen, Germany) and an equal number of samples from healthy individuals. This panel was primarily used for development of the assay. The clinical study panel on the other hand was a secondary one comprising of 100 samples and was used to evaluate the performance of the assay. Reference viral load values were also assigned to this panel using artus HIV-1 RG RT-PCR kit (Qiagen, Germany).
Most commercial HIV-1 viral load assays are designed and optimized for the detection of viruses found in North America and Europe. As a result, these assays either under-quantify or fail to amplify some of the HIV-1 non-B subtypes [6–8]. This observation motivated us to identify the gag gene from Indian subtype C HIV-1 viruses as the target to develop this SYBR Green chemistry based HIV-1 viral load estimation assay. The linearity of our assay ranged from 50 HIV-1 RNA copies to 107 HIV-1 RNA copies/ml of plasma sample and had a lower limit of detection of 50 HIV-1 RNA copies/ml.
The normalized measure of dispersion of the probability distribution of data generated from inter assay precision testing and expressed as coefficient of variation was found to increase as the values came closer to the detection limit. This was found to be acceptable since the linear range of detection was wide and at par with standard quantitative real time PCR assays [9–11].
The sensitivity of the assay was found to be 100%. No amplification in 25 clinical samples from HIV-1 sero negative reference panel indicated 100% specificity. This finding was further consolidated by dissociation curve analysis of all the sero positive reference panel samples. Sharp peaks at 85.5°C were detected in all sero positive reference panel samples and was absent in samples from healthy individuals. The results obtained from clinical study panel indicated a high level of concordance between the viral loads generated using the commercial kit and the assay described in this study.
The poor sensitivity of two commercial assays [HIV RNA Real-TM Quant (Sacace Biotechnologies S.r.l, Italy) and HIV-1 Real Time RT PCR kit (Life River, Shanghai ZJ Biotech, China) respectively] shown in this study may be due to presence of nucleotide mismatches within the primer and probe binding regions of HIV-1 genome. We believe that suboptimal attention towards HIV-1 subtype C specific sequences originating from India was the primary reason for such results. This finding is in line with data presented by Drexler et al. which demonstrated that HIV-1 positive samples which were reported negative by three commercial assays were shown to have a HIV-1 viral load of >5,000 copies/ml when tested using a home brew real time PCR protocol [7].
To evaluate the performance of the assay in a resource limited setting we used the Wilcoxon signed-rank test (similar to the paired student’s t-test) as a non-parametric statistical hypothesis test to compare the two related samples (plasma samples stored in -20°C and at room temperature respectively) and ascertain whether their population mean ranks differed. Results indicated that there were no significant differences in viral loads detected in both the groups (Wilcoxon signed rank test, p = 0.105) thus confirming the conclusions drawn from similar studies undertaken elsewhere [12]. In resource limiting setting like India cold chain facility is not always available for transportation of plasma samples from collection centers to the reference laboratories. Stability of the samples at ambient temperature is therefore important for proper clinical management of the patients.
Conclusions
There has been a significant increase in sensitivity of HIV-1 RNA quantification due to systematic involvement of advanced molecular techniques over the years. However, enhanced cost and demand for greater reagent and instrument sophistication is a necessary evil that needs to be traded off to achieve high level of accuracy and consistency. This is therefore a compelling reason for exploring areas where economy and ease of work can be linked to accuracy, sensitivity and specificity in HIV-1 RNA quantification. To address these issues we developed and evaluated the performance of SYBR Green chemistry based HIV-1 viral load detection assay. The significance of this study is high for a developing country like India which has a high rate of HIV-1 infection. Some commercial assays originating from Europe and China with poor attention to India-specific HIV-1 Subtype C genome sequences generated sub optimal data in our study and therefore carry a risk of generating error prone data with potential to mislead therapeutic decisions. In these contexts the assay described in this study specifically designed for Indian Subtype C viruses is useful, economical and reliable for routine monitoring of HIV-1 viral load in resource limited settings.
Methods
Clinical sample collection and processing
The reference panel comprised of blood samples collected from 25 each of HIV-1 sero-positive (drug naïve) and sero-negative (with no AIDS related risk factors) individuals from India in K2-EDTA vacutainer tubes (Becton Dickinson, San Diego, California, USA) after obtaining written consent from the donors. The status of HIV-1 infection was confirmed by (a) Tridot rapid assay (J. Mitra & Co. Ltd., New Delhi, India) (b) multiple enzyme-linked immunosorbent assays (Biochem Immunosystems, Montreal, Canada) and (c) western blot analysis (LAV Blot HIV-1, Bio- Rad, France). The viral load for each of these samples was determined using a US FDA-approved kit (artus HIV-1 RG RT-PCR kit; Qiagen, Germany) and used as reference concentration.
