Abstract
Sirt1 is the most evolutionarily conserved mammalian sirtuin. It plays a vital role in the regulation of metabolism, stress responses, genome stability, and ultimately aging. Although much attention has focused on the identification of the cellular targets and functional networks controlled by Sirt1, the mechanisms that regulate Sirt1 activity by biological stimuli have only recently begun to emerge. As an enzyme, the activity of Sirt1 can be controlled by the availability of its substrates, post-translational modifications, interactions with other proteins, or changes in its expression levels. In this review, we briefly review the ways and means by which the activity of Sirt1 is fine-tuned under different conditions.
Keywords: Sirt1, energy metabolism, aging, nutrients, metabolic sensor
Sirt1 at the forefront of sirtuin research
Since the discovery of the first silent mating-type information regulation 2 (Sir2), or sirtuin, in yeast a mere quarter century ago, there has been a stampede of research to elucidate the biological functions of the entire sirtuin family. Sirt1 is the most evolutionarily conserved sirtuin among the seven mammalian homologs, and it has been shown to play crucial roles in mammalian health and disease and is often associated with the most complex physiological processes, including metabolism, cancer, and aging. The wide- and far-reaching effects of Sirt1 are a natural consequence of its capacity to regulate the activity of key cellular players through its enzymatic activity. Therefore, not surprisingly, the activity of Sirt1 itself can be adjusted in response to biological stimuli. Here, we briefly review the ways and means by which the activity of Sirt1 is fine-tuned. We first summarize the mechanisms that regulate Sirt1 activity without altering its expression and then describe the mechanisms that affect Sirt1 activity by altering its expression.
Regulation of Sirt1 activity without altering its expression levels
Regulation through NAD+
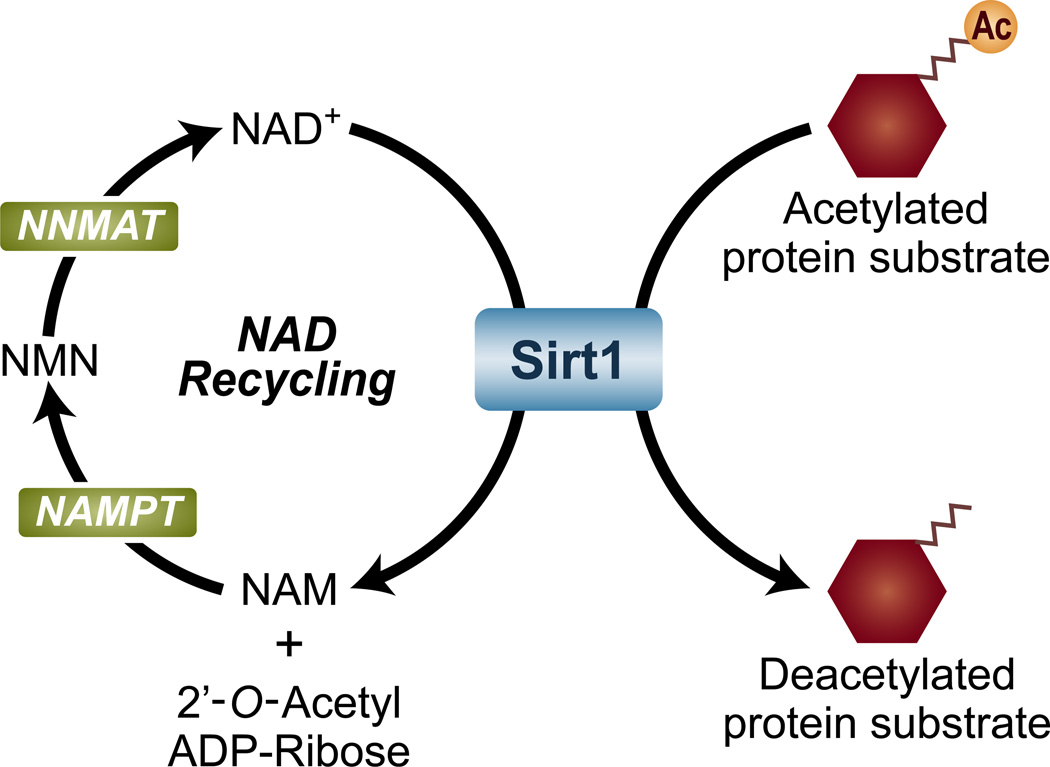
Like all enzymes, the catalytic activity of Sirt1 is dependent on its substrates: acetylated proteins and NAD+. The Sirt1 enzymatic reaction catalyzes the deacetylation of protein targets by hydrolyzing NAD+ and transferring the lysine-bound acetyl group to the 2′-OH position of ADP-ribose, thereby generating a protein with a deacetylated lysine residue, 2′-O-Acetyl-ADPR, and nicotinamide (Figure 1) [1, 2]. Although NAD+ has countless functions, it is best known for its role in redox reactions, where it readily accepts electrons to generate NADH, which then gives off electrons to re-form NAD+ [reviewed in [3]]. Because of this unique property, NAD+ is extensively used in energy harvesting reactions inside cells, most notably the electron transport chain. As a result, the cellular energy status and the levels of NAD+ are inherently coupled.
Figure 1. The enzymatic reaction catalyzed by Sirt1.
Sirt1 catalyzes the deacetylation of several proteins by consuming nicotinamide adenine dinucleotide (NAD+), generating nicotinamide (NAM) and 2′-O-Acetyl-ADP-Ribose. NAM is recycled back into NAD+ by the enzymes nicotinamide phosphorybosyltransferase (NAMPT), nicotinamide mononucleotide adenylyltransferase (NMNAT), and the nicotinamide mononucleotide (NMN) intermediate.
The discovery that Sirt1 exclusively utilizes NAD+ as a co-substrate raised the possibility that its activity is also intrinsically linked to the energy status of the cell. In this scenario, a low-energy status would increase Sirt1 activity by increasing the levels of NAD+, whereas a high-energy status would decrease Sirt1 activity by decreasing the levels of NAD+ [2]. An ample body of research has largely found this to be the case. For instance, increased NAD+ levels and Sirt1 activity have been observed during low-energy conditions, such as fasting, caloric restriction, and exercise, in a variety of mammalian tissues [4–10]. By contrast, energy-rich, high- fat diet feeding has been found to reduce the NAD+ levels, thereby decreasing the activity of Sirt1 [11–14].
In addition to fluctuations in NAD+ availability in response to the energy status, changes in the biosynthesis of NAD+ can also alter NAD+ levels and Sirt1 activity. Mammalian cells have the capacity to synthesize NAD+ from tryptophan (de novo pathway), nicotinic acid (Preiss-Halder pathway), or nicotinamide (salvage pathway), the latter representing the main source of cellular NAD+ [reviewed in [15]]. Not surprisingly, enzymes and intermediate compounds involved in the NAD+ biosynthesis have been shown to alter Sirt1 activity in an NAD+-dependent manner. For instance, increasing the activity of the enzyme nicotinamide phosphoribosyltransferase (Nampt), which performs the rate-limiting step in the salvage pathway, concomitantly enhances the levels of NAD+ and activity of Sirt1 [reviewed in [16]]; conversely, decreasing the activity of Nampt reduces NAD+ levels and Sirt1 activity [reviewed in [16]]. Likewise, the treatment of cells or animals with nicotinamide mononucleotide, the product of Nampt and an NAD+ precursor, elevates NAD+ levels and Sirt1 activity [reviewed in [16]].
