1. Introduction
Detection of chemical and biological agents plays a fundamental role in biomedical, forensic and environmental sciences1–4 as well as in anti bioterrorism applications.5–7 The development of highly sensitive, cost effective, miniature sensors is therefore in high demand which requires advanced technology coupled with fundamental knowledge in chemistry, biology and material sciences.8–13
In general, sensors feature two functional components: a recognition element to provide selective/specific binding with the target analytes and a transducer component for signaling the binding event. An efficient sensor relies heavily on these two essential components for the recognition process in terms of response time, signal to noise (S/N) ratio, selectivity and limits of detection (LOD).14,15 Therefore, designing sensors with higher efficacy depends on the development of novel materials to improve both the recognition and transduction processes. Nanomaterials feature unique physicochemical properties that can be of great utility in creating new recognition and transduction processes for chemical and biological sensors15–27 as well as improving the S/N ratio by miniaturization of the sensor elements.28
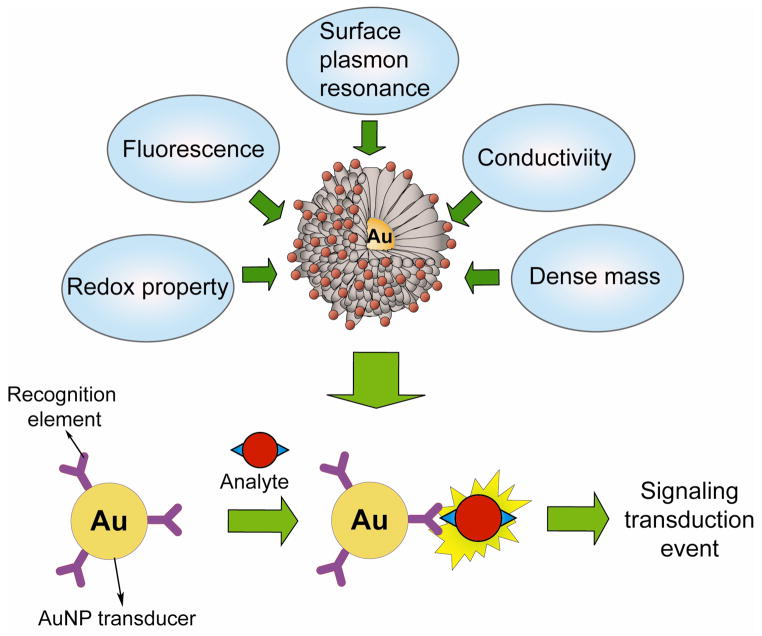
Gold nanoparticles (AuNPs) possess distinct physical and chemical attributes that make them excellent scaffolds for the fabrication of novel chemical and biological sensors (Figure 1).29–36 First, AuNPs can be synthesized in a straightforward manner and can be made highly stable. Second, they possess unique optoelectronic properties. Third, they provide high surface-to-volume ratio with excellent biocompatibility using appropriate ligands.30 Fourth, these properties of AuNPs can be readily tuned varying their size, shape and the surrounding chemical environment. For example, the binding event between recognition element and the analyte can alter physicochemical properties of transducer AuNPs, such as plasmon resonance absorption, conductivity, redox behavior, etc. that in turn can generate a detectable response signal. Finally, AuNPs offer a suitable platform for multi-functionalization with a wide range of organic or biological ligands for the selective binding and detection of small molecules and biological targets.30–32,36 Each of these attributes of AuNPs has allowed researchers to develop novel sensing strategies with improved sensitivity, stability and selectivity. In the last decade of research, the advent of AuNP as a sensory element provided us a broad spectrum of innovative approaches for the detection of metal ions, small molecules, proteins, nucleic acids, malignant cells, etc. in a rapid and efficient manner.37
Figure 1.
Physical properties of AuNPs and schematic illustration of an AuNP-based detection system.
In this current review, we have highlighted the several synthetic routes and properties of AuNPs that make them excellent probes for different sensing strategies. Furthermore, we will discuss various sensing strategies and major advances in the last two decades of research utilizing AuNPs in the detection of variety of target analytes including metal ions, organic molecules, proteins, nucleic acids, and microorganisms.
2. Synthesis and Surface Functionalization
Numerous preparative methods for gold nanoparticles have been reported, including both “top-down” (physical manipulation) and “bottom-up” (chemical transformation) approaches.30 During the last two decades, considerable effort has been devoted to synthesis of AuNPs, focusing on control over their size, shape, solubility, stability and functionality. It is worth noting that the term colloid and cluster are frequently used interchangeably; the former generally refers to particles having diameter more than 10 nm, while the latter commonly refers to smaller particles.
2.1. Citrate and Related Particle Preparation Methods
The scientific synthesis of colloidal gold can be traced back to Michael Faraday’s work in 1857, in which the gold hydrosols were prepared by reduction of an aqueous solution of chloroaurate with phosphorus dissolved in carbon disulfide.38 Later in 1951, Turkevich developed one of the most popular approaches for the synthesis of AuNPs, using citrate reduction of HAuCl4 in water.39 In this method, citric acid acts as both reducing and stabilizing agent and provides AuNPs in diameters of 20 nm. Further studies by Frens’ group enabled control of AuNPs size by varying the feed ratio of gold salt to sodium citrate.40 The kinetics of the Turkevich process was provided by Chow and Zukoski.41 Detailed studies and evolution of the Turkevich reaction have been reported and employed in numerous applications.42–48
2.2. The Brust-Schiffrin Method for Thiol-protected AuNPs
After Mulvaney’s initial attempt of stabilizing AuNPs with alkanethiols,49 a significant breakthrough in the field of AuNPs synthesis was achieved by Brust and Schiffrin in 1994. They reported a two-phase synthetic strategy, (the Brust-Schiffrin method), utilizing strong thiol-gold interactions to protect AuNPs with thiol ligands (Figure 2). In this method, AuCl4− is transferred from aqueous phase to toluene using the surfactant tetraoctylammonium bromide (TOAB) and reduced by sodium borohydride (NaBH4) with dodecanethiol.50 On addition of NaBH4, a quick color change from orange to deep brown takes place in organic phase. The AuNPs are generated in toluene with controlled diameters in the range 1.5 to 5 nm. These thiol-protected AuNPs feature superior stability due to strong thiol-gold interaction and they can be easily handled, characterized and functionalized. The nanoparticles can be thoroughly dried and then redispersed in organic solvents without any aggregation or decomposition. Various reaction conditions, such as gold/thiol ratio, temperature, and reduction rate, can be used to tune the particle size.51 Immediate quenching after reduction or use of sterically bulky ligands gives a higher portion of small core NPs (≤ 2 nm).52–56 With the translation of this synthesis into single-phase system, 57–59 many modifications have been reported to obtain functional thiols-stabilized AuNPs. Isolable, water-soluble gold clusters protected by monolayers of tiopronin have been generated with an average core size of 1.8 nm.60 Arenethiol ligands generate relatively larger and thermally less stable AuNPs than the alkanethiol protected clusters.53 All-aromatic monolayer-protected clusters with potential value for enhanced rates of electron transfer can be synthesized by differential extraction of the poly-anionic products using benzenethiolates.61 Alkylthiosulfates (Bunte salts) can be used as ligand precursors to synthesize thiol-stabilized AuNPs62 and water soluble AuNPs at a larger size as well.63,64 AuNPs of mixed monolayer65,66 and single-component monolayer67 of thymine moieties have been prepared for assembly studies via molecular recognition. A recent study on the identification and quantification of the precursor species in the Brust-Schiffrin method was also reported.68 Superhydride69, hexadecylaniline (inter alia)70, organometallic reagents (2-propylmagnesium bromide)71, 9-borabicyclo[3.3.1]nonane (9-BBN)72 and glutathione73 have been used as alternative reagents to NaBH4 for the reduction of Au (III) in synthesis of thiol-protected AuNPs.
Figure 2.
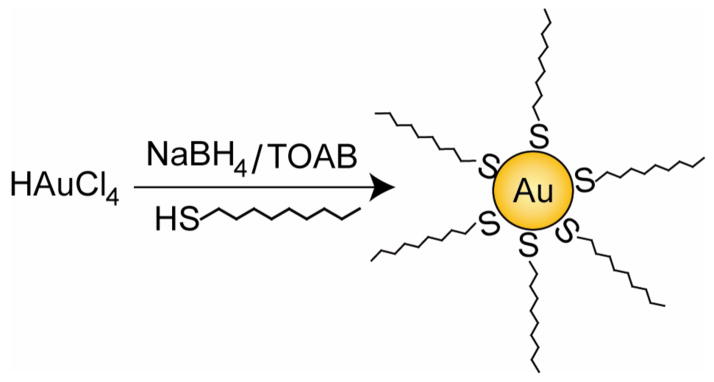
Brust-Schiffrin method for two-phase synthesis of AuNPs by reduction of gold salts in presence of external thiol ligands.
2.3. Place Exchange Method to form Mixed Monolayer AuNPs
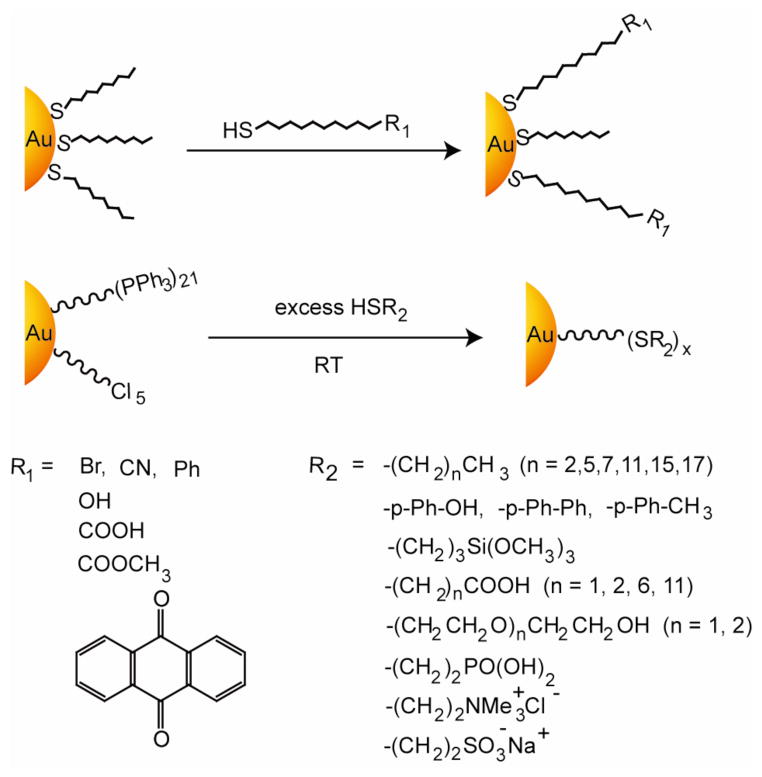
Place exchange, i.e. substitution of thiol ligands by different thiols was reported by Murray et al.54,74,75 This versatile technique introduces chemical functionality onto AuNPs monolayer in a divergent fashion. In this method, the initially anchored thiol ligands are exchanged in by free thiol ligands (Figure 3, top). The reaction time and the feed ratio of the functional ligands control the loading efficiency onto AuNPs surface. Moreover, introducing two or more functional ligands during the place exchange reaction can provide mixed monolayer protected AuNPs for synergistic applications. The ligands on the AuNPs surface also interact with each other, leading to a rigid monolayer76 and show a certain level of intra-monolayer mobility, providing optimization of the interaction with the analytes.77 Under appropriate conditions, ligands can also slowly hop between NPs.78 Place exchange is efficient for ultra-small 1.1 nm diameter phenylethanethiolate-stabilized AuNPs.79,80 Therefore, a range of functional groups can be employed in the synthesis of AuNPs by the use of functionalized thiols to place exchange with phosphine stabilized nanoparticles.(Figure 3, bottom).81,82
Figure 3.
General scheme for place exchange reaction for alkanethiol-protected AuNPs using functionalized thiols, giving examples of particles that can be rapidly generated.
2.4. Physical Methods for Particle Modification
Physical methods enable further manipulation of the structure and hence properties of AuNPs. Thermolysis,83,84 digestive ripening85,86 and conventional ripening87,88 have significantly reduced average particle size and polydispersity and triggered formation of superlattices in 2D and 3D. UV and laser irradiation are another parameter that can modify particle structure.89–94 Ultrasonic fields provide an approach to control the reduction rate of AuCl4− in aqueous solutions and therefore affect core sizes with the intensity of the ultrasound and the reactor position. 95–100 Control of the particle size can also be provided by radiolysis.101–103 Once formed, size-exclusion chromatography can separate suspended AuNPs by shape and size.104
2.5. Polymer Stabilized AuNPs
Polymer-stabilized AuNPs were first reported by Helcher in 1718, though of course the characterization was rather limited.105 Polymers commonly used for stabilization include poly(N-vinylpyrrolidone) (PVP),106–108 poly(ethylene glycol) (PEG),109–112 poly(4-vinylpyridine),113–115 poly(vinyl alcohol) (PVA),116 poly(vinyl methyl ether) (PVME),117 chitosan,118 polyethyleneimine (PEI),119 poly(diallyl dimethylammonium chloride) (PDDA),120 poly(methyl methacrylate) (PMMA),121,122 polystyrene-block polymers,123,124 poly(dG)-poly(dC),125 and poly(N-isopropylacrylamide) (PNIPam).126,127
There are four strategies widely used for creating polymer-functionalized nanoparticles. 1) The “grafting from” approach consists of polymer chain growth from small initiators attached to AuNPs.128–131 This technique generally provides a very dense polymer brush. Frequently used methods include living radical polymerization (LRP)121 and surface-initiated atom-transfer radical polymerization (SI-ATRP).113,132 2) “Grafting to” enables one-pot synthesis of AuNPs stabilized by sulfur-containing polymers,133–139 and generally produces a sparser coverage.134 3) Physisorption using block copolymer micelles (nanoreactors), water-soluble polymers, or star block copolymers.140–146 4) “Post-modification of pre-formed AuNPs”. In this method, AuNPs are generated in the first stage through conventional methods such as Brust-Schiffrin method, followed by the exchange or modification with polymers. 147,148
2.6. Other Capping Ligands
While most AuNP functionalization has been done using thiol/thiolated ligands, a variety of other ligands have been used to passivate and functionalize AuNPs. Other sulfur-containing ligands include disulfides,149–156 multivalent (and hence more stable) di-157 and trithiols,158,159 thioethers160,161 xanthates,162 and resorcinarene tetrathiols.163 Iodine can be used to oxidize and decompose these thiol-stabilized AuNPs.164 Amine-capped AuNPs were reported using primary amines.165 Self-assembled gold(I) amine precursors including [AuCl(NH2R)] (R=C8H17, C12H25 and C16H33) yield AuNPs upon decomposition through air exposure or in tetrahydrofuran (THF).166,167 Laurylamine (LAM) or octadecylamine (ODA) has been used to generate monodispersed particles.168 Oleyl amine (OLA),169 aromatic amines,170 amino acids,171–175 diamines,176 tetraoctylammonium177, porphyrins178 and hyperbranched polyethylenimine179 have been used as reducing/capping agents in synthesis of AuNPs, while a direct one-pot synthesis of amine-stabilized AuNPs using 3-(trimethoxysilylpropyl)diethylenetriamine have been reported.180 The controlled synthesis of AuNPs in quaternary ammonium ionic liquids by simple heating has been developed recently,181 and piperazine derivatives have been used as reducing/capping agents.182 Tri-n-octylphosphine oxide (TOPO), has been used in the presence of stabilizer octadecylamine.183,184 Studies using phosphine,185,186 carboxylate ligands,187–190 lactic acid,191 and hydroquinone192 as stabilizing ligands have also been documented.
3. Physical Properties
3.1. Size-dependent Electronic and Optoelectronic Properties
AuNPs possess quantum size effect that leads to discrete electron transition energy levels. For example, hexanethiol-functionalized AuNPs (Au147, d = 1.62 nm) display 15 redox states at room temperature,193 demonstrating that AuNPs can possess molecule-like redox properties.194 Moreover, the quantized capacitance charging behavior of AuNPs can be tuned by external ligands, magnetic fields and electrolyte ions, leading to potential applications in electronic devices and electrochemical labels.195,196
In addition to their redox activity, AuNPs feature a surface plasmon band (SPB).197 The SPB is a result from the collective oscillation of the conduction electrons due to the resonant excitation by the incoming photons. This SPB is absent in both small AuNPs (< 2 nm diameter) and bulk gold. The nature of this surface plasmon resonance (SPR) was elucidated by Mie in 1908.198 The SPR is influenced not only by size, but also by solvent, ligand, core charge, and temperature. Murray and co-workers has observed spectral shift induced by solvent refractive index changes that are consistent with Mie theory.199 The refractive index dependence on solvent and ligand can alter the optical thickness, which can be used for detecting impurities due to different refractive indexes of gold oxide and gold chloride.200 The core charge, as mentioned above, is influential in determining SPB energy, causing shifts to higher energy with excess electronic charge and to lower one with electron deficiency.199,201–203 The mechanism of the dominant electronic dephasing was proposed to be that only electron-electron interactions were involved rather than electron-photon coupling.204 However, experimental data from femtosecond light scattering confirmed the occurrence of both processes in the individual AuNPs.205 Furthermore, the SPR frequency is sensitive to the proximity of other nanoparticles. Therefore, the aggregation of nanoparticles results in significant red-shifting (from ~520 nm to ~650 nm) and broadening in the SPB, changing the solution color from red to blue due to the interparticle plasmon coupling.206,207 This phenomenon has made AuNPs an attractive candidate for colorimetric sensors (vide infra).
A rough estimation of the nanoparticle concentration can be made based on the number of gold atoms and the molar extinction coefficient of colloidal gold (~4000 M−1 cm−1/gold atom).208 The extinction coefficients of AuNPs with different sizes and stabilizing ligands have been obtained experimentally.209 For example, AuNPs with 20 nm diameter have a molar extinction coefficient of 1×109 M−1 cm−1, substantially higher than the organic dyes, indicating that AuNPs offer a superior light collecting efficiency.
3.2. Fluorescence Quenching
Early fluorescence studies on AuNPs focused on fluorescent ligands, such as pyrenyl,210 polyoctylthiophenyl,211 fluorenyl,212 and other probes.213–216 Photoluminescence was reported later for water-soluble AuNPs.217 AuNPs, however, also show enhancement in fluorescence at appropriate fluorophore-to-metal distances on solid substrates.218 It is believed that this phenomenon is due to the reflected far-field radiation from the fluorophore back onto itself.
Fluorescence quenching is a commonly observed consequence when fluorophores are appended onto AuNPs. The resonant energy transfer has led to applications in biophotonics219 and materials science.220–222 This quenching occurs when the emission spectrum overlaps with the gold surface plasmon band.223–225 Both radiative and non-radiative rates are particle dependant, with higher efficiencies of quenching occurring with small nanoparticles.226–229 A long-range molecular ruler, termed as nanosurface energy transfer (NSET), was demonstrated for breaking the “FRET barrier”.230 NSET is similar to FRET but is capable of measuring nearly 2-fold greater distances, making nanoparticles smaller than 2 nm of particular interest in bionanotechnology.231,232 Another process that quenches fluorophores is photoinduced electron transfer (PET) to nanoparticles that can be modulated by charging/discharging the gold core.233,234
4. Colorimetric Sensing
The aggregation of AuNPs of appropriate sizes (d > 3.5 nm) induces interparticle surface plasmon coupling, resulting in a visible color change from red to blue at nanomolar concentrations.207 The color change during AuNP aggregation (or redispersion of an aggregate) provides a practical platform for absorption-based colorimetric sensing of any target analyte that directly or indirectly triggers the AuNP aggregation or redispersion.16,235,236
4.1. Detection of Metal Ions
4.1.1. Alkali and Alkaline Earth Metal Ions
AuNP-based colorimetric sensing for metal ions generally requires the incorporation of chelating agents onto the nanoparticle surface. The presence of analyte ion induces the nanoparticle aggregation by forming multidentate interparticle complexes with the chelating ligand (Figure 4). For example, AuNPs carrying 15-crown-5 moieties have been fabricated for the colorimetric detection of potassium ions (K+) via formation of a 2:1 sandwich complex between 15-crown-5 moiety and K+.237 Most attractively, this sensor system showed micromolar recognition and colorimetric response towards K+ even in the presence of physiologically important cations, such as Li+, Cs+, NH4+, Mg2+, Ca2+, and excess Na+. Later the performance of this sensor system was improved by bifunctionalizing the AuNPs with thioctic acid and crown thiols.238 The increased rate of K+ recognition from this system has been attributed to a cooperative effect that allows crown moiety to be preorganized by the negatively charged carboxylate moiety of the thioctic acid for K+ recognition. Utilizing this principle the analogous detection of Na+ in urine has been achieved by incorporating 12-crown-4 onto the AuNP surface together with the thioctic acid.238 In a similar fashion Li+ has been detected by utilizing phenanthroline-functionalized 4 nm AuNPs,239 and lactose-functionalized 16 nm AuNPs have been used to detect Ca2+ 240 through metal ion-mediated carbohydrate-carbohydrate interactions.
Figure 4.
Schematic depiction of red-to-blue color colorimetric sensing of metal ions using chelating ligands.
4.1.2. Heavy Metal Ions
Heavy metal ions such as Pb2+, Cd2+, and Hg2+ pose significant public health hazards. Hupp et al. have reported a simple colorimetric technique for the sensing of aqueous heavy metal ions utilizing 11-mercaptoundecanoic acid (MUA)-functionalized 13 nm AuNPs.241 The color change (red to blue) is driven by heavy-metal ion chelation process where the surface carboxylates act as metal ion receptors. Colorimetric response was observed from Pb2+, Cd2+, and Hg2+ (≥400 μM), whereas Zn2+ displays no response to this assay process. The ion driven aggregation process was also detected using the enhanced hyper-Rayleigh scattering (HRS) response from the nanoparticle aggregates, leading to a more sensitive (25 μM) detection of Pb2+ ion. Chang and coworkers have significantly improved the selectivity and sensitivity of this system by fine tuning of the buffer composition and monolayer structure,242 achieving 100 nM sensitivity. A colorimetric sensor for Pb2+ was developed by Chen and coworkers that utilized mixed monolayer-protected AuNPs carrying both carboxylate and 15-crown-5 functionalities.243 In this system, initial aggregates of AuNPs were formed due to hydrogen bonding interactions between carboxylic acid residues in a methanol/water solvent system. Addition of Pb2+ disrupts the hydrogen-bonded assembly by associating with crown ether moiety and generating an electrostatic repulsion between the AuNPs, resulting in a blue-to-red color change. This system showed high sensitivity and selectivity over other metal ions including Cd2+ and Hg2+. Colorimetric detection of Cu2+ and Hg2+ has been achieved using AuNPs decorated with cysteine and peptide functionality.244,245 Quaternary ammonium-functionalized AuNPs have been utilized by Liu et al. to devise a simple colorimetric sensor for Hg2+ at room temperature, with the abstraction of AuNP stabilizing thiols by Hg2+ inducing aggregation.246 Mirkin and coworkers have employed DNA-functionalized AuNPs for the selective and sensitive detection of Hg2+.247 This thiolated-DNA based detection system relies on the thymidine-Hg2+-thymidine coordination chemistry and the melting temperature (Tm) of the nanoparticle aggregates. For the assay, AuNPs were functionalized with two different thiolated-DNA sequences (designated as probe 1 and probe 2 in Figure 5). When mixed together, probe 1 and probe 2 form aggregates with lower Tm due to T-T mismatches in their base sequence. Presence of Hg2+ in the system raises the Tm of the AuNP aggregates through selectively coordinating with the T-T mismatches to form stable T-Hg2+-T base pairs, providing detection down to 100 nM Hg2+.247 However, this method requires an electronic heating element coupled with the sensor system for careful monitoring of Tm during the detection process. To avoid this need for heating during the read-out process Liu and coworkers have optimized the number of T units in the DNA strands so that the system operates at ambient temperature.248 Chang and coworkers have reported a visual Hg2+ sensing method based on Hg2+-induced conformation change of a T-rich single-stranded DNA (ssDNA).249 Recently, simple thymine functionalized AuNPs have been employed for the colorimetric detection of Hg2+ ion.250 Specific interaction of Hg2+ with thymines residues from two AuNPs induces aggregation process and corresponding color change. Mirkin and coworkers have reported the discrimination of cysteine from other amino acids utilizing T-Hg2+-T coordination chemistry and employing the DNA based probes.251 AuNPs modified with 5,5′-dithiobis (2-nitrobenzoic acid) were also used for the detection of trace levels Cr3+ (99.6 ppb) in the presence of 15 other metal ions in aqueous solution.252
Figure 5.
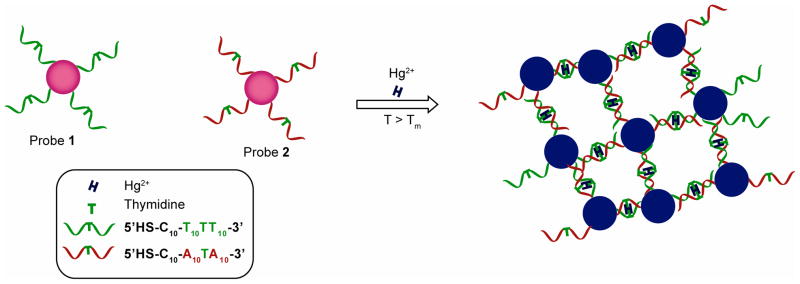
Colorimetric detection of Hg2+ using DNA functionalized AuNPs exploiting thymidine- Hg2+-thymidine coordination chemistry.
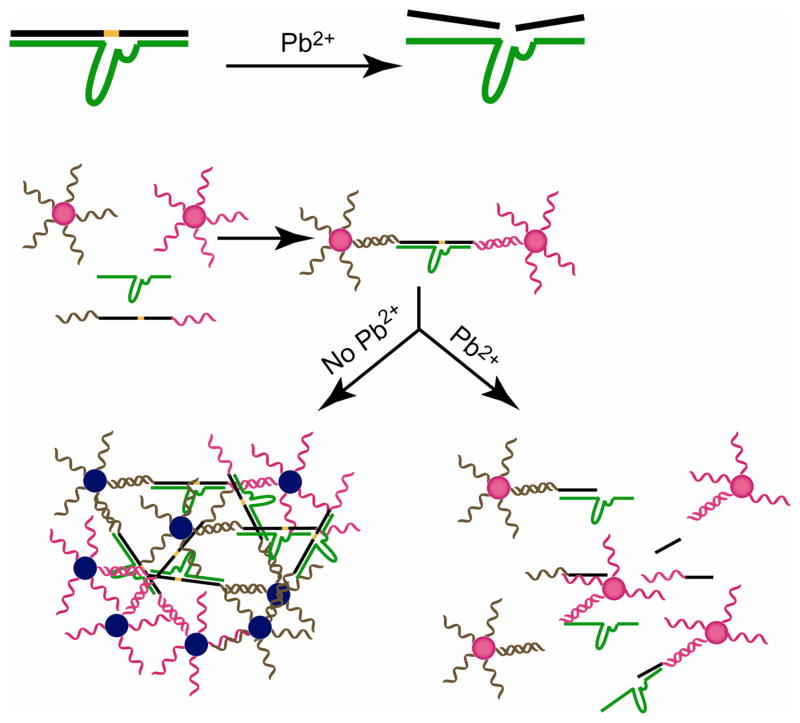
DNAzymes are DNA-based catalysts.253–258 Liu and Lu have fabricated highly selective lead biosensors using DNAzyme-directed assembly of AuNPs,259–262 allowing the tuning of sensitivity over several orders of magnitude. These DNAzymes were obtained through the combinatorial method systematic evolution of ligands by exponential enrichment (SELEX). In their sensor design, a DNAzyme specific to the Pb2+ ion was chosen as the target recognition element and DNA-functionalized AuNPs were used as the signal transducer element. The Pb2+-specific DNAzyme is comprised of an enzyme strand and a substrate strand. In the presence of Pb2+ ion, the enzyme strand carries out catalytic reactions involving hydrolytic cleavage of the substrate strand. When incubated with DNAzyme, the DNA-functionalized AuNPs form blue-colored assemblies through Watson-Crick base pairing as shown in Figure 6. The DNAzyme is activated in the presence of Pb2+ in the solution. Activated DNAzyme cleaves the substrate strand to dissemble the AuNPs, resulting in a blue-to-red color change. The sensor was capable of detecting Pb2+ concentration of 100 nM. More importantly, other divalent metal ions such as Ca2+, Co2+, Ni2+, and Cd2+ did not induce any color change. Optimization to control the AuNP orientation in the assemblies allowed fast detection of Pb2+ (< 10 min) at ambient temperature.262 Later, a highly selective and sensitive colorimetric sensor for uranyl (UO22+) based on AuNP and UO22+ selective DNAzyme has been reported using both labeled and label-free methods.263
Figure 6.
Pb2+ directed assembly of AuNPs by the DNAzymes resulting in detection.
4.1.3. Other Metal Ions
Hutchison and coworkers have developed a sensitive and selective trivalent lanthanide (Ln3+) ions sensor based upon tetramethylmalonamide (TMMA) functionalized AuNPs.264 The presence of Ln3+ ions in the AuNP solution initiates AuNP cross-linking and concomitant red to blue color change through the formation of 2:1 TMMA-Ln3+ chelating complex. An immediate colorimetric response to the Ln3+ ions was detected, with sensitivity down to ~50 nM for Eu3+ and Sm3+. Recently, Wang and coworkers have devised a colorimetric method for detecting Al3+ based on pentapeptide (CALNN) functionalized AuNPs.265 The affinity of Al3+ towards the functional carboxylic group of the pentapeptide induces AuNP aggregation and color change. This assay was successfully applied for sensing of the Al3+ on living cellular surfaces under physiological conditions.
4.2. Detection of Anions
Numerous efforts have been devoted to the development of sensor system for anionic species.266–268 Designing recognition motifs for anions is challenging due to their lower charge to radius ratio, pH sensitivity, wide range of geometries, and solvent dependent binding affinity and selectivity.268 Kubo et al. have attached isothiouronium groups onto AuNP surface and demonstrated sensing of oxanions such as AcO−, HPO42−, and malonate in 10% (v/v) H2O-MeOH solution.269 Colorimetric sensing of hydrophilic anions (e.g. dihydrogen phosphate) has been achieved in dichloromethane using AuNPs with phenyl urea anion binders.270 AuNPs coated with ethylene glycol-appended isothiouronium units were used to detect F− in water using 3-nitrophenylboronic acid as a mediator at pH 5.5.271 Ionic liquid functionalized AuNPs have been applied for anion sensing.272,273 For example, Itoh et al. have used ionic liquids based on the imidazolium cation for colorimetric sensing of I− and PF6−.272 AuNPs coated with thioglucose groups have been used by Watanabe et al. to sense fluoride anions over a relatively narrow concentration range (20 ~ 40 mM) in water,274 with high selectivity against other anions such as Cl−, Br−, I−, AcO−, and NO3−. Ahn et al. have reported selective sensing of trans-dicarboxylates such as fumarate (one of the key components generated in the Krebs cycle) over its cis-isomer, maleate utilizing AuNPs functionalized with an anion-recognition motif.275,276 Utilizing this system, discrimination of benzene-1,4-dicarboxylate from its isomers benzene-1,2- and benzene-1,3-dicarboxylate in water was also possible.276
Fast and sensitive detection of CN− is important for environmental monitoring and the evaluation of food safety.277,278 Han et al. reported a colorimetric detection method for cyanide anions in aqueous solution employing adenosine triphosphate-stabilized AuNPs and a Cu2+–phenanthroline complex as the receptor unit.279 In their sensing ensemble, exposure of CN− to Cu2+–phenanthroline complex induced a decomplexation process to generate free phenanthroline, which subsequently caused the ATP-stabilized AuNPs to aggregate resulting in color change. Utilizing this system 10−5 M CN− was detected in neutral aqueous solution. Han et al. have reported an AuNP-embedded plasticized poly(vinyl chloride) (PVC) membrane for sensing of iodide anions in the presence of F−, Cl−, Br−, N3−, NO2−, NO3−and CH3COO−.280 Yuan et al. have employed AuNPs decorated with a zinc (II) dipicolylamine-phosphate binding group for the colorimetric detection of bis-phosphorylated peptides.281 Li et al. have utilized a “click” reaction coupled with the AuNP probes for the visual detection of ascorbic acid.282 Mirkin et al. have employed the Griess reaction (coupling of sulfanilamide and naphthylethylenediamine by nitrite) for the AuNP-based colorimetric detection of nitrite.283
4.3. Detection of Small Organic Molecules
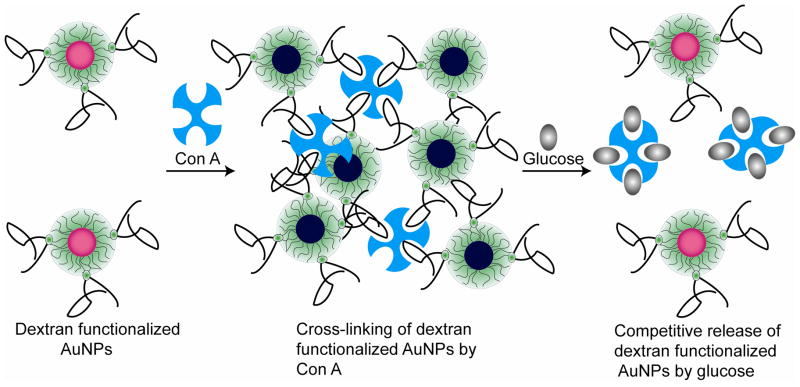
Geddes et al. have demonstrated a competitive colorimetric glucose assay using assemblies of concanavalin A (Con A) and dextran-functionalized AuNPs.284,285 As shown in Figure 7, multivalent binding with Con A crosslinks dextran-coated nanoparticles. The presence of glucose in the system competitively binds Con A, releasing the dextran-coated AuNPs that can be readily monitored by either conventional UV/Vis spectrometry284 or wavelength-ratiometric resonance light scattering with a glucose dynamic sensing range of 1 – 40 mM.285 Due to the wide detection range, this system can potentially be useful to diagnose the blood glucose level in healthy people (3 – 8 mM) and in diabetics (2 – 40 mM). Molecularly imprinted polymers (MIP) with embedded AuNPs (Au-MIP) have been used by Sugimoto et al. as a colorimetric sensor for adrenaline.286 In the absence of adrenaline, the shrunken MIP gel provides close proximity of AuNPs. Adrenaline swells the MIP gel and causes a blue shift in the plasmon absorption band of the immobilized AuNPs, with a dynamic range from 5 μM to 2 mM. AuNPs surface functionalized with water-soluble copolymers [poly(N-n-isopropylacrylamide-co-acryloyldiethyletriamine)] have been employed by Uehare et al. for the detection of glutathione.287 In their system, the AuNP aggregates form upon mixing the AuNP solution with the polymer. Addition of glutathione results in spontaneous disassembly of the aggregates with concomitant colorimetric response. AuNPs conjugated with thermoresponsive co-polymers have been utilized for the colorimetric sensing of thiol compounds.288,289 Recently, cysteamine-modified AuNPs have been employed for the colorimetric detection of melamine290 and 2,4,6-Trinitrotoluene (TNT)291 in real world matrices.
Figure 7.
Colorimetric glucose sensing via the dissociation of Con A-aggregated dextran coated AuNPs. The presence of Con A crosslinks dextran-coated AuNPs with concomitant blue-shift in SPR. The addition of glucose diminishes the Con A-AuNPs interaction releasing the individual dextran-coated AuNPs.
Aptamers are single-stranded oligonucleic acid-based binding molecules that are generated by SELEX, an in vitro selection process.292 These functional DNA or RNA structures can bind a wide variety of targets with high affinity and specificity. Aptamer-based analytical methods have been used with the AuNP-based platform for colorimetric detection of small organic molecules.293–299 For example, an effective colorimetric adenosine sensor was designed by Lu and coworkers.293 As illustrated in Figure 8, the sensor is composed of nanoparticle aggregates containing three functional components: two kinds of ssDNA-modified AuNPs and a linker DNA molecule that carries adenosine aptamer. Initially, AuNPs and the linker DNA were suspended in solution to generate purple AuNPs. In the AuNP aggregation process, the linker DNA molecule pairs respectively with two ssDNA-functionalized AuNPs where a part of adenosine aptamer also takes part in the DNA hybridization process. When adenosine is present in the system, the aptamer changes its structure to bind with adenosine. This adenosine binding process results in the disassembly of the AuNP aggregates with a concomitant blue-to-red color change. Utilizing this system, adenosine was detected in concentrations from 0.3 to 2 mM. To demonstrate the generality of this system, Lu and coworkers have further constructed a colorimetric sensor for cocaine employing a cocaine aptamer.293 Interestingly, the use of a mixture of different aptamers provided smart materials with cooperative responses to adenosine and cocaine.294 The generality of the approach has been further demonstrated by introducing a third aptamer/nanoparticle component (responsive to potassium ions300) with this system. Lu and coworkers have also immobilized both adenosine and cocaine aptamer-linked nanoparticle aggregates onto a lateral flow device, resulting a more sensitive “dipstick” test which can be performed in complex sample matrices such as human blood serum.299 Stone et al. have reported an AuNP-based colorimetric detection system for theophylline using a theophylline recognizing aptamer.298 Recently, an aptamer-based colorimetric biosensor for Ochratoxin A (OTA) has been reported by Marty et al.296 Highly specific target recognition property of aptazymes has been utilized to devise colorimetric sensor for small organic molecules.301 For example, Lu et al. have devised an adenosine biosensor based on the adenosine recognizing aptazyme-directed assembly of AuNPs.302 RNA aptazyme-tethered AuNP was employed by Ogawa et al. for developing sensing system to visually detect ligands of a cleavase-like RNA aptazyme at room temperature.303 Gu et al. have employed a highly specific oxytetracycline binding ssDNA aptamer to discriminate oxytetracycline from other tetracyclines, such as doxycycline and tetracycline.304
Figure 8.
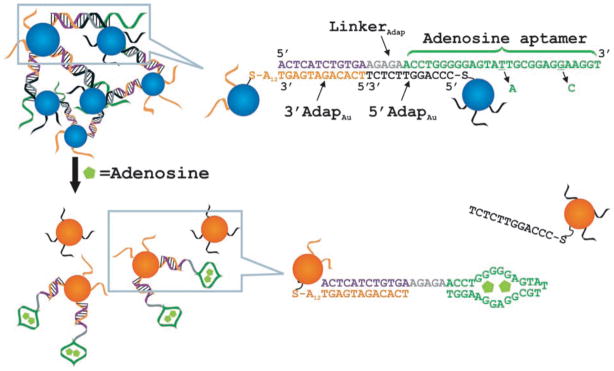
Schematic representation of colorimetric sensing of adenosine using aptamer linked AuNPs (left panel). The mutated linker with two mutations denoted by short black arrows was used as a control. Reprinted with permission from Angew. Chem. Int. Ed. (Ref 293). Copyright 2006 Wiley-VCH Verlag & Co. KGaA, Weinheim.
4.4. Detection of Oligonucleotides
Detection of genetic mutations provides crucial target for diagnostics,305,306 while nucleic acid detection can be used to identify microbial pathogens. Fluorescent and radioactive detection readout methods (e.g. PCR, RT-PCR, northern blot, southern blot, and high density microarrays) are the conventional techniques for the detection of oligonucleotides.307–310 AuNP-based colorimetric assays have been demonstrated to be a highly competitive technology for oligonucleotide targets.311,312
DNA-meditated AuNP assembly was demonstrated by Mirkin in 1996.313 Fabrication of AuNPs functionalized with thiolated DNA strand allowed researchers to tailor the properties of the nanoparticle probes according to the assay method. This discovery has stimulated extensive use of oligonucleotide-directed AuNP aggregation for colorimetric detection of oligonucleotides311,314–321 and fabrication of structured assemblies.322 In this appraoch, two ssDNA-modified AuNP probes were used for colorimetric detection of target oligonucleotides. The base sequences in the AuNP probes are designed so they are complementary to both ends of the target oligonucleotides (Figure 9). AuNP aggregation with concomitant color change is triggered by the presence of target oligonucleotides as a result of hybridization of DNA strand. Highly specific base-pairing of DNA strands coupled with the intense absorptivity of AuNPs enables the sub-picomolar quantitative colorimetric detection of oligonucleotides.317 Maeda et al. have shown that the aggregation of DNA-functionalized AuNPs can also be induced by hybridization of target DNA that does not cross-link the AuNPs,323 with an unusual sensitivity of this system towards single-base mismatches. Rothberg et al. have shown that citrate-stabilized AuNPs can discern ssDNA and double-stranded DNA (dsDNA) at the level of 100 fmol through simple electrostatic interactions.324 Mismatches even at the level of single base-pair have been easily detected through this way. A nearly “universal” sensing strategy employing an ssDNA probe, unmodified AuNPs, and a positively charged, water-soluble conjugated polyelectrolyte has been demonstrated by Xia et al. to detect a broad range of targets including DNA, proteins, small molecules, and inorganic ions.325
Figure 9.
