Abstract
ABC-F proteins have evaded functional characterization even though they comprise one of the most widely distributed branches of the ATP-binding cassette (ABC) superfamily. Herein, we demonstrate that YjjK, the most prevalent eubacterial ABC-F protein, gates ribosome entry into the translation elongation cycle through a nucleotide-dependent interaction sensitive to ATP/ADP ratio. Accordingly, we rename this protein Energy-dependent Translational Throttle A (EttA). We determined the crystal structure of Escherichia coli EttA and used it to design mutants for biochemical studies, including enzymological assays of the initial steps of protein synthesis. These studies suggest that EttA may regulate protein synthesis in energy-depleted cells, which have a low ATP/ADP ratio. Consistent with this inference, ΔettA cells exhibit a severe fitness defect in long-term stationary phase. These studies demonstrate that an ABC-F protein regulates protein synthesis via a novel mechanism sensitive to cellular energy status.
Keywords: Protein synthesis, translational regulation, ABC-F protein family, YjjK, ATP/ADP ratio, stationary phase fitness, X-ray crystallography
Most genomes encode multiple ABC superfamily1 proteins. They are named after stereotyped ATP-binding cassettes (ABCs), which share characteristic sequence motifs involved in ATP hydrolysis. The Walker A and B motifs participate in binding and hydrolyzing the β and γ phosphates of ATP and are shared with a larger group of mechanically active enzymes that includes the F1 and AAA+ ATPases and the superfamily I and II helicases2. However, the C motif or Signature Sequence, with consensus LSGGQ, is found exclusively in ABC ATPases1. These residues drive a mechanical powerstroke involving formation of an “ATP-sandwich dimer”3–5 in which the LSGGQ from one subunit reciprocally encapsulates the ribose and triphosphate of an ATP molecule bound to the Walker motifs in the other subunit.
While transporters in the ABC superfamily (ABC transporters) represent the most common molecular architecture used to couple transmembrane transport to ATP hydrolysis6, the superfamily also includes soluble proteins performing diverse biochemical functions. These include UvrA7 and Rad508, which function in DNA repair, and also eEF39 and ABCE1 (RLI1)10–13, which are translation factors. ABCE1 binds to the ribosomal aminoacyl-tRNA binding (A) site in eukaryotic and archaeal post-termination complexes to assist ribosome recycling. The eEF3 protein has been proposed to stimulate the release of deacylated tRNAs from the tRNA exit (E) site of ribosomes14,15 and, more recently, to assist recycling of yeast post-termination ribosomal complexes16. ABC-F17,18 and RbbA19,20 proteins also interact with ribosomes, but their exact biochemical functions remain uncharacterized.
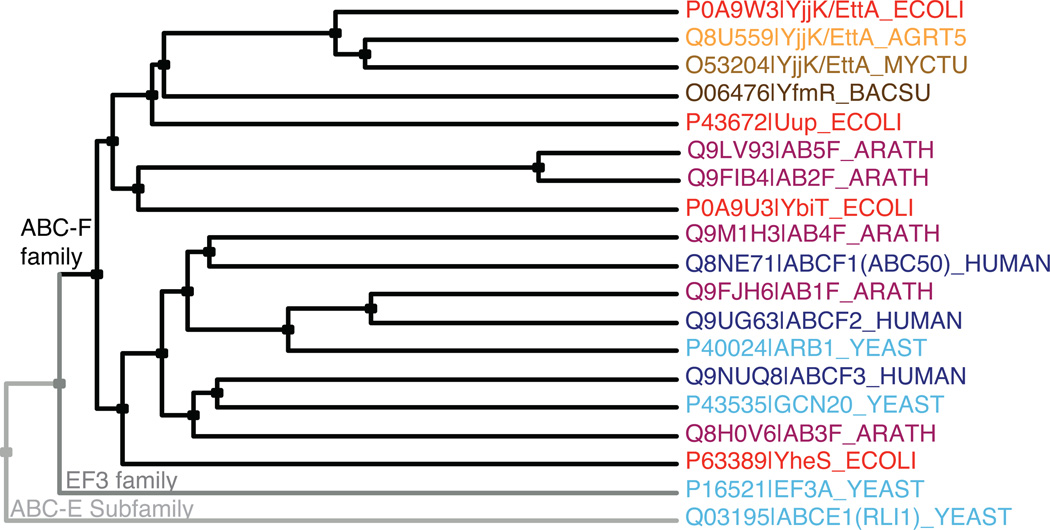
ABC-F proteins (ABC-Fs) comprise the most pervasively distributed soluble-protein family within the ABC superfamily. Multiple ABC-F family members are encoded in all eukaryotic and most eubacterial genomes21, including three in humans, two in Saccharomyces cerevisiae, five in Arabidopsis thaliana, and four in E. coli (Fig. 1 and Supplementary Fig. 1). ABC-Fs have two tandem ABCs separated by an ~80 residue linker in a single polypeptide chain. PFAM22 identifies this linker as a conserved domain (PF12848 or ABC_tran_2) distinct from the ATPase domains (PF00005 or ABC_tran). PF12848 is found in other proteins with diverse organizations generally including at least one ABC domain. However, it is not found in ABC-E or eEF3, which instead contain different domains12,15 not found in ABC-Fs. Moreover, although ABC-Fs show stronger sequence similarity to eEF3 than to other soluble ABC proteins (Fig. 1 and Supplementary Fig. 1), eEF3 is more closely related to several ABC transporters than to ABC-Fs. Therefore, ABC-Fs represent a distinct phylogenetic lineage that probably evolved independently from the other soluble ABC protein families and have a different biochemical function.
Figure 1.
ABC-F phylogeny. Cladogram produced using CLUSTAL-Ω and labeled with Swissprot species codes, which shows two bacterial orthologs of EttA (from A. tumefaciens and M. tuberculosis), one bacterial paralog of EttA (YfmR from B. subtillis), two non-ABC-F family proteins containing tandem ABC domains (eEF3 and ABCE1 from S. cerevisiae), and all ABC-F proteins from E. coli (red), S. cerevisiae (cyan), A. thaliana (purple), and H. sapiens (blue). Note that all of these ABC-F proteins, but neither eEF3 nor ABCE1, contain the PF12848 domain in addition to tandem ABC domains.
Despite their ubiquitous distribution, no ABC-F protein has had its exact function elucidated, although some seem to play a role in protein synthesis. The N-terminal domain of GCN20, a yeast ABC-F, modulates a ribosome-associated kinase that regulates translation upon amino-acid starvation23,24. However, this domain is not found in other ABC-F families, and the established activity of GCN20 does not require its ABC domains. ARB1, the second yeast ABC-F, is essential and impairs ribosome biogenesis upon depletion25. Human ABC50 (ABC-F1) influences translation initiation at an internal ribosome entry site (IRES) in vitro; consistent with a broader role in translation initiation, a hydrolysis-deficient mutant of ABC50 causes polysome depletion in vivo18. In contrast, the E. coli ABC-F Uup, has been proposed to function in DNA recombination26,27. Nothing is known about the functions of the other E. coli ABC-Fs28, which were given provisional names YbiT, YheS, and YjjK during annotation of the E. coli K12 genome (Fig. 1 and Supplementary Fig. 1).
We set out to elucidate the biochemical and physiological functions of E. coli YjjK, using a combination of structural, enzymological, and genetic methods. This protein has orthologs in more than half of eubacteria, making it the most widespread of the >20 phylogenetically distinct ABC-F classes in eubacteria. We propose renaming this protein Energy-dependent Translational Throttle A (EttA) based on our results presented below, which demonstrate that it is a translation factor that gates ribosome entry into the translation elongation cycle through a nucleotide-dependent interaction sensitive to ATP/ADP ratio.
RESULTS
Crystal structure of E. coli EttA
We solved the X-ray crystal structure of E. coli EttA using single-wavelength anomalous diffraction from selenomethionine-labeled protein. This nucleotide-free structure, the first determined for any ABC-F protein, was refined at 2.4 Å resolution to a free R-factor of 18.3% (Table 1, Figs. 2a-b, and Supplementary Figs. 1–4). The asymmetric unit contains a domain-swapped dimer with only minor deviations from 2-fold symmetry (Fig. 2a). Purified EttA participates in a slowly reversible monomer-dimer equilibrium (Supplementary Fig. 5a) that favors the monomer at the ~7–20 µM concentration measured in vivo29 but the dimer at the ~240 µM concentration used for crystallization. In vitro translation assays presented below suggest that the monomer form of EttA regulates protein synthesis, because it is active at a 3 µM concentration at which the monomer predominates in solution (Supplementary Fig. 5a). This inference is confirmed by the results in the accompanying paper30, which reports the cryogenic electron microscopy (cryo-EM) structure of a functional complex of EttA with 70S ribosomes that was generated using equivalent in vitro translation reactions. In the domain-swapped dimer of EttA (Fig. 2a), ABC1 from one protomer interacts with ABC2 from the other protomer. The cryo-EM structure of EttA30 indicates that this ABC1-ABC2 complex (Fig. 2b), comprising half of the dimer structure, represents the active form of EttA.
Table 1.
Data collection and refinement statistics a
| YjjK/EttA | |
|---|---|
| Data collection | |
| Space group | P21 |
| Cell dimensions | |
| a, b, c (Å) | 45.4, 233.5, 54.1 |
| α, β, γ (°) | 90.0, 91.3, 90.0 |
| Resolution (Å) | 50.0 – 2.4 (2.44 – 2.40) |
| Rsym | 14.3% (57.7%) |
| I / σI | 14.1 (2.0) |
| Completeness (%) | 93.7% (83.2%) for I > −σI |
| Redundancy | 6.5 (3.2) |
| Refinement | |
| Resolution (Å) | 50.0 – 2.40 (2.46 – 2.40) |
| No. reflections | 41106 (2328) |
| Rwork / Rfree | 18.3% (24.0%) / 24.3% (32.0%) |
| Model contents | |
| Residues in protomer A | 1–133, 140–281, 286–548 |
| Residues in protomer B | 3–131, 140–279, 283–540 |
| Organic ions and molecules | 1 citrate, 1 triethyleneglycol, 9 glycerol |
| Inorganic ions | 11 sulfates |
| No. atoms | 8904 |
| Protein | 8376 (23 alternate conformations) |
| Ligand/ion | 132 |
| Water | 396 |
| Ramachandran distribution | |
| Most favored | 98.3% |
| Additionally allowed | 1.7% |
| B-factors (Å2) | 35.3 (Wilson 27.69) |
| Protein | 35.3 |
| Ligand/ion | 50.7 |
| Water | 31.7 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.006 |
| Bond angles (°) | 1.23 |
Data collection statistics correspond to a dataset derived from a single crystal as described in the text. Values in parentheses are for the highest-resolution shell. Data collection statistics come from SCALEPACK, while other statistics come from PHENIX.
