Abstract
It has emerged that palindrome-mediated genomic instability generates DNA-based rearrangements. The presence of palindromic AT-rich repeats (PATRRs) at the translocation breakpoints suggested a palindrome-mediated mechanism in the generation of several recurrent constitutional rearrangements: the t(11;22), t(17;22) and t(8;22). To date, all reported PATRR mediated translocations include the PATRR on chromosome 22 (PATRR22) as a translocation partner. Here, the constitutional rearrangement, t(3;8)(p14.2;q24.1), segregating with renal cell carcinoma in two families, is examined. The chromosome 8 breakpoint lies in PATRR8 in the first intron of the RNF139 (TRC8) gene while the chromosome 3 breakpoint is located in an AT-rich palindromic sequence in intron 3 of the FHIT gene (PATRR3). Thus, the t(3;8) is the first PATRR-mediated, recurrent, constitutional translocation that does not involve PATRR22. Furthermore, similar to the t(11;22) and t(8;22), we detect de novo translocations involving PATRR3 in normal sperm. The breakpoint on chromosome 3 is in proximity to FRA3B, the most common fragile site in the human genome and a site of frequent deletions in tumor cells. However, the lack of involvement of PATRR3 sequence in numerous FRA3B-related deletions suggests that there are several different DNA sequence based etiologies responsible for chromosome 3p14.2 genomic rearrangements.
Keywords: Palindrome, PATRR, translocation, FRA3B, renal cell carcinoma
Introduction
Multiple types of repetitive sequence are abundant in the human genome. They are capable of forming various non-B DNA structures, which frequently induce genomic instability and rearrangements (1–4). Palindromic sequences or inverted repeats represent an unstable DNA motif because they can induce unusual stem-loop DNA structures (5, 6). Such repetitive sequences are often responsible for inducing spontaneously arising genomic rearrangements such as recombination or deletions in model organisms (7–9).
Palindromic AT-rich repeat (PATRR)-mediated translocations are an extensively described form of repetitive DNA instability. Previously, recurrent translocations at 22q11 including the constitutional t(11;22)(q23;q11), t(8;22)(q24;q11), and t(17;22)(q11;q11) have been reported. The presence of PATRRs at 22q11, as well as within the relevant 11q23, 8q24 and 17q11 regions, suggests a PATRR-mediated etiology for these recurrent constitutional translocations with 22q11. These translocations have been reported in unrelated families and the breakpoints in almost all translocation carriers are localized at the center of the PATRR on both chromosomes (10–15). Similarly, a t(1;22)(q21;q11), t(9;22)(p21;q11) and a t(4;22)(q35;q11), all non-recurrent translocations, were reported as PATRR mediated (16–18). To date, all PATRR-mediated translocations have involved the PATRR on chromosome 22 (PATRR22). Therefore, PATRR22 represents a hotspot for constitutional translocations (19). However, in this manuscript, the first PATRR mediated translocation that does not involve PATRR22 is described. The constitutional t(3;8)(p14.2;q24.2) has previously been associated with the inheritance of renal cell carcinoma in two unrelated families (20, 21). t(3;8) balanced carriers have breakpoints in the 5’ untranslated region (UTR) of FHIT on chromosome 3 and in the sterol sensing domain of RNF139 TRC8) on chromosome 8 (21, 22). Although it was known that the short arm of chromosome 3 is altered by deletions or translocations in renal cell carcinoma, until this study, the t(3;8) breakpoints were uncharacterized at the nucleotide level.
Previously, it was known that the t(3;8) breakpoint on chromosome 3 either coincides with or is close to FRA3B, the most common fragile site in the human genome but the precise distance between the 3p translocation breakpoint and the fragile site was not known (23–26). Common fragile sites are specific chromosomal regions that preferentially exhibit chromosome instability visible on mitotic chromosomes as non-random gaps, constrictions or breaks following partial inhibition of DNA synthesis. The gaps coincide with regions of deletions, and intra- and inter-chromosomal recombination (27, 28). Numerous heterozygous and homozygous deletions in various human cancers and precancerous lesions involve FRA3B, spanning FHIT introns 3 to 7 (29). Thus, it has been suggested that the presence of FRA3B may predispose to the t(3;8) translocation as well as to FRA3B deletions (30).
Here we characterize the t(3;8) translocation breakpoint junctions and confirm that this is a palindrome mediated translocation with a breakpoint localized to a newly described PATRR on chromosome 3 (PATRR3). PATRR3 and its surrounding sequence in translocation carriers and normal human samples have been examined and characterized. We also report detection of de novo translocations mediated by PATRR3 in sperm samples from normal males. Although the PATRR3 sequence is capable of adopting secondary structure, FRA3B deletions do not encompass PATRR3. These analyses contribute to an understanding of the mechanisms responsible for PATRR induced genomic rearrangements. These studies indicate that PATRR3-mediated translocations appear to be unrelated to FRA3B deletions, which lie in close proximity of, but distal to PATRR3.
Materials and Methods
Sequence analysis of t(3;8) junction fragments and the breakpoint region
Control samples were collected through the clinical cytogenetics laboratory at the Children’s Hospital of Philadelphia with proper institutional review board (IRB) approval and patient consent or they were purchased from the Coriell Institute for Medical Research. Genomic DNA was extracted from blood, lymphoblast or fibroblast cell lines using the PureGene DNA purification kit (QIAGEN). The patients varied in ethnicity to include 12 Caucasians, 2 African Americans, 2 bi-racial individuals, and 1 Asian. The human-mouse chromosome 3-only somatic cell hybrid cell line (GM11713) was obtained from Coriell Mutant Cell Repository. To narrow the location of the breakpoints, PCRs were performed to analyze sequences located on the derivative human chromosome 3 in a somatic cell hybrid. der(3)t(3;8) and der(8)t(3;8) specific PCRs were performed as described in previous articles (31, 32) differing only in primer selection. PCR products were purified by ExoSap-IT (GE Healthcare) and sequenced bi-directionally by an ABI Prism Sequencer 3730 (Applied Biosystems by Life Technologies). The resulting sequences were aligned with PATRR3 and PATRR8 sequences. PCR primers are listed in Table 1.
Table 1.
