Abstract
Stress impacts negatively on plant growth and crop productivity, caicultural production worldwide. Throughout their life, plants are often confronted with multiple types of stress that affect overall cellular energy status and activate energy-saving responses. The resulting low energy syndrome (LES) includes transcriptional, translational, and metabolic reprogramming and is essential for stress adaptation. The conserved kinases sucrose-non-fermenting-1-related protein kinase-1 (SnRK1) and target of rapamycin (TOR) play central roles in the regulation of LES in response to stress conditions, affecting cellular processes and leading to growth arrest and metabolic reprogramming. We review the current understanding of how TOR and SnRK1 are involved in regulating the response of plants to low energy conditions. The central role in the regulation of cellular processes, the reprogramming of metabolism, and the phenotypic consequences of these two kinases will be discussed in light of current knowledge and potential future developments.
Keywords: energy signaling, TOR, SnRK1, bZIP, T6P, stress, metabolism
INTRODUCTION
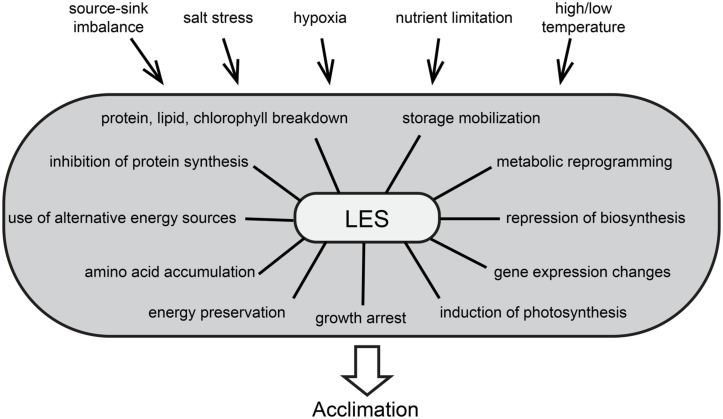
Suboptimal growth conditions related to temperature, light, water supply, and soil characteristics are among the most limiting factors for crop yield worldwide. Fruit and seeds constitute about 75% of world crop production (Liu et al., 2013a). They rely on the supply of carbohydrates from photosynthetic source tissues to sustain growth and development (Rolland et al., 2006) and this fine-tuned balance can be severely disturbed at different levels under biotic and abiotic stress conditions (Lemoine et al., 2013). In general, stress conditions cause the alteration of a set of processes and biochemical reactions. These changes can be encompassed by the term low energy syndrome (LES) and play a major role in the adaptation to stress conditions (Figure 1). In this review we summarize recent advances in the understanding of LES and the different signaling pathway components from energy deficiency toward adaptation, focusing on Arabidopsis thaliana as a plant model.
FIGURE 1.
The low energy syndrome (LES) is a collection of phenotypical consequences of stress condition and drives plant acclimation to environmental changes. The figure aims at depicting all causes and consequences described so far, but LES can be induced by one or multiple stress conditions and the acclimation process can include only a subset of the depicted outcomes.
THE LOW ENERGY SYNDROME IS PART OF STRESS ADAPTATION
Controlling energy homeostasis is a challenge for all organisms, as they must constantly sense and integrate internal and external signals to optimize growth and development, often under suboptimal conditions (Polge and Thomas, 2007). This is particularly critical for plants as they are sessile organisms, so it is very interesting to understand how they have evolved to overcome these constraints and how they respond differently to stresses when compared to other organisms. Their adaptation and survival depend on their capacity to efficiently manage energy resources in all tissues, and to coordinate energy consumption and preservation (Baena-González et al., 2007; Baena-González, 2010). Stress conditions affect source and sink tissues differently. In plants exposed to stress, sink organs like seeds or tubers often suffer from reduced sugar import and are impaired in biomass production (Pinheiro et al., 2001; Cuellar-Ortiz et al., 2008). Accordingly, stress factors like nutrient limitation, hypoxia, excess of salt, and low or high temperatures were discussed to impair fruit and seed development by interfering with the source-sink balance (Gibon et al., 2002; Geigenberger, 2003; Bailey-Serres et al., 2012; Lemoine et al., 2013; Liu et al., 2013b). Furthermore, stress conditions often affect photosynthesis and respiration in source leaves and this can accentuate the source-sink imbalance. The resulting energy deprivation was suggested to be common to most types of stress and to trigger specific responses (Baena-González and Sheen, 2008), leading to the massive alteration of cellular processes, referred to as LES. This includes growth arrest and metabolic reprogramming, comprising the repression of biosynthetic activities and sugar storage, as well as the induction of catabolic processes, photosynthesis, and sugar remobilization (Paul and Pellny, 2003; Gibon et al., 2006). In addition, the expression of thousands of genes is altered (Usadel et al., 2008). These genes were described to form a network that regulates plant metabolism under stress conditions for the purpose of energy preservation (Avin-Wittenberg et al., 2012). At the same time, metabolic reprogramming favors catabolic processes of molecules other than carbohydrates, resulting in protein, lipid and chlorophyll breakdown (Contento et al., 2004; Thimm et al., 2004). Accordingly, metabolomics data show an increase in amino acids coming from protein degradation (Caldana et al., 2011), which may contribute to sustain levels of TCA cycle intermediates (Araújo et al., 2012). Furthermore, translation rates decrease dramatically (Kawaguchi et al., 2003, 2004; Branco-Price et al., 2005, 2008; Nicolaï et al., 2006; Mustroph et al., 2009), although often without alteration of specific mRNA levels, which allows a rapid recovery after removal of the stress (Piques et al., 2009; Juntawong and Bailey-Serres, 2012).
These massive alterations on all cellular levels that comprise LES occur during many different stress situations and are therefore discussed as central processes necessary for adaptation. Even though LES involves a collection of phenotypic outcomes, their interconnection and regulation remain to be fully described. Generally, stress adaptation involves both universal and stress specific reactions, indicating that plants perceive multiple stresses and transduces the signal through pathways which may cross-talk at various levels (Chinnusamy et al., 2004). A number of signaling pathways are involved in the regulation of energy utilization and can be linked to the adaptation to stress conditions. Key players of energy signaling are the evolutionary conserved protein kinases sucrose-non-fermenting-1-related protein kinase-1 (SnRK1) and target of rapamycin (TOR). They are proposed to be antagonists in the coordination of energy consumption and preservation (Robaglia et al., 2012) and the balance of their activities might be essential to the regulation of LES in stress adaptation.
SnRK1 IS A METABOLIC SENSOR KINASE
Sucrose-non-fermenting-1-related protein kinase-1 is a metabolic sensor that can decode energy deficiency signals and induce an extensive metabolic reprogramming. This is mediated by a number of transcription factors and downstream targets that start an energy-saving program at several levels, including transcription, translation or direct phosphorylation of targets (Baena-González et al., 2007). SnRK1 is the plant homolog of the yeast sucrose non-fermenting-1 (SNF1) and the animal AMP-activated protein kinase (AMPK; Halford et al., 2004). SNF1-related protein kinases show close to 50% identity, rising to 65% for the kinase domains (Polge and Thomas, 2007). Their primary role is the integration of nutrient availability, stress signals, and energy expenditure, to be able to activate the required adaptations for homeostasis and survival (Halford and Hardie, 1998; Hardie et al., 1998; Ghillebert et al., 2011). Plants contain two other subfamilies, SnRK2 and SnRK3. They are less similar to SNF1 and AMPK and unique to plants (Halford et al., 2004), and are also involved in plant responses to several stresses (Coello et al., 2011). SnRK2 is involved in ABA signaling, responses to cold, and was shown to improve drought tolerance when overexpressed (Fujita et al., 2009; Halford and Hey, 2009; Yoshida et al., 2014). The SnRK3 family includes SOS2 (salt overly sensitive 2), involved in conferring salt tolerance (Liu et al., 2000). There is no evidence of redundancy between the different SnRK families and SnRK2 and SnRK3 do not complement the yeast snf1Δ deletion mutant growth phenotype (Hrabak et al., 2003). It is clear that they cannot fulfill the role of SnRK1 (Halford and Hey, 2009), even though there is some similarity in target recognition (Zhang et al., 2008). It was suggested that SnRK2 and SnRK3 arose in plants by duplication of SnRK1 and then diverged rapidly during plant evolution to meet new needs related to networks linking stress and ABA signaling with metabolic signaling (Halford and Hey, 2009).
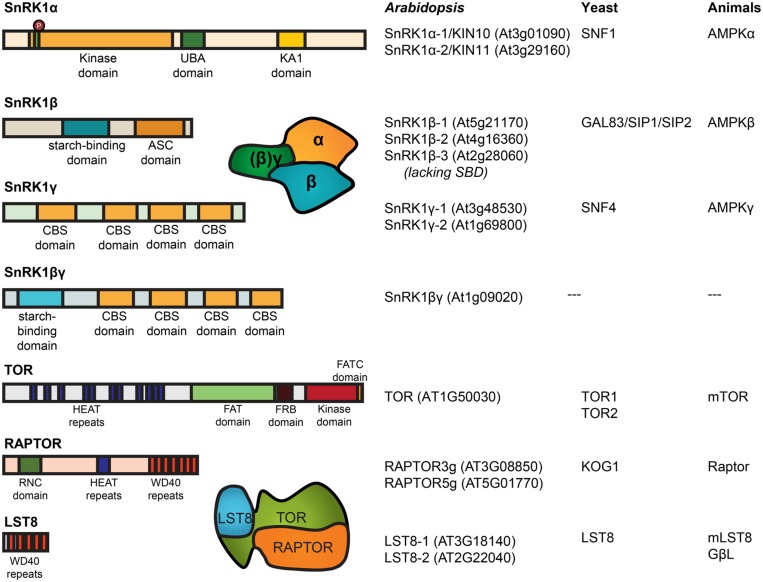
The SnRK1/SNF1/AMPK kinases typically function as heterotrimeric complexes and require a catalytic α-subunit, KIN10 and KIN11 in plants, and regulatory β and γ subunits (Figure 2). The non-catalytic subunits are also conserved among the SnRK1/SNF1/AMPK complex. They are likely involved in substrate recognition, subcellular localization, and regulation of the complex activity (Polge and Thomas, 2007; Ghillebert et al., 2011). Interestingly, the Arabidopsis AKINβ1, AKINβ2, and AKINβ3 have markedly different expression patterns, which suggests a level of regulation based on interactions targeting the β subunits in response to different signals (Bouly et al., 1999; Gissot et al., 2004).
