Abstract
Background
Tigecycline resistance in Acinetobacter baumannii is primarily acquired through overexpression of the AdeABC efflux pump. Besides AdeRS, other two-component regulatory systems (TCSs) involving the regulation of this transporter have not been clarified.
Results
In this study, we found that the TCS genes baeR and baeS are co-transcribed and function as stress responders under high osmotic conditions. The baeSR and adeAB genes showed increased transcription in both the laboratory-induced and clinical tigecycline-resistant strains compared with the wild-type strain. The deletion of baeR in the ATCC 17978 strain led to 67–73% and 68% reduction in adeA and adeB expression, respectively, with a resultant 2-fold decrease in the tigecycline minimal inhibition concentration (MIC). In contrast, the overexpression of baeR resulted in a doubled tigecycline MIC, with a more than 2-fold increase in adeA and adeB expression. The influence of baeR knockout on adeAB gene expression can also be observed in the laboratory-induced tigecycline-resistant strain. A time-kill assay showed that the baeR deletion mutant showed an approximate 1-log10 reduction in colony forming units (CFUs) relative to the wild-type strain when the tigecycline concentration was 0.25 μg/mL throughout the assay period. The wild-type phenotype could be restored by trans-complementation with pWH1266-kan r -baeR. Increasing the tigecycline concentration to 0.5 μg/mL produced an even more marked 4.7-log10 reduction in CFUs of the baeR deletion mutant at 8 h, while only a 2.1-log10 reduction was observed for the wild-type strain.
Conclusions
Taken together, these data show for the first time that the BaeSR TCS influences the tigecycline susceptibility of A. baumannii through the positive regulation of the resistance-nodulation-division efflux pump genes adeA and adeB.
Keywords: Acinetobacter baumannii, Tigecycline, Two-component regulatory system, Efflux pumps
Background
Acinetobacter baumannii has emerged as a major cause of nosocomial infections, especially in intensive care units [1]. Both its ability to acquire resistant determinants and to adapt to harsh environments has made A. baumannii a successful pathogen [2]. A. baumannii has high rates of resistance to many available antibiotics in clinical practice. For example, imipenem-resistant A. baumannii constituted > 50% of a worldwide collection of clinical samples between 2005 and 2009 [3]. A Taiwanese surveillance report of antimicrobial resistance in 2000 found that 73% of A. baumannii isolates collected from 21 medical centers and regional hospitals were ceftazidime-resistant [4]. Therefore, there are only a few effective anti-Acinetobacter drugs currently available, including polymyxins and tigecycline [5]. Tigecycline is the first drug from the glycylcycline class, a new class of antibiotics derived from tetracycline [6]. Tigecycline acts as a protein synthesis inhibitor by binding to the 30S ribosomal subunit, and thus blocking entry of the tRNA into the A site of the ribosome during translation. Although tigecycline has an expanded spectrum of antibacterial activity, previous studies have shown that tigecycline resistance has emerged in A. baumannii. Resistance in these strains is associated with multidrug efflux systems, especially the overexpression of the adeABC genes, which encode an efflux pump [7,8]. The AdeABC pump belongs to the resistance-nodulation-division (RND) family, which has a three-component structure [9].
Bacterial two-component systems (TCSs) play an important role in the regulation of adaptation to and signal transduction of environmental stimuli, including stress conditions [10]. TCSs are typically composed of a membrane-localized sensor with histidine kinase activity and a cytoplasmic response regulator (RR). Generally, upon sensing environmental changes, signaling begins via autophosphorylation of the sensor protein at a conserved histidine residue. The phosphate is then transferred to an aspartic acid residue in the so-called receiver domain of the corresponding RR. Phosphorylation may induce conformational changes in RRs, which alters their DNA- binding properties, thus modulating downstream gene expression [11]. Importantly, the roles of TCSs in the regulation of antimicrobial resistance have recently been documented in several species of bacteria [12-14]. Additionally, the AdeS-AdeR TCS controls genes encoding the AdeABC pump in A. baumannii[15]. AdeS is a sensor kinase, whereas AdeR is an RR. Point mutations in AdeS and AdeR, or a truncation of AdeS due to an ISAba1 insertion, may be related to the overexpression of AdeABC, which leads to multidrug resistance [15,16]. However, the existence of adeABC-overexpressing mutants without any mutations in adeRS[7] and the low expression of adeABC in a clinical strain of A. baumannii with the ISAbaI insertion in the adeRS operon [16] suggest that the regulation of adeABC gene expression is complicated, and other regulatory mechanisms may be involved.
BaeSR is a TCS and is one of the five extracytoplasmic response pathways in Escherichia coli. BaeSR detects environmental signals and responds by altering the bacterial envelope [17]. The main function of the Bae response is to upregulate efflux pump expression in response to specific envelope-damaging agents [18]. Indole, flavonoids, and sodium tungstate have been shown to be novel inducers of the BaeSR response [18,19]. In Salmonella typhimurium, one of the physiological roles of BaeR is to respond to stresses that specifically damage MdtA, leading to an induction of MdtA transport and the removal of the toxic agent (e.g., tungstate waste) from the cell [19]. In TolC mutants or efflux mutants of E. coli, the overexpression of spy, which encodes a periplasmic chaperone, depends on the BaeRS and CpxARP stress response systems [20]. A genome-wide analysis of E. coli gene expression showed that BaeR overproduction activates genes involved in multidrug transport, flagellum biosynthesis, chemotaxis, and maltose transport [21]. Furthermore, BaeSR is also able to activate the transcription of the yegMNOB (mdtABCD) transporter gene cluster in E. coli and increases its resistance to novobiocin and deoxycholate [22]. Because there is a potential similarity in the biological functions of mdtABCD in E. coli and adeABC in A. baumannii, we here explore the role of BaeSR in the regulation of the transporter gene adeAB in A. baumannii and report the positive regulation of these factors, which leads to increased tigecycline resistance.
Results
Sequence analysis of the AdeAB efflux pump and the BaeR/BaeS TCS
A search of the GenBank database (http://www.ncbi.nlm.nih.gov/genbank) revealed that, similar to other strains of A. baumannii, the ATCC 17978 strain contains sequences encoding the AdeABC-type RND efflux pump. There are two adeA genes (A1S_1751 and A1S_1752) and one adeB gene (A1S_1750) in the genome; however, no adeC gene was found. AdeB is a transmembrane component with two conserved domains: the hydrophobe/amphiphile efflux-1 (HAE1) family signature and a domain conserved within the protein export membrane protein SecD_SecF. Both AdeA proteins are inner membrane fusion proteins with biotin-lipoyl-like conserved domains. We designated A1S_1751 as AdeA1 and A1S_1752 as AdeA2 for differentiation.
The A. baumannii ATCC 17978 gene A1S_2883 encoded a protein of 228 amino acids. Sequence alignments of A. baumannii A1S_2883 with BaeR homologs in other bacteria showed that A1S_2883 shared 64.6% similarity with BaeR of E. coli str. K-12 substr. MG1655 and 65.2% similarity with BaeR of Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 (Figure 1A). In addition, protein analysis using Prosite (http://prosite.expasy.org/) predicted that A. baumannii A1S_2883 contained a response regulatory domain at amino acid residues 3 to 115 and a phosphorylation site at amino acid residue 51 (aspartate). Therefore, the role of A1S_2883 may be similar to that of BaeR in other bacterial species; thus, we have designated A1S_2883 as BaeR in A. baumannii.
Figure 1.
Sequence alignment of BaeR and BaeS from Acinetobacter baumannii ATCC 17978 and other bacteria. (A) Sequence alignments of A. baumannii A1S_2883 with BaeR homologs in other bacteria revealed that A1S_2883 shares 64.6% similarity with BaeR of Escherichia coli and 65.2% similarity with BaeR of Salmonella LT2. (B) A1S_2884 shares 48.1% similarity with BaeS of E. coli and 46.3% similarity with BaeS of Salmonella LT2. REC and trans_reg_C are two conserved domains of BaeR, while HAMP, HisKA, and HATPase_c are conserved domains of BaeS. AB17978: A. baumannii ATCC 17978, ABAYE: A. baumannii AYE, E. coli: E. coli str. K-12 substr. MG1655, SalmonellaLT2: Salmonella enterica subsp. enterica serovar Typhimurium str. LT2.