Plasma were separated from the blood samples within 4 hours of collection and stored in a -20°C freezer. Details of the study participants are described in Table 2.
Table 2.
Characteristics of reference study panel
| Sero positive reference panel (n = 25) | Sero negative reference panel (n = 25) | ||||
|---|---|---|---|---|---|
| Sample ID | Age | Gender | Sample ID | Age | Gender |
| DBL001 | 28 | Male | DBN001 | 29 | Male |
| DBL002 | 39 | Female | DBN002 | 24 | Female |
| DBL003 | 44 | Male | DBN003 | 21 | Male |
| DBL004 | 21 | Male | DBN004 | 46 | Male |
| DBL005 | 47 | Male | DBN005 | 35 | Male |
| DBL006 | 24 | Female | DBN006 | 39 | Female |
| DBL007 | 27 | Female | DBN007 | 42 | Female |
| DBL008 | 35 | Male | DBN008 | 31 | Male |
| DBL009 | 45 | Male | DBN009 | 36 | Male |
| DBL010 | 54 | Male | DBN010 | 45 | Female |
| DBL011 | 19 | Female | DBN011 | 26 | Male |
| DBL012 | 28 | Male | DBN012 | 21 | Female |
| DBL013 | 34 | Female | DBN013 | 37 | Male |
| DBL014 | 31 | Male | DBN014 | 41 | Male |
| DBL015 | 25 | Female | DBN015 | 28 | Male |
| DBL016 | 42 | Male | DBN016 | 37 | Female |
| DBL017 | 47 | Male | DBN017 | 35 | Male |
| DBL018 | 29 | Female | DBN018 | 32 | Female |
| DBL019 | 24 | Male | DBN019 | 27 | Male |
| DBL020 | 27 | Female | DBN020 | 26 | Female |
| DBL021 | 29 | Female | DBN021 | 29 | Female |
| DBL022 | 36 | Female | DBN022 | 38 | Male |
| DBL023 | 34 | Female | DBN023 | 39 | Female |
| DBL024 | 31 | Male | DBN024 | 34 | Male |
| DBL025 | 50 | Male | DBN025 | 38 | Male |
The clinical study panel comprised of blood samples collected from 100 HIV-1 sero-positive individuals. The viral load for each of these samples was determined using a US FDA-approved kit (artus HIV-1 RG RT-PCR kit; Qiagen, Germany) and used for analysis and comparison.
The study program was duly approved by a bio-safety and bio-ethics committee (Approval number SNGL/2012/GY06) overseeing ethical aspect of such research programs within the organization.
Oligonucleotide primer design
In order to avoid primer mismatch the most conserved region of HIV-1 gag gene was chosen for designing of the oligonucleotide primers. This region was identified by analyzing several full length HIV-1 gag gene nucleotide sequences submitted from India and obtained from NCBI GenBank database (http://www.ncbi.nlm.nih.gov/genbank) [13]. The sequences were aligned with the help of multiple sequence alignment program ClustalW v.2 [14] and conserved regions identified were used to manually design PCR primers. Five different sets of forward and reverse primers were designed, their reaction kinetics as well as self-complimentarity checked using Primer Express 3.0 program (Life Technologies, USA) and experimentally evaluated for their efficiency in amplifying target DNA.
Construction of reference standards
For construction of reference standards, the HIV-1 gag gene PCR fragment generated using the newly designed PCR primers was cloned in the pCR4-TOPO vector (Life Technologies. USA) following manufacturer’s instructions. The recombinant plasmid was purified and identity of the cloned insert confirmed by double strand DNA sequencing using the BigDye Terminator v3.1 Cycle Sequencing Kit and ABI PRISM® 3500 Dx Genetic Analyzer (Applied Biosystems, USA).
In order to generate quantization standard, the recombinant plasmid was serial diluted and calibrated against reference standards of the US FDA approved artus HIV-1 RG RT-PCR kit (Qiagen, Germany). A seven dilution series of the reference standard was prepared with copy numbers as follows: 101, 102, 103, 104, 105, 106 and 107 HIV-1 viral RNA copies/ml.
RNA extraction and reverse transcription–real time PCR
HIV-1 RNA was extracted from plasma samples using a QIAamp Viral RNA mini kit according to manufacturer’s instructions (Qiagen, Germany). SYBR Green I - based one-step real time quantitative RT-PCR amplification was performed using a StepOne Plus Real Time PCR System (Applied Biosystems, USA). Fifty μL of a reaction mixture comprised of 25 μL of 2X QuantiFast SYBR Green RT-PCR master mix (Qiagen, Germany) which contained HotStarTaq DNA Polymerase, 0.5 μL 100X QuantiFast RT Mix, 20 μL of RNA and each primers (forward and reverse) at a final concentration of 5 pmoles per reaction.
