Abstract
During the initial stages of the base excision DNA repair (BER) pathway, DNA glycosylases are responsible for locating and removing the majority of endogenous oxidative base lesions. The bifunctional formamidopyrimidine DNA glycosylase (Fpg) and endonuclease VIII (Nei) are members of the Fpg/Nei family, one of the two families of glycosylases that recognize oxidized DNA bases, the other being the HhH/GPD (or Nth) superfamily. Structural and biochemical developments over the past decades have led to novel insights into the mechanism of damage recognition by the Fpg/Nei family of enzymes. Despite the overall structural similarity among members of this family, these enzymes exhibit distinct features that make them unique. This review summarizes the current structural knowledge of the Fpg/Nei family members, emphasizes their substrate specificities, and describes how these enzymes search for lesions.
Keywords: Base excision repair; bifunctional-DNA glycosylase; lesion recognition, structure and substrates; substrate specificity
I. Introduction
Escherichia coli formidopyrimidine (Fapy) DNA glycosylase (Fpg) was originally discovered in Tomas Lindahl’s laboratory as a DNA glycosylase that removes methylFapyG from alkylated DNA (1). The E. coli gene for Fpg was subsequently cloned (2) and the protein further characterized in a number of laboratories (3-6). E. coli mutM mutants were identified in Jeffrey Miller’s laboratory as mutators that gave rise to G→T transversions (7). When the MutM protein was subsequently purified, it was found to be identical to Fpg (8). Following these initial findings there were a number of biochemical studies showing that 8-oxoguanine (8-oxoG) was also a substrate for Fpg and that Fpg preferred 8-oxoG over methylFapyG (9,10). Because of this substrate preference and because guanine is the most readily oxidized DNA base, the conclusion was drawn that 8-oxoguanine was the biologically relevant substrate for Fpg. These studies led to the formulation of the GO model for 8-oxoG repair (11) which proposed that when guanine is oxidized to 8-oxoguanine, it is removed by Fpg. If 8-oxoG is not removed prior to replication, A is often inserted opposite the 8-oxoG by DNA polymerases (12-15). If this occurs, the A can be removed by another glycosylase called MutY (16). The GO model also included MutT that removes 8-oxoguanine nucleoside triphosphates from the nucleotide pool by hydrolyzing them to 8-oxodGMP (17). Taken together these data supported the idea that 8-oxoguanine is a biologically important, potentially mutagenic oxidative DNA lesion. However recent studies have shown that unmethylated FapyG is also a good substrate for Fpg (18,19) and like 8-oxoG, A can also be incorporated opposite FapyG (20,21) and the incorporated A can be removed by MutY (22). FapyG, which is formed from the same adduct radical as 8-oxoG (23), appears to be responsible for a substantial number of mutations originally attributed to 8-oxoG and thus is also a biologically relevant substrate (24).
E. coli nei (endonuclease VIII) was originally discovered in the Wallace laboratory as an activity that recognizes oxidized pyrimidines (25,26). The gene was cloned and the protein sequence was shown to be very similar to that of Fpg (27). nei mutants had little or no phenotype, but, when coupled with an nth mutation, they were mutators leading to C→T transitions (27). The nth gene encodes endonuclease III which also recognizes oxidized pyrimidines with a substrate specificity that substantially overlaps that of Nei (for reviews see (28,29)).
It was not until the twenty-first century and the sequencing of the human genome that in silico analysis allowed the Wallace, Mitra, and Seeberg laboratories to identify, clone and characterize three Fpg/Nei homologs in mammalian cells, the so-called Neil1 (nei-like), Neil2 and Neil3 proteins (30-34). Mouse Neil1 and Neil3 were also found in mice nullizygous for nth (35). The substrate specificities of human NEIL1 and NEIL2 have been well-characterized (30-34,36-40). In addition, NEIL1 forms specific interactions with a number of replication proteins and is cell cycle regulated (41-44). Thus, it has been proposed that NEIL1 acts as a cow catcher ahead of the replication fork, eliminating potentially mutagenic lesions (42-44). NEIL2 prefers lesions in single-stranded DNA over duplex DNA and interacts with a number of transcription factors including RNA polymerase II and has been suggested to act in transcription-coupled repair (45). Although attempts had been made to determine the activity of NEIL3 (33,46,47), it has only been recently that NEIL3 has been purified and characterized (24,48) and its glycosylase activity shown to be similar to that of NEIL2 (24). In mice, Neil3 is present during embryonic development (49) and was found in brain stem cells (49,50). In humans, expression of NEIL3 has only been observed in thymus (51).
II. Fpg/Nei Phylogeny
Sequence alignments of members of the Fpg/Nei family of glycosylases indicate that they share many structural and biochemical features (34). Some of the hallmark motifs of this family include conserved residues in the helix-two-turns-helix motif (H2TH), a zinc finger motif, and a common catalytic mechanism involving either an N-terminal proline (for example in NEIL1 and NEIL2) or a valine residue (as in human NEIL3 and the giant mimivirus Nei2 (MvNei2)) as the active site nucleophile. Despite these commonalities, each glycosylase prefers a different spectrum of oxidative lesions. Moreover, some of these subfamilies have changed significantly in sequence from their common ancestor, making it difficult to infer the evolution of these enzymes.
Phylogenetic analysis and functional studies of the Fpg/Nei family indicate that in Actinobacteria alone, six gene clades occur, two within the Nei proteins and four within the Fpg clade (52). The plant and fungi clade is clearly part of the Fpg family while within metazoans, Neil2 and Neil3 form their own clade separate from Neil1. The Neil1 protein, like members of the plant and fungi Fpg/Nei proteins, does not have the canonical zinc finger, but possesses a “zincless-finger” motif, which lacks the four characteristic cysteine residues that coordinate a zinc ion. This motif superimposes well with the zinc finger domains of EcoNei and EcoFpg, despite the absence of sequence homology. In contrast to Neil1, both Neil2 and Neil3 possess a zinc finger domain: the former contains a C-H-C-C-type zinc finger whereas the latter has a RanBP-type zinc finger very similar to the one found in bacterial Fpg. Some shared conserved structural features suggest that the zincless fingers evolved independently of the zinc finger motifs. Recent evidence suggests that the Neil2 and Neil3 proteins evolved from a common ancestor while Neil1 evolved separately (Barrantes-Reynolds, unpublished data).
We speculate that horizontal gene transfer, a common occurrence in bacteria, seems to be a likely event in the initial evolution of EcoNei proteins from a common ancestor which contained at least one Fpg/Nei homolog and exhibited features similar to EcoFpg (53). Vertical evolution may have been responsible for the transfer of an early Fpg/Nei gene to early eukaryotes in which these Fpg/Nei homologs led to the diversification of the Fpg/Nei proteins in higher eukaryotes (34,52,53).
III. Fpg/Nei Structures
A. Introduction
Over the past decade, there has been a significant increase in the number of crystal structures of Fpg/Nei glycosylases (54-69). The advent of techniques such as reductive cross-linking using sodium borohydride has played an essential role in trapping stable protein-DNA complexes for the purposes of crystallization and to elucidate the mechanism and role of these intricate enzymes ((54-56,64-66,70,71) and for reviews see (70,71)). Other approaches successfully used to produce stable glycosylase/DNA complexes include the generation of site-directed mutants of active site residues to abolish catalysis and the use of non-cleavable substrates such as tetrahydrofuran (THF) that mimics an abasic (AP) site (60,72) and non-cleavable cyclopentane FapyG (cFapyG) (68). A summary of all the currently available crystal structures of the Fpg/Nei family of glycosylases and their substrate preferences is listed in Table 1. Crystal structures of Fpg proteins from various bacterial species like Thermus thermophilus (Tth) Fpg (without DNA)(54), Escherichia coli (EcoFpg) (55), Geobacillus stearothermophilus Fpg (BstFpg) (57,59,64-66) and Lactococcus lactis Fpg (LlaFpg) (58,60,63,68,69) complexed with DNA substrates have been determined. The Fpg/DNA complexes include Schiff base intermediates, non-covalent complexes with AP-site analogs, and recognition or end-product complexes. Although the structure of EcoNei as a Schiff base intermediate in a complex with DNA was solved (56), it wasn’t until recently that the unliganded structure of EcoNei was determined which revealed a unique and interesting interdomain global conformational change upon DNA binding (62). Furthermore, the structures of unliganded human NEIL1 (61), unliganded MvNei1 and MvNei1 in a complex with THF were subsequently obtained (67). The first crystal structures of an Nei bound to damaged bases were recently reported: MvNeil1 was captured in a complex with DNA containing either thymine glycol (Tg) or 5-hydroxyuracil (5-OHU). 104
Table 1.
