Abstract
Background/Aims
Despite numerous studies on the relation of genetic polymorphisms with irritable bowel syndrome (IBS), the results still remain inconclusive. The aim of this study was to assess the possible association between SLC6A4 serotonin transporter gene linked polymorphic region (5-HTTLPR), ADRA2A −1291C>G, GNB3 825C>T, CCK1R intron 779T>C and TRPV1 945G>C polymorphisms and IBS based on Rome III criteria in Korea.
Methods
Study subjects were prospectively recruited from visitors to Seoul National University Bundang Hospital between July 2009 and January 2014. Ninety-nine IBS patients and 171 healthy controls were enrolled. Polymorphisms of above-mentioned 5 genes were genotyped. Serum serotonin from 101 participants was measured by ELISA and compared according to SLC6A4 5-HTTLPR polymorphisms and IBS subtypes.
Results
Regarding SLC6A4 5-HTTLPR polymorphism, L/L genotype was significantly associated with the total IBS, constipation predominant IBS (IBS-C) and mixture of diarrhea and constipation IBS (IBS-M) (adjusted OR: 4.35, 95% CI: 1.04–16.67; adjusted OR: 11.11, 95% CI: 1.69–50.00 and adjusted OR: 5.56, 95% CI: 1.05–33.33, respectively). Carrying ADRA2A −1291G allele was significantly associated with total IBS and diarrhea predominant IBS (adjusted OR: 3.37, 95% CI: 1.16–9.77 and adjusted OR: 5.64, 95% CI: 1.18–27.01, respectively). IBS-C patients showed reduced level of serum serotonin compared to controls and patients with diarrhea predominant IBS (50.2 ng/mL vs. 69.0 ng/mL and 92.9 ng/mL, P = 0.017 and P = 0.001, respectively).
Conclusions
Genetic polymorphisms of SLC6A4 5-HTTLPR and ADRA2A −1291C>G could be one of the pathophysiological factors of IBS in Korea. Reduced serum serotonin shown in the IBS-C group suggested a role of serotonin in IBS, but large study is needed for confirming genotypic difference in serum serotonin level.
Keywords: Irritable bowel syndrome; Polymorphism, genetic; Receptors, adrenergic; Polymorphism, single nucleotide; Serotonin
Introduction
Irritable bowel syndrome (IBS) is one of the most common gastrointestinal functional disorder.1 The prevalence of IBS widely ranges between 10% and 20% of the population according to Western studies.2,3 This has been reported to induce abdominal pain or discomfort chronically, and impair quality of life (QOL), raising enormous healthcare expenditure.4,5
Although the etiology of IBS remains largely unknown, motility disturbances, visceral hypersensitivity, genetic variation, psychological distress, altered bacterial flora and immune dysregulation, have been postulated as a possible candidate.6,7 IBS has been also shown a familial clustering, but not in manner of major Mendelian inheritance. Rather, it has been considered as a complex genetic disorder, in which the multiple genetic variants on several genes contribute the development of this condition. Up to now, more than 60 genes have been evaluated to determine whether specific genetic variants may be associated with IBS.7 Among these genetic variants, the genetic polymorphisms of serotonin transporter gene linked polymorphic region (5-HTTLPR) have been most widely evaluated in IBS.8,9 The serotonin transporter protein (SERT) is encoded by a single gene (SLC6A4) on chromosome17q11, and 44 bp insertion/deletion in 5-HTTLPR creates a short (S) and a long (L) allele which show the different transcriptional efficiency.10 In addition to 5-HTTLPR polymorphism, with respect to colonic motility and sensation, the polymorphisms of G-protein β3 (GNB3) 825C>T, alpha 2A adrenergic receptor, ADRA2A −1291C>G, cholecystokinin receptor 1 (CCK1R) intron 779T>C and transient receptor potential ion channel of the vanilloid type 1 (TRPV1) 945G>C were recently investigated.11–15
The genetic variation in the adrenergic receptor such as ADRA2A −1291C>G has been proposed as a mechanism for the modification of motor and sensory functions in IBS patients.16,17
Another protein of interest is the G-protein. Common GNB3-825C>T polymorphism has been believed to be associated with enhanced G-protein activation,18 and there are several studies which indicate a significant association with functional dyspepsia.19,20 Meanwhile, the possible relation between cholecystokinin (CCK) and IBS was first reported by Harvey et al21 who had observed intravenous injection of CCK caused increase in colonic motility of patients with IBS. Finally, TRPV1 is a member of a sensory ion channel superfamily. This receptor is expressed in the gastrointestinal tract and responsible for gastrointestinal chemo-/mechano-sensation, nociception and hyperalgesia. Increased TRPV1 nerve fibers have been observed in the colonic mucosa of IBS patients and its up-regulation may contribute to generation of IBS symptoms.22 However, despite large amount of studies, it is still unclear whether these genetic polymorphisms play a certain role in IBS.
From this background, the aim of the present study was to explore the association between above-mentioned 5 genetic polymorphisms and IBS based on the Rome III criteria in Korean population. In addition, since there were rare studies which demonstrated the relationship between the serotonin level in blood and SLC6A4 polymorphism of SERT-promoter (SERT-P) in IBS, we aimed to compare the serum level of serotonin among various IBS subtypes and different genotypes of SLC6A4 5-HTTLPR polymorphism.
Materials and Methods
Study Subjects
Case and control subjects were prospectively recruited among the patients who visited the gastroenterologic clinic or health promotion center of Seoul National University Bundang Hospital between July 2009 and January 2014. All subjects underwent examinations including blood tests, abdominal imaging and upper endoscopy to evaluate the presence of any organic disease. If there were indications or patient’s wish, colonoscopy was performed additionally. All of them completed a validated bowel disease questionnaire, which was translated from the original Bowel Disease Questionnaire (BDQ) into Korean (Korean BDQ).23 The questionnaire contained 56 gastrointestinal symptom-related items which based on the Rome III criteria. Socio-demographic status, past medical history, smoking, alcohol habit, marital status, educational level and employment status were also included.
Patients who met IBS on the Rome III criteria were consecutively enrolled in the patient group1; IBS was defined as recurrent abdominal pain or discomfort for at least 3 days per month in the past 3 months with at least 2 of the following; improvement with defecation, onset associated with a change in frequency of stool, and onset associated with a change in form of stool. Patients scored their usual defecations according to the Bristol Stool Form Scale (BSFS). Stools described as 1 or 2 on BSFS were considered as hard or lumpy stools, and stools described as 6 or 7 on BSFS were considered as loose (mushy) or watery stools. Four subtypes of IBS were defined according to the predominant stool pattern; we defined diarrhea predominant IBS (IBS-D) as having loose (mushy) or watery stool for ≥ 25% of the time and hard or lumpy stools for < 25% of the time. We defined constipation predominant IBS (IBS-C) as having hard or lumpy stools for ≥ 25% of time and loose (mushy) or watery stools for < 25% of time. mixture of diarrhea and constipation IBS (IBS-M) was defined as having hard or lumpy stools for ≥ 25% of time and having loose (mushy) or watery stools for ≥ 25% of time. Unsubtyped IBS (IBS-U) was defined as stool abnormalities which did not meet criteria for either IBS-C, IBS-D or IBS-M.24
The healthy check-up subjects without any gastrointestinal symptoms were consecutively enrolled as healthy controls. Exclusion criteria included history of inflammatory bowel disease; abdominal operations (except appendectomy); pregnant or lactating women; suffering from severe systemic disease including malignancy; and those with hepatic, biliary, pancreatic, small bowel or large bowel disorders, psychiatric disorder requiring medication.
All participants were biologically unrelated native Koreans and provided informed consent. The study protocol was approved by the Ethics Committee of Seoul National University Bundang Hospital (B-0906/077-008).
