Abstract
Small-cell lung cancer (SCLC) is a subtype of lung cancer with poor prognosis. To identify accurate predictive biomarkers and effective therapeutic modalities, we focus on a long noncoding RNA, Hox transcript antisense intergenic RNA (HOTAIR), and investigated its expression, cellular functions, and clinical relevance in SCLC. In this study, HOTAIR expression was assessed in 35 surgical SCLC samples and 10 SCLC cell lines. The efficacy of knockdown of HOTAIR by siRNA transfection was evaluated in SBC-3 cells in vitro, and the gene expression was analyzed using microarray. HOTAIR was expressed highly in pure, rather than combined, SCLC (P = 0.012), that the subgroup with high expression had significantly more pure SCLC (P = 0.04), more lymphatic invasion (P = 0.03) and more relapse (P = 0.04) than the low-expression subgroup. The knockdown of HOTAIR in SBC-3 cells led to decreased proliferation activity and decreased invasiveness in vitro. Gene expression analysis indicated that depletion of HOTAIR resulted in upregulation of cell adhesion-related genes such as ASTN1, PCDHA1, and mucin production-related genes such as MUC5AC, and downregulation of genes involved in neuronal growth and signal transduction including NTM and PTK2B. Our results suggest that HOTAIR has an oncogenic role in SCLC and could be a prognostic biomarker and therapeutic target.
Keywords: HOTAIR, invasiveness, lincRNA, proliferation, small-cell lung cancer
Introduction
Lung cancer is a leading cause of cancer death worldwide 1. Small-cell lung cancer (SCLC) is an aggressive subtype, characterized by a neuroendocrine nature, which represents ∼15% of all newly diagnosed lung cancers 2. SCLC patients have a poor prognosis compared with non–small-cell lung cancers (NSCLCs) due to more rapid growth and more frequent recurrence. Although the survival of NSCLC patients has been significantly improved by targeted chemotherapy, there are currently no targeted drugs effective against SCLC. To identify accurate predictive biomarkers and to develop effective therapeutic modalities, elucidation of molecular mechanisms underlying the rapid growth and the high propensity for relapse of SCLC is essential.
Advances in experimental technology have been applied to studies of malignant tumors including SCLC 3. In particular, application of modern genetic profiling technology to the study of noncoding RNAs has revealed a crucial role for these molecules in tumor cell regulation. Similar to short regulatory noncoding RNAs (ncRNAs), such as microRNAs, many long intergenic ncRNAs (lincRNAs) have been found to be important by functioning as the interface between DNA and specific chromatin remodeling activities 3–7. These lincRNAs are involved in diverse cellular processes, including cell-cycle regulation, immune surveillance, and stem cell pluripotency.
Hox transcript antisense intergenic RNA (HOTAIR) is one of the few biologically well-studied lincRNAs 8,9. Previous studies have demonstrated that HOTAIR is transcribed from HoxC gene as an antisense transcript, and binds polycomb repressive complex 2 (PRC2) and LSD1-CoREST-REST complex as scaffolds, leading to catalyzing trimethylation of H3K27 and spontaneous demethylation of H3K4, and to repressing transcription of HoxD genes 9. REST (RE1 silencing transcriptional factor, also called neuron-restrictive silencer factor) and its corepressors negatively regulate neurogenesis and contribute to the maintenance of pluripotency of neural cells 10, whereas LSD1 (lysin-specific demethylase 1) regulates neural stem cell proliferation 11. In relation to DNA methylation, EZH2, a compartment of PRC2, directly interacts with DNA methyltransferases (DNMT1, DNMT3A and DNMT3B). This interaction is necessary for maintenance of DNA methylation and stable repression of specific genes, including many tumor suppressors 12. In fact, 20% of the lincRNAs have been shown to associate with PRC2. The homeobox-containing genes as targets of HOTAIR are a family of transcriptional regulators encoding DNA-binding homeodomains involved in the control of normal development 4,5. Also, aberrant expression of homeobox genes is associated with both morphological abnormalities and carcinogenesis 6,7. Moreover, a most recent study suggested that the role of HOTAIR in tumorigenesis occurs through triggering epithelial-to-mesenchymal transition (EMT) and acquiring stemness and its maintenance 13.
Although HOTAIR and its association in cancer metastasis and prognosis of diverse cancers have been suggested in several studies 14–22, its functions in SCLC remain unclear. In this study, we investigated the role of HOTAIR for cellular proliferation and patients' prognosis to develop a biomarker and a new target for therapy of SCLC.
Materials and Methods
Clinical samples and cell lines
Between January 1995 and December 2010, 3460 patients with primary lung cancer underwent surgery at the Cancer Institute Hospital of Japanese Foundation for Cancer Research (JFCR), Tokyo, Japan. Since SCLC is usually inoperable, only 55 (1.6%) cases had been diagnosed as SCLC by expert pathologists using hematoxylin and eosin (H&E) staining, based on the WHO classification 23.
Due to inadequate amounts of viable cancer cells, 20 cases were excluded from the study leaving 35 cases. Basis on TNM classification of malignant tumors 7th edition, all cases were staged. Specimens were snap-frozen in liquid nitrogen typically within 15 min after removal and stored at −80°C. Written informed consent for research was obtained from all patients, and our institutional review board approved the study plan. We collected clinicopathological details including neoadjuvant and adjuvant chemotherapy (NAC and AC, respectively), and listed them in Table 1.
Table 1.
