Abstract
Imaging and evaluation of the optic nerve head (ONH) plays an essential part in the detection and clinical management of glaucoma. The morphological characteristics of ONHs vary greatly from person to person and this variability means it is difficult to quantify them in a standardized way. We developed and evaluated a feature extraction approach using shift-invariant wavelet packet and kernel principal component analysis to quantify the shape features in ONH images acquired by scanning laser ophthalmoscopy (Heidelberg Retina Tomograph [HRT]). The methods were developed and tested on 1996 eyes from three different clinical centers. A shape abnormality score (SAS) was developed from extracted features using a Gaussian process to identify glaucomatous abnormality. SAS can be used as a diagnostic index to quantify the overall likelihood of ONH abnormality. Maps showing areas of likely abnormality within the ONH were also derived. Diagnostic performance of the technique, as estimated by ROC analysis, was significantly better than the classification tools currently used in the HRT software – the technique offers the additional advantage of working with all images and is fully automated.
OCIS codes: (100.2960) Image analysis; (100.4993) Pattern recognition, Baysian processors; (100.7410) Wavelets; (150.1835) Defect understanding; (170.4470) Ophthalmology; (170.5755) Retina scanning; (170.4580) Optical diagnostics for medicine
1. Introduction
Glaucoma is a chronic, slowly progressive optic neuropathy that can lead to irreversible sight loss. Some amalgamation of biomechanical and schematic factors acting on optic nerve head (ONH) tissue cause retinal ganglion cell (RGC) dysfunction in glaucoma [1, 2]. Classification of abnormality in the ONH is therefore central to the clinical diagnosis and monitoring of glaucoma. To date, characterization of ONH has, for example, been limited to simple measures of size of the ONH or the volume of neuroretinal rim area comprising the RGC axons [3]. Yet, ONH geometry is more complicated than this. It is thought that application of geometric morphometric methods, or some analysis of shape features, would facilitate better assessment of the ONH and likely improve classification and detection of abnormality [4].
The confocal scanning laser ophthalmoscope provides reproducible three-dimensional images of the ONH and surrounding area. It is an established retinal imaging technology, described in detail elsewhere [5], and is now widely used in eye clinics. The technology is typified by the commercially available Heidelberg Retina Tomograph (HRT; Heidelberg Engineering, Inc. Heidelberg, Germany) which produces both reflectance and topographic (ONH surface shape) images of the ONH and the surrounding areas (Fig. 1) [6]. For clinical quantification, the information contained in the image is often reduced to a single ONH parameter such as the size of the neuroretinal rim area or depth of ‘cupping’ in relation to the size of the ONH. These ‘size features’ are amenable to clinical understanding and simple quantification [7–10]. However, this approach is wasteful since, for example, the topographic image includes approximately 65000 pixels, each one representing a surrogate measure of the topographic height of the ONH or retinal area at the corresponding location. Moreover, current methods do not attempt to characterize any distinguishing spatial or morphometric features that might be suggestive of abnormality or otherwise. The main idea described in this paper is to take advantage of these features in the images in order to better detect glaucomatous damage in the ONH.
Fig. 1.

An example of HRT topographic (a) and reflectance (b) images. The ONH is conventionally divided into six sectors indicated on the reflectance image.
The most widely used diagnostic tool on the HRT is the Moorfields Regression Analysis (MRA) [6]. By comparing the measured neuroretinal rim area to normative limits, globally and in six separate sectors (Fig. 1), the MRA classifies an ONH as “within normal limits”, “borderline”, or “outside normal limits”. This method corrects for the well-established relationship between ONH size and rim area: larger ONHs tend to have larger rim areas [11–13]. Simple size features of the ONH are also used by other methods in the software adopting linear discriminant functions and other machine-learning classifiers [14–16]. In order to quantify the size of ONH and rim area, the MRA requires manual delineation (contouring) of the ONH by connecting points on ONH margin that are subjectively selected by the operator/clinician. This process inevitably introduces measurement variability, which in turn adversely affects the diagnostic performance of MRA [17].
A three-dimensional model of the topographical image has been incorporated into another diagnostic tool available in the HRT software [18] yielding a metric called the glaucoma probability score (GPS). The GPS uses two measures of peripapillary retinal nerve fiber layer shape (horizontal and vertical curvature) and three measures of optic nerve head shape (cup size, cup depth and rim steepness) for input into a relevance vector machine classifier that estimates the probability of having ONH damage consistent with glaucoma. GPS has the advantage over the MRA of not requiring manual contouring of ONH margin. GPS demonstrates similar diagnostic performance to MRA, but a number of ONHs cannot be classified by GPS when the topographical surface cannot be approximated by the modeled surface [19]. In short, the model cannot be used in some ONHs meaning that a percentage of eyes cannot be classified. Moreover, in healthy eyes, large ONHs are more often classified with abnormality than are medium-sized or small ONHs, whilst in eyes with glaucoma, both MRA and GPS are less sensitive in small ONHs than in large ones [15, 19–22]. In the Nottingham regression analysis [23], the effect of disc size was neutralized, allowing the algorithm to perform equally over the normal range of disc areas. A statistical correction to the MRA, using quantile regression [24] has recently been described to improve the specificity of the diagnostic precision in large ONHs but neither of these improvements has yet to be adopted in HRT software.
Whilst much work has been done to characterize size-related parameters of the ONH, little attempt has been made to identify characteristics about the shape of an ONH including, for example, ONHs that are tilted or crowded by vessel structure. Moreover, although a subjectively selected feature offers transparent methods conforming to clinical understanding, they may not reflect the most intrinsic information about ONHs due to the limited knowledge about them. Therefore, this study proposed an automated feature extraction method to objectively quantify the shape characteristics of ONHs. The method was developed and tested on images from 1996 eyes. Estimates of diagnostic performance of the technique were compared with MRA and GPS.
2. Data sets
The study used data sets of HRT images acquired from three different clinical centers: Royal Eye Hospital in Manchester, UK [25], QEII Eye Care Centre of Queen Elizabeth II Health Sciences Centre in Halifax, Canada [15] and Queen's Medical Centre, Nottingham, UK [26, 27]. The Manchester and Halifax data sets were used for developing feature extraction and diagnostic methods, whilst the Nottingham data set was used for independent validation of these methods.
