Abstract
Recent advances that allow us to collect more data on DNA sequences and metabolites have increased our understanding of connections between the intestinal microbiota and metabolites, at a whole-systems level. We can also now better study the effects of specific microbes on specific metabolites. Here, we review how the microbiota determines levels of specific metabolites, how the metabolite profile develops in infants, and prospects for assessing a person’s physiological state based on their microbes and/or metabolites. Although data acquisition technologies have improved, computational challenges to integrating data from multiple levels remain formidable; developments in this area will significantly improve our ability to interpret current and future datasets.
Introduction
Rapid advances in sequencing technologies over the past decade have allowed researchers worldwide to assess how the intestinal microbiome affects human health1. Humans develop symbiotic relationships with microbes at a young age2. Factors such as the environment3, proximity to other humans and animals4, diet5, 6, genetics7, and temporal variation8 affect the assemblage of microbes on our skin, in our mouths, and in our guts9, 10. Our microbiota has been compared to a previously unknown organ in terms of its effects; it has extensive metabolic capabilities, and carries ~150-fold more genes than the human genome. Microbes provide the host with a range of otherwise inaccessible metabolic capabilities11.
Unlike the human genome, the microbiome is relatively plastic. It can be rapidly altered through factors such as diet6, drugs, probiotics, and microbially produced metabolites. Deliberate alterations in the microbiota and/or microbiome can therefore affect health. The intestinal microbiota is viewed increasingly as an important target of pharmacologic agents—specific microbes have been shown to deactivate or activate specific xenobiotics, which can alter the effects of different therapeutic agents12. The systems-level effects of the entire microbial community on the whole metabolite repertoire are just beginning to be understood.
Metabolomics and metabolite profiling analyses have been widely used to identify disease biomarkers. For example, quantification of triglycerides, glucose, and cholesterol in the blood can be used to determine the risk of heart disease. Similarly, the first microbiome studies sought to identify taxa that correlated with disease, physiological state, drug use, or dietary intake. However, not all exposures can alter the composition of the microbial community or its gene content; some can affect gene expression13, 14.
Humanized mice (created by transplanting human fecal microbiota into the mouse gut) have metabolomes distinct from those of conventionally raised mice15. This observation indicates that different gut microbes can produce changes in metabolites throughout their host. This shift in focus from determining “who is there” toward understanding “what are they doing” drives current studies of the human microbiota. Metabolomic studies will allow us to move from observing patterns to understanding mechanisms.
Metabolomic analyses also help researchers to understand the effects of rare taxa, and taxa with genomic variations that affect function. Organisms are considered to be of the same species if they have greater than 97% identity in the 16S rRNA gene. However, genomes from the same species can have large differences in DNA sequences outside the 16S rRNA gene. Importantly, they often have different sets of gene clusters that regulate production of specialized metabolites (e.g. antibiotics, virulence factors, siderophores, etc.) and the composition of the microbial communities, as well as encode many antibiotic resistance genes16. Rasko et al. determined that among 17 Escherichia coli isolates, the average genome size of a single isolate was 5020 nucleotides (nt), although the pan-genome was ~13,000 nt17. Furthermore, rare taxa might have a large effect on the overall community metabolome if they have important metabolic activities, perhaps acting as keystone species.
Although definitions of what constitutes a core microbiome in terms of membership is elusive, there does seem to be at least a core functional profile for the gut microbiota.10 Identifying biologically important variations against this core remains a challenge. Metabolomic analyses provide a partial picture of metabolism rather than the potential for metabolism, and the expression of this core set of functions can change with alterations in available substrates, such as xenobiotics, even if the microbial species membership and abundance remain constant13. We review the intimate connections among animal hosts, their microbiota, and the metabolites produced by either one.
Different microbial communities metabolize xenobiotic agents and dietary components in different ways to produce variable effects on many tissues in the host, including the brain18 (Figure 1). We discuss general metabolomic technologies and their implementation for study of human health, assess cases in which changes in gut microbiota alter host metabolic profiles, examine the ways in which gut microbiota process xenobiotics and nutritional inputs, and examine the analytical limitations of associating microbial abundances with metabolic profiles.