Comparisons of clinicopathological factors of all SCLC patients enrolled (n = 35) and those with high- and low expression of HOTAIR
| All cases, examined (n = 35) | Cases with high-HOTAIR expression (n = 12) | Cases with low-HOTAIR expression (n = 23) | |||||
|---|---|---|---|---|---|---|---|
| Factors | N | % | N | % | N | % | P value |
| Age (mean ± SD) | 65.8 ± 6.60 | 63.3 ± 6.70 | 67.1 ± 6.30 | 0.10 | |||
| Gender | |||||||
| M | 25 | 71.4 | 9 | 75.0 | 16 | 69.6 | 0.53 |
| F | 10 | 28.6 | 3 | 25.0 | 7 | 30.4 | |
| Cumulative smoking (pack-years) | 52 ± 31 | 52 ± 19 | 52 ± 36 | 0.96 | |||
| Chemotherapy type | |||||||
| NAC − AC− | 3 | 8.60 | 1 | 8.33 | 2 | 8.70 | |
| NAC + AC− | 3 | 8.60 | 0 | 0 | 3 | 13.0 | |
| NAC − AC+ | 23 | 65.7 | 8 | 66.7 | 15 | 65.2 | |
| NAC + AC+ | 6 | 17.1 | 3 | 25.0 | 3 | 13.0 | |
| Regimen | |||||||
| NAC(n = 9) | CDDP + VP16 (8) | 22.9 | 3 | 25.0 | 5 | 21.7 | |
| CDDP + DOC (1) | 2.90 | 0 | 0 | 1 | 4.35 | ||
| AC(n = 29) | CDDP + VP161 (22) | 62.9 | 10 | 83.3 | 12 | 52.2 | |
| CBDCA + VP162 (4) | 11.4 | 0 | 0 | 4 | 17.4 | ||
| Mix; 1 and 2 (3) | 8.60 | 1 | 8.33 | 2 | 8.70 | ||
| Histological type (SCLC) | |||||||
| Pure | 24 | 68.6 | 11 | 91.7 | 13 | 56.5 | 0.04* |
| Combined | 11 | 31.4 | 1 | 8.33 | 10 | 43.5 | |
| With Ad (5) | 45.5 | 0 | 0 | 5 | 50.0 | ||
| With LCC (2) | 18.2 | 0 | 0 | 2 | 20.0 | ||
| With LCNEC (4*) | 36.4 | 1* | 100 | 2 | 20.0 | ||
| With others (1) | 9.10 | 0 | 0 | 1 | 10.0 | ||
| *include combined case (with both LCNEC/Ad) | *include combined case (with both LCNEC/Ad) | ||||||
| Operation (SCLC) | |||||||
| Partial resection | 3 | 8.60 | 1 | 8.33 | 2 | 8.70 | |
| Segmentectomy | 0 | 0 | 0 | 0 | 0 | 0 | |
| Lobectomy (1 lobe) | 27 | 77.1 | 10 | 83.3 | 17 | 73.9 | |
| Lobectomy + partial resection | 1 | 2.90 | 1 | 8.33 | 0 | 0 | |
| Lobectomy (2 lobes) | 2 | 5.70 | 0 | 0 | 2 | 8.70 | |
| Pneumectomy | 2 | 5.70 | 0 | 0 | 2 | 8.70 | |
| Pathological stage | |||||||
| 1a | 11 | 31.4 | 1 | 8.33 | 10 | 43.5 | |
| 1b | 4 | 11.4 | 2 | 16.7 | 2 | 8.70 | |
| 2a | 7 | 20.0 | 3 | 25.0 | 4 | 17.4 | |
| 2b | 2 | 5.70 | 0 | 0 | 2 | 8.70 | |
| 3a | 10 | 28.6 | 6 | 50.0 | 4 | 17.4 | |
| 3b | 1 | 2.90 | 0 | 0 | 1 | 4.35 | |
| 4 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Invasion (microscopic) | |||||||
| Vascular invasion | 29 | 82.9 | 9 | 75.0 | 20 | 87.0 | 0.33 |
| Lymphatic invasion | 20 | 57.1 | 10 | 83.3 | 10 | 43.5 | 0.03* |
| Relapse | 15 | 42.9 | 8 | 66.7 | 7 | 30.4 | 0.04* |
| Brain (6) | 40.0 | 4 | 50.0 | 2 | 28.6 | ||
| Lung (2) | 13.3 | 2 | 25.0 | 0 | 0 | ||
| Mediastinal LNs (4) | 26.7 | 3 | 37.5 | 1 | 14.3 | ||
| Stomach (1) | 6.70 | 0 | 0 | 1 | 14.3 | ||
| Liver (3) | 20.0 | 1 | 12.5 | 2 | 28.6 | ||
| Adrenal gl. (2) | 5.70 | 1 | 12.5 | 1 | 14.3 | ||
| Others (pleural eff.) (1) | 6.70 | 0 | 0 | 1 | 14.3 | ||
| Survival | |||||||
| Alive | 19 | 54.3 | 5 | 41.7 | 14 | 60.9 | |
| Dead | 16 | 45.7 | 7 | 58.3 | 9 | 39.1 | |
| Lung cancer (11) | 68.7 | 6 | 85.7 | 5 | 55.6 | ||
| Other malignancy (2) | 12.5 | 1 | 14.3 | 1 | 11.1 | ||
| Other disease (2) | 12.5 | 0 | 0 | 2 | 22.2 | ||
| Unknown (1) | 6.30 | 0 | 0 | 1 | 11.1 | ||
| DSS (mean ± SD) | 45.3 ± 35.7 mos. | 38.1 ± 27.2 mos | 49.0 ± 39.5 mos | ||||
| RFS/DFS (SCLC) (mean ± SD) | 40.9 ± 38.5 mos. | 30.8 ± 30.7 mos | 46.3 ± 41.6 mos | ||||
SCLC, small-cell lung cancer; smoking index, a product of number of cigarettes per day by duration in years; NAC, neoadjuvant chemotherapy; AC, adjuvant chemotherapy; CDDP, cisplatin; CBDCA, carboplatin; VP-16, etoposide; DOC, docetaxel; Ad, adenocarcinoma; LCC, large cell carcinoma; LCNEC, large cell neuroendocrine carcinoma; LNs, lymph nodes; DSS, disease-specific survival; RFS, relapse-free survival; DFS, disease-free survival.