FIGURE 2.
Domain structure and nomenclature of Arabidopsis SnRK1 and TOR subunits. SnRK1 structures include the conserved phosphorylation sites on T-loop of the α-subunit. The α-subunit contains the kinase domain, together with an auto-inhibitory (UBA) domain and a kinase associated (KA1) domain where the interaction with the ß-subunit takes place. The β-subunit (except in SnRK1ß3) contains a starch-binding domain (Ávila-Castañeda et al., 2014) and binds to the γ-subunit at the association with the SNF1 complex (ASC) domain. The plant-specific βγ-subunit might take the place described for the γ-subunit in mammals and yeast, containing multiple cystathionine β-synthase (CBS) domains. TOR contain two FAT domains (FRAP, ATM, and TRAP) probably constituting the active center, a PI3K kinase domain, a FRB domain (FKB12-rapamycin binding) for interaction with the inhibitor FKB12, and a number of HEAT repeats [huntingtin, elongation factor 3 (EF3), protein phosphatase 2A (PP2A), TOR1] for the interaction with RAPTOR. Next to HEAT repeats, RAPTOR contains a RNC domain (raptor N-terminal conserved/putative caspase domain) and a number of WD40 repeat domains (Hay and Sonenberg, 2004). Nomenclature as described before (Robaglia et al., 2012).
The activity of SnRK1 depends on phosphorylation in the highly conserved T-loop by upstream kinases (Sugden et al., 1999). In Arabidopsis, the protein kinases SnRK1-activating kinase 1 and 2 (AtSnAK1 and AtSnAK2) were shown to complement a yeast triple kinase mutant by restoring SNF1 upstream kinase activity (Shen and Hanley-Bowdoin, 2006). In addition, they phosphorylate non-truncated AtSnRK1 catalytic subunits in vitro, making them putative candidates as SnRK1 physiological upstream kinases (Crozet et al., 2010). Even though it is known that SnRK1 activation requires phosphorylation, it has not been clarified how it is affected by the cellular energy level. In contrast to the mammalian AMPK, SnRK1 is not allosterically activated by AMP, but it was shown that T-loop dephosphorylation and the resulting inactivation of the kinase are inhibited by low concentrations of AMP (Sugden et al., 1999). Furthermore, SnRK1 activity is modulated by specific phosphatases. Two clade A type 2C protein phosphatases (PP2C) were recently shown to dephosphorylate and inactivate SnRK1 through interaction with the catalytic subunit (Rodrigues et al., 2013).
The activity of SnRK1 is also inhibited by trehalose-6-phosphate (T6P). The association between trehalose metabolism and sugar-sensing in plants has recently become more evident (Tsai and Gazzarrini, 2014). Despite its role as a carbon source and in stress protection in resurrection plants, fungi, bacteria, and non-vertebrate animals (Elbein et al., 2003; Paul et al., 2008), the amount of trehalose in the majority of plants is too low to perform this function. It was suggested that trehalose has a major role on metabolism, growth, and development, acting as a signal of sugar availability (Schluepmann et al., 2003; Ramon and Rolland, 2007; Gómez et al., 2010). Trehalose is synthesized from UDP-glucose and glucose-6P via the intermediate T6P in a two-step pathway involving trehalose phosphate synthase (TPS) and trehalose phosphate phosphatase (TPP), and degraded by trehalase (Paul et al., 2008). T6P has a distinctive role in metabolic signaling, and class II TPSs, that include AtTPS5-11, are targets of phosphorylation by SnRK1 (Glinski and Weckwerth, 2005). AtTPS5 is induced by sugars and repressed by starvation (Schluepmann et al., 2004), while the opposite is true for AtTPS8-10 (Osuna et al., 2007).
Trehalose-6-phosphate inhibits the catalytic activity of SnRK1 in vitro at physiological concentrations, causing expression changes of KIN10 marker genes consistent with an inactivation of SnRK1 (Zhang et al., 2009), while Arabidopsis seedlings overexpressing SnRK1 show a glucose-hypersensitive phenotype (Cho et al., 2012), similar to seedlings with low T6P (Schluepmann et al., 2003). However, even though T6P inhibits the growth of Arabidopsis seedlings, it does not inhibit SnRK1 catalytic activity in extracts of mature leaves (Zhang et al., 2009), suggesting that an intermediary factor is needed for SnRK1 inhibition by T6P. Recently, it was suggested that T6P and SnRK1 might act through different, but interacting signaling pathways (Lunn et al., 2014) and can play antagonistic roles during stress responses (O’Hara et al., 2013). This is particularly important in stress conditions that negatively affect carbon levels, leading to an activation of starvation responses through SnRK1. For example, T6P levels are much lower in rosettes harvested in the dark and in carbon-starved seedlings (Lunn et al., 2006; Carillo et al., 2013; Yadav et al., 2014). However, stress conditions do not necessarily lead to carbon depletion. For example, under moderate drought or cold stress, a wide range of carbohydrates accumulate in Arabidopsis (Muller et al., 2011), including abundant sugars such as hexoses and sucrose, other sugars such as trehalose or mannitol, amino acids, organic acids, structural C-rich compounds like cellulose, among others. In grapevine, sucrose and T6P contents increase in response to chilling (Fernandez et al., 2012). T6P may therefore play a role in the inhibition of SnRK1 under conditions where carbon sources are not limited (Lunn et al., 2014). The cross-talk between SnRK1 and T6P when growth is limited by sink capacity was recently studied by varying temperature and nutrient supply to induce sink limitation, and feed sucrose and glucose at physiological levels (Nunes et al., 2013). In these conditions, T6P responds specifically to sucrose, even at different growth rates. Moreover, there was a strong correlation between T6P- and SnRK1-regulated gene expression, but not between T6P and relative growth rate. It appears that SnRK1 marker gene expression is related to T6P content regardless of the growth outcome, but further investigations will hopefully elucidate the relationship between SnRK1 and T6P.
SnRK1 REGULATES STRESS RESPONSES UPON LOW ENERGY
The activation of SnRK1 initiates massive transcriptional changes, possibly by affecting a number of transcription factors (Baena-González et al., 2007). The gene expression profile mediated by the SnRK1 subunit KIN10 is positively correlated with the one induced by deprivation of sugar and carbon, and negatively correlated with that controlled by sugars. This places KIN10 as a regulator of gene expression upon starvation and stress conditions. The most prominent KIN10-activated genes represent a variety of major catabolic pathways, including degradation of cell wall, starch, sucrose, amino acid, lipid, and proteins which provide alternative sources of energy and metabolites. Additionally, a large set of genes involved in energy-consuming ribosome biogenesis and anabolism are repressed.
From the possible mechanisms by which SnRK1 affects transcription, the members of the S1 class of the basic leucine zipper (bZIP) transcription factors are probably the best described (Figure 3). They belong to a large family of several classes in eukaryotes (Reinke et al., 2013) and function as homo-or heterodimers, which increases their potential for regulation (Jakoby et al., 2002; Corrêa et al., 2008; Schütze et al., 2008). From the bZIP family, bZIP1, bZIP11, and bZIP53 were proposed to mediate some of the transcriptional changes induced by the SnRK1 signaling pathway (Baena-González et al., 2007) and could be linked to the regulation of LES. In the presence of sucrose and glucose, the transcript levels of bZIP1 and bZIP53 decrease, and energy availability also seems to affect the phenotypes of their mutants. bZIP53 overexpression results in reduced plant size, delayed bolting and expression of seed-specific genes in leaves (Alonso et al., 2009). bZIP1 knockout plants were shown to grow faster than wild type on medium lacking glucose (Kang et al., 2010), while a plant overexpressing bZIP1 showed a stronger starvation response indicated by faster leaf-yellowing in extended night conditions (Dietrich et al., 2011). Under ambient growth conditions, bZIP1 gene expression is limited to sink tissue like pollen or young leaves (Weltmeier et al., 2009), but transcript levels were shown to be increased in source leaves after sugar starvation induced by extended night (Dietrich et al., 2011), while bZIP11 transcript levels increase in the presence of glucose and sucrose (Rook et al., 1998). On the other hand, bZIP11 translation is repressed by sucrose mediated by an upstream open reading frame (Wiese et al., 2004; Rahmani et al., 2009). This decrease is observed at physiological sucrose levels in most tissues, restricting bZIP11 activity to conditions of low energy availability (Wiese et al., 2004). Transgenic plants overexpressing bZIP11 show reduced plant size, seed production, viability and a wide effect on gene regulation and metabolism as demonstrated by microarray analysis of these plants (Hanson et al., 2008).
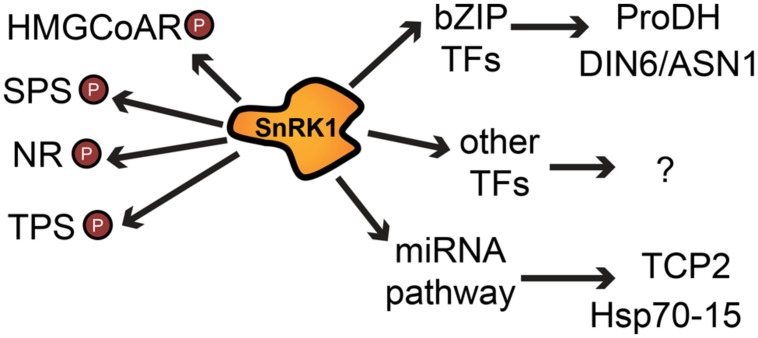
FIGURE 3.
Simplified summary of SnRK1 effects on cellular processes by direct phosphorylation of target proteins and by alteration of mRNA levels of many genes via transcription factors or the miRNA machinery. Arrows indicate a positive or negative regulatory effect; P denotes phosphorylation. Abbreviations: HMGCoAR, 3-hydroxy-3- methylglutaryl-coenzyme A reductase; SPS, sucrose phosphate synthase; NR, nitrate reductase; TPS, trehalose phosphate synthase; bZIP TF, basic leucine zipper transcription factor; ProDH, proline dehydrogenase; DIN6/ASN1, dark inducible 6/asparagine synthetase 1; TCP2, teosinte branched 1, cycloidea and PCF transcription factor 2; Hsp70–15, heat shock protein 70–15.