Modifying the rate at which NAD+ is synthesized can influence the activity of Sirt1, but so can modifying the rate at which NAD+ is destroyed. Poly ADP-ribose polymerases, or PARPs, are enzymes that create polymers of ADP-ribose at the expense of NAD+ [reviewed in [17]]. When active, these enzymes exhibit an astonishing appetite for NAD+ and can rapidly reduce or even deplete it in the cell [reviewed in [18]]. Not surprisingly, ablating Parp1 or Parp2, two of the most ubiquitous PARPs, elevates NAD+ levels and promotes the activity of Sirt1 [19, 20]. In addition, through its powerful NAD-hydrolyzing activity, the multifunctional enzyme cluster of differentiation 38 (CD38) can also influence cellular NAD levels and module the activity of Sirt1 [21]. Taken together, the levels of NAD+, an essential substrate for Sirt1, have a profound effect on the regulation of Sirt1.
Post-translational modifications
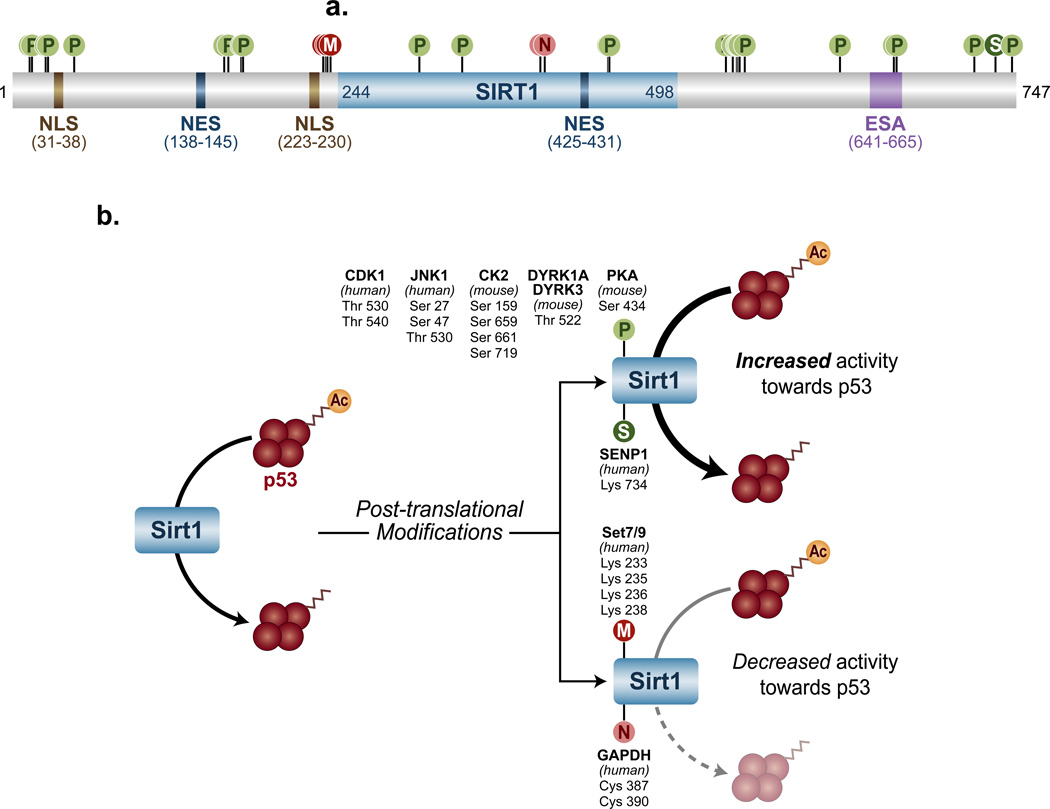
Post-translational modifications (PTMs) are known to impact the activity of a multitude of enzymes, and Sirt1 is no exception. Here, we describe several of them and their influence on Sirt1 activity and function (Figure 2).
Figure 2. Sirt1 post-translational modifications and their effects.
(a) The human protein Sirt1 contains several domains, including NLSs (nuclear localization signals), NESs (nuclear export signals), the ESA (essential for Sirt1 activity), and an enzymatic core (indicated in light blue) from residues 244 through 498. Residue numbers for each domain are shown in parentheses beneath each domain name. In addition, Sirt1 is also subject to a variety of post-translational modifications, including phosphorylation (P), methylation (M), nitrosylation (N), and SUMOylation (S). (b) Phosphorylation and SUMOylation (above, right), can increase the deacetylase activity of Sirt1 towards p53, whereas methylation and nitrosylation (below, right) decrease this activity.
Regulation through phosphorylation(s)
Protein phosphorylation is one the most common biological approaches of protein regulation. It involves the covalent addition of a phosphate group by kinases to serine, threonine, tyrosine, or in rare cases, histidine residues. Early mass spectrometry experiments detected at least 13 serine/threonine phosphorylation sites in the N- and C-terminal domains of Sirt1, and it is likely that many more exist [22]. Studies on the impact of these modifications have only recently begun, but the initial results have already yielded valuable information about how they influence Sirt1 activity and function. JUN N-terminal kinase 1 (JNK1), for instance, is capable of phosphorylating Sirt1 at Ser 27 and 47, and Thr 530, particularly under stressful cellular conditions. Intriguingly, these phosphorylations appear to increase the deacetylase activity of Sirt1 towards one of its substrates, histone H3, but have no effect towards another substrate, p53. This suggests that some or all of these PTMs can alter the activity of Sirt1 in a substrate-specific manner [23]. Physiologically, the JNK1-Sirt1 signaling pathway has recently been found to protect the heart from oxidative stress [24].
In addition to JNK1, other kinases can also alter the activity of Sirt1 with important biological effects. Two pro-survival dual specificity tyrosine phosphorylation-regulated kinases (DYRKs), DYRK1A and DYRK3, have been shown to phosphorylate murine Sirt1 Thr 522 (homologous to human Thr 530) in response to genotoxic stress [25, 26]. This PTM substantially enhances Sirt1 deacetylase activity towards acetylated p53 independently of NAD+, and protects cells from genotoxic stress-induced apoptosis [25]. Intriguingly, this phosphorylation is transient; it peaks within one hour and lasts less than two hours after the initial genotoxic or heat stress. Therefore, it appears that the primary biological role of this PTM is to delay apoptosis, providing cells extra time to repair before committing to suicide.
Casein kinase II (CK2) phosphorylates Ser 154, 649, 651, and 683 of murine Sirt1, and Ser 659 and 661 of human Sirt1 [27–29]. These PTMs increase the substrate affinity and therefore activity of Sirt1 towards acetylated p53 and p65 (a main component of the pro-inflammatory Nuclear Factor Kappa B (NFκB) complex)[28]. Like Sirt1 phosphorylation at Thr 522, these PTMs increase cellular survival after genotoxic treatment [27, 28].
Additionally, the cyclin B/cyclin-dependent kinase 1 (CDK1) complex also phosphorylates Sirt1 Thr 530 and Ser 540 [22]. However, although it is still unclear how these PTMs influence the activity of Sirt1, mutating these residues disrupts cell cycle progression and slows cellular proliferation, indicating that at least some of the downstream targets of Sirt1 are affected [22, 30].