Genotyping
Genomic DNA from blood samples was isolated using a commercially available kit (QIAamp DNA blood mini kit, QIAGEN Inc., Valencia, CA, USA) following the manufacturer’s instructions. The polymerase chain reaction (PCR)-based restriction fragment length polymorphism (RFLP) assay was performed in each DNA sample to detect the presence of the long (L) or short (S) allele in the SLC6A4 gene (5-HTTLPR), using a Perkin Elmer model 9600 (Perkin Elmer, Norwalk, CT, USA). The forward primer was 5’-TCCTCCGCTTTGGCGCCTCTTCC-3’, and the reverse primer was 5’-TGGGGGTTGCAGGGGAGATCCTG-3’. The 469 bp fragment of the 5-HTTLPR polymorphism was designated “S” and the 512-bp fragment was designated “L”. The genotyping of CCK1R intron-779T>C was also performed using PCR-RFLP assay. The forward primer for CCK1R intron-779 T>C was 5’-CTGTTCACTTGAGGAGCTTTG-3’, and the reverse primer was 5’-TTAGAAGCTGACCTCCAACATGG-3’. The PCR product was digested with PstI, and the T allele yielded a DNA fragment of 264/480 bp, while the C allele yielded a DNA fragment of 744 bp.
The GNB3 825C>T, ADRA2A −1291C>G, and TRPV1 945G>C single nucleotide polymorphisms (SNP) were determined using 5′ exonuclease TaqMan genotyping assays on an ABI StepOnePlus Real time PCR System, according to the manufacturer's instructions (Applied Biosystems, Foster City, CA, USA). The predesigned primer and probe sets were ordered at http://www.appliedbiosystems.com/ (GNB3 825C>T assay ID number: C___2184734_10; ADRA2A −1291C>G assay ID number: C___7611979_10; TRPV1 945G>C assay ID number: C___1093688_20) and used according to their protocols.
The genotyping results of SLC6A4 5-HTTLPR, GNB3 825C>T, ADRA2A −1291C>G, CCK1R intron 779T>C, and TRPV1 945G>C were confirmed by direct sequencing with ABI version 3.1 Sequence Analysis software (Applied Biosystems).
Measurement of Serum Serotonin
Serum serotonin levels were measured to evaluate their association with SLC6A4 polymorphism of SERT-P. Blood samples were obtained after an overnight fast of 8 hours. The blood was centrifuged and then serum was frozen at the temperature of −70°C.25 The total serotonin estimation was done using a commercially available ELISA kit (IBL International GmbH, Hamburg, Germany). The serum level of serotonin was expressed in ng/mL.
Statistical Methods
Hardy-Weinberg equilibrium of each gene allele in healthy controls was assessed by Chi-square test. Continuous and categorical demographic data were compared by Student t test and Chi-square test, respectively. The logistic regression analysis was used to estimate the OR and 95% CI for IBS groups. Age, sex and body mass index (BMI) were included as covariates in each of the logistic regression model. Both additive (e.g., each C/C, C/T and T/T for GNB3) and dominant/recessive (e.g., C/C vs. C/T + T/T and C/C + C/T vs. T/T for GNB3) models were evaluated in all the analyses. Serum serotonin level was analyzed by Mann-Whitney U test. Statistical analysis was performed using the SPSS for Windows version 19.0 (SPSS Inc., Chicago, IL, USA). P < 0.05 was considered as the level of significance.
Results
Subject Characteristics
Ninety-nine IBS patients who met the Rome III criteria and 171 healthy controls were enrolled in the present study. The demographic and clinical symptom data are shown in Tables 1 and 2, respectively. Ninety-four (55.0%) subjects of 171 controls and 74 (71.7%) of 99 IBS patients underwent colonoscopy. One and 3 subjects, respectively, received sigmoidoscopy in each group.
Table 1.
Characteristics of Healthy Controls and Patients With Irritable Bowel Syndrome
| Controls (n = 171) | IBS, total (n = 99) | IBS-D (n = 51) | IBS-C (n = 13) | IBS-M (n = 35) | |
|---|---|---|---|---|---|
| Age (mean ± SD, yr) | 55.7 ± 11.9 | 45.4 ± 13.6a | 45.0 ± 14.3a | 48.9 ± 10.0 | 44.9 ± 14.0a |
| Female (n [%]) | 86 (50.3) | 61 (61.6) | 29 (56.9) | 10 (76.9) | 22 (62.9) |
| BMI (mean ± SD, kg/m2) | 23.6 ± 3.3 | 22.5 ± 3.3b | 22.2 ± 3.0b | 24.3 ± 4.2 | 22.3 ± 3.1 |
| Smoking (n [%]) | |||||
| Current/ Ex-smoker | 70 (40.9) | 38 (38.4) | 21 (41.2) | 4 (30.8) | 13 (37.1) |
| Never smoker | 101 (59.1) | 61 (61.6) | 30 (58.8) | 9 (69.2) | 22 (62.9) |
| Alcoholc (n [%]) | |||||
| ≥ 35 g/wk | 43 (25.1) | 21 (21.6) | 12 (23.5) | 5 (38.5) | 4 (11.4) |
| < 35 g/wk | 51 (29.9) | 43 (43.4) | 20 (39.2) | 3 (23.1) | 20 (57.1) |
| Stopped/Never | 77 (45.0) | 35 (35.4) | 19 (37.3) | 5 (38.5) | 11 (31.4) |
| Marital status (n [%]) | |||||
| Single | 12 (7.0) | 21 (21.2)d | 13 (25.5)d | 1 (7.7) | 7 (20.0)d |
| Married | 139 (81.3) | 71 (71.7) | 36 (70.6) | 9 (69.2) | 26 (74.3) |
| Divorced/Separated/Widowed | 20 (11.7) | 7 (7.1) | 2 (3.9) | 3 (23.1) | 2 (5.7) |
| Education (n [%]) | |||||
| Middle or below | 32 (18.7) | 13 (13.1) | 4 (7.8) | 5 (38.5) | 4 (11.4) |
| High School | 52 (30.4) | 24 (24.2) | 14 (27.5) | 3 (23.1) | 5 (20.0) |
| University or beyond | 87 (50.9) | 62 (62.6) | 33 (64.7) | 5 (38.5) | 23 (68.6) |
| Employment status (n [%]) | |||||
| Employed | 112 (65.5) | 62 (62.6) | 32 (62.7) | 8 (61.5) | 22 (62.9) |
| Unemployed | 10 (5.8) | 5 (5.1) | 3 (5.9) | 1 (7.7) | 1 (2.9) |
| Student/Housewife | 49 (28.7) | 32 (32.3) | 15 (31.4) | 4 (30.8) | 12 (34.3) |
| Spicy food (n [%]) | 134 (78.4) | 72 (72.7) | 32 (62.7)e | 11 (84.6) | 29 (82.9) |
P < 0.001 and
P < 0.05, compared to healthy controls by ANOVA or Student t test;
The amount of alcohol in a bottle of Korean So-ju is considered approximately 70 g;
Single individuals were more likely to be in IBS group (IBS-D and IBS-M) compared to control (Chi-square test, P < 0.05);
Patients with IBS-D ingest spicy food less frequently than controls (Chi-square test, P < 0.05).
IBS, irritable bowel syndrome; IBS-D, diarrhea predominant IBS; IBS-C, constipation predominant IBS; IBS-M, mixture of diarrhea and constipation IBS.
Table 2.
Bowel Symptoms in Irritable Bowel Syndrome Subgroups According to the ROME III Criteria
| Symptoms | IBS-D (n = 51) | IBS-C (n = 13) | IBS-M (n = 35) | P-value | ||
|---|---|---|---|---|---|---|
|
| ||||||
| IBS-D vs. IBS-C | IBS-D vs. IBS-M | IBS-C vs. IBS-M | ||||
| Stool form (BSFS)a | 5.5 (3.0–7.0) | 3.0 (1.0–5.0)a | 5.0 (2.0–6.0) | < 0.001 | < 0.001 | 0.002 |
| Bowel movement (per week) | 11.5 (3.5–28.0) | 3.5 (1.0–19.0) | 6.5 (2.0–19.0) | 0.004 | 0.004 | 0.265 |
| Abdominal pain severityb | 3.0 (1.0–5.0) | 3.0 (1.0–5.0) | 3.0 (2.0–5.0) | 0.566 | 0.203 | 0.809 |
| Straining (n [%]) | 6 (11.8) | 9 (69.2) | 9 (25.7) | < 0.001 | 0.094 | 0.009 |
| Feeling of incomplete emptying (n [%]) | 7 (13.7) | 8 (61.5) | 13 (37.1) | 0.001 | 0.012 | 0.130 |
| Sensation that stool cannot be passed (n [%]) | 28 (54.9) | 12 (92.3) | 27 (77.1) | 0.022 | 0.035 | 0.410 |
| Manual removal of stool (n [%]) | 2 (3.9) | 4 (30.8) | 6 (17.1) | 0.013 | 0.038 | 0.425 |
| Urgency (n [%]) | 8 (15.7) | 1 (7.7) | 6 (17.1) | 0.672 | 0.857 | 0.656 |
| Nausea (n [%]) | 22 (43.1) | 8 (61.5) | 22 (62.9) | 0.235 | 0.072 | 0.999 |
| Vomiting (n [%]) | 8 (15.7) | 4 (30.8) | 10 (28.6) | 0.243 | 0.149 | 0.999 |
| Bloating (n [%]) | 38 (74.5) | 9 (69.2) | 30 (85.7) | 0.732 | 0.210 | 0.228 |
Observed most often;
1: very mild, 2: mild, 3: moderate, 4: severe, 5: very severe.
