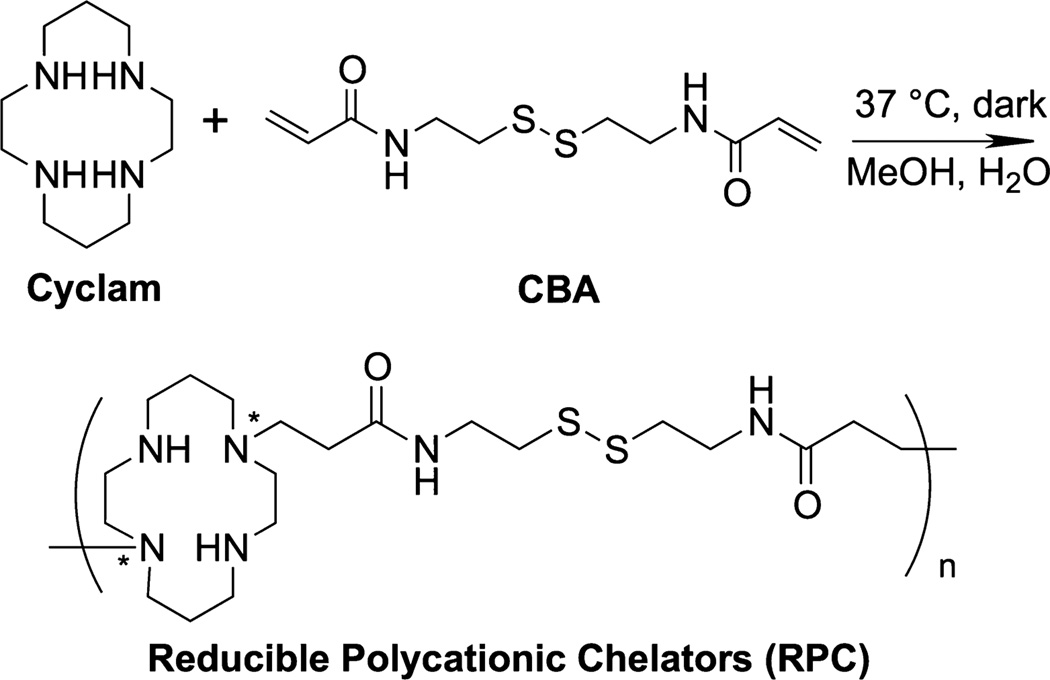
Abstract
A series of reducible polycationic copper chelators (RPCs) based on 1,4,8,11-tetraazacyclotetradecane (cyclam) were synthesized by Michael addition. Molecular weight of the polycations was controlled by reaction stoichiometry and reaction conditions, resulting in polymers with molecular weights ranging from 4400 to 13 800. The cyclam moieties in the polycations retained their ability to form complexes with Cu(II). The presence of disulfide bonds in the polycations resulted in substantially lower cytotoxicity than control 25 kDa poly-(ethyleneimine). RPC as well as their complexes with Cu(II) exhibited high transfection activity in vitro. The reported polycationic Cu(II) chelates represent promising nucleic acid delivery vectors with potential for future theranostic applications.
INTRODUCTION
Polyplexes based on bioreducible polycations are among the most investigated biodegradable systems in gene delivery. The main benefits of bioreducible polycations include reduced toxicity and, compared with hydrolytically degradable polycations, better spatial control of disassembly and release of DNA that is localized predominantly to the cytoplasm and nucleus.1–3 The intracellular degradation is also a known contributing factor to the decreased toxicity.2,4 Improved spatial control of polyplex disassembly is known to enhance transfection of multiple types of nucleic acids, including plasmid DNA, mRNA, siRNA, and shRNA in a number of cancer cell lines.5–8 Bioreducible polyplexes take advantage of the strong reducing intracellular environment mediated by small redox molecules like glutathione (GSH) and intracellular protein thiols.9–12 After the polyplexes are endocytosed into cells and enter into the cytoplasm or nucleus, the bioreducible polycations are broken down by thiol-disulfide exchange reactions, and the nucleic acids are released intracellularly. In addition to intracellular reduction, interactions of the polycation disulfides with cell surface thiols have been implicated in an increased cellular uptake of the bioreducible polyplexes.13
Cyclam (1,4,8,11-tetraazacyclotetradecane) is a well-known macrocyclic ligand with four secondary amines that forms highly stable complexes with virtually all transition metal ions. Macrocyclic chelators and their metal complexes have numerous biomedical applications, including positron emission tomography (PET) and magnetic resonance imaging (MRI).14–16 The in vivo stability of the macrocyclic metal chelates is significantly enhanced over acyclic chelators such as ethylenediaminetetraacetic acid (EDTA).14 Derivatives of cyclam and its relative cyclen (1,4,7,10-tetraazacyclododecane) have been the subject of intensive research in the development of PET imaging probes using Cu radioisotopes.15,17–19 Benefits of cyclams in PET imaging include high complexation affinity and high stability, which substantially benefit the systemic delivery of these imaging probes.14 At the same time, 64Cu shows great promise in PET imaging and targeted radiotherapy due to its half life (t1/2 = 12.7 h), decay characteristics (β+ (19%); β− (40%)), and easy large-scale production.14,20 Antibody-conjugated macrocyclic chelators have been reported to complex with 67Cu(II) (t1/2 = 62 h; β− (100%)) due to their high in vivo stability.14,21 Complexes of 64Cu with bicyclam compound AMD3100 have been shown to be effective and stable PET imaging agents suitable for tracking CXCR4 positive tumors in vivo.22,23 The ability to form metal complexes combined with the cationic nature at physiological pH make cyclam a suitable building block for designing nucleic acid delivery vectors with the possibility of easy labeling for simultaneous in vivo PET imaging. To the best of our knowledge, there are no previous reports of cyclam-based polycations used in gene delivery, although several reports describe incorporation of cyclen in gene delivery systems but their metal chelates were not described.24–28
The goal of this study was to explore the possibility of using cyclam as the building block of reducible polycationic chelators (RPCs) as a novel type of theranostic vectors for combined gene delivery and future PET imaging. In this initial study, we report the synthesis and effect of Cu(II) complexation on physicochemical properties of RPC/DNA polyplexes and their gene delivery performance in vitro.
MATERIALS AND METHODS
Materials
N,N′-Cystaminebisacrylamide (CBA) was obtained from Polysciences (Warrington, PA). Cyclam was purchased from Alfa Aesar (Ward Hill, MA). Heparin (sodium salt) and 25 kDa branched poly(ethyleneimine) (PEI) were purchased from Sigma-Aldrich (St. Louis, MO). Plasmid DNA, gWiz high-expression luciferase (gWiz-Luc) containing luciferase reporter gene, was from Aldevron (Fargo, ND). Dulbecco’s modified eagle medium (DMEM), Dulbecco’s phosphate-buffered saline (PBS), fetal bovine serum (FBS), l-glutamine, and penicillin-streptomycin (Pen-Strep) solution were from Thermo Scientific (Waltham, MA). All other reagents and chemicals were obtained from Fisher Scientific or VWR International unless otherwise noted.
Synthesis and Characterization of RPC
RPC were synthesized by direct Michael addition polymerization of different molar ratios of cyclam and CBA. Calculated amounts of cyclam and CBA were weighted and dissolved in methanol/water mixture (v/v 7:3). Polymerization was allowed to proceed in the dark at 37 °C for 24−48 h, and the reaction was stopped before gelation occurred. The reaction mixture was then added dropwise to excess of 1.25 M HCl in ethanol so that pH of the mixture was kept around 3. The resulting precipitated RPC•HCl was then isolated by centrifugation and washed twice with ethanol to remove excess HCl, and the product was dried in vacuum. The polymers were then dissolved in water and further purified by dialysis against water for 2 days (MWCO 1000 for RPC/3 and MWCO 3500 for RPC/2 and 1.8). The polymers were then lyophilized after dialysis.
