Abstract
Pre-mRNA splicing is a critical step in eukaryotic gene expression, which involves removal of noncoding intron sequences from pre-mRNA and ligation of the remaining exon sequences to make a mature message. Splicing is carried out by a large ribonucleoprotein complex called the spliceosome. Since the first description of the pre-mRNA splicing reaction in the 1970s, elegant genetic and biochemical studies have revealed that the enzyme that catalyzes the reaction, the spliceosome, is an exquisitely dynamic macromolecular machine, and its RNA and protein components undergo highly ordered, tightly coordinated rearrangements in order to carry out intron recognition and splicing catalysis. Studies using the genetically tractable unicellular eukaryote budding yeast (Saccharomyces cerevisiae) have played an instrumental role in deciphering splicing mechanisms. In this chapter, we discuss how yeast genetics has been used to deepen our understanding of the mechanism of splicing and explore the potential for future mechanistic insights using S. cerevisiae as an experimental tool.
Keywords: Pre-mRNA Splicing, Saccharomyces cerevisiae, Yeast genetics, Synthetic lethality, Temperature-sensitive (ts) screening, Suppressor screening, DExD/H-box protein, SGA analysis, E-MAP
1 Introduction
Eukaryotic genes are often interrupted by noncoding intron sequences. In order to achieve proper gene expression, introns are removed from the pre-mRNA and the remaining exon sequences are ligated to produce a mature messenger RNA. This process, “pre-messenger RNA splicing” is carried out by an evolutionary conserved, ~3 MDa ribonucleoprotein complex called the spliceosome which is composed of 5 snRNAs and over 100 associated proteins [1, 2]. As is suggested by the functional conservation of the spliceosome, splicing is a crucial aspect of gene expression for all eukaryotic cells—from the unicellular eukaryote S. cerevisiae to mammalian cells. For example, it is estimated that 90 % of human genes undergo splicing, and although introns are less prevalent in S. cerevisiae (found in ~6 % of genes) >30 % of the total mature messages in yeast are derived from intron containing genes [3].
The spliceosome is a dynamic ribonucleoprotein machine. It assembles in a stepwise manner onto the nascent transcript, recognizes splice site sequences in the RNA via RNA–RNA and RNA–protein interactions, and configures into a catalytically active structure. The dynamic, ATP-driven rearrangements of the spliceosome are intricately coordinated to ensure precise cleavage and ligation of exons. Characterization of the precise nature and timing of these spliceosomal rearrangements and the proteins that direct them have been central challenges for researchers. In light of the strong functional conservation of the spliceosome, classical yeast genetics using the experimentally tractable model eukaryote S. cerevisiae has proven to be a powerful tool for identifying the components of the splicing machinery and elucidating their mechanisms of action. The approaches employed include a variety of screens, e.g., temperature-sensitive (ts)/cold-sensitive (cs), enhancer (e.g., synthetic lethality), and suppressor screens, all of which have led the way to identification of genes and characterization of proteins that are involved in splicing.
In this chapter, we discuss how S. cerevisiae has been used to study pre-mRNA splicing. We describe how temperature-sensitive mutant screens have revealed components of the splicing machinery. We also describe how suppressor screens have allowed a detailed characterization of RNA and protein interactions that guide intron recognition and catalysis. Finally, we describe low- and high-throughput methods such as Synthetic Genetic Array (SGA) and Epistatic MiniArray Profile (E-MAP) analyses used to identify functional interactions between splicing components and discuss how such data are interpreted.
1.1 Pre-mRNA Splicing and the “Awesome Power of Yeast Genetics”
Genetic manipulation of S. cerevisiae has been used with great effect to understand the roles of conserved genomic sequences as well as the functional relationships among genes or sets of genes. There are numerous reasons why yeast has become a favorite model organism for genetic analyses. For one, despite being a eukaryote, yeast share the technical advantages with bacteria of rapid growth, ease of mutagenesis, and ease of long-term archival storage by freezing. Moreover, transformed DNA can be integrated into the genome via homologous recombination, thus allowing efficient gene knockout and mutation. An important feature of S. cerevisiae that underlies its genetic tractability is the fact that it exists stably as both haploid and diploid cells, and the haploid product of meiosis can be isolated via microdissection of a tetrad ascus. Finally, yeast serves as an extremely useful model organism for understanding the basic mechanisms of pre-mRNA splicing because much of the molecular machinery involved in gene expression can be generalized to multicellular eukaryotic organisms. Genetic strategies such as mutation, deletion, or genetic depletion of factors associated with splicing have substantially contributed to understanding the mechanism of splicing, and the insights gleaned about pre-mRNA splicing beautifully illustrate the often alluded to “awesome power of yeast genetics [4].”
