Abstract
RNA splicing is the cellular process that has only recently been found to be an important target for various cancers. Among the spliceosome genes that are involved in cancers, SF3B1 is most frequently mutated. Recurrent mutation in codon 625 has been found in uveal melanoma, but this mutation has not been identified in cutaneous melanoma. We used whole-exome sequencing to explore the mutational landscape of 295 melanoma samples, 231 of which are cutaneous melanoma. Out of these cutaneous melanoma samples, we found 2 samples with R625 mutation in SF3B1 gene. The results were validated by Sanger sequencing. We conclude that SF3B1 R625 mutation does occur in cutaneous melanoma, although with a low frequency (~1%).
Keywords: Melanoma, SF3B1, Cancer genetics, Whole-exome sequencing
Introduction
Recent high throughput sequencing of cancer genomes has led to new discoveries of mutations in cellular processes that were not previously known to play a causal role in cancer. One such processes is RNA-splicing (for review see [1]). Among the recently discovered spliceosome genes that are involved in cancers, SF3B1 is the most frequently mutated. SF3B1 mutations are found with high frequency in myelodysplastic syndromes (MDS) and chronic myelogenous leukemia (CLL); it is also mutated in solid tumors such as lung adenocarcinomas, breast cancer, and pancreatic cancer. In uveal melanoma, recurrent mutations at codon 625 of SF3B1 have been identified [2–4].
SF3B1 encodes subunit one of the splicing factor 3b protein complex, which is part of the U2 small nuclear RNAs (snRNP) that binds pre-mRNA upstream of the intron’s branch site. SF3B1 mutations usually occur within the 22 HEAT repeats in the C-terminal region. Codon R625 has the highest mutation frequency in uveal melanoma [2–4], whereas the mutation hotspot is at K700 for MDS and CLL.
While multiple groups identified SF3B1 R625 mutations in uveal melanoma [2–4], none identified this mutation in cutaneous melanoma [5]. We have sequenced the whole exome of a cohort of 295 melanoma samples, 231 of which are cutaneous melanoma [6]. We report here that although the frequency is low, SF3B1 R625 mutation does occur in cutaneous melanoma.
Material and methods
Sample selection
For details of sample selection, see reference [6].
Exome sequencing and data analysis
For details of exome sequencing and data analysis, see reference [6].
Sanger sequencing
The following primers were used to amplify the region of the SF3B1 R625 for validation by Sanger sequencing: F1: 5′-CCTCGTGGTCATTGAACCGC -3′; B1: 5′-ACTTCTAAGATGTGGCAAGATGGC -3′
Results
Our cohort contains all samples described in an earlier publication [6] with additional samples (Table 1). The whole-exome sequencing of 295 melanoma samples identified 5 with mutations in SF3B1 R625 (Table 2). Out of these five samples, two (YUBOO and G2306T) are uveal melanoma, two (YUPAO and YUGAFFE) are cutaneous melanoma, and one sample for which the location of the primary lesion is unknown (YUKAY). Three samples have p.R625H mutation, while the other two have p. R625C mutation (amino acids are numbered based on GenBank accession NM_012433.2). The alignments of reads from whole-exome sequencing around the mutations convincingly show that the mutations are authentic, even though the mutation frequency is low. The whole-exome sequencing results were validated by Sanger sequencing, as shown in Figure 1. The mutation peaks in both samples are weak when compared with the reference peaks. These mutation peaks, however, correlate with the mutation frequencies detected in the whole-exome sequencing. For the cutaneous sample YUPAO (shown in the top of Figure 1), there are total 251 reads mapped to this position, with the number of reads for each base as: A=0, C=211 (84%, 80+, 131−), G=0, T=40 (16%, 15+, 25−). The “+” and “−” signs indicate the numbers of reads in forward and reverse strands respectively. SF3B1 is on the reverse strand of hg19 reference genome and the bases shown here are the original counts on hg19 forward strand. For the uveal sample YUBOO (shown in the bottom of Figure 1), there are total 137 reads, with the number of reads for each base as: A=0, C=112(82%, 39+, 73−), G=0, T=25 (18%, 9+, 16−). The other samples with mutations in SF3B1 R625 show similar mutation frequencies. The low mutation frequencies might reflect the presence of stroma in the samples. The matching normal DNAs available for four of the five samples were all wild-type, indicating the somatic origin SF3B1 R625.
Table 1.
Samples types and their metastatic status
| Tumor type | Primary | Metastasis | Total |
|---|---|---|---|
| Cutaneous | 66 | 165 | 231 |
| Mucosal | 3 | 7 | 10 |
| Ocular | 20 | 5 | 25 |
| Unknown | 0 | 29 | 29 |
| Total | 89 | 206 | 295 |
Table 2.