P < 0.05.
Ten SCLC cell lines (COLO-668, COR-L51, COR-L88, DMS-79, DMS-53, Lu-134A, MS-1, SBC-3, SBC-5, and SBC-1), one adenocarcinoma cell line (A549) and a normal lung cell line (MRC-5), derived from embryonic normal lung tissue, were used. The former four lines were obtained from the European Collection of Cell Cultures, and the other were from Japanese Collection of Research Bio-resources or the RIKEN Bio Resource Center. All cells were maintained in the Dulbecco's modified Eagle's medium (DMEM) supplemented with 2 mmol/L l-alanyl-l-glutamine solution, 1.1% antibiotic-antimycotic mixed stock solution (Nacalai Tesque, Kyoto, Japan) and 10% fetal bovine serum (FBS), at 37°C, 5% CO2 incubator.
Primary-cultured normal bronchial epithelial (NBE) cells were obtained from fresh surgical materials. Emergence and proliferation of bronchial epithelial cells around the samples without any proliferation of fibroblasts and contamination was confirmed by microscopy.
RNA preparation, reverse transcription, and quantitative real-time polymerase chain reaction
Total RNA from tissues and cells were extracted using the RNeasy mini kit or RNeasy mini kit plus (Qiagen, Tokyo, Japan). cDNAs were generated from 30 ng of total RNA. The resulting cDNA was subjected to a 45-cycle polymerase chain reaction (PCR) amplification step followed by quantitative real-time PCR (qRT-PCR) using the LightCycler 480 SYBR Green I Master protocol (Roche Applied Science, Indianapolis, IN). Triplicates were run for each gene for each sample as previously 16. Based on previous studies 24, the amount of HOTAIR RNA was normalized to that of beta-actin (ACTB) in tissue samples and xenografts, and to Glyceraldehydes-3-phosphate dehydrogenase (GAPDH) in cell lines and normal cells. The HOTAIR/ACTB ratio in 35 SCLC and 15 noncancerous lung tissues randomly chosen from the 35 patients were analyzed by qRT-PCR. Tumors were divided into two groups with high- and low expression based on HOTAIR/ACTB ratios, using receiver operating characteristic (ROC) curve analysis. The primer sequences are listed in Table S1.
HOTAIR expression of SCLC cell lines as well as control cells
We assessed HOTAIR expression in above cell lines and normal controls, normalizing to GAPDH. To define high- and low-expression groups, we used the level of normal controls, that is, the cut-off level of high expression was defined as above those of normal controls.
SCLC cell xenografts
We examined HOTAIR expression in xenografts as well 25. Four-week-old male nude mice Crlj:CD1-Foxn1NU with ICR background were purchased from Charles River Laboratories, Japan, housed at the animal care facility of our institute and kept under standard temperature, humidity, and timed-lighting conditions and provided mouse chow and water ad libitum. The five SCLC cell lines with high expression (final concentration, 1.0 × 107 cells/0.3 mL phosphate buffered saline [PBS] each) were injected directly into three sites of the abdominal subcutaneous adipose tissue of the mice in 0.3 mL of sterile PBS. Developed tumors were immediately frozen in liquid nitrogen and stored at −80°C until use.
RNA interference
Cells were transfected with 20 nmol/L small interfering (si)RNAs targeting HOTAIR, using Lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA) as per manufacturer's directions. Transfection efficiency was assessed using a fluorescence microscope (Leica-DMIRE2) following 72 h incubation after transfection of labeled positive control; BLOCK-iT Alexa Fluor Red Fluorescent Oligo (Invitrogen) according to manufacturer's procedures. Twenty nmol/L (final concentration) of siRNAs and 1.5 μL of Lipofectamine RNAiMAX in a total volume of 101.5 μL were used for transfection of SBC-3 cells in 24-well based analysis (transduction efficiency: 100%, knockdown efficiency: 50%). We transfected #1–3 siHOTAIR as previously 8,9,14 to SBC-3 cells. After 72 h, total RNAs were collected for qRT-PCR analysis. Primer sequences are listed in Table S1.
Cell proliferation assay and matrigel invasion assay
For cell proliferation assays, 4.0 × 104 cells were plated in triplicate on 24-well plates containing DMEM medium with 10% FBS, 1% antibiotics, and glutamine solution. Subsequently, the cell number was calculated using the Trypan-blue staining and an automated cell counter (TC10 Bio-Rad Laboratories, Tokyo, Japan) after transfection with siRNA for 24, 48, 72, and 96 h.
A matrigel invasion assay was performed using the Biocoat Matrigel Invasion Chamber (BD) according to the manufacturer's protocols. In brief, 4.0 × 104 cells were plated in the upper insert chamber in serum-free medium. The bottom chamber contained DMEM medium with 10% FBS as an inducer of invasion. After 48 h, the bottom of the insert chamber was fixed and stained with Diff-Quick staining (Sysmex, Kobe, Japan). Cells on the dissected stained membrane were counted under a microscope. Each membrane was divided into four quadrants and the sum of all four quadrants was calculated. Each matrigel invasion assay was performed in triplicate, and compared with migration assays using control insert membranes.
Comprehensive gene expression analysis using microarrays
Total RNAs from #1siHOTAIR-transfected SBC-3 cells and control cells (siGFP-transfected cells), were extracted using RNeasy mini kit Plus (Qiagen) and hybridized to the microarrays, Sure Print G3 Human GE 8 × 60K microarrays (Agilent Technologies, Santa Clara, CA), according to the manufacturer's instructions. Subsequently, data analysis was carried out using the GeneSpring GX 12 software (Agilent Technologies), with a stringency of P < 0.1 and a twofold or more change using gene ontology analysis.