Finally, a serine residue located at the highly conserved core domain of Sirt1, Ser 434, has recently been shown to be a phosphorylation target of the cyclic AMP/Protein kinase A (cAMP/PKA)-signaling pathway [31]. This PTM rapidly enhances the intrinsic deacetylase activity of Sirt1 independently of cellular NAD+ levels, stimulating fatty acid oxidation and energy expenditure in response to several stimuli.
Regulation through methylation
Methylation of lysine or arginine is another PTM that can influence the biological activities of proteins. Sirt1 is methylated at Lys 233, 235, 236, and 238 by the lysine methyltransferase SET domain containing 7 (Setd7 or Set7/9), but the roles of these PTMs are still unclear. In vitro, these PTMs do not alter Sirt1 deacetylase activity per se, but the interaction of Sirt1 and Set7/9 is able to prevent Sirt1 from binding to p53. This raises the possibility that Sirt1 methylation promotes the Sirt1-Set7/9 interaction, thereby reducing the Sirt1-p53 interaction. Thus, methylation could modify Sirt1 activity by changing the affinity of Sirt1 towards its protein substrates [30].
Regulation through SUMOylation
SUMOylation is the reversible covalent addition of the relatively small (~12 kDa) Small Ubiquitin like-Modifier (SUMO) protein to lysine residues. Human Sirt1 is SUMOylated at Lys 734, and Lys 734 point mutants exhibit severely reduced deacetylase activity towards p53, suggesting that SUMOylation potentiates the activity of Sirt1. The deSUMOylating enzyme sentrin-specific protease (SENP) is capable of interacting with SUMOylated Sirt1 and removing this PTM. Consistently, knocking down SENP increases the SUMOylation levels of Sirt1 and cellular resistance to stress-induced apoptosis, a trait often associated with increased Sirt1 activity [32]. However, the lysine 734-sumoylation site is not conserved in mouse Sirt1, raising the possibility that Sirt1 activity is differentially regulated in different species.
Regulation through nitrosylation
Nitrosylation is yet another PTM that affects the activity of Sirt1 [33]. It involves the covalent incorporation of a nitric oxide moiety into thiol groups [reviewed in [34]]. S-nitrosylated glyceraldehyde-3-phosphate dehydrogenase (SNO-GAPDH) appears to be responsible for the nitrosylation of Sirt1 at Cys 387 and 390. These residues are localized in the catalytic core of Sirt1, therefore, it is not surprising that their nitrosylation reduces the deacetylase activity of Sirt1, at least in relation to peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α), a widely reported protein substrate of Sirt1 [33].
Regulation through protein-protein interactions
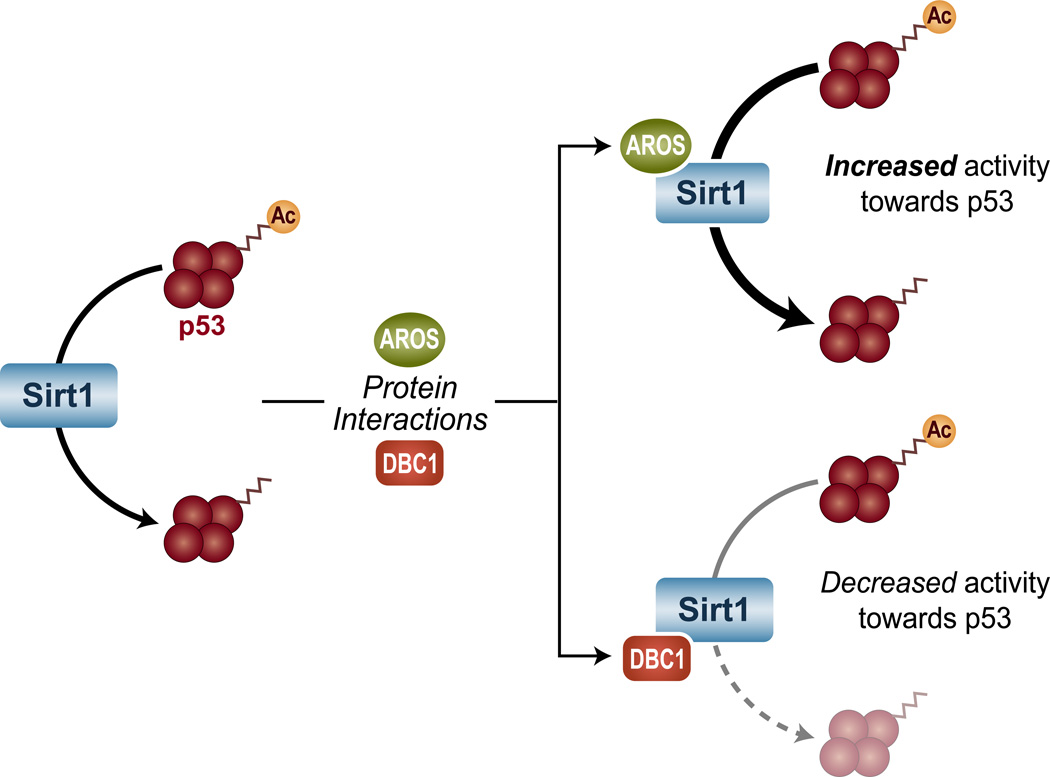
Several transient protein-protein interactions also play key roles in the regulation of Sirt1 (Figure 3). For instance, interaction with active regulator of Sirt1 (AROS) enhances the activity Sirt1 towards acetylated p53. AROS appears to bind to the N terminus of Sirt1, but little is known about how this interaction increases the Sirt1 activity [35].
Figure 3. Protein interactions alter Sirt1 activity.
Sirt1 association with AROS (Active Regulator of Sirt1) increases the deacetylase activity of Sirt1 towards acetylated p53, whereas Sirt1 binding to DBC1 (Deleted in Breast Cancer 1) leads to decreased Sirt1 activity.
In contrast to AROS, deleted in breast cancer 1 (DBC1) is a negative regulator of Sirt1 [36]. The discovery of the ‘essential for Sirt1 activity’ (ESA) domain within Sirt1 protein has been crucial to understanding how DBC1 impairs the activity of Sirt1. The ESA domain is a 25 amino acid region in the C terminus o f Sirt1 that intramolecularly interacts with the Sirt1 deacetylase core to activate its enzymatic activity. DBC1 also interacts with the Sirt1 deacetylase core, potentially displacing ESA, decreasing the accessibility of the core domain to protein substrates, and thereby inactivating Sirt1 [37]. Interestingly, cellular stress results in phosphorylation of DBC1 at Thr 454, which appears to create a second binding site for Sirt1 to augment the DBC1-Sirt1 interaction [38]. In cells, expression of DBC1 results in a marked inhibition of the Sirt1 deacetylase activity, as judged by the acetylation levels of three of its substrates: p53, forkhead box O1 (FOXO1), and FOXO3. Consistently, knocking down DBC1 increases the ability of cells to tolerate genotoxic stress in a Sirt1-dependent manner [36, 38, 39]. Furthermore, deletion of DBC1 in mice leads to increased Sirt1 activity in several tissues, protecting animals from high-fat diet-induced liver steatosis and inflammation [39].
In addition to endogenous cellular factors, foreign proteins can also control the activity of Sirt1. Similar to DBC1, the Tat protein encoded by the human immunodeficiency virus (HIV) binds the Sirt1 catalytic core and impairs its ability to deacetylate p65 [40, 41]. Unlike the interaction between Sirt1 and DBC1, however, the Tat-Sirt1 binding did not decrease the ability of Sirt1 to interact with acetyl-p65, suggesting that allosteric changes rather than substrate access are responsible for the decrease in Sirt1 activity [41].