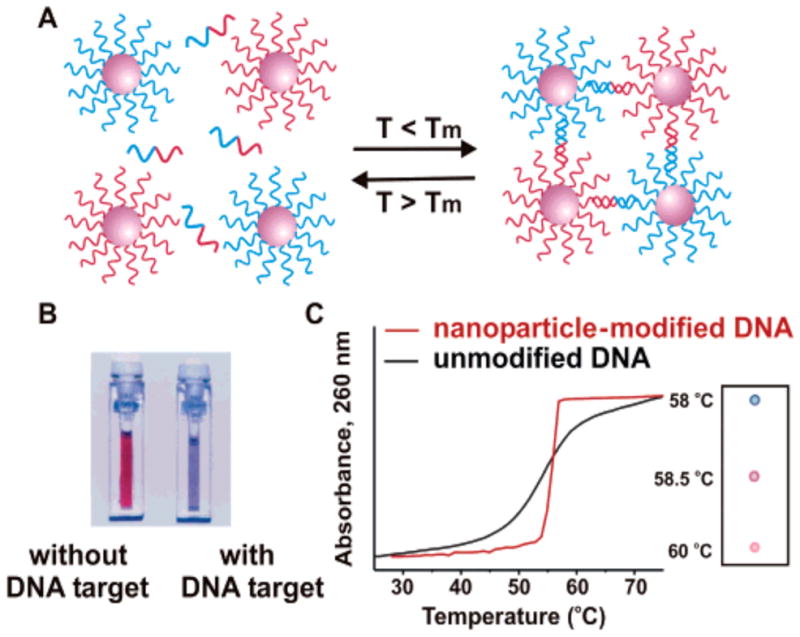
Aggregation of oligonucleotide AuNPs in presence of complementary target DNA (A), leading to change in color of solution from red to blue (B). Reprinted with permission from Science and Chem. Rev. (Ref 314 and Ref 16). Copyright 1997 American Association for the Advancement of Science and 2005 American Chemical Society.
Another important application of oligonucleotide-directed AuNP assembly is for the colorimetric screening and triplex DNA binders.326,327 Screening of triplex DNA binders uses three components: two sets of AuNPs functionalized with non-complementary ssDNA strand and a free strand of ssDNA that can form triplex structure with the DNA (Figure 10).326 At room temperature, no aggregation of ssDNA functionalized AuNPs takes place due to the low stability of the triplex structure. However, the presence of appropriate triplex DNA binders (e.g. benzo[e]pyridoindole and coralyne (CORA)) stabilizes the triplex structure, inducing the AuNP aggregation and corresponding color change. The simplicity of this approach makes a convenient and high-throughput method for identifying triplex binders from large combinatorial libraries. Oligonucleotide-directed AuNP assembly has also been applied for determining the relative binding strengths of molecules to duplex DNA by colorimetric assay, providing a strategy for identifying DNA binding drugs.328,329
Figure 10.
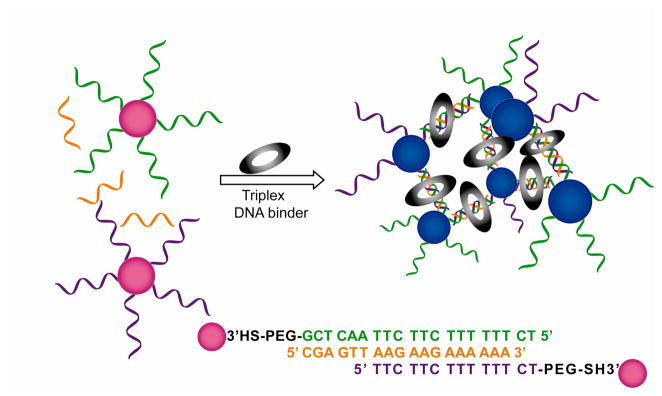
Schematic illustration of structure and color change of DNA functionalized AuNPs in presence of triplex DNA binders.
4.5. Detection of Proteins
Many disease states (e.g. cancer) are often associated with the presence of certain biomarker proteins and/or irregular protein concentrations. AuNPs have been successfully applied for colorimetric detection of proteins. A diverse range of carbohydrate functionalized AuNPs have been prepared for the detection of carbohydrate binding proteins.330–340 For example, the aggregation of β-D-lactopyranoside (Lac)-functionalized AuNPs has been utilized by Kataoka et al. for the detection of Recinus communis agglutinin (RCA120).338 The degree of colloidal aggregation was proportional to the protein concentration, thus allowing this method to be useful in quantitative detection of lectin. Significantly, a high sensitivity of lectin detection (lectin concentration of 1 ppm), has been achieved with this system.338 Later, the density of Lac moiety on the particle surface has been modulated for controlling the concentration range of lectin detection.339 The investigation showed that a critical Lac density (>20%) is required to trigger lectin-induced aggregation. The protein-directed assembly of gold glyconanoparticles has also been developed for facile and sensitive identification of protein-protein interactions. In an interesting study, binding partners of Con A have been identified by utilizing the assemblies of Con A and mannose-modified AuNPs, since the protein-protein interactions disrupt the initial nanoparticle-protein assemblies.341 Similarly, a series of gluco- and manno-oligosaccharide-functionalized AuNPs have been used to sense carbohydrate-protein using the lectin Con A.342 Recently, a novel fluorescent based competition binding assay was also reported by Wang et al. to measure the binding affinity of glyconanoparticles (AuNPs conjugated with underivatized mono-, oligo-, and polysaccharides) with model protein (lectin Con A).343 Lactose-stabilized gold glyconanoparticles have been employed by Russell et al. to measure calcium ion-mediated carbohydrate-carbohydrate interactions.240 The controlled aggregation of lactose-stabilized ~16 nm AuNPs has been harnessed to obtain colorimetric detection of cholera toxin at nanomolar levels.344 AuNPs functionalized with a series of synthetic sugar probes have been utilized by Uzawa et al. for discriminating ricin toxin.345 By utilizing biotin-functionalized AuNPs deposited on glass substrates, label-free optical methods to study biomolecular interactions in real time has been developed by Chilkoti et al.346,347 Likewise, 15 nm sialic acid functionalized AuNPs have been employed to the optical detection of JC virus VLPs through sialic acid recognition.348
In an aptamer-based strategy, AuNP probes carrying platelet-derived growth factors (PDGFs)-specific aptamers have been employed by Chang et al. to detect PDGFs at nanomolar concentrations.349 Additionally, by conducting a competitive binding assay, this aptamer-AuNP-PDGF assembly has been used to detect PDGF receptors.349 Dong et al. have reported an even simpler aptamer-based colorimetric protein sensing method using unmodified AuNP probes.350 In their sensor design, unmodified AuNPs were initially stabilized with thrombin-binding aptamers. In the presence of thrombin, the aptamers fold into a G-quadruplex/duplex structure due to the aptamer-protein recognition. As the apatamers fold, their relatively rigid structure induces AuNP aggregation with a detection limit of 0.83 nM. Likewise, glass surfaces have been modified with thrombin-specific aptamer to achieve thrombin sensing.351 Since thrombin includes two binding sites for the aptamer, this process leads to a “sandwich” complex. Later, this method was extended by further enlargement of the immobilized AuNPs in a growth solution containing HAuCl4, CTAB, and NADH.352 This process initiates the SPR coupling interactions between the adjacent AuNPs, and provided sensitivity limit of 2 nM for thrombin. A general antigen-antibody interaction has also been applied for the AuNP aggregation-based immunoassay for proteins.353 Utilizing this method, Rosenzweig et al. have demonstrated a detection limit of 2 μg/mL of anti-protein in serum samples.354
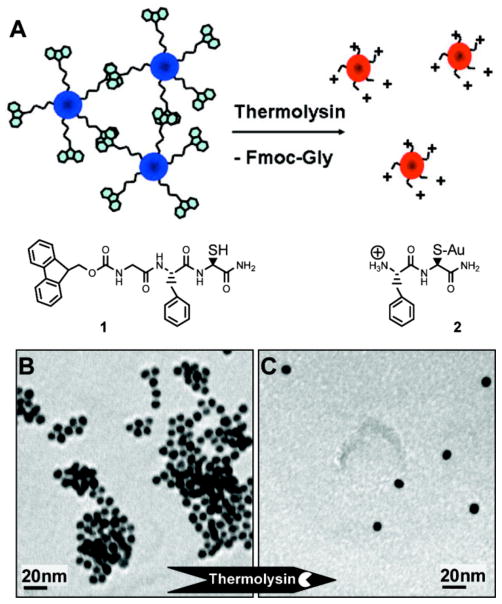
Dithiols cross-link AuNPs,355 with dithiol-functionalized peptides affording a useful platform for colorimetric detection of proteases.356 Scrimin et al. have designed two C- and N-terminal cysteinyl derivatives of peptide substrates specific to thrombin and lethal factor.357 For their assay, the peptides were first treated with the analytes. Subsequently, the solution was incubated with citrate-stabilized 12 nm AuNPs. Nanoparticle aggregation was induced by the intact peptides in the absence of target proteases, whereas the protease-cleaved peptides do not bridge the AuNPs. Later, Stevens et al. further simplified this two-stage approach by employing AuNPs decorated with Fmoc-protected peptides that bear a cysteine anchor.358 Presence of thermolysin in the system cleaved the peptide ligands, leading to AuNPs dispersion in the solution along with a blue-to-red color change (Figure 11). An impressive sensitivity of 90 zg mL−1 (less than 380 molecules) has been reported from this system. Based on the enzymatic cleavage of DNA molecules, Mirkin et al. have developed a real-time colorimetric screening method for endonuclease activity by using DNA-mediated AuNP assemblies.359 Simultaneous determination of the efficiencies of endonuclease inhibitors (e.g. molecules with DNA-binding ability) has been achieved utilizing the colorimetric endonuclease-inhibition assay. Similarly, detection of kinase,360 phosphatases,361,362 β-lactamase,363 and aminopeptidase364 along with the screening of their activity have been achieved utilizing the enzyme-triggered AuNP assembly/disassembly approach. Zare et al. have demonstrated a colorimetric sensor for protein conformational changes by utilizing AuNP probes.365
Figure 11.
Schematic illustration of thermolysin triggered dispersion of AuNP assemblies (A), TEM images of AuNPs (functionalized with 1) before (B) and after (C) addition of thermolysin and generation of 2. Reprinted with permission from J. Am. Chem. Soc. (Ref 358). Copyright 2007 American Chemical Society.
5. Fluorescence-based Sensing
5.1. FRET-based Detection of Metal Ions and Small Molecules
AuNPs can serve as excellent fluorescence quenchers for FRET-based assays223 due to their extraordinary high molar extinction coefficients and broad energy bandwidth.366 Murray and coworkers have reported a FRET based assay for the detection of various metal ions.367 Electrostatic complexation of anionic tiopronin-coated AuNPs and [Ru (bpy)3]3+, a well-known cationic fluorophore, results in fluorescence quenching of [Ru (bpy)3]3+. The complexes then can be dissociated by addition of electrolytes and the fluorescence of [Ru (bpy)3]3+ restored. AuNP FRET quenching has been utilized in Hg2+ sensing.368 Chang et al. reported that selectivity of the optimized Rhodamine B-abosrbed AuNPs system for Hg2+ is 50 times higher than that of other metal ions (e.g. Pb2+, Cd2+, Co2+) with a detection limit of 2.0 ppb. A FRET based Cu2+ ion sensor has been developed using bispyridyl perylene bridged AuNPs by Zhu and collaborators.369 In the absence of the Cu2+ ion, the fluorescence of the bispyridyl perylene on AuNPs is quenched. The Cu2+ ions then replace the bispyridyl perylene from the AuNPs, restoring fluorescence. Thomas et al. have used lanthanide complexes of bipyridine-functionalized AuNPs as phosphorescent sensors for alkaline earth metal ions and transition metal ions.370
Besides detecting metal ions, FRET based AuNP assays have been utilized for sensing small organic molecules. Chang et al. have used AuNPs in which Nile red non-covalently attached to AuNPs for sensing thiols at submicromolar levels.371 As another example, Tang et al. have designed a FRET based cholesterol sensor by using β-cyclodextrin (β-CD) functionalized AuNPs.372 Inclusion of fluorescein (FL) into cavity of CD on AuNPs causes complexation of AuNP-CD-FL construct, resulting in fluorescence quenching of FL through FRET. In the presence of cholesterol, FL inside CD is replaced by the cholesterol due to their higher binding affinity to CD, restoring FL fluorescence for detection of cholesterol at nanomolar concentrations. FRET based AuNP assays have also been reported for detecting homocysteine.373,374 Due to strong fluorescent properties of AuNP smaller than 1.2 nm, AuNPs have been used to detect metal ions and proteins, using aggregation induced quenching or enhanced fluorescence of AuNPs.375–378
5.2. AuNP-based Molecular Beacons
Hairpin FRET-based systems for sensing DNA have been created by labeling molecular beacons with AuNPs.212 As shown in Figure 12, the nucleic acid probe conjugated with the organic dye is self-complementary, forming the hairpin structure on AuNP with effective FRET fluorescence quenching. The hairpin structure changes to rodlike through complementary hybridization with the target DNA, resulting in an increase in fluorescence of the dye. By employing similar principle, Nie et al. have shown that single stranded oligonucleotide-functionalized AuNPs with fluorophore-termini can assemble into a constrained arch like conformation.379 In this conformation, the fluorophore is efficiently quenched by AuNP due to close donor and acceptor distance. Upon binding with the target DNA, the constrained conformation opens and the fluorophore is separated from the AuNP, affording fluorescence turn-on. AuNPs based FRET assay have also been used to monitor the cleavage of DNA by nucleases.231
Figure 12.
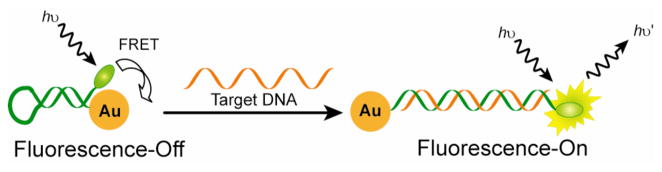
Schematic illustration of DNA detection, showing the conformational changes of dye-oligonucleotide-AuNP conjugates before and after hybridization with the target DNA.
Mirkin et al. have developed AuNP probes, (nano-flares) that are designed to detect and quantify intracellular analytes e.g. mRNA in cells.380–382 Hybridization of dye-terminated DNA reporter sequences with oligonucleotide-functionalized AuNPs quenches fluorescence of the reporter. The presence of a target then displaces and releases the reporter from AuNPs by constructing more stable duplex between the target and the oligonucleotide on AuNPs. Bai et al. have fabricated a FRET based AuNP assay to identify organic molecules that stabilize G-quadruplexes.383 Initially, AuNPs carrying fluorescein-tagged probe DNA quench the fluorescence of the probe. Upon specific binding of a target organic molecule, intramolecular folding of the linear probe DNA into G-quadruplexes formation increases the distance between the AuNP and the probe DNA with concomitant enhancement of fluorescence. Fan et al. have reported multicolor fluorescent AuNP-based molecular beacons to detect target molecular analytes.384 In their system, the multicolor dye-labeled aptamers are duplexed with DNA probes on AuNPs through complementary hybridization, resulting in fluorescence quenching of the dyes. In the presence of target molecules, the dye-labeled aptamer-target molecule binding separates the duplex, leading to fluorescence recovery of the dyes. FRET based AuNP assay labeled with fluorescence probes has also been reported to detect Hg2+.385
5.3. Sensors based on FRET between QDs and AuNPs
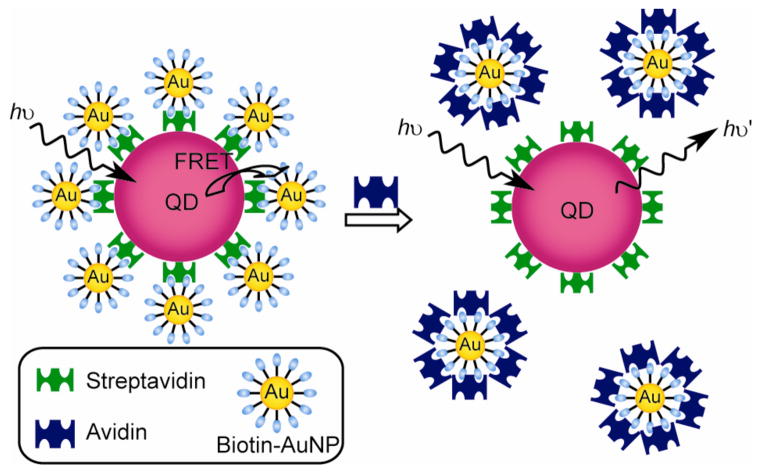
Semiconductor quantum dots (QDs) have been utilized for FRET-based AuNPs assays for detection of proteins, utilizing the high efficiency and stability of these fluorophores.386 Malvin et al. have reported a fluorescent competitive assay for DNA identification using QDs and AuNPs, where AuNPs were assembled with CdSe QDs through short complementary DNA strands, causing fluorescence quenching of the CdSe QDs.387 Addition of complementary oligonucleotides then displaces the AuNP-DNA from the QD-DNA, resulting in QD fluorescence restoration. Similarly, Kim et al. have fabricated an enzyme inhibition assay using biotinylated AuNPs and streptavidin-coated QDs as a FRET donor-acceptor couple (Figure 13).388 The biotinylated AuNPs specifically bind with the streptavidin-functionalized QDs forming quenched assemblies. The presence of avidin then releases the biotinylated AuNPs from QDs through a competitive binding with the biotinylated AuNPs. The authors have also demonstrated an approach for the rapid and simple detection of protein glycosylation by using dextran-functionalized QDs and Con A-coated AuNPs.389
Figure 13.
Competitive inhibition assay for the detection of avidin using QD-AuNP conjugates.
Yu et al. have used assembly of Con A conjugated QDs and β-CD-modified AuNPs for determination of glucose in serum.390 In practice, β-CD-modified AuNP is displaced by glucose due to its higher binding affinity to β-CD than Con A, resulting in release of Con A-conjugated QDs and recovery of QD fluorescence. Guo et al. have designed an inhibition assay for identification of Pb2+ based on the modulation in FRET efficiency between QDs and AuNPs.391 Initially, the positively charged QDs form FRET donor-acceptor assemblies with negatively charged AuNPs by electrostatic interaction. The presence of Pb2+ aggregates AuNPs via an ion-templated chelation, inhibiting the FRET process, with a detection limit of 30 ppb of Pb2+. Lu et al. have reported that simultaneous colorimetric and fluorescent detection of adenosine and cocaine in a single solution with QD-encoded aptamer sensors.392
5.4. “Chemical Nose” Approach for the Detection of Proteins, Pathogens and Mammalian Cells
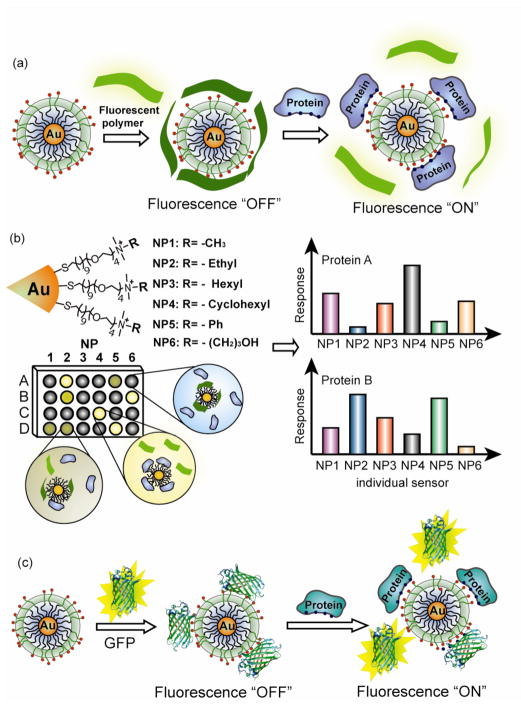
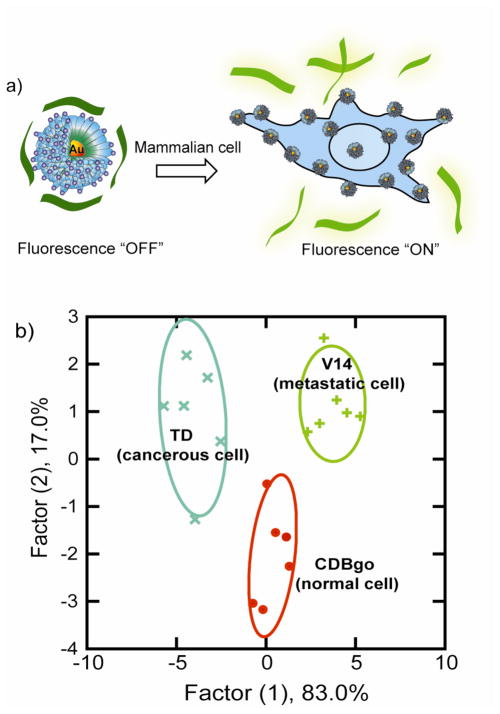
The above FRET based assays employ specific interactions. Array-based sensing, i.e. “chemical nose/tongue” strategies provides a useful alternative that uses differential selective interactions to generate patterns that can be used to identify analytes or changes in complex mixtures.8 Recently, Rotello et al. have developed a protein sensor using chemical nose technology.393 The prototype sensor array was generated using six cationic AuNPs of differing structures and an anionic poly (p-phenyleneethynylene) (PPE) polymer. As illustrated in Figure 14a, electrostatic complexation of AuNPs and polymer results in fluorescence quenching of the polymer (fluorescence “OFF”) through energy transfer. Addition of protein analytes then disrupts the quenched polymer-AuNPs complexes via competitive binding, causing fluorescence recovery of the polymer (fluorescence “ON”). The protein-nanoparticle interactions are differential,394,395 leading to a fingerprint fluorescence response pattern for individual proteins (Figure 14b) that was characterized using linear discriminate analysis (LDA). By employing the same principle, an AuNP-green fluorescent protein construct was used to detect and identify proteins at 500 nM in undiluted human serum (~1 mM overall protein concentration)(Figure 14c).396
Figure 14.
Schematic illustration of ‘chemical nose’ sensor array based on AuNP-fluorescent polymer/GFP conjugates. (a) The competitive binding between protein and quenched polymer-AuNP complexes leads to the restoration of fluorescence (b) The combination of an array of sensors generates fingerprint response patterns for individual proteins. (c) The competitive binding between protein and nanoparticle-GFP complexes leads to fluorescence light-up.
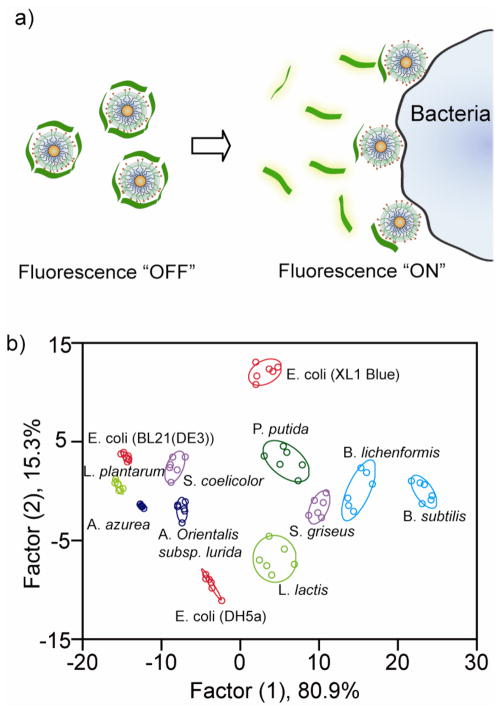
Analogous AuNP-conjugated polymer systems have been used to detect and identify bacteria.397 Three cationic AuNPs and one anionic PPE polymer were used to generate the sensor. Presence of bacteria disrupts the initially quenched assemblies leading to fluorescence restoration of PPE. From the distinct fluorescence response patterns, the sensor array was capable of identifying 12 bacteria including both Gram-positive (e.g. A. azurea, B. subtilis) and Gram-negative (e.g. E. coli, P. putida) species, as well as three different strains of E. coli (Figure 15).
Figure 15.
Array-based sensing of bacteria. a) Displacement assay between bacteria and the AuNP-PPE complex. b) Canonical score plot for the first two factors of simplified fluorescence response patterns obtained with NP-PPE assembly arrays against bacteria (95% confidence ellipses shown). Reprinted with permission from Angew. Chem. Int. Ed. (Ref 397). Copyright 2008 Wiley-VCH Verlag & Co. KGaA.
The AuNP-conjugated polymer systems have been used for rapid and effective differentiation between normal, cancerous, and metastatic cells.398–400 The fluorescence responses analyzed by LDA were capable of distinguishing (i) different cell types; (ii) normal, cancerous and metastatic human breast cells; and (iii) isogenic normal, cancerous and metastatic murine epithelial cell lines (Figure 16).
Figure 16.
a) Schematic depiction of the fluorophore-displacement cell detection array. Displacement of quenched fluorescent polymer by a cell with consequent restoration of polymer fluorescence. b) Canonical score plot for the first two factors of simplified fluorescence response patterns obtained with AuNP-conjugated polymer assembly arrays against different mammalian cell types. Reprinted with permission with permission from Proc. Natl. Acad. Sci. U.S.A. (Ref 398). Copyright 2009 National Academy of Sciences.
Rotello et al. have also reported an enzyme-AuNP sensor array for detecting proteins in which the sensitivity is amplified via enzymatic activity.401 In this system, cationic AuNPs are combined with β-galactosidase (β-Gal) through electrostatic interaction, inhibiting the β-Gal enzymatic activity. Addition of analyte proteins releases the β-Gal from AuNPs and restores the β-Gal activity, providing an amplified readout of the binding event (Figure 17).
Figure 17.
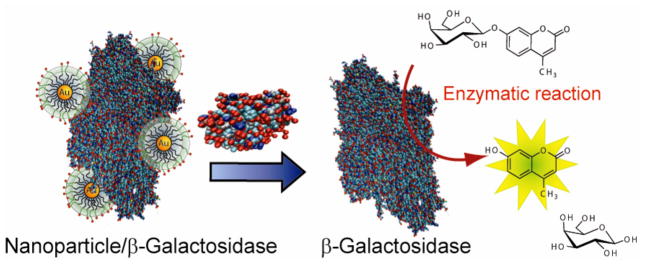
A schematic representation of the sensor system comprised of β-galactosidase (β-Gal) and cationic AuNPs. β-Gal is displaced from the β-Gal/AuNP complex by protein analytes, restoring the catalytic activity of β-Gal towards the fluorogenic substrate 4-methylumbelliferyl-β-D-galactopyranoside, resulting in an amplified signal for detection. Reprinted with permission from J. Am. Chem. Soc. (Ref 401). Copyright 2010 American Chemical Society.
6. Electrical and Electrochemical Sensing
AuNPs feature excellent conductivity, high surface area and catalytic properties402 making them excellent materials for electrochemical 15,403–418 In this section, we will summarize the use of AuNPs for electrocatalytic and electrochemical sensing.
6.1. Vapor Sensing
The electronic properties of self-assembled films of monolayer-protected AuNPs can be varied by tuning the particle size, interparticle separation, surface functionality, and chemical environments.419 Chemiresistors are solid-state devices that rely on this sensitivity through changes in electrical resistance upon interaction with a chemical species. Over the past decade, there have been a number of chemiresistor vapor sensors based on thiol functionalized AuNPs.420–426 For example, Wohltjen and Snow have fabricated a chemiresistor by deposition of a thin film of octanethiol-coated AuNPs (d ~ 2 nm) onto an interdigitated microelectrode.427 A rapid decrease in the conductance due to film swelling was observed in presence of toluene and tetrachloroethylene, with a detection limit of ca. 1 ppm. Later, Shen and coworkers have shown that aromatic-functionalized AuNPs exhibited different sensory responses depending on the nature of the terminal functionality (OH, CH3, NH2, COOH) of aromatic thiols.428,429
Vossmeyer et al. have systematically investigated the sensing of toluene and tetrachloroethylene using films consisting of dodecylamine-stabilized AuNPs and α,ω-dithiols with different chain lengths (C6, C9, C12, C16).430 At a given concentration of toluene, it was observed that the resistance responses increase exponentially with increase of –CH2 units. This effect was attributed to the augmentation of sorption sites with increasing ligand length. Zhong and coworkers have proved the correlation between the vapor-response sensitivity and the interparticle spacing properties.146,431 Recently, Wieczorek and coworkers successfully detected dissolved organic analytes using a thin film of hexanethiol protected AuNPs inkjet-printed onto microelectrodes.432 On exposure to toluene, dichloromethane, and ethanol dissolved in 1 M KCl solution, an increase in impedance at 1 Hz was observed with detection limits of 0.1, 10, and 3000 ppm, respectively. Further studies by this group revealed that morphology, ionic strength and hydrophobic-hydrophilic character of nanoparticle film play an essential role in sensing.433,434
Mixed monolayer surfaces of AuNPs have been used to develop ‘electronic-tongue’ type sensor arrays by varying the ratios of the different ligands on the nanoparticle surface. 435,436 For example, AuNPs of two different thiol ligands have been fabricated by Kim et al.436 that showed different chemical selectivities and produced rapid and reversible responses towards the vapors of 1-propanol, acetone and cyclohexane. AuNP-dendrimer composites have also been explored in vapor sensing.437–441 In these layer-by-layer (LBL) self-assemblies, the AuNPs provide the conductive film material while the dendrimers serve to cross-link the nanoparticles and to provide sites for the selective sorption of analyte molecules. An interesting bioconjugate material has been synthesized by simple reaction of the spider silk with aqueous chloroauric acid.442 The environment-dependent expansion/contraction phenomenon of spider-silk modulates electron transport between nanoparticles, differentiating the polarity of alcohol vapors (from methanol to butanol) by distinct conductivity changes.
6.2. Electronic AuNP Sensors Employing Macrocyclic Complexation
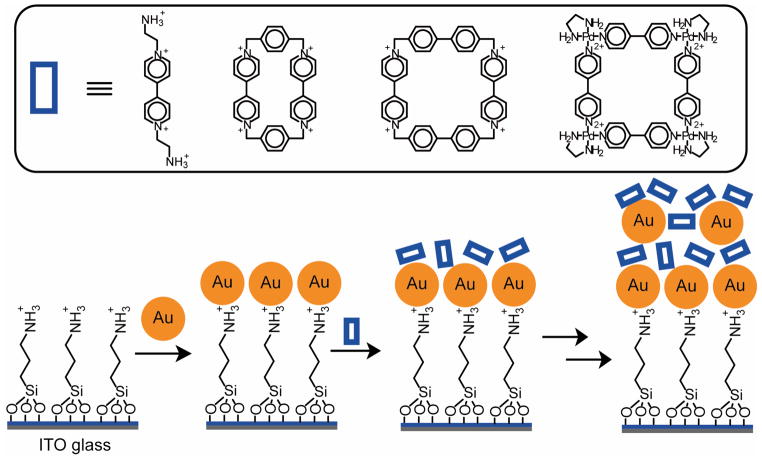
The synergistic combination of electroactive AuNPs and macrocyclic compounds provides useful sensor systems.443–446 Willner and coworkers have constructed nanostructured assemblies via electrostatic cross-linking of citrate-stabilized AuNPs (12±1 nm) and oligocationic cyclophanes (molecular squares). The assembly process was repeated in a stepwise manner to attain LBL assembly of anionic AuNPs and oligocationic cyclophanes (Figure 18). For sensing, the bipyridinium cyclophanes serve as π-acceptors447 for the association of π-donor substrates such as hydroquinone in their cavities, generating an electrochemical response.448 The sensitivity of the resulting sensor can be tuned via the number of assembled layers on the conductive surface.443 The binding affinity between the macrocycles and the analytes determines the selectivity of the electrodes, with cyclobis(paraquat-p-phenylene) cyclophane responding to hydroquinone, while the enlarged cyclophane cyclobis(paraquat-p-biphenylene) responds only to dihydroxymethyl ferrocene.446 Sensing studies with anionic π-donor analytes such as 3,4-dihydroxyphenylacetic acid and an acyclic crosslinker N,N′-diaminoethyl-4,4′-bipyridinium were also performed.445 Willner and coworkers have also developed a sensing interface by assembling a film consisting of polyethyleneimine, AuNPs and cyclobis(paraquat-p-phenylene) on the Al2O3 insulating layer of an ion-sensitive field-effect transistors.449,450 This device is able to sense charged analytes that are attached to the cyclophane, regardless of their redox activity. Detection of adrenaline was accomplished by measuring either the source-drain current or the gate-source voltage, with a detection limit of 1×10−6 M.450
Figure 18.
Electroactive multilayers formed by the simultaneous deposition of anionic AuNPs and oligocationic cyclophanes. The AuNPs provide excellent conductive surfaces, while the macrocycles serve as π-acceptors to bind with π-donor analytes (e.g. hydroquinone), generating electrochemical responses.
6.3. AuNPs as Platforms for Electrocatalyic and Electrochemical Sensors
AuNPs feature catalytic activity that results from their large surface area-to-volume ratio and their interface-dominated properties.451–454 AuNPs can decrease the overpotentials of many electroanalytical reactions and maintain the reversibility of redox reactions.455 Numerous approaches such as electrostatic interaction,443–446 electrochemical deposition,456–460 and mixing with components in a composite electrode matrix,461 have been applied to deposit AuNPs on electrode surfaces. For example, AuNPs have been used as electrochemical enhancers for electrogenerated chemiluminescence (ECL) sensors.462–464
6.3.1. Detection of Small Molecules
AuNPs has been used for enhanced electrochemical detection of numerous small molecules465–477 including glucose,478–487 dopamine,488–493 uric acid,494–500 ascorbic acid,496–498,501–504 epinephrine,500,505–508 bisphenol A,509 nitrite,510–513 etc. Identification of several phenolic compounds;514 e.g. catechol,515 and aliphatic dicarboxylic acids; oxalic, succinic, malic, and tartaric516 were also reported. For example, Wang and coworkers have electrocatalytically detected epinephrine using a self-assembled dithiothreitol(DTT)-dodecanethiol(DDT)-Au colloid modified gold electrode. The electrode reaction of epinephrine is significantly improved at the nano-Au electrode, providing a detection limit of 60 nM.506 Recently, Luczak reported a voltammetric sensor for detection of norepinephrine using AuNPs, cystamine (CA) and 3, 3′-dithiodipropionic acid (DTDPA) modified gold electrodes. Moreover, the system is able to detect norepinephrine in presence of interferents ascorbic and uric acids.517 Further, Zhang et al. have reported the superior electrocatalytic acitivity of positively charged AuNPs over negatively charged AuNPs using 4-dimethylaminopyridine (DMAP) coated AuNPs/L-cysteine film on gold electrode. Compared with electrodes modified by negatively charged AuNPs/L-cysteine, or L-cysteine alone, the electrode modified by the positively charged AuNPs/L-cysteine exhibited enhanced electrochemical behavior toward the oxidation of biomolecules such as ascorbic acid, dopamine and hydrogen peroxide.518
6.3.2. Detection of Toxic Chemicals and Drugs
AuNP-based electrodes have been used to detect toxic ions such as arsenic,519–524 mercury,519,525–528 antimony,529 and chromium530,531. Several groups also reported high catalytic activity of AuNP-modified electrodes for electrocatalytic oxidation and detection of carbon monoxide,532,533 nitric oxide,534–539 and hydrazine.540–544 Hydrogen peroxide (H2O2) was detected using AuNP decorated electrodes through enzymatic,545–555 non-enzymatic,556–560 and microfluidic electrochemical561 approaches. Various pesticides, e.g. atrazine,562 methyl parathion,563,564 paraoxon ethyl,565 carbofuran,564 phoxim;564 and different drugs such as paracetamol,504,566 atenolol,567 prednisolone,568 ethamsylate,569 were also detected using AuNP-modified electrodes. Recently, Raj has detected isoniazid, a popular anti-tuberculosis drug, by chemisorbing 70–100 nm AuNPs on a sol-gel-derived 3D silicate network with a detection sensitivity of 0.1 nM.570
6.3.3. Detection of Mammalian Cells using AuNP-modified Electrodes
AuNPs/chitosan nanocomposite gels were used for electrochemical monitoring of adhesion, proliferation, and apoptosis of cells on electrodes. Living cells immobilized on glassy carbon electrode (GCE) demonstrated an irreversible voltammetric response and enhanced the electron-transfer resistance with a limit of detection of 8.71× 102 cells/mL.571 Later, K562 leukemia cells were immobilized onto a microporous cellulose membrane modified with AuNPs and the effectiveness of anti-tumor drug methotrexate effect was monitored through the electrochemical response from cells.572 Similarly, living pancreatic adenocarcinoma cells were immobilized on a composite electrode using AuNPs and carbon paste and used to determine the cytotoxic effect of antitumor drug adriamycin.573 Recently, de la Escosura-Muňiz et al. have developed an electrocatalytic platform/sensor for the specific identification of tumor cells. In their system, molecules on cell surfaces are recognized by antibodies conjugated with AuNPs (Figure 19) with catalytic hydrogen reduction used to provide cell detection.574
Figure 19.
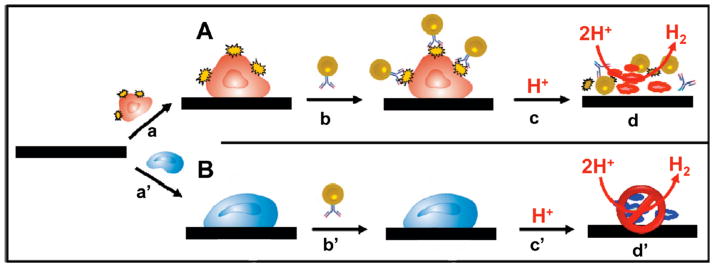
Specific identification of (A) Tumoral Cell Line (HMy2) expressing surface HLA-DR molecules compared to a (B) PC-3 Cell Line that is negative to this marker using AuNP-conjugated antibody coupled with an electrochemical sensor. The specific binding of AuNPs with tumor cells catalyses hydrogen evolution in the acidic environment, generating electrochemical responses. Reprinted with permission from Anal. Chem. (Ref 574). Copyright 2009 Americal Chemical Society.
6.4. AuNP-based Electrochemical Enzymatic Biosensors
6.4.1. AuNPs act as “Electron Wires” facilitating Direct Electron Transfer
The direct electrical communication of redox enzymes with electrodes is a useful strategy for biochemical sensoring.575 However, the redox center of most oxidoreductases is electrically insulated by the protein. AuNP-based electrodes acting as “electron wires” can facilitate direct electron transfer between redox proteins and bulk electrode materials, thus allowing electrochemical sensing without redox mediators,576–585 exploiting the physical properties of AuNPs.586–589 One practical method to develop AuNP-based enzyme electrodes uses immobilization of the enzyme on AuNPs deposited directly on the surface of the bulk electrode.590 Different strategies have been applied to immobilize enzymes onto AuNPs, such as direct attachment by the use of cysteine,591 via thiol linkers,592 and through covalent bonds.593 The co-deposition of redox enzymes and AuNPs on the electrode surfaces reduces the insulating effect of protein shell providing an excellent biosensor platform594,595 to detect various biomolecules.596–602 Moreover, adsorption of biomolecules onto AuNPs surface can preserve their bioactivity using biocompatible AuNPs.603,604 Again, the immobilization of enzymes onto AuNPs can increase their turnover rates,605–607 enhancing sensitivity. Willner and coworkers have constructed a gold nanoparticle based bioelectrocatalytic system with highly efficient electrical contact of glucose oxidase (GOx) with the electrode support. In their system, electron transfer between the enzyme active sites and the electrode support was facilitated through the reconstitution of apo-glucose oxidase (apo-GOx) on a 1.4 nm AuNP-functionalized with the cofactor flavin adenine dinucleotide (FAD) (Figure 20).608 The resulting AuNP-reconstituted enzyme electrodes featured high electron-transfer turnover rate of ~5000 s−1(seven-fold higher than that of native GOx). This approach was further explored to pyrroloquinoline quinine (PQQ)-dependent enzymes by the reconstitution of apo-glucose dehydrogenase (GDH) on a PQQ-functionalized AuNP-modified gold electrode.609 Kerman et al. have demonstrated a streptavidin-coated AuNP-based sensing strategy to monitor protein phosphorylation.610
Figure 20.