Samples with SF3B1 R625 mutation
| Sample name | Chromosome position (chr2, hg19/GRCh37) | Nucleotide change | Amino acid change | BRAF V600 mutation | NRAS Q61 mutation | Tumor type/origin/tumor location | Primary or metastatic |
|---|---|---|---|---|---|---|---|
| YUBOO | 198267483 | C > T | R625H | - | - | uveal/uvea/head/neck | metastatic |
| YUKAY | 198267483 | C > T | R625H | - | - | unknown/unknown/trunk | metastatic |
| YUPAO | 198267483 | C > T | R625H | - | - | acral/acral/extremity | metastatic |
| YUGAFFE | 198267484 | G > A | R625C | V600Ka | - | cutaneous/head/neck/lymph node | metastatic |
| G2306T | 198267484 | G > A | R625C | - | - | uveal/uvea/uvea | primary |
double mutations in the same codon with chr7:140453136 A>T and chr7:140453137 C>T
Figure 1.
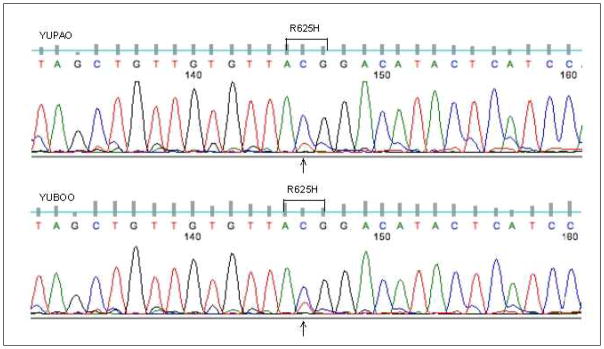
Sequencing chromatograms for two samples showing the SF3B1 mutation at R625 position. On the top is cutaneous sample YUPAO and on the bottom is uveal sample YUBOO.
One melanoma sample in this cohort has BRAF V600 mutation (YUGAFFE) and none has NRAS mutation (Table 2).
Other SF3B1 non-synonymous somatic mutations were also present as listed in Table 3 (amino acids are numbered based on GenBank accession NM_012433.2).
Table 3.
Additional SF3B1 non-synonymous somatic mutations.
| position | Nucleotide change | Amino acid change | Sample names | Sample type/origin/tumor location | Primary or metastatic |
|---|---|---|---|---|---|
| 198273258 | G > A | R318*-R | YUKLAB | cutaneous/unknown/trunk | metastatic |
| 198267432 | G > A | P642L-P | YULAN | cutaneous/head/neck/lymph node | metastatic |
| 198267360 | T > G | K666K-T | YUGISMO | cutaneous/trunk/trunk | metastatic |
| 198266795 | C > T | A713A-T | YUPAT | cutaneous/trunk/lung | metastatic |
| 198266497 | G > C | P780P-R | G2310T | uveal/uvea/uvea | primary |
| 198262755 | G > A | R1074C-R | YUSEN | cutaneous/head/neck/trunk | metastatic |
| 198260927 | G > T | A1131A-D | YUEGO | cutaneous/head/neck/trunk | metastatic |
| 198257761 | T > C | M1231M-V | YUKNOLL | unknown/unknown/liver | metastatic |
| 198257176 | G > A | H1256H-Y | YUBEL | unknown/unknown/lymph node | metastatic |
Discussion
The report of mutations in codon 625 of SF3B1 in uveal melanoma [2–4] prompted further investigation regarding the presence of this mutation in cutaneous melanoma [5]. The published study examined 85 cutaneous melanoma samples using direct Sanger sequencing [5]. The cohort included 22 superficial spreading, 24 acral-lentiginous, 36 nodular, and 3 lentigo-maligna melanomas. The mutation was not detected in 81 samples (four samples failed or were ambiguous in the sequencing). In our larger cohort, we found five samples with SF3B1 R625 mutations, two of which are cutaneous melanoma and one of unknown origin, showing that the mutation does occur in this type of melanoma, although at low frequency.
We also noticed that most of our samples that have SF3B1 R625 mutations are metastatic melanoma. This is in contrast to previous finding that SF3B1 R625 mutations are rare in metastatic tumors and are associated with better prognosis [2, 4, 7]. Harbour et al did not detect any SF3B1 mutation in their five distant metastatic uveal melanoma [2], and Griewank et al found one out of 26 metastatic uveal melanoma samples [7]. The difference might be due to sample bias, since there are more metastatic than primary tumors in our cohort. The other possibility is that the SF3B1 mutations might play different roles in uveal and cutaneous melanomas. SF3B1 mutations have been found to be associated with different prognosis in different type of cancers. The SF3B1 mutations in CLL are associated with poorer prognosis [8], while in MDS the mutations are associated with better prognosis [9].