The polymers were analyzed by 1H NMR to confirm the completion of the reaction and polymer composition. Weight- (Mw) and number-average (Mn) molecular weights as well as polydispersity index (PDI) were determined by size exclusion chromatography (SEC) using Viscotek GPCmax chromatography system consisting of an autosampler, a pump, a CTO-10ASVP Shimadzu column oven, a refractive index detector, a low- and right-angle light scattering detector, and OmniSEC software for chromatographic data analysis/storage (Malvern Instruments, U.K.). The columns used in series were single-pore AquaGel columns (cat. no. PAA-202 and PAA-203) from PolyAnalytik (London, ON, Canada). Sodium acetate buffer (0.3 M, pH 5) was used as an eluent at a flow rate of 0.3 mL/min.
Determination of Degree of Branching
The degree of branching was estimated using a degradative method with LC-MS/MS detection following a published procedure.29 Two mg of each RPC was dissolved in 1 mL of sodium phosphate buffer (20 mM, pH 7.2) before adding 3.75 mg tris(2-carboxyethyl) phosphine (TCEP) as a reducing agent to cleave all disulfide bonds. The mixture was then stirred at room temperature for 1 h, followed by the addition of 2.5 mg of N-ethylmaleimide (NEM) to cap the free thiols. The reaction was then kept at room temperature for an additional 2 h. The resulting NEM-derivatized fragments were analyzed by AQUITY UPLC TQD system (Waters, MA). Chromatographic analysis was performed using ACQUITY UPLC BEH ShieldRP18 column (2.1 × 100 mm, 1.7 μm). A gradient solvent system consisted of 0.1% formic acid (FA) in acetonitrile (solvent B) and 0.1% FA in water (solvent A). The gradient was increased from 5 to 95% solvent B over 3 min at a flow rate of 0.2 mL/min. Spectra were acquired using single ion recording (SIR) mode for m/z = 457.46, 713.60, 969.79, and 1225.79. The ratio of degraded fragments of terminal (T), linear (L), single-branched (S) and double-branched (D), cyclam units was determined by peak integrations in LC-MS/MS chromatograms. The relative degree of branching (DB) was calculated using the following equation for AB3 type polymers:30
Formation of RPC Complexes with Cu(II)
Cu(II) complexes of RPC were formed by incubating polymer solutions with CuCl2 in 0.1 M sodium acetate buffer (pH 6.0) for 1 h at room temperature. Complex formation was evaluated from the changes in absorption spectra (400−800 nm) measured by Synergy 2 Microplate Reader (BioTek, VT).
Ethidium Bromide (EtBr) Exclusion Assay
The ability of RPC polycations to condense gWiz-Luc DNA was determined by EtBr exclusion assay by measuring the changes in EtBr/DNA fluorescence. DNA solution at a concentration of 20 μg/mL in 10 mM HEPES buffer (pH 7.4) was mixed with EtBr (1 μg/mL) and fluorescence was measured and set to 100% using an excitation wavelength of 540 nm and an emission wavelength of 590 nm. Fluorescence readings were then taken following a stepwise addition of a polycation solution, and the condensation curve for each polycation was constructed.
Preparation and Characterization of RPC Polyplexes
gWiz-Luc DNA solution in 10 mM HEPES (pH 7.4) was prepared to give a DNA concentration of 20 μg/mL in the polyplexes. Polyplexes were formed by adding a predetermined volume of polymer to achieve the desired polycation/DNA weight/weight (w/w) ratio and mixed by vigorous vortexing for 10 s. Polyplexes were further allowed to stand for 30 min prior to use. The determination of hydrodynamic diameters and zeta potentials of polyplexes was performed by dynamic light scattering. Results were expressed as mean ± standard deviation (S.D.) of 3−10 experimental runs.
Agarose Gel Electrophoresis
The disassembly of the polyplexes was examined by agarose gel electrophoresis using a previously published protocol.5 In brief, gWiz-Luc DNA polyplexes were incubated under indicated conditions of different concentrations of heparin with or without a reducing agent (either GSH or DTT) at 37 °C for 1 h. Samples were then loaded onto a 0.8% agarose gel containing 0.5 μg/mL EtBr and run for 75 min at 120 V in 0.5× Tris/Borate/EDTA (TBE) running buffer. The gel was then visualized under UV. The intensity of the bands corresponding to free DNA was analyzed by ImageJ.
Cell Culture
Human breast cancer cell line MDA-MB-231 was a kind gift from Dr. Jing Li, Karmanos Cancer Institute (Detroit, MI). The cells were maintained in RPMI 1640 medium supplemented with 10% FBS. Murine melanoma cell line B16F10 and human hepatocellular carcinoma cell line Hep G2 were purchased from ATCC (Manassas, VA). B16F10 cells were maintained in DMEM media supplemented with 10% FBS and Hep G2 cells were maintained in MEM media supplemented with 10% FBS. All the cells were cultured at 37 °C in 5% CO2 atmosphere.
Transfection of DNA Polyplexes
All transfection experiments were conducted in 48-well plates with cells at logarithmic growth phase following a previously published protocol.4 Cells were seeded at a density of 40 000 cells/well 24 h prior to transfection. On the day of transfection, cells were incubated with the polyplexes (DNA conc. 2.35 μg/mL) in 170 μL of serum-free or 10% FBS-containing media. After 4 h of incubation, polyplexes were completely removed, and the cells were cultured in complete culture medium for 24 h prior to measuring luciferase expression. The medium was discarded, and the cells were lysed in 100 μL of 0.5× cell culture lysis reagent buffer (Promega, Madison, WI) for 30 min. To measure the luciferase content, we automatically injected 100 μL of 0.5 mM luciferin solution into each well of 20 μL of cell lysate, and the luminescence was integrated over 10 s using Synergy 2 Microplate Reader (BioTek). Total cellular protein in the cell lysate was determined by the Bicinchoninic acid protein assay using calibration curve constructed with standard bovine serum albumin solutions (Pierce, Rockford, IL). Transfection activity was expressed as relative light units (RLU)/mg cellular protein ± SD of quadruplicate samples.
Cytotoxicity of Polycations
The toxicity of polycations was evaluated by MTS assay in MDA-MB-231 and Hep G2 cells. The cells were plated into 96-well microplates at a density of 20 000 cells/well. After 24 h, culture medium was replaced by 150 μL of serial dilutions of a polymer in serum-supplemented medium, and the cells were incubated for 24 h. Polymer solutions were aspirated and replaced by a mixture of 100 μL serum-free media and 20 μL of MTS reagent (CellTiter 96 aqueous non-radioactive cell proliferation assay, Promega). After 2 h of incubation, the absorbance was measured spectrophotometrically in Synergy 2 Microplate Reader (BioTek) at a wavelength of 490 nm. The relative cell viability (%) was calculated as [A]sample/[A]untreated × 100%. The IC50 values were calculated as polymer concentration, which inhibits the growth of 50% of cells relative to untreated cells. The IC50 values were calculated based on “log(inhibitor) vs response − absolute IC50” curve fitting procedure in GraphPad Prism, with constrains of Fifty = 50, Top = 100, and a formula Y = Bottom + (Top − Bottom)/(1 + 10^(LogIC50-X)*HillSlope + log((Top − Bottom)/(Fifty − Bottom) − 1))).
Statistical Analysis
Significant differences between two groups were determined using Student’s t test, and differences among multiple groups were determined using analysis of variance (ANOVA). A p value <0.05 was considered to be statistically significant in all cases.