The Manchester and Halifax data set contains images of one eye from each of 164 visually healthy people and 252 patients with a clinical diagnosis of glaucoma. These data are described in detail elsewhere [24, 25]. In addition 692 eyes from people attending clinics at the same centers (Manchester and Halifax) were included in this data set for developing our methods. These eyes either had an equivocal clinical diagnosis of glaucoma or were from people attending clinics as glaucoma suspects. The Nottingham data set includes eyes from 806 elderly visually healthy people who had been assessed as part of the Bridlington Eye Assessment Project (BEAP (2002-5)), a large cross-sectional community-based study on eye disease in the elderly in the UK [26, 27]. This data set was artificially enriched with a sample of 82 eyes from 82 patients with a clinical diagnosis of glaucoma; the ratio of cases to controls in this validation data set was kept at approximately ten to one in order to at least mimic the situation of using the HRT in a case finding or primary care situation. In all cases the patients were given a diagnosis of primary open angle glaucoma by a glaucoma specialist made from an established reference test (visual fields) [28] and other clinical indicators – the diagnosis was not based on ONH abnormally alone. But it should be noted that different visual field instruments and criteria were used in the three centers. The HRT image quality in all data sets was quantified by mean pixel height standard deviation (MPHSD), a standard measure of image quality used by HRT devices [29]. All images were required to have MPHSD <50µm in order to be included in the study.
The patient data were collected in accordance with the tenets of the Declaration of Helsinki from studies that had research ethics committee approval with all participants giving informed consent. Data were anonymized and transferred to a secure database held at City University London.
3. ONH shape feature extraction
Both topographic and reflectance images (Fig. 1) of each eye were first rescaled so that the pixel size of the images is 10 × 10 microns. A region of 256-by-256 pixels around the image center was used for analysis. Smaller images were padded by replicating pixels on edges.
Features of ONHs were extracted in two steps. First, topography and reflectance images were individually processed by a shift-invariant wavelet packet to extract the spatial-frequency features. Second, the wavelet features were then adaptively selected by the kernel principal component analysis (KPCA) to further reduce the dimensionality of extracted features.
3.1 Adaptive shift-invariant wavelet packet feature extraction
A wavelet transform expresses a general function as a series of wavelets. In particular, it expresses a signal as a linear combination of a particular set of functions by shifting and dilating one single function named the mother wavelet. This methodology is widely used in applications such as image de-noise, image compression, feature extraction and classification [30–35]. The decomposition of the signal leads to a set of wavelet coefficients that are spatial-frequency localized. The wavelet transform for discrete signal and digital image processing can be efficiently implemented with the Mallat algorithm [36]. In a discrete wavelet transform (DWT) for a 2D image, the image is convolved with complementary low- and high-pass filters corresponding to the mother wavelet; the convolution with the low-pass filter forms the approximation coefficients, and the convolution with the high-pass filters leads to detail coefficients on the horizontal, vertical and diagonal. The approximation and detail coefficients are then down-sampled to be half size, and will be referred to here as approximation and detail images. The approximation image can be successively decomposed to higher levels with lower frequency components at larger scales. Note that the detail images are not further decomposed in conventional DWT.
The down-sampling in DWT significantly reduces the number of coefficients and forms optimally sparse and efficient representation of images. However, it also causes DWT to be shift variant: a small shift of the image greatly perturbs the wavelet coefficients [37]. It has been reported that shift variance complicates wavelet analysis and algorithms have to cope with a wide range of possible wavelet coefficient patterns caused by image shift [38, 39]. Therefore, our method adopted an adaptive shift-invariant wavelet packet decomposition [35] that generates shift-invariance and non-redundant wavelet coefficients.
The wavelet packet is a generalization of DWT in which not only the approximation image, but also the detail images from the lower level of DWT are decomposed. This forms a quad-tree of decomposition shown in Fig. 2 with the original image at the root. To achieve the shift-invariance, a redundant set of wavelet packet coefficients for shifted images is computed. These coefficients are redundant because they include the same information as those in the non-shifted image. The redundancy is then eliminated by averaging the corresponding coefficients of shifted images to form a shift-invariant wavelet packet representation [35]. As an example, in Fig. 2, when decomposing the approximation image AA, it is shifted by the combination of zero or one pixel on the horizontal and vertical. The wavelet coefficients of shifted AA are averaged to form the approximation AAA, and details AAH, AAV, AAD.
Fig. 2.
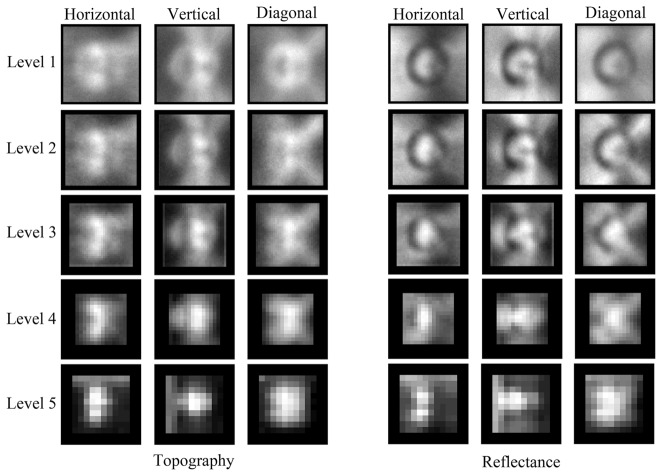
Illustration of adaptive shift-invariant wavelet packet. The value of Shannon entropy is indicated on the branches of each wavelet decomposition with the format of ‘lower level entropy: summation of higher level entropy’. The crosses indicate that the decomposition is prevented due to the increased information cost.
The wavelet packet, compared with the wavelet transform, still produces significantly more coefficients due to the decomposition of detail images. The information cost was used to adaptively select the coefficients that should be further decomposed [35, 40]. In particular, Shannon entropy (zero order entropy of the frequency of pixel intensity) was computed for an image to be decomposed and is compared with the sum of Shannon entropy of the four decomposition images. If the information cost of the current image is less than the sum of information cost of decomposition images, it will not be further decomposed; otherwise, it will be decomposed until the information cost increases or a maximum level (five in this study) is reached. In Fig. 2, the information cost of each image and the sum of information cost in the decomposition images are given; the abandoned decomposition (due to increased information cost) is indicated by crosses. Two uniform decomposition trees are then selected for topography and reflectance images respectively according to the criteria that a branch of the quad-tree is kept only if the information cost is decreased by the corresponding decomposition in more than 50% of the images in the training data set.
In each uniform quad-tree, the coefficients in the remaining decomposition were further selected. Note that no coefficient on the border with the width of mother wavelet was selected because these coefficients could not be reliably calculated by convolution. All approximation coefficients at the highest level (leaf approximation) in the quad-tree were selected. The selection of detail coefficients was carried out in two steps. First, an adaptive histogram-based thresholding [41] was used to select the coefficients with the largest magnitudes in each detail image. Second, the number of topography, or reflectance images, that select each detail coefficient in the first step was counted, and the feature coefficients chosen were those ranked to be in the top 3% of the most selected coefficients in the quad-tree. Note that the coefficient selection was carried out independently for topography and reflectance images.