Primer sequence
| Column1 | Primer | Sequence 5' to 3' |
|---|---|---|
| Surround PATRR3 | FHITi3-4F | GTTCCCCTTGAAATCACTGC |
| FHITi3-4R | AGGTTACCAAAGTGATCAAACC | |
| Surround PATRR3 | FHITi3-5F | CACAAGGCTCACCACTAATCG |
| FHITi3-5R | GCCGCTAAAACAATTCTTCC | |
| Surround PATRR8 | RNF139i1-1F | TTATTTGTCTATCTGATGCCTTCC |
| RNF139i1-1R | CATGGAAGGTAACAAGAAAATGG | |
| Surround PATRR8 | RNF139i1-2F | TTAGTGGCCCATTTTCTTGG |
| RNF139i1-2R | CAGTAGACGCATTTCACAATCC | |
| PATRR3 | PATRR3-386F | GCACCCTGAAGGCTACTTGTTAAAG |
| PATRR3+411R | AACTGGGCTGGACCTCTTTTGGAAC | |
| PATRR3 Nested PCR | PATRR3 NestFw | AACTGGGCTGGACCTCTTTTGGAAC |
| PATRR3 NestRv | GGCGCAAAAAAAARAAAGATATATGATATG | |
| der(3)t(3;8) | PATRR3+411R | AACTGGGCTGGACCTCTTTTGGAAC |
| PATRR8+267R | CATTTAAGTGATGACTCTGTCCAGGG | |
| der(8)t(3;8) | PATRR8-512F | GATTACATATGGCATCTGGTAGGCTG |
| PATRR3-386F | GCACCCTGAAGGCTACTTGTTAAAG | |
| der(3)t(3;22) | PATRR3+411R | AACTGGGCTGGACCTCTTTTGGAAC |
| JF22 | CCTCCAACGGATCCATACT | |
| der(22)t(3;22) | JF22 | CCTCCAACGGATCCATACT |
| PATRR3-386F | GCACCCTGAAGGCTACTTGTTAAAG | |
| der(3)t(3;11) | PATRR3+411R | AACTGGGCTGGACCTCTTTTGGAAC |
| PATRR11-216F | GAGAGTAAAGAAATAGTTCAGAAAGG | |
| der(11)t(3;11) | PATRR11+212R | CCACAGACTCATTCATGGAACC |
| PATRR3-386F | GCACCCTGAAGGCTACTTGTTAAAG |
PATRR3 genotyping
For analysis of PATRR3 polymorphisms, genomic DNA was extracted from buccal cells or blood samples as described above. The PATRR3 region was amplified by PCR with the use of KOD Xtreme™Hot Start DNA Polymerase (EMD4Biosciences). PCR cycles were as follows: 94°C for 2.5min, 35 cycles of at 98°C for 10sec and 63°C for 5min, followed by a final extension at 63°C for 10min. PCR products were separated by agarose gel electrophoresis, then gel slices were purified by QIAquick Gel Extraction Kit (QIAGEN). Nested PCR was performed on the gel purified DNA to determine the size of the PATRR3. The PCR and nested PCR primers are listed in Table 1. The products of nested PCR reactions were sequenced directly using an ABI Prism Sequencer 3730 (Applied Biosystems by Life Technologies).
Detection of PATRR3 involved de novo translocation in normal males
Semen samples, acquired with IRB approval, were from individual anonymous donors with various genotypes of PATRR3. Testis samples were acquired, with IRB approval, from the Cooperative Human Tissue Network. Genomic DNA was extracted from semen or testis samples and translocation-specific PCRs were performed as above. Primers for detection of PATRR-mediated translocations have been previously described and are listed in Table 1 (15, 32). Multiple batches of 100ng sperm DNA each containing 33,000 haploids were amplified. The frequency of translocation events was calculated in the following manner. The number of positive PCR reactions per total number of reactions was counted. The frequency was calculated on the basis that the probability of a positive PCR reaction corresponds to a total sum of a binomial series of the translocation frequency calculated using the equation q=1-(1-p1/n as described previously (31).
SNP arrays for detection FRA3B deletions
Genomic DNA was extracted from cultured fibroblasts or peripheral blood. DNA quality was assessed by a Nano Drop Spectrometer and gel electrophoresis. SNP array analysis was performed using the Illumina Quad610 genotyping bead chip (Illumina, San Diego, CA, USA) (33). Structural aberrations and copy number differences were visually detected using Illumina’s BeadStudio software. The average resolution across the genome for the Illumina platform is 4kb per SNP. In order to be confident of a duplication or deletion 10kb is a minimum requirement. The B allele frequency and logR ratio were used to determine copy number changes.
