Abstract
The Trithorax and Polycomb groups of chromatin regulators are critical for cell-lineage specification during normal development; functions that often become deregulated during tumorigenesis. As an example, oncogenic fusions of the Trithorax-related protein MLL can initiate aggressive leukemias by altering the transcriptional circuitry governing hematopoietic cell differentiation, a process that is known to require additional epigenetic pathways to implement. Here we used shRNA screening to identify chromatin regulators uniquely required in a mouse model of MLL-fusion acute myeloid leukemia, which revealed a role for the Polycomb Repressive Complex 2 (PRC2) in maintenance of this disease. shRNA-mediated suppression of PRC2 subunits Eed, Suz12, or Ezh1/Ezh2 led to proliferation-arrest and differentiation of leukemia cells, with a minimal impact on growth of several non-transformed hematopoietic cell lines. The requirement for PRC2 in leukemia is partly due to its role in direct transcriptional repression of genes that limit the self-renewal potential of hematopoietic cells, including Cdkn2a. In addition to implicating a role for PRC2 in the pathogenesis of MLL-fusion leukemia, our results suggest, more generally, that Trithorax and Polycomb group proteins can cooperate with one another to maintain aberrant lineage programs in cancer.
Keywords: chromatin, leukemia, epigenetics, MLL, PRC2
Introduction
Cellular identity in multicellular organisms is reinforced by master-regulatory transcription factors in concert with chromatin modifying activities. A major epigenetic regulatory axis maintaining the “ON” or “OFF” state of transcription is composed of the Trithorax- and Polycomb-groups of chromatin regulators, respectively (for reviews see 1, 2). First discovered in Drosophila based on their antagonistic regulation of homeotic phenotypes (3, 4), Trithorax and Polycomb group genes have emerged as key regulators of transcriptional programs underlying embryonic development, tissue homeostasis, and the pathogenesis of several human diseases, including cancer (5). Trithorax and Polycomb group proteins possess diverse regulatory activities directed toward chromatin, including lysine methyltransferase, ubiquitin ligase, chromatin remodeling ATPase, as well as a host of histone-binding modules (1, 2).
Polycomb Repressive Complex 2 (PRC2) mediates gene silencing through catalysis of histone H3K27 methylation (6–8). PRC2 is minimally comprised of two essential non-catalytic subunits, Eed and Suz12, as well as one of two SET domain-containing methyltransferase subunits, Ezh1 or Ezh2 (6–9). Recruitment of PRC2 generally occurs at CpG-rich promoter sequences in the genome, mediated through an assortment of protein-protein and protein-RNA interactions to establish localized domains of H3K27 methylation (10, 11). This histone mark serves as a docking site for other Polycomb complexes, which exert a repressive effect on transcription (6). A key function of PRC2 in mammals is to regulate stem cell function, where it can promote self-renewal through direct repression of pro-differentiation genes (12–14). Additionally, several lines of evidence link the function of PRC2 to the pathogenesis of human cancer. Ezh2 is overexpressed in many different malignancies and mutations that elevate its tri-methyltransferase activity are found in subtypes of lymphoma, together suggesting a pro-tumorigenic role for this complex (5, 15–17). However, Ezh2 loss-of-function mutations have also been observed in myelodysplastic syndrome (MDS), suggesting a tumor suppressor function in certain cellular contexts (18, 19). Interestingly, Ezh2 loss-of-function mutations are rarely observed in primary acute myeloid leukemia (AML), suggesting that a role for PRC2 in myeloid cancer might be highly dependent on cellular and genetic context (Ross Levine, personal communication).
In mammals, a major class of Trithorax-group genes belongs to the Mixed Lineage Leukemia (MLL) subfamily. MLL encodes a histone H3K4 methyltransferase essential for hematopoietic development through maintenance of transcription of various genes, most notably HOX clusters (20). Mutant forms of MLL also act as potent oncogenes in AML pathogenesis, which are associated with chemotherapy-resistant disease (reviewed in 21). MLL can often be mutated via chromosomal translocation, where the N-terminal fragment of MLL is fused to the C-terminus of one of over 50 known partner genes, with AF9 being the most common in AML (22). MLL-AF9 acts in a gain-of-function manner via aberrant recruitment of AF9-interacting proteins to normal MLL target genes, resulting in transcriptional hyperactivation. The biological consequence of MLL-AF9 expression is a blockade of myeloid maturation and an enduring state of self-renewal. Coupling of MLL-AF9 expression with activating mutations in the MAP kinase-signaling pathway (e.g. NRASG12D), is thought to be sufficient for leukemic transformation (23, 24). As one of the only known examples of a proto-oncogene chromatin regulator, MLL-fusion leukemia represents a paradigm for understanding causality between epigenetic alterations and cancer pathogenesis.
To better define the repertoire of epigenetic regulators specifically required for maintaining MLL-fusion AML, we systematically compared the effects of suppressing chromatin-regulators in a mouse model of MLL-AF9;NrasG12D AML versus non-transformed hematopoietic cell lines. Surprisingly, our results identify PRC2 as a highly-specific requirement for the maintenance of this disease. Inhibition of PRC2 results in differentiation of leukemia cells due to anomalous upregulation of specific growth-inhibitory genes, including the Cdkn2a gene products p16Ink4a and p19Arf. Together, these findings reveal an unexpected collaboration between the activating function of Trithorax and the repressive function of Polycomb group proteins to execute an aberrant self-renewal program in AML.
