Abstract
The M2 protein of influenza viruses forms an acid-activated tetrameric proton channel. We used solid-state nuclear magnetic resonance spectroscopy to determine the structure and functional dynamics of the pH-sensing and proton-selective histidine-37 in M2 bound to a cholesterol-containing virus-envelope-mimetic membrane so as to better understand the proton conduction mechanism. In the high-pH closed state, the four histidines form an edge-face π-stacked structure, preventing the formation of a hydrogen-bonded water chain to conduct protons. In the low-pH conducting state, the imidazoliums hydrogen-bond extensively with water and undergo microsecond ring reorientations with an energy barrier greater than 59 kilojoules per mole. This barrier is consistent with the temperature dependence of proton conductivity, suggesting that histidine-37 dynamically shuttles protons into the virion. We propose a proton conduction mechanism in which ring-flip–assisted imidazole deprotonation is the rate-limiting step.
Proton transport in synthetic materials is mediated either solely by hydrogen-bonded (H-bonded) water, as in hydrated ionic polymers (1), or solely by titratable heterocycles, such as imidazoles tethered to the backbone of anhydrous polymers (2). In comparison, the conduction mechanism of biological proton channels in cell membranes is more complex because both water and titratable protein sidechains are usually present (3). The influenza M2 protein forms a tetrameric proton channel that is important for the virus life cycle (4). Activated below pH 6, the M2 channel conducts 10 to 10,000 protons per second (5, 6). The pH-sensing and proton-selective residue is a single histidine, His37, in the transmembrane (TM) domain (7). 15N chemical shifts of His37 in 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC)/dimyristoyl phosphatidylglycerol (DMPG) bilayers indicated that the four histidines titrate with pKas of 8.2, 8.2, 6.3, and <5.0 (where Ka is the acid dissociation constant); thus, the third protonation event is responsible for channel activation (8). However, the precise role of His37 in proton conduction is still debated. Two models have been proposed. In the “shutter” model, the pore at His37 is enlarged through electrostatic repulsion among the imidazoliums, permitting a continuous H-bonded water chain over which protons hop by means of the Grotthuss mechanism (9, 10). The rate-limiting step is proton-hopping across three or four charged imidazoliums, with a calculated energy barrier of 29 to 42 kJ/mol (9). In the “shuttle” model, His37 actively participates in proton relay through protonation and deprotonation. Tautomerization or ring flips reestablish the original conformation required for the next proton relay (11). The rate-limiting step in this model is the His37 conformational change.
Although high-resolution structures of the M2 TM domain (M2TM) in detergents at high and low pH have been reported (12, 13), the His37 sidechain conformations differed in these structures, and sidechain dynamics and water interactions were not probed. Further, detergent molecules can perturb the packing of weakly bound membrane protein complexes; thus, the structures may not accurately reflect the chemistry of the imidazoles in the lipid membrane.
To elucidate the proton conduction mechanism of M2, we used solid-state nuclear magnetic resonance (NMR) to determine the structure and dynamics of His37 in M2TM reconstituted into a cholesterol-rich virus-envelope-mimetic lipid membrane (14, 15). Extensive data yielded the His37 protonation state, tautomerization, rotameric conformation, sidechain dynamics, and hydrogen bonding from pH 8.5 to 4.5. Here, we focus on pH 8.5 for the closed channel and pH 4.5 for the conducting channel. M2TM exhibits acid-activated and amantadine-sensitive proton currents similar to the intact protein (16) and fully assembles into four-helix bundles (17) in the viral membrane with immobilized backbones (14), which allowed His37 sidechain motion to be elucidated.
Histidine 15N and 13C chemical shifts are exquisitely sensitive to the protonation state and tautomeric structure of imidazoles. Deprotonation increases the 15N chemical shift by ~80 parts per million (ppm) (18), and Cγ/Cδ2 chemical shifts also systematically depend on the imidazole structure (19). Two-dimensional (2D) 13C-13C and 15N-13C correlation spectra of His37-labeled M2TM revealed only neutral imidazoles at pH 8.5. The Nε2-protonated τ-tautomer and Nδ1-protonated π-tautomer exist at a~3:1 ratio (Fig. 1), with slow or no exchange at ambient temperature (fig. S1). Inter-tautomer Cδ2(τ)-Cγ(π) and Cδ2(τ)-Cδ2(π) cross peaks (Fig. 1A) indicate that both tautomers exist in each channel. The ~25% fraction of the π-tautomer is much higher than in small imidazole-containing compounds (18), suggesting stabilization of the protonated Nδ1(π) through hydrogen bonding (20).