A. baumannii ATCC 17978 gene A1S_2884 encodes a protein of 487 amino acids. Sequence alignments showed that A1S_2884 shared 48.1% similarity with BaeS of E. coli str. K-12 substr. MG1655 and 46.3% similarity with BaeS of Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 (Figure 1B). Protein analysis using Prosite predicted that A. baumannii A1S_2884 contains a HAMP (histidine kinase, adenylyl cyclase, methyl-accepting protein, and phosphatase) and a histidine kinase domain at amino acid residues 214 to 266 and 274 to 487, respectively. In addition, the histidine at residue 277 is predicted to be a phosphorylation site. Therefore, the role of A1S_2884 may be similar to that of BaeS in other bacterial species; thus, A1S_2884 is designated as BaeS in A. baumannii.
Co-transcription of baeR and baeS as an operon
Although the BaeSR TCS has been characterized in E. coli[21,22], the biological functions of BaeSR in A. baumannii have not been revealed. Genome analysis of A. baumannii ATCC 17978 shows that the coding sequences of baeR (A1S_2883) and baeS (A1S_2884) are arranged sequentially, suggesting that the two genes may be co-transcribed as an operon. To test this hypothesis, reverse transcription-polymerase chain reaction (RT-PCR) was performed using the primers baeS-co F and baeS-co R (Table 1). As shown in Figure 2A, a 793-bp DNA fragment covering the junction between both baeR and baeS was amplified by RT-PCR. We concluded that baeR and baeS are co-transcribed as a single operon in A. baumannii ATCC 17978.
Table 1.
Oligonucleotides used in this study
| Primer name | Sequence (5′ to 3′) a |
|---|---|
|
baeS-co_F |
CGCGTAGTACAGGTGGAACA |
|
baeR-co_R |
TCCACTCATGACGGTTACCA |
|
kan
r
-BamHI_F |
ATATGGATCCCCGGAATTGCCAGCTGGGGC |
|
kan
r
-KpnI_R |
ATATGGTACCTCAGAAGAACTCGTCAAGAA |
|
baeR-up-SalI_F |
TTAAGTCGACCGCCCGATTATGGTCAATAG |
|
baeR-up-BamHI_R |
TATAGGATCCGCTTACTTCGAACCCAGCAG |
|
baeR-dw-KpnI_F |
ATCGGGTACCTTGCTTAGAAAAGTTATGCT |
|
baeR-dw-SacI_R |
AAATGAGCTCATGCTTTAGGGGTGGCTTCT |
|
baeR-up-check_R |
CTTTCCCAGTGGTGGTTACG |
|
baeR-dw-check_R |
GACGGACGTGGCTTACTCAT |
| pWH1266 check_F |
TGCCACCTGACGTCTAAGAA |
| pWH1266 check_R |
TCATACACGGTGCCTGACTG |
|
sacB_F |
AGTTTTGTTCAGCGGCTTGT |
|
sacB_R |
GGTCAGGTTCAGCCACATTT |
|
baeR_F |
GGATGGTTTAACGATTTGCC |
|
baeR_R |
TCCACTCATGACGGTTACCA |
|
baeS_F |
CTTTCCCAGTGGTGGTTACG |
|
baeS_R |
GAGCCAAGTCTGCCAAATCT |
|
baeS probe_F |
CCTCAATACTGGTGAAACCA |
|
baeS probe_R |
TCCCCCAATCATGATAAACG |
|
rpoB_F |
AGTCACGCGAAGTTGAAGGT |
|
rpoB_R |
GCGGTATGGAGTTTCCAAGA |
| 16 s rRNA_F |
GTAGCTTGCTACTGGACCTAG |
| 16 s rRNA_R |
CATACTCTAGCTCACCAGTATCG |
|
Kan-2
r
-EcoRI_F |
AATAGAATTCACATCTCAACCATCATCG |
|
Kan-2
r
-EcoRI_R |
AATAGAATTCCATCTCAACCCTGAAGC |
|
baeR-XhoI_F |
TTAACTCGAGCATGTTTCATGATGGTC |
|
baeR-XbaI_R |
TAGCTCTAGATTATTCTTCTGGATATTCG |
| qbaeS_F |
GCCATTCAGCAGCATTCTTTC |
| qbaeS_R |
ATTTACCGTTCCCGCATCTG |
| qbaeR_F |
TGACAGCACGTACCGAAGAAA |
| qbaeR_R |
CATAATCATCTGCCCCCATGT |
| qadeB_F |
ACAAGACCGCGCTAACTTAGGT |
| qadeB_R |
TGCCATTGCCATAAGTTCATCT |
| qadeA1_F |
CCTCAAGCGCTATTGGTTCCT |
| qadeA1_R |
CCTGAGGCTCGCCACTGA |
| qadeA2_F |
TTTGAGGCCGATGTAAATAGCA |
| qadeA2_R |
GTCTTGCCACCTCAGCTTCAG |
| q16s rRNA_F |
AGCATTTCGGATGGGAACTTTA |
| q16s rRNA_R | GTCGTCCCCGCCTTCCT |
aRestriction sites introduced into the primers are highlighted using bold fonts and described in the primer name.
Figure 2.
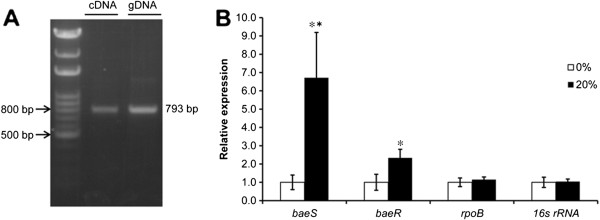
Evaluation of baeR and baeS expression. (A) The co-transcription of baeR and baeS was determined by agarose gel electrophoresis of the product obtained by reverse transcription polymerase chain reaction (RT-PCR). Lane 2 (cDNA) and 3 (genomic DNA) reveal a 793-bp DNA fragment covering the junction between both the baeR and baeS genes. (B) The relative transcript levels of baeR and baeS, as determined by RT-PCR, under different osmolarity conditions. The cells were grown on Luria-Bertani (LB) agar with or without 20% sucrose (37°C, 220 rpm). 16S rRNA and rpoB genes were used as controls. The expression levels of baeR and baeS were 2.3- and 6.7-fold higher in cells experiencing osmotic stress than those in cells grown without sucrose. The results are displayed as the means ± SD from four independent experiments. *, P < 0.05; **, P < 0.01.
Transcription of baeR and baeS under normal and stressed conditions
TCSs are commonly involved in stress responses in bacteria. Because no previous studies have explored the function of A1S_2883 and A1S_2884, we began by testing the response of both genes to high osmotic conditions to determine if they have functions that are similar to those of their BaeSR counterparts in other bacteria. To determine whether A. baumannii baeSR participates in the stress response, the relative levels of baeR and baeS transcription were detected in cells grown in Luria-Bertani (LB) agar (37°C, 220 rpm) with or without 20% sucrose. RT-PCR analysis showed that the expression levels of baeR and baeS were 2.3- and 6.7-fold higher in cells exposed to osmotic stress compared with cells grown without sucrose (Figure 2B). This result suggested that the BaeSR TCS in A. baumannii was involved in cellular adaptation to stress conditions such as high osmolarity.