Various mother wavelets including daubechies (db1-db6), symlets (sym2-sym6) and biorthogonal (bio1.1, bio1.3, bio1.5, bio2.2, bio2.4, bio3.1, bio3.3) were considered [42, 43]; those that performed best in the diagnostic model (described below) in 10-fold cross validation on the training data set were used in the final model. The validation data set (Nottingham) was not used at all in this process.
3.2 Kernel principal component analysis dimensionality reduction
There are far fewer wavelet coefficients than the number of pixels in the 256 × 256 image, but there is still redundancy that can be eliminated by examining the spatial covariance among wavelet coefficients. Kernel principal component analysis (KPCA) [44] was used to project wavelet coefficients to a non-linear low-dimensional hyper-surface. Handling non-linearity has made KPCA a useful tool for image understanding [45, 46].
The selected wavelet coefficients of the topography and reflectance images were input into a KPCA model with a non-normalized isotropic Gaussian kernel. KPCA was carried out by using Eigen-decomposition of the kernel matrix [44]. The principal components with the largest variance in the feature space were selected and a feature vector for an image was formed by projection onto the selected principal components. The features from topography and reflectance images were then combined by a principal component analysis to form the final features for the ONH. Only the images in the training data set were used to establish the principal components in KPCA. The parameters in the kernel and the number of principal components were selected such that they gave the best diagnostic performance in 10-fold cross validation on the training data set.
4. Validation and experiments
In order to demonstrate that the extracted features reflect the intrinsic information about the ONH, they were used to construct a diagnostic model that quantifies the uncertainty of glaucomatous abnormality. In turn this was used to infer the local configuration of abnormality in the ONHs.
4.1 Shape abnormality score (SAS)
A Gaussian process (GP) [47, 48] model was used to classify each ONH into healthy and glaucomatous categories. This diagnostic model, like GPS, outputs an overall probability score that we term the shape abnormality score (SAS). This score takes a value between 0 and 1, where 0 means definitely healthy and 1 indicates definitely glaucomatous.
Given a M-dimensional feature vector of a ONH and its corresponding diagnosis label y with if is unhealthy and if is healthy, the likelihood of y was modeled by a cumulative Gaussian response function :
| (1) |
where is a latent function with a GP prior. The response function in (1) compresses the value from with arbitrary range to be between 0 and 1. Let denote the M-dimensional feature vectors of N ONHs in the training data set, consists of latent functions of corresponding and contains labels of corresponding . Similarly, is a feature vector of a test ONH with latent function . A GP over defines a joint Gaussian distribution so the prior and posterior are also normal distributions. The covariance matrix of the GP has elements being non-normalized Gaussian kernel with automatic relevance determination parameters.
In order to make a prediction for , the first step is to calculate the distribution of latent variable given the training data:
| (2) |
and then evaluate the probability of label for
| (3) |
The optimization for kernel parameters was made by maximizing the marginal likelihood of training data:
| (4) |
Because in (1) and in (2) are not known distributions in an exponential family, the integrations in (2)-(4) are not analytically tractable. Therefore, expectation propagation [49, 50] was used to approximate with a normal distribution by a standard model reduction technique (moment matching); this enables analytical integration in (2)-(4). The GP algorithm started with estimating kernel parameters by maximizing the approximated log marginal likelihood (4) using 100 iterations of the scaled conjugate gradient algorithm [51, 52]. With optimized covariance function parameters, the SAS score for a test ONH with feature was given by (3) under an expectation propagation approximation.
The diagnostic model was developed on the Manchester and Halifax data sets and then independently tested on the Nottingham data set. The statistical sensitivity and false positive rate were evaluated and the diagnostic performance was assessed by the receiver operating characteristic (ROC) analysis. Ten thousand bootstrap iterations of the validation data were carried out to estimate the 95% confidence interval (CI) of the area under ROC (AuROC) curve. Moreover, because methods with high false positive rate (low specificity) are not clinically useful in diagnostic tests for glaucoma, a partial AuROC curve with false positive rate ≤15% was also evaluated. The partial AuROC curve was normalized by dividing it by 0.15.
4.2 Localization of abnormality
SAS provides an overall index about the ‘integrity’ of the ONH but glaucomatous damage often happens at discrete locations in the ONH area [10]. Therefore, estimating the spatial feature of ONH abnormality is also essential for clinical diagnosis of glaucomatous loss. For example, MRA and GPS analyses identify glaucomatous abnormality in six sectors (Fig. 1) of the ONH, which may not be adequate resolution to describe the ‘local’ configuration of abnormality. We therefore derive a pixel level localization of abnormality from the SAS score.
Ideally, localization of abnormality in the ONH area can be achieved by comparing the ONH with its ‘healthy status’. A reference healthy ONH is however seldom available because patients will typically present with abnormality, making the direct comparison impossible. We propose a method of estimating a reference healthy ONH in a ‘backwards fashion’ by searching for an ONH that scores a SAS of 0 by continuously deforming the features of the measured ONH. The deformation is achieved by iteratively minimizing approximated (3) with respect to feature . The minimization starts from the features of the actual measured ONH and uses a gradient descent to search for a new feature that reduces SAS in each iteration. The iteration stops when SAS is close to 0 (below in implementation). The output feature corresponds to the reference healthy ONH.
The features of both measured and reference healthy ONHs were then converted back to the original image space. This is achieved by reversing the feature extraction procedures. In particular, reverse PCA first converts the feature vector of ONH back to feature vectors for topography and reflectance images. A pre-image technique of KPCA [53] is then used to convert the feature vector of topography image to wavelet packet coefficients, which are then transformed to the topography image by the inverse wavelet packet. The difference between the reconstructed topography images of the measured and reference healthy ONH can then be calculated and presented as local abnormality map on pixel level.
The analytical methods described were implemented in MATLAB R2013a (MathWorks Inc., Natick, MA). An executable version of this code is freely available from the authors. All GPS and MRA analyses were conducted on the images using the HRT‐III software and results were transferred to a database in order to make statistical comparisons with the SAS results.
5. Results
5.1 ONH feature extraction
The sym4 mother wavelet, when applied on both topography and reflectance images, led to the best diagnostic performance in cross validation on the training data set. The following results for feature extraction are therefore all based on the results from the sym4 wavelet.