Results
Characterization of the t(3;8) breakpoints
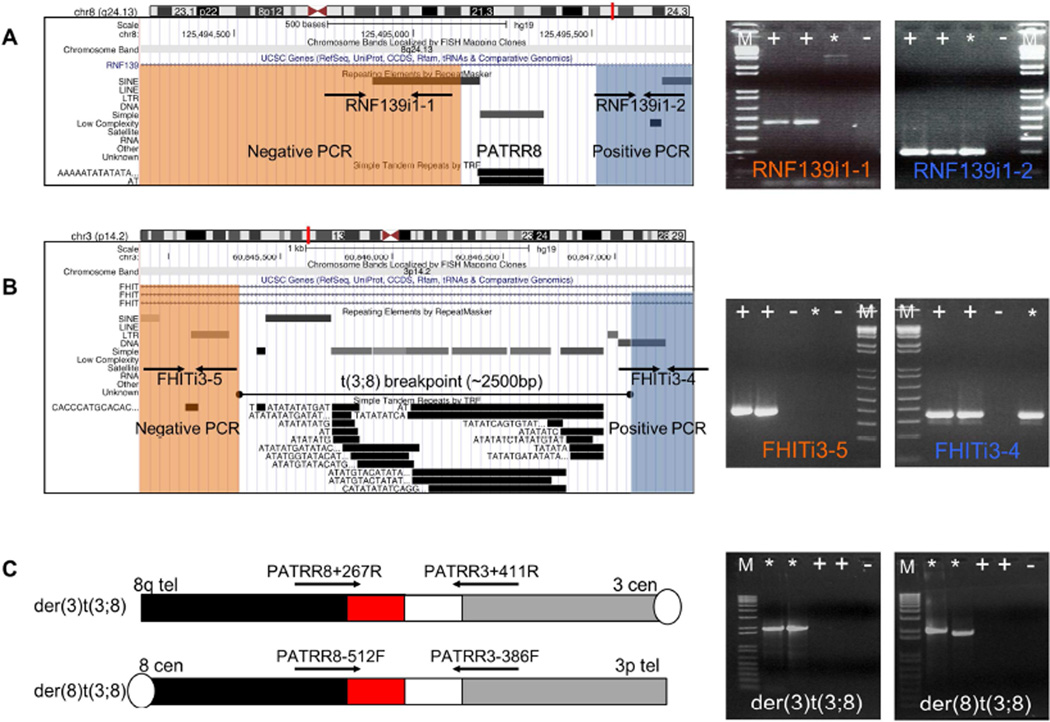
Previously, the breakpoint locations of the t(3;8) were narrowed to the third intron of the FHIT gene located on chromosome 3 and the first intron of the RNF139 gene on chromosome 8 (21, 22) (Supplementary figure 1). Since a PATRR on chromosome 8 (PATRR8), the breakpoint of the recurrent t(8;22), is also located in the first intron of RNF139 we hypothesized that the breakpoint of the t(3;8) might also reside in PATRR8. To further characterize the breakpoint, we utilized DNA from a somatic cell hybrid containing the human der(3)t(3;8). We used multiple sets of PCR primers in the vicinity of PATRR8 (Figure 1A) and in FHIT intron 3 (Figure 1B) to narrow the chromosome breakpoint regions to a 0.6 kb region on the derivative chromosome 8 and a 2.6 kb region of chromosome 8 on the der(3) that included PATRR8. Interestingly, the chromosome 3 breakpoint region contained an AT rich region, suggesting that this region may also contain a PATRR.
Figure 1. Determination of the breakpoints of the t(3;8).
(A) t(3;8) breakpoint mapping on chromosome 8 with DNA from der(3)t(3;8) somatic cell hybrid. On the left is the ideogram of chromosome 8 with a screenshot from the UCSC genome browser below in the region containing the PATRR in the RNF 139 gene. On the right are the relevant PCRs. (B) t(3;8) breakpoint mapping on chromosome 3 with DNA from der(3)t(3;8) somatic cell hybrid. On the left is the ideogram of chromosome 3 with a screenshot from the UCSC genome browser below in the region containing PATRR3 in the FHIT gene. On the right are the relevant PCRs. Lane +, Healthy control; Lane *, der(3)t(3;8) hybrid cell; Lane -, Negative control; Lane M, 1kb+ladder. Sequences surrounding the breakpoints are visualized in the UCSC genome browser. (C) PCR for detection of the breakpoint region of the t(3;8) translocation in a balanced carrier. Primer locations are shown as arrows above the chromosomes. The black bar indicates chromosome 8q, the red bar the chromosome 8 PATRR, the white bar indicates the chromosome 3 PATRR and the grey bar chromosome 3p.. Lane +, Healthy control; Lane *, t(3;8) carrier; Lane −, Negative control; Lane M, 1kb+ladder.
Cell lines derived from the originally reported t(3;8) translocation subjects were used to determine the translocation breakpoints (21, 22). Despite being unrelated, translocations segregating in these two families have grossly the same breakpoints. Translocation-specific PCR and DNA sequencing were performed as previously described (31) (Figure 1C). DNA sequences from carriers in both families demonstrated almost identical breakpoints in the PATRR8 region. The chromosome 3 breakpoint was reconstructed using junction fragment sequences because intact sequence was not present in the reference genomes available. Since access to the individuals in whom the t(3;8) originally arose is unavailable for further analysis we cannot rule out the possibility that the reconstructed PATRR sequence is incorrect and has been altered by DNA resection during translocation. Nonetheless, this reconstructed breakpoint includes a novel PATRR sequence, which consists of VNTRs and (AT)n sequences. The reconstructed PATRR3 from the carriers differ from one another and are about 750bp (AB690558) and 600bp (AB690559) in length. They are predicted to have symmetrical hairpin structures by the M-fold software program (http://mfold.rna.albany.edu/?q=mfold) (34) (Supplementary Figure 2A). We suggest that these palindromic sequences contribute to the genomic instability of PATRR3, leading to unusual DNA structures.
The t(3;8) translocation breakpoint on chromosome 3 is either localized in or close to the FRA3B region, which has been associated with deletions in various tumor cells (22, 30, 35). To confirm that the t(3;8) did not induce copy number changes in the FRA3B region, we performed a SNP array using DNA from each of the translocation carriers. This analysis did not identify copy number alterations at either 3p14 in the FRA3B region or at the chromosome 8 breakpoint region (Supplementary Figure 3). Thus, as is the case with other PATRR-mediated translocations, the t(3;8)s appear to be balanced translocations that do not result in a gain or loss of genetic material (14, 15, 36).
Polymorphisms of PATRR3 sequences and their structures
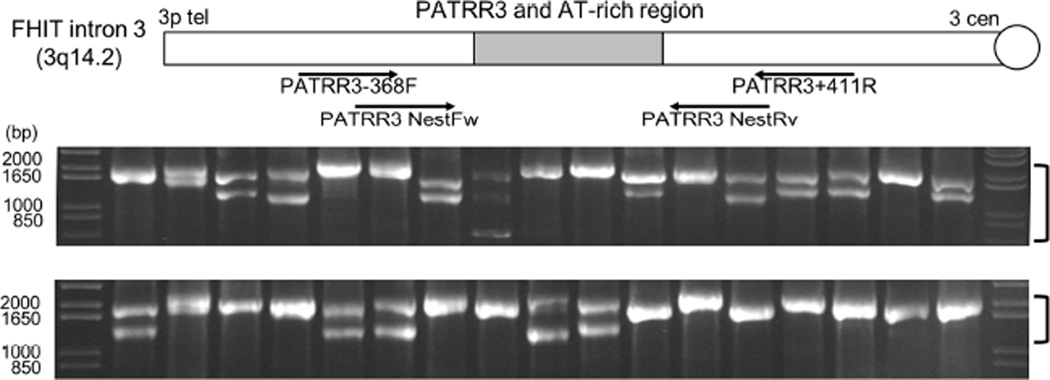
In order to further characterize PATRR3 sequence variation, we performed extensive genotyping using DNA from several human ethnic groups. As reported previously, PATRRs are often polymorphic in size (15, 32, 37, 38). PCR results reveal that PATRR3 is hypervariable in length among individuals, and ranges from 600 to 2030bp (Figure 2). Sequence analysis demonstrates that all of the samples have different fragment lengths, despite the fact that different alleles appear to be the same length by gel electrophoresis. We found only one symmetrical PATRR3 (S-PATRR3), while all other alleles contain an asymmetrical PATRR3 (AS-PATRR3). The potential secondary structure of each of the PATRR3 sequences was predicted using the M-fold software program (34) (Supplementary Figure 2B). The S-PATRR3 is comprised of a symmetrical long hairpin structure, while AS-PATRR3s appear to have shorter hairpin structures of various lengths. The number and/or orientation of the VNTRs and (AT)n length contribute to differences in size and secondary structure forming propensity.
