Abstract
Cellular diversity and architectural complexity create barriers to understanding the function of the mammalian central nervous system (CNS) at a molecular level. To address this problem, we recently developed a methodology that provides the ability to profile the entire translated mRNA complement of any genetically defined cell population. This methodology, which we termed translating ribosome affinity purification, or TRAP, combines cell-type-specific transgene expression with affinity purification of translating ribosomes. TRAP can be used to study the cell-type-specific mRNA profiles of any genetically defined cell type, and has been successfully used to date in organisms ranging from D. melanogaster to mice and human cultured cells. Unlike other methodologies that rely upon micro-dissection, cell panning, or cell sorting, the TRAP methodology bypasses the need for tissue fixation or single-cell suspensions (and potential artifacts these treatments introduce), and reports on mRNAs in the entire cell body. This protocol provides a step-by-step guide to implementing the TRAP methodology, which takes two days to complete once all materials are in hand.
Keywords: polyribosome, polysome, mRNA, purification, translating ribosome affinity purification, cell-type-specific, gene expression, mRNA translation
INTRODUCTION
The advent of microarray mRNA expression profiling and RNA sequencing has made possible the simultaneous interrogation of the mRNA expression profiles of all genes in a genome. However, these techniques have not been easily applied to the study of cells that reside in complex tissues. In tissues such as the mammalian brain, the high level of cellular heterogeneity, combined with close anatomical intermixing of diverse cell types, complicates analysis of gene expression data derived from whole tissue samples. Thus, observed gene expression profiles cannot be attributed to any particular cell type, and any changes that are not unidirectional in all cells may not be detected, due to averaging of information across different cell types in the target tissue. Conversely, techniques such as in situ hybridization and immunohistochemistry afford single-cell resolution, but cannot be routinely applied at a genome-wide scale. We recently developed a methodology that we termed translating ribosome affinity purification, which allows the interrogation of the entire translated messenger RNA (mRNA) complement of any genetically defined cell type.
Overview of TRAP
TRAP utilizes both indirect tagging of mRNAs and cell-type-specific genetic targeting of transgene expression (Figure 1). Indirect tagging of mRNAs is achieved by the incorporation of an affinity tag, such as enhanced green fluorescent protein (EGFP), on the large ribosomal subunit protein L10a. EGFP-tagged ribosomes not only assist in cell visualization in tissue, but tagged ribosomes can also be affinity purified with EGFP antibodies. Cell-type-specific expression of EGFP-L10a is accomplished by driving expression of the transgene under regulatory elements known to direct cell-type-specific expression. If ribosomes are maintained on the mRNAs that they are translating upon tissue harvest, purification of the cell-type-specific tagged ribosomes will also yield cell-type-specific translated mRNAs. Translated mRNAs thus purified can subsequently be analyzed by any of the common methods used to study RNA expression, including Northern blot, quantitative PCR (qPCR), microarray, or RNA sequencing. We have successfully used TRAP purified mRNA as input into qPCR, microarray, and RNA sequencing reactions (for examples, see references 17 and 18).
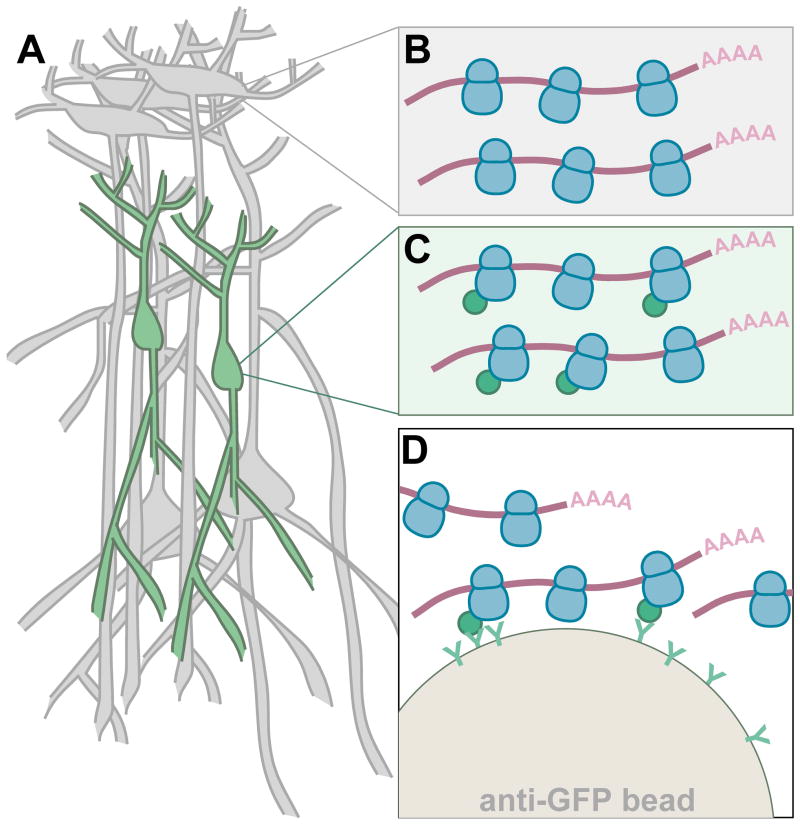
Figure 1. The Translating Ribosome Affinity Purification (TRAP) strategy.
(A) The cell type of interest is targeted with appropriate genetic elements to express the EGFP-L10a transgene. Translating polyribosomes (polysomes) originating from non-targeted cells (grey cells, B) do not have an EGFP tag on their ribosomes, while those originating from targeted cells (green cells, C) do. Lysis of all cells releases both tagged and non-tagged polysomes. Only the latter are captured on an anti-GFP affinity matrix (D), which can be used for purification of the cell-type-specific mRNA associated with tagged polysomes.
Alternative Approaches
Several molecular profiling methods have been developed to study the genome-wide expression profiles of cells in complex tissues, including methods that rely upon the isolation of cells of interest after tissue dissociation. Such methods include fluorescence-activated cell sorting (FACS) of dissociated neurons1–7, manual sorting of fluorescent cells8–10, and immunopanning of dissociated cells with or without FACS7,11. Other methods have relied upon the identification of cell types of interest within sectioned tissue, without tissue dissociation. These methods include laser-capture microdissection12,13 and aspiration of patched cells after electrophysiological recording14,15. While each of these methods has unique advantages, they all suffer from the inability to profile cells in situ, without the introduction of experimental noise caused by tissue fixation, tissue dissociation, tissue incubation ex vivo, or high amplification of only single-cell mRNA content. Further, as these methods require dissociation of tissue or extraction of cells from tissue, they produce expression profiles that reflect only the mRNA content of the cell soma, not the entire cell.