Fig. 1.
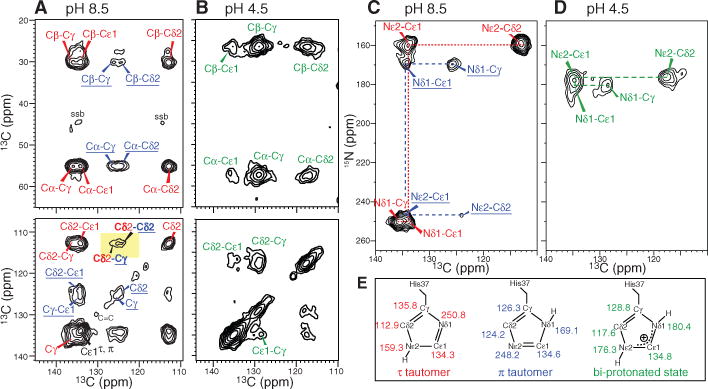
15N and 13C chemical shifts of His37-labeled M2TM in viral membranes reveal pH-dependent imidazole protonation state and tautomeric structures. (A and B) 2D 13C-13C correlation spectra, (A) pH 8.5 and (B) pH 4.5. The τ- and π-tautomer peaks are assigned in red and blue, and the charged His37 peaks are in green. (C and D) 2D 15N-13C correlation spectra, (C) pH 8.5 and (D) pH 4.5. (E) Summary of the imidazole chemical shifts.
At pH 4.5, both Nδ1 and Nε2 exhibit protonated chemical shifts (170 to 180 ppm); no unprotonated signal was observed at 250 ppm (fig. S1), indicating that the neutral species is below the detection limit (≤5%). The charged imidazoliums showed much larger linewidths than the neutral species (table S1), indicating broader conformational distribution of the protein at low pH.
We probed interhelical packing of the His37 tetrad through χ1- and χ2-dependent backbone-sidechain distances. The Cα-Nδ1 distance constrains the χ2 torsion angle, whereas the Cδ2-Nα distance constrains both χ1 and χ2 angles. At both pH, the Cα-Nδ1 distance was 3.9 Å (Fig. 2 and fig. S2), which ruled out the +60° and −60° χ2 rotamers and was consistent only with the 180° rotamer. Similar experiments yielded a Cδ2-Nα distance of 4.4 to 4.9 Å (fig. S3), which ruled out the χ1 = −60° rotamer. Thus, the high-pH τ-tautomer adopts the tt rotamer, which is consistent with interhelical His37-Trp41 (21) and Trp41-Trp41 distances (17). At pH 4.5, the Cα-Nδ1 (3.9 Å), Cα-Nε2 (4.4 Å), and Cδ2-Nα (>4.5 Å) distances similarly indicated the tt conformation (Fig. 2B and fig. S3).
Fig. 2.
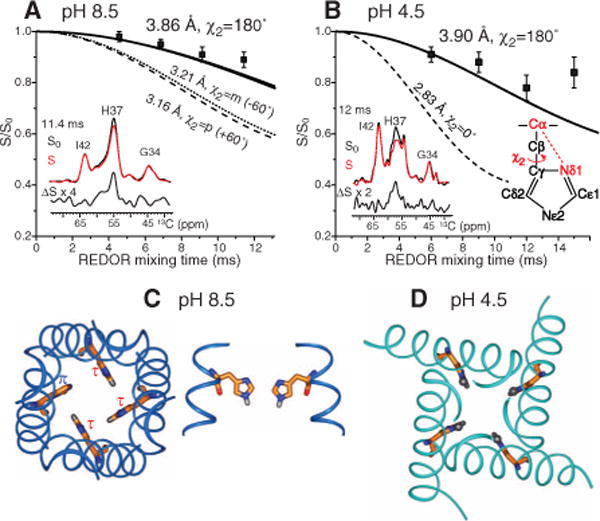
His37 rotameric conformation from Cα-Nδ1 distances. (A) pH 8.5 data, with representative rotational-echo double-resonance control (S0), dephased (S), and difference (∆S) spectra. The 3.9 Å distance indicates χ2 = 180°. (B) pH 4.5 data, showing a similar distance and χ2 angle. (C) Top and side views of the His37 tetrad in the tt rotamer in the high-pH structure [Protein Data Bank (PDB) number 2KQT] (22). (D) Top view of the His37 tetrad in the tt rotamer in the low-pH structure (PDB number 3C9J) (13).
