Abstract
An unresolved question about GPCR function is the role of membrane components in receptor stability and activation. In particular, cholesterol is known to affect the function of membrane proteins, but the details of its effect on GPCRs are still elusive. Here, we describe how cholesterol modulates the behavior of the TM1-TM2-TM7-helix 8(H8) functional network that comprises the highly conserved NPxxY(x)5,6F motif, through specific interactions with the receptor. The inferences are based on the analysis of microsecond length molecular dynamics (MD) simulations of rhodopsin in an explicit membrane environment. Three regions on the rhodopsin exhibit the highest cholesterol density throughout the trajectory: the extracellular end of TM7, a location resembling the high-density sterol area from the electron microscopy data; the intracellular parts of TM1, TM2, and TM4, a region suggested as the cholesterol binding site in the recent X-ray crystallography data on β2-adrenergic GPCR; and the intracellular ends of TM2-TM3, a location that was categorized as the high cholesterol density area in multiple independent 100 ns MD simulations of the same system. We found that cholesterol primarily affects specific local perturbations of the helical TM domains such as the kinks in TM1, TM2, and TM7. These local distortions, in turn, relate to rigid-body motions of the TMs in the TM1-TM2-TM7-H8 bundle. The specificity of the effects stems from the nonuniform distribution of cholesterol around the protein. Through correlation analysis we connect local effects of cholesterol on structural perturbations with a regulatory role of cholesterol in the structural rearrangements involved in GPCR function.
Keywords: GPCR, membrane, atomistic simulations, signaling, allosteric behavior
INTRODUCTION
Cholesterol (Chol) is an essential component of mammalian cell membranes and is critical for membrane organization, dynamics, and function.1–5 Distributed nonuniformly between cellular organelles,3 compartments of mem-branes,4,6–10 and even between leaflets of the lipid bilayer,11 cholesterol not only influences biophysical properties of a fluid lipid matrix, but also is implicated in the stabilization and functioning of membrane proteins. Among these is the largest family of integral membrane proteins, G-protein-coupled receptors (GPCRs)12,13 that play crucial roles in modulating cellular response to a variety of physical or chemical stimuli. As a result, they are also of outstanding pharmaceutical interest. Sharing the seven-transmembrane (7-TM) segment design first observed for the light receptor, rhodopsin14–17 and more recently for β2-adrenergic (β2-AR) 18–20 and β1-adrenergic 21 receptors, GPCRs exist in a dynamic equilibrium between various inactive and active intermediate states, and become ligand-activated by switching between these states.22–28 Once in the active form, GPCRs interact with their G protein partners, triggering events that ultimately lead to a signaling cascade.23
Cholesterol has long been suspected to regulate GPCR activity, and the recent suggestion of special cholesterol-enriched membrane compartments that selectively partition certain membrane proteins while excluding others, has unambiguously established Chol as a major effector in stabilization and functioning of GPCRs. The consequences of Chol-receptor interactions have been explored extensively for rhodopsin.29–33 An increased amount of Chol in the retinal rod cells has been shown to stabilize the inactive state of rhodopsin in those cells, resulting in reduced signaling efficiency. In a similar fashion, depletion of cholesterol from the membranes of neonatal cardiac myocytes altered the signaling behavior of rhodopsin-like β2-AR receptor34,35: in Chol-depleted cells β2-AR coupled more strongly to a partnering G protein, compared to untreated cells. Further, cholesterol has been implicated in regulating signaling of the receptors for oxytocin, serotonin1A, and galanin.12,13,36–38
The structural changes associated with GPCR activation have been characterized as functional microdomains (FMs) that are conserved across the GPCR family (see for example, Refs. 22, 28 and 39). These FMs work as modular units that respond to changes triggered in the receptor conformation activation signals and operate as activation switches throughout the structure.40,41 The local conformational rearrangements associated with GPCR activation involve various structural perturbations in the transmembrane helices (TMs) such as bending and twisting motions enabled by specialized features such as proline kinks or glycine pairs.42 Interestingly, these elements of the intramolecular signaling path can be modulated even in the absence of a ligand, for instance by point mutations resulting in constitutively active mutant constructs.43–45 Cholesterol can affect these functionally important modes of motion in GPCRs, either through direct interactions with proteins 13,46or indirectly, through its ability to alter the biophysical properties of the lipid bilayer,4 but the details remain elusive.
The interest in the significance of specific Chol-membrane protein interactions has been rekindled recently by several structural studies showing cholesterol interacting with GPCRs. Using electron microscopy to explore dimeric arrangements of rhodopsin and its photointermediate product metarhodopsin I (meta-I), Schertler and coworkers 47,48 identified a cloud from a single cholesterol molecule positioned between antiparallel dimers of rhodopsin; the Chol is situated between the extracellular (EC) part of TM6-TM7 of one rhodopsin molecule and the intracellular (IC) part of TM4 of a neighboring GPCR molecule. More recently, the X-ray crystallographic structure of the β2-AR receptor obtained by Kobilkas’ group18 associates three cholesterol molecules with the β2-AR receptor monomer. One Chol is positioned near the crystallographic dimeric interface at TM1-helix 8 (H8), and the other two are situated on the intracellular (IC) side of the TM bundle near TM1, TM2, TM3, and TM4 (TM1-TM4 region). The latter cloud of cholesterol has been observed in the latest different crystal structure of β2-AR as well.49 Because the cholesterol molecules in the TM1-TM4 region appear not to be involved in the crystal packing, it has been argued that this region on the receptor is a specific cholesterol-binding site of the β2-AR GPCR.49
These experimental observations offered strong evidence about GPCR-Chol interactions, but it is still unclear what role they play in stabilizing the receptor and regulating its function. Molecular dynamics (MD) simulations have proven to be an invaluable way to clarify such issues and the results have significantly improved our understanding about dynamics in GPCR-membrane systems, 50–57 and how different membrane components may influence the stability of the GPCR.50–58 In particular, the packing of physiologically important polyunsaturated docosahexaenoic acid (DHA) lipid chains and cholesterol has been explored in a series of independent 100-ns MD simulations of rhodopsin in 2:2:1 1-stearoyl-2-docosahexaenoyl-sn-Glycero-3-phosphocholine (SDPC) lipid/ 1-stearoyl-2-docosahexaenoyl-sn-Glycero-3-phosphoethanolamine (SDPE) lipid/cholesterol membrane.55 Unlike Chol, the DHA fatty acids are known to stabilize the active state of the rhodopsin and enhance the kinetics of the photocycle.31,32 These modeling studies revealed that overall packing of cholesterol against the rhodopsin was somewhat weaker compared to the tighter arrangement of DHA chains around the protein. Thus, the results suggested that DHA and not cholesterol was involved in the strongest direct interactions with the protein. Still, several groups of residues that preferentially interact with tightly packed cholesterol molecules were identified, most notably the amino acid sequence 3.20–3.27 at the extracellular (EC) side of TM3.
These MD simulations improved our knowledge about mechanisms of the GPCR-lipid interactions, but their relatively short time-scales cannot be expected to reveal the consequences of the observed packing of different membrane components on the local structural perturbations of the helices, and on the dynamics of FMs. In general, short time-scale limitations inherent to the common simulations of the atomistic models (≤100 ns) have not allowed the exploration of detailed modulatory effects of specific lipid-receptor interactions that require long equilibration times. Because the local motions associated with these FMs are critical to the intramolecular signaling cascade, for receptor stability, and for partitioning among various conformational states related to function, it is essential to identify their relation to the membrane environment. Such molecular-level details require long-time dynamics simulations of the receptor-membrane system. To this end, we have evaluated the dynamic and structural characteristics of cholesterol-protein interactions from the trajectory of a fully atomistic 1.6 µs MD simulations of rhodopsin embedded in 2:2:1 SDPC/SDPE/ Cholesterol membrane. We describe the findings indicating that cholesterol, through specific interactions with the receptor, modulates the behavior of a critical FM— the IC side of TM1-TM2-TM7-H8 network. This region includes the highly conserved NPxxY(x)5,6F sequence connecting the TM7 with the juxtamembrane helix, H8. The FM has been extensively studied by means of mutagenesis,59–62 computational simulation,63 crystallography,14–17 metal ion site engineering,64 and other bio-physical techniques.65–67 The results have revealed the crucial role of the interactions in the NPxxY(x)5,6F motif for the activation process.63 Further, significant rigid-body motions on the IC end of TM1-TM2-TM7-H8 bundle upon rhodopsin activation have been observed 65–65 —including movement of the IC portion of TM7 away from TM1 by 2–4 Å,65 and outward displacement of TM2 by 3 Å relative to H8.66 The local dynamic changes in this region are so distinctive that they have been proposed as specific markers of the state of activation of GPCR structures.63
Results from the simulations presented in this work indicate that the dynamics of cholesterol around the rhodopsin relate to the activation elements in TM1-TM2-TM7-H8 bundle of the receptor, namely, to the relative angular motion between TM7 and H8, and to relative movement of TM1 and TM7. Interestingly, Chol influences these rigid-body motions indirectly, through the helical kink parameters in TM1, TM2, and TM7. In particular, we find that the presence of cholesterol near Val1.58, Tyr2.63, and Pro7.38 residues correlates with changes in the kink angle values in the TM1, TM2, and TM7, respectively. Local distortions in TM1 and TM2, in turn, are linked to the shortening of the distances between TM1 and TM7 and between TM7 and H8.
Based on these findings, we conclude that cholesterol molecules at the extracellular end of the TM2-TM3 bundle and at the intracellular side of the TM1-TM2-TM4 network stabilize TM1-TM2-TM7-H8 functional microdomain of rhodopsin through direct interactions with the receptor.