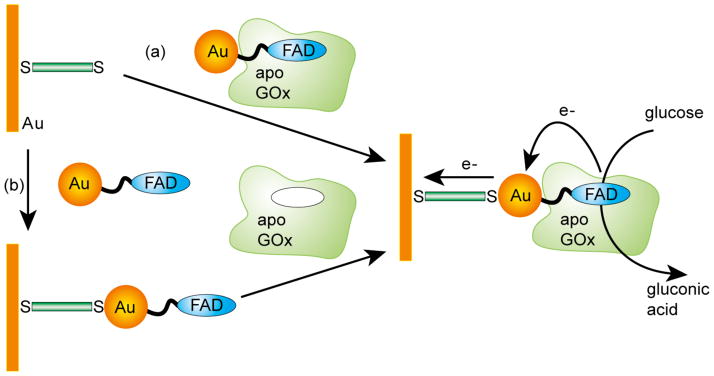
Fabrication of GOx electrode by the reconstitution of GOx on a FAD-functionalized AuNP : (a) The adsorption of AuNP-reconstituted of apo-GOx to a dithiol monolayer assembled on a gold electrode and (b) the adsorption of FAD-AuNPs to a dithiol-modified gold electrode followed by the reconstitution of apo-GOx onto functional AuNPs. Reprinted with permission from Science (Ref 608). Copyright 2003 American Association for the Advancement of Science.
Hemoglobin (Hb) has been extensively studied as a redox protein for direct electron transfer to AuNP-modified electrodes. Hb has very slow electron transfer rate to bulk electrodes, however, AuNPs can greatly enhance the electron transfer between Hb and electrodes with611 or without redox mediator.612–622 Yuan has used this system for amperometric sensing of H2O2 using Hb immobilized on multiwall carbon nanotubes (MWCNT)/AuNPs623 and onto AuNPs/MWCNT/chitosan composite matrices624 on GCE surface. Amperometric nitrite sensors have used Hb immobilized on AuNP-modified screen printed electrode625 and onto one-dimensional AuNP assemblies.626 Similarly, direct electrochemistry of myoglobin (Mb), an oxygen transporter in muscle tissues, was used to detect H2O2 using a variety of AuNP-modified electrodes.627–631 Also, an electrochemical H2O2 biosensor was created by immobilizing Mb and colloidal AuNPs onto Nafion-modified GCE. The immobilized Mb exhibited excellent electrocatalytic response to the reduction of H2O2 with a detection limit of 0.5 μM.632
Direct electron transfer to electrodes of P450 enzymes, CYP2B6633 and CYP11A1634 were applied to detect drugs and cholesterol respectively. Similarly, the electrocatalytic activity of cytochrome c immobilized on AuNP-modified electrodes635,636,637 was used to sense H2O2. LBL assembly methods based on electrostatic interaction have been employed to interface redox proteins by facilitating direct electron transfer.638,639 For example, multilayer films of GOx/AuNPs on gold electrodes using cysteamine as a covalent cross-linker were prepared by LBL technique. The bioelectrocatalytic response was directly correlated to the number of deposited bilayers.640 Chen et al. have reported a H2O2 biosensor following the similar approach based on HRP immobilization on an LBL assembly of films of AuNPs and toluidine blue that responded rapidly to H2O2 with a detection limit of 70 nM.641 Zhu and coworkers have fabricated the composite C@SiO2 with AuNPs (AuNPs–C@SiO2) by LBL assembly technique to sense H2O2.642 Cobalt hexacyanoferrate-modified AuNPs were alternated with poly(vinylsulfonic acid) layers on indium tin oxide (ITO) electrodes and used as a platform for immobilization of GOx in the presence of bovine serum albumin (BSA) using glutaraldehyde as a cross-linker. This hybrid electrode successfully measured amperometric response of glucose at 0.0 V vs saturated calomel electrode (SCE).643
6.4.2. Enzyme Biosensors using AuNPs Composite Electrode Matrices
The incorporation of nanomaterials into composite electrode matrices presents another approach to enzyme biosensors, providing low background currents, straightforward surface generation, and the ability to incorporate of different substances into the bulk electrode matrix. AuNP-based composite electrode matrices have been used to detect phenol,461 hypoxanthine,644 H2O2,645,646 atenolol,647 glucose,648 etc. Similarly, tyrosinase biosensors consisting of composite graphite-Teflon electrodes modified with AuNPs have been developed by Pingarrón et al. to detect different alkyl- and chlorophenols. The presence of AuNPs in the composite matrix increased the kinetics of the enzyme reaction and the electrochemical reduction of o-quinones at the electrodes, thus allowing nanomolar detection of phenolic compounds.649
AuNPs are often conjugated to other nanomaterials to improve their binding efficiency on electrode matrices.650–653 Electrodes fetauring AuNPs conjugated with carbon nanotubes (CNT) provide excellent electrocatalytic ability654 enabling electrochemical biosensors for detection of glucose,655–659 cytochrome c,660 tryptophan,661 hydroxylamine,662 bisphenol A,663 etc. For example, an electrochemical sensor platform was constructed by covalent integration of AuNPs and CNTs onto a poly(thionine) modified GCE. Using HRP, the synergistic effect of the combined matrix in the presence of redox polymer mediator provided faster electron transfer and higher enzyme immobilization efficiency for detection of H2O2..664
6.4.3. AuNP/Polymer Matrices for Novel Electrochemical Biosensors
Electropolymerization provides a strategy for biomolecule immobilization on the electrode surfaces in the presence of AuNPs. For example, Au-polypyrrole,665 Au-polyaniline,666 and AuNP-(3-mercaptopropyl)-trimethoxysilane (MPS) nanocomposite667 bioelectrodes have been fabricated to detect glucose as an analyte. AuNPs were assembled onto an AgCl@polyaniline nanocomposite-modified GCE, providing an amperometric glucose biosensor based on GOx. The hybrid electrode system showed superior electrocatalytic activity and reproducibility, detecting glucose at 4 pM.668 Similarly, an AuNP/polyaniline/AgCl/gelatin matrix has been successfully used for glucose biosensing.669 A H2O2 biosensor has been created by electropolymerization of p-aminobenzene sulfonic acid using cyclic voltammetry. In this approach, AuNPs were assembled in an interface containing amine groups of thionine followed by HRP adsorption and the resulting biosensor responded to H2O2 with a detection limit 0.64 μM.670 Zhu et al. have reported the use of an AuNP/nafion/polythionine/gelatin matrix on Pt disk electrodes to immobilize HRP for H2O2 sensing.671 Recently, a surface molecular self-assembly strategy for molecular imprinting in electropolymerized polyaminothiophenol (PATP) membranes at the surface of AuNPs modified GCE was reported for the electrochemical detection of the pesticide chlorpyrifos (CPF). 672
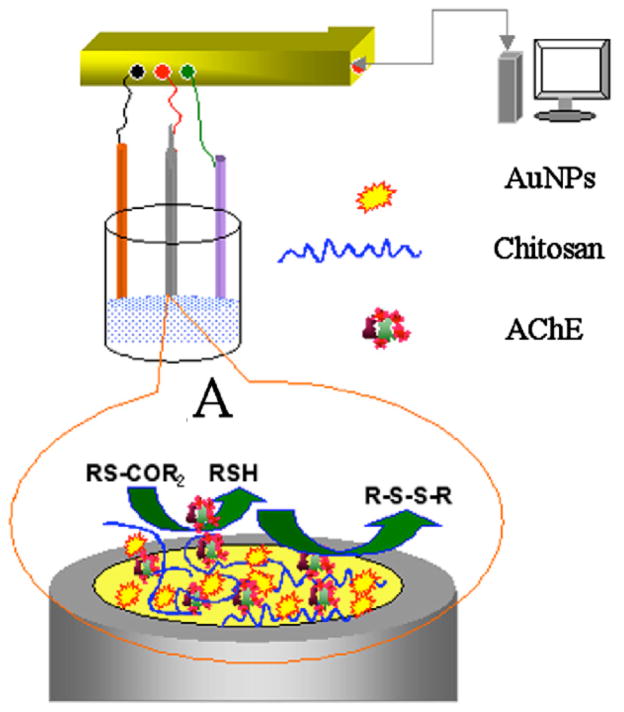
Several groups have used the biopolymer chitosan to immobilize enzymes due to its biocompatibility and high mechanical strength. Using this approach, amperometric biosensors have been fabricated including GOx673–678 to detect glucose, HRP598,679–685 to detect H2O2, tyrosinase686 to detct phenolic compounds, acetylcholinesterase (AChE) to measure drug sensitivity687 or malathion activity688 were developed. A novel in situ interfacing of AuNPs with a chitosan hydrogel was achieved by one-step electrochemical deposition of tetrachloroauric (III) acid and chitosan on gold electrodes (Figure 21).689 The deposited interface showed excellent biocompatibility and stability. With AChE as a model enzyme, rapid amperometric sensing of the pesticides malathion and monocrotophos was achieved with a detection limit of 1 ng/mL.
Figure 21.
Acetylcholinesterase enzyme biosensor constructs using AuNPs/chitosan hydrogel matrices for the detection of pestisides. A) Megascopic interface of AChE/Chitosan–AuNP modified gold electrode. Reprinted with permission from J. Electroanal. Chem. (Ref 689). Copyright 2007 Elsevier B.V.
6.5. AuNP-based Electrochemical Detection of Oligonucleotides
The unique electronic/electrochemical properties of AuNPs provide an alternative platform to optical sensing of oligonucleotides,690–698 providing an efficient tool for immobilizing DNA on electrodes699 as well as a label to signal the hybridization event.700–702 For example, an oligonucleotide with a mercaptohexyl group at the 5 -phosphate end was attached onto an AuNP-modified gold electrode, increasing nucleic acid loading efficiency to 1 × 1014 molecules/cm2, ~10 times higher than a bare gold electrode.703 A variety of AuNP-based DNA sensing strategies (Figure 22) have been developed, including AuNP dissolution by acid,704–708 direct detection of AuNP/DNA conjugates anchored onto sensor surfaces,709–711 and AuNPs as carriers for other electroactive labels.712–717 Sensing enhancement by precipitation of silver718–726 and gold727 onto AuNPs labels have been used to achieve amplified signals. Recently, Bonanni et al. have used streptavidin-coated AuNPs to provide impedimetric signal amplification for detecting DNA hybridization events. The probe oligomer was adsorbed on a graphite/epoxy composite electrode and the impedance measurement was performed using a ferrocyanide/ferricyanide redox marker. The coating with streptavidin favoured the rapid formation of conjugates with biotinylated target DNA hybrid with a limit of detection 11.8 pM.728
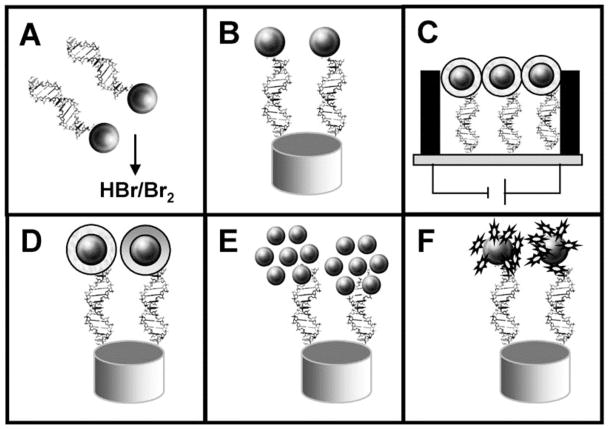
Figure 22.
Schematic procedure of the different strategies used for the integration of AuNPs into DNA sensing systems: (A) previous dissolving of AuNPs by using HBr/Br2 mixture followed by Au(III) ions detection, (B) direct detection of AuNPs anchored onto the surface of the genosensor, (C) conductometric detection, (D) enhancement with silver or gold followed by detection, (E) AuNPs as carriers of other AuNPs, (F) AuNPs as carriers of other electroactive labels. Reprinted with permission from Electroanalysis (Ref 691). Copyright 2007 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim.
DNA sensing has likewise been performed using AuNP-DNA conjugates on GCEs using methylene blue (MB) as an electroactive label and differential pulse voltammetry (DPV). The resulting system enhanced the response signal during immobilization and hybridization by increasing the density of redox active sites.729 Fang and coworkers have immobilized an oligonucleotide with a mercaptohexyl group at the 5 -phosphate end onto 16 nm AuNPs self-assembled on a cystamine-modified gold electrode. The modified electrode immobilized 10-fold greater quantities of ssDNA than planar gold electrodes.730
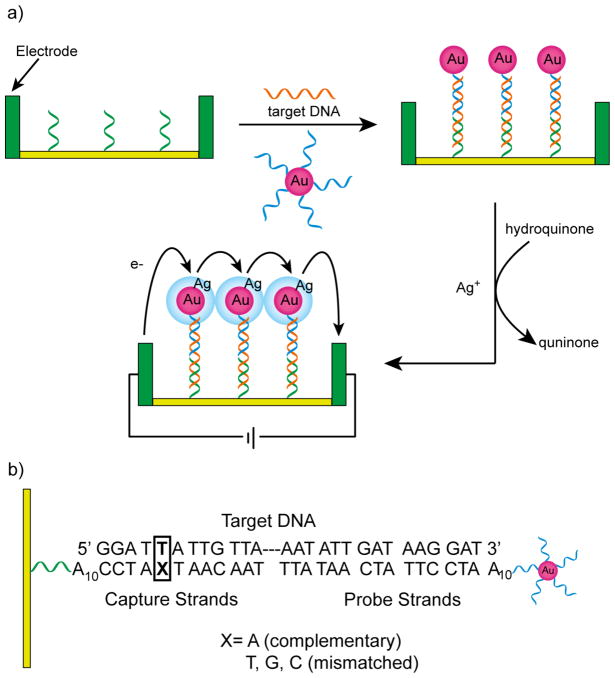
AuNPs can change the conductivity across a microelectrode gap, providing highly sensitive electronic detection of DNA hybridization.731,732 Mirkin and coworkers have reported an electronic DNA detection method where a short capture oligonucleotide was immobilized between electrodes in a microelectrode array with 20 μm gaps. Using a three-component sandwich approach, hybridized target DNA and AuNPs functionalized with oligonucleotides were bound between the electrodes leads followed by silver deposition onto AuNPs to enhance conductivity (Figure 23). Using this method, a sensitivity of 500 fM has been achieved with a point mutation selectivity factor of 105:1 in target DNA.732 Following the same principle, Urban et al. have studied the changes of resistance across the microelectrode gap resulting from AuNP-labeled DNA in a parallel array readout system.733 Additionally Diessel and coworkers have further demonstrated the utility of this strategy for the detection of single-nucleotide polymorphism.734
Figure 23.
a) Schematic illustration of electrical detection of DNA based on “sandwich” hybridization with DNA functionalized AuNPs followed by silver enhancement. b) Sequences of capture, target, and probe DNA strands. Reproduced with permission from Science (Ref 732). Copyright 2002 American Association for the Advancement of Science.
The redox properties of AuNPs have been used as electrochemical labels for DNA detection. For example, Ozsoz et al. have developed a sandwich-type electrochemical genosensor where the labeled target was captured by probe strands immobilized onto a graphite electrode and hybridization detected by electrochemical gold oxidation.735 The response is enhanced due to the many oxidizable gold atoms in each nanoparticle label, with the detection limit as low as 0.78 fmol DNA for PCR amplicons. Similarly, another design was used to determine a specific binding event between E. coli ssDNA binding protein (SSB) and single-stranded oligonucleotides conjugated to AuNPs. SSB was adhered onto a SAM of single-stranded oligonucleotide modified AuNPs, and the resulting Au-tagged SSB was used as the hybridization label. Binding of Au tagged SSB to the probe was monitored through Au oxidation signal change, providing a detection limit of 2.17 pM target DNA.736 Willner et. al. reported an amplified electrochemical DNA detection method using aggregation of AuNPs on electrodes.737 In their sensor, MB acts as a specific electrochemical indicator for the dsDNA aggregation of the AuNPs, with the AuNP assemblies facilitating the electrical communication of the intercalated MB with the electrodes with a detection limit of 0.1 pM for a 27-mer DNA. More recently, a new strategy for the label-free electrochemical DNA detection has been developed using an AuNP/poly(neutral red) modified electrode that transduces hybridization through current decrease, with a detection limit of 4.2 pM.738
In addition to DNA recognition, AuNP-DNA-modified electrodes have been used to study the interaction of DNA with small molecules. For example, a DNA-modified electrode was fabricated by self-assembling (3-Aminopropyl) trimethoxysilane and AuNPs and electrochemically immobilizing DNA onto ITO electrode. This DNA-modified electrode was used to detect the synthetic abortifacient, mifepristone with a detection limit of 200 nM.739 Likewise, interfacial interactions between immobilized DNA probes and DNA binding drugs were investigated using impedance spectroscopy. Use of AuNP functionalized substrates resulted in detection limits of 5 nM for nogalamycin, 10 nM for mythramycin, and 40 nM for netropsin, 15–40-fold lower compared to flat gold substrates.740 A norepinephrine biosensor was designed using an electrodeposited DNA membrane doped with AuNPs with a detection limit of 5 nM.741
6.6 AuNP-based Electrochemical Immunosensors
Electrochemical immunosensors based on AuNPs enhance the electrochemical signal transduction of the binding event between antigen and antibody, providing a better surface for maintaining immunoreagent stability upon immobilization.742
6.6.1. Detection of Viral Surface Antigens & Others
Antibody immobilization onto AuNP-modified electrodes has been used to detect Hepatitis B virus surface antigen (HBsAg) with detection limits of 50 ng/L743 and 7.8 ng/mL.744 Similarly, HBsAg detection was performed by electrostatic adsorption of the corresponding antibody onto AuNP/tris(2,2-bipyridyl)cobalt (III) multilayer films,745 AuNPs entrapped on polyvinyl butyral/nafion film coated PtE surface,746 cysteamine and AuNP modified gold electrodes.747 Other studies have used a nanoporous gold electrode with HRP labeled secondary antibody–AuNPs bioconjugates,748 by self-assembling AuNPs to a thiol-containing sol-gel network and assembling hepatitis B surface antibody on to the surface of AuNPs749,750 and by immobilizing hepatitis B surface antibody on a platinum disk electrode based on an AuNP/nafion/gelatin matrix.751 An alternate strategy involving AuNPs and HRP modified gold electrode was used to build an amperometric immunosensor for HBsAg (Figure 24). The sensor was comparable to a BSA-blocked immunosensor in terms of linearity and sensitivity and detected HBsAg as low as 0.85 ng/mL.752 Recently, a label-free amperometric immunosensor based on chitosan-branched ferrocene (CS-Fc) and AuNPs was developed to detect HBsAg. The decrease of amperometric signal corresponding to specific antigen-antibody binding was proportional to HBsAg concentration with a detection limit 0.016 ng/mL.753 A copper-enhanced AuNP tag provided improved electrochemical performance for detecting HBsAg with a detection limit of 87 pg/mL.754
Figure 24.
Schematic illustration of the stepwise immunosensor fabrication process to detect HBsAg: (a) formation of nafion monolayer; (b) adsorption of thionine; (c) formation of AuNP-monolayer; (d) HBsAb loading; (e) blocking with HRP. Reprinted with permission from Anal. Chim. Acta (Ref 752). Copyright 2005 Elsevier B.V.
Diphtheria antigen was detected through diphtheria antibody immobilization on PtE modified with AuNPs755,756 and silica/silver/AuNPs757 in polyvinyl butyral matrices. A modified approach using a mixture of AuNPs and silica nanoparticles provided higher sensitivity, better reproducibility, and long-term stability of the system.758 A label-free amperometric immunosensor based on multi-layer assembly of polymerized o-phenylenediamine and AuNPs was used to detect Japanese B encephalitis vaccine with a detection limit of 6×10−9 log pfu/mL (pfu/mL is plaque forming unit).759
Electrochemical AuNP-based immunosensors have been used to detect a wide array of small molecule analytes, including carcinogenic substances (Aflatoxin B1,760,761 Ochratoxin A,762,763 naphthalene,764 paraoxon),765 herbicides (atrazine,766,767 picloram768), and hormones (human chorionic gonadotrophin hormone (pregnancy test marker),769–774 progesterone,775,776 phytohormone abscisic acid,777 17β-estradiol,778,779 human growth hormone780). Furthermore, detection of different proteins such as transferrin,781,782 human serum albumin,783,784 thrombin,785 hemoglobin,786 protein A,787 and apolipoprotein A-I788 has also been successfully achieved.
6.6.2. Detection of Biomarkers for Cancer and Other Disease States
α-Fetoprotein (AFP) is a tumor-marker for metastatic cancer. Several groups have used AuNP-based immunosensors to detect AFP by immobilizing AFP antibodies onto AuNP-decorated thionine/nafion membranes,789,790 through electro-deposition of AuNPs and prussian blue on ITO electrode,791 using carbon paste electrode constructed with an ionic liquid and AuNPs,792 with AFP-antibody functionalized AuNPs,793 using chitosan-AuNP composite film on a GCE,794 using films of MWCNT/DNA/thionine/AuNPs,795 using 1,1′-bis-(2-mercapto)-4,4′-bipyridinium dibromide functionalized AuNPs onto GCE,796 and by immobilizing AFP-antigen onto GCE modified with AuNPs and a CNT doped chitosan film.797 AuNPs and enzyme amplification have been applied to develop an immunosensor to detect AFP based microelectrodes and microwells systems fabricated by SU-8 photoresist on silicon wafer with a detection limit of 5 ng/mL.798
Another biomarker, carcinoma antigen 125 (CA125) was electrochemically detected by immobilizing anti-CA125 on a thionine and AuNP-modified carbon paste electrode.799 Anti-CA125 was later immobilized on AuNPs stabilized with a cellulose acetate membrane on a GCE surface. Using o-phenylenediamine and H2O2 as enzyme substrates, the sensor provided a competitive immunoassay format to detect CA125, with HRP-labeled CA125 antibody as a tracer.800 Similarly, various antigens were successfully detected by several groups using AuNP-based immunosensors, e.g. carbohydrate antigen 19-9,801 carbohydrate antigen 125,802 prostate-specific antigen,803–807 and mammary cancer 15-3 antigen.808
A wide range of additional biomarkers have been detected besides cancer biomarkers, including cholera toxin,809 interleukin-6,810–812 vascular endothelial growth factors,813 Annexin II and MUC5AC antigens,814 Schistosoma japonicum antigen,815,816 Salmonella typhi antigen,817,818 foodborne pathogen Escherichia coli O157:H7,819 osteoprotegerin,820 protein markers for acute myocardial infarction (AMI) e.g. myoglobin, cardiac troponin complex and the MB isoform of the enzyme creatine kinase,821 dust mite allergen Der f2,822 carcinoembryonic antigen (CEA),823–837 etc. Recently, Ho et al. have reported an AuNP-based electrochemical sandwich immunosensor for α-Enolase (ENO1) antigen, a potential diagnostic marker for lung cancer. The immunosensor operates through physisorption of anti-ENO1 monoclonal antibody on polyethylene glycol-modified disposable screen-printed electrodes followed by using anti-ENO1-tagged AuNP congregate bioprobes as electrochemical signal probes (Figure 25) with a detection limit as low as 11.9 fg (equivalent to 5 μL of a 2.38 pg/mL solution).838 Recently, Ju and coworkers have developed a disposable reagentless electrochemical immunoassay array for multianalyte determination by immobilizing AuNPs modified with HRP-labeled antibodies on screen printed electrodes. Chitosan/sol-gel was used to trap the corresponding antigens of the analytes from the sample solutions.839,840 Upon formation of immunocomplex, the direct electrochemical signal of the HRP decreased due to increasing steric blocking.
Figure 25.
Schematic representation of the operation of the electrochemical immunosensor for the detection of ENO1. Reprinted with permission from Anal. Chem. (Ref 838). Copyright 2010 American Chemical Society.
6.6.3. Detection of Immunoglobulins
Immunoglobulins are important biomarkers for a wide variety of disease states. Numerous examples of determining human immunoglobulin G (IgG)841–856 as well as immunoglobulin E (IgE)857 via AuNP-based electrochemical immunosensors can be found in literature. For example, Velev and Kaler designed a conductivity immunoassay system that used silver metallization to enhance sensitivity.858 Mao et al. have reported an electrochemical method based on copper precipitation onto AuNP tags to detect human IgG and anodic stripping voltammetry (ASV) reaching a detection limit of 0.5 ng/mL.859 Pioneering work using AuNPs as electrochemical labels for voltammetric detection of proteins were performed by Costa-García et al.860,861 and Limoges et al.862 For example, Limoges and coworkers reported an electrochemical immunoassay for IgG using AuNP-labeled antibodies and ASV. In this approach, the AuNP-labeled antibody forms sandwich complexes with the goat IgG target and the immobilized antibody. After removal of the unbound labeled antibody, AuNPs were dissolved in an acidic bromine-bromide solution to release gold ions that were electrochemically detected, providing an IgG detection limit of 3 pM,862 competitive with ELISA. Likewise, human IgG has been detected by ASV using cyclic accumulation of AuNPs. In this sensor, the probe antibody in the sandwich complexes is labeled with dethiobiotin and avidin-AuNPs. The alternating treatment of the system with biotin solution and avidin-AuNPs resulted in cyclic accumulation of AuNPs. The detection limit of this method was 0.1 ng/mL of human IgG.863 Recently, Merkoçi and coworkers reported an electrocatalytic silver-enhanced metalloimmunoassay using AuNPs as labels and microparamagnetic beads (MBs) as platforms for primary antibody immobilization to detect human IgG with a very low detection limit of 23 fg/mL (Figure 26).864 Finally, based on the catalytic effect of AuNPs on the electroreduction of silver ions, Huang and coworkers have reported the sensitivity enhancement for an electrochemical immunoassay by the autocatalytic deposition of Au3+ onto AuNPs. By coupling the autocatalytic deposition with square-wave stripping voltammetry, the model analyte rabbit IgG could be determined quantitatively with a detection limit 1.6 fM.865
Figure 26.
(A) AuNP conjugation with anti-human IgG; (B) analytical procedure for the sandwich type assay and the obtaining of the analytical signal based on the catalytic effect of AuNPs on the silver electrodeposition. Reprinted with permission from Biosens. Bioelectron. (Ref 864). Copyright 2009 Elsevier B.V
The rather analogous mouse IgG was identified with a detection limit of 1 ng/mL using a reagentless amperometric immunosensor based on AuNPs on a nafion modified GCE with 3,3′,5,5′-tetramethylbenzidine (TMB) as redox mediator.866 A polymer-membrane based potentiometric IgG sensing method using silver enlargement of AuNPs tracers was reported by Chumbimuni-Torres et al. to detect mouse IgG with a detection limit of ~12.5 pmol in the 50 μL sample.867 A novel DNA-free ultrasensitive sandwich-type heterogeneous electrochemical immunosensor using AuNP nanocatalyst labels was reported by Das et al.868 As illustrated in Figure 27a partially ferrocenyltethered dendrimer (Fc-D) was immobilized to the ITO electrode by covalent bonding between dendrimer amines and carboxylic acids of a phosphonate self-assembled monolayer. Unreacted amines of Fc-D were modified with biotin groups to allow the specific binding of streptavidin. Then, biotinylated antibodies were immobilized to the streptavidin-modified ITO electrode. An IgG-nanocatalyst conjugate was also prepared via direct adsorption of IgG on 10 nm AuNPs. Mouse IgG and prostate specific antigen (PSA) were chosen as target analytes (Figure 27b).868 The signal amplification was achieved by the catalytic reduction of p-nitrophenol (NP) to p-aminophenol (AP) using gold AuNP catalyst labels as well as through the NaBH4 mediated chemical reduction of p-quinone imine (QI). This DNA-free method can detect 1 fg/mL of PSA, comparable to that of the bio-barcode assay.869 An extension of this model using magnetic beads for easy magnetic separation and immunoreactions leads to greater signal amplification by AuNPs, detecting mouse IgG as low as 7 aM using CV and 0.7 aM using DPV measurements.870
Figure 27.
(a) Schematic representation of the preparation of an immunosensing layer. (b) Schematic view of electrochemical detection of mouse IgG or PSA. Reprinted with permission from J. Am. Chem. Soc. (Ref 868). Copyright 2006 American Chemical Society.
7. AuNP-based Surface Plasmon Resonance Sensors
The interaction of light at the surface of the noble metal film excites surface electromagnetic waves and sets them to resonate with incident light wave, resulting in the absorption of the light. This phenomenon is known as surface plasmon resonance (SPR)871,872 and depends on the refractive index of the interfacial region. Metal nanoparticles such as gold227,873–875 and silver876,877 exhibit localized surface plasmon resonance (LSPR) at specific incident wavelengths, generating strong light scattering and the appearance of intense surface plasmon absorption bands. The intensity and the frequency of the absorption band is a characteristic of the particular metal nanoparticles and highly dependent on their size, shape as well as the surrounding environment.878–882 Using this phenomenon many LSPR-based chemical and biological sensors have been developed.14,309,883–895 Although numerous biosensors have been developed using silver nanoparticles896–903 we will focus on AuNP based sensors.894,904–915
7.1. Sensors based on Change in LSPR Absorption of AuNPs
The general principle behind LSPR-based sensors is the wavelength shift in the LSPR spectrum arising from local dielectric changes caused by analyte adsorption. LSPR assays have been conducted both in solution phase916–918 and on surfaces coated with nanoparticle monolayers.919–922 In examples of the former, absorption maxima of LSPR was red-shifted when AuNPs functionalized with monoclonal antibodies interacted with analytes.923,924 Moreover, the wavelength shift was found to be proportional to the amount of ligands.924
Most AuNP-based SPR sensors, however, have been fabricated by immobilizing nanoparticles onto surface.925,926 The introduction of AuNPs onto the sensing surface provides an effective way to increase the sensitivity of SPR sensors owing to the high dielectric constants of AuNPs and the electromagnetic coupling between AuNPs and the metal film on the surface.927 For example, a gold film-coated chip was used to detect dopamine in nanomolar concentration by immobilizing an MIP gel with embedded AuNPs.928 Various substrates such as quartz, optical fibers, ITO glass, sol-gel matrix, etc. have been used for supports for AuNPs, allowing the detection of numerous analytes such as human serum albumin,929 BSA,930,931 human IgG,932 streptavidin,933 interleukin-1β,934 propanethiol,935 etc. Recently, AuNPs encapsulated by hydroxyl/thiol-functionalized fourth generation PAMAM dendrimer were immobilized onto maleimide terminated SAMs to detect insulin.936 The resulting AuNP modified dendrimer surface provided high stability and enhanced sensitivity with a detection limit of 0.5 pM. The sensor was further testified by analyzing human serum samples from normal and diabetic patients with good correlation to standard methods.936
The aggregation of AuNPs leads to an alteration in surface absorption band that gives rise to a visible color change. Exploting this principle, Mirkin et al. have developed a colorimetric sensor for DNA hybridization assay using oligonucletide functionalized AuNPs both in dispersions as well as on surface (See Section 4.4). Other SPR based sensors using aggregation of AuNPs have been reported to detect proteins (via antigen-antibody or biotin streptavidin interaction),354,937,938 lectin,331,338 etc.
7.2. AuNP-mediated SPR Signal Amplification
AuNPs have been used to enhance the signals of the propagating SPR spectroscopic signals to increase sensor sensitivity.939–943 The signal amplification was explained by the electronic coupling interaction of the propagating surface plasmons with localized surface plasmons of AuNPs944,945 and depends on various factors such as size, shape, and the distance from the metal generating SPR.946,947
7.2.1. Sensing of Proteins
Protein sensing through antigen-antibody interaction can be detected exploting AuNP-amplified SPR phenomena. For example, Natan et al. have reported an AuNP-enhanced SPR immunosensing system using either antigen or secondary antibody functionalized AuNPs as signal enhancers.904 In an example of this sandwich strategy, a gold film coated with γ-chain and Fc specific monoclonal goat antihuman IgG (α-h-IgG(Fc)) generates a small plasmon shift upon addition of human IgG and the second free antibody. The plasmon shift, however, increases 28-fold times compared to an unamplified assay when the secondary free antibody is replaced by an electrostatic conjugate between AuNPs and α-h-IgG(Fc). Using this method picomolar detection of human IgG has been achieved. Similarly, several competitive and sandwich immunoassays have been developed using AuNP-enhanced SPR signals to detect human tissue inhibitor of metalloproteinases-2,948 anti-glutamic acid decarboxylase antibody,949 allergen,950 TNT,951 human IgE,952 and testosterone.953 The sensitivity of these assays can be enhanced using fluorescence-labeled antibodies decorated with AuNPs, leading to the technique called localized surface plasmon resonance coupled fluorescence fiber optic sensor.954–957
7.2.2. Sensing of Oligonucleotides
The sensitivity of oligonucleotide detection can be improved by using AuNP-amplified SPR.958,959 Keating et al. have developed a sandwich approach where 12-mer oligonucleotides were first linked covalently onto a gold substrate followed by hybridization of one half of the target DNA molecules. Then, a sequence complementary to the other half of the target was added with or without tagging of AuNPs. The AuNP-tagged surface demonstrated a 10-fold increase in angle shift concomitant with a 1000-fold improvement in sensitivity and a ~10 pM detection limit for the target 24-mer oligonucleotide.960 For example, Zhou et al. have shown that an intermediate carboxylated dextran layer between gold film and the immobilized DNA molecules effectively eliminates the nonspecific adsorption of oligonucleotide functionalized AuNPs, resulting femtomolar level of detection for 39-mer DNA (Figure 28).961 In a representative study, real time multicolor DNA detection has been achieved exploiting AuNP-amplified diffraction, where ssDNA-modified AuNPs and micropatterned chemoresponsive diffraction gratings were used to interrogate simultaneously at multiple laser wavelengths.962
Figure 28.
Schematic representation of sandwich DNA detection assay via AuNP-mediated SPR signal amplification. The SPR measurements were carried out by injecting the oligonucleotide functionalized AuNPs into the flow cell housing sensors covered with various duplexes or capture probes. The intermediate dextran layer reduces the nonspecific adsorption of AuNPs, improving detection sensitivity. Adapted from Ref 961.
7.3. Sensors based on AuNP Plasmon Resonance Scattering
In addition to changes in LSPR absorption, the plasmon resonance scattering phenomena of AuNPs provides a useful tool for sensor design.197,963 The plasmon scattering of 36 nm diameter AuNPs is 10–100 times stronger than dyes or quantum dots. Exploiting this nanoscale phenomena, several groups have developed immunoassays to detect human IgG,964 kanamycin,965 and lysozyme in human urine.966 For example, Ren and coworkers have developed a highly selective and sensitive homogeneous immunoassay and DNA hybridization assay using plasmon scattering of single AuNP probe. The sandwich immunoassay was used to detect cancer biomarkers such as CEA, AFP in femtomolar range, and aptamer recognition for thrombin as low as 2.72 pM.967 Recently, Ling et al. have reported an LSPR light scattering sensor for Ag+ with unmodified AuNPs exploiting the specific recognition property of Ag+ with a cytosine-cytosine mismatch base pair. The addition of Ag+ removes the oligonucleotide from the AuNP surface causing aggregation concomitant with dramatic increment of LSPR scattering intensity. The LSPR light scattering intensity was proportional to concentration of Ag+, with a limit of detection of 62 nM.968 El Sayed et al. have demonstrated a biosensor technique using SPR scattering images and SPR absorption spectra from anti-EGFR functionalized AuNPs for the diagnosis of oral epithelial cancer cells in vitro.969 The anti-EGFR functionalized AuNPs bind 600% stronger to oral malignant cells HOC 313 clone 8 and HSC 3 than normal cell HaCaT, resulting in a sharper SPR absorption band with a red shift (Figure 29).
Figure 29.
SPR light scattering images to distinguish between normal cells (left panel, HaCaT) and cancerous cells (middle & right panel, HOC and HSC) after incubation with anti-EGFR conjugated AuNPs. The anti-EGFR conjugated AuNPs bind specifically to the surface of cancer cells resulting in a sharper SPR band with a red shifted maxima. Reproduced with permission from Nano Lett. (Ref 969). Copyright 2005 American Chemical Society.
8. Surface Enhanced Raman Scattering (SERS)-based Sensing
The physical phenomenon behind the Raman spectroscopy is the inelastic scattering of photons by a molecule having quantized vibrational level/signature.970 Raman scattering is sensitive to different vibrational modes and consequently can provide a “fingerprint” of the target molecules.971 However, the direct application of this technique in sensitive detection and identification of analyte molecules is severely restricted owing to the low efficiency of inelastic photon scattering by molecules leading to a weak signal.972 The inherent limitation of low scattering intensity arises from the fact that the Raman scattering cross-sections for molecules are usually small, typically 10−30–10−25 cm2/molecule, 10 to 15 orders of magnitude smaller than that of fluorescence cross section. In the presence of plasmonic nanoparticles or rough metal surfaces, however, the Raman scattering intensity from a molecule can be enhanced by up to 1014 order of magnitudes.973–977 This phenomenon has been attributed to a local electromagnetic field enhancement induced by the plasmon resonance of surfaces and is called surface enhanced Raman scattering (SERS).978–981 The field enhancement is dependent on the size, shape, orientation, and aggregation of the nanoparticle. The large enhancement in detectable signal coupled with the unique molecular fingerprints generated has made SERS a powerful tool for the multiplex detection of analytes, with the ability to achieve a detection of single molecule level.976,977
8.1. Detection of Small Organic Molecules
AuNPs have been utilized for the SERS based detection of small organic molecules such as explosives.982–985 For example, Ray et al. have used AuNPs modified with cysteine as label-free SERS probe for highly selective and sensitive recognition of TNT.982 Cysteine modified AuNPs undergo aggregation in the presence of TNT due to the formation of donor-acceptor Meisenheimer complex between TNT and cysteine. The resulting ‘hot spots’ for enhancement of the Raman signal provided a sensitivity of 2 pM level. Kneipp et al. have shown that SERS nanosensor made from AuNP nanoaggregates and 4-mercaptobenzoic acid attached as a reporter can be used for monitoring changes in local pH of the cellular compartments of living cells.986
8.2. Detection of Oligonucleotides
Mirkin et al. have utilized AuNP probes labeled with oligonucleotides and Raman-active dyes for the SERS-based multiplexed detection of DNA and RNA targets (Figure 30).987 To detect the presence of specific target DNA strands, a three-component sandwich assay in a microarray format was used. For the assay, dye-labeled AuNP probes were captured by the target oligonucleotide strands, followed by silver enhancement, generating detectable SERS signals exclusively from the Raman dyes immobilized on the particles, with a limit of detection of 20 fM. More importantly, this method was able to discriminate single nucleotide polymorphisms relating to six different viruses. Irudayaraj et al. have incorporated non-fluorescent Raman tags onto DNA-conjugated AuNP (~30 nm) probes for the SERS detection of DNA.988 In their system, the surface coverage of the Raman tags on the AuNP surface has been modulated to control the intensity of Raman signal from the probes. Simultaneous identification of upto eight probes with a detection limit of ~100 nM without further metal enhancement was achieved. A DNA sensor using SERS from AuNP aggregates formed via DNA photoligation has been described by Maenosono et al.989 In their system, SERS signals arise after hybridization from the Raman-active molecules present in the NP aggregates. Liu et al. have reported a sequence-specific DNA detection using SERS spectroscopic fingerprint from thiol-oligonucleotide-modified ZnO/Au nanocomposite probes.990 Nie et al. have developed a SERS beacons using DNA conjugated AuNP probe.991 Signals from this AuNP-based SERS beacons can be turned on and off by biomolecular binding and dissociation events. Using both distance dependent SERS enhancement of the isolated AuNPs and the “hot spots” created by two conjugated AuNPs, Hu et al. have developed a novel nano-junction based biosensor for the detection of sub-attomolar HIV-1 DNA.992 Johnson et al. have demonstrated an indirect capture model assay for SERS-based detection of DNA using AuNPs.993 SERS based approach-utilizing AuNP probes for identifying and quantifying multiple gene segments extracted from cells has been described by Irudayaraj et al.994 Multiple AuNP-on-wire systems as a SERS sensing platform for viral DNA have been developed by Kang and coworkers.995 Their particle-on-wire sensor provided SERS signals only in the presence of target DNA, leading to the detection of DNA concentration ranging from 10 pM to 10 nM. Raman dye labeled AuNP probes have also been applied for SERS based in vivo and in vitro imaging.996–1001
Figure 30.