Placing the His37 rotamer into the different backbone structures at low pH and high pH revealed substantially different packing of the imidazole tetrad (Fig. 2, C and D). In the closed channel (22), the major τ-tautomers pack in an edge-face fashion in which each Cε1-Hε1 bond points to the center of the neighboring ring. The packing is tight, with a nearest-neighbor Cε1-Nε2 distance of ~4.9 Å, which is consistent with inter-tautomer cross peaks and suggests aromatic CH-π interaction (23). The high density of π-electrons should repel water oxygens and orient them in opposite directions across the tetrad, thus disabling proton conduction. The tetrad dimension is still possible for metal-ion coordination (24), which may explain Cu2+ inhibition of M2 (25). The minor π-tautomer can readily maintain the fourfold symmetry by adopting tt or t0 rotamer. In comparison, in the low-pH structure (13) the imidazoliums show no edge-face stacking and leave a much wider pore.
The considerable packing difference suggests that His37 sidechains may be immobilized at high pH but dynamic at low pH. To test this hypothesis, we measured one-bond Cγ-Nδ1 and Cδ2-Hδ2 dipolar couplings at physiological temperature. The viral membrane immobilized the protein backbone, giving Nα-H and Cα-Hα order parameters of 1.0 (figs. S4 and S5) (14), thus isolating potential sidechain motion. Fast motions scale the couplings by an order parameter (S) that reflects the motional amplitude. At pH 8.5, we obtained rigid-limit Cγ-Nδ1 and Cε1-Nδ1 couplings (1.15 kHz and 1.39 Å) and a rigid-limit Cδ2-Hδ2 coupling (23.9 kHz and 1.08 Å) (Fig. 3, A and B), confirming immobilization of the neutral imidazoles by edge-face stacking. However, at pH 4.5 the Cγ-Nδ1 and Cδ2-Hδ2 couplings are scaled by a factor of 0.85 and 0.80, respectively (Fig. 3C), indicating sidechain motion. The availability of two-order parameters constrained the geometry of the imidazolium motion. The most likely motional axis is the Cβ-Cγ bond (26). Uniaxial rotation is ruled out because it predicts a very small SCγ–Nδ1 of 0.06 because of the 57° angle of the Cγ-Nδ1 bond to the Cβ-Cγ axis. The well-known 180° ring flip motion is also ruled out because it has little effect on the Cδ2-Hδ2 coupling (SCδ2–Hδ2 = 0.94) (table S2). The near-invariance of the Cδ2-Hδ2 coupling to 180° ring flips also rules out a scenario in which some imidazoliums undergo ring flips whereas others remain static (fig. S6). Instead, analysis of the S dependence on χ2 angles shows that only two-site jumps with a χ2 change of 45° satisfies both the Cγ-Nδ1 and Cδ2-H order parameters (fig. S7). Given the average χ2 of 180° at low temperature, the most likely instantaneous χ2 angles are about 160° and −155° (Fig. 3D).
Fig. 3.
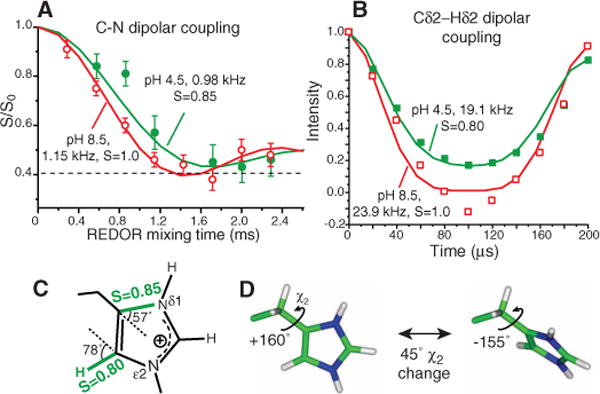
His37 sidechains reorient at low pH but remain static at high pH at physiological temperature. (A) 303 K 13C-15N dipolar couplings. At pH 8.5, a 1:1 combination of Cγ-Nδ1 and Cε1-Nδ1 couplings reaches the rigid limit. At pH 4.5, the dominant Cγ-Nδ1 coupling is motionally averaged. (B) Cδ2-Hδ2 coupling at 308 K is motionally averaged at pH 4.5 but in the rigid limit at pH 8.5. (C) Measured order parameters at pH 4.5. (D) Two-site jump of imidazolium at low pH. A 45° reorientation around the Cβ-Cγ bond fits the observed order parameters.