IBS, irritable bowel syndrome; IBS-D, diarrhea predominant IBS; IBS-C, constipation predominant IBS; IBS-M, mixture of diarrhea and constipation IBS; BSFS, Bristol Stool Form Scale.
IBS patients were classified into IBS-D (n = 51), IBS-C (n = 13) and IBS-M (n = 35) according to their predominant stool form (Table 2). In this study, 2 patients with BSFS 3 and 5 were grouped into IBS-C since they took laxative and they demonstrated severe straining and feeling of incomplete evacuation. Each of them had L/S and S/S genotypes. Five patients experiencing alternating periods of both IBS-C and IBS-D during the total follow-up period were included in the IBS-M group.
With regard to demographic characteristics, controls were older than the total patients with IBS (55.7 ± 11.9 years vs. 45.4 ± 13.6 years, P < 0.001) (Table 1). The BMI of control group was higher than that of total patients with IBS (23.6 ± 3.3 kg/m2 vs. 22.5 ± 3.3 kg/m2, P = 0.012). The mean age of patients with IBS-D and IBS-M, respectively, was lower than the mean age of healthy controls (45.0 ± 14.3 years and 44.9 ± 14.0 years vs. 55.7 ± 11.9 years, all P < 0.001). The mean BMI of IBS-D was lower than that of controls (22.2 ± 3.0 kg/m2 vs. 23.6 ± 3.3 kg/m2, P = 0.032). IBS-D or IBS-M patients were more likely to be single compared to control (25.5% and 20.0% vs. 7.0%, P = 0.001 and P = 0.040, respectively). Subjects in IBS-D group were found to ingest spicy food less frequently than controls (78.4% and 62.7%, P = 0.024). However, there were no significant differences in other variables including gender, smoking, alcohol, education and employment status.
As for clinical symptoms including bowel habit, IBS-D patients showed more frequent bowel movement than IBS-C and IBS-M patients (11.5 vs. 3.5 and 6.5, all P = 0.004) (Table 2). Patients with IBS-C experienced straining during defecation more frequently than those with IBS-D or IBS-M. Furthermore, patients with IBS-C and IBS-M was more likely to have feeling of incomplete defecation, sensation that the stool cannot be passed when having a bowel movement and needs to press on or around bottom to remove stool compared to those with IBS-D. However, there were no significant differences in the severity of abdominal pain or discomfort, urgency, nausea, vomiting and bloating.
Association Between 5 Genetic Polymorphisms and Irritable Bowel Syndrome
The genotypic distributions, crude ORs and adjusted ORs by age, sex and BMI of the 5 polymorphisms are summarized in Table 3. All of the polymorphisms in healthy controls were found to be in Hardy-Weinberg equilibrium (P > 0.05).
Table 3.
Distribution of the Genotypes in Healthy Controls and Patients With Irritable Bowel Syndrome
| Controls (n = 171) | IBS, total (n = 99) | OR (95% CI) | P-value | Adjusted OR (95% CI) | P-value | |
|---|---|---|---|---|---|---|
| SLC6A4 5-HTTLPR (n [%]) | ||||||
| S/S (addictive) | 123 (73.2) | 63 (63.6) | 1 (reference) | 1 (reference) | ||
| L/S | 41 (24.4) | 28 (28.3) | 1.33 (0.76–2.35) | 0.321 | 1.01 (0.54–1.91) | 0.966 |
| L/L | 4 (2.4) | 8 (8.1) | 3.91 (1.13–13.47) | 0.031 | 4.38 (1.04–18.53) | 0.045 |
| L carrier (dominant) | 45 (26.8) | 36 (36.4) | 1.56 (0.92–2.66) | 0.101 | 1.24 (0.69–2.24) | 0.475 |
| L/L (recessive) | 4 (2.4) | 8 (8.1) | 3.57 (1.05–12.5) | 0.041 | 4.35 (1.04–16.67) | 0.043 |
| ADRA2A −1291C>G (n [%]) | ||||||
| C/C (addictive) | 23 (13.4) | 5 (5.1) | 1 (reference) | 1 (reference) | ||
| C/G | 74 (43.3) | 46 (46.4) | 2.86 (1.02–8.05) | 0.047 | 3.59 (1.19–10.84) | 0.023 |
| G/G | 74 (43.3) | 48 (48.5) | 2.98 (1.06–8.38) | 0.038 | 3.17 (1.06–9.54) | 0.040 |
| G carrier (dominant) | 148 (86.6) | 94 (94.9) | 2.92 (1.07–7.95) | 0.036 | 3.37 (1.16– 9.77) | 0.025 |
| G/G (recessive) | 74 (43.3) | 48 (48.5) | 1.23 (0.75–2.04) | 0.407 | 1.10 (0.64–1.89) | 0.732 |
| GNB3 825C>T (n [%]) | ||||||
| C/C (addictive) | 40 (23.4) | 26 (26.3) | 1 (reference) | 1 (reference) | ||
| C/T | 81 (47.4) | 46 (46.4) | 0.87 (0.47–1.61) | 0.666 | 0.90 (0.46–1.77) | 0.764 |
| T/T | 50 (29.2) | 27 (27.3) | 0.83 (0.42–1.64) | 0.593 | 0.87 (0.42–1.83) | 0.720 |
| T carrier (dominant) | 131 (76.6) | 73 (73.7) | 0.86 (0.48–1.52) | 0.597 | 0.89 (0.48–1.66) | 0.717 |
| T/T (recessive) | 50 (29.2) | 27 (27.3) | 0.91 (0.52–1.56) | 0.730 | 0.93 (0.51–1.69) | 0.822 |
| CCK1R intron 779T>C (n [%]) | ||||||
| T/T (addictive) | 95 (55.5) | 50 (50.5) | 1 (reference) | 1 (reference) | ||
| T/C | 67 (39.2) | 40 (40.4) | 1.13 (0.67–1.91) | 0.635 | 1.04 (0.59–1.83) | 0.889 |
| C/C | 9 (5.3) | 9 (9.1) | 1.90 (0.71–5.09) | 0.202 | 1.54 (0.48–4.94) | 0.465 |
| C carrier (dominant) | 76 (44.5) | 49 (49.5) | 1.23 (0.75–2.01) | 0.423 | 1.10 (0.64–1.89) | 0.742 |
| C/C (recessive) | 9 (5.3) | 9 (9.1) | 1.79 (0.69–4.76) | 0.230 | 1.52 (0.49–4.76) | 0.473 |
| TRPV1 945G>C (n [%]) | ||||||
| G/G (addictive) | 42 (25.8) | 26 (26.8) | 1 (reference) | 1 (reference) | ||
| G/C | 71 (43.5) | 48 (49.5) | 1.09 (0.59–2.01) | 0.777 | 1.23 (0.62–2.42) | 0.551 |
| C/C | 50 (30.7) | 23 (23.7) | 0.74 (0.37–1.49) | 0.402 | 0.92 (0.43–1.97) | 0.829 |
| C carrier (dominant) | 121 (74.2) | 71 (73.2) | 0.95 (0.54–1.68) | 0.854 | 1.10 (0.59–2.08) | 0.760 |
| C/C (recessive) | 50 (30.7) | 23 (23.7) | 0.70 (0.40–1.25) | 0.228 | 0.81 (0.43–1.49) | 0.496 |
IBS, irritable bowel syndrome; 5-HTTLPR, serotonin transporter gene linked polymorphic region.