METHODS
MD simulation details
The 1.6 µs MD simulations of a rhodopsin molecule (PDB code 1U1968,69) embedded in a lipid bilayer of 49 SDPC lipid, 50 SDPE lipid, and 24 cholesterol (Chol) molecules hydrated by 7400 TIP3 waters, 14 Na, and 16 Cl ions, involved a total of 43,222 atoms. Details of the system setup and equilibration have been reported elsewhere.55,56,58 In short, simulations were performed in the dark state at 310 K temperature in the NVE ensemble using the Program BlueMatter,70 with a rectilinear 56.5 × 79.2 × 95.5434 Å period box. The CHARMM27 force field was used for the protein,71 and refined CHARMM parameters for saturated chain,72 polyunsaturated chain,73 and cholesterol 74 were utilized to describe membrane components. Construction and equilibration were performed under CHARMM version 27,75 as detailed elsewhere.55,56 Long-range electrostatic interactions were calculated using the particle-particle-particle-mesh Ewald summation technique,76,77 with a 1283 grid for the fast Fourier transform, a charge-interpolation distance of 4 mesh points, and the Ewald a value set to 0.35 Å−1 .Real-space electrostatics and repulsion-dispersion were smoothly truncated at 10 Å. All bonds containing hydrogen were constrained to their equilibrium values using the RATTLE algorithm,78 allowing us to run dynamics with a 2 fs time-step using the velocity Verlet integrator79
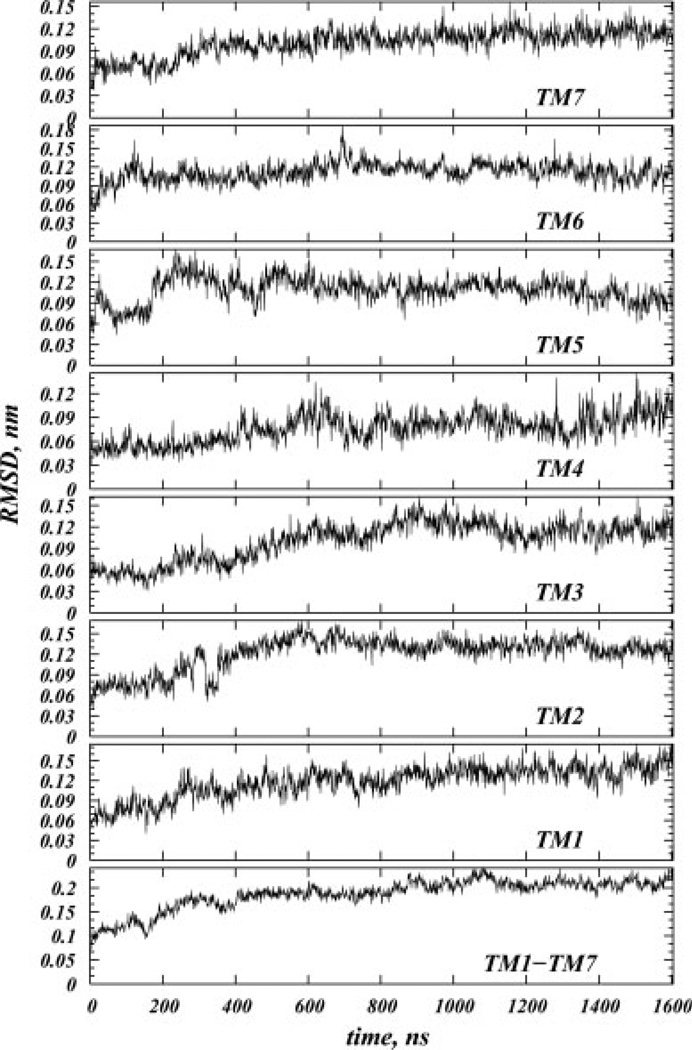
The root-mean-square deviations (RMSDs) versus time for the Cα atoms of the entire TM bundle of the protein relative to its initial X-ray crystallographic structure, as well as RMSDs for the individual helices are shown in Figure 1. The system appears to reach steady state at ~600 ns, when RMSD of the TM bundle reaches a plateau value of ~0.2 nm. Similar pattern is observed for the individual helices.
Figure 1.
Root-mean-square deviation (RMSD) versus time for the Cα atoms of the transmembrane helical bundle (TM1-TM7) and for the Cα atoms of individual TM helices.
Rhodopsin residue numbering and calculation of helical kink parameters
The GPCR residue numbering scheme follows the generic numbering system defined by Ballesteros and Weinstein. According to this scheme, the most conserved residue in each TM is assigned a number 50, and then a pair of numbers (A1.A2) are used to describe amino acid residues in TMs. A1 refers to the TM number and A2 denotes position of the amino acid relative to the most conserved residue in the TM, with numbers decreasing toward the N-terminus and increasing toward the C-terminus.
To describe local distortions on TMs during the MD simulation caused by proline or consecutive glycine residues,42 we calculated bend and face shift angles around the following residues on TMs: Pro1.48, Gly2.56, Gly3.35, Pro4.59, Pro5.50, Pro6.50, and two Pro7.38 and Pro7.50 residues on TM7. The bend angle, which measures the extent of helical kink, is defined as the angle between pre-kink and post-kink parts in a TM, and the face shift angle describes the distortion that causes helix to twist in such a way that amino acids previously facing a same side of the helix are now shifted and positioned on different sides of the TM81.
To quantify the changes in the helix distortion parameters throughout the MD trajectory, we used the ProKink package 81 in the publicly available software Simulaid.82 Details about the geometric definitions and the computational protocol implemented in ProKink can be found in Ref. 81. In short, rhodopsin snapshots at different time-points were fitted onto the starting reference structure of the protein, and the Cα atom of the Pro residue in the relevant TM was positioned at the cartesian origin. To calculate the bend angle of a helix, for each trajectory frame the coordinate system was rotated around the axis passing through the preproline helical segment until the long axes of the post and preproline parts were in the same plane. From this orientation, the bend or kink angle was measured as the angle between the axes of the two parts of the helix. To obtain the face shift angle, for each snapshot the postproline segment which included Pro Cα atom was rotated so that both pre and postproline helical parts shared a long axis.81 The face shift angle was then calculated as the angle between projections of two vectors onto the plane perpendicular to the long axis: these vectors are the on connecting the Pro Cα atom with the cartesian origin, and the average vector connecting the Cα atoms of the (i-3) and (i-4) amino acids with the origin.
Analysis of spatial distribution of cholesterol around rhodopsin
The spatial distribution of cholesterol around rhodopsin in the MD simulations was calculated as the 3D spatial distribution function (SDF) of Chol oxygen atoms O1 around the protein (see Fig. 1). The SDF is defined as74,83–86:
where N(r,θ,φ) is the number of O1 atoms in the volume element ΔV = r2 sinθΔrΔθΔφ, ρ is the number density of O1, taken as the ratio of number of O1 atoms to the volume of the simulation box, and Nframes denotes number of trajectory snapshots analyzed.
To calculate the SDF, the unit cell was divided into small volume elements of Δr = 1 Å, Δθ = 1°, and Δφ = 1° for each trajectory frame, and rhodopsin snapshots at different time-points were aligned against the starting reference structure of the protein. Å histogram of O1 atoms was then collected over Nframes snapshots, and the SDF was obtained from Eq. (1).
In principle, the SDF may depend on the choice of atoms (or group of atoms) used to build the histogram. To test the sensitivity of our results on atom selection, we performed the SDF calculations using center-of-mass of each cholesterol molecule rather than O1 atoms, and obtained results similar to that when only O1 atoms were used.
Correlation analysis
To quantify concerted motions in rhodopsin during dynamics, we performed correlation analysis on the time-evolution data of different variables, with special emphasis on the helical kink parameters and the TM1-TM2-TM7-H8 bundle. To this end, we followed the time-sequence of m = 10 selected variables that included proline kink and face-shift angles in TM1, TM2, TM6, and TM7, the distance between TM1 and TM7 both from the intracelluar (d{TM1-TM7 IC}) and the extracellular (d{TM1-TM7 EC}) sides, the angle between TM7 and H8 (α{TM7-H8}), and the distance between Cys7.63 and Leu2.39 residues (d{Cys7.63-Leu2.39}).
We first studied pair-wise correlations between all the variables, constructing the matrix of coefficients of determination, R2 using Spearman’s rank correlation test (see for instance Ref. 87). In this method, given Nframes pairs of observations, (xi, yi), first the xi and yi values separately are assigned a rank, and then the corresponding difference, di between the xi and yi ranks is found for each pair. The R2 is then defined as:
Because it uses rankings, Spearman’s method eliminates the sensitivity of the correlation test to the function linking the pairs of values and thus is preferred over parametric tests when no α priori knowledge exists on the functional relationship between xi and yi pairs.
After establishing pairwise correlations, we grouped variables with similar R2 values into clusters, with the purpose of identifying a (larger) set of dynamic quantities that evolved during MD simulation in a similar fashion. To this end, we used hierarchical cluster analysis87 on the m × m matrix of R2 values. In this technique the “distance” between two samples in m-dimensional space is defined based on the difference between their R2 values; samples with the shortest distance (thus the most similar ones), are grouped in the same limb of a tree. Groups of samples that are distinctly different are placed in other limbs. Created in such a manner, the resulting cluster dendrogram reveals concerted motions in rhodopsin and describes allosteric features of the protein during dynamics.
For all the statistics analyses we used open source software R.87 The level of confidence for calculated R2 values was assessed by measuring corresponding P-values.87 All the correlations discussed in this work were found to be within 95% confidence interval.
RESULTS AND DISCUSSION
We start by describing the distribution of cholesterol molecules around rhodopsin and identify three regions on the protein where Chol maintains close contact during microsecond dynamics. We then detail how cholesterol at these locations modulates the helical kinks on TM1, TM2, and TM7 that are involved in the rigid-body movements associated with the activation pathway in the TM1-TM2-TM7-H8 bundle.
Long time-scale dynamics of cholesterol around rhodopsin
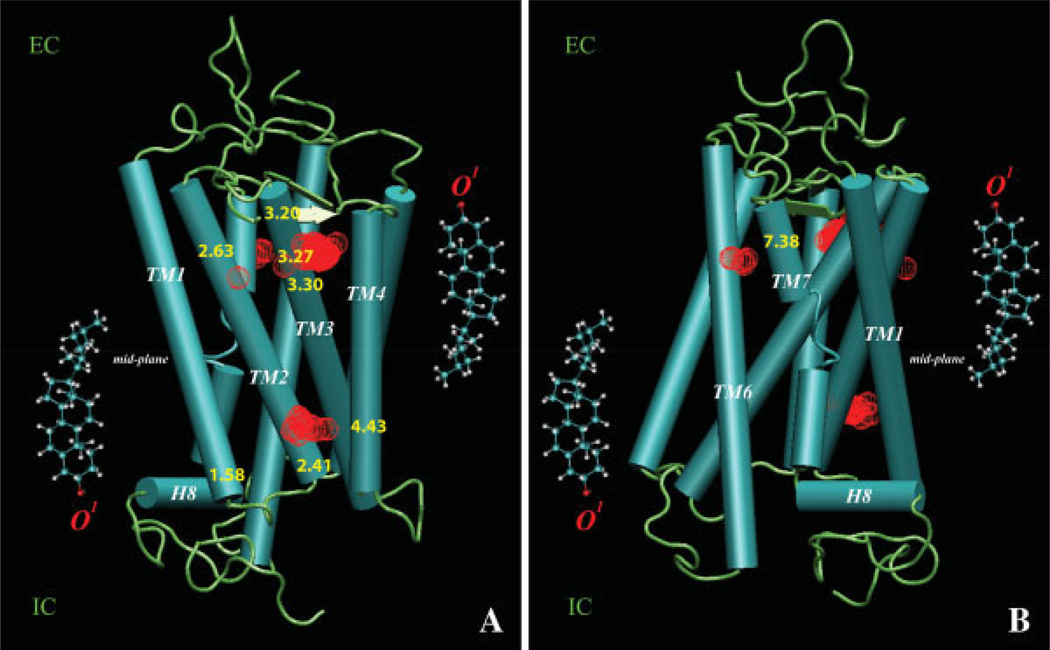
Figure 2 illustrates the average spatial distribution of cholesterol around rhodopsin during the 1.6 µs trajectory, and the positions that exhibit highest density of cholesterols. The high-density regions of Chol O1 atoms (red spheres) were identified by the locations of the 3D SDF peaks and are shown in Figure 2 superimposed on the rhodopsin structure at t = 0 ns (rendered as cartoon).
Figure 2.