Construction of baeR deletion mutants and baeR-reconstituted strains
To further study the role of the BaeSR TCS in A. baumannii, in-frame deletion mutants of baeR were generated using the method of Sugawara et al. [23]. The successful construction of baeR deletion mutants was verified by PCR (Additional file 1: Figure S1B), RT-PCR (Additional file 2: Figure S2), and Southern blot assays (Additional file 3: Figure S3B). To generate the baeR-reconstituted strain, pWH1266-kan r -baeR was introduced into the baeR deletion mutant (AB1026; Table 2) by electroporation. The baeR-reconstituted strain was designated AB1027. In addition, pWH1266-kan r -baeR was also introduced into the wild-type strain to generate the strain AB1028. Successful construction of the AB1027 and AB1028 strains was verified by RT-PCR. The expression of baeR was comparable in the wild-type and the baeR-reconstituted AB1027 strains, whereas baeR was overexpressed in AB1028 relative to the wild-type strain (data not shown).
Table 2.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant feature(s) | Source or reference | |
|---|---|---|---|
|
A. baumannii strains |
ATCC 17978 |
Wild-type strain |
ATCC |
| |
AB1026 (ΔbaeR::kan
r
) |
Derived from ATCC 17978. baeR mutant obtained by kan
r
gene replacement |
This study |
| |
AB1027 |
AB1026 baeR::pWH1266 |
This study |
| |
AB1028 |
ATCC 17978 baeR::pWH1266 |
This study |
| |
AB1029 |
ATCC 17978 kan:: pWH1266 |
This study |
| |
ABtc |
Induced tigecycline resistant ATCC 17978 |
This study |
| |
ABtcm (ΔbaeR::kan
r
) |
Derived from ABtc. baeR mutant obtained by kan
r
gene replacement |
This study |
| |
ABhl1 |
Tigecycline resistant clinical isolate |
This study |
|
E. coli strains |
XL1 blue |
recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F’ proAB lacIqZΔM15 Tn10 (Tetr)] |
Stratagene |
| |
S17-1 (ATCC 47055) |
thi pro hsdR hsdM recA[RP42-Tc::Mu- Km::Tn7 (TprSmr)Tra+] |
ATCC |
| Plasmids |
pEX18Tc |
Suicide vector containing sacB, Tcr |
40 |
| |
pSFS2A |
Containing kan
r
, an FRT site, FLP1, and CaSAT1 as a SAT1 flipper |
41 |
| |
pEX18Tc-Δbae::kan
r
|
pEX18Tc containing baeR upstream and downstream fragments joined by a kan
r
cassette |
This study |
| |
pWH1266 (ATCC 77092) |
E. coli-A. baumannii shuttle cloning vector, containing Ampr, Tetr |
43 |
| |
pC2HP |
Provided kan
r
for pWH1266 |
42 |
| |
pWH1266-kan
r
|
pWH1266 containing kan
r
|
This study |
| pWH1266-kan r -baeR | pWH1266-kan r containing baeR | This study | |
Minimal inhibitory concentration (MIC) determination
To correlate BaeR with tigecycline susceptibility, the MIC of tigecycline was determined. For A. baumannii ATCC 17978, the MIC of tigecycline was 0.5 μg/mL. However, the MIC of tigecycline for the baeR deletion mutant was 0.25 μg/mL; baeR reconstitution restored the MIC to the wild-type level (MIC 0.5 μg/mL). Moreover, the overexpression of baeR in AB1028 raised the MIC of tigecycline to 1 μg/mL. The introduction of pWH1266 alone did not affect the MIC of tigecycline, whereas the MICs obtained with the induced tigecycline-resistant strain ABtc and the clinical tigecycline-resistant strain ABhl1 were 256 and 16 μg/mL, respectively. These results indicate that BaeR is closely related to the tigecycline susceptibility of A. baumannii.
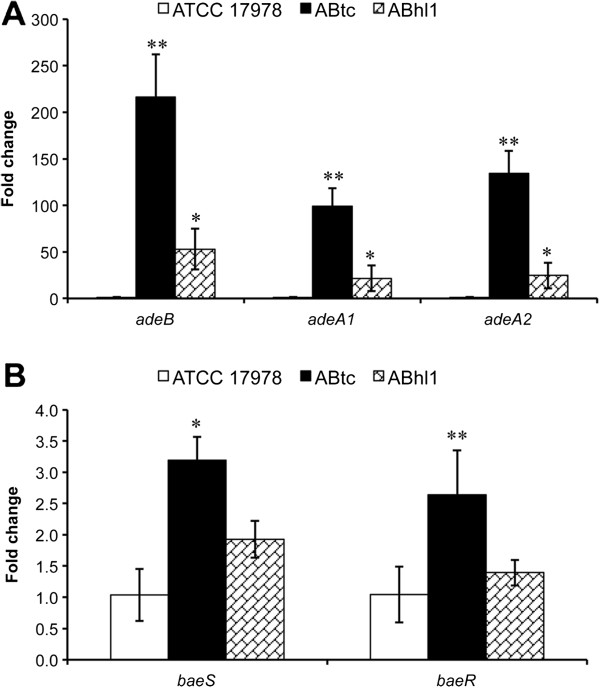
Expression of the adeAB and baeSR genes in strains with different levels of tigecycline resistance
To further decipher the role of the BaeSR TCS and AdeAB in tigecycline resistance, we analyzed gene expression in the wild-type A. baumannii strain ATCC 17978 as well as the ABtc and ABhl1 strains. The quantitative real-time PCR (qRT-PCR) results showed that the expression levels of adeB were 216- and 53-fold higher in ABtc and ABhl1, respectively, than in the wild-type strain. ABtc and ABhl1 also showed increased transcription of adeA1 and adeA2, although it was less marked than that of adeB (Figure 3A). The expression levels of baeS and baeR in ABtc increased 3.19 and 2.64-fold, respectively, compared with the wild-type strain, whereas those in ABhl1 only increased 1.93 and 1.39-fold, respectively (Figure 3B). Overall, the combination of the qRT-PCR results with the MIC assay above suggest that both BaeSR and AdeAB are involved in the tigecycline resistance of A. baumannii.
Figure 3.
Transcript levels of the adeA, adeB, baeR, and baeS genes in A. baumanniistrains. ABtc and ABhl1 are laboratory-induced and clinically isolated tigecycline-resistant strains, respectively. The corresponding tigecycline minimum inhibitory concentrations (MICs) of ATCC 17978, ABtc, and ABhl1 were 0.5, 256, and 16 μg/mL, respectively. Gene expression was detected by quantitative real-time PCR (qRT-PCR). (A) qRT-PCR showed that the expression levels of adeB in ABtc and ABhl1 were 216- and 53-fold higher than those in the wild-type strain, respectively. The adeA1 expression levels in ABtc and ABhl1 were 99- and 22-fold higher than those in the wild-type strain, respectively, whereas the adeA2 expression levels in ABtc and ABhl1 were 134- and 25-fold higher. (B) The expression levels of baeS and baeR in ABtc increased 3.19 and 2.64 times, respectively, compared with the wild-type strain, whereas those in ABhl1 only increased 1.93 and 1.39 times, respectively. 16S rRNA gene was used as a control. The results are displayed as the means ± SD from four independent experiments. *, P < 0.05; **, P < 0.01.