1.2 Identification of Temperature-Sensitive Mutations in Pre-mRNA Processing Factors
The first temperature-sensitive mutant screening study in S. cerevisiae was performed by Leland Hartwell in 1967 [5]. Hartwell took advantage of the small genome of S. cerevisiae and its ability to exist as both a haploid and a diploid cell to study the dominance or recessiveness of mutations and their complementation. Cells were exposed to mutagen, evaluated for their ability to grow at 23 °C, but not at 36 °C, and then analyzed by their abilities to produce RNA. A set of ts mutants screened in this study fell into ten complementation groups and were named RNA2–RNA11, as these mutants showed inhibited production of ribosomal protein gene mRNA [6] and turned out to be defective in splicing [7]. Almost 20 years later, these mutants were renamed prp (pre-RNA processing) mutants, and studies from John Abelson’s laboratory showed that many of these PRP-encoded (PRP2–PRP11) gene products were involved in and essential for pre-mRNA splicing in vitro [8]. Additional ts prp mutants were isolated and screened by Northern blot analysis using an ACT1 intron probe, which allowed analysis of the levels of actin pre-mRNA, the intron lariat intermediate, and the excised lariat product [9]. Subsequent work from Christine Guthrie’s lab identified cold-sensitive (cs) mutants involved in splicing [10]. All of these early studies laid the groundwork for genetic analysis of yeast splicing [10]. Despite the progress that has been made toward identifying the genes and their products involved in pre-mRNA splicing, there remain many questions about the roles of these splicing factors, which can be addressed genetically. In particular, conditional alleles of essential genes can provide insights into the functions of essential components of the spliceosome, their interactions within the spliceosome, and interactions with other gene expression machineries. Moreover, mutagenesis of cells containing known gene mutations or deletions can be used to screen for functional interactions between components of the splicing machinery or between splicing factors and proteins involved in other gene expression processes, as will be discussed below.
2 Materials
Haploid S. cerevisiae strains (e.g., W303, S288C, or one of the BY strains derived from S288C [11]; markers may be any that are suitable for further genetics and biochemical study, such as auxotrophic or drug-resistance markers).
YPD liquid media.
YPD plates.
Sodium phosphate buffer pH 7.0.
Ethyl methanesulfonate (EMS).
5 % sodium thiosulfate (autoclaved).
3 Methods
3.1 EMS Mutagenesis: Generating Temperature-Sensitive Mutants to Study Splicing (See Note 1)
Grow the wild-type haploid strains to stationary phase in 50 ml of YPD medium.
Centrifuge cells and resuspend the pellet in 0.1 M sodium phosphate buffer pH 7.0 at a density of about 108 cells/ml (OD600 of 1.0 is ~3 × 107 cells/ml). Transfer cells to a glass culture tube.
At this point, save an aliquot of cells without EMS to compare cell survival (see Note 2). Then treat the cells with 3 % EMS at 30 °C for 60 min with agitation.
Pellet cells and remove EMS. Be sure to dispose of this in a designated EMS waste container.
Dilute the mutagenized cells 40-fold into 5 % sodium thiosulfate to inactivate the EMS. Spin down cells, remove supernatant, and repeat the inactivation step.
Wash the cells twice with sterile water. If the cells are clumpy, a brief vortexing will help facilitate uniform spreading.
Spread the cells onto YPD plates and incubate at the permissive temperature (23 °C) for a few days until there are ~200 colonies per plate (see Note 3).
Replica-plate the cells from the petri plates grown under permissive conditions onto fresh YPD plates and incubate at 37 °C. Store the 23 °C control plates.
Compare the original 23 °C plate and the 37 °C replicate-plate to identify colonies which show poor or no growth at 37 °C. The colonies that are identified should be restreaked onto fresh YPD plates and grown at 23 °C and 37 °C to retest growth (and decrease the likelihood of false positives).
Carry out an initial phenotypic analysis. To study the pre- mRNA splicing defects in these mutants a variety of functional assays have been used including Northern blots, primer extension, or in vitro splicing of a prototypical intron-containing gene, such as ACT1.
As mutant analysis can be complicated by the presence of multiple mutations, it is important to backcross mutants with an appropriate untreated WT strain of opposite mating type.
Check the temperature sensitivity of diploid strains by plating on the YPD plates and incubate at 23 and 37 °C, respectively (see Note 4).
For a detailed discussion of screen saturation, i.e., knowing when “enough is enough,” see ref. 12.