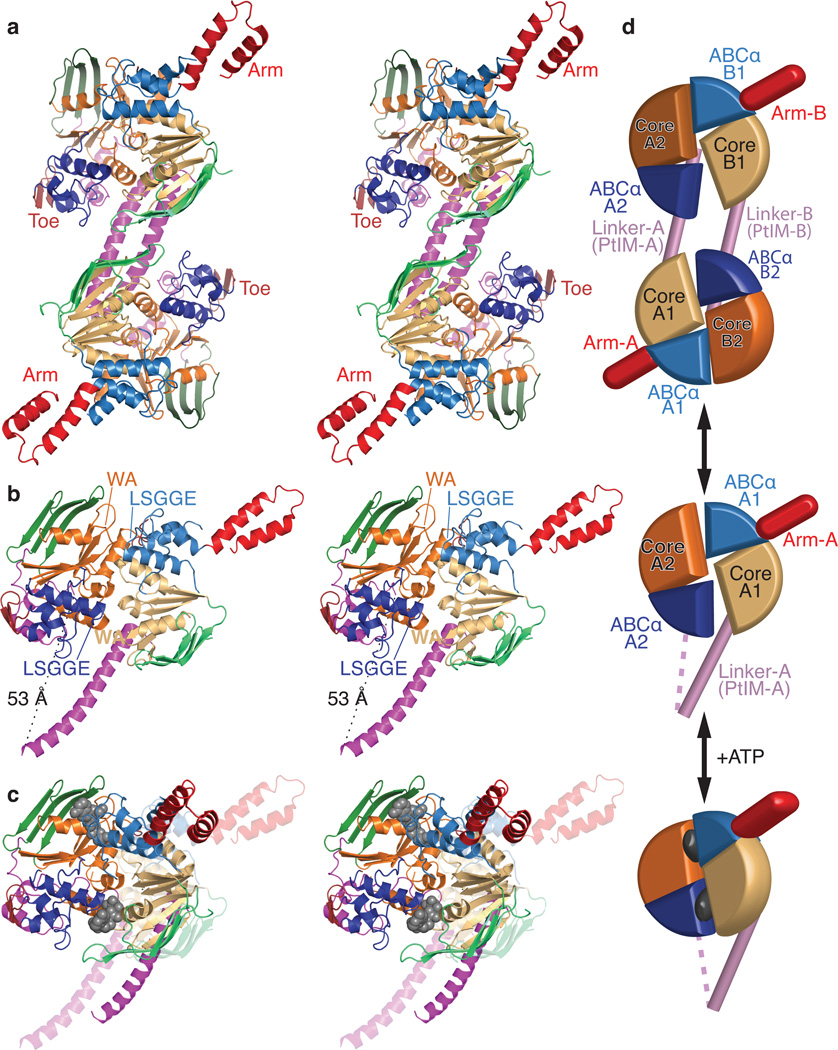
Figure 2.
Crystal structure of E. coli EttA. (a) Stereopair showing the nucleotide-free EttA dimer in the asymmetric unit (Table 1). The ABC domains in each protomer are colored lighter (ABC1) and darker (ABC2) shades of similar colors (green for ABCβ, tan-orange for F1-like core, and blue for ABCα subdomains, red for the arm and toe motifs, and magenta for the PtIM30). (b) Equivalently colored stereopair showing a magnified view of one interacting ABC1-ABC2 domain pair in the EttA dimer (generated by deleting 1-286 in protomer A and 278-555 in protomer B), which provides a model for the nucleotide-free conformation of the EttA monomer. Labels indicate the Walker A (WA) motif in the Fl-like core and the LSGGE signature sequence in the ABCα subdomain. The Walker B motif (Φ4DE, with Φ being any hydrophobe and terminating in catalytic base) is located between the WA and LSGGE motifs within each ABC. (c) Stereopair showing models for the nucleotide-free (translucent colors) and ATP-bound (solid colors) conformations of the EttA monomer superimposed via least-squares alignment of ABC2. The nucleotide-free conformation represents one ABC1-ABC2 domain pair from the crystallographically observed EttA dimer (panel b), while the ATP-bound conformation was modeled using rigid-body rotations to align the crystallographically observed nucleotide-free conformations of ABC1 and ABC2 to the two protomers in the ATP-sandwich dimer of the E171Q mutant of MJ0796; see “Structural Superposition” in Online Methods for details. (d) Schematics of the EttA dimer (top), nucleotide-free monomer (middle), and modeled ATP-bound monomer (bottom) colored as above.
The tandem ABC domains in EttA (ABC1 in lighter and ABC2 in darker colors) are canonical in structure except for one insertion of substantial size in each domain (Figs. 2a-b and Supplementary Figs. 1–2). These insertions, dubbed the “arm” in ABC1 and the “toe” in ABC2, occur in the loop after the first of the three α-helices in the ABCα subdomain (blue), which is the primary site of transmembrane-domain contact in ABC transporters. The arm is a 45-residue α-helical hairpin spanning amino acids 95–139 (lighter red), while the toe is a 12-residue antiparallel β-hairpin spanning amino acids 414–423 (darker red). Based on their structural uniqueness and location, we hypothesized that these structures mediate important functional interactions, an inference verified below.
Minor structural variations are observed at two other sites in the ABC domains, at the C-terminus of the ABCβ subdomain and in the segment preceding the LSGGQ motif, which are frequent sites of structural diversity in ABC proteins31. The catalytic motifs in EttA are canonical, with two exceptions. The first is the lack of an aromatic residue in most EttA orthologs at the C-terminus of the first β-strand in the ABCβ subdomain of ABC1, at a position where an aromatic residue typically stacks with the adenine base of ATP (Supplementary Fig. 1). The second is the substitution of glutamate for glutamine in the LSGGQ motifs in ABC1 of all othologs and ABC2 of most of them (i.e., making their sequences LSGGE). Less conservative substitutions are observed in ABC1 in some other ABC-F family members (Supplementary Fig. 1). Structural superposition demonstrates that ABC1 and ABC2 of EttA are slightly more closely related to each other than to other ABC domains, but that they are not more closely related to eEF3 and ABCE1 than to several transmembrane transporters (Supplementary Table 1).
As previously observed in other nucleotide-free ABC superfamily structures32,33, ABC1 and ABC2 of EttA interact in an “open” conformation in which their ATP-binding sites are both positioned in a deep groove at their mutual interface. However, the Walker A and LSGGE motifs are too far apart to tightly encapsulate ATP in the inter-ABC interface. Their F1-like ATP-binding cores would need to rotate by 44° (as modeled in Fig. 2c and Supplementary Fig. 3a) to bring them into the closed, catalytically active ATP-sandwich dimer conformation adopted by ABC domains upon binding ATP3–5. Furthermore, within each domain, the ABCα subdomain is rotated away from its ATP-binding core by 18–20° compared to the canonical ATP-binding conformation (Supplementary Fig. 3b-c). The observed deviations from this conformation are all characteristic of nucleotide-free ABC domain structures34,35.
The 81-residue linker between the ABC domains in EttA (magenta) is a unique feature of ABC-Fs that PFAM22 identifies as conserved domain PF12848. We designate it as the “P-site tRNA-interaction motif” (PtIM) based on the cryo-EM structure of ribosome-bound EttA30, which shows a monomer of EttA making extensive interactions with the ribosomal tRNA exit (E) site and an initiator tRNA in the ribosomal peptidyl-tRNA binding (P) site30. The first half of the PtIM forms an ~50 Å long extension of the C-terminal α-helix in the ATP-binding core of ABC1. In the crystal structure of EttA, the second half of the PtIM forms a pair of shorter α-helices that pack onto ABC2 (Fig. 2a-b and Supplemental Fig. 4a). These α-helices are followed by 7-residues (residues 311–317) that pack into a deep groove between the ABCα and F1-like core subdomains of ABC2 on the surface opposite its interface with ABC1. Possible functional implications of this interaction are described in Supplemental Fig. 4b.
Approximately 3,500 Å2 of solvent-accessible surface area per protomer is buried in the interface of the domain-swapped dimer of EttA in the asymmetric unit of its crystal structure (Fig. 2a). One-quarter of this interface (~920 Å2) comes from a reciprocal packing interaction between the first α-helix in the PtIM, which prevents it from adopting the α-helical hairpin configuration that interacts with ribosomes30 (Supplementary Fig. 4). A single rigid-body rotation simultaneously brings the ATP-binding cores of both ABC1-ABC2 domain pairs into the canonical ATP-sandwich conformation (Supplementary Fig. 3d). This result suggests that the EttA dimer might be able to bind four ATP molecules cooperatively, although experimental evidence of such cooperativity has not yet been obtained. This dimer could represent an inactive form that buffers the active monomer pool at high EttA concentrations, but further investigation will be required to understand the significance of the dimer.
EttA-EQ2 stops cell growth by inhibiting protein synthesis
Guided by the crystal structure, we designed two EttA mutants for physiological studies. EttA-EQ2 contains dual glutamate-to-glutamine substitutions in the catalytic bases following the Walker B motifs in both ABC domains (Glu188 and Glu470). Based on results with other ABC ATPases5,33,36, these substitutions prevent ATP hydrolysis and trap EttA in its ATP-bound conformation. The other designed mutant, EttA-Δarm, deletes the arm motif, a unique structural feature in ABC1 of ABC-Fs. We used a tightly controlled arabinose-dependent promoter to induce expression of EttA-EQ2 in E. coli MG1655 cells, which results in arrest of growth either in the absence (Fig. 3a) or presence (unpublished) of a ΔettA mutation deleting the endogenous gene. In contrast, induction of wild-type EttA (WT-EttA), EttA-Δarm, or EttA-Δarm-EQ2 has no effect on growth. Abrogation of the toxicity of EttA-EQ2 upon deletion of the arm supports our inference that this motif contributes to ABC-F function.
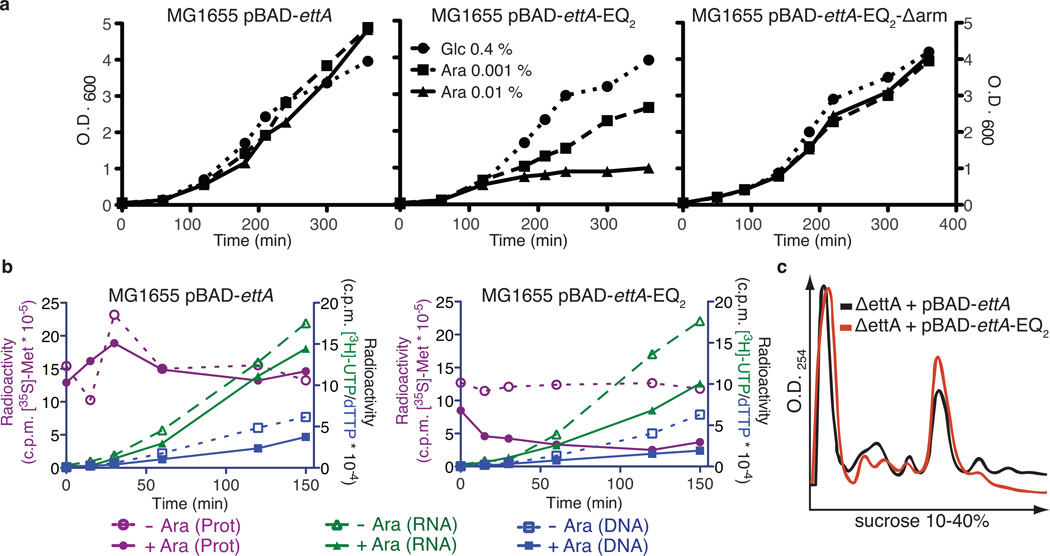
Figure 3.