Spatial distribution of cholesterol around rhodopsin: Two views (A, B) of the high-density regions of Chol O1 atoms indicated by red mesh, revealed through the locations of the 3D SDF peaks and superimposed on the rhodopsin structure at t = 0 ns. Panel A highlights densities around TM1, TM2, TM3, and TM4; Panel B shows densities close to position Pro7.38 of TM7. For clarity, both panels also show the locations of cholesterol molecules on two leaflets of a typical lipid membrane with respect to the bilayer mid-plane (shown in horizontal line); the position of Chol O1 atoms used in the SDF calculations is indicated in red. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com]
Figure 2 reveals that during the microsecond time-scale dynamics the distribution of cholesterol is not uniform around the protein and that more Chol associates with rhodopsin at the extracellular end. In particular, our results identify three regions of the protein that are extensively involved in interactions with Chol.
Cholesterol at the extracellular ends of TM2-TM3
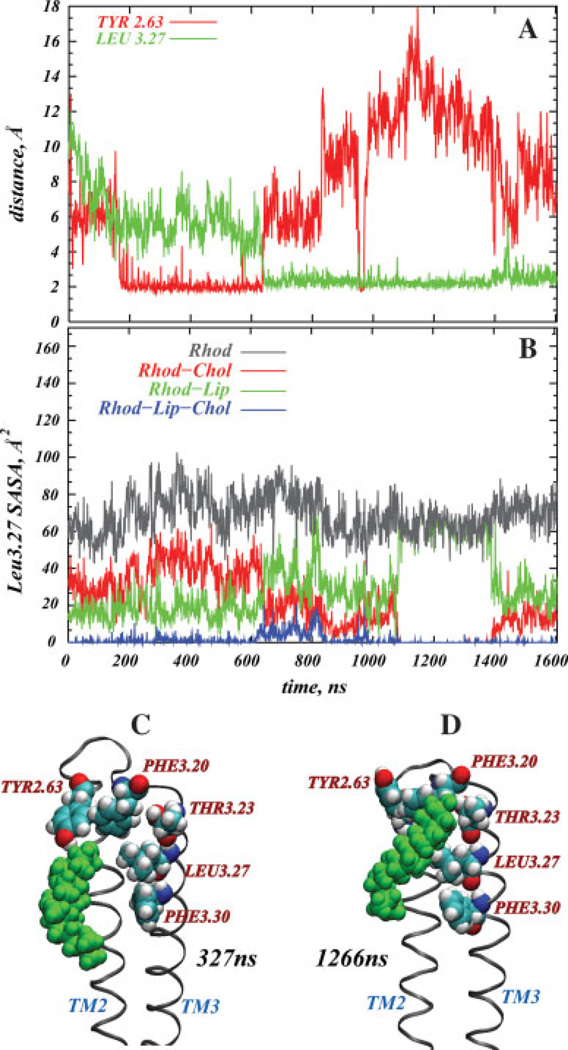
A single cholesterol molecule populates the high density area at the EC sides of TM2 and TM3, in the proximity of Tyr2.63 and near Phe3.30, Leu3.27, Thr3.23, and Phe3.20 of TM3 [Fig. 1(A)]. Interestingly, this region has been identified as a high-sterol area in series of independent 100-ns MD simulations of the same system.55 Figure 3 shows the time-evolution of the distance from Tyr2.63 and Leu3.27 to the nearest cholesterol molecule (panel A), and the time-sequence of the solvent-accessible surface area (SASA)88 of Leu3.27 (panel B). The two snapshots in Figure 3 depict cholesterol around the TM2-TM3 bundle at the 327 ns (C) and 1266 ns (D) time-points. The SASA calculations were done with naccess 2.1.1.89 To assess how different membrane components affect solvent accessibility of Leu3.27, we calculated SASA first considering only protein molecular surface [gray in Fig. 3(B)]; then, we repeat calculations for the molecular surfaces of the protein and all the lipids (green), the protein and all the cholesterols (red), and the total SASA (blue), taking into consideration the molecular surfaces of rhodopsin and all the lipid membrane components, that is, SDPC, SDPE, and cholesterol [Fig. 3(B)].
Figure 3.
Cholesterol at the extracellular (EC) ends of TM2 and TM3. Panel A shows the time-evolution of the minimum distance from Tyr2.63 and Leu3.27 to the nearest cholesterol molecule measured as the distances between the nearest atom pair; Panel B shows the time-sequence of the solvent-accessible surface area (SASA) of the Leu3.27 residue. The SASA is calculated in several ways, that is, considering only protein molecular surface (gray), the molecular surfaces of the protein and all the lipids (green), the molecular surfaces of the protein and all the cholesterols (red), and the total SASA (blue), obtained by taking into consideration molecular surfaces of the rhodopsin and all the lipid membrane components, that is, SDPC, SDPE, and cholesterol. Panels C and D depict the positions of cholesterol (in green) near the TM2-TM3 bundle at 327 and 1266 ns time-points, respectively. Key residues on the protein are shown in space-fill. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com]
Figure 3(A) reveals that after first 200 ns of simulations, cholesterol enters the proximity of Tyr2.63 and forms a complex [see Fig. 3(C)] that persists for more than 400 ns, until cholesterol moves away from Tyr2.63 and engages in strong interactions with residues on TM3 (compare panels A and D). In particular, Chol interacts with Phe3.20 and Thr3.23 through its polar hydroxyl group, and at the same time with Leu3.27 and Phe3.30 through its ring and tail atoms. This contact with TM3 continues for the last ~1 µs of the trajectory. Near the 970 ns time-point, the Chol is equidistant from Tyr2.63 and Leu3.27, and during the 1.4–1.45 µs interval, it moves away somewhat from Leu3.27 and toward Tyr2.63.
To test if the cholesterol is engaged in hydrogen bonding interactions with any of the residues in this area, we performed additional quantitative analysis on Chol OH group orientation around hydrogen donor and acceptor moieties on Tyr2.63 and Thr3.23, the residues closest to cholesterol hydroxyl during the simulation. The interaction energies between Chol OH and Tyr2.63 and Thr3.23 were calculated (data not shown). We found that as Chol enters the proximity of Tyr2.63 [200–600 ns interval, see Fig. 3(A)], cholesterol O1 atom orients itself at ~160° angle with respect to Tyr2.63 hydroxyl group, and we calculate the interaction energy between Chol OH and Tyr2.63 to be ~12 kJ/mol. Notably, the angle and energy values track the distance plot [Fig. 3(A)] faithfully which suggests the direct involvment of hydrogen bond in the proximity of Chol to rhodopsin. We observe analogous behavior of the same cholesterol molecule between 1.0 and 1.6 µs interval around Thr3.23 residue. Thus, we conclude that the Chol O1 atom is involved in hydrogen bonding interactions with Tyr2.63 residue during 200–600 ns time-span and with Thr3.23 residue between 1.0 and 1.6 µs interval.
Remarkably, when Chol engages residues on TM3, it also brings the bulky Tyr2.63 and Phe3.20 side-chains into closer proximity to each other [see Fig. 3(D)]. Strong aromatic-aromatic interaction between Tyr2.63 and Phe3.20, along with cholesterol forming dynamic complexes with Tyr2.63, Phe3.30, Leu3.27, Thr3.23, and Phe3.20, result in formation of a small pocket for a single Chol molecule. This pocket remains populated by cholesterol during the microsecond dynamics, as the bulky side-chains of Tyr and Phe restrict cholesterol’s motion to the area of the cavity.
This point is further illustrated in Figure 3(B), which details the effects of different membrane components on the solvent accessibility of Leu3.27, and reveals that the total SASA of Leu3.27 (Rhod-Lip-Chol in panel B) is low during the entire simulation. The plots illustrate that the SASA of Leu3.27 calculated only for the molecular surface of the protein (Rhod) is distinctly higher than the total SASA, indicating that Leu3.27 is in contact with lipids and cholesterol throughout the trajectory. During the first ~600 ns, Leu3.27 solvent accessibility is limited mostly by lipids, as the SASA profile for rhodopsin and lipid molecular surfaces (Rhod-Lip) is lower than the SASA obtained for the protein and cholesterol molecular surfaces (Rhod-Chol). But, in the second part of the simulation, as the Chol starts interacting with TM3 residues, the cholesterol influence on the SASA becomes stronger than that of lipids, as the Rhod-Chol curve on Figure 3(B) falls below Rhod-Lip graph at ~600 ns time-point. Thus, from 600 ns onwards cholesterol displaces nearby SDPC and/or SDPE lipids from the vicinity of Leu3.27. Furthermore, panel B reveals that the SASA profiles for Rhod-Lip-Chol and Rhod-Chol coincide with each other during 1.1–1.4 µs time interval, and the solvent accessibilities measured for Rho and Rhod-Lip are identical within the same time-span. From these observations we infer that the cholesterol completely fills up the pocket during the 1.1–1.4 µs time period.
Cholesterol around the intracellular ends of TM1-TM2-TM4: comparison to the crystallographic structure of the β2-adrenergic receptor
In the second high-density region identified from the SDF analysis, a single Chol is situated near the IC ends of TM1, TM2, and TM4 [Fig. 2(A)]. Interestingly, this high-density area resembles the cholesterol concentration detected in recent X-ray crystallographic structures on β2-AR GPCR.18,49 These studies have identified two Chol molecules at the cytoplasmic end, inside the groove formed by TM1, TM2, TM3, and TM4 of β2-AR, and suggested that the presence of Chol in this area stabilizes the receptor primarily by enhancing helical packing at the IC end of the TM1-TM2-TM3-TM4 bundle, and by increasing the thermal stability of the protein.49
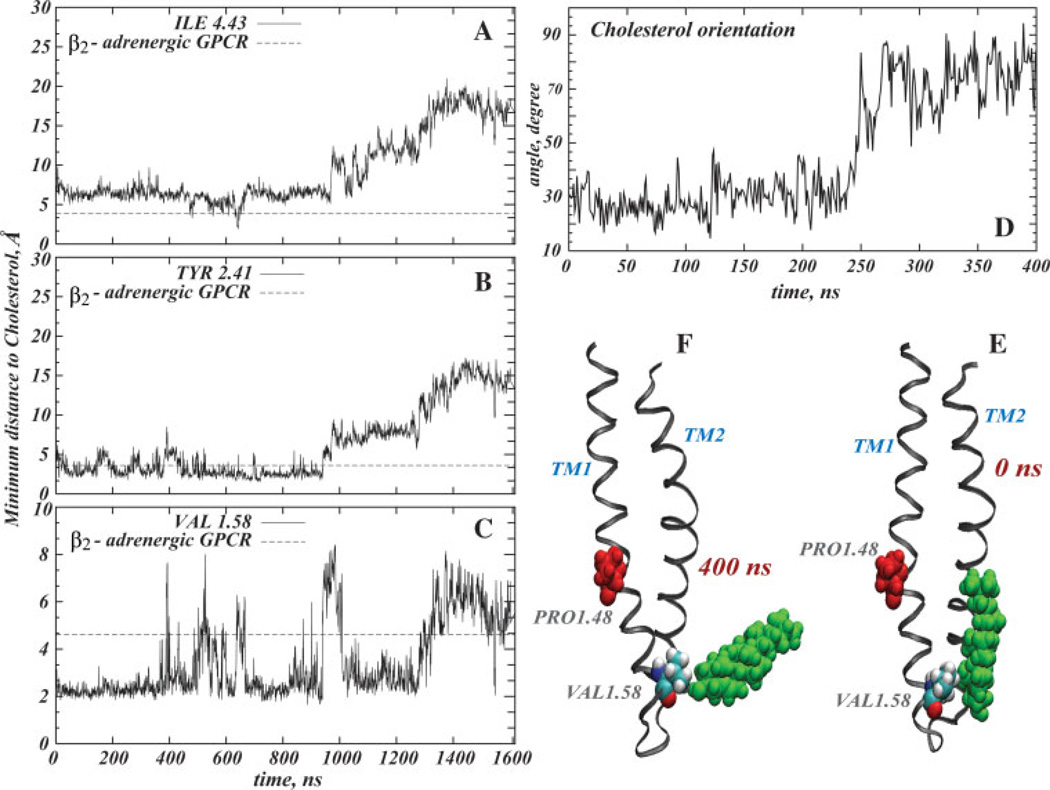
Figure 4 shows the time-evolution of the distance from Val1.58, Tyr2.41, and Ile4.43 residues to the nearest Chol molecule. These three residues correspond to the positions closest to the cholesterol molecules in the (β2-adrenergic GPCR X-ray crystallographic structure,18 and the dotted line indicates the corresponding crystallographic distances. Panel D shows the change in the orientation of the cholesterol with respect to TM1 during the first 400 ns of dynamics, expressed as the angle between cholesterol ring plane and the vector describing the direction of the cytoplasmic segment of TM1. Also shown in Figure 4 are two snapshots, at t = 0 ns (E) and t = 400 ns (F), of cholesterol near the IC ends of TM1-TM2.