Several gene expression studies identified putative target genes of these transcription factors and there is considerable overlap between their targets and genes regulated by SnRK1. bZIP11 targets include several genes associated with LES, involved in the regulation of trehalose and other minor regulatory carbohydrates, such as myo-inositol and raffinose (Ma et al., 2011). bZIP11 also induces GDH1 and GDH2, genes encoding glutamate dehydrogenase (Hanson et al., 2008). The double mutant gdh1gdh2 was shown to be more susceptible to extended night, likely due to the role of these enzymes in amino acid degradation (Miyashita and Good, 2008a,b). Other genes involved in amino acid metabolism were shown to be regulated by bZIP transcription factors. Members of the S1 class specifically activate the G-box containing promoter of the SnRK1 regulated gene DIN6/ASN1 (Baena-González et al., 2007). They were also shown to activate gene expression of proline dehydrogenase (ProDH; Satoh et al., 2004) by binding to G-boxes contained in the promoter region (Weltmeier et al., 2006; Dietrich et al., 2011). While ProDH is thought to be involved in stress recovery (Weltmeier et al., 2006), the accumulation of asparagine and other amino acids during dark-induced starvation was proposed to result from protein degradation in order to provide an alternative to carbon as energy source (Caldana et al., 2011; Dietrich et al., 2011).
Basic leucine zipper transcription factors mediate many, but not all SnRK1 effects on transcription. It remains to be studied which factors mediate the first, direct effect on gene expression and which genes are regulated by secondary mechanisms. Further studies will hopefully reveal a number of additional transcription factors involved in SnRK1 regulation of gene expression.
Sucrose-non-fermenting-1-related protein kinase-1 also affects enzymes by direct phosphorylation. For example, it inhibits the activity of HMG-CoA reductase, the rate-limiting step in sterol synthesis (Clarke and Hardie, 1990; Mackintosh et al., 1992). Two other enzymes were shown to be substrates of SnRK1, sucrose phosphate synthase (SPS), and nitrate reductase (NR; McMichael et al., 1995; Douglas et al., 1997) which are key biosynthetic enzymes involved in the control of nitrogen assimilation and sucrose synthesis (Figure 3).
In addition to transcriptional changes and direct phosphorylation, SnRK1 was recently shown to activate a miRNA pathway (Confraria et al., 2013). Some of the candidate miRNA targets can be connected to the SnRK1 pathway and miRNAs can therefore be placed as components of the SnRK1 signaling pathway, as they regulate mRNA targets and possibly tune down specific cellular processes during the stress response (Figure 3). Most of the affected genes correspond to genes related to ribosomal proteins (RPs) and translation, which is in accordance with the role of SnRK1 as a repressor of biosynthetic processes and as a modulator of energy metabolism (Baena-González et al., 2007; Baena-González and Sheen, 2008). In animals there seems to be a link between miRNAs and metabolism: AMPK activation was recently reported to induce the differential accumulation of multiple miRNAs (Liu et al., 2013a), suggesting that miRNAs could be possible common elements in diverse organisms for restoring homeostasis following stress (Confraria et al., 2013). A recent paper supports this view by showing a strong connection between the regulation of miRNA expression and glucose-mediated regulatory responses (Duarte et al., 2013). These recent findings make it clear that SnRK1 mode of action goes beyond direct phosphorylation or modulation of transcription and promise new discoveries to come on the interplay between multiple pathways in the regulation of the LES.
THE TOR KINASE IS INVOLVED IN LES
Another key component in this network is the serine/threonine kinase TOR. TOR kinase genes are present in every eukaryote genome analyzed so far and they share 40–60% sequence identity (De Virgilio and Loewith, 2006; Wullschleger et al., 2006). These large proteins are well described for their central roles in the energy signaling pathways of yeast, mammals and plants (Wullschleger et al., 2006; Robaglia et al., 2012). TOR is activated, in both yeast and mammals, by high amino acid levels, but inactivated under amino acid starvation (Jewell et al., 2013). In plants, TOR activity has been linked to cell and organ size, seed yield, and stress resistance (Ren et al., 2012) and it was suggested to play a role in the regulation of carbon partitioning and growth (Zhang et al., 2013). Furthermore, it has been shown that TOR and SnRK1 interact closely and act in opposite ways in the regulation of nutrient-driven processes like autophagy (Robaglia et al., 2012).
In yeast and mammals, TOR functions in two complexes with distinct functions, TOR complex 1 (TORC1) and TORC2, characterized by different interaction partners. Of these, only subunits of TORC1 (TOR, LST8/GbetaL and KOG1/RAPTOR) are present in the Arabidopsis genome (Figure 2; van Dam et al., 2011). Mutations that disrupt TOR or RAPTOR genes were shown to be embryo lethal (Menand et al., 2002; Deprost et al., 2005). In addition, the low sensitivity of the Arabidopsis TOR toward rapamycin (Xiong and Sheen, 2012; Caldana et al., 2013) makes the study of TOR signaling in plants more difficult than in other organisms, where the inhibitor was extensively used to study TOR functions (Wullschleger et al., 2006). Alternative approaches were developed including the expression of a yeast FKBP12 protein which confers rapamycin sensitivity (Sormani et al., 2007; Ren et al., 2012), modulation of TOR expression by overexpressor or RNA interference constructs (Deprost et al., 2007; Ren et al., 2011) and the expression of an inducible artificial microRNA targeting TOR (Caldana et al., 2013).
The Arabidopsis TOR promoter is active in root and apical primary meristems, embryo and endosperm, but not in source leaves or differentiated cells (Menand et al., 2002). TOR acts on cell cycle control in Arabidopsis root meristems by directly phosphorylating the transcription factor E2Fa that regulates S-phase gene expression (Xiong et al., 2013). It was proposed as a potential integrator of cell cycle, cell expansion and cytoplasmic growth (Sablowski and Carnier Dornelas, 2014) and could thus be responsible for the activation of growth in the meristems in response to sugars provided by the photosynthetic source tissues (Xiong et al., 2013). Furthermore, TOR inhibition leads to a reduction of the length of the root meristematic zone and the division zone therein (Montané and Menand, 2013). Recently, TOR was shown to be activated by the growth hormone auxin and is involved in the regulation of translation of auxin responsive genes (Bögre et al., 2013; Schepetilnikov et al., 2013). Accordingly, reduction of TOR expression in Arabidopsis results in severe growth arrest with plants displaying decreased cell size, whereas TOR overexpression leads to an increase in shoot and root growth (Deprost et al., 2007).
TOR complex 1 was described to be involved in the control of transcription, protein synthesis, and autophagy in yeast and mammals (Martin and Hall, 2005; De Virgilio and Loewith, 2006; Wang and Proud, 2011). In Arabidopsis, massive transcriptional changes induced by glucose in seedlings are dependent on TOR signaling. The genes activated by this signaling pathway are involved in amino acid synthesis, translation, glycolysis and the TCA cycle, cell wall synthesis and modification, whereas genes involved in protein and amino acid degradation and autophagy regulation were downregulated (Xiong et al., 2013). The role of TOR signaling in the induction of biosynthesis and the repression of catabolic pathways was underlined by RNA sequencing and microarray analysis studying gene expression changes in response to TOR inactivation (Ren et al., 2012; Caldana et al., 2013). These transcriptional changes were shown to be accompanied by an increase in starch content (Moreau et al., 2012) and an accumulation of organic and amino acids (Ren et al., 2012; Caldana et al., 2013), as well as a decrease in galactinol and raffinose levels (Moreau et al., 2012). This led to the conclusion that TOR downregulation mimics starvation (Caldana et al., 2013) and strengthens the importance of the TOR pathway in starvation responses.
Furthermore, the TOR kinase was implicated in sugar signaling pathways to control translation. Protein synthesis is a very energy demanding process and therefore needs to be tightly regulated in function of the cellular energy availability. In mammals and yeast, TORC1 affects the level of rRNA (Claypool et al., 2004; Mayer et al., 2004; Li et al., 2006a; Tsang et al., 2010) and RP gene expression (Jorgensen et al., 2004; Martin et al., 2004; Rudra and Warner, 2004), as well as cap-dependent translation (Ma and Blenis, 2009) and scanning along structured 5′-UTRs (Meyuhas and Dreazen, 2009).
The tight regulation of translation according to energy availability also occurs in plants (Pal et al., 2013; Lastdrager et al., 2014). For some of the well described pathway components, like the phosphorylation of 4E-BP, evidence in plants is lacking (Van Der Kelen et al., 2009; Ren et al., 2011). Nevertheless, plants expressing RNAi constructs for TOR or its positive downstream effector TAP46 displayed a significant decrease in polysomal loading and in protein synthesis (Ahn et al., 2011). The transcription of RP genes is induced after sugar treatment and seems to be dependent on the diurnal cycle (Bläsing et al., 2005; Usadel et al., 2008; Baerenfaller et al., 2012) and glucose induction of RP gene expression was shown to depend on TOR activity (Xiong et al., 2013). Furthermore, TOR overexpression was shown to induce rRNA production and ChIP experiments showed that TOR binds directly to the 45S promoter region (Ren et al., 2011). TOR mediated ribosomal protein S6 kinase (S6K) activity was proposed to be important to maintain eukaryotic translation initiation factor 3 subunit H (eIF3h) phosphorylation, which is needed for translation reinitiation and thus for translation of uORF containing mRNAs (Schepetilnikov et al., 2013). Starvation inhibits the TOR kinase and therefore allows energy costly ribosome biogenesis and translation processes to be reduced in growth limiting conditions (Ma and Blenis, 2009; Robaglia et al., 2012).