Influence of the BaeSR TCS on adeAB efflux pump expression
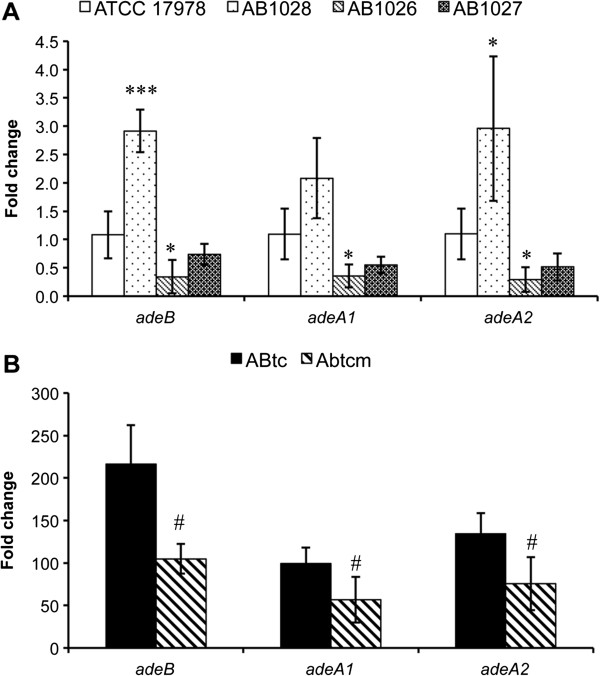
To understand whether baeR influenced the tigecycline MIC by affecting the adeAB efflux pump gene, the expression of adeA1, adeA2, and adeB in ATCC 17978, AB1026, AB1027, and AB1028 was analyzed by qRT-PCR. The expression levels of adeB, adeA1, and adeA2 in AB1028 were approximately 2.9-, 2.1-, and 3-fold higher, respectively, than those in ATCC 17978, while the deletion of baeR from the wild-type strain decreased the expression levels of these three pump genes by 68.3%, 67.3%, and 73.5%, respectively (Figure 4A). The decreased expression of the pump genes can be partially restored by baeR reconstitution (Figure 4A). To determine the impact of baeR deletion on adeR expression, RT-PCR was also performed. No differences in adeR expression were observed between AB1026 and the wild-type strain (data not shown). Overall, these findings suggest that BaeR upregulates the expression of adeAB genes.
Figure 4.
Transcript levels of the adeA and adeB genes in different strains of A. baumannii. AB1026, AB1027, and AB1028 are the baeR deletion mutant, baeR reconstitution, and wild-type with baeR overexpression strains, respectively. ABTcm is the baeR deletion mutant of ABtc, which was a laboratory-induced tigecycline-resistant strain. The relative expression of adeB, adeA1, and adeA2 was determined by qRT-PCR. (A) The expression levels of adeB, adeA1, and adeA2 in AB1028 were approximately 2.9-, 2.1-, and 3-fold higher, respectively, than those in ATCC 17978, while the deletion of baeR in the wild-type strain decreased the expression levels of these three pump genes by 68.3%, 67.3%, and 73.5%, respectively. The decreased expression of the pump genes can be partially restored by baeR reconstitution. (B) The expression levels of adeB, adeA1, and adeA2 in ABtcm were 51.5%, 42.7%, and 43.7% lower, respectively, than those in ABtc. 16S rRNA gene was used as a control. The results are displayed as the means ± SD from three independent experiments. *, P < 0.05; ***, P < 0.001. #, P < 0.05 between ABtc and ABtcm.
Expression analysis of adeAB in induced tigecycline-resistant A. baumannii and its baeR mutant
To further confirm the role of baeR in the tigecycline resistance of A. baumannii via the AdeAB efflux pump, a baeR deletion mutant of ABtc (ABtcm) was constructed and adeAB expression was analyzed by qRT-PCR. The expression levels of adeB, adeA1, and adeA2 in ABtcm were 51.5, 42.7%, and 43.7% lower, respectively, than those in ABtc (Figure 4B). These data confirmed the contribution of BaeR to the regulation of AdeAB, which is essential to tigecycline resistance in A. baumannii.
Time-kill assay
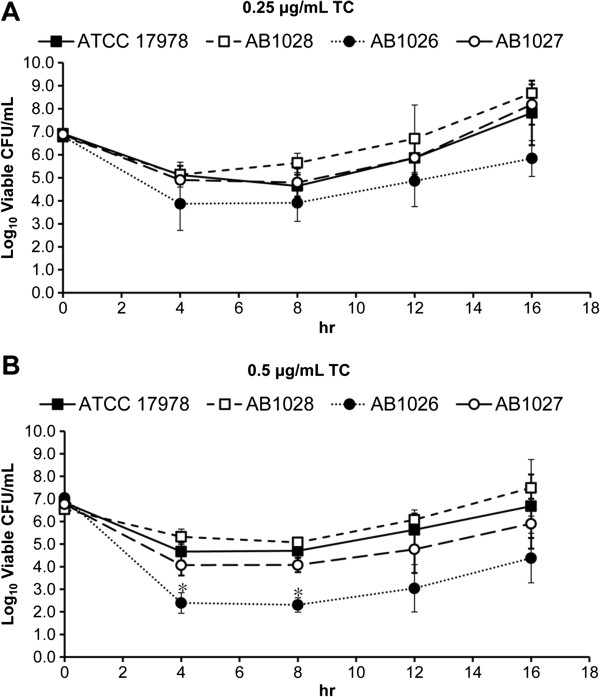
To further compare the effects of BaeR on tigecycline susceptibility, time-kill assays were performed using ATCC 17978, AB1026, AB1027, and AB1028. There were no differences in the surviving colony forming units (CFUs) among these four strains when tigecycline was not added to the LB agar. In the presence of 0.25 μg/mL tigecycline, all tested strains had similar surviving CFU curves; the lowest value was observed at 4 h, which was followed by regrowth (Figure 5A). Additionally, AB1026 showed a greater reduction in CFUs than the wild-type strain (e.g., 2.9-log10 versus 1.8-log10 reduction, respectively, at 4 h) throughout the assay period, which could be restored by baeR reconstitution. Increasing the tigecycline concentration to 0.5 μg/mL produced an even more marked 4.7-log10 reduction in the CFUs of AB1026 at 8 h, which was followed by regrowth. In contrast, a smaller reduction (2.1-log10 reduction at 8 h) was observed for the wild-type strain (Figure 5B). However, baeR reconstitution did not fully restore the ability of AB1026 to resist 0.5 μg/mL tigecycline. AB1028 showed a slightly smaller reduction in CFUs than the wild-type strain in the presence of 0.25 and 0.5 μg/mL tigecycline. Therefore, the time-kill assay indicates that the BaeSR TCS plays a role in the tigecycline susceptibility of A. baumannii.
Figure 5.
Time-kill assays for ATCC 17978, AB1026, AB1027, and AB1028 with 0.25 μg/mL (A) and 0.5 μg/mL (B) tigecycline. In the presence of 0.25 μg/mL tigecycline, all tested strains showed similar surviving colony forming unit (CFU) curves, in which the lowest value occurred at 4 h and was followed by regrowth. AB1026 had a greater CFU reduction than the wild-type strain throughout the assay period, which could be restored by baeR reconstitution. Increasing the tigecycline concentration to 0.5 μg/mL resulted in a marked 4.7-log10 CFU reduction for AB1026 at 8 h, which was followed by regrowth, whereas a smaller reduction was observed for the wild-type strain. baeR reconstitution did not fully restore the ability of AB1026 to resist 0.5 μg/mL tigecycline. AB1028 had a slight, though not significant, CFU reduction compared to the wild-type strain in the presence of 0.25 or 0.5 μg/mL tigecycline. Viable counts represented by CFUs were determined at time 0 and at 4, 8, 12, and 16 h after inoculation. A time-kill curve was constructed for each strain. The results are displayed as the means ± SD from three independent experiments. *, P < 0.05.
Discussion
Previous studies that investigated the regulation of AdeABC efflux pumps in A. baumannii primarily focused on the AdeRS TCS, which is located upstream of the adeABC operon and is transcribed in the opposite direction [15]. Several point mutations in adeR or adeS have been proposed as the major cause of AdeABC efflux pump overexpression, including a threonine-to-methionine substitution at position 153 [15], a glycine-to-aspartate mutation at position 30 [24], an alanine-to-valine substitution at position 94 of AdeS [25], or a proline-to-leucine substitution at position 116 of AdeR [15]. However, the effect of AdeR or AdeS mutations on the expression of AdeABC is not always consistent. Different tigecycline MICs were observed in two transformed strains with the same mutations in the DNA-binding domain of the AdeR protein [16]. adeABC-overexpressing mutants that did not carry any mutations in adeRS compared with their isogenic parents were also reported [7,25]. Another mechanism leading to the overexpression of AdeABC involves the transposition of an ISAba1 copy into adeS[15], which stimulates AdeR to interact with and activate the adeABC promoter [16]. In contrast to the results of the above-mentioned studies of AdeRS, four imipenem-resistant A. baumannii strains carrying adeB but lacking adeRS were identified by Hou et al. [26], suggesting that another regulatory mechanism may be involved. Henry et al. reported that BaeSR was associated with the increased expression of the multidrug resistance-associated efflux pump genes macAB-tolC and adeIJK in their transcriptional analysis of lipopolysaccharide-deficient A. baumannii 19606R [27]. Therefore, the role of BaeSR in the expression of the AdeABC efflux pump deserves investigation. Our data demonstrate that BaeSR influences the tigecycline susceptibility of A. baumannii ATCC 17978 through its positive regulation of the transcription of transporter genes adeA and adeB. This result supported the possibility that other TCSs aside from AdeRS may be involved in the regulation of the AdeABC efflux pump in A. baumannii.