3.2 Genetic Screens Identify Key Splice Site Sequences and the Proteins That Recognize Them
The discovery of introns in the late 1970s immediately raised the question of what sequence elements led to their removal. To address this question, a chimeric construct was made in which the S. cerevisiae ACT1 intron sequence was subjected to random mutagenesis and fused upstream of the HIS4 gene sequence. As a consequence, the His4 protein product was only generated through precise splicing of the actin intron. This construct was transformed into cells deleted for the endogenous HIS4 gene so that splicing of this actin-HIS4 construct was required for the cells to grow on media containing the histidine precursor histidinol [13, 14]. Colonies that showed defective growth in histidinol-containing media were further analyzed to identify the sequences in the intron that were required for different steps of the splicing reaction [13]. For example, a mutant actin-HIS4 construct in which the branch-point (BP) was mutated from TACTAAC to TACTACC [15] caused accumulation of the unspliced pre-mRNA in vivo, which was not sufficient for growth in media containing histidinol [13, 14].
The use of the chimeric ACT1-HIS4 construct proved to be a powerful tool for identifying RNAs and proteins that recognize introns—both directly and indirectly. For example, it was shown that compensatory mutations in U2 snRNA that restored base pairing between the snRNA and the TACTAAC sequence in the ACT1 intron could also restore growth on histidinol and splicing [16], thus demonstrating that base pairing between U2 snRNA and the branchpoint was critical for splicing. Similar compensatory mutation experiments demonstrated U1 base pairing with the 5′ splice site [17]. Around the same time, a trans-acting factor involved in branchpoint recognition was also identified. Growth of cells harboring the ACT1-HIS4 construct that was mutated at the branchpoint was assessed to identify spontaneous suppressors that had acquired the ability to grow on media containing histidinol. This led to the identification of an extragenic suppressor that improved the splicing of the mutant actin-HIS4 construct but decreased the splicing efficiency of the wild-type intron [15]. This suppressor was named rna16-1, and was later characterized as prp16-1, an allele of the PRP16 gene which encodes an essential splicing factor Prp16 and contains ATPase activity [18]. Shortly thereafter numerous mutant branch site suppressors were identified that all mapped to the region of Prp16 responsible for its ATPase activity [19].
3.3 Suppressor Screens Reveal the Fundamental Role of DExD/H-Box Proteins in Splicing
A remarkable feature of splicing is that no new phosphodiester bonds are formed in the two catalytic reactions; nonetheless, splicing is an ATP-dependent reaction. In recent years, there has been a growing appreciation for the role of a family of proteins found to be important in numerous gene expression reactions—the DExD/H-box family of proteins, so named for the presence of a conserved motif in the protein (D (Asp)-E (Glu)-A (Ala)-D (Asp)). Eight such DExD/H-box proteins have been shown to play roles throughout the splicing cycle [20]. Whereas these proteins show RNA binding activity, RNA-dependent ATPase activity, and, similar to the DNA helicases, some nucleic acid unwinding activity, the substrates of these proteins and/or their mechanisms of action have remained elusive. Nonetheless, some of the strongest indications of their roles in splicing have come from yeast genetics.
For example, important insights into the activity of one of these DExD/H proteins, Prp16 (introduced above), were gleaned in a screen to identify suppressors of a cold-sensitive allele prp16-302. This genetic screen revealed that deletion of the gene encoding a component of the Prp19p-associated complex (NTC), ISY1, suppressed the growth defect associated with the ATPase-deficient prp16-302 mutant [21]. This work also demonstrated that the reduced fidelity of branch site recognition seen in the prp16-302 mutant could be suppressed by an ISY1 deletion. These observations are consistent with a growing appreciation for the role of DExD/H-box proteins in maintaining the fidelity of splicing. Specifically, mutations in the DExD/H-box proteins and/or interacting partners are able to affect the use of nonconsensus splice sites, (i.e., splicing fidelity), indicating that the ATP-dependent activities of DExD/H-box proteins are required for proper spliceosome rearrangements that are required to maintain splicing fidelity [22, 23].
Each of the DExD/H-box proteins is encoded by an essential gene. Hence, temperature-sensitive or cold-sensitive alleles have been isolated for each—PRP16, PRP5, SUB2, PRP28, BRR2, PRP2, PRP22, and PRP43—in order to analyze their protein functions [20]. Moreover, genetic screens have identified suppressors of cs and ts mutants of each of these, which have greatly informed our understanding of the functions of DExD/H-box proteins in splicing, including crucial roles in the fidelity of splicing [20, 22, 23].