INTERACTION BETWEEN SnRK1 AND TOR SIGNALING
The TOR and SnRK1 signaling pathways have emerged as crucial in regulating the perception and responses to nutrient and energy levels. The TOR kinase is activated in favorable nutritional and energy conditions, while SnRK1 is stimulated upon nutrient and energy starvation. It is becoming increasingly clear that they act in opposite ways in the regulation of nutrient-driven processes, such as autophagy. For example, SnRK1 induces autophagy to promote recycling of cytosolic components in response to situations where C or N metabolites are in short supply. Conversely, TOR restrains autophagy in energy-replete conditions and is involved in the regulation of N assimilation and in the synthesis of C metabolites like starch or raffinose (Robaglia et al., 2012). They both regulate many similar processes in the context of LES (Figure 4) and massively affect the transcription of a number of genes. By comparing the expression of genes targeted by SnRK1/KIN10 (Baena-González et al., 2007) and TOR (Xiong et al., 2013) in two available transcriptional datasets (data was acquired from protoplast and seedlings, respectively), we found that there was a significant overlap in the genes affected by both TOR and SnRK1. More than half of the genes (294 out of 507) described as KIN10 upregulated target genes were found to be downregulated by glucose in a TOR-dependent manner. Interestingly, 47 genes which were oppositely affected by TOR and SnRK1 are annotated to encode RPs or proteins related to translation. Furthermore, a similar proportion of genes found to be downregulated by KIN10 (260 of 515 genes) were among the putative upregulated targets of the TOR kinase, including genes annotated to be involved in amino acid metabolism and involved in carbohydrate metabolism. This underlines the hypothesis that TOR and SnRK1 act antagonistically in the regulation of central processes such as translation and carbohydrate and amino acid metabolism, a role that is likely to be crucial during the establishment of stress responses.
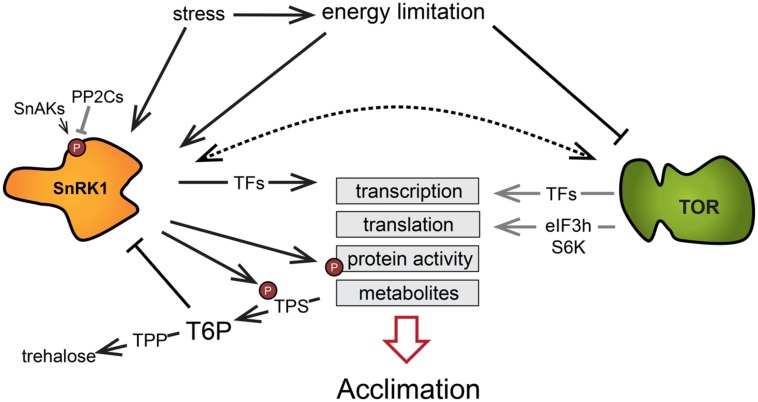
FIGURE 4.
The low energy syndrome involves processes regulated by TOR and SnRK1 central to the adaptation to energy limitation. The kinases SnRK1 and TOR are affected by energy availability and mediate transcription, translation, enzyme activity, and the accumulation of metabolites in order to drive adaptation to different conditions. Gray arrows indicate processes shut down in response to energy limitation and the dashed arrow denotes a probable interaction between these two signaling pathways. Abbreviations: SnAK, SnRK1-activating kinase; PP2C, type 2C protein phosphatase; TPS, trehalose phosphate synthase; T6P, trehalose 6-phosphate; TPP, trehalose phosphate phosphatase; eIF3h, eukaryotic translation initiation factor 3 subunit H; S6K, ribosomal protein S6 kinase.
Target of rapamycin and SnRK1 are both evolutionary conserved and have been shown to interact in mammalian systems. AMPK is described to regulate the mTOR complex 1 in different ways, by phosphorylating components of the mTOR signaling pathway (Xu et al., 2012). The phosphorylation of the TSC2/TSC1 complex by AMPK leads to the inactivation of Rheb, a GTPase that activates mTORC1. Furthermore, AMPK has been described to directly regulate the Raptor subunit of the mTORC1 complex (Inoki et al., 2012). mTOR and AMPK have opposite roles in the regulation of autophagy by targeting different phosphorylation sites of ULK1 (Kim et al., 2011). Under nutrient limitation, AMPK activates ULK1 and autophagy, whereas mTORC1 inactivates ULK1 in nutrient-rich conditions (Inoki et al., 2012). In plants, some of the described factors are present, including the Arabidopsis TCTP that was proposed as a regulator of Rheb in TOR signaling (Berkowitz et al., 2008) and the AMPK phosphorylation site in Raptor (Robaglia et al., 2012). However, their role in the interaction between the TOR and SnRK1 signaling pathways remains to be further analyzed. Additionally, many of the described genes are missing in Arabidopsis, such as Rheb itself, TSC1, and TSC2 (van Dam et al., 2011). Their functions were taken over by other factors, indicating that plants evolved energy signaling pathways different from mammals or yeast that remain to be fully described (Xiong and Sheen, 2014).
TOWARD A BETTER UNDERSTANDING OF LES
The studies discussed above underline the central role of the energy signaling network composed of TOR, SnRK1, bZIP transcription factors, and T6P in the control of growth and development. Exposure of plants to conditions that challenge their energy homeostasis results in significant metabolic reprogramming to prevent damage to cells, tissue, and organs. LES involves various changes in transcription, translation, enzymatic activities, and metabolite levels ultimately aiming at the adaptation to energy deprivation (Figure 4). Amongst others, this leads to protein, lipid, and chlorophyll breakdown (Contento et al., 2004; Thimm et al., 2004), while in parallel, carbon utilization is inhibited, which, taken together, severely affects growth (Gibon et al., 2004).
Even though much is already known about the LES network, there are still many open questions. More experimental data is needed to unravel unknown mechanisms behind LES, but several experimental limitations may be restraining new discoveries. For example, most studies were conducted in protoplasts, whole seedlings or plant tissue without distinguishing source and sink organs. Since the interplay of molecules is complex and coordinated in both time and space, increasing spatial resolution of metabolites with the use of better techniques, like subcellular fractionation (Nägele and Heyer, 2013), is necessary to enable a better understanding of regulation of stress conditions by allowing tissue-, cell-, and compartment specific analyses of metabolic changes. Furthermore, the pleiotropic nature of SnRK1 and TOR limits the use of mutants as mutations result in undesired effects that can hardly be precluded. The use of constitutive promoters for overexpression leads to the loss of information about the localization of the expression of a certain gene. This information is very important as many of the discussed genes are differentially expressed between source or sink tissues, which likely impacts on their role and function. It would thus be useful to develop additional mutants, especially including organ and temporal specific promoters, to overcome these current issues. The use of inducible systems is of high interest for both SnRK1 and TOR, but particularly for the latter since knock-out mutants are not available due to lethality. Hence, the uses of inducible artificial microRNAs are a valuable option to down-regulate its expression.
The study of networks like LES requires the analysis of complex datasets. It is important to realize that the challenge is not only the acquisition of more and better experimental data, but especially to be able to integrate it to facilitate its interpretation via methods often referred to as systems biology or omics-based approaches. To improve upon current approaches, theoretical methods of uni- and multivariate statistics and mathematical modeling have been developed which now allow large scale analysis of biological networks (Nägele and Weckwerth, 2012). Several problems related to plant science are being addressed using these approaches, including responses to stress, (Yamaguchi-Shinozaki and Shinozaki, 2006), plant defense (Li et al., 2006b) and the identification of new players, such as transcription factors (Hirai et al., 2007).
Low energy signaling is a complex network that integrates multiple cellular and environmental signals and comprises several cellular changes. To avoid limitation in the interpretation of the biological events, it is important to study this pathway on all possible levels, comprising the gene, transcript, protein, and metabolite levels. Limitations can arise, for example, from the consideration of only transcriptomic data. With this type of data, differences in transcription and RNA stability cannot be distinguished. Also, changes in transcript levels often do not reflect changes in protein or metabolic levels and little information is obtained at this level. Substantial contributions have been made mainly concerning the integration of transcript and metabolite data for A. thaliana (Hirai et al., 2004, 2005; Tohge et al., 2005). By combining data from different molecular levels, a better understanding of pathway regulation being affected under low energy conditions and contributing to the acclimatory response will be significantly promoted. One type of systems biology approach is based on the collection of data from different platforms followed by data driven integration using advanced statistical models to study the dynamic interactions between components (Yuan et al., 2008; Fukushima et al., 2009; Keurentjes, 2009). Another more targeted approach where a specific regulatory model involving all known molecular players is built uses mathematical modeling to advance understanding of biological processes (Pokhilko et al., 2010, 2012, 2013; Gould et al., 2013; Seaton et al., 2014). The interaction between predictive models and experimental confirmation can be very effective, as having a more directed approach in a given experiment could enable the achievement of faster and more targeted results. Furthermore, unknown components of the studied system can be discovered (Dalchau et al., 2011). Some of the components that comprise the LES network may still be unknown, but this type of approach could provide very useful information especially in the connection between TOR and SnRK1.
The use of systems biology is limited by the availability of data and requires generalization, simplification, and assumptions. However, it clearly has great potential to increasingly contribute to the understanding of biological networks, including the LES pathway, in combination with other approaches that involve cell biology, biochemistry, or genetics. TOR and SnRK1 are related in the processes they regulate and are activated under opposite conditions, but so far there is no indication in plants that they or their targets interact directly to optimize the activation and repression of certain processes. While in mammals AMPK phosphorylates the TSC2/TSC1 complex and Raptor, TSC2 and the AMPK phosphorylation site in Raptor are missing in Arabidopsis and it is not yet clear whether TOR and SnRK1 pathways interact in a similar way. It is possible that despite their connection, TOR down-regulation is just another way to limit energy use, independent from SnRK1. However, given the complexity of the network and the interconnections seen so far, not only in plants but also in other systems, it seems more likely that additional links or factors are yet to be unraveled. It is crucial to establish if there is a direct interaction between TOR and SnRK1, or if they act independently. In this context, a better understanding of their (sub-)cellular localization could provide insights to their mode of action and possible interaction. Protein interaction studies may uncover novel interactions between known LES components or even unravel new components of the network. Their role in the metabolic reprogramming induced by energy deprivation may then be tested by metabolomics studies which directly give information about changes in the concentrations of central metabolite pools. A multidisciplinary approach on various levels of cellular and organismal organization is needed to be able to draw a comprehensive picture about low energy induced metabolic reprogramming.
AUTHOR CONTRIBUTIONS
Filipa Tomé is first author, Thomas Nägele is second author, Magdalena Gamm is last author, and all other authors contributed equally to the work.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Johannes Hanson and Matthew Hannah for critical reading and helpful comments. Furthermore, we are grateful to Elena Baena-González, Christina Chaban, Wolfgang Dröge-Laser, Berend Snel, Markus Teige, Jesus V. Carbajosa, Wolfram Weckwerth, and Ulrike Zentgraf for their ideas and support. All authors are part of the Marie Curie ITN project MERIT, Grant Agreement number 264474, from where financial support was obtained. The authors would like to thank the reviewers for their comments which helped to improve this manuscript.