Most A. baumannii strains have an RND efflux pump, AdeABC, which has a three-component structure with AdeB forming the transmembrane component, AdeA forming the inner membrane fusion protein, and AdeC forming the outer membrane protein [9]. However, according to the NCBI GenBank database, A. baumannii ATCC 17978 lacks an adeC gene but has two adeA genes and one adeB gene. A. baumannii AYE, A. baumannii ACICU, A. baumannii ATCC 19606, and A. baumannii TYTH-1 all possess an AdeC-like outer membrane protein. Marchand et al. constructed a clinical A. baumannii strain with an inactivated adeC. This derivative mutant displayed resistance to the various substrates of the AdeABC pump that was similar to that of the wild-type strain, indicating that adeC is not essential for resistance [15]. Because adeC was not found in 41% of the clinical isolates carrying adeRS-adeAB in one study [28], it is reasonable to deduce that AdeAB could recruit another outer membrane protein to form a functional tripartite complex [29].
The first description of tigecycline non-susceptibility was reported by Peleg et al. [7]. These authors found that the efflux pump inhibitor phenyl-arginine-β-naphthylamide could cause a four-fold reduction in the MIC of tigecycline in two tigecycline-non-susceptible isolates. The qRT-PCR results showed 40-fold and 54-fold increases in adeB expression in these two isolates compared to that observed in a tigecycline-susceptible isolate. Their finding is consistent with our comparison of tigecycline MICs and expression levels of AdeAB among the wild-type, ABhl1, and ABtc strains. Despite the important role of AdeABC in antibiotic resistance, this efflux pump operon is cryptic in natural isolates of A. baumannii[15,30]. Antibiotic exposure, including exposure to tigecycline, could induce pump overexpression, resulting in drug resistance [29]; this was observed in our ABtc strain. Furthermore, there was a statistically significant linear relationship between log-transformed adeA expression values and log-transformed MICs of tigecycline in clinical isolates of the A. calcoaceticus-A. baumannii complex, indicating that the overexpression of the AdeABC efflux pump is a prevalent mechanism for this resistance phenotype [31].
The modest increase in AdeAB pump gene expression in AB1028 relative to the wild-type strain may have been due to the overexpression of BaeSR. However, because ABtcm had only moderately reduced adeB, adeA1, and adeA2 expression levels relative to ABtc, we proposed that control mechanisms aside from BaeSR, such as sequence changes in adeR or adeS, were responsible for the overexpression of these pump genes. The regulators that are involved in efflux gene expression are either local or global regulators [32]. One of the most well-studied examples is the AcrAB-TolC system of E. coli[33]. This system is under the control of the local repressor gene acrR, which negatively regulates the transcription of acrAB. On the other hand, global stress conditions are assumed to result in the generation of global transcription regulators. These regulators are unlikely to be MarA, SoxS, or Rob, but could be their homologs. Such regulators increase the transcription of not only acrAB but also acrR, which functions as a secondary modulator to repress acrAB. Fernando et al. demonstrated that the transcription patterns of both adeB and adeJ are cell density-dependent and similar, indicating a role for global regulatory mechanisms in the expression of these genes in A. baumannii[34]. Two-component regulatory systems mediate the adaptive responses of bacterial cells to a broad range of environmental stimuli [35]. In this study, qRT-PCR analysis of baeSR expression under high sucrose conditions suggested that this TCS was involved in the regulation related to this stress condition. Therefore, we propose that BaeSR, which functions as an envelope stress response system to external stimuli, also influences the transcription of adeAB in A. baumannii by functioning as a regulator of global transcription. Meanwhile, the well-described adeR is an example of a local regulator that activates adeABC expression [15,16]. However, the relationship between BaeSR and AdeRS must be further clarified. Because the expression of adeRS was only marginally increased in the baeSR deletion mutants in this study, we assume that the crosstalk between these TCSs might be absent or only very weak. The question of whether other TCSs are involved in the regulation of the AdeABC efflux pump and how they interact in A. baumannii merits further investigation.
Conclusions
In this study, we showed for the first time that the BaeSR TCS influences the tigecycline susceptibility of A. baumannii by positively regulating the RND efflux pump genes adeA and adeB. However, whether BaeSR can also contribute to tigecycline resistance through other transporter genes, such as macAB-tolC and adeIJK, is not yet clear, and related studies are underway. Overall, this finding highlights the complexity of AdeABC transporter regulation and could be a starting point for understanding the role of TCSs in the antimicrobial susceptibility of bacteria.
Methods
Bacterial strains, plasmids, growth conditions, and antibiotic susceptibility testing
The bacterial strains and plasmids used in this study are listed in Table 2. The cells were grown at 37°C in LB broth and agar. To determine the MIC, a broth microdilution method was used according to the 2012 CLSI guidelines [36]. Briefly, bacteria were inoculated into 1 mL cation-adjusted Mueller-Hinton broth (CAMHB) (Sigma-Aldrich, St. Louis, MO) containing different concentrations of tigecycline (Pfizer, Collegeville, PA) to reach ≈ 5 × 105 CFU/mL, and the cultures were incubated at 37°C for 24 h. The lowest tigecycline concentration that completely inhibited bacterial growth was defined as the MIC, and growth was determined by unaided eyes and by measuring optical densities (ODs) using a spectrophotometer. On the basis of the report published by Pachón-Ibáñez et al., the provisional MIC breakpoints for tigecycline are ≤2, 4, and ≥8 μg/mL to designate susceptible, intermediate, and resistant strains, respectively [37].
DNA manipulation
Plasmid DNA was prepared with the FavorPrep™ Plasmid DNA Extraction Mini Kit (Favorgen, Ping-Tung, Taiwan). A. baumannii genomic DNA was extracted as described previously [38]. PCR amplification of the DNA was performed in a Thermo Hybaid PXE 0.2 HBPX02 Thermal Cycler (Thermo Scientific, Redwood, CA), using ProTaq™ DNA Polymerase (Protech, Taipei, Taiwan) or the KAPA HiFi™ PCR Kit (Kapa Biosystems, Boston, MA). DNA fragments were extracted from agarose gels and purified using the GeneKlean Gel Recovery & PCR CleanUp Kit (MDBio, Inc., Taipei, Taiwan). Nucleotide sequences of the PCR products were verified using an ABI 3730XL DNA Analyzer (Applied Biosystems, South San Francisco, CA).