Results and Discussion
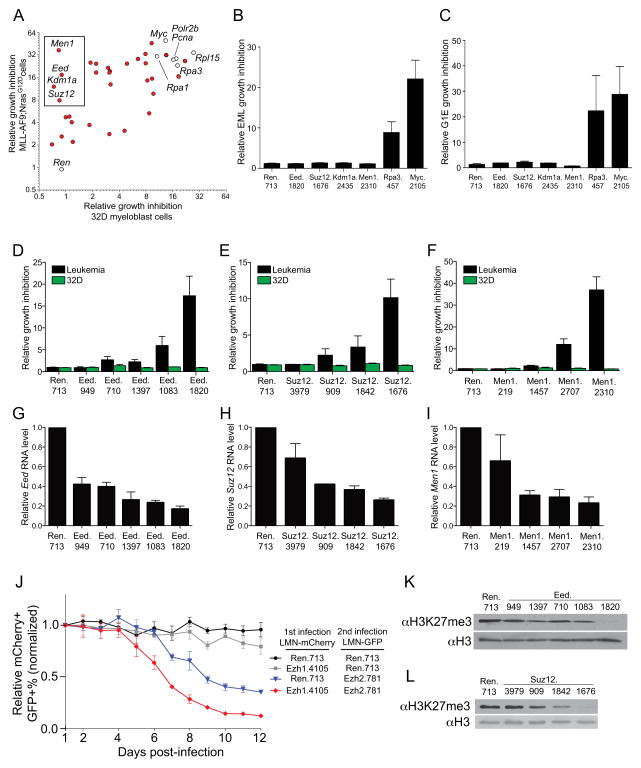
We previously reported an shRNA screening strategy for identifying epigenetic regulators needed for growth of a murine model of MLL-AF9;NrasG12D-driven AML, which revealed the bromodomain protein Brd4 as a dependency in this disease (25). This screen identified several additional chromatin regulators as necessary for leukemia growth, however the disease-specificity and in vivo relevance were unexplored (25). To address this, we compared the impact of shRNAs capable of suppressing growth of leukemia with their effect on growth of 32D, an immortalized, non-leukemic myeloid cell line. A number of positive control shRNAs targeting Polr2b, Rpl15, Myc, Rpa1, Rpa3, and Pcna were included, which all suppressed growth comparably in both 32D and leukemia contexts (Figure 1A). The most leukemia-specific growth requirement identified was Men1, encoding Menin, a known cofactor for MLL-AF9 with a well-established and highly-specific maintenance role in this disease (26, 27) (Figure 1A). Based on the criteria of potent leukemia growth inhibition with neutral effects in 32D, our analysis identified 3 additional candidates, Eed, Suz12, and Kdm1a (Figure 1A). shRNAs targeting these genes also did not affect growth of non-transformed G1E and EML hematopoietic cell lines (28, 29), confirming their leukemia-specific requirement (Figures 1B, C). To rule out off-target effects, we retrieved all shRNAs designed to target Eed, Suz12, Men1, and Kdm1a from the original library and compared their knockdown efficiency with their relative impact on AML proliferation (Figures 1D–I, Supplementary Figure 2). In all cases the degree of knockdown correlated with the level of growth inhibition, suggesting that the observed effects with shRNA were due to knockdown of their predicted target.
Figure 1. RNAi screen identifies Eed and Suz12 as unique requirements for growth of MLL-AF9;NrasG12D leukemia.
(A) Scatter-plot comparison of the relative growth inhibition conferred by LMN-shRNAs in MLL-AF9;NrasG12D leukemia and 32D myeloblasts. All shRNAs evaluated were identified from a pooled negative-selection shRNA screen reported previously (25). MLL-AF9;NrasG12D leukemia or 32D myeloblasts were transduced with individual LMN-shRNA vectors (MSCV-miR30-shRNA-PGK-NeoR-IRES-GFP), followed by measurement of the GFP-percentage at day 2 and day 12 post-infection using a Guava Easycyte (Millipore). Growth inhibition was calculated as the ratio of the GFP% measured at day 2 to day 12 of partially transduced cell populations. Since leukemia and 32D cells grow at comparable rates in vitro (Supplementary Figure 1), relative GFP-depletion is a suitable assay for comparing growth effects in each line. Control shRNAs are indicated with white circles. Box indicates shRNAs with leukemia-specific growth inhibition. (B–F) Relative growth inhibition conferred by indicated LMN-shRNAs in EML, G1E, leukemia, and 32D cell lines, calculated as in (A) (n = 3). (G–I) Quantitative reverse transcription PCR measuring knockdown efficiency in 32D myeloblasts cells following transduction with LMN-shRNAs and selection with G418. Measurements were normalized to Gapdh, with the relative mRNA level in the cells with control Ren shRNA set to 1 (n = 3). (J) Relative change double-transduced cell percentage following co-transduction with indicated LMN-shRNAs linked to either GFP or mCherry reporters. The results were normalized to the GFP+/mCherry+ percentage measured at day 1, set to 1 (n = 3). (K, L) H3K27me3 Western blotting of acid extracted histones prepared from 32D cells transduced with the indicated LMN-shRNA following G418 selection. The levels of total histone H3 serve as a loading control. A representative experiment of three replicates is shown. All error bars represent s.e.m.