Expression of EttA-EQ2 causes trans-dominant toxicity in vivo due to inhibition of protein synthesis. (a) Graphs showing OD600 profiles during expression of EttA variants in E. coli MG1655 in LB medium at 37 °C. Cells harboring pBAD-ettA, pBAD-ettA-EQ2, or pBAD-ettA-EQ2-Δarm plasmids were grown overnight in LB with 0.4% (w/v) glucose (Glc) to repress EttA expression and then diluted 1:100 into the same medium or alternatively one containing 0.001-0.01% (w/v) arabinose (Ara) to induce increasing levels of expression. (b) Graphs showing results from experiments using radiolabeled precursors to characterize the influence of expressing EttA variants on protein, RNA, and DNA synthesis in vivo. MG1655 cells harboring pBAD-ettA or pBAD-ettA-EQ2 plasmids were grown at 37 °C in M9 glycerol minimal medium to OD600 ~0.2 prior to induction of EttA expression using 0.2% (w/v) Ara at zero time on these graphs. RNA or DNA were labeled by adding [3H]UTP (green) or [3H]dTTP (blue), respectively, to the cultures at the same time as the inducer, while protein was labeled at the indicated time points by subjecting an aliquot of the culture to a 1 minute pulse with [35S]methionine (red). Cells were spotted onto a Whatman 3MM filter and washed with trichloroacetic acid (TCA) before scintillation counting of the radioactivity incorporated into polymers. (c) Plots of sucrose gradient profiles of polysomes from MG1655 ΔettA cells harboring pBAD-ettA or pBAD-ettA-EQ2 plasmids induced with 0.1% Ara for 30 minutes after reaching an OD600 of 0.6.
In vivo pulse-chase experiments using radiolabeled substrates for protein synthesis, RNA transcription, or DNA replication demonstrate that EttA-EQ2 induction rapidly inhibits protein synthesis (Fig. 3b). The slower and weaker inhibition of RNA and DNA synthesis suggests that these effects are secondary to inhibition of protein synthesis. Indeed, purified EttA-EQ2, but neither WT-EttA, EttA-Δarm, nor EttA-Δarm-EQ2, inhibits in vitro translation of a luciferase reporter mRNA (Supplementary Fig. 6a-b). Immunoblot analyses of fractions from sucrose density gradient ultracentrifugation of ribosomes from E. coli MG1655 cells (Supplementary Fig. 2b) show that endogenous WT-EttA co-fractionates with both 70S ribosomes (monosomes) and poly-ribosomes (polysomes). Equivalent analyses conducted 30 min after induction of EttA-EQ2 reveals a decrease in polysomes relative to monosomes. These observations suggest that the ATP-bound conformation of EttA, as trapped by the EQ2 mutations, inhibits protein synthesis after formation of the 70S ribosomal initiation complex (70S IC) but prior to its entry into the elongation cycle (Fig. 3c). In vitro translation experiments on a single mRNA using radiolabeled [35S]methionine support this conclusion (Supplementary Fig. 6c).
EttA-EQ2•ATP traps ribosomes after formation of the first peptide bond
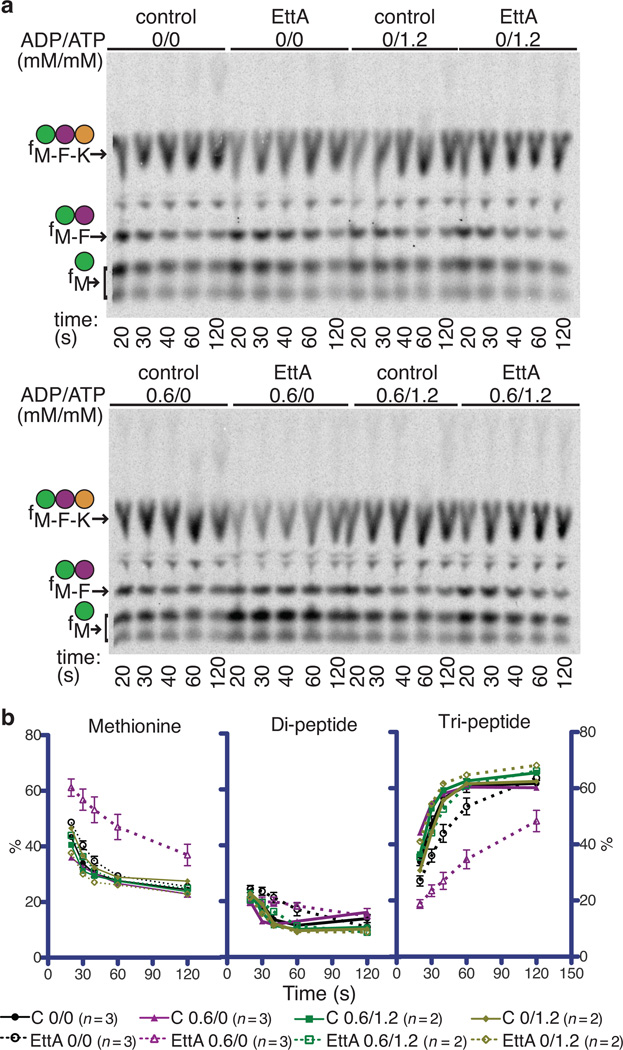
We employed a purified in vitro translation system37 to demonstrate that EttA-EQ2 specifically inhibits protein synthesis after formation of the first peptide bond (Fig. 4 and Supplementary Fig. 7). We pre-formed a 70S IC by incubating translation initiation factors 1, 2, and 3 with 70S ribosomes, then adding mRNA followed by [35S]fMet-tRNAfMet. The model mRNA, previously used for enzymological studies of ribosome-catalyzed protein synthesis37,38, contains a Shine-Delgarno sequence, initial codons encoding an fMet-Phe-Lys-Glu (fMFKE) tetrapeptide, and 16 additional codons to fill the ribosomal mRNA-binding channel. After 70S IC formation, we conducted translation elongation reactions in a buffer containing 0.6 mM ATP in addition to 1 mM GTP, the latter nucleotide being required for proper function of elongation factors EF-Tu and EF-G. Reaction products were analyzed using electrophoretic thin layer chromatography (eTLC), which separates unreacted [35S]fMet amino acid substrate and di-, tri-, and tetrapeptide products39.
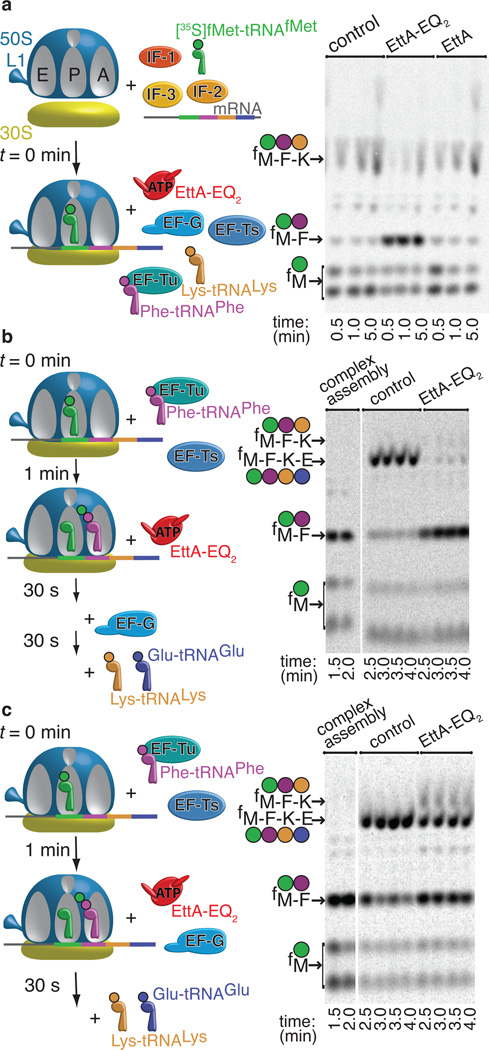
Figure 4.
EttA-EQ2 inhibits translation after formation of the first peptide bound. Minimum in vitro translation assays were performed as explained in the schematics on the left at 37 °C in the presence of 0.5 mM ATP, 1.0 mM GTP, and a phophoenolpyruvate-based energy-regenerating system. Reaction products were analyzed by electrophoretic thin-layer chromatography (eTLC) and autoradiography (right). (a) After 70S IC formation, either buffer or 2.5 µM WT-EttA or EttA-EQ2 was added in parallel with the elongation factors, Phe-tRNAPhe and Lys-tRNALys. (b) After formation of the 70S IC and subsequent addition of EF-Tu, EF-Ts, and Phe-tRNAPhe to drive synthesis of the first peptide bond, either buffer or EttA-EQ2 was added 1 minute later, and the reaction was allowed to proceed for 30 seconds prior to the addition of EF-G, Lys-tRNALys, and Glu-tRNAGlu to enable tetrapeptide synthesis. (c) Same protocol as panel b, but to determine whether EF-G and EttA-EQ2 kinetically compete with one another, EF-G was added in parallel with buffer or EttA-EQ2 at 1 minute after addition of EF-Tu, EF-Ts, and Phe-tRNAPhe. Thirty seconds later, Lys-tRNALys and Glu-tRNAGlu were added to enable tetrapeptide synthesis. Reactions were conducted in Polymix Buffer (3.5 mM Mg(OAc)2, 100 mM KCl, 5 mM NH4OAc, 0.5 mM Ca(OAc)2, 0.1 mM EDTA, 1 mM spermidine, 5 mM putrescine, 6 mM 2-mercaptoethanol, 50 mM Tris-OAc, pH 6.9) using an mRNA template directing synthesis of an fMet-Phe-Lys-Glu (fMFKE) tetrapeptide.
Fig. 4a shows tripeptide synthesis in reactions initiated by adding a mixture containing EF-Tu, EF-Ts, Phe-tRNAPhe, and Lys-tRNALys to the pre-formed 70S IC and subsequently adding EF-G. When EttA is omitted or WT-EttA is added at the same time as EF-G, an fMFK tripeptide is synthesized efficiently before the translating ribosome stalls at the fourth codon due to the absence of a cognate Glu-tRNAGlu (Fig. 4a). In contrast, addition of EttA-EQ2 at the same time as EF-G strongly inhibits translation elongation after formation of the first peptide bond, resulting in a reduction in fMFK tripeptide yield and accumulation of fMF dipeptide (Fig. 4a). This observation reveals that EttA-EQ2, which should be locked in the ATP-bound conformation, blocks translation after the first aminoacyl-tRNA has been incorporated into the A site and participated in peptide-bond formation at the peptidyl-transferase center (PTC), but prior to a second round of peptide-bond formation. The same result is obtained when EttA-EQ2 is added prior to 70S IC formation (Supplementary Fig. 7a), demonstrating that 70S IC formation is not inhibited by EttA-EQ2.