Advantages and Limitations of TRAP
We have previously described a protocol for purification of TRAP-tagged ribosome-mRNA complexes from mouse brain tissue16,17. As described in these studies, critical steps for successful purification of translating ribosomes include: immediate homogenization of target tissue; stabilization of translating ribosomes on intact mRNA by inclusion of magnesium, cycloheximide, and RNase inhibitors in purification solutions; solubilization of rough endoplasmic reticulum-bound ribosomes under non-denaturing conditions; and the use of an affinity matrix that has low background RNA binding. A limitation of the TRAP methodology as originally published was the low yield of mRNA, which necessitated using a large amount of starting material from transgenic animals. Recent optimizations to the TRAP methodology include an increase in RNA yields, greater signal-to-noise ratios, and the ability to freeze tissue before performing TRAP purifications, optimizations that are due to a change in the affinity matrix and RNA extraction conditions18–21.
TRAP has the advantage that is does not require the fixation or dissociation of tissue, or the extraction of cells from tissue for the capture of cell-type-specific mRNA. This allows for the in situ profiling of an entire cell’s mRNA translation profile. This advantage gives TRAP a higher degree of sensitivity than other methods. Further, the TRAP transgene labels the cell type of interest with EGFP, thus allowing for visualization in immunohistochemical or electrophysiological studies. Another advantage of TRAP versus other gene expression profiling methodologies is that it reveals the translated mRNA content of a cell, which will more closely match the protein content than will the total RNA gene expression profile. If the true total RNA profile is desired, another methodology may be preferable to TRAP. A limitation of the TRAP methodology as originally published was the need to generate transgenic animal lines for each cell type of interest. However, this potential limitation has been reduced for mouse studies with the recent generation of several conditional TRAP mouse lines22,23. Nevertheless, a genetic element is always needed to drive cell-type-specific expression of the TRAP transgene in the cell type of interest. In cases where this is not possible (e.g. human brain tissue studies), other methods such as laser capture micro-dissection may be preferable, even if they have less sensitivity.
Applications
Mouse studies
As originally described, cell-type-specific TRAP expression in mice was achieved by the use of cell-type-specific genetic targeting driven by Bacterial Artificial Chromosomes (BACs). BACs are capable of carrying up to approximately 200 kilobases of DNA, a large enough segment of DNA to ensure that the regulatory elements of most genes are included with the transgene to be targeted24–26. Several options now exist to achieve TRAP expression in mice: an existing BAC-TRAP transgenic mouse line can be obtained16,17; a new BAC-TRAP transgenic mouse line can be constructed27; viral transduction of a conditional TRAP construct can be used in conjunction with one of many publically available Cre driver lines28,29, or a conditional TRAP mouse line22,23 can be used in combination with a Cre driver line. In cases where investigators wish to target rare cell populations, BAC-TRAP lines may be of most use, as such lines will drive the highest level of TRAP transgene expression due to integration of multiple copies of the transgene in the genome (unpublished observations, M. Heiman and N. Heintz).
Studies in cultured cells
The TRAP transgene has been successfully used to study translation in cultured, transfected human cells using a constitutive promoter16. We have also succesfully used Amaxa nucleofection, as well as adenoviral and lentiviral transduction to express the TRAP transgene in primary mouse neurons (M. Heiman, unpublished observations). TRAP could be employed to study specific cell types in mixed cultures by placing the TRAP transgene under a compact cell-type-specific promoter, or by using cells taken from transgenic, cell-type-specific Cre-expresing mice (in combination with a conditional TRAP construct).
Studies in other species
TRAP and related ribosomes-tagging approaches have now also been successfully used to study cell-type-specific gene expression in Drosophila30, plants31, Xenopus32, and zebrafish19. Readers are directed to the appropriate references for details on genetic elements needed to achieve cell-type-specific expression in these species. Two important considerations in adapting TRAP to other species is to make sure that codon usage for the EGFP coding sequence is species-optimized, and that the ribosomal protein orthologous to mouse large ribosomal protein L10a is used in constructing the TRAP transgene. In addition, the volumes listed in the protocol below were optimized for working with mouse brain tissue (typically 50–200mg), and thus the number listed may need to be scaled up or down if the amount of starting material is larger or smaller, respectively.
Downstream Applications
RNA purified according to the TRAP protocol is of high purity and can be used directly for various downstream applications, including Northern blotting, qPCR, microarray analysis, and RNA sequencing. An important consideration when choosing a downstream application is the TRAP yield of RNA, which will depend on the abundance of the cell type of study.
Experimental design
Polysome stabilization during cell and tissue collection
Stabilization of the polysome complex is essential to collect mRNAs that accurately reflect the translational state of the cell type of interest. Polysomes can be stabilized by the addition of cycloheximide and magnesium33, and by maintaining the samples at 4°C. Another key step in successfully stabilizing polysomes after cell lysis is to control endogenous RNase activity. Neuronal polysomes have been reported to be particularly sensitive to degradation by RNase33. As blood is a major source of RNase contamination34, a quick wash of the dissected tissue to remove blood is useful to control RNase levels. Maintaining the samples at 4°C, and the addition of recombinant RNase inhibitors to the lysis buffer, will also help to minimize RNase activity. High levels of RNase activity that are not controlled by these precautions can be controlled by the addition of 0.75 mg/ml heparin to all solutions35. However, we do not routinely use heparin in our preparations due to the subsequent need to perform lithium chloride precipitation of the purified RNA in order to prevent inhibition of downstream enzymes by heparin36. If there is doubt regarding the integrity of polysomes in a lysate preparation, polysome profiles should be analyzed after zonal centrifugation, as previously described17,37 and shown for reference in Figure 2.
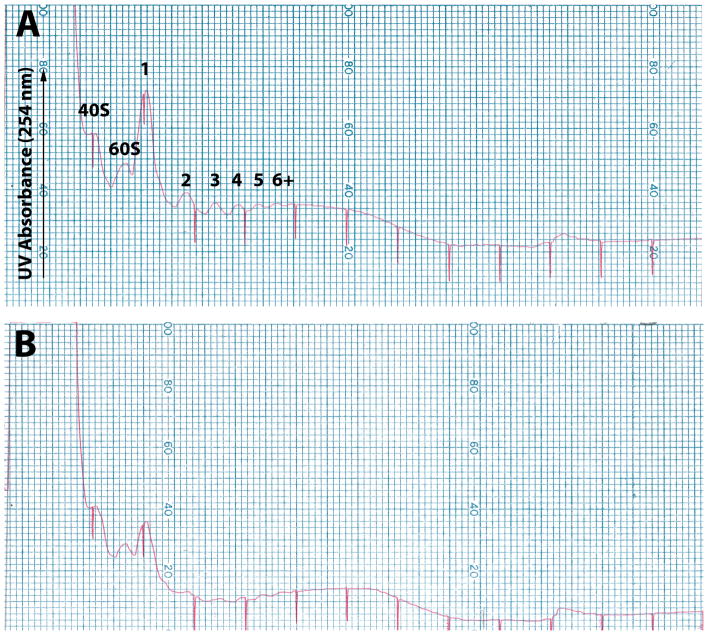
Figure 2. Representative polyribosome profiles from fresh and frozen striatal tissue.