The restricted nature of the imidazolium ring reorientation may result from the symmetric low pH across the bilayer in the NMR samples because the imidazoles may not need to substantially reorient to be deprotonated and reprotonated. When a proton concentration gradient exists, such as in the virus membrane, full ring flips may occur. The motion must be much faster than 104 s−1 to average the Cδ2-Hδ2 coupling. Temperature-dependent Cδ2-Hδ2 couplings from 308 to 243 K indicated that the imidazolium was frozen by 263 K but fully mobile at 293 K (fig. S8). Using a lower-limit of 50 kHz for the 293 K rate and an upper-limit of 3 kHz for the 263 K rate, we obtained an energy barrier of >59 kJ/mol, which is consistent with the 50 to 120 kJ/mol reported for imidazole motions in synthetic proton conductors (27). M2 proton conductivities differ by 14-fold between 18° and 37°C at pH 5.7 (5), indicating an energy barrier of 104 kJ/mol. Thus, the barrier of imidazolium motion is consistent with the functional data, whereas the barrier for water-mediated proton hopping (29 to 42 kJ/mol) is not (9), suggesting that His37 ring reorientation is directly involved in proton transport, as in the “shuttle” model. Imidazole motion was also observed at the physiological pH of 6.0 and 5.2, at which the channel first opened and both charged and neutral histidines were present (fig. S8) (7, 8). Thus, His37 motion appears to be an intrinsic property of the spacious conducting channel, although its precise amplitudes and rates may vary with pH.
Water is still necessary for delivering protons to the imidazoles before they can be relayed to the virus interior. Thus, water-His37 hydrogen bonding is implied in the shuttle model. We probed H-bond formation by measuring imidazole N-H and C-H dipolar couplings at 243 K, at which the sidechain was frozen. H-bond formation stretches the N-H and C-H bonds from their covalent lengths (1.03 Å and 1.10 Å), thus weakening dipolar couplings (28). At pH 8.5, the protonated Nδ1(π) showed a significantly stretched N-H bond of 1.08 Å, indicating hydrogen bonding and explaining the π-tautomer stabilization. Even the unprotonated Nδ1(τ) showed a sizeable coupling of 2.1 kHz, suggesting a nearest-proton distance of 1.8 Å and a weak Nδ1…H-O H-bond. In contrast, the protonated Nε2(τ) exhibited an unstretched bond length of 1.03 Å (11.1 kHz) (Fig. 4A), despite the presence of a small amount of water in the H37-W41 region on the basis of Nε2(τ)-water cross peaks in 2D 15N-1H correlation spectra (fig. S9). At pH 4.5, the combined Nδ1/Nε2 peak showed a reduced N-H coupling of 8.8 kHz, indicating a stretched bond of 1.11 Å. Given the spaciousness of the low-pH pore, the H-bond acceptors cannot be another imidazolium. Trp41-His37 aromatic interaction may partly contribute to Nε2-H bond stretching (10, 29), but we propose the most likely cause for N-H bond elongation at low pH is H-bond with frozen water, which is more abundant in the low-pH channel than the high-pH channel, as shown with spin diffusion NMR (30) and molecular dynamics simulations (9, 31).
Fig. 4.
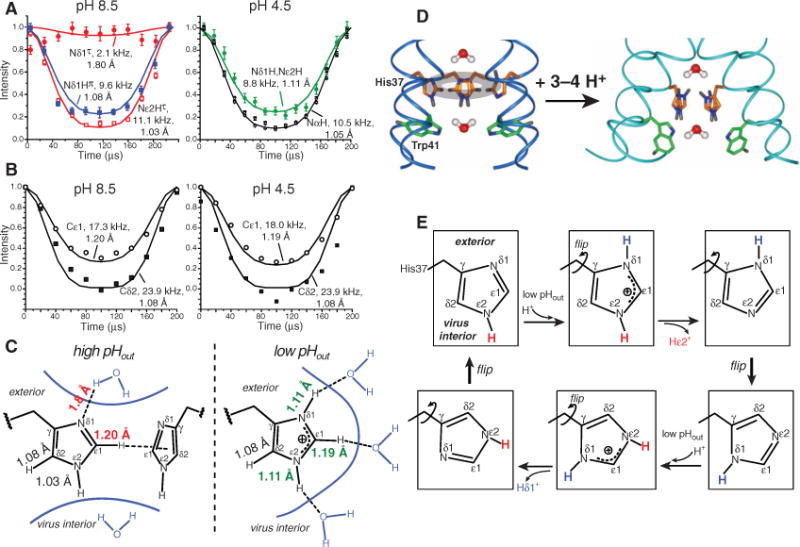
Charged His37 H-bonds with water and undergoes ring flips to relay protons. (A) N-H dipolar couplings at 243 K. At pH 8.5, Nε2(τ) is not H-bonded, whereas Nδ1(π) is. The unprotonated Nδ1(τ) shows a weak H-bond. At pH 4.5, both nitrogens show weak couplings and bond stretching. (B) C-H dipolar couplings of Cδ2 and Cε1 at 243 K. The Cε1-Hε1 bond is stretched, whereas Cδ2-Hδ2 is not. (C) Imidazole bond lengths and H-bond networks at high and low pH. (D) His37 structure and dynamics and proposed water orientations across the tetrad. Trp41 may interact with His37 at high pHout. (E) Proposed imidazole structural changes in a cycle in which multiple ring reorientations mediate the transfer of two protons.