Figure 2. Genotyping Human PATRR3 by PCR to determine PATRR3 polymorphisms.
Agarose gel electrophoresis of nested PCR products from 17 normal human samples. Primer locations for PCR and nested PCR are depicted below the chromosome. Bracket beside the gel indicates the PATRR3 product(s).
Translocation specific PCR using normal human sperm
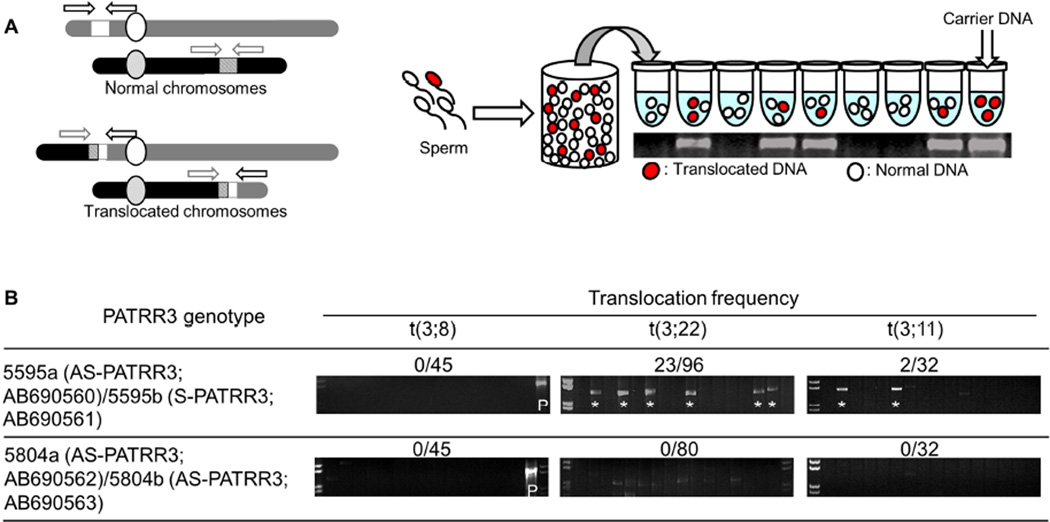
We have previously demonstrated that de novo t(11;22)s occur frequently in sperm from normal healthy males (31). Recently, we also identified de novo PATRR-mediated t(8;22)s as well as t(8;11)s by a similar PCR method, suggesting that a proportion of constitutional translocations result from a palindrome-mediated mechanism in meiosis (15). Here, we tried to detect de novo occurrences of PATRR3-involved translocations by PCR in sperm from normal healthy males with various PATRR3 alleles (Figure 3A). First, we performed der(3)t(3;8) and der(8)t(3;8) translocation-specific PCR in sperm samples. However neither the der(3)t(3;8) nor der(8)t(3;8) were observed in any of our samples (Translocation frequency <3.4×10−7). Despite the fact that t(3;22) and t(3;11) translocations have not been reported in the literature, t(3;22) and the t(3;11) were detected (Figure 3B) at a low translocation frequency (≤10−6) in a sperm sample derived from a heterozygous S-PATRR3 carrier. Sequence analysis revealed that all of the translocations appear to originate from the S-PATRR3 allele (5595b). Sperm samples from carriers of AS-PATRR3s (5804) on both chromosomes did not produce any de novo PATRR3 related translocations, or produced such a small number of de novo translocations that they were below the sensitivity of this PCR assay. However de novo t(11;22) and t(8;22) translocations from these individuals could be detected by PCR. Thus, given the caveat that we only found one individual with an S-PATTR3 allele, the occurrence of de novo PATRR3 involved translocations appears to be dependent upon the presence of PATRR3 symmetry.
Figure 3. Detection of de novo PATRR3 involved translocations.
(A) Strategy of translocation-specific PCR. Arrowheads indicate each relevant primer. Diagram of the strategy used for estimation of translocation frequency by PCR. Genomic DNA was isolated from sperm samples. Translocation-specific PCR was performed using multiple batches of template DNA. Additional details of the methodology are described in Materials and Methods. The gel images show representative PCR results derived from sperm DNA samples. (B) Translocation frequency of PATRR3-related PCR. Lane *, PCR positive; Lane P, t(3;8) carrier. PCR products from 200ng of sperm DNA.