RESULTS AND DISCUSSION
Synthesis and Characterization of RPC
A series of three RPCs was synthesized by direct copolymerization of cyclam and CBA. The polymerization was achieved by Michael addition of different molar ratios of a disulfide containing bisacrylamide CBA and cyclam (Scheme 1). Because cyclam contains four secondary amines available for the polymerization, it is important to identify reaction conditions to avoid gel formation. We first optimized the polymerization condition by testing different temperatures, solvents, and feeding ratios of the two monomers. We found that the polymerization is easy to control without rapid gel formation when conducted in methanol/water (v/v 7:3) at 37 °C. Changing the cyclam/CBA stoichiometry allowed us to control degree of branching of the synthesized polymers. Molar ratio of cyclam/CBA > 0.9 was needed to avoid gel formation within 24 h. In this study, three RPC polymers were obtained using molar feeding ratio of cyclam/CBA molar ratios of 1.5, 1, and 0.9, corresponding to amine/acrylamide functional group ratios of 3, 2, and 1.8, respectively. The reaction conditions for each RPC polymer are summarized in Table 1.
Scheme 1.
Synthesis of RPCa
a*Exact location of the CBA attachment could not be determined. Any of the secondary amines are susceptible to modification.
Table 1.
Synthesis and Characterization of RPC Polycations
| feeding ratio (amine to acrylamide) |
feeding ratio (cyclam to CBA) |
reaction time (h) |
final cyclam % |
Mn | Mw | PDI | degree of branching |
|
|---|---|---|---|---|---|---|---|---|
| RPC/3 | 3 | 1.5 | 48 | 56.6 | 3100 | 4400 | 1.41 | 0.07 |
| RPC/2 | 2 | 1 | 48 | 49.6 | 12 000 | 13 700 | 1.14 | 0.24 |
| RPC/1.8 | 1.8 | 0.9 | 24 | 47.8 | 12 900 | 13 800 | 1.07 | 0.26 |
The RPC polymers were analyzed by 1H NMR to confirm completion of the reaction by disappearance of the characteristic CBA acrylamide peaks (5.5 to 6.5 ppm). 1H NMR was also used to determine the composition of RPC. On the basis of the integration of the signature peaks (4H) from cyclam (1.8 to 2.0 ppm) and the signature peaks (4H) from CBA (3.4 to 3.6 ppm) (Figure 1A), the final molar content of cyclam in each RPC polymer product was determined, and the results are summarized in Table 1. The molecular weights and PDI determined by SEC are also listed in Table 1. RPC/3 had the lowest Mw of 4400, whereas the RPC/2 and 1.8 showed similar Mw values of 13 700 and 13 800, respectively. The SEC data also showed a decrease in PDI with decreasing cyclam content. However, the low PDI values for RPC/2 and 1.8 might be underestimated considering the nature of the reaction and the limitations of the SEC column to separate fully low-molecular-weight branched polycations.
Figure 1.
Characterization of RPC polycations. (A) Typical 1H NMR (D2O) spectrum (RPC/2). (B) Chemical structures and typical LC-MS/MS spectrum of reduced RPC/2 fragments.
Because there are four secondary amines on the cyclam ring available for reaction and quick gel formation at feeding ratio (amine to acrylamide) below 1.8 was observed, it was expected that branching occurs and varies depending on the feeding ratio of the two monomers. To estimate the degree of branching of the polycations, we broke down disulfide bonds in RPC by a thiol-free reducing agent TCEP, followed by capping the thiol group using NEM to form a stable thioether bond. The degradation fragments were then analyzed by LC-MS/MS. The chemical structure of all possible fragments and their mass after ionization (m/z) are listed in Figure 1B. All four possible degradation products could be found in all three RPC polymers. Figure 1C shows an example of the LC-MS/MS spectrum for RPC/2. Calculated degrees of branching of RPC polycations are listed in Table 1. The results show that RPC/3 exhibited the least branched structure when compared with the other two polycations. No significant difference in the degree of branching was found between RPC/2 and RPC/1.8.
Copper(II) Complexation of RPC Polymers
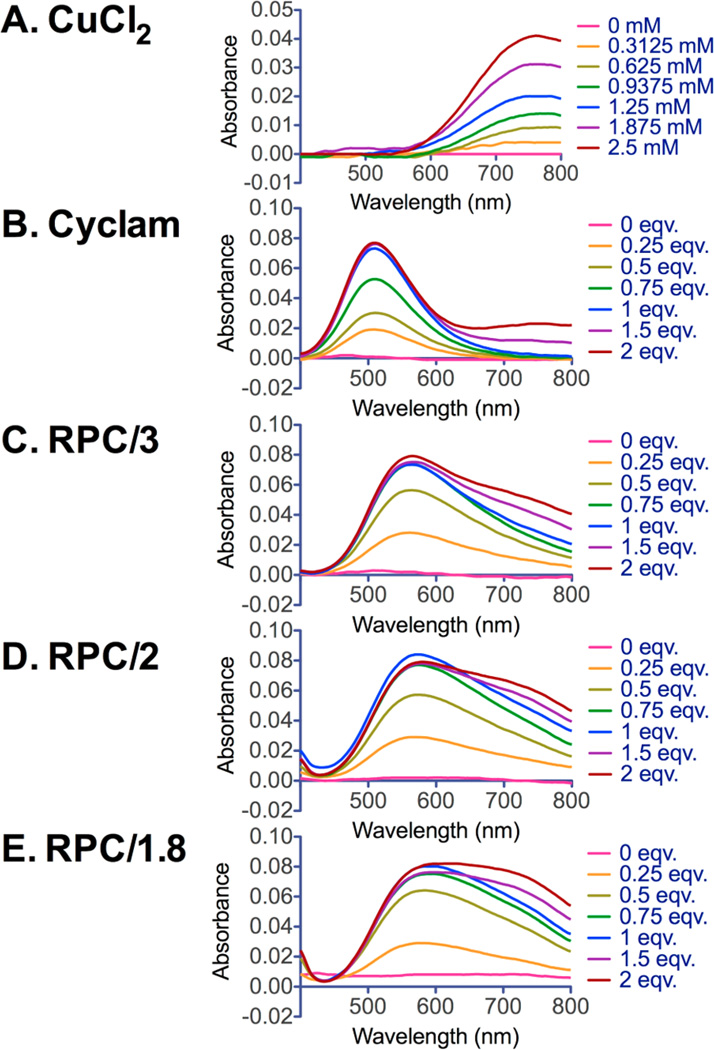
To explore the potential of the RPC polymers for PET imaging, we used nonradioactive CuCl2 in this preliminary study to evaluate if the cyclam moiety in RPC retains its ability to form complexes with Cu(II) (Scheme 2). Cu(II) complexes with RPC were easily prepared in acetate buffer and characterized by UV−vis spectroscopy (Figure 2). Cyclam was used as a positive control, and CuCl2 was used as a negative control for monitoring free uncomplexed Cu(II). The results show that for cyclam (Figure 2B) titration with CuCl2 led to the appearance of a peak with a maximum at 510 nm that reached a maximum absorbance when 1 equiv of CuCl2 was added. The sharp peak at the equivalent point indicates the formation of a well-defined uniform cyclam−metal complex.31 The addition of excess Cu(II) gives absorbance above 600 nm, which corresponds to the unbound CuCl2. All three RPC polymers (Figure 2C−E) showed absorption maximum at λmax = 570 nm that reached maximum absorbance at a nearly equimolar cyclam/Cu ratio, indicating that cyclam units in the RPC are sterically accessible to Cu(II) complexation. However, Cu(II) complexes of RPC exhibited broader peaks with λmax shifted to higher wavelengths, suggesting altered Cu(II) complexation ability of RPC caused by the compositional heterogeneity of the cyclam moieties in the polymers. RPC/1.8 exhibited the broadest absorption peak among the three polymers, a likely consequence of its highest degree of branching. Overall, all RPC polymers have retained their ability to form Cu(II) complexes, which makes them promising theranostic agents for future use in gene delivery combined with PET imaging.