3.4 Enhancer and Suppressor Screens Identify Functional Interactions Between Components of the Spliceosome
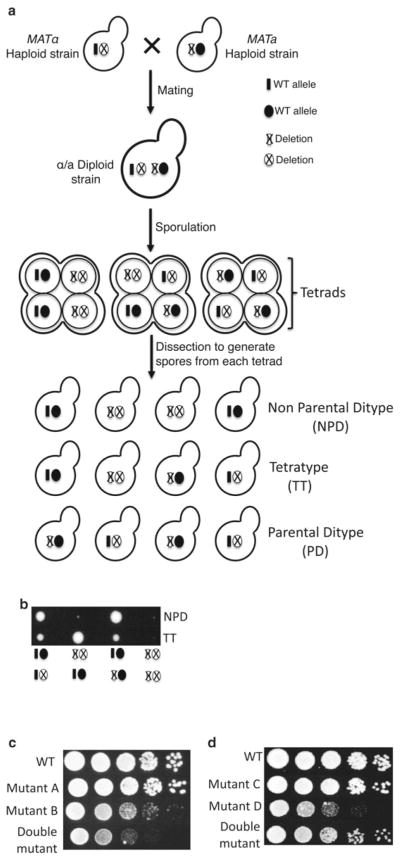
Genetic screens such as those described above highlight the power of yeast genetics to identify functional interactions between proteins and/or protein and RNA. Applying basic yeast genetics, deletion (or mutation) of one gene can be combined with deletion (or mutation) of another gene such that the double mutant cells lead to a phenotypic modulation (Fig. 1). The degree of phenotypic modulation may allow one to predict or understand the functionality of the gene(s).
Fig. 1.
Genetic interaction analysis in the yeast Saccharomyces cerevisiae. (a) Schematic diagram shows how classical genetic interaction studies are performed between yeast mutant strains. The first is the mating of two haploid strains in which the gene of interest is deleted and replaced by a selectable marker. The mating will generate a heterozygous diploid strain, and sporulation of the diploid strain will generate haploid spores clustered in a 4-spore ascus. Microdissection of each tetrad generates four haploid cells that can grow to form colonies and can be genotyped to identify the combination of WT and mutant alleles. Segregation of mutant alleles leads to three classes of tetrads: nonparental ditype (NPD), tetratype (TT), and parental ditype (PD). (b) Representative picture of a tetrad dissection plate. Each row represents a tetrad, which shows growth from each spore. The genotype of each spore is depicted below the panel. The genotype of the spores can be determined by growing the cells under selective conditions to identify the presence of selectable markers associated with the mutations (e.g., geneticin-resistance). Alternatively, genomic DNA can be isolated from the cells, and PCR can be performed using primers that specifically identify the mutations of interest. (c and d) Show representative pictures of a dilution growth assay comparing WT, single mutants, and double mutants. Cells were grown at 30 °C to the same OD600 ~0.5 and then tenfold serial dilutions were spotted on YPD plates. Panel (c) shows the synthetic growth defect phenotype of double mutant cells (bottom row), which grow more slowly than either of the single mutants (middle two rows ). The dilution assay in panel (d) shows the suppressor phenotype of the double mutant cells (bottom row ), which grow better than single mutants (middle two rows)
To assess genetic interactions, two haploid mutants (such as nonessential deletions) of opposite mating types can be mated to generate a diploid strain, and each mutation can be followed using a selectable marker. For example, through the yeast deletion project, a near complete collection of gene-deletion mutants has been generated (available through Open Biosystems) such that for each strain one gene has been precisely deleted [24]. Each gene deletion is marked by the kanMX gene, and the knockout can be followed by growth on media containing the aminoglycoside antibiotic geneticin. Diploid yeast cells undergo meiosis and unlinked genes will assort independently. When the strain is induced to sporulate by growth under nitrogen and carbon deprivation conditions, a 4-spore tetrad ascus is formed that can be dissected (Fig. 1a, b) using a micromanipulator. In this way, individual spores, representing each of the four products of meiosis can form colonies. Then the double mutants can be identified and then compared to each parent and a wild-type strain. This approach has been effectively used to directly query the relationship between two specific genes in “directed” genetic interactions studies.
When the spores containing double mutants show an enhanced negative phenotype, such as becoming more sick than the parents or inviable, this phenotype is referred to as a negative genetic interaction: a synthetic growth defect or synthetic lethality, respectively (Fig. 1b, c). In general, synthetic lethality reflects an interaction that is essential for viability, and synthetic sickness represents an interaction that is important for viability. Since a remarkable 80 % of yeast genes can be individually deleted in haploid cells and the cells remain viable, it is possible to analyze synthetic interactions in null alleles. Furthermore, conditional and hypomorphic alleles can be used to assess genetic interactions involving essential genes. The observation of synthetic lethality, particularly between null alleles, is generally interpreted to mean that the products of the two genes may be involved in synergistic functional pathways, may contribute to a complex, or may have activities that, in WT cells, buffer one another [25–27]. In other words, the presence of one gene allows the cells to tolerate loss of function of another gene that is essential or important for viability. The use of conditional and hypomorphic alleles has provided a powerful tool for identifying genes encoding products involved in the same pathway. An example of this is when combined mutations in two components of a complex weaken interactions within the complex sufficiently to diminish its function below the threshold for viability. Analysis of synthetic lethality has revealed important interactions between proteins involved in the same pathway such as translocation to the golgi, as well as those involved in functionally redundant or overlapping pathways such as DNA replication and DNA repair [25].