Adjusted OR, sex, age and BMI adjusted ORs vs. 171 healthy controls, by multiple logistic regression model.
When the 5 kinds of genotypic frequency were compared between controls and total IBS patients, the frequency of the C/C genotype of ADRA2A was significantly lower in the IBS group than that in the control group (5.1% vs. 13.4%, P < 0.05).
In logistic regression models, SLC6A4 5-HTTLPR L/L genotype was associated with IBS (adjusted OR: 4.38, 95% CI: 1.04–18.53, P = 0.045 and adjusted OR: 4.35, 95% CI: 1.04–16.67, P = 0.043) in both addictive and recessive models (Table 3). Each of C/G and C/C genotype and G carrier of ADRA2A were also significantly associated with increased susceptibility to IBS (adjusted OR: 3.59, 95% CI: 1.19–10.84, P = 0.023; adjusted OR: 3.17, 95% CI: 1.06–9.54, P = 0.040 and adjusted OR: 3.37, 95% CI: 1.16–9.77, P = 0.025, respectively). However, no significant differences were found in other 3 genetic polymorphisms between total IBS patients and control group.
When the genotypic distribution among subtypes of IBS and controls was analyzed, significant association was observed between SLC6A4 5-HTTLPR L/L genotype and IBS-C in both addictive and recessive models (adjusted OR: 10.63, 95% CI: 1.57–71.94, P = 0.015 and adjusted OR: 11.11, 95% CI: 1.69–50.00, P = 0.012, respectively) (Table 4). Similar to this result, significant association was detected for L homozygotes in IBS-M in both addictive and recessive models (adjusted OR: 6.75, 95% CI: 1.23–37.07, P = 0.028 and adjusted OR: 5.56, 95% CI: 1.05–33.33, P = 0.043, respectively).
Table 4.
Distribution of the Genotypes in Irritable Bowel Syndrome Subtypes
| Controls (n = 171) | IBS-D (n = 51) | IBS-C (n = 13) | IBS-M (n = 35) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|||||||
| n (%) | n (%) | OR (95% CI) | Adjusted OR (95% CI) | n (%) | OR (95% CI) | Adjusted OR (95% CI) | n (%) | OR (95% CI) | Adjusted OR (95% CI) | |
| SLC6A4 5-HTTLPR | ||||||||||
| S/S (addictive) | 123 (73.2) | 38 (74.5) | 1 (reference) | 1 (reference) | 7 (53.8) | 1 (reference) | 1 (reference) | 18 (51.4) | 1 (reference) | 1 (reference) |
| L/S | 41 (24.4) | 12 (23.5) | 0.95 (0.45–1.98) | 0.74 (0.32–1.71) | 3 (23.1) | 1.29 (0.32–5.20) | 0.97 (0.23–4.16) | 13 (37.1) | 2.17 (0.98–4.80) | 1.65 (0.70–3.87) |
| L/L | 4 (2.4) | 1 (2.0) | 0.81 (0.09–7.46) | 0.30 (0.03–3.29) | 3 (23.1) | 13.18 (2.46–70.68)a | 10.63 (1.57–71.94)b | 4 (11.5) | 6.83 (1.57–29.77)b | 6.75 (1.23–37.07)b |
| L carrier (dominant) | 45 (26.8) | 13 (25.5) | 0.94 (0.46–1.91) | 0.68 (0.30–1.53) | 6 (46.2) | 2.34 (0.75–7.35) | 1.71 (0.51–5.73) | 17 (48.6) | 2.58 (1.23–5.44) | 2.01 (0.90–4.47) |
| L/L (recessive) | 4 (2.4) | 1 (2.0) | 0.82 (0.09–7.69) | 0.32 (0.03–3.57) | 10 (76.9) | 12.50 (2.44–50.00)a | 11.11 (1.69–50.00)b | 31 (88.5) | 5.26 (1.25–20.00)b | 5.56 (1.05–33.33)b |
| ADRA2A −1291C>G | ||||||||||
| C/C (addictive) | 23 (13.4) | 2 (3.9) | 1 (reference) | 1 (reference) | 2 (15.4) | 1 (reference) | 1 (reference) | 1 (2.9) | 1 (reference) | 1 (reference) |
| C/G | 74 (43.3) | 26 (51.0) | 4.04 (0.89–18.34) | 6.39 (1.29–31.68)b | 5 (38.5) | 0.78 (0.14–4.28) | 0.72 (0.12–4.30) | 15 (42.9) | 4.66 (0.58–37.23) | 8.40 (0.95–74.07) |
| G/G | 74 (43.3) | 23 (45.1) | 3.57 (0.78–16.32) | 4.90 (0.98–24.60) | 6 (46.1) | 0.93 (0.18–4.94) | 0.86 (0.16–4.80) | 19 (54.2) | 5.91 (0.75–46.55) | 7.64 (0.90–64.64) |
| G carrier (dominant) | 148 (86.6) | 49 (96.1) | 3.81 (0.87–16.74) | 5.64 (1.18–27.01)b | 11 (84.6) | 0.86 (0.18–4.11) | 0.80 (0.16–4.06) | 34 (97.1) | 5.28 (0.69–40.49) | 7.91 (0.96–65.37) |
| G/G (recessive) | 74 (43.3) | 23 (45.1) | 1.08 (0.57–2.00) | 1.02 (0.51–2.04) | 7 (53.9) | 1.12 (0.36–3.45) | 1.10 (0.34–3.57) | 16 (45.8) | 1.56 (0.75–3.23) | 1.30 (0.36–3.45) |
| GNB3 825C>T | ||||||||||
| C/C (addictive) | 40 (23.4) | 19 (37.3) | 1 (reference) | 1 (reference) | 2 (15.4) | 1 (reference) | 1 (reference) | 5 (14.3) | 1 (reference) | 1 (reference) |
| C/T | 81 (47.4) | 21 (41.1) | 0.55 (0.26–1.13) | 0.53 (0.24–1.18) | 6 (46.1) | 1.48 (0.29–7.67) | 1.19 (0.22–6.42) | 19 (54.3) | 1.88 (0.65–5.39) | 2.11 (0.68–6.54) |
| T/T | 50 (29.2) | 11 (21.6) | 0.46 (0.20–1.09) | 0.46 (0.18–1.16) | 5 (38.5) | 2.00 (0.37–10.86) | 1.63 (0.29–9.28) | 11 (31.4) | 1.76 (0.57–5.48) | 1.96 (0.58–6.61) |
| T carrier (dominant) | 131 (76.6) | 32 (62.7) | 0.51 (0.26–1.00) | 0.50 (0.24–1.05) | 11 (84.6) | 1.68 (0.36–7.89) | 1.36 (0.28–6.62) | 30 (85.7) | 1.83 (0.67–5.03) | 2.05 (0.69–6.05) |
| T/T (recessive) | 50 (29.2) | 11 (21.6) | 0.67 (0.32–1.41) | 0.67 (0.30–1.49) | 8 (61.5) | 1.51 (0.47–4.76) | 1.43 (0.43–4.76) | 24 (68.6) | 1.11 (0.51–2.44) | 1.14 (0.49–2.63) |
| CCK1R intron 779T>C | ||||||||||
| T/T (addictive) | 95 (55.5) | 24 (47.1) | 1 (reference) | 1 (reference) | 4 (30.8) | 1 (reference) | 1 (reference) | 22 (62.9) | 1 (reference) | 1 (reference) |
| T/C | 67 (39.2) | 20 (39.2) | 1.18 (0.60–2.31) | 1.06 (0.51–2.18) | 9 (69.2) | 3.19 (0.94–10.79) | 2.53 (0.72–8.86) | 11 (31.4) | 0.71 (0.32–1.56) | 0.72 (0.31–1.67) |
| C/C | 9 (5.3) | 7 (13.7) | 3.08 (1.04–9.11) | 2.17 (0.59–8.01) | 0 | – | – | 2 (5.7) | 0.96 (0.19–4.76) | 1.09 (0.19–6.09) |
| C carrier (dominant) | 76 (44.5) | 27 (52.9) | 1.41 (0.75–2.63) | 1.18 (0.59–2.35) | 9 (69.2) | 2.81 (0.83–9.49) | 2.30 (0.66–8.01) | 13 (37.1) | 0.74 (0.35–1.56) | 0.76 (0.34–1.69) |
| C/C (recessive) | 9 (5.3) | 7 (13.7) | 2.86 (1.01–8.33) | 2.13 (0.60–7.69) | 13 (100) | - | 33 (94.3) | 1.09 (0.23–5.26) | 1.20 (0.22–6.67) | |
| TRPV1 945G>C | ||||||||||