Schematic representation of three component sandwich assay for SERS-based oligonucleotide detection. Reproduced with permission from Science (Ref 987). Copyright 2002 American Association for the Advancement of Science.
8.3. Detection of Proteins
AuNPs have been widely used in SERS based immunoassays of proteins.1002–1009 For example, Grubisha et al. reported an immunoassay using 30 nm AuNPs functionalized with a monolayer of an intrinsically strong Raman scatterer (5-thiol-2-nitrobenzoate) followed by a layer of covalently linked antibodies.1004 In this system, a sandwich assay format based on monoclonal antibodies has been used for the detection of free PSA. The sensitivity from this system has been drastically enhanced due to their unique senor design, which not only minimizes the separation between label and particle surface but also maximizes the number of labels on each particle. The detection limit of this immunoassay was ~1 pg mL−1 in human serum. Kim et al. have demonstrated a label-free detection system for avidin via avidin-mediated self-assembly between biotinylated Au nanowires and biotinylated AuNPs. The avidin induced self-assembly process created ‘hot spots’ between the nanowire and AuNPs that strongly enhanced the Raman signal.1010 An aptasensor based on the formation of SERS ‘hot spots’ between nanowires and AuNPs has also been used to detect human α-thrombin in serum.1011 Hu et al. have used AuNPs functionalized with thrombin specific aptamer for the detection of thrombin.1012 Their electrostatic interaction based assay approach provided a detection limit of 20 pM. Mirkin et al. have used AuNPs functionalized with either protein ligands or antibodies and Raman dyes to perform multiplexed screening of protein-small molecule interactions and protein-protein interactions in a microarray format.1013 Reich et al. have employed DNA-bridged AuNP assemblies for the detection of protein-DNA interactions via SERS.1014 Self-assembly of peptide functionalized AuNPs has been used for the SERS based detection of proteases.1015 Wang et al. demonstrated a microarray approach based on SERS for the detection of peptide–protein or protein–antibody interactions.1016 A SERS-based assay technique was developed by Gu et al. to detect the activity of alkaline phosphatase enzyme at ultralow concentrations.1017 SERS based rapid screening of pathogenic bacteria has also been achieved utilizing AuNPs.1018,1019
9. AuNPs in Quartz Crystal Microbalance-based Sensing
Quartz crystal microbalances (QCM) are ultrasensitive piezoelectric devices that use frequency changes on a quartz crystal resonator to monitor changes in mass arising from analyte binding. This technique has been widely used to sense chemical vapors, microorganisms, and DNA probes due to its high sensitivity and label-free detection.1020–1026 The incorporation of AuNPs into QCM-based sensing systems1027 can enhance detection sensitivity by their high surface area and by serving as a “mass enhancer” by amplifying the frequency changes.1028 On the small molecule front, AuNPs and AuNP-dendrimer composite behaving as sorptive materials have been utilized to detect chemical vapors.1029–1032
9.1. Detection of Oligonucleotides
Several groups have reported QCM-based oligonucleotide sensors incorporating AuNPs.1033 For example, Duan and coworkers have shown the introduction of AuNPs greatly enhances the immobilization capacity and the detection limit for oligonucleotide sensing using QCM.1034 In their findings, the immobilization of ~12 nm diameter AuNPs onto a gold coated QCM resulted in easier attachment of thiol containing ssDNA, resulting in higher sensitivity upon addition of target oligonucleotides. Sandwich-based approaches using ssDNA-modified AuNPs, can significantly improve the detection limit, where one end of the target oligonucleotide hybridizes with the immobilized ssDNA (recognition element) while the other end hybridizes with AuNPs (mass enhancer).1035–1043 It should be noted, however, the nonspecific adsorption of AuNPs to the gold film of QCM should be avoided to prevent the overestimation of signal amplification.1044 On the other hand, catalytic deposition of gold films onto amplifier AuNPs has been proved to enhance the sensitivity of the QCM approach for DNA detection, and a detection limit of 1 fM has been achieved.1045 Microcantilever-based DNA sensors are a parallel method that monitors mass changes on much smaller platforms. The sensor element, however, in comparison to QCM, is at least 100 times smaller, providing a high density sensor array for multiplexed detection. Using the sandwich approach together with AuNP-mediated amplification on a microcantilever, Dravid et al. obtained a detection limit of 23 pM for a 30-mer DNA.1046
9.2. Detection of Proteins
Several sandwich type assays on QCM using AuNPs have been reported for immunosensing of proteins.1047–1052 For example, detection of streptavidin on a QCM has been achieved using AuNPs as a signal amplifier.1053 The experimental set up involves the immobilization of biotinylated BSA on the gold surface of the QCM electrode. Addition of streptavidin generates a small frequency change that is amplified by incubation with biotin functionalized AuNP providing a detection limit of 1 ng/mL. A similar approach has been described by Kim et al. to detect C-reactive protein.1054 To attain further sensitivity Su et al. developed a QCM biosensor using primary AuNP-amplified sandwich immunoassay with silver enhancement technique. The AuNP-promoted silver reduction and the deposition of silver improved the detection of human IgG with two fold of magnitudes.1055 Other QCM-based biosensors have also been reported for the detection of pathogens,1056–1058 antisperm antibody,1059 human carcinoma cells,1060 lectin,1061 and dengue virus1062 exploiting analogous AuNP-enhanced signal amplification.
10. AuNP-based Bio-Barcode Assays
A multiplexed and ultrasensitive detection for proteins and nucleic acids has been developed by Mirkin et al. using an AuNP-based bio-barcode assay to amplify the target molecules.16,1063,1064 The bio-barcode assay was first utilized for identifying PSA.869 As illustrated in Figure 31a, the target of interest PSA was captured by targeting antibodies immobilized on magnetic microparticles. An AuNP encoding double-stranded barcode DNAs and PSA targeting antibodies then bound with the magnetic microparticle. After magnetic separation of the complexes, thermal dehybridization of the barcode DNAs on AuNPs was carried out to afford the single-stranded free barcode DNA and the ssDNA encoded AuNPs. The free barcode ssDNAs were analyzed by using the PCR amplification to determine the presence of PSA, providing a limit of detection of 3 aM. When the ssDNA-encoded AuNP probes were analyzed by using chip-based hybridization followed by silver amplification, PSA was detected at 30 aM.869
Figure 31.
AuNP-based bio-barcode assay of (a) proteins and (b) DNA
Exploiting the same principle, an AuNP-based bio-barcode assay to detect DNA has also been reported.1065–1067 As shown in Figure 31b specific ssDNA replaced the antibodies on magnetic microparticle in the protein detection system. Upon complementary binding with the target DNA, both magnetic microparticle and bio-barcoded AuNPs form sandwich assemblies. The magnetic separation of the sandwich assemblies followed by thermal dehybridization released the free bar-code DNA for analysis. Scanometric detection of this method provides 500 zeptomolar sensitivity, comparable to many PCR-based approaches.1066 The authors also demonstrated that the bio-barcoded amplification method can be used for identifying multiple DNA targets and bacterial genomic DNA.1068
The bio-barcode assay has been applied to detect specific targets for many diseases.1069,1070 The bio-barcode approach has also been used to measure the concentration of amyloid-β-derived diffusible ligand (ADDL), a soluble pathogenic Alzheimer’s disease marker in the cerebrospinal fluid (CSF).1071 In this work, the bio-barcode assay was used to detect ADDL in CSF at clinically relevant concentrations (< 1 pM) that were not detectable by conventional ELISA or blotting assay. Mirkin et al. also reported a bio-barcode assay for detecting human telomerase, a biomarker for cancer diagnosis.1072,1073 Recently, AuNP-based bio-barcode assay has been conducted to detect PSA in the serum of patients who have experienced radical prostatectomy for prostate cancer.1074 The assay is ~ 300 times more sensitive than commercial immunoassays.
The AuNP-based bio-barcode assay has been employed for multiplexed detection of protein cancer markers by using a mixture of different bio-barcoded AuNP probes.1075,1076 During the detection process, the released barcode DNA molecules can be specifically immobilized onto DNA functionalized surfaces followed by hybridization with ssDNA-AuNP and silver amplification. The multiplexed barcode assay can detect the target markers at low-femtomolar concentrations in serum.
An analogous fluorophore labeled bio-barcode amplification method simplifies the detection process of the previously reported bio-barcode AuNPs assay by eliminating the requirement for barcode sorting.1077 Quantification of the assay is analyzed by measuring the fluorescence intensity of the released fluorophore-labeled barcode DNA. When the assay was tested with PSA, a detection limit of 300 aM was obtained without enzymatic amplification or microarray analysis. Wolff et al. have reported fluorescent DNA barcode-based assay for detecting avian influenza virus with PCR-like sensitivity.1078
A colorimetric bio-barcode amplification assay has been developed by Groves et al. for the protein detection.1079,1080 In this system, the released barcode DNAs serve as a bridging agent for two ssDNA-functionalized AuNPs, inducing aggregation of the AuNPs and consequent red-to-blue color change to detect target molecules. The authors showed that cytokine, a biomarker for immunodeficiency-related diseases was detected upto 30 aM concentration, three orders of magnitude more sensitive than conventional cytokine detection assays.
11. Concluding Remarks
The physical properties and ease of synthesis and functionalization make AuNPs a versatile platform for chemical and biological sensors. The ease of functionalization provides facile attachment of both specific and selective recognition elements. The optoelectronic and physical properties of AuNPs then enable a diverse array of transduction processes. Advanced nanodiagnostic techniques have paved the way for affording easy, rapid, low-cost, and multiplexed identification of biomarkers. However, optimization of parameters for the sensing systems is still required to meet the demands of clinical diagnostics. In particular, the development of efficient sensors to detect analytes in complex biological fluids such as urine, serum, and blood remains a challenge. Additionally, efficient detection of disease often requires multiplex analysis, an issue where the ease of functionalization can enhance both the recognition process as well as provide building blocks for further miniaturization and hence an increased number of recognition elements.. Taken together, AuNPs provide an exceptional platform for the creation of nanosensors. As described in this review, there have been numerous sensor systems fashioned to date of both fundamental and clinical importance. The fundamental advantages of AuNPs have generated an exponential increase in their use in sensing that will continue to revolutionize the field of diagnostics for years to come.
Acknowledgments
We acknowledge the support of the N.I.H. (EB012246, GM07717) and the NSF (CHE-0808945, VR), MRSEC facilities, and the Center for Hierarchical Manufacturing (DMI-0531171).
References
- 1.Diamond D. Principles of Chemical and Biological Sensors. John Wiley & Sons, Inc; New York, NY: 1998. [Google Scholar]
- 2.Sadik OA, Land WH, Wang J. Electroanalysis. 2003;15:1149–1159. [Google Scholar]
- 3.El-Sherif M, Bansal L, Yuan JM. Sensors. 2007;7:3100–3118. doi: 10.3390/s7123100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sapsford KE, Bradburne C, Detehanty JB, Medintz IL. Mater Today. 2008;11:38–49. [Google Scholar]
- 5.Burnworth M, Rowan SJ, Weder C. Chem Eur J. 2007;13:7828–7836. doi: 10.1002/chem.200700720. [DOI] [PubMed] [Google Scholar]
- 6.Paddle BM. Biosens Bioelectron. 1996;11:1079–1113. doi: 10.1016/0956-5663(96)82333-5. [DOI] [PubMed] [Google Scholar]
- 7.Russell AJ, Berberich JA, Drevon GE, Koepsel RR. Annu Rev Biomed Eng. 2003;5:1–27. doi: 10.1146/annurev.bioeng.5.121202.125602. [DOI] [PubMed] [Google Scholar]
- 8.Miranda OR, Creran B, Rotello VM. Curr Opin Chem Biol. 2010;14:728–736. doi: 10.1016/j.cbpa.2010.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De M, Ghosh PS, Rotello VM. Adv Mater. 2008;20:4225–4241. [Google Scholar]
- 10.Anslyn EV, Rotello VM. Curr Opin Chem Biol. 2010;14:683–684. doi: 10.1016/j.cbpa.2010.08.024. [DOI] [PubMed] [Google Scholar]
- 11.Cote GL, Lec RM, Pishko MV. IEEE Sens J. 2003;3:251–266. [Google Scholar]
- 12.Sanchez C, Julian B, Belleville P, Popall M. J Mater Chem. 2005;15:3559–3592. [Google Scholar]
- 13.Thomas SW, Joly GD, Swager TM. Chem Rev. 2007;107:1339–1386. doi: 10.1021/cr0501339. [DOI] [PubMed] [Google Scholar]
- 14.Anker JN, Hall WP, Lyandres O, Shah NC, Zhao J, Van Duyne RP. Nat Mater. 2008;7:442–453. doi: 10.1038/nmat2162. [DOI] [PubMed] [Google Scholar]
- 15.Shipway AN, Katz E, Willner I. Chemphyschem. 2000;1:18–52. doi: 10.1002/1439-7641(20000804)1:1<18::AID-CPHC18>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 16.Rosi NL, Mirkin CA. Chem Rev. 2005;105:1547–1562. doi: 10.1021/cr030067f. [DOI] [PubMed] [Google Scholar]
- 17.Agasti SS, Rana S, Park MH, Kim CK, You CC, Rotello VM. Adv Drug Delivery Rev. 2010;62:316–328. doi: 10.1016/j.addr.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alivisatos P. Nat Biotechnol. 2004;22:47–52. doi: 10.1038/nbt927. [DOI] [PubMed] [Google Scholar]
- 19.Asefa T, Duncan CT, Sharma KK. Analyst. 2009;134:1980–1990. doi: 10.1039/b911965p. [DOI] [PubMed] [Google Scholar]
- 20.Cheng MMC, Cuda G, Bunimovich YL, Gaspari M, Heath JR, Hill HD, Mirkin CA, Nijdam AJ, Terracciano R, Thundat T, Ferrari M. Curr Opin Chem Biol. 2006;10:11–19. doi: 10.1016/j.cbpa.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 21.Pandey P, Datta M, Malhotra BD. Anal Lett. 2008;41:159–209. [Google Scholar]
- 22.Parak WJ, Gerion D, Pellegrino T, Zanchet D, Micheel C, Williams SC, Boudreau R, Le Gros MA, Larabell CA, Alivisatos AP. Nanotechnology. 2003;14:R15–R27. [Google Scholar]
- 23.Niemeyer CM. Angew Chem Int Ed. 2001;40:4128–4158. doi: 10.1002/1521-3773(20011119)40:22<4128::AID-ANIE4128>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 24.Tansil NC, Gao ZQ. Nano Today. 2006;1:28–37. [Google Scholar]
- 25.Welser K, Adsley R, Moore BM, Chan WC, Aylott JW. Analyst. 2011;136:29–41. doi: 10.1039/c0an00429d. [DOI] [PubMed] [Google Scholar]
- 26.West JL, Halas NJ. Curr Opin Biotechnol. 2000;11:215–217. doi: 10.1016/s0958-1669(00)00082-3. [DOI] [PubMed] [Google Scholar]
- 27.Khanna VK. Defence Sci J. 2008;58:608–616. [Google Scholar]
- 28.Sheehan PE, Whitman LJ. Nano Lett. 2005;5:803–807. doi: 10.1021/nl050298x. [DOI] [PubMed] [Google Scholar]
- 29.Boisselier E, Astruc D. Chem Soc Rev. 2009;38:1759–1782. doi: 10.1039/b806051g. [DOI] [PubMed] [Google Scholar]
- 30.Daniel MC, Astruc D. Chem Rev. 2004;104:293–346. doi: 10.1021/cr030698+. [DOI] [PubMed] [Google Scholar]
- 31.Haick H. J Phys D: Appl Phys. 2007;40:7173–7186. [Google Scholar]
- 32.Zayats M, Baron R, Popov I, Willner I. Nano Lett. 2005;5:21–25. doi: 10.1021/nl048547p. [DOI] [PubMed] [Google Scholar]
- 33.Zhao W, Brook MA, Li YF. ChemBioChem. 2008;9:2363–2371. doi: 10.1002/cbic.200800282. [DOI] [PubMed] [Google Scholar]
- 34.Bunz UHF, Rotello VM. Angew Chem Int Ed. 2010;49:3268–3279. doi: 10.1002/anie.200906928. [DOI] [PubMed] [Google Scholar]
- 35.Sperling RA, Rivera Gil P, Zhang F, Zanella M, Parak WJ. Chem Soc Rev. 2008;37:1896–1908. doi: 10.1039/b712170a. [DOI] [PubMed] [Google Scholar]
- 36.Radwan SH, Azzazy HME. Expert Rev Mol Diagn. 2009;9:511–524. doi: 10.1586/erm.09.33. [DOI] [PubMed] [Google Scholar]
- 37.Wilson R. Chem Soc Rev. 2008;37:2028–2045. doi: 10.1039/b712179m. [DOI] [PubMed] [Google Scholar]
- 38.Faraday M. Philos Trans R Soc London. 1857;147:145–181. [Google Scholar]
- 39.Turkevich J, Stevenson PC, Hillier J. Discuss Faraday Soc. 1951:55–75. [Google Scholar]
- 40.Frens G. Nature: Phys Sci. 1973;241:20–22. [Google Scholar]
- 41.Chow MK, Zukoski CF. J Colloid Interface Sci. 1994;165:97–109. [Google Scholar]
- 42.Ji XH, Song XN, Li J, Bai YB, Yang WS, Peng XG. J Am Chem Soc. 2007;129:13939–13948. doi: 10.1021/ja074447k. [DOI] [PubMed] [Google Scholar]
- 43.Kimling J, Maier M, Okenve B, Kotaidis V, Ballot H, Plech A. J Phys Chem B. 2006;110:15700–15707. doi: 10.1021/jp061667w. [DOI] [PubMed] [Google Scholar]
- 44.Nguyen DT, Kim DJ, So MG, Kim KS. Adv Powder Technol. 2010;21:111–118. [Google Scholar]
- 45.Polte J, Ahner TT, Delissen F, Sokolov S, Emmerling F, Thunemann AF, Kraehnert R. J Am Chem Soc. 2010;132:1296–1301. doi: 10.1021/ja906506j. [DOI] [PubMed] [Google Scholar]
- 46.Pong BK, Elim HI, Chong JX, Ji W, Trout BL, Lee JY. J Phys Chem C. 2007;111:6281–6287. [Google Scholar]
- 47.Uppal MA, Kafizas A, Ewing MB, Parkin IP. New J Chem. 2010;34:2906–2914. [Google Scholar]
- 48.Uppal MA, Kafizas A, Lim TH, Parkin IP. New J Chem. 2010;34:1401–1407. [Google Scholar]
- 49.Giersig M, Mulvaney P. Langmuir. 1993;9:3408–3413. [Google Scholar]
- 50.Brust M, Walker M, Bethell D, Schiffrin DJ, Whyman R. J Chem Soc, Chem Commun. 1994:801–802. [Google Scholar]
- 51.Hostetler MJ, Wingate JE, Zhong CJ, Harris JE, Vachet RW, Clark MR, Londono JD, Green SJ, Stokes JJ, Wignall GD, Glish GL, Porter MD, Evans ND, Murray RW. Langmuir. 1998;14:17–30. [Google Scholar]
- 52.Chen SW. Langmuir. 1999;15:7551–7557. [Google Scholar]
- 53.Chen SW, Murray RW. Langmuir. 1999;15:682–689. [Google Scholar]
- 54.Hostetler MJ, Green SJ, Stokes JJ, Murray RW. J Am Chem Soc. 1996;118:4212–4213. [Google Scholar]
- 55.Ingram RS, Hostetler MJ, Murray RW. J Am Chem Soc. 1997;119:9175–9178. [Google Scholar]
- 56.Templeton AC, Wuelfing MP, Murray RW. Acc Chem Res. 2000;33:27–36. doi: 10.1021/ar9602664. [DOI] [PubMed] [Google Scholar]
- 57.Brust M, Fink J, Bethell D, Schiffrin DJ, Kiely C. J Chem Soc, Chem Commun. 1995:1655–1656. [Google Scholar]
- 58.Kanaras AG, Kamounah FS, Schaumburg K, Kiely CJ, Brust M. Chem Commun. 2002:2294–2295. doi: 10.1039/b207838b. [DOI] [PubMed] [Google Scholar]
- 59.Zheng M, Li ZG, Huang XY. Langmuir. 2004;20:4226–4235. doi: 10.1021/la035981i. [DOI] [PubMed] [Google Scholar]
- 60.Templeton AC, Chen SW, Gross SM, Murray RW. Langmuir. 1999;15:66–76. [Google Scholar]
- 61.Price RC, Whetten RL. J Am Chem Soc. 2005;127:13750–13751. doi: 10.1021/ja053968+. [DOI] [PubMed] [Google Scholar]
- 62.Shon YS, Gross SM, Dawson B, Porter M, Murray RW. Langmuir. 2000;16:6555–6561. [Google Scholar]
- 63.Shon YS, Wuelfing WP, Murray RW. Langmuir. 2001;17:1255–1261. [Google Scholar]
- 64.Lohse SE, Dahl JA, Hutchison JE. Langmuir. 2010;26:7504–7511. doi: 10.1021/la904306a. [DOI] [PubMed] [Google Scholar]
- 65.Boal AK, Ilhan F, DeRouchey JE, Thurn-Albrecht T, Russell TP, Rotello VM. Nature. 2000;404:746–748. doi: 10.1038/35008037. [DOI] [PubMed] [Google Scholar]
- 66.Itoh H, Tahara A, Naka K, Chujo Y. Langmuir. 2004;20:1972–1976. doi: 10.1021/la0359777. [DOI] [PubMed] [Google Scholar]
- 67.Zhou JF, Beattie DA, Sedev R, Ralston J. Langmuir. 2007;23:9170–9177. doi: 10.1021/la700449f. [DOI] [PubMed] [Google Scholar]
- 68.Goulet PJG, Lennox RB. J Am Chem Soc. 2010;132:9582–9584. doi: 10.1021/ja104011b. [DOI] [PubMed] [Google Scholar]
- 69.Yee CK, Jordan R, Ulman A, White H, King A, Rafailovich M, Sokolov J. Langmuir. 1999;15:3486–3491. [Google Scholar]
- 70.Ascencio JA, Perez M, Jose-Yacaman M. Surf Sci. 2000;447:73–80. [Google Scholar]
- 71.Sugie A, Hatta T, Kanie K, Muramatsu A, Mori A. Chem Lett. 2009;38:562–563. [Google Scholar]
- 72.Sardar R, Shumaker-Parry JS. Chem Mater. 2009;21:1167–1169. [Google Scholar]
- 73.Negishi Y, Takasugi Y, Sato S, Yao H, Kimura K, Tsukuda T. J Am Chem Soc. 2004;126:6518–6519. doi: 10.1021/ja0483589. [DOI] [PubMed] [Google Scholar]
- 74.Templeton AC, Hostetler MJ, Kraft CT, Murray RW. J Am Chem Soc. 1998;120:1906–1911. [Google Scholar]
- 75.Hostetler MJ, Templeton AC, Murray RW. Langmuir. 1999;15:3782–3789. [Google Scholar]
- 76.Jadzinsky PD, Calero G, Ackerson CJ, Bushnell DA, Kornberg RD. Science. 2007;318:430–433. doi: 10.1126/science.1148624. [DOI] [PubMed] [Google Scholar]
- 77.Boal AK, Rotello VM. J Am Chem Soc. 2000;122:734–735. [Google Scholar]
- 78.Zachary M, Chechik V. Angew Chem Int Ed. 2007;46:3304–3307. doi: 10.1002/anie.200604070. [DOI] [PubMed] [Google Scholar]
- 79.Donkers RL, Lee D, Murray RW. Langmuir. 2004;20:1945–1952. doi: 10.1021/la0497494. [DOI] [PubMed] [Google Scholar]
- 80.Wang W, Murray RW. Langmuir. 2005;21:7015–7022. doi: 10.1021/la0508700. [DOI] [PubMed] [Google Scholar]
- 81.Woehrle GH, Warner MG, Hutchison JE. J Phys Chem B. 2002;106:9979–9981. [Google Scholar]
- 82.Woehrle GH, Brown LO, Hutchison JE. J Am Chem Soc. 2005;127:2172–2183. doi: 10.1021/ja0457718. [DOI] [PubMed] [Google Scholar]
- 83.Leff DV, Ohara PC, Heath JR, Gelbart WM. J Phys Chem. 1995;99:7036–7041. doi: 10.1103/PhysRevLett.75.3466. [DOI] [PubMed] [Google Scholar]
- 84.Tao AR, Habas S, Yang PD. Small. 2008;4:310–325. [Google Scholar]
- 85.Prasad BLV, Stoeva SI, Sorensen CM, Klabunde KJ. Langmuir. 2002;18:7515–7520. [Google Scholar]
- 86.Prasad BLV, Stoeva SI, Sorensen CM, Klabunde KJ. Chem Mater. 2003;15:935–942. [Google Scholar]
- 87.Zhong CJ, Zhang WX, Leibowitz FL, Eichelberger HH. Chem Commun. 1999:1211–1212. [Google Scholar]
- 88.Maye MM, Zheng WX, Leibowitz FL, Ly NK, Zhong CJ. Langmuir. 2000;16:490–497. [Google Scholar]
- 89.Kurita H, Takami A, Koda S. Appl Phys Lett. 1998;72:789–791. [Google Scholar]
- 90.Sau TK, Pal A, Jana NR, Wang ZL, Pal T. J Nanopart Res. 2001;3:257–261. [Google Scholar]
- 91.Mossmer S, Spatz JP, Moller M, Aberle T, Schmidt J, Burchard W. Macromolecules. 2000;33:4791–4798. [Google Scholar]
- 92.Meltzer S, Resch R, Koel BE, Thompson ME, Madhukar A, Requicha AAG, Will P. Langmuir. 2001;17:1713–1718. [Google Scholar]
- 93.Mallick K, Wang ZL, Pal T. J Photochem Photobiol, A. 2001;140:75–80. [Google Scholar]
- 94.Sobhan MA, Withford MJ, Goldys EM. Langmuir. 2010;26:3156–3159. doi: 10.1021/la903088e. [DOI] [PubMed] [Google Scholar]
- 95.Chen W, Cai WP, Liang CH, Zhang LD. Mater Res Bull. 2001;36:335–342. [Google Scholar]
- 96.Chen W, Cai WP, Zhang L, Wang GZ, Zhang LD. J Colloid Interface Sci. 2001;238:291–295. doi: 10.1006/jcis.2001.7525. [DOI] [PubMed] [Google Scholar]
- 97.Niidome Y, Hori A, Sato T, Yamada S. Chem Lett. 2000:310–311. [Google Scholar]
- 98.Pol VG, Gedanken A, Calderon-Moreno J. Chem Mater. 2003;15:1111–1118. [Google Scholar]
- 99.Reed JA, Cook A, Halaas DJ, Parazzoli P, Robinson A, Matula TJ, Grieser F. Ultrason Sonochem. 2003;10:285–289. doi: 10.1016/S1350-4177(03)00093-2. [DOI] [PubMed] [Google Scholar]
- 100.Zhou Y, Wang CY, Zhu YR, Chen ZY. Chem Mater. 1999;11:2310–2312. [Google Scholar]
- 101.Dawson A, Kamat PV. J Phys Chem B. 2000;104:11842–11846. [Google Scholar]
- 102.Gachard E, Remita H, Khatouri J, Keita B, Nadjo L, Belloni J. New J Chem. 1998;22:1257–1265. [Google Scholar]
- 103.Henglein A, Meisel D. Langmuir. 1998;14:7392–7396. [Google Scholar]
- 104.Wei GT, Liu FK, Wang CRC. Anal Chem. 1999;71:2085–2091. doi: 10.1021/ac990044u. [DOI] [PubMed] [Google Scholar]
- 105.Helcher HH. Aurum Potabile oder Gold Tinstur. J. Herbord Klossen; Breslau and Leipzig: 1718. [Google Scholar]
- 106.Meguro K, Nakamura Y, Hayashi Y, Torizuka M, Esumi K. Bull Chem Soc Jpn. 1988;61:347–350. [Google Scholar]
- 107.Carotenuto G. Appl Organomet Chem. 2001;15:344–351. [Google Scholar]
- 108.Seoudi R, Fouda AA, Elmenshawy DA. Physica B. 2010;405:906–911. [Google Scholar]
- 109.Hayat MA. Colloidal Gold, Principles, Methods and Applications. Academic Press; New York: 1989. [Google Scholar]
- 110.Oh E, Susumu K, Goswami R, Mattoussi H. Langmuir. 2010;26:7604–7613. doi: 10.1021/la904438s. [DOI] [PubMed] [Google Scholar]
- 111.Susumu K, Mei BC, Mattoussi H. Nat Protoc. 2009;4:424–436. doi: 10.1038/nprot.2008.247. [DOI] [PubMed] [Google Scholar]
- 112.Tracy JB, Kalyuzhny G, Crowe MC, Balasubramanian R, Choi JP, Murray RW. J Am Chem Soc. 2007;129:6706–6707. doi: 10.1021/ja071042r. [DOI] [PubMed] [Google Scholar]
- 113.Li DX, He Q, Cui Y, Li JB. Chem Mater. 2007;19:412–417. [Google Scholar]
- 114.Bohrisch J, Wendler U, Jaeger W. Macromol Rapid Commun. 1997;18:975–982. [Google Scholar]
- 115.Harnish B, Robinson JT, Pei ZC, Ramstrom O, Yan MD. Chem Mater. 2005;17:4092–4096. [Google Scholar]
- 116.Pucci A, Bernabo M, Elvati P, Meza LI, Galembeck F, Leite CAD, Tirelli N, Ruggeri G. J Mater Chem. 2006;16:1058–1066. [Google Scholar]
- 117.Bhattacharjee RR, Chakraborty M, Mandal TK. J Phys Chem B. 2006;110:6768–6775. doi: 10.1021/jp056675b. [DOI] [PubMed] [Google Scholar]
- 118.Huang HZ, Yang XR. Biomacromolecules. 2004;5:2340–2346. doi: 10.1021/bm0497116. [DOI] [PubMed] [Google Scholar]
- 119.Sun XP, Dong SJ, Wang EK. Polymer. 2004;45:2181–2184. [Google Scholar]
- 120.Gole A, Murphy CJ. Chem Mater. 2005;17:1325–1330. [Google Scholar]
- 121.Mandal TK, Fleming MS, Walt DR. Nano Lett. 2002;2:3–7. [Google Scholar]
- 122.Yilmaz E, Suzer S. Appl Surf Sci. 2010;256:6630–6633. [Google Scholar]
- 123.Spatz JP, Mossmer S, Moller M. Angew Chem Int Ed. 1996;35:1510–1512. [Google Scholar]
- 124.Spatz JP, Roescher A, Moller M. Adv Mater. 1996;8:337–340. [Google Scholar]
- 125.Zikich D, Borovok N, Molotsky T, Kotlyar A. Bioconjugate Chem. 2010;21:544–547. doi: 10.1021/bc900527a. [DOI] [PubMed] [Google Scholar]
- 126.Zhu MQ, Wang LQ, Exarhos GJ, Li ADQ. J Am Chem Soc. 2004;126:2656–2657. doi: 10.1021/ja038544z. [DOI] [PubMed] [Google Scholar]
- 127.Morones JR, Frey W. J Nanopart Res. 2010;12:1401–1414. [Google Scholar]
- 128.Ohno K, Koh K, Tsujii Y, Fukuda T. Macromolecules. 2002;35:8989–8993. [Google Scholar]
- 129.Raula J, Shan J, Nuopponen M, Niskanen A, Jiang H, Kauppinen EI, Tenhu H. Langmuir. 2003;19:3499–3504. [Google Scholar]
- 130.Kim DJ, Kang SM, Kong B, Kim WJ, Paik HJ, Choi H, Choi IS. Macromol Chem Phys. 2005;206:1941–1946. [Google Scholar]
- 131.Nuss S, Bottcher H, Wurm H, Hallensleben ML. Angew Chem Int Ed. 2001;40:4016–4018. doi: 10.1002/1521-3773(20011105)40:21<4016::AID-ANIE4016>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 132.Yoon KR, Ramaraj B, Lee SM, Kim DP. Surf Interface Anal. 2008;40:1139–1143. [Google Scholar]
- 133.Wuelfing WP, Gross SM, Miles DT, Murray RW. J Am Chem Soc. 1998;120:12696–12697. [Google Scholar]
- 134.Corbierre MK, Cameron NS, Lennox RB. Langmuir. 2004;20:2867–2873. doi: 10.1021/la0355702. [DOI] [PubMed] [Google Scholar]
- 135.Shan J, Nuopponen M, Jiang H, Kauppinen E, Tenhu H. Macromolecules. 2003;36:4526–4533. [Google Scholar]
- 136.Kim BJ, Bang J, Hawker CJ, Kramer EJ. Macromolecules. 2006;39:4108–4114. [Google Scholar]
- 137.Suzuki D, Kawaguchi H. Langmuir. 2005;21:8175–8179. doi: 10.1021/la0504356. [DOI] [PubMed] [Google Scholar]
- 138.Hussain I, Graham S, Wang ZX, Tan B, Sherrington DC, Rannard SP, Cooper AI, Brust M. J Am Chem Soc. 2005;127:16398–16399. doi: 10.1021/ja055321v. [DOI] [PubMed] [Google Scholar]
- 139.Huang HM, Chang CY, Liu IC, Tsai HC, Lai MK, Tsiang RCC. J Polym Sci, Part A: Polym Chem. 2005;43:4710–4720. [Google Scholar]
- 140.Shan J, Tenhu H. Chem Commun. 2007:4580–4598. doi: 10.1039/b707740h. [DOI] [PubMed] [Google Scholar]
- 141.Kim F, Connor S, Song H, Kuykendall T, Yang PD. Angew Chem Int Ed. 2004;43:3673–3677. doi: 10.1002/anie.200454216. [DOI] [PubMed] [Google Scholar]
- 142.Salvati R, Longo A, Carotenuto G, De Nicola S, Pepe GP, Nicolais L, Barone A. Appl Surf Sci. 2005;248:28–31. [Google Scholar]
- 143.Ah CS, Yun YJ, Park HJ, Kim WJ, Ha DH, Yun WS. Chem Mater. 2005;17:5558–5561. [Google Scholar]
- 144.Chen Y, Gu X, Nie CG, Jiang ZY, Xie ZX, Lin CJ. Chem Commun. 2005:4181–4183. doi: 10.1039/b504911c. [DOI] [PubMed] [Google Scholar]
- 145.Hoppe CE, Lazzari M, Pardinas-Blanco I, Lopez-Quintela MA. Langmuir. 2006;22:7027–7034. doi: 10.1021/la060885d. [DOI] [PubMed] [Google Scholar]
- 146.Wang LY, Kariuki NN, Schadt M, Mott D, Luo J, Zhong CJ, Shi XJ, Zhang C, Hao WB, Lu S, Kim N, Wang JQ. Sensors. 2006;6:667–679. [Google Scholar]
- 147.Corbierre MK, Cameron NS, Sutton M, Mochrie SGJ, Lurio LB, Ruhm A, Lennox RB. J Am Chem Soc. 2001;123:10411–10412. doi: 10.1021/ja0166287. [DOI] [PubMed] [Google Scholar]
- 148.Kang YJ, Taton TA. Angew Chem Int Ed. 2005;44:409–412. doi: 10.1002/anie.200461119. [DOI] [PubMed] [Google Scholar]