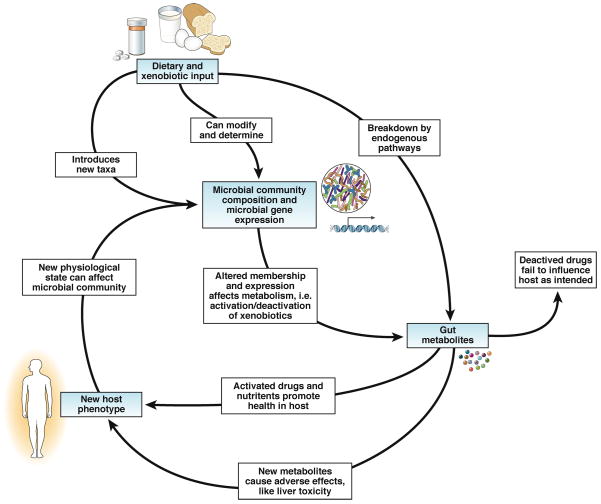
Figure 1. Interactions Among Host, Microbiota, and Metabolites.
In this simplified model, the gut microbiota metabolize substrate inputs from the host including diet and xenobiotics into metabolites that can enter the host’s bloodstream and affect the host peripherally. For example, therapeutic drugs can be inactivated, reducing their efficacy. Alternatively, drugs may converted to derivatives with non-target and possibly toxic effects. Changes in these input substrates, therefore, change the reservoir of available microbial substrates and alter the metabolomic profile of the gut, yielding variable effects on the host. The new host phenotype can, in turn, have a feedback effect on the microbial community.
Metabolomics in Assessment of Metabolic Status
Metabolomic studies analyze complex systems, including the repertoire of small-molecule metabolites in the gut, using high-throughput analytical methods. Mass spectrometry and nuclear magnetic resonance spectroscopy allow robust and sensitive identification of metabolites produced by microbes and host cells, in samples such as feces, urine, and tissue (see comprehensive reviews in 19, 20). These tools allow researchers to determine the effects that treatments or perturbations have on the host’s metabolic profile, by analyzing the presence and quantity of thousands of metabolites simultaneously. Although it is a challenge to assign spectral features, spectral networking platforms,21, 22 aided by open-source metabolome databases such as HMDB23, METLIN24, LIPIDS MAPS25, MassBank26, and NIST,27 allow for faster identification and annotation of known and unknown metabolites28. By comparing pre- and post-perturbation metabolomic profiles using multivariate statistics, metabolites that are significantly affected by experimental variables can be identified and placed into the larger context of how the host was affected overall.
Effects of the Microbiome on the Metabolome
Metabolomic analyses allow for the metabolism of the gut microbiota to be directly compared with metabolic outcomes in the host. Wikoff et al.29 directly tested the effect of gut microbiota on the host by comparing the plasma metabolomic profile, obtained via untargeted mass spectrometry, between germ-free and conventionally raised mice. They found that concentrations of more than 10% of all metabolites detected in the plasma differed by at least 50% between mice with and without gut microbes. Furthermore, many metabolites were detected only in serum from conventionally raised mice (not germ-free mice). For example, serum levels of tryptophan decreased 40% in serum from conventional mice compared to germ-free mice—likely due to the presence of bacteria that produce tryptophanases29.
Another detailed study evaluated the systemic effects of probiotics, prebiotics, and their combination (termed ‘synbiotics’) in initially germ-free mice colonized with a combination of microbes representing those found in a human infant (Bacteroides distasonis, Clostridium perfringens, Escherichia coli, Bifidobacterium breve, Bifidobacterium longum, Staphylococcus aureus, and Staphylococcus epidermidis)30. Dietary supplementation with the probiotic Lactobacillus rhamnosus NCC4007 and the prebiotic galactosyl-oligosaccharides significantly altered the relative proportions of the 7-member community, and led to systemic changes in the metabolic profiles of different tissues from the mice. For example, a prebiotic increased proportions of B breve, B longum, and B distasonis; decreased proportions of E coli and C perfringens; and altered lipid metabolism by reducing plasma levels of glucose and hepatic levels of triglycerides. Probiotics also had systemic effects, lowering plasma levels of lipoprotein, hepatic levels of glutamine, and glycogen levels. Overall, prebiotics significantly altered the metabolome in the plasma, urine, feces, liver, pancreas, renal cortex, renal medulla, and adrenal glands; probiotics produced differences in all these compartments except the pancreas.