Summary of current crystal structures of the Fpg/Nei glycosylases
| Protein | Mutation | Complex | Substrate:Opposite base |
Resolution (Å) |
PDB ID/ References |
Substrate Specificity |
|---|---|---|---|---|---|---|
|
| ||||||
| TthFpg | - | - | 1.90 | 1EE8 (54) | 8-oxoG, FapyG, Me-FapyG, FapyA (54) | |
|
| ||||||
| EcoFpg | - | SBI | - | 2.10 | 1K82 (55) | Sp = 8-oxoG, FapyG, Me-FapyG, FapyA, and
Gh (6,18,72,79,89,101-103) |
|
| ||||||
| BstFpg | - | SBI | - | 1.70 | 1L1Z (57) | 8-oxoG, FapyG, Me-FapyG, FapyA (57,59,64-66) |
| - | SBI | - | 2.40 | 1L2B | ||
| - | RC | rAb:C | 1.80 | 1L1T | ||
| - | RC | rAb:T | 2.20 | 1L2C | ||
| - | RC | rAb:G | 2.00 | 1L2D | ||
|
| ||||||
| E3Q | LRC | 8-oxoG:C | 2.34 | 1R2Y (59) | ||
|
| ||||||
| E3Q | LRC | DHU:C | 1.63 | 1R2Z (64) | ||
| E3Q-DXL | CC1 | 8-oxoG:C | 2.35 | 2F5S | ||
| E3Q-DXL | CC2 | 8-oxoG:C | 2.35 | 2F5Q | ||
| E3Q-DXL | IC1 | A:T | 2.00 | 2F5N | ||
| E3Q-DXL | IC2 | A:T | 2.00 | 2F5P | ||
| E3Q-DXL | IC3 | G:C | 2.05 | 2F5O | ||
|
| ||||||
| E3Q-DXL | LRC3 | 8-oxoG:C | 1.85 | 3GPY (65) | ||
| Δ220-235-DXL | EC3 | 8-oxoG:C | 1.89 | 3GO8 | ||
| V222P-DXL | EC3 | 8-oxoG:C | 2.05 | 3GP1 | ||
| T224P-DXL | EC3 | 8-oxoG:C | 2.15 | 3GPP | ||
| Δ220-235-DXL | EC4 | 8-oxoG:C | 1.62 | 3GPU | ||
| Δ220-235-DXL | IC4 | G:C | 1.78 | 3GPX | ||
| E3Q-DXL | LRC5 | 8-oxoG:C | 1.70 | 3GQ4 | ||
| Δ220-235-DXL | EC5 | 8-oxoG:C | 1.83 | 3GQ3 | ||
| V222P-DXL | IC5 | G:C | 1.90 | 3GQ5 | ||
|
| ||||||
| N174C-DXL | XGC | G:C | 2.60 | 3JR4 (66) | ||
| N174C-DXL | LRC | 8-oxoG:C | 1.70 | 3JR5 | ||
|
| ||||||
| LlaFpg | P1G | RC | Pr:C | 2.55 | 1KFV (58) | 8-oxoG, FapyG, Me-FapyG, FapyA, Hyd (58,60,63,68,69) |
|
| ||||||
| ΔP1 | LRC | cFapydG:C | 1.80 | 1TDZ (60) | ||
|
| ||||||
| P1G | RC | Pr:C | 1.90 | 1NNJ (63) | ||
| P1G | RC | THF:C | 1.90 | 1PJJ | ||
| - | RC | Pr:C | 1.90 | 1PJI | ||
| - | RC | THF:C | 1.95 | 1PM5 | ||
|
| ||||||
| - | RC | cFapydG:C | 1.95 | 1XC8 (68) | ||
| - | RC | N7-Benzyl-FapyG:C | 1.90 | 3C58 | ||
|
| ||||||
| - | LRC | cHyd:C | 1.80 | 2XZF (69) | ||
| - | DPC | cHyd:C | 1.80 | 2XZU | ||
|
| ||||||
| EcoNei | SBI | - | 1.42 | 1K3W (56) | Tg, 5-OHU, 5-OHC, DHT, DHU, Sp,
Gh (34,72,76,81) |
|
| SBI | - | 1.25 | 1K3X | |||
|
| ||||||
| - | - | - | 2.80 | 1Q39 (62) | ||
| E2A | - | - | 2.30 | 1Q3C | ||
| R252A | - | - | 2.05 | 1Q3B | ||
|
| ||||||
| NEIL1 | ΔC-terminal56 | - | - | 2.10 | 1TDH (61) | Sp=Gh>Tg>DHU>5-OHU>5-OHC>DHT>FapyG=FapyA>>8-oxoG (30,36,39,44,61,83,86) |
|
| ||||||
| MvNei1 | - | RC | THF:C | 2.20 | 3A46 (67) | Sp=Gh>Tg>5-OHU>5-OHC>DHT=DHU>>8-oxoG (39,67,82) |
| - | - | 2.60 | 3A42 | |||
| - | - | 2.30 | 3A45 | |||
Schiff-base intermediate: SBI
Disulfide cross-linking: DXL
Recognition complex: RC
Lesion Recognition Complex: LRC
DNA-protein covalent: DPC
Interrogation Complex: IC
Control Complex: CC
Encounter Complex: EC
Extrahelical G complex: XGC
5-hydroxy-5-methylhydantoin: Hyd or cHyd, where c refers to a carbanucleoside
Overall, the structures of the Fpg and Nei proteins are similar, with a distinct 2-domain architecture connected by a flexible hinge region (Figure 1A and B using EcoFpg and EcoNei as examples) (55,56,72). In general, the N-terminal region is predominantly β-sheet rich and is composed of a β-sandwich flanked by α-helices. The C-terminal domain comprises α-helices, two of which form a conserved H2TH motif, as well as two anti-parallel β-strands that fold into a zinc finger motif. These signature motifs are characteristic of both Fpg and Nei subfamilies. The zinc finger and H2TH motifs have been shown to be absolutely required for Fpg to bind to DNA (72-74). In addition to structural similarity, the members of this superfamily exhibit a similar multi-step catalytic mechanism that generally involves a nucleophilic attack at the C1′ position of the target nucleotide by an N-terminal proline residue (in the case of Fpg, Nei, and NEIL1) (75,76). A comparison of these structures is further discussed below.
Figure 1. Overall structural comparison between EcoFpg and EcoNei.
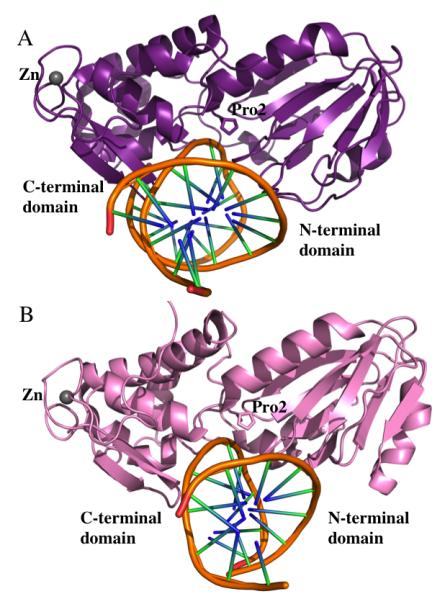
A. EcoFpg bound to DNA. Both and N- and C-terminal domains are colored in purple (PDB ID 1K82) (55). B. EcoNei bound to DNA. N- and C-terminal domains are shown in pale pink. (IK3W from the PDB) (56). Zinc atoms are shown as gray spheres in both cases and the DNA is displayed as a ribbon. PyMol was used to generate the Figures (DeLano Scientific, The PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC.).
B. Substrate Preference
Bacterial Fpg proteins characteristically excise oxidized purines, whereas bacterial Nei and the Neil proteins excise oxidized pyrimidines (for reviews see (28,29,34,71,72)). Interestingly, the Fpg proteins share similar substrate specificity with Ogg whereas the substrate specificity of the Nei family members overlaps with that of the Nth family. However, all Fpg/Nei family members share sequence homology. Glycosylases vary in their discrimination of the bases opposite the lesion as well as their preference for the nature of the DNA, i.e. single-stranded DNA, double-stranded DNA, or bubble-containing substrates. Fpg primarily exhibits a substrate preference for purines such as 8-oxoG and FapyG, albeit oxidized pyrimidines are also removed (77,78) whereas bacterial Nei and the rest of the eukaryotic family members such as NEIL1 and NEIL2 recognize a wider array of substrates.