Since Eed and Suz12 both encode integral subunits of PRC2 (7, 8, 30, 31), we next investigated the role of other components in this complex. The methyltransferase subunit of PRC2 can be either Ezh1 or its close homolog Ezh2, with both genes being required to maintain the total cellular content of H3K27 methylation (9). While individual shRNAs targeting Ezh1 or Ezh2 displayed only weak or no growth inhibitory effects (data not shown), co-expression of the two most potent Ezh1 and Ezh2 shRNAs together led to synergistic inhibition of MLL-leukemia proliferation, indicating a requirement for the PRC2 catalytic subunit (Figure 1J, Supplementary Figure 3). Importantly, knockdown of Eed or Suz12 alone was sufficient to deplete total H3K27me3 levels, consistent with prior reports of these core subunits being essential for PRC2 activity (30, 31) (Figure 1K, L). Interestingly, there appears to be a threshold of approximately 80% knockdown of Eed or Suz12 that results in severe depletion of total H3K27me3 levels and ensuing effects on leukemia growth (Figure 1G, H, K, L). Together, these data suggest that MLL-AF9;NrasG12D leukemia is reliant on PRC2 for rapid proliferation in culture.
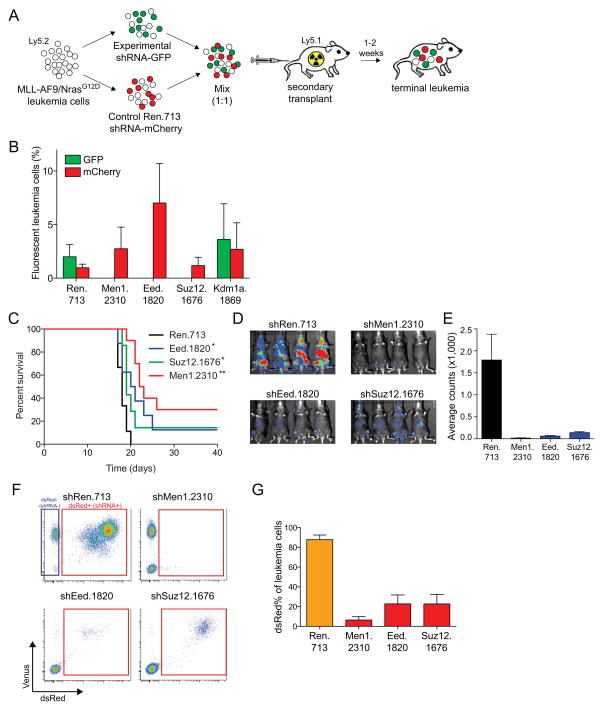
We next examined whether the leukemia-specific requirements observed in vitro were also relevant in vivo. First, we performed a competitive transplant assay where MLL-AF9;NrasG12D leukemia cells transduced with experimental or control shRNAs were mixed and evaluated for their relative ability to initiate disease in secondary recipient mice (Figure 2A). Leukemia cells transduced with shRNAs targeting Men1, Eed or Suz12 were outcompeted by control cells during leukemia growth in vivo, as indicated by the depletion of GFP positivity following 10 days of disease expansion (Figure 2B). Interestingly, while Kdm1a knockdown inhibited leukemia proliferation in vitro, this level of knockdown had a negligible impact on proliferation in vivo (Figure 2B). This observation was confirmed with multiple independent Kdm1a shRNAs (data not shown), suggesting the requirement for this specific gene might be augmented under tissue culture conditions.
Figure 2. Suppression of Eed or Suz12 impairs leukemia progression in vivo.
(A) Schematic of two-color competition assay measuring impact of LMN-shRNAs on leukemia expansion in vivo. MLL-AF9;NrasG12D leukemia cultures were transduced with either experimental shRNAs from the LMN vector, or with a Ren.713 control shRNA expressed from LMN-mCherry. Experimental and control cultures were mixed at 1:1 GFP:mCherry ratio followed by transplantation of 1 × 106 cells into secondary recipients on day 2 post-infection, prior to any depletion of GFP+ cells in vitro. Upon reaching terminal disease state (~15 days), as indicated by moribund appearance and whole-body bioluminescent signal, mice were sacrificed and bone marrow was collected for flow cytometry evaluation. Gating was performed on donor-derived leukemia populations (CD45.2+) and the ratio of GFP:mCherry was measured using a LSRII flow cytometer. (B) Percentage of GFP and mCherry positivity within donor-derived leukemia cells (CD45.2+) derived from bone marrow at the terminal disease endpoint (~15 days following transplant). Each shRNA group included 5 mice. (C–D) Tet-on competent MLL-AF9;NrasG12D leukemia cultures were retrovirally transduced with TRPMV-Neo constructs (pSIN-TRE-dsRed-miR30-shRNA-PGK-Venus-IRES-NeoR) followed by G418 selection (1 mg/ml for 6 days). Transduced cells were then transplanted into secondary recipient animals, followed by initiation of doxycycline administration after 1–2 days. Mice were monitored thereafter by bioluminescent imaging (IVIS Spectrum system; Caliper LifeSciences) and for differences in overall survival. Bone marrow from leukemic mice at terminal disease stage, was analyzed by flow cytometry for the percentage of dsRed+/shRNA+ cells in donor-derived (CD45.2+) leukemia populations. Each shRNA group contained 8–10 mice. (C) Kaplan-Meier survival curves of mice transplanted with Tet-On competent MLL-AF9;NrasG12D leukemia cells transduced with the indicted shRNAs in the TRMPV-Neo vector (32). Statistical significance was calculated by using log-rank test comparing to shRen control group; *, P<0.05; **,P<0.0001. (D) Representative bioluminescent imaging of leukemia disease burden at day 15 post-transplant. (E) Quantification of bioluminescent imaging shown in (D). Mean values were calculated from 4 replicate mice. (F) Flow cytometry analysis of donor-derived (Cd45.2+) leukemia cells in terminally-diseased mice from (C). The gate shown includes shRNA+/dsRed+ cells. Representative plots are shown. (G) Quantitation of results shown in (F) (n = 6 to n = 9). All error bars shown represent s.e.m. All mouse experiments included in this work were approved by The Cold Spring Harbor Animal Care and Use Committee.