Genomic Analysis of the FRA3B region
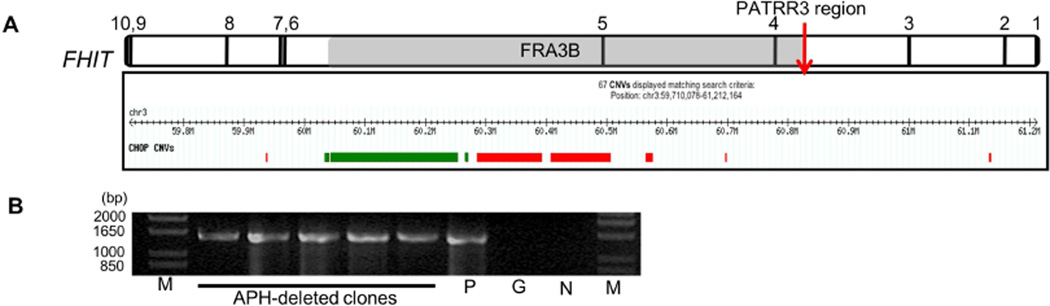
Sequence analysis using the M-fold program indicates that PATRR3s are predicted to adopt secondary structures, with the potential for impeding replication fork progression (Supplementary figure 2). FRA3B related deletions, which are induced by aphidicolin (APH), appear to be located in the t(3;8) translocation breakpoint region (39). Furthermore, APH-induced deletions have been detected in the PATRR on chromosome 11 (PATRR11) (40). These observations suggested that PATRR3 sequences might form unusual structures and induce recurrent FRA3B deletions. To determine whether PATRR3 sequence is involved in FRA3B deletions, we analyzed human samples with deletions in the vicinity of FRA3B and DNA from a chromosome 3-only human-mouse somatic cell hybrid with an intact FRA3B region. The somatic cell hybrid contains one normal human chromosome 3 and was not initially treated with APH. First, using array data derived from 2067 healthy controls (41), we assessed copy number variation in the FRA3B region. Many deletions in the FRA3B region were observed, yet none appeared to include PATRR3. Recurrent deletions were concentrated in intron 5 of the FHIT gene (Figure 4A). In a previous study, Durkin et al., using the same chromosome 3 human-mouse somatic cell hybrid, established that deletions in the FRA3B region are induced upon APH-mediated replication stress (29). Using the hybrid clones containing APH induced deletions, PCR was performed to determine whether the PATRR3 region is retained. All clones retain PATRR3 (Figure 4B). Of note, the PATRR3 sequence of the human-mouse somatic cell hybrid is AS-PATRR3 (AB690564). Despite the AS-PATRR3 allele type, many FRA3B deletions were seen. These results indicate that deletions of FRA3B do not include PATRR3, suggesting that PATRR3 sequence might not induce the FRA3B deletions.
Figure 4. Copy number variation in the FHIT gene.
(A) FRA3B deletions in 2067 samples from healthy individuals are illustrated. Deletions are depicted under the genomic position. Green bars indicate recurrent deletions. Red bars indicate unique deletions. In total, 61 deletions were found in the FHIT gene. Among them, 55 deletions were not unique (green bars 2+6+47). Indicated by the red bars are the unique deletions. (B) PCR for PATRR3 region in APH-induced FRA3B deletions. Lane P, Chromosome 3 hybrid DNA (Non APH-treated); Lane M, 1kb+ladder; Lane G, mouse genomic DNA; Lane N, Negative control.
Discussion
In this study, a recurrent, constitutional PATRR-mediated translocation, which does not involve PATRR22, has been examined in two unrelated families. This translocation, the t(3;8)(p14.2;q24.1), has previously been associated with hereditary renal cell carcinoma in these individuals and their translocation carrier relatives. Intriguingly, the breakpoint sequence of chromosome 8 in the t(3;8) is in the same region as the recurrent t(8;22) breakpoint (15). Also the breakpoint of chromosome 3 is localized at the center of a novel AT-rich palindromic sequence (Figure 1A, B). Neither translocation carrier demonstrates DNA deletions surrounding the breakpoint regions (Supplementary Figure 3). Collectively, these findings indicate that the t(3;8) is another constitutional, recurrent and balanced PATRR-mediated translocation. Reconstructing the original chromosome 3 region from the der(3) and der(8) junction fragments reveals that PATRR3 consists of a combination of many VNTRs and (AT)n sequences that form an inverted repeat. In the general population, PATRR3 varies in size and sequence. The majority of human PATRR3 alleles are asymmetric, and appear to have arisen either by deletion of a symmetrical allele or by transmission with minor nucleotide substitutions (42).
The frequency of de novo PATRR3-related translocations is relatively low, as compared to t(11;22) and t(8;22) translocations (15, 32). Despite the possibility of PATRR3 having the longest palindromic sequence, de novo translocations are rarely seen (43). We have observed that PATRR3 has several large mismatches between its proximal and distal arms and a relatively low AT-content (about 75%) as compared with other PATRRs. The AT-content of a PATRR greatly affects its predicted propensity to form secondary structures. A low melting temperature would be expected to permit double-stranded DNA to denature more readily, allowing the subsequent formation of a single-stranded folded structure (44, 45). Thus, as a result of its lower AT content, PATRR3 might be less likely to form a cruciform structure than other PATRRs. Consequently de novo PATRR3-related translocations may be produced infrequently in sperm (46).
De novo translocations between PATRR3 and other PATRRs [t(3;22) and t(3;11)] were seen as PCR products only in the sperm sample carrying the symmetrical PATRR3 allele, even though neither of these translocations has been reported in the literature. In contrast, de novo t(3;8)s have not been detected in sperm, despite the fact that several balanced translocation carriers have been reported. Prior to the present t(3;8) findings, all of the PATRR-mediated translocations had included PATRR22 as a partner (43, 47). This suggests that there may be a hierarchy of susceptibility to translocation that is likely based on the configuration of both of the partner PATRRs.
In fact, numerous t(11;22) translocation carriers have been reported (10, 36, 48, 49). Therefore it appears that the most translocation vulnerable PATRR is PATRR22, with the next most susceptible being the PATRR11. PATRR11 and 22 related translocations are observed at a high frequency in sperm samples (32, 38). Similarly, t(8;22) translocation carriers have been reported in at least 12 unique cases, and the t(8;22) arises in sperm at a frequency of approximately 2×10−6. The hypothetical t(8;11) rearrangement is also identified in sperm, albeit infrequently (15). Previous studies indicate that PATRR8 is less susceptible to translocation than are PATRR11 and 22, which seems to mirror the experience related to occurrence of PATRR3 related translocations. It supports the observation that de novo t(3;8) translocations did not take place or were produced so infrequently that they were below the sensitivity of the assay. The low allele frequency of the S-PATRR3 may also explain why only two PATRR3-related translocations have been reported to date. Similarly, the t(17;22) translocation has not been detected in sperm from healthy individuals (<5×10−6) (37). From these results, we hypothesize that PATRR3 and PATRR17 are stable and less susceptible to translocation. Alternatively, spatial proximity between PATRR3 and other relevant PATRR containing chromosomes may not occur (50).