| G/G (addictive) | 42 (25.8) | 10 (20.4) | 1 (reference) | 1 (reference) | 4 (30.8) | 1 (reference) | 1 (reference) | 12 (34.3) | 1 (reference) | 1 (reference) |
| G/C | 71 (43.5) | 25 (51.0) | 1.48 (0.65–3.38) | 1.99 (0.77–5.14) | 7 (53.8) | 1.04 (0.29–3.75) | 1.26 (0.32–4.93) | 16 (45.7) | 0.79 (0.34–1.83) | 0.76 (0.30–1.90) |
| C/C | 50 (30.7) | 14 (28.6) | 1.18 (0.47–2.92) | 1.60 (0.57–4.48) | 2 (15.4) | 0.42 (0.07–2.41) | 0.47 (0.08–2.81) | 7 (20.0) | 0.49 (0.18–1.34) | 0.56 (0.19–1.65) |
| C carrier (dominant) | 121 (74.2) | 39 (79.6) | 1.35 (0.62–2.95) | 1.83 (0.75–4.48) | 9 (69.2) | 0.78 (0.23–2.67) | 0.90 (0.25–3.26) | 23 (65.7) | 0.67 (0.31–1.45) | 0.68 (0.29–1.59) |
| C/C (recessive) | 50 (30.7) | 14 (28.6) | 0.90 (0.45–1.82) | 0.99 (0.46–2.13) | 11 (84.6) | 0.41 (0.09–1.92) | 0.41 (0.08–2.00) | 28 (80.0) | 0.56 (0.23–1.37) | 0.66 (0.26–1.69) |
IBS, irritable bowel syndrome; IBS-D, diarrhea predominant IBS; IBS-C, constipation predominant IBS; IBS-M, mixture of diarrhea and constipation IBS; 5-HTTLPR, serotonin transporter gene linked polymorphic region.
Adjusted OR, sex- age- and BMI adjusted ORs vs. 171 healthy controls, by multiple logistic regression model.
P < 0.01;
P < 0.05; Bold characteristics represent statistically significance.
With respect to ADRA2A −1291C>G polymorphism, patients with C/G genotype and G carrier were at higher risk of IBS-D (adjusted OR: 6.39, 95% CI: 1.29–31.68, P = 0.023 and adjusted OR: 5.64, 95% CI: 1.18–27.01, P = 0.030, respectively).
However, in other genetic polymorphisms such as GNB3 825C>T, CCK1R intron 779T>C and TRPV1 945G>C, there were no significant differences between controls and each IBS subtype. These results were not changed when those with IBS-M and IBS-C were combined into one group and compared with the control group.
Furthermore, there were no differences in the frequency of any symptoms described in Table 2 among patients with different genotypes of genetic polymorphism which studied in the present study.
Association of Irritable Bowel Syndrome Subtypes, SLC6A4 Genotypes and Serum Level of Serotonin
Serum was collected from 101 participants who had agreed to the blood sampling. Among them, 96 samples were analyzed since other 5 specimens were extremely outlied. As a result, 49 controls and 47 IBS patients were analyzed (Table 5). Regarding the genotypic distribution, 51 subjects with S/S, 36 subjects with L/S and 9 subjects with L/L genotype were included in this analysis.
Table 5.
Serum Level of Serotonin According to Irritable Bowel Syndrome Subtypes and SLC6A4 Serotonin Transporter Gene Linked Polymorphic Region Genotypes
| Variable | Numbera | Serum serotonin (median [range], ng/mL) | P-valueb | P-valuec |
|---|---|---|---|---|
| Symptom Groups | ||||
| Control | 49 | 69.0 (32.7–150.8) | ||
| IBS, total | 47 | 69.8 (24.5–269.1) | 0.684 | |
| IBS-D | 21 | 92.9 (39.2–227.6) | 0.025 | |
| IBS-C | 11 | 50.2 (25.7–146.3) | 0.017 | 0.001 |
| IBS-M | 15 | 53.7 (24.5–269.2) | 0.096 | 0.003 |
| IBS-C + IBS-M | 26 | 51.9 (24.5–269.5) | 0.010 | < 0.001 |
| SLC6A4 5-HTTLPR | ||||
| S/S | 51 | 78.5 (25.7–150.8) | ||
| L/S | 36 | 67.1 (24.5–269.2) | 0.593 | |
| L/L | 9 | 55.2 (34.5–144.8) | 0.100 | |
| L/S + L/L | 45 | 65.6 (24.5–269.2) | 0.299 | |
| S/S + L/S | 87 | 74.3 (24.5–269.2) | 0.143 |
Five outliers were excluded among 101 samples which were measured by ELISA,
Compared with the control group or S/S genotype of SLC6A4 5-HTTLPR by Mann-Whitney U test,
Compared with the IBS-D by Mann-Whitney U test.
IBS, irritable bowel syndrome; IBS-D, diarrhea predominant IBS; IBS-C, constipation predominant IBS; IBS-M, mixture of diarrhea and constipation IBS; 5-HTTLPR, serotonin transporter gene linked polymorphic region.
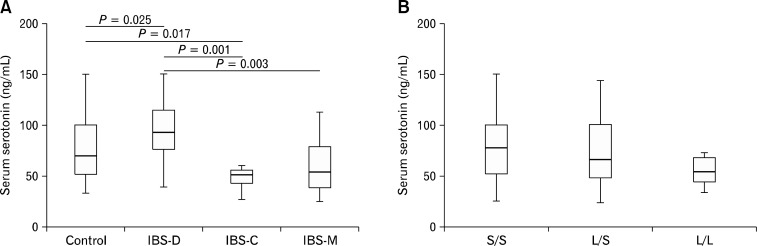
When the serum serotonin level was compared between control and total IBS patients, no significance difference was detected (69.0 ng/mL vs. 69.8 ng/mL, P = 0.684). In analyses among controls and the IBS subtypes, IBS-D showed marginally elevated serum serotonin level (69.0 ng/mL vs. 92.9 ng/mL, P = 0.025). Patients with IBS-C showed markedly reduced serum level of serotonin compared to controls and IBS-D (50.2 ng/mL vs. 69.0 ng/mL and 92.9 ng/mL, P = 0.017 and P = 0.001, respectively) (Table 5 and Figure A). In addition, serum level of IBS-M was significantly lower than that of IBS-D (53.7 ng/mL vs. 92.9 ng/mL, P = 0.003). When IBS-C and IBS-M were combined into one group, serum serotonin level of “IBS-C and IBS-M” was lower than those of control and IBS-D (51.9 ng/mL vs. 69.0 ng/mL and 92.9 ng/mL, P = 0.010 and P < 0.001, respectively) (Table 5).
Figure.
Serum level of serotonin (A) patients with constipation predominant IBS (IBS-C) showed significantly reduced level of serum serotonin compared to controls and patients with diarrhea predominant IBS (IBS-D), while those with IBS-D revealed highest level among other subtypes and control. (B) There was no significant difference in serum serotonin level among different SLC6A4 serotonin transporter gene linked polymorphic region (5-HTTLPR) genotypes. The whiskers in the box and whiskers plot represent the 10th and 90th percentiles. IBS-M, mixture of diarrhea and constipation IBS.
However, when the relationship between serum serotonin level and SLC6A4 5-HTTLPR genotypes was assessed, serum level of serotonin of those with S/S was not significantly different from that of L homozygotes (78.5 ng/mL vs. 55.2 ng/mL, P = 0.100) (Table 5 and Figure B). Because of small sample size, further subgroup analysis regarding both IBS subtypes and genotypes was not conducted.