Recently, it was determined that EcoFpg is more efficient at removing spiroiminodihydantoin (Sp), a further oxidation product of 8-oxoG, from double-stranded DNA substrates than 8-oxoG itself (79,80). EcoNei, like endonuclease III, recognizes Tg, dihydrothymine (DHT), β-ureidoisobutyric acid and urea residues (for reviews see (28,29,34,72,76). EcoNei can also recognize 5-hydroxycytosine (5-OHC), 5-hydroxyuracil (5-OHU) and uracil glycol (81). MvNei1 and NEIL1 share substrate preferences for oxidized pyrimidines in duplex DNA and also recognize and process lesions from single-stranded DNA (82,83). Although 8-oxoG is not a preferred substrate for NEIL1, its further oxidation products guanidinohydantoin (Gh) and Sp are both excellent substrates for these enzymes when paired opposite C rather than A (39,40,83). The NEIL1 protein also excises Tg, 5,6-dihydrouracil (DHU), FapyA and FapyG, as well as 5-OHU, 5-OHC, and oxanine (30,31,84-86). Bacterial Fpg (87,88) and all the eukaryotic members of the Fpg/Nei family recognize lesions in single-stranded DNA (31,36,83). NEIL2 and MmuNeil3 prefer to excise lesions present in single-stranded, bubble or forked DNA structures over duplex DNA (24,36,39,42-44,48).
C. Comparison of Structures of the Fpg/Nei Family
As mentioned above, there are currently crystal structures of Fpg proteins from four bacterial species, namely TthFpg, EcoFpg, BstFpg, and LlaFpg (54,55,57-60,63-66,68,69). All four proteins share the same domain structure and considerable sequence homology. Structures of intermediates covalently linked to duplex DNA indicate that the DNA binds to the enzyme in a positively charged groove that runs roughly orthogonal to the DNA axis (55,59). Bacterial Fpg binds DNA in the minor groove and the damaged base is extruded through the major groove. The DNA appears to be severely kinked at the lesion point (~66o roll angle in the case of EcoFpg (55)) upon enzyme binding thereby allowing the extruded base to be positioned in the active site for catalysis. The minor groove is widened considerably at the lesion site, however, the rest of the DNA duplex surrounding the lesion retains canonical B-form (55,59). Upon nucleotide eversion, three highly conserved residues in the bacterial Fpg proteins namely Met74, Arg109 and Phe111 (in EcoFpg) fill the void that is created and stabilize the opposite base (Figure 2A) (55). Met74 is part of the β4/5 loop and occupies the position of the extruded base by entering through the minor groove while Arg109 and Phe111 are part of a loop connecting strands β7 and β8. Phe111 is wedged between the base opposite the lesion (a cytosine) and the neighboring base, and causes unstacking of these bases leading to the severe kinking of the DNA. Additionally, Arg109 forms H-bonds with the opposite base leading to discrimination against A as the opposite base (55,89).
Figure 2. Specific interactions between EcoFpg and DNA.
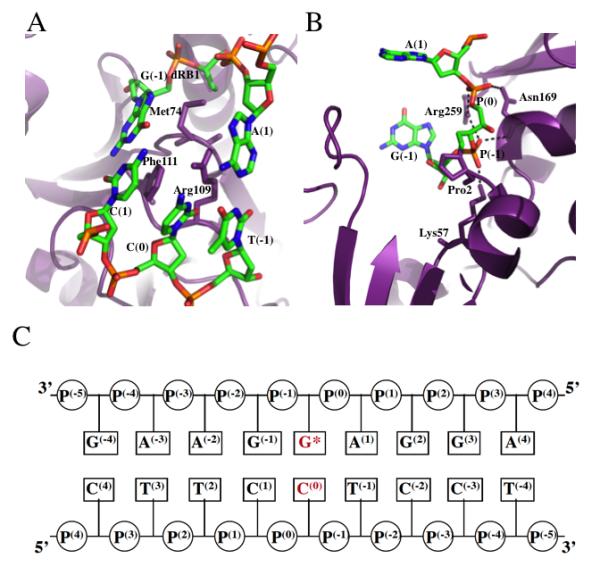
A. Triad of void-filling residues Met74, Phe111 and Arg109 that intercalate into the DNA causing severe kinking at the site of the damage. B. Interaction of conserved residues Lys57, Asn169 and Arg259 with DNA phosphates surrounding the ring-opened deoxyribitol moiety (dRb1) (PDB ID code 1K82 (55)). C. DNA sequence context present in the crystal structure of EcoFpg bound to DNA indicating the typical nomenclature used to describe the phosphates and the bases surrounding the lesion. The lesion is indicated by G* while C(0) is the opposite base, both of which are indicated in red lettering.
The analogous residues in MvNei1 (Leu84, Arg114, and Phe116) (61), human NEIL1 (Met81, Arg118, and Phe120) (67) and Arabidopsis thaliana Fpg (AthFpg) (Met78, Arg126 and Phe128) (Stephanie Duclos, Pierre Aller, Pawel Jaruga, Miral Didzaroglu, Susan S. Wallace and Sylvie Doubie, manuscript submitted to DNA Repair) are similar to those seen in the bacterial Fpg proteins which discriminate against A as a base opposite the lesion (89). In contrast, EcoNei inserts three consecutive residues Gln69, Leu70 and Tyr71 into the void created upon base extrusion (56). The three residues are located on a loop connecting β4 and β5. Tyr71 is wedged between the orphaned base and its 3′ neighbor and stabilizes the severely kinked DNA. The void-filling residues are lacking in MmuNeil3, which appears to be related to the preference of this enzyme for single-stranded DNA (Minmin Liu, Kayo Imamujra, Sylvie Dolublie and Susan S. Wallace, manuscript in preparation).
In addition to the void-filling residues, the Fpg/Nei proteins contain an absolutely conserved Lys residue (Lys57 in EcoFpg) and a conserved Asn (Asn169 in EcoFpg), which is part of the H2TH motif (55). Lys 57 forms salt-bridges with P−1 and P−2 while Asn169 forms bonds through backbone and side-chain amides to P−1 and P0 of the DNA (Figure 2B and C). Another highly conserved residue in the Fpg/Nei family of proteins is Arg259 (in EcoFpg), which is part of the zinc finger motif and is involved in the formation of salt bridges with the phosphodiester backbone (55) (Figure 2B). In the bacterial Fpg and eukaryotic Fpg/Nei proteins, there is no gross conformational change in the overall domain structure upon DNA binding (55,57,59,64,72). On the other hand, the side-chains of the void-filling residues and conserved residues in the hallmark motifs, show small changes. The development of disulfide-crosslinking techniques used to study the structure of BstFpg bound to lesion-containing DNA and undamaged DNA indicate that the enzyme possesses intrahelical recognition of the damage and can detect the subtle differences between the damaged base and its undamaged counterpart even at an initial encounter ((64,65) and see below). Comparing 8-oxoG with guanine in DNA suggests that the enzyme induces a local conformational change in the DNA backbone in which the sugar pucker (C2′-endo) adopts a different conformation (C4′-exo) to prevent a steric clash between the 8-oxo group of 8-oxoG and the C2′ of the sugar (64).
A loop region (called the αF-β9/10 loop) in the α-helical C-terminal domain of the Fpg proteins is presumed to be involved in lesion recognition. In the unliganded structure of TthFpg, this lesion recognition loop is ordered (54), but in structures of BstFpg bound to DNA containing an AP site, the density for this loop disappears suggesting that this region is disordered (57). In the presence of lesion-containing DNA in complex with catalytically inactive enzyme, the density for this loop resurfaces indicating conformational mobility upon catalysis (59,64,65). This loop plays a key role in the recognition of 8-oxoG: a projection from the loop wraps around the damaged base forming an extensive network of hydrogen bonds (59). This same loop was shown to wrap around FapyG in the LlaFpg structure (60). The major difference between the binding of Fpg to 8-oxoG and FapyG lies in the fact that binding in the extrahelical base-binding pocket of Fpg for the former lesion occurs in the syn conformation whereas FapyG is in the anti conformation. However, despite the difference in base conformations, a similar type of interaction exists between the main-chain carbonyl carbons of conserved residues S218 (in LlaFpg) and S221 (in BstFpg), which bind to the protonated N1 and N7 of FapyG and 8-oxoG, respectively. Similarly, the conserved I220 (in LlaFpg) and the analogous V223 (in BstFpg) use their main-chain group for hydrogen bonding with the carbonyl moiety at position 6 of both lesions (60).