Post-mitochondrial supernatants (prepared as described in this protocol) were prepared from freshly dissected striatal tissue (A) or tissue that had been flash-frozen and stored for four months at −80°C (B), subjected to sucrose-gradient zonal centrifugation as previously described17 and gradient fractions were collected while UV absorbance at 254 nm was monitored. An increase in 254 nm absorbance indicates an increase in RNA content in the fraction. Both fresh and frozen samples were prepared from young adult TRAP mice. Small ribosomal subunit (40S), large ribosomal subunit (60S), monosome (1) and polysome (2, 3, 4, 5, 6+) peaks are indicated. Red ticks indicate gradient fractions. Animal experiments were reviewed and approved by the Rockefeller University Institutional Animal Care and Use Committee.
Expected RNA integrity and yields
Most of the RNA purified by TRAP will be ribosomal RNA (rRNA), and thus any analysis should show mostly purification of this RNA species, although messenger RNA (mRNA, the desired species in most cases) and transfer RNA (tRNA) are also purified. The integrity (lack of degradation) of mRNA can be indirectly estimated by assessing the quality of the rRNA. Using an Agilent Bioanalyzer, we routinely obtain RNA integrity numbers (RINs) of 9.5–10 (out of a maximum 10) from cultured cells, and 7.5–10 (out of a maximum of 10) from brain tissue extracted from unperfused, adult mice. Poor RNA integrity is caused by inefficient control of endogenous, or the addition of exogenous, RNase activity. Expected total RNA yields will vary based upon: the number of cells expressing the TRAP transgene; the expression level of the TRAP transgene; as well as the size and translational state of the cell types expressing the TRAP transgene. Thus actively growing, large cells, expressing high levels of EGFP-L10a will yield more total RNA than post-mitotic, small cells, expressing low levels of EGFP-L10a. As a comparison metric: for 0.7×106 labeled spiny projection neurons in 30 mg of one mouse striatum, with the transgene driven by a moderate-strength driver (Drd1a), we often recover ~50ng of polysomal RNA21.
Effects of freezing tissue on polyribosome integrity and mRNA yields
Optimal polysome integrity, and thus TRAP RNA yields, is obtained from fresh tissue that is immediately homogenized in lysis buffer that will stabilize polysomes. Our studies indicate that approximately half of the monosomes, as well as the polysome aggregates, are lost after flash freezing of tissue in liquid nitrogen, storage at −80°C, and thawing coincident with homogenization (Figure 2). Nevertheless, studies that require the use of large numbers of transgenic animals, or that animals be sacrificed after defined periods of drug exposure are impractical to perform with fresh tissue only. On average, our total RNA yields from TRAP frozen tissue samples are approximately half (46.2%) of that normally obtained from fresh tissue, as predicted from the results of the polysome profiles (Figure 2). We suggest that all samples in an experiment be processed in the same way (fresh or frozen), to avoid experimental artifacts; in particular, we predict that the loss of TRAP yield upon freezing may result in a loss of representation of rare messages, when comparing to fresh tissue.
Experimental group design
Before starting a TRAP study, investigators should ensure that they have the relevant licenses for animal research, and that their proposed TRAP study has been reviewed and approved by the investigator’s Institutional Animal Care and Use Committee. Investigators should adhere to the ARRIVE guidelines for reporting animal experiments38.
Feasibility pilot experiment To determine if a particular tissue source is feasible for use in TRAP studies, purifications should be run from TRAP-expressing and control (non-TRAP expressing) animals to determine the background level of RNA purification from the prospective tissue source. The authors typically use 1 TRAP transgenic and 1 non-transgenic animal each for 3 different affinity matrix concentrations (see Box 1) to determine background when using a new tissue source. As long as there is a difference in amount of RNA purified from these two sources, a TRAP study can be conducted. Brain tissue regions that are very rich in myelin tend to have the highest background RNA levels.
Statistical power analysis pilot experiment While the particulars of each investigator’s scientific hypothesis will direct the TRAP study design (e.g. sex, drug treatment, genetic perturbation, age of animals), nevertheless a second pilot experiment, with 3 TRAP-expressing biological replicates collected under the same purification conditions, should be used to determine the sample size that will be needed in the full study. Results from this pilot study will not only inform the investigators as to expected RNA yields, but should also be used to perform a statistical power analysis based upon the number of genes and hypotheses to be interrogated in downstream applications. It is common for 6–10 biological replicates to be needed for adequate statistical power in genome-wide expression studies.
Collecting experimental groups As with any experiment that will amplify RNA, it is essential to process paired TRAP samples in downstream amplification steps at the same time to avoid amplification artifacts (e.g. processing for microarray or RNA sequencing analysis). Optimally, all samples are harvested, purified, and amplified together. However, in experiments that make use of a large number of experimental groups and biological replicates, this is often impractical. For maximum TRAP sensitivity, at minimum each set of samples that will be directly compared at the raw data level (e.g. vehicle and drug treatment) should be purified and amplified together.
Box 1. Preparation of the affinity matrix ● TIMING 2–2.5 h.
Each purification will require: 300 μl Streptavidin MyOne T1 Dynabeads, 120 μl Biotinylated Protein L (1 μg/μl in 1x PBS), and 50 μg each of GFP antibodies 19C8 and 19F7 (100 μg total antibody).
For feasibility pilot experiments (see Experimental Design section), half and double the matrix component amounts, keeping ratios the same, can also be tried. Investigators should keep altering the amounts, keeping ratios the same, until an optimal amount of matrix is found that captures all tagged message. For example, if the amounts listed above are optimal for a new cell type, one would expect to see a halving of TRAP yield with half the amount of matrix used in TRAP purifications -- and no detectable purification of RNA from the non-TRAP control with either concentration. (We typically use a 1.5-fold excess of optimal concentration in our TRAP experiments, to account for matrix pipetting error.) Such a pilot experiment is recommended because the amount of affinity matrix needed will vary by cell type, as the abundance of the target cell type, its translational state, its size, and the local tissue background RNA binding levels are all characteristics that will vary between different investigators’ TRAP experiments. The amounts listed above were empirically determined to ensure complete binding of all epitope in a relatively abundant cell type (e.g. 0.7×106 spiny projection neurons in 30 mg striatal tissue), but rare cells may need a fraction of the amounts listed here. For such rare cell types, it is advisable to use the minimum amount of matrix needed to reduce background RNA binding.
Upon receipt, record the two antibodies’ concentrations, as they will vary by batch. If the antibodies arrive unfrozen, mix each tube gently and aliquot each antibody into single experiment aliquots (to be used within a week), snap-freeze aliquots in liquid nitrogen, and store the aliquots at −80°C. If the antibodies arrive frozen, store immediately at −80°C; they should be thawed on ice and aliquoted before or at first use. On the day of use, thaw aliquot to be used that day on ice, spin tubes at maximum speed (>13,000 x g) in microcentrifuge for 10 minutes, 4 °C, and take supernatants (antibody) to new tubes. Add sodium azide as needed. Antibody can be kept at 4 °C for a few days. If interval between IPs is longer than this, aliquot, snap-freeze in liquid nitrogen, and store in single-use aliquots at −80 °C.
Resuspend the Streptavidin MyOne T1 Dynabeads thoroughly in the original bottle by gentle hand mixing.