Figure 4.
Cholesterol at the intracellular (IC) ends of TM1, TM2, and TM4. Minimum distances from Ile4.43 (A), Tyr2.41 (B), and Val1.58 (C) residues to the nearest cholesterol (see definition in Fig. 3) are plotted as a function of time in the simulation. For comparison, the distances from the corresponding residues of the β2-adrenergic receptor to nearest cholesterol in the X-ray crystallographic structure,18 are indicated by a dotted line. Panel D plots the change in the orientation of the cholesterol with respect to TM1 during the first 400 ns of simulations. Two snapshots, at t = 0 ns (E) and t = 400 ns (F), show cholesterol (in green) near the IC ends of TM1-TM2. The Val1.58 residue is depicted in space-fill, and Pro1.48 is rendered in red. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com]
Panels A–C in Figure 4 show that during the first microsecond of dynamics the distances from Val1.58, Tyr2.41, and Ile4.43 to the nearest Chol are in a good agreement with the crystallographic data for the (β2-adrenergic GPCR. Later, cholesterol moves away from TM2 and TM4, but still maintains contact with TM1. Divergence from TM2 and TM4 helices at the latter stages of the trajectory can be explained by the finding that the interaction of the Chol with Val1.58 is stronger than with any other residue in this area. Hence, on average the cholesterol spends more time around IC end of TM1.
Interestingly, in the initial ~200–400 ns time interval Chol tilts away from TM1 [see Fig. 4(D–F)] and pulls Val1.58 with it. This motion results in tilting of the IC segment of TM1 around the Pro1.48 kink, as seen from Figure 4(F) and will be discussed further below, in relation to the proline kink distortions.
Note, that Ref. 18 reports on a third Chol molecule in the vicinity of the IC end of TM7 and H8. On the microsecond scales of the MD simulations, we did not find any cholesterol molecule near TM7 and H8. Thus, it is reasonable to suspect that this cholesterol is involved in stabilizing the observed crystallographic dimeric arrangement of β2-AR receptors.
Cholesterol interacting with Pro7.38
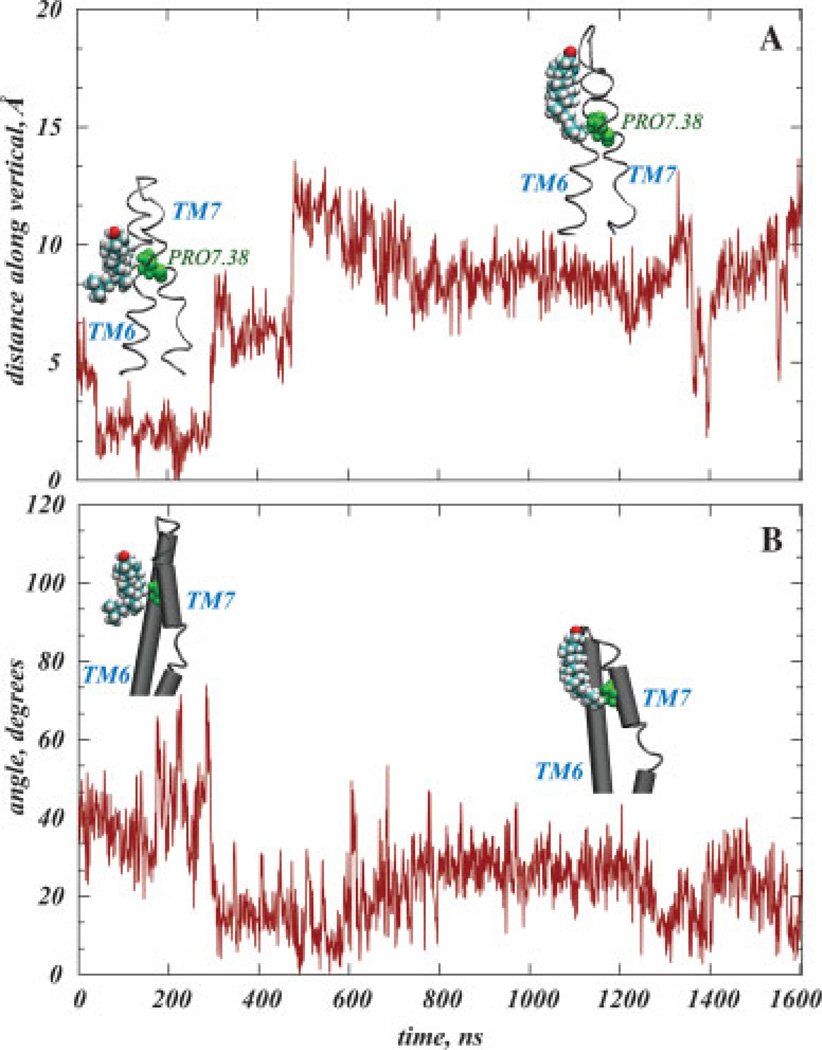
The third area of cholesterol high-density identified from the SDF calculations is at the EC end of TM7, near the Pro7.38 residue [Fig. 2(B)]. Notably, this region corresponds to the a sterol cloud detected in the electron microscopy experiments that position Chol around EC parts of TM6-TM7.47,48 Figure 5 shows the dynamics of cholesterol inside this area in the simulations; panel A plots the change with time of the relative position of Pro7.38 and the nearest cholesterol molecule calculated from differences in the locations along the membrane normal of Chol O and Pro7.38 Cα atoms. Panel B shows how the relative orientation of cholesterol changes with respect to the EC fragment of TM7 during simulation, computed from the angle between the cholesterol ring plane and the vector directed along the EC segment of TM7.
Figure 5.
Cholesterol interacting with TM7 Pro7.38. The time-sequence in panel (A) is for the z-directional distance between the cholesterol oxygen and the Cα atom of Pro7.38. Panel (B) shows the angle between the cholesterol ring and the axis of the extracellular segment of TM7 as a function of trajectory time. Cholesterol is shown in space-fill, and Pro7.38 in green. The TM6-TM7 bundle of rhodopsin is shown as ribbons (A), or as cylinders (B). [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com]
In the initial stages of the simulation, cholesterol is situated with its polar group close to Pro7.38 [Fig. 5(A)], and appears to be tilted along the vertical. After ~300 ns, Chol moves in the direction of the hydrophilic interface, closer to the extracellular loop (ECL) 3 that connects TM6 and TM7, but maintains contact with the Pro7.38 residue through its carbon tail atoms. In this process, the cholesterol tilt changes so that its ring becomes nearly parallel to the extracellular segment of TM7 [Fig. 5(B)].
Relation to proline kink distortions in the TM helices
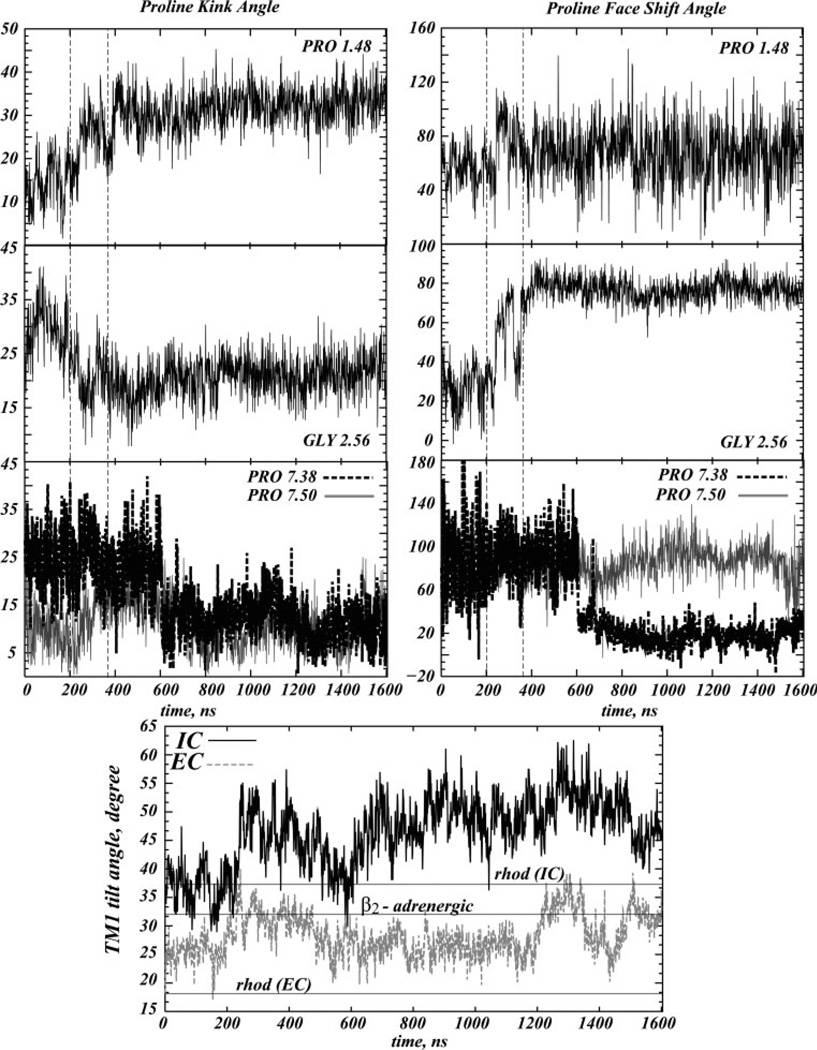
Figure 6 shows the time-sequences of changes in the bend angle and face shift parameters of the proline kinks81 in TM1, TM2, and TM7, and also details the dynamics of the tilt angle with respect to the membrane normal at the IC and the EC segments of TM1 (bottom panel). For comparison, the bottom panel shows the values of the IC and the EC segment tilts in the TM1 from the X-ray crystallographic structure of rhodopsin68,69 (horizontal lines at 37° and 18°), as well as the overall tilt in the TM1 helix from the crystallographic structure of β2-AR GPCR18 (horizontal line at 32°). Note that, unlike rhodopsin, β2-AR does not contain Pro in TM1. Thus, this helix in β2-AR is not distorted due to kinks. Consequently, its orientation in Figure 6 is described by a single value representing the tilt of the entire TM1 helix with respect to the vertical.