Regulation by small chemicals
Like the vast majority of enzymes, the activity of Sirt1 can also be modified by small chemicals [reviewed in [42, 43]]. Sirt1 activity is associated with decreased apoptosis, a feat likely accomplished by the deacetylation and inactivation of p53, therefore Sirt1 inhibitors could be useful for the treatment of cancers [reviewed in [42]]. Increased Sirt1 activity, conversely, has been implicated in the prevention and treatment of metabolic disorders as well as several aging-associated diseases [reviewed in [44]]. Therefore, Sirt1 activators are promising compounds for the treatment of a wide variety of conditions, most notably the metabolic syndrome [reviewed in [43]].
The most prominent among Sirt1-activating compounds is the polyphenol resveratrol, a small chemical that was initially proposed as a broad activator of sirtuins nearly a decade ago. But although resveratrol can promote the activity of Sirt1, it is still inconclusive and hotly debated whether its effects are direct or indirect [reviewed in [45, 46]]. Resveratrol also targets many other enzymes and can act as a powerful antioxidant on its own, making its effects difficult to interpret [reviewed in [47]]. Therefore, it is evident that the resveratrol-Sirt1 relationship is more complex than initially proposed and that more research is needed to fully elucidate the mechanism(s) by which resveratrol stimulates Sirt1 activity. Several potential Sirt1-activating artificial compounds have also been developed [reviewed in [43]]. Like resveratrol, however, there is much debate as to whether they act directly on Sirt1 or mediate their biological effects through other means [reviewed in [46]].
Unlike the muddy field of Sirt1 activators, there is more clarity with regards to Sirt1 inhibitors. Among them, sirtinol was the first compound reported to inhibit the activity of sirtuins [48]. Its mechanism of action is still unclear, but sirtinol can bind directly to Sirt1 and inhibit its enzymatic activity Sirt1 with an IC50 of about 40 µM [49]. More recently, however, many sirtinol-related inhibitors with far greater potency have been developed [50]. Splitomicin and its derivatives constitute yet another family of sirtuin inhibitors [51, 52]. In contrast to sirtinol, splitomicin is a poor Sirt1 inhibitor [40, 53], but derivative compounds that have much greater inhibitory potency have since been synthesized [54]. One such inhibitor is HR73, which exhibits an IC50 towards Sirt1 that is lower than 5 µM [40]. Other compounds capable of inhibiting Sirt1 have also been developed in the past few years and include: EX-527 (IC50 ~38 nM towards Sirt1) [49], tenovins (IC50 ~20 µM for tenovin-6 towards Sirt1) [55], and suramin (IC50 ~0.3 µM towards Sirt1) [56]. Nicotinamide, another commonly used sirtuin inhibitor, is a natural precursor of NAD+ and the by-product of the Sirt1 enzymatic reaction. However, with an IC50 of about 85 µM towards Sirt1, and a concentration of less than 1 µM in mammalian serum, nicotinamide is unlikely to actually impair Sirt1 function in vivo [49, 57–59]. Nonetheless, it is possible that the concentration of nicotinamide could be far greater in the immediate vicinity of Sirt1 and, if so, it could at least partially inhibit Sirt1 and generate a negative feedback loop, a common regulatory feature of enzymes. It should be noted that many of these Sirt1 inhibitors not only inhibit other sirtuins but also several unrelated enzymes, making it difficult to sort out their Sirt1-specific effects.
As discussed above, the levels of NAD, a substrate of Sirt1, post-translational modifications of Sirt1, protein-protein interaction with Sirt1, and small chemicals have been found to modulate Sirt1 activity independently of Sirt1 transcriptional changes. The overall activity level of Sirt1, however, is also subject to its own expression levels. Next, we discuss mechanisms that alter Sirt1 expression levels.
Modulation of Sirt1 activity by altering its expression levels
Regulation by transcription factors
Sirt1 activity is not only controlled by the accessibility of its substrates but also by its expression levels (Figure 4). Interestingly, a synergistic system seems to have evolved, whereby the levels of factors that impact the activity of Sirt1 coordinately shift with Sirt1 expression levels. For instance, low-energy conditions that favor increased Sirt1 activity through higher levels of NAD+ also tend to increase Sirt1 expression [4, 6, 60, 61]. As a result, it is often impossible to determine the exact contribution of Sirt1 expression towards its in vivo activity levels. Nonetheless, here we explore several factors and conditions can influence the expression levels and activity of Sirt1.
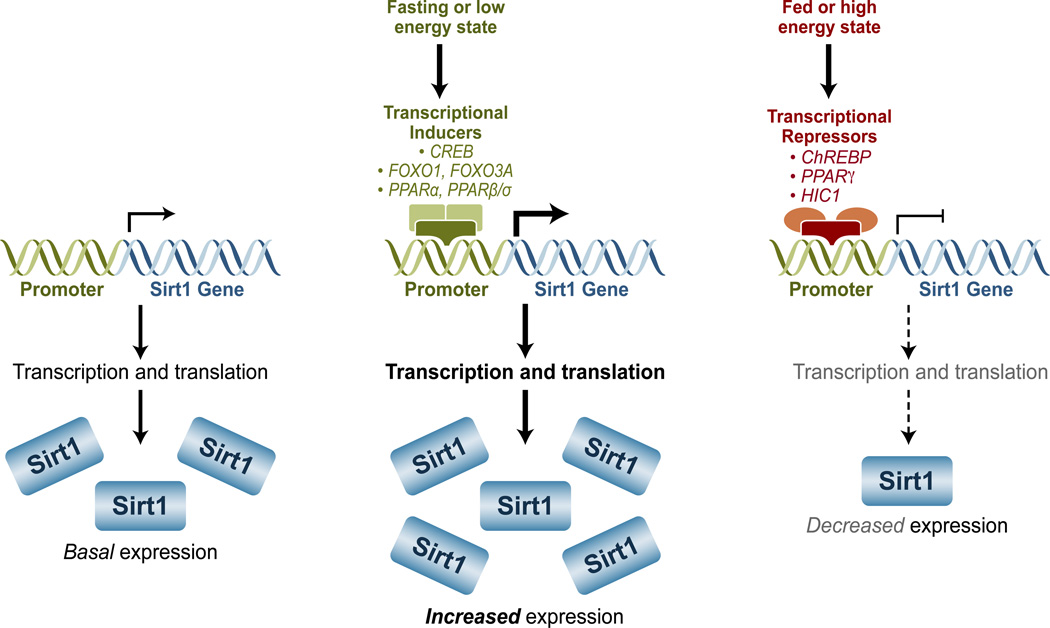
Figure 4. Transcriptional regulation Sirt1.
In a low energy state, several transcription factors, like CREB (cyclic AMP response-element-binding protein), FOXO1 (forkhead box O1), FOXO3A, PPARα (Peroxisome proliferator-activated receptors alpha), and PPARβ/σ, can increase and the expression of Sirt1; in a high energy state, ChREBP (carbohydrate response-element-binding protein), PPARγ, and HIC1 (hypermethylated in cancer protein 1) can repress Sirt1 expression.