Interestingly, another study that evaluated the effects of probiotics and prebiotics in adults found that neither significantly affected proportions of microbes in fecal samples, but RNA sequencing data showed altered expression of microbial genes that control carbohydrate metabolism14. It is possible that the relatively simpler communities that reside in infants are more susceptible to probiotic and prebiotic manipulation than the more diverse and complex communities found in adults. Prebiotics and probiotics might therefore have the largest effects when administered early in life. However, this hypothesis requires testing in animal models.
The dietary components that escape digestion in the upper gastrointestinal tract provide most of the substrates for the intestinal microbiota. Fermentation of carbohydrates by the intestinal microbiota leads to the production of short-chain fatty acids (SCFA) such as butyrate, propionate, and acetate. Studies have shown that patients with inflammatory bowel diseases such as ulcerative colitis have fewer butyrate producing bacteria (e.g., Roseburia hominis and Faecalibacterium prausnitzii) in their intestine, resulting in lower levels of butyrate31, 32. In addition to butyrate, propionate can potentiate de novo generation of T-regulatory cells in the peripheral immune system. Modulation of butyrate- and propionate-producing microbes might therefore be used to treat inflammatory bowel diseases such as ulcerative colitis. However, the anti-inflammatory mechanisms of butyrate and other SCFA remain poorly defined.
Predictive Microbial Metagenomes
Metagenomic information can been used to determine how metabolism is affected by different disease states. Studies of obesity have shown that individuals with increased adiposity have lower microbial diversity than lean individuals 33, 34. The more-diverse microbiota of lean individuals contains significantly higher proportions of microbes correlated with anti-inflammatory responses, such as Faecalibacterium prausnitzii. The less-diverse microbiota of obese individuals contains higher proportions of Bacteroides sp. and Ruminococcus gnavus, each of which could have inflammatory effects33. Gene content analysis of these groups revealed the less-diverse microbiota appeared to produce lower levels of butyrate, have increased potential for production of hydrogen sulfide, and have reduced capability for management of oxidative stress. One poorly understood aspect of the microbiome, and its potential to produce a variety of metabolites, is whether microbial diversity itself has protective effects for the host, or whether low diversity is a side effect of specific disorders (rather than a cause)35. This relationship can best be resolved in humans by prospective longitudinal studies.
Although it would be ideal to obtain metabolomic and metagenomic data for every sample for which a 16S amplicon profile has been collected, these techniques are currently far more expensive than 16S amplicon profiling. Fully matched datasets are therefore prohibitively expensive and time consuming to produce. However, recent advances in software, including Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt)36, that exploit the strong association between phylogeny and function now allow researchers to estimate the metabolomic functional profile of a community using 16S amplicon sequences. Briefly, PICRUST takes a phylogenetic tree where the gene profile of a subset of nodes is known, and then uses ancestral-state reconstruction to estimate the functional gene content for other uncharacterized nodes. PICRUSt was able to make strong predictions (average Spearman r = 0.82) for inferred metagenomes from 16S marker genes, compared against fully sequenced metagenomes obtained from the Human Microbiome Project.
Another powerful computational tool is Predicted Relative Metabolic Turnover (PRMT), which uses gene number to predict the relative consumption and production of metabolites in a system; it can be used for modeling and hypothesis generation37. Tools such as PICRUSt and PRMT could be cost-effective methods to determine whether additional resources should be used for more comprehensive metabolic profiling and metagenomic sequencing. However, findings must be validated with matched datasets, to assess the limits of their performance.