Figure 6.
Helix kink parameters for TM1 (calculated around Pro1.48), TM2 (around Gly2.56), and TM7 (around Pro7.38 and Pro7.50), and dynamics in the tilt angle of the IC and the EC segments of TM1 (bottom panel). For comparison, the bottom panel shows the values of the IC and the EC segment tilts in the TM1 from the X-ray crystallographic structure of rhodopsin68,69 (horizontal lines at 37° and 18°), as well as the overall tilt in the TM1 helix from the crystallographic structure of β2-AR GPCRcherezov18 (horizontal line at 32°).
Figure 6 reveals major interrelated changes in kink distortions in TM1 and TM2 helices occurring in the ~200–400 ns interval. Thus, in this time interval the bend angle in TM1 increased by ~20°, whereas the bend angle in TM2 diminished approximately to the same extent, and the face shift angle at Gly2.56 increased by ~60°. Interestingly, these structural perturbations in TM1 and TM2 remain unchanged from 400 ns onwards.
Notably, comparison of the data from Figure 3 to the time-evolution of the kink parameters in TM2 reveals that the bend and face-shift angles around Gly2.56 start to change when the cholesterol at the EC sides of TM2-TM3 engages in interactions with Tyr2.63, around 200 ns into the trajectory. During the next 1.4 is the Chol not only establishes strong contacts with Tyr2.63, Phe3.30, Leu3.27, Thr3.23, and Phe3.20 residues, but also stabilizes the Tyr2.63-Phe3.20 complex, as described earlier. Strong Tyr2.63-Phe3.20 and Tyr2.63-Chol interactions result in motion of the EC segment of TM2 toward TM3 (data not shown). As a consequence, the TM2 helix kinks and twists around the Gly2.56-Gly2.57 residue pair (Fig. 6). These findings suggest that the helical distortions on TM2 are modulated by cholesterol interactions with the Tyr2.63, Phe3.30, Leu3,27, Thr3.23, and Phe3.20 residues.
Comparison of the results from Figure 4 to the perturbations in TM1 suggests that the dynamics of Chol around the IC ends of TM1-TM2-TM4 relate to the observed change in kink angle on TM1. Thus, the Chol near the IC side of TM1 tilts and pulls the neighboring Val1.58 residue away from the helical bundle during the same time-interval in which the higher proline kink value is established in TM1, that is, between 200 and 400 ns [compare Figs. 4(D) and 6].
We note that around 600 ns time-point the system fluctuates through a state in which the IC and the EC ends of TM1 have similar tilts of ~32° away from the vertical axis (Fig. 6, bottom panel), a value that coincides with the overall tilt of the TM1 helix in the β2-AR X-ray structure (horizontal line). Remarkably, it is the same time-interval, around 600 ns, when the relative cholesterol positioning from Val1.58, Tyr2.41, and Ile4.43 in the dynamic trajectory correlates the best with the corresponding crystallographic distances in β2-AR, as described in Figure 4, further supporting a relationship between the position of cholesterol around the IC ends of TM1-TM2-TM4 bundle and local distortions in the TM1.
Interestingly, the magnitude and overall behavior of the kink distortions in TM1 and TM2 are very similar. The inter-helix interactions between TM1 and TM2 are likely stabilized by the aromatic interactions present throughout the simulation between the Phe1.49 and Phe2.55 residues that are adjacent to Pro1.48 and Gly2.56, respectively.
To relate cholesterol dynamics around Pro7.38 to local helical distortions on TM7, we contrast the results from Figure 5 to the time-sequences of kink and face-shift angles in TM7 around Pro7.38. The comparison reveals that the ~20° decrease in the bend angle and the ~80° reduction in the face-shift angle around Pro7.38 in TM7 occurred at about the same time as Chol stabilized its position relative to Pro7.38, at ~600 ns. Thus, the initially bent and twisted EC segment of TM7 around Pro7.38 became elongated and untangled when cholesterol moved away, suggesting that TM7 helical kink distortions near Pro7.38 are modulated by direct interactions with cholesterol.
Figure 6 shows that the IC fragment of TM7 is relatively stable against local perturbations around the Pro7.50 residue that is part of the highly conserved NPxxY(x)5,6F motif; the kink parameters around Pro7.50 remain relatively unchanged, although in the ~200–400 ns time-interval TM7 actually experiences a change in the kink of ~5° around Pro7.50, that is related to interactions between Asn7.49 and Tyr7.53 within the NPxxY(x)5,6F sequence (the distance between these residues shortens by 0.5 Å during the same period of time—data not shown). Overall, the time plot indicates significant preservation of the structural stability of this functionally important microdomain during the simulations, and very few distortions in this IC end of TM7.
Since cholesterol is generally considered to stabilize GPCRs, and our results clearly implicate it in the modulation of local distortions of selected TM segments such as the proline kinks that were suggested to play important roles in receptor function,42 it is intriguing to determine the relation between the observed perturbations associated with cholesterol and the larger scale rigid-body motions in the TM1-TM2-TM7-H8 network that are related to receptor activation.62
Relation to the dynamics of the TM1-TM2-TM7-H8 bundle
The dynamics of the TM1-TM2-TM7-H8 bundle are illustrated in Figure 7 where the distance between the IC ends of TM1 and TM7 is shown to decrease by 1 Å, and the distance on the EC side the TM1-TM7 to be shortened by 3 Å. From Figure 7(C) we infer as well that the angle between TM7 and H8 has decreased from near perpendicular to ~77° in a motion that is related to relative movements of Asn7.49 and Tyr7.53 residues within NPxxY(x)5,6F motif: shortening of Asn7.49-Tyr7.53 distance by 0.5 Å along with strong Tyr7.53-Phe7.60 interactions results in rigid-body motion of H8 toward TM7, a rearrangement that was shown to relate to receptor activation.62
Figure 7.
Dynamics in the TM1-TM2-TM7-H8 bundle. A: Change in distances between extracellular and intracellular ends of TM1 and TM7. B: The time evolution of the distance between the Cα atoms of Cys7.63 and Leu2.39. C: The time-sequence of the angle between TM7 and H8. For comparison, on each panel we show same quantities measured in X-ray crystallographic structure of rhodopsin.68,69 Note that the X-ray data is taken from the initial structure of the simulation at t = 0 time–point.
Comparison of these results to Figure 6 suggests that the large scale motions between TM1 and TM7 and between TM7 and H8 may be correlated with the local perturbations in TM1 and TM2: from Figure 7, d{TM1-TM7 EC} and α{TM7-H8} change in the ~200–400 ns time interval, the same time interval when major variations occur in the kink parameters in TM1 and TM2, as discussed in the previous section. The relation between the helical deformations on TM2 and relative TM7-H8 motions is noteworthy, as the connection between TM7-H8 bundle and TM2 has been suggested in the recent modeling work on activation of rhodopsin.62 This study has proposed that the magnitude of TM7-TM2 interactions can be modulated by the mutations at 7.53 and 7.60 positions, which in turn contribute to the final phenotype of the receptor. Here we observe that the shortening of the TM7-H8 distance is related to straightening and twisting of TM2. Importantly, these local perturbations are due to direct cholesterol-rhodopsin interactions.
At the IC end of the TM1-TM7-H8 bundle we find Val1.56 and Thr1.53 residues interacting strongly with the Phe7.60 residue in the NPxxY(x)5,6F sequence (data not shown), contributing to the overall stability of the inactive-like conformation of the TM1-TM7-H8 functional microdomain: Asn7.49-Tyr7.53 and Tyr7.53-Phe7.60 modulate the dynamics of the TM7-H8 bundle, and additional Phe7.60 interactions with Val1.56 and Thr1.53 further stabilize the TM1-TM7-H8 network. These contacts on the IC ends of TM1-TM7 may also play a role in regulating distortions in TM1 by preventing an extreme kink due to the steric effect of the interaction with cholesterol on the cytoplasmic-end residues of TM1.
Not too surprisingly, the dynamics within TM1-TM2-TM7-H8 bundle relate to identified active-like elements —the distance between Cys7.63 residue on H8 and Leu2.39 residue on TM2 has increased by 1–1.5 Å, as seen from Figure 7(B), suggesting that cytoplasmic end of TM2 moved away from H8 somewhat.65 Such findings are in line with the common understanding that GPCRs are in dynamic equilibrium between various inactive- and active-like states.62 Importantly, comparison of Figure 6 with the dynamics between Cys7.63 and Leu2.39 residues suggests that the change in d{Cys7.63-Leu2.39} may be linked to the helical frustrations on TM1 and TM2 as well.
To address quantitatively the possible relations between the local distortions on TMs and the observed rigid-body motions in TM1-TM2-TM7-H8 bundle, we performed correlation analysis on the time-evolution data of the structural helical perturbations from Figure 6 and the dynamic variables from Figure 7, and the results are described below.