Statistical analysis
Continuous datasets were compared using an independent t-test between two groups, and categorical datasets were analyzed by the chi-square test. Significance of difference between two or more groups was estimated with the Mann–Whitney U-test or the Kruskal–Wallis test, as appropriate. Disease-specific survival (DSS) was defined by death only from SCLC. Relapse-free survival (RFS) was defined by metastasis as the first recurrence event. In this study, because second primary cancers were not found in all subjects, RFS is de facto disease-free survival (DFS). DSS and RFS/DFS curves were plotted according to the Kaplan–Meier method with the Cox–Mantel log-rank test applied for comparison. Univariate and multivariate analyses by the Cox proportional hazard method were performed. All differences were considered statistically significant at the level of P < 0.05, and there being a tendency at a level of P < 0.10. SPSS 19.0 (IBM Corporation, Somers, NY) was used for statistical analyses.
Results
Clinicopathological profiles of 35 SCLC cases
We assessed 35 surgically removed SCLC tumors and 15 lung tissues. The patients were mostly male, the average age was 65.8 years, and cumulative smoking was over 50 pack-years. Nine cases undertook chemotherapy before surgery, typically four courses of platinum (cisplatin [CDDP] or carboplatin [CBDCA]) plus etoposide (VP-16), and 26 (74%) cases did not have chemotherapy. Histologically, 69% (n = 24) were pure SCLC and vascular invasions were often observed (83%, n = 29). As expected, there were many stage I cases (n = 15, 43%). Generally, although SCLC patients once improved after chemotherapy, relapses frequently occur, resulting in poor prognosis. During the period of observation, 43% (n = 15) had relapses and 46% (n = 16) died. DSS and RFS/DFS were 45.3 ± 35.7 months and 40.9 ± 38.5 months, respectively.
HOTAIR expressions and clinicopathological factors in SCLC
We assessed HOTAIR expression levels in 35 tumors and 15 normal tissues by qRT-PCR normalized to the ACTB. HOTAIR levels between tumor tissues (n = 35) and normal tissues (n = 15) were not different significantly when analyzed across all cases (P = 0.992 [Mann-Whitney U test]; Fig. 1A). When compared between pure-SCLC tumor tissues (n = 24) and combined-SCLC tumor tissues (n = 11), HOTAIR expression was significantly higher in pure SCLC (P = 0.009; Fig. 1B). For pure-SCLC cases, HOTAIR had a tendency to be expressed more in tumor tissues (n = 24) than normal tissues (n = 13) (P = 0.092; Fig. 1C). Since chemotherapy prior to surgery might have some effect on cellular nature, analysis was limited to cases without such treatment (pure-NAC(−) cases), resulting in significantly higher expression in tumor (n = 18) than normal tissues (n = 9) (P = 0.012; Fig. 1D).
Figure 1.
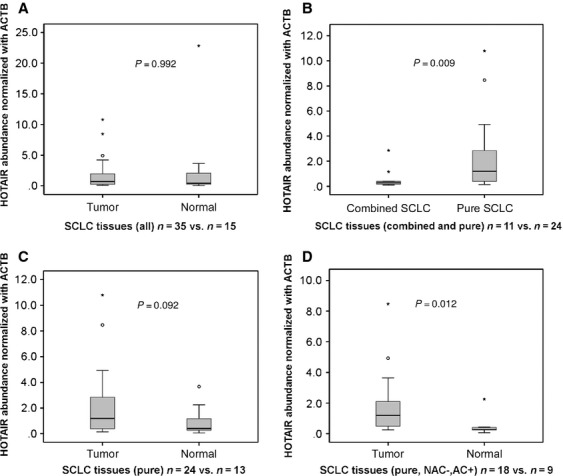
HOTAIR expression levels assessed by quantitative RT-PCR in 35 SCLC and normal tissues. Comparisons between all the tumors (n = 35) and normal tissues (n = 15) (A), pure (n = 24) and combined (n = 11) SCLC (B) (P = 0.009), pure SCLC and normal (C), and between pure SCLC with adjuvant chemotherapy and no neoadjuvant chemotherapy (n = 18) and normal (D) (P = 0.012) were made. HOTAIR was highly expressed in pure SCLC than combined SCLC and normal tissues.
We divided 35 subjects by HOTAIR expression into two groups: high expression (n = 12) and low expression (n = 23), according to a HOTAIR/ACTB ratio of 1.368 in tumor tissues, obtained by the ROC method (Fig. 2A). The high-expression group contained significantly more pure SCLC (P = 0.04), more lymphatic invasion (P = 0.03), and more relapse (P = 0.04) than the low-expression group (Table 1). To perform survival analysis, we focused only on cases with pure SCLC, without NAC and with AC (n = 8 in high-expression group and n = 10 in low-expression group). The high-expression group tended to have lower survival for RFS/DFS (P = 0.086), but not for DSS (P = 0.263) (Fig. 2B and C). Univariate analyses of DSS in all the subjects revealed that there were tendencies toward poor prognosis in the cases with relapse and pathological stages II or higher (P = 0.058, 0.099, respectively, Table S2). Multivariate analyses of DSS demonstrated that AC (P = 0.005) and pathological stages (P = 0.02) were significant prognostic factors (Table S2). However, HOTAIR expression was not a prognostic factor by either uni- or multivariate analyses. In regard to RFS/DFS, multivariate analyses showed that AC and pathological stages were significant prognostic factors (P = 0.004 and 0.021, respectively) and there was a tendency to poor prognosis in the cases with high-HOTAIR expression (P = 0.071) as indicated in Table S2.
Figure 2.