RNA isolation, RT-PCR, and qRT-PCR
For total RNA isolation, A. baumannii ATCC 17978 was grown overnight in LB broth (37°C, 220 rpm, 16 h) to reach an OD600 of approximately 6.5. The overnight cultures were sub-cultured at a 1:100 dilution in 25 mL fresh LB medium. The cells were grown to mid-log phase and harvested by centrifugation at 4°C. The cell pellets were resuspended in 200 μL ice-cold RNA extraction buffer (0.1 M Tris-Cl [pH 7.5], 0.1 M LiCl, 0.01 M ethylenediaminetetraacetic acid [pH 8.0], 5% sodium dodecyl sulfate [SDS], 2% β-mercaptoethanol), and 200 μL ice-cold phenol-chloroform-isoamyl alcohol (PCIA [25:24:1], pH 4.5) was added and vortexed for 2 min. The supernatants were then collected by centrifugation, added to 200 μL ice-cold PCIA, and mixed well. This step was repeated three times. Then, RNA was precipitated with ethanol at -80°C overnight and collected by centrifugation at maximum speed for 5 min. The RNA pellets were dissolved in 25–100 μL diethylpyrocarbonate-treated water. DNA was removed using Ambion® TURBO™ DNase (Life Technologies, Grand Island, NY), and cDNA was synthesized by reverse transcription using High-Capacity cDNA Reverse Transcriptase Kits (Applied Biosystems). The cDNAs were used in PCR reactions with different primers (Table 1).
qRT-PCR was carried out with a StepOne™ Real-Time PCR System (Life Technologies). The primers used for qRT-PCR are listed in Table 1. Briefly, each 20-μL reaction mixture contained 25 ng cDNA, 10 μL Power SYBR green PCR master mix (Life Technologies), and 300 nM each forward and reverse primer. The reactions were performed with 1 cycle at 95°C for 10 min followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. The 16S rRNA transcript was used as an endogenous control for the qRT-PCR. The data were analyzed using StepOne v2.1 software (Life Technologies).
Induction of tigecycline resistance
To induce tigecycline resistance, serial passaging was performed as previously described [39] with some modifications. Briefly, on day 1, 3 mL of LB broth containing tigecycline at the MIC was inoculated with A. baumannii (passage 1), and the cultures were incubated at 37°C with shaking (220 rpm). On day 3, 30 μL of the culture was transferred to 3 mL of LB broth containing tigecycline at 8× the MIC (passage 2), and the cultures were again incubated at 37°C with shaking (220 rpm). On day 5, 30 μL of the culture was transferred into LB broth containing tigecycline at 16× the MIC (passage 3), and the cultures were again incubated at 37°C with shaking (220 rpm). This passaging was repeated on day 7 (passage 4). On day 9, aliquots (3 mL) of the cultures were mixed with 10% glycerol and stored at -80°C until use. Daily passaging in tigecycline-free LB was conducted for 30 days for both ATCC 17978 and the clinical strain.
Construction of baeR deletion mutants and baeR reconstituted strains
To assess the contribution of BaeR to the regulation of tigecycline resistance, baeR deletion mutants of A. baumannii ATCC 17978 were constructed as previously described [23] with some modifications. The suicide vector pEX18Tc [40] was first cloned with a 953-bp DNA fragment carrying a kanamycin resistance cassette, which was PCR-amplified from the pSFS2A plasmid [41], to generate pEX18Tc-kan r . DNA fragments carrying the upstream and downstream regions of the baeR gene, referred to as baeR-up and baeR-dw, were independently amplified by PCR using the primer pairs baeR-up-SalI-F and baeR-up-BamHI-R or baeR-dw-KpnI-F and baeR-dw-SacI-R (Table 1). The baeR-up fragment (1,119 bp) was digested with SalI and BamHI enzymes, whereas the baeR-dw fragment (1,120 bp) was digested with KpnI and SacI enzymes (Additional file 4: Figure S4A). Both enzyme-digested DNA fragments were then independently cloned into the corresponding restriction sites of pEX18Tc-kan r , generating pEX18Tc-kan r -baeR-flanking. The resultant plasmid was then transformed into the E. coli S17-1 strain using the standard CaCl2/heat shock method [38]. Then, trans-conjugation was performed between E. coli S17-1 donor cells and A. baumannii ATCC 17978 recipient cells to transfer and integrate pEX18Tc-kan r -baeR-flanking into the chromosome of ATCC 17978 (Additional file 4: Figure S4B). By growing the ATCC 17978 conjugate cells on LB agar containing 10% sucrose, the cells were able to resolve the suicide plasmid pEX18Tc (Additional file 4: Figure S4C). Sucrose-resistant colonies were examined to verify that they had the kanamycin-resistant phenotype as a result of plasmid eviction. The absence of the baeR gene sequence in the genome was verified by PCR and RT-PCR and further confirmed by Southern blot hybridization.
To reconstitute the baeR gene in the baeR-deleted mutants, a DNA fragment carrying the entire baeR gene sequence was generated by PCR using the genomic DNA of A. baumannii ATCC 17978 as the template. Briefly, a kanamycin resistance cassette was first amplified from the pC2HP vector [42] and cloned into the E. coli/Acinetobacter shuttle vector pWH1266 [43,44] (Additional file 5: Figure S5A and S5B). Subsequently, the baeR DNA fragment was cloned into the XbaI/XhoI restriction sites (Additional file 5: Figure S5C). The plasmid was transformed into the wild-type strain and the baeR deletion mutants by electroporation, thus creating the baeR-overexpressing strain and complemented mutant strains, respectively. The overexpression and baeR-reconstituted strains were selected on LB agar containing 10 μg/mL tetracycline and were further verified by PCR (Additional file 5: Figure S5D) and RT-PCR (Additional file 2: Figure S2).
Southern blot hybridization
Southern blot analysis was performed as reported in a previous publication [45]. Genomic DNA was extracted, and approximately 10 μg was digested with BclI overnight at 50°C. The DNA was then separated on a 0.8% agarose gel containing 1:10,000 SYBR Safe gel stain (Invitrogen, Grand Island, NY), transferred onto a positively charged nylon membrane (Pall Corporation, Port Washington, NY) via the alkaline transfer method [38], and fixed by baking at 80°C for 2 h. The membrane was hybridized with an [α-32P] dCTP-labeled baeS probe (Additional file 3: Figure S3A) using prehybridization buffer (6× saline sodium citrate [SSC; 1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 5× Denhardt’s reagent, 0.5% SDS, 100 μg/mL salmon sperm DNA, and 50% formamide) at 42°C overnight. The membrane was then washed and visualized by autoradiography.
Time-kill assay
The time-kill assays were carried out in duplicate as previously described [46] with some modifications. Briefly, cells were grown to log phase and sub-cultured into 10 mL CAMHB broth without (control) or with tigecycline (0.25 or 0.5 μg/mL) to a cell density of approximately 5 × 105 CFU/mL. The cultures were incubated in an ambient atmosphere at 37°C. At different time points (0, 4, 8, 12, and 16 h) after inoculation, 0.1 mL of the culture was removed from each tube and 10-fold serially diluted. Then, 25 μL of each diluted cell suspension was spotted onto LB agar in duplicate. Viable cell counts were determined, the duplicates were averaged, and the data were plotted.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
MFL conceived the study design and drafted the manuscript. YYL performed the laboratory work, including the mutant construction and complementation, gene expression, and time-kill assays. HWL carried out the MIC determinations. CYL participated in the overall design of this study and assisted in writing the manuscript. All authors have read and approved the final manuscript.
Supplementary Material
Verification of the baeR deletion mutants. (A) Diagram of the baeR gene and deletion mutant verification using appropriate primers. (B) Successful baeR gene fragment deletion was deduced based on a change in the PCR band size from 4539 bp to 4884 bp.
Southern blot analysis. (A) Genomic DNA from the baeR deletion mutant and the parental strain was digested by BclI. The location of the specific DNA probe is shown. (B) The bands corresponding to 6.7-kb and 2.8-kb fragments are indicated. Four independent clones of AB1026 are included.
Construction of the baeR deletion mutant. (A) A single crossover between pEX18Tc containing baeR upstream and downstream sequences joined by a kan r cassette and the ATCC 17978 chromosome. (B) Two mechanisms by which the plasmid can integrate into the chromosome are diagrammed. (C) The suicide plasmid was excised by 10% sucrose counter-selection and selection of the in-frame baeR deletion strain with kanamycin.
Shuttle vector pWH1266 and verification of pWH1266 introduction into different strains of Acinetobacter baumannii.(A) pWH1266. (B) pWH1266 with kanamycin cassette insertion. (C)baeR insertion into the XbaI/XhoI restriction sites in pWH1266. (D) Successful baeR gene fragment insertion into the kanamycin cassette was deduced based on a change in the PCR band size from 1375 bp to 983 bp. AB1027, AB1028, and AB1029 represent the baeR reconstituted strain, the baeR-overexpressing strain, and the A. baumannii ATCC 17978 strain with pWH1266, respectively.
baeR gene expression in different A. baumannii strains as determined by reverse transcription polymerase chain reaction. No baeR expression could be observed in AB1026. AB1027 was the baeR-reconstituted strain derived from AB1026, which had a baeR expression level similar to that of the wild-type strain. AB1028 and AB1029 represent the baeR-overexpressing strain and A. baumannii ATCC 17978 with pWH1266, respectively.