Discussion
The present study revealed an association between L/L genotype of SERT-P polymorphism and IBS, especially IBS-C and IBS-D and an association between the non-C genotype in ADRA2A −1291C>G polymorphism and total IBS and IBS-D. However, GNB3 825C>T, CCK1R intron 779T>C and TRPV1 945G>C polymorphism did not show significant differences between controls and total IBS, nor each IBS subtype. In the analysis of serum serotonin, while increased level of serotonin was detected in the IBS-D patients compared to controls, reduced level of serotonin in IBS-C patients was shown as compared with controls and IBS-D patients. However, no significant difference was found in serum serotonin level among different genotypes.
Serotonin is the best studied neurotransmitter in IBS. The facts that most of serotonin is synthesized in the intestine and the presence of many kinds of receptors within the intestinal wall involving gastrointestinal secretion, motility, and visceral perception implies that serotonin or serotonin signaling pathway may contribute to IBS.17 In particular, SERT play an important role in modulating the level of serotonin,26 and excessive transcription of SERT in those with L allele would show high reuptake activity of serotonin and subsequently, reducing it in the synaptic cleft.27
There are studies similar with our result about SERT polymorphism. Recently, the subgroup analysis in the meta-analysis of 12 studies demonstrated that Asian with S/S or L/S showed significantly reduced risk of IBS.28 Regarding the association with IBS subtypes, the significant association between L/L genotype and IBS-C was shown in Chinese study.29 Similarly to our result, in another recent meta-analysis with 3,443 IBS cases and 3,359 controls of 25 studies, the L homozygote was demonstrated to be a risk factor for IBS-C development. Moreover, L allele and L/L genotype were significantly associated with increased IBS-C risk in the East Asian population.30
The genotypic association between the L/L genotype and not only IBS-C but also IBS-M may result from possible misclassification of IBS-M or spectrum disorder of IBS itself. IBS-M is a highly heterogeneous due to range of symptoms that are similar to those in IBS-D and IBS-C,31 this would cause a large proportion of patients remains IBS-M. Although this entity were not well studied currently, some studies has reported IBS-M is more similar to IBS-C than IBS-D based on similarities in stool frequency, consistency, psychological symptoms and a higher likelihood of transition between these 2 subtypes longitudinally.32,33 In the present study, it appears that clinical symptoms of IBS-M tend to more resemble IBS-C than IBS-D. Therefore, there might be a considerable movement from IBS-C to IBS-M, but despite several times’ review of medical chart including history of medication such as laxative and antidiarrheal or longitudinal symptom change, 35 IBS-M patients in the present study could not be subclassified into either of IBS-D or IBS-C. Further study for characterization of this mixed bowel pattern group is warranted.
However, there are other studies reporting a significant association between IBS-D and the S/S genotype.34,35 An Indian study demonstrated that the frequency of SLC6A4 S/S genotype was higher in IBS-D than IBS-C, IBS-M and controls.36 These genotypic results were different from our study, but taking the serotonin level and its association on subtypes into consideration, they might be in line with our study at least in part. Serotonin in rectal mucosa was markedly elevated in IBS-D patients compared to IBS-M and IBS-C patients and in S/S carriers compared with those in L/S and L/L genotypes.36 Houghton et al37 previously showed increased serotonin in the plasma of IBS-D patients compared with control. Although the relation between the serotonin level in plasma and mucosa remained uncertain, these results indicate that serotonin could be associated with colonic transit or stool form and SLC6A4 5-HTTLPR polymorphism might regulate this level.
However, contrary to the present study, another Indian study showed the significantly higher frequency of the S/S genotype of the SERT-P polymorphism in male IBS-C patients.38
With regard to the relation of ADRA2A −1291C>G polymorphism on IBS, in the present study, a significant association between carrying G allele and IBS-D was demonstrated (adjusted OR: 5.64; 95% CI: 1.18–27.01; P = 0.030) (Table 3). In a recent study conducted in India, the frequencies of non-C genotypes of ADRA2A were significantly higher than that of C/C genotypes between IBS-D and control groups (G/G + C/G vs. C/C; OR: 2.08, 95% CI: 1.06–4.07).14 Although how this SNP had an influence on the expression of α2A-adrenergic receptor (α2A AR) has not been proved yet, ADRA2A −1291C>G polymorphism in the promoter region could be related with loss of function. A previous experimental study demonstrated that gastrointestinal transit of α2A AR-knockout mice was doubled compared to wild type and intraperitoneally injected medetomidine (α2A agonist) slowed gastrointestinal transit in wild type but not α2A AR-knockout mice.39 There is an another report that adrenoreceptors modulate antisecretory action of intestine and the impairment in the α2A ARs may cause a decrease in absorption of ions and water leading to diarrhea.40 Nevertheless, on the contrary to the present study, Kim et al11 showed an association between ADRA2A − 1291C>G polymorphism and IBS-C.
Significant relationship between GNB3 825C>T and functional dyspepsia was shown.19,20 However, in terms with IBS, most Caucasian and one Korean studies showed no significant association.9,12,41,42 Only one Greek study showed significant association between T homozygotes (or T allele) and IBS.43 In the present study, we could not find any significant association between this SNP and IBS, either.
To our knowledge, as for the association between CCK1R intron 779T>C and IBS, only one study has been published. In the study which was conducted in Korea, a significant association of CCK1R intron 779 T>C polymorphism with “IBS-C and IBS-M” was reported (OR, 2.43).13 However, in the present study, no association with IBS or subtypes was shown.
It has been reported that TRPV1 945G>C polymorphism increased the expression level of mRNA and protein, in vitro.44 The increased TRPV1 expression and hypersensitivity to capsaicin have been reported in IBS.45 From these backgrounds, it is conceivable that the SNP of the gene could contribute to development of IBS, but no significant association was detected between them in the present study. This result is consistent with previously published data.46,47
As the explanation for the contradictory results among these genetic association studies, racial or regional differences have been most widely addressed as a possible reason. Discrepancy in genotype distribution of SLC6A4 5-HTTLPR between Caucasian and Asian patients was well documented.17,18,48 Nonetheless, the genotypic distributions of most polymorphisms in controls of this study were similar to those of other studies conducted in Korea, Japan and China lending strength to the validity of our result particularly in Asian countries.13,29,35,42,46,47 Secondly, the difficulty in determining IBS subtypes could contribute to different results of the genetic association studies in IBS. This may come from variable diagnostic criteria for IBS,49 inevitable limitation of symptom-based diagnosis or lack in stability of symptom over time. There might be also a possibility of misclassification since underlying biological mechanisms could be different in spite of symptom similarity of people grouped in the same category. Furthermore, IBS is a complex disease not caused by a single gene. Therefore, gene-gene or gene-environment interaction could lead to different results.
We also have to admit the small sample size of overall patients, especially IBS-C in this study may have limited its accuracy to detect differences between the groups. Although the proportion of IBS-C seems widely variable in Asia,50 the recent study which was conducted in Korea based on ROME III criteria reported that the proportion of IBS-C was only 12.0%.23 When the necessary sample size for detecting the significant association was calculated based on the observed genotypic frequencies (with a power of 80% and a level of significance of 5% in unmatched case-control [1:1]; OR, 2.0), the smallest number of sample size among the 5 polymorphisms was approximately 150 in control and total IBS group, respectively. However, the sample size of IBS patients in the present study was far less than the expected value and this inevitably reduced statistical power. As a result, only in the subgroup analyses with IBS-C, the statistical power of SLC6A4 5-HTTLPR was above 80%. Therefore, the possibility of type II error cannot be excluded in each polymorphism analysis, although there were some studies which demonstrated the genetic association with modest sample size.12,13,29 Larger studies such as multi-center studies are needed for the accurate estimation of the genotypic risk. We did not perform the correction for multiple testing in order to reduce the likelihood that real effects would be missed. We also corrected age, sex and BMI by multiple logistic regression models not 1:1 age- and sex-matching in order to maintain current sample size, but this might cause incomplete correction. Another limitation was that we measured serotonin level in only 101 subjects due to difficulty in obtaining agreement and we used serum than rectal mucosa, which did not reflect focal level of this neurotransmitter of where symptoms arise. Consequently, the result in this study should be interpreted cautiously, and larger scale study is needed to confirm this.