The αF-β9/10 loops of bacterial Fpgs are functionally similar and are of comparable length (~27 residues) and conformation. In contrast, in the eukaryotic members of the family, which do not recognize 8-oxoG, this putative lesion-recognition loop is generally shorter or even missing as in the case of NEIL1 (61), AthFpg (Stephanie Duclos, Pierre Aller, Pawel Jaruga, Miral Didzaroglu, Susan S. Wallace and Sylvie Doublie, Manuscript submitted to DNA Repair) and MmuNeil3 (Liu et. al., manuscript in preparation). A superposition of BstFpg bound to DNA (containing 8-oxoG:C, (59)) with the unliganded human NEIL1 (61) and the MvNei1 enzymes illustrates that this loop wraps around the lesion only in the case of BstFpg (Figure 3). In contrast, in the case of NEIL1, the loop is replaced by an α-helix; the loop is shorter in MvNei1, and in both cases this segment is unable to wrap around the lesion (Figure 3). These data are consistent with the fact that 8-oxoG is not a good substrate for NEIL1 or any of the eukaryotic and mimivirus enzymes that are missing this loop. In fact, deletion of the αF-β9/10 loop in EcoFpg yielded a variant that retains catalytic ability on oxidized pyrimidines and FapyG, but not 8-oxoG, implying that this loop is important for stabilizing 8-oxoG and not the other lesions (Stephanie Duclos, Pierre Aller, Pawel Jaruga, Miral Didzaroglu, Susan S. Wallace and Sylvie Doublie, Manuscript submitted to DNA Repair).
Figure 3. Superposition of BstFpg (E3Q mutant, green) bound to DNA containing 8-oxoG with human NEIL1 (orange) and MvNei1 (beige).
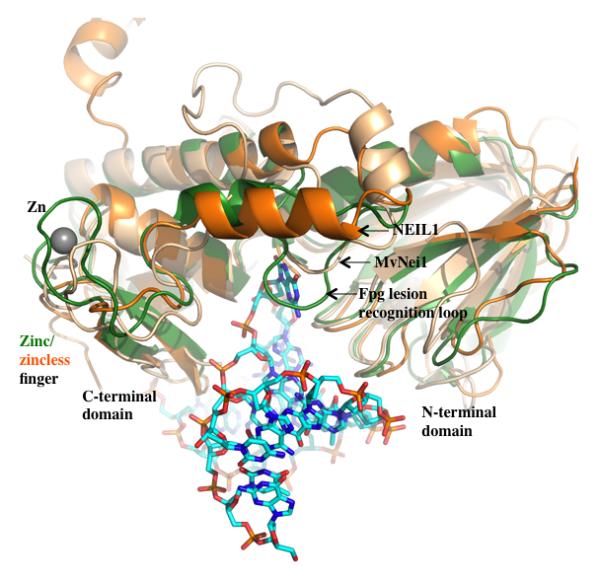
Overall the three proteins superimpose well with an RMSD of 1.1 Å – 1.3 Å upon aligning C-α of analogous residues from the NEIL1 and MvNei1 with BstFpg (performed using COOT (100) and Superimpose (M. Rould, personal communication)). Key differences among the three enzymes are the zinc/zinc-less finger in Fpg vs. NEIL1 and MvNei1, and the presence of the lesion-recognition loop in Fpg. The 8-oxoG containing DNA is displayed as a ball and stick model. (PDB ID codes for the BstFpg protein, NEIL1 and MvNei1 are 1R2Y, 1TDH and 3A42 respectively).
The bacterial Nei proteins are composed of members that share several characteristics with Fpg but also provide some unique variations. The crystal structure of EcoNei reveals a similar structural fold and conservation of motifs present in the Fpg proteins (56,62). A striking difference between the unliganded and DNA-bound EcoNei structures is a large conformational change of about 50 degrees between the N- and C-terminal domains. This is the only DNA glycosylase to date that has been reported to display a DNA-induced global conformational change, in which the glycosylase transforms from an elongated “open” form to a “closed” DNA-bound form (56,62). This conformational change was not observed for the MvNei1 protein upon DNA binding (67).
The structure of human NEIL1 reveals the presence of a structural motif composed of two antiparallel β-strands that mimic the zinc finger fold. This motif superimposes well with the zinc finger of EcoNei and the bacterial Fpg proteins (59,61) (Figure 3). However, the canonical Cys residues, and the loops connecting the β-strands of the zinc finger are missing in NEIL1, which prevents the coordination of a zinc atom (Figure 3). This motif termed “zincless-finger” contains a highly conserved Arg277 residue which, when mutated, significantly diminishes glycosylase activity (56,61). This zincless motif is also harbored by MvNei1 (61,67), and the plant and fungal Fpg glycosylases (Stephanie Duclos, Pierre Aller, Pawel Jaruga, Miral Didzaroglu, Susan S. Wallace and Sylvie Doublie, Manuscript submitted to DNA Repair).
In summary, members of the Fpg/Nei family are structurally similar, but display significant variations in conserved domains/motifs involved in DNA interactions. One of the main differences between EcoNei and the bacterial Fpg proteins is the composition of the void-filling, intercalation triad. In EcoNei, all three residues are located on the same β4/β5 loop and are consecutive, i.e. Gln69, Leu70 and Tyr71. In bacterial Fpg proteins, NEIL1, MvNei1 and AthFpg as mentioned above, the residues that constitute the triad reside in two different loops. Another difference between EcoFpg and the eukaryotic family members lies in the lesion-recognition loop located in the C-terminal domain of both proteins. In bacterial Fpg, the damaged base is everted from the DNA helix and is enveloped in a deep pocket, which is capped by the lesion recognition loop at one end (Figure 3). This loop is missing in EcoNei and the eukaryotic members for which a structure exists, including NEIL1, AthFpg and Neil3 and appears to be required for excising 8-oxoG. The vast repertoire of substrates of the Fpg/Nei family members and their different preferences for opposite bases and DNA substrates (single-stranded, double-stranded, or bubble DNA substrates) warrants further structural and biochemical scrutiny.
IV. Glycosylases Search for Lesions
It has long been a question in the field as to how DNA glycosylases locate the lesions they recognize in a sea of undamaged bases. This issue is complicated by the fact that a glycosylase flips out the damage from the DNA helix into its active site pocket in order to perform its enzymatic function. Furthermore, glycosylases do not use biochemical energy and rely on thermal energy so that lesions are found through random collisions between the glycosylase and the DNA molecule. Because of this, three-dimensional diffusion is considered to be too inefficient to account for the number of lesions the glycosylase must excise. Glycosylases are thought to bind to a non-specific site on the DNA molecule and slide along the DNA by one-dimensional diffusion until the enzyme finds the lesion or disassociates from DNA. There have been a number of hypotheses proposed for the lesion search itself. One model suggests that the glycosylase binds to an extruded DNA lesion and then moves along the DNA testing every single base (90). This appears to be unlikely since both kinetics (91) and single-molecule studies (92,93) have shown that glycosylases scan DNA close to diffusion limits making it thermodynamically impossible to sequentially extrude and examine every base. In the second model, the DNA glycosylase traps a randomly extruded damaged base. This extrusion is more likely with lesions since hydrogen bonding and stacking interactions would be altered compared to the normal bases. This appears to be the mechanism used by uracil DNA glycosylase (94). In the third model, glycosylases slide along the DNA molecule and are able to recognize their particular substrate by specific interactions between the glycosylase and the DNA molecule. This model has been suggested by structural studies (64,65) and by a recent single-molecule study (95).
Two groups have attempted to address the question of how glycosylases search for a lesion using single-molecule approaches (92,93,95). In the first study (92), human OGG1 (oxoguanine DNA glycosylase, a member of the HhH superfamily) labeled with Cy3, was observed to undergo one-dimensional sliding along DNA that was stretched by shear flow. A similar diffusive motion was observed with BstFpg. These same authors showed that the one-dimensional diffusion constants measured were consistent with the glycosylases diffusing along the DNA helix in a rotational manner (93). In a recent study from our laboratory (95), quantum dot-labeled E. coli Fpg, Nei, and Nth were imaged in the absence of flow. In this study the glycosylases were shown to diffuse along the DNA with a broad distribution of rates that ranged over two orders of magnitude. This broad distribution was common to all three glycosylases suggesting that both the Fpg/Nei family and HhH superfamily scan using a similar mechanism. When the diffusive behavior was analyzed further, the three glycosylases were shown to exhibit a continuum of motion that was in keeping with rotational diffusion along the DNA molecule and that ranged from a slow, subdiffusive to a faster, unrestricted diffusive behavior.