Scheme 2.
Structure of the Complex of RPC with Copper(II)
Figure 2.
Cu(II) complexation of RPC polymers (C−E). Cyclam (B) was used as positive control and CuCl2 (A) was used to monitor free Cu(II). The absorption spectra were obtained by UV−vis spectroscopy after 1 h of incubation with CuCl2.
We next tested transfection activity of the RPC polycations and their Cu(II) complexes. Conformational changes caused by binding with transition-metal ions like Cu(II)32 increase rigidity of the cyclam ring due to locked positions of the secondary amines. We therefore hypothesized that the likely increased rigidity will affect DNA binding ability and polyplex formation of RPC. In addition to unusual sequence of protonation constants in cyclam (pK1 11.6, pK2 10.6, pK3 1.61 and pK4 2.42),33 complexation with Cu(II) is likely to increase the acidity of the amines in the cyclam ring and thus alter the overall protonation of the RPC. All of these effects are likely to influence the electrostatic interactions during the formation of DNA polyplexes, cytotoxicity of RPC, stability of the polyplexes against disassembly, and ultimately transfection activity of the polyplexes. Finally, there is a potential for increased toxicity of the Cu(II) complexes of RPC due to adverse effects of the heavy metal ion itself. Hence, we conducted a comprehensive evaluation of the physical properties and biological activity of the RPC polycations as well as their Cu(II) complexes. We have selected RPC with 50 and 100% of the cyclam moieties complexed with Cu(II) and compared the results obtained with copper-free RPC.
DNA Condensation
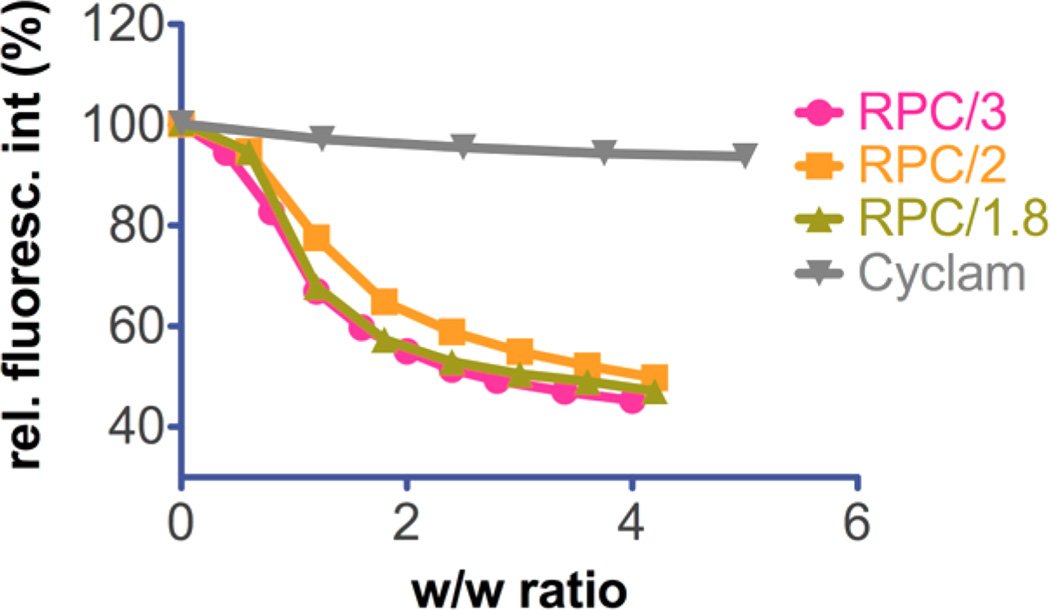
To test the DNA condensation capability of RPC and their Cu(II) complexes, we conducted EtBr exclusion assay (Figure 3). All RPCs were able to condense DNA fully at w/w > 2, as indicated by no further fluorescence decrease beyond this ratio, whereas the cyclam monomer failed to condense DNA into polyplexes. All three PRCs displayed similar condensation curves of typical sigmoidal shape. Condensation curves for the Cu(II)-complexed RPC polymers could not be reliably obtained because of the observed fluorescence quenching by the copper present. Therefore, the effect of Cu(II) complexation on improved binding of RPC could not be determined by this method.
Figure 3.
DNA condensation of RPC polymers and cyclam monomer by EtBr exclusion assay.
Particle Size and Zeta Potential of DNA Polyplexes
The physical properties of DNA/polycation complexes have crucial effect on both in vitro and in vivo biological performance. Polyplexes with appropriate size and surface charge greatly benefit the cellular uptake and gene transfer ability. Hydrodynamic size and zeta-potential of polyplexes were measured by DLS before every in vitro test. Representative particle size and zeta-potential data of three RPC polyplexes prepared at different w/w ratios are summarized in Table 2. The sizes of RPC/DNA polyplexes fell into a broad range of 80−205 nm. All polyplexes were positively charged with zeta potential ranging from 13 to 29 mV. No significant increase in the surface charge of the polyplexes was observed above the equivalent w/w ratio 2. Low zeta potential that was found in RPC/3 polyplexes prepared at lower w/w ratio is probably due to the low molecular weight of the polycation.
Table 2.
Size and Zeta-Potential of RPC/DNA Polyplexes at Different w/w Ratios.
| [−] Cu(II) |
[+] 50% Cu(II) |
[+] 100% Cu(II) |
|||||
|---|---|---|---|---|---|---|---|
| polymer | w/w | size (nm) | zeta potential (mV) | size (nm) | zeta potential (mV) | size (nm) | zeta potential (mV) |
| RPC/3 | 5 | 102 ± 2 | 13 ± 1 | ||||
| 10 | 124 ± 7 | 14 ± 2 | |||||
| 15 | 111 ± 3 | 17 ± 5 | 92 ± 1 | 18 ± 1 | 114 ± 1 | 16 ± 3 | |
| 20 | 114 ± 3 | 15 ± 3 | |||||
| 25 | 125 ± 2 | 17 ± 3 | |||||
| RPC/2 | 5 | 119 ± 2 | 21 ± 4 | ||||
| 10 | 125 ± 2 | 26 ± 2 | |||||
| 15 | 205 ± 9 | 16 ± 2 | 136 ± 6 | 26 ± 6 | 75 ± 0 | 17 ± 2 | |
| 20 | 167 ± 3 | 17 ± 2 | |||||
| 25 | 163 ± 3 | 16 ± 3 | |||||
| RPC/1.8 | 5 | 87 ± 2 | 16 ± 2 | ||||
| 10 | 90 ± 3 | 23 ± 4 | |||||
| 15 | 121 ± 3 | 29 ± 3 | 82 ± 10 | 20 ± 3 | 69 ± 2 | 19 ± 2 | |
| 20 | 81 ± 3 | 20 ± 2 | |||||
| 25 | 80 ± 1 | 17 ± 2 | |||||
After complexation with 50 and 100% Cu(II), no significant difference in surface charge of the polyplexes was observed. However, Cu(II)-complexed RPC/2 and RPC/1.8 polyplexes showed a decreasing trend in the hydrodynamic sizes with increasing metal complexation. This trend was not found in the Cu(II)-complexed RPC/3 polyplexes. The effect of copper complexation appears to be more pronounced in the polycations with higher degree of branching.