Alternatively, the double mutant products of a genetic cross could have less severe effect on growth than the single mutants alone, and this phenotype is referred to as a positive interaction or genetic suppression (Fig. 1d). Such a phenotype in the double mutant suggests several interesting possibilities that can be further assessed experimentally. A positive genetic interaction often identifies genes acting antagonistically in the same pathway [26, 27]. For example, deletion of the U2 snRNP protein Cus2 suppresses the lethality associated with mutation of the ATP binding domain of the DExD/H protein Prp5. Cus2 is thought to negatively regulate the formation of Stem IIa of the snRNA to ensure proper timing of this spliceosomal rearrangement and U2 snRNP interaction with the branchpoint. The data suggest that one of the functions of the Prp5 ATPase activity is to displace Cus2, thus allowing the U2 snRNA to adopt the IIa conformation [28, 29]. Positive genetic interactions may also be observed when two gene products physically interact. In this case, a mutant form of the gene may generate a product with decreased functionality, but the protein produced by a suppressor mutation in another gene can associate with and correct the function. Finally, the phenotype caused by a mutation could allow a cell to bypass some defect caused by the first mutation. In such a case the genes are likely to be in separate pathways.
Although the classical, directed genetic interaction studies can uncover important functional relationships that can lead to testable hypotheses, it is important to note that further molecular and biochemical experiments are usually necessary to decipher the molecular functions of the genes. An instructive example of this is provided by the analysis of positive and negative genetic interactions between the yeast cap binding complex and the histone H2B deubiquitylation machinery [30].
3.5 High-Throughput Methods to Identify Genetic Interactions
Classical genetic screens and “low-throughput” genetic interaction studies have proven to be extremely useful for understanding the organization of molecular pathways in yeast. Furthermore, the availability of yeast deletion and mutant collections, in which each nonessential gene in the genome is deleted (with molecular bar-codes at either end of the deletion cassette to allow its identification) or in which essential genes are modified to alter expression, e.g., Decreased Abundance by mRNA Perturbation or DAmP [31], has made it possible to understand, at a global level, the cellular functions of gene(s) in the context of a biological network. Large-scale genetic interaction studies have been particularly powerful for identifying and analyzing genes that encode multifunctional proteins and act in multiple cellular pathways [27]. Moreover, these methods utilizing yeast strain collections allow a systematic, quantitative assessment of interactions within and between networks. Here we describe two such tools for global analysis of genetic interactions—SGA analysis [32, 33] and quantitative interactions mapping via E-MAP [27].
3.6 SGA and E-MAP Analyses in Yeast (Saccharomyces cerevisiae)
SGA and E-MAP approaches allow systematic, unbiased, quantitative, and comprehensive methodologies for constructing a predictive network of genes, which are functionally related or distinct [27, 33, 34]. Focusing first on SGA analysis, this approach enables the systematic generation of double mutants in order to reveal genome-wide synthetic genetic interactions by using a combination of genetic methodologies and robotic devices. A detailed description of the tools and reagents used in such analysis are nicely described elsewhere [32]. Briefly a “query” strain can be crossed to an ordered array of ~5,000 viable gene-deletion mutants and ~1,000 essential genes with conditional mutations of the opposite mating type [32, 33]. After selection for diploid cells, sporulation, and selection of meiotic progeny, the double mutant cells in the arrays can be transferred to selective plates and photographed using high-resolution digital imaging. The yeast colony sizes of the two individual mutants and the double mutants are compared in order to obtain measures of fitness and genetic interactions. Using this approach, Charles Boone’s lab performed SGA analysis in S. cerevisiae on a genome-wide scale which involved ~1,700 “query” mutants and generated ~170,000 genetic interactions [35, 36].
SGA analysis produces a large “genetic landscape” of the cell, and data from this network can be used to predict the function of the particular gene depending on the genetic interactions it shows. The interactions can then be clustered to reveal functionally related genes that exist in the same protein complex or pathway, much like the clustering of genes in a microarray. If a gene of unknown function shows similar genetic interactions with genes that make up a functional “cluster” this can provide an indication of the cellular activity of the gene’s product. Moreover, genetic interactions between two clusters can reveal how they are functionally related [35, 36]. It is important to note that one limitation to interpreting the published genetic interactions data sets is the lack of validation for most of the reported interactions.