None of the approximation coefficients needed to be further decomposed in the wavelet packet because the decomposition increased the information cost in over 50% of the training images. Therefore, the shift-invariant wavelet packet produced the same number of coefficients as that of ordinary wavelet decomposition. The decomposition was carried out to a maximum of five levels for topography and reflectance images. The number of topography or reflectance images that select each detail coefficient is demonstrated in Fig. 3. The 3% (1615) most commonly selected coefficients from the topography and reflectance images were chosen to be processed by KPCA.
Fig. 3.
Illustrating the number of images that select each detail coefficient in the topography and reflectance images. Brighter pixels indicate more important coefficients selected by a larger number of images. The contrast is normalized in each image for visualization purpose.
The KPCA was carried out using different scale parameters in Gaussian kernels and various numbers of most significant topography and reflectance features were used to search for the optimal feature space that provides the best diagnostic performance on the training data set. Twenty-six topography features and three reflectance features from the KPCA were selected. These features account for 83% and 32% variance in topography and reflectance wavelet features respectively. The PCA that combines these topography and reflectance features produced twenty-two features that accounted for 95% variance of twenty-nine topography and reflectance features.
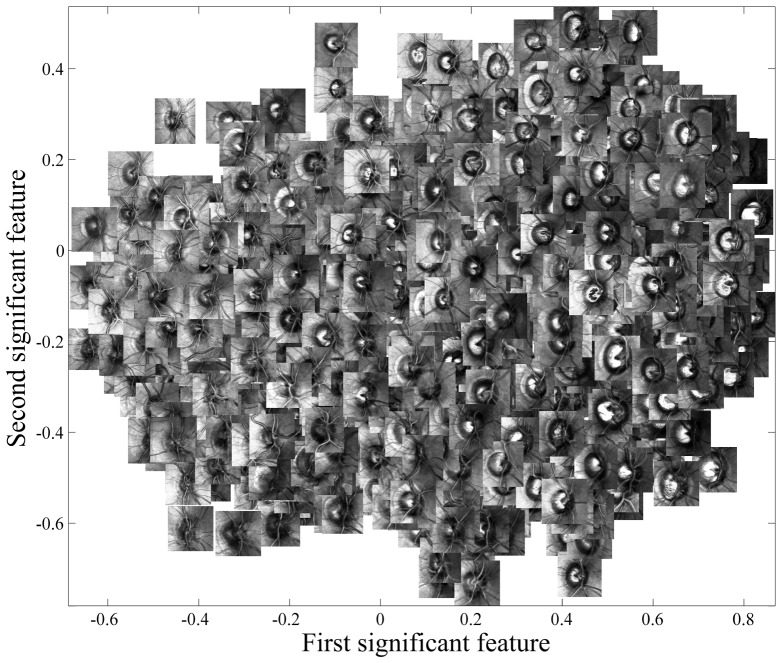
As an illustration, the two most significant features of ONHs are presented in a plot in Fig. 4 with corresponding ONH images superimposed. These features, although objectively extracted, indicate important morphological characteristics of the ONHs. For instance, the ONHs on the left side of this plot tend to be smaller and more crowded by vessel structure, than those on the right. Note that many images with appearance of peripapillary atrophy [54] are grouped in the top-right of the feature space. Of course these features were extracted automatically without the knowledge of diagnosis.
Fig. 4.
The scatter plot of the first two most significant features of the ONH.
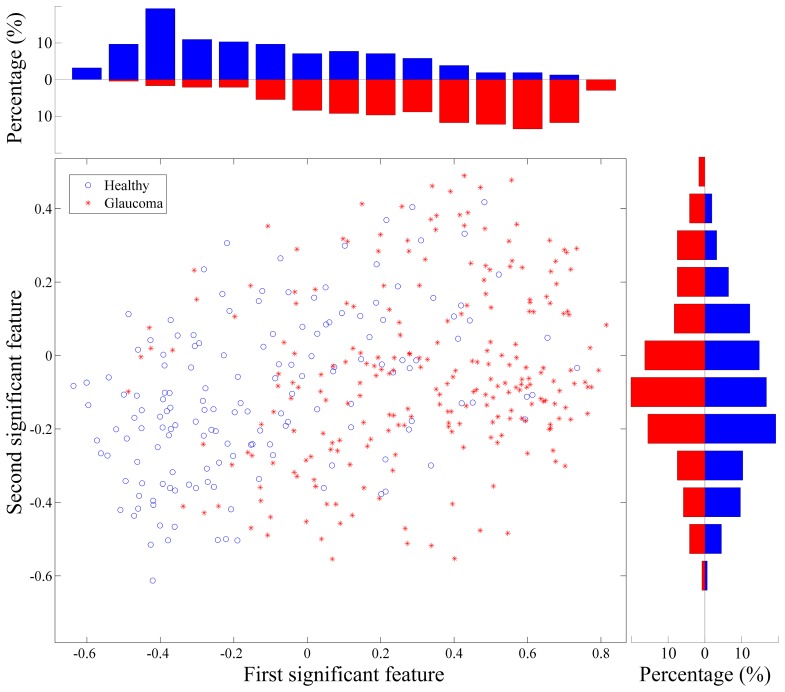
Figure 5 shows the same scatter plot but only with the images of eyes having a clinical diagnosis. Note that the clinical diagnosis is only for visualization purpose and is not used in the feature extraction. It can be observed that in each feature, the glaucoma and healthy images distribute differently but still have overlapping. The two individual features, once combined in a two-dimensional space show better evidence of separation between ONHs from eyes with a clinical diagnosis of glaucoma (red symbols towards top-right) and healthy eyes (blue symbols towards bottom-left). The classification model acts intuitively in the same manner: it ‘aggregates’ the separation in individual features and carries out classification in multivariate feature space.
Fig. 5.
The scatter plot of the first and second most significant features from those images with a diagnosis in the training data set. The axes from KPCA and PCA are in arbitrary units. The histogram of glaucoma (red) and healthy (blue) data in each feature are shown on the top and right of the scatter plot. Note that the diagnosis information was unknown during feature extraction and it is only displayed here for visualization and demonstrative purpose.
5.2 Diagnostic performance
The diagnostic performance of SAS was compared with that of MRA and GPS classification made directly from HRT software on an independent data set (Nottingham) not used in the model development. ROC curves were calculated for the three classifiers in Fig. 6.
Fig. 6.

ROC curve for SAS, MRA and GPS analysis. The unity line corresponds to random classification.
The MRA gives an overall global classification of the ONH as ‘within normal limits’, ‘outside normal limits’ or on ‘borderline’ and does not output a continuous score about abnormality. Therefore, the ROC curve of MRA cannot be produced as a continuous line. Instead, two points on the ROC curve were calculated when the ONHs on borderline were classified as glaucoma or visually healthy respectively and were plotted in Fig. 6. MRA is more sensitive but less specific when the ONHs on borderline are classified as abnormal.