We used variations in the assay protocol to pinpoint the step at which EttA-EQ2 inhibits the elongation cycle. To test whether inhibition occurs before the first round of EF-G-catalyzed translocation40,41 on the mRNA template, we varied the order of addition of the components needed to elongate the fMF dipeptide (Figs. 4b-c and Supplementary 7b), which accumulates in a reaction that proceeds for 1 min in the absence of EF-G and Lys-tRNALys (leftmost lanes in Fig. 4b-c). Subsequent addition of EF-G together with Lys-tRNALys and Glu-tRNAGlu results in extension of the fMF dipeptide into an fMFKE tetrapeptide (center lanes in Fig. 4b-c), demonstrating that the fMF dipeptide product remains covalently attached to tRNAPhe in the A site of the ribosomal pre-translocation (PRE) complex. Addition of EttA-EQ2 prior to EF-G/Lys-tRNALys/Glu-tRNAGlu shows almost complete inhibition of the extension of the fMF dipeptide (right lanes in Fig. 4b). In contrast, much weaker inhibition is observed when EttA-EQ2 is added simultaneously with (right lanes in Fig. 4c) or subsequent to (Supplementary Fig. 7b) EF-G/Lys-tRNALys/Glu-tRNAGlu. These results demonstrate that EttA-EQ2 and EF-G kinetically compete for interaction with the ribosomal pre-translocation complex carrying deacylated tRNAfMet in the P site and fMF-tRNAPhe in the A site.
Remarkably, fMFKE tetrapeptide synthesis reactions do not show accumulation of fMFK tripeptide, even when ~50% of fMFKE synthesis is inhibited by EttA-EQ2 (Figs. 4c and Supplementary Fig. 7b). Therefore, although it strongly inhibits extension of the fMF dipeptide into an fMFK tripeptide, EttA-EQ2 does not significantly inhibit extension of the fMFK tripeptide into an fMFKE tetrapeptide. These observations demonstrate that EttA-EQ2 is specific for ribosomal complexes that have cleared the initiation stage of protein synthesis but have not yet undergone the first round of EF-G-catalyzed translocation40,41.
EttA prevents peptide-bond formation in the presence of ADP
We further varied the assay protocol to evaluate whether WT-EttA’s activity is influenced by alterations in ATP/ADP ratio, a parameter that tracks cellular energy supply. WT-EttA, like most ABC ATPases42, interacts in an approximately equivalent manner with adenine and guanine nucleotides (unpublished), while the essential GTPase translation factors are specific for guanine43. Therefore, we reduced the concentration of GTP used in our in vitro translations from 1 mM to 300 µM to enable addition of physiologically relevant concentrations of ADP and ATP to produce significant variations in the ratio of nucleotide triphosphates (NTPs) to nucleotide diphosphates (NDPs). (Note that we use the term ATP/ADP ratio in this manuscript as shorthand for the NTP/NDP ratio, because these ratios track one another in E. coli44–48).
WT-EttA produces a small, but reproducible, stimulation of fMFK formation in tripeptide synthesis assays in 300 µM GTP and 1.2 mM ATP (top right in Fig. 5a). In contrast, WT-EttA produces an appreciable kinetic inhibition of fMK dipeptide and fMFK tripeptide formation in equivalent assays in the presence of the same concentration of GTP but with 0.6 mM ADP substituted for 1.2 mM ATP (bottom left in Fig. 5a-b). Because dipeptide synthesis must precede tripeptide synthesis, and WT-EttA kinetically inhibits both, we infer that the protein inhibits fMF dipeptide synthesis in the presence of ADP. These results contrast with those presented above demonstrating that, in the presence of ATP, EttA-EQ2 allows fMF dipeptide synthesis while specifically inhibiting fMFK tripeptide synthesis (Fig. 4). Analysis of our cryo-EM structure30 confirms that ribosome-bound EttA-EQ2 is trapped in an ATP-bound conformation in the presence of ATP. Therefore, the contrasting results observed in our in vitro translation experiments conducted with WT-EttA•ADP compared to EttA-EQ2•ATP indicate important differences in the functional interactions of EttA with translating ribosomes depending on the relative concentrations of ADP vs. ATP.
Figure 5.
WT-EttA inhibits synthesis of the first peptide bond at low ATP/ADP ratio. (a) Room temperature in vitro translations with or without 0.6 mM ADP and 1.2 mM ATP were analyzed by eTLC. Reactions, conducted as in Fig. 4a but with the 70S IC desalted in Polymix Buffer, contained 0.3 mM GTP, 0.6 µM 70S ribosomes, and when indicated 3.5 µM of an EttA variant added in parallel with the elongation factors, Phe-tRNAPhe, and Lys-tRNALys. (b) Quantification of products in the autoradiograms in panel a using ImageQuant software, with error bars representing standard error of the mean.
This inference is supported by single-molecule fluorescence resonance energy transfer (smFRET) experiments showing modest but statistically significant differences in the influence of EttA on the structure and dynamics of the ribosomal L1 stalk in the presence of ADP vs. ATP (Supplementary Fig. 8). These experiments employed a donor fluorophore at the base of the L1 stalk and an acceptor fluorophore at its apical tip (smFRETL1-L9)49. In the presence of ATP, EttA-EQ2 increases the mean FRET efficiency (EFRET), suggesting a decrease in mean donor-acceptor separation consistent with our cryo-EM structure30. WT-EttA produces a similar but smaller increase in EFRET in the presence of ATP, presumably reflecting a mixed population of free and EttA-bound 70S ICs due to transient interaction of ATP-bound EttA prior to dissociation induced by ATP hydrolysis. In contrast, in the presence of ADP, WT-EttA produces a small decrease in EFRET, suggesting an increase in mean donor-acceptor separation. This change in EFRET in the opposite direction from what is observed in the presence of ATP demonstrates that EttA modulates the structure or dynamics of the L1 stalk differently in the presence of ATP vs. ADP.
Importantly, inhibition by WT-EttA in the presence of 0.6 mM ADP is relieved when 1.2 mM ATP is simultaneously included in in vitro translation reactions (bottom right in Fig. 5a). Therefore, the ADP/ATP ratio controls WT-EttA activity, and a super-stoichiometric ratio of ATP relieves ADP-dependent inhibition of protein synthesis by EttA. These observations suggest that an elevated cellular ADP/ATP ratio, as found in energy-depleted cells44,50, will cause EttA to stabilize 70S ICs in a “hibernating” conformation that prevents committing metabolic resources to synthesis of incomplete proteins. This hypothesis, based on our in vitro enzymological studies, suggests that EttA could play a significant role in controlling protein synthesis in stationary-phase cells, in which the rates of protein synthesis and cell growth51–54 decline due to depletion of nutritional and energetic resources.
ΔettA impairs fitness in long-term stationary phase
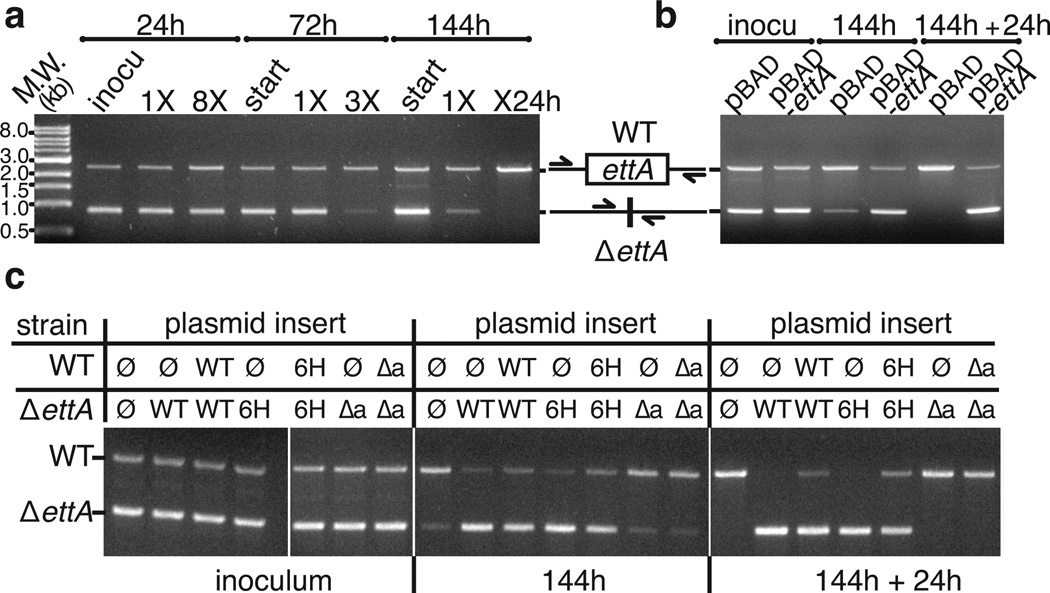
Consistent with this hypothesis, Western blots demonstrate that EttA expression increases in stationary phase (Supplementary Fig. 5c), when there is a declining ATP/ADP ratio44,50,53. Increasing expression should promote formation of EttA-bound, hibernating 70S ICs poised to rapidly resume protein synthesis when energy, in the form of ATP, becomes available again. Therefore, we investigated whether EttA influences fitness when growth in fresh LB medium is resumed out of stationary phase. Indeed, ΔettA E. coli exhibits a progressively more severe competitive disadvantage as residency in stationary phase is extended from one to six days (Fig. 6a) prior to restarting growth. This defect is complemented by expression of WT-EttA or His6-EttA (Fig. 6d-c) but not EttA-Δarm (Fig. 6c). The parallel effects of the Δarm mutation in abrogating the inhibition of in vitro translation by EttA-EQ2 and in eliminating the in vivo fitness advantage conferred by WT-EttA supports the hypothesis that this advantage derives from the functional interaction of EttA with ribosomes.
Figure 6.
WT and His6-EttA promote survival in long-term stationary phase. Agarose gels are shown that visualize PCR products quantifying the relative population of wild-type vs. ΔettA cells in competitive fitness assays in LB at 37 °C. The chromosomal region flanking ettA by 400 basepairs was amplified from total DNA in mixed cultures. (a) Starting cultures containing a 1:1 mixture of overnights from the individual strains were grown for 24, 72, or 144 hours prior to re-inoculation into fresh medium and re-growth for the same period of time. Eight growth cycles were performed for the 24-hour culture, three for the 72-hour culture, and two for the 144-hour culture, which was re-grown for an additional 24 hours prior to analysis. (b) Mixed cultures of the ΔettA strain containing pBAD or pBAD-ettA plasmids were grown for 144 hours, prior to re-inoculation for an additional 24 hours. Immunoblotting analysis with anti-EttA antibody (unpublished) demonstrates a roughly physiological level of expression from the pBAD-ettA plasmid under these growth conditions (i.e., without supplementation with glucose to repress expression from the arabinose promoter controlling expression of EttA). (c) Results from equivalent complementation experiments performed on mixed cultures of the ΔettA and wild-type strains containing pBAD plasmid with different inserts (ø: no insert, WT: WT-EttA, 6H: His6-EttA, Δa: EttA-Δarm).
DISCUSSION
Our biochemical results demonstrate that EttA, the most widely distributed ABC-F protein among eubacteria, is a novel translation factor that controls the progression of 70S ICs into the translation elongation cycle using a mechanism sensitive to the ATP/ADP ratio. We also present genetic experiments showing that knockout of the ettA gene produces a severe fitness defect in E. coli in long-term stationary phase (Fig. 6). This observation supports the hypothesis that EttA contributes to regulating the commitment of metabolic resources to protein synthesis and preventing the synthesis of incomplete proteins in energy-depleted cells.