Contributor Information
Ming-Feng Lin, Email: c9977@ms27.hinet.net.
Yun-You Lin, Email: s110481@hotmail.com.
Hui-Wen Yeh, Email: vivian3929@yahoo.com.tw.
Chung-Yu Lan, Email: cylan@life.nthu.edu.tw.
Acknowledgements
This study was supported by a grant from the National Taiwan University Hospital, Chu-Tung Branch. The authors also thank Dr. Kia-Chih Chang (Tzu Chi University, Taiwan) for providing the clinical A. baumannii strains and Dr. Ming-Li Liou (Yuanpei University, Taiwan) for providing the wild-type strain. We also thank Jeng-Yi Chen for his technical assistance.
References
- Fournier PE, Richet H. The epidemiology and control of Acinetobacter baumannii in health care facilities. Clin Infect Dis. 2006;42(5):692–699. doi: 10.1086/500202. [DOI] [PubMed] [Google Scholar]
- Perez F, Hujer AM, Hujer KM, Decker BK, Rather PN, Bonomo RA. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother. 2007;51(10):3471–3484. doi: 10.1128/AAC.01464-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes RE, Farrell DJ, Sader HS, Jones RN. Comprehensive assessment of tigecycline activity tested against a worldwide collection of Acinetobacter spp. (2005–2009) Diagn Microbiol Infect Dis. 2010;68(3):307–311. doi: 10.1016/j.diagmicrobio.2010.07.003. [DOI] [PubMed] [Google Scholar]
- Lauderdale TL, Clifford McDonald L, Shiau YR, Chen PC, Wang HY, Lai JF, Ho M. The status of antimicrobial resistance in Taiwan among gram-negative pathogens: the Taiwan surveillance of antimicrobial resistance (TSAR) program, 2000. Diagn Microbiol Infect Dis. 2004;48(3):211–219. doi: 10.1016/j.diagmicrobio.2003.10.005. [DOI] [PubMed] [Google Scholar]
- Gordon NC, Wareham DW. Multidrug-resistant Acinetobacter baumannii: mechanisms of virulence and resistance. Int J Antimicrob Agents. 2010;35(3):219–226. doi: 10.1016/j.ijantimicag.2009.10.024. [DOI] [PubMed] [Google Scholar]
- Rose WE, Rybak MJ. Tigecycline: first of a new class of antimicrobial agents. Pharmacotherapy. 2006;26(8):1099–1110. doi: 10.1592/phco.26.8.1099. [DOI] [PubMed] [Google Scholar]
- Peleg AY, Adams J, Paterson DL. Tigecycline Efflux as a Mechanism for Nonsusceptibility in Acinetobacter baumannii. Antimicrob Agents Chemother. 2007;51(6):2065–2069. doi: 10.1128/AAC.01198-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzin A, Keeney D, Bradford PA. AdeABC multidrug efflux pump is associated with decreased susceptibility to tigecycline in Acinetobacter calcoaceticus-Acinetobacter baumannii complex. J Antimicrob Chemother. 2007;59(5):1001–1004. doi: 10.1093/jac/dkm058. [DOI] [PubMed] [Google Scholar]
- Vila J, Marti S, Sanchez-Cespedes J. Porins, efflux pumps and multidrug resistance in Acinetobacter baumannii. J Antimicrob Chemother. 2007;59(6):1210–1215. doi: 10.1093/jac/dkl509. [DOI] [PubMed] [Google Scholar]
- Alm E, Huang K, Arkin A. The evolution of two-component systems in bacteria reveals different strategies for niche adaptation. PLoS Comput Biol. 2006;2(11):e143. doi: 10.1371/journal.pcbi.0020143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AH, Stock AM. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem Sci. 2001;26(6):369–376. doi: 10.1016/S0968-0004(01)01852-7. [DOI] [PubMed] [Google Scholar]
- Sun S, Negrea A, Rhen M, Andersson DI. Genetic analysis of colistin resistance in Salmonella enterica serovar Typhimurium. Antimicrob Agents Chemother. 2009;53(6):2298–2305. doi: 10.1128/AAC.01016-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishii R, Takei M. Relationship between the expression of ompF and quinolone resistance in Escherichia coli. J Infect Chemother. 2009;15(6):361–366. doi: 10.1007/s10156-009-0716-6. [DOI] [PubMed] [Google Scholar]
- Barrow K, Kwon DH. Alterations in two-component regulatory systems of phoPQ and pmrAB are associated with polymyxin B resistance in clinical isolates of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2009;53(12):5150–5154. doi: 10.1128/AAC.00893-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand I, Damier-Piolle L, Courvalin P, Lambert T. Expression of the RND-type efflux pump AdeABC in Acinetobacter baumannii is regulated by the AdeRS two-component system. Antimicrob Agents Chemother. 2004;48(9):3298–3304. doi: 10.1128/AAC.48.9.3298-3304.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun JR, Perng CL, Chan MC, Morita Y, Lin JC, Su CM, Wang WY, Chang TY, Chiueh TS. A truncated AdeS kinase protein generated by ISAba1 insertion correlates with tigecycline resistance in Acinetobacter baumannii. PLoS ONE. 2012;7(11):e49534. doi: 10.1371/journal.pone.0049534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bury-Mone S, Nomane Y, Reymond N, Barbet R, Jacquet E, Imbeaud S, Jacq A, Bouloc P. Global analysis of extracytoplasmic stress signaling in Escherichia coli. PLoS Genet. 2009;5(9):e1000651. doi: 10.1371/journal.pgen.1000651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leblanc SK, Oates CW, Raivio TL. Characterization of the induction and cellular role of the BaeSR two-component envelope stress response of Escherichia coli. J Bacteriol. 2011;193(13):3367–3375. doi: 10.1128/JB.01534-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appia-Ayme C, Patrick E, Sullivan MJ, Alston MJ, Field SJ, AbuOun M, Anjum MF, Rowley G. Novel inducers of the envelope stress response BaeSR in Salmonella Typhimurium: BaeR is critically required for tungstate waste disposal. PLoS ONE. 2011;6(8):e23713. doi: 10.1371/journal.pone.0023713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosner JL, Martin RG. Reduction of cellular stress by TolC-dependent efflux pumps in Escherichia coli indicated by BaeSR and CpxARP activation of spy in efflux mutants. J Bacteriol. 2013;195(5):1042–1050. doi: 10.1128/JB.01996-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino K, Honda T, Yamaguchi A. Genome-wide analyses of Escherichia coli gene expression responsive to the BaeSR two-component regulatory system. J Bacteriol. 2005;187(5):1763–1772. doi: 10.1128/JB.187.5.1763-1772.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranova N, Nikaido H. The BaeSR Two-Component Regulatory System Activates Transcription of the yegMNOB (mdtABCD) Transporter Gene Cluster in Escherichia coli and Increases Its Resistance to Novobiocin and Deoxycholate. J Bacteriol. 2002;184(15):4168–4176. doi: 10.1128/JB.184.15.4168-4176.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara E, Nikaido H. OmpA is the principal nonspecific slow porin of Acinetobacter baumannii. J Bacteriol. 2012;194(15):4089–4096. doi: 10.1128/JB.00435-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne S, Guigon G, Courvalin P, Perichon B. Screening and quantification of the expression of antibiotic resistance genes in Acinetobacter baumannii with a microarray. Antimicrob Agents Chemother. 2010;54(1):333–340. doi: 10.1128/AAC.01037-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornsey M, Ellington MJ, Doumith M, Thomas CP, Gordon NC, Wareham DW, Quinn J, Lolans K, Livermore DM, Woodford N. AdeABC-mediated efflux and tigecycline MICs for epidemic clones of Acinetobacter baumannii. J Antimicrob Chemother. 2010;65(8):1589–1593. doi: 10.1093/jac/dkq218. [DOI] [PubMed] [Google Scholar]