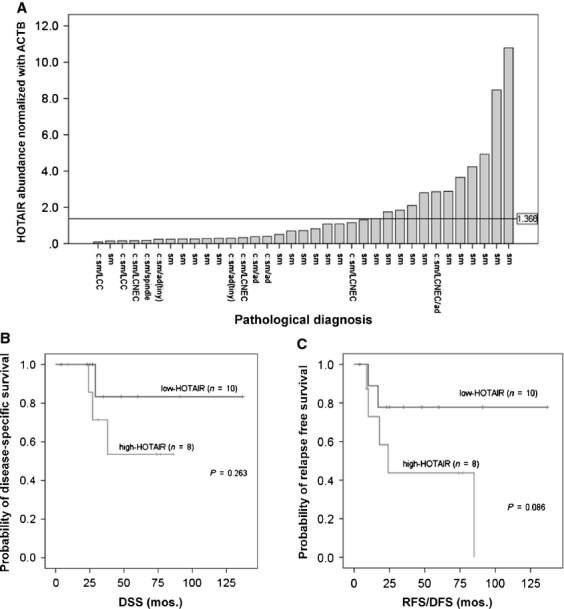
HOTAIR expression levels in SCLC and survival curves by HOTAIR expression. (A) HOTAIR expression in 35 SCLC and a reference level (HOTAIR/ACTB ratio = 1.368). (B and C) The Kaplan–Meier DSS curves and RFS/DFS curves, respectively, according to HOTAIR levels in pure-SCLC cases with adjuvant chemotherapy and no neoadjuvant chemotherapy (n = 18). Note that there was a tendency to poor prognosis in the high-expression group for the RFS/DFS of these groups (P = 0.086). For abbreviations, see text.
Expression of HOTAIR in SCLC cells and their xenografts
We next examined HOTAIR expression in cell lines and normal cells. Normal cells had no expression. There were five SCLC cell lines (COR-L51, COR-L88, DMS-79, Lu-134A, and SBC-3) whose expression was significantly higher than normal, whereas the other five cell lines and the adenocarcinoma line showed the same low-level expression as normal cells. The SBC-3 cells expressed particularly highly (Fig. 3A). We made xenografts from these five cell lines and assessed HOTAIR expression using RT-PCR as normalized to ACTB. Although Lu-134A and DMS-79 lines expressed significantly highly compared with states of culture, it was SBC-3 that expressed most highly (Fig. 3B).
Figure 3.
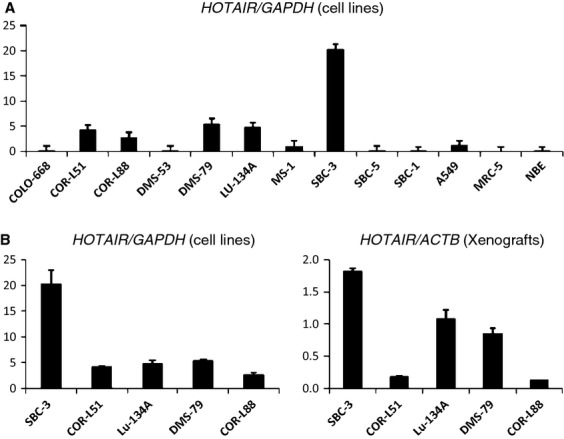
Expression of HOTAIR in cell lines and xenografts of SCLC. (A) Relative expression levels normalized to GAPDH. The SBC-3 line showed particularly high expression. (B) Comparisons in five SCLC cell lines with higher expression than normal cells. For abbreviations, see text.
Consequently, we selected SBC-3 cells for further analyses. The SBC-3 line is an adherent cell line, others are not, and therefore RNAi was successful.
Depletion of HOTAIR by siRNA leading to decreased proliferation and invasiveness
#1–3 siRNAs against HOTAIR gene and siGFP were conformed as described 8,9,14. All three siRNAs worked well and the gene expression was reduced significantly (Fig. 4A). In the proliferation experiments, however, depletion of HOTAIR by #1 siHOTAIR in SBC-3 cells dramatically decreased its proliferation ability whereas transfection of #2 or #3 siHOTAIR did not (Fig. 4B). Also, in the invasion assay, #1 siHOTAIR successfully reduced matrix invasiveness as compared with controls (siGFP transfection), whereas #2–3 siRNA did not (Fig. 4C).
Figure 4.
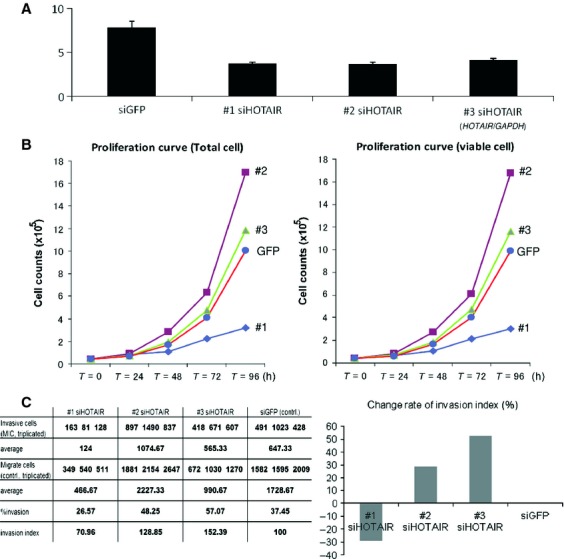
HOTAIR abundance, cellular proliferation and invasiveness in SBC-3 cells with transfection of siRNA and of siGFP. (A) The graph showed the HOTAIR/GAPDH ratio. #1–3 siHOTAIRs successfully reduced the expression levels. (B), #1 siHOTAIR-transfected cells reduced their ability of proliferation after 72 h, although #2, #3 siHORAIR did not reduce. (C) Invasiveness was assessed by % invasion and invasion index according to manufactures' protocol. In #1 siHOTAIR-transfected cells, almost 30% reduction of matrigel invasiveness was seen, whereas #2 and #3 siHOTAIR did not reduce invasiveness.
Gene ontology analysis in siHOTAIR-transfected cells
We tested whether HOTAIR depletion by siRNA affected the pattern of gene transcriptions, especially in genes related to proliferation and invasiveness targeted by siHOTAIR. We succeeded in the transfection experiments using the SBC-3 cell line and subsequently performed gene expression analysis. Raw data of two samples (#1 siHOTAIR-transfected cells and siGFP-transfected cells) were deposited in the GEO database (GSE43877). We identified significantly altered 110 genes. Ontology analyses demonstrated that many of these genes were related to cell adhesion, proliferation, and mucin formation.