Correlation analysis of dynamics structural perturbations
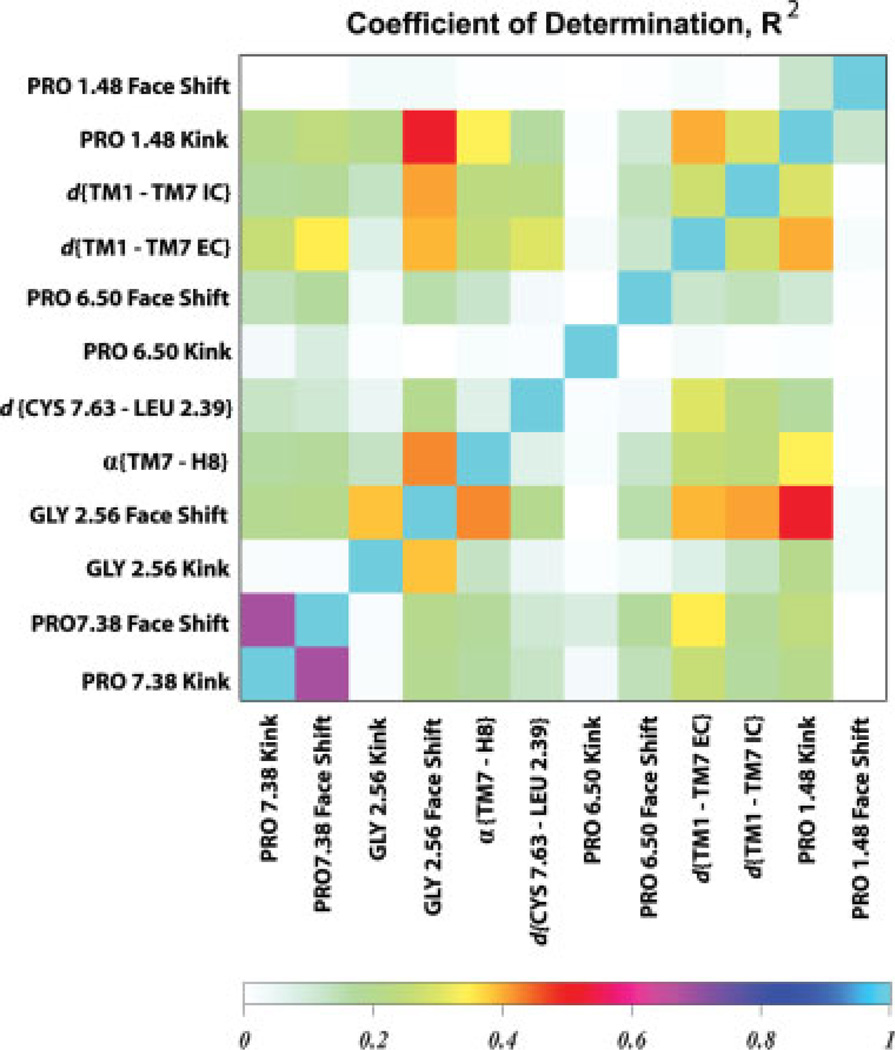
Figure 8 details pairwise correlations between the selected dynamic variables in a matrix representation of the coefficient of determination, R2 (see details in Methods). In this symmetric graph, the color code describes the strength of the correlations (both positive and negative) between the pairs, “1” indicating perfect correlation (either positive or negative), and “0” representing no correlation between variables. The dynamic quantities in the matrix include the structural perturbations discussed in Figures 6 and 7 as well as the proline bend and face-shift angles on TM6 around the highly conserved Pro6.50 residue.
Figure 8.
Coefficient of determination, R2, matrix calculated for selected variables including proline kink parameters in TM1, TM2, TM6, and TM7, and dynamic quantities from Figure 6: d{TM1-TM7 IC}, d{TM1-TM7 EC}, α{TM7-H8}, and d{Cys7.63-Leu2.39}. The color code represents the strength of individual pair-wise correlations where “1” indicates perfect correlation (either positive or negative), and “0” represents no correlation between variables. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com]
Figure 8 confirms the existence of correlations between several pairs of variables. The bend and face-shift angles around Pro7.38 exhibit the strongest correlation of all parameter pairs shown in the plot. On the other hand, the bend and face-shift angles around Gly2.56 as well as the proline kink angle around Pro1.48 appear to be correlated with the largest number of other dynamic variables in this set. In particular, the plot suggests that the local perturbations around Gly2.56 and Pro1.48 are related to α{TM7-H8} and d{TM1-TM7}.
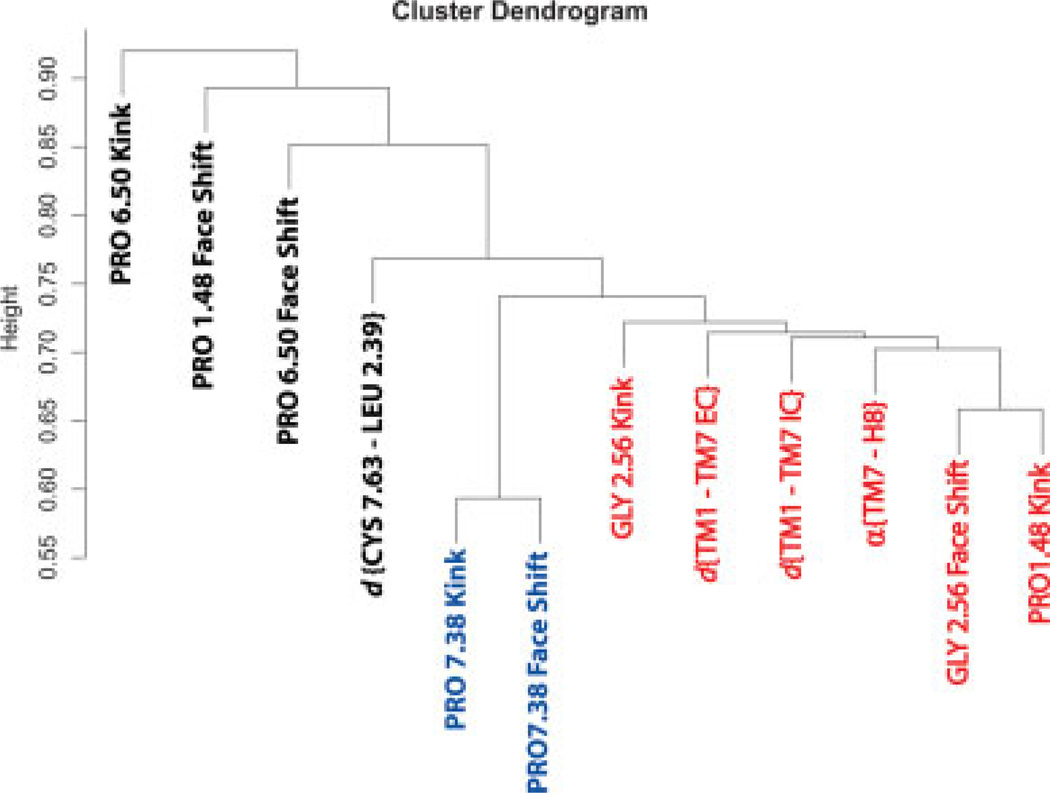
A cluster dendrogram of these pairwise correlation data (see Methods) is presented in Figure 9 showing the results of the clustering analysis on the R2 matrix. In this tree, the variables with the most similar R2 are grouped in the same limb of a tree, and the ones with distinctly different R2 values are placed in other limbs. The vertical scale of the dendrogram is in arbitrary units, such that the two variables joined at 0.5 height are perfectly correlated and therefore ideally similar, whereas a 0.5–1.0 length connection indicates pairs of increasingly dissimilar variables. Thus, the similarity of any two quantities on Figure 9 can be judged by tracing the line joining them: if this line traverses a short distance in the vertical direction, then the variables can be regarded as similar; if, on the other hand, the line connecting the pair is relatively long, then the variables can be treated as dissimilar. Following this rule, we observe that the bend and face-shift angles around Pro7.38 are located on the bottom of the tree, joined at around 0.57, which confirms the correlation identified in Figure 8 and serves as a positive control of the tree construction. Similarly, the Pro6.50 Kink and Pro1.48 Face Shift represent negative controls on the dendrogram, since these dynamic quantities do not seem to correlate with other variables in the R2 matrix (see Fig. 8).
Figure 9.
Cluster tree (dendrogram) constructed from the variables used in Figure 7. The vertical scale is such that 0.5 denotes perfect correlation and thus complete similarity of two variables, and 1.0 represents complete dissimilarity. The dendrogram contains a large cluster, highlighted in red color, which includes variables describing local distortion in TM1, TM2, and rigid-body movements of TM1, TM7, and H8. As expected from the R2 analysis, the Pro7.38 kink parameters correlate very strongly with each other and are the most similar variables, thus serving here as a positive control. Proline kink parameters in TM6, not correlated to any variables, represent a negative control. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com]
Remarkably, the cluster tree in Figure 9 contains a limb (shown in red color), consisting of data describing perturbations around Gly2.56 and Pro1.48 as well as the rigid-body motions inside TM1-TM2-TM7-H8 bundle. Quantitative identification of this large connected set establishes the existence of strong correlations between the local distortions on TM1 and TM2 and relative motions of TM7-H8 and TM1-TM7. With that the cluster dendrogram vividly illustrates the allosteric features of the TM1-TM2-TM7-H8 bundle, as large-scale helix displacements inside the bundle are modulated by local structural perturbations on TM1 and TM2. Because our data revealed that specific perturbations are related to direct contacts between cholesterol and the protein, our results emphasize the role of specific Chol-rhodopsin contacts in modulating the stability and interactions of the TM1-TM2-TM7-H8 functional network, and the allosteric nature of these interactions that are known to relate to receptor function as detailed earlier.
CONCLUSIONS
Based on the analysis of the microsecond length MD simulations of rhodopsin in an explicit membrane environment, we described a detailed mechanism by which cholesterol, through direct and structurally specific contacts with the rhodopsin, modulates the behavior of the TM1-TM2-TM7-H8 functional network. This network is known to be essential for GPCR activation (see Refs. 63, 65–67). Thus, we showed that cholesterol molecules at the EC ends of TM2-TM3 bundle (near Tyr2.63, Phe3.30, Leu3.27, Thr3.23, and Phe3.20 residues), at the IC sides of TM1-TM2-TM4 (near Val1.58, Tyr2.41, and Ile4.43), and near the EC end of TM7 (near Pro7.38) affect primarily a set of specific local perturbations of the TM domains, such as the kinks in TM1, TM2, and TM7, respectively. Remarkably, the local distortions in TM1 and TM2, in turn, relate to shortening of the distance between TM1 and TM7, and reduce the angle between TM7 and H8—motions that have been shown to be elements of the activation dynamics of GPCRs. Based on these results, we propose that the specific interactions of cholesterol with Tyr2.63, Phe3.30, Leu3.27, Thr3.23, and Phe3.20 residues as well as with Val1.58 have important physiological roles both in stabilizing GPCR structure, and in modulating function-related allosteric behavior.
ACKNOWLEDGMENTS
The authors thank Drs. Jason R. Banfelder and Lucy Skrabanek for insightful discussions on correlation analysis.
Grant sponsor: National Institutes of Health; Grant numbers: P01 DA012923, P01 DA012408; Grant sponsor: Computational Resources of the HRH Prince Alwaleed Bin Talal Bin Abdulaziz Alsaud Institute for Computational Biomedicine at Weill Medical College of Cornell University.