Moreover, while extremely powerful, global analysis of genetic interactions among randomly chosen gene pairs, as is the case with SGA analysis, yields mostly neutral interactions. In fact, only 0.5 % queried interactions show negative or positive interactions between the genes [35, 36]. With this in mind, E-MAP analysis was designed to explore genetic networks among genes that are likely to be involved in the same or similar functions [27]. Like SGA, E-MAP explores large-scale genetic interactions and involves genetic methodologies, robotic tools, and quantitative analysis of double mutants, and similar to SGA, E-MAP also generates a large amount of genetic interaction network data. Initially, E-MAPs were designed to measure pairwise interactions between rationally selected sets of genes increased the frequency of detecting genetic interactions, thus providing a deeper data set for analysis of specific pathways. Importantly, the computational analyses employed in E-MAP studies allowed the identification of positive interactions not strong enough to be observed in the early SGA studies (although with time enhanced computational tools have increased the sensitivity of SGA analyses as well). E-MAP analyses have been performed to study genetic interaction between genes involved in the same pathway such as chromatin assembly pathways [27], between kinases and their substrates [37], and components of the secretory pathway [38]. Applying high-density, targeted, pathway analysis via quantitative E-MAP can uncover the function of an unknown gene or known genes with unknown function. Nonetheless, it is important that the genetic interactions identified through these approaches be validated.
Interestingly, E-MAP analysis of genes acting in RNA processing pathways have revealed genetic interactions between the components of the complexes involved in RNA processing as well as significant genetic “crosstalk” between complexes. For example, positive interactions have been shown between cytoplasmic RNA biogenesis and mitochondrial RNA biogenesis, whereas negative interactions have been shown between genes involved in mRNA splicing, mRNA export, and the nuclear export. The E-MAP approach has suggested a new role for 19S proteasome subunit, Sem1/Dss1 in mRNA splicing and mRNA export [39]. E-MAP analysis also suggested that the SR-like protein Npl3 interacts with both the splicing machinery and the histone H2B deubiquitylation machinery [40, 41]. Subsequent experiments have supported this dual role for Npl3 in pre-mRNA splicing and the coupling of RNA splicing with histone H2B ubiquitylation [41].
3.7 Using Yeast Genetics to Study Complex Networks: Insights into Coordination of Gene Expression Processes
There is growing evidence that spliceosome assembly occurs co-transcriptionally (Merkhofer and Johnson, Chapter 6). As both the processes of transcription and splicing are multicomponent assembly processes, genetic interaction studies play an extremely useful role in uncovering the coordination between these two reactions. High-throughput and directed genetic studies have been performed between transcription factors and splicing factors to understand the crosstalk between transcription and splicing, for example [30, 39, 41–44]. One of the first such examples of how genetic analyses can inform understanding of mechanism comes from directed genetic studies showing synthetic interactions between the chromatin modifying enzyme, Gcn5, and components of the U2 snRNP [42]. Subsequent studies revealed that the dynamics of histone acetylation affect the recruitment of the U2 snRNP to pre-mRNA [43]. Large-scale genetic analysis using SGA analysis and E-MAP will almost certainly continue to provide important insights into the interconnections between RNA processing, transcription, and chromatin modification.
Acknowledgments
We would like to thank Drs. Kristin Patrick (UCSF) and Lorraine Pillus (UCSD) for critical reading of the manuscript. Funding was provided by the National Institutes of General Medical Sciences (GM085764) and the National Science Foundation (MCB-1051921).
Footnotes
Caution: EMS is a strong mutagen and must be used in a fume hood. All the glassware must be rinsed with 5 % sodium thiosulfate to inactivate EMS.
Calibrate the survival efficiency by treating cells with EMS for varying amounts of time, keeping all other parameters same, to achieve approximately 10–30 % survival.
Adding fiduciary marks to this plate and the empty plates onto which the cells will be replica-plated will allow easier alignment of the colonies following replica-plating.
Diploid strains that do not show temperature sensitivity indicate recessive mutations, while diploid strains that show temperature sensitivity indicate dominant mutations.