As described earlier, members of the Fpg/Nei family of DNA glycosylases have three void-filling residues that are inserted into the DNA helix and aid in flipping out the damaged base, and as well stabilize the DNA helix (55,56,59,64-67). The HhH superfamily uses a similar mechanism (96,97). Interestingly, a crystal structure of BstFpg crosslinked to undamaged DNA revealed that one of these void-filling residues, a phenylalanine, was found to be wedged into the helix occupying a position analogous to its position in the Fpg complex bound to damage-containing DNA (64) (Figure 2A). These data, together with kinetics data (98,99), suggest that the phenylalanine may be acting as a wedge that scans for deformability of the base pair such as in the sugar pucker. Interestingly, when the corresponding E. coli wedge residue, Phe111, was mutated to an alanine, there was a significant increase in the mean diffusion constant compared to the wild-type protein (95). Moreover, the diffusive properties characteristic of wild-type were altered, that is, the slow, subdiffusive population of glycosylases was selectively lost. Similar results were observed when the analogous residues in Nei and Nth were mutated (Dunn et al. unpublished observations) suggesting that the slow subdiffusive glycosylases are those interrogating the DNA for damages. Taken together, the data support the idea that the Fpg/Nei family of DNA glycosylases diffuse one-dimensionally along the DNA molecule with diffusion constants that are consistent with rotation around the DNA molecule, presumably in the minor groove where they bind. It also appears that at least part of the glycosylase search mechanism may be accomplished by insertion of a particular wedge residue that senses the topography of the minor groove and pauses either to check for damage at random locations or in response to subtle deformations of the DNA helix.
V. Concluding Remarks
Advances in the structural biology and biochemistry of glycosylases have led to a better understanding of how these complex enzymes recognize and excise damaged bases. Based on current in vitro studies, we can speculate on the mechanisms of specific lesion recognition. However, despite the vast knowledge gained, several unanswered questions still remain. For instance, we know that the Fpg/Nei family members recognize a broad range of substrates but it is not clear how these enzymes discriminate among each of these lesions and how they distinguish these from undamaged bases. Moreover, as some glycosylases are active at different times in the cell cycle and interact with a number of protein partners, how are these enzymes involved in processes such as DNA replication or transcription? Additionally, it remains difficult to classify certain members of the Fpg/Nei family under a specific subfamily. For example, even though members of the Fpg/Nei family of proteins are structurally similar, some elements such as the intercalation triad present in NEIL1 and MvNei1 suggest that these members could be classified under the Fpg sub-family contrary to the Nei sub-family after which they were originally named. Many aspects of phylogenetic characterization, lesion recognition, substrate specificities and the biological functions of this glycosylase family still remain to be elucidated.
Acknowledgements
We would like to thank Minmin Liu, Drs. Scott Kathe, Stephanie Duclos, and Ramiro Barrantes-Reynolds for helpful discussions. This work was supported by NIH/NCI P01 CA098993 (S.S. Wallace, PI).
REFERENCES
- 1.Chetsanga CJ, Lindahl T. Release of 7-methylguanine residues whose imidazole rings have been opened from damaged DNA by a DNA glycosylase from Escherichia coli. Nucleic Acids Res. 1979;6:3673–3684. doi: 10.1093/nar/6.11.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boiteux S, O’Connor TR, Laval J. Formamidopyrimidine-DNA glycosylase of Escherichia coli: cloning and sequencing of the fpg structural gene and overproduction of the protein. EMBO J. 1987;6:3177–3183. doi: 10.1002/j.1460-2075.1987.tb02629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chetsanga CJ, Lozon M, Makaroff C, Savage L. Purification and characterization of Escherichia coli formamidopyrimidine-DNA glycosylase that excises damaged 7-methylguanine from deoxyribonucleic acid. Biochemistry. 1981;20:5201–5207. doi: 10.1021/bi00521a016. [DOI] [PubMed] [Google Scholar]
- 4.Breimer LH. Enzymatic excision from gamma-irradiated polydeoxyribonucleotides of adenine residues whose imidazole rings have been ruptured. Nucleic Acids Res. 1984;12:6359–6367. doi: 10.1093/nar/12.16.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Connor TR, Laval J. Physical association of the 2,6-diamino-4-hydroxy-5N-formamidopyrimidine-DNA glycosylase of Escherichia coli and an activity nicking DNA at apurinic/apyrimidinic sites. Proc Natl Acad Sci U S A. 1989;86:5222–5226. doi: 10.1073/pnas.86.14.5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boiteux S, O’Connor TR, Lederer F, Gouyette A, Laval J. Homogeneous Escherichia coli FPG protein. A DNA glycosylase which excises imidazole ring-opened purines and nicks DNA at apurinic/apyrimidinic sites. J Biol Chem. 1990;265:3916–3922. [PubMed] [Google Scholar]
- 7.Cabrera M, Nghiem Y, Miller JH. mutM, a second mutator locus in Escherichia coli that generates G.C----T.A transversions. J Bacteriol. 1988;170:5405–5407. doi: 10.1128/jb.170.11.5405-5407.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Michaels ML, Pham L, Cruz C, Miller JH. MutM, a protein that prevents G.C----T.A transversions, is formamidopyrimidine-DNA glycosylase. Nucleic Acids Res. 1991;19:3629–3632. doi: 10.1093/nar/19.13.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tchou J, Kasai H, Shibutani S, Chung MH, Laval J, Grollman AP, Nishimura S. 8-oxoguanine (8-hydroxyguanine) DNA glycosylase and its substrate specificity. Proc Natl Acad Sci U S A. 1991;88:4690–4694. doi: 10.1073/pnas.88.11.4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tchou J, Bodepudi V, Shibutani S, Antoshechkin I, Miller J, Grollman AP, Johnson F. Substrate specificity of Fpg protein. Recognition and cleavage of oxidatively damaged DNA. J Biol Chem. 1994;269:15318–15324. [PubMed] [Google Scholar]
- 11.Michaels ML, Miller JH. The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine) J Bacteriol. 1992;174:6321–6325. doi: 10.1128/jb.174.20.6321-6325.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brieba LG, Eichman BF, Kokoska RJ, Doublie S, Kunkel TA, Ellenberger T. Structural basis for the dual coding potential of 8-oxoguanosine by a high-fidelity DNA polymerase. EMBO J. 2004;23:3452–3461. doi: 10.1038/sj.emboj.7600354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shibutani S, Takeshita M, Grollman AP. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- 14.Moriya M, Ou C, Bodepudi V, Johnson F, Takeshita M, Grollman AP. Site-specific mutagenesis using a gapped duplex vector: a study of translesion synthesis past 8-oxodeoxyguanosine in E. coli. Mutat Res. 1991;254:281–288. doi: 10.1016/0921-8777(91)90067-y. [DOI] [PubMed] [Google Scholar]
- 15.Wood ML, Dizdaroglu M, Gajewski E, Essigmann JM. Mechanistic studies of ionizing radiation and oxidative mutagenesis: genetic effects of a single 8-hydroxyguanine (7-hydro-8-oxoguanine) residue inserted at a unique site in a viral genome. Biochemistry. 1990;29:7024–7032. doi: 10.1021/bi00482a011. [DOI] [PubMed] [Google Scholar]