To further confirm that Eed and Suz12 play an important role during leukemia progression in vivo, we made use of doxycycline (dox)-inducible shRNA vectors to deliver knockdown following disease engraftment into recipient animals (32). As compared to controls, dox-induced Men1, Eed, and Suz12 knockdown led to inhibition of leukemia expansion in vivo associated with a statistically significant, albeit modest, survival benefit (Figure 2C, D). Similar to results described above, as the leukemia expanded in vivo, cells expressing Men1, Eed, or Suz12 shRNA were outcompeted by shRNA-negative cells, as indicated by depletion of shRNA/dsRed-positivity within the terminal leukemia burden (Figure 2F, G). These experiments further support PRC2 being required for rapid leukemia expansion in vivo and not simply for disease engraftment.
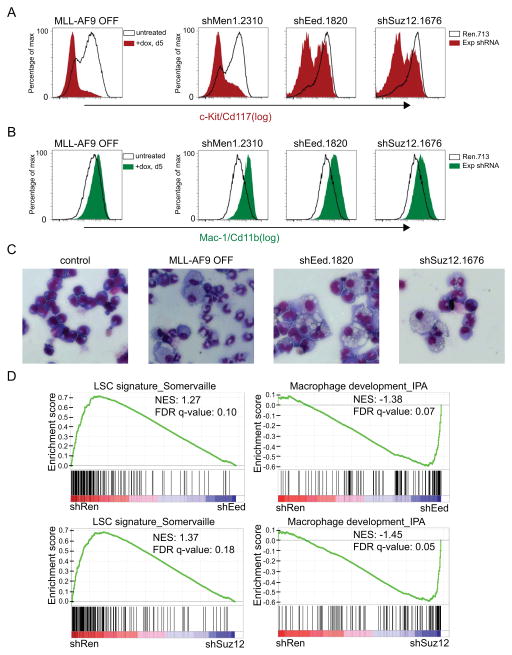
A hallmark of MLL-fusion AML is the inappropriate blockade of myeloid differentiation and aberrant self-renewal capacity present within leukemia stem cell subpopulations, which can be prospectively identified as a c-Kit/Cd117 expressing subpopulation (33, 34). Based on the known role of PRC2 in supporting self-renewal in other stem-cell contexts (12–14), we considered whether a similar function might be relevant in leukemia. Indeed, shRNA-mediated suppression of Eed or Suz12 in MLL-AF9;NrasG12D leukemia cells led to a marked decrease in c-Kit levels (Figure 3A), paralleled by an upregulation of the myeloid differentiation marker Mac-1 (Figure 3B), as well as a morphologic transition toward a differentiated macrophage-like appearance (Figure 3C). As a control, we verified that a similar differentiation-like phenotype was induced upon suppression of MLL-AF9 or Menin, consistent with prior findings (26, 35, 36) (Figure 3A–C). Finally, we performed expression microarrays of leukemia cells transduced with Eed or Suz12 shRNA. Gene Set Enrichment Analysis (GSEA) of these data revealed a systematic downregulation of genes known to be highly expressed in leukemia stem cells and upregulation of macrophage-specific genes (37) (Figure 3D). Based on these findings, we conclude that PRC2, like MLL-AF9, is required to enforce the blockade of myeloid differentiation present in leukemia.
Figure 3. Suppression of Eed or Suz12 results in differentiation of MLL-AF9;NrasG12D leukemia cells.
(A, B) Flow cytometry analysis of cell surface levels of c-Kit/Cd117 and Mac-1/Cd11b. MLL-AF9 was suppressed as a positive control using a TET-off system (35), following 5 days of 1 ug/ml doxycycline treatment. Untreated TET-off leukemia cells were used a negative control. Men1, Eed, and Suz12 LMN-shRNAs were transduced into MLL-AF9;NrasG12D leukemia cultures, with cell-surface staining and flow-cytometry analysis performed on day 7 post-infection. Since the average infection efficiency was ~ 20%, shRNA+/GFP+ cells were compared to shRNA-/GFP- within each culture as an internal negative control. (C) Light microscopy of May–Grunwald/Giemsa-stained MLL-AF9;NrasG12D leukemia cells under the same experimental condition used in (A, B), except G418 was administered to LMN-transduced cells following infection to select for shRNA+/GFP+ cells. Imaging was performed using X40 objective. Representative images of three biological replicates are shown. (D) GSEA of microarray data obtained from leukemia cells transduced with Eed and Suz12 LMN-shRNAs, 5 days post-infection/G418 selection. NES, normalized enrichment score; FDR q-val, false discovery rate q-value. All error bars shown represent s.e.m.