REFERENCES
- Ahn C. S., Han J.-A., Lee H.-S., Lee S., Pai H.-S. (2011). The PP2A regulatory subunit Tap46, a component of the TOR signaling pathway, modulates growth and metabolism in plants. Plant Cell 23 185–209 10.1105/tpc.110.074005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso R., Oñate-Sánchez L., Weltmeier F., Ehlert A., Diaz I., Dietrich K., et al. (2009). A pivotal role of the basic leucine zipper transcription factor bZIP53 in the regulation of Arabidopsis seed maturation gene expression based on heterodimerization and protein complex formation. Plant Cell 21 1747–1761 10.1105/tpc.108.062968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo W. L., Nunes-Nesi A., Nikoloski Z., Sweetlove L. J., Fernie A. R. (2012). Metabolic control and regulation of the tricarboxylic acid cycle in photosynthetic and heterotrophic plant tissues. Plant Cell Environ. 35 1–21 10.1111/j.1365-3040.2011.02332.x [DOI] [PubMed] [Google Scholar]
- Ávila-Castañeda A., Gutiérrez-Granados N., Ruiz-Gayosso A., Sosa-Peinado A., Martínez-Barajas E., Coello P. (2014). Structural and functional basis for starch binding in the SnRK1 subunits AKINß2 and AKINßγ. Front. Plant Sci. 5:1–8 10.3389/fpls.2014.00199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avin-Wittenberg T., Tzin V., Angelovici R., Less H., Galili G. (2012). Deciphering energy-associated gene networks operating in the response of Arabidopsis plants to stress and nutritional cues. Plant J. 70 954–966 10.1111/j.1365-313X.2012.04926.x [DOI] [PubMed] [Google Scholar]
- Baena-González E. (2010). Energy signaling in the regulation of gene expression during stress. Mol. Plant 3 300–313 10.1093/mp/ssp113 [DOI] [PubMed] [Google Scholar]
- Baena-González E., Rolland F., Thevelein J. M., Sheen J. (2007). A central integrator of transcription networks in plant stress and energy signaling. Nature 448 938–942 10.1038/nature06069 [DOI] [PubMed] [Google Scholar]
- Baena-González E., Sheen J. (2008). Convergent energy and stress signaling. Trends Plant Sci. 13 474–482 10.1016/j.tplants.2008.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baerenfaller K., Massonnet C., Walsh S., Baginsky S., Bühlmann P., Hennig L., et al. (2012). Systems-based analysis of Arabidopsis leaf growth reveals adaptation to water deficit. Mol. Syst. Biol. 8:606 10.1038/msb.2012.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey-Serres J., Fukao T., Gibbs D. J., Holdsworth M. J., Lee S. C., Licausi F., et al. (2012). Making sense of low oxygen sensing. Trends Plant Sci. 17 129–138 10.1016/j.tplants.2011.12.004 [DOI] [PubMed] [Google Scholar]
- Berkowitz O., Jost R., Pollmann S., Masle J. (2008). Characterization of TCTP, the translationally controlled tumor protein, from Arabidopsis thaliana. Plant Cell 20 3430–3447 10.1105/tpc.108.061010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bläsing O., Gibon Y., Günther M., Höhne M., Morcuende R., Osuna D., et al. (2005). Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. Plant Cell 17 3257–3281 10.1105/tpc.105.035261.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bögre L., Henriques R., Magyar Z. (2013). TOR tour to auxin. EMBO J. 32 1069–1071 10.1038/emboj.2013.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouly J., Gissot L., Lessard P., Kreis M., Thomas M. (1999). Arabidopsis thaliana proteins related to the yeast SIP and SNF4 interact with AKINa1, an SNF1-like protein kinase. Plant J. 18 541–550 10.1046/j.1365-313X.1999.00476.x [DOI] [PubMed] [Google Scholar]
- Branco-Price C., Kaiser K. A., Jang C. J., Larive C. K., Bailey-Serres J. (2008). Selective mRNA translation coordinates energetic and metabolic adjustments to cellular oxygen deprivation and reoxygenation in Arabidopsis thaliana. Plant J. 56 743–755 10.1111/j.1365-313X.2008.03642.x [DOI] [PubMed] [Google Scholar]
- Branco-Price C., Kawaguchi R., Ferreira R. B., Bailey-Serres J. (2005). Genome-wide analysis of transcript abundance and translation in Arabidopsis seedlings subjected to oxygen deprivation. Ann. Bot. 96 647–660 10.1093/aob/mci217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldana C., Degenkolbe T., Cuadros-Inostroza A., Klie S., Sulpice R., Leisse A., et al. (2011). High-density kinetic analysis of the metabolomic and transcriptomic response of Arabidopsis to eight environmental conditions. Plant J. 67 869–884 10.1111/j.1365-313X.2011.04640.x [DOI] [PubMed] [Google Scholar]
- Caldana C., Li Y., Leisse A., Zhang Y., Bartholomaeus L., Fernie A. R., et al. (2013). Systemic analysis of inducible target of rapamycin mutants reveal a general metabolic switch controlling growth in Arabidopsis thaliana. Plant J. 73 897–909 10.1111/tpj.12080 [DOI] [PubMed] [Google Scholar]
- Carillo P., Feil R., Gibon Y., Satoh-Nagasawa N., Jackson D., Bläsing O. E., et al. (2013). A fluorometric assay for trehalose in the picomole range. Plant Methods 9:21 10.1186/1746-4811-9-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V., Schumaker K., Zhu J.-K. (2004). Molecular genetic perspectives on cross-talk and specificity in abiotic stress signaling in plants. J. Exp. Bot. 55 225–236 10.1093/jxb/erh005 [DOI] [PubMed] [Google Scholar]
- Cho Y.-H., Hong J.-W., Kim E.-C., Yoo S.-D. (2012). Regulatory functions of SnRK1 in stress-responsive gene expression and in plant growth and development. Plant Physiol. 158 1955–1964 10.1104/pp.111.189829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke P. R., Hardie D. G. (1990). Regulation of HMG-CoA reductase: identification of the site phosphorylated by the AMP-activated protein kinase in vitro and in intact rat liver. EMBO J. 9 2439–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claypool J. A., French S. L., Johzuka K., Eliason K., Vu L., Dodd J. A., et al. (2004). Tor pathway regulates Rrn3p-dependent recruitment of yeast RNA polymerase I to the promoter but does not participate in alteration of the number of active genes. Mol. Biol. Cell 15 946–956 10.1091/mbc.E03 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coello P., Hey S. J., Halford N. G. (2011). The sucrose non-fermenting-1-related (SnRK) family of protein kinases: potential for manipulation to improve stress tolerance and increase yield. J. Exp. Bot. 62 883–893 10.1093/jxb/erq331 [DOI] [PubMed] [Google Scholar]
- Confraria A., Martinho C., Elias A., Rubio-Somoza I., Baena-González E. (2013). miRNAs mediate SnRK1-dependent energy signaling in Arabidopsis. Front. Plant Sci. 4:197 10.3389/fpls.2013.00197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contento A. L., Kim S., Bassham D. C. (2004). Transcriptome profiling of the response of Arabidopsis suspension culture cells to Suc starvation. Plant Physiol. 135 2330–2347 10.1104/pp.104.044362.2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrêa L. G. G., Riaño-Pachón D. M., Schrago C. G., dos Santos R. V., Mueller-Roeber B., Vincentz M. (2008). The role of bZIP transcription factors in green plant evolution: adaptive features emerging from four founder genes. PLoS ONE 3:e2944 10.1371/journal.pone.0002944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crozet P., Jammes F., Valot B., Ambard-Bretteville F., Nessler S., Hodges M., et al. (2010). Cross-phosphorylation between Arabidopsis thaliana sucrose nonfermenting 1-related protein kinase 1 (AtSnRK1) and its activating kinase (AtSnAK) determines their catalytic activities. J. Biol. Chem. 285 12071–12077 10.1074/jbc.M109.079194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuellar-Ortiz S. M., De La Paz Arrieta-Montiel M., Acosta-Gallegos J., Covarrubias A. (2008). Relationship between carbohydrate partitioning and drought resistance in common bean. Plant Cell Environ. 31 1399–1409 10.1111/j.1365-3040.2008.01853.x [DOI] [PubMed] [Google Scholar]
- Dalchau N., Baek S. J., Briggs H. M., Robertson F. C., Dodd A. N., Gardner M. J., et al. (2011). The circadian oscillator gene GIGANTEA mediates a long-term response of the Arabidopsis thaliana circadian clock to sucrose. Proc. Natl. Acad. Sci. U.S.A. 108 5104–5109 10.1073/pnas.1015452108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deprost D., Truong H.-N., Robaglia C., Meyer C. (2005). An Arabidopsis homolog of RAPTOR/KOG1 is essential for early embryo development. Biochem. Biophys. Res. Commun. 326 844–850 10.1016/j.bbrc.2004.11.117 [DOI] [PubMed] [Google Scholar]
- Deprost D., Yao L., Sormani R., Moreau M., Leterreux G., Nicolaï M., et al. (2007). The Arabidopsis TOR kinase links plant growth, yield, stress resistance and mRNA translation. EMBO Rep. 8 864–870 10.1038/sj.embor.7401043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Virgilio C., Loewith R. (2006). The TOR signaling network from yeast to man. Int. J. Biochem. Cell Biol. 38 1476–1481 10.1016/j.biocel.2006.02.013 [DOI] [PubMed] [Google Scholar]
- Dietrich K., Weltmeier F., Ehlert A., Weiste C., Stahl M., Harter K., et al. (2011). Heterodimers of the Arabidopsis transcription factors bZIP1 and bZIP53 reprogram amino acid metabolism during low energy stress. Plant Cell 23 381–395 10.1105/tpc.110.075390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas P., Pigaglio E., Ferrer A., Halford N. G., MacKintosh C. (1997). Three spinach leaf nitrate reductase-3-hydroxy-3-methylglutaryl-CoA reductase kinases that are regulated by reversible phosphorylation and/or Ca2+ ions. Biochem. J. 325 101–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte G. T., Matiolli C. C., Pant B. D., Schlereth A., Scheible W.-R., Stitt M., et al. (2013). Involvement of microRNA-related regulatory pathways in the glucose-mediated control of Arabidopsis early seedling development. J. Exp. Bot. 64 4301–4312 10.1093/jxb/ert239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbein A. D., Pan Y. T., Pastuszak I., Carroll D. (2003). New insights on trehalose: a multifunctional molecule. Glycobiology 13 17R–27R 10.1093/glycob/cwg047 [DOI] [PubMed] [Google Scholar]