PATRR3 is located in or in proximity to FRA3B, in which frequent deletions and translocations are seen in various tumor cells (29, 30, 51). Therefore we investigated the possibility of a relationship between PATRR3 instability and FRA3B deletions in normal human samples and in APH-treated human-mouse somatic cell hybrids. Many deletions were observed in proximity to FRA3B, but they did not include the PATRR3 region. Recurrent deletions were centered in intron 5 of the FHIT gene, which is approximately 600kb away from PATRR3. From these results, we conclude that PATRR3 appears to be directly involved in PATRR3-related translocations, but not in the majority of FRA3B deletions. This suggests that PATRR instability may be limited to vulnerability to translocation and not enhanced susceptibility to deletion. In general, common fragile sites, like FRA3B, are prone to breakage under certain culture conditions or as a result of treatment with specific agents (28). This instability has been thought to be sequence driven as a result of being prone to forming unusual secondary structures (52, 53) and a paucity of activated replication origins in FRA3B following replication stress (54). Common fragile sites, including FRA3B, are comprised of relatively AT-rich sequences (23, 55). Therefore they are likely to form unusual secondary DNA structures, which can impede DNA replication and induce genomic rearrangements (24, 53). Despite the fact that PATRRs appear capable of forming hairpin secondary structures (Supplementary figure 2), PATRR3 does not appear to be directly involved in the FRA3B deletions, at least in those we examined. This result corroborates a recent opinion that FRA3B fragility does not rely on fork slowing or stalling caused by secondary structures, but on a paucity of replication initiation events in lymphoblast cell lines (54).
In conclusion, we have identified a PATRR on chromosome 3 that is involved in a recurrent palindrome-mediated translocation. In addition, the occurrence of de novo t(3;11)s and t(3;22)s in normal healthy male sperm indicate that PATRR-mediated translocations can also involve chromosome 3. This indicates that PATRR mediated translocations are not unique to chromosome 22, but represent a universal pathway to chromosomal rearrangement. Analysis of chromosome 3p rearrangements shows that the PATRR3 sequence does not appear to be directly involved at the site of FRA3B deletions, suggesting a non-PATRR mechanism for fragile site instability. Thus, genomic rearrangements on 3p, in the PATRR and FRA3B regions, appear to have several different etiologies.
Supplementary Material
Arrowheads indicate the breakpoint of the t(3;8)(p14;q24). Shaded grey bar indicates the FRA3B region. Exons are shown as horizontal numbered bars.
(A) M-fold analysis of the secondary structure of PATRR3 reconstructions derived from the t(3;8) translocation carriers. Arrows show translocation breakpoints. (B) M-fold analysis of the secondary structure of PATRR3 reconstructions. Sequence surrounding the PATRR3 region is capable of forming a hairpin structures.
Log R ratio for individual probes on chromosomes 3 (top) and chromosome 8 (bottom) are illustrated as individual blue dots. There are no significant copy number changes. These plots were generated using GenomeStudio software (Illumina).
Acknowledgements
The authors wish to thank Laura K. Conlin and Sharon E. Plon for providing technical assistance and invaluable reagents. These studies were supported by CA039926, from the National Cancer Institute (B.S.E.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health. The studies were also supported by funds from the Charles E.H. Upham Chair in Pediatrics (B.S.E.). One of the authors, (T.K.), was supported by a JSPS Postdoctoral Fellowship for Research Abroad.
This manuscript is dedicated to the memory of Rosaline Willis (mother of B.S.E.). Her support and encouragement enabled the maturation of this research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Takema Kato, Email: takema@fujita0hu.ac.jp.
Colleen P. Franconi, Email: franconic@email.chop.edu.
Molly B. Sheridan, Email: msherid3@jhmi.edu.
April M. Hacker, Email: hacker@email.chop.edu.
Hidehito Inagakai, Email: hinagaki@fujita-hu.ac.jp.
Thomas W. Glover, Email: glover@umich.edu.
Martin F. Arlt, Email: arltm@umich.edu.
Harry A. Drabkin, Email: drabkin@musc.edu.
Robert M. Gemmill, Email: gemmill@musc.edu.
Hiroki Kurahashi, Email: kura@fujita-hu.ac.jp.
Beverly S. Emanuel, Email: beverly@mail.med.upenn.edu.
References
- 1.Bacolla A, Wojciechowska M, Kosmider B, et al. The involvement of non-B DNA structures in gross chromosomal rearrangements. DNA Repair (Amst) 2006;5:1161–1170. doi: 10.1016/j.dnarep.2006.05.032. [DOI] [PubMed] [Google Scholar]
- 2.Wang G, Vasquez KM. Non-B DNA structure-induced genetic instability. Mutat Res. 2006;598:103–119. doi: 10.1016/j.mrfmmm.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 3.Zhao J, Bacolla A, Wang G, et al. Non-B DNA structure-induced genetic instability and evolution. Cell Mol Life Sci. 2010;67:43–62. doi: 10.1007/s00018-009-0131-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wells RD. Non-B DNA conformations, mutagenesis and disease. Trends Biochem Sci. 2007;32:271–278. doi: 10.1016/j.tibs.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 5.Leach DR. Long DNA palindromes, cruciform structures, genetic instability and secondary structure repair. Bioessays. 1994;16:893–900. doi: 10.1002/bies.950161207. [DOI] [PubMed] [Google Scholar]
- 6.Lewis SM, Cote AG. Palindromes and genomic stress fractures: bracing and repairing the damage. DNA Repair (Amst) 2006;5:1146–1160. doi: 10.1016/j.dnarep.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 7.Akgun E, Zahn J, Baumes S, et al. Palindrome resolution and recombination in the mammalian germ line. Mol Cell Biol. 1997;17:5559–5570. doi: 10.1128/mcb.17.9.5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collick A, Drew J, Penberth J, et al. Instability of long inverted repeats within mouse transgenes. EMBO J. 1996;15:1163–1171. [PMC free article] [PubMed] [Google Scholar]