Metabolomic Profiles of Infants
Changes to the microbiome and immune system during infancy may have lasting effects, such as in contributing to the development of allergies14, 38, 39. Distinct changes in the microbiota occur during the first 2 years of life, and correlate with changes in environment, diet—these can be tracked by studying changes in infants’ fecal metabolomes. A study that followed infants at risk for celiac disease showed that the metabolomes of infants less than 6 months old were dominated by sugars, including lactose and glucose. However, after 6 months, their metabolomes shifted, increasing concentrations of amino acids and SCFA. Principal coordinates analysis showed that the metabolome of infants at 2 years of age resembles more closely that of adults, due to increased levels of acetate and butyrate40. These findings are supported by 16S amplicon studies showing that the infant microbiota comes to resemble that of adults from the same community at 2 years of age 41. It is also apparent that the intestinal microbiota of infants is specifically adapted to metabolize the infant’s earliest nutrient source, breast milk. Specific Bifidobacterium species have genomes enriched in genes that regulate processing of human milk-derived oligosaccharides. These might have a competitive advantage that places them among the first colonizers of the human intestine42.
Xenobiotic Metabolism
In addition to diet-derived macronutrients, the microbes residing in the gastrointestinal tract may be exposed to a variety of xenobiotic compounds (antibiotics, other drugs, and diet-derived bioactive compounds). Because the gut microbiome encodes so many enzymes with different activities, it is not surprising that many of xenobiotic compounds are often metabolized by the gut microbiota. It has been at least 40 years since we began to appreciate the contribution of microbes to xenobiotic metabolism43–45. However, we are only beginning to uncover the mechanisms of this process. Adding to the complexity of these interactions, xenobiotics can also modulate the expression and activity of the gut microbiome13. Metabolites of microbial origin may interfere with host metabolism of xenobiotics, and diet-derived nutrients can regulate microbial metabolism of xenobiotics.
One of the first studies to provide detailed evidence for the interaction between the gut microbiota and metabolism of xenobiotics came from Clayton et al. in 200946. Their study leveraged a powerful metabolomic analysis pipeline to correlate the presence of the microbial metabolite, p-cresol, with a reduction in the ratio of sulfonated to glucuronidated acetaminophen. Increased p-cresol production reduces the capacity of the liver to properly metabolize this widely used analgesic drug, presumably because p-cresol competes with sulfotransferase 46. Subsequent studies from this group showed that metabolites of microbial origin could modulate expression and activities of a range of host enzymes, including those of major xenobiotic-metabolizing cytochrome enzymes47. These seminal observations are beginning to lay the foundation for a metagenomic approach to selection of therapy based on microbial and host metabolism.
In addition to its interactions with metabolite production, the gut microbiota can also have a more-direct role in xenobiotic metabolism, by catalyzing a multitude of reactions that influence the fate of these compounds. Recent reviews have summarized the many processes by which microbes metabolize xenobiotics12, 48, 49. Although these activities are largely catalogued, there are only a few for which the exact mechanisms are being characterized. For example, it has been known for decades that the cardiac drug digoxin can be inactivated by Eggerthella lenta, a common gut bacterium within the Actinobacteria50. Researchers have recently identified a cytochrome-encoding operon that is upregulated by digoxin and other cardiac glycosides and is unique to strains capable of inactivating digoxin. Inactivation of digoxin was blocked by increasing dietary protein intake by mice mono-associated with E lenta51, likely due to the inhibitory effect of arginine52.
Wallace et al. studied how the microbiota can determine the effects of the colorectal cancer drug irinotecan. Enzymes produced by microbes have long been known to deconjugate an irinotecan metabolite in the gut, causing inflammation, diarrhea, and anorexia. After a successful screen for a small-molecule inhibitor of the microbial β-glucuronidase enzyme that mediates this deconjugation, Wallace et al. showed that the side effects of irinotecan could be greatly reduced by co-administration with this β glucuronidase inhibitor53. Interestingly, recent studies show that the presence of the microbiota increases the efficacy of chemotherapeutic drugs, indicating that the microbiota have previously unappreciated, but integral roles in mediating responses to these drugs 54, 55.