References
- 1.Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol Cell. 2003;12(1):5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- 2.Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136(4):701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 3.Ares M, Jr, Grate L, Pauling MH. A handful of intron-containing genes produces the lion’s share of yeast mRNA. RNA. 1999;5(9):1138–1139. doi: 10.1017/s1355838299991379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sherman F. An introduction to the genetics and molecular biology of the yeast Saccharomyces cerevisiae. In: Meyers RA, editor. The encyclopedia of molecular biology and molecular medicine. VCH Publisher; Weinheim, Germany: 1997. pp. 302–325. [Google Scholar]
- 5.Hartwell LH. Macromolecule synthesis in temperature-sensitive mutants of yeast. J Bacteriol. 1967;93(5):1662–1670. doi: 10.1128/jb.93.5.1662-1670.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hartwell LH, McLaughlin CS, Warner JR. Identification of ten genes that control ribosome formation in yeast. Mol Gen Genet. 1970;109(1):42–56. doi: 10.1007/BF00334045. [DOI] [PubMed] [Google Scholar]
- 7.Rosbash M, Harris PK, Woolford JL, Jr, et al. The effect of temperature-sensitive RNA mutants on the transcription products from cloned ribosomal protein genes of yeast. Cell. 1981;24(3):679–686. doi: 10.1016/0092-8674(81)90094-5. [DOI] [PubMed] [Google Scholar]
- 8.Lustig AJ, Lin RJ, Abelson J. The yeast RNA gene products are essential for mRNA splicing in vitro. Cell. 1986;47(6):953–963. doi: 10.1016/0092-8674(86)90810-x. [DOI] [PubMed] [Google Scholar]
- 9.Vijayraghavan U, Company M, Abelson J. Isolation and characterization of pre- mRNA splicing mutants of Saccharomyces cerevisiae. Genes Dev. 1989;3(8):1206–1216. doi: 10.1101/gad.3.8.1206. [DOI] [PubMed] [Google Scholar]
- 10.Noble SM, Guthrie C. Identification of novel genes required for yeast pre-mRNA splicing by means of cold-sensitive mutations. Genetics. 1996;143(1):67–80. doi: 10.1093/genetics/143.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brachmann CB, Davies A, Cost GJ, et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14(2):115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 12.Hawley RS, Walker MY. Advanced genetic analysis: finding meaning in a genome. Blackwell Publishing; Malden, MA: 2003. [Google Scholar]
- 13.Parker R, Guthrie C. A point mutation in the conserved hexanucleotide at a yeast 5′ splice junction uncouples recognition, cleavage, and ligation. Cell. 1985;41(1):107–118. doi: 10.1016/0092-8674(85)90065-0. [DOI] [PubMed] [Google Scholar]
- 14.Vijayraghavan U, Parker R, Tamm J, et al. Mutations in conserved intron sequences affect multiple steps in the yeast splicing pathway, particularly assembly of the spliceosome. EMBO J. 1986;5(7):1683–1695. doi: 10.1002/j.1460-2075.1986.tb04412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Couto JR, Tamm J, Parker R, et al. A trans-acting suppressor restores splicing of a yeast intron with a branch point mutation. Genes Dev. 1987;1(5):445–455. doi: 10.1101/gad.1.5.445. [DOI] [PubMed] [Google Scholar]
- 16.Parker R, Siliciano PG, Guthrie C. Recognition of the TACTAAC box during mRNA splicing in yeast involves base pairing to the U2-like snRNA. Cell. 1987;49(2):229–239. doi: 10.1016/0092-8674(87)90564-2. [DOI] [PubMed] [Google Scholar]
- 17.Siliciano PG, Guthrie C. 5′ splice site selection in yeast: genetic alterations in base- pairing with U1 reveal additional requirements. Genes Dev. 1988;2(10):1258–1267. doi: 10.1101/gad.2.10.1258. [DOI] [PubMed] [Google Scholar]
- 18.Schwer B, Guthrie C. PRP16 is an RNA-dependent ATPase that interacts transiently with the spliceosome. Nature. 1991;349(6309):494–499. doi: 10.1038/349494a0. [DOI] [PubMed] [Google Scholar]
- 19.Burgess SM, Guthrie C. A mechanism to enhance mRNA splicing fidelity: the RNA-dependent ATPase Prp16 governs usage of a discard pathway for aberrant lariat intermediates. Cell. 1993;73(7):1377–1391. doi: 10.1016/0092-8674(93)90363-u. [DOI] [PubMed] [Google Scholar]
- 20.Chang TH, Tung L, Yeh FL, et al. Functions of the DExD/H-box proteins in nuclear pre-mRNA splicing. Biochim Biophys Acta. 2013;1829(8):764–774. doi: 10.1016/j.bbagrm.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 21.Villa T, Guthrie C. The Isy1p component of the NineTeen complex interacts with the ATPase Prp16p to regulate the fidelity of pre-mRNA splicing. Genes Dev. 2005;19(16):1894–1904. doi: 10.1101/gad.1336305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Semlow DR, Staley JP. Staying on message: ensuring fidelity in pre-mRNA splicing. Trends Biochem Sci. 2012;37(7):263–273. doi: 10.1016/j.tibs.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koodathingal P, Staley JP. Splicing fidelity: DEAD/H-box ATPases as molecular clocks. RNA Biol. 2013;10(7) doi: 10.4161/rna.25245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Winzeler EA, Shoemaker DD, Astromoff A, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285(5429):901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 25.Hartman JL, Garvik B, Hartwell L. Principles for the buffering of genetic variation. Science. 2001;291(5506):1001–1004. doi: 10.1126/science.291.5506.1001. [DOI] [PubMed] [Google Scholar]