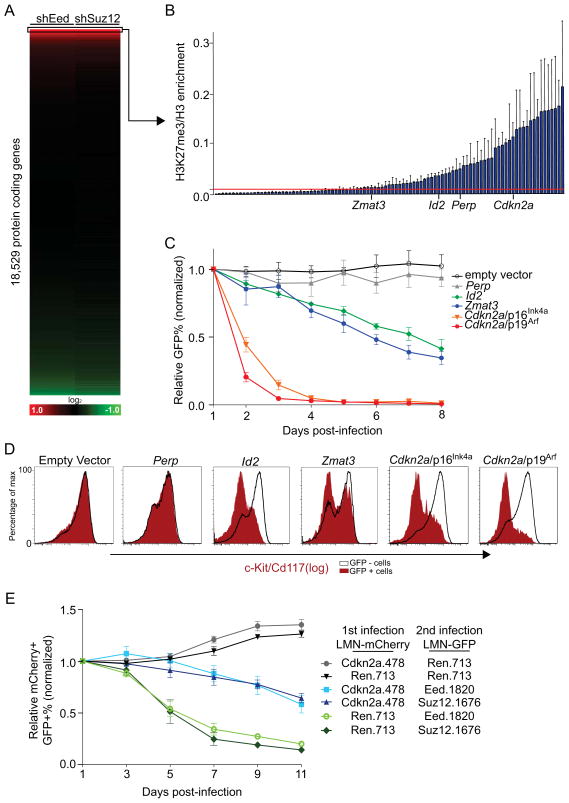
We next sought to identify the direct repression targets of PRC2 relevant to its leukemia-maintenance function. To this end, we analyzed the expression microarray data described above to identify 149 genes displaying >2-fold upregulation following both Eed and Suz12 knockdown (Figure 4A). This group of candidate PRC2-targets was further reduced to 97 based on the presence of a CpG island near the promoter region, a sequence feature strongly linked with PRC2 recruitment in the mammalian genome (11). We next used ChIP-qPCR to further discriminate direct from indirect PRC2 targets by measuring levels of H3K27me3 in the vicinity of the promoter region, which identified 60 genes with significant enrichment (Figure 4B). We surveyed the published literature pertaining to these genes for evidence of any known role in either a) promoting myeloid differentiation b) negatively regulating stem cell self-renewal or c) inhibiting the cell-cycle or cell survival. Using these criteria, we identified Id2 (38), Perp (39), Zmat3 (40), and Cdkn2a (41) as candidates for further evaluation (Supplementary Figure 4).
Figure 4. PRC2 directly represses transcription of genes that restrict self-renewal potential of leukemia cells.
(A) Expression microarray heatmap representing the fold-change of all protein-coding genes, comparing Ren.713 to Eed.1820 (left) or Ren.713 to Suz12.1676 (right). Genes are ranked in both columns based on the level of fold-change in the Eed dataset. Box indicates >2-fold upregulated genes. (B) ChIP-qPCR was performed in MLL-AF9;NrasG12D leukemia cells using anti-H3 or anti-H3K27me3 antibody (n = 3). Primers were designed in the vicinity of the transcription start site of each gene indicted. Enrichment was calculated as the ratio of H3K27me3 to H3 recovery, normalized to input DNA. The red line represents a 10-fold enrichment threshold above background. All PRC2 target genes identified here are listed in supplementary figure 8. (C) Relative change GFP-positive percentage following retroviral transduction of MLL-AF9;NrasG12D leukemia cells with indicated MSCV-GFP cDNA overexpression construct. Results were normalized to the GFP percentage measured at day 1 post-transduction, set to 1 (n = 3). (D) Flow cytometry analysis of c-Kit/Cd117 levels in experiment shown in (C) after either 7 days post-infection (for Perp, Id2 and Zmat3) or after 2 days post-infection (for p16Ink4a and p19ARF) (n = 3). A representative experiment of three biological replicates is shown. (E) Relative change in cell populations co-transduced with the indicated shRNAs linked to GFP and mCherry reporters using the LMN vector. The results were normalized to the percentage of GFP/mCherry double positive cells measured at day 1, set to 1 (n = 5). All error bars shown represent s.e.m.
To determine whether repression of these genes might be required for leukemia growth, we first examined the impact of retroviral overexpression of each in MLL-AF9;NrasG12D leukemia cells. Overexpression of all of the candidate genes but Perp led to growth inhibition, with the two protein products of the Cdkn2a locus, p16Ink4a and p19Arf displaying the greatest potency (Figure 4C). Using flow cytometry, we found that overexpression of Id2, Zmat3, p16Ink4a, or p19Arf all led to depletion of c-Kit-positive cells from leukemia cultures (Figure 4D), resembling the phenotype observed upon inhibition of PRC2 (Figure 3A). While the collective repression of multiple downstream targets could account for the entirety of a PRC2-requirement in leukemia, we considered that a substantial contribution might be mediated by Cdkn2a repression, based on the potent phenotypes observed upon p16Ink4a and p19Arf overexpression. To evaluate this, we employed a validated Cdkn2a shRNA to suppress levels of both p16Ink4a and p19Arf and evaluated whether this influenced the PRC2-requirement for leukemia growth (42) (Supplementary Figure 5). Indeed, shRNA knockdown of Cdkn2a rendered leukemia cells less dependent on Eed and Suz12 for rapid growth (Figure 4E), indicating that at least one role for PRC2 in MLL-fusion AML is to maintain low levels of p16Ink4a and p19Arf to permit leukemia proliferation.