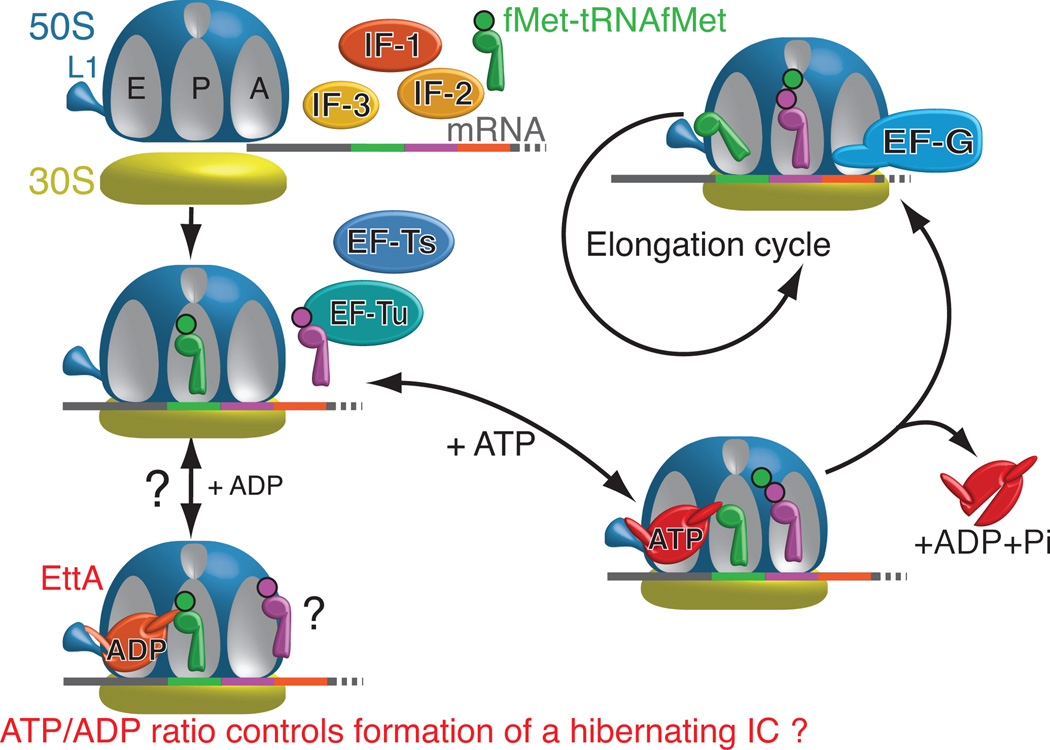
Our results, combined with those in the accompanying paper30 and published work on ABC ATPases3–5, support a straightforward model for the interaction of ATP-bound EttA with the 70S IC (Fig. 7), although several alternative models outlined below could explain the more complex influence of ADP on this interaction. The ATP hydrolysis cycle of ABC ATPases, like that of other NTPases, involves orderly progression through a series of conformational states coupled to ATP binding, ATP hydrolysis, and release of the products (ADP and inorganic phosphate). As observed for other ABC ATPases3–5, EttA’s two ABC domains adopt an open conformation in the absence of bound nucleotide, as visualized in our nucleotide-free X-ray crystal structure of WT-EttA (Fig. 1a-b) in which the ATP-binding site in each ABC domain faces the other ABC domain without directly contacting it (Fig. 1a-b). Binding of two ATP molecules to these sites closes the interface between the ABC domains to produce a more compact conformation with greatly increased affinity for the 70S IC, as visualized in the cryo-EM structure of ATP-bound EttA-EQ2 reported in the accompanying paper30. This structure shows EttA bound in the E site of the ribosome, where its arm motif contacts the L1 stalk of the large ribosomal subunit and its PtIM interacts with the acceptor stem of a P site-bound deacylated initiator tRNAfMet. The small but reproducible stimulation of dipeptide synthesis by WT-EttA in the presence of ATP (Fig. 5) suggests that ATP-bound EttA stabilizes the ribosome in a conformation that promotes peptide-bond formation in the peptidyl-transferase center. Interaction with the ribosome, in turn, stimulates ATP hydrolysis by EttA (Supplementary Fig. 5b), and this reaction triggers release of EttA from the ribosome and entry of the ribosome into the translation elongation cycle. By blocking ATP hydrolysis, the EQ2 mutations in EttA trap the protein in the otherwise transient ATP-bound state and consequently block its release from the ribosome. In WT-EttA, transient electrostatic forces generated during ATP hydrolysis may accelerate this release process. Once engaged in the translation elongation cycle, the ribosome becomes resistant to the rebinding of EttA (Fig 4 and Supplementary Fig. 6c), presumably either due to EttA having reduced affinity for elongator tRNAs compared to the initiator tRNAfMet in the P site or due to deacylated tRNAs passing through and blocking the E site as they exit the translating ribosome.
Figure 7.
Schematic model of EttA function based on the results presented here and in the companion paper30. In the presence of ADP, EttA inhibits formation of the first peptide bound (Fig. 5b), which may be mediated by stabilization of the 70S IC in a hibernating state by ADP-bound EttA. (See Discussion). In contrast, ATP-bound EttA stimulates the formation of the first peptide bond by the ribosome and then, concomitantly with ATP hydrolysis, dissociates from the ribosome, thereby allowing it to enter the elongation cycle.
Our data show that WT-EttA has a qualitatively different effect on translation in the presence of ADP compared to either WT-EttA or EttA-EQ2 in the presence of ATP (Fig. 5 and Fig. 4). In the presence of ADP, WT-EttA inhibits synthesis of the first peptide bond by the 70S IC rather than promoting or trapping the product of this reaction, as observed in the presence of ATP for WT-EttA and EttA-EQ2, respectively. Several models could explain this alternative activity in the presence of ADP compared to ATP. One possibility is that ADP interacts with the ribosome to alter its interaction with EttA, while an alternative possibility is that ADP binds to one or both of the ATPase active sites in EttA, resulting in an altered conformation that still binds to the 70S IC but stabilizes it in a conformation inhibiting rather than promoting formation of the first peptide. There are several possible explanations for the different behavior of EttA upon binding ADP directly compared to the post-hydrolysis complex with ADP formed following the binding of ATP. A related mechanistic issue concerns whether there is functional asymmetry between the two ATPase active sites in EttA. These issues are addressed in the Supplementary Notes.
Additional studies will be required to understand how EttA interacts with other cellular systems regulating protein synthesis in stationary-phase. A key contributor is likely to be the coupled reductions in GTP/GDP and ATP/ADP ratios in energy-depleted cells (a phenomenon mediated by the phosphotransferase activities of nucleoside diphosphate kinase and adenylate kinase45–48). GDP exerts strong feedback inhibition of most of the essential GTPase translation factors55, and this effect will reduce the rates of both initiation and elongation in energy-depleted cells. This baseline metabolic effect should amplify the activity of EttA and the other proteins that modulate protein translation in stressed and energy-depleted cells. These include some toxin-antitoxin systems56,57, the Ribosomal Silencing Factor58 (RsfA) protein, and the Ribosome Modulation Factor (RMF) protein59,60. Toxin-antitoxin systems improve survival under stress conditions by inhibiting critical physiological processes including protein synthesis57. RsfA, which has a phylogenetic distribution as broad as EttA, inhibits translation in stationary phase by preventing the joining of the large and small ribosomal subunits to form the 70S IC58. RMF, which has a narrow phylogenetic distribution that is limited to proteobacteria, drives dimerization of 70S ribosomes in stationary phase to form inactive 100S di-ribosome complexes59,60. While experiments focused on each of these factors individually have shown that they can contribute to controlling protein synthesis in energy-depleted cells, the manner in which they interact under differ metabolic and environmental conditions is not understood.
The biochemical properties of EttA raise intriguing possibilities for regulation of protein synthesis in response to such environmental variations. The observation that EttA targets a 70S IC poised to translate a bound mRNA suggests that it could act preferentially on mRNAs encoding specific target proteins18, while specificity seems unlikely for the other factors that regulate protein synthesis in energy-depleted cells. If EttA does have such specificity, its activity inhibiting entry into the translational elongation cycle at high ADP concentration (Fig. 5) can attenuate the expression of specific proteins under conditions of energy depletion while simultaneously preparing them for rapid synthesis when energy levels return to normal. Such targeted “hibernation” activity would enable EttA and potentially other ABC-Fs18 to influence cellular fitness not only under conditions of energy deprivation but also in an anticipatory manner upon resumption of growth. ABC-Fs could thereby provide a powerful mechanism for differential control of the translation of specific proteins not only under conditions of growth limitation but also at the time of growth re-initiation. In this context, we note that, in the presence of ATP, E. coli YbiT-EQ2 interacts with ribosomes in vitro in a similar manner to EttA-EQ2 (unpublished).
Our results establish a technical foundation for broader and deeper studies of ABC-F proteins. The fact that these proteins have evaded detailed functional characterization until now, despite their great phylogenetic prevalence and diversity, suggests that substantial gaps remain in understanding the physiology and systems biology of protein synthesis.
ONLINE METHODS
The online Supplementary Note documents protein-purification, crystallization, methods used in this paper and methods used in the experiments presented in Supplementary Information.
Bacterial strains
Standard E. coli strains for cloning (DH5α) and protein expression (BL21(λDE3) and B834) were obtained from commercial vendors. Strains MG1655 (sequenced WT strain) and FB21853 (MG1655 yjjK::Tn5) were purchased from the E. coli Genome Project at the University of Wisconsin (www.genome.wisc.edu). All other strains were purchased from the E. coli Genetic Stock Center at Yale University 61 or constructed in the course of these studies. Genetic and physiological assays were performed using E. coli K12 strain MG1655 or derivates. Because we found that strain FB21853 is not isogenic to the sequenced WT strain MG1655, we re-constructed MG1655 yjjK::Tn5 by P1 transduction62 of the yjjK::Tn5 locus from strain FB21853 into the sequenced WT strain MG1655. The resulting strain, designated ettA::Tn5, was used for the early phases of the work reported in this paper. The interruption of the ettA gene in this strain was verified by Western blot with an antibody against the EttA protein (raised as described below). We also built a strain deleted of yjjK/ettA that do not carry any antibiotic resistance, using the procedure developed by Datsenko and Wanner63. Briefly, the strain deleted for yjjK in the Keio collection64 (JW4354-1, CGSC# 11108) was used as a template to generate the PCR product to mutate the MG1655 strain by amplification of yjjK/ettA locus with primers 400 pb upstream and downstream of the locus. This PCR product was electroporated in the MG1655 strain carrying the pKD46, the transformed strain was cured of the pKD46 plasmid and the insertion of the PCR product in the genome at the good locus was verified by PCR. The positive strain was cured of the pKD46 plasmid and transformed with the FLP helper plasmid pCP20. The resulting colonies were screened for the flip-out of the kanamycin marker by PCR. The verified strain was cured of the pCP20 plasmid and used for the fitness experiment. This strain is referred to as ΔettA. All the constructs were verified by PCR and sequencing of the modified locus.