Despite aforementioned limitations, the present study attempted to evaluate the association of various polymorphisms in a study population. Especially for CCK1R intron 779T>C and TRPV1 945G>C polymorphisms, there are very few associational studies regarding IBS around the world. Furthermore, this is one of the rare reports which tried to show functional relationship between SLC6A4 5-HTTLPR polymorphism and blood serotonin level as a causal factor of IBS.
In summary, we have demonstrated that SLC6A4 5-HTTLPR L/L genotype was associated with IBS-C and IBS-M and low serum serotonin level was related with IBS-C, while patients with IBS-D showed elevated serotonin level. There was also an association between ADRA2A −1291G allele and IBS, especially IBS-D. With respect to GNB3 825C>T, CCK1R intron 779T>C, and TRPV1 945G>C polymorphisms, there seems to be no significant associations with IBS.
In conclusion, the genetic polymorphism of SLC6A4 5-HTTLPR and ADRA2A −1291C>G might be one of the pathophysiological factors of IBS in Korea. Serum serotonin level may be related with colonic transit, but whether the genotypic difference of SERT-P increases the risk for IBS via modulating serotonin level was needed to be proven. Furthermore, since IBS is a heterogeneous and multifactorial disease, more comprehensive approach including multiple genes and gene-environment interaction should be considered.
Acknowledgments
The authors thank Division of Statistics in Medical Research Collaborating Center at Seoul National University Bundang Hospital for statistical analyses.
Footnotes
Financial support: This work was supported by the National Research Foundation of Korea (NRF) grant for the Global Core Research Center (GCRC) funded by the Korean government (MSIP) (No. 2011-0030001).
Conflicts of interest: None.
Author contributions: Yoon Jin Choi interpreted data and revised the manuscript; Sung Wook Hwang analyzed data and drafted the article; Nayoung Kim designed this study, supervised preparing manuscript; Ji Hyun Park genotyped the genetic polymorphisms and measured serum serotonin level; Jane C Oh checked the contents and edited English; Dong Ho Lee advised design and supervised manuscript.
References
- 1.Drossman DA. The functional gastrointestinal disorders and the Rome III process. Gastroenterology. 2006;130:1377–1390. doi: 10.1053/j.gastro.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 2.Saito YA, Schoenfeld P, Locke III GR., 3rd The epidemiology of irritable bowel syndrome in North America: a systematic review. Am J Gastroenterol. 2002;97:1910–1915. doi: 10.1111/j.1572-0241.2002.05913.x. [DOI] [PubMed] [Google Scholar]
- 3.Locke GR., 3rd The epidemiology of functional gastrointestinal disorders in North America. Gastroenterol Clin North Am. 1996;25:1–19. doi: 10.1016/s0889-8553(05)70362-9. [DOI] [PubMed] [Google Scholar]
- 4.Tack J, Masaoka T, Janssen P. Functional dyspepsia. Curr Opin Gastroenterol. 2011;27:549–557. doi: 10.1097/MOG.0b013e32834b7ca8. [DOI] [PubMed] [Google Scholar]
- 5.Kaji M, Fujiwara Y, Shiba M, et al. Prevalence of overlaps between GERD, FD and IBS and impact on health-related quality of life. J Gastroenterol Hepatol. 2010;25:1151–1156. doi: 10.1111/j.1440-1746.2010.06249.x. [DOI] [PubMed] [Google Scholar]
- 6.Ford AC, Talley NJ. Irritable bowel syndrome. BMJ. 2012;345:e5836. doi: 10.1136/bmj.e5836. [DOI] [PubMed] [Google Scholar]
- 7.Saito YA. The role of genetics in IBS. Gastroenterolo Clin North Am. 2011;40:45–67. doi: 10.1016/j.gtc.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Tilburg MA, Whitehead WE. New paradigm for studying genetic contributions to irritable bowel syndrome. Dig Dis Sci. 2012;57:2484–2486. doi: 10.1007/s10620-012-2370-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saito YA, Larson JJ, Atkinson EJ, et al. The Role of 5-HTT LPR and GNβ3 825C> T polymorphisms and gene–environment interactions in irritable bowel syndrome (IBS) Dig Dis Sci. 2012;57:2650–2657. doi: 10.1007/s10620-012-2319-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heils A, Teufel A, Petri S, et al. Allelic variation of human serotonin transporter gene expression. J Neurochem. 1996;66:2621–2624. doi: 10.1046/j.1471-4159.1996.66062621.x. [DOI] [PubMed] [Google Scholar]
- 11.Kim HJ, Camilleri M, Carlson PJ, et al. Association of distinct α2 adrenoceptor and serotonin transporter polymorphisms with constipation and somatic symptoms in functional gastrointestinal disorders. Gut. 2004;53:829–837. doi: 10.1136/gut.2003.030882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saito YA, Locke G, 3rd, Zimmerman JM, et al. A genetic association study of 5-HTT LPR and GNβ3 C825T polymorphisms with irritable bowel syndrome. Neurogastroenterol Motil. 2007;19:465–470. doi: 10.1111/j.1365-2982.2007.00905.x. [DOI] [PubMed] [Google Scholar]
- 13.Park SY, Rew JS, Lee SM, et al. Association of CCK1 receptor gene polymorphisms and irritable bowel syndrome in Korean. J Neurogastroenterol Motil. 2010;16:71–76. doi: 10.5056/jnm.2010.16.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sikander A, Rana SV, Sharma SK, et al. Association of alpha 2A adrenergic receptor gene (ADRA2A) polymorphism with irritable bowel syndrome, microscopic and ulcerative colitis. Clinica Chimica Acta. 2010;411:59–63. doi: 10.1016/j.cca.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 15.Tahara T, Shibata T, Nakamura M, et al. Homozygous TRPV1 315C influences the susceptibility to functional dyspepsia. J Clin Gastroenterol. 2010;44:e1–e7. doi: 10.1097/MCG.0b013e3181b5745e. [DOI] [PubMed] [Google Scholar]
- 16.Lario S, Calls J, Cases A, Oriola J, Torras A, Rivera F. MspI identifies a biallelic polymorphism in the promoter region of the α2A-adrenergic receptor gene. Clin Genet. 1997;51:129–130. [PubMed] [Google Scholar]
- 17.Camilleri M, Katzka DA. Irritable bowel syndrome: methods, mechanisms, and pathophysiology. Genetic epidemiology and pharmacogenetics in irritable bowel syndrome. Am J Physiol Gastrointest Liver Physiol. 2012;302:G1075–G1084. doi: 10.1152/ajpgi.00537.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oshima T, Toyoshima F, Nakajima S, Fukui H, Watari J, Miwa H. Genetic factors for functional dyspepsia. J Gastroenterol Hepatol. 2011;26(suppl 3):83–87. doi: 10.1111/j.1440-1746.2011.06639.x. [DOI] [PubMed] [Google Scholar]
- 19.Van Lelyveld N, Linde JT, Schipper M, Samsom M. Candidate genotypes associated with functional dyspepsia. Neurogastroenterol Motil. 2008;20:767–773. doi: 10.1111/j.1365-2982.2008.01102.x. [DOI] [PubMed] [Google Scholar]
- 20.Holtmann G, Siffert W, Haag S, et al. G-protein β3 subunit 825 CC genotype is associated with unexplained (functional) dyspepsia. Gastroenterology. 2004;126:971–979. doi: 10.1053/j.gastro.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 21.Harvey RF, Read AE. Effect of cholecystokinin on colonic motility and symptoms in patients with the irritable-bowel syndrome. The Lancet. 1973;301:1–3. doi: 10.1016/s0140-6736(73)91219-1. [DOI] [PubMed] [Google Scholar]