- 9.Gordenin DA, Lobachev KS, Degtyareva NP, et al. Inverted DNA repeats: a source of eukaryotic genomic instability. Mol Cell Biol. 1993;13:5315–5322. doi: 10.1128/mcb.13.9.5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edelmann L, Spiteri E, Koren K, et al. AT-rich palindromes mediate the constitutional t(11;22) translocation. Am J Hum Genet. 2001;68:1–13. doi: 10.1086/316952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kurahashi H, Shaikh TH, Hu P, et al. Regions of genomic instability on 22q11 and 11q23 as the etiology for the recurrent constitutional t(11;22) Hum Mol Genet. 2000;9:1665–1670. doi: 10.1093/hmg/9.11.1665. [DOI] [PubMed] [Google Scholar]
- 12.Gotter AL, Nimmakayalu MA, Jalali GR, et al. A palindrome-driven complex rearrangement of 22q11.2 and 8q24.1 elucidated using novel technologies. Genome Res. 2007;17:470–481. doi: 10.1101/gr.6130907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kehrer-Sawatzki H, Haussler J, Krone W, et al. The second case of a t(17;22) in a family with neurofibromatosis type 1: sequence analysis of the breakpoint regions. Hum Genet. 1997;99:237–247. doi: 10.1007/s004390050346. [DOI] [PubMed] [Google Scholar]
- 14.Kurahashi H, Shaikh T, Takata M, et al. The constitutional t(17;22): another translocation mediated by palindromic AT-rich repeats. Am J Hum Genet. 2003;72:733–738. doi: 10.1086/368062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sheridan MB, Kato T, Haldeman-Englert C, et al. A palindrome-mediated recurrent translocation with 3, 1 meiotic nondisjunction: the t(8;22)(q24.13;q11.21) Am J Hum Genet. 2010;87:209–218. doi: 10.1016/j.ajhg.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gotter AL, Shaikh TH, Budarf ML, et al. A palindrome-mediated mechanism distinguishes translocations involving LCR-B of chromosome 22q11.2. Hum Mol Genet. 2004;13:103–115. doi: 10.1093/hmg/ddh004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nimmakayalu MA, Gotter AL, Shaikh TH, et al. A novel sequencebased approach to localize translocation breakpoints identifies the molecular basis of a t(4;22) Hum Mol Genet. 2003;12:2817–2825. doi: 10.1093/hmg/ddg301. [DOI] [PubMed] [Google Scholar]
- 18.Tan X, Anzick SL, Khan SG, et al. Chimeric Negative Regulation of p14ARF and TBX1 by a t(9;22) Translocation Associated with Melanoma, Deafness, and DNA Repair Deficiency. Hum Mutat. 2013 doi: 10.1002/humu.22354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kurahashi H, Inagaki H, Hosoba E, et al. Molecular cloning of a translocation breakpoint hotspot in 22q11. Genome Res. 2007;17:461–469. doi: 10.1101/gr.5769507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cohen AJ, Li FP, Berg S, et al. Hereditary renal-cell carcinoma associated with a chromosomal translocation. N Engl J Med. 1979;301:592–595. doi: 10.1056/NEJM197909133011107. [DOI] [PubMed] [Google Scholar]
- 21.Poland KS, Azim M, Folsom M, et al. A constitutional balanced t(3;8)(p14;q24.1) translocation results in disruption of the TRC8 gene and predisposition to clear cell renal cell carcinoma. Genes Chromosomes Cancer. 2007;46:805–812. doi: 10.1002/gcc.20466. [DOI] [PubMed] [Google Scholar]
- 22.Gemmill RM, West JD, Boldog F, et al. The hereditary renal cell carcinoma 3;8 translocation fuses FHIT to a patched-related gene, TRC8. Proc Natl Acad Sci U S A. 1998;95:9572–9577. doi: 10.1073/pnas.95.16.9572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boldog F, Gemmill RM, West J, et al. Chromosome 3p14 homozygous deletions and sequence analysis of FRA3B. Hum Mol Genet. 1997;6:193–203. doi: 10.1093/hmg/6.2.193. [DOI] [PubMed] [Google Scholar]
- 24.Mimori K, Druck T, Inoue H, et al. Cancer-specific chromosome alterations in the constitutive fragile region FRA3B. Proc Natl Acad Sci U S A. 1999;96:7456–7461. doi: 10.1073/pnas.96.13.7456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mulvihill DJ, Wang YH. Two breakpoint clusters at fragile site FRA3B form phased nucleosomes. Genome Res. 2004;14:1350–1357. doi: 10.1101/gr.2304404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilke CM, Guo SW, Hall BK, et al. Multicolor FISH mapping of YAC clones in 3p14 and identification of a YAC spanning both FRA3B and the t(3;8) associated with hereditary renal cell carcinoma. Genomics. 1994;22:319–326. doi: 10.1006/geno.1994.1390. [DOI] [PubMed] [Google Scholar]
- 27.Arlt MF, Durkin SG, Ragland RL, et al. Common fragile sites as targets for chromosome rearrangements. DNA Repair (Amst) 2006;5:1126–1135. doi: 10.1016/j.dnarep.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 28.Durkin SG, Glover TW. Chromosome fragile sites. Annu Rev Genet. 2007;41:169–192. doi: 10.1146/annurev.genet.41.042007.165900. [DOI] [PubMed] [Google Scholar]
- 29.Durkin SG, Ragland RL, Arlt MF, et al. Replication stress induces tumor-like microdeletions in FHIT/FRA3B. Proc Natl Acad Sci U S A. 2008;105:246–251. doi: 10.1073/pnas.0708097105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Glover TW, Coyle-Morris JF, Li FP, et al. Translocation t(3;8)(p14.2;q24.1) in renal cell carcinoma affects expression of the common fragile site at 3p14(FRA3B) in lymphocytes. Cancer Genet Cytogenet. 1988;31:69–73. doi: 10.1016/0165-4608(88)90013-1. [DOI] [PubMed] [Google Scholar]
- 31.Kurahashi H, Emanuel BS. Unexpectedly high rate of de novo constitutional t(11;22) translocations in sperm from normal males. Nat Genet. 2001;29:139–140. doi: 10.1038/ng1001-139. [DOI] [PubMed] [Google Scholar]
- 32.Kato T, Inagaki H, Yamada K, et al. Genetic variation affects de novo translocation frequency. Science. 2006;311:971. doi: 10.1126/science.1121452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gunderson KL, Steemers FJ, Lee G, et al. A genome-wide scalable SNP genotyping assay using microarray technology. Nat Genet. 2005;37(5):549–554. doi: 10.1038/ng1547. [DOI] [PubMed] [Google Scholar]