Calculate the amount of Dynabeads required based on the ratios above. Note that affinity matrix to be used in one particular experiment should be aliquoted to samples from a common source: either prepare all matrix in one larger tube, or prepare batches in smaller tubes and combine into a larger tube for mixing prior to aliquoting. As recommended by the manufacturer, throughout all manipulations, keep the volume of beads close to the original volume from the source bottle.
-
Transfer the beads to be used to a tube(s) and collect on magnet (30–60 seconds).
?TROUBLESHOOTING
Wash the beads with 1x PBS one time (1 ml for all washes if in a 1.5-ml tube).
Collect the beads on the magnet and resuspend in the appropriate volume of 1x PBS (original bead volume minus volume of biotinylated Protein L to be added).
Incubate the beads with biotinylated Protein L in 1x PBS (aim for 1 ml total volume if using a 1.5-ml tube) for 35 minutes at room temperature using gentle end-over-end mixing in a tube rotator.
Collect the Protein L-coated beads on the magnet.
Wash the coated beads 5 times with 1x PBS containing 3% (weight/volume) IgG, Protease-free BSA.
Proceed to antibody binding in low-salt buffer, binding 50 μg each of 19C8 and 19F7 (100 μg total, in 1 ml total volume) for 1 hour using gentle end-over-end rotation in a tube rotator. Do not vortex affinity matrix after antibody binding.
| After antibody binding, wash beads 3 times with low-salt buffer. After washing, resuspend the beads in a volume of low-salt buffer such that each IP will receive an aliquot of the components listed above -- beads/Protein L/Ab (the affinity matrix in ratios listed above) -- in a 200 μl final aliquot volume. ■ PAUSE POINT Once prepared, the affinity matrix can be used immediately, or can be stored for up to two weeks at 4°C with the addition of 0.02% sodium azide. If pre-prepared affinity matrix is stored in sodium azide, it should be washed three times quickly in low-salt buffer before use. Pre-prepared affinity matrix may be difficult to resuspend quickly, and may be carefully resuspended by gentle agitation overnight on a tube rotator. Do not vortex affinity matrix after antibody binding.
RNase-free technique
RNase contamination on lab equipment can be widespread in a lab not currently conducting RNA work. Additionally, RNase is a very stable enzyme. Before starting, it is thus important to set up an RNase-free work zone. Use RNase decontaminating reagents such as RNase-Zap (Ambion) to decontaminate work surfaces and equipment that may be contaminated with RNase. Certified RNase-free plasticware and reagents, as well as aerosol-resistant tips, should be used whenever possible. Wear and change gloves often, as well as whenever common use lab equipment that may have RNase contamination is touched.
Material availability
Plasmids for use in making TRAP transgenic mice and published TRAP mouse lines are available from The Rockefeller University by request. Monoclonal GFP antibodies 19C8 and 19F7 are available for purchase from the Memorial Sloan-Kettering Monoclonal Antibody Facility. The TRAP transgene sequence as used in our studies is supplied below in the MATERIALS section.
MATERIALS
REAGENTS
Absolutely RNA Nanoprep kit (Agilent #400753)
Biotinylated Protein L, recombinant, purified (Fisher Scientific # PI-29997) ◆ CRITICAL preparation of the affinity matrix using biotinylated Protein L, GFP antibodies, and magnetic Dynabeads takes a few hours, and can be performed up to two weeks before the actual experiment (Box 1)
BSA, Bovine Serum Albumin, IgG and protease-free (Jackson ImmunoResearch #001-000-162)
Cycloheximide (Sigma # C7698) !CAUTION Very toxic, dangerous for the environment. Collect all waste for proper disposal.
DHPC, 1,2-diheptanoyl-sn-glycero-3-phosphocholine (Avanti Polar Lipids 850306P-200 mg) ◆ CRITICAL Be sure to order powdered form in the 200mg size or larger, since smaller sizes often come in non-resealable glass ampules. Store powder at −20°C and thaw to room temperature (24°C) before opening to reconstitute.
DTT, DL-Dithiothreitol (Sigma # D9779) !CAUTION Harmful irritant
Ethanol (Sigma # #E7023)
GFP antibodies (Memorial-Sloan Kettering Monoclonal Antibody Facility; clone names: Htz-GFP-19F7 and Htz-GFP-19C8, bioreactor supernatant purity) ◆ CRITICAL preparation of the affinity matrix using biotinylated Protein L, GFP antibodies, and magnetic Dynabeads takes a few hours, and can be performed up to two weeks before the actual experiment (Box 1) ◆ CRITICAL For TRAP purifications, these monoclonal antibodies are superior to commercially available ones, and thus should not be substituted. Different lots of antibodies will have different antibody concentrations, but even when normalized for concentration, different lots may have slightly different functional antibody concentration if lots have been freeze-thawed. We recommend ordering a large enough lot to cover most planned purifications, and to save the antibodies in single-experiment aliquots. If additional lots are needed, a side-by-side comparison for efficacy (based on TRAP yield) should be performed with old versus new lots.
Glucose (Sigma # G7528)
HBSS, Hank’s Balanced Salt Solution, 10X (Invitrogen # 14065-056)
HEPES, 1M, pH 7.3, RNase-free (Affymetrix #16924)
Magnesium chloride, MgCl2, 1M, RNase-free (Applied Biosystems #AM9530G)
Methanol (Sigma # 322415)
NP-40 (chemical name: Nonylphenyl Polyethylene Glycol), 10%, 10 × 5 ml ampules (AG Scientific # P1505)
Nuclease-free water (Applied Biosystems #AM9939)
Phosphate buffered saline, PBS, 10x, RNase-free (Applied Biosystems #AM9625)
Potassium chloride, KCl, 2 M, RNase-free (Applied Biosystems #AM9640G)
Protease inhibitor tablets, Mini-complete, EDTA-free (Roche #11836170001) ◆ CRITICAL It is essential that no EDTA be used in any of the solutions prepared in this protocol. EDTA will chelate magnesium, leading to dissociation of polysomes.
Quant-it RiboGreen Assay (Invitrogen # R11490)
RNase Zap Wipes (Applied Biosystems #AM9786)
RNasin, recombinant (Promega # N2515)
Sodium azide (Sigma # S2002) !CAUTION Very toxic and dangerous for the environment; contact with acids generates a very toxic gas. Collect and dispose of waste properly.
Sodium bicarbonate, NaHCO3 (Sigma # S6297)
Streptavidin MyOne T1 Dynabeads (Invitrogen # 65601) ◆ CRITICAL STEP preparation of the affinity matrix using biotinylated Protein L, GFP antibodies, and magnetic Dynabeads takes a few hours, and can be performed up to two weeks before the actual experiment (Box 1)
Sulfolane (Sigma # #T22209-100G) for use with the Absolutely RNA Nanoprep Kit (Agilent #400753)
Superasin (Applied Biosystems # AM2694)
The TRAP transgene as originally described17 is a fusion of the EGFP coding sequence (taken from GenBank U57606) to mouse ribosomal protein Rpl10a (GenBank BC083346). This TRAP transgene sequence was assembled as follows: EGFP coding sequence minus its stop codon, linker sequence coding for SGRTQISSSSFEF, followed by the Rpl10a coding sequence minus its first M-encoding codon.