A long and growing list of transcription factors can mediate changes in the expression of Sirt1 (Figure 4). In response to fasting, for instance, cyclic AMP response-element-binding protein (CREB), a transcription factor whose activation is mediated by protein kinase A (PKA) in response to low nutrient availability, induces the expression of Sirt1 [60, 62]. Conversely, the carbohydrate response-element-binding protein (ChREBP) responds to re-feeding and represses Sirt1 [60]. Unlike CREB, which acts through several CREB-binding sites distributed throughout the Sirt1 gene, ChREBP works through a single ChREBP-response element located in the Sirt1 promoter [60].
Members of the FOXO family of transcription factors also regulate the expression of Sirt1. FOXO1 induces Sirt1 expression by binding to FOXO1-response elements in the Sirt1 promoter [63, 64]. Interestingly, Sirt1 can also deacetylate FOXO1 and increase its transcriptional activity, suggesting a positive feedback loop between Sirt1 and FOXO1 [64]. FOXO3A, by contrast, translocates into the nucleus under low-energy conditions, where it interacts with p53 at p53 response-elements in the Sirt1 promoter to activate Sirt1 [63]. FOXO3A is also a target of the Sirt1 deacetylase activity, but unlike FOXO1, Sirt1 can either activate or inhibit the transcriptional activity of FOXO3A, depending on environmental stimuli [65, 66].
Peroxisome proliferator-activated receptors (PPARs) are nuclear receptors that function as transcription factors and are also able to regulate the expression levels of Sirt1. PPARα can increase Sirt1 expression levels during a 24-hour fast in mice, presumably through a PPAR-responsive element (PPRE) present in the Sirt1 promoter [6]. Unlike FOXOs, there is no evidence that PPARα is a deacetylation target of Sirt1, but Sirt1 can enhance the activity of PPARα through its co-activators, suggesting a positive feedback loop [67]. PPARβ/σ is yet another transcription factor capable of increasing the expression of Sirt1 [68]. Its actions seem to be mediated by Sp1, another positive regulator of Sirt1 expression [68]. PPARγ, by contrast, represses the Sirt1 gene, presumably by directly interacting with the Sirt1 promoter [69]. In addition, because PPARγ is a deacetylation target of Sirt1, its negative effects on Sirt1 suggest the existence of a negative PPARγ-Sirt1 feedback loop [69].
Finally, the hypermethylated in cancer protein 1 (HIC1) can negatively regulate the expression of Sirt1 [70]. The repression exerted by HIC1 is dependent on CtBP (carboxyl-terminal-binding protein), an NAD+/NADH redox sensor, consistent with low Sirt1 expression observed during abundant energy conditions [71]. Interestingly, like PPARγ, Sirt1 is able to interact with and deacetylate HIC1, thereby reducing its inhibitory actions and suggesting the existence of yet another negative feedback loop [72].
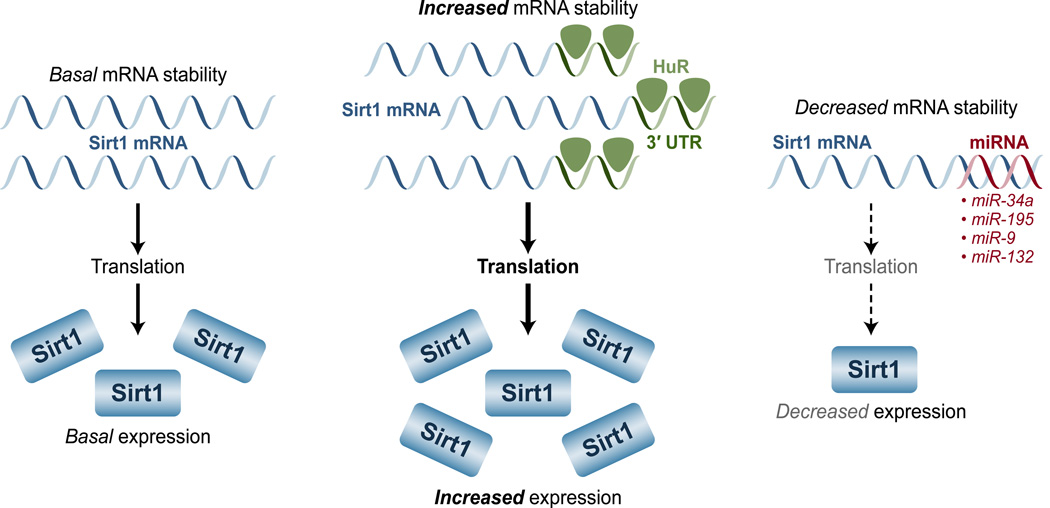
Regulation by RNA stability
The abundance of Sirt1 is not only controlled at the transcriptional level, but also by post-transcriptional events, such as RNA stability. In this regard, the Hu antigen R (HuR) plays a major role in stabilizing the Sirt1 mRNA transcript (Figure 5). In the presence of HuR, Sirt1 mRNA exhibits a half-life more than 8 hours, but in its absence, the Sirt1 mRNA half-life declines to only about 1 hour; such a decline inevitably leads to lower Sirt1 expression and activity [73]. HuR stabilizes the Sirt1 mRNA transcript by binding with high affinity to three HuR RNA-recognition motifs located at the 3’-untranslated region of the Sirt1 mRNA [73]. This stabilizing interaction, however, is regulated by certain physiological conditions. During oxidative stress, for instance, the checkpoint kinase 2 (Chk2) phosphorylates HuR at multiple residues and promotes its dissociation from the Sirt1 mRNA, leading to a reduction in the expression levels and activity of Sirt1 [73].
Figure 5. Factors that alter Sirt1 mRNA stability can alter the levels of Sirt1.
HuR interacts with the 3’-untranslated region of the Sirt1 mRNA, thereby increasing its stability and Sirt1 protein levels; miRNAs such miR-34a, -195, -9, and -132, target Sirt1mRNA and leads to its degradation, which results in decreased levels of Sirt1 expression.
Regulation by miRNA
Sirt1 expression and its activity are also under the influence of several microRNAs (miRNAs) (Figure 5). These short RNA molecules (~22 nucleotides) bind target mRNAs and are capable of suppressing their translation or reducing their stability. Several miRNAs target Sirt1 mRNA and subsequently decrease its expression levels [reviewed in [74]]. miR-34a was the first miRNA that was shown to regulate Sirt1, therefore it is the most studied. Not surprisingly, miR-34a has been found to have wide-ranging effects in cells, tissues, and organs. For instance, it has been implicated in apoptosis and several cancers [reviewed in [74]]. The effects of miR-34a on many cancers are not unexpected, considering that Sirt1 controls the activity of p53 through deacetylation [reviewed in [75]]. Similarly, miR-34a also affects the cardiovascular system by influencing angiogenesis and cellular senescence in a Sirt1-dependent manner [76, 77]. Finally, miR-34a has also been linked to metabolism. In the livers of obese (ob/ob) mice or wild-type fed a high-fat diet, the expression of miR-34a expression increases, while that of Sirt1 decreases, suggesting that miR-34a might control the expression of Sirt1 during these conditions [78]. The farnesoid X receptor (FXR), a major regulator of bile acid metabolism, also appears to influence the expression of Sirt1 through miR-34a [78].