Among the upregulated genes, there were those related to cell adhesion such as ASTN1 (astrotactin 1), PCDHA1 and 10 (protocadherin-alpha [Pcdha] 1 and 10), and CLDN11 (Claudin-11), and genes involved in mucin production including MUC5AC and MUC4. In addition, ECM2, coding an extracellular matrix protein, and NRP2, coding a transmembrane protein interacting with vascular endothelial growth factor, were among the top 15 upregulated genes (Table S3). The top 15 downregulated genes include NTM, neurotrimin, encoding a protein that may promote neurite outgrowth and adhesion, PTK2B, protein tyrosine kinase 2 beta (also known as Pyk2), which functions in the activation of MAPK signaling pathways, and CTNNA2, catenin (cadherin-associated protein) alpha 2, expression of which is important for maintaining a subset of neurons. In addition, SIRPG (signal regulatory protein gamma), a member of the immunoglobulin superfamily, known to be involved in the negative regulation of receptor tyrosine kinase-coupled signaling processes, and ITGB8 (Integrin beta-8), coding a member of the integrin beta chain family, are downregulated. Generally, integrin complexes mediate cell–cell and cell–extracellular matrix interactions and play a role in human airway epithelial proliferation.
Discussion
Among lincRNAs, HOTAIR is one of the most remarkable because of its relevance to metastases in common cancers such as breast and colon cancers. Here, we first demonstrated that HOTAIR was expressed in pure SCLC and higher expression was significantly related to lymphatic invasion and relapse. Multivariate analyses demonstrated that HOTAIR expression correlated with RFS. In vitro experiments demonstrated that half of SCLC cell lines expressed HOTAIR at higher levels than normal cells and that, using SBC-3 cells, knockdown of HOTAIR decreased proliferative activity and cellular invasiveness with altered expression of cell adhesion-related genes.
Accumulating reports suggest that HOTAIR is related to poorer prognosis of tumors. Several studies have shown a relationship between high HOTAIR expression and the poorer prognosis of diverse cancers 14–22, but there are few studies that reveal the importance of Hox-related genes and/or HOTAIR in SCLC because of the scarcity of fresh tissue samples. In fact, almost all evidence obtained for alterations of expression in Hox-related genes in SCLC to date was based on cell lines and their xenografts 26–29. To our knowledge, this is the first report on HOTAIR expression in primary SCLC tissues. In particular, we have shown that HOTAIR expression is significantly higher in SCLC tumors than normal tissues, considering induction chemotherapy (Fig. 1D), and that high expression of HOTAIR is relevant for relapse of SCLC. This tumor type is special for lung cancer, but proved to share similar characteristics of HOTAIR-related cellular regulations with ordinary carcinomas including those of breast and colon. Furthermore, our knockdown experiments of HOTAIR demonstrated that its depletion in SBC-3 cells caused altered expression of genes involved in neural cell adhesion, proliferation, and mucin formation although the findings are based on one cell line.
It is interesting, although not surprising, that HOTAIR correlates with SCLC proliferation and invasion with expression of genes implicated in cell adhesion and mucin production. In fact, depletion of HOTAIR upregulates genes implicated with cellular adhesion and mucin production including ASTN1, PCDHA1, and MUC5AC. The roles of each of these genes have been elucidated to some extant: ASTN1, is required for appropriate and timely migration of cerebellar granule cells 30; PCDHA1 belongs to the cadherin superfamily and mediate the formation and maintenance of specific synaptic connections 31; MUC5AC, whose expression of protein and mRNA is significantly decreased in gastric cancer tissue 32. On the other hand, the depletion of HOTAIR downregulates some genes implicated in tumor invasion and proliferation; NTM is expressed in fetal brain at higher levels than that in mature brain and is more highly expressed in nervous tumors than that in normal brain tissues 33; PTK2B mediates cell proliferation and invasiveness in HCC cells by upregulation of the c-Src and ERK/MAPK-signaling pathway, and is related to progression and metastasis in breast cancer, together with focal adhesion kinase 34,35. Genes with fold change values of 10 or more were including genes mainly relevant to neural development (ASTN1, PCDHA1, MUC5AC, and NTM). Our data suggest that HOTAIR mainly regulates the expression of genes related to neural development in SCLC cells.
Possible limitations of our analysis and interpretation are as follows: First, our analyses were based on surgically resectable cases representing a very minor population. In fact, several reports indicated that patients with surgically resectable SCLC had much better prognosis and showed 5-year survival rate 30–70% 36–38, and we were not able to show significantly different survival between the groups with low- and high-HOTAIR expression (Fig. 2B and C). However, if we only used material from inoperable cases, in other words, the majority of SCLC patients, almost all patients were of extended disease, and as a result, we had almost no patients with good prognosis. Certainly, we need more cases with good prognosis to obtain significant difference of prognosis between low- and high-HOTAIR groups. Second, we succeeded in RNAi experiments using only the SBC-3 cells. As mentioned above, the SBC-3 cell line was established from a bone metastasis and not from a primary site. It is known that HOTAIR expression in metastatic sites was higher than in primary sites for breast cancer 14. Possibly, high expression in the SBC-3 cells may be due to its metastatic nature, rather than its neuroendocrine nature. Further analysis using other cell lines is warranted although RNAi experiments will be a challenge.