- 16.Michaels ML, Cruz C, Grollman AP, Miller JH. Evidence that MutY and MutM combine to prevent mutations by an oxidatively damaged form of guanine in DNA. Proc Natl Acad Sci U S A. 1992;89:7022–7025. doi: 10.1073/pnas.89.15.7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maki H, Sekiguchi M. MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature. 1992;355:273–275. doi: 10.1038/355273a0. [DOI] [PubMed] [Google Scholar]
- 18.Boiteux S, Gajewski E, Laval J, Dizdaroglu M. Substrate specificity of the Escherichia coli Fpg protein (formamidopyrimidine-DNA glycosylase): excision of purine lesions in DNA produced by ionizing radiation or photosensitization. Biochemistry. 1992;31:106–110. doi: 10.1021/bi00116a016. [DOI] [PubMed] [Google Scholar]
- 19.Krishnamurthy N, Haraguchi K, Greenberg MM, David SS. Efficient removal of formamidopyrimidines by 8-oxoguanine glycosylases. Biochemistry. 2008;47:1043–1050. doi: 10.1021/bi701919u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patro JN, Wiederholt CJ, Jiang YL, Delaney JC, Essigmann JM, Greenberg MM. Studies on the replication of the ring opened formamidopyrimidine, Fapy.dG in Escherichia coli. Biochemistry. 2007;46:10202–10212. doi: 10.1021/bi700628c. [DOI] [PubMed] [Google Scholar]
- 21.Wiederholt CJ, Greenberg MM. Fapy.dG instructs Klenow exo(-) to misincorporate deoxyadenosine. J Am Chem Soc. 2002;124:7278–7279. doi: 10.1021/ja026522r. [DOI] [PubMed] [Google Scholar]
- 22.Wiederholt CJ, Delaney MO, Pope MA, David SS, Greenberg MM. Repair of DNA containing Fapy.dG and its beta-C-nucleoside analogue by formamidopyrimidine DNA glycosylase and MutY. Biochemistry. 2003;42:9755–9760. doi: 10.1021/bi034844h. [DOI] [PubMed] [Google Scholar]
- 23.Dizdaroglu M, Jaruga P, Birincioglu M, Rodriguez H. Free radical-induced damage to DNA: mechanisms and measurement. Free Radic Biol Med. 2002;32:1102–1115. doi: 10.1016/s0891-5849(02)00826-2. [DOI] [PubMed] [Google Scholar]
- 24.Liu M, Bandaru V, Bond JP, Jaruga P, Zhao X, Christov PP, Burrows CJ, Rizzo CJ, Dizdaroglu M, Wallace SS. The mouse ortholog of NEIL3 is a functional DNA glycosylase in vitro and in vivo. Proc Natl Acad Sci U S A. 2010;107:4925–4930. doi: 10.1073/pnas.0908307107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Melamede RJ, Hatahet Z, Kow YW, Ide H, Wallace SS. Isolation and characterization of endonuclease VIII from Escherichia coli. Biochemistry. 1994;33:1255–1264. doi: 10.1021/bi00171a028. [DOI] [PubMed] [Google Scholar]
- 26.Jiang D, Hatahet Z, Melamede RJ, Kow YW, Wallace SS. Characterization of Escherichia coli endonuclease VIII. J Biol Chem. 1997;272:32230–32239. doi: 10.1074/jbc.272.51.32230. [DOI] [PubMed] [Google Scholar]
- 27.Jiang D, Hatahet Z, Blaisdell JO, Melamede RJ, Wallace SS. Escherichia coli endonuclease VIII: cloning, sequencing, and overexpression of the nei structural gene and characterization of nei and nei nth mutants. J Bacteriol. 1997;179:3773–3782. doi: 10.1128/jb.179.11.3773-3782.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wallace SS. In: Oxidative Stress and the Molecular Biology of Antioxidant Defenses. Scandalios J, Oxidative Stress and the Molecular Biology of Antioxidant Defenses, editor. Cold Spring Harbor Press; Cold Spring Harbor: 1997. pp. 49–90. [Google Scholar]
- 29.Wallace SS. Enzymatic processing of radiation-induced free radical damage in DNA. Radiat Res. 1998;150:S60–79. [PubMed] [Google Scholar]
- 30.Bandaru V, Sunkara S, Wallace SS, Bond JP. A novel human DNA glycosylase that removes oxidative DNA damage and is homologous to Escherichia coli endonuclease VIII. DNA Repair (Amst) 2002;1:517–529. doi: 10.1016/s1568-7864(02)00036-8. [DOI] [PubMed] [Google Scholar]
- 31.Hazra TK, Izumi T, Boldogh I, Imhoff B, Kow YW, Jaruga P, Dizdaroglu M, Mitra S. Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc Natl Acad Sci U S A. 2002;99:3523–3528. doi: 10.1073/pnas.062053799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hazra TK, Kow YW, Hatahet Z, Imhoff B, Boldogh I, Mokkapati SK, Mitra S, Izumi T. Identification and characterization of a novel human DNA glycosylase for repair of cytosine-derived lesions. J Biol Chem. 2002;277:30417–30420. doi: 10.1074/jbc.C200355200. [DOI] [PubMed] [Google Scholar]
- 33.Morland I, Rolseth V, Luna L, Rognes T, Bjoras M, Seeberg E. Human DNA glycosylases of the bacterial Fpg/MutM superfamily: an alternative pathway for the repair of 8-oxoguanine and other oxidation products in DNA. Nucleic Acids Res. 2002;30:4926–4936. doi: 10.1093/nar/gkf618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wallace SS, Bandaru V, Kathe SD, Bond JP. The enigma of endonuclease VIII. DNA Repair (Amst) 2003;2:441–453. doi: 10.1016/s1568-7864(02)00182-9. [DOI] [PubMed] [Google Scholar]
- 35.Takao M, Kanno S, Kobayashi K, Zhang QM, Yonei S, van der Horst GT, Yasui A. A back-up glycosylase in Nth1 knock-out mice is a functional Nei (endonuclease VIII) homologue. J Biol Chem. 2002;277:42205–42213. doi: 10.1074/jbc.M206884200. [DOI] [PubMed] [Google Scholar]
- 36.Dou H, Mitra S, Hazra TK. Repair of oxidized bases in DNA bubble structures by human DNA glycosylases NEIL1 and NEIL2. J Biol Chem. 2003;278:49679–49684. doi: 10.1074/jbc.M308658200. [DOI] [PubMed] [Google Scholar]
- 37.Katafuchi A, Nakano T, Masaoka A, Terato H, Iwai S, Hanaoka F, Ide H. Differential specificity of human and Escherichia coli endonuclease III and VIII homologues for oxidative base lesions. J Biol Chem. 2004;279:14464–14471. doi: 10.1074/jbc.M400393200. [DOI] [PubMed] [Google Scholar]
- 38.Jaruga P, Birincioglu M, Rosenquist TA, Dizdaroglu M. Mouse NEIL1 protein is specific for excision of 2,6-diamino-4-hydroxy-5-formamidopyrimidine and 4,6-diamino-5-formamidopyrimidine from oxidatively damaged DNA. Biochemistry. 2004;43:15909–15914. doi: 10.1021/bi048162l. [DOI] [PubMed] [Google Scholar]
- 39.Hailer MK, Slade PG, Martin BD, Rosenquist TA, Sugden KD. Recognition of the oxidized lesions spiroiminodihydantoin and guanidinohydantoin in DNA by the mammalian base excision repair glycosylases NEIL1 and NEIL2. DNA Repair (Amst) 2005;4:41–50. doi: 10.1016/j.dnarep.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 40.Krishnamurthy N, Zhao X, Burrows CJ, David SS. Superior removal of hydantoin lesions relative to other oxidized bases by the human DNA glycosylase hNEIL1. Biochemistry. 2008;47:7137–7146. doi: 10.1021/bi800160s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wiederhold L, Leppard JB, Kedar P, Karimi-Busheri F, Rasouli-Nia A, Weinfeld M, Tomkinson AE, Izumi T, Prasad R, Wilson SH, et al. AP endonuclease-independent DNA base excision repair in human cells. Mol Cell. 2004;15:209–220. doi: 10.1016/j.molcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 42.Dou H, Theriot CA, Das A, Hegde ML, Matsumoto Y, Boldogh I, Hazra TK, Bhakat KK, Mitra S. Interaction of the human DNA glycosylase NEIL1 with proliferating cell nuclear antigen. The potential for replication-associated repair of oxidized bases in mammalian genomes. J Biol Chem. 2008;283:3130–3140. doi: 10.1074/jbc.M709186200. [DOI] [PubMed] [Google Scholar]