Similar C-H coupling measurements revealed that the Cδ2-Hδ2 bond was unstretched (1.08 Å) at either pH, whereas the Cε1-Hε1 bond was stretched to 1.20 Å at pH 8.5 and 1.19 Å at pH 4.5 (Fig. 4B). The latter may be attributed to CH-π interactions at high pH and H-bond with water at low pH. The imidazole Cε1-Hε1 bond is known to be prone to elongation because of its acidic nature (32), although the large magnitude of stretching observed here is surprising and requires further investigation.
These bond lengths reveal an extensive H-bond network that covers three sides of the imidazolium at low pH (Fig. 4C), creating a continuous H-bonded chain. Similar to the histidine in the catalytic triad of serine proteases, Cε1 hydrogen bonding may facilitate Nε2 deprotonation by evenly distributing the positive charge and increasing Nε2 electronegativity (26, 33). At high pH, the H-bond network is incomplete, excluding Nε2, which we attribute to the opposing water orientation and possible His37-Trp41 interactions (Fig. 4D).
Taken together, these data suggest the following mechanism for proton gating and conduction by M2 (Fig. 4, D and E). At high pHout, the neutral imidazoles form tightly packed electron-rich CH-π stacks, preventing the formation of a H-bonded water chain. The outward-facing Nδ1(π) H-bonds with water, whereas the inward-facing Nε2 does not. Lowering pHout protonates Nδ1, resulting in several imidazoliums per channel, which repel each other and cause backbone conformational changes that widen the pore (34, 35). More water permeates this region (30), establishing a H-bonded chain that includes His37. The larger pore frees the imidazoliums to undergo microsecond ring reorientations. We propose a proton conduction mechanism in which imidazolium deprotonation is facilitated by Cε1-Hε1 hydrogen bonding and continuous ring flips achieve the dual purpose of properly aligning the charged imidazolium with the C-terminal water molecules so as to cause proton transfer and then pointing the unprotonated nitrogen to the low-pH extracellular side to be reprotonated. Our data indicate that the highest energy barrier of this process is the imidazolium motion, which may account for the temperature dependence of M2 proton conductance, possibly in combination with an additional small barrier for proton transfer (5). This dynamically assisted proton transfer model is consistent with the observed deuterium isotope effect, whose magnitude also suggested a mixed H-bonded chain with dissimilar elements (6). Thus, the present data strongly suggest that His37 is actively involved in proton conduction by M2. The structural information obtained here is largely invisible to conventional high-resolution techniques and demonstrates the ability of solid-state NMR to elucidate functionally important membrane protein dynamics and chemistry.