Plasmids
The gene coding for EttA (YjjK) was amplified by PCR using MG1655 genomic DNA as a template with 5’ primer containing the NcoI restriction site, and 6 codons coding for His in front of the initiator GTG codon which was replaced by an ATG codon. The 3’ primer used for this PCR had the stop codon of yjjK followed by an XhoI restriction site. This PCR product was cloned into the pBAD/Myc-HisA (invitrogen) using the restriction enzyme NcoI and XhoI (Fermentas). The resulting plasmid was called pBAD-His6-ettA. For the plasmid pBAD-ettA, which expressed the native protein without tag, the same procedure was used but with a 5’ primer that does not contain 6 codon for histidine. The plasmid expressing the EttA-E188Q mutant was made by QuikChange II Site-Directed Mutagenesis (Agilent Technologies) using primers that replaced the codon of the Glutamate 188 with a Glutamine and used the pBAD-ettA plasmid as template. The resulting plasmid was verified and named pBAD-ettA-E188Q. The plasmid expressing the EttA-EQ2 was made using the same technique with primers, which replaced the codon of the Glutamate 470 with a Glutamine and the pBAD-ettA-E188Q as template; the resulting plasmid was named pBAD-ettA-EQ2. The deletion of the Arm domain (plasmid pBAD-ettA-Δarm) was also done by QuickChange using the pBAD-ettA as template and primer designed to substitute the three residue sequence GGS for residues 96 to 141 in the native EttA sequence (EVVNALKRLDEVYALYADPDADFDKLAAEQGRLEEIIQAHDGHN LN). The plasmid pBAD-ettA-EQ2-Δarm was created using the same approach, but with the plasmid pBAD-ettA-EQ2 as template. The same constructs, expressing a His-tagged protein, were made similarly, but using the pBAD-His6-ettA as starting plasmid. For structure determination, the yjjK/ettA gene was inserted into vector pET28c (EMD Biosciences) at the NcoI and XhoI restriction sites so as to express the full-length protein with no additional tags or amino acids. All the plasmids were verified by DNA sequencing.
Bacterial growth media
Bacteria were cultivated in LB medium (Affymetrix/USB). Ampicillin was added at 100 µg/ml for cultures harboring pBAD based plasmids. Kanamycin was added at 25 µg/ml for the mutant construct.
Estimation of EttA concentration in vivo
The quantitative proteomics study of Lu et al.29 reports the concentration of EttA (YjjK) to be 2167 molecules/cell during exponential growth in glucose minimal medium, which corresponds to 7 µM protomer (assuming an average cell volume65 of 4.96 × 10−16 L). We have verified by western-blot analysis that the expression level of EttA is similar in exponential phase in glucose medium minimal or LB (unpublished). The western-blot data presented in Supplementary Figure 5c shows that EttA expression increases after 24 hours of growth, to a level ~3-fold higher than in exponential phase. Therefore, based on the calibration described above, EttA concentration in stationary phase is ~21µM.
Crystallization, X-ray data collection, and structure determination
Crystals of EttA (either native or selenomethionie derivatized) regularly exhibited streaked and highly mosaic diffraction. Out of hundreds of crystals screened, a single selenomethionine crystal showed diffraction convincingly beyond 3 Å using a rotating anode X-ray source. Data from this crystal were collected on NSLS beamline X12C using a Brandeis-B4 detector, a Nonius/Bruker diffractometer (c. 1999), an ambient temperature of 130K, and a wavelength corresponding to maximum f” as measured by an online fluorescence scan (0.97961Å). A total of 529 frames of 1° oscillation images were processed with DENZO and merged with SCALEPACK using the “no merge original index” and “scale anomalous” options66. The resulting dataset was highly redundant and complete to a limiting resolution of 2.4 Å (see Table 1) and, based on a solvent content of 50%, was expected to contain two EttA protomers per asymmetric unit. The resulting dataset was analyzed with the “SAD” option in SOLVE version 2.0367 using a limiting resolution of 3.0 Å. The anomalous signal-to-noise ratio for this dataset was estimated at only 0.64. Nonetheless, SOLVE identified 18 selenium sites that obeyed 2-fold rotational non-crystallographic symmetry (NCS). However, the resulting electron density maps were uninterpretable. Inversion of the site pattern and recalculation of phases followed by extensive solvent flattening in RESOLVE68 did produce an interpretable electron density map. The protein model was built by hand using O69 and initially refined in CNS70 using standard procedures along with NCS restraints. Further iterative refinement and rebuilding were carried out using PHENIX71 and COOT72, respectively. Refinement in PHENIX was carried out using the same set of “free” reflections as had been used in CNS, but NCS restraints were not applied. The final model of EttA contained 1065 protein residues in two chains (6 alternate conformations), 11 sulfate ions, 1 citrate ion, 1 triethyleneglycol molecule, 9 molecules of glycerol and 396 waters. The model refined to R/R-free values of 18.3% and 24.3% respectively with no Ramachandran outliers and excellent geometry throughout (see Table 1). The two molecules are related to one another by a rotation of 180 about an axis parallel to the crystallographic C axis. The two protomers differ only slightly when superimposed with an RMSD of 1.23 Å for 95% of common atoms. Structural analysis was carried out using CCP473 and the Uppsala Software Factory suites74. Structure figures for EttA were produced in PYMOL. Coordinates for E. coli EttA are deposited in the Protein Data Bank under accession code 4FIN.
Structural Superposition for Fig. 2c
Alignments were based on least-squares superposition of the ABCβ and F1-like core subdomains in one ABC domain of EttA with one corresponding regions of one protomer in the MJ0706 ATP-sandwich dimer5. ATP molecules from the MJ0796 dimer are shown in gray space-filling representation.
In vivo assays of DNA, RNA, and protein synthesis
MG1655 cells harboring pBAD-ettA or pBAD-ettA-EQ2 plasmids were grown at 37 °C in M9 glycerol minimal medium with 0.1 mg/ml of amino acid (minus Met and Cys) to OD600 ~0.2 prior to induction of EttA expression using 0.2% L-arabinose (at zero time on these graphs). Control cultures were done the same way without L-arabinose. [35S]methionine incorporation was carried out using the protocol of Hirashima and Inouye75. At each time point 1 ml of each culture was briefly incubated with 6 µl of [35S]methionine for 1 min. The reaction was stopped by adding 300 µl of cold methionine (0.1mg/ml). A 50 µl volume of this sample was applied on a Whatman 3MM filter. The filters were immediately washed with a solution of 10% trichloroacetic acid (TCA) and 0.5 µg/ml of methionine, boiled for 30 min and then washed 3 times with fresh cold TCA. Finally, the filters were rinsed with acetone and dried before radioactivity was determined with a scintillation counter. The incorporation of [methyl-3H]thymidine and [methyl-3H]uracil incorporation were carried out using the protocol of Christensen-Dalsgaard and Gerdes76. After induction (time=0), 1 ml of each culture was incubated at 37° C with 50 µl of [methyl-3H]thymidine, or [methyl-3H]uracil. At each time point, 50 µl were put on Whatman 3MM filter. The filters were immediately washed with a solution of 10% TCA and 0.5 µg/ml of dTTP or UTP, and then washed 3 times with fresh cold 10% TCA. Finally, the filters were rinsed with 95% ethanol and dried before radioactivity was determined with a scintillation counter.
Minimum purified in vitro translation assay with eTLC detection
All the components and proteins were prepared and purified exactly as described in the method of Fei et al.37. The [35S]fMet-tRNAfMet was prepared with the same protocol, but with the methionine replaced by 3 µM of [35S]Methionine (Perkin Elmer) and quenched 5 min after the beginning of the reaction with 16 µM of cold methionine. Estimation of aminoacylation and formylation yields was assessed by hydrophobic interaction chromatography37. The Glu-tRNAGlu was prepared as the other aa-tRNA37. The Glu-tRNA-synthethase was prepared as described by Shimizu et al.77. All the minimum purified in vitro translation assays were done in Polymix Buffer (50 mM Tris-OAc, pH 6.9, 100 mM KCL, 5 mM NH4OAc, 0.5 mM Ca(OAc)2, 0.1 mM EDTA, 1 mM spermidine, 5 mM putrescine, 3.5 mM Mg(OAc)2, 6 mM 2-mercaptoethanol) with 0.3 µM [35S]fMet-tRNAfMet using the pT7gp32.1-20 mRNA template (described in Supplementary Information).
The experiments presented in Figure 4 and Supplementary Figure 7 were performed using the standard procedure37 which includes a GTP-regenerating system. Because it was not possible to use this protocol for experiments conducted using WT-EttA at different concentrations of ATP and ADP, the GTP-regenerating system was omitted, and the GTP concentration was adjusted to a final concentration of 0.3 mM. For all the minimum purified in vitro translation assays, the reaction products were analyzed on eTLC after hydrolysis of the product with 0.2 M of KOH and separation of the products by eTLC using the method described by Youngman, et al.39 A 0.5 µl volume of each sample was spotted onto TLC-cellulose (EMD Chemicals) plates, dried, and separated by electrophoresis in pyridine acetate buffer, pH 2.8 (20% glacial acetic acid and 0.06% f pyridine) at 1200 V for 20 min.
Standard in vitro minimum purified translation assays using the GTP-regenerating system were performed at 37 °C with an mRNA pT7gp32.1-20 at 1.7 µM, [35S]fMet-tRNAfMet (0.3 µM), 70S ribosome (0.45 µM), the initiation factors (~ 0.5 µM each), the corresponding aa-tRNA (0.7 µM) and the elongation factors (EF-Tu 2, EF-Ts 1, EF-G 1.5 µM). The reactions were performed in the presence or absence of WT-EttA or EttA-EQ2 (2.5 µM) with ATP (0.5 mM) at different steps of the reaction. The reaction was assembled in sequential order: First, the 70S IC was assembled by incubation of the 70S ribosome and initiation factors (Ifs) 1, 2 and 3 in Polymix Buffer with GTP for 10 min at 37°C. Second, the mRNA was added and the reaction was incubated for another 10 min at 37°C. Third, [35S]fMet-tRNAfMet was added and another 10 min of incubation at 37°C took place. Finally, the 70S IC was kept on ice for at least 10 min before being used for the elongation reactions. The reactions were assembled as described in the Figure 3 legend. EF-G and ternary complex were prepared with the GTP-regenerating buffer as described by Fei et al.37.
Standard in vitro minimum purified translation assays without the GTP-regenerating system were run using an equivalent protocol but with the following changes: After the formation of the 70S IC the reaction was buffer exchanged in Polymix Buffer without GTP using a Zeba spin column (Thermo Scientific). The resulting GTP-free 70S IC was aliquoted and stored at −80°C. The reactions were run using this 70S IC with nearly the same conditions as before. However, the 70S ribosome was adjusted at 0.6 µM and GTP at 0.3 mM. All the reactions were done at room temperature to slow down the process. The reactions were run in the presence or absence of combinations of ATP (1.2 mM) and ADP (0.6 mM). After adding the 70S IC, either buffer (control) or WT-EttA, (EttA) (3.5 µM) was added in parallel with the elongation factors Phe-tRNAPhe and Lys-tRNALys.