It is important to note that the inbuilt GPS was not able to reliably process 97 out of 888 test ONHs (~11%) due to algorithm failure and could not produce classification in at least one sector. The total AuROC, normalized partial AuROC and their 95% CI of SAS and GPS are given in Table 1. The AuROC of MRA are not calculated as the sensitivity and specificity could not be continuously estimated for MRA. The median (95% CI) difference of AuROC curve between SAS and GPS is 0.10 (0.05, 0.16). The bootstrap technique for estimating the differences assumes that the data is related (different classifiers are applied to the same data). It therefore indicates a statistically significant difference in the AuROC curves. The median (95% CI) difference of the partial AuROC between SAS and GPS was 0.16 (0.09, 0.20). Moreover, at a specificity of 85% (the highest specificity for MRA) the sensitivities (95% CI) of MRA, GPS and SAS are shown in Table 1. The sensitivity of SAS is significantly better than those of MRA and GPS.
Table 1. Statistics (median [95% CI]) of ROC analysis.
| Total AuROC | Normalized partial AuROC | Sensitivities at specificity of 85% | |
|---|---|---|---|
| SAS | 0.89 (0.83, 0.93) | 0.59 (0.50, 0.68) | 77% (73%, 83%) |
| GPS | 0.80 (0.75, 0.87) | 0.42 (0.32, 0.51) | 61% (55%, 66%) |
| MRA | − | − | 65% (61%, 69%) |
It is worth noting that MRA and GPS are based on the topography of the ONH only. Conversely, SAS uses features from both topography and reflectance images of ONHs. Although only three reflectance features were selected in SAS, they provide an improved diagnostic performance. This is shown in the ROC curve in Fig. 7. The reflectance features, are relatively poor at identifying glaucomatous abnormality on their own but are able to ‘boost’ the diagnostic performance with combined features especially in the region with low false positive rate.
Fig. 7.

ROC curve of SAS derived from the topography, reflectance and combined features.
4.7 Localization of abnormality
With the proposed technique, local abnormality in the ONH was represented by the difference between the reference healthy topography and the measured topography; it is presented as a heat map superimposed on the reflectance image. ONHs from ten eyes from the validation data set, all with a clinical diagnosis of glaucoma, are shown in Fig. 8; these images are demonstrated with the local abnormality map showing the sites of likely change in the ONH. Examples (i) and (ii) show typical damage on the superior and inferior regions of relatively large and small ONHs respectively. Example (iii) shows an ONH with advanced and diffused defect and (iv) shows an example with local loss. It is noteworthy that in examples (v) to (x), MRA indicates neuroretinal rim area not to be outside normal limits for the overall or any of the sectorial classification. Examples (v) and (vi) show moderate or at least small distortions highlighted by red clusters despite low overall SAS values. Examples (vii) to (x) show small, crowded or tilted ONHs that are also misdiagnosed by MRA analysis (false negatives).
Fig. 8.
Examples of localized abnormality maps for ten eyes with a clinical diagnosis of glaucoma. The topography, reflectance and abnormality map are displayed for each ONH. SAS scores are given in the lower left corner of each topography image. Difference between the measured topography and reference healthy topography is represented by a heat map. The transparency of the abnormality heat map corresponds to the amount of change and a scale is shown on the margin of the abnormality map.
6. Discussion
We have presented a new method for detecting abnormality in optic nerve head images acquired using scanning laser tomography (HRT). The technique uses a feature extraction analysis based on application of a shift-invariant wavelet packet and KPCA. In contrast to the methods used on HRT software the technique derives objective morphological characteristics of ONHs that can be used to derive a probability of abnormality (SAS). This analysis can be performed without the manual contouring of ONH margin as required by the MRA and should lead to more repeatable and reproducible classification, although we have not tested that here. Our technique is advantageous because it means an ONH can be interpreted automatically without any user or expert processing. It has been widely reported that the contouring process leads to high levels of intra-observer variability [17, 55]. This is also an advantage of the GPS that is available in the commercial software. However, the shape model that underpins the calculation of GPS often fails to fit the measured data - this results in an algorithm failure and the software cannot produce a classification decision. In our validation experiment a significant number of eye (11%) could not be processed by the GPS; this figure is consistent with the literature [19].
Our results provide some statistical evidence that SAS has better overall diagnostic precision than MRA and GPS when we considered differences in AuROC curves. Also SAS appeared to offer better estimates of sensitivity to detect a glaucomatous ONH at higher levels of specificity. This is encouraging from a clinical point of view because any detection tool for glaucoma must maintain reasonable sensitivity at high specificity because of the prevalence of the disease. Moreover, SAS requires no manual input and the classification algorithm never fails to produce a result - these are clear practical advantages for our method compared to those techniques currently used by the HRT software. Of course, glaucoma diagnosis and management does not rely on assessment of the ONH alone: information on risk factors, intraocular pressure and visual fields are also used. The HRT cannot be used in isolation to diagnose glaucoma. Our method has simply been shown to be statistically better than the techniques currently used by the commercially available software on the HRT. Moreover, whether the improvement by the proposed method translates to clinically significant gains in the diagnostic precision of classifying abnormal ONHs must be the subject of a further study that must follow appropriate standards [56]. For example, in the current study we defined glaucomatous ONHs to be from eyes with a clinical diagnosis of glaucoma that may have varied across the three centers.
Our method took advantage of building a model from a large number of eyes from different clinical centers. It is well established that if modeling and testing is done on the same data then model estimates of effects will be overly optimistic. Our validation experiment used a data set that was completely independent of that used to develop the model. This data was used to compare relative diagnostic performance between our new method and the diagnostic tools available on the HRT software. An even larger test data set could be used to further validate the differences we have seen in our experiment.
MRA and GPS classify neuroretinal rim area at six pre-defined sectors. Our technique can yield an abnormality map that provides pixel level quantification of distortion or abnormality of the ONH. The procedures involved in the feature extraction analysis are invertible, meaning that an ONH in the feature space can be converted back to the original image space by a reverse wavelet packet and pre-image technique. The clinical relevance of examining the ONH at this level of resolution with these images will need to be tested but we speculate that a clinician might usefully interpret these localized abnormalities in conjunction with other images or changes in the visual field. Moreover, this method can potentially be used as a tool for quantifying change in time series measurements of ONHs. Currently the so-called topographic change analysis (TCA) [57, 58] is used for this purpose in the HRT software. TCA compares the most recently acquired images with a set of baseline images, with change flagged at super-pixels. Our method could be adapted similarly and as good baseline images are acquired then these could be used instead of the reference healthy ONH that our feature space method realizes.