Supplementary Material
Footnotes
References and Notes
- 1.Mauritz KA, Moore RB. Chem Rev. 2004;104:4535. doi: 10.1021/cr0207123. [DOI] [PubMed] [Google Scholar]
- 2.Bureekaew S, et al. Nat Mater. 2009;8:831. doi: 10.1038/nmat2526. [DOI] [PubMed] [Google Scholar]
- 3.Nagle JF, Morowitz HJ. Proc Natl Acad Sci USA. 1978;75:298. doi: 10.1073/pnas.75.1.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cady SD, Luo WB, Hu F, Hong M. Biochemistry. 2009;48:7356. doi: 10.1021/bi9008837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin TI, Schroeder C. J Virol. 2001;75:3647. doi: 10.1128/JVI.75.8.3647-3656.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mould JA, et al. J Biol Chem. 2000;275:8592. doi: 10.1074/jbc.275.12.8592. [DOI] [PubMed] [Google Scholar]
- 7.Wang C, Lamb RA, Pinto LH. Biophys J. 1995;69:1363. doi: 10.1016/S0006-3495(95)80003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu J, et al. Proc Natl Acad Sci USA. 2006;103:6865. [Google Scholar]
- 9.Chen H, Wu Y, Voth GA. Biophys J. 2007;93:3470. doi: 10.1529/biophysj.107.105742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okada A, Miura T, Takeuchi H. Biochemistry. 2001;40:6053. doi: 10.1021/bi0028441. [DOI] [PubMed] [Google Scholar]
- 11.Pinto LH, et al. Proc Natl Acad Sci USA. 1997;94:11301. doi: 10.1073/pnas.94.21.11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schnell JR, Chou JJ. Nature. 2008;451:591. doi: 10.1038/nature06531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stouffer AL, et al. Nature. 2008;451:596. doi: 10.1038/nature06528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo W, Cady SD, Hong M. Biochemistry. 2009;48:6361. doi: 10.1021/bi900716s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Materials and methods are available as supporting material on Science Online.
- 16.Ma C, et al. Proc Natl Acad Sci USA. 2009;106:12283. [Google Scholar]
- 17.Luo W, Mani R, Hong M. J Phys Chem. 2007;111:10825. doi: 10.1021/jp073823k. [DOI] [PubMed] [Google Scholar]
- 18.Munowitz M, Bachovchin WW, Herzfeld J, Dobson CM, Griffin RG. J Am Chem Soc. 1982;104:1192. [Google Scholar]
- 19.Henry B, Tekely P, Delpuech JJ. J Am Chem Soc. 2002;124:2025. doi: 10.1021/ja011638t. [DOI] [PubMed] [Google Scholar]
- 20.Bachovchin WW, Roberts JD. J Am Chem Soc. 1978;100:8041. [Google Scholar]
- 21.Nishimura K, Kim S, Zhang L, Cross TA. Biochemistry. 2002;41:13170. doi: 10.1021/bi0262799. [DOI] [PubMed] [Google Scholar]
- 22.Cady SD, et al. Nature. 2010;463:689. doi: 10.1038/nature08722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Waters ML. Curr Opin Chem Biol. 2002;6:736. doi: 10.1016/s1367-5931(02)00359-9. [DOI] [PubMed] [Google Scholar]
- 24.Brinen LS, Willett WS, Craik CS, Fletterick RJ. Biochemistry. 1996;35:5999. doi: 10.1021/bi9530200. [DOI] [PubMed] [Google Scholar]
- 25.Gandhi CS, et al. J Biol Chem. 1999;274:5474. doi: 10.1074/jbc.274.9.5474. [DOI] [PubMed] [Google Scholar]
- 26.Ash EL, et al. Proc Natl Acad Sci USA. 2000;97:10371. doi: 10.1073/pnas.97.19.10371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fischbach I, Spiess HW, Saalwachter K, Goward GR. J Phys Chem. 2004;108:18500. [Google Scholar]
- 28.Song XJ, Rienstra CM, McDermott AE. Magn Reson Chem. 2001;39(S1):S30. [Google Scholar]
- 29.Tang Y, Zaitseva F, Lamb RA, Pinto LH. J Biol Chem. 2002;277:39880. doi: 10.1074/jbc.M206582200. [DOI] [PubMed] [Google Scholar]
- 30.Luo W, Hong M. J Am Chem Soc. 2010;132:2378. doi: 10.1021/ja9096219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yi M, Cross TA, Zhou HX. J Phys Chem B. 2008;112:7977. doi: 10.1021/jp800171m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scheiner S, Kar T, Pattanayak J. J Am Chem Soc. 2002;124:13257. doi: 10.1021/ja027200q. [DOI] [PubMed] [Google Scholar]
- 33.Derewenda ZS, Derewenda U, Kobos PM. J Mol Biol. 1994;241:83. doi: 10.1006/jmbi.1994.1475. [DOI] [PubMed] [Google Scholar]
- 34.Khurana E, et al. Proc Natl Acad Sci USA. 2009;106:1069. doi: 10.1073/pnas.0811720106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yi M, Cross TA, Zhou HX. Proc Natl Acad Sci USA. 2009;106:13311. doi: 10.1073/pnas.0906553106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.We thank S Li for discussions This work was supported by NSF grant MCB-543473 and NIH grant GM088204.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