Finally, although the knockdown efficacy was similar in #1–3 siHOTAIRs, suppression of cell proliferation and invasion were induced only by #1 siHOTAIR. This might be because the time to onset of gene silencing only by #1 siRNA was appropriate, and because the sequence conditions of the three siRNAs were different. In particular for the latter point, following four sequence conditions were suggested to give rise to highly effective RNAi in mammalian cells 39,40: (1) the 5′ antisense-strand (AS) end, A or U; (2) the 5′ sense-strand (SS) end, G or C; (3) the 5′-terminal one third of AS, A/U-rich; and (4) a long G/C stretch, absent from the 5′-terminal two thirds of SS. Considering these conditions as compared among #1–3 siHOTAIRs, only #1 siHOTAIR met the four conditions at the same time.
siHOTAIR may represent a new therapeutic target for SCLC, in particular, of its metastatic phase. Although several kinds of pharmaceutical agents are available for SCLC treatment, HOTAIR therapy may be useful after the development of multidrug resistance. However, we have to bear in mind that biological, pathological, and clinical evidence for SCLC on HOTAIR is very limited and that our knowledge of homeobox genes, the target genes of HOTAIR, is very sparse. Further analysis of HOTAIR expression in systemic tissues of both adults and fetuses followed by further cell line experiments and in vivo analyses on tissue effects by siRNA administration using a more number of siRNA will be required.
Acknowledgments
Parts of this study were supported financially by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan, grants from the Japan Society for the Promotion of Science, the Ministry of Health, Labour and Welfare, and the Princess Takamatsu Cancer Research Fund. We thank Mime Kobayashi and Tamiko Minamisawa, Division of Protein Engineering, The JFCR Cancer Institute, for their help in using fluorescence microscope, and Takeshi Fujiwara, The JFCR Genome Center, for his help in using web-based databases.
Conflict of Interest
Yuichi Ishikawa is supported by research grants from SONY corporation and Daiichi-Sankyo Co. Ltd.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Table S1. Primer sequences of RT-PCR for HOTAIR quantification as well as GAPDH and ACTB as controls. Also, primers for RNAi experiments are shown.
Table S2. Univariate and multivariate analysis of DSS and RFS/DFS.
Table S3.HOTAIR expression mainly associated with genes contributes to cell adhesion.
References
- 1.Field JK, Duffy SW. Lung cancer screening: the way forward. Br. J. Cancer. 2008;99:557–562. doi: 10.1038/sj.bjc.6604509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Puglisi M, Dolly S, Faria A, Myerson JS, Popat S, O'Brien ME. Treatment options for small cell lung cancer – do we have more choice? Br. J. Cancer. 2010;102:629–638. doi: 10.1038/sj.bjc.6605527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.D'Angelo SP, Pietanza MC. The molecular pathogenesis of small cell lung cancer. Cancer Biol. Ther. 2010;10:1–10. doi: 10.4161/cbt.10.1.12045. [DOI] [PubMed] [Google Scholar]
- 4.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 5.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 6.Tsai MC, Spitale RC, Chang HY. Long intergenic noncoding RNAs: New links in cancer progression. Cancer Res. 2011;71:3–7. doi: 10.1158/0008-5472.CAN-10-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pauli A, Rinn JL, Schier AF. Non-coding RNAs as regulators of embryogenesis. Nat. Rev. Genet. 2011;12:136–149. doi: 10.1038/nrg2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human Hox loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 11.Sun G, Alzayady K, Stewart R, Ye P, Yang S, Li W, et al. Histone demethylase LSD1 regulates neural stem cell proliferation. Mol. Cell. Biol. 2010;30:1997–2005. doi: 10.1128/MCB.01116-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sauvageau M, Sauvageau G. Polycomb group proteins: multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell. 2010;7:299–313. doi: 10.1016/j.stem.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alves CP, Fonseca AS, Muys BR, de Barros E, Lima Bueno R, Bürger MC. The lincRNA Hotair is required for epithelial-to-mesenchymal transition and stemness maintenance of cancer cells lines. Stem Cells. 2013;31:2827–2832. doi: 10.1002/stem.1547. [DOI] [PubMed] [Google Scholar]
- 14.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang Z, Zhou L, Wu LM, Lai MC, Xie HY, Zhang F, et al. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann. Surg. Oncol. 2011;18:1243–1250. doi: 10.1245/s10434-011-1581-y. [DOI] [PubMed] [Google Scholar]
- 16.Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 17.Li X, Wu Z, Mei Q, Li X, Guo M, Fu X, et al. Long non-coding RNA HOTAIR, a driver of malignancy, predicts negative prognosis and exhibits oncogenic activity in oesophageal squamous cell carcinoma. Br. J. Cancer. 2013;109:2266–2278. doi: 10.1038/bjc.2013.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang L, Zhang W, Su B, Yu B. Long noncoding RNA HOTAIR is associated with motility, invasion, and metastatic potential of metastatic melanoma. Biomed. Res. Int. 2013;2013:251098. doi: 10.1155/2013/251098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu ZY, Yu QM, Du YA, Yang LT, Dong RZ, Huang L, et al. Knockdown of long non-coding RNA HOTAIR suppresses tumor invasion and reverses epithelial-mesenchymal transition in gastric cancer. Int. J. Biol. Sci. 2013;9:587–597. doi: 10.7150/ijbs.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lv XB, Lian GY, Wang HR, Song E, Yao H, Wang MH. Long noncoding RNA HOTAIR is a prognostic marker for esophageal squamous cell carcinoma progression and survival. PLoS ONE. 2013;8:e63516. doi: 10.1371/journal.pone.0063516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nie Y, Liu X, Qu S, Song E, Zou H, Gong C. Long non-coding RNA HOTAIR is an independent prognostic marker for nasopharyngeal carcinoma progression and survival. Cancer Sci. 2013;104:458–464. doi: 10.1111/cas.12092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakagawa T, Endo H, Yokoyama M, Abe J, Tamai K, Tanaka N, et al. Large noncoding RNA HOTAIR enhances aggressive biological behavior and is associated with short disease-free survival in human non-small cell lung cancer. Biochem. Biophys. Res. Commun. 2013;436:319–324. doi: 10.1016/j.bbrc.2013.05.101. [DOI] [PubMed] [Google Scholar]