- 43.Hegde ML, Theriot CA, Das A, Hegde PM, Guo Z, Gary RK, Hazra TK, Shen B, Mitra S. Physical and functional interaction between human oxidized base-specific DNA glycosylase NEIL1 and flap endonuclease 1. J Biol Chem. 2008;283:27028–27037. doi: 10.1074/jbc.M802712200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Theriot CA, Hegde ML, Hazra TK, Mitra S. RPA physically interacts with the human DNA glycosylase NEIL1 to regulate excision of oxidative DNA base damage in primer-template structures. DNA Repair (Amst) 2010;9:643–652. doi: 10.1016/j.dnarep.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Banerjee D, Mandal SM, Das A, Hegde ML, Das S, Bhakat KK, Boldogh I, Sarkar PS, Mitra S, Hazra TK. Preferential repair of oxidized base damage in the transcribed genes of mammalian cells. J Biol Chem. 2011;286:6006–6016. doi: 10.1074/jbc.M110.198796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takao M, Kanno S, Shiromoto T, Hasegawa R, Ide H, Ikeda S, Sarker AH, Seki S, Xing JZ, Le XC, et al. Novel nuclear and mitochondrial glycosylases revealed by disruption of the mouse Nth1 gene encoding an endonuclease III homolog for repair of thymine glycols. EMBO J. 2002;21:3486–3493. doi: 10.1093/emboj/cdf350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krokeide SZ, Bolstad N, Laerdahl JK, Bjoras M, Luna L. Expression and purification of NEIL3, a human DNA glycosylase homolog. Protein Expr Purif. 2009;65:160–164. doi: 10.1016/j.pep.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 48.Takao M, Oohata Y, Kitadokoro K, Kobayashi K, Iwai S, Yasui A, Yonei S, Zhang QM. Human Nei-like protein NEIL3 has AP lyase activity specific for single-stranded DNA and confers oxidative stress resistance in Escherichia coli mutant. Genes Cells. 2009;14:261–270. doi: 10.1111/j.1365-2443.2008.01271.x. [DOI] [PubMed] [Google Scholar]
- 49.Hildrestrand GA, Neurauter CG, Diep DB, Castellanos CG, Krauss S, Bjoras M, Luna L. Expression patterns of Neil3 during embryonic brain development and neoplasia. BMC Neurosci. 2009;10:45. doi: 10.1186/1471-2202-10-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rolseth V, Runden-Pran E, Luna L, McMurray C, Bjoras M, Ottersen OP. Widespread distribution of DNA glycosylases removing oxidative DNA lesions in human and rodent brains. DNA Repair (Amst) 2008;7:1578–1588. doi: 10.1016/j.dnarep.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Torisu K, Tsuchimoto D, Ohnishi Y, Nakabeppu Y. Hematopoietic tissue-specific expression of mouse Neil3 for endonuclease VIII-like protein. J Biochem. 2005;138:763–772. doi: 10.1093/jb/mvi168. [DOI] [PubMed] [Google Scholar]
- 52.Kathe SD, Barrantes-Reynolds R, Jaruga P, Newton MR, Burrows CJ, Bandaru V, Dizdaroglu M, Bond JP, Wallace SS. Plant and fungal Fpg homologs are formamidopyrimidine DNA glycosylases but not 8-oxoguanine DNA glycosylases. DNA Repair (Amst) 2009;8:643–653. doi: 10.1016/j.dnarep.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pumo DE, Barrantes-Reynolds R, Kathe SD, Wallace SS, Bond JP. In: Genome Instability and Transgenerational Effects. Kovalchuk I, Kovalchuk O, editors. Nova Science Publishers, Inc.; New York: 2010. pp. 71–88. [Google Scholar]
- 54.Sugahara M, Mikawa T, Kumasaka T, Yamamoto M, Kato R, Fukuyama K, Inoue Y, Kuramitsu S. Crystal structure of a repair enzyme of oxidatively damaged DNA, MutM (Fpg), from an extreme thermophile, Thermus thermophilus HB8. EMBO J. 2000;19:3857–3869. doi: 10.1093/emboj/19.15.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gilboa R, Zharkov DO, Golan G, Fernandes AS, Gerchman SE, Matz E, Kycia JH, Grollman AP, Shoham G. Structure of formamidopyrimidine-DNA glycosylase covalently complexed to DNA. J Biol Chem. 2002;277:19811–19816. doi: 10.1074/jbc.M202058200. [DOI] [PubMed] [Google Scholar]
- 56.Zharkov DO, Golan G, Gilboa R, Fernandes AS, Gerchman SE, Kycia JH, Rieger RA, Grollman AP, Shoham G. Structural analysis of an Escherichia coli endonuclease VIII covalent reaction intermediate. EMBO J. 2002;21:789–800. doi: 10.1093/emboj/21.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fromme JC, Verdine GL. Structural insights into lesion recognition and repair by the bacterial 8-oxoguanine DNA glycosylase MutM. Nat Struct Biol. 2002;9:544–552. doi: 10.1038/nsb809. [DOI] [PubMed] [Google Scholar]
- 58.Serre L, Pereira de Jesus K, Boiteux S, Zelwer C, Castaing B. Crystal structure of the Lactococcus lactis formamidopyrimidine-DNA glycosylase bound to an abasic site analogue-containing DNA. EMBO J. 2002;21:2854–2865. doi: 10.1093/emboj/cdf304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fromme JC, Verdine GL. DNA lesion recognition by the bacterial repair enzyme MutM. J Biol Chem. 2003;278:51543–51548. doi: 10.1074/jbc.M307768200. [DOI] [PubMed] [Google Scholar]
- 60.Coste F, Ober M, Carell T, Boiteux S, Zelwer C, Castaing B. Structural basis for the recognition of the FapydG lesion (2,6-diamino-4-hydroxy-5-formamidopyrimidine) by formamidopyrimidine-DNA glycosylase. J Biol Chem. 2004;279:44074–44083. doi: 10.1074/jbc.M405928200. [DOI] [PubMed] [Google Scholar]
- 61.Doublie S, Bandaru V, Bond JP, Wallace SS. The crystal structure of human endonuclease VIII-like 1 (NEIL1) reveals a zincless finger motif required for glycosylase activity. Proc Natl Acad Sci U S A. 2004;101:10284–10289. doi: 10.1073/pnas.0402051101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Golan G, Zharkov DO, Feinberg H, Fernandes AS, Zaika EI, Kycia JH, Grollman AP, Shoham G. Structure of the uncomplexed DNA repair enzyme endonuclease VIII indicates significant interdomain flexibility. Nucleic Acids Res. 2005;33:5006–5016. doi: 10.1093/nar/gki796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pereira de Jesus K, Serre L, Zelwer C, Castaing B. Structural insights into abasic site for Fpg specific binding and catalysis: comparative high-resolution crystallographic studies of Fpg bound to various models of abasic site analogues-containing DNA. Nucleic Acids Res. 2005;33:5936–5944. doi: 10.1093/nar/gki879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Banerjee A, Santos WL, Verdine GL. Structure of a DNA glycosylase searching for lesions. Science. 2006;311:1153–1157. doi: 10.1126/science.1120288. [DOI] [PubMed] [Google Scholar]
- 65.Qi Y, Spong MC, Nam K, Banerjee A, Jiralerspong S, Karplus M, Verdine GL. Encounter and extrusion of an intrahelical lesion by a DNA repair enzyme. Nature. 2009;462:762–766. doi: 10.1038/nature08561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Qi Y, Spong MC, Nam K, Karplus M, Verdine GL. Entrapment and structure of an extrahelical guanine attempting to enter the active site of a bacterial DNA glycosylase, MutM. J Biol Chem. 2010;285:1468–1478. doi: 10.1074/jbc.M109.069799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Imamura K, Wallace SS, Doublie S. Structural characterization of a viral NEIL1 ortholog unliganded and bound to abasic site-containing DNA. J Biol Chem. 2009;284:26174–26183. doi: 10.1074/jbc.M109.021907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Coste F, Ober M, Le Bihan YV, Izquierdo MA, Hervouet N, Mueller H, Carell T, Castaing B. Bacterial base excision repair enzyme Fpg recognizes bulky N7-substituted-FapydG lesion via unproductive binding mode. Chem Biol. 2008;15:706–717. doi: 10.1016/j.chembiol.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 69.Le Bihan YV, Angeles Izquierdo M, Coste F, Aller P, Culard F, Gehrke TH, Essalhi K, Carell T, Castaing B. 5-Hydroxy-5-methylhydantoin DNA lesion, a molecular trap for DNA glycosylases. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Verdine GL, Norman DP. Covalent trapping of protein-DNA complexes. Annu Rev Biochem. 2003;72:337–366. doi: 10.1146/annurev.biochem.72.121801.161447. [DOI] [PubMed] [Google Scholar]
- 71.Fromme JC, Banerjee A, Verdine GL. DNA glycosylase recognition and catalysis. Curr Opin Struct Biol. 2004;14:43–49. doi: 10.1016/j.sbi.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 72.Zharkov DO, Shoham G, Grollman AP. Structural characterization of the Fpg family of DNA glycosylases. DNA Repair (Amst) 2003;2:839–862. doi: 10.1016/s1568-7864(03)00084-3. [DOI] [PubMed] [Google Scholar]
- 73.O’Connor TR, Graves RJ, de Murcia G, Castaing B, Laval J. Fpg protein of Escherichia coli is a zinc finger protein whose cysteine residues have a structural and/or functional role. J Biol Chem. 1993;268:9063–9070. [PubMed] [Google Scholar]