TRAP-expressing cells can be obtained by transfecting or transducing transformed or primary cells with the TRAP transgene sequence under the control of a suitable promoter. Alternatively, TRAP expressing primary cells can be harvested from TRAP transgenic mice (see below) for culture in vitro.
TRAP transgenic mice (Available from The Rockefeller University by request) !CAUTION Only use mice after proper training by and registration with the Institution Animal Care and Use Committee (IACUC) at your institution
EQUIPMENT
2100 Electrophoresis Bioanalyzer with Nanochips and Picochips (Agilent #G2939AA, # 5067-1511, # 5067-1513)
Cell scrapers (Sarstedt #83.1832)
Homogenizers (Fisher Scientific #K8855100020)
Magnet (will depend on purification scale; samples in 1.5-ml tubes can be concentrated on a DynaMag-2, Invitrogen #123-21D)
Minicentrifuge (Fisher Scientific # 05-090-100, or equivalent)
Nanodrop 2000C Spectrophotometer (Thermo Scientific #ND-2000C)
Refrigerated centrifuge (Eppendorf #5430R, or equivalent with rotor for 1.5-ml microcentrifuge tubes)
RNase-free 1.5-ml microcentrifuge tubes (Applied Biosystems #AM12450)
RNase-free 50-ml conical tubes (Applied Biosystems #AM12501)
RNase-free 1000 μl filter tips (Rainin # RT-1000F)
RNase-free 200 μl filter tips (Rainin # RT-200F)
RNase-free 20 μl filter tips (Rainin # RT-20F)
Rotor for homogenizers (Yamato #LT-400D, or equivalent)
Tube rotator, Labquake brand (Thermo Fisher #13-687-12Q, or equivalent)
REAGENT SETUP
Cell-lysis buffer
Mix 20 mM HEPES KOH (pH 7.3), 150 mM KCl, 10 mM MgCl2, and 1% NP-40 in RNase-free water. Store at 4 °C for up to several months. To an aliquot, immediately before use, add EDTA-free protease inhibitors (one mini tablet per 10 ml), 0.5 mM DTT, 100 μg/ml cycloheximide, and 10 μl/ml rRNasin and Superasin. ◆ CRITICAL Brain ribosomes (monosomes) and polyribosomes (polysomes) require relatively high levels of MgCl2 (10–12 mM) for optimal purification33.
Cycloheximide
In a clean plastic weigh boat, weigh out and resuspend 100 mg cycloheximide in 1ml methanol. Store at 4 °C for up to one day.
DHPC
Reconstitute in RNase-free water to 300 mM. Warm powder to room temperature before reconstitution. Powder needs to sit at room temperature with occasional vortexing for ~30 minutes to fully go into solution. Once reconstituted in water use up to 7 days later, stored at 4°C in its glass packaging. Do not store in plastic.
Dissection buffer
Mix 1x HBSS, 2.5 mM HEPES-KOH (pH 7.3), 35 mM glucose, and 4 mM NaHCO3 in RNase-free water. Store at 4 °C for several months. To an aliquot, immediately before use, add 100 μg/ml cycloheximide.
DTT
Reconstitute DTT to 1M in water, filter sterilize, and store at −20 °C in single-use aliquots.
High-salt buffer
Mix 20 mM HEPES KOH (pH 7.3), 350 mM KCl, 10 mM MgCl2, and 1% NP-40 in RNase-free water. Store at 4 °C for up to several months. To an aliquot, immediately before use, add 0.5 mM DTT and 100 μg/ml cycloheximide.
Low-salt buffer
See recipe for cell-lysis buffer above; however, there is no need to add RNase inhibitors to the low-salt buffer if it is not used for cell lysis. Thus to an aliquot, immediately before use, add 0.5 mM DTT and 100 μg/ml cycloheximide.
Protein L
Reconstitute as recommended by the manufacturer. Once reconstituted, extra Protein L should be aliquoted in single-use aliquots and stored at −80 °C.
Tissue-lysis buffer
Mix 20 mM HEPES KOH (pH 7.4), 150 mM KCl, and 10 mM MgCl2, in RNase-free water. Store at 4 °C for several months. To an aliquot, immediately before use, add EDTA-free protease inhibitors, 0.5 mM DTT, 100 μg/ml cycloheximide, and 10 μl/ml rRNasin and Superasin.
PROCEDURE
◆ CRITICAL It is important to prepare reagents, including the affinity matrix (Box 1), before starting, in order to process cells or tissue as quickly as possible.
-
1|
Lysate Preparation ● TIMING 30–60 min for either option
Lysates can be prepared from either cultured cells (Option A) or from tissue (Option B). The differences in procedure are driven mainly by the fact that detergent can be used to lyse cells in culture, but should be avoided in tissue lysis (as we believe that the presence of detergent can cause myelin or fat to aggregate with polysomes).
CRITICAL STEP Always maintain samples at 4°C.
Option A Cell Culture Lysate Preparation
To cell culture media add 100μg/ml of cycloheximide, and return plates to incubator for 15 min.
Wash the cells 3 times on ice with 10 mls 1X PBS containing 100 μg/ml cycloheximide, aspirating as much PBS as possible after each wash.
Add 1 ml ice-cold cell lysis buffer to each 10-cm plate.
Incubate on ice for 10 minutes.
Scrape cells and cell debris from plate surface with cell scrapers and transfer to a pre-chilled homogenizer
Homogenize in cold room, or at room temperature by placing the homogenizer in a 50-ml conical tube with ice. Homogenize samples in a motor-driven Teflon-glass homogenizer, at 900 r.p.m., with 12 full strokes. First insert the Teflon pestle into the glass tube until the solution submerges the entire Teflon pestle, start to stir at 300 r.p.m., and then raise speed slowly to 900 r.p.m. Lower the glass tube but do not let the Teflon pestle rise to air-solution interface, because it will produce significant aeration, which may lead to protein denaturation. Alternatively, cultured cell lysis can be performed by vigorous pipetting up and down ten times with a P1000 pipette tip (without the creation of bubbles), although best lysis is obtained using a homogenizer.
-
Transfer lysate to a chilled microcentrifuge tube and centrifuge at 2,000 x g for 10 minutes at 4 °C. Lysate volume should be similar to that added to tissue culture dish.
?TROUBLESHOOTING
-
Transfer the supernatants (S2) to a new, chilled microcentrifuge tube and add to each sample 1/9 volume (e.g. 111 μl to 1000 μl) of 300 mM DHPC (DHPC reconstituted in water).
?TROUBLESHOOTING
Mix by quick hand inversion and incubate on ice for 5 minutes.
Centrifuge at 20,000 x g for 10 minutes at 4°C.
Take the resulting supernatant, S20, to new, chilled tubes and proceed to immunopurification (IP, step 2). Lysates can be stored on ice for several hours while additional samples are collected, before proceeding to Step 2. A small aliquot of S20 (e.g. 1% total volume) can be saved at this point to compare to enrichment of transcripts in purified material by downstream assays.
Option B Tissue Lysate Preparation
-
Remove brain tissue from mice
◆ CRITICAL STEP Be sure that all procedures for handling mice are registered with your Institutional Animal Care and Use Committee (IACUC).