Different cancers also have different predominate mutations in SF3B1. In hematological, breast and pancreatic cancers codon K700 mutations predominate, whereas in uveal melanoma the R625 codon mutations predominate. We did not detect any K700 mutation in our cohort. It seems that SF3B1 plays different roles in the biology of different cancers. Although now it is clear that SF3B1, as well as other genes involved in RNA splicing, plays causal role in cancer, the exact effect of the mutations on protein functions and splicing patterns are still needed to be fully elucidated by further experiments.
We detected mutations in BAP1 in both sun-exposed and uveal melanoma [6]. Another gene, EIF1AX, which encodes eukaryotic translation initiation factor 1A (eIF1A), was also recently found to be frequently mutated in uveal melanoma [3]. Interestingly, in our cohort we also found EIF1AX mutations in both uveal and cutaneous melanomas. We found 6 mutations in 25 uveal melanomas (~24%) and 5 mutations in 231 cutaneous melanomas (~2%). As discovered previously in uveal melanomas [3], the nonsynonymous EIF1AX mutations are clustered around the N terminus of the protein for both cutaneous and uveal melanomas in our cohort (data not shown). It would be interesting to see if interactions between the different cellular machinery encoded by these two genes play a role in uveal and cutaneous melanoma biology.
Acknowledgments
This work was supported by the Yale SPORE in Skin Cancer funded by the National Cancer Institute grant number 1 P50 CA121974 (principal investigator, R.H.), the Melanoma Research Alliance (a Team award to R.H. and, M.K.), Gilead Sciences, Inc. (R.H.) and a generous gift from Roz and Jerry Meyer (R.H).
Contributor Information
Yong Kong, Email: yong.kong@yale.edu.
Michael Krauthammer, Email: michael.krauthammer@yale.edu.
Ruth Halaban, Email: ruth.halaban@yale.edu.
References
- 1.Garraway LA, Lander ES. Lessons from the cancer genome. Cell. 2013;153:17–37. doi: 10.1016/j.cell.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 2.Harbour JW, Roberson ED, Anbunathan H, Onken MD, Worley LA, Bowcock AM. Recurrent mutations at codon 625 of the splicing factor SF3B1 in uveal melanoma. Nat Genet. 2013;45:133–135. doi: 10.1038/ng.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martin M, Masshofer L, Temming P, Rahmann S, Metz C, Bornfeld N, van de Nes J, Klein-Hitpass L, Hinnebusch AG, Horsthemke B, et al. Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nat Genet. 2013 doi: 10.1038/ng.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Furney SJ, Pedersen M, Gentien D, Dumont AG, Rapinat A, Desjardins L, Turajlic S, Piperno-Neumann S, de la Grange P, Roman-Roman S, et al. SF3B1 mutations are associated with alternative splicing in uveal melanoma. Cancer Discov. 2013 doi: 10.1158/2159-8290.CD-13-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schilling B, Bielefeld N, Sucker A, Hillen U, Zimmer L, Schadendorf D, Zeschnigk M, Griewank KG. Lack of SF3B1 R625 mutations in cutaneous melanoma. Diagn Pathol. 2013;8:87. doi: 10.1186/1746-1596-8-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krauthammer M, Kong Y, Ha BH, Evans P, Bacchiocchi A, McCusker JP, Cheng E, Davis MJ, Goh G, Choi M, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet. 2012;44:1006–1014. doi: 10.1038/ng.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Griewank KG, van de Nes J, Schilling B, Moll I, Sucker A, Kakavand H, Haydu LE, Asher M, Zimmer L, Hillen U, et al. Genetic and clinico-pathologic analysis of metastatic uveal melanoma. Mod Pathol. 2013 doi: 10.1038/modpathol.2013.138. [DOI] [PubMed] [Google Scholar]
- 8.Quesada V, Conde L, Villamor N, Ordonez GR, Jares P, Bassaganyas L, Ramsay AJ, Bea S, Pinyol M, Martinez-Trillos A, et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat Genet. 2012;44:47–52. doi: 10.1038/ng.1032. [DOI] [PubMed] [Google Scholar]
- 9.Papaemmanuil E, Cazzola M, Boultwood J, Malcovati L, Vyas P, Bowen D, Pellagatti A, Wainscoat JS, Hellstrom-Lindberg E, Gambacorti-Passerini C, et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med. 2011;365:1384–1395. doi: 10.1056/NEJMoa1103283. [DOI] [PMC free article] [PubMed] [Google Scholar]