- 149.Shelley EJ, Ryan D, Johnson SR, Couillard M, Fitzmaurice D, Nellist PD, Chen Y, Palmer RE, Preece JA. Langmuir. 2002;18:1791–1795. [Google Scholar]
- 150.Hasan M, Bethell D, Brust M. J Am Chem Soc. 2002;124:1132–1133. doi: 10.1021/ja0120577. [DOI] [PubMed] [Google Scholar]
- 151.Porter LA, Ji D, Westcott SL, Graupe M, Czernuszewicz RS, Halas NJ, Lee TR. Langmuir. 1998;14:7378–7386. doi: 10.1021/la980870i. [DOI] [PubMed] [Google Scholar]
- 152.Yonezawa T, Yasui K, Kimizuka N. Langmuir. 2001;17:271–273. [Google Scholar]
- 153.Manna A, Chen PL, Akiyama H, Wei TX, Tamada K, Knoll W. Chem Mater. 2003;15:20–28. [Google Scholar]
- 154.Torigoe K, Esumi K. J Phys Chem B. 1999;103:2862–2866. [Google Scholar]
- 155.Letsinger RL, Elghanian R, Viswanadham G, Mirkin CA. Bioconjugate Chem. 2000;11:289–291. doi: 10.1021/bc990152n. [DOI] [PubMed] [Google Scholar]
- 156.Rouhana LL, Jaber JA, Schlenoff JB. Langmuir. 2007;23:12799–12801. doi: 10.1021/la702151q. [DOI] [PubMed] [Google Scholar]
- 157.Resch R, Baur C, Bugacov A, Koel BE, Echternach PM, Madhukar A, Montoya N, Requicha AAG, Will P. J Phys Chem B. 1999;103:3647–3650. [Google Scholar]
- 158.Li Z, Jin RC, Mirkin CA, Letsinger RL. Nucleic Acids Res. 2002;30:1558–1562. doi: 10.1093/nar/30.7.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Tan YW, Li YF, Zhu DB. Langmuir. 2002;18:3392–3395. [Google Scholar]
- 160.Maye MM, Chun SC, Han L, Rabinovich D, Zhong CJ. J Am Chem Soc. 2002;124:4958–4959. doi: 10.1021/ja025724k. [DOI] [PubMed] [Google Scholar]
- 161.Li XM, de Jong MR, Inoue K, Shinkai S, Huskens J, Reinhoudt DN. J Mater Chem. 2001;11:1919–1923. [Google Scholar]
- 162.Tzhayik O, Sawant P, Efrima S, Kovalev E, Klug JT. Langmuir. 2002;18:3364–3369. [Google Scholar]
- 163.Balasubramanian R, Kim B, Tripp SL, Wang XJ, Lieberman M, Wei A. Langmuir. 2002;18:3676–3681. [Google Scholar]
- 164.Sun L, Crooks RM, Chechik V. Chem Commun. 2001:359–360. [Google Scholar]
- 165.Leff DV, Brandt L, Heath JR. Langmuir. 1996;12:4723–4730. [Google Scholar]
- 166.Gomez S, Philippot K, Colliere V, Chaudret B, Senocq F, Lecante P. Chem Commun. 2000:1945–1946. [Google Scholar]
- 167.Heath JR, Knobler CM, Leff DV. J Phys Chem B. 1997;101:189–197. [Google Scholar]
- 168.Kumar A, Mandal S, Selvakannan PR, Pasricha R, Mandale AB, Sastry M. Langmuir. 2003;19:6277–6282. doi: 10.1021/la034209c. [DOI] [PubMed] [Google Scholar]
- 169.Aslam M, Fu L, Su M, Vijayamohanan K, Dravid VP. J Mater Chem. 2004;14:1795–1797. [Google Scholar]
- 170.Newman JDS, Blanchard GJ. Langmuir. 2006;22:5882–5887. doi: 10.1021/la060045z. [DOI] [PubMed] [Google Scholar]
- 171.Bhargava SK, Booth JM, Agrawal S, Coloe P, Kar G. Langmuir. 2005;21:5949–5956. doi: 10.1021/la050283e. [DOI] [PubMed] [Google Scholar]
- 172.Selvakannan PR, Mandal S, Phadtare S, Pasricha R, Sastry M. Langmuir. 2003;19:3545–3549. doi: 10.1021/la034209c. [DOI] [PubMed] [Google Scholar]
- 173.Selvakannan P, Mandal S, Phadtare S, Gole A, Pasricha R, Adyanthaya SD, Sastry M. J Colloid Interface Sci. 2004;269:97–102. doi: 10.1016/s0021-9797(03)00616-7. [DOI] [PubMed] [Google Scholar]
- 174.Leontowich AFG, Calver CF, Dasog M, Scott RWJ. Langmuir. 2010;26:1285–1290. doi: 10.1021/la902465b. [DOI] [PubMed] [Google Scholar]
- 175.Mandal S, Selvakannan P, Phadtare S, Pasricha R, Sastry M. Proc Indian Acad Sci Chem Sci. 2002;114:513–520. [Google Scholar]
- 176.Selvakannan PR, Kumar PS, More AS, Shingte RD, Wadgaonkar PP, Sastry M. Langmuir. 2004;20:295–298. doi: 10.1021/la0350352. [DOI] [PubMed] [Google Scholar]
- 177.Isaacs SR, Cutler EC, Park JS, Lee TR, Shon YS. Langmuir. 2005;21:5689–5692. doi: 10.1021/la050656b. [DOI] [PubMed] [Google Scholar]
- 178.Kotiaho A, Lahtinen R, Efimov A, Lehtivuori H, Tkachenko NV, Kanerva T, Lemmetyinen H. J Photochem Photobiol, A. 2010;212:129–134. [Google Scholar]
- 179.Duan HW, Nie SM. J Am Chem Soc. 2007;129:2412–2413. doi: 10.1021/ja067727t. [DOI] [PubMed] [Google Scholar]
- 180.Zhu HG, Pan ZW, Hagaman EW, Liang CD, Overbury SH, Dai S. J Colloid Interface Sci. 2005;287:360–365. doi: 10.1016/j.jcis.2005.01.106. [DOI] [PubMed] [Google Scholar]
- 181.Huang W, Chen SM, Liu YS, Fu HY, Wu GZ. Nanotechnology. 2011;22:025602. doi: 10.1088/0957-4484/22/2/025602. [DOI] [PubMed] [Google Scholar]
- 182.Das AK, Raj CR. J Colloid Interface Sci. 2011;353:506–511. doi: 10.1016/j.jcis.2010.09.080. [DOI] [PubMed] [Google Scholar]
- 183.Green M, O’Brien P. Chem Commun. 2000:183–184. [Google Scholar]
- 184.Fleming DA, Williams ME. Langmuir. 2004;20:3021–3023. [PubMed] [Google Scholar]
- 185.Weare WW, Reed SM, Warner MG, Hutchison JE. J Am Chem Soc. 2000;122:12890–12891. [Google Scholar]
- 186.Moores A, Goettmann F, Sanchez C, Le Floch P. Chem Commun. 2004:2842–2843. doi: 10.1039/b412553c. [DOI] [PubMed] [Google Scholar]
- 187.Yamamoto M, Nakamoto M. Chem Lett. 2003;32:452–453. [Google Scholar]
- 188.Shakeri-Zadeh A, Ghasemifard M, Mansoor GA. Physica E. 2010;42:1272–1280. [Google Scholar]
- 189.Sarkar A, Shukla SP, Adhikari S, Mukherjee T. Int J Nano Technol. 2010;7:1027–1037. [Google Scholar]
- 190.Wang WX, Chen QF, Jiang C, Yang DZ, Liu XM, Xu SK. Colloid Surface A. 2007;301:73–79. [Google Scholar]
- 191.Yin XJ, Chen SG, Wu AG. Micro Nano Lett. 2010;5:270–273. [Google Scholar]
- 192.Sirajuddin, Mechler A, Torriero AAJ, Nafady A, Lee CY, Bond AM, O’Mullane AP, Bhargava SK. Colloids Surf, A. 2010;370:35–41. [Google Scholar]
- 193.Quinn BM, Liljeroth P, Ruiz V, Laaksonen T, Kontturi K. J Am Chem Soc. 2003;125:6644–6645. doi: 10.1021/ja0349305. [DOI] [PubMed] [Google Scholar]
- 194.Antonello S, Holm AH, Instuli E, Maran F. J Am Chem Soc. 2007;129:9836–9837. doi: 10.1021/ja071191+. [DOI] [PubMed] [Google Scholar]
- 195.Schmid G, Simon U. Chem Commun. 2005:697–710. doi: 10.1039/b411696h. [DOI] [PubMed] [Google Scholar]
- 196.Subramaniam C, Pradeep T, Chakrabarti J. Phys Rev Lett. 2005;95:164501. doi: 10.1103/PhysRevLett.95.164501. [DOI] [PubMed] [Google Scholar]
- 197.Jain PK, Lee KS, El-Sayed IH, El-Sayed MA. J Phys Chem B. 2006;110:7238–7248. doi: 10.1021/jp057170o. [DOI] [PubMed] [Google Scholar]
- 198.Mie G. Ann Phys. 1908;25:377–445. [Google Scholar]
- 199.Templeton AC, Pietron JJ, Murray RW, Mulvaney P. J Phys Chem B. 2000;104:564–570. [Google Scholar]
- 200.Swanson NL, Billard BD. Nanotechnology. 2003;14:353–357. [Google Scholar]
- 201.Link S, Mohamed MB, El-Sayed MA. J Phys Chem B. 1999;103:3073–3077. [Google Scholar]
- 202.Yan BH, Yang Y, Wang YC. J Phys Chem B. 2003;107:9159–9159. [Google Scholar]
- 203.Rechberger W, Hohenau A, Leitner A, Krenn JR, Lamprecht B, Aussenegg FR. Opt Commun. 2003;220:137–141. [Google Scholar]
- 204.Link S, El-Sayed MA. J Phys Chem B. 1999;103:4212–4217. [Google Scholar]
- 205.Itoh T, Asahi T, Masuhara H. Appl Phys Lett. 2001;79:1667–1669. [Google Scholar]
- 206.Su KH, Wei QH, Zhang X, Mock JJ, Smith DR, Schultz S. Nano Lett. 2003;3:1087–1090. [Google Scholar]
- 207.Srivastava S, Frankamp BL, Rotello VM. Chem Mater. 2005;17:487–490. [Google Scholar]
- 208.Link S, Wang ZL, El-Sayed MA. J Phys Chem B. 1999;103:3529–3533. [Google Scholar]
- 209.Liu XO, Atwater M, Wang JH, Huo Q. Colloids Surf, B. 2007;58:3–7. doi: 10.1016/j.colsurfb.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 210.Thomas KG, Kamat PV. J Am Chem Soc. 2000;122:2655–2656. [Google Scholar]
- 211.Xu P, Yanagi H. Chem Mater. 1999;11:2626–2628. [Google Scholar]
- 212.Dubertret B, Calame M, Libchaber AJ. Nat Biotechnol. 2001;19:365–370. doi: 10.1038/86762. [DOI] [PubMed] [Google Scholar]
- 213.Sarathy KV, Narayan KS, Kim J, White JO. Chem Phys Lett. 2000;318:543–548. [Google Scholar]
- 214.Makarova OV, Ostafin AE, Miyoshi H, Norris JR, Meisel D. J Phys Chem B. 1999;103:9080–9084. [Google Scholar]
- 215.Gu T, Whitesell JK, Fox MA. Chem Mater. 2003;15:1358–1366. [Google Scholar]
- 216.Dulkeith E, Morteani AC, Niedereichholz T, Klar TA, Feldmann J, Levi SA, van Veggel F, Reinhoudt DN, Moller M, Gittins DI. Phys Rev Lett. 89:2002. 203002. doi: 10.1103/PhysRevLett.89.203002. [DOI] [PubMed] [Google Scholar]
- 217.Zheng J, Zhang CW, Dickson RM. Phys Rev Lett. 2004;93:077402. doi: 10.1103/PhysRevLett.93.077402. [DOI] [PubMed] [Google Scholar]
- 218.Lakowicz JR. Anal Biochem. 2005;337:171–194. doi: 10.1016/j.ab.2004.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Imahori H, Fukuzumi S. Adv Mater. 2001;13:1197–1199. [Google Scholar]
- 220.Imahori H, Arimura M, Hanada T, Nishimura Y, Yamazaki I, Sakata Y, Fukuzumi S. J Am Chem Soc. 2001;123:335–336. doi: 10.1021/ja002838s. [DOI] [PubMed] [Google Scholar]
- 221.Lakowitca JR. Principles of Fluorescence Spectroscopy. Klumer; New York: 1999. [Google Scholar]
- 222.Wilcoxon JP, Martin JE, Parsapour F, Wiedenman B, Kelley DF. J Chem Phys. 1998;108:9137–9143. [Google Scholar]
- 223.Sapsford KE, Berti L, Medintz IL. Angew Chem Int Ed. 2006;45:4562–4589. doi: 10.1002/anie.200503873. [DOI] [PubMed] [Google Scholar]
- 224.Gersten J, Nitzan A. J Chem Phys. 1981;75:1139–1152. [Google Scholar]
- 225.Oh E, Hong MY, Lee D, Nam SH, Yoon HC, Kim HS. J Am Chem Soc. 2005;127:3270–3271. doi: 10.1021/ja0433323. [DOI] [PubMed] [Google Scholar]
- 226.Bigioni TP, Whetten RL, Dag O. J Phys Chem B. 2000;104:6983–6986. [Google Scholar]
- 227.Link S, El-Sayed MA. Int Rev Phys Chem. 2000;19:409–453. [Google Scholar]
- 228.Mohamed MB, Volkov V, Link S, El-Sayed MA. Chem Phys Lett. 2000;317:517–523. [Google Scholar]
- 229.Dulkeith E, Ringler M, Klar TA, Feldmann J, Javier AM, Parak WJ. Nano Lett. 2005;5:585–589. doi: 10.1021/nl0480969. [DOI] [PubMed] [Google Scholar]
- 230.Yun CS, Javier A, Jennings T, Fisher M, Hira S, Peterson S, Hopkins B, Reich NO, Strouse GF. J Am Chem Soc. 2005;127:3115–3119. doi: 10.1021/ja043940i. [DOI] [PubMed] [Google Scholar]
- 231.Ray PC, Fortner A, Darbha GK. J Phys Chem B. 2006;110:20745–20748. doi: 10.1021/jp065121l. [DOI] [PubMed] [Google Scholar]
- 232.Jennings TL, Singh MP, Strouse GF. J Am Chem Soc. 2006;128:5462–5467. doi: 10.1021/ja0583665. [DOI] [PubMed] [Google Scholar]
- 233.Kamat PV, Barazzouk S, Hotchandani S. Angew Chem Int Ed. 2002;41:2764–2767. doi: 10.1002/1521-3773(20020802)41:15<2764::AID-ANIE2764>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 234.Thomas KG, Kamat PV. Acc Chem Res. 2003;36:888–898. doi: 10.1021/ar030030h. [DOI] [PubMed] [Google Scholar]
- 235.Liu RR, Liew RS, Zhou H, Xing BG. Angew Chem Int Ed. 2007;46:8799–8803. doi: 10.1002/anie.200702773. [DOI] [PubMed] [Google Scholar]
- 236.Jiang Y, Zhao H, Lin YQ, Zhu NN, Ma YR, Mao LQ. Angew Chem Int Ed. 2010;49:4800–4804. doi: 10.1002/anie.201001057. [DOI] [PubMed] [Google Scholar]
- 237.Lin SY, Liu SW, Lin CM, Chen CH. Anal Chem. 2002;74:330–335. doi: 10.1021/ac0156316. [DOI] [PubMed] [Google Scholar]
- 238.Lin SY, Chen CH, Lin MC, Hsu HF. Anal Chem. 2005;77:4821–4828. doi: 10.1021/ac050443r. [DOI] [PubMed] [Google Scholar]
- 239.Obare SO, Hollowell RE, Murphy CJ. Langmuir. 2002;18:10407–10410. [Google Scholar]
- 240.Reynolds AJ, Haines AH, Russell DA. Langmuir. 2006;22:1156–1163. doi: 10.1021/la052261y. [DOI] [PubMed] [Google Scholar]
- 241.Kim YJ, Johnson RC, Hupp JT. Nano Lett. 2001;1:165–167. [Google Scholar]
- 242.Huang CC, Chang HT. Chem Commun. 2007:1215–1217. doi: 10.1039/b615383f. [DOI] [PubMed] [Google Scholar]
- 243.Lin SY, Wu SH, Chen CH. Angew Chem Int Ed. 2006;45:4948–4951. doi: 10.1002/anie.200600771. [DOI] [PubMed] [Google Scholar]
- 244.Yang WR, Gooding JJ, He ZC, Li Q, Chen GN. J Nanosci Nanotechnol. 2007;7:712–716. [PubMed] [Google Scholar]
- 245.Si S, Kotal A, Mandal TK. J Phys Chem C. 2007;111:1248–1255. [Google Scholar]
- 246.Liu DB, Qu WS, Chen WW, Zhang W, Wang Z, Jiang XY. Anal Chem. 2010;82:9606–9610. doi: 10.1021/ac1021503. [DOI] [PubMed] [Google Scholar]
- 247.Lee JS, Han MS, Mirkin CA. Angew Chem Int Ed. 2007;46:4093–4096. doi: 10.1002/anie.200700269. [DOI] [PubMed] [Google Scholar]
- 248.Xue XJ, Wang F, Liu XG. J Am Chem Soc. 2008;130:3244–3245. doi: 10.1021/ja076716c. [DOI] [PubMed] [Google Scholar]
- 249.Liu CW, Hsieh YT, Huang CC, Lin ZH, Chang HT. Chem Commun. 2008:2242–2244. doi: 10.1039/b719856f. [DOI] [PubMed] [Google Scholar]
- 250.Liu XJ, Cheng XH, Bing T, Fang CL, Shangguan DH. Anal Sci. 2010;26:1169–1172. doi: 10.2116/analsci.26.1169. [DOI] [PubMed] [Google Scholar]
- 251.Lee JS, Ulmann PA, Han MS, Mirkin CA. Nano Lett. 2008;8:529–533. doi: 10.1021/nl0727563. [DOI] [PubMed] [Google Scholar]
- 252.Dang YQ, Li HW, Wang B, Li L, Wu YQ. ACS Appl Mater Interfaces. 2009;1:1533–1538. doi: 10.1021/am9001953. [DOI] [PubMed] [Google Scholar]
- 253.Joyce GF. Annu Rev Biochem. 2004;73:791–836. doi: 10.1146/annurev.biochem.73.011303.073717. [DOI] [PubMed] [Google Scholar]
- 254.Achenbach JC, Chiuman W, Cruz RPG, Li Y. Curr Pharm Biotechnol. 2004;5:321–336. doi: 10.2174/1389201043376751. [DOI] [PubMed] [Google Scholar]
- 255.Breaker RR. Nat Biotechnol. 1997;15:427–431. doi: 10.1038/nbt0597-427. [DOI] [PubMed] [Google Scholar]
- 256.Breaker RR. Science. 2000;290:2095–2096. doi: 10.1126/science.290.5499.2095. [DOI] [PubMed] [Google Scholar]
- 257.Lu Y. Chem Eur J. 2002;8:4589–4596. [PubMed] [Google Scholar]
- 258.Sen D, Geyer CR. Curr Opin Chem Biol. 1998;2:680–687. doi: 10.1016/s1367-5931(98)80103-8. [DOI] [PubMed] [Google Scholar]
- 259.Liu JW, Lu Y. J Am Chem Soc. 2003;125:6642–6643. doi: 10.1021/ja034775u. [DOI] [PubMed] [Google Scholar]
- 260.Liu JW, Lu Y. J Am Chem Soc. 2004;126:12298–12305. doi: 10.1021/ja046628h. [DOI] [PubMed] [Google Scholar]
- 261.Liu JW, Lu Y. Chem Mater. 2004;16:3231–3238. [Google Scholar]
- 262.Liu J, Lu Y. J Am Chem Soc. 2005;127:12677–12683. doi: 10.1021/ja053567u. [DOI] [PubMed] [Google Scholar]
- 263.Lee JH, Wang ZD, Liu JW, Lu Y. J Am Chem Soc. 2008;130:14217–14226. doi: 10.1021/ja803607z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Lisowski CE, Hutchison JE. Anal Chem. 2009;81:10246–10253. doi: 10.1021/ac902271t. [DOI] [PubMed] [Google Scholar]
- 265.Li XK, Wang JN, Sun LL, Wang ZX. Chem Commun. 2010;46:988–990. doi: 10.1039/b920135a. [DOI] [PubMed] [Google Scholar]
- 266.Schmidtchen FP, Berger M. Chem Rev. 1997;97:1609–1646. doi: 10.1021/cr9603845. [DOI] [PubMed] [Google Scholar]
- 267.Martinez-Manez R, Sancenon F. Chem Rev. 2003;103:4419–4476. doi: 10.1021/cr010421e. [DOI] [PubMed] [Google Scholar]
- 268.Beer PD, Gale PA. Angew Chem Int Ed. 2001;40:486–516. [PubMed] [Google Scholar]
- 269.Kubo Y. Tetrahedron Lett. 2005;46:4369–4372. [Google Scholar]
- 270.Kado S, Furui A, Akiyama Y, Nakahara Y, Kimura K. Anal Sci. 2009;25:261–265. doi: 10.2116/analsci.25.261. [DOI] [PubMed] [Google Scholar]
- 271.Minami T, Kaneko K, Nagasaki T, Kubo Y. Tetrahedron Lett. 2008;49:432–436. [Google Scholar]
- 272.Itoh H, Naka K, Chujo Y. J Am Chem Soc. 2004;126:3026–3027. doi: 10.1021/ja039895g. [DOI] [PubMed] [Google Scholar]
- 273.Garcia-Etxarri A, Aizpurua J, Molina-Aldareguia J, Marcilla R, Pomposo JA, Mecerreyes D. Front Phys China. 2010;5:330–336. [Google Scholar]
- 274.Watanabe S, Seguchi H, Yoshida K, Kifune K, Tadaki T, Shiozaki H. Tetrahedron Lett. 2005;46:8827–8829. [Google Scholar]
- 275.Youk KS, Kim KM, Chatterjee A, Ahn KH. Tetrahedron Lett. 2008;49:3652–3655. [Google Scholar]
- 276.Chatterjee A, Oh DJ, Kim KM, Youk KS, Ahn KH. Chem Asian J. 2008;3:1962–1967. doi: 10.1002/asia.200800233. [DOI] [PubMed] [Google Scholar]
- 277.Muir GD. Hazards in the Chemical Laboratory. 2. The Chemical Society; London: 1977. [Google Scholar]
- 278.Baskin SIB, TG . Medical Aspects of Chemical and Biological Warfare. TMM Publications; Washington: 1997. [Google Scholar]
- 279.Kim MH, Kim S, Jang HH, Yi S, Seo SH, Han MS. Tetrahedron Lett. 2010;51:4712–4716. [Google Scholar]
- 280.Lee KY, Kim DW, Heo J, Kim JS, Yang JK, Cheong GW, Han SW. Bull Korean Chem Soc. 2006;27:2081–2083. [Google Scholar]
- 281.Zhang SH, Wang J, Han LA, Li CG, Wang W, Yuan Z. Sens Actuat B Chem. 2010;147:687–690. [Google Scholar]
- 282.Zhang YF, Li BX, Xu CL. Analyst. 2010;135:1579–1584. doi: 10.1039/c0an00056f. [DOI] [PubMed] [Google Scholar]
- 283.Daniel WL, Han MS, Lee JS, Mirkin CA. J Am Chem Soc. 2009;131:6362–6363. doi: 10.1021/ja901609k. [DOI] [PubMed] [Google Scholar]
- 284.Aslan K, Lakowicz JR, Geddes CD. Anal Biochem. 2004;330:145–155. doi: 10.1016/j.ab.2004.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Aslan K, Lakowicz JR, Geddes CD. Anal Chem. 2005;77:2007–2014. doi: 10.1021/ac0484880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Matsui J, Akamatsu K, Nishiguchi S, Miyoshi D, Nawafune H, Tamaki K, Sugimoto N. Anal Chem. 2004;76:1310–1315. doi: 10.1021/ac034788q. [DOI] [PubMed] [Google Scholar]
- 287.Uehara N, Ookubo K, Shimizu T. Langmuir. 2010;26:6818–6825. doi: 10.1021/la100460w. [DOI] [PubMed] [Google Scholar]
- 288.Shimada T, Ookubo K, Komuro N, Shimizu T, Uehara N. Langmuir. 2007;23:11225–11232. doi: 10.1021/la700664u. [DOI] [PubMed] [Google Scholar]
- 289.Okubo K, Shimada T, Shimizu T, Uehara N. Anal Sci. 2007;23:85–90. doi: 10.2116/analsci.23.85. [DOI] [PubMed] [Google Scholar]
- 290.Liang XS, Wei HP, Cui ZQ, Deng JY, Zhang ZP, You XY, Zhang XE. Analyst. 2011;136:179–183. doi: 10.1039/c0an00432d. [DOI] [PubMed] [Google Scholar]
- 291.Jiang Y, Zhao H, Zhu NN, Lin YQ, Yu P, Mao LQ. Angew Chem Int Ed. 2008;47:8601–8604. doi: 10.1002/anie.200804066. [DOI] [PubMed] [Google Scholar]
- 292.Bunka DHJ, Stockley PG. Nat Rev Microbiol. 2006;4:588–596. doi: 10.1038/nrmicro1458. [DOI] [PubMed] [Google Scholar]
- 293.Liu JW, Lu Y. Angew Chem Int Ed. 2006;45:90–94. doi: 10.1002/anie.200502589. [DOI] [PubMed] [Google Scholar]
- 294.Liu JW, Lu Y. Adv Mater. 2006;18:1667–1671. [Google Scholar]
- 295.Liu J, Lu Y. Nat Protoc. 2006;1:246–252. doi: 10.1038/nprot.2006.38. [DOI] [PubMed] [Google Scholar]
- 296.Cheng Y, Yong W, Marty JL, Xiurong Y. Biosens Bioelectron. 2010;26:2724–2727. doi: 10.1016/j.bios.2010.09.032. [DOI] [PubMed] [Google Scholar]
- 297.Liu XP, Zhou ZH, Zhang LL, Tan ZY, Shen GL, Yu RQ. Chin J Chem. 2009;27:1855–1859. [Google Scholar]
- 298.Chavez JL, Lyon W, Kelley-Loughnane N, Stone MO. Biosens Bioelectron. 2010;26:23–28. doi: 10.1016/j.bios.2010.04.049. [DOI] [PubMed] [Google Scholar]
- 299.Liu JW, Mazumdar D, Lu Y. Angew Chem Int Ed. 2006;45:7955–7959. doi: 10.1002/anie.200603106. [DOI] [PubMed] [Google Scholar]
- 300.Wang LH, Liu XF, Hu XF, Song SP, Fan CH. Chem Commun. 2006:3780–3782. doi: 10.1039/b607448k. [DOI] [PubMed] [Google Scholar]
- 301.Zhao WA, Chiuman W, Brook MA, Li YF. Chembiochem. 2007;8:727–731. doi: 10.1002/cbic.200700014. [DOI] [PubMed] [Google Scholar]
- 302.Liu JW, Lu Y. Anal Chem. 2004;76:1627–1632. doi: 10.1021/ac0351769. [DOI] [PubMed] [Google Scholar]
- 303.Ogawa A. Bioorg Med Chem Lett. 2011;21:155–159. doi: 10.1016/j.bmcl.2010.11.048. [DOI] [PubMed] [Google Scholar]
- 304.Kim YS, Kim JH, Kim IA, Lee SJ, Jurng J, Gu MB. Biosens Bioelectron. 2010;26:1644–1649. doi: 10.1016/j.bios.2010.08.046. [DOI] [PubMed] [Google Scholar]
- 305.Ludwig JA, Weinstein JN. Nat Rev Cancer. 2005;5:845–856. doi: 10.1038/nrc1739. [DOI] [PubMed] [Google Scholar]
- 306.Cairns P. Nat Rev Cancer. 2007;7:531–543. doi: 10.1038/nrc2170. [DOI] [PubMed] [Google Scholar]
- 307.Lien K-Y, Lee G-B. Analyst. 2010;135:1499–1518. doi: 10.1039/c000037j. [DOI] [PubMed] [Google Scholar]
- 308.Marti AA, Jockusch S, Stevens N, Ju JY, Turro NJ. Acc Chem Res. 2007;40:402–409. doi: 10.1021/ar600013q. [DOI] [PubMed] [Google Scholar]
- 309.Sassolas A, Leca-Bouvier BD, Blum LJ. Chem Rev. 2008;108:109–139. doi: 10.1021/cr0684467. [DOI] [PubMed] [Google Scholar]
- 310.Kolpashchikov DM. Chem Rev. 2010;110:4709–4723. doi: 10.1021/cr900323b. [DOI] [PubMed] [Google Scholar]
- 311.Thaxton CS, Georganopoulou DG, Mirkin CA. Clin Chim Acta. 2006;363:120–126. doi: 10.1016/j.cccn.2005.05.042. [DOI] [PubMed] [Google Scholar]
- 312.Sato K, Hosokawa K, Maeda M. Anal Sci. 2007;23:17–20. doi: 10.2116/analsci.23.17. [DOI] [PubMed] [Google Scholar]
- 313.Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. Nature. 1996;382:607–609. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- 314.Elghanian R, Storhoff JJ, Mucic RC, Letsinger RL, Mirkin CA. Science. 1997;277:1078–1081. doi: 10.1126/science.277.5329.1078. [DOI] [PubMed] [Google Scholar]
- 315.Storhoff JJ, Elghanian R, Mucic RC, Mirkin CA, Letsinger RL. J Am Chem Soc. 1998;120:1959–1964. [Google Scholar]
- 316.Storhoff JJ, Lucas AD, Garimella V, Bao YP, Muller UR. Nat Biotechnol. 2004;22:883–887. doi: 10.1038/nbt977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Reynolds RA, Mirkin CA, Letsinger RL. J Am Chem Soc. 2000;122:3795–3796. [Google Scholar]
- 318.Cao YC, Jin RC, Thaxton S, Mirkin CA. Talanta. 2005;67:449–455. doi: 10.1016/j.talanta.2005.06.063. [DOI] [PubMed] [Google Scholar]
- 319.Li HX, Rothberg LJ. J Am Chem Soc. 2004;126:10958–10961. doi: 10.1021/ja048749n. [DOI] [PubMed] [Google Scholar]
- 320.Chakrabarti R, Klibanov AM. J Am Chem Soc. 2003;125:12531–12540. doi: 10.1021/ja035399g. [DOI] [PubMed] [Google Scholar]
- 321.Li JH, Chu X, Liu YL, Jiang JH, He ZM, Zhang ZW, Shen GL, Yu RQ. Nucleic Acids Res. 2005;33:9. doi: 10.1093/nar/gni163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Feldkamp U, Niemeyer CM. Angew Chem Int Ed. 2006;45:1856–1876. doi: 10.1002/anie.200502358. [DOI] [PubMed] [Google Scholar]
- 323.Sato K, Hosokawa K, Maeda M. J Am Chem Soc. 2003;125:8102–8103. doi: 10.1021/ja034876s. [DOI] [PubMed] [Google Scholar]
- 324.Li HX, Rothberg L. Proc Natl Acad Sci USA. 2004;101:14036–14039. doi: 10.1073/pnas.0406115101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Xia F, Zuo XL, Yang RQ, Xiao Y, Kang D, Vallee-Belisle A, Gong X, Yuen JD, Hsu BBY, Heeger AJ, Plaxco KW. Proc Natl Acad Sci USA. 2010;107:10837–10841. doi: 10.1073/pnas.1005632107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Han MS, Lytton-Jean AKR, Mirkin CA. J Am Chem Soc. 2006;128:4954–4955. doi: 10.1021/ja0606475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Chen CE, Song GT, Yang XJ, Ren JS, Qu XG. Biochimie. 2010;92:1416–1421. doi: 10.1016/j.biochi.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 328.Han MS, Lytton-Jean AKR, Oh BK, Heo J, Mirkin CA. Angew Chem Int Ed. 2006;45:1807–1810. doi: 10.1002/anie.200504277. [DOI] [PubMed] [Google Scholar]
- 329.Hurst SJ, Han MS, Lytton-Jean AKR, Mirkin CA. Anal Chem. 2007;79:7201–7205. doi: 10.1021/ac071253e. [DOI] [PubMed] [Google Scholar]
- 330.Schofield CL, Haines AH, Field RA, Russell DA. Langmuir. 2006;22:6707–6711. doi: 10.1021/la060288r. [DOI] [PubMed] [Google Scholar]
- 331.Watanabe S, Yoshida K, Shinkawa K, Kumagawa D, Seguchi H. Colloid Surface B. 2010;81:570–577. doi: 10.1016/j.colsurfb.2010.07.061. [DOI] [PubMed] [Google Scholar]
- 332.Narain R, Housni A, Gody G, Boullanger P, Charreyre MT, Delair T. Langmuir. 2007;23:12835–12841. doi: 10.1021/la702378n. [DOI] [PubMed] [Google Scholar]
- 333.Nakamura-Tsuruta S, Kishimoto Y, Nishimura T, Suda Y. J Biochem. 2008;143:833–839. doi: 10.1093/jb/mvn038. [DOI] [PubMed] [Google Scholar]
- 334.Schofield CL, Mukhopadhyay B, Hardy SM, McDonnell MB, Field RA, Russell DA. Analyst. 2008;133:626–634. doi: 10.1039/b715250g. [DOI] [PubMed] [Google Scholar]
- 335.Barrientos AG, de la Fuente JM, Jimenez M, Solis D, Canada FJ, Martin-Lomas M, Penades S. Carbohydr Res. 2009;344:1474–1478. doi: 10.1016/j.carres.2009.04.029. [DOI] [PubMed] [Google Scholar]
- 336.Thygesen MB, Sorensen KK, Clo E, Jensen KJ. Chem Commun. 2009:6367–6369. doi: 10.1039/b911676a. [DOI] [PubMed] [Google Scholar]
- 337.Chuang YJ, Zhou XC, Pan ZW, Turchi C. Biochem Biophys Res Commun. 2009;389:22–27. doi: 10.1016/j.bbrc.2009.08.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Otsuka H, Akiyama Y, Nagasaki Y, Kataoka K. J Am Chem Soc. 2001;123:8226–8230. doi: 10.1021/ja010437m. [DOI] [PubMed] [Google Scholar]
- 339.Takae S, Akiyama Y, Otsuka H, Nakamura T, Nagasaki Y, Kataoka K. Biomacromolecules. 2005;6:818–824. doi: 10.1021/bm049427e. [DOI] [PubMed] [Google Scholar]
- 340.Toyoshima M, Miura Y. J Polym Sci Polym Chem. 2009;47:1412–1421. [Google Scholar]
- 341.Tsai CS, Yu TB, Chen CT. Chem Commun. 2005:4273–4275. doi: 10.1039/b507237a. [DOI] [PubMed] [Google Scholar]
- 342.Halkes KM, de Souza AC, Maljaars CEP, Gerwig GJ, Kamerling JP. Eur J Org Chem. 2005:3650–3659. [Google Scholar]
- 343.Wang X, Ramstrom O, Yan MD. Anal Chem. 2010;82:9082–9089. doi: 10.1021/ac102114z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Schofield CL, Field RA, Russell DA. Anal Chem. 2007;79:1356–1361. doi: 10.1021/ac061462j. [DOI] [PubMed] [Google Scholar]
- 345.Uzawa H, Ohga K, Shinozaki Y, Ohsawa I, Nagatsuka T, Seto Y, Nishida Y. Biosens Bioelectron. 2008;24:923–927. doi: 10.1016/j.bios.2008.07.049. [DOI] [PubMed] [Google Scholar]
- 346.Nath N, Chilkoti A. Anal Chem. 2002;74:504–509. doi: 10.1021/ac015657x. [DOI] [PubMed] [Google Scholar]
- 347.Nath N, Chilkoti A. Anal Chem. 2004;76:5370–5378. doi: 10.1021/ac049741z. [DOI] [PubMed] [Google Scholar]
- 348.Niikura K, Nagakawa K, Ohtake N, Suzuki T, Matsuo Y, Sawa H, Ijiro K. Bioconjugate Chem. 2009;20:1848–1852. doi: 10.1021/bc900255x. [DOI] [PubMed] [Google Scholar]
- 349.Huang CC, Huang YF, Cao ZH, Tan WH, Chang HT. Anal Chem. 2005;77:5735–5741. doi: 10.1021/ac050957q. [DOI] [PubMed] [Google Scholar]
- 350.Wei H, Li BL, Li J, Wang EK, Dong SJ. Chem Commun. 2007:3735–3737. doi: 10.1039/b707642h. [DOI] [PubMed] [Google Scholar]
- 351.Pavlov V, Xiao Y, Shlyahovsky B, Willner I. J Am Chem Soc. 2004;126:11768–11769. doi: 10.1021/ja046970u. [DOI] [PubMed] [Google Scholar]
- 352.Xiao Y, Pavlov V, Levine S, Niazov T, Markovitch G, Willner I. Angew Chem Int Ed. 2004;43:4519–4522. doi: 10.1002/anie.200460608. [DOI] [PubMed] [Google Scholar]
- 353.Dykman LA, Bogatyrev VA, Khlebtsov BN, Khlebtsov NG. Anal Biochem. 2005;341:16–21. doi: 10.1016/j.ab.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 354.Thanh NTK, Rosenzweig Z. Anal Chem. 2002;74:1624–1628. doi: 10.1021/ac011127p. [DOI] [PubMed] [Google Scholar]
- 355.Brust M, Bethell D, Schiffrin DJ, Kiely CJ. Adv Mater. 1995;7:795–797. [Google Scholar]
- 356.You CC, Arvizo RR, Rotello VM. Chem Commun. 2006:2905–2907. doi: 10.1039/b605508g. [DOI] [PubMed] [Google Scholar]
- 357.Guarise C, Pasquato L, De Filippis V, Scrimin P. Proc Natl Acad Sci USA. 2006;103:3978–3982. doi: 10.1073/pnas.0509372103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Laromaine A, Koh LL, Murugesan M, Ulijn RV, Stevens MM. J Am Chem Soc. 2007;129:4156–4157. doi: 10.1021/ja0706504. [DOI] [PubMed] [Google Scholar]
- 359.Xu XY, Han MS, Mirkin CA. Angew Chem Int Ed. 2007;46:3468–3470. doi: 10.1002/anie.200605249. [DOI] [PubMed] [Google Scholar]
- 360.Wang ZX, Levy R, Fernig DG, Brust M. J Am Chem Soc. 2006;128:2214–2215. doi: 10.1021/ja058135y. [DOI] [PubMed] [Google Scholar]
- 361.Choi Y, Ho NH, Tung CH. Angew Chem Int Ed. 2007;46:707–709. doi: 10.1002/anie.200603735. [DOI] [PubMed] [Google Scholar]
- 362.Zhao WA, Chiuman W, Lam JCF, Brook MA, Li YF. Chem Commun. 2007:3729–3731. doi: 10.1039/b705335e. [DOI] [PubMed] [Google Scholar]
- 363.Jiang TT, Liu RR, Huang XF, Feng HJ, Teo WL, Xing BG. Chem Commun. 2009:1972–1974. doi: 10.1039/b818853j. [DOI] [PubMed] [Google Scholar]
- 364.Uehara N, Fujita M, Shimizu T. Anal Sci. 2009;25:267–273. doi: 10.2116/analsci.25.267. [DOI] [PubMed] [Google Scholar]
- 365.Chah S, Hammond MR, Zare RN. Chem Biol. 2005;12:323–328. doi: 10.1016/j.chembiol.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 366.Jain PK, El-Sayed IH, El-Sayed MA. Nano Today. 2007;2:18–29. [Google Scholar]
- 367.Huang T, Murray RW. Langmuir. 2002;18:7077–7081. [Google Scholar]
- 368.Huang C-C, Chang H-T. Anal Chem. 2006;78:8332–8338. doi: 10.1021/ac061487i. [DOI] [PubMed] [Google Scholar]
- 369.He XR, Liu HB, Li YL, Wang S, Li YJ, Wang N, Xiao JC, Xu XH, Zhu DB. Adv Mater. 2005;17:2811–2815. [Google Scholar]
- 370.Ipe BI, Yoosaf K, Thomas KG. J Am Chem Soc. 2006;128:1907–1913. doi: 10.1021/ja054347j. [DOI] [PubMed] [Google Scholar]
- 371.Chen S-J, Chang H-T. Anal Chem. 2004;76:3727–3734. doi: 10.1021/ac049787s. [DOI] [PubMed] [Google Scholar]
- 372.Zhang N, Liu YY, Tong LL, Xu KH, Zhuo LH, Tang B. Analyst. 2008;133:1176–1181. doi: 10.1039/b803226b. [DOI] [PubMed] [Google Scholar]
- 373.Lin JH, Chang CW, Tseng WL. Analyst. 2010;135:104–110. doi: 10.1039/b916511h. [DOI] [PubMed] [Google Scholar]
- 374.Huang CC, Tseng WL. Anal Chem. 2008;80:6345–6350. doi: 10.1021/ac8006973. [DOI] [PubMed] [Google Scholar]
- 375.Huang CC, Yang Z, Lee KH, Chang HT. Angew Chem Int Ed. 2007;46:6824–6828. doi: 10.1002/anie.200700803. [DOI] [PubMed] [Google Scholar]
- 376.Huang CC, Chen CT, Shiang YC, Lin ZH, Chang HT. Anal Chem. 2009;81:875–882. doi: 10.1021/ac8010654. [DOI] [PubMed] [Google Scholar]
- 377.Chen CT, Chen WJ, Liu CZ, Chang LY, Chen YC. Chem Commun. 2009:7515–7517. doi: 10.1039/b916919a. [DOI] [PubMed] [Google Scholar]
- 378.Chen WB, Tu XJ, Guo XQ. Chem Commun. 2009:1736–1738. doi: 10.1039/b820145e. [DOI] [PubMed] [Google Scholar]
- 379.Maxwell DJ, Taylor JR, Nie SM. J Am Chem Soc. 2002;124:9606–9612. doi: 10.1021/ja025814p. [DOI] [PubMed] [Google Scholar]