-
Perform rapid hand dissection of desired tissue region and place tissue into ice-cold dissection buffer quickly to wash.
■ PAUSE POINT Dissected tissue can be stored for several years at −80°C if immediately flash frozen in liquid nitrogen upon dissection. To freeze tissue, immediately upon dissection: wash tissue in dissection buffer for 2 seconds; transfer to a clean, empty microcentrifuge tube; cap tube and fully immerse tube immediately into liquid nitrogen; transfer frozen tubes to storage location at −80°C, without allowing tubes to warm above −80°C. When ready to use tissue, without allowing the tissue to thaw, use cold forceps to transfer frozen tissue piece(s) quickly into a cold homogenizer containing pre-chilled tissue lysis buffer and IMMEDIATELY homogenize, such that tissue thaws as it is being homogenized. !CAUTION Handle liquid nitrogen with proper cryogenic protective equipment and only use tubes that are resistant to liquid nitrogen.
Transfer tissue to a pre-chilled homogenizer on ice that contains tissue-lysis buffer. Homogenize approximately 25–50 mg tissue per 1ml of tissue-lysis buffer. The homogenizer listed in the Equipment section is ideal for 1ml volumes, but similar models of Teflon-glass homogenizers are available for smaller or larger volumes (scale according to a 25–50 mg:1ml of tissue-lysis buffer ratio). ◆ CRITICAL STEP If more than one dissection is being performed, homogenize each sample as it is dissected.
-
Homogenize in cold room, or at room temperature by placing the homogenizer in a 50-ml conical tube with ice. Homogenize samples in a motor-driven Teflon-glass homogenizer, at 900 r.p.m., with 12 full strokes. First insert the Teflon pestle into the glass tube until the solution submerges the entire Teflon pestle, start to stir at 300 r.p.m., and then raise speed slowly to 900 r.p.m. Lower the glass tube but do not let the Teflon pestle rise to air-solution interface, because it will produce significant aeration, which may lead to protein denaturation.
?TROUBLESHOOTING
Transfer the lysate into a pre-chilled microcentrifuge tube on ice.
Prepare a post-nuclear supernatant (S2) by centrifugation at 4°C, 10 minutes, 2,000 x g
Transfer S2 to a new, pre-chilled microcentrifuge tube on ice
Add 1/9 sample volume of 10% NP-40 to S2 (final concentration = 1%); mix gently by hand inversion of tube. Pulse centrifuge the sample in a minifuge to collect the liquid at the bottom of the tube.
-
Add 1/9 sample volume of 300 mM DHPC (final concentration = 30 mM); mix gently by hand inversion and incubate on ice for 5 minutes.
?TROUBLESHOOTING
Prepare post-mitochondrial supernatant (S20) by centrifugation at 4°C, 10 minutes, 20,000 x g
Take S20 to a new, pre-chilled microcentrifuge tube and proceed immediately to immunopurification (IP, step 2). Lysates can be stored on ice for several hours while additional samples are collected, before proceeding to Step 2. A small aliquot of S20 (e.g. 1% total volume) can be saved at this point to compare to enrichment of transcripts in purified material by downstream assays. If so, we recommend incubating this S20 aliquot at 4 °C for the same length of time as the TRAP immunopurification (16–18 h) before freezing the aliquot at −80 °C, to ensure all samples are equally incubated.
Immunopurification (IP) ● TIMING 18–20 h
-
2|
Thoroughly resuspend the pre-prepared affinity matrix ((Box 1). If pre-prepared affinity matrix was stored in sodium azide, it should be washed three times quickly in low-salt buffer before use.
-
3|
Add 200 μl of freshly resuspended beads to each S20 sample (~1000 μl)
CRITICAL STEP Always resuspend the affinity matrix thoroughly by gentle pipetting immediately before use.
?TROUBLESHOOTING
-
4|
Incubate at 4 °C for 16–18 hours with gentle end-over-end mixing in a tube rotator. This incubation time is longer than described in our original method17, and reflects recent optimizations to the purification scheme that serve to increase RNA yield18–21 (10-fold for rare cell types).
-
5|
After incubation, collect the beads with a magnet (well-chilled in an ice bucket). Use a minifuge to spin down beads from caps in between each wash. The whole or an aliquot of the unbound fraction can be saved at −80 °C at this point to compare to enrichment of transcripts in purified material by downstream assays.
-
6|
Resuspend beads in 1000 μl of high-salt buffer and collect with magnet as above. ◆ CRITICAL STEP All washes should be performed by careful pipetting that avoids the introduction of bubbles. After beads are visibly resuspended, beads should be mixed by pipetting at least four more times. High background can result from insufficient bead resuspension during washes.
-
7|
Repeat wash 3 times (1000 μl high-salt buffer each time, total 4 washes).
-
8|
After the fourth wash with high-salt buffer, remove all remaining wash buffer, remove tubes from magnet, and warm tubes to room temperature. Resuspend the beads in 100 μl Nanoprep Lysis Buffer with β-ME (use Lysis Buffer from the Stratagene Absolutely RNA Nanoprep kit or equivalent), vortex, incubate for 10 minutes at room temperature, remove the RNA (now in Nanoprep Lysis Buffer) from the beads with the magnet, and proceed immediately to RNA clean-up, following kit manufacturer’s instructions. ■ PAUSE POINT Eluted RNA (in Lysis Buffer and removed from beads) can be kept frozen in Lysis Buffer with β-ME at −80 °C before column clean-up. Warm to room temperature upon thawing, before resuming purification with kit.
◆ CRITICAL STEP Other buffers from alternate RNA purification kits may be used, but the buffer used to release bound RNA from affinity matrix must contain the denaturant guanidine thiocyanate.
◆ CRITICAL STEP Guanidine thiocyanate can form crystals at low temperatures. Be sure to extract and clean up RNA at room temperature to avoid crystallization and carry-through to downstream applications.
RNA clean-up and quantitation ● TIMING 4–6 h
-
9|
Follow the Agilent Nanoprep kit, or similar kit, manufacturer’s instructions for RNA clean-up (including the optional DNase digestion and the optional 2 RNA elutions with elution buffer heated to 60 °C, all optional steps per kit instructions). RNA purified in this fashion is of high enough purity for use in most downstream applications, and our studies have indicated that further acidic phenol purification of these samples does not improve the purity of the samples (as judged by spectrophotometer readings and use in downstream amplification reactions, e.g. reverse transcription).
◆CRITICAL STEP After column purification at room temperature, return samples to ice and keep on ice at all times; purified RNA should be kept at −80°C for long-term storage periods.
◆CRITICAL STEP Perform RNase-free DNase digestion at this step if it is required for downstream applications.
■PAUSE POINT After column purification, RNA can be stored at −80°C for several years.
-
10|
To assay the integrity of the RNA, analyze 1 μl of each sample on a Bioanalyzer 2100 using an RNA Pico/Nano chip (follow Agilent’s protocol for running chips). PicoChip assay qualitative range: 200–5000 pg/μl; NanoChip assay qualitative range: 5–500 ng/μl.