- Fernandez O., Vandesteene L., Feil R., Baillieul F., Lunn J. E., Clément C. (2012). Trehalose metabolism is activated upon chilling in grapevine and might participate in Burkholderia phytofirmans induced chilling tolerance. Planta 236 355–369 10.1007/s00425-012-1611-4 [DOI] [PubMed] [Google Scholar]
- Fujita Y., Nakashima K., Yoshida T., Katagiri T., Kidokoro S., Kanamori N., et al. (2009). Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol. 50 2123–2132 10.1093/pcp/pcp147 [DOI] [PubMed] [Google Scholar]
- Fukushima A., Kusano M., Redestig H., Arita M., Saito K. (2009). Integrated omics approaches in plant systems biology. Curr. Opin. Chem. Biol. 13 532–538 10.1016/j.cbpa.2009.09.022 [DOI] [PubMed] [Google Scholar]
- Geigenberger P. (2003). Response of plant metabolism to too little oxygen. Curr. Opin. Plant Biol. 6 247–256 10.1016/S1369-5266(03)00038-4 [DOI] [PubMed] [Google Scholar]
- Ghillebert R., Swinnen E., Wen J., Vandesteene L., Ramon M., Norga K., et al. (2011). The AMPK/SNF1/SnRK1 fuel gauge and energy regulator: structure, function and regulation. FEBS J. 278 3978–3990 10.1111/j.1742-4658.2011.08315.x [DOI] [PubMed] [Google Scholar]
- Gibon Y., Bläsing O. E., Palacios-Rojas N., Pankovic D., Hendriks J. H. M., Fisahn J., et al. (2004). Adjustment of diurnal starch turnover to short days: depletion of sugar during the night leads to a temporary inhibition of carbohydrate utilization, accumulation of sugars and post-translational activation of ADP-glucose pyrophosphorylase in the followin. Plant J. 39 847–862 10.1111/j.1365-313X.2004.02173.x [DOI] [PubMed] [Google Scholar]
- Gibon Y., Usadel B., Blaesing O. E., Kamlage B., Hoehne M., Trethewey R., et al. (2006). Integration of metabolite with transcript and enzyme activity profiling during diurnal cycles in Arabidopsis rosettes. Genome Biol. 7:R76 10.1186/gb-2006-7-8-r76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibon Y., Vigeolas H., Tiessen A., Geigenberger P., Stitt M. (2002). Sensitive and high throughput metabolite assays for inorganic pyrophosphate, ADPGlc, nucleotide phosphates, and glycolytic intermediates based on a novel enzymic cycling system. Plant J. 30 221–235 10.1046/j.1365-313X.2001.01278.x [DOI] [PubMed] [Google Scholar]
- Gissot L., Polge C., Bouly J., Lemaitre T., Kreis M., Thomas M. (2004). AKINβ3, a plant specific SnRK1 protein, is lacking domains present in yeast and mammals non-catalytic β-subunits. Plant Mol. Biol. 56 747–759 10.1007/s11103-004-5111-1 [DOI] [PubMed] [Google Scholar]
- Glinski M., Weckwerth W. (2005). Differential multisite phosphorylation of the trehalose-6-phosphate synthase gene family in Arabidopsis thaliana: a mass spectrometry-based process for multiparallel peptide library phosphorylation analysis. Mol. Cell. Proteomics 4 1614–1625 10.1074/mcp.M500134-MCP200 [DOI] [PubMed] [Google Scholar]
- Gómez L. D., Gilday A., Feil R., Lunn J. E., Graham I. A. (2010). AtTPS1-mediated trehalose 6-phosphate synthesis is essential for embryogenic and vegetative growth and responsiveness to ABA in germinating seeds and stomatal guard cells. Plant J. 64 1–13 10.1111/j.1365-313X.2010.04312.x [DOI] [PubMed] [Google Scholar]
- Gould P. D., Ugarte N., Domijan M., Costa M., Foreman J., Macgregor D., et al. (2013). Network balance via CRY signaling controls the Arabidopsis circadian clock over ambient temperatures. Mol. Syst. Biol. 9:650 10.1038/msb.2013.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halford N. G., Hardie D. G. (1998). SNF1-related protein kinases: global regulators of carbon metabolism in plants? Plant Mol. Biol. 37 735–748 10.1023/A:1006024231305 [DOI] [PubMed] [Google Scholar]
- Halford N. G., Hey S. J. (2009). Snf1-related protein kinases (SnRKs) act within an intricate network that links metabolic and stress signaling in plants. Biochem. J. 419 247–259 10.1042/BJ20082408 [DOI] [PubMed] [Google Scholar]
- Halford N. G., Hey S., Jhurreea D., Laurie S., McKibbin R. S., Zhang Y., et al. (2004). Highly conserved protein kinases involved in the regulation of carbon and amino acid metabolism. J. Exp. Bot. 55 35–42 10.1093/jxb/erh019 [DOI] [PubMed] [Google Scholar]
- Hanson J., Hanssen M., Wiese A., Hendriks M. M., Smeekens S. (2008). The sucrose regulated transcription factor bZIP11 affects amino acid metabolism by regulating the expression of ASPARAGINE SYNTHETASE1 and PROLINE DEHYDROGENASE2. Plant J. 53 935–949 10.1111/j.1365-313X.2007.03385.x [DOI] [PubMed] [Google Scholar]
- Hardie D. G., Carling D., Carlson M. (1998). The AMP-activated/SNF1 protein kinase subfamily: metabolic sensors of the eukaryotic cell? Annu. Rev. Biochem. 67 821–855 10.1146/annurev.biochem.67.1.821 [DOI] [PubMed] [Google Scholar]
- Hay N., Sonenberg N. (2004). Upstream and downstream of mTOR. Genes Dev. 18 1926–1945 10.1101/gad.1212704 [DOI] [PubMed] [Google Scholar]
- Hirai M. Y., Klein M., Fujikawa Y., Yano M., Goodenowe D. B., Yamazaki Y., et al. (2005). Elucidation of gene-to-gene and metabolite-to-gene networks in Arabidopsis by integration of metabolomics and transcriptomics. J. Biol. Chem. 280 25590–25595 10.1074/jbc.M502332200 [DOI] [PubMed] [Google Scholar]
- Hirai M. Y., Sugiyama K., Sawada Y., Tohge T., Obayashi T., Suzuki A., et al. (2007). Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 104 6478–6483 10.1073/pnas.0611629104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai M. Y., Yano M., Goodenowe D. B., Kanaya S., Kimura T., Awazuhara M., et al. (2004). Integration of transcriptomics and metabolomics for understanding of global responses to nutritional stresses in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 101 10205–10210 10.1073/pnas.0403218101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrabak E. M., Chan C. W. M., Gribskov M., Harper J. F., Choi J. H., Halford N., et al. (2003). The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 132 666–680 10.1104/pp.102.011999.666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K., Kim J., Guan K.-L. (2012). AMPK and mTOR in cellular energy homeostasis and drug targets. Annu. Rev. Pharmacol. Toxicol. 52 381–400 10.1146/annurev-pharmtox-010611-134537 [DOI] [PubMed] [Google Scholar]
- Jakoby M., Weisshaar B., Dröge-Laser W., Vicente-Carbajosa J., Tiedemann J., Kroj T., et al. (2002). bZIP transcription factors in Arabidopsis. Trends Plant Sci. 7 106–111 10.1016/S1360-1385(01)02223-3 [DOI] [PubMed] [Google Scholar]
- Jewell J. L., Russell R. C., Guan K.-L. (2013). Amino acid signaling upstream of mTOR. Nat. Rev. Mol. Cell Biol. 14 133–139 10.1038/nrm3522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen P., Rupeš I., Sharom J. R., Schneper L., Broach J. R., Tyers M. (2004). A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 18 2491–2505 10.1101/gad.1228804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juntawong P., Bailey-Serres J. (2012). Dynamic light regulation of translation status in Arabidopsis thaliana. Front. Plant Sci. 3:66 10.3389/fpls.2012.00066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S. G., Price J., Lin P.-C., Hong J. C., Jang J.-C. (2010). The Arabidopsis bZIP1 transcription factor is involved in sugar signaling, protein networking, and DNA binding. Mol. Plant 3 361–373 10.1093/mp/ssp115 [DOI] [PubMed] [Google Scholar]
- Kawaguchi R., Girke T., Bray E. A., Bailey-Serres J. (2004). Differential mRNA translation contributes to gene regulation under non-stress and dehydration stress conditions in Arabidopsis thaliana. Plant J. 38 823–839 10.1111/j.1365-313X.2004.02090.x [DOI] [PubMed] [Google Scholar]
- Kawaguchi R., Williams A. J., Bray E. A., Bailey-Serres J. (2003). Water-deficit-induced translational control in Nicotiana tabacum. Plant Cell Environ. 26 221–229 10.1046/j.1365-3040.2003.00952.x [DOI] [Google Scholar]
- Keurentjes J. J. B. (2009). Genetical metabolomics: closing in on phenotypes. Curr. Opin. Plant Biol. 12 223–230 10.1016/j.pbi.2008.12.003 [DOI] [PubMed] [Google Scholar]
- Kim J., Kundu M., Viollet B., Guan K.-L. (2011). AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 13 132–141 10.1038/ncb2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lastdrager J., Hanson J., Smeekens S. (2014). Sugar signals and the control of plant growth and development. J. Exp. Bot. 65 799–807 10.1093/jxb/ert474 [DOI] [PubMed] [Google Scholar]
- Lemoine R., La Camera S., Atanassova R., Dédaldéchamp F., Allario T., Pourtau N., et al. (2013). Source-to-sink transport of sugar and regulation by environmental factors. Front. Plant Sci. 4:272 10.3389/fpls.2013.00272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Tsang C. K., Watkins M., Bertram P. G., Zheng X. F. S. (2006a). Nutrient regulates Tor1 nuclear localization and association with rDNA promoter. Nature 442 1058–1061 10.1038/nature05020 [DOI] [PubMed] [Google Scholar]
- Li S., Assmann S. M., Albert R. (2006b). Predicting essential components of signal transduction networks: a dynamic model of guard cell abscisic acid signaling. PLoS Biol. 4:e312 10.1371/journal.pbio.0040312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Ishitani M., Halfter U., Kim C. S., Zhu J. K. (2000). The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc. Natl. Acad. Sci. U.S.A. 97 3730–3734 10.1073/pnas.060034197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Liu W., Ying H., Zhao W., Zhang H. (2013a). Analysis of microRNA expression profile induced by AICAR in mouse hepatocytes. Gene 512 364–372 10.1016/j.gene.2012.09.118 [DOI] [PubMed] [Google Scholar]