REFERENCES
- 1.Yeagle PL. The biophysics and cell biology of cholesterol: an hypothesis for the essential role of cholesterol in mammalian cells. In: Finegold L, editor. Cholesterol in membrane models. Boca Raton, FL: CRC Press; 1999. pp. 1–12. [Google Scholar]
- 2.Liscum L, Underwood KW. Intracelluarl cholesterol transport and compartmentation. J Biol Chem. 1995;270:15443–15446. doi: 10.1074/jbc.270.26.15443. [DOI] [PubMed] [Google Scholar]
- 3.Liscum L, Munn NJ. Intracellular cholesterol transport. Biocim Bio-phys Acta. 1999;1438:19–37. doi: 10.1016/s1388-1981(99)00043-8. [DOI] [PubMed] [Google Scholar]
- 4.Simons K, Ikonen E. How cells handle cholesterol. Science. 2000;290:1721–1726. doi: 10.1126/science.290.5497.1721. [DOI] [PubMed] [Google Scholar]
- 5.Mouritsen OG, Zuckermann MJ. What’s so special about cholesterol? Lipids. 2004:1101–1113. doi: 10.1007/s11745-004-1336-x. [DOI] [PubMed] [Google Scholar]
- 6.Rukmini R, Rawat SS, Biswas SC, Chattopadhyay A. Cholesterol organization in membranes at low concentrations: effects of curvature stress and membrane thickness. Biophys J. 2001;81:2122–2134. doi: 10.1016/S0006-3495(01)75860-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schroeder F, Woodford JK, Kavecansky J, Wood WG, Joiner C. Cholesterol domains in biological membranes. Mol Membr Biol. 1995;21:113–119. doi: 10.3109/09687689509038505. [DOI] [PubMed] [Google Scholar]
- 8.Schroeder F, Frolov AA, Murphy EJ, Atshaves BP, Jefferson JR, Pu L, Wood WG, Foxworth WB, Kier AB. Recent advances in membrane cholesterol domain dynamics and intracellular cholesterol trafficking. Proc Soc Exp Biol Med. 1996;213:150–177. doi: 10.3181/00379727-213-44047. [DOI] [PubMed] [Google Scholar]
- 9.Simons K, Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 10.Xu X, London E. The effect of sterol structure on membrane lipid domains reveals how cholesterol can induce lipid domain formation. Biochemistry. 2000;39:843–849. doi: 10.1021/bi992543v. [DOI] [PubMed] [Google Scholar]
- 11.Schroeder F, Nemecz G, Wood WG, Joiner C, Morrot G, AyrautJar-rier M, Devaux PF. Transmembrane distribution of sterol in the human erythrocyte. Biochim Biophys Acta. 1991;1066:181–192. doi: 10.1016/0005-2736(91)90185-b. [DOI] [PubMed] [Google Scholar]
- 12.Pucadyil TJ, Chattapodhyay A. Role of cholesterol in the function and organization of G-protein coupled receptors. Prog Lip Res. 2006;45:295–333. doi: 10.1016/j.plipres.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 13.Burger K, Gimpl G, Fahrenholz F. Regulation of receptor function by cholesterol. Cell Mol Life Sci. 2000;57:1577–1592. doi: 10.1007/PL00000643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Crystal structure of rhodopsin: a G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 15.Teller DC, Okada T, Behnke CA, Palczewski K, Stenkamp RE. Advance in determination of a high-resolution three-dimensional structure of rhodopsin, a model of G-protein-coupled receptors (GPCRs) Biochemistry. 2001;40:7761–7772. doi: 10.1021/bi0155091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okada T, Fujiyosni Y, Silow M, Navarro J, Nadau EM, Shichida Y. Functional role of internal water moleculer in rhodopsin revealed by X-ray crystallography. Proc Natl Acad Sci USA. 2002;99:5982–5987. doi: 10.1073/pnas.082666399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salom D, Lodowski DT, Stenkamp RE, Le Trong I, Golczak M, Jastrzebska B, Harris T, Ballesteros JA, Palczewski K. Crystal structure of a photoactivated deprotonated intermediate of rhodopsin. Proc Natl Acad Sci USA. 2006;103:16123–16128. doi: 10.1073/pnas.0608022103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HK, Kuhn P, Weis WI, Kobilka BK, Steven RC. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rasmussen SG, Choi HK, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, Burghammer M, Ratnala VR, Sanishvili R, Fischetti RF, Schertler GF, Weis WI, Kobilka BK. Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 20.Rosenbaum DM, Cherezov V, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, CHoi HJ, Yao XJ, Weis WI, Stevens RC, Kobilka BK. GPCR engineering yields high-resolution structural insights into beta2-adrenergic receptor function. Science. 2007;318:1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- 21.Warne A, Serrano-Vega MJ, Baker JG, Moukhametzianov R, Edwards PC, Henderson R, Leslie AGW, Tate CG, Schertler GFX. Structure of the beta1-adrenergic G protein-coupled receptor. Nature. 2008;454:486–491. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Visiers I, Ballesterol JA, Weinstein H. Three-dimensional representations of G-protein-coupled receptor structures and mechanisms. Methods Enzymol. 2002;343:329–371. doi: 10.1016/s0076-6879(02)43145-x. [DOI] [PubMed] [Google Scholar]
- 23.Kobilka BK. G protein coupled receptor structure and activation. Biochim Biophys Acta. 2007;1768:794–807. doi: 10.1016/j.bbamem.2006.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kobilka BK, Deupi X. Conformational complexity of G-protein-coupled receptors. Trends Pharmacol Sci. 2007;28:397–406. doi: 10.1016/j.tips.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 25.Hubbell WL, Altenbach C, Hubbell CM, Khorana HG. Rhodopsin structure, dynamics and activation: a perspective from crystallography, site-directed spin labeling, sulfhydryl reactivity, and disulfide crosslinking. Adv Protein Chem. 2003;63:243–290. doi: 10.1016/s0065-3233(03)63010-x. [DOI] [PubMed] [Google Scholar]
- 26.Bartfai T, Benovic J, Bockaert J, Bond RA, Bouvier M, Christopoulos A, Civelli O, Devi LA, George SR, Inui A, Kobilka BK, Leurs R, Neubig R, Pin JP, Quirion R, Roques BP, Sakmar TP, Seifert R, Stenkamp RE, Strange PG. The state of GPCR research in 2004. Nat Rev Drug Discov. 2004;3:577–626. doi: 10.1038/nrd1458. [DOI] [PubMed] [Google Scholar]
- 27.Kristiansen K. Molecular mechanisms of ligand binding, signaling and regulation within the superfamily of G-protein-coupled receptors: molecular modeling and mutagenesis approaches to receptor structure and function. Pharmacol Ther. 2004;103:21–80. doi: 10.1016/j.pharmthera.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 28.Filizola M, Weinstein H. The structure and dynamics of GPCR oligomers: a new focus in models of cell-signaling mechanisms and drug design. Curr Opin Drug Discov Dev. 2005;8:577–584. [PubMed] [Google Scholar]
- 29.Niu SL, Mitchell DC, Litman BJ. Manipulation of cholesterol levels in rod disk membranes by methyl-beta-cyclodextrin. Effects on receptor activation. J Biol Chem. 2002;277:20139–20145. doi: 10.1074/jbc.M200594200. [DOI] [PubMed] [Google Scholar]
- 30.Polozova A, Litman BJ. Cholesterol dependent recruitment of di22:6-PC by a G-protein coupled receptor into lateral domains. Biophys J. 2000;79:2632–2643. doi: 10.1016/S0006-3495(00)76502-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mitchell DC, Niu SL, Litman BJ. Optimization of receptor-G protein coupling by bilayer lipid composition. I. Kinetics of rhodopsin-transducin binding. JBC. 2001;276:42801–42806. doi: 10.1074/jbc.M105772200. [DOI] [PubMed] [Google Scholar]
- 32.Niu SL, Mitchell DC, Litman BJ. Optimization of receptor-G protein coupling by bilayer lipid composition. II. Formation of metarhodopsin II-transducin complex. JBC. 2001;276:42807–42811. doi: 10.1074/jbc.M105778200. [DOI] [PubMed] [Google Scholar]
- 33.Benett MP, Mitchell DC. Regulation of membrane proteins by dietary lipids: effects of cholesterol and DHA-containing phospholipids on rhodopsin stability and function. Biophys J. 2008;95:1206–1216. doi: 10.1529/biophysj.107.122788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiang Y, Rybin VO, Steinberg SF, Kobilka B. Caveolar localization dictates physiologic signaling of beta 2-adrenoceptors in neonatal cardiac myocytes. J Biol Chem. 2002;277:34280–34286. doi: 10.1074/jbc.M201644200. [DOI] [PubMed] [Google Scholar]
- 35.Whorton MR, Bokoch MP, Rasmussen SGF, Huang B, Zare RN, Kobilka B, Sunahara RK. A monomeric G protein-coupled receptor isolated in a high-density lipoprotein particle efficiently activates its G protein. Proc Natl Acad Sci USA. 2007;104:7682–7687. doi: 10.1073/pnas.0611448104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gimpl G, Fahrenholz F. Cholesterol as stabilizer of the oxytocin receptor. Biochim Biophys Acta. 2002;1564:384–392. doi: 10.1016/s0005-2736(02)00475-3. [DOI] [PubMed] [Google Scholar]
- 37.Gimpl G, Burger K, Fahrenholz F. Cholesterol as modulator of receptor function. Biochemistry. 1997;36:10959–10974. doi: 10.1021/bi963138w. [DOI] [PubMed] [Google Scholar]
- 38.Pang L, Graziano M, Wang S. Membrane cholesterol modulates galanin-GalR2 interaction. Biochemistry. 1999;38:12003–12011. doi: 10.1021/bi990227a. [DOI] [PubMed] [Google Scholar]
- 39.Weinstein H. Hallucinogen actions on 5-HT receptors reveal distinct mechanisms of activation and signaling by G protein-coupled receptors. AAPS J. 2006;7:E871–E884. doi: 10.1208/aapsj070485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smit MJ, Vischer HF, Bakker RA, Jongejan A, Timmerman H, Pardo L, Leurs R. Pharmacogenomic and structural analysis of constitutive G protein-coupled receptor activity. Annu Rev Pharmacol Toxicol. 2007;47:53–87. doi: 10.1146/annurev.pharmtox.47.120505.105126. [DOI] [PubMed] [Google Scholar]
- 41.Bhattacharya S, Hall SE, Li H, Vaidehi N. Ligand stabilized conformational states of human {beta}2 adrenergic receptor: insight into G protein coupled receptor activation. Biophys J. 2008;94:2027–2042. doi: 10.1529/biophysj.107.117648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sansom MSP, Weinstein H. Hinges, swivels and switches: the role of prolines in signaling via transmembrane alpha-helices. Trends Pharmacol Sci. 2000;21:445–451. doi: 10.1016/s0165-6147(00)01553-4. [DOI] [PubMed] [Google Scholar]
- 43.Seifert R, Wnezel-Seifert K. Constitutive activity of G-protein-coupled receptors: cause of disease and common property of wild-type receptors. Naunyn Schmiedebergs Arch Pharmacol. 2002;366:381–416. doi: 10.1007/s00210-002-0588-0. [DOI] [PubMed] [Google Scholar]
- 44.Han M, Smith SO, Sakmar TP. Consitutive activation of opsin by mutation of methionine 257 on transmembrane helix 6. Biochemistry. 1998;37:8253–8261. doi: 10.1021/bi980147r. [DOI] [PubMed] [Google Scholar]