Effect of Reduction on Stability of Polyplexes against Polyelectrolyte Exchange
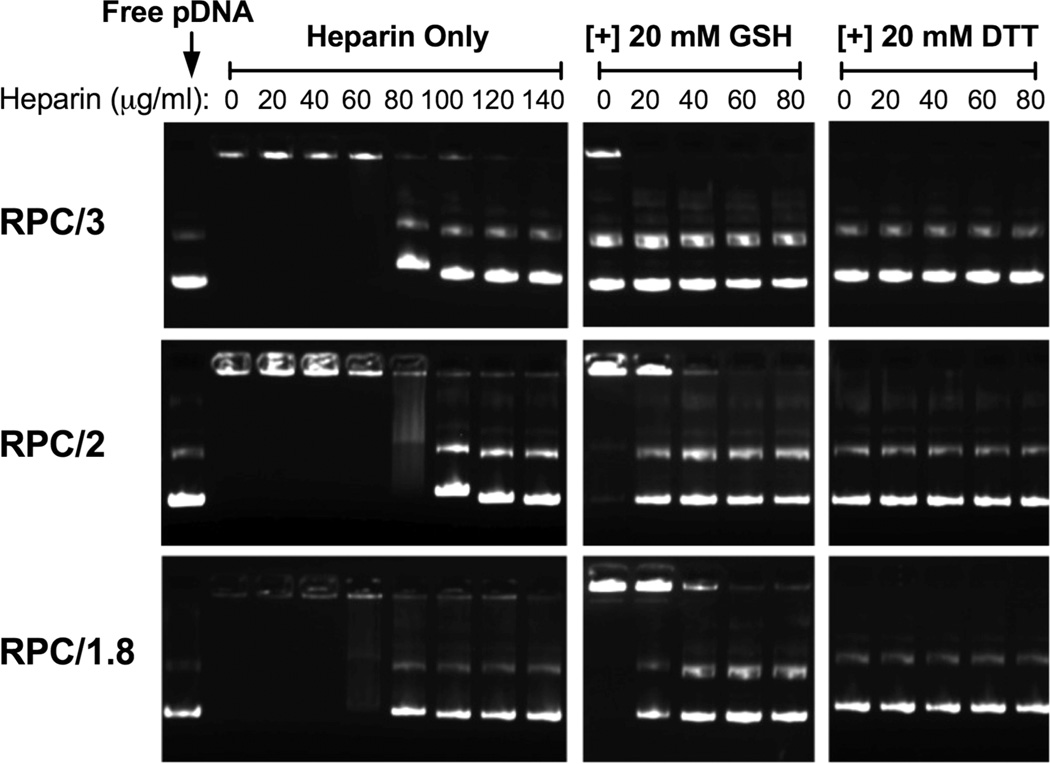
The effect of RPC disulfide reduction on the resistance of the polyplexes against polyelectrolyte exchange with a competing polyanion heparin was evaluated by agarose gel electrophoresis (Figure 4). In the absence of a reducing agent, the polyplexes showed first signs of destabilization at a heparin concentration of 80 μg/mL. Under reducing conditions of 20 mM GSH selected to mimic the redox status in the cell nuclei,34 PRC/3 polyplexes disassembled and DNA was released without involvement of heparin. Polyplexes formed with PRC of more branched structure did not release DNA, as readily as suggested by the fact that 20 mM GSH alone was not sufficient to destabilize the polyplexes. However, the addition of a low concentration of heparin (40 μg/mL) caused complete DNA release (disappearance of fluorescent signal in the well) from RPC/2 polyplexes, and a higher concentration of heparin was required to release DNA fully from RPC/1.8 polyplexes. Replacing GSH with a stronger reducing agent DTT resulted in complete DNA release from all three polyplexes without the presence of heparin. This result suggests that the differences in degree of the polycation branching affect the sensitivity to reduction-triggered disassembly of polyplexes, with the least branched polycations exhibiting the highest sensitivity to reduction.
Figure 4.
DNA release from polyplexes by agarose gel electrophoresis with heparin ∓20 mM GSH or DTT. All RPC/DNA polyplexes were prepared at w/w 5.
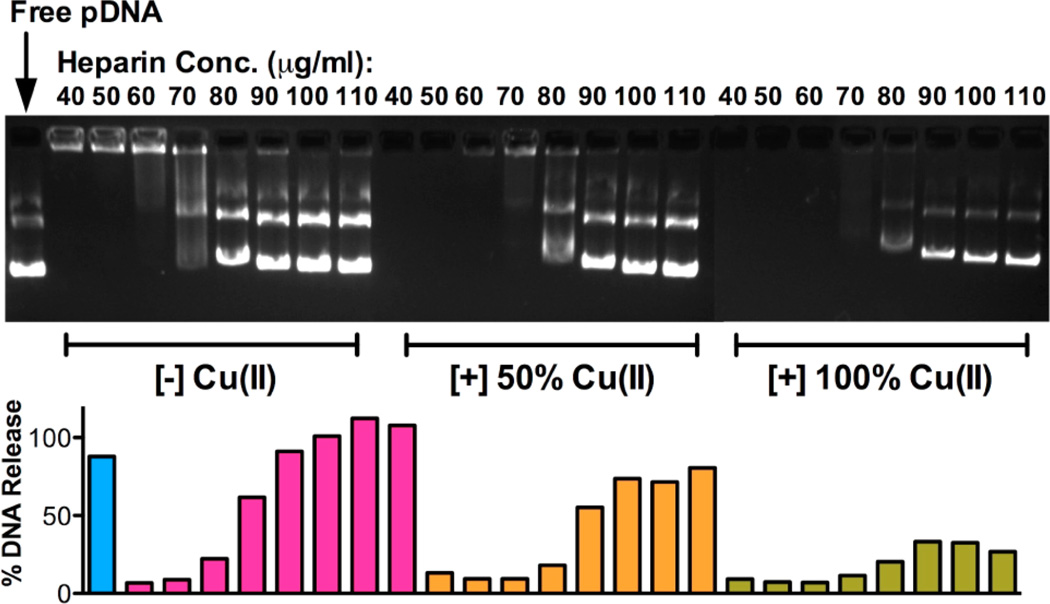
The heparin stability of polyplexes of RPC complexed with Cu(II) was tested using the same gel electrophoresis method. Figure 5 shows the result of RPC/1.8 polyplexes as an example. Cu(II)-free RPC/1.8 polyplexes started to show signs of disassembly and traces of DNA release at heparin concentration 70 μg/mL and complete DNA release above 80 μg/mL. For polyplexes prepared with RPC/1.8 complexed with Cu(II), only partial DNA release was observed at heparin concentration of 80 μg/mL (~80% for 50% Cu(II) polyplexes and ~30% for 100% Cu(II) polyplexes). This result suggests that the presence of Cu(II) increases the stability of polyplexes against polyanion exchanges. However, this might not benefit gene delivery efficiency of these RPC polyplexes due to increased difficulty in polyplex disassembly and DNA release.
Figure 5.
Stability of Cu(II) complexes of RPC/1.8 against heparin disassembly. The amount of DNA released from the polyplexes was quantified by analyzing the fluorescence intensity of the bands.