In vivo fitness assays
MG1655 and the corresponding ΔettA stain were grown overnight separately in LB at 37°C and 250 rpm. Ampicillin was added at 100 µg/ml for complementation experiments, which were conduced with cells harboring either the pBAD, pBAD-ettA, pBAD-His6-ettA, or pBAD-ettA-Δarm plasmid. The overnight cultures were mixed together in a ratio of 1:1 based on OD600 and diluted 100-fold into fresh LB. At the indicated times (24, 72, or 144 hours), these cultures were diluted 1000-fold into fresh medium. This serial regrowth procedure was repeated for the number of times indicated in the figure. For the 144-hour experiment, the growth was continued after the second re-start for only 24 hours. All of the regrowth experiments were performed in triplicate with independent inocula. For PCR analysis, a 100 µl aliquot of each culture was centrifuged at 6000 rpm for 5 min, and washed in 1 ml of Phosphate Buffered Saline (PBS) buffer, and the resulting pellets were stored at -20 °C. The pellets were resuspended in 200 µl of milliQ water, and 0.5 µl of the resulting solution was added to 30 µl of Gotaq PCR reaction mix (Promega) with primers designed to hybridize 400 pb upstream and downstream of the ettA gene. After 20 cycles of PCR amplification (95 °C for 30 sec, 55 °C for 30 sec and 72 °C for 2 min), the products were separated on a 1% TBE agarose gel that was stained with ethidium bromide. The gel was imaged on a UV transilluminator using a camera configured to avoid saturation. The PCR assay was calibrated by analyzing immediately after mixing samples containing varying ratios of WT and ΔettA cells. The calibration procedure demonstrated that that the assay detects ΔettA cells with somewhat higher sensitivity, meaning that it provides a conservative estimate of the degree of depletion of the ΔettA cells. The most important results from this PCR-based assays of co-culture content were verified by plating on LB-agar cells from one 8×24 hour restart experiment and using colony PCR to determine the genotype of 10 of the resulting single colonies. This assay showed that that nine colonies contained WT cells while only one contained delta-EttA cells.
Analytical gel-filtration and static light-scattering analyses
Protein samples were injected onto a Shodex 804 column (Showa Denko, Tokyo, Japan) running at 4 °C in 150 mM NaCl, 5% (v/v) glycerol, 20 mM Tris-Cl, pH 7.2. The column effluent was monitored using static-light scattering (Dawn) and refractive index (Optilab) detectors from Wyatt Technologies.
Immunochemistry
Polyclonal rabbit antiserum was generated by Invitrogen’s EvoQuest division using purified EttA as an antigen. After protein separation on a 10 % SDS-PAGE gel, electrotransfer onto nitrocellulose, and blocking with 5% blotting grade non-fat dry milk (Bio-Rad), immunoblots were incubated with a 1:20,000 dilution of antiserum from a terminal bleed and developed with the ECL system (GE Biosciences) using horseradish peroxidase-conjugated goat anti-rabbit secondary antibodies (NA9340V, GE Biosciences). Pre-immune serum did not show any immunoreactivity at the molecular weight of EttA. The specificity of the EttA antiserum was verified using ettA::tn5 knockout which did not show any immunoreactivity at the molecular weight of EttA in blots of whole cells cultured in LB medium (Supplementary Figure 5c); the isogenic control strain MG1655 showed EttA immunoreactivity similar to that observed from strain DH5α. The antibody was also affinity purified against EttA using the affinity purification of polyclonal antisera described by Levin78. The western-blot presented in Supplementary Figure 5c was incubated with a 1:2,000 dilution of affinity purified anti-EttA antibody, developed with a donkey anti-rabbit secondary antibodies conjugate to IRDye 680 (926-32223, Li-cor) and scanned on an Odyssey CLx scanner (Li-cor).
Polysome analyses
Polysomes were isolated from WT strain MG1655 using the freeze-thaw-lysozyme lysis method of Ron et al.79 with 0.1 mg/ml chloramphenicol added to the growth medium 10 min prior to harvesting and to the cell lysis buffers. They were separated on 10% - 40% (w/v) sucrose gradients in a buffer containing 10 mM Mg-OAc, 20 mM Tris-OAc, pH 7.6, NH4OAc 100 mM. The gradients were spun in a SW40Ti rotor at 40,000 rpm for 2 hours prior to manual fractionation using a Brandel Model 184 fractionator. Fractions were analyzed using SDS-PAGE followed by immunoblotting with anti-EttA antiserum. In the control experiment, 0.2 mg/ml RNase A (Sigma-Aldrich) was added to the polysome preparation prior to loading on the gradient. Polysomes were isolated from the strains over-expressing EttA or EttA-EQ2 using the same protocol, but the starting strains were MG1655 ΔettA cells harboring pBAD-ettA or pBAD-ettA-EQ2 plasmids. After reaching an OD600 of 0.6, cells were induced with 0.1% (w/v) L-arabinose for 10, 20, 30, or 40 min. The sucrose density-gradient profile in figure 3c, which shows complete depletion of polysomes in the pBAD-ettA-EQ2 cells at the 30 min time point, is representative of the results of three independent replicate experiments.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by US National Science Foundation grants to J.F.H. (0424043) and R.L.G. (MCB CAREER 0644262), a Burroughs Wellcome Fund award to R.L.G. (CABS 1004856), a Canadian Institutes of Health Research grant to H.-J.W. (MOP 114938), a grant from the US National Institutes of Health (NIH) Protein Structure Initiative to the Northeast Structural Genomics Consortium (GM074958), and NIH grants to R.L.G. (GM084288) and to J.F. (GM29169 and GM55440). M.T.E. was supported by the NIH Training Program in Molecular Biophysics at Columbia University (T32 GM008281). J.F. is an Investigator supported by the Howard Hughes Medical Institute. The authors thank J. Hurley and N. Woychik of the University of Medicine and Dentistry of the State of New Jersey for assistance with in vivo radiolabelling, A. Tzagoloff for sharing equipment, and the members of the Hunt and Gonzalez laboratories for advice and technical assistance.
Footnotes
Accession codes. Coordinates of the X-ray structure of EttA have been deposited in RCSB Protein Data Bank (PDB) under accession number 4FIN.
AUTHOR CONTRIBUTIONS
P.C.M.S. determined the crystal structure and performed the polysome analysis on WT-EttA, J.J.F. performed the ATPase measurements. G.B., with assistance from A.J.T., performed the other biochemical and genetic studies. W.N. performed the smFRET experiments. M.T.E. provided training and reagents for in vitro translation assays and eTLC analysis of in vitro translation products. G.B., P.C.M.S, H.-J.W., R.L.G., and J.F.H. designed the experiments. G.B., P.C.M.S., B.C., J.F., R.L.G., and J.F.H. conceived the research program and wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Davidson AL, Dassa E, Orelle C, Chen J. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol Mol Biol Rev. 2008;72:317–364. doi: 10.1128/MMBR.00031-07. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cavanaugh LF, Palmer AG, 3rd, Gierasch LM, Hunt JF. Disorder breathes life into a DEAD motor. Nat Struct Mol Biol. 2006;13:566–569. doi: 10.1038/nsmb0706-566. [DOI] [PubMed] [Google Scholar]
- 3.Jones PM, George AM. Subunit interactions in ABC transporters: towards a functional architecture. FEMS Microbiol Lett. 1999;179:187–202. doi: 10.1111/j.1574-6968.1999.tb08727.x. [DOI] [PubMed] [Google Scholar]
- 4.Hopfner KP, et al. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell. 2000;101:789–800. doi: 10.1016/s0092-8674(00)80890-9. [DOI] [PubMed] [Google Scholar]
- 5.Smith PC, et al. ATP binding to the motor domain from an ABC transporter drives formation of a nucleotide sandwich dimer. Mol Cell. 2002;10:139–149. doi: 10.1016/s1097-2765(02)00576-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holland IB, Blight MA. ABC-ATPases, adaptable energy generators fuelling transmembrane movement of a variety of molecules in organisms from bacteria to humans. J Mol Biol. 1999;293:381–399. doi: 10.1006/jmbi.1999.2993. [DOI] [PubMed] [Google Scholar]
- 7.Jaciuk M, Nowak E, Skowronek K, Tanska A, Nowotny M. Structure of UvrA nucleotide excision repair protein in complex with modified DNA. Nat Struct Mol Biol. 2011;18:191–197. doi: 10.1038/nsmb.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lammens K, et al. The Mre11:Rad50 structure shows an ATP-dependent molecular clamp in DNA double-strand break repair. Cell. 2011;145:54–66. doi: 10.1016/j.cell.2011.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Skogerson L, Wakatama E. A ribosome-dependent GTPase from yeast distinct from elongation factor 2. Proc Natl Acad Sci U S A. 1976;73:73–76. doi: 10.1073/pnas.73.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khoshnevis S, et al. The iron-sulphur protein RNase L inhibitor functions in translation termination. EMBO Rep. 2010;11:214–219. doi: 10.1038/embor.2009.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pisarev AV, et al. The role of ABCE1 in eukaryotic posttermination ribosomal recycling. Mol Cell. 2010;37:196–210. doi: 10.1016/j.molcel.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barthelme D, et al. Ribosome recycling depends on a mechanistic link between the FeS cluster domain and a conformational switch of the twin-ATPase ABCE1. Proc Natl Acad Sci U S A. 2011;108:3228–3233. doi: 10.1073/pnas.1015953108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Becker T, et al. Structural basis of highly conserved ribosome recycling in eukaryotes and archaea. Nature. 2012;482:501–506. doi: 10.1038/nature10829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kamath A, Chakraburtty K. Role of yeast elongation factor 3 in the elongation cycle. J Biol Chem. 1989;264:15423–15428. [PubMed] [Google Scholar]
- 15.Andersen CB, et al. Structure of eEF3 and the mechanism of transfer RNA release from the E-site. Nature. 2006;443:663–668. doi: 10.1038/nature05126. [DOI] [PubMed] [Google Scholar]
- 16.Kurata S, et al. Ribosome recycling step in yeast cytoplasmic protein synthesis is catalyzed by eEF3 and ATP. Proc Natl Acad Sci U S A. 2010;107:10854–10859. doi: 10.1073/pnas.1006247107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tyzack JK, Wang X, Belsham GJ, Proud CG. ABC50 interacts with eukaryotic initiation factor 2 and associates with the ribosome in an ATP-dependent manner. J Biol Chem. 2000;275:34131–34139. doi: 10.1074/jbc.M002868200. [DOI] [PubMed] [Google Scholar]
- 18.Paytubi S, et al. ABC50 promotes translation initiation in mammalian cells. J Biol Chem. 2009;284:24061–24073. doi: 10.1074/jbc.M109.031625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kiel MC, Aoki H, Ganoza MC. Identification of a ribosomal ATPase in Escherichia coli cells. Biochimie. 1999;81:1097–1108. doi: 10.1016/s0300-9084(99)00352-1. [DOI] [PubMed] [Google Scholar]
- 20.Babu M, et al. Ribosome-dependent ATPase interacts with conserved membrane protein in Escherichia coli to modulate protein synthesis and oxidative phosphorylation. PLoS One. 2011;6:e18510. doi: 10.1371/journal.pone.0018510. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Kerr ID. Sequence analysis of twin ATP binding cassette proteins involved in translational control, antibiotic resistance, and ribonuclease L inhibition. Biochem Biophys Res Commun. 2004;315:166–173. doi: 10.1016/j.bbrc.2004.01.044. [DOI] [PubMed] [Google Scholar]