- 74.Tchou J, Michaels ML, Miller JH, Grollman AP. Function of the zinc finger in Escherichia coli Fpg protein. J Biol Chem. 1993;268:26738–26744. [PubMed] [Google Scholar]
- 75.Tchou J, Grollman AP. The catalytic mechanism of Fpg protein. Evidence for a Schiff base intermediate and amino terminus localization of the catalytic site. J Biol Chem. 1995;270:11671–11677. doi: 10.1074/jbc.270.19.11671. [DOI] [PubMed] [Google Scholar]
- 76.Kropachev KY, Zharkov DO, Grollman AP. Catalytic mechanism of Escherichia coli endonuclease VIII: roles of the intercalation loop and the zinc finger. Biochemistry. 2006;45:12039–12049. doi: 10.1021/bi060663e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hatahet Z, Kow YW, Purmal AA, Cunningham RP, Wallace SS. New substrates for old enzymes. 5-Hydroxy-2′-deoxycytidine and 5-hydroxy-2′-deoxyuridine are substrates for Escherichia coli endonuclease III and formamidopyrimidine DNA N-glycosylase, while 5-hydroxy-2′-deoxyuridine is a substrate for uracil DNA N-glycosylase. J Biol Chem. 1994;269:18814–18820. [PubMed] [Google Scholar]
- 78.D’Ham C, Romieu A, Jaquinod M, Gasparutto D, Cadet J. Excision of 5,6-dihydroxy-5,6-dihydrothymine, 5,6-dihydrothymine, and 5-hydroxycytosine from defined sequence oligonucleotides by Escherichia coli endonuclease III and Fpg proteins: kinetic and mechanistic aspects. Biochemistry. 1999;38:3335–3344. doi: 10.1021/bi981982b. [DOI] [PubMed] [Google Scholar]
- 79.Guo Y, Bandaru V, Jaruga P, Zhao X, Burrows CJ, Iwai S, Dizdaroglu M, Bond JP, Wallace SS. The oxidative DNA glycosylases of Mycobacterium tuberculosis exhibit different substrate preferences from their Escherichia coli counterparts. DNA Repair (Amst) 2010;9:177–190. doi: 10.1016/j.dnarep.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Luo W, Muller JG, Rachlin EM, Burrows CJ. Characterization of spiroiminodihydantoin as a product of one-electron oxidation of 8-Oxo-7,8-dihydroguanosine. Org Lett. 2000;2:613–616. doi: 10.1021/ol9913643. [DOI] [PubMed] [Google Scholar]
- 81.Purmal AA, Lampman GW, Bond JP, Hatahet Z, Wallace SS. Enzymatic processing of uracil glycol, a major oxidative product of DNA cytosine. J Biol Chem. 1998;273:10026–10035. doi: 10.1074/jbc.273.16.10026. [DOI] [PubMed] [Google Scholar]
- 82.Bandaru V, Zhao X, Newton MR, Burrows CJ, Wallace SS. Human endonuclease VIII-like (NEIL) proteins in the giant DNA Mimivirus. DNA Repair (Amst) 2007;6:1629–1641. doi: 10.1016/j.dnarep.2007.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhao X, Krishnamurthy N, Burrows CJ, David SS. Mutation versus repair: NEIL1 removal of hydantoin lesions in single-stranded, bulge, bubble, and duplex DNA contexts. Biochemistry. 2010;49:1658–1666. doi: 10.1021/bi901852q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dong L, Meira LB, Hazra TK, Samson LD, Cao W. Oxanine DNA glycosylase activities in mammalian systems. DNA Repair (Amst) 2008;7:128–134. doi: 10.1016/j.dnarep.2007.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nakano T, Katafuchi A, Shimizu R, Terato H, Suzuki T, Tauchi H, Makino K, Skorvaga M, Van Houten B, Ide H. Repair activity of base and nucleotide excision repair enzymes for guanine lesions induced by nitrosative stress. Nucleic Acids Res. 2005;33:2181–2191. doi: 10.1093/nar/gki513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yeo J, Goodman RA, Schirle NT, David SS, Beal PA. RNA editing changes the lesion specificity for the DNA repair enzyme NEIL1. Proc Natl Acad Sci U S A. 2010;107:20715–20719. doi: 10.1073/pnas.1009231107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ishchenko AA, Koval VV, Fedorova OS, Douglas KT, Nevinsky GA. Structural requirements of double and single stranded DNA substrates and inhibitors, including a photoaffinity label, of Fpg protein from Escherichia coli. J Biomol Struct Dyn. 1999;17:301–310. doi: 10.1080/07391102.1999.10508363. [DOI] [PubMed] [Google Scholar]
- 88.Ishchenko AA, Bulychev NV, Maksakova GA, Johnson F, Nevinsky GA. Single-stranded oligodeoxyribonucleotides are substrates of Fpg protein from Escherichia coli. IUBMB Life. 1999;48:613–618. doi: 10.1080/713803570. [DOI] [PubMed] [Google Scholar]
- 89.Zaika EI, Perlow RA, Matz E, Broyde S, Gilboa R, Grollman AP, Zharkov DO. Substrate discrimination by formamidopyrimidine-DNA glycosylase: a mutational analysis. J Biol Chem. 2004;279:4849–4861. doi: 10.1074/jbc.M310262200. [DOI] [PubMed] [Google Scholar]
- 90.Verdine GL, Bruner SD. How do DNA repair proteins locate damaged bases in the genome? Chem Biol. 1997;4:329–334. doi: 10.1016/s1074-5521(97)90123-x. [DOI] [PubMed] [Google Scholar]
- 91.Porecha RH, Stivers JT. Uracil DNA glycosylase uses DNA hopping and short-range sliding to trap extrahelical uracils. Proc Natl Acad Sci U S A. 2008;105:10791–10796. doi: 10.1073/pnas.0801612105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Blainey PC, van Oijen AM, Banerjee A, Verdine GL, Xie XS. A base-excision DNA-repair protein finds intrahelical lesion bases by fast sliding in contact with DNA. Proc Natl Acad Sci U S A. 2006;103:5752–5757. doi: 10.1073/pnas.0509723103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Blainey PC, Luo G, Kou SC, Mangel WF, Verdine GL, Bagchi B, Xie XS. Nonspecifically bound proteins spin while diffusing along DNA. Nat Struct Mol Biol. 2009;16:1224–1229. doi: 10.1038/nsmb.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cao C, Jiang YL, Stivers JT, Song F. Dynamic opening of DNA during the enzymatic search for a damaged base. Nat Struct Mol Biol. 2004;11:1230–1236. doi: 10.1038/nsmb864. [DOI] [PubMed] [Google Scholar]
- 95.Dunn AR, Kad NM, Nelson SR, Warshaw DM, Wallace SS. Single Qdot-labeled glycosylase molecules use a wedge amino acid to probe for lesions while scanning along DNA. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bruner SD, Norman DP, Verdine GL. Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature. 2000;403:859–866. doi: 10.1038/35002510. [DOI] [PubMed] [Google Scholar]
- 97.Fromme JC, Verdine GL. Structure of a trapped endonuclease III-DNA covalent intermediate. EMBO J. 2003;22:3461–3471. doi: 10.1093/emboj/cdg311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kuznetsov NA, Zharkov DO, Koval VV, Buckle M, Fedorova OS. Reversible chemical step and rate-limiting enzyme regeneration in the reaction catalyzed by formamidopyrimidine-DNA glycosylase. Biochemistry. 2009;48:11335–11343. doi: 10.1021/bi901100b. [DOI] [PubMed] [Google Scholar]
- 99.Koval VV, Kuznetsov NA, Ishchenko AA, Saparbaev MK, Fedorova OS. Real-time studies of conformational dynamics of the repair enzyme E. coli formamidopyrimidine-DNA glycosylase and its DNA complexes during catalytic cycle. Mutat Res. 2010;685:3–10. doi: 10.1016/j.mrfmmm.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 100.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 101.Michaels ML, Tchou J, Grollman AP, Miller JH. A repair system for 8- oxo-7,8-dihydrodeoxyguanine. Biochemistry. 1992;31:10964–10968. doi: 10.1021/bi00160a004. [DOI] [PubMed] [Google Scholar]
- 102.Zharkov DO, Ishchenko AA, Douglas KT, Nevinsky GA. Recognition of damaged DNA by Escherichia coli Fpg protein: insights from structural and kinetic data. Mutat Res. 2003;531:141–156. doi: 10.1016/j.mrfmmm.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 103.Hamm ML, Gill TJ, Nicolson SC, Summers MR. Substrate specificity of Fpg (MutM) and hOGG1, two repair glycosylases. J Am Chem Soc. 2007;129:7724–7725. doi: 10.1021/ja0716453. [DOI] [PubMed] [Google Scholar]