- 26.Boone C, Bussey H, Andrews BJ. Exploring genetic interactions and networks with yeast. Nat Rev Genet. 2007;8(6):437–449. doi: 10.1038/nrg2085. [DOI] [PubMed] [Google Scholar]
- 27.Collins SR, Roguev A, Krogan NJ. Quantitative genetic interaction mapping using the E-MAP approach. Methods Enzymol. 2010;470:205–231. doi: 10.1016/S0076-6879(10)70009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perriman R, Ares M., Jr ATP can be dispensable for prespliceosome formation in yeast. Genes Dev. 2000;14(1):97–107. [PMC free article] [PubMed] [Google Scholar]
- 29.Perriman R, Barta I, Voeltz GK, et al. ATP requirement for Prp5p function is determined by Cus2p and the structure of U2 small nuclear RNA. Proc Natl Acad Sci USA. 2003;100(24):13857–13862. doi: 10.1073/pnas.2036312100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hossain MA, Claggett JM, Nguyen T, et al. The cap binding complex influences H2B ubiquitination by facilitating splicing of the SUS1 pre-mRNA. RNA. 2009;15(8):1515–1527. doi: 10.1261/rna.1540409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Breslow DK, Cameron DM, Collins SR, et al. A comprehensive strategy enabling high-resolution functional analysis of the yeast genome. Nat Methods. 2008;5(8):711–718. doi: 10.1038/nmeth.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tong AH, Boone C. Synthetic genetic array analysis in Saccharomyces cerevisiae. Methods Mol Biol. 2006;313:171–192. doi: 10.1385/1-59259-958-3:171. [DOI] [PubMed] [Google Scholar]
- 33.Baryshnikova A, Costanzo M, Dixon S, et al. Synthetic genetic array (SGA) analysis in Saccharomyces cerevisiae and Schizosaccharomyces pombe. Methods Enzymol. 2010;470:145–179. doi: 10.1016/S0076-6879(10)70007-0. [DOI] [PubMed] [Google Scholar]
- 34.Tong AH, Evangelista M, Parsons AB, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294(5550):2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- 35.Tong AH, Lesage G, Bader GD, et al. Global mapping of the yeast genetic interaction network. Science. 2004;303(5659):808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- 36.Baryshnikova A, Costanzo M, Kim Y, et al. Quantitative analysis of fitness and genetic interactions in yeast on a genome scale. Nat Methods. 2010;7(12):1017–1024. doi: 10.1038/nmeth.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fiedler D, Braberg H, Mehta M, et al. Functional organization of the S. cerevisiae phosphorylation network. Cell. 2009;136(5):952–963. doi: 10.1016/j.cell.2008.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schuldiner M, Collins SR, Thompson NJ, et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123(3):507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 39.Wilmes GM, Bergkessel M, Bandyopadhyay S, et al. A genetic interaction map of RNA-processing factors reveals links between Sem1/Dss1-containing complexes and mRNA export and splicing. Mol Cell. 2008;32(5):735–746. doi: 10.1016/j.molcel.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kress TL, Krogan NJ, Guthrie C. A single SR-like protein, Npl3, promotes pre-mRNA splicing in budding yeast. Mol Cell. 2008;32(5):727–734. doi: 10.1016/j.molcel.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moehle EA, Ryan CJ, Krogan NJ, et al. The yeast SR-like protein Npl3 links chromatin modification to mRNA processing. PLoS Genet. 2012;8(11):e1003101. doi: 10.1371/journal.pgen.1003101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gunderson FQ, Johnson TL. Acetylation by the transcriptional coactivator Gcn5 plays a novel role in co-transcriptional spliceosome assembly. PLoS Genet. 2009;5(10):e1000682. doi: 10.1371/journal.pgen.1000682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gunderson FQ, Merkhofer EC, Johnson TL. Dynamic histone acetylation is critical for cotranscriptional spliceosome assembly and spliceosomal rearrangements. Proc Natl Acad Sci USA. 2011;108(5):2004–2009. doi: 10.1073/pnas.1011982108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hossain MA, Chung C, Pradhan SK, et al. The yeast cap binding complex modulates transcription factor recruitment and establishes proper histone H3K36 trimethylation during active transcription. Mol Cell Biol. 2013;33(4):785–799. doi: 10.1128/MCB.00947-12. [DOI] [PMC free article] [PubMed] [Google Scholar]