The nucleotide sequence of the primers used in this study is as follows: 5′- ACATCAAGCAGCCATGCAAAT - 3′ (forward) and 5′- TACTAGTAGTTCCTGCTATGTC - 3′ (reverse). ROX dye was used as a passive reference to normalize fluorescent signals. The thermal cycling profile comprised of 30 minutes of reverse transcription at 45°C, 5 minutes of heating at 95°C to activate HotStart Taq DNA Polymerase followed by 45 cycles of PCR amplification each comprising of 95°C for 15 seconds, 55°C for 15 seconds, and 72°C for 30 seconds. Data collection was done at 72°C of each cycle. Following amplification, a melting curve (dissociation curve) analysis was performed to verify the authenticity of the amplified products by checking their specific melting temperatures (Tm). In each batch all reference standards as well as samples were tested in triplicate and appropriate negative as well as reagent controls were included to exclude the possibility of cross contamination during the setting up of reactions.
Intra- and inter-assay validation
For intra- and inter-assay validation 25 HIV-1 sero-positive samples were tested 10 times each in different batches independently and in triplicate including the reference standards and controls and their coefficient of variation was calculated.
Sensitivity and specificity
For finding limit of detection of the assay 10, 30, 50, 100 and 500 copies/ml of serially diluted standards were tested in triplicate in 20 independent experiments. In order to measure the specificity of the assay all known HIV-1 sero-negative samples (Table 2) were subjected to real time PCR amplification followed by comparison of the melting-temperature data with known positives as well as no-template control (NTC) samples.
Comparing sensitivity of detection against two commercial HIV-1 copy number detection assays
A panel of 90 plasma samples which was a part of the study panel and with HIV-1 viral copy number ranging from 5,000–25,000 HIV-1 RNA copies/mL as estimated using artus HIV-1 RG RT-PCR kit (Qiagen, Germany) and confirmed as sub type C using method described by Siddappa et al. [15] were simultaneously subjected to quantitative detection of HIV-1 RNA using (a) HIV RNA Real-TM Quant (Sacace Biotechnologies S.r.l, Italy) (b) HIV-1 Real Time RT PCR kit (Life River, Shanghai ZJ Biotech, China) and (c) assay method developed in this study. All RT-PCR reactions were set in duplicate in two separate thermal cyclers of identical make (StepOne Plus Real Time PCR System; Applied Biosystems, USA) in order to minimize the influence of experimental errors.
Effect of storage time and temperature on stability of plasma samples
Plasma samples of 10 patients with known viral load were collected and two aliquots were made from each of them. One set of plasma were stored at -20°C and the other at 25°C. RNA was extracted from both set of samples at day seven and their viral load estimated in triplicate using the method described in this study.
Statistical analysis
The linearity of the assay was determined by linear regression method using the MedCalc v.12.7.2 (Acacialaan 22, B-8400 Ostend, Belgium) program. Sensitivity of the assay was expressed as the percentage of samples that scored positive using the protocol described in this study divided by the total number of samples that scored positive using an US -FDA approved commercial assay. Specificity of the assay was calculated as the number of negative samples scored using the present study divided by the total number of known negative samples obtained from healthy donors and was expressed in percentage. Performance of the assay was measured by doing a comparative analysis with artus HIV-1 RG RT-PCR kit (Qiagen, Germany) and applying the Spearman correlation coefficient formula (http://www.wessa.net/rankcorr.wasp). The effect of ambient temperature on the stability of HIV-1 RNA was analyzed using the Wilcoxon signed rank test [16] and the box and a whisker plot was created using BioStat program (AnalystSoft Inc, USA).
Acknowledgments
The authors are grateful to all the study participants for agreeing to provide their valuable clinical samples for this study.
Footnotes
Competing interests
The authors declare no competing interests.
Authors' contributions
AA and PS carried out the viral RNA extraction and real time PCR reactions. HP generated the recombinant plasmids used in this study. AA and SV analyzed the amplification data and advised on the resource population. RNM and MW assisted in drafting the manuscript. PNM and SV conceptualized the work and drafted the complete manuscript. All authors read and approved the final manuscript.
Contributor Information
Arpan Acharya, Email: juarpan@yahoo.co.in.
Salil Vaniawala, Email: salil@sngenelab.com.
Parth Shah, Email: parth.shah@supratechlabs.com.
Harsh Parekh, Email: harshparekh88@gmail.com.
Rabindra Nath Misra, Email: rabindranath.misra@yahoo.in.
Minal Wani, Email: minal.wani@dpu.edu.in.
Pratap N Mukhopadhyaya, Email: pnm.istra@yahoo.com.