Our findings support a role for PRC2 in MLL-AF9;NrasG12D AML to prevent differentiation and sustain aberrant self-renewal. As such, this study illustrates how Trithorax and Polycomb group regulators can together support a pathologic cellular state in cancer. The repressive function of PRC2, in this case directed towards genes that restrict self-renewal like Cdkn2a, acts complementary to MLL-AF9, which directly activates expression of genes that promote self-renewal like HoxA9. Importantly, inhibition of PRC2 has only a minimal impact on the expression of well-established direct target genes of MLL-AF9, which also harbor minimal levels of H3K27 methylation at their promoters (Supplementary Figure 6), suggesting it is the segregation of each pathway to distinct target sites in the genome which can enable a complementary function in this disease. This is in stark contrast to the antagonism generally seen between Trithorax and Polycomb proteins when they intersect at common target genes. It should be noted that the presence of cooperating mutations in NRAS might also influence the collaborative actions of MLL-AF9 and PRC2 seen in this experimental system. While NRAS mutations are among the most common mutations that co-occur with MLL-translocations (24), the presence of alternative cooperating mutations (e.g. FLT3ITD) might impose a different degree of PRC2 dependency, an issue that could be addressed in the future by examining other genetically-engineered models of MLL-fusion leukemia.
The Polycomb Repressive Complex 1 (PRC1) subunit Cbx8 has recently been reported to interact directly with AF9, thereby supporting the function of MLL-AF9 in leukemogenesis (43). In this case, Cbx8 functions independently of the repressive PRC1 complex, acting instead to support trans-activation by MLL-AF9 (43). While this mechanism is entirely distinct from the repressive function of PRC2 described here, both studies highlight collaboration between Trithorax and Polycomb proteins in the setting of cancer, which may represent an emerging model of epigenetic dysfunction driving tumorigenesis.
A recurring theme in the study of cancer epigenetics is the duality of many chromatin regulators in displaying both tumor-promoting and tumor-suppressing activities, depending on cellular context. For example, the inactivating mutations of Ezh2 seen in MDS suggest a tumor-suppressor function in this disease (18, 19), in contrast to the tumor-maintenance role in MLL-fusion AML described here and that observed previously in several human AML cell lines (44, 45). The repressive landscape of H3K27me3 is known to be both plastic as well as heritable in nature, mediated by the ability of Eed to recruit PRC2 to sites of preexisting H3K27me3 (46, 47). Based on these epigenetic properties, we speculate that myeloid cells can select for, and thereafter maintain, a global configuration of repressive H3K27me3 that complements the specific genetic-lesions driving leukemic transformation. Consistent with such a model, we see significant differences in levels of H3K27me3 at many promoters when comparing MLL-fusion leukemia to 32D cells (Supplementary Figure 7), suggesting variable patterns of PRC2 recruitment in different myeloid contexts. Additionally, an important feature of the mouse model used here is that an extremely aggressive disease is initiated by high-level expression of two potent oncogenes, a context that would be expected to differ demonstrably from the evolution of MDS (48). Hence, it is a possibility that PRC2 plays a maintenance role in primary AML by buffering against stress induced as a secondary consequence of potent oncogenes (41). This is further suggested by our identification of Cdkn2a as a key downstream repression target of PRC2, a finding also seen in a prior study examining PRC2 function in human AML cell lines (45). Nevertheless, since a knockout of Ezh2 in adult mouse bone marrow does not significantly interfere with hematopoietic output, the possibility exists for PRC2 to be a therapeutic-target in MLL-leukemia (49). Elucidating how such epigenetic dependencies emerge and evolve during tumorigenesis will likely have a direct impact on the clinical implementation of novel epigenetics-based therapies, which are currently under intense development as anti-cancer drug candidates.
Supplementary Material
Acknowledgments
We thank J. Simon, E. Earl and L. Bianco for support with mouse work; S. Hearn for microscopy support; G. Hannon laboratory for support of shRNA screening methodology; and G. Blobel for comments on the manuscript. C.R.V., J.S., E.W., and M.T. were supported by the Don Monti Memorial Research Foundation, Laurie Strauss Leukemia Foundation, Sass Foundation, Edward P. Evans Foundation, and F.M. Kirby Foundation for research support. J.Z. was supported by a research fellowship from the German Research Foundation (DFG) and by the Andrew Seligson Memorial Clinical Fellowship at CSHL; A.R.R. was supported by an NIH traineeship and the Barbara McClintock fellowship. S.W.L. is supported by a Specialized Center of Research (SCOR) grant from the Leukemia and Lymphoma Society of America, a Cancer Target Discovery and Development (CTD2) grant from the National Cancer Institute, and by the Howard Hughes Medical Institute.
Footnotes
Conflict of interest statement
The authors declare no conflict of interest.