Transfection Efficiency
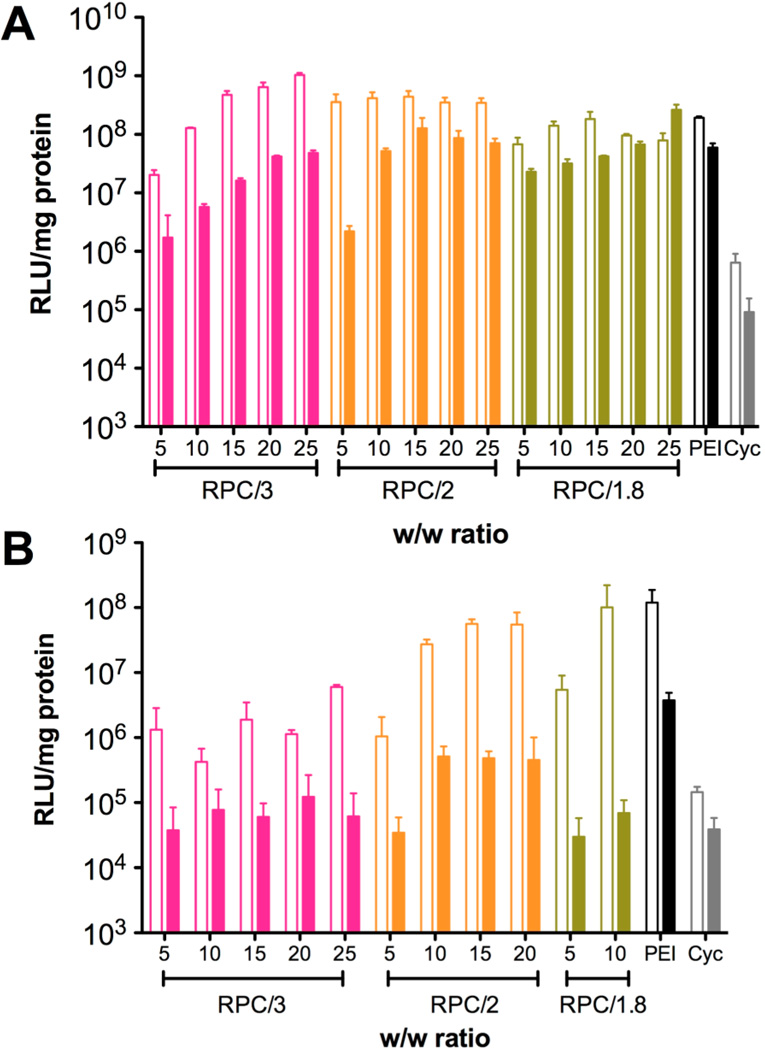
In vitro transfection activity of RPC polyplexes at different w/w ratios was evaluated in murine melanoma cell line B16F10 and human breast cancer cell line MDA-MB-231 (Figure 6). PEI polyplexes at w/w 1.2 (N/P 10) were used as a positive control. The results showed that in B16F10 cells, all RPC polyplexes exhibited high transfection activity comparable to PEI above a certain w/w ratio, whereas the cyclam monomer failed to mediate any significant transfection. The effect of w/w ratio on transfection activity was especially pronounced in PRC/3 polyplexes, which might be attributed to the low molecular weight of the polymer and high sensitivity to disulfide reduction. Serum usually has a negative effect on in vitro transfection activity due to the interaction of positively charged polyplexes with serum proteins like albumin.35–37 Here the influence of serum on transfection of RPC/1.8 polyplexes was not as significant as the negative effect observed for RPC/3 and RPC/2 polyplexes, especially when the polyplexes were prepared at high w/w ratio. In MDAMB-231 cells, the transfection activity of the RPC polymers was not as high as in B16F10 cells. Nevertheless, RPC/2 and RPC/1.8 still showed transfection activity comparable to PEI in serum-free condition. Cytotoxicity of RPC polyplexes was significantly higher in MDA-MB-231 cells than in B16F10 cells, and thus the transfection results for RPC/2 polyplexes prepared above w/w 20 and RPC/1.8 polyplexes prepared above w/w 10 are not included in Figure 6B.
Figure 6.
Transfection efficiency of RPC polyplexes prepared using different w/w ratio in (A) B16F10 cells and (B) MDA-MB-231 cells. All transfection experiments were conducted in serum-free (empty bars) and 10% FBS-containing medium (solid bars) during polyplex incubation.
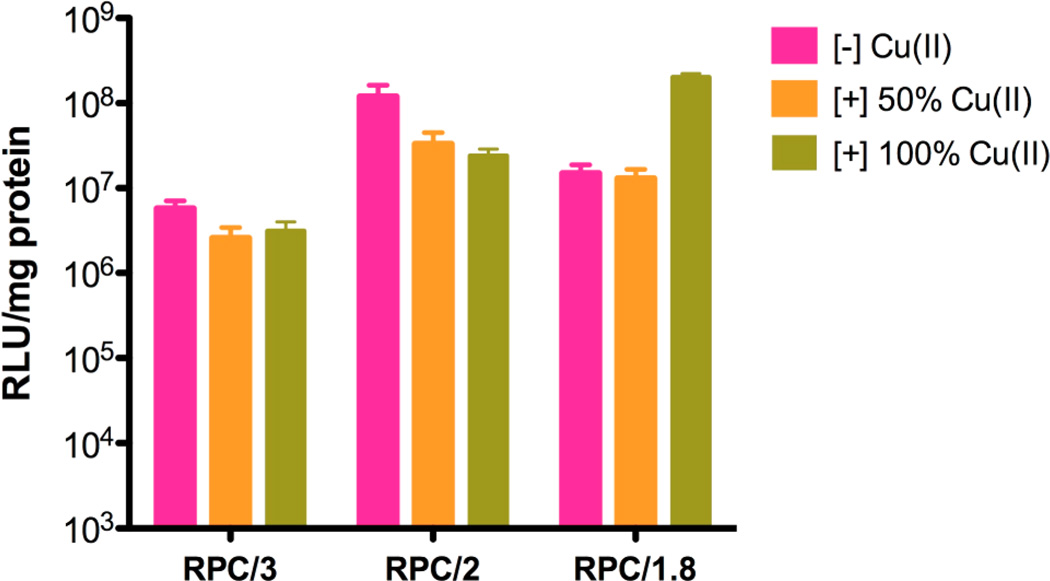
The in vitro luciferase transfection efficiency of 50 and 100% Cu(II)-complexed RPC polymers was evaluated as above using B16F10 cells in the presence of 10% serum. As shown in Figure 7, for RPC/3 and RPC/2 polyplexes, the transfection efficiency of both 50% Cu(II) and 100%(II) polyplexes showed a slight decrease (two- to five-fold), which might be attributed to the increased resistance of the copper-containing polyplexes against disassembly, as discussed above. However, transfection activity of RPC/1.8 complexed with 100% Cu(II) increased 20-fold when compared with Cu(II)-free polyplexes. Because the properties of RPC/2 and RPC/1.8 are similar, the most likely explanation for the observed transfection increase is the higher toxicity (decreased protein content in the cell lysate) of the copper complexes of RPC/1.8, as described below.
Figure 7.
Transfection efficiency of Cu(II) complexes of RPC polymers in B16F10 cells in the presence of serum. The RPC and Cu(II)-complexed RPC polyplexes were prepared at w/w 15.