- 380.Seferos DS, Giljohann DA, Hill HD, Prigodich AE, Mirkin CA. J Am Chem Soc. 2007;129:15477–15479. doi: 10.1021/ja0776529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Zheng D, Seferos DS, Giljohann DA, Patel PC, Mirkin CA. Nano Lett. 2009;9:3258–3261. doi: 10.1021/nl901517b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Prigodich AE, Seferos DS, Massich MD, Giljohann DA, Lane BC, Mirkin CA. ACS Nano. 2009;3:2147–2152. doi: 10.1021/nn9003814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Jin Y, Li HY, Bai JY. Anal Chem. 2009;81:5709–5715. doi: 10.1021/ac900482p. [DOI] [PubMed] [Google Scholar]
- 384.Zhang J, Wang LH, Zhang H, Boey F, Song SP, Fan CH. Small. 2010;6:201–204. doi: 10.1002/smll.200901012. [DOI] [PubMed] [Google Scholar]
- 385.Wang H, Wang YX, Jin JY, Yang RH. Anal Chem. 2008;80:9021–9028. doi: 10.1021/ac801382k. [DOI] [PubMed] [Google Scholar]
- 386.Medintz IL, Clapp AR, Mattoussi H, Goldman ER, Fisher B, Mauro JM. Nat Mater. 2003;2:630–638. doi: 10.1038/nmat961. [DOI] [PubMed] [Google Scholar]
- 387.Dyadyusha L, Yin H, Jaiswal S, Brown T, Baumberg JJ, Booy FP, Melvin T. Chem Commun. 2005:3201–3203. doi: 10.1039/b500664c. [DOI] [PubMed] [Google Scholar]
- 388.Oh E, Hong M-Y, Lee D, Nam S-H, Yoon HC, Kim H-S. J Am Chem Soc. 2005;127:3270–3271. doi: 10.1021/ja0433323. [DOI] [PubMed] [Google Scholar]
- 389.Oh E, Lee D, Kim YP, Cha SY, Oh DB, Kang HA, Kim J, Kim HS. Angew Chem Int Ed. 2006;45:7959–7963. doi: 10.1002/anie.200601948. [DOI] [PubMed] [Google Scholar]
- 390.Tang B, Cao LH, Xu KH, Zhuo LH, Ge JH, Li QF, Yu LJ. Chem Eur J. 2008;14:3637–3644. doi: 10.1002/chem.200701871. [DOI] [PubMed] [Google Scholar]
- 391.Wang X, Guo XQ. Analyst. 2009;134:1348–1354. doi: 10.1039/b822744f. [DOI] [PubMed] [Google Scholar]
- 392.Liu JW, Lee JH, Lu Y. Anal Chem. 2007;79:4120–4125. doi: 10.1021/ac070055k. [DOI] [PubMed] [Google Scholar]
- 393.You CC, Miranda OR, Gider B, Ghosh PS, Kim IB, Erdogan B, Krovi SA, Bunz UHF, Rotello VM. Nat Nanotechnol. 2007;2:318–323. doi: 10.1038/nnano.2007.99. [DOI] [PubMed] [Google Scholar]
- 394.You CC, De M, Han G, Rotello VM. J Am Chem Soc. 2005;127:12873–12881. doi: 10.1021/ja0512881. [DOI] [PubMed] [Google Scholar]
- 395.De M, You CC, Srivastava S, Rotello VM. J Am Chem Soc. 2007;129:10747–10753. doi: 10.1021/ja071642q. [DOI] [PubMed] [Google Scholar]
- 396.De M, Rana S, Akpinar H, Miranda OR, Arvizo RR, Bunz UHF, Rotello VM. Nat Chem. 2009;1:461–465. doi: 10.1038/nchem.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Phillips RL, Miranda OR, You CC, Rotello VM, Bunz UHF. Angew Chem Int Ed. 2008;47:2590–2594. doi: 10.1002/anie.200703369. [DOI] [PubMed] [Google Scholar]
- 398.Bajaj A, Miranda OR, Kim IB, Phillips RL, Jerry DJ, Bunz UHF, Rotello VM. Proc Natl Acad Sci USA. 2009;106:10912–10916. doi: 10.1073/pnas.0900975106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Bajaj A, Miranda OR, Phillips R, Kim IB, Jerry DJ, Bunz UHF, Rotello VM. J Am Chem Soc. 2010;132:1018–1022. doi: 10.1021/ja9061272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Bajaj A, Rana S, Miranda OR, Yawe JC, Jerry DJ, Bunz UHF, Rotello VM. Chem Sci. 2010;1:134–138. [Google Scholar]
- 401.Miranda OR, Chen HT, You CC, Mortenson DE, Yang XC, Bunz UHF, Rotello VM. J Am Chem Soc. 2010;132:5285–5289. doi: 10.1021/ja1006756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Katz E, Willner I, Wang J. Electroanalysis. 2004;16:19–44. [Google Scholar]
- 403.Guo SJ, Wang EK. Anal Chim Acta. 2007;598:181–192. doi: 10.1016/j.aca.2007.07.054. [DOI] [PubMed] [Google Scholar]
- 404.Pumera M, Sanchez S, Ichinose I, Tang J. Sens Actuat B Chem. 2007;123:1195–1205. [Google Scholar]
- 405.Willner I, Willner B. Pure Appl Chem. 2002;74:1773–1783. [Google Scholar]
- 406.Lucarelli F, Tombelli S, Minunni M, Marrazza G, Mascini M. Anal Chim Acta. 2008;609:139–159. doi: 10.1016/j.aca.2007.12.035. [DOI] [PubMed] [Google Scholar]
- 407.Shipway AN, Lahav M, Willner I. Adv Mater. 2000;12:993–998. [Google Scholar]
- 408.Pingarron JM, Yanez-Sedeno P, Gonzalez-Cortes A. Electrochim Acta. 2008;53:5848–5866. [Google Scholar]
- 409.Wang J. Analyst. 2005;130:421–426. doi: 10.1039/b414248a. [DOI] [PubMed] [Google Scholar]
- 410.Hernandez-Santos D, Gonzalez-Garcia MB, Garcia AC. Electroanalysis. 2002;14:1225–1235. [Google Scholar]
- 411.Wang J. Anal Chim Acta. 2003;500:247–257. [Google Scholar]
- 412.Erdem A. Talanta. 2007;74:318–325. doi: 10.1016/j.talanta.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 413.Fritzsche W, Taton TA. Nanotechnology. 2003;14:R63–R73. doi: 10.1088/0957-4484/14/12/R01. [DOI] [PubMed] [Google Scholar]
- 414.Kerman K, Saito M, Yamamura S, Takamura Y, Tamiya E. TrAC, Trends Anal Chem. 2008;27:585–592. [Google Scholar]
- 415.Yanez-Sedeno P, Pingarron JM. Anal Bioanal Chem. 2005;382:884–886. doi: 10.1007/s00216-005-3221-5. [DOI] [PubMed] [Google Scholar]
- 416.Merkoci A. FEBS J. 2007;274:310–316. doi: 10.1111/j.1742-4658.2006.05603.x. [DOI] [PubMed] [Google Scholar]
- 417.Kerman K, Kobayashi M, Tamiya E. Meas Sci Technol. 2004;15:R1–R11. [Google Scholar]
- 418.Mao X, Liu GD. J Biomed Nanotechnol. 2008;4:419–431. [Google Scholar]
- 419.Zamborini FP, Leopold MC, Hicks JF, Kulesza PJ, Malik MA, Murray RW. J Am Chem Soc. 2002;124:8958–8964. doi: 10.1021/ja025965s. [DOI] [PubMed] [Google Scholar]
- 420.Ahn H, Chandekar A, Kang B, Sung C, Whitten JE. Chem Mater. 2004;16:3274–3278. [Google Scholar]
- 421.Im J, Chandekar A, Whitten JE. Langmuir. 2009;25:4288–4292. doi: 10.1021/la900016u. [DOI] [PubMed] [Google Scholar]
- 422.Joseph Y, Peic A, Chen XD, Michl J, Vossmeyer T, Yasuda A. J Phys Chem C. 2007;111:12855–12859. [Google Scholar]
- 423.Ibanez FJ, Zamborini FP. ACS Nano. 2008;2:1543–1552. doi: 10.1021/nn800109q. [DOI] [PubMed] [Google Scholar]
- 424.Joseph Y, Guse B, Vossmeyer T, Yasuda A. J Phys Chem C. 2008;112:12507–12514. [Google Scholar]
- 425.Guo JL, Pang PF, Cai QY. Sens Actuat B Chem. 2007;120:521–528. [Google Scholar]
- 426.Ibanez FJ, Gowrishetty U, Crain MM, Walsh KM, Zamborini FP. Anal Chem. 2006;78:753–761. doi: 10.1021/ac051347t. [DOI] [PubMed] [Google Scholar]
- 427.Wohltjen H, Snow AW. Anal Chem. 1998;70:2856–2859. [Google Scholar]
- 428.Evans SD, Johnson SR, Cheng YL, Shen T. J Mater Chem. 2000;10:183–188. [Google Scholar]
- 429.Zhang HL, Evans SD, Henderson JR, Miles RE, Shen TH. Nanotechnology. 2002;13:439–444. [Google Scholar]
- 430.Joseph Y, Besnard I, Rosenberger M, Guse B, Nothofer H-G, Wessels JM, Wild U, Knop-Gericke A, Su D-S, Schlögl R, Yasuda A, Vossmeyer T. J Phys Chem B. 2003;107:7406–7413. [Google Scholar]
- 431.Wang LY, Shi XJ, Kariuki NN, Schadt M, Wang GR, Rendeng Q, Choi J, Luo J, Lu S, Zhong CJ. J Am Chem Soc. 2007;129:2161–2170. doi: 10.1021/ja0673074. [DOI] [PubMed] [Google Scholar]
- 432.Raguse B, Chow E, Barton CS, Wieczorek L. Anal Chem. 2007;79:7333–7339. doi: 10.1021/ac070887i. [DOI] [PubMed] [Google Scholar]
- 433.Chow E, Herrmann J, Barton CS, Raguse B, Wieczorek L. Anal Chim Acta. 2009;632:135–142. doi: 10.1016/j.aca.2008.10.070. [DOI] [PubMed] [Google Scholar]
- 434.Raguse B, Barton CS, Muller KH, Chow E, Wieczorek L. J Phys Chem C. 2009;113:15390–15397. [Google Scholar]
- 435.Chow E, Gengenbach TR, Wieczorek L, Raguse B. Sens Actuat B Chem. 2010;143:704–711. [Google Scholar]
- 436.Kim YJ, Yang YS, Ha SC, Cho SM, Kim YS, Kim HY, Yang H, Kim YT. Sens Actuat B Chem. 2005;106:189–198. [Google Scholar]
- 437.Krasteva N, Besnard I, Guse B, Bauer RE, Müllen K, Yasuda A, Vossmeyer T. Nano Lett. 2002;2:551–555. [Google Scholar]
- 438.Krasteva N, Fogel Y, Bauer RE, Mullen K, Matsuzawa N, Yasuda A, Vossmeyer T. Adv Funct Mater. 2007;17:881–888. [Google Scholar]
- 439.Krasteva N, Guse B, Besnard I, Yasuda A, Vossmeyer T. Sens Actuat B Chem. 2003;92:137–143. [Google Scholar]
- 440.Vossmeyer T, Guse B, Besnard I, Bauer RE, Mullen K, Yasuda A. Adv Mater. 2002;14:238–242. [Google Scholar]
- 441.Park MH, Ofir Y, Samanta B, Rotello VM. Adv Mater. 2009;21:2323. [Google Scholar]
- 442.Singh A, Hede S, Sastry M. Small. 2007;3:466–473. doi: 10.1002/smll.200600413. [DOI] [PubMed] [Google Scholar]
- 443.Shipway AN, Lahav M, Blonder R, Willner I. Chem Mater. 1999;11:13–15. [Google Scholar]
- 444.Lahav M, Gabai R, Shipway AN, Willner I. Chem Commun. 1999:1937–1938. [Google Scholar]
- 445.Lahav M, Shipway AN, Willner I. J Chem Soc, Perkin Trans 2. 1999:1925–1931. [Google Scholar]
- 446.Lahav M, Shipway AN, Willner I, Nielsen MB, Stoddart JF. J Electroanal Chem. 2000;482:217–221. [Google Scholar]
- 447.Odell B, Reddington MV, Slawin AMZ, Spencer N, Stoddart JF, Williams DJ. Angew Chem Int Ed. 1988;27:1547–1550. [Google Scholar]
- 448.Blonder R, Sheeney L, Willner I. Chem Commun. 1998:1393–1394. [Google Scholar]
- 449.Kharitonov AB, Shipway AN, Katz E, Willner I. Rev Anal Chem. 1999;18:255–260. doi: 10.1021/ac990997s. [DOI] [PubMed] [Google Scholar]
- 450.Kharitonov AB, Shipway AN, Willner I. Anal Chem. 1999;71:5441–5443. doi: 10.1021/ac990997s. [DOI] [PubMed] [Google Scholar]
- 451.Thompson DT. Nano Today. 2007;2:40–43. [Google Scholar]
- 452.Valden M, Lai X, Goodman DW. Science. 1998;281:1647–1650. doi: 10.1126/science.281.5383.1647. [DOI] [PubMed] [Google Scholar]
- 453.Haruta M, Date M. Appl Catal A. 2001;222:427–437. [Google Scholar]
- 454.Turner M, Golovko VB, Vaughan OPH, Abdulkin P, Berenguer-Murcia A, Tikhov MS, Johnson BFG, Lambert RM. Nature. 2008;454:981–983. doi: 10.1038/nature07194. [DOI] [PubMed] [Google Scholar]
- 455.Li YY, Schluesener HJ, Xu SQ. Gold Bull. 2010;43:29–41. [Google Scholar]
- 456.Li Y, Shi GQ. J Phys Chem B. 2005;109:23787–23793. doi: 10.1021/jp055256b. [DOI] [PubMed] [Google Scholar]
- 457.Guerin S, Attard GS. Electrochem Commun. 2001;3:544–548. [Google Scholar]
- 458.Ueda M, Dietz H, Anders A, Kneppe H, Meixner A, Plieth W. Electrochim Acta. 2002;48:377–386. [Google Scholar]
- 459.Plieth W, Dietz H, Anders A, Sandmann G, Meixner A, Weber M, Kneppe H. Surf Sci. 2005;597:119–126. [Google Scholar]
- 460.Lin TH, Hung WH. J Electrochem Soc. 2009;156:D45–D50. [Google Scholar]
- 461.Liu SQ, Yu JH, Ju HX. J Electroanal Chem. 2003;540:61–67. [Google Scholar]
- 462.Cui H, Dong YP. J Electroanal Chem. 2006;595:37–46. [Google Scholar]
- 463.Sun XP, Du Y, Dong SJ, Wang EK. Anal Chem. 2005;77:8166–8169. doi: 10.1021/ac051476+. [DOI] [PubMed] [Google Scholar]
- 464.Zhang LH, Xu ZA, Sun XP, Dong SJ. Biosens Bioelectron. 2007;22:1097–1100. doi: 10.1016/j.bios.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 465.Jena BK, Raj CR. Electroanalysis. 2007;19:816–822. [Google Scholar]
- 466.Feng KJ, Sun CH, Kang Y, Chen JW, Jiang JH, Shen GL, Yu RQ. Electrochem Commun. 2008;10:531–535. [Google Scholar]
- 467.Goyal RN, Oyama M, Singh SP. Electroanalysis. 2007;19:575–581. [Google Scholar]
- 468.Mora L, Hernandez P, Vicente J, Galan F, Hernandez L. Electroanalysis. 2008;20:2084–2089. [Google Scholar]
- 469.Liu YJ, Nie LH, Tao WY, Yao SZ. Electroanalysis. 2004;16:1271–1278. [Google Scholar]
- 470.Cubukcu M, Timur S, Anik U. Talanta. 2007;74:434–439. doi: 10.1016/j.talanta.2007.07.039. [DOI] [PubMed] [Google Scholar]
- 471.Yang SL, Qu LB, Li G, Yang R, Liu CC. J Electroanal Chem. 2010;645:115–122. [Google Scholar]
- 472.Zhang J, Wang YQ, Lv RH, Xu L. Electrochim Acta. 2010;55:4039–4044. [Google Scholar]
- 473.Cheng H, Chen CD, Zhang SA. Anal Sci. 2009;25:1221–1225. doi: 10.2116/analsci.25.1221. [DOI] [PubMed] [Google Scholar]
- 474.Kannan P, John SA. Anal Chim Acta. 2010;663:158–164. doi: 10.1016/j.aca.2010.01.045. [DOI] [PubMed] [Google Scholar]
- 475.Wei XH, Wang F, Yin YM, Liu QY, Zou LN, Ye BX. Analyst. 2010;135:2286–2290. doi: 10.1039/c0an00256a. [DOI] [PubMed] [Google Scholar]
- 476.Liu D, Luo P, Sun WW, Zhang LP, Wang Z. Anal Biochem. 2010;404:14–20. doi: 10.1016/j.ab.2010.04.023. [DOI] [PubMed] [Google Scholar]
- 477.Han L, Hang XL. Electroanalysis. 2009;21:124–129. [Google Scholar]
- 478.Kurniawan F, Tsakova V, Mirsky VM. Electroanalysis. 2006;18:1937–1942. [Google Scholar]
- 479.Li Y, Song YY, Yang C, Xia XH. Electrochem Commun. 2007;9:981–988. [Google Scholar]
- 480.Jena BK, Raj CR. Chem Eur J. 2006;12:2702–2708. doi: 10.1002/chem.200501051. [DOI] [PubMed] [Google Scholar]
- 481.Sun YY, Bai Y, Yang WW, Sun CQ. Electrochim Acta. 2007;52:7352–7361. [Google Scholar]
- 482.Chen M, Diao GW. Talanta. 2009;80:815–820. doi: 10.1016/j.talanta.2009.07.068. [DOI] [PubMed] [Google Scholar]
- 483.Liu XY, Zeng XD, Mai NN, Liu Y, Kong B, Li YH, Wei WZ, Luo SL. Biosens Bioelectron. 2010;25:2675–2679. doi: 10.1016/j.bios.2010.04.045. [DOI] [PubMed] [Google Scholar]
- 484.Jia F, Shan CS, Li FH, Niu L. Biosens Bioelectron. 2008;24:945–950. doi: 10.1016/j.bios.2008.07.057. [DOI] [PubMed] [Google Scholar]
- 485.Wang JW, Wang LP, Di JW, Tu YF. Sens Actuat B Chem. 2008;135:283–288. [Google Scholar]
- 486.Zheng BZ, Qian L, Yuan HY, Xiao D, Yang XP, Paau MC, Choi MMF. Talanta. 2010;82:177–183. doi: 10.1016/j.talanta.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 487.Labib M, Hedstrom M, Amin M, Mattiasson B. Anal Chim Acta. 2010;659:194–200. doi: 10.1016/j.aca.2009.11.028. [DOI] [PubMed] [Google Scholar]
- 488.Raj CR, Okajima T, Ohsaka T. J Electroanal Chem. 2003;543:127–133. [Google Scholar]
- 489.Li MG, Gao F, Yang P, Wang L, Fang B. Surf Sci. 2008;602:151–155. [Google Scholar]
- 490.Zhang L, Jiang X. J Electroanal Chem. 2005;583:292–299. [Google Scholar]
- 491.Hu GZ, Zhang DP, Wu WL, Yang ZS. Colloids Surf B. 2008;62:199–205. doi: 10.1016/j.colsurfb.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 492.Huang X, Li YX, Wang P, Wang L. Anal Sci. 2008;24:1563–1568. doi: 10.2116/analsci.24.1563. [DOI] [PubMed] [Google Scholar]
- 493.Li J, Lin XQ. Sens Actuat B Chem. 2007;124:486–493. [Google Scholar]
- 494.Zhang SJ, Xu ML, Zhang YZ. Electroanalysis. 2009;21:2607–2610. [Google Scholar]
- 495.Raj CR, Ohsaka T. J Electroanal Chem. 2003;540:69–77. [Google Scholar]
- 496.Hu GZ, Ma YG, Guo Y, Shao SJ. Electrochim Acta. 2008;53:6610–6615. [Google Scholar]
- 497.Kannan P, John SA. Anal Biochem. 2009;386:65–72. doi: 10.1016/j.ab.2008.11.043. [DOI] [PubMed] [Google Scholar]
- 498.Kalimuthu P, John SA. J Electroanal Chem. 2008;617:164–170. [Google Scholar]
- 499.Lu LP, Lin XQ. Anal Sci. 2004;20:527–530. doi: 10.2116/analsci.20.527. [DOI] [PubMed] [Google Scholar]
- 500.Li J, Lin XQ. Anal Chim Acta. 2007;596:222–230. doi: 10.1016/j.aca.2007.05.057. [DOI] [PubMed] [Google Scholar]
- 501.Wang JL, Wang F, Zou XQ, Xu ZA, Dong SJ. Electrochem Commun. 2007;9:343–347. [Google Scholar]
- 502.Sivanesan A, Kannan P, John SA. Electrochim Acta. 2007;52:8118–8124. [Google Scholar]
- 503.Ragupathy D, Gopalan AI, Lee KP. Sens Actuat B Chem. 2010;143:696–703. [Google Scholar]
- 504.Nair SS, John SA, Sagara T. Electrochim Acta. 2009;54:6837–6843. [Google Scholar]
- 505.Jin BK, Zhang H. Anal Lett. 2002;35:1907–1918. [Google Scholar]
- 506.Wang L, Bai JY, Huang PF, Wang HJ, Zhang LY, Zhao YQ. Electrochem Commun. 2006;8:1035–1040. [Google Scholar]
- 507.Luczak T. Electrochim Acta. 2009;54:5863–5870. [Google Scholar]
- 508.Zhang H, Gui XQ, Xu Y, Jin BK. Chin Chem Lett. 2002;13:153–156. [Google Scholar]
- 509.Yin HS, Zhou YL, Ai SY, Han RX, Tang TT, Zhu LS. Microchim Acta. 2010;170:99–105. [Google Scholar]
- 510.Zhao ML, Ni DD, Wang JW, Di JW, Tu YF. Chin J Anal Chem. 2008;36:1729–1731. [Google Scholar]
- 511.Li J. Chin J Chem. 2009;27:2373–2378. [Google Scholar]
- 512.Liu Y, Gu HY. Microchim Acta. 2008;162:101–106. [Google Scholar]
- 513.Jiang YN, Luo HQ, Li NB. Int J Environ Anal Chem. 2007;87:295–306. [Google Scholar]
- 514.Sanz VC, Mena ML, Gonzalez-Cortes A, Yanez-Sedeno P, Pingarron JM. Anal Chim Acta. 2005;528:1–8. [Google Scholar]
- 515.Su L, Mao LQ. Talanta. 2006;70:68–74. doi: 10.1016/j.talanta.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 516.El-Cheick FM, Rashwan FA, Mahmoud HA, El-Rouby M. J Solid State Electrochem. 2010;14:1425–1443. [Google Scholar]
- 517.Luczak T. Electroanalysis. 2009;12:1539–1549. [Google Scholar]
- 518.Zhang LY, Yuan R, Chai YQ, Li XL. Anal Chim Acta. 2007;596:99–105. doi: 10.1016/j.aca.2007.05.050. [DOI] [PubMed] [Google Scholar]
- 519.Jena BK, Raj CR. Anal Chem. 2008;80:4836–4844. doi: 10.1021/ac071064w. [DOI] [PubMed] [Google Scholar]
- 520.Song YS, Muthuraman G, Chen YZ, Lin CC, Zen JM. Electroanalysis. 2006;18:1763–1770. [Google Scholar]
- 521.Dai X, Compton RG. Electroanalysis. 2005;17:1325–1330. [Google Scholar]
- 522.Baron R, Sljukic B, Salter C, Crossley A, Compton RG. Russ J Phys Chem A. 2007;81:1443–1447. [Google Scholar]
- 523.Xiao L, Wildgoose GG, Compton RG. Anal Chim Acta. 2008;620:44–49. doi: 10.1016/j.aca.2008.05.015. [DOI] [PubMed] [Google Scholar]
- 524.Majid E, Hrapovic S, Liu YL, Male KB, Luong JHT. Anal Chem. 2006;78:762–769. doi: 10.1021/ac0513562. [DOI] [PubMed] [Google Scholar]
- 525.Zhu ZQ, Su YY, Li J, Li D, Zhang J, Song SP, Zhao Y, Li GX, Fan CH. Anal Chem. 2009;81:7660–7666. doi: 10.1021/ac9010809. [DOI] [PubMed] [Google Scholar]
- 526.Dominguez-Renedo O, Alonso-Lomillo MA, Ferreira-Goncalves L, Arcos-Martinez MJ. Talanta. 2009;79:1306–1310. doi: 10.1016/j.talanta.2009.05.043. [DOI] [PubMed] [Google Scholar]
- 527.Xu H, Zeng LP, Xing SJ, Shi GY, Xian YZ, Lin LT. Electrochem Commun. 2008;10:1839–1843. [Google Scholar]
- 528.Abollino O, Giacomino A, Malandrino M, Piscionieri G, Mentasti E. Electroanalysis. 2008;20:75–83. [Google Scholar]
- 529.Renedo OD, Martinez MJA. Anal Chim Acta. 2007;589:255–260. doi: 10.1016/j.aca.2007.02.069. [DOI] [PubMed] [Google Scholar]
- 530.Liu BZ, Lu LY, Wang M, Zi YQ. J Chem Sci. 2008;120:493–498. [Google Scholar]
- 531.Liu GD, Lin YY, Wu H, Lin Y. Environ Sci Technol. 2007;41:8129–8134. doi: 10.1021/es071726z. [DOI] [PubMed] [Google Scholar]
- 532.Maye MM, Lou YB, Zhong CJ. Langmuir. 2000;16:7520–7523. [Google Scholar]
- 533.Diao P, Zhang DF, Guo M, Zhang Q. J Catal. 2007;250:247–253. [Google Scholar]
- 534.Milsom EV, Novak J, Oyama M, Marken F. Electrochem Commun. 2007;9:436–442. [Google Scholar]
- 535.Gu HY, Yu AM, Yuan SS, Chen HY. Anal Lett. 2002;35:647–661. [Google Scholar]
- 536.Kannan P, John SA. Electrochim Acta. 2010;55:3497–3503. [Google Scholar]
- 537.Varatharajan S, Kumar KS, Berchmans S, Amutha R, Kiruthiga PV, Devi KP. Analyst. 2010;135:2348–2354. doi: 10.1039/c0an00091d. [DOI] [PubMed] [Google Scholar]
- 538.Zhu M, Liu M, Shi GY, Xu F, Ye XY, Chen JS, Jin LT, Jin JY. Anal Chim Acta. 2002;455:199–206. [Google Scholar]
- 539.Yu AM, Liang ZJ, Cho JH, Caruso F. Nano Lett. 2003;3:1203–1207. [Google Scholar]
- 540.Jena BK, Raj CR. J Phys Chem C. 2007;111:6228–6232. [Google Scholar]
- 541.Maduraiveeran G, Ramaraj R. Electrochem Commun. 2007;9:2051–2055. [Google Scholar]
- 542.Baron R, Sljukic B, Salter C, Crossley A, Compton RG. Electroanalysis. 2007;19:1062–1068. [Google Scholar]
- 543.Li J, Lin XQ. Sens Actuat B Chem. 2007;126:527–535. [Google Scholar]
- 544.Yi QF, Yu WQ. J Electroanal Chem. 2009;633:159–164. [Google Scholar]
- 545.Yin HS, Ai SY, Shi WJ, Zhu LS. Sens Actuat B Chem. 2009;137:747–753. [Google Scholar]
- 546.Xu XX, Liu SQ, Ju HX. Sensors. 2003;3:350–360. [Google Scholar]
- 547.Xiao Y, Ju HX, Chen HY. Anal Chim Acta. 1999;391:73–82. [Google Scholar]
- 548.Xu SY, Peng B, Han XZ. Biosens Bioelectron. 2007;22:1807–1810. doi: 10.1016/j.bios.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 549.Wang L, Wang EK. Electrochem Commun. 2004;6:225–229. [Google Scholar]
- 550.Li XL, Wu J, Gao N, Shen GL, Yu RQ. Sens Actuat B Chem. 2006;117:35–42. [Google Scholar]
- 551.Xu SY, Han XZ. Biosens Bioelectron. 2004;19:1117–1120. doi: 10.1016/j.bios.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 552.Wang JW, Wang LP, Di JW, Tu YF. Talanta. 2009;77:1454–1459. doi: 10.1016/j.talanta.2008.09.034. [DOI] [PubMed] [Google Scholar]
- 553.Zheng Y, Lin XQ. Chin J Anal Chem. 2008;36:604–608. [Google Scholar]
- 554.Patolsky F, Gabriel T, Willner I. J Electroanal Chem. 1999;479:69–73. [Google Scholar]
- 555.Xu SY, Tu GL, Peng B, Han XZ. Anal Chim Acta. 2006;570:151–157. doi: 10.1016/j.aca.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 556.Liu AP, Dong WJ, Liu EJ, Tang WH, Zhu JQ, Han JC. Electrochim Acta. 2010;55:1971–1977. [Google Scholar]
- 557.Qiu JD, Peng HZ, Liang RP, Li J, Xia XH. Langmuir. 2007;23:2133–2137. doi: 10.1021/la062788q. [DOI] [PubMed] [Google Scholar]
- 558.Pandey PC, Singh B. Biosens Bioelectron. 2008;24:842–848. doi: 10.1016/j.bios.2008.07.016. [DOI] [PubMed] [Google Scholar]
- 559.Li YL, Zhang J, Zhu H, Yang F, Yang XR. Electrochim Acta. 2010;55:5123–5128. [Google Scholar]
- 560.Hung CC, Wen TC, Wei Y. Mater Chem Phys. 2010;122:392–396. [Google Scholar]
- 561.Chikkaveeraiah BV, Liu HY, Mani V, Papadimitrakopoulos F, Rusling JF. Electrochem Commun. 2009;11:819–822. doi: 10.1016/j.elecom.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 562.Kim GY, Shim J, Kang MS, Moon SH. J Hazard Mater. 2008;156:141–147. doi: 10.1016/j.jhazmat.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 563.Kang TF, Wang F, Lu LP, Zhang Y, Liu TS. Sens Actuat B Chem. 2010;145:104–109. [Google Scholar]
- 564.Yin HS, Ai SY, Xu J, Shi WJ, Zhu LS. J Electroanal Chem. 2009;637:21–27. [Google Scholar]
- 565.Upadhyay S, Rao GR, Sharma MK, Bhattacharya BK, Rao VK, Vijayaraghavan R. Biosens Bioelectron. 2009;25:832–838. doi: 10.1016/j.bios.2009.08.036. [DOI] [PubMed] [Google Scholar]
- 566.Goyal RN, Gupta VK, Oyama M, Bachheti N. Electrochem Commun. 2005;7:803–807. [Google Scholar]
- 567.Goyal RN, Gupta VK, Oyama M, Bachheti N. Electrochem Commun. 2006;8:65–70. [Google Scholar]
- 568.Goyal RN, Oyama M, Bachheti N, Singh SP. Bioelectrochemistry. 2009;74:272–277. doi: 10.1016/j.bioelechem.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 569.Yang GJ, Qu XL, Shen M, Wang CY, Qu QS, Hu XY. Sens Actuat B Chem. 2007;128:258–265. [Google Scholar]
- 570.Jena BK, Raj CR. Talanta. 2010;80:1653–1656. doi: 10.1016/j.talanta.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 571.Ding L, Hao C, Xue YD, Ju HX. Biomacromolecules. 2007;8:1341–1346. doi: 10.1021/bm061224y. [DOI] [PubMed] [Google Scholar]
- 572.Yan F, Chen J, Ju HX. Electrochem Commun. 2007;9:293–298. [Google Scholar]
- 573.Du D, Liu SL, Chen J, Ju HX, Lian HZ, Li JX. Biomaterials. 2005;26:6487–6495. doi: 10.1016/j.biomaterials.2005.03.048. [DOI] [PubMed] [Google Scholar]
- 574.de la Escosura-Muniz A, Sanchez-Espinel C, Diaz-Freitas B, Gonzalez-Fernandez A, Maltez-da Costa M, Merkoci A. Anal Chem. 2009;81:10268–10274. doi: 10.1021/ac902087k. [DOI] [PubMed] [Google Scholar]
- 575.Ghindilis AL, Atanasov P, Wilkins E. Electroanalysis. 1997;9:661–674. [Google Scholar]
- 576.Brown KR, Fox AP, Natan MJ. J Am Chem Soc. 1996;118:1154–1157. [Google Scholar]
- 577.Wang L, Wang EK. Electrochem Commun. 2004;6:49–54. [Google Scholar]
- 578.Murata K, Suzuki M, Kajiya K, Nakamura N, Ohno H. Electrochem Commun. 2009;11:668–671. [Google Scholar]
- 579.Xiao Y, Ju HX, Chen HY. Anal Biochem. 2000;278:22–28. [Google Scholar]
- 580.Zhao JG, Henkens RW, Stonehuerner J, Odaly JP, Crumbliss AL. J Electroanal Chem. 1992;327:109–119. [Google Scholar]
- 581.Li F, Feng Y, Wang Z, Yang LM, Zhuo LH, Tang B. Biosens Bioelectron. 2010;25:2244–2248. doi: 10.1016/j.bios.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 582.Xiang C, Zou Y, Sun LX, Xu F. Sens Actuat B Chem. 2009;136:158–162. [Google Scholar]
- 583.Liu Y, Yuan R, Chai YQ, Tang DP, Dai JY, Zhong X. Sens Actuat B Chem. 2006;115:109–115. [Google Scholar]
- 584.Xu Q, Cai WY, Zhu JJ. Chem Lett. 2005;34:832–833. [Google Scholar]
- 585.Guo SF, Wang LH, Lu TH, Ding XL, Huang XH. J Electroanal Chem. 2010;644:80–84. [Google Scholar]
- 586.Liu SQ, Leech D, Ju HX. Anal Lett. 2003;36:1–19. [Google Scholar]
- 587.Zhao S, Zhang K, Bai Y, Yang WW, Sun CQ. Bioelectrochemistry. 2006;69:158–163. doi: 10.1016/j.bioelechem.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 588.Du D, Ding JW, Cai J, Zhang JM, Liu L. Talanta. 2008;74:1337–1343. doi: 10.1016/j.talanta.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 589.Zhang H, Hu N. Biosens Bioelectron. 2007;23:393–399. doi: 10.1016/j.bios.2007.04.018. [DOI] [PubMed] [Google Scholar]
- 590.Zhao JG, Daly OJP, Henkens RW, Stonehuerner J, Crumbliss AL. Biosens Bioelectron. 1996;11:493–502. [Google Scholar]
- 591.Andreescu S, Luck LA. Anal Biochem. 2008;375:282–290. doi: 10.1016/j.ab.2007.12.035. [DOI] [PubMed] [Google Scholar]
- 592.Lei CX, Hu SQ, Gao N, Shen GL, Yu RQ. Bioelectrochemistry. 2004;65:33–39. doi: 10.1016/j.bioelechem.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 593.Zhang SX, Wang N, Yu HJ, Niu YM, Sun CQ. Bioelectrochemistry. 2005;67:15–22. doi: 10.1016/j.bioelechem.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 594.Crumbliss AL, Perine SC, Stonehürner J, Tubergen KR, Zhao JG, Henkens RW. Biotech Bioeng. 1992;40:483–490. doi: 10.1002/bit.260400406. [DOI] [PubMed] [Google Scholar]
- 595.Shulga O, Kirchhoff JR. Electrochem Commun. 2007;9:935–940. [Google Scholar]
- 596.Bharathi S, Nogami M. Analyst. 2001;126:1919–1922. doi: 10.1039/b105318n. [DOI] [PubMed] [Google Scholar]
- 597.Zhou N, Wang J, Chen T, Yu ZG, Li GX. Anal Chem. 2006;78:5227–5230. doi: 10.1021/ac0605492. [DOI] [PubMed] [Google Scholar]
- 598.Lin JH, Zhang LJ, Zhang SS. Anal Biochem. 2007;370:180–185. doi: 10.1016/j.ab.2007.06.021. [DOI] [PubMed] [Google Scholar]
- 599.Cai WY, Xu Q, Zhao XN, Zhu JH, Chen HY. Chem Mater. 2006;18:279–284. [Google Scholar]
- 600.Wang XY, Zhong H, Lv Y, Chen HY. Chem Lett. 2003;32:1054–1055. [Google Scholar]
- 601.Zhang JJ, Zhu JJ. Sci China, Ser B. 2009;52:815–820. [Google Scholar]
- 602.Mena ML, Yanez-Sedeno P, Pingarron JM. Anal Biochem. 2005;336:20–27. doi: 10.1016/j.ab.2004.07.038. [DOI] [PubMed] [Google Scholar]
- 603.Gole A, Dash C, Ramakrishnan V, Sainkar SR, Mandale AB, Rao M, Sastry M. Langmuir. 2001;17:1674–1679. [Google Scholar]
- 604.Gole A, Vyas S, Phadtare S, Lachke A, Sastry M. Colloids Surf B. 2002;25:129–138. [Google Scholar]
- 605.You CC, Agasti SS, De M, Knapp MJ, Rotello VM. J Am Chem Soc. 2006;128:14612–14618. doi: 10.1021/ja064433z. [DOI] [PubMed] [Google Scholar]
- 606.Pandey P, Singh SP, Arya SK, Gupta V, Datta M, Singh S, Malhotra BD. Langmuir. 2007;23:3333–3337. doi: 10.1021/la062901c. [DOI] [PubMed] [Google Scholar]
- 607.Ahirwal GK, Mitra CK. Sensors. 2009;9:881–894. doi: 10.3390/s90200881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 608.Xiao Y, Patolsky F, Katz E, Hainfeld JF, Willner I. Science. 2003;299:1877–1881. doi: 10.1126/science.1080664. [DOI] [PubMed] [Google Scholar]
- 609.Zayats M, Katz E, Baron R, Willner I. J Am Chem Soc. 2005;127:12400–12406. doi: 10.1021/ja052841h. [DOI] [PubMed] [Google Scholar]
- 610.Kerman K, Chikae M, Yamamura S, Tamiya E. Anal Chim Acta. 2007;588:26–33. doi: 10.1016/j.aca.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 611.Ma LP, Yuan R, Chai YQ, Chen SH, Ling SJ. Bioprocess Biosyst Eng. 2009;32:537–544. doi: 10.1007/s00449-008-0275-8. [DOI] [PubMed] [Google Scholar]
- 612.Zhang L, Jiang XU, Wang EK, Dong SJ. Biosens Bioelectron. 2005;21:337–345. doi: 10.1016/j.bios.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 613.Fang B, Wang GF, Yang XH, Zha QQ, Zhang WZ, Kan XW. Anal Lett. 2004;37:2911–2924. [Google Scholar]
- 614.Ruo Y, ShuRui C, Yaqin C, FengXian G, Oing Z, MingYu T, ZhongQiang T, Yi X. Sci China, Ser B. 2007;50:620–628. [Google Scholar]
- 615.Yang G, Yuan R, Chai YQ. Colloids Surf B. 2008;61:93–100. doi: 10.1016/j.colsurfb.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 616.Liu Y, Jiang QY, Lu SY, Zhang Y, Gu HY. Appl Biochem Biotechnol. 2009;152:418–427. doi: 10.1007/s12010-008-8238-8. [DOI] [PubMed] [Google Scholar]
- 617.Guo HL, Liu DY, Yu XD, Xia XH. Sens Actuat B Chem. 2009;139:598–603. [Google Scholar]
- 618.Tang MY, Chen SH, Yuan R, Chai YQ, Gao FX, Xie Y. Anal Sci. 2008;24:487–491. doi: 10.2116/analsci.24.487. [DOI] [PubMed] [Google Scholar]
- 619.Li JX, Zhou LH, Han X, Liu HL. Sens Actuat B Chem. 2008;135:322–326. [Google Scholar]
- 620.Gu HY, Yu AM, Chen HY. J Electroanal Chem. 2001;516:119–126. [Google Scholar]
- 621.Han XJ, Cheng WL, Zhang ZL, Dong SJ, Wang EK. Biochim Biophys Acta. 2002;1556:273–277. doi: 10.1016/s0005-2728(02)00372-9. [DOI] [PubMed] [Google Scholar]
- 622.Feng JJ, Xu JJ, Chen HY. J Electroanal Chem. 2005;585:44–50. [Google Scholar]
- 623.Chen SH, Yuan R, Chai YQ, Zhang LY, Wang N, Li XL. Biosens Bioelectron. 2007;22:1268–1274. doi: 10.1016/j.bios.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 624.Chen SH, Ynan R, Chai YQ, Yin B, Xu Y. Electroanalysis. 2008;20:2141–2147. [Google Scholar]
- 625.Xu XX, Liu SQ, Li B, Ju HX. Anal Lett. 2003;36:2427–2442. [Google Scholar]
- 626.Hong JM, Dai ZH. Sens Actuat B Chem. 2009;140:222–226. [Google Scholar]
- 627.Zhang JD, Oyama M. J Electroanal Chem. 2005;577:273–279. [Google Scholar]
- 628.Liu SQ, Ju HX. Electroanalysis. 2003;15:1488–1493. [Google Scholar]
- 629.Cao W, Wei CM, Hu JB, Li QL. Electroanalysis. 2008;20:1925–1931. [Google Scholar]
- 630.Zhang H, Hu NF. J Phys Chem B. 2007;111:10583–10590. doi: 10.1021/jp0741556. [DOI] [PubMed] [Google Scholar]