References
- 1.Schuettengruber B, Martinez AM, Iovino N, Cavalli G. Trithorax group proteins: switching genes on and keeping them active. Nat Rev Mol Cell Biol. 2011;12(12):799–814. doi: 10.1038/nrm3230. [DOI] [PubMed] [Google Scholar]
- 2.Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009 Oct;10(10):697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 3.Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978 Dec 7;276(5688):565–70. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- 4.Kennison JA, Tamkun JW. Dosage-dependent modifiers of polycomb and antennapedia mutations in Drosophila. Proc Natl Acad Sci U S A. 1988 Nov;85(21):8136–40. doi: 10.1073/pnas.85.21.8136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bracken AP, Helin K. Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nat Rev Cancer. 2009 Nov;9(11):773–84. doi: 10.1038/nrc2736. [DOI] [PubMed] [Google Scholar]
- 6.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002 Nov 1;298(5595):1039–43. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 7.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002 Nov 15;16(22):2893–905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, et al. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002 Oct 18;111(2):197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- 9.Shen X, Liu Y, Hsu YJ, Fujiwara Y, Kim J, Mao X, et al. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell. 2008 Nov 21;32(4):491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007 Jun 29;129(7):1311–23. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008 Oct;4(10):e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006 May 18;441(7091):349–53. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 13.Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006 Apr 21;125(2):301–13. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ezhkova E, Pasolli HA, Parker JS, Stokes N, Su IH, Hannon G, et al. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell. 2009 Mar 20;136(6):1122–35. doi: 10.1016/j.cell.2008.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002 Oct 10;419(6907):624–9. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 16.Yap DB, Chu J, Berg T, Schapira M, Cheng SW, Moradian A, et al. Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood. 2011 Feb 24;117(8):2451–9. doi: 10.1182/blood-2010-11-321208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wigle TJ, Knutson SK, Jin L, Kuntz KW, Pollock RM, Richon VM, et al. The Y641C mutation of EZH2 alters substrate specificity for histone H3 lysine 27 methylation states. FEBS Lett. 2011 Oct 3;585(19):3011–4. doi: 10.1016/j.febslet.2011.08.018. [DOI] [PubMed] [Google Scholar]
- 18.Ernst T, Chase AJ, Score J, Hidalgo-Curtis CE, Bryant C, Jones AV, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet. 2010 Aug;42(8):722–6. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- 19.Nikoloski G, Langemeijer SM, Kuiper RP, Knops R, Massop M, Tonnissen ER, et al. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat Genet. 2010 Aug;42(8):665–7. doi: 10.1038/ng.620. [DOI] [PubMed] [Google Scholar]
- 20.Yu BD, Hess JL, Horning SE, Brown GA, Korsmeyer SJ. Altered Hox expression and segmental identity in Mll-mutant mice. Nature. 1995 Nov 30;378(6556):505–8. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]
- 21.Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007 Nov;7(11):823–33. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- 22.Ayton PM, Cleary ML. Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins. Oncogene. 2001 Sep 10;20(40):5695–707. doi: 10.1038/sj.onc.1204639. [DOI] [PubMed] [Google Scholar]
- 23.Gilliland DG, Jordan CT, Felix CA. The molecular basis of leukemia. Hematology Am Soc Hematol Educ Program. 2004:80–97. doi: 10.1182/asheducation-2004.1.80. [DOI] [PubMed] [Google Scholar]
- 24.Zuber J, Radtke I, Pardee TS, Zhao Z, Rappaport AR, Luo W, et al. Mouse models of human AML accurately predict chemotherapy response. Genes Dev. 2009 Apr 1;23(7):877–89. doi: 10.1101/gad.1771409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011 Oct 27;478(7370):524–8. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yokoyama A, Somervaille TC, Smith KS, Rozenblatt-Rosen O, Meyerson M, Cleary ML. The menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis. Cell. 2005 Oct 21;123(2):207–18. doi: 10.1016/j.cell.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 27.Maillard I, Chen YX, Friedman A, Yang Y, Tubbs AT, Shestova O, et al. Menin regulates the function of hematopoietic stem cells and lymphoid progenitors. Blood. 2009 Feb 19;113(8):1661–9. doi: 10.1182/blood-2009-01-135012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsai S, Bartelmez S, Sitnicka E, Collins S. Lymphohematopoietic progenitors immortalized by a retroviral vector harboring a dominant-negative retinoic acid receptor can recapitulate lymphoid, myeloid, and erythroid development. Genes Dev. 1994 Dec 1;8(23):2831–41. doi: 10.1101/gad.8.23.2831. [DOI] [PubMed] [Google Scholar]
- 29.Weiss MJ, Keller G, Orkin SH. Novel insights into erythroid development revealed through in vitro differentiation of GATA-1 embryonic stem cells. Genes Dev. 1994 May 15;8(10):1184–97. doi: 10.1101/gad.8.10.1184. [DOI] [PubMed] [Google Scholar]