Computational Challenges to Discovering Correlations
Identifying statistically meaningful patterns in metabolite contingency tables (tables recording the abundance of each metabolite count in each sample) is straightforward in theory but often conducted with mathematically unfounded techniques in practice. For instance, analysis of variance and Student t test methods are frequently used to identify significant differences in abundances of metabolites among sample groups without establishing that the underlying data meet the distribution requirements. Normality, equality of variance, and homogenous population characteristics are required for proper calculations of statistical significance (either P values or false discovery rates). Although non-parametric tests can be substituted to deal with the non-normality of the data, these approaches still does not resolve 2 fundamental computational challenges: extraction of biologically significant results from the mass of statistically significant results and the fact that multivariate biological data are typically normalized to a sum—the simplex constraints this imposes violate the Euclidean-space models assumed by most test statistics (see below).
The most widely applied method to reduce biologically irrelevant, but statistically significant, results is to remove features (taxa, KEGG Orthology groups, Enzyme Commission numbers, etc.) from the contingency table prior to testing on the basis of a metric that assigns expected biological relevance to a feature. This ‘metric’ is usually as simple as overall table abundance (e.g. remove feature i if i is less than 1% of all observations) or overall sample representation (e.g. remove feature i if i is in less than 20% of samples). This filtering approach is motivated by the intuition that more widely shared features will be more biologically important, and has the additional attraction of reducing the severity of multiple hypothesis test correction factors. Unfortunately, although widespread, this approach has not yet been systematically benchmarked or evaluated for sensitivity, specificity, or even false discovery control, particularly in fields combining microbiome and metabolomic datasets.
A complementary approach to identifying differential representation of features among groups is to look for interactions among features via co-occurrence analysis (Figure 2). Traditional co-occurrence detection methods including Spearman or Pearson correlation between feature vectors are not reliable when the data are ‘compositional’ (i.e. lie in a simplex rather than Euclidean space)56, 57. Because compositionality is a feature of much -omics data (16S amplicon surveys are inherently compositional because normalization for unequal sampling effort in any contingency table introduces compositionality), methods such as ‘SparCC’ and ‘CoNet’ have been developed to capture true correlations. Although these methods are well-founded in mathematics, have been benchmarked and validated in only limited circumstances and their performance has not yet been characterized for metabolomic data in general.
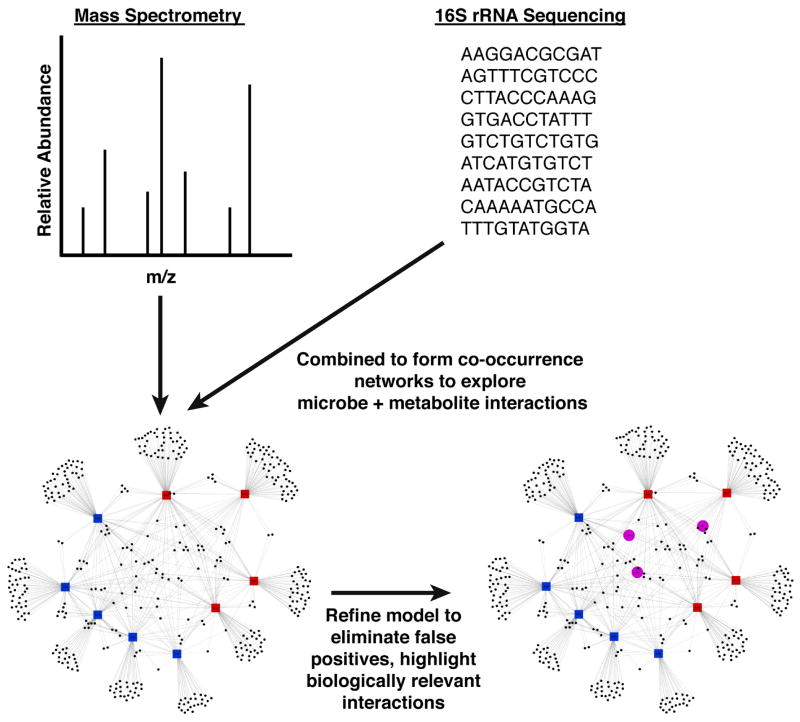
Figure 2. Exploring the Interactions Between Metabolomics and the Microbiome.