- 631.Zhang H, Lu HY, Hu NF. J Phys Chem B. 2006;110:2171–2179. doi: 10.1021/jp055301f. [DOI] [PubMed] [Google Scholar]
- 632.Yang WW, Li YC, Bai Y, Sun CQ. Sens Actuat B Chem. 2006;115:42–48. [Google Scholar]
- 633.Liu SQ, Peng L, Yang XD, Wu YF, He L. Anal Biochem. 2008;375:209–216. doi: 10.1016/j.ab.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 634.Shumyantseva VV, Carrara S, Bavastrello V, Riley DJ, Bulko TV, Skryabin KG, Archakov AI, Nicolini C. Biosens Bioelectron. 2005;21:217–222. doi: 10.1016/j.bios.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 635.Ju HX, Liu SQ, Ge BX, Lisdat F, Scheller FW. Electroanalysis. 2002;14:141–147. [Google Scholar]
- 636.Li SQ, Xia J, Liu CY, Cao W, Hu JB, Li QL. J Electroanal Chem. 2009;633:273–278. [Google Scholar]
- 637.Xiang CL, Zou YJ, Sun LX, Xu F. Electrochem Commun. 2008;10:38–41. [Google Scholar]
- 638.Hoshi T, Sagae N, Daikuhara K, Anzai J. Talanta. 2007;71:644–647. doi: 10.1016/j.talanta.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 639.Hoshi T, Sagae N, Daikuhara K, Takahara K, Anzai JI. Mater Sci Eng, C. 2007;27:890–894. [Google Scholar]
- 640.Yang WW, Wang JX, Zhao S, Sun YY, Sun CQ. Electrochem Commun. 2006;8:665–672. [Google Scholar]
- 641.Chen SH, Yuan R, Chai YQ, Xu L, Wang N, Li XN, Zhang LY. Electroanalysis. 2006;18:471–477. [Google Scholar]
- 642.Wang YY, Chen XJ, Zhu JJ. Electrochem Commun. 2009;11:323–326. [Google Scholar]
- 643.Crespilho FN, Ghica ME, Florescu M, Nart FC, Oliveira ON, Brett CMA. Electrochem Commun. 2006;8:1665–1670. [Google Scholar]
- 644.Agui L, Manso J, Yanez-Sedeno P, Pingarron JM. Sens Actuat B Chem. 2006;113:272–280. [Google Scholar]
- 645.Lei CX, Hu SQ, Shen GL, Yu RQ. Talanta. 2003;59:981–988. doi: 10.1016/S0039-9140(02)00641-0. [DOI] [PubMed] [Google Scholar]
- 646.Liu SQ, Ju HX. Anal Biochem. 2002;307:110–116. doi: 10.1016/s0003-2697(02)00014-3. [DOI] [PubMed] [Google Scholar]
- 647.Behpour M, Honarmand E, Ghoreishi SM. Bull Korean Chem Soc. 2010;31:845–849. [Google Scholar]
- 648.Liu SQ, Ju HX. Biosens Bioelectron. 2003;19:177–183. doi: 10.1016/s0956-5663(03)00172-6. [DOI] [PubMed] [Google Scholar]
- 649.Carralero V, Mena ML, Gonzalez-Cortes A, Yanez-Sedeno P, Pingarron JM. Biosens Bioelectron. 2006;22:730–736. doi: 10.1016/j.bios.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 650.Li JL, Han T, Wei NN, Du JY, Zhao XW. Biosens Bioelectron. 2009;25:773–777. doi: 10.1016/j.bios.2009.08.026. [DOI] [PubMed] [Google Scholar]
- 651.Wei YY, Li Y, Zhang ND, Shi GY, Jin LT. Ultrason Sonochem. 2010;17:17–20. doi: 10.1016/j.ultsonch.2009.06.017. [DOI] [PubMed] [Google Scholar]
- 652.Wang HC, Wang XS, Zhang XQ, Qin X, Zhao ZX, Miao ZY, Huang N, Chen Q. Biosens Bioelectron. 2009;25:142–146. doi: 10.1016/j.bios.2009.06.022. [DOI] [PubMed] [Google Scholar]
- 653.Kang Q, Yang LX, Cai QY. Bioelectrochemistry. 2008;74:62–65. doi: 10.1016/j.bioelechem.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 654.Liu Y, Wang MK, Zhao F, Guo ZH, Chen HJ, Dong SJ. J Electroanal Chem. 2005;581:1–10. [Google Scholar]
- 655.Manso J, Mena ML, Yanez-Sedeno P, Pingarron J. J Electroanal Chem. 2007;603:1–7. [Google Scholar]
- 656.Liu Y, Wu S, Ju HX, Xu L. Electroanalysis. 2007;19:986–992. [Google Scholar]
- 657.Wang Y, Wei WZ, Liu XY, Zeng XD. Mater Sci Eng, C. 2009;29:50–54. [Google Scholar]
- 658.Wu BY, Hou SH, Yin F, Zhao ZX, Wang YY, Wang XS, Chen Q. Biosens Bioelectron. 2007;22:2854–2860. doi: 10.1016/j.bios.2006.11.028. [DOI] [PubMed] [Google Scholar]
- 659.Manesh KM, Kim JH, Santhosh P, Gopalan AY, Lee KP, Kang HD. J Nanosci Nanotechnol. 2007;7:3365–3372. doi: 10.1166/jnn.2007.836. [DOI] [PubMed] [Google Scholar]
- 660.Wu YH, Hu SS. Colloids Surf B. 2005;41:299–304. doi: 10.1016/j.colsurfb.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 661.Guo YJ, Guo SJ, Fang YX, Dong SJ. Electrochim Acta. 2010;55:3927–3931. [Google Scholar]
- 662.Bui MPN, Pham XH, Han KN, Li CA, Lee EK, Chang HJ, Seong GH. Electrochem Commun. 2010;12:250–253. [Google Scholar]
- 663.Tu XM, Yan LS, Luo XB, Luo SL, Xie QJ. Electroanalysis. 2009;21:2491–2494. [Google Scholar]
- 664.Feng H, Wang H, Zhang Y, Yan B, Shen G, Yu RQ. Anal Sci. 2007;23:235–239. doi: 10.2116/analsci.23.235. [DOI] [PubMed] [Google Scholar]
- 665.Njagi J, Andreescu S. Biosens Bioelectron. 2007;23:168–175. doi: 10.1016/j.bios.2007.03.028. [DOI] [PubMed] [Google Scholar]
- 666.Xian YZ, Hu Y, Liu F, Xian Y, Wang HT, Jin LT. Biosens Bioelectron. 2006;21:1996–2000. doi: 10.1016/j.bios.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 667.Zhong X, Yuan R, Chai YQ, Liu Y, Dai JY, Tang DP. Sens Actuat B Chem. 2005;104:191–198. [Google Scholar]
- 668.Yan W, Feng XM, Chen XJ, Hou WH, Zhu JJ. Biosens Bioelectron. 2008;23:925–931. doi: 10.1016/j.bios.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 669.Ozdemir C, Yeni F, Odaci D, Timur S. Food Chem. 2010;119:380–385. [Google Scholar]
- 670.Gao FX, Yuan R, Chai YQ, Chen SH, Cao SR, Tang MY. J Biochem Bioph Methods. 2007;70:407–413. doi: 10.1016/j.jbbm.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 671.Zhu Q, Yuan R, Chai YQ, Zhuo Y, Zhang Y, Li XL, Wang N. Anal Lett. 2006;39:483–494. [Google Scholar]
- 672.Xie CG, Li HF, Li SQ, Wu J, Zhang ZP. Anal Chem. 2010;82:241–249. doi: 10.1021/ac901860t. [DOI] [PubMed] [Google Scholar]
- 673.Luo XL, Xu JJ, Du Y, Chen HY. Anal Biochem. 2004;334:284–289. doi: 10.1016/j.ab.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 674.Wu BY, Hou SH, Yin F, Li J, Zhao ZX, Huang JD, Chen Q. Biosens Bioelectron. 2007;22:838–844. doi: 10.1016/j.bios.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 675.Xue MH, Xu Q, Zhou M, Zhu JJ. Electrochem Commun. 2006;8:1468–1474. [Google Scholar]
- 676.Du Y, Luo XL, Xu JJ, Chen HY. Bioelectrochemistry. 2007;70:342–347. doi: 10.1016/j.bioelechem.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 677.Shan CS, Yang HF, Han DX, Zhang QX, Ivaska A, Niu L. Biosens Bioelectron. 2010;25:1070–1074. doi: 10.1016/j.bios.2009.09.024. [DOI] [PubMed] [Google Scholar]
- 678.Zeng XD, Li XF, Xing L, Liu XY, Luo SL, Wei WZ, Kong B, Li YH. Biosens Bioelectron. 2009;24:2898–2903. doi: 10.1016/j.bios.2009.02.027. [DOI] [PubMed] [Google Scholar]
- 679.Luo XL, Xu JJ, Zhang Q, Yang GJ, Chen HY. Biosens Bioelectron. 2005;21:190–196. doi: 10.1016/j.bios.2004.07.029. [DOI] [PubMed] [Google Scholar]
- 680.Tangkuaram T, Ponchio C, Kangkasomboon T, Katikawong P, Veerasai W. Biosens Bioelectron. 2007;22:2071–2078. doi: 10.1016/j.bios.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 681.Zhao XJ, Mai ZB, Kang XH, Zou XY. Biosens Bioelectron. 2008;23:1032–1038. doi: 10.1016/j.bios.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 682.Xu Q, Mao C, Liu NN, Zhu JJ, Sheng J. Biosens Bioelectron. 2006;22:768–773. doi: 10.1016/j.bios.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 683.Wang JF, Yuan R, Chai YQ, Li WJ, Fu P, Min LG. Colloids Surf B. 2010;75:425–431. doi: 10.1016/j.colsurfb.2009.09.015. [DOI] [PubMed] [Google Scholar]
- 684.Li WJ, Yuan R, Chai YQ, Zhou L, Chen SH, Li N. J Biochem Bioph Methods. 2008;70:830–837. doi: 10.1016/j.jprot.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 685.Lin JH, Qu W, Zhang SS. Anal Biochem. 2007;360:288–293. doi: 10.1016/j.ab.2006.10.030. [DOI] [PubMed] [Google Scholar]
- 686.Liu ZM, Wang H, Yang Y, Yang HF, Hu SQ, Shen GL, Yu RQ. Anal Lett. 2004;37:1079–1091. [Google Scholar]
- 687.Du D, Chen SZ, Cai J, Song DD. J Electroanal Chem. 2007;611:60–66. [Google Scholar]
- 688.Du D, Ding JW, Cai J, Zhang AD. Sens Actuat B Chem. 2007;127:317–322. [Google Scholar]
- 689.Du D, Ding JW, Cai J, Zhang AD. J Electroanal Chem. 2007;605:53–60. [Google Scholar]
- 690.Drummond TG, Hill MG, Barton JK. Nat Biotechnol. 2003;21:1192–1199. doi: 10.1038/nbt873. [DOI] [PubMed] [Google Scholar]
- 691.Castaneda MT, Alegret S, Merkoci A. Electroanalysis. 2007;19:743–753. [Google Scholar]
- 692.Moller R, Fritzsche W. Curr Pharm Biotechnol. 2007;8:274–285. doi: 10.2174/138920107782109967. [DOI] [PubMed] [Google Scholar]
- 693.Fang YZ, Xu Y, He PG. J Biomed Nanotechnol. 2005;1:276–285. [Google Scholar]
- 694.Selvaraju T, Das J, Jo K, Kwon K, Huh CH, Kim TK, Yang H. Langmuir. 2008;24:9883–9888. doi: 10.1021/la801828a. [DOI] [PubMed] [Google Scholar]
- 695.Ma HY, Zhang LP, Pan Y, Zhang KY, Zhang YZ. Electroanalysis. 2008;20:1220–1226. [Google Scholar]
- 696.Zhang ZL, Pang DW, Yuan H, Cai RX, Abruna H. Anal Bioanal Chem. 2005;381:833–838. doi: 10.1007/s00216-004-2972-8. [DOI] [PubMed] [Google Scholar]
- 697.Zhang KI, Ma HY, Zhang LP, Zhang YZ. Electroanalysis. 2008;20:2127–2133. [Google Scholar]
- 698.Fu YZ, Yuan R, Xu L, Chai YQ, Zhong X, Tang DP. Biochem Eng J. 2005;23:37–44. [Google Scholar]
- 699.Lin XQ, Miao Q, Jin BK. Chin Chem Lett. 1999;10:157–160. [Google Scholar]
- 700.Yang J, Yang T, Feng YY, Jiao K. Anal Biochem. 2007;365:24–30. doi: 10.1016/j.ab.2006.12.039. [DOI] [PubMed] [Google Scholar]
- 701.Liu SF, Li YF, Li JR, Jiang L. Biosens Bioelectron. 2005;21:789–795. doi: 10.1016/j.bios.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 702.Lin XQ, Zheng SJ, Miao Q, Jin BK. Anal Lett. 2002;35:1373–1385. [Google Scholar]
- 703.Wu ZS, Jiang JH, Shen GL, Yu RQ. Human Mutation. 2007;28:630–637. doi: 10.1002/humu.20487. [DOI] [PubMed] [Google Scholar]
- 704.Authier L, Grossiord C, Brossier P, Limoges B. Anal Chem. 2001;73:4450–4456. doi: 10.1021/ac0103221. [DOI] [PubMed] [Google Scholar]
- 705.Wang J, Xu DK, Kawde AN, Polsky R. Anal Chem. 2001;73:5576–5581. doi: 10.1021/ac0107148. [DOI] [PubMed] [Google Scholar]
- 706.Wang J, Polsky R, Xu DK. Langmuir. 2001;17:5739–5741. [Google Scholar]
- 707.Wang J, Xu DK, Polsky R. J Am Chem Soc. 2002;124:4208–4209. doi: 10.1021/ja0255709. [DOI] [PubMed] [Google Scholar]
- 708.Kawde AN, Wang J. Electroanalysis. 2004;16:101–107. [Google Scholar]
- 709.Pumera M, Castaneda MT, Pividori MI, Eritja R, Merkoci A, Alegret S. Langmuir. 2005;21:9625–9629. doi: 10.1021/la051917k. [DOI] [PubMed] [Google Scholar]
- 710.Castaneda MT, Merkoci A, Pumera M, Alegret S. Biosens Bioelectron. 2007;22:1961–1967. doi: 10.1016/j.bios.2006.08.031. [DOI] [PubMed] [Google Scholar]
- 711.Kerman K, Saito M, Morita Y, Takamura Y, Ozsoz M, Tamiya E. Anal Chem. 2004;76:1877–1884. doi: 10.1021/ac0351872. [DOI] [PubMed] [Google Scholar]
- 712.Wang J, Li JH, Baca AJ, Hu JB, Zhou FM, Yan W, Pang DW. Anal Chem. 2003;75:3941–3945. doi: 10.1021/ac0344079. [DOI] [PubMed] [Google Scholar]
- 713.Baca AJ, Zhou FM, Wang J, Hu JB, Li JH, Wang JX, Chikneyan ZS. Electroanalysis. 2004;16:73–80. [Google Scholar]
- 714.Jin Y, Lu W, Hu JQ, Yao X, Li JH. Electrochem Commun. 2007;9:1086–1090. [Google Scholar]
- 715.Wang JX, Zhu X, Tu QY, Guo Q, Zarui CS, Momand J, Sun XZ, Zhou FM. Anal Chem. 2008;80:769–774. doi: 10.1021/ac0714112. [DOI] [PubMed] [Google Scholar]
- 716.Zhang J, Song SP, Zhang LY, Wang LH, Wu HP, Pan D, Fan CH. J Am Chem Soc. 2006;128:8575–8580. doi: 10.1021/ja061521a. [DOI] [PubMed] [Google Scholar]
- 717.Wang J, Zhang SJ, Zhang YZ. Anal Biochem. 2010;396:304–309. doi: 10.1016/j.ab.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 718.Li LL, Cai H, Lee TMH, Barford J, Hsing IM. Electroanalysis. 2004;16:81–87. [Google Scholar]
- 719.Wang MJ, Sun CY, Wang LY, Ji XH, Bai YB, Li TJ, Li JH. J Pharm Biomed Anal. 2003;33:1117–1125. doi: 10.1016/s0731-7085(03)00411-4. [DOI] [PubMed] [Google Scholar]
- 720.Cai H, Wang YQ, He PG, Fang YH. Anal Chim Acta. 2002;469:165–172. [Google Scholar]
- 721.Lee TMH, Li LL, Hsing IM. Langmuir. 2003;19:4338–4343. [Google Scholar]
- 722.Cai H, Shang C, Hsing IM. Anal Chim Acta. 2004;523:61–68. [Google Scholar]
- 723.Lee TMH, Cai H, Hsing IM. Electroanalysis. 2004;16:1628–1631. [Google Scholar]
- 724.Cai H, Wang YQ, He PG, Fang YZ. Chem J Chinese U. 2003;24:1390–1394. [Google Scholar]
- 725.Lee TMH, Cai H, Hsing IM. Analyst. 2005;130:364–369. doi: 10.1039/b413143f. [DOI] [PubMed] [Google Scholar]
- 726.Hanaee H, Ghourchian H, Ziaee AA. Anal Biochem. 2007;370:195–200. doi: 10.1016/j.ab.2007.06.029. [DOI] [PubMed] [Google Scholar]
- 727.Rochelet-Dequaire M, Limoges B, Brossier P. Analyst. 2006;131:923–929. doi: 10.1039/b603963d. [DOI] [PubMed] [Google Scholar]
- 728.Bonanni A, Esplandiu MJ, del Valle M. Electrochim Acta. 2008;53:4022–4029. [Google Scholar]
- 729.Kang JW, Li XN, Wu GF, Wang ZH, Lu XQ. Anal Biochem. 2007;364:165–170. doi: 10.1016/j.ab.2007.01.037. [DOI] [PubMed] [Google Scholar]
- 730.Cai H, Xu C, He PG, Fang YZ. J Electroanal Chem. 2001;510:78–85. [Google Scholar]
- 731.Moller R, Csaki A, Kohler JM, Fritzsche W. Langmuir. 2001;17:5426–5430. [Google Scholar]
- 732.Park SJ, Taton TA, Mirkin CA. Science. 2002;295:1503–1506. doi: 10.1126/science.1067003. [DOI] [PubMed] [Google Scholar]
- 733.Urban M, Moller R, Fritzsche W. Rev Sci Instrum. 2003;74:1077–1081. [Google Scholar]
- 734.Diessel E, Grothe K, Siebert HM, Warner BD, Burmeister J. Biosens Bioelectron. 2004;19:1229–1235. doi: 10.1016/j.bios.2003.11.013. [DOI] [PubMed] [Google Scholar]
- 735.Ozsoz M, Erdem A, Kerman K, Ozkan D, Tugrul B, Topcuoglu N, Ekren H, Taylan M. Anal Chem. 2003;75:2181–2187. doi: 10.1021/ac026212r. [DOI] [PubMed] [Google Scholar]
- 736.Kerman K, Morita Y, Takamura Y, Ozsoz M, Tamiya E. Anal Chim Acta. 2004;510:169–174. [Google Scholar]
- 737.Li D, Yan Y, Wieckowska A, Willner I. Chem Commun. 2007:3544–3546. doi: 10.1039/b704731b. [DOI] [PubMed] [Google Scholar]
- 738.Zhang KY, Zhang YZ. Electroanalysis. 2010;22:673–679. [Google Scholar]
- 739.Xu JH, Zhu JJ, Zhu YL, Gu K, Chen HY. Anal Lett. 2001;34:503–512. [Google Scholar]
- 740.Li CZ, Liu YL, Luong JHT. Anal Chem. 2005;77:478–485. doi: 10.1021/ac048672l. [DOI] [PubMed] [Google Scholar]
- 741.Lu LP, Wang SQ, Lin XQ. Anal Chim Acta. 2004;519:161–166. [Google Scholar]
- 742.de la Escosura-Muniz A, Parolo C, Merkoci A. Mater Today. 2010;13:17–27. [Google Scholar]
- 743.Wang MJ, Wang LY, Wang G, Ji XH, Bai YB, Li TJ, Gong SY, Li JH. Biosens Bioelectron. 2004;19:575–582. doi: 10.1016/s0956-5663(03)00252-5. [DOI] [PubMed] [Google Scholar]
- 744.Tang DP, Yuan R, Chai YQ, Dai JY, Zhong X, Liu Y. Bioelectrochemistry. 2004;65:15–22. doi: 10.1016/j.bioelechem.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 745.Tang DP, Yuan R, Chai YQ, Fu YZ, Dai JY, Liu Y, Zhong X. Biosens Bioelectron. 2005;21:539–548. doi: 10.1016/j.bios.2004.11.024. [DOI] [PubMed] [Google Scholar]
- 746.Ruo Y, Tang DP, Chai YQ, Zhang LY, Liu Y, Zhong X, Dai JY. Sci China, Ser B. 2005;48:49–57. [Google Scholar]
- 747.Bin He Y, Luo HQ, Li NB. Biosens Bioelectron. 2007;22:2952–2957. doi: 10.1016/j.bios.2006.11.031. [DOI] [PubMed] [Google Scholar]
- 748.Ding CF, Li H, Hu KC, Lin JM. Talanta. 2010;80:1385–1391. doi: 10.1016/j.talanta.2009.09.040. [DOI] [PubMed] [Google Scholar]
- 749.Tang DP, Yuan R, Chai YQ, Zhong X, Liu Y, Dai JY. Clin Biochem. 2006;39:309–314. doi: 10.1016/j.clinbiochem.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 750.Liang RP, Qiu HD, Cai PX. Anal Chim Acta. 2005;534:223–229. [Google Scholar]
- 751.Tang DP, Yuan R, Chai YQ, Zhong X, Liu Y, Dai JY, Zhang LY. Anal Biochem. 2004;333:345–350. doi: 10.1016/j.ab.2004.06.035. [DOI] [PubMed] [Google Scholar]
- 752.Zhuo Y, Yuan R, Chai YQ, Zhang Y, Li XL, Zhu Q, Wang N. Anal Chim Acta. 2005;548:205–210. [Google Scholar]
- 753.Qiu JD, Liang RP, Wang R, Fan LX, Chen YW, Xia XH. Biosens Bioelectron. 2009;25:852–857. doi: 10.1016/j.bios.2009.08.048. [DOI] [PubMed] [Google Scholar]
- 754.Shen GY, Zhang Y. Anal Chim Acta. 2010;674:27–31. doi: 10.1016/j.aca.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 755.Tang DP, Yuan R, Chai YQ, Zhang LY, Zhong X, Liu Y, Dai JY. Sens Actuat B Chem. 2005;104:199–206. [Google Scholar]
- 756.Tang DP, Yuan R, Chai YQ, Zhong X, Liu Y, Dai JY. Biochem Eng J. 2004;22:43–49. doi: 10.1016/j.ab.2004.06.035. [DOI] [PubMed] [Google Scholar]
- 757.Tang DP, Yuan R, Chai YQ, Fu YZ. Electrochem Commun. 2005;7:177–182. [Google Scholar]
- 758.Tang DP, Ren JJ. Electroanalysis. 2005;17:2208–2216. [Google Scholar]
- 759.Yuan R, Zhang LY, Li QF, Chai YQ, Cao SR. Anal Chim Acta. 2005;531:1–5. [Google Scholar]
- 760.Liu Y, Qin ZH, Wu XF, Jiang H. Biochem Eng J. 2006;32:211–217. [Google Scholar]
- 761.Sharma A, Matharu Z, Sumana G, Solanki PR, Kim CG, Malhotra BD. Thin Solid Films. 2010;519:1213–1218. [Google Scholar]
- 762.Bone L, Vidal JC, Duato P, Castillo JR. Anal Methods. 2010;2:335–341. [Google Scholar]
- 763.Liu XP, Deng YJ, Jin XY, Chen LG, Jiang JH, Shen GL, Yu RQ. Anal Biochem. 2009;389:63–68. doi: 10.1016/j.ab.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 764.Zhang Y, Zhuang HS. Chin J Anal Chem. 2010;38:153–157. [Google Scholar]
- 765.Hu SQ, Xie JW, Xu QH, Rong KT, Shen GL, Yu RQ. Talanta. 2003;61:769–777. doi: 10.1016/S0039-9140(03)00368-0. [DOI] [PubMed] [Google Scholar]
- 766.Valera E, Ramon-Azcon J, Sanchez FJ, Marco MP, Rodriguez A. Sens Actuat B Chem. 2008;134:95–103. [Google Scholar]
- 767.Valera E, Muniz D, Rodriguez A. Microelectron Eng. 2010;87:167–173. [Google Scholar]
- 768.Tang L, Zeng GM, Shen GL, Li YP, Zhang Y, Huang DL. Environ Sci Technol. 2008;42:1207–1212. doi: 10.1021/es7024593. [DOI] [PubMed] [Google Scholar]
- 769.Chen J, Tang JH, Yan F, Ju HX. Biomaterials. 2006;27:2313–2321. doi: 10.1016/j.biomaterials.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 770.Idegami K, Chikae M, Kerman K, Nagatani N, Yuhi T, Endo T, Tamiya D. Electroanalysis. 2008;20:14–21. [Google Scholar]
- 771.Chen J, Yan F, Tan F, Ju HX. Electroanalysis. 2006;18:1696–1702. [Google Scholar]
- 772.Chai R, Yuan R, Chai YQ, Ou CF, Cao SR, Li XL. Talanta. 2008;74:1330–1336. doi: 10.1016/j.talanta.2007.08.046. [DOI] [PubMed] [Google Scholar]
- 773.Yang GM, Chang YB, Yang H, Tan L, Wu ZS, Lu XX, Yang YH. Anal Chim Acta. 2009;644:72–77. doi: 10.1016/j.aca.2009.04.021. [DOI] [PubMed] [Google Scholar]
- 774.Chen J, Yan F, Du D, Wu J, Ju HX. Electroanalysis. 2006;18:670–676. [Google Scholar]
- 775.Carralero V, Gonzalez-Cortes A, Yanez-Sedeno P, Pingarron JM. Anal Chim Acta. 2007;596:86–91. doi: 10.1016/j.aca.2007.05.046. [DOI] [PubMed] [Google Scholar]
- 776.Carralero V, Gonzalez-Cortes A, Yanez-Sedeno P, Pingarron JM. Electroanalysis. 2007;19:853–858. [Google Scholar]
- 777.Wang RZ, Li YW, Li Q, Shen GL, Xiao LT. Anal Lett. 2009;42:2893–2904. [Google Scholar]
- 778.Liu XQ, Wong DKY. Talanta. 2009;77:1437–1443. doi: 10.1016/j.talanta.2008.09.027. [DOI] [PubMed] [Google Scholar]
- 779.Liu XQ, Duckworth PA, Wong DKY. Biosens Bioelectron. 2010;25:1467–1473. doi: 10.1016/j.bios.2009.10.047. [DOI] [PubMed] [Google Scholar]
- 780.Rezaei B, Khayamian T, Majidi N, Rahmani H. Biosens Bioelectron. 2009;25:395–399. doi: 10.1016/j.bios.2009.07.026. [DOI] [PubMed] [Google Scholar]
- 781.Yin TJ, Wei WZ, Yang L, Gao XH, Gao YP. Sens Actuat B Chem. 2006;117:286–294. [Google Scholar]
- 782.Hu SQ, Xie ZM, Lei CX, Shen GL, Yu RQ. Sens Actuat B Chem. 2005;106:641–647. [Google Scholar]
- 783.Ahirwal GK, Mitra CK. Biosens Bioelectron. 2010;25:2016–2020. doi: 10.1016/j.bios.2010.01.029. [DOI] [PubMed] [Google Scholar]
- 784.Kim JH, Cho JH, Cha GS, Lee CW, Kim HB, Paek SH. Biosens Bioelectron. 2000;14:907–915. doi: 10.1016/s0956-5663(99)00063-9. [DOI] [PubMed] [Google Scholar]
- 785.Rahman MA, Son JI, Won MS, Shim YB. Anal Chem. 2009;81:6604–6611. doi: 10.1021/ac900285v. [DOI] [PubMed] [Google Scholar]
- 786.Qu L, Xia SH, Bian C, Sun JZ, Han JH. Biosens Bioelectron. 2009;24:3419–3424. doi: 10.1016/j.bios.2008.07.077. [DOI] [PubMed] [Google Scholar]
- 787.Lin CC, Chen LC, Huang CH, Ding SJ, Chang CC, Chang HC. J Electroanal Chem. 2008;619:39–45. [Google Scholar]
- 788.Zhang S, Huang F, Liu BH, Ding JJ, Xu X, Kong JL. Talanta. 2007;71:874–881. doi: 10.1016/j.talanta.2006.05.081. [DOI] [PubMed] [Google Scholar]
- 789.Zhuo Y, Yuan R, Chai YQ, Tang DP, Zhang Y, Wang N, Li XL, Zhu Q. Electrochem Commun. 2005;7:355–360. [Google Scholar]
- 790.Zhuo Y, Yuan R, Chai YQ, Zhang Y, Li XL, Wang N, Zhu QA. Sens Actuat B Chem. 2006;114:631–639. [Google Scholar]
- 791.Li Y, Liang WB, Fang LC, Huang H, Deng J, Zheng JS. J Chem Sci. 2009;121:1069–1076. [Google Scholar]
- 792.Ding CF, Zhao F, Ren R, Lin JM. Talanta. 2009;78:1148–1154. doi: 10.1016/j.talanta.2009.01.036. [DOI] [PubMed] [Google Scholar]
- 793.Tan L, Chen YQ, Yang H, Shi Y, Si JF, Yang GM, Wu ZS, Wang P, Lu XX, Bai HP, Yang YH. Sens Actuat B Chem. 2009;142:316–320. [Google Scholar]
- 794.Liu YX, Yuan R, Chai YQ, Hong CL, Guan S. Bioprocess Biosyst Eng. 2010;33:613–618. doi: 10.1007/s00449-009-0385-y. [DOI] [PubMed] [Google Scholar]
- 795.Ran XQ, Yuan R, Chai YQ, Hong CL, Qian XQ. Colloids Surf B. 2010;79:421–426. doi: 10.1016/j.colsurfb.2010.05.012. [DOI] [PubMed] [Google Scholar]
- 796.Liang WB, Yi WJ, Li SH, Yuan R, Chen A, Chen S, Xiang GM, Hu CM. Clin Biochem. 2009;42:1524–1530. doi: 10.1016/j.clinbiochem.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 797.Lin JH, He CY, Zhang LJ, Zhang SS. Anal Biochem. 2009;384:130–135. doi: 10.1016/j.ab.2008.09.033. [DOI] [PubMed] [Google Scholar]
- 798.Xu YY, Chao BA, Chen SF, Xia SH. Anal Chim Acta. 2006;561:48–54. [Google Scholar]
- 799.Tang DP, Yuan R, Chai YQ. Anal Chim Acta. 2006;564:158–165. [Google Scholar]
- 800.Wu LN, Chen J, Du D, Ju HX. Electrochim Acta. 2006;51:1208–1214. [Google Scholar]
- 801.Du D, Xu XX, Wang SF, Zhang AD. Talanta. 2007;71:1257–1262. doi: 10.1016/j.talanta.2006.06.028. [DOI] [PubMed] [Google Scholar]
- 802.Fu XH. Electroanalysis. 2007;19:1831–1839. [Google Scholar]
- 803.Kim DJ, Lee NE, Park JS, Park IJ, Kim JG, Cho HJ. Biosens Bioelectron. 2010;25:2477–2482. doi: 10.1016/j.bios.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 804.Liu Y. Thin Solid Films. 2008;516:1803–1808. [Google Scholar]
- 805.Liu ZY, Yuan R, Chai YQ, Zhuo Y, Hong CL. Acta Chim Sinica. 2009;67:637–644. [Google Scholar]
- 806.Rusling JF, Sotzing G, Papadimitrakopoulosa F. Bioelectrochemistry. 2009;76:189–194. doi: 10.1016/j.bioelechem.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 807.Mani V, Chikkaveeraiah BV, Patel V, Gutkind JS, Rusling JF. ACS Nano. 2009;3:585–594. doi: 10.1021/nn800863w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 808.Tang DP, Yuan R, Chai YQ. Biosens Bioelectron. 2007;22:1116–1120. doi: 10.1016/j.bios.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 809.Loyprasert S, Hedstrom M, Thavarungkul P, Kanatharana P, Mattiasson B. Biosens Bioelectron. 2010;25:1977–1983. doi: 10.1016/j.bios.2010.01.020. [DOI] [PubMed] [Google Scholar]
- 810.Liang KZ, Mu WJ. Anal Chim Acta. 2006;580:128–135. doi: 10.1016/j.aca.2006.07.068. [DOI] [PubMed] [Google Scholar]
- 811.Liang KZ, Mu WJ, Huang MY, Yu ZX, Lai QK. Electroanalysis. 2006;18:1505–1510. [Google Scholar]
- 812.Munge BS, Krause CE, Malhotra R, Patel V, Gutkind JS, Rusling JF. Electrochem Commun. 2009;11:1009–1012. doi: 10.1016/j.elecom.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 813.Kim GI, Kim KW, Oh MK, Sung YM. Biosens Bioelectron. 2010;25:1717–1722. doi: 10.1016/j.bios.2009.12.015. [DOI] [PubMed] [Google Scholar]
- 814.Kim DM, Noh HB, Park DS, Ryu SH, Koo JS, Shim YB. Biosens Bioelectron. 2009;25:456–462. doi: 10.1016/j.bios.2009.08.007. [DOI] [PubMed] [Google Scholar]
- 815.Lei CX, Gong FC, Shen GL, Yu RQ. Sens Actuat B Chem. 2003;96:582–588. [Google Scholar]
- 816.Chu X, Xiang ZF, Fu X, Wang SP, Shen GL, Yu RQ. J Immunol Methods. 2005;301:77–88. doi: 10.1016/j.jim.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 817.Dungchai W, Siangproh W, Chaicumpa W, Tongtawe P, Chailapakul O. Talanta. 2008;77:727–732. [Google Scholar]
- 818.Yang GJ, Huang JL, Meng WJ, Shen M, Jiao XA. Anal Chim Acta. 2009;647:159–166. doi: 10.1016/j.aca.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 819.Lin YH, Chen SH, Chuang YC, Lu YC, Shen TY, Chang CA, Lin CS. Biosens Bioelectron. 2008;23:1832–1837. doi: 10.1016/j.bios.2008.02.030. [DOI] [PubMed] [Google Scholar]
- 820.Singh K, Rahman MA, Son JI, Kim KC, Shim YB. Biosens Bioelectron. 2008;23:1595–1601. doi: 10.1016/j.bios.2008.01.016. [DOI] [PubMed] [Google Scholar]
- 821.Piras L, Reho S. Sens Actuat B Chem. 2005;111:450–454. [Google Scholar]
- 822.Huang HZ, Ran PX, Liu ZG. Bioelectrochemistry. 2007;70:257–262. doi: 10.1016/j.bioelechem.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 823.Li XL, Yuan R, Chai YQ, Zhang LY, Zhuo Y, Zhang Y. J Biotechnol. 2006;123:356–366. doi: 10.1016/j.jbiotec.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 824.An HZ, Yuan RT, Tang DP, Chai YQ, Li N. Electroanalysis. 2007;19:479–486. [Google Scholar]
- 825.Huang KJ, Niu DJ, Xie WZ, Wang W. Anal Chim Acta. 2010;659:102–108. doi: 10.1016/j.aca.2009.11.023. [DOI] [PubMed] [Google Scholar]
- 826.Ou CF, Yuan R, Chai YQ, Tang MY, Chai R, He XL. Anal Chim Acta. 2007;603:205–213. doi: 10.1016/j.aca.2007.08.052. [DOI] [PubMed] [Google Scholar]
- 827.Zhong ZY, Wu W, Wang D, Wang D, Shan JL, Qing Y, Zhang ZM. Biosens Bioelectron. 2010;25:2379–2383. doi: 10.1016/j.bios.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 828.Liu ZY, Yuan R, Chai YQ, Zhuo Y, Hong CL, Yang X. Sens Actuat B Chem. 2008;134:625–631. [Google Scholar]
- 829.Li N, Zhao HW, Yuan R, Peng KF, Chai YQ. Electrochim Acta. 2008;54:235–241. [Google Scholar]
- 830.Shi YT, Yuan R, Chai YQ, He XL. Electrochim Acta. 2007;52:3518–3524. [Google Scholar]
- 831.Shi YT, Yuan R, Chai YQ, Tang MY, He XL. J Electroanal Chem. 2007;604:9–16. [Google Scholar]
- 832.Zhuo Y, Yu RJ, Yuan R, Chai YQ, Hong CL. J Electroanal Chem. 2009;628:90–96. [Google Scholar]
- 833.He XL, Yuan R, Chai YQ, Shi YT. J Biochem Bioph Methods. 2008;70:823–829. doi: 10.1016/j.jbbm.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 834.Song ZJ, Yuan R, Chai YQ, Yin B, Fu P, Wang JF. Electrochim Acta. 2010;55:1778–1784. [Google Scholar]
- 835.Yuan YR, Yuan R, Chai YQ, Zhuo Y, Miao XM. J Electroanal Chem. 2009;626:6–13. [Google Scholar]
- 836.Tang H, Chen JH, Nie LH, Kuang YF, Yao SZ. Biosens Bioelectron. 2007;22:1061–1067. doi: 10.1016/j.bios.2006.04.027. [DOI] [PubMed] [Google Scholar]
- 837.Zhang TT, Yuan R, Chai YQ, Liu KG, Ling SJ. Microchim Acta. 2009;165:53–58. [Google Scholar]
- 838.Ho JAA, Chang HC, Shih NY, Wu LC, Chang YF, Chen CC, Chou C. Anal Chem. 2010;82:5944–5950. doi: 10.1021/ac1001959. [DOI] [PubMed] [Google Scholar]
- 839.Wu J, Yan F, Zhang XQ, Yan YT, Tang JH, Ju HX. Clin Chem. 2008;54:1481–1488. doi: 10.1373/clinchem.2007.102350. [DOI] [PubMed] [Google Scholar]
- 840.Wu J, Yan YT, Yan F, Ju HX. Anal Chem. 2008;80:6072–6077. doi: 10.1021/ac800905k. [DOI] [PubMed] [Google Scholar]
- 841.Wu ZS, Li JS, Luo MH, Shen GL, Yu RQ. Anal Chim Acta. 2005;528:235–242. [Google Scholar]
- 842.Mao X, Jiang JH, Huang Y, Shen GL, Yu RQ. Sens Actuat B Chem. 2007;123:198–203. [Google Scholar]
- 843.Chen ZP, Peng ZF, Zhang P, Jin XF, Jiang JH, Zhang XB, Shen GL, Yu RQ. Talanta. 2007;72:1800–1804. doi: 10.1016/j.talanta.2007.02.020. [DOI] [PubMed] [Google Scholar]
- 844.Chen H, Jiang JH, Huang Y, Deng T, Li JS, Shen GL, Yu RQ. Sens Actuat B Chem. 2006;117:211–218. [Google Scholar]
- 845.Chu X, Fu X, Chen K, Shen GL, Yu RQ. Biosens Bioelectron. 2005;20:1805–1812. doi: 10.1016/j.bios.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 846.Fu YZ, Yuan R, Tang DP, Chai YQ, Xu L. Colloids Surf B. 2005;40:61–66. doi: 10.1016/j.colsurfb.2004.10.022. [DOI] [PubMed] [Google Scholar]
- 847.Carralero V, Gonzalez-Cortes A, Yanez-Sedeno P, Pingarron JM. Anal Lett. 2008;41:244–259. [Google Scholar]
- 848.Leng CA, Lai GS, Yan F, Ju HX. Anal Chim Acta. 2010;666:97–101. doi: 10.1016/j.aca.2010.03.060. [DOI] [PubMed] [Google Scholar]
- 849.Cui RJ, Huang HP, Yin ZZ, Gao D, Zhu JJ. Biosens Bioelectron. 2008;23:1666–1673. doi: 10.1016/j.bios.2008.01.034. [DOI] [PubMed] [Google Scholar]
- 850.Liu MQ, Jiang JH, Feng YL, Huang Y, Shen GL, Yu RQ. Chin J Anal Chem. 2010;38:258–262. [Google Scholar]
- 851.Huang KJ, Niu DJ, Sun JY, Zhu XL, Zhu JJ. Anal Bioanal Chem. 2010;397:3553–3561. doi: 10.1007/s00216-010-3868-4. [DOI] [PubMed] [Google Scholar]
- 852.Tian D, Duan C, Wang W, Li N, Zhang H, Cui H, Lu Y. Talanta. 2009;78:399–404. doi: 10.1016/j.talanta.2008.11.037. [DOI] [PubMed] [Google Scholar]
- 853.Tian DY, Duan CF, Wang W, Cui H. Biosens Bioelectron. 2010;25:2290–2295. doi: 10.1016/j.bios.2010.03.014. [DOI] [PubMed] [Google Scholar]
- 854.Marcon L, Melnyk O, Stievenard D. Biosens Bioelectron. 2008;23:1185–1188. doi: 10.1016/j.bios.2007.10.027. [DOI] [PubMed] [Google Scholar]
- 855.Liu GD, Lin YH. J Nanosci Nanotechnol. 2005;5:1060–1065. doi: 10.1166/jnn.2005.178. [DOI] [PubMed] [Google Scholar]
- 856.Ambrosi A, Castaneda MT, Killard AJ, Smyth MR, Alegret S, Merkoci A. Anal Chem. 2007;79:5232–5240. doi: 10.1021/ac070357m. [DOI] [PubMed] [Google Scholar]
- 857.Chen CR, Liu DY, Wu ZS, Luo QM, Shen GL, Yu RQ. Electrochem Commun. 2009;11:1869–1872. [Google Scholar]
- 858.Velev OD, Kaler EW. Langmuir. 1999;15:3693–3698. [Google Scholar]
- 859.Mao X, Jiang JH, Luo Y, Shen GL, Yu RQ. Talanta. 2007;73:420–424. doi: 10.1016/j.talanta.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 860.Garcia MBG, Garcia AC. Bioelectrochem Bioenerg. 1995;38:389–395. [Google Scholar]