Cytotoxicity
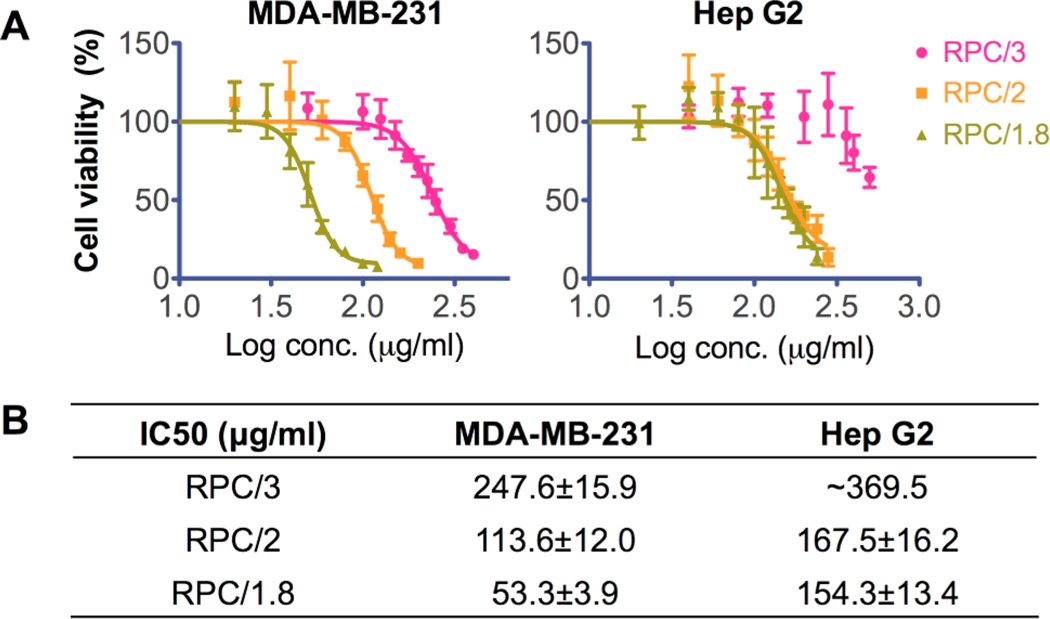
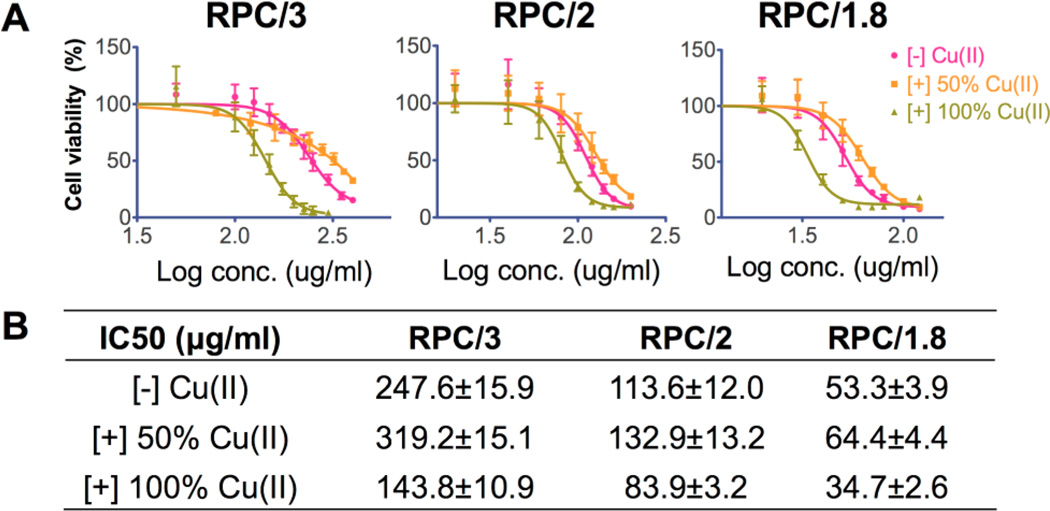
It is well recognized that bioreducible polycations exhibit significantly decreased cytotoxicity compared with nonreducible counterparts.2 Here the in vitro cytotoxicity of RPC polymers was evaluated by MTS assay in human breast cancer MDA-MB-231 cells because we found in the previous transfection experiments that these cells were more sensitive to the polycations. The cytotoxicity of RPC was also tested in human liver hepatocellular carcinoma Hep G2 cells, which is a well-recognized cell line model used for in vitro predictive toxicity screening38,39 (Figure 8). IC50 values of each polymer in these two cell lines are summarized in Figure 8B. The cytotoxicity of RPC polymers was significantly decreased compared with 25 kD PEI in MDA-MB-231 cells (IC50 14 μg/mL40). The cytotoxicity of RPC was even lower in Hep G2 cells compared with PEI (IC50 18 μg/mL41), suggesting a potentially decreased hepatic toxicity, which might benefit future in vivo applications. In both cell lines, RPC with higher molecular weight and higher degree of branching showed increased toxicity. Interestingly, in MDA-MB-231 cells, RPC/2 showed two-fold higher IC50 than RPC/1.8, whereas little difference was observed in Hep G2 cells, suggesting that the cytotoxicity of these polycations is cell-type-dependent.
Figure 8.
Cytotoxicity of RPC polycations in (A) MDA-MB-231 cells and Hep G2 cells by MTS assay. Calculated IC50 values of RPC are listed in the table (B).
The cytotoxicity of RPC complexed with Cu(II) was evaluated by MTS assay in MDA-MB-231 cells as above (Figure 9). For all the three polycations, 100% Cu(II)-complexed RPC polymers had significantly increased cytotoxicity compared with copper-free RPC. This is most likely due to increased damage to the cell membrane due to higher rigidity of the RPC complexed with copper. No toxicity was observed when the cells were treated with equivalent concentrations of free copper in the form of CuCl2. Interestingly, RPC complexed with 50% Cu(II) had significantly lower cytotoxicity compared with Cu(II)-free RPC in the case of all three polycations. The reason for this observation remains unclear but the fact that toxicity of the Cu(II) complexes is highly dependent on the degree of metal complexation needs to be taken into account when selecting appropriate doses of 64Cu(II) for future PET imaging.
Figure 9.
(A) Cell viability curves of Cu(II) complexes of RPC polycations in MDA-MB-231 cells by MTS assay. (B) IC50 values calculated from data in (A).
CONCLUSIONS
A new type of bioreducible cyclam-based polymeric chelators for combined gene delivery and potential PET imaging was developed. These RPC polycations could be easily synthesized by one-step Michael addition reaction, and they showed low cytotoxicity and high transfection efficiency in vitro. Transfection activity and cytotoxicity of RPC complexed with Cu(II) depend on the degree of Cu(II) complexation. The capability of RPC to form Cu(II) complexes makes them promising theranostic candidates for future use in gene delivery combined with PET imaging.
ACKNOWLEDGMENTS
This work was supported in part by a grant from the Office of Vice President for Research, Wayne State University and in part by NIH grant CA109711.
Footnotes
The authors declare no competing financial interest.