- Liu Y.-H., Offler C. E., Ruan Y.-L. (2013b). Regulation of fruit and seed response to heat and drought by sugars as nutrients and signals. Front. Plant Sci. 4:282 10.3389/fpls.2013.00282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunn J. E., Delorge I., Figueroa C. M., Van Dijck P., Stitt M. (2014). Trehalose metabolism in plants. Plant J. 10.1111/tpj.12509 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Lunn J. E., Feil R., Hendriks J. H. M., Gibon Y., Morcuende R., Osuna D., et al. (2006). Sugar-induced increases in trehalose 6-phosphate are correlated with redox activation of ADPglucose pyrophosphorylase and higher rates of starch synthesis in Arabidopsis thaliana. Biochem. J. 397 139–148 10.1042/BJ20060083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Hanssen M., Lundgren K., Hernández L., Delatte T., Ehlert A., et al. (2011). The sucrose-regulated Arabidopsis transcription factor bZIP11 reprograms metabolism and regulates trehalose metabolism. New Phytol. 191 733–745 10.1111/j.1469-8137.2011.03735.x [DOI] [PubMed] [Google Scholar]
- Ma X. M., Blenis J. (2009). Molecular mechanisms of mTOR-mediated translational control. Nat. Rev. Mol. Cell Biol. 10 307–318 10.1038/nrm2672 [DOI] [PubMed] [Google Scholar]
- Mackintosh R. W., Davies S. P., Clarke P. R., Weekes J., Gillespie J. G., Gibb B. J., et al. (1992). Evidence for a protein kinase cascade in higher plants 3-hydroxy-3-methylglutaryl-CoA reductase kinase. Eur. J. Biochem 209 923–931 10.1111/j.1432-1033.1992.tb17364.x [DOI] [PubMed] [Google Scholar]
- Martin D. E., Hall M. N. (2005). The expanding TOR signaling network. Curr. Opin. Cell Biol. 17 158–166 10.1016/j.ceb.2005.02.008 [DOI] [PubMed] [Google Scholar]
- Martin D. E., Soulard A., Hall M. N. (2004). TOR regulates ribosomal protein gene expression via PKA and the Forkhead transcription factor FHL1. Cell 119 969–979 10.1016/j.cell.2004.11.047 [DOI] [PubMed] [Google Scholar]
- Mayer C., Zhao J., Yuan X., Grummt I. (2004). mTOR-dependent activation of the transcription factor TIF-IA links rRNA synthesis to nutrient availability. Genes Dev. 18 423–434 10.1101/gad.285504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMichael R. W., Bachmann M., Huber S. C. (1995). Spinach leaf sucrose-phosphate synthase and nitrate reductase are phosphorylated/inactivated by multiple protein kinases in vitro. Plant Physiol. 1081077–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menand B., Desnos T., Nussaume L., Berger F., Bouchez D., Meyer C., et al. (2002). Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proc. Natl. Acad. Sci. U.S.A. 99 6422–6427 10.1073/pnas.092141899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyuhas O., Dreazen A. (2009). Ribosomal protein S6 kinase from TOP mRNAs to cell size. Prog. Mol. Biol. Transl. Sci. 90 109–153 10.1016/S1877-1173(09)90003-5 [DOI] [PubMed] [Google Scholar]
- Miyashita Y., Good A. G. (2008a). Glutamate deamination by glutamate dehydrogenase plays a central role in amino acid catabolism in plants. Plant Signal. Behav. 3 842–843 10.1093/jxb/erm340.842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita Y., Good A. G. (2008b). NAD(H)-dependent glutamate dehydrogenase is essential for the survival of Arabidopsis thaliana during dark-induced carbon starvation. J. Exp. Bot. 59 667–680 10.1093/jxb/erm340 [DOI] [PubMed] [Google Scholar]
- Montané M.-H., Menand B. (2013). ATP-competitive mTOR kinase inhibitors delay plant growth by triggering early differentiation of meristematic cells but no developmental patterning change. J. Exp. Bot. 64 4361–4374 10.1093/jxb/ert242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau M., Azzopardi M., Clément G., Dobrenel T., Marchive C., Renne C., et al. (2012). Mutations in the Arabidopsis homolog of LST8 / GbL, a partner of the target of rapamycin kinase, impair plant growth, flowering, and metabolic adaptation to long days. Plant Cell 24 463–481 10.1105/tpc.111.091306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller B., Pantin F., Génard M., Turc O., Freixes S., Piques M., et al. (2011). Water deficits uncouple growth from photosynthesis, increase C content, and modify the relationships between C and growth in sink organs. J. Exp. Bot. 62 1715–1729 10.1093/jxb/erq438 [DOI] [PubMed] [Google Scholar]
- Mustroph A., Zanetti M. E., Jang C. J. H., Holtan H. E., Repetti P. P., Galbraith D. W., et al. (2009). Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106 18843–18848 10.1073/pnas.0906131106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nägele T., Heyer A. G. (2013). Approximating subcellular organisation of carbohydrate metabolism during cold acclimation in different natural accessions of Arabidopsis thaliana. New Phytol. 198 777–787 10.1111/nph.12201 [DOI] [PubMed] [Google Scholar]
- Nägele T., Weckwerth W. (2012). Mathematical modeling of plant metabolism—from reconstruction to prediction. Metabolites 2 553–566 10.3390/metabo2030553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolaï M., Roncato M. A., Canoy A. S., Rouquié D., Sarda X., Freyssinet G., et al. (2006). Large-scale analysis of mRNA translation states during sucrose starvation in Arabidopsis cells identifies cell proliferation and chromatin structure as targets of translational control. Plant Physiol. 141 663–673 10.1104/pp.106.079418.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes C., O’Hara L. E., Primavesi L. F., Delatte T. L., Schluepmann H., Somsen G. W., et al. (2013). The trehalose 6-phosphate/SnRK1 signaling pathway primes growth recovery following relief of sink limitation. Plant Physiol. 162 1720–1732 10.1104/pp.113.220657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hara L. E., Paul M. J., Wingler A. (2013). How do sugars regulate plant growth and development? New insight into the role of trehalose-6-phosphate. Mol. Plant 6 261–274 10.1093/mp/sss120 [DOI] [PubMed] [Google Scholar]
- Osuna D., Usadel B., Morcuende R., Gibon Y., Bläsing O. E., Höhne M., et al. (2007). Temporal responses of transcripts, enzyme activities and metabolites after adding sucrose to carbon-deprived Arabidopsis seedlings. Plant J. 49 463–491 10.1111/j.1365-313X.2006.02979.x [DOI] [PubMed] [Google Scholar]
- Pal S. K., Liput M., Piques M., Ishihara H., Obata T., Martins M. C. M., et al. (2013). Diurnal changes of polysome loading track sucrose content in the rosette of wild-type Arabidopsis and the starchless pgm mutant. Plant Physiol. 162 1246–1265 10.1104/pp.112.212258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul M. J., Pellny T. K. (2003). Carbon metabolite feedback regulation of leaf photosynthesis and development. J. Exp. Bot. 54 539–547 10.1093/jxb/erg052 [DOI] [PubMed] [Google Scholar]
- Paul M. J., Primavesi L. F., Jhurreea D., Zhang Y. (2008). Trehalose metabolism and signaling. Annu. Rev. Plant Biol. 59 417–441 10.1146/annurev.arplant.59.032607.092945 [DOI] [PubMed] [Google Scholar]
- Pinheiro C., Chaves M. M., Ricardo C. P. (2001). Alterations in carbon and nitrogen metabolism induced by water deficit in the stems and leaves of Lupinus albus L. J. Exp. Bot. 52 1063–1070 10.1093/jexbot/52.358.1063 [DOI] [PubMed] [Google Scholar]
- Piques M., Schulze W. X., Höhne M., Usadel B., Gibon Y., Rohwer J., et al. (2009). Ribosome and transcript copy numbers, polysome occupancy and enzyme dynamics in Arabidopsis. Mol. Syst. Biol. 5:314 10.1038/msb.2009.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokhilko A., Fernández A. P., Edwards K. D., Southern M. M., Halliday K. J., Millar A. J. (2012). The clock gene circuit in Arabidopsis includes a repressilator with additional feedback loops. Mol. Syst. Biol. 8:574 10.1038/msb.2012.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokhilko A., Hodge S. K., Stratford K., Knox K., Edwards K. D., Thomson A. W., et al. (2010). Data assimilation constrains new connections and components in a complex, eukaryotic circadian clock model. Mol. Syst. Biol. 6:416 10.1038/msb.2010.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokhilko A., Mas P., Millar A. J. (2013). Modelling the widespread effects of TOC1 signaling on the plant circadian clock and its outputs. BMC Syst. Biol. 7:23 10.1186/1752-0509-7-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polge C., Thomas M. (2007). SNF1/AMPK/SnRK1 kinases, global regulators at the heart of energy control? Trends Plant Sci. 12 20–28 10.1016/j.tplants.2006.11.005 [DOI] [PubMed] [Google Scholar]
- Rahmani F., Hummel M., Schuurmans J., Wiese-Klinkenberg A., Smeekens S., Hanson J. (2009). Sucrose control of translation mediated by an upstream open reading frame-encoded peptide. Plant Physiol. 150 1356–1367 10.1104/pp.109.136036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramon M., Rolland F. (2007). Plant development: introducing trehalose metabolism. Trends Plant Sci. 12 185–188 10.1016/j.tplants.2007.03.007 [DOI] [PubMed] [Google Scholar]
- Reinke A. W., Baek J., Ashenberg O., Keating A. E. (2013). Networks of bZIP protein-protein interactions diversified over a billion years of evolution. Science 340 730–734 10.1126/science.1233465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren M., Qiu S., Venglat P., Xiang D., Feng L., Selvaraj G., et al. (2011). Target of rapamycin regulates development and ribosomal RNA expression through kinase domain in Arabidopsis. Plant Physiol. 155 1367–1382 10.1104/pp.110.169045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren M., Venglat P., Qiu S., Feng L., Cao Y., Wang E., et al. (2012). Target of rapamycin signaling regulates metabolism, growth, and life span in Arabidopsis. Plant Cell 24 4850–4874 10.1105/tpc.112.107144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robaglia C., Thomas M., Meyer C. (2012). Sensing nutrient and energy status by SnRK1 and TOR kinases. Curr. Opin. Plant Biol. 15 301–307 10.1016/j.pbi.2012.01.012 [DOI] [PubMed] [Google Scholar]