- Hou PF, Chen XY, Yan GF, Wang YP, Ying CM. Study of the correlation of imipenem resistance with efflux pumps AdeABC, AdeIJK, AdeDE and AbeM in clinical isolates of Acinetobacter baumannii. Chemotherapy. 2012;58(2):152–158. doi: 10.1159/000335599. [DOI] [PubMed] [Google Scholar]
- Henry R, Vithanage N, Harrison P, Seemann T, Coutts S, Moffatt JH, Nation RL, Li J, Harper M, Adler B, Boyce JD. Colistin-resistant, lipopolysaccharide-deficient Acinetobacter baumannii responds to lipopolysaccharide loss through increased expression of genes involved in the synthesis and transport of lipoproteins, phospholipids, and poly-beta-1,6-N-acetylglucosamine. Antimicrob Agents Chemother. 2012;56(1):59–69. doi: 10.1128/AAC.05191-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemec A, Maixnerova M, van der Reijden TJ, van den Broek PJ, Dijkshoorn L. Relationship between the AdeABC efflux system gene content, netilmicin susceptibility and multidrug resistance in a genotypically diverse collection of Acinetobacter baumannii strains. J Antimicrob Chemother. 2007;60(3):483–489. doi: 10.1093/jac/dkm231. [DOI] [PubMed] [Google Scholar]
- Coyne S, Courvalin P, Perichon B. Efflux-mediated antibiotic resistance in Acinetobacter spp. Antimicrob Agents Chemother. 2011;55(3):947–953. doi: 10.1128/AAC.01388-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnet S, Courvalin P, Lambert T. Resistance-nodulation-cell division-type efflux pump involved in aminoglycoside resistance in Acinetobacter baumannii strain BM4454. Antimicrob Agents Chemother. 2001;45(12):3375–3380. doi: 10.1128/AAC.45.12.3375-3380.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzin A, Immermann FW, Bradford PA. RT-PCR and statistical analyses of adeABC expression in clinical isolates of Acinetobacter calcoaceticus-Acinetobacter baumannii complex. Microb Drug Resist. 2010;16(2):87–89. doi: 10.1089/mdr.2009.0131. [DOI] [PubMed] [Google Scholar]
- Wieczorek P, Sacha P, Hauschild T, Zorawski M, Krawczyk M, Tryniszewska E. Multidrug resistant Acinetobacter baumannii–the role of AdeABC (RND family) efflux pump in resistance to antibiotics. Folia Histochem Cytobiol. 2008;46(3):257–267. doi: 10.2478/v10042-008-0056-x. [DOI] [PubMed] [Google Scholar]
- Ma D, Alberti M, Lynch C, Nikaido H, Hearst JE. The local repressor AcrR plays a modulating role in the regulation of acrAB genes of Escherichia coli by global stress signals. Mol Microbiol. 1996;19(1):101–112. doi: 10.1046/j.1365-2958.1996.357881.x. [DOI] [PubMed] [Google Scholar]
- Fernando D, Kumar A. Growth phase-dependent expression of RND efflux pump- and outer membrane porin-encoding genes in Acinetobacter baumannii ATCC 19606. J Antimicrob Chemother. 2012;67(3):569–572. doi: 10.1093/jac/dkr519. [DOI] [PubMed] [Google Scholar]
- Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute. Methods for dilution antimicrobial susceptibility test for bacteria that grow aerobically-approved standard-ninth edition, M07-A9., vol. M07-A9. Wayne PA: CLSI; 2012. [Google Scholar]
- Pachon-Ibanez ME, Jimenez-Mejias ME, Pichardo C, Llanos AC, Pachon J. Activity of tigecycline (GAR-936) against Acinetobacter baumannii strains, including those resistant to imipenem. Antimicrob Agents Chemother. 2004;48(11):4479–4481. doi: 10.1128/AAC.48.11.4479-4481.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell W. Molecular cloning: a laboratory manual. 3. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Li J, Rayner CR, Nation RL, Owen RJ, Spelman D, Tan KE, Liolios L. Heteroresistance to colistin in multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother. 2006;50(9):2946–2950. doi: 10.1128/AAC.00103-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene. 1998;212(1):77–86. doi: 10.1016/S0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- Reuss O, Vik A, Kolter R, Morschhauser J. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene. 2004;341:119–127. doi: 10.1016/j.gene.2004.06.021. [DOI] [PubMed] [Google Scholar]
- Stynen B, Van Dijck P, Tournu H. A CUG codon adapted two-hybrid system for the pathogenic fungus Candida albicans. Nucleic Acids Res. 2010;38(19):e184. doi: 10.1093/nar/gkq725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunger M, Schmucker R, Kishan V, Hillen W. Analysis and nucleotide sequence of an origin of DNA replication in Acinetobacter calcoaceticus and its use for Escherichia coli shuttle plasmids. Gene. 1990;87(1):45–51. doi: 10.1016/0378-1119(90)90494-C. [DOI] [PubMed] [Google Scholar]
- Liou ML, Soo PC, Ling SR, Kuo HY, Tang CY, Chang KC. The sensor kinase BfmS mediates virulence in Acinetobacter baumannii. J Microbiol Immunol Infect. in press. [DOI] [PubMed]
- Hsu PC, Yang CY, Lan CY. Candida albicans Hap43 is a repressor induced under low-iron conditions and is essential for iron-responsive transcriptional regulation and virulence. Eukaryot Cell. 2011;10(2):207–225. doi: 10.1128/EC.00158-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantar C, Di Chiara M, Nicola F, Relloso S, Smayevsky J. Comparative in vitro bactericidal activity between cefepime and ceftazidime, alone and associated with amikacin, against carbapenem-resistant Pseudomonas aeruginosa strains. Diagn Microbiol Infect Dis. 2000;37(1):41–44. doi: 10.1016/S0732-8893(99)00156-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Verification of the baeR deletion mutants. (A) Diagram of the baeR gene and deletion mutant verification using appropriate primers. (B) Successful baeR gene fragment deletion was deduced based on a change in the PCR band size from 4539 bp to 4884 bp.
Southern blot analysis. (A) Genomic DNA from the baeR deletion mutant and the parental strain was digested by BclI. The location of the specific DNA probe is shown. (B) The bands corresponding to 6.7-kb and 2.8-kb fragments are indicated. Four independent clones of AB1026 are included.
Construction of the baeR deletion mutant. (A) A single crossover between pEX18Tc containing baeR upstream and downstream sequences joined by a kan r cassette and the ATCC 17978 chromosome. (B) Two mechanisms by which the plasmid can integrate into the chromosome are diagrammed. (C) The suicide plasmid was excised by 10% sucrose counter-selection and selection of the in-frame baeR deletion strain with kanamycin.
Shuttle vector pWH1266 and verification of pWH1266 introduction into different strains of Acinetobacter baumannii.(A) pWH1266. (B) pWH1266 with kanamycin cassette insertion. (C)baeR insertion into the XbaI/XhoI restriction sites in pWH1266. (D) Successful baeR gene fragment insertion into the kanamycin cassette was deduced based on a change in the PCR band size from 1375 bp to 983 bp. AB1027, AB1028, and AB1029 represent the baeR reconstituted strain, the baeR-overexpressing strain, and the A. baumannii ATCC 17978 strain with pWH1266, respectively.
baeR gene expression in different A. baumannii strains as determined by reverse transcription polymerase chain reaction. No baeR expression could be observed in AB1026. AB1027 was the baeR-reconstituted strain derived from AB1026, which had a baeR expression level similar to that of the wild-type strain. AB1028 and AB1029 represent the baeR-overexpressing strain and A. baumannii ATCC 17978 with pWH1266, respectively.