Both metabolomics and high-throughput sequencing produce a wealth of information. Visualizing the interactions between these highly multivariate datasets is important for elucidating relationships. In this example tripartite network, the large blue nodes represent samples, which are connected to red diamonds (metabolites) with red edges, and connected to black circles (OTUs) with black lines. The closer an OTU node or a metabolite node is to a sample node, the larger the relative abundance of that metabolite or that OTU in that sample. Therefore, OTUs and metabolites that are close together in the network tend to be found in the same samples (and this suggests, but does not conclusively prove, that the metabolite may be produced by that OTU). The tripartite network also demonstrates which metabolites and OTUs are shared by samples, and which metabolites and OTUs are unique to a given sample. As discussed in this review, methods are being developed to help separate out biologically important associations from amongst many statistically significant ones. Once identified, we can visualize how biologically important metabolites are controlled by the interaction between host and microbiome.
Conclusion
The overall diversity and plasticity of the gut microbiota, in comparison to our human genomes, provides exciting new prospects for personalized medicine—particularly for studies to determine the mechanisms by which microbes affect production of metabolites from drugs and diet. Although there is much work to be done, especially in terms of computational methods, the experimental frameworks of metabolomics and microbial community analysis that have emerged should allow for rapid host characterization followed by subsequent analyses of clinical potential.
Acknowledgments
FUNDING: LKU is supported by the National Institutes of Health Signaling and Cellular Recognition Training Grant (T32 GM08759). HJH is supported by the Canadian Institutes of Health Research (MFE-112991). JV and LR are supported by National Research Initiative Grant 2009-55200-05197 from the USDA National Institute for Food and Agriculture. This work was supported in part by the Howard Hughes Medical Institute.
Footnotes
COI: No conflicts of interest exist.
References
- 1.Tringe SG, Hugenholtz P. A renaissance for the pioneering 16S rRNA gene. Curr Opin Microbiol. 2008;11:442–6. doi: 10.1016/j.mib.2008.09.011. [DOI] [PubMed] [Google Scholar]
- 2.Dominguez-Bello MG, Blaser MJ, Ley RE, et al. Development of the human gastrointestinal microbiota and insights from high-throughput sequencing. Gastroenterology. 2011;140:1713–9. doi: 10.1053/j.gastro.2011.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ursell LK, Van Treuren W, Metcalf JL, et al. Replenishing our defensive microbes. Bioessays. 2013;35:810–7. doi: 10.1002/bies.201300018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song SJ, Lauber C, Costello EK, et al. Cohabiting family members share microbiota with one another and with their dogs. Elife. 2013;2:e00458. doi: 10.7554/eLife.00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu GD, Chen J, Hoffmann C, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–8. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.David LA, Maurice CF, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2013 doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benson AK, Kelly SA, Legge R, et al. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc Natl Acad Sci U S A. 2010;107:18933–8. doi: 10.1073/pnas.1007028107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caporaso JG, Lauber CL, Costello EK, et al. Moving pictures of the human microbiome. Genome Biology. 2011;12:R50. doi: 10.1186/gb-2011-12-5-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costello EK, Lauber CL, Hamady M, et al. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–7. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Human Microbiome Project C. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–14. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haiser HJ, Turnbaugh PJ. Is it time for a metagenomic basis of therapeutics? Science. 2012;336:1253–5. doi: 10.1126/science.1224396. [DOI] [PubMed] [Google Scholar]