- 22.Punta M, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vazquez de Aldana CR, Marton MJ, Hinnebusch AG. GCN20, a novel ATP binding cassette protein, and GCN1 reside in a complex that mediates activation of the eIF-2 alpha kinase GCN2 in amino acid-starved cells. EMBO J. 1995;14:3184–3199. doi: 10.1002/j.1460-2075.1995.tb07321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sattlegger E, Hinnebusch AG. Polyribosome binding by GCN1 is required for full activation of eukaryotic translation initiation factor 2{alpha} kinase GCN2 during amino acid starvation. J Biol Chem. 2005;280:16514–16521. doi: 10.1074/jbc.M414566200. [DOI] [PubMed] [Google Scholar]
- 25.Dong J, Lai R, Jennings JL, Link AJ, Hinnebusch AG. The novel ATP-binding cassette protein ARB1 is a shuttling factor that stimulates 40S and 60S ribosome biogenesis. Mol Cell Biol. 2005;25:9859–9873. doi: 10.1128/MCB.25.22.9859-9873.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hopkins JD, Clements M, Syvanen M. New class of mutations in Escherichia coli (uup) that affect precise excision of insertion elements and bacteriophage Mu growth. J Bacteriol. 1983;153:384–389. doi: 10.1128/jb.153.1.384-389.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murat D, Bance P, Callebaut I, Dassa E. ATP hydrolysis is essential for the function of the Uup ATP-binding cassette ATPase in precise excision of transposons. J Biol Chem. 2006;281:6850–6859. doi: 10.1074/jbc.M509926200. [DOI] [PubMed] [Google Scholar]
- 28.Murat D, Goncalves L, Dassa E. Deletion of the Escherichia coli uup gene encoding a protein of the ATP binding cassette superfamily affects bacterial competitiveness. Res Microbiol. 2008;159:671–677. doi: 10.1016/j.resmic.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 29.Lu P, Vogel C, Wang R, Yao X, Marcotte EM. Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat Biotechnol. 2007;25:117–124. doi: 10.1038/nbt1270. [DOI] [PubMed] [Google Scholar]
- 30.Chen B, et al. EttA regulates translation by binding to the ribosomal E site and restricting ribosome-tRNA dynamics. doi: 10.1038/nsmb.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zaitseva J, Jenewein S, Jumpertz T, Holland IB, Schmitt L. H662 is the linchpin of ATP hydrolysis in the nucleotide-binding domain of the ABC transporter HlyB. EMBO J. 2005;24:1901–1910. doi: 10.1038/sj.emboj.7600657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karcher A, Schele A, Hopfner KP. X-ray structure of the complete ABC enzyme ABCE1 from Pyrococcus abyssi. J Biol Chem. 2008;283:7962–7971. doi: 10.1074/jbc.M707347200. [DOI] [PubMed] [Google Scholar]
- 33.Oldham ML, Chen J. Crystal structure of the maltose transporter in a pretranslocation intermediate state. Science. 2011;332:1202–1205. doi: 10.1126/science.1200767. [DOI] [PubMed] [Google Scholar]
- 34.Diederichs K, et al. Crystal structure of MalK, the ATPase subunit of the trehalose/maltose ABC transporter of the archaeon Thermococcus litoralis. EMBO J. 2000;19:5951–5961. doi: 10.1093/emboj/19.22.5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karpowich N, et al. Crystal structures of the MJ1267 ATP binding cassette reveal an induced-fit effect at the ATPase active site of an ABC transporter. Structure. 2001;9:571–586. doi: 10.1016/s0969-2126(01)00617-7. [DOI] [PubMed] [Google Scholar]
- 36.Vergani P, Lockless SW, Nairn AC, Gadsby DC. CFTR channel opening by ATP-driven tight dimerization of its nucleotide-binding domains. Nature. 2005;433:876–880. doi: 10.1038/nature03313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fei J, et al. A highly purified, fluorescently labeled in vitro translation system for single-molecule studies of protein synthesis. Methods Enzymol. 2010;472:221–259. doi: 10.1016/S0076-6879(10)72008-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yusupova GZ, Yusupov MM, Cate JH, Noller HF. The path of messenger RNA through the ribosome. Cell. 2001;106:233–241. doi: 10.1016/s0092-8674(01)00435-4. [DOI] [PubMed] [Google Scholar]
- 39.Youngman EM, Brunelle JL, Kochaniak AB, Green R. The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release. Cell. 2004;117:589–599. doi: 10.1016/s0092-8674(04)00411-8. [DOI] [PubMed] [Google Scholar]
- 40.Fei J, Kosuri P, MacDougall DD, Gonzalez RL., Jr Coupling of ribosomal L1 stalk and tRNA dynamics during translation elongation. Mol Cell. 2008;30:348–359. doi: 10.1016/j.molcel.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 41.Agrawal RK, Heagle AB, Penczek P, Grassucci RA, Frank J. EF-G-dependent GTP hydrolysis induces translocation accompanied by large conformational changes in the 70S ribosome. Nat Struct Biol. 1999;6:643–647. doi: 10.1038/10695. [DOI] [PubMed] [Google Scholar]
- 42.Aleksandrov AA, Cui L, Riordan JR. Relationship between nucleotide binding and ion channel gating in cystic fibrosis transmembrane conductance regulator. J Physiol. 2009;587:2875–2886. doi: 10.1113/jphysiol.2009.170258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramakrishnan V. Ribosome structure and the mechanism of translation. Cell. 2002;108:557–572. doi: 10.1016/s0092-8674(02)00619-0. [DOI] [PubMed] [Google Scholar]
- 44.Buckstein MH, He J, Rubin H. Characterization of nucleotide pools as a function of physiological state in Escherichia coli. J Bacteriol. 2008;190:718–726. doi: 10.1128/JB.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Glembotski CC, Chapman AG, Atkinson DE. Adenylate energy charge in Escherichia coli CR341T28 and properties of heat-sensitive adenylate kinase. J Bacteriol. 1981;145:1374–1385. doi: 10.1128/jb.145.3.1374-1385.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu Q, Inouye M. Adenylate kinase complements nucleoside diphosphate kinase deficiency in nucleotide metabolism. Proc Natl Acad Sci U S A. 1996;93:5720–5725. doi: 10.1073/pnas.93.12.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bernard MA, Ray NB, Olcott MC, Hendricks SP, Mathews CK. Metabolic functions of microbial nucleoside diphosphate kinases. J Bioenerg Biomembr. 2000;32:259–267. doi: 10.1023/a:1005537013120. [DOI] [PubMed] [Google Scholar]
- 48.Walton GM, Gill GN. Nucleotide regulation of protein synthesis. Methods Enzymol. 1979;60:578–590. doi: 10.1016/s0076-6879(79)60055-1. [DOI] [PubMed] [Google Scholar]
- 49.Fei J, Richard AC, Bronson JE, Gonzalez RL., Jr Transfer RNA-mediated regulation of ribosome dynamics during protein synthesis. Nat Struct Mol Biol. 2011;18:1043–1051. doi: 10.1038/nsmb.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tran QH, Unden G. Changes in the proton potential and the cellular energetics of Escherichia coli during growth by aerobic and anaerobic respiration or by fermentation. Eur J Biochem. 1998;251:538–543. doi: 10.1046/j.1432-1327.1998.2510538.x. [DOI] [PubMed] [Google Scholar]
- 51.Swedes JS, Sedo RJ, Atkinson DE. Relation of growth and protein synthesis to the adenylate energy charge in an adenine-requiring mutant of Escherichia coli. J Biol Chem. 1975;250:6930–6938. [PubMed] [Google Scholar]
- 52.Jewett MC, Miller ML, Chen Y, Swartz JR. Continued protein synthesis at low [ATP] and [GTP] enables cell adaptation during energy limitation. J Bacteriol. 2009;191:1083–1091. doi: 10.1128/JB.00852-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chapman AG, Fall L, Atkinson DE. Adenylate energy charge in Escherichia coli during growth and starvation. J Bacteriol. 1971;108:1072–1086. doi: 10.1128/jb.108.3.1072-1086.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gaal T, Bartlett MS, Ross W, Turnbough CL, Gourse RL. Transcription Regulation by Initiating NTP Concentration: rRNA Synthesis in Bacteria. Science. 1997;278:2092–2097. doi: 10.1126/science.278.5346.2092. [DOI] [PubMed] [Google Scholar]
- 55.Walton GM, Gill GN. Regulation of ternary (Met-tRNAf - GTP - eukaryotic initiation factor 2) protein synthesis initiation complex formation by the adenylate energy charge. Biochim Biophys Acta. 1976;418:195–203. doi: 10.1016/0005-2787(76)90069-1. [DOI] [PubMed] [Google Scholar]
- 56.Schifano JM, et al. Mycobacterial toxin MazF-mt6 inhibits translation through cleavage of 23S rRNA at the ribosomal A site. Proc Natl Acad Sci U S A. 2013;110:8501–8506. doi: 10.1073/pnas.1222031110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamaguchi Y, Park JH, Inouye M. Toxin-antitoxin systems in bacteria and archaea. Annu Rev Genet. 2011;45:61–79. doi: 10.1146/annurev-genet-110410-132412. [DOI] [PubMed] [Google Scholar]
- 58.Hauser R, et al. RsfA (YbeB) proteins are conserved ribosomal silencing factors. PLoS Genet. 2012;8:e1002815. doi: 10.1371/journal.pgen.1002815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Polikanov YS, Blaha GM, Steitz TA. How hibernation factors RMF, HPF, and YfiA turn off protein synthesis. Science. 2012;336:915–918. doi: 10.1126/science.1218538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamagishi M, et al. Regulation of the Escherichia coli rmf gene encoding the ribosome modulation factor: growth phase- and growth rate-dependent control. EMBO J. 1993;12:625–630. doi: 10.1002/j.1460-2075.1993.tb05695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Berlyn MB, Letovsky S. Genome-related datasets within the E. coli Genetic Stock Center database. Nucleic Acids Res. 1992;20:6143–6151. doi: 10.1093/nar/20.23.6143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Miller JH, editor. Short course in bacterial genetics a laboratory manual and handbook for Escherichia coli and related bacteria. Plainview, N.Y.: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 63.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baba T, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100050. 2006 0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Neidhardt FC, Curtiss R. Escherichia coli and Salmonella : cellular and molecular biology. Washington, D.C.: ASM Press; 1996. [Google Scholar]
- 66.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 67.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Terwilliger TC. Maximum-likelihood density modification using pattern recognition of structural motifs. Acta Crystallogr. D Biol. Crystallogr. 2001;57:1755–1762. doi: 10.1107/S0907444901013737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jones TA, Zou J-Y, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 70.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D. Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 71.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 74.Kleywegt GJ. Quality control and validation. Methods Mol Biol. 2007;364:255–272. doi: 10.1385/1-59745-266-1:255. [DOI] [PubMed] [Google Scholar]
- 75.Hirashima A, Inouye M. Specific biosynthesis of an envelope protein of Escherichia coli. Nature. 1973;242:405–407. doi: 10.1038/242405a0. [DOI] [PubMed] [Google Scholar]
- 76.Christensen-Dalsgaard M, Gerdes K. Two higBA loci in the Vibrio cholerae superintegron encode mRNA cleaving enzymes and can stabilize plasmids. Mol Microbiol. 2006;62:397–411. doi: 10.1111/j.1365-2958.2006.05385.x. [DOI] [PubMed] [Google Scholar]
- 77.Shimizu Y, et al. Cell-free translation reconstituted with purified components. Nat Biotechnol. 2001;19:751–755. doi: 10.1038/90802. [DOI] [PubMed] [Google Scholar]
- 78.Levin PA. 6 Light microscopy techniques for bacterial cell biology. In: Philippe Sansonetti AZ, editor. Methods in Microbiology. Vol. Volume 31. Academic Press; 2002. pp. 115–132. [Google Scholar]
- 79.Ron EZ, Kohler RE, Davis BD. Polysomes extracted from Escherichia coli by freeze-thaw-lysozyme lysis. Science. 1966;153:1119–1120. doi: 10.1126/science.153.3740.1119. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.