References
- 1.‘Global report: UNAIDS report on the global AIDS epidemic 2012’. 2012. [Google Scholar]
- 2.Gazzard B. British HIV Association (BHIVA) guidelines for the treatment of HIV-infected adults with antiretroviral therapy. HIV Med. 2005;6(2):1–61. doi: 10.1111/j.1468-1293.2005.0311b.x. [DOI] [PubMed] [Google Scholar]
- 3.Holguin A, Lopez M, Molinero M, Soriano V. Performance of three commercial viral load assays, versant human immunodeficiency virus type 1 (HIV-1) RNA bDNA v3.0, cobas AmpliPrep/cobas TaqMan HIV-1, and NucliSens HIV-1 EasyQ v1.2, testing HIV-1 non-B subtypes and recombinant variants. J Clin Microbiol. 2008;46:2918–2923. doi: 10.1128/JCM.02414-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knudtson KL, Adams PS, Grove DS, Hollingshead DJ, Hunter TC, Shipley GL. The ABRF NARG real-time PCR survey 2007. Taking the pulse of the quantitative PCR field. 2007. [Google Scholar]
- 5.Brumme CJ, Swenson LC, Wynhoven B, YIP B, Skinner S, Lima VD, Montaner JSG, Harrigan PR. Technical and regulatory shortcomings of the TaqMan version 1 HIV viral load assay. PLoS One. 2012;7(8):e43882. doi: 10.1371/journal.pone.0043882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alaeus A, Lidman K, Sonnerborg A, Albert J. Subtype specific problems with quantification of plasma HIV-1 RNA. AIDS. 1997;11:859–865. doi: 10.1097/00002030-199707000-00004. [DOI] [PubMed] [Google Scholar]
- 7.Drexler JF, De Souza Luna LK, Pedroso C, Pedral-Sampaio DB, Queiroz ATL, Brites C, Netto EM, Drosten C. Rates of and reasons for failure of commercial human immunodeficiency virus type 1 viral load assays in Brazil. J Clin Microbiol. 2007;45:2061–2063. doi: 10.1128/JCM.00136-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Korn K, Weissbrich B, Henke-Gendo C, Heim A, Jauer CM, Taylor N, Eberle J. Single-point mutations causing more than 100-fold underestimation of human immunodeficiency virus type 1 (HIV-1) load with the cobas TaqMan HIV-1 real-time PCR assay. J Clin Microbiol. 2009;47:1238–1240. doi: 10.1128/JCM.02204-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mehta N, Trzmielina S, Nonyane B, Eliot MN, Lin R, Foulkes AS, McNeal K, Ammann A, Eulalievyolo V, Sullivan JL, Luzuriaga K, Somasundaran M. Low-cost HIV-1 diagnosis and quantification in dried blood spots by real time PCR. PLoS One. 2009;4(6):e5819. doi: 10.1371/journal.pone.0005819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fibriani A, Farah N, Kusumadewi I, Pas SD, Van Crevel R, Van Der Ven A, Boucher CA, Schutten M. Low cost HIV-1 quantitative RT-PCR assay in resource-limited settings: improvement and implementation. J Virol Methods. 2012;185(1):118–123. doi: 10.1016/j.jviromet.2012.06.015. [DOI] [PubMed] [Google Scholar]
- 11.Gibellini D, Vitone F, Gori E, Placa ML, Re MC. Quantitative detection of human immunodeficiency virus type 1 (HIV-1) viral load by SYBR green real-time RT-PCR technique in HIV-1 seropositive patients. J Virol Methods. 2004;115(2):183–189. doi: 10.1016/j.jviromet.2003.09.030. [DOI] [PubMed] [Google Scholar]
- 12.Holodniy M, Rainen L, Herman S, Yen-Lieberman B. Stability of plasma human immunodeficiency virus load in VACUTAINER PPT plasma preparation tubes during overnight shipment. J Clin Microbiol. 2000;38(1):323–326. doi: 10.1128/jcm.38.1.323-326.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benson DA, Cavanaugh M, Clark K, Karsch-mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2013;41:D36–D42. doi: 10.1093/nar/gks1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larkin MA, Blackshields G, Brown NP, Chenna R, Mcgettigan PA, Mcwilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 15.Siddappa NB, Dash PK, Mahadevan A, Jayasuryan N, Hu F, Dice B, Keefe R, Satish KS, Satish B, Sreekanthan K, Chatterjee R, Venu K, Satishchandra P, Ravi V, Shankar SK, Shankarappa R, Ranga U. Identification of subtype C human immunodeficiency virus type 1 by subtype-specific PCR and its use in the characterization of viruses circulating in the southern parts of India. J Clin Microbiol. 2004;42(6):2742–2751. doi: 10.1128/JCM.42.6.2742-2751.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frank W. Individual comparisons by ranking methods. Biom Bull. 1945;1(6):80–83. doi: 10.2307/3001968. [DOI] [Google Scholar]