- 13.Maurice CF, Haiser HJ, Turnbaugh PJ. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell. 2013;152:39–50. doi: 10.1016/j.cell.2012.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McNulty NP, Yatsunenko T, Hsiao A, et al. The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Science translational medicine. 2011;3:106ra106. doi: 10.1126/scitranslmed.3002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marcobal A, Kashyap PC, Nelson TA, et al. A metabolomic view of how the human gut microbiota impacts the host metabolome using humanized and gnotobiotic mice. ISME J. 2013;7:1933–43. doi: 10.1038/ismej.2013.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Penders J, Stobberingh EE, Savelkoul PH, et al. The human microbiome as a reservoir of antimicrobial resistance. Front Microbiol. 2013;4:87. doi: 10.3389/fmicb.2013.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rasko DA, Rosovitz MJ, Myers GS, et al. The pangenome structure of Escherichia coli: comparative genomic analysis of E. coli commensal and pathogenic isolates. J Bacteriol. 2008;190:6881–93. doi: 10.1128/JB.00619-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cryan JF, Dinan TG. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci. 2012;13:701–12. doi: 10.1038/nrn3346. [DOI] [PubMed] [Google Scholar]
- 19.Dettmer K, Aronov PA, Hammock BD. Mass spectrometry-based metabolomics. Mass Spectrom Rev. 2007;26:51–78. doi: 10.1002/mas.20108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Slupsky CM. Nuclear magnetic resonance-based analysis of urine for the rapid etiological diagnosis of pneumonia. Expert Opin Med Diagn. 2011;5:63–73. doi: 10.1517/17530059.2011.537653. [DOI] [PubMed] [Google Scholar]
- 21.Rath CM, Alexandrov T, Higginbottom SK, et al. Molecular analysis of model gut microbiotas by imaging mass spectrometry and nanodesorption electrospray ionization reveals dietary metabolite transformations. Anal Chem. 2012;84:9259–67. doi: 10.1021/ac302039u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watrous J, Roach P, Alexandrov T, et al. Mass spectral molecular networking of living microbial colonies. Proc Natl Acad Sci U S A. 2012;109:E1743–52. doi: 10.1073/pnas.1203689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wishart DS, Jewison T, Guo AC, et al. HMDB 3. 0--The Human Metabolome Database in 2013. Nucleic Acids Res. 2013;41:D801–7. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith CA, O’Maille G, Want EJ, et al. METLIN: a metabolite mass spectral database. Ther Drug Monit. 2005;27:747–51. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- 25.Sud M, Fahy E, Cotter D, et al. LMSD: LIPID MAPS structure database. Nucleic Acids Res. 2007;35:D527–32. doi: 10.1093/nar/gkl838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horai H, Arita M, Kanaya S, et al. MassBank: a public repository for sharing mass spectral data for life sciences. J Mass Spectrom. 2010;45:703–14. doi: 10.1002/jms.1777. [DOI] [PubMed] [Google Scholar]
- 27.Stein SE. Chemical substructure identification by mass spectral library searching. J Am Soc Mass Spectrom. 1995;6:644–55. doi: 10.1016/1044-0305(95)00291-K. [DOI] [PubMed] [Google Scholar]
- 28.Yang JY, Sanchez LM, Rath CM, et al. Molecular networking as a dereplication strategy. J Nat Prod. 2013;76:1686–99. doi: 10.1021/np400413s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wikoff WR, Anfora AT, Liu J, et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A. 2009;106:3698–703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin FP, Sprenger N, Yap IK, et al. Panorganismal gut microbiome-host metabolic crosstalk. J Proteome Res. 2009;8:2090–105. doi: 10.1021/pr801068x. [DOI] [PubMed] [Google Scholar]
- 31.Machiels K, Joossens M, Sabino J, et al. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut. 2013 doi: 10.1136/gutjnl-2013-304833. [DOI] [PubMed] [Google Scholar]
- 32.Wang W, Chen L, Zhou R, et al. Increased Proportion of Bifidobacterium and the Lactobacillus group and Loss of Butyrate-producing Bacteria in Inflammatory Bowel Disease. J Clin Microbiol. 2013 doi: 10.1128/JCM.01500-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Le Chatelier E, Nielsen T, Qin J, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–6. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- 34.Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–4. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fang S, Evans RM. Microbiology: Wealth management in the gut. Nature. 2013;500:538–9. doi: 10.1038/500538a. [DOI] [PubMed] [Google Scholar]