- 23.Travis WD, Brambilla E, Muller-Hermelink HK, Harris CC. Pathology and genetics: tumours of the lung, pleura, thymus and heart. Vol. 1. Lyon, France: IARC; 2004. pp. 31–34. [Google Scholar]
- 24.Nguewa PA, Agorreta J, Blanco D, Lozano MD, Gomez-Roman J, Sanchez BA, et al. Identification of importin 8 (IPO8) as the most accurate reference gene for the clinicopathological analysis of lung specimens. BMC Mol. Biol. 2008;9:103. doi: 10.1186/1471-2199-9-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Daniel VC, Marchionni L, Hierman JS, Rhodes JT, Devereux WL, Rudin CM, et al. A primary xenograft model of small-cell lung cancer reveals irreversible changes in gene expression imposed by culture in vitro. Cancer Res. 2009;69:3364–3373. doi: 10.1158/0008-5472.CAN-08-4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tiberio C, Barba P, Magli MC, Arvelo F, Le Chevalier T, Poupon MF, et al. Hox gene expression in human small-cell lung cancers xenografted into nude mice. Int. J. Cancer. 1994;58:608–615. doi: 10.1002/ijc.2910580426. [DOI] [PubMed] [Google Scholar]
- 27.Lechner JF, Fugaro JM, Wong Y, Pass HI, Harris CC, Belinsky SA. Perspective: cell differentiation theory may advance early detection of and therapy for lung cancer. Radiat. Res. 2001;155:235–238. doi: 10.1667/0033-7587(2001)155[0235:pcdtma]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 28.Cantile M, Franco R, Tschan A, Baumhoer D, Zlobec I, Schiavo G, et al. HoxD13 expression across 79 tumor tissue types. Int. J. Cancer. 2009;125:1532–1541. doi: 10.1002/ijc.24438. [DOI] [PubMed] [Google Scholar]
- 29.Grier DG, Thompson A, Kwasniewska A, McGonigle GJ, Halliday HL, Lappin TR. The pathophysiology of Hox genes and their role in cancer. J. Pathol. 2005;205:154–171. doi: 10.1002/path.1710. [DOI] [PubMed] [Google Scholar]
- 30.Adams NC, Tomoda T, Cooper M, Dietz G, Hatten ME. Mice that lack astrotactin have slowed neuronal migration. Development. 2002;129:965–972. doi: 10.1242/dev.129.4.965. [DOI] [PubMed] [Google Scholar]
- 31.Wu Q, Maniatis T. A striking organization of a large family of human neural cadherin-like cell adhesion genes. Cell. 1999;97:779–790. doi: 10.1016/s0092-8674(00)80789-8. [DOI] [PubMed] [Google Scholar]
- 32.Shi D, Qiu XM, Bao YF. Effects of Helicobacter pylori infection on MUC5AC protein expression in gastric cancer. Future Oncol. 2013;9:115–120. doi: 10.2217/fon.12.172. [DOI] [PubMed] [Google Scholar]
- 33.Liu J, Li G, Peng X, Liu B, Yin B, Tan X, et al. The cloning and preliminarily functional analysis of the human neurotrimin gene. Sci. China C Life Sci. 2004;47:158–164. doi: 10.1360/03yc0072. [DOI] [PubMed] [Google Scholar]
- 34.Sun CK, Man K, Ng KT, Ho JW, Lim ZX, Cheng Q, et al. Proline-rich tyrosine kinase 2 (Pyk2) promotes proliferation and invasiveness of hepatocellular carcinoma cells through c-Src/ERK activation. Carcinogenesis. 2008;29:2096–2105. doi: 10.1093/carcin/bgn203. [DOI] [PubMed] [Google Scholar]
- 35.Behmoaram E, Bijian K, Jie S, Xu Y, Darnel A, Bismar TA, et al. Focal adhesion kinase-related proline-rich tyrosine kinase 2 and focal adhesion kinase are co-overexpressed in early-stage and invasive ErbB-2-positive breast cancer and cooperate for breast cancer cell tumorigenesis and invasiveness. Am. J. Pathol. 2008;173:1540–1550. doi: 10.2353/ajpath.2008.080292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lim E, Belcher E, Yap YK, Nicholson AG, Goldstraw P. The role of surgery in the treatment of limited disease small cell lung cancer: time to reevaluate. J. Thorac. Oncol. 2008;3:1267–1271. doi: 10.1097/JTO.0b013e318189a860. [DOI] [PubMed] [Google Scholar]
- 37.Schreiber D, Rineer J, Weedon J, Vongtama D, Wortham A, Kim A, et al. Survival outcomes with use of surgery in limited-stage small cell lung cancer: should its role be re-evaluated? Cancer. 2010;116:1350–1357. doi: 10.1002/cncr.24853. [DOI] [PubMed] [Google Scholar]
- 38.Inoue M, Sawabata N, Okumura M. Surgical intervention for small-cell lung cancer: what is the surgical role? Gen. Thorac. Cardiovasc. Surg. 2012;60:401–405. doi: 10.1007/s11748-012-0072-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ui-Tei K, Naito Y, Takahashi F, Haraguchi T, Ohki-Hamazaki H, Juni A, et al. Guidelines for the selection of highly effective siRNA sequences for mammalian and chick RNA interference. Nucleic Acids Res. 2004;32:936–948. doi: 10.1093/nar/gkh247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ui-Tei K, Naito Y, Saigo K. Essential notes regarding the design of functional siRNAs for efficient mammalian RNAi. J. Biomed. Biotechnol. 2006;2006:65052. doi: 10.1155/JBB/2006/65052. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Primer sequences of RT-PCR for HOTAIR quantification as well as GAPDH and ACTB as controls. Also, primers for RNAi experiments are shown.
Table S2. Univariate and multivariate analysis of DSS and RFS/DFS.
Table S3.HOTAIR expression mainly associated with genes contributes to cell adhesion.