There are some technical limitations to our method because of limitations in the images themselves. For instance, the convolution involved in the wavelet packet cannot be reliably estimated on the border of the image. With the image size (256-by-256 pixel), the maximum level of wavelet packet is set as five levels because a wider mother wavelet in the higher level of wavelet packet makes an unreliable border in convolution too large with regard to the image size.
In summary we have proposed an analytical method for extracting topographic features that can be used for classifying ONH abnormality. The method proposed is wholly adaptable to other types of imaging of the ONH and is not restricted to those images captured by HRT. For instance, the method of ‘reversing’ the damage to infer the ONH in its healthy status could be applied to the ONH topographic images acquired by the rapidly developing spectral-domain optical coherence tomography (SD-OCT), forming a classification and abnormality localization method for the SD-OCT ONH images. Moreover, recent anatomic findings with SD-OCT have challenged the basis and accuracy of current methods for neuroretinal rim evaluation [59, 60]. Identification of the Bruch’s membrane opening as a marker for the outer border of neuroretinal rim suggests a clinical step-change in how the ONH topography can be delineated using SD-OCT [61]. The algorithms for this improved image acquisition of the ONH using the SD-OCT are just becoming clinically available. Our analytical methods have the potential to be used with these reconstructions of the ONH and this awaits further study.
Acknowledgment
This study was funded by the UK Guide Dogs for the Blind Association. HZ was supported by a Research Fellow Award from the National Institute for Health Research, National Health Service, United Kingdom. The views expressed in this publication are those of the authors and not necessarily those of the National Health Service, the National Institute for Health Research or the Department of Health. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We thank Paul Artes (Dalhousie University, Halifax, Canada) and David Henson (University of Manchester) for providing data for this study. We also thank Neil O’Leary (Dalhousie University, Halifax, Canada) for help with preparing some of the data.
References and links
- 1.Jonas J. B., Budde W. M., Panda-Jonas S., “Ophthalmoscopic evaluation of the optic nerve head,” Surv. Ophthalmol. 43(4), 293–320 (1999). 10.1016/S0039-6257(98)00049-6 [DOI] [PubMed] [Google Scholar]
- 2.Burgoyne C. F., Downs J. C., Bellezza A. J., Suh J. K., Hart R. T., “The optic nerve head as a biomechanical structure: a new paradigm for understanding the role of IOP-related stress and strain in the pathophysiology of glaucomatous optic nerve head damage,” Prog. Retin. Eye Res. 24(1), 39–73 (2005). 10.1016/j.preteyeres.2004.06.001 [DOI] [PubMed] [Google Scholar]
- 3.Sigal I. A., Flanagan J. G., Tertinegg I., Ethier C. R., “3D morphometry of the human optic nerve head,” Exp. Eye Res. 90(1), 70–80 (2010). 10.1016/j.exer.2009.09.013 [DOI] [PubMed] [Google Scholar]
- 4.Sanfilippo P. G., Cardini A., Hewitt A. W., Crowston J. G., Mackey D. A., “Optic disc morphology--rethinking shape,” Prog. Retin. Eye Res. 28(4), 227–248 (2009). 10.1016/j.preteyeres.2009.05.004 [DOI] [PubMed] [Google Scholar]
- 5.Webb R. H., Hughes G. W., Delori F. C., “Confocal scanning laser ophthalmoscope,” Appl. Opt. 26(8), 1492–1499 (1987). 10.1364/AO.26.001492 [DOI] [PubMed] [Google Scholar]
- 6.Wollstein G., Garway-Heath D. F., Hitchings R. A., “Identification of early glaucoma cases with the scanning laser ophthalmoscope,” Ophthalmology 105(8), 1557–1563 (1998). 10.1016/S0161-6420(98)98047-2 [DOI] [PubMed] [Google Scholar]
- 7.Mardin C. Y., Jünemann A. G., “The diagnostic value of optic nerve imaging in early glaucoma,” Curr. Opin. Ophthalmol. 12(2), 100–104 (2001). 10.1097/00055735-200104000-00004 [DOI] [PubMed] [Google Scholar]
- 8.Kesen M. R., Spaeth G. L., Henderer J. D., Pereira M. L., Smith A. F., Steinmann W. C., “The Heidelberg Retina Tomograph vs clinical impression in the diagnosis of glaucoma,” Am. J. Ophthalmol. 133(5), 613–616 (2002). 10.1016/S0002-9394(02)01395-8 [DOI] [PubMed] [Google Scholar]
- 9.Kamal D. S., Viswanathan A. C., Garway-Heath D. F., Hitchings R. A., Poinoosawmy D., Bunce C., “Detection of optic disc change with the Heidelberg retina tomograph before confirmed visual field change in ocular hypertensives converting to early glaucoma,” Br. J. Ophthalmol. 83(3), 290–294 (1999). 10.1136/bjo.83.3.290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strouthidis N. G., Garway-Heath D. F., “New developments in Heidelberg retina tomograph for glaucoma,” Curr. Opin. Ophthalmol. 19(2), 141–148 (2008). 10.1097/ICU.0b013e3282f4450b [DOI] [PubMed] [Google Scholar]
- 11.Garway-Heath D. F., Hitchings R. A., “Quantitative evaluation of the optic nerve head in early glaucoma,” Br. J. Ophthalmol. 82(4), 352–361 (1998). 10.1136/bjo.82.4.352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caprioli J., Miller J. M., “Optic disc rim area is related to disc size in normal subjects,” Arch. Ophthalmol. 105(12), 1683–1685 (1987). 10.1001/archopht.1987.01060120081030 [DOI] [PubMed] [Google Scholar]
- 13.Jonas J. B., Gusek G. C., Naumann G. O., “Optic disc, cup and neuroretinal rim size, configuration and correlations in normal eyes,” Invest. Ophthalmol. Vis. Sci. 29(7), 1151–1158 (1988). [PubMed] [Google Scholar]