?TROUBLESHOOTING
-
11|
Quantify samples on a small-volume spectrophotometer (e.g. Nanodrop, ThermoScientific). For precise quantification of low-concentration samples (<10 ng/μl), run a RiboGreen (or similar) fluorescence-based assay. Use of a spectrophotometer to quantitate samples that have been column-purified and are of a concentration of less than ~10 ng/μl is not recommended, as debris from the columns leads to inaccurate readings (silica shed from the column scatters light).
■ PAUSE POINT After quantitation and check of integrity, RNA can be kept at −80°C for several years.
● TIMING
Box 1, Preparation of the affinity matrix: 2–2.5 h
Step 1, Lysate Preparation: 30–60 min
Steps 2–8, Immunopurification 18–20 h
Steps 9–11, RNA clean-up and quantitation, 4- 6 h
?TROUBLESHOOTING
Troubleshooting advice can be found in Table 1.
Table 1.
Troubleshooting table
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| Box 1, step 3 | Unequal amount of beads appear in equivalent aliquots | Beads not fully resuspended before being aliquoted | Mix beads well by swirling bottle and and manual pipetting. Ensure that no bead clumps exist in bottle or are seen on pipette tip during aliquoting step. |
| Step 1, option B, iv | Tissue is difficult to homogenize | Too much tissue is used for volume of lysis buffer chosen | Use a Teflon-glass homogenizer and volume of lysis buffer that keeps a ratio of about 25–50 mg tissue: 1 ml tissue lysis buffer. |
| Step 1, option A, vii | Lysate volume is much larger than amount of cell lysis buffer used | Insufficient aspiration of wash solution before cell lysis | Ensure, with a vacuum line, that all wash buffer is removed from the dish surface before lysis buffer is added. |
| Step 1, option A, viii AND option B, ix | DHPC is difficult to resuspend | Insufficient time has passed for complete hydration | Reconstitute DHPC with water to 300mM; it will need to sit at room temperature with occasional vortexing for ~30 minutes to fully go into solution. Once reconstituted in water use it up to 7 days later, stored at 4°C in a glass bottle. Do not store in plastic. |
| 3 | Affinity matrix is difficult to resuspend | Affinity matrix has been stored at 4°C for an extended period of time | On a tube rotator, rotate tube containing matrix gently for several hours until beads are completely resuspended. |
| 10 | RNA is degraded | RNase contamination | Use only RNase-free reagents, use aerosol-resistant filter pipette tips, change gloves often, and keep samples on ice at all times. If working with tissue, homogenize tissue sample immediately after collection and wash tissue before homogenization to remove blood. If working with cultured cells, ensure no mycoplasma or anti-viral response is present: both of these conditions may elevate endogenous RNase levels dramatically in cultured cells. |
| 10 | RNA yields are low | Inefficient immunoaffinity purification caused by: RNase contamination; EDTA is present in buffers; inefficient cell/tissue lysis |
Ensure polysome integrity is maintained by adding cycloheximide, MgCl2, and RNase inhibitors to all solutions, and by keeping samples on ice at all times. Ensure that protease inhibitors used do not have EDTA. Ensure that a tight-fitting homogenizer similar to the one listed in the Equipment section is used. Use a volume of lysis buffer that keeps a ratio of about 25–50 mg tissue: 1 ml tissue lysis buffer. Save aliquots of and perform western blots on each step of the purification. Use a non-mouse GFP antibody for western blot analysis: efficiency of purifications can be checked by running western blots (against EGFP) with immunopurification input, unbound, and immunopurified samples. |
| 10 | RNA is not of high enough concentration for use in downstream applications | RNA is diluted during clean-up | Concentrate RNA samples using a vacuum concentrator, with no or low heat settings. |
ANTICIPATED RESULTS
With the stabilization of ribosomes on mRNA and inhibition of RNase activity, all mRNAs associated with EGFP-tagged ribosomes should be recovered. Absolute yields will vary depending upon the nature of the target material, as well as the strength of the regulatory elements that drive the expression of the TRAP transgene. However, for comparison, from 0.7×106 labeled spiny projection neurons in 30 mg of one mouse striatum, with the transgene driven by a moderate-strength driver (Drd1a) we often recover ~50ng of polysomal RNA21. Purified RNA integrity from cultured cells is in the 9.5–10 range, and 7.5–10 for RNA purified from brain tissue. These numbers reflect Agilent’s Bioanalyzer RIN scoring system; analysis of samples on agarose gels should reveal sharp, undegraded bands at approximately 5 and 2 kilobases (kb) corresponding to 28S and 18S rRNA, respectively. The TRAP co-purified mRNA can usually not be visualized directly by these methods, due to its small contribution to the total RNA amount. If obtained from the procedure outlined above, undegraded TRAP RNA that is immunopurified and column-purified is of high enough quality for immediate use in many downstream applications, including qPCR, microarray analysis, RNA sequencing, or Northern blotting, without the need for further purification.
Acknowledgments
We acknowledge members of the Greengard, Heintz, and R. Darnell labs for helpful discussions, advice, and feedback. This work was supported by grants from The Picower Foundation, The Simons Foundation, and NIMH award MH090963 to P.G.; The Simons Foundation, The Howard Hughes Medical Institute, and NIMH award MH090963 to N.H.
Footnotes
AUTHOR CONTRIBUTION STATEMENT
M.H., R. K., and R.F. assembled the step-by-step protocol; M.H., N.H., and P.G. wrote the paper.
COMPETING FINANCIAL INTERESTS
The authors have no competing financial interests.