REFERENCES
- 1.Manickam DS, Hirata A, Putt DA, Lash LH, Hirata F, Oupicky D. Biomaterials. 2008;29:2680–2688. doi: 10.1016/j.biomaterials.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen J, Wu C, Oupicky D. Biomacromolecules. 2009;10:2921–2927. doi: 10.1021/bm900724c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin C, Zhong Z, Lok MC, Jiang X, Hennink WE, Feijen J, Engbersen JF. J. Controlled Release. 2006;116:130–137. doi: 10.1016/j.jconrel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 4.Read ML, Singh S, Ahmed Z, Stevenson M, Briggs SS, Oupicky D, Barrett LB, Spice R, Kendall M, Berry M, Preece JA, Logan A, Seymour LW. Nucleic Acids Res. 2005;33:e86. doi: 10.1093/nar/gni085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manickam DS, Li J, Putt DA, Zhou QH, Wu C, Lash LH, Oupicky D. J. Controlled Release. 2010;141:77–84. doi: 10.1016/j.jconrel.2009.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rahbek UL, Howard KA, Oupicky D, Manickam DS, Dong M, Nielsen AF, Hansen TB, Besenbacher F, Kjems J. J. Gene Med. 2008;10:81–93. doi: 10.1002/jgm.1120. [DOI] [PubMed] [Google Scholar]
- 7.Kim J, Nam HY, Kim TI, Kim PH, Ryu J, Yun CO, Kim SW. Biomaterials. 2011;32:5158–5166. doi: 10.1016/j.biomaterials.2011.03.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beloor J, Choi CS, Nam HY, Park M, Kim SH, Jackson A, Lee KY, Kim SW, Kumar P, Lee SK. Biomaterials. 2012;33:1640–1650. doi: 10.1016/j.biomaterials.2011.11.008. [DOI] [PubMed] [Google Scholar]
- 9.Jiang XM, Fitzgerald M, Grant CM, Hogg PJ. J. Biol. Chem. 1999;274:2416–2423. doi: 10.1074/jbc.274.4.2416. [DOI] [PubMed] [Google Scholar]
- 10.Donoghue N, Hogg PJ. Methods Enzymol. 2002;348:76–86. doi: 10.1016/s0076-6879(02)48628-4. [DOI] [PubMed] [Google Scholar]
- 11.Gilbert HF. J. Biol. Chem. 1997;272:29399–29402. doi: 10.1074/jbc.272.47.29399. [DOI] [PubMed] [Google Scholar]
- 12.Hansen RE, Roth D, Winther JR. Proc. Natl. Acad. Sci. U. S. A. 2009;106:422–427. doi: 10.1073/pnas.0812149106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li J, Manickam DS, Chen J, Oupicky D. Eur. J. Pharm. Sci. 2012;46:173–180. doi: 10.1016/j.ejps.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun X, Wuest M, Weisman GR, Wong EH, Reed DP, Boswell CA, Motekaitis R, Martell AE, Welch MJ, Anderson CJ. J. Med. Chem. 2002;45:469–477. doi: 10.1021/jm0103817. [DOI] [PubMed] [Google Scholar]
- 15.Boswell CA, Regino CA, Baidoo KE, Wong KJ, Milenic DE, Kelley JA, Lai CC, Brechbiel MW. Bioorg. Med. Chem. 2009;17:548–552. doi: 10.1016/j.bmc.2008.11.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murugesa S, Shetty SJ, Srivastava TS, Noronha OP, Samuel AM. Appl. Radiat. Isot. 2001;55:641–646. doi: 10.1016/s0969-8043(01)00113-0. [DOI] [PubMed] [Google Scholar]
- 17.Jin ZH, Furukawa T, Galibert M, Boturyn D, Coll JL, Fukumura T, Saga T, Dumy P, Fujibayashi Y. Nucl. Med. Biol. 2011;38:529–540. doi: 10.1016/j.nucmedbio.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 18.Leung K. Molecular Imaging and Contrast Agent Database (MICAD) Bethesda, MD: NCBI; 2004. [PubMed] [Google Scholar]
- 19.Sun X, Kim J, Martell AE, Welch MJ, Anderson CJ. Nucl. Med. Biol. 2004;31:1051–1059. doi: 10.1016/j.nucmedbio.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 20.Philpott GW, Schwarz SW, Anderson CJ, Dehdashti F, Connett JM, Zinn KR, Meares CF, Cutler PD, Welch MJ, Siegel BA. J. Nucl. Med. 1995;36:1818–1824. [PubMed] [Google Scholar]
- 21.Cole WC, DeNardo SJ, Meares CF, McCall MJ, DeNardo GL, Epstein AL, O’Brien HA, Moi MK. Int. J. Radiat. Appl. Instrum. Part B. 1986;13:363–368. doi: 10.1016/0883-2897(86)90011-5. [DOI] [PubMed] [Google Scholar]
- 22.Nimmagadda S, Pullambhatla M, Stone K, Green G, Bhujwalla ZM, Pomper MG. Cancer Res. 2010;70:3935–3944. doi: 10.1158/0008-5472.CAN-09-4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacobson O, Weiss ID, Szajek L, Farber JM, Kiesewetter DO. Bioorg. Med. Chem. 2009;17:1486–1493. doi: 10.1016/j.bmc.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang QD, Ren J, Ou WJ, Fu Y, Cai MQ, Zhang J, Zhu W, Yu XQ. Chem. Biol. Drug Des. 2012;79:879–887. doi: 10.1111/j.1747-0285.2012.01355.x. [DOI] [PubMed] [Google Scholar]
- 25.Huang QD, Zhong GX, Zhang Y, Ren J, Fu Y, Zhang J, Zhu W, Yu XQ. PLoS One. 2011;6:e23134. doi: 10.1371/journal.pone.0023134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu JL, Ma QP, Huang QD, Yang WH, Zhang J, Wang JY, Zhu W, Yu XQ. Eur. J. Med. Chem. 2011;46:4133–4141. doi: 10.1016/j.ejmech.2011.06.015. [DOI] [PubMed] [Google Scholar]
- 27.Huang QD, Ou WJ, Chen H, Feng ZH, Wang JY, Zhang J, Zhu W, Yu XQ. Eur. J. Pharm. Biopharm. 2011;78:326–335. doi: 10.1016/j.ejpb.2011.03.017. [DOI] [PubMed] [Google Scholar]
- 28.Xiang YZ, Feng ZH, Zhang J, Liao YL, Yu CJ, Yi WJ, Zhu W, Yu XQ. Org. Biomol. Chem. 2010;8:640–647. doi: 10.1039/b914877a. [DOI] [PubMed] [Google Scholar]
- 29.Christensen LV, Chang CW, Kim WJ, Kim SW, Zhong Z, Lin C, Engbersen JF, Feijen J. Bioconjugate Chem. 2006;17:1233–1240. doi: 10.1021/bc0602026. [DOI] [PubMed] [Google Scholar]
- 30.Holter D, Burgath A, Frey H. Acta Polym. 1997;48:30–35. [Google Scholar]
- 31.Lo AT, Salam NK, Hibbs DE, Rutledge PJ, Todd MH. PLoS One. 2011;6:e17446. doi: 10.1371/journal.pone.0017446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paisey SJ, Sadler PJ. Chem. Commun. (Cambridge U. K.) 2004:306–307. doi: 10.1039/b312752b. [DOI] [PubMed] [Google Scholar]
- 33.Hancock RD, Motekaitis RJ, Mashishi J, Cukrowski I, Reibenspies JH, Martell AE. J. Chem. Soc. Perkin Trans. . 1996;2:1925–1929. [Google Scholar]
- 34.Soundara Manickam D, Oupicky D. J. Drug Targeting. 2006;14:519–526. doi: 10.1080/10611860600834409. [DOI] [PubMed] [Google Scholar]
- 35.Li S, Wang Y, Wang S, Zhang J, Wu SF, Wang BL, Zhu W, Yu XQ. Bioorg. Med. Chem. 2012;20:1380–1387. doi: 10.1016/j.bmc.2012.01.016. [DOI] [PubMed] [Google Scholar]
- 36.Zheng M, Zhong Z, Zhou L, Meng F, Peng R, Zhong Z. Biomacromolecules. 2012;13:881–888. doi: 10.1021/bm2017965. [DOI] [PubMed] [Google Scholar]
- 37.Dong Y, Li J, Wu C, Oupicky D. Pharm. Res. 2010;27:1927–1938. doi: 10.1007/s11095-010-0197-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mersch-Sundermann V, Knasmuller S, Wu XJ, Darroudi F, Kassie F. Toxicology. 2004;198:329–340. doi: 10.1016/j.tox.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 39.Bokhari M, Carnachan RJ, Cameron NR, Przyborski SA. J. Anat. 2007;211:567–576. doi: 10.1111/j.1469-7580.2007.00778.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y, Wang LS, Goh SH, Yang YY. Biomacromolecules. 2007;8:1028–1037. doi: 10.1021/bm061051c. [DOI] [PubMed] [Google Scholar]
- 41.Wang Y, Gao S, Ye WH, Yoon HS, Yang YY. Nat. Mater. 2006;5:791–796. doi: 10.1038/nmat1737. [DOI] [PubMed] [Google Scholar]