- 14.Mardin C. Y., Hothorn T., Peters A., Jünemann A. G., Nguyen N. X., Lausen B., “New Glaucoma Classification Method Based on Standard Heidelberg Retina Tomograph Parameters by Bagging Classification Trees,” J. Glaucoma 12(4), 340–346 (2003). 10.1097/00061198-200308000-00008 [DOI] [PubMed] [Google Scholar]
- 15.Ford B. A., Artes P. H., McCormick T. A., Nicolela M. T., LeBlanc R. P., Chauhan B. C., “Comparison of data analysis tools for detection of glaucoma with the heidelberg retina tomograph,” Ophthalmology 110(6), 1145–1150 (2003). 10.1016/S0161-6420(03)00230-6 [DOI] [PubMed] [Google Scholar]
- 16.Zangwill L. M., Chan K., Bowd C., Hao J., Lee T.-W., Weinreb R. N., Sejnowski T. J., Goldbaum M. H., “Heidelberg Retina Tomograph Measurements of the Optic Disc and Parapapillary Retina for Detecting Glaucoma Analyzed by Machine Learning Classifiers,” Invest. Ophthalmol. Vis. Sci. 45(9), 3144–3151 (2004). 10.1167/iovs.04-0202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iester M., Mariotti V., Lanza F., Calabria G., “The effect of contour line position on optic nerve head analysis by Heidelberg Retina Tomograph,” Eur. J. Ophthalmol. 19(6), 942–948 (2009). [DOI] [PubMed] [Google Scholar]
- 18.Swindale N. V., Stjepanovic G., Chin A., Mikelberg F. S., “Automated Analysis of Normal and Glaucomatous Optic Nerve Head Topography Images,” Invest. Ophthalmol. Vis. Sci. 41(7), 1730–1742 (2000). [PubMed] [Google Scholar]
- 19.Coops A., Henson D. B., Kwartz A. J., Artes P. H., “Automated Analysis of Heidelberg Retina Tomograph Optic Disc Images by Glaucoma Probability Score,” Invest. Ophthalmol. Vis. Sci. 47(12), 5348–5355 (2006). 10.1167/iovs.06-0579 [DOI] [PubMed] [Google Scholar]
- 20.Hawker M. J., Vernon S. A., Ainsworth G., “Specificity of the Heidelberg Retina Tomograph’s diagnostic algorithms in a normal elderly population: the Bridlington Eye Assessment Project,” Ophthalmology 113(5), 778–785 (2006). 10.1016/j.ophtha.2005.10.068 [DOI] [PubMed] [Google Scholar]
- 21.Saito H., Tsutsumi T., Araie M., Tomidokoro A., Iwase A., “Sensitivity and specificity of the Heidelberg Retina Tomograph II Version 3.0 in a population-based study: the Tajimi Study,” Ophthalmology 116(10), 1854–1861 (2009). 10.1016/j.ophtha.2009.03.048 [DOI] [PubMed] [Google Scholar]
- 22.Iester M., Mikelberg F. S., Drance S. M., “The effect of optic disc size on diagnostic precision with the Heidelberg retina tomograph,” Ophthalmology 104(3), 545–548 (1997). 10.1016/S0161-6420(97)30277-2 [DOI] [PubMed] [Google Scholar]
- 23.Hawker M. J., Vernon S. A., Tattersall C. L., Dua H. S., “Linear regression modeling of rim area to discriminate between normal and glaucomatous optic nerve heads: the Bridlington Eye Assessment Project,” J. Glaucoma 16(4), 345–351 (2007). 10.1097/IJG.0b013e3180618d72 [DOI] [PubMed] [Google Scholar]
- 24.Artes P. H., Crabb D. P., “Estimating Normative Limits of Heidelberg Retina Tomograph Optic Disc Rim Area with Quantile Regression,” Invest. Ophthalmol. Vis. Sci. 51(1), 355–361 (2010). 10.1167/iovs.08-3354 [DOI] [PubMed] [Google Scholar]
- 25.Kwartz A. J., Henson D. B., Harper R. A., Spencer A. F., McLeod D., “The effectiveness of the Heidelberg Retina Tomograph and laser diagnostic glaucoma scanning system (GDx) in detecting and monitoring glaucoma,” Health Technol. Assess. 9(46), 1–132 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Vernon S. A., Hawker M. J., Ainsworth G., Hillman J. G., Macnab H. K., Dua H. S., “Laser Scanning Tomography of the Optic Nerve Head in a Normal Elderly Population: The Bridlington Eye Assessment Project,” Invest. Ophthalmol. Vis. Sci. 46(8), 2823–2828 (2005). 10.1167/iovs.05-0087 [DOI] [PubMed] [Google Scholar]
- 27.Hawker M. J., Vernon S. A., Tattersall C. L., Dua H. S., “Detecting glaucoma with RADAAR: the Bridlington Eye Assessment Project,” Br. J. Ophthalmol. 90(6), 744–748 (2006). 10.1136/bjo.2005.082115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henson D. B., Visual Fields, 2nd ed (Oxford University Press, 2000). [Google Scholar]
- 29.Strouthidis N. G., White E. T., Owen V. M., Ho T. A., Hammond C. J., Garway-Heath D. F., “Factors affecting the test-retest variability of Heidelberg retina tomograph and Heidelberg retina tomograph II measurements,” Br. J. Ophthalmol. 89(11), 1427–1432 (2005). 10.1136/bjo.2005.067298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vetterli M., “Wavelets, approximation, and compression,” IEEE Signal Process. Mag. 18, 59–73 (2010). [Google Scholar]
- 31.Starck J.-L., Murtagh F., “Astronomical image and signal processing: looking at noise, information and scale,” IEEE Signal Process. Mag. 18(2), 30–40 (2001). 10.1109/79.916319 [DOI] [Google Scholar]
- 32.Skodras A., Christopoulos C., Ebrahimi T., “The JPEG 2000 still image compression standard,” IEEE Signal Process. Mag. 18(5), 36–58 (2001). 10.1109/79.952804 [DOI] [Google Scholar]
- 33.Chang T., Kuo C. J., “Texture analysis and classification with tree-structured wavelet transform,” IEEE Trans. Image Process. 2(4), 429–441 (1993). 10.1109/83.242353 [DOI] [PubMed] [Google Scholar]
- 34.Laine A., Fan J., “Texture Classification by Wavelet Packet Signatures,” IEEE Trans. Pattern Anal. Mach. Intell. 15(11), 1186–1191 (1993). 10.1109/34.244679 [DOI] [Google Scholar]
- 35.Pun C.-M., Lee M.-C., “Extraction of Shift Invariant Wavelet Features for Classification of Images with Different Sizes,” IEEE Trans. Pattern Anal. Mach. Intell. 26(9), 1228–1233 (2004). 10.1109/TPAMI.2004.67 [DOI] [PubMed] [Google Scholar]