Aside from miR-34a, many other miRNAs have also been found to target Sirt1. In cardiomyocytes, for instance, miR-195 is induced by the fatty acid palmitate to inhibit Sirt1, and miR-199a represses both Sirt1 and hypoxia-inducible factor (HIF)-1alpha under hypoxic conditions. Both of these miRNAs can promote apoptosis in a Sirt1-dependent manner [79, 80]. Yet another miRNA, miR-9, targets Sirt1 in pancreatic beta cells; Sirt1 is a regulator of insulin secretion, therefore miR-9 has the potential to play a major role in glucose metabolism [81, 82]. Finally, miR-132 appears to regulate Sirt1 in adipose tissue and, therefore, might play a role in the metabolic syndrome by influencing secretion of pro-inflammatory chemokines in a Sirt1-dependent manner [83].
Regulation through ubiquitylation
Ubiquitin is an 8.5-kDa protein present in all eukaryotic cells. Although it has many functions, ubiquitin is mainly associated with protein degradation through its covalent attachment to proteins as a PTM called ubiquitylation. Once a protein is mono-ubiquitylated, multiple other ubiquitins are often subsequently added to create a sizeable poly-ubiquitin chain. Ultimately, proteins that are ubiquitylated in this manner are rapidly degraded by the proteasome. Thus, ubiquitylation can be thought of as a cellular tag that signals when to degrade specific proteins.
Like many proteins, the expression of Sirt1 can be controlled by ubiquitylation. Upon phosphorylation at Ser 47 by JNK1, Sirt1 is rapidly ubiquitylated and degraded. Interestingly, this phosphorylation increases the activity of Sirt1 in the short-term, but, by promoting the ubiquitylation and subsequent degradation of Sirt1, decreases the expression of Sirt1 in the long-term [84]. Consistently, in an animal model of obesity, which leads to the persistent activation of JNK1, the expression of Sirt1 is reduced [24]. Ubiquitylation, therefore, provides yet another viable avenue to regulate Sirt1.
As discussed above, the expression of Sirt1 expression is dependent on transcription factors like CREB, ChREBP, FOXO1, FOXO3, several PPARs, and HIC1. In addition, Sirt1 expression can also be modulated by several miRNAs that affect the stability of its mRNA, and by ubiquitylation, which shortens the half-life of the Sirt1 protein.
Concluding remarks
As one of the most conserved mammalian sirtuins, Sirt1 plays essential roles in many biological processes. Nature has evolved a variety of strategies to tightly modulate the activity of Sirt1 in response to different stimuli. First, as an enzyme, the activity of Sirt1 can be directly regulated by substrate availability, PTMs, interacting protein partners, or small molecule activators or re pressors. A second approach involves the alteration of Sirt1 expression levels through transcription factors, RNA binding proteins, miRNAs, or the ubiquitin-proteasome system. Interestingly, both of these approaches seem to act in concert to bring about the desired level of Sirt1 activity. It is clear that Sirt1 is temporally and spatially regulated in response to various environmental cues, and that the dysregulation of this master regulator can lead to a multiple pathological conditions.
Box 1. Outstanding questions.
How do the different Sirt1 post-translational modifications crosstalk to one another?
What is more important for the regulation of Sirt1: the activity of each Sirt1 molecule, or the overall level of Sirt1 expression?
Why are there two systems (activity and expression) that regulate Sirt1?
Under what conditions is each system (activity and expression) more or less important?
What is impact of each Sirt1 regulatory mechanism in terms of health and disease?
Acknowledgements
We thank Drs. John Cidlowski, Aaron Jetten, Brant Hamel, and members of the Li lab for critical reading of the manuscript. The work related to this article was supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences to X.L. (Z01 ES102205).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Tanner KG, et al. Silent information regulator 2 family of NAD- dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(26):14178–14182. doi: 10.1073/pnas.250422697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Imai S, et al. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403(6771):795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 3.Houtkooper RH, et al. The secret life of NAD+: an old metabolite controlling new metabolic signaling pathways. Endocrine reviews. 2010;31(2):194–223. doi: 10.1210/er.2009-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodgers JT, et al. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434(7029):113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 5.Chen D, et al. Tissue-specific regulation of SIRT1 by calorie restriction. Genes & development. 2008;22(13):1753–1757. doi: 10.1101/gad.1650608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hayashida S, et al. Fasting promotes the expression of SIRT1, an NAD+ -dependent protein deacetylase, via activation of PPARalpha in mice. Molecular and cellular biochemistry. 2010;339(1–2):285–292. doi: 10.1007/s11010-010-0391-z. [DOI] [PubMed] [Google Scholar]
- 7.Graham TE, Saltin B. Estimation of the mitochondrial redox state in human skeletal muscle during exercise. Journal of applied physiology. 1989;66(2):561–566. doi: 10.1152/jappl.1989.66.2.561. [DOI] [PubMed] [Google Scholar]
- 8.Chabi B, et al. Relationship between Sirt1 expression and mitochondrial proteins during conditions of chronic muscle use and disuse. Journal of applied physiology. 2009;107(6):1730–1735. doi: 10.1152/japplphysiol.91451.2008. [DOI] [PubMed] [Google Scholar]
- 9.Canto C, et al. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458(7241):1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Canto C, et al. Interdependence of AMPK and SIRT1 for metabolic adaptation to fasting and exercise in skeletal muscle. Cell metabolism. 2010;11(3):213–219. doi: 10.1016/j.cmet.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoshino J, et al. Nicotinamide mononucleotide, a key NAD(+) intermediate, treats the pathophysiology of diet- and age-induced diabetes in mice. Cell metabolism. 2011;14(4):528–536. doi: 10.1016/j.cmet.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim HJ, et al. Metabolomic analysis of livers and serum from high-fat diet induced obese mice. Journal of proteome research. 2011;10(2):722–731. doi: 10.1021/pr100892r. [DOI] [PubMed] [Google Scholar]
- 13.Kendrick AA, et al. Fatty liver is associated with reduced SIRT3 activity and mitochondrial protein hyperacetylation. The Biochemical journal. 2011;433(3):505–514. doi: 10.1042/BJ20100791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tao R, et al. Hepatic FoxOs regulate lipid metabolism via modulation of expression of the nicotinamide phosphoribosyltransferase gene. The Journal of biological chemistry. 2011;286(16):14681–14690. doi: 10.1074/jbc.M110.201061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Belenky P, Bogan KL, Brenner C. NAD+ metabolism in health and disease. Trends in biochemical sciences. 2007;32(1):12–19. doi: 10.1016/j.tibs.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 16.Imai S. A possibility of nutriceuticals as an anti-aging intervention: activation of sirtuins by promoting mammalian NAD biosynthesis. Pharmacological research : the official journal of the Italian Pharmacological Society. 2010;62(1):42–47. doi: 10.1016/j.phrs.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gibson BA, Kraus WL. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nature reviews. Molecular cell biology. 2012 doi: 10.1038/nrm3376. [DOI] [PubMed] [Google Scholar]