- 30.Pasini D, Bracken AP, Jensen MR, Lazzerini Denchi E, Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004 Oct 13;23(20):4061–71. doi: 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Montgomery ND, Yee D, Chen A, Kalantry S, Chamberlain SJ, Otte AP, et al. The murine polycomb group protein Eed is required for global histone H3 lysine-27 methylation. Curr Biol. 2005 May 24;15(10):942–7. doi: 10.1016/j.cub.2005.04.051. [DOI] [PubMed] [Google Scholar]
- 32.Zuber J, McJunkin K, Fellmann C, Dow LE, Taylor MJ, Hannon GJ, et al. Toolkit for evaluating genes required for proliferation and survival using tetracycline-regulated RNAi. Nat Biotechnol. 2010 Jan;29(1):79–83. doi: 10.1038/nbt.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Somervaille TC, Cleary ML. Identification and characterization of leukemia stem cells in murine MLL-AF9 acute myeloid leukemia. Cancer Cell. 2006 Oct;10(4):257–68. doi: 10.1016/j.ccr.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 34.Krivtsov AV, Twomey D, Feng Z, Stubbs MC, Wang Y, Faber J, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006 Aug 17;442(7104):818–22. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- 35.Zuber J, Rappaport AR, Luo W, Wang E, Chen C, Vaseva AV, et al. An integrated approach to dissecting oncogene addiction implicates a Myb-coordinated self-renewal program as essential for leukemia maintenance. Genes Dev. 2011 Aug 1;25(15):1628–40. doi: 10.1101/gad.17269211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zeisig BB, Milne T, Garcia-Cuellar MP, Schreiner S, Martin ME, Fuchs U, et al. Hoxa9 and Meis1 are key targets for MLL-ENL-mediated cellular immortalization. Mol Cell Biol. 2004 Jan;24(2):617–28. doi: 10.1128/MCB.24.2.617-628.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Somervaille TC, Matheny CJ, Spencer GJ, Iwasaki M, Rinn JL, Witten DM, et al. Hierarchical maintenance of MLL myeloid leukemia stem cells employs a transcriptional program shared with embryonic rather than adult stem cells. Cell Stem Cell. 2009 Feb 6;4(2):129–40. doi: 10.1016/j.stem.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nigten J, Breems-de Ridder MC, Erpelinck-Verschueren CA, Nikoloski G, van der Reijden BA, van Wageningen S, et al. ID1 and ID2 are retinoic acid responsive genes and induce a G0/G1 accumulation in acute promyelocytic leukemia cells. Leukemia. 2005 May;19(5):799–805. doi: 10.1038/sj.leu.2403699. [DOI] [PubMed] [Google Scholar]
- 39.Attardi LD, Reczek EE, Cosmas C, Demicco EG, McCurrach ME, Lowe SW, et al. PERP, an apoptosis-associated target of p53, is a novel member of the PMP-22/gas3 family. Genes Dev. 2000 Mar 15;14(6):704–18. [PMC free article] [PubMed] [Google Scholar]
- 40.Hellborg F, Qian W, Mendez-Vidal C, Asker C, Kost-Alimova M, Wilhelm M, et al. Human wig-1, a p53 target gene that encodes a growth inhibitory zinc finger protein. Oncogene. 2001 Sep 6;20(39):5466–74. doi: 10.1038/sj.onc.1204722. [DOI] [PubMed] [Google Scholar]
- 41.Williams RT, Sherr CJ. The INK4-ARF (CDKN2A/B) locus in hematopoiesis and BCR-ABL-induced leukemias. Cold Spring Harb Symp Quant Biol. 2008;73:461–7. doi: 10.1101/sqb.2008.73.039. [DOI] [PubMed] [Google Scholar]
- 42.Dickins RA, McJunkin K, Hernando E, Premsrirut PK, Krizhanovsky V, Burgess DJ, et al. Tissue-specific and reversible RNA interference in transgenic mice. Nat Genet. 2007 Jul;39(7):914–21. doi: 10.1038/ng2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tan J, Jones M, Koseki H, Nakayama M, Muntean AG, Maillard I, et al. CBX8, a Polycomb Group Protein, Is Essential for MLL-AF9-Induced Leukemogenesis. Cancer Cell. 2011 Nov 15;20(5):563–75. doi: 10.1016/j.ccr.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Villa R, Pasini D, Gutierrez A, Morey L, Occhionorelli M, Vire E, et al. Role of the polycomb repressive complex 2 in acute promyelocytic leukemia. Cancer Cell. 2007 Jun;11(6):513–25. doi: 10.1016/j.ccr.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 45.Fiskus W, Wang Y, Sreekumar A, Buckley KM, Shi H, Jillella A, et al. Combined epigenetic therapy with the histone methyltransferase EZH2 inhibitor 3-deazaneplanocin A and the histone deacetylase inhibitor panobinostat against human AML cells. Blood. 2009 Sep 24;114(13):2733–43. doi: 10.1182/blood-2009-03-213496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ, 3rd, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009 Oct 8;461(7265):762–7. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A, et al. A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol. 2008 Nov;10(11):1291–300. doi: 10.1038/ncb1787. [DOI] [PubMed] [Google Scholar]
- 48.Bejar R, Stevenson K, Abdel-Wahab O, Galili N, Nilsson B, Garcia-Manero G, et al. Clinical effect of point mutations in myelodysplastic syndromes. N Engl J Med. 2011 Jun 30;364(26):2496–506. doi: 10.1056/NEJMoa1013343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mochizuki-Kashio M, Mishima Y, Miyagi S, Negishi M, Saraya A, Konuma T, et al. Dependency on the polycomb gene Ezh2 distinguishes fetal from adult hematopoietic stem cells. Blood. 2011 Dec 15;118(25):6553–61. doi: 10.1182/blood-2011-03-340554. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.