- 36.Mallat S. G., “A Theory for Multiresolution Signal Decomposition: The Wavelet Representation,” IEEE Trans. Pattern Anal. Mach. Intell. 11(7), 674–693 (1989). 10.1109/34.192463 [DOI] [Google Scholar]
- 37.Selesnick I. W., Baraniuk R. G., Kingsbury N. G., “The dual-tree complex wavelet transform,” IEEE Signal Process. Mag. 22(6), 123–151 (2005). 10.1109/MSP.2005.1550194 [DOI] [Google Scholar]
- 38.Dragotti P. L., Vetterli M., “Wavelet footprints: theory, algorithms, and applications,” IEEE Trans. Signal Process. 51(5), 1306–1323 (2003). 10.1109/TSP.2003.810296 [DOI] [Google Scholar]
- 39.Kingsbury N., “Image processing with complex wavelets,” Philos. Trans. R. Soc., A 357(1760), 2543–2560 (1999). 10.1098/rsta.1999.0447 [DOI] [Google Scholar]
- 40.Coifman R. R., Wickerhauser M. V., “Entropy-based algorithms for best basis selection,” IEEE Trans. Inf. Theory 38(2), 713–718 (1992). 10.1109/18.119732 [DOI] [Google Scholar]
- 41.Ridler T. W., Calvard S., “Picture Thresholding Using an Iterative Selection Method,” IEEE Trans. Syst. Man Cybern. 8(8), 630–632 (1978). 10.1109/TSMC.1978.4310039 [DOI] [Google Scholar]
- 42.Chui C. K., An introduction to wavelets (Academic Press Professional, Inc., 1992). [Google Scholar]
- 43.Daubechies I., Ten lectures on wavelets (Society for Industrial and Applied Mathematics, 1992). [Google Scholar]
- 44.Schölkopf B., Smola A., Müller K.-R., “Nonlinear Component Analysis as a Kernel Eigenvalue Problem,” Neural Comput. 10(5), 1299–1319 (1998). 10.1162/089976698300017467 [DOI] [Google Scholar]
- 45.Yang J., Frangi A. F., Yang J. Y., Zhang D., Jin Z., “KPCA plus LDA: a Complete Kernel Fisher Discriminant Framework for Feature Extraction and Recognition,” IEEE Trans. Pattern Anal. Mach. Intell. 27(2), 230–244 (2005). 10.1109/TPAMI.2005.33 [DOI] [PubMed] [Google Scholar]
- 46.Jung K., Kim K., Kim H. J., “Face recognition using kernel principal component analysis,” IEEE Sig. Proc. Lett. 9(2), 40–42 (2002). 10.1109/97.991133 [DOI] [Google Scholar]
- 47.Williams C. K. I., “Regression with Gaussian Processes,” in Mathematics of Neural Networks: Models, Algorithms and Applications, (Kluwer, 1995) [Google Scholar]
- 48.Williams C. K. I., Barber D., “Bayesian classification with Gaussian processes,” IEEE Trans. Pattern Anal. Machine Intelligence 20(12), 1342–1351 (1998). 10.1109/34.735807 [DOI] [Google Scholar]
- 49.Minka T. P., “A family of algorithms for approximate bayesian inference,” (Massachusetts Institute of Technology, 2001). [Google Scholar]
- 50.Rasmussen C. E., Williams C. K. I., Gaussian Processes for Machine Learning (MIT Press, 2006). [Google Scholar]
- 51.Møller M. F., “A scaled conjugate gradient algorithm for fast supervised learning,” Neural Netw. 6(4), 525–533 (1993). 10.1016/S0893-6080(05)80056-5 [DOI] [Google Scholar]
- 52.Hestenes M. R., Stiefel E., “Methods of conjugate gradients for solving linear systems,” J. Res. Natl. Bur. Stand. 49(6), 409–436 (1952). 10.6028/jres.049.044 [DOI] [Google Scholar]
- 53.Kwok J. T., Tsang I. W., “The pre-image problem in kernel methods,” IEEE Trans. Neural Netw. 15(6), 1517–1525 (2004). 10.1109/TNN.2004.837781 [DOI] [PubMed] [Google Scholar]
- 54.Park K. H., Park S. J., Lee Y. J., Kim J. Y., Caprioli J., “Ability of peripapillary atrophy parameters to differentiate normal-tension glaucoma from glaucomalike disk,” J. Glaucoma 10(2), 95–101 (2001). 10.1097/00061198-200104000-00005 [DOI] [PubMed] [Google Scholar]
- 55.Hawker M. J., Ainsworth G., Vernon S. A., Dua H. S., “Observer agreement using the Heidelberg retina tomograph: the Bridlington Eye Assessment Project,” J. Glaucoma 17(4), 280–286 (2008). 10.1097/IJG.0b013e31815c3a33 [DOI] [PubMed] [Google Scholar]
- 56.Bossuyt P. M., Reitsma J. B., Bruns D. E., Gatsonis C. A., Glasziou P. P., Irwig L. M., Moher D., Rennie D., de Vet H. C., Lijmer J. G., Standards for Reporting of Diagnostic Accuracy , “The STARD statement for reporting studies of diagnostic accuracy: explanation and elaboration,” Ann. Intern. Med. 138(1), 1–12 (2003). 10.7326/0003-4819-138-1-200301070-00010 [DOI] [PubMed] [Google Scholar]
- 57.Chauhan B. C., Blanchard J. W., Hamilton D. C., LeBlanc R. P., “Technique for detecting serial topographic changes in the optic disc and peripapillary retina using scanning laser tomography,” Invest. Ophthalmol. Vis. Sci. 41(3), 775–782 (2000). [PubMed] [Google Scholar]
- 58.Bowd C., Balasubramanian M., Weinreb R. N., Vizzeri G., Alencar L. M., O’Leary N., Sample P. A., Zangwill L. M., “Performance of confocal scanning laser tomograph Topographic Change Analysis (TCA) for assessing glaucomatous progression,” Invest. Ophthalmol. Vis. Sci. 50(2), 691–701 (2008). 10.1167/iovs.08-2136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Strouthidis N. G., Yang H., Reynaud J. F., Grimm J. L., Gardiner S. K., Fortune B., Burgoyne C. F., “Comparison of Clinical and Spectral Domain Optical Coherence Tomography Optic Disc Margin Anatomy,” Invest. Ophthalmol. Vis. Sci. 50(10), 4709–4718 (2009). 10.1167/iovs.09-3586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reis A. S., Sharpe G. P., Yang H., Nicolela M. T., Burgoyne C. F., Chauhan B. C., “Optic disc margin anatomy in patients with glaucoma and normal controls with spectral domain optical coherence tomography,” Ophthalmology 119(4), 738–747 (2012). 10.1016/j.ophtha.2011.09.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chauhan B. C., Burgoyne C. F., “From clinical examination of the optic disc to clinical assessment of the optic nerve head: a paradigm change,” Am. J. Ophthalmol. 156(2), 218–227 (2013). 10.1016/j.ajo.2013.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]