- 36.Langille MG, Zaneveld J, Caporaso JG, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814–21. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Larsen N, Vogensen FK, van den Berg FW, et al. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS ONE. 2010;5:e9085. doi: 10.1371/journal.pone.0009085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heederik D, von Mutius E. Does diversity of environmental microbial exposure matter for the occurrence of allergy and asthma? J Allergy Clin Immunol. 2012;130:44–50. doi: 10.1016/j.jaci.2012.01.067. [DOI] [PubMed] [Google Scholar]
- 39.Blaser M. Antibiotic overuse: Stop the killing of beneficial bacteria. Nature. 2011;476:393–4. doi: 10.1038/476393a. [DOI] [PubMed] [Google Scholar]
- 40.Sellitto M, Bai G, Serena G, et al. Proof of concept of microbiome-metabolome analysis and delayed gluten exposure on celiac disease autoimmunity in genetically at-risk infants. PLoS One. 2012;7:e33387. doi: 10.1371/journal.pone.0033387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yatsunenko T, Rey FE, Manary MJ, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–7. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sela DA, Chapman J, Adeuya A, et al. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc Natl Acad Sci U S A. 2008;105:18964–9. doi: 10.1073/pnas.0809584105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scheline RR. Metabolism of foreign compounds by gastrointestinal microorganisms. Pharmacol Rev. 1973;25:451–523. [PubMed] [Google Scholar]
- 44.Goldman P, Peppercorn MA, Goldin BR. Metabolism of drugs by microorganisms in the intestine. Am J Clin Nutr. 1974;27:1348–55. doi: 10.1093/ajcn/27.11.1348. [DOI] [PubMed] [Google Scholar]
- 45.Goldman P. Biochemical pharmacology of the intestinal flora. Annu Rev Pharmacol Toxicol. 1978;18:523–39. doi: 10.1146/annurev.pa.18.040178.002515. [DOI] [PubMed] [Google Scholar]
- 46.Clayton TA, Baker D, Lindon JC, et al. Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:14728–14733. doi: 10.1073/pnas.0904489106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Claus SP, Ellero SL, Berger B, et al. Colonization-induced host-gut microbial metabolic interaction. MBio. 2011;2:e00271–10. doi: 10.1128/mBio.00271-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sousa T, Paterson R, Moore V, et al. The gastrointestinal microbiota as a site for the biotransformation of drugs. Int J Pharm. 2008;363:1–25. doi: 10.1016/j.ijpharm.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 49.Haiser HJ, Turnbaugh PJ. Developing a metagenomic view of xenobiotic metabolism. Pharmacol Res. 2013;69:21–31. doi: 10.1016/j.phrs.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saha JR, Butler VP, Jr, Neu HC, et al. Digoxin-inactivating bacteria: identification in human gut flora. Science. 1983;220:325–7. doi: 10.1126/science.6836275. [DOI] [PubMed] [Google Scholar]
- 51.Haiser HJ, Gootenberg DB, Chatman K, et al. Predicting and manipulating cardiac drug inactivation by the human gut bacterium Eggerthella lenta. Science. 2013;341:295–8. doi: 10.1126/science.1235872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sperry JF, Wilkins TD. Arginine, a growth-limiting factor for Eubacterium lentum. J Bacteriol. 1976;127:780–4. doi: 10.1128/jb.127.2.780-784.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wallace BD, Wang H, Lane KT, et al. Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science. 2010;330:831–5. doi: 10.1126/science.1191175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Iida N, Dzutsev A, Stewart CA, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013;342:967–70. doi: 10.1126/science.1240527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Viaud S, Saccheri F, Mignot G, et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science. 2013;342:971–6. doi: 10.1126/science.1240537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Friedman J, Alm EJ. Inferring correlation networks from genomic survey data. PLoS Comput Biol. 2012;8:e1002687. doi: 10.1371/journal.pcbi.1002687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Faust K, Sathirapongsasuti JF, Izard J, et al. Microbial co-occurrence relationships in the human microbiome. PLoS Comput Biol. 2012;8:e1002606. doi: 10.1371/journal.pcbi.1002606. [DOI] [PMC free article] [PubMed] [Google Scholar]