- Rodrigues A., Adamo M., Crozet P., Margalha L., Confraria A., Martinho C., et al. (2013). ABI1 and PP2CA phosphatases are negative regulators of Snf1-related protein kinase1 signaling in Arabidopsis. Plant Cell 1–15 10.1105/tpc.113.114066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland F., Baena-Gonzalez E., Sheen J. (2006). Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu. Rev. Plant Biol. 57 675–709 10.1146/annurev.arplant.57.032905.105441 [DOI] [PubMed] [Google Scholar]
- Rook F., Gerrits N., Kortstee A., van Kampen M., Borrias M., Weisbeek P., et al. (1998). Sucrose-specific signaling represses translation of the Arabidopsis ATB2 bZIP transcription factor gene. Plant J. 15 253–263 10.1046/j.1365-313X.1998.00205.x [DOI] [PubMed] [Google Scholar]
- Rudra D., Warner J. R. (2004). What better measure than ribosome synthesis? Genes Dev. 18 2431–2436 10.1101/gad.1256704 [DOI] [PubMed] [Google Scholar]
- Sablowski R., Carnier Dornelas M. (2014). Interplay between cell growth and cell cycle in plants. J. Exp. Bot. 65 2703–2714 10.1093/jxb/ert354 [DOI] [PubMed] [Google Scholar]
- Satoh R., Fujita Y., Nakashima K., Shinozaki K., Yamaguchi-Shinozaki K. (2004). A novel subgroup of bZIP proteins functions as transcriptional activators in hypoosmolarity-responsive expression of the ProDH gene in Arabidopsis. Plant Cell Physiol. 45 309–317 10.1093/pcp/pch036 [DOI] [PubMed] [Google Scholar]
- Schepetilnikov M., Dimitrova M., Mancera-Martínez E., Geldreich A., Keller M., Ryabova L. (2013). TOR and S6K1 promote translation reinitiation of uORF-containing mRNAs via phosphorylation of eIF3h. EMBO J. 32 1087–1102 10.1038/emboj.2013.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluepmann H., Dijken A., Van Aghdasi M., Wobbes B., Paul M. (2004). Trehalose mediated growth inhibition of Arabidopsis seedlings is due to trehalose-6-phosphate accumulation. Plant Physiol. 135 879–890 10.1104/pp.104.039503.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluepmann H., Pellny T., van Dijken A., Smeekens S., Paul M. (2003). Trehalose 6-phosphate is indispensable for carbohydrate utilization and growth in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 100 6849–6854 10.1073/pnas.1132018100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schütze K., Harter K., Chaban C. (2008). Post-translational regulation of plant bZIP factors. Trends Plant Sci. 13 247–255 10.1016/j.tplants.2008.03.002 [DOI] [PubMed] [Google Scholar]
- Seaton D. D., Ebenhöh O., Millar A. J., Pokhilko A. (2014). Regulatory principles and experimental approaches to the circadian control of starch turnover. J. R. Soc. Interface 11:20130979 10.1098/rsif.2013.0979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W., Hanley-Bowdoin L. (2006). Geminivirus infection up-regulates the expression of two Arabidopsis protein kinases related to yeast SNF1- and mammalian AMPK-activating kinases. Plant Physiol. 142 1642–1655 10.1104/pp.106.088476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sormani R., Yao L., Menand B., Ennar N., Lecampion C., Meyer C., et al. (2007). Saccharomyces cerevisiae FKBP12 binds Arabidopsis thaliana TOR and its expression in plants leads to rapamycin susceptibility. BMC Plant Biol. 7:26 10.1186/1471-2229-7-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugden C., Crawford R. M., Halford N. G., Hardie D. G. (1999). Regulation of spinach SNF1-related (SnRK1) kinases by protein kinases and phosphatases is associated with phosphorylation of the T loop and is regulated by 5′-AMP. Plant J. 19 433–439 10.1046/j.1365-313X.1999.00532.x [DOI] [PubMed] [Google Scholar]
- Thimm O., Bläsing O., Gibon Y., Nagel A., Meyer S., Krüger P., et al. (2004). Mapman: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37 914–939 10.1111/j.1365-313X.2004.02016.x [DOI] [PubMed] [Google Scholar]
- Tohge T., Nishiyama Y., Hirai M. Y., Yano M., Nakajima J., Awazuhara M., et al. (2005). Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J. 42 218–235 10.1111/j.1365-313X.2005.02371.x [DOI] [PubMed] [Google Scholar]
- Tsai A. Y.-L., Gazzarrini S. (2014). Trehalose-6-phosphate and SnRK1 kinases in plant development and signaling: the emerging picture. Front. Plant Sci. 5:119 10.3389/fpls.2014.00119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang C. K., Liu H., Zheng X. F. S. (2010). mTOR binds to the promoters of RNA polymerase I- and III- transcribed genes. Cell Cycle 9 953–957 10.4161/cc.9.5.10876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B., Bläsing O. E., Gibon Y., Retzlaff K., Höhne M., Günther M., et al. (2008). Global transcript levels respond to small changes of the carbon status during progressive exhaustion of carbohydrates in Arabidopsis rosettes. Plant Physiol. 146 1834–1861 10.1104/pp.107.115592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dam T. J. P., Zwartkruis F. J. T., Bos J. L., Snel B. (2011). Evolution of the TOR pathway. J. Mol. Evol. 73 209–220 10.1007/s00239-011-9469-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Kelen K., Beyaert R., Inzé D., De Veylder L. (2009). Translational control of eukaryotic gene expression. Crit. Rev. Biochem. Mol. Biol. 44 143–168 10.1080/10409230902882090 [DOI] [PubMed] [Google Scholar]
- Wang X., Proud C. G. (2011). mTORC1 signaling: what we still don’t know. J. Mol. Cell Biol. 3 206–220 10.1093/jmcb/mjq038 [DOI] [PubMed] [Google Scholar]
- Weltmeier F., Ehlert A., Mayer C. S., Dietrich K., Wang X., Schütze K., et al. (2006). Combinatorial control of Arabidopsis proline dehydrogenase transcription by specific heterodimerisation of bZIP transcription factors. EMBO J. 25 3133–3143 10.1038/sj.emboj.7601206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weltmeier F., Rahmani F., Ehlert A., Dietrich K., Schütze K., Wang X., et al. (2009). Expression patterns within the Arabidopsis C/S1 bZIP transcription factor network: availability of heterodimerization partners controls gene expression during stress response and development. Plant Mol. Biol. 69 107–119 10.1007/s11103-008-9410-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese A., Elzinga N., Wobbes B., Smeekens S. (2004). A conserved upstream open reading frame mediates sucrose-induced repression of translation. Plant Cell 16 1717–1729 10.1105/tpc.019349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger S., Loewith R., Hall M. N. (2006). TOR signaling in growth and metabolism. Cell 124 471–484 10.1016/j.cell.2006.01.016 [DOI] [PubMed] [Google Scholar]
- Xiong Y., McCormack M., Li L., Hall Q., Xiang C., Sheen J. (2013). Glucose-TOR signaling reprograms the transcriptome and activates meristems. Nature 496 181–186 10.1038/nature12030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Sheen J. (2012). Rapamycin and glucose-target of rapamycin (TOR) protein signaling in plants. J. Biol. Chem. 287 2836–2842 10.1074/jbc.M111.300749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Sheen J. (2014). TOR signaling networks in plant growth and metabolism. Plant Physiol. 164 499–512 10.1104/pp.113.229948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Ji J., Yan X.-H. (2012). Cross-talk between AMPK and mTOR in regulating energy balance. Crit. Rev. Food Sci. Nutr. 52 373–381 10.1080/10408398.2010.500245 [DOI] [PubMed] [Google Scholar]
- Yadav U. P., Ivakov A., Feil R., Duan G. Y., Walther D., Giavalisco P., et al. (2014). The sucrose-trehalose 6-phosphate (Tre6P) nexus: specificity and mechanisms of sucrose signaling by Tre6P. J. Exp. Bot. 65 1051–1068 10.1093/jxb/ert457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K., Shinozaki K. (2006). Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 57 781–803 10.1146/annurev.arplant.57.032905.105444 [DOI] [PubMed] [Google Scholar]
- Yoshida T., Fujita Y., Maruyama K., Mogami J., Todaka D., Shinozaki K., et al. (2014). Four Arabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic-acid signaling in response to osmotic stress. Plant. Cell Environ. 10.1111/pce.12351 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J. S., Galbraith D. W., Dai S. Y., Griffin P., Stewart C. N. (2008). Plant systems biology comes of age. Trends Plant Sci. 13 165–171 10.1016/j.tplants.2008.02.003 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Andralojc P. J., Hey S. J., Primavesi L. F., Specht M., Koehler J., et al. (2008). Arabidopsis sucrose non-fermenting-1-related protein kinase-1 and calcium-dependent protein kinase phosphorylate conserved target sites in ABA response element binding proteins. Ann. Appl. Biol. 153 401–409 10.1111/j.1744-7348.2008.00302.x [DOI] [Google Scholar]
- Zhang Y., Persson S., Giavalisco P. (2013). Differential regulation of carbon partitioning by the central growth regulator Target of Rapamycin (TOR). Mol. Plant 6 1731–1733 10.1093/mp/sst094 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Primavesi L. F., Jhurreea D., Andralojc P. J., Mitchell R. A C., Powers S. J., et al. (2009). Inhibition of SNF1-related protein kinase1 activity and regulation of metabolic pathways by trehalose-6-phosphate. Plant Physiol. 149 1860–1871 10.1104/pp.108.133934 [DOI] [PMC free article] [PubMed] [Google Scholar]