- 45.Kim JM, Altenbach C, Kono M, Oprian DD, Hubbell WL, Khorana HG. Structural origins of constitutive activation in rhodopsin: role of the K296/E113 salt bridge. Proc Natl Acad Sci USA. 2004;101:12508–12513. doi: 10.1073/pnas.0404519101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee AG. How lipids affect the activities of integral membrane proteins. Biochim Biophys Acta. 2004;1666:62–87. doi: 10.1016/j.bbamem.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 47.Ruprecht JJ, Mielke T, Vogel T, Villa C, Schertler GFX. Electron crystallography reveals the structure of metarhodopsin I. EMBO J. 2004;23:3609–3620. doi: 10.1038/sj.emboj.7600374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schertler GFX. Structure of rhodopsin and the metarhodopsin I photointermediate. Curr Opin Struct Biol. 2005;15:408–415. doi: 10.1016/j.sbi.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 49.Hanson MA, Cherezov V, Griffith MT, Roth CB, Jaakola VP, Chien EYT, Velasquez J, Kuhn P, Stevens RC. A specific cholesterol binding site is established by the 2.8 Å structure of the human β2-adrenergic receptor. Structure. 2008;16:897–905. doi: 10.1016/j.str.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huber T, Botelho AV, Beyer K, Brown MF. Membrane model for the G-protein-coupled receptor rhodopsin: hydrophobic interface and dynamical structure. Biophys J. 2004;86:2078–2100. doi: 10.1016/S0006-3495(04)74268-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jang H, Crozier PS, Stevens MJ, Woolf TB. How environment supports a state: molecular dynamics simulations of two states in bacteriorhodopsin suggest lipid and water compensation. Biophys J. 2004;87:129–145. doi: 10.1529/biophysj.104.039602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saam J, Tajkhorshid E, Hayashi S, Schulten K. Molecular dynamics investigation of primary photoinduced events in the activation of rhodopsin. Biophys J. 2002;83:3097–3112. doi: 10.1016/S0006-3495(02)75314-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lemaitre V, Yeagle P, Watts A. Molecular dynamics simulations of retinal in rhodopsin: from the dark-adapted state towards lumirhodopsin. Biochemistry. 2005;44:12667–12680. doi: 10.1021/bi0506019. [DOI] [PubMed] [Google Scholar]
- 54.Filizola M, Wang SX, Weinstein H. Dynamic models of G-protein coupled receptors dimers: indications of asymmetry in the rhodopsin dimer from molecular dynamics simulations in a POPC bilayer. JCAMD. 2006;20:405–416. doi: 10.1007/s10822-006-9053-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grossfield A, Feller SE, Pitman MC. A role for direct interactions in the modulation of rhodopsin by omega-3 polyunsaturated lipids. Proc Natl Acad Sci USA. 2006;103:4888–4893. doi: 10.1073/pnas.0508352103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pitman MC, Grossfield A, Suits F, Feller SE. Role of cholesterol and polyunsaturated chains in lipid-protein interactions: molecular dynamics simulation of rhodopsin in a realistic membrane environment. J Am Chem Soc. 2005;127:4576–4577. doi: 10.1021/ja042715y. [DOI] [PubMed] [Google Scholar]
- 57.Feller SE, Gawrisch K, Woolf TM. Rhodopsin exhibits a preference for solvation by polyunsaturated docosohexanoic acid. JACS. 2003;125:4434–4435. doi: 10.1021/ja0345874. [DOI] [PubMed] [Google Scholar]
- 58.Grossfield A, Pitman MC, Feller SE, Soubias O, Gawrisch K. Internal hydration increases during activation of the G-protein-coupled receptro rhodopsin. JMB. 2008;381:478–486. doi: 10.1016/j.jmb.2008.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fritze O, Filipek S, Kuksa V, Palczewski K, Hofmann KP, Ernst OP. Role of the conserved NPxxY(x)5,6F motif in the rhodopsin ground state and during activation. Proc Natl Acad Sci USA. 2003;100:2290–2295. doi: 10.1073/pnas.0435715100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Prioleau C, Visiers I, Ebersole BJ, Weinstein H, Sealfon SC. Conserved helix 7 tyrosine acts as a multistate conformational switch in the 5HT2C receptor. JBC. 2002;277:36577–36584. doi: 10.1074/jbc.M206223200. [DOI] [PubMed] [Google Scholar]
- 61.Kalatskaya I, Schüssler S, Blaukat A, Müller-Esterl W, Jochum M, Proud D, Faussner A. Mutation of tyrosine in the conserved NPXXY sequence leads to constitutive phosphorylation and inter-nalization, but not signaling, of the human B2 Bradykinin receptor. JBC. 2004;279:31268–31276. doi: 10.1074/jbc.M401796200. [DOI] [PubMed] [Google Scholar]
- 62.Anavi-Goffer S, Fleischer D, Hurst DP, Lynch DL, Barnett-Norris J, Shi S, Lewis DL, Mukhapadhyay S, Howlett AC, Reggio PH, Abood ME. Helix 8 Leu in the CB1 Cannabinoid receptor contributes to selective signal transduction mechanisms. JBC. 2007;282:25100–25112. doi: 10.1074/jbc.M703388200. [DOI] [PubMed] [Google Scholar]
- 63.Han DS, Wang SX, Weinstein H. Active state-like conformational elements in the β2-AR and a photoactivated intermediate of rho-dopsin identified by dynamic properties of GPCRs. Biochemistry. 2008;47:7317–7321. doi: 10.1021/bi800442g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Elling CE, Thirstrup K, Holst B, Schwartz TW. Conversion of agonist site to metal-ion chelator site in β2-adrenergic receptor. Proc Natl Acad Sci USA. 1999;96:12322–12327. doi: 10.1073/pnas.96.22.12322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Altenbach C, Cai K, Klein-Seetharaman J, Khorana HG, Hubbell WL. Structure and function in rhodopsin: mapping light-dependent changes in distance between residue 65 in helix TM1 and residues in the sequence 306-319 at the cytoplasmic end of helix TM7 and in helix H8. Biochemistry. 2001;40:15483–15492. doi: 10.1021/bi011546g. [DOI] [PubMed] [Google Scholar]
- 66.Altenbach C, Cai K, Klein-Seetharaman J, Khorana HG, Hubbell WL. Structure and function in rhodopsin: mapping light-dependent changes in distance between residue 316 in helix 8 and residues in the sequence 60–75, covering the cytoplasmic end of helices TM1 and TM2 and their connection loop CL1. Biochemistry. 2001;40:15493–15500. doi: 10.1021/bi011545o. [DOI] [PubMed] [Google Scholar]
- 67.Altenbach C, Kusnetzow AK, Ernst OP, Hofmann KP, Hubbell WL. High-resolution distance mapping in rhodopsin reveals the pattern of helix movement due to activation. Proc Natl Acad Sci USA. 2008;105:7439–7444. doi: 10.1073/pnas.0802515105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. http://www.rcsb.org.pdb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Okada T, Sugihara M, Bondar AN, Elstner M, Entel P, Buss V. The reitnal conformation and its environment in Rhodopsin in light of new 2.2Å crystal structure. J Mol Biol. 2004;342:571–583. doi: 10.1016/j.jmb.2004.07.044. [DOI] [PubMed] [Google Scholar]
- 70.Fitch BG, Germain RS, Mendell M, Pitera J, Pitman M, Rayshub-skiy A, Sham Y, Suits F, Swope WC, Ward TJC, Zhestkov Y, Zhou R. Blue Matter, an application framework for molecular simulations on Blue Gene. J Para Distrib Comp. 2003;63:759–773. [Google Scholar]
- 71.MacKerell AD, Jr, Bashford D, Bellot M, Dunbrack R, Jr, Evanseck J, Field M, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau F, Mattos C, Michnick S, Ngo T, Nguyen D, Prodhom B, Reiher W, III, Roux B, Schlenkrich M, Smith J, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 72.Klauda JB, Brooks BR, MacKerell AD, Jr, Venable RM, Pastor RW. Ab initio study pm the torsional surface of alkanes and its effect on molecular dimulations of alkanes and a DPPC bilayer. J Phys Chem B. 2005;109:5300–5311. doi: 10.1021/jp0468096. [DOI] [PubMed] [Google Scholar]
- 73.Feller SE, Gawrisch K, MacKerell AD., Jr Polyunsaturated fatty acids in lipid bilayers: intrinsic and environmental contributions to their unique physical properties. J Am Chem Soc. 2002;124:318–326. doi: 10.1021/ja0118340. [DOI] [PubMed] [Google Scholar]
- 74.Pitman M, Suits F, MacKerell AD, Jr, Feller SE. Molecular-level organization of saturated and polyunsaturated fatty acids in phosphatidylcholine bilayer containing cholesterol. Biochemistry. 2002;43:15318–15328. doi: 10.1021/bi048231w. [DOI] [PubMed] [Google Scholar]
- 75.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. CHARMM: a program for macromolecular energy minimization and dynamics calculations. J Comput Chem. 1983;4:187–217. [Google Scholar]
- 76.Hockney RW, Eastwood JW. Computer Simulation using particles. New York: McGraw-Hill; 1981. [Google Scholar]
- 77.Pollock E, Glosli J. Comments on P3M, FMM, and the Ewald method for large periodic Coulombic systems. Comput Phys Commun. 1981;95:93–110. [Google Scholar]
- 78.Andersen HC. Rattle: A “velocity” version of the shake algorithm for molecular dynamics calculations. J Comput Phys. 1983;52:24–34. [Google Scholar]
- 79.Swope WC, Andersen HC, Berens PH, Wilson KR. A computer simulation method for the calculation of equilibrium constants for the formation of physical clusters of molecules: application to small water clusters. J Chem Phys. 1982;6:637–649. [Google Scholar]
- 80.Ballesteros JA, Weinstein H. Integrated methods for the construction of three dimensional models and computational probing of structure function relations in G-protein coupled receptors. Meth Neurosci. 1995;25:366–428. [Google Scholar]
- 81.Visiers I, Braunheim BB, Weinstein H. Prokink: a protocol for numerical evaluation of helix distortions by proline. Protein Eng. 2000;13:603–606. doi: 10.1093/protein/13.9.603. [DOI] [PubMed] [Google Scholar]
- 82. http://atlas.physbio.mssm.edu/~mezei/simulaid/
- 83.Kusalik PG, Svishchev IM. The spatial structure in liquid water. Science. 1994;265:1219–1221. doi: 10.1126/science.265.5176.1219. [DOI] [PubMed] [Google Scholar]
- 84.Kulin´ska K, Kulin´ski T, Lyubartsev A, Laaksonen A, Adamiak RW. Spatial distribution functions as a tool in the analysis of ribonucleic acids hydration—molecular dynamics studies. Comp Chem. 2000;24:451–457. doi: 10.1016/s0097-8485(99)00085-6. [DOI] [PubMed] [Google Scholar]
- 85.Pandit SA, Jakobsson A, Scott HL. Simulation of the early stages of nano-domain formation in mixed bilayers of sphingomyelin, cholesterol, and dioleylphosphatidylcholine. Biophys J. 2004;87:3312–3322. doi: 10.1529/biophysj.104.046078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pitman MC, Suits F, MacKerell AD, Jr, Feller SE. Molecular-level organization of saturated and polyunsaturated fatty acids in a phosphatidylcholine bilayer containing cholesterol. Biochemistry. 2004;43:15318–15328. doi: 10.1021/bi048231w. [DOI] [PubMed] [Google Scholar]
- 87.Crawley MJ. The R book. England: Wiley; 2007. [Google Scholar]
- 88.Lee B, Richards FM. The interpretation of protein structures: estimation of static accessibility. J Mol Biol. 1971;55:379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- 89.Hubbard SJ, Thornton JM. “NACCESS,” Computer Program. University College London: Department of Biochemistry and Molecular Biology; 1993. [Google Scholar]