- 34.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohta M, Inoue H, Cotticelli MG, et al. The FHIT gene, spanning the chromosome 3p14.2 fragile site and renal carcinoma-associated t(3;8) breakpoint, is abnormal in digestive tract cancers. Cell. 1996;84:587–597. doi: 10.1016/s0092-8674(00)81034-x. [DOI] [PubMed] [Google Scholar]
- 36.Kurahashi H, Shaikh TH, Zackai EH, et al. Tightly clustered 11q23 and 22q11 breakpoints permit PCR-based detection of the recurrent constitutional t(11;22) Am J Hum Genet. 2000;67:763–768. doi: 10.1086/303054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Inagaki H, Ohye T, Kogo H, et al. Palindromic AT-rich repeat in the NF1 gene is hypervariable in humans and evolutionarily conserved in primates. Hum Mutat. 2005;26:332–342. doi: 10.1002/humu.20228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tong M, Kato T, Yamada K, et al. Polymorphisms of the 22q11.2 breakpoint region influence the frequency of de novo constitutional t(11;22)s in sperm. Hum Mol Genet. 2010;19:2630–2637. doi: 10.1093/hmg/ddq150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Glover TW, Stein CK. Chromosome breakage and recombination at fragile sites. Am J Hum Genet. 1988;43:265–273. [PMC free article] [PubMed] [Google Scholar]
- 40.Kurahashi H, Inagaki H, Kato T, et al. Impaired DNA replication prompts deletions within palindromic sequences, but does not induce translocations in human cells. Hum Mol Genet. 2009;18:3397–3406. doi: 10.1093/hmg/ddp279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shaikh TH, Gai X, Perin JC, et al. High-resolution mapping and analysis of copy number variations in the human genome: a data resource for clinical and research applications. Genome Res. 2009;19:1682–1690. doi: 10.1101/gr.083501.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kato T, Inagaki H, Kogo H, et al. Two different forms of palindrome resolution in the human genome: deletion or translocation. Hum Mol Genet. 2008;17:1184–1191. doi: 10.1093/hmg/ddn008. [DOI] [PubMed] [Google Scholar]
- 43.Kato T, Kurahashi H, Emanuel BS. Chromosomal translocations and palindromic AT-rich repeats. Curr Opin Genet Dev. 2012;22:221–228. doi: 10.1016/j.gde.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bowater R, Aboul-ela F, Lilley DM. Large-scale stable opening of supercoiled DNA in response to temperature and supercoiling in (A + T)-rich regions that promote low-salt cruciform extrusion. Biochemistry. 1991;30:11495–11506. doi: 10.1021/bi00113a003. [DOI] [PubMed] [Google Scholar]
- 45.Dayn A, Malkhosyan S, Duzhy D, et al. Formation of (dA-dT)n cruciforms in Escherichia coli cells under different environmental conditions. J Bacteriol. 1991;173:2658–2664. doi: 10.1128/jb.173.8.2658-2664.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kato T, Inagaki H, Tong M, et al. DNA secondary structure is influenced by genetic variation and alters susceptibility to de novo translocation. Mol Cytogenet. 2011;4:18. doi: 10.1186/1755-8166-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kurahashi H, Inagaki H, Ohye T, et al. Palindrome-mediated chromosomal translocations in humans. DNA Repair (Amst) 2006;5:1136–1145. doi: 10.1016/j.dnarep.2006.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tapia-Paez I, Kost-Alimova M, Hu P, et al. The position of t(11;22)(q23;q11) constitutional translocation breakpoint is conserved among its carriers. Hum Genet. 2001;109:167–177. doi: 10.1007/s004390100560. [DOI] [PubMed] [Google Scholar]
- 49.Carter MT, St Pierre SA, Zackai EH, et al. Phenotypic delineation of Emanuel syndrome (supernumerary derivative 22 syndrome): Clinical features of 63 individuals. Am J Med Genet A. 2009;149A:1712–1721. doi: 10.1002/ajmg.a.32957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ashley T, Gaeth AP, Inagaki H, et al. Meiotic recombination and spatial proximity in the etiology of the recurrent t(11;22) Am J Hum Genet. 2006;79:524–538. doi: 10.1086/507652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fang JM, Arlt MF, Burgess AC, et al. Translocation breakpoints in FHIT and FRA3B in both homologs of chromosome 3 in an esophageal adenocarcinoma. Genes Chromosomes Cancer. 2001;30:292–298. doi: 10.1002/1098-2264(2000)9999:9999<::aid-gcc1095>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 52.Lukusa T, Fryns JP. Human chromosome fragility. Biochim Biophys Acta. 2008;1779:3–16. doi: 10.1016/j.bbagrm.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 53.Mishmar D, Rahat A, Scherer SW, et al. Molecular characterization of a common fragile site (FRA7H) on human chromosome 7 by the cloning of a simian virus 40 integration site. Proc Natl Acad Sci U S A. 1998;95:8141–8146. doi: 10.1073/pnas.95.14.8141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Letessier A, Millot GA, Koundrioukoff S, et al. Cell-type-specific replication initiation programs set fragility of the FRA3B fragile site. Nature. 2011;470:120–123. doi: 10.1038/nature09745. [DOI] [PubMed] [Google Scholar]
- 55.Shiraishi T, Druck T, Mimori K, et al. Sequence conservation at human and mouse orthologous common fragile regions, FRA3B/FHIT and Fra14A2/Fhit. Proc Natl Acad Sci U S A. 2001;98:5722–5727. doi: 10.1073/pnas.091095898. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Arrowheads indicate the breakpoint of the t(3;8)(p14;q24). Shaded grey bar indicates the FRA3B region. Exons are shown as horizontal numbered bars.
(A) M-fold analysis of the secondary structure of PATRR3 reconstructions derived from the t(3;8) translocation carriers. Arrows show translocation breakpoints. (B) M-fold analysis of the secondary structure of PATRR3 reconstructions. Sequence surrounding the PATRR3 region is capable of forming a hairpin structures.
Log R ratio for individual probes on chromosomes 3 (top) and chromosome 8 (bottom) are illustrated as individual blue dots. There are no significant copy number changes. These plots were generated using GenomeStudio software (Illumina).