- 861.Gonzalez-Garcia MB, Fernandez-Sanchez C, Costa-Garcia A. Biosens Bioelectron. 2000;15:315–321. doi: 10.1016/s0956-5663(00)00074-9. [DOI] [PubMed] [Google Scholar]
- 862.Dequaire M, Degrand C, Limoges B. Anal Chem. 2000;72:5521–5528. doi: 10.1021/ac000781m. [DOI] [PubMed] [Google Scholar]
- 863.Mao X, Jiang J, Chen J, Huang Y, Shen G, Yu R. Anal Chim Acta. 2006;557:159–163. [Google Scholar]
- 864.de la Escosura-Muniz A, Costa MMD, Merkoci A. Biosens Bioelectron. 2009;24:2475–2482. doi: 10.1016/j.bios.2008.12.028. [DOI] [PubMed] [Google Scholar]
- 865.Liao KT, Huang HJ. Anal Chim Acta. 2005;538:159–164. [Google Scholar]
- 866.Wu Y, Zheng JW, Li Z, Zhao YR, Zhang Y. Biosens Bioelectron. 2009;24:1389–1393. doi: 10.1016/j.bios.2008.07.075. [DOI] [PubMed] [Google Scholar]
- 867.Chumbimuni-Torres KY, Dai Z, Rubinova N, Xiang Y, Pretsch E, Wang J, Bakker E. J Am Chem Soc. 2006;128:13676–13677. doi: 10.1021/ja065899k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 868.Das J, Aziz MA, Yang H. J Am Chem Soc. 2006;128:16022–16023. doi: 10.1021/ja0672167. [DOI] [PubMed] [Google Scholar]
- 869.Nam JM, Thaxton CS, Mirkin CA. Science. 2003;301:1884–1886. doi: 10.1126/science.1088755. [DOI] [PubMed] [Google Scholar]
- 870.Selvaraju T, Das J, Han SW, Yang H. Biosens Bioelectron. 2008;23:932–938. doi: 10.1016/j.bios.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 871.Tudos AJ, Schasfoort RBM. Handbook of Surface Plasmon Resonance. RSC Publishing; 2008. [Google Scholar]
- 872.Love JC, Estroff LA, Kriebel JK, Nuzzo RG, Whitesides GM. Chem Rev. 2005;105:1103–1169. doi: 10.1021/cr0300789. [DOI] [PubMed] [Google Scholar]
- 873.Eustis S, El-Sayed MA. Chem Soc Rev. 2006;35:209–217. doi: 10.1039/b514191e. [DOI] [PubMed] [Google Scholar]
- 874.Mustafa DE, Yang TM, Xuan Z, Chen SZ, Tu HY, Zhang AD. Plasmonics. 2010;5:221–231. [Google Scholar]
- 875.Guerrero E, Munoz-Marquez MA, Garcia MA, Crespo P, Fernandez-Pinel E, Hernando A, Fernandez A. Nanotechnology. 2008;19:175701. doi: 10.1088/0957-4484/19/17/175701. [DOI] [PubMed] [Google Scholar]
- 876.Haes AJ, Zou SL, Schatz GC, Van Duyne RP. J Phys Chem B. 2004;108:109–116. doi: 10.1021/jp051178g. [DOI] [PubMed] [Google Scholar]
- 877.Haes AJ, Van Duyne RP. J Am Chem Soc. 2002;124:10596–10604. doi: 10.1021/ja020393x. [DOI] [PubMed] [Google Scholar]
- 878.Underwood S, Mulvaney P. Langmuir. 1994;10:3427–3430. [Google Scholar]
- 879.Mulvaney P. Langmuir. 1996;12:788–800. [Google Scholar]
- 880.Burda C, Chen XB, Narayanan R, El-Sayed MA. Chem Rev. 2005;105:1025–1102. doi: 10.1021/cr030063a. [DOI] [PubMed] [Google Scholar]
- 881.Lee KS, El-Sayed MA. J Phys Chem B. 2006;110:19220–19225. doi: 10.1021/jp062536y. [DOI] [PubMed] [Google Scholar]
- 882.Zhang XM, Zhang JH, Wang HA, Hao YD, Zhang X, Wang TQ, Wang YN, Zhao R, Zhang H, Yang B. Nanotechnology. 2010;21:465702. doi: 10.1088/0957-4484/21/46/465702. [DOI] [PubMed] [Google Scholar]
- 883.Homola J, Yee SS, Gauglitz G. Sens Actuat B Chem. 1999;54:3–15. [Google Scholar]
- 884.Aslan K, Lakowicz JR, Geddes CD. Curr Opin Chem Biol. 2005;9:538–544. doi: 10.1016/j.cbpa.2005.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 885.Hutter E, Fendler JH. Adv Mater. 2004;16:1685–1706. [Google Scholar]
- 886.Haes AJ, Van Duyne RP. Anal Bioanal Chem. 2004;379:920–930. doi: 10.1007/s00216-004-2708-9. [DOI] [PubMed] [Google Scholar]
- 887.Ray PC. Chem Rev. 2010;110:5332–5365. doi: 10.1021/cr900335q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 888.Homola J. Chem Rev. 2008;108:462–493. doi: 10.1021/cr068107d. [DOI] [PubMed] [Google Scholar]
- 889.Stewart ME, Anderton CR, Thompson LB, Maria J, Gray SK, Rogers JA, Nuzzo RG. Chem Rev. 2008;108:494–521. doi: 10.1021/cr068126n. [DOI] [PubMed] [Google Scholar]
- 890.Willets KA, Van Duyne RP. Annu Rev Phys Chem. 2007;58:267–297. doi: 10.1146/annurev.physchem.58.032806.104607. [DOI] [PubMed] [Google Scholar]
- 891.Yonzon CR, Stuart DA, Zhang XY, McFarland AD, Haynes CL, Van Duyne RP. Talanta. 2005;67:438–448. doi: 10.1016/j.talanta.2005.06.039. [DOI] [PubMed] [Google Scholar]
- 892.Yguerabide J, Yguerabide EE. Anal Biochem. 1998;262:137–156. doi: 10.1006/abio.1998.2759. [DOI] [PubMed] [Google Scholar]
- 893.Jin RC, Cao YC, Hao EC, Metraux GS, Schatz GC, Mirkin CA. Nature. 2003;425:487–490. doi: 10.1038/nature02020. [DOI] [PubMed] [Google Scholar]
- 894.Kealley CS, Arnold MD, Porkovich A, Cortie MB. Sens Actuat B Chem. 2010;148:34–40. [Google Scholar]
- 895.Jain PK, Huang X, El-Sayed IH, El-Sayad MA. Plasmonics. 2007;2:107–118. [Google Scholar]
- 896.Fan MK, Thompson M, Andrade ML, Brolo AG. Anal Chem. 2010;82:6350–6352. doi: 10.1021/ac101495m. [DOI] [PubMed] [Google Scholar]
- 897.McFarland AD, Van Duyne RP. Nano Lett. 2003;3:1057–1062. [Google Scholar]
- 898.Haes AJ, Van Duyne RP. Expert Review of Molecular Diagnostics. 2004;4:527–537. doi: 10.1586/14737159.4.4.527. [DOI] [PubMed] [Google Scholar]
- 899.Yonzon CR, Jeoungf E, Zou SL, Schatz GC, Mrksich M, Van Duyne RP. J Am Chem Soc. 2004;126:12669–12676. doi: 10.1021/ja047118q. [DOI] [PubMed] [Google Scholar]
- 900.Lai T, Hou QN, Yang HA, Luo XG, Xi MR. Acta Biochim Biophys Sin. 2010;42:787–792. doi: 10.1093/abbs/gmq085. [DOI] [PubMed] [Google Scholar]
- 901.Endo T, Shibata A, Yanagida Y, Higo Y, Hatsuzawa T. Mater Lett. 2010;64:2105–2108. [Google Scholar]
- 902.Filippo E, Serra A, Manno D. Sens Actuat B Chem. 2009;138:625–630. [Google Scholar]
- 903.Serra A, Filippo E, Re M, Palmisano M, Vittori-Antisari M, Buccolieri A, Manno D. Nanotechnology. 2009;20:165501. doi: 10.1088/0957-4484/20/16/165501. [DOI] [PubMed] [Google Scholar]
- 904.Lyon LA, Musick MD, Natan MJ. Anal Chem. 1998;70:5177–5183. doi: 10.1021/ac9809940. [DOI] [PubMed] [Google Scholar]
- 905.Cheng SF, Chau LK. Anal Chem. 2003;75:16–21. doi: 10.1021/ac020310v. [DOI] [PubMed] [Google Scholar]
- 906.Raschke G, Kowarik S, Franzl T, Sonnichsen C, Klar TA, Feldmann J, Nichtl A, Kurzinger K. Nano Lett. 2003;3:935–938. [Google Scholar]
- 907.Chen CH, Tsao TC, Li WY, Shen WC, Cheng CW, Tang JL, Jen CP, Chau LK, Wu WT. Microsyst Technol. 2010;16:1207–1214. [Google Scholar]
- 908.Chau LK, Lin YF, Cheng SF, Lin TJ. Sens Actuat B Chem. 2006;113:100–105. [Google Scholar]
- 909.Ye J, Bonroy K, Nelis D, Frederix F, D’Haen J, Maes G, Borghs G. Colloid Surface A. 2008;321:313–317. [Google Scholar]
- 910.Uechi I, Yamada S. Anal Bioanal Chem. 2008;391:2411–2421. doi: 10.1007/s00216-008-2121-x. [DOI] [PubMed] [Google Scholar]
- 911.Park KH, Kim S, Yang SM, Park HG. J Nanosci Nanotechnol. 2009;9:1374–1378. doi: 10.1166/jnn.2009.c160. [DOI] [PubMed] [Google Scholar]
- 912.Kajiura M, Nakanishi T, Iida H, Takada H, Osaka T. J Colloid Interface Sci. 2009;335:140–145. doi: 10.1016/j.jcis.2009.03.016. [DOI] [PubMed] [Google Scholar]
- 913.Gao SY, Koshizaki N, Tokuhisa H, Koyama E, Sasaki T, Kim JK, Ryu J, Kim DS, Shimizu Y. Adv Funct Mater. 2010;20:78–86. [Google Scholar]
- 914.Wang L, Li T, Du Y, Chen CG, Li L, Zhou M, Dong SJ. Biosens Bioelectron. 2010;25:2622–2626. doi: 10.1016/j.bios.2010.04.027. [DOI] [PubMed] [Google Scholar]
- 915.Wang TJ, Lin WS. Appl Phys Lett. 2006;89:173903. [Google Scholar]
- 916.Watanabe S, Sonobe M, Arai M, Tazume Y, Matsuo T, Nakamura T, Yoshida K. Chem Commun. 2002:2866–2867. doi: 10.1039/b205751d. [DOI] [PubMed] [Google Scholar]
- 917.Hone DC, Haines AH, Russell DA. Langmuir. 2003;19:7141–7144. [Google Scholar]
- 918.Fahnestock KJ, Manesse M, McIlwee HA, Schauer CL, Boukherroub R, Szunerits S. Analyst. 2009;134:881–886. doi: 10.1039/b817140h. [DOI] [PubMed] [Google Scholar]
- 919.Kalyuzhny G, Vaskevich A, Schneeweiss MA, Rubinstein I. Chem Eur J. 2002;8:3850–3857. doi: 10.1002/1521-3765(20020902)8:17<3849::AID-CHEM3849>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 920.Kalyuzhny G, Schneeweiss MA, Shanzer A, Vaskevich A, Rubinstein I. J Am Chem Soc. 2001;123:3177–3178. doi: 10.1021/ja005703v. [DOI] [PubMed] [Google Scholar]
- 921.Deng JJ, Song Y, Wang YA, Di JW. Biosens Bioelectron. 2010;26:615–619. doi: 10.1016/j.bios.2010.07.014. [DOI] [PubMed] [Google Scholar]
- 922.Tokareva I, Minko S, Fendler JH, Hutter E. J Am Chem Soc. 2004;126:15950–15951. doi: 10.1021/ja044575y. [DOI] [PubMed] [Google Scholar]
- 923.Englebienne P. Analyst. 1998;123:1599–1603. doi: 10.1039/a804010i. [DOI] [PubMed] [Google Scholar]
- 924.Englebienne P, Van Hoonacker A, Verhas M. Analyst. 2001;126:1645–1651. [Google Scholar]
- 925.Prabhakar A, Mukherji S. Lab Chip. 2010;10:3422–3425. doi: 10.1039/c005253a. [DOI] [PubMed] [Google Scholar]
- 926.Zhu SL, Zhang JB, Yue LYL, Hartono D, Liu AQ. Adv Mater Res. 2009;74:95–98. [Google Scholar]
- 927.Wang J. Small. 2005;1:1036–1043. doi: 10.1002/smll.200500214. [DOI] [PubMed] [Google Scholar]
- 928.Matsui J, Akamatsu K, Hara N, Miyoshi D, Nawafune H, Tamaki K, Sugimoto N. Anal Chem. 2005;77:4282–4285. doi: 10.1021/ac050227i. [DOI] [PubMed] [Google Scholar]
- 929.Frederix F, Friedt JM, Choi KH, Laureyn W, Campitelli A, Mondelaers D, Maes G, Borghs G. Anal Chem. 2003;75:6894–6900. doi: 10.1021/ac0346609. [DOI] [PubMed] [Google Scholar]
- 930.Azzam EMS, Bashir A, Shekhah O, Alawady ARE, Birkner A, Grunwald C, Woll C. Thin Solid Films. 2009;518:387–391. [Google Scholar]
- 931.Kim HM, Jin SM, Lee SK, Kim MG, Shin YB. Sensors. 2009;9:2334–2344. doi: 10.3390/s90402334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 932.Wang J, Wang LY, Sun Y, Zhu XN, Cao YB, Wang XH, Zhang HQ, Song DQ. Colloids Surf B. 2010;75:520–525. doi: 10.1016/j.colsurfb.2009.09.029. [DOI] [PubMed] [Google Scholar]
- 933.Lin YB, Zou Y, Mo YY, Guo JP, Lindquist RG. Sensors. 2010;10:9397–9406. doi: 10.3390/s101009397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 934.Chiang CY, Hsieh ML, Huang KW, Chau LK, Chang CM, Lyu SR. Biosens Bioelectron. 2010;26:1036–1042. doi: 10.1016/j.bios.2010.08.047. [DOI] [PubMed] [Google Scholar]
- 935.Briglin SM, Gao T, Lewis NS. Langmuir. 2004;20:299–305. doi: 10.1021/la0351717. [DOI] [PubMed] [Google Scholar]
- 936.Frasconi M, Tortolini C, Botre F, Mazzei F. Anal Chem. 2010;82:7335–7342. doi: 10.1021/ac101319k. [DOI] [PubMed] [Google Scholar]
- 937.Connolly S, Fitzmaurice D. Adv Mater. 1999;11:1202–1205. [Google Scholar]
- 938.Shenton W, Davis SA, Mann S. Adv Mater. 1999;11:449–452. [Google Scholar]
- 939.Fu E, Ramsey SA, Yager P. Anal Chim Acta. 2007;599:118–123. doi: 10.1016/j.aca.2007.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 940.Ko S, Park TJ, Kim HS, Kim JH, Cho YJ. Biosens Bioelectron. 2009;24:2592–2597. doi: 10.1016/j.bios.2009.01.030. [DOI] [PubMed] [Google Scholar]
- 941.Siiman O, Burshteyn A. J Phys Chem B. 2000;104:9795–9810. [Google Scholar]
- 942.Manera MG, Spadavecchia J, Taurino A, Rella R. J Opt. 2010;12:035003. [Google Scholar]
- 943.Hutter E, Pileni MP. J Phys Chem B. 2003;107:6497–6499. [Google Scholar]
- 944.Roy D, Fendler J. Adv Mater. 2004;16:479–508. [Google Scholar]
- 945.Hutter E, Cha S, Liu JF, Park J, Yi J, Fendler JH, Roy D. J Phys Chem B. 2001;105:8–12. [Google Scholar]
- 946.Hutter E, Fendler JH, Roy D. J Phys Chem B. 2001;105:11159–11168. doi: 10.1021/jp052764c. [DOI] [PubMed] [Google Scholar]
- 947.Chah S, Hutter E, Roy D, Fendler JH, Yi J. Chem Phys. 2001;272:127–136. [Google Scholar]
- 948.Pieper-Furst U, Stocklein WFM, Warsinke A. Anal Chim Acta. 2005;550:69–76. [Google Scholar]
- 949.Cao C, Sim SJ. Biosens Bioelectron. 2007;22:1874–1880. doi: 10.1016/j.bios.2006.07.021. [DOI] [PubMed] [Google Scholar]
- 950.Huang HZ, Ran PX, Liu ZG. Sens Actuat B Chem. 2008;131:417–423. [Google Scholar]
- 951.Kawaguchi T, Shankaran DR, Kim SJ, Matsumoto K, Toko K, Miura N. Sens Actuat B Chem. 2008;133:467–472. [Google Scholar]
- 952.Wang JL, Munir A, Li ZH, Zhou HS. Biosens Bioelectron. 2009;25:124–129. doi: 10.1016/j.bios.2009.06.016. [DOI] [PubMed] [Google Scholar]
- 953.Mitchell JS, Lowe TE. Biosens Bioelectron. 2009;24:2177–2183. doi: 10.1016/j.bios.2008.11.018. [DOI] [PubMed] [Google Scholar]
- 954.Chang YF, Chen RC, Lee YJ, Chao SC, Su LC, Li YC, Chou C. Biosens Bioelectron. 2009;24:1610–1614. doi: 10.1016/j.bios.2008.08.019. [DOI] [PubMed] [Google Scholar]
- 955.Hsieh BY, Chang YF, Ng MY, Liu WC, Lin CH, Wu HT, Chou C. Anal Chem. 2007;79:3487–3493. doi: 10.1021/ac0624389. [DOI] [PubMed] [Google Scholar]
- 956.Huang JC, Chang YF, Chen KH, Su LC, Lee CW, Chen CC, Chen YMA, Chou C. Biosens Bioelectron. 2009;25:320–325. doi: 10.1016/j.bios.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 957.Chang YF, Wang SF, Huang JC, Su LC, Yao L, Li YC, Wu SC, Chen YMA, Hsieh JP, Chou C. Biosens Bioelectron. 2010;26:1068–1073. doi: 10.1016/j.bios.2010.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 958.Yamaguchi A, Juodkazis S, Matsuo S, Misawa H. Chem Lett. 2002:190–191. [Google Scholar]
- 959.Fang SP, Lee HJ, Wark AW, Corn RM. J Am Chem Soc. 2006;128:14044–14046. doi: 10.1021/ja065223p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 960.He L, Musick MD, Nicewarner SR, Salinas FG, Benkovic SJ, Natan MJ, Keating CD. J Am Chem Soc. 2000;122:9071–9077. [Google Scholar]
- 961.Yao X, Li X, Toledo F, Zurita-Lopez C, Gutova M, Momand J, Zhou FM. Anal Biochem. 2006;354:220–228. doi: 10.1016/j.ab.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 962.Bailey RC, Nam JM, Mirkin CA, Hupp JT. J Am Chem Soc. 2003;125:13541–13547. doi: 10.1021/ja035479k. [DOI] [PubMed] [Google Scholar]
- 963.Du BA, Li ZP, Liu CH. Angew Chem Int Ed. 2006;45:8022–8025. doi: 10.1002/anie.200603331. [DOI] [PubMed] [Google Scholar]
- 964.Jans H, Liu X, Austin L, Maes G, Huo Q. Anal Chem. 2009;81:9425–9432. doi: 10.1021/ac901822w. [DOI] [PubMed] [Google Scholar]
- 965.Wang XY, Zou MJ, Xu X, Lei R, Li KA, Li N. Anal Bioanal Chem. 2009;395:2397–2403. doi: 10.1007/s00216-009-3134-9. [DOI] [PubMed] [Google Scholar]
- 966.Wang XY, Xu Y, Xu XA, Hu K, Xiang MH, Li LM, Liu F, Li N. Talanta. 2010;82:693–697. doi: 10.1016/j.talanta.2010.05.034. [DOI] [PubMed] [Google Scholar]
- 967.Xie C, Xu FG, Huang XY, Dong CQ, Ren JC. J Am Chem Soc. 2009;131:12763–12770. doi: 10.1021/ja903873n. [DOI] [PubMed] [Google Scholar]
- 968.Wu CK, Xiong C, Wang LJ, Lan CC, Ling LS. Analyst. 2010;135:2682–2687. doi: 10.1039/c0an00201a. [DOI] [PubMed] [Google Scholar]
- 969.El-Sayed IH, Huang XH, El-Sayed MA. Nano Lett. 2005;5:829–834. doi: 10.1021/nl050074e. [DOI] [PubMed] [Google Scholar]
- 970.Smith E, Dent G. Modern Raman Spectroscopy—A Practical Approach. Wiley; Chichester: 2005. [Google Scholar]
- 971.Kneipp K, Kneipp H, Itzkan I, Dasari RR, Feld MS. Chem Rev. 1999;99:2957. doi: 10.1021/cr980133r. [DOI] [PubMed] [Google Scholar]
- 972.Banholzer MJ, Millstone JE, Qin LD, Mirkin CA. Chem Soc Rev. 2008;37:885–897. doi: 10.1039/b710915f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 973.Fleischm M, Hendra PJ, McQuilla Aj. Chem Phys Lett. 1974;26:163–166. [Google Scholar]
- 974.Jeanmaire DL, Vanduyne RP. J Electroanal Chem. 1977;84:1–20. [Google Scholar]
- 975.Albrecht MG, Creighton JA. J Am Chem Soc. 1977;99:5215–5217. [Google Scholar]
- 976.Nie SM, Emory SR. Science. 1997;275:1102–1106. doi: 10.1126/science.275.5303.1102. [DOI] [PubMed] [Google Scholar]
- 977.Kneipp K, Wang Y, Kneipp H, Perelman LT, Itzkan I, Dasari R, Feld MS. Phys Rev Lett. 1997;78:1667–1670. [Google Scholar]
- 978.Stockman MI. Surface-Enhanced Raman Scattering: Physics and Applications. 2006;103 [Google Scholar]
- 979.Schatz GC, Young MA, Van Duyne RP. Surface-Enhanced Raman Scattering: Physics and Applications. 2006;103 [Google Scholar]
- 980.Moskovits M. Surface-Enhanced Raman Scattering: Physics and Applications. 2006;103 [Google Scholar]
- 981.Otto A. J Raman Spectrosc. 2006;37:937–947. [Google Scholar]
- 982.Dasary SSR, Singh AK, Senapati D, Yu HT, Ray PC. J Am Chem Soc. 2009;131:13806–13812. doi: 10.1021/ja905134d. [DOI] [PubMed] [Google Scholar]
- 983.Gupta S, Agrawal M, Uhlmann P, Simon F, Oertel U, Stamm M. Macromolecules. 2008;41:8152–8158. [Google Scholar]
- 984.Chang SH, Ko HH, Singamaneni S, Gunawidjaja R, Tsukruk VV. Anal Chem. 2009;81:5740–5748. doi: 10.1021/ac900537d. [DOI] [PubMed] [Google Scholar]
- 985.Ko H, Chang S, Tsukruk VV. ACS Nano. 2009;3:181–188. doi: 10.1021/nn800569f. [DOI] [PubMed] [Google Scholar]
- 986.Kneipp J, Kneipp H, Wittig B, Kneipp K. J Phys Chem C. 2010;114:7421–7426. [Google Scholar]
- 987.Cao YC, Jin R, Mirkin CA. Science. 2002;297:1536–1540. doi: 10.1126/science.297.5586.1536. [DOI] [PubMed] [Google Scholar]
- 988.Sun L, Yu CX, Irudayaraj J. Anal Chem. 2007;79:3981–3988. doi: 10.1021/ac070078z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 989.Thuy NTB, Yokogawa R, Yoshimura Y, Fujimoto K, Koyano M, Maenosono S. Analyst. 2010;135:595–602. doi: 10.1039/b919969a. [DOI] [PubMed] [Google Scholar]
- 990.Liu YC, Zhong MY, Shan GY, Li YJ, Huang BQ, Yang GL. J Phys Chem B. 2008;112:6484–6489. doi: 10.1021/jp710399d. [DOI] [PubMed] [Google Scholar]
- 991.Qian XM, Zhou X, Nie SM. J Am Chem Soc. 2008;130:14934–14935. doi: 10.1021/ja8062502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 992.Hu J, Zheng PC, Jiang JH, Shen GL, Yu RQ, Liu GK. Analyst. 2010;135:1084–1089. doi: 10.1039/b920358c. [DOI] [PubMed] [Google Scholar]
- 993.Harpster MH, Zhang H, Sankara-Warrier AK, Ray BH, Ward TR, Kollmar JP, Carron KT, Mecham JO, Corcoran RC, Wilson WC, Johnson PA. Biosens Bioelectron. 2009;25:674–681. doi: 10.1016/j.bios.2009.05.020. [DOI] [PubMed] [Google Scholar]
- 994.Sun L, Irudayaraj J. J Phys Chem B. 2009;113:14021–14025. doi: 10.1021/jp908225f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 995.Kang T, Yoo SM, Yoon I, Lee SY, Kim B. Nano Lett. 2010;10:1189–1193. doi: 10.1021/nl1000086. [DOI] [PubMed] [Google Scholar]
- 996.Maiti KK, Dinish US, Fu CY, Lee JJ, Soh KS, Yun SW, Bhuvaneswari R, Olivo M, Chang YT. Biosens Bioelectron. 2010;26:398–403. doi: 10.1016/j.bios.2010.07.123. [DOI] [PubMed] [Google Scholar]
- 997.Willets KA. Anal Bioanal Chem. 2009;394:85–94. doi: 10.1007/s00216-009-2682-3. [DOI] [PubMed] [Google Scholar]
- 998.Qian XM, Peng XH, Ansari DO, Yin-Goen Q, Chen GZ, Shin DM, Yang L, Young AN, Wang MD, Nie SM. Nat Biotechnol. 2008;26:83–90. doi: 10.1038/nbt1377. [DOI] [PubMed] [Google Scholar]
- 999.Kneipp J, Kneipp H, Rice WL, Kneipp K. Anal Chem. 2005;77:2381–2385. doi: 10.1021/ac050109v. [DOI] [PubMed] [Google Scholar]
- 1000.Kneipp K, Haka AS, Kneipp H, Badizadegan K, Yoshizawa N, Boone C, Shafer-Peltier KE, Motz JT, Dasari RR, Feld MS. Appl Spectrosc. 2002;56:150–154. [Google Scholar]
- 1001.Tang HW, Yang XB, Kirkham J, Smith DA. Anal Chem. 2007;79:3646–3653. doi: 10.1021/ac062362g. [DOI] [PubMed] [Google Scholar]
- 1002.Ni J, Lipert RJ, Dawson GB, Porter MD. Anal Chem. 1999;71:4903–4908. doi: 10.1021/ac990616a. [DOI] [PubMed] [Google Scholar]
- 1003.Driskell JD, Uhlenkamp JM, Lipert RJ, Porter MD. Anal Chem. 2007;79:4141–4148. doi: 10.1021/ac0701031. [DOI] [PubMed] [Google Scholar]
- 1004.Grubisha DS, Lipert RJ, Park HY, Driskell J, Porter MD. Anal Chem. 2003;75:5936–5943. doi: 10.1021/ac034356f. [DOI] [PubMed] [Google Scholar]
- 1005.Han XX, Kitahama Y, Itoh T, Wang CX, Zhao B, Ozaki Y. Anal Chem. 2009;81:3350–3355. doi: 10.1021/ac802553a. [DOI] [PubMed] [Google Scholar]
- 1006.Wang GF, Park HY, Lipert RJ. Anal Chem. 2009;81:9643–9650. doi: 10.1021/ac901711f. [DOI] [PubMed] [Google Scholar]
- 1007.Manimaran M, Jana NR. J Raman Spectrosc. 2007;38:1326–1331. [Google Scholar]
- 1008.Lin CC, Yang YM, Chen YF, Yang TS, Chang HC. Biosens Bioelectron. 2008;24:178–183. doi: 10.1016/j.bios.2008.03.035. [DOI] [PubMed] [Google Scholar]
- 1009.Stevenson R, Ingram A, Leung H, McMillan DC, Graham D. Analyst. 2009;134:842–844. doi: 10.1039/b902174d. [DOI] [PubMed] [Google Scholar]
- 1010.Kang T, Yoon I, Kim J, Hee H, Kim B. Chem Eur J. 2010;16:1351–1355. doi: 10.1002/chem.200901708. [DOI] [PubMed] [Google Scholar]
- 1011.Wang YL, Lee K, Irudayaraj J. Chem Commun. 2010;46:613–615. doi: 10.1039/b919607b. [DOI] [PubMed] [Google Scholar]
- 1012.Hu J, Zheng PC, Jiang JH, Shen GL, Yu RQ, Liu GK. Anal Chem. 2009;81:87–93. doi: 10.1021/ac801431m. [DOI] [PubMed] [Google Scholar]
- 1013.Cao YC, Jin RC, Nam JM, Thaxton CS, Mirkin CA. J Am Chem Soc. 2003;125:14676–14677. doi: 10.1021/ja0366235. [DOI] [PubMed] [Google Scholar]
- 1014.Bonham AJ, Braun G, Pavel I, Moskovits M, Reich NO. J Am Chem Soc. 2007;129:14572–14573. doi: 10.1021/ja0767837. [DOI] [PubMed] [Google Scholar]
- 1015.Maher RC, Maier SA, Cohen LF, Koh L, Laromaine A, Dick JAG, Stevens MM. J Phys Chem C. 2010;114:7231–7235. [Google Scholar]
- 1016.Li T, Liu DJ, Wang ZX. Biosens Bioelectron. 2009;24:3335–3339. doi: 10.1016/j.bios.2009.04.033. [DOI] [PubMed] [Google Scholar]
- 1017.Ruan CM, Wang W, Gu BH. Anal Chem. 2006;78:3379–3384. doi: 10.1021/ac0522106. [DOI] [PubMed] [Google Scholar]
- 1018.Huang PJ, Tay LL, Tanha J, Ryan S, Chau LK. Chem Eur J. 2009;15:9330–9334. doi: 10.1002/chem.200901397. [DOI] [PubMed] [Google Scholar]
- 1019.Rule KL, Vikesland PJ. Environ Sci Technol. 2009;43:1147–1152. doi: 10.1021/es801531t. [DOI] [PubMed] [Google Scholar]
- 1020.Marx KA. Biomacromolecules. 2003;4:1099–1120. doi: 10.1021/bm020116i. [DOI] [PubMed] [Google Scholar]
- 1021.Fritz J. Analyst. 2008;133:855–863. doi: 10.1039/b718174d. [DOI] [PubMed] [Google Scholar]
- 1022.Milburn C, Zhou J, Bravo O, Kumar C, Soboyejo WO. J Biomed Nanotechnol. 2005;1:30–38. [Google Scholar]
- 1023.Wu GH, Datar RH, Hansen KM, Thundat T, Cote RJ, Majumdar A. Nat Biotechnol. 2001;19:856–860. doi: 10.1038/nbt0901-856. [DOI] [PubMed] [Google Scholar]
- 1024.Zhou XC, Huang LQ, Li SFY. Biosens Bioelectron. 2001;16:85–95. doi: 10.1016/s0956-5663(00)00136-6. [DOI] [PubMed] [Google Scholar]
- 1025.Oshima K, Nakajima H, Takahashi S, Kera Y, Shimomura M, Miyauchi S. Sens Actuat B Chem. 2005;105:473–478. [Google Scholar]
- 1026.Hwang KS, Lee SM, Kim SK, Lee JH, Kim TS. Annu Rev Anal Chem. 2009;2:77–98. doi: 10.1146/annurev-anchem-060908-155232. [DOI] [PubMed] [Google Scholar]
- 1027.Saya D, Nicu L, Guirardel M, Tauran Y, Bergaud C. Rev Sci Instrum. 2004;75:3010–3015. [Google Scholar]
- 1028.Mao XL, Yang LJ, Su XL, Li YB. Biosens Bioelectron. 2006;21:1178–1185. doi: 10.1016/j.bios.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 1029.Grate JW, Nelson DA, Skaggs R. Anal Chem. 2003;75:1868–1879. doi: 10.1021/ac0206364. [DOI] [PubMed] [Google Scholar]
- 1030.Grate JW. Anal Chem. 2003;75:6759–6759. doi: 10.1021/ac030280b. [DOI] [PubMed] [Google Scholar]
- 1031.Han L, Daniel DR, Maye MM, Zhong CJ. Anal Chem. 2001;73:4441–4449. doi: 10.1021/ac0104025. [DOI] [PubMed] [Google Scholar]
- 1032.Krasteva N, Fogel Y, Bauer RE, Mullen K, Joseph Y, Matsuzawa N, Yasuda A, Vossmeyer T. Adv Funct Mater. 2007;17:881–888. [Google Scholar]
- 1033.Han SB, Lin JQ, Satjapipat M, Baca AJ, Zhou FM. Chem Commun. 2001:609–610. [Google Scholar]
- 1034.Lin L, Zhao HQ, Li JR, Tang JA, Duan MX, Jiang L. Biochem Biophys Res Commun. 2000;274:817–820. doi: 10.1006/bbrc.2000.3233. [DOI] [PubMed] [Google Scholar]
- 1035.Zhou XC, O’Shea SJ, Li SFY. Chem Commun. 2000:953–954. [Google Scholar]
- 1036.Patolsky F, Ranjit KT, Lichtenstein A, Willner I. Chem Commun. 2000:1025–1026. [Google Scholar]
- 1037.Willner I, Patolsky F, Weizmann Y, Willner B. Talanta. 2002;56:847–856. doi: 10.1016/s0039-9140(01)00658-0. [DOI] [PubMed] [Google Scholar]
- 1038.Nie LB, Yang Y, Li S, He NY. Nanotechnology. 2007;18:305501. [Google Scholar]
- 1039.Liu T, Tang J, Han MM, Jiang L. Biochem Biophys Res Commun. 2003;304:98–100. doi: 10.1016/s0006-291x(03)00531-x. [DOI] [PubMed] [Google Scholar]
- 1040.Liu T, Tang J, Jiang L. Biochem Biophys Res Commun. 2004;313:3–7. doi: 10.1016/j.bbrc.2003.11.098. [DOI] [PubMed] [Google Scholar]
- 1041.Tao L, Ji’an T, Han MM, Jiang L. Chin Sci Bull. 2003;48:873–875. [Google Scholar]
- 1042.Liu SF, Li JR, Jiang L. Colloid Surface A. 2005;257–58:57–62. [Google Scholar]
- 1043.Li SH, Li XC, Zhang JP, Zhang YK, Han JH, Jiang L. Colloid Surface A. 2010;364:158–162. [Google Scholar]
- 1044.Liu T, Tang J, Jiang L. Biochem Biophys Res Commun. 2002;295:14–16. doi: 10.1016/s0006-291x(02)00628-9. [DOI] [PubMed] [Google Scholar]
- 1045.Weizmann Y, Patolsky F, Willner I. Analyst. 2001;126:1502–1504. doi: 10.1039/b106613g. [DOI] [PubMed] [Google Scholar]
- 1046.Su M, Li SU, Dravid VP. Appl Phys Lett. 2003;82:3562–3564. [Google Scholar]
- 1047.Tang DQ, Zhang DJ, Tang DY, Ai H. J Immunol Methods. 2006;316:144–152. doi: 10.1016/j.jim.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 1048.Jla XE, Xie QJ, Zhang YY, Yao SZ. Anal Sci. 2007;23:689–696. doi: 10.2116/analsci.23.689. [DOI] [PubMed] [Google Scholar]
- 1049.Jin XY, Jin XF, Chen LG, Jiang JH, Shen GL, Yu RQ. Biosens Bioelectron. 2009;24:2580–2585. doi: 10.1016/j.bios.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 1050.Fu YC, Xie QJ, Jia XE, Xu XH, Meng WH, Yao SZ. J Electroanal Chem. 2007;603:96–106. [Google Scholar]
- 1051.Ding YJ, Wang H, Li JS, Shen GL, Yu RQ. Chem J Chinese U. 2005;26:222–226. [Google Scholar]
- 1052.Chu X, Zhao ZL, Shen GL, Yu RQ. Sens Actuat B Chem. 2006;114:696–704. [Google Scholar]
- 1053.Kim NH, Baek TJ, Park HG, Seong GH. Anal Sci. 2007;23:177–181. doi: 10.2116/analsci.23.177. [DOI] [PubMed] [Google Scholar]
- 1054.Kim N, Kim DK, Cho YJ. Curr Appl Phys. 2010;10:1227–1230. [Google Scholar]
- 1055.Su XD, Li SFY, O’Shea SJ. Chem Commun. 2001:755–756. [Google Scholar]
- 1056.Xia H, Wang F, Huang Q, Huang JF, Chen M, Wang J, Yao CY, Chen QH, Cai GR, Fu WL. Sensors. 2008;8:6453–6470. doi: 10.3390/s8106453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1057.Wang LJ, Wei QS, Wu CS, Hu ZY, Ji J, Wang P. Chin Sci Bull. 2008;53:1175–1184. [Google Scholar]
- 1058.Chen SH, Wu VCH, Chuang YC, Lin CS. J Microbiol Methods. 2008;73:7–17. doi: 10.1016/j.mimet.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 1059.Shen GY, Tan SZ, Nie HG, Shen GL, Yu RQ. J Immunol Methods. 2006;313:11–19. doi: 10.1016/j.jim.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 1060.Ma ZF, Wu JL, Zhou TH, Chen ZH, Dong YG, Tang JT, Sui SF. New J Chem. 2002;26:1795–1798. [Google Scholar]
- 1061.Lyu YK, Lim KR, Lee BY, Kim KS, Lee WY. Chem Commun. 2008:4771–4773. doi: 10.1039/b807438k. [DOI] [PubMed] [Google Scholar]
- 1062.Chen SH, Chuang YC, Lu YC, Lin HC, Yang YL, Lin CS. Nanotechnology. 2009;20:215501. doi: 10.1088/0957-4484/20/21/215501. [DOI] [PubMed] [Google Scholar]
- 1063.Nam JM, Park SJ, Mirkin CA. J Am Chem Soc. 2002;124:3820–3821. doi: 10.1021/ja0178766. [DOI] [PubMed] [Google Scholar]
- 1064.Hill HD, Mirkin CA. Nat Protoc. 2006;1:324–336. doi: 10.1038/nprot.2006.51. [DOI] [PubMed] [Google Scholar]
- 1065.Stoeva SI, Lee JS, Thaxton CS, Mirkin CA. Angew Chem Int Ed. 2006;45:3303–3306. doi: 10.1002/anie.200600124. [DOI] [PubMed] [Google Scholar]
- 1066.Nam JM, Stoeva SI, Mirkin CA. J Am Chem Soc. 2004;126:5932–5933. doi: 10.1021/ja049384+. [DOI] [PubMed] [Google Scholar]
- 1067.Li J, Song SP, Liu XF, Wang LH, Pan D, Huang Q, Zhao Y, Fan CH. Adv Mater. 2008;20:497–500. [Google Scholar]
- 1068.Hill HD, Vega RA, Mirkin CA. Anal Chem. 2007;79:9218–9223. doi: 10.1021/ac701626y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1069.Goluch ED, Nam JM, Georganopoulou DG, Chiesl TN, Shaikh KA, Ryu KS, Barron AE, Mirkin CA, Liu C. Lab Chip. 2006;6:1293–1299. doi: 10.1039/b606294f. [DOI] [PubMed] [Google Scholar]
- 1070.Liu MY, Jia CP, Huang YY, Lou XH, Yao SH, Jin QH, Zhao JL, Xiang JQ. Analyst. 2010;135:327–331. doi: 10.1039/b916629g. [DOI] [PubMed] [Google Scholar]
- 1071.Georganopoulou DG, Chang L, Nam JM, Thaxton CS, Mufson EJ, Klein WL, Mirkin CA. Proc Natl Acad Sci USA. 2005;102:2273–2276. doi: 10.1073/pnas.0409336102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1072.Zheng GF, Daniel WL, Mirkin CA. J Am Chem Soc. 2008;130:9644–9645. doi: 10.1021/ja803035p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1073.Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PLC, Coviello GM, Wright WE, Weinrich SL, Shay JW. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 1074.Thaxton CS, Elghanian R, Thomas AD, Stoeva SI, Lee JS, Smith ND, Schaeffer AJ, Klocker H, Horninger W, Bartsch G, Mirkin CA. Proc Natl Acad Sci USA. 2009;106:18437–18442. doi: 10.1073/pnas.0904719106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1075.Stoeva SI, Lee JS, Smith JE, Rosen ST, Mirkin CA. J Am Chem Soc. 2006;128:8378–8379. doi: 10.1021/ja0613106. [DOI] [PubMed] [Google Scholar]
- 1076.Kim D, Daniel WL, Mirkin CA. Anal Chem. 2009;81:9183–9187. doi: 10.1021/ac9018389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1077.Oh BK, Nam JM, Lee SW, Mirkin CA. Small. 2006;2:103–108. doi: 10.1002/smll.200500260. [DOI] [PubMed] [Google Scholar]
- 1078.Cao C, Dhumpa R, Bang DD, Ghavifekr Z, Hogberg J, Wolff A. Analyst. 2010;135:337–342. doi: 10.1039/b916821b. [DOI] [PubMed] [Google Scholar]
- 1079.Nam JM, Wise AR, Groves JT. Anal Chem. 2005;77:6985–6988. doi: 10.1021/ac0513764. [DOI] [PubMed] [Google Scholar]
- 1080.Nam JM, Jang KJ, Groves JT. Nat Protoc. 2007;2:1438–1444. doi: 10.1038/nprot.2007.201. [DOI] [PubMed] [Google Scholar]