- 18.Virag L. Structure and function of poly(ADP-ribose) polymerase-1: role in oxidative stress-related pathologies. Current vascular pharmacology. 2005;3(3):209–214. doi: 10.2174/1570161054368625. [DOI] [PubMed] [Google Scholar]
- 19.Bai P, et al. PARP-2 regulates SIRT1 expression and whole-body energy expenditure. Cell metabolism. 2011;13(4):450–460. doi: 10.1016/j.cmet.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bai P, et al. PARP-1 inhibition increases mitochondrial metabolism through SIRT1 activation. Cell metabolism. 2011;13(4):461–468. doi: 10.1016/j.cmet.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chini EN. CD38 as a regulator of cellular NAD: a novel potential pharmacological target for metabolic conditions. Current pharmaceutical design. 2009;15(1):57–63. doi: 10.2174/138161209787185788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sasaki T, et al. Phosphorylation regulates SIRT1 function. PloS one. 2008;3(12):e4020. doi: 10.1371/journal.pone.0004020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nasrin N, et al. JNK1 phosphorylates SIRT1 and promotes its enzymatic activity. PloS one. 2009;4(12):e8414. doi: 10.1371/journal.pone.0008414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vinciguerra M, et al. mIGF-1/JNK1/SirT1 signaling confers protection against oxidative stress in the heart. Aging cell. 2012;11(1):139–149. doi: 10.1111/j.1474-9726.2011.00766.x. [DOI] [PubMed] [Google Scholar]
- 25.Guo X, et al. DYRK1A and DYRK3 promote cell survival through phosphorylation and activation of SIRT1. The Journal of biological chemistry. 2010;285(17):13223–13232. doi: 10.1074/jbc.M110.102574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo X, et al. The NAD(+)-dependent protein deacetylase activity of SIRT1 is regulated by its oligomeric status. Scientific reports. 2012;2:640. doi: 10.1038/srep00640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dixit D, et al. Inhibition of Casein kinase-2 induces p53-dependent cell cycle arrest and sensitizes glioblastoma cells to tumor necrosis factor (TNFalpha)-induced apoptosis through SIRT1 inhibition. Cell death & disease. 2012;3:e271. doi: 10.1038/cddis.2012.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kang H, et al. CK2 is the regulator of SIRT1 substrate-binding affinity, deacetylase activity and cellular response to DNA-damage. PloS one. 2009;4(8):e6611. doi: 10.1371/journal.pone.0006611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zschoernig B, Mahlknecht U. Carboxy-terminal phosphorylation of SIRT1 by protein kinase CK2. Biochemical and biophysical research communications. 2009;381(3):372–377. doi: 10.1016/j.bbrc.2009.02.085. [DOI] [PubMed] [Google Scholar]
- 30.Liu X, et al. Methyltransferase Set7/9 regulates p53 activity by interacting with Sirtuin 1 (SIRT1) Proceedings of the National Academy of Sciences of the United States of America. 2011;108(5):1925–1930. doi: 10.1073/pnas.1019619108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gerhart-Hines Z, et al. The cAMP/PKA pathway rapidly activates SIRT1 to promote fatty acid oxidation independently of changes in NAD(+) Molecular cell. 2011;44(6):851–863. doi: 10.1016/j.molcel.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang Y, et al. SIRT1 sumoylation regulates its deacetylase activity and cellular response to genotoxic stress. Nature cell biology. 2007;9(11):1253–1262. doi: 10.1038/ncb1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kornberg MD, et al. GAPDH mediates nitrosylation of nuclear proteins. Nature cell biology. 2010;12(11):1094–1100. doi: 10.1038/ncb2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakamura T, Lipton SA. Emerging Role of Protein-Protein Transnitrosylation in Cell Signaling Pathways. Antioxidants & redox signaling. 2012 doi: 10.1089/ars.2012.4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim EJ, et al. Active regulator of SIRT1 cooperates with SIRT1 and facilitates suppression of p53 activity. Molecular cell. 2007;28(2):277–290. doi: 10.1016/j.molcel.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 36.Kim JE, Chen J, Lou Z. DBC1 is a negative regulator of SIRT1. Nature. 2008;451(7178):583–586. doi: 10.1038/nature06500. [DOI] [PubMed] [Google Scholar]
- 37.Kang H, et al. Peptide switch is essential for Sirt1 deacetylase activity. Molecular cell. 2011;44(2):203–213. doi: 10.1016/j.molcel.2011.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuan J, et al. Regulation of SIRT1 activity by genotoxic stress. Genes & development. 2012;26(8):791–796. doi: 10.1101/gad.188482.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Escande C, et al. Deleted in breast cancer-1 regulates SIRT1 activity and contributes to high-fat diet-induced liver steatosis in mice. The Journal of clinical investigation. 2010;120(2):545–558. doi: 10.1172/JCI39319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pagans S, et al. SIRT1 regulates HIV transcription via Tat deacetylation. PLoS biology. 2005;3(2):e41. doi: 10.1371/journal.pbio.0030041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kwon HS, et al. Human immunodeficiency virus type 1 Tat protein inhibits the SIRT1 deacetylase and induces T cell hyperactivation. Cell host & microbe. 2008;3(3):158–167. doi: 10.1016/j.chom.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alcain FJ, Villalba JM. Sirtuin inhibitors. Expert opinion on therapeutic patents. 2009;19(3):283–294. doi: 10.1517/13543770902755111. [DOI] [PubMed] [Google Scholar]
- 43.Alcain FJ, Villalba JM. Sirtuin activators. Expert opinion on therapeutic patents. 2009;19(4):403–414. doi: 10.1517/13543770902762893. [DOI] [PubMed] [Google Scholar]
- 44.Herranz D, Serrano M. SIRT1: recent lessons from mouse models. Nature reviews. Cancer. 2010;10(12):819–823. doi: 10.1038/nrc2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu Y, et al. The controversial links among calorie restriction, SIRT1, and resveratrol. Free radical biology & medicine. 2011;51(2):250–256. doi: 10.1016/j.freeradbiomed.2011.04.034. [DOI] [PubMed] [Google Scholar]
- 46.Schmidt C. GSK/Sirtris compounds dogged by assay artifacts. Nature biotechnology. 2010;28(3):185–186. doi: 10.1038/nbt0310-185. [DOI] [PubMed] [Google Scholar]
- 47.Nakata R, Takahashi S, Inoue H. Recent advances in the study on resveratrol. Biological & pharmaceutical bulletin. 2012;35(3):273–279. doi: 10.1248/bpb.35.273. [DOI] [PubMed] [Google Scholar]
- 48.Grozinger CM, et al. Identification of a class of small molecule inhibitors of the sirtuin family of NAD-dependent deacetylases by phenotypic screening. The Journal of biological chemistry. 2001;276(42):38837–38843. doi: 10.1074/jbc.M106779200. [DOI] [PubMed] [Google Scholar]
- 49.Peck B, et al. SIRT inhibitors induce cell death and p53 acetylation through targeting both SIRT1 and SIRT2. Molecular cancer therapeutics. 2010;9(4):844–855. doi: 10.1158/1535-7163.MCT-09-0971. [DOI] [PubMed] [Google Scholar]
- 50.Mai A, et al. Design, synthesis, and biological evaluation of sirtinol analogues as class III histone/protein deacetylase (Sirtuin) inhibitors. Journal of medicinal chemistry. 2005;48(24):7789–7795. doi: 10.1021/jm050100l. [DOI] [PubMed] [Google Scholar]