References
- 1.St John PA, Kell WM, Mazzetta JS, Lange GD, Barker JL. Analysis and isolation of embryonic mammalian neurons by fluorescence-activated cell sorting. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1986;6:1492–1512. doi: 10.1523/JNEUROSCI.06-05-01492.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang S, Roy NS, Benraiss A, Goldman SA. Promoter-based isolation and fluorescence-activated sorting of mitotic neuronal progenitor cells from the adult mammalian ependymal/subependymal zone. Developmental neuroscience. 2000;22:167–176. doi: 10.1159/000017437. 17437. [DOI] [PubMed] [Google Scholar]
- 3.Tomomura M, Rice DS, Morgan JI, Yuzaki M. Purification of Purkinje cells by fluorescence-activated cell sorting from transgenic mice that express green fluorescent protein. The European journal of neuroscience. 2001;14:57–63. doi: 10.1046/j.0953-816x.2001.01624.x. [DOI] [PubMed] [Google Scholar]
- 4.Arlotta P, et al. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45:207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 5.Lobo MK, Karsten SL, Gray M, Geschwind DH, Yang XW. FACS-array profiling of striatal projection neuron subtypes in juvenile and adult mouse brains. Nature neuroscience. 2006;9:443–452. doi: 10.1038/nn1654. [DOI] [PubMed] [Google Scholar]
- 6.Marsh ED, Minarcik J, Campbell K, Brooks-Kayal AR, Golden JA. FACS-array gene expression analysis during early development of mouse telencephalic interneurons. Developmental neurobiology. 2008;68:434–445. doi: 10.1002/dneu.20602. [DOI] [PubMed] [Google Scholar]
- 7.Cahoy JD, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sugino K, et al. Molecular taxonomy of major neuronal classes in the adult mouse forebrain. Nature neuroscience. 2006;9:99–107. doi: 10.1038/nn1618. [DOI] [PubMed] [Google Scholar]
- 9.Hempel CM, Sugino K, Nelson SB. A manual method for the purification of fluorescently labeled neurons from the mammalian brain. Nature protocols. 2007;2:2924–2929. doi: 10.1038/nprot.2007.416. [DOI] [PubMed] [Google Scholar]
- 10.Okaty BW, Miller MN, Sugino K, Hempel CM, Nelson SB. Transcriptional and electrophysiological maturation of neocortical fast-spiking GABAergic interneurons. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2009;29:7040–7052. doi: 10.1523/JNEUROSCI.0105-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barres BA, Silverstein BE, Corey DP, Chun LL. Immunological, morphological, and electrophysiological variation among retinal ganglion cells purified by panning. Neuron. 1988;1:791–803. doi: 10.1016/0896-6273(88)90127-4. [DOI] [PubMed] [Google Scholar]
- 12.Luo L, et al. Gene expression profiles of laser-captured adjacent neuronal subtypes. Nature medicine. 1999;5:117–122. doi: 10.1038/4806. [DOI] [PubMed] [Google Scholar]
- 13.Yao F, et al. Microarray analysis of fluoro-gold labeled rat dopamine neurons harvested by laser capture microdissection. Journal of neuroscience methods. 2005;143:95–106. doi: 10.1016/j.jneumeth.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 14.Surmeier DJ, Song WJ, Yan Z. Coordinated expression of dopamine receptors in neostriatal medium spiny neurons. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1996;16:6579–6591. doi: 10.1523/JNEUROSCI.16-20-06579.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toledo-Rodriguez M, et al. Correlation maps allow neuronal electrical properties to be predicted from single-cell gene expression profiles in rat neocortex. Cereb Cortex. 2004;14:1310–1327. doi: 10.1093/cercor/bhh092. [DOI] [PubMed] [Google Scholar]
- 16.Doyle JP, et al. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell. 2008;135:749–762. doi: 10.1016/j.cell.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heiman M, et al. A translational profiling approach for the molecular characterization of NS cell types. Cell. 2008;135:738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mellen M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151:1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tryon RC, Pisat N, Johnson SL, Dougherty JD. Development of translating ribosome affinity purification for zebrafish. Genesis. 2013;51:187–192. doi: 10.1002/dvg.22363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stanley S, et al. Profiling of Glucose-Sensing Neurons Reveals that GHRH Neurons Are Activated by Hypoglycemia. Cell metabolism. 2013;18:596–607. doi: 10.1016/j.cmet.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 21.Heiman M, et al. Molecular Adaptations of Striatal Spiny Projection Neurons During Levodopa-Induced Dyskinesia. Proceedings of the National Academy of Sciences of the United States of America. 2014 doi: 10.1073/pnas.1401819111. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hupe M, Li MX, Gertow Gillner K, Adams RH, Stenman JM. Evaluation of TRAP-sequencing technology with a versatile conditional mouse model. Nucleic acids research. 2013 doi: 10.1093/nar/gkt995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou P, et al. Interrogating translational efficiency and lineage-specific transcriptomes using ribosome affinity purification. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:15395–15400. doi: 10.1073/pnas.1304124110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heintz N. BAC to the future: the use of bac transgenic mice for neuroscience research. Nature reviews. Neuroscience. 2001;2:861–870. doi: 10.1038/35104049. [DOI] [PubMed] [Google Scholar]
- 25.Shizuya H, et al. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:8794–8797. doi: 10.1073/pnas.89.18.8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shizuya H, Kouros-Mehr H. The development and applications of the bacterial artificial chromosome cloning system. The Keio journal of medicine. 2001;50:26–30. doi: 10.2302/kjm.50.26. [DOI] [PubMed] [Google Scholar]
- 27.Gong S, Kus L, Heintz N. Rapid bacterial artificial chromosome modification for large-scale mouse transgenesis. Nature protocols. 2010;5:1678–1696. doi: 10.1038/nprot.2010.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gong S, et al. Targeting Cre recombinase to specific neuron populations with bacterial artificial chromosome constructs. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2007;27:9817–9823. doi: 10.1523/JNEUROSCI.2707-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nature neuroscience. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomas A, et al. A versatile method for cell-specific profiling of translated mRNAs in Drosophila. PloS one. 2012;7:e40276. doi: 10.1371/journal.pone.0040276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mustroph A, Juntawong P, Bailey-Serres J. Isolation of plant polysomal mRNA by differential centrifugation and ribosome immunopurification methods. Methods Mol Biol. 2009;553:109–126. doi: 10.1007/978-1-60327-563-7_6. [DOI] [PubMed] [Google Scholar]
- 32.Watson FL, et al. Cell type-specific translational profiling in the Xenopus laevis retina. Developmental dynamics : an official publication of the American Association of Anatomists. 2012;241:1960–1972. doi: 10.1002/dvdy.23880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zomzely CE, Roberts S, Gruber CP, Brown DM. Cerebral protein synthesis. II Instability of cerebral messenger ribonucleic acid-ribosome complexes. The Journal of biological chemistry. 1968;243:5396–5409. [PubMed] [Google Scholar]
- 34.Neuwelt EA, Boguski MS, Frank JJ, Procter-Appich K, Levy CC. Possible sites of origin of human plasma ribonucleases as evidenced by isolation and partial characterization of ribonucleases from several human tissues. Cancer research. 1978;38:88–93. [PubMed] [Google Scholar]
- 35.Gauthier D, Ven Murthy MR. Efficacy of RNase inhibitors during brain polysome isolation. Neurochemical research. 1987;12:335–339. doi: 10.1007/BF00993241. [DOI] [PubMed] [Google Scholar]
- 36.Jung R, Lubcke C, Wagener C, Neumaier M. Reversal of RT-PCR inhibition observed in heparinized clinical specimens. BioTechniques. 1997:23, 24, 26, 28. doi: 10.2144/97231bm03. [DOI] [PubMed] [Google Scholar]
- 37.McQuillen K, Roberts RB, Britten RJ. Synthesis of Nascent Protein by Ribosomes in Escherichia Coli. Proceedings of the National Academy of Sciences of the United States of America. 1959;45:1437–1447. doi: 10.1073/pnas.45.9.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kilkenny C, Browne WJ, Cuthill IC, Emerson M, Altman DG. Improving bioscience research reporting: The ARRIVE guidelines for reporting animal research. Journal of pharmacology & pharmacotherapeutics. 2010;1:94–99. doi: 10.4103/0976-500X.72351. [DOI] [PMC free article] [PubMed] [Google Scholar]
Key references
- Heiman M, et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135:738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellen M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151:1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiman M, et al. Molecular Adaptations of Striatal Spiny Projection Neurons During Levodopa-Induced Dyskinesia. Proceedings of the National Academy of Sciences of the United States of America. 2014 doi: 10.1073/pnas.1401819111. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]