- 22.Akbar A, Yiangou Y, Facer P, Walters JR, Anand P, Ghosh S. Increased capsaicin receptor TRPV1-expressing sensory fibres in irritable bowel syndrome and their correlation with abdominal pain. Gut. 2008;57:923–929. doi: 10.1136/gut.2007.138982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noh YW, Jung HK, Kim SE, Jung SA. Overlap of erosive and non-erosive reflux diseases with functional gastrointestinal disorders according to Rome III criteria. J Neurogastroenterol Motil. 2010;16:148–156. doi: 10.5056/jnm.2010.16.2.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Longstreth GF, Thompson WG, Chey WD, Houghton LA, Mearin F, Spiller RC. Functional bowel disorders. Gastroenterology. 2006;130:1480–1491. doi: 10.1053/j.gastro.2005.11.061. [DOI] [PubMed] [Google Scholar]
- 25.Wiśniewska-Jarosińska M, Harasiuk A, Walecka-Kapica E, Pawłowicz M, Stec-Michalska K, Chojnacki J. Postprandial secretion of serotonin and melatonin in patients with functional dyspepsia. Clin Exp Med Lett. 2009;50:165–168. [Google Scholar]
- 26.Kim DY, Camilleri M. Serotonin: a mediator of the brain-gut connection. Am J Gastroenterol. 2000;95:2698–2709. doi: 10.1111/j.1572-0241.2000.03177.x. [DOI] [PubMed] [Google Scholar]
- 27.Lesch KP, Bengel D, Heils A, et al. Association of anxiety-related traits with a polymorphism in the serotonin transporter gene regulatory region. Science. 1996;274:1527–1531. doi: 10.1126/science.274.5292.1527. [DOI] [PubMed] [Google Scholar]
- 28.Areeshi MY, Haque S, Panda AK, Mandal RK. A serotonin transporter gene (SLC6A4) polymorphism is associated with reduced risk of irritable bowel syndrome in American and Asian population: a meta-analysis. PLoS One. 2013;8:e75567. doi: 10.1371/journal.pone.0075567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y, Nie Y, Xie J, et al. The association of serotonin transporter genetic polymorphisms and irritable bowel syndrome and its influence on tegaserod treatment in Chinese patients. Dig Dis Sci. 2007;52:2942–2949. doi: 10.1007/s10620-006-9679-y. [DOI] [PubMed] [Google Scholar]
- 30.Zhang ZF, Duan ZJ, Wang LX, Yang D, Zhao G, Zhang L. The serotonin transporter gene polymorphism (5-HTTLPR) and irritable bowel syndrome: a meta-analysis of 25 studies. BMC Gastroenterol. 2014;14:23. doi: 10.1186/1471-230X-14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Su AM, Shih W, Presson AP, Chang L. Characterization of symptoms in irritable bowel syndrome with mixed bowel habit pattern. Neurogastroenterol Motil. 2014;26:36–45. doi: 10.1111/nmo.12220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Drossman DA, Morris CB, Hu Y, et al. A prospective assessment of bowel habit in irritable bowel syndrome in women: defining an alternator. Gastroenterology. 2005;128:580–589. doi: 10.1053/j.gastro.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 33.Weinland SR, Morris CB, Hu Y, Leserman J, Bangdiwala SI, Drossman DA. Characterization of episodes of irritable bowel syndrome using ecological momentary assessment. Am J Gastroenterol. 2011;106:1813–1820. doi: 10.1038/ajg.2011.170. [DOI] [PubMed] [Google Scholar]
- 34.Yeo A, Boyd P, Lumsden S, et al. Association between a functional polymorphism in the serotonin transporter gene and diarrhoea predominant irritable bowel syndrome in women. Gut. 2004;53:1452–1458. doi: 10.1136/gut.2003.035451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park JM, Choi MG, Park JA, et al. Serotonin transporter gene polymorphism and irritable bowel syndrome. Neurogastroenterol Motil. 2006;18:995–1000. doi: 10.1111/j.1365-2982.2006.00829.x. [DOI] [PubMed] [Google Scholar]
- 36.Kumar S, Ranjan P, Mittal B, Ghoshal UC. Serotonin transporter gene (SLC6A4) polymorphism in patients with irritable bowel syndrome and healthy controls. J Gastrointestin Liver Dis. 2012;21:31–38. [PubMed] [Google Scholar]
- 37.Houghton LA, Atkinson W, Whitaker RP, Whorwell PJ, Rimmer MJ. Increased platelet depleted plasma 5-hydroxytryptamine concentration following meal ingestion in symptomatic female subjects with diarrhoea predominant irritable bowel syndrome. Gut. 2003;52:663–670. doi: 10.1136/gut.52.5.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sikander A, Rana SV, Sinha SK, et al. Serotonin transporter promoter variant: analysis in Indian IBS patients and control population. J Clin Gastroenterol. 2009;43:957–961. doi: 10.1097/MCG.0b013e3181b37e8c. [DOI] [PubMed] [Google Scholar]
- 39.Scheibner J, Trendelenburg AU, Hein L, Starke K, Blandizzi C. α2-Adrenoceptors in the enteric nervous system: a study in α2A-adrenoceptor-deficient mice. Br J Pharmacol. 2002;135:697–704. doi: 10.1038/sj.bjp.0704512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu L, Coupar IM. Role of α2-adrenoceptors in the regulation of intestinal water transport. Br J Pharmacol. 1997;120:892–898. doi: 10.1038/sj.bjp.0700958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Andresen V, Camilleri M, Kim HJ, et al. Is there an association between GNβ3-C825T genotype and lower functional gastrointestinal disorders? Gastroenterology. 2006;130:1985–1994. doi: 10.1053/j.gastro.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 42.Kim HG, Lee KJ, Lim SG, Jung JY, Cho SW. G-protein beta3 subunit C825T polymorphism in patients with overlap syndrome of functional dyspepsia and irritable bowel syndrome. J Neurogastroenterol Motil. 2012;18:205–210. doi: 10.5056/jnm.2012.18.2.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Markoutsaki T, Karantanos T, Gazouli M, Anagnou NP, Ladas SD, Karamanolis DG. Serotonin transporter and G protein beta3 subunit gene polymorphisms in Greeks with irritable bowel syndrome. Dig Dis Sci. 2011;56:3276–3280. doi: 10.1007/s10620-011-1726-7. [DOI] [PubMed] [Google Scholar]
- 44.Xu H, Tian W, Fu Y, Oyama TT, Anderson S, Cohen DM. Functional effects of nonsynonymous polymorphisms in the human TRPV1 gene. Am J Physiol Renal Physiol. 2007;293:F1865–F1876. doi: 10.1152/ajprenal.00347.2007. [DOI] [PubMed] [Google Scholar]
- 45.Li X, Cao Y, Wong RK, Ho KY, Wilder‐Smith CH. Visceral and somatic sensory function in functional dyspepsia. Neurogastroenterol Motil. 2013;25:246, e165. doi: 10.1111/nmo.12044. [DOI] [PubMed] [Google Scholar]
- 46.Song YA, Park SY, Park YL, et al. Association between single nucleotide polymorphisms of the transient receptor potential vanilloid 1 (TRPV-1) gene and patients with irritable bowel syndrome in Korean populations. Acta Gastroenterol Belg. 2012;75:222–227. [PubMed] [Google Scholar]
- 47.Toya Y, Chiba T, Sugai T, Habano W, Suzuki K. Association between brain-gut peptide polymorphisms and irritable bowel syndrome. J Gastroenterol Hepatol Res. 2013;2:576–580. [Google Scholar]
- 48.van Kerkhoven LA, Laheij RJ, Jansen JB. Meta-analysis: a functional polymorphism in the gene encoding for activity of the serotonin transporter protein is not associated with the irritable bowel syndrome. Aliment Pharmacol Ther. 2007;26:979–986. doi: 10.1111/j.1365-2036.2007.03453.x. [DOI] [PubMed] [Google Scholar]
- 49.Ersryd A, Posserud I, Abrahamsson H, Simrén M. Subtyping the irritable bowel syndrome by predominant bowel habit: Rome II versus Rome III. Aliment Pharmacol Ther. 2007;26:953–961. doi: 10.1111/j.1365-2036.2007.03422.x. [DOI] [PubMed] [Google Scholar]
- 50.Chang FY, Lu CL, Chen TS. The current prevalence of irritable bowel syndrome in Asia. J Neurogastroenterol Motil. 2010;16:389–400. doi: 10.5056/jnm.2010.16.4.389. [DOI] [PMC free article] [PubMed] [Google Scholar]