- 51.Bedalov A, et al. Identification of a small molecule inhibitor of Sir2p. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(26):15113–15118. doi: 10.1073/pnas.261574398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hirao M, et al. Identification of selective inhibitors of NAD+-dependent deacetylases using phenotypic screens in yeast. The Journal of biological chemistry. 2003;278(52):52773–52782. doi: 10.1074/jbc.M308966200. [DOI] [PubMed] [Google Scholar]
- 53.Posakony J, et al. Inhibitors of Sir2: evaluation of splitomicin analogues. Journal of medicinal chemistry. 2004;47(10):2635–2644. doi: 10.1021/jm030473r. [DOI] [PubMed] [Google Scholar]
- 54.Freitag M, et al. Synthesis and biological activity of splitomicin analogs targeted at human NAD(+)-dependent histone deacetylases (sirtuins) Bioorganic & medicinal chemistry. 2011;19(12):3669–3677. doi: 10.1016/j.bmc.2011.01.026. [DOI] [PubMed] [Google Scholar]
- 55.Lain S, et al. Discovery, in vivo activity, and mechanism of action of a small-molecule p53 activator. Cancer cell. 2008;13(5):454–463. doi: 10.1016/j.ccr.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trapp J, et al. Structure-activity studies on suramin analogues as inhibitors of NAD+-dependent histone deacetylases (sirtuins) ChemMedChem. 2007;2(10):1419–1431. doi: 10.1002/cmdc.200700003. [DOI] [PubMed] [Google Scholar]
- 57.Bernofsky C. Physiology aspects of pyridine nucleotide regulation in mammals. Molecular and cellular biochemistry. 1980;33(3):135–143. doi: 10.1007/BF00225285. [DOI] [PubMed] [Google Scholar]
- 58.Catz P, et al. Simultaneous determination of myristyl nicotinate, nicotinic acid, and nicotinamide in rabbit plasma by liquid chromatography-tandem mass spectrometry using methyl ethyl ketone as a deproteinization solvent. Journal of chromatography. B, Analytical technologies in the biomedical and life sciences. 2005;829(1–2):123–135. doi: 10.1016/j.jchromb.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 59.Jacobson EL, et al. Evaluating the role of niacin in human carcinogenesis. Biochimie. 1995;77(5):394–398. doi: 10.1016/0300-9084(96)88152-1. [DOI] [PubMed] [Google Scholar]
- 60.Noriega LG, et al. CREB and ChREBP oppositely regulate SIRT1 expression in response to energy availability. EMBO reports. 2011;12(10):1069–1076. doi: 10.1038/embor.2011.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Heilbronn LK, et al. Glucose tolerance and skeletal muscle gene expression in response to alternate day fasting. Obesity research. 2005;13(3):574–581. doi: 10.1038/oby.2005.61. [DOI] [PubMed] [Google Scholar]
- 62.Fusco S, et al. A role for neuronal cAMP responsive-element binding (CREB)-1 in brain responses to calorie restriction. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(2):621–626. doi: 10.1073/pnas.1109237109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nemoto S, Fergusson MM, Finkel T. Nutrient availability regulates SIRT1 through a forkhead-dependent pathway. Science. 2004;306(5704):2105–2108. doi: 10.1126/science.1101731. [DOI] [PubMed] [Google Scholar]
- 64.Xiong S, et al. FoxO1 mediates an autofeedback loop regulating SIRT1 expression. The Journal of biological chemistry. 2011;286(7):5289–5299. doi: 10.1074/jbc.M110.163667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brunet A, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303(5666):2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 66.Motta MC, et al. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116(4):551–563. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- 67.Purushotham A, et al. Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell metabolism. 2009;9(4):327–338. doi: 10.1016/j.cmet.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Okazaki M, et al. PPARbeta/delta regulates the human SIRT1 gene transcription via Sp1. Endocrine journal. 2010;57(5):403–413. doi: 10.1507/endocrj.k10e-004. [DOI] [PubMed] [Google Scholar]
- 69.Han L, et al. SIRT1 is regulated by a PPAR{gamma}-SIRT1 negative feedback loop associated with senescence. Nucleic acids research. 2010;38(21):7458–7471. doi: 10.1093/nar/gkq609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen WY, et al. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell. 2005;123(3):437–448. doi: 10.1016/j.cell.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 71.Zhang Q, et al. Metabolic regulation of SIRT1 transcription via a HIC1:CtBP corepressor complex. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(3):829–833. doi: 10.1073/pnas.0610590104. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 72.Dehennaut V, et al. Molecular dissection of the interaction between HIC1 and SIRT1. Biochemical and biophysical research communications. 2012;421(2):384–388. doi: 10.1016/j.bbrc.2012.04.026. [DOI] [PubMed] [Google Scholar]
- 73.Abdelmohsen K, et al. Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Molecular cell. 2007;25(4):543–557. doi: 10.1016/j.molcel.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yamakuchi M. MicroRNA Regulation of SIRT1. Frontiers in physiology. 2012;3:68. doi: 10.3389/fphys.2012.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yamakuchi M, Lowenstein CJ. MiR-34, SIRT1 and p53: the feedback loop. Cell cycle. 2009;8(5):712–715. doi: 10.4161/cc.8.5.7753. [DOI] [PubMed] [Google Scholar]
- 76.Ito T, Yagi S, Yamakuchi M. MicroRNA-34a regulation of endothelial senescence. Biochemical and biophysical research communications. 2010;398(4):735–740. doi: 10.1016/j.bbrc.2010.07.012. [DOI] [PubMed] [Google Scholar]
- 77.Zhao T, Li J, Chen AF. MicroRNA-34a induces endothelial progenitor cell senescence and impedes its angiogenesis via suppressing silent information regulator 1. American journal of physiology. Endocrinology and metabolism. 2010;299(1):E110–E116. doi: 10.1152/ajpendo.00192.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lee J, et al. A pathway involving farnesoid X receptor and small heterodimer partner positively regulates hepatic sirtuin 1 levels via microRNA-34a inhibition. The Journal of biological chemistry. 2010;285(17):12604–12611. doi: 10.1074/jbc.M109.094524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhu H, et al. MicroRNA-195 promotes palmitate-induced apoptosis in cardiomyocytes by down-regulating Sirt1. Cardiovascular research. 2011;92(1):75–84. doi: 10.1093/cvr/cvr145. [DOI] [PubMed] [Google Scholar]
- 80.Rane S, et al. Downregulation of miR-199a derepresses hypoxia-inducible factor-1alpha and Sirtuin 1 and recapitulates hypoxia preconditioning in cardiac myocytes. Circulation research. 2009;104(7):879–886. doi: 10.1161/CIRCRESAHA.108.193102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Moynihan KA, et al. Increased dosage of mammalian Sir2 in pancreatic beta cells enhances glucose-stimulated insulin secretion in mice. Cell metabolism. 2005;2(2):105–117. doi: 10.1016/j.cmet.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 82.Ramachandran D, et al. Sirt1 and mir-9 expression is regulated during glucose-stimulated insulin secretion in pancreatic beta-islets. The FEBS journal. 2011;278(7):1167–1174. doi: 10.1111/j.1742-4658.2011.08042.x. [DOI] [PubMed] [Google Scholar]
- 83.Strum JC, et al. MicroRNA 132 regulates nutritional stress-induced chemokine production through repression of SirT1. Molecular endocrinology. 2009;23(11):1876–1884. doi: 10.1210/me.2009-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gao Z, et al. Sirtuin 1 (SIRT1) protein degradation in response to persistent c-Jun N-terminal kinase 1 (JNK1) activation contributes to hepatic steatosis in obesity. The Journal of biological chemistry. 2011;286(25):22227–22234. doi: 10.1074/jbc.M111.228874. [DOI] [PMC free article] [